GC is the fourth most commonly diagnosed malignancy
and the second leading cause of cancer-related death with a total
of 989,600 new stomach cancer cases and 738,000 deaths according to
global cancer statistics (1).
Surgery is the dominant treatment of GC, however, >80% of
diagnoses occur at the middle to late stage of the disease and some
of the patients miss the opportunity for surgery (2,3),
thus highlighting an urgent need for novel biomarkers and improved
therapies. Multiple studies have shown that GC is the outcome of
missregulation of many related genes including oncogenes and tumor
suppressor genes (4). Accumulating
data strongly suggest that deregulation of FBPs, which mediates the
degradation of many regulatory proteins, contributes to the
initiation and promotion of cancer. In this review, we summarize
the literature on the FBPs involved in GC, focusing on the function
and mechanism of each related F-box protein (FBP) in GC.
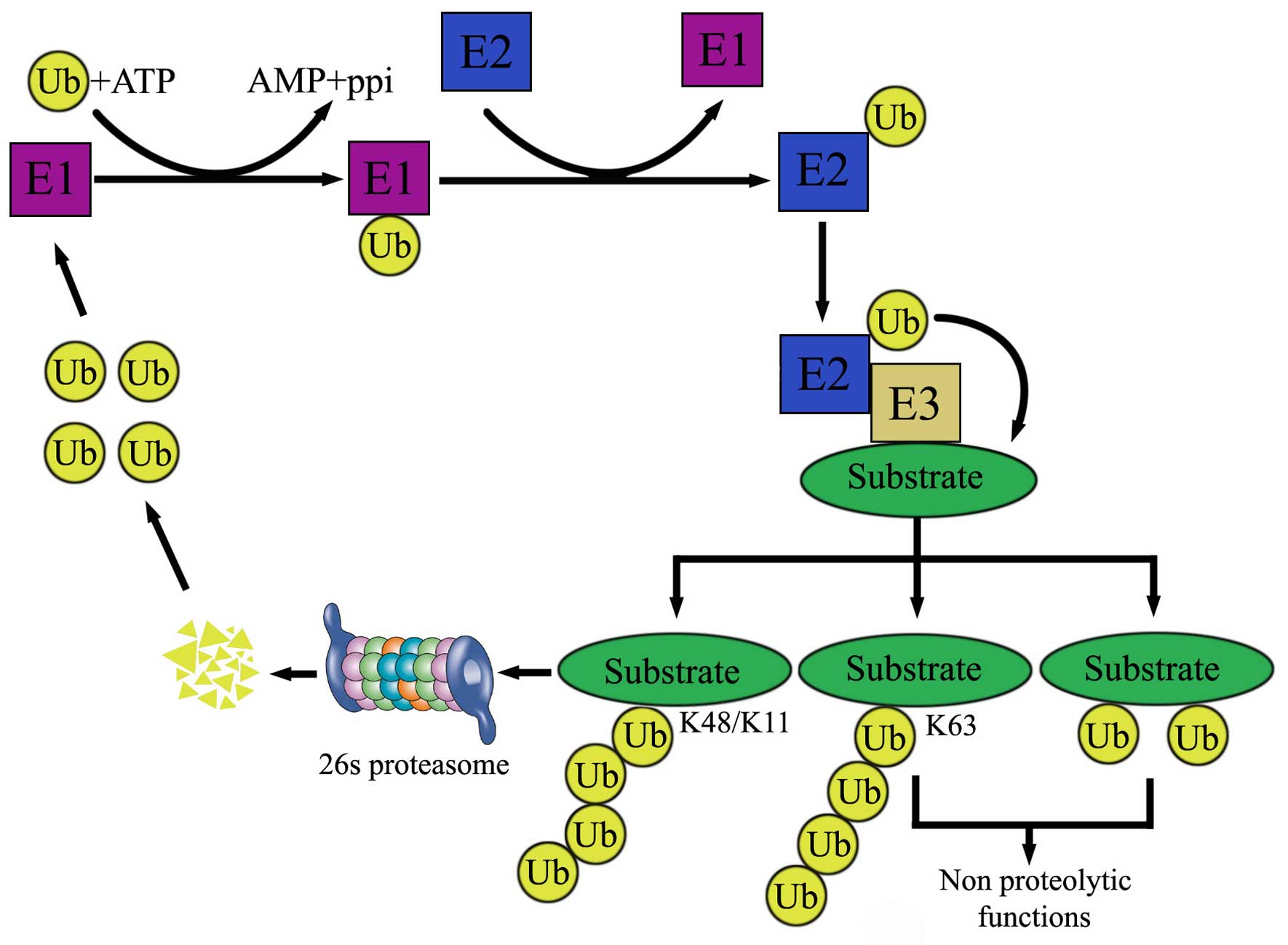
Protein degradation is essential for the removal of
excessive proteins. Two major protein degradation systems exist in
cells, the authophagy-lysosome and UPS. Approximately 80–90% of
intracellular proteins are degraded though UPS. Ubiquitination is a
process in which ubiquitin moieties are covalently attached to a
substrate through an enzymatic cascade. The proteasome and
deubiquitinases (DUBs) are essential components of this system.
Depending on ubiquitin-ubiquitin linkage, polyubiquitinated
proteins can either be activated through K63 linkage, or recognized
and degraded by the 26S proteasome (through K48 linkage). Formation
of a ubiquitin Lys48 chain on the ɛ-NH2 group of a substrate
internal Lys residue (polyubiquitination) can target the substrate
for degradation by the 26S proteasome in an ATP-dependent manner
(Fig. 1).
Ubiquitin attachment to substrates is accomplished
by the coordinated activity of three enzymes (Fig. 1): ubiquitin-activating enzyme (E1),
ubiquitin conjugating enzyme (E2) and ubiquitin-protein ligase
(E3). The degradation of proteins by the UPS is mainly comprised of
two steps. The first step is linking a ubiquitin molecule to E1 via
a high-energy thiol ester at carboxyl terminal glycine residue in
an ATP and Mg2+-dependent manner. Next, E2 accepts the
activated ubiquitin molecule from E1 and with help from an E3
ubiquitin ligase-transfers it to the lysine residue of a target
protein. Then the ubiquitin proteins are recognized and degraded by
26 proteasomes to several small peptides.
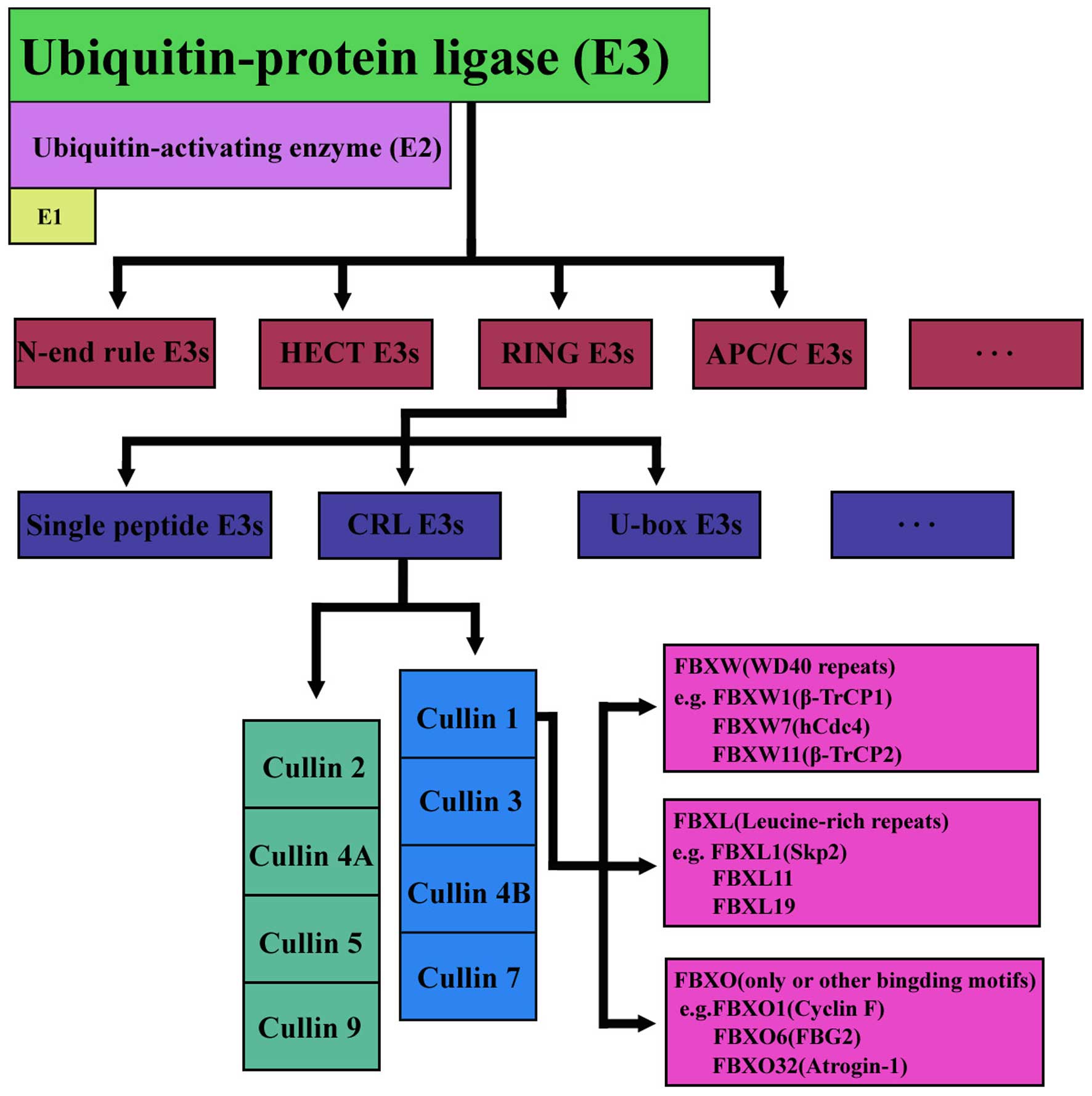
F-box is a widespread protein motif of ~40–50 amino
acids and it functions as a site of protein-protein interaction.
FBPs can be organized into three categories on the basis of the
presence of recognizable domains beyond the F-box domain, those
with WD 40 repeats (FBXW), leucine-rich repeats (FBXL), or other
domain-containing proteins (FBXO) (10). In humans, ~69 FBPs have been
identified so far. The FBXW family is composed of 10 proteins, all
members containing WD40 repeat domains. FBXL family donates each
protein group harboring an F-box and leucine-rich domains,
comprising 22 proteins. The remaining 37 F-box proteins were named
FBXO family (Fig. 2). FBXO is an
abbreviation for F-box and other domains. These F-box proteins
often have conserved homology domains that were either recognized
or are not present in a large number of F-box proteins. Currently,
emerging experimental and clinical data has begun to reveal some
interesting biological functions on FBXO proteins. The abnormal
activation of F-box proteins were found in many human cancers, but
the underlying mechanism was not completely clarified. Below we
discuss each related F-box protein, including its targeting
substrates and affected biologic processes in GC.
FBPs exert antitumoral or promoting effect depending
on the specific substrates they degrade. FBPs function as
oncoproteins when overexpressed if their substrates are tumor
suppressors or as tumor suppressors if their substrates are
oncoproteins. A FBP has more than one substrate, for the same
reason, one substrate can be degraded by different FBPs. Three FBPs
(SKP2, FBXO6 and FBXO32) have shown potential oncogenic role in
GC.
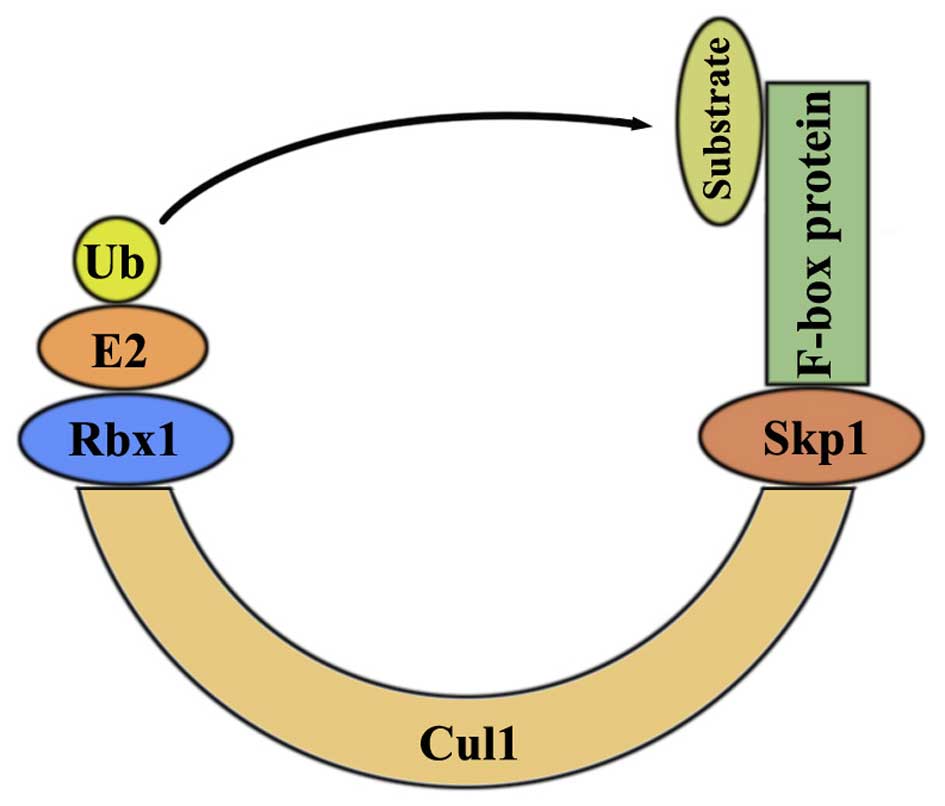
SKP2 was characterized as an Skp1-binding protein to
regulate cell cycle progression through ubiquitin mediated
proteolysis (11). It is regulated
by a network of proteins including cyclins, cyclin-dependent
kinases (CDKs) (12) and CDK
inhibitors (CKIs) (13).
Burgeoning amounts of literature strongly implies that SKP2 plays
an oncogenic role in human cancers. Firstly, SKP2 was found to be
frequently overexpressed in various human cancers (14–17)
and it was associated with poor prognosis as well as tumor
metastasis (18). Secondly, most
of its downstream substrates were tumor suppressor proteins
including p27 and p21 (19).
Thirdly, SKP2 acetylation could promote its oncogenic function by
regulating the downstream targets such as p21 and FOXO1, and
upregulated the acetylation of SKP2 promoted cell growth (20,21).
Fourthly, SKP2 genes were associated with the increased cell
cytotoxicity induced by drugs like platinum (22). Recently, research has shown that
SKP2 predicts poor prognosis and maintains a cancer stem cell pool
(23–25). Moreover, SKP2 has been proven to be
linked with radiotherapy and chemotherapy resistance (26,27).
Collectively, these studies suggest that SKP2 may be an ideal
therapeutic target for human cancer.
In a study of 98 cases of human GC, it was found
that overexpression of SKP2 at the mRNA level was associated with
poor OS (28). The overexpression
of SKP2 in gastric carcinoma cells decreased the level of p27,
increased cell growth rate, rendered cancer cells more resistant to
actinomycin D-induced apoptosis and increased their invasion
potential (28). SKP2 protein was
proved to be negatively associated with p27 in gastric carcinoma
and was positively associated with differentiation degree, vessel
invasion and lymph node metastasis (29). Other regulators of SKP2 also exist.
COX-2 contributed to the expression of SKP2 and poor survival in
human gastric carcinomas (30).
E2F-1 reversed the multidrug resistance by downregulating the
expression of SKP2 in gastric cell line SGC7901 (31). Mechanistic investigation using the
in vitro cell culture models was performed to validate the
critical role of SKP2-p27 axis in the growth and invasion of
SGC7901. E2F-1 is one of DNA-binding protein in E2F family (E2F-1
to E2F-6), and it has the potential to function as an oncogene
(32). Wei et al (33) discovered that the knockdown of SKP2
expression suppressed the ability of GC cell line MGC803 to form
tumors and to metastasize to the lungs of mice. In contrast,
overexpression of Skp2 promoted tumorigenesis of GC in mice.
Proper post-translational modification of the
substrates are often required for their interaction with respective
FBPs (37). The old paradigm of
phosphorylation-directed substrate recognition by FBPs may still be
dominant, but it is no longer absolute. FBXO6 can recognize degrons
that are modified by glycosylation or the addition of mannose
oligo-saccharides. For instance, FBXO6 binds to a glycosylated
degron in T cell receptor α-chain 17. FBXO6, also named Fbs2, is
involved in the endoplasmic reticulum-associated degradation (ERAD)
pathway by mediating the ubiquitination of glycoproteins (38). FBXO6 transcript is widely expressed
in various tissues (39), which
was reported to be involved in neuroblastoma tumorigenesis
(40). Merry et al
(41) showed an inverse
correlation of FBXO6 and Chk1 protein in breast tumor tissues, and
the low expression of FBXO6 causing Chk1 accumulation increases
tumor cells resistance to chemotherapy drug irinotecan. Chk1 is a
key protein kinase in the replication checkpoint. FBXO6 mediates
the Chk1 ubiquitination and degradation, thus terminating the
checkpoint (42). Inhibition of
FBXO6 gene expression increased the biological effects of cadmium
toxicity (43). Zhang et al
(44) reported that FBXO6
accelerated the growth and proliferation of GC cells and normal
gastric cells. These findings shed new light on the tumor-promoting
role of FBXO6. However, FBXO6 did not affect apoptosis and invasion
of GC cells or normal gastric cells. One of the possible
explanation is that FBXO6 has little influence on the key genes
concerned with apoptosis and invasion. FBXO6 might exert many
functions of cells with other FBPs participating in the metabolism,
underscoring the need for further studies aimed at clarifying the
specific role of FBXO6 in GC.
Numerous FBPs including FBXW7, β-TrCP, FBXL5 and
FBXO31, have been shown to be tumor suppressors in GC. Frequent
inactivating mutations or downregulated expression of these E3
ubiquitin ligases have been detected in GC. Besides mutation and
gene copy loss, epigenetic alteration also contributed to
inactivation of these tumor suppressors. FBPs with tumor suppressor
activity in GC are discussed in detail below.
FBXW7 has been implicated as a tumor suppressor gene
in various tumors. Frequent inactivation of FBXW7 by deletions or
point mutations occurs in cancers. It is found that hCDC4 mutations
exist not only in early, but also advanced GC. In agreement, the
study by Milne et al showed similar results (72). Yokobori et al (73) for the first time revealed the
relationship between low expression of FBXW7 and other tumor
suppressor genes. Their results showed that both p53 mutation and
low FBXW7 expression were correlated with poor prognosis in
clinical GC cases. FBXW7 was demonstrated to be regulated by miRNA.
miR-223 in GC cell line SGC7901 were able to decrease FBXW7 mRNA
expression and reduced cellular apoptosis and increased
proliferation and invasion in vitro (74). Eto et al (75) reported that miR-223/FBXW7 pathway
affected the GC sensitivity to the biological targeted drug
trastuzumab.
Human β-TrCP, was first identified in 1998 as a
binding partner of HIV-1 Vpu protein in a yeast two-hybrid
screening. The role of β-TrCP in cancers is tissue-dependent.
β-TrCP mainly acts as an oncoprotein, but in some situations as a
tumor suppressor, depending on the function of the targeted
substrates. Two main substrates of β-TrCP are IκB in the NF-κB
pathway and β-catenin in Wnt pathway (76,77).
IκB, inhibitor of NF-κB, functions as a tumor suppressor. β-catenin
has been identified to be upregulated in various types of human
cancer, and is always correlated with poor prognosis and short
survival (78,79). IκB and β-catenin exert antagonistic
functions, thus making it hard for them to be ideal drug targets.
The development of β-TrCP inhibitors might be a feasible
therapeutic approach for NF-κB-associated human disease. β-TrCP
also participates in cell adhesion and migration (80). β-TrCP is a member of the SCF family
with both oncogenic and tumor suppressor properties. Taken
together, β-TrCP might have a greater role as an oncogenic protein
than as a tumor suppressor in cancer.
Most examples of F-box protein degradation are
linked to the cell cycle, but the regulation of FBXL5 is unique in
this regard. SKP1-CUL1-FBXL5 mediates the degradation of iron
regulatory protein 2 (IRP2) under conditions of high iron levels in
eukaryotic cells (83,84). IRP2 is a key event for the
maintenance of an appropriate intracellular concentration of iron.
Liver-specific deletion of FBXL5 resulted in deregulation of both
hepatic and systemic iron homeostasis, leading to the development
of steatohepatitis (85).
Gradually, the role of FBXL5 in cancer is being clarified. Dragoi
et al (86) demonstrated
that depletion of FBXL5 did not alter the level of E-cadherin mRNA
significantly but increased E-cadherin protein level. These results
suggested that FBXL5 affected E-cadherin expression at the
post-transcriptional level. FBXL5 was associated with
epithelial-to-mesenchymal transition (EMT) and metastatic prowess.
FBXL5 negatively modulated human single-strand DNA binding protein
1 (hSSB1) for destruction to regulate DNA damage response in lung
cancer cell lines and clinical lung cancer samples (87). FBXL5 overexpression in GC cells
mediated the ubiquitination and degradation of cortactin (88). Cortactin was proved to be involved
in the regulation of cell migration and cell-cell junction
(89) and was reported to increase
cancer cells migration and invasion (90). Further studies are required to
reveal the exact molecular mechanisms and signaling pathways
through which FBXL5 modulates GC cell migration and invasion.
FBXO31, is located in chromosome 16q24.3, and plays
a crucial role in DNA damage response (91,92),
neuronal morphogenesis (93) and
tumorigenesis (94–97). FBXO31 was first identified as a
candidate tumor suppressor in breast cancer (97). A well-characterized substrate of
FBXO31 is cyclin D1, which was identified in melanoma cell line
SK-MEL-28 (91). However, a study
from Kogo et al showed conflicting results in esophageal
squamous cell carcinoma (ESCC) (95). They found that the expression level
of FBXO31 was positively associated with tumor invasion depth and
clinical stage. They revealed concordant expression rather than
inverse correlation between cyclin D1 and FBXO31 in ESCC cells,
which indicated that cyclin D1 was not a candidate substrate in
ESCC. Therefore, the expression and function of FBXO31 is still
controversial. In this context, Zhang et al (96) detected the expression of FBXO31 in
GC. They demonstrated that the expression of FBXO31 was
significantly decreased in GC (82/90) and the decreased expression
of FBXO31 was associated with tumor size, tumor infiltration,
clinical grade and patients’ prognosis in GC. Furthermore, Zhang
et al (96) found that
FBXO31 could inhibit colony formation and induces G1 phase arrest
in GC cells possibly by targeting cyclin D1 through the
ubiquitin-proteasome pathway. This result is consistent with Santra
et al (91). Furthermore,
overexpression of FBXO31 dramatically inhibited xenograft tumor
growth in nude mice. Zhang et al (96) highlighted the regulatory mechanism
of miRNAs to FBXO31, they first revealed that the increased
expression of miR-20a and miR-17 in GC inhibited the expression of
FBXO31. In spite of all the work on FBXO31, the expression profile,
clinical significance, biological functions and the regulation
mechanism of FBXO31 in different cancers is still unclear. It is
necessary to investigate other mechanism, such as DNA methylation,
loss of heterozygosity (LOH) contributing to the downregulation in
GC tissue.
The UPS is an important mechanism regulating protein
degradation. Extensive study of UPS has led to a better
understanding of the molecular mechanism by which E3 ligases
regulate cellular processes and of how their deregulations
contribute to GC. FBPs, as the main component of E3 ligases, have a
critical role in digestive system cancers (10). Substantial current research is
focused on identifying new FBPs and the substrates that binds to
each FBP. Only PBPs, SKP2, FBXW7 and βTRCP, showed strong clinical
relevance by virtue of its ability to regulate key substrates in
the initiation and development of GC, but more research is needed
to apply the results to clinic. According to our experimental
results, the expression of FBXO31 in ESCC has been proved to be
related to drug resistance in vitro. Therefore, FBPs have
great potential as biomarkers to monitor clinical indexes.
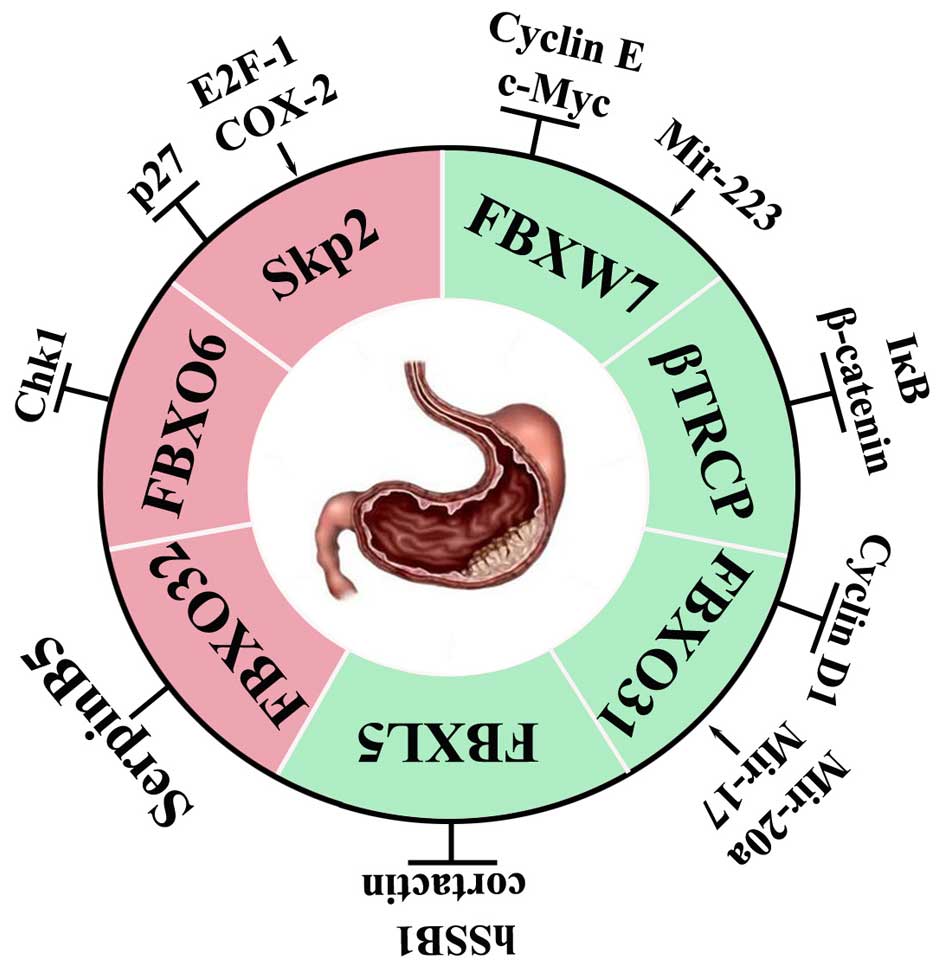
However, all the current discovered functions are
just the tip of the iceberg and the range of the FBP-dependent
processes will increase. SKP2, the authentic oncoprotein, has shown
to be involved in helicobacter pylori related chronic
gastritis. However, whether patients with FBXW7 mutation are more
likely to be infected with helicobacter pylori is still
unknown. FBXL5, FBXO6, FBXO31 and FBXO32 have begun to show their
roles in participation in the initiation and progression of GC.
Unfortunately, the specific roles of these proteins in GC still
need to be further investigated. Future study will be directed to
functional characterization of all 69 F-box proteins and their
corresponding substrates to thoroughly elucidate the biological
processes in GC. Here we have provided a useful conceptual
framework for understanding the complex FBPs related to GC
(Fig. 4). The recent therapeutic
success of the proteasome inhibitor bortezomib has expedited the
efforts to develop inhibitors of other components in the UPS. FBPs,
such as those described in this review, are promising targets or
biomarkers for GC in theory, however, FBPs suffers from several
challenges. For example, one F-box protein can promote the
degradation of either oncogenes or tumor suppressor genes at the
same cell, the therapeutic outcome of inhibitors has to be cell
context-dependent. Therefore, a better mechanistic understanding of
the many uncharacterized FBPs both with regard to their substrates
and the posttranslational modifications will be a hotpot of future
research.
J.G. and J.-R.H. are grateful to the Fundamental
Research Funds for the Central Universities of Central South
University (No. 2015zzts119).
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng L, Wu C, Xi P, Zhu M, Zhang L, Chen
S, Li X, Gu J and Zheng Y: The survival and the long-term trends of
patients with gastric cancer in Shanghai, China. BMC Cancer.
14:3002014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu HS and Xiao HS: MicroRNAs as potential
biomarkers for gastric cancer. World J Gastroenterol.
20:12007–12017. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yakirevich E and Resnick MB: Pathology of
gastric cancer and its precursor lesions. Gastroenterol Clin North
Am. 42:261–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yen H-CS, Xu Q, Chou DM, Zhao Z and
Elledge SJ: Global protein stability profiling in mammalian cells.
Science. 322:918–923. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao Y and Sun Y: Cullin-RING Ligases as
attractive anti-cancer targets. Curr Pharm Des. 19:3215–3225. 2013.
View Article : Google Scholar
|
|
7
|
Deshaies RJ and Joazeiro CA: RING domain
E3 ubiquitin ligases. Annu Rev Biochem. 78:399–434. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reed SI: Ratchets and clocks: The cell
cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol.
4:855–864. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peters J-M: The anaphase promoting
complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell
Biol. 7:644–656. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gong J, Lv L and Huo J: Roles of F-box
proteins in human digestive system tumors (Review). Int J Oncol.
45:2199–2207. 2014.PubMed/NCBI
|
|
11
|
Bai C, Sen P, Hofmann K, Ma L, Goebl M,
Harper JW and Elledge SJ: SKP1 connects cell cycle regulators to
the ubiquitin proteolysis machinery through a novel motif, the
F-box. Cell. 86:263–274. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morgan DO: Principles of CDK regulation.
Nature. 374:131–134. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sherr CJ and Roberts JM: Inhibitors of
mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149–1163.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hershko D, Bornstein G, Ben-Izhak O,
Carrano A, Pagano M, Krausz MM and Hershko A: Inverse relation
between levels of p27(Kip1) and of its ubiquitin ligase subunit
Skp2 in colorectal carcinomas. Cancer. 91:1745–1751. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fukuchi M, Masuda N, Nakajima M, Fukai Y,
Miyazaki T, Kato H and Kuwano H: Inverse correlation between
expression levels of p27 and the ubiquitin ligase subunit Skp2 in
early esophageal squamous cell carcinoma. Anticancer Res.
24B:777–783. 2004.
|
|
16
|
Yang G, Ayala G, De Marzo A, Tian W,
Frolov A, Wheeler TM, Thompson TC and Harper JW: Elevated Skp2
protein expression in human prostate cancer: Association with loss
of the cyclin-dependent kinase inhibitor p27 and PTEN and with
reduced recurrence-free survival. Clin Cancer Res. 8:3419–3426.
2002.PubMed/NCBI
|
|
17
|
Traub F, Mengel M, Lück HJ, Kreipe HH and
von Wasielewski R: Prognostic impact of Skp2 and p27 in human
breast cancer. Breast Cancer Res Treat. 99:185–191. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li JQ, Wu F, Masaki T, Kubo A, Fujita J,
Dixon DA, Beauchamp RD, Ishida T, Kuriyama S and Imaida K:
Correlation of Skp2 with carcinogenesis, invasion, metastasis, and
prognosis in colorectal tumors. Int J Oncol. 25:87–95.
2004.PubMed/NCBI
|
|
19
|
Hershko DD: Oncogenic properties and
prognostic implications of the ubiquitin ligase Skp2 in cancer.
Cancer. 112:1415–1424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Inuzuka H, Gao D, Finley LW, Yang W, Wan
L, Fukushima H, Chin YR, Zhai B, Shaik S, Lau AW, et al:
Acetylation-dependent regulation of Skp2 function. Cell.
150:179–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Inuzuka H, Zhong J, Liu P, Sarkar
FH, Sun Y and Wei W: Identification of acetylation-dependent
regulatory mechanisms that govern the oncogenic functions of Skp2.
Oncotarget. 3:1294–1300. 2012.PubMed/NCBI
|
|
22
|
da Silva GN, de Camargo EA, Sávio AL and
Salvadori DM: MRE11A and SKP2 genes are associated with the
increased cytotoxicity induced by the synergistic effects of
cisplatin and gemcitabine in bladder cancer cells. Mol Biol Rep.
41:4613–4621. 2014.PubMed/NCBI
|
|
23
|
Wang J, Huang Y, Guan Z, Zhang JL, Su HK,
Zhang W, Yue CF, Yan M, Guan S and Liu QQ: E3-ligase Skp2 predicts
poor prognosis and maintains cancer stem cell pool in
nasopharyngeal carcinoma. Oncotarget. 5:5591–5601. 2014.PubMed/NCBI
|
|
24
|
Pascal LE and Wang Z: Virtual drug design:
Skp1-Skp2 inhibition targets cancer stem cells. Asian J Androl.
15:717–718. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chan CH, Morrow JK, Li CF, Gao Y, Jin G,
Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, et al: Pharmacological
inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem
cell traits and cancer progression. Cell. 154:556–568. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Totary-Jain H, Sanoudou D, Dautriche CN,
Schneller H, Zambrana L and Marks AR: Rapamycin resistance is
linked to defective regulation of Skp2. Cancer Res. 72:1836–1843.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang XC, Tian LL, Tian J and Jiang XY:
Overexpression of SKP2 promotes the radiation resistance of
esophageal squamous cell carcinoma. Radiat Res. 177:52–58. 2012.
View Article : Google Scholar
|
|
28
|
Masuda TA, Inoue H, Sonoda H, Mine S,
Yoshikawa Y, Nakayama K, Nakayama K and Mori M: Clinical and
biological significance of S-phase kinase-associated protein 2
(Skp2) gene expression in gastric carcinoma: Modulation of
malignant phenotype by Skp2 overexpression, possibly via p27
proteolysis. Cancer Res. 62:3819–3825. 2002.PubMed/NCBI
|
|
29
|
Ma XM, Liu Y, Guo JW, Liu JH and Zuo LF:
Relation of overexpression of S phase kinase-associated protein 2
with reduced expression of p27 and PTEN in human gastric carcinoma.
World J Gastroenterol. 11:6716–6721. 2005.
|
|
30
|
Honjo S, Kase S, Osaki M, Ardyanto TD,
Kaibara N and Ito H: COX-2 correlates with F-box protein, Skp2
expression and prognosis in human gastric carcinoma. Int J Oncol.
26:353–360. 2005.PubMed/NCBI
|
|
31
|
Yan LH, Wang XT, Yang J, Kong FB, Lian C,
Wei WY, Luo W, Xie YB and Xiao Q: Reversal of multidrug resistance
in gastric cancer cells by E2F-1 downregulation in vitro and in
vivo. J Cell Biochem. 115:34–41. 2014. View Article : Google Scholar
|
|
32
|
Trimarchi JM and Lees JA: Sibling rivalry
in the E2F family. Nat Rev Mol Cell Biol. 3:11–20. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wei Z, Jiang X, Liu F, Qiao H, Zhou B,
Zhai B, Zhang L, Zhang X, Han L, Jiang H, et al: Downregulation of
Skp2 inhibits the growth and metastasis of gastric cancer cells in
vitro and in vivo. Tumour Biol. 34:181–192. 2013. View Article : Google Scholar
|
|
34
|
Eguchi H, Herschenhous N, Kuzushita N and
Moss SF: Helicobacter pylori increases proteasome-mediated
degradation of p27(kip1) in gastric epithelial cells. Cancer Res.
63:4739–4746. 2003.PubMed/NCBI
|
|
35
|
Kim SS, Meitner P, Konkin TA, Cho YS,
Resnick MB and Moss SF: Altered expression of Skp2, c-Myc and p27
proteins but not mRNA after H. pylori eradication in chronic
gastritis. Mod Pathol. 19:49–58. 2006. View Article : Google Scholar
|
|
36
|
Kim SS, Cho YS, Kim HK, Shin OR, Chae HS,
Choi MG and Chung IS: The effect of rosiglitazone on the cell
proliferation and the expressions of p27 and skp2 in helicobacter
pylori infected human gastric epithelial cells. Korean J
Gastroenterol. 55:225–231. 2010.(In Korean). View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Skaar JR, Pagan JK and Pagano M:
Mechanisms and function of substrate recruitment by F-box proteins.
Nat Rev Mol Cell Biol. 14:369–381. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yoshida Y, Chiba T, Tokunaga F, Kawasaki
H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K, et al:
E3 ubiquitin ligase that recognizes sugar chains. Nature.
418:438–442. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshida Y, Tokunaga F, Chiba T, Iwai K,
Tanaka K and Tai T: Fbs2 is a new member of the E3 ubiquitin ligase
family that recognizes sugar chains. J Biol Chem. 278:43877–43884.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Janoueix-Lerosey I, Novikov E, Monteiro M,
Gruel N, Schleiermacher G, Loriod B, Nguyen C and Delattre O: Gene
expression profiling of 1p35-36 genes in neuroblastoma. Oncogene.
23:5912–5922. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Merry C, Fu K, Wang J, Yeh IJ and Zhang Y:
Targeting the checkpoint kinase Chk1 in cancer therapy. Cell Cycle.
9:279–283. 2010. View Article : Google Scholar
|
|
42
|
Zhang YW, Brognard J, Coughlin C, You Z,
Dolled-Filhart M, Aslanian A, Manning G, Abraham RT and Hunter T:
The F box protein Fbx6 regulates Chk1 stability and cellular
sensitivity to replication stress. Mol Cell. 35:442–453. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hwang GW, Du K, Takahashi T and Naganuma
A: Inhibition of F-box protein FBXO6 gene expression by RNA
interference enhances cadmium toxicity in HEK293 cells. J Toxicol
Sci. 36:847–849. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang L, Hou Y, Wang M, Wu B and Li N: A
study on the functions of ubiquitin metabolic system related gene
FBG2 in gastric cancer cell line. J Exp Clin Cancer Res. 28:782009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gomes MD, Lecker SH, Jagoe RT, Navon A and
Goldberg AL: Atrogin-1, a muscle-specific F-box protein highly
expressed during muscle atrophy. Proc Natl Acad Sci USA.
98:14440–14445. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mastrocola R, Reffo P, Penna F,
Tomasinelli CE, Boccuzzi G, Baccino FM, Aragno M and Costelli P:
Muscle wasting in diabetic and in tumor-bearing rats: Role of
oxidative stress. Free Radic Biol Med. 44:584–593. 2008. View Article : Google Scholar
|
|
47
|
Costelli P, Muscaritoli M, Bossola M,
Penna F, Reffo P, Bonetto A, Busquets S, Bonelli G, Lopez-Soriano
FJ, Doglietto GB, et al: IGF-1 is downregulated in experimental
cancer cachexia. Am J Physiol Regul Integr Comp Physiol.
291:R674–R683. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
D’Orlando C, Marzetti E, François S,
Lorenzi M, Conti V, di Stasio E, Rosa F, Brunelli S, Doglietto GB,
Pacelli F, et al: Gastric cancer does not affect the expression of
atrophy-related genes in human skeletal muscle. Muscle Nerve.
49:528–533. 2014. View Article : Google Scholar
|
|
49
|
Bonetto A, Penna F, Aversa Z, Mercantini
P, Baccino FM, Costelli P, Ziparo V, Lucia S, Rossi Fanelli F and
Muscaritoli M: Early changes of muscle insulin-like growth factor-1
and myostatin gene expression in gastric cancer patients. Muscle
Nerve. 48:387–392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zou Z, Anisowicz A, Hendrix MJ, Thor A,
Neveu M, Sheng S, Rafidi K, Seftor E and Sager R: Maspin, a serpin
with tumor-suppressing activity in human mammary epithelial cells.
Science. 263:526–529. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lei KF, Liu BY, Wang YF, Chen XH, Yu BQ,
Guo Y and Zhu ZG: SerpinB5 interacts with KHDRBS3 and FBXO32 in
gastric cancer cells. Oncol Rep. 26:1115–1120. 2011.PubMed/NCBI
|
|
52
|
Hartwell LH, Mortimer RK, Culotti J and
Culotti M: Genetic control of the cell division cycle in yeast: V.
Genetic analysis of cdc mutants. Genetics. 74:267–286.
1973.PubMed/NCBI
|
|
53
|
Sionov RV, Netzer E and Shaulian E:
Differential regulation of FBXW7 isoforms by various stress
stimuli. Cell Cycle. 12:3547–3554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Crusio KM, King B, Reavie LB and Aifantis
I: The ubiquitous nature of cancer: The role of the SCF(Fbw7)
complex in development and transformation. Oncogene. 29:4865–4873.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Matsumoto A, Onoyama I and Nakayama KI:
Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or
p53-dependent manner. Biochem Biophys Res Commun. 350:114–119.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Welcker M, Orian A, Grim JE, Eisenman RN
and Clurman BE: A nucleolar isoform of the Fbw7 ubiquitin ligase
regulates c-Myc and cell size. Curr Biol. 14:1852–1857. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Minella AC, Welcker M and Clurman BE: Ras
activity regulates cyclin E degradation by the Fbw7 pathway. Proc
Natl Acad Sci USA. 102:9649–9654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yada M, Hatakeyama S, Kamura T, Nishiyama
M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K and
Nakayama KI: Phosphorylation-dependent degradation of c-Myc is
mediated by the F-box protein Fbw7. EMBO J. 23:2116–2125. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hoeck JD, Jandke A, Blake SM, Nye E,
Spencer-Dene B, Brandner S and Behrens A: Fbw7 controls neural stem
cell differentiation and progenitor apoptosis via Notch and c-Jun.
Nat Neurosci. 13:1365–1372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rocher-Ros V, Marco S, Mao JH, Gines S,
Metzger D, Chambon P, Balmain A and Saura CA: Presenilin modulates
EGFR signaling and cell transformation by regulating the ubiquitin
ligase Fbw7. Oncogene. 29:2950–2961. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Inuzuka H, Shaik S, Onoyama I, Gao D,
Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al:
SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for
ubiquitylation and destruction. Nature. 471:104–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tan M, Zhao Y, Kim SJ, Liu M, Jia L,
Saunders TL, Zhu Y and Sun Y: SAG/RBX2/ROC2 E3 ubiquitin ligase is
essential for vascular and neural development by targeting NF1 for
degradation. Dev Cell. 21:1062–1076. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fukushima H, Matsumoto A, Inuzuka H, Zhai
B, Lau AW, Wan L, Gao D, Shaik S, Yuan M, Gygi SP, et al: SCF(Fbw7)
modulates the NFκB signaling pathway by targeting NFκB2 for
ubiquitination and destruction. Cell Rep. 1:434–443. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Busino L, Millman SE, Scotto L, Kyratsous
CA, Basrur V, O’Connor O, Hoffmann A, Elenitoba-Johnson KS and
Pagano M: Fbxw7α- and GSK3-mediated degradation of p100 is a
pro-survival mechanism in multiple myeloma. Nat Cell Biol.
14:375–385. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lochab S, Pal P, Kapoor I, Kanaujiya JK,
Sanyal S, Behre G and Trivedi AK: E3 ubiquitin ligase Fbw7
negatively regulates granulocytic differentiation by targeting
G-CSFR for degradation. Biochim Biophys Acta. 1833:2639–2652. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tu K, Yang W, Li C, Zheng X, Lu Z, Guo C,
Yao Y and Liu Q: Fbxw7 is an independent prognostic marker and
induces apoptosis and growth arrest by regulating YAP abundance in
hepatocellular carcinoma. Mol Cancer. 13:1102014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Huhn S, Bevier M, Pardini B, Naccarati A,
Vodickova L, Novotny J, Vodicka P, Hemminki K and Försti A:
Colorectal cancer risk and patients’ survival: Influence of
polymorphisms in genes somatically mutated in colorectal tumors.
Cancer Causes Control. 25:759–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Brim H, Abu-Asab MS, Nouraie M, Salazar J,
Deleo J, Razjouyan H, Mokarram P, Schaffer AA, Naghibhossaini F and
Ashktorab H: An integrative CGH, MSI and candidate genes
methylation analysis of colorectal tumors. PLoS One. 9:e821852014.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Babaei-Jadidi R, Li N, Saadeddin A,
Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R,
Abuzinadah M, Davis H, et al: FBXW7 influences murine intestinal
homeostasis and cancer, targeting Notch, Jun, and DEK for
degradation. J Exp Med. 208:295–312. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Davis H, Lewis A, Behrens A and Tomlinson
I: Investigation of the atypical FBXW7 mutation spectrum in human
tumours by conditional expression of a heterozygous propellor tip
missense allele in the mouse intestines. Gut. 63:792–799. 2014.
View Article : Google Scholar :
|
|
71
|
Grim JE, Knoblaugh SE, Guthrie KA, Hagar
A, Swanger J, Hespelt J, Delrow JJ, Small T, Grady WM, Nakayama KI,
et al: Fbw7 and p53 cooperatively suppress advanced and
chromosomally unstable intestinal cancer. Mol Cell Biol.
32:2160–2167. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Milne AN, Leguit R, Corver WE, Morsink FH,
Polak M, de Leng WW, Carvalho R and Offerhaus GJ: Loss of CDC4/
FBXW7 in gastric carcinoma. Cell Oncol. 32:347–359. 2010.
|
|
73
|
Yokobori T, Mimori K, Iwatsuki M, Ishii H,
Onoyama I, Fukagawa T, Kuwano H, Nakayama KI and Mori M:
p53-altered FBXW7 expression determines poor prognosis in gastric
cancer cases. Cancer Res. 69:3788–3794. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li J, Guo Y, Liang X, Sun M, Wang G, De W
and Wu W: MicroRNA-223 functions as an oncogene in human gastric
cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol.
138:763–774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Eto K, Iwatsuki M, Watanabe M, Ishimoto T,
Ida S, Imamura Y, Iwagami S, Baba Y, Sakamoto Y, Miyamoto Y, et al:
The sensitivity of gastric cancer to trastuzumab is regulated by
the miR-223/FBXW7 pathway. Int J Cancer. 136:1537–1545. 2015.
View Article : Google Scholar
|
|
76
|
Shirane M, Hatakeyama S, Hattori K and
Nakayama K and Nakayama K: Common pathway for the ubiquitination of
IkappaBalpha, IkappaBbeta, and IkappaBepsilon mediated by the F-box
protein FWD1. J Biol Chem. 274:28169–28174. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Spiegelman VS, Slaga TJ, Pagano M,
Minamoto T, Ronai Z and Fuchs SY: Wnt/beta-catenin signaling
induces the expression and activity of betaTrCP ubiquitin ligase
receptor. Mol Cell. 5:877–882. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang N, Wei P, Gong A, Chiu WT, Lee HT,
Colman H, Huang H, Xue J, Liu M, Wang Y, et al: FoxM1 promotes
β-catenin nuclear localization and controls Wnt target-gene
expression and glioma tumorigenesis. Cancer Cell. 20:427–442. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Mokkapati S, Niopek K, Huang L, Cunniff
KJ, Ruteshouser EC, deCaestecker M, Finegold MJ and Huff V:
β-catenin activation in a novel liver progenitor cell type is
sufficient to cause hepatocellular carcinoma and hepatoblastoma.
Cancer Res. 74:4515–4525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM
and Zhou BP: Stabilization of snail by NF-kappaB is required for
inflammation-induced cell migration and invasion. Cancer Cell.
15:416–428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Saitoh T and Katoh M: Expression profiles
of betaTRCP1 and betaTRCP2, and mutation analysis of betaTRCP2 in
gastric cancer. Int J Oncol. 18:959–964. 2001.PubMed/NCBI
|
|
82
|
Kim CJ, Song JH, Cho YG, Kim YS, Kim SY,
Nam SW, Yoo NJ, Lee JY and Park WS: Somatic mutations of the
beta-TrCP gene in gastric cancer. APMIS. 115:127–133. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Vashisht AA, Zumbrennen KB, Huang X,
Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M,
Sangfelt O, et al: Control of iron homeostasis by an iron-regulated
ubiquitin ligase. Science. 326:718–721. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Salahudeen AA, Thompson JW, Ruiz JC, Ma
HW, Kinch LN, Li Q, Grishin NV and Bruick RK: An E3 ligase
possessing an iron-responsive hemerythrin domain is a regulator of
iron homeostasis. Science. 326:722–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Moroishi T, Nishiyama M, Takeda Y, Iwai K
and Nakayama KI: The FBXL5-IRP2 axis is integral to control of iron
metabolism in vivo. Cell Metab. 14:339–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Dragoi AM, Swiss R, Gao B and Agaisse H:
Novel strategies to enforce an epithelial phenotype in mesenchymal
cells. Cancer Res. 74:3659–3672. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chen ZW, Liu B, Tang NW, Xu YH, Ye XY, Li
ZM, Niu XM, Shen SP, Lu S and Xu L: FBXL5-mediated degradation of
single-stranded DNA-binding protein hSSB1 controls DNA damage
response. Nucleic Acids Res. 42:11560–11569. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Cen G, Ding HH, Liu B and Wu WD: FBXL5
targets cortactin for ubiquitination-mediated destruction to
regulate gastric cancer cell migration. Tumour Biol. 35:8633–8638.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wu H and Parsons JT: Cortactin, an
80/85-kilodalton pp60src substrate, is a filamentous actin-binding
protein enriched in the cell cortex. J Cell Biol. 120:1417–1426.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
MacGrath SM and Koleske AJ: Cortactin in
cell migration and cancer at a glance. J Cell Sci. 125:1621–1626.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Santra MK, Wajapeyee N and Green MR: F-box
protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest
after DNA damage. Nature. 459:722–725. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Jia L and Sun Y: F-box proteins FBXO31 and
FBX4 in regulation of cyclin D1 degradation upon DNA damage.
Pigment Cell Melanoma Res. 22:518–519. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Vadhvani M, Schwedhelm-Domeyer N,
Mukherjee C and Stegmüller J: The centrosomal E3 ubiquitin ligase
FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS
One. 8:e575302013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Huang HL, Zheng WL, Zhao R, Zhang B and Ma
WL: FBXO31 is down-regulated and may function as a tumor suppressor
in hepatocellular carcinoma. Oncol Rep. 24:715–720. 2010.PubMed/NCBI
|
|
95
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: FBXO31 determines poor prognosis in esophageal squamous
cell carcinoma. Int J Oncol. 39:155–159. 2011.PubMed/NCBI
|
|
96
|
Zhang X, Kong Y, Xu X, Xing H, Zhang Y,
Han F, Li W, Yang Q, Zeng J, Jia J, et al: F-box protein FBXO31 is
down-regulated in gastric cancer and negatively regulated by miR-17
and miR-20a. Oncotarget. 5:6178–6190. 2014.PubMed/NCBI
|
|
97
|
Kumar R, Neilsen PM, Crawford J, McKirdy
R, Lee J, Powell JA, Saif Z, Martin JM, Lombaerts M, Cornelisse CJ,
et al: FBXO31 is the chromosome 16q24.3 senescence gene, a
candidate breast tumor suppressor, and a component of an SCF
complex. Cancer Res. 65:11304–11313. 2005. View Article : Google Scholar : PubMed/NCBI
|