Introduction
Natural killer cells (NK cells), a subset of
approximately 5–15% of the peripheral blood lymphocyte population,
based on the expression levels of surface markers, are classified
into two subpopulations with different functions,
CD3−CD56dimCD16+ (90%) and
CD3− CD56brightCD16− (10%)
(1–4). NK cells typically lack
antigen-specific cell surface receptors (5), consistent with the fact that NK cells
do not require tumor-specific antigen recognition when killing
tumor cells. This advantage of NK cells compared with T and B cells
has led many scientists to study the immunotherapeutic effects of
NK cells (6,7). In recent years, NK cell-based
adoptive immunotherapy to treat cancer has become increasingly
important (6,8–10).
However, progress in the development of this therapy is hindered by
the lack of a feasible method to expand large numbers of highly
cytotoxic NK cells ex vivo (11). Previous studies have examined
various methods to amplify NK cells. Using a starting population of
CD56+-selected PBMCs stimulated with cytokines alone,
Klingemann et al gained only 5- to 40-fold NK expansion
(12), whereas Berg et al
used the same starting population but added EBV-LCL as feeder cells
and obtained 300- to 930-fold NK expansion in 15 days, with 98% NK
purity (13). Other studies have
utilized unseparated PBMCs as the initial expanded population, but
typically also include feeder cells, such as the K562 cell line
expressing IL-15 and 4-1BBL on the surface (14), and K562 cell line with
membrane-bound IL-21 (15),
achieving tens of thousands of times greater expansion of NK cells
with nearly 80% purity. In addition, Carlens et al obtained
193-fold expansion, but only 55% NK purity, using unseparated PBMCs
stimulated with cytokines alone without feeder cells (16).
4-1BBL, also known as CD137L or TNFSF9, a member of
the tumor necrosis factor ligand superfamily, is expressed in B
cells, dendritic cells, activated T cells and macrophages. This
protein binds to the receptor 4-1BB, generating a co-stimulatory
signal for T cell activation and expansion (17,18).
Although 4-1BB is typically expressed on activated but not resting
T-cells (19,20), this receptor has also been detected
on NK cells (21). The functional
domain of 4-1BBL is located in the extracellular domain (22,23),
and soluble 4-1BBL has no activity compared with the natural
membrane-bound form (24).
Interleukin-21 (IL-21) has been recently identified, and the
functional effects of this protein on the immune system have been
recently studied (25). IL-21, a
secreted protein, mediates the proliferation, differentiation and
antitumorigenicity of cells via binding to a composite receptor
comprising the private receptor IL-21R, which is also expressed on
a variety of B, T, NK cell lines, and the common receptor γ-chain,
which is also shared by IL-2, IL-4, IL-7, IL-9, IL-13 and IL-15
(26–29). Wang et al used these two
receptor molecules, which were expressed on the feeder cells, to
stimulate the expansion of NK cells and achieved significant cell
expansion (30). However, a
disadvantage of this method is the effective removal of the feeder
cells from the activated NK cells.
In the present study, we introduced a novel strategy
involving solid phase recombinant human 4-1BBL and IL-21 to expand
NK cells using the PBMCs from healthy donors, and this method does
not require separation from other cells. Here, the recombinant
human extracellular domains of 4-1BBL and IL-21 were, respectively
expressed in E. coli as BirA-tagged recombinant proteins.
After biotinylation, the two protein factors were bound to
streptavidin-labeled Dynabeads to constitute the objective
irritant. This objective irritant was used to stimulate human PBMCs
three times in 21 days, achieving considerable NK fold-expansion,
high purity, and nearly 100% potent cytotoxicity of NK cells.
Moreover, the advantage of removing the beads co-cultured with NK
cells using a magnetic separation rack prevents the contamination
of other components of the co-culture system, such as feeder
cells.
Materials and methods
Materials and reagents
The restriction enzymes, including BamHI,
NdeI, XhoI, and Pfu DNA polymerase used in the
present study were purchased from Tiangen Biotech Co., Ltd.
(Beijing, China). The T4 DNA ligase and the ligase buffer were
purchased from Fermentas. All primers used in the PCR experiments
were synthesized at Invitrogen Co., Ltd. (Shanghai, China), and the
recombinant plasmids were also sequenced at Invitrogen. The protein
marker was purchased from GenStar Biosolutions Co., Ltd. (Beijing,
China). Streptavidin was purchased from Tianjin Heowns, Biochemical
Technology Co., Ltd. (Tianjin, China). Dynabeads® M-280
Streptavidin was purchased from Invitrogen Co., Ltd. (Shanghai,
China). X-VIVO™ 15 medium was purchased from Sartorius Stedim
Biotech GmbH. The FITC-conjugated anti-human CD3 monoclonal
antibody was purchased from Biolegend, and APC-conjugated
anti-human CD56 monoclonal antibody was purchased from Miltenyi
Biotec. Human AB serum was purchased from Andygene Co. (USA). Fetal
bovine serum (FBS) was purchased from Molecular Devices (USA).
Plasmids, strains, cells and cell
lines
The plasmids pGEX-6p-1 and pET-28a and E.
coli strains DH5α and BL21 (DE3) were obtained from our
laboratory. We isolated PBMCs from the concentrated human leukocyte
blood of healthy donors provided at the Beijing Red Cross Blood
Center. The cell line K562 transformed into luciferase plasmid
(K562-luc cells) was also obtained from our laboratory.
Plasmid constructs
To generate the plasmid construct
pGEX-6p-1-h4-1BBL-BirA-tag, we fused the BirA-tag gene to the
N-terminus of the 184-amino acid extracellular domain of human
4-1BBL. There is a (GGGGS)3 linker between h4-1BBL and BirA-tag.
Overlap PCR was used for target gene amplification, and we inserted
the target gene between the BamHI and XhoI
restriction sites in the plasmid pGEX-6p-1. In addition, we
amplified the human IL-21 gene without the signal peptide and
generated a C-terminal BirA-tagged fusion protein, with a linker
bridging IL-21 and the BirA-tag, as described above. The target
gene was also amplified using overlap PCR, however, the target gene
was inserted between the NdeI and XhoI restriction
sites in pET-28a to generate the recombinant plasmid
pET-28a-hIL-21-BirA-tag. The primers used to PCR amplify the target
genes are shown in Table I.
 | Table IPrimers used to PCR amplify the
target genes. |
Table I
Primers used to PCR amplify the
target genes.
| h4-1BBL-BirA-tag
(660 bp) |
| RP1 | 5′-CCGCTCGAGTTATTCCGACCTCGGTGAAGGGAGTCCGG-3′ |
| FP2 |
5′-AGCGGCGGCGGGGGCAGTGGAGGAGGGGGATCACGCGAGGGTCCCGAGCTTTCG-3′ |
| FP3 |
5′-AGGCCCAGAAGATCGAGTGGCACGGAGGCGGAGGTAGCGGCGGCGGGGGCAGTGGAGG-3′ |
| FP4 | 5′-CGCGGATCCGGTCTGAACGACATATTTGAGGCCCAGAAGATCGAGTGGCACGG-3′ |
| hIL-21-BirA-tag
(504 bp) |
| FP5 |
5′-GAATTCCATATGCAAGATCGCCACATGATTAGAATGC-3′ |
| RP6 |
5′-CGAGCCCCCGCCGCCCGAACCCCCACCACCGGAATCTTCACTTCCGTGTGTTC-3′ |
| RP7 |
5′-GAAGATGTCGTTGAGGCCCGATCCCCCTCCTCCCGAGCCCCCGCCGCCCGAACC-3′ |
| RP8 | 5′-CCGCTCGAGTTAGTGCCATTCGATTTTTTGTGCTTCGAAGATGTCGTTGAGGCC-3′ |
| BirA-tag |
5′-GGTCTGAACGACATATTTGAGGCCCAGAAGATCGAGTGGCAC-3′ |
| Linker |
5′-GGTGGTGGGGGTTCGGGCGGCGGGGGCTCGGGAGGAGGGGGATCG-3′ |
Expression and purification of
protein
To express the proteins h4-1BBL and hIL-21, the
recombinant plasmids pGEX-6p-1-h4-1BBL-BirA-tag and
pET-28a-hIL-21-BirA-tag were transformed into E. coli strain
BL21 (DE3) through heat shock transformation. To express h4-1BBL,
the cells were grown for 8 h at 37°C from a single colony in 5 ml
LB medium supplemented with 50 μg/ml of ampicillin. Subsequently,
the cells were diluted 100-fold into 300 ml of fresh medium
(supplemented as above) and 40-fold into two liters of fresh medium
(supplemented as above) and incubated at 37°C with shaking at 200
rpm until reaching an OD600 of 0.6. Protein expression was induced
upon the addition of 1 mM IPTG. The cells were grown at 25°C for 5
h and subsequently harvested through centrifugation. To express
hIL-21, the single colony propagation was performed as described
for h4-1BBL, but the medium was supplemented with 50 μg/ml of
kanamycin. When the OD600 reached 0.8, the culture was induced with
1 mM IPTG and subsequently incubated overnight at 37°C. The protein
expression was examined using 12% sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE).
To purify h4-1BBL-BirA-tag protein, the harvested
cells were lysed using ultrasonication. The GST-h4-1BBL fusion
protein was harvested from the supernatant using Glutathione
Sepharose 4 Fast Flow according to the manufacturer’s instructions.
The GST-tag was cleaved using precision protease (PSP) on the
column at 4°C overnight, and the target protein was purified
through fast protein liquid chromatography (FPLC). To purify
hIL-21-BirA-tag protein, we obtained the inclusion bodies after
lysing the cells, and refolded the proteins using the rapid
dilution method. Subsequently, we concentrated the protein from the
refolding buffer using Ultrafiltration concentration tubes (10
kDa), followed by FPLC.
Protein biotinylation
Several solutions were used for protein
biotinylation: biotin-protein ligase (3 mg/ml BirA enzyme);
Solution A (0.5 M bicine buffer, pH 8.3); Solution B (100 mM ATP,
100 mM MgOAc, and 200 mM biotin); extra D-biotin (500 mM biotin);
pepstatin (2 mg/ml in DMSO) and leupeptin (2 mg/ml in
dH2O). The biotinylation reaction system included 700 μl
of target protein (1–2 mg), 100 μl Solution A, 100 μl Solution B,
100 μl extra D-biotin, 50 μl BirA enzyme, 2 μl pepstatin and 2 μl
leupeptin. The reaction was incubated overnight at 25°C. For
biotinylated h4-1BBL-BirA-tag (h4-1BBL-BirA-tag-biotin) protein was
directly purified through FPLC. However, the biotinylated
hIL-21-BirA-tag (hIL-21-BirA-tag-biotin) protein was purified using
Ni-NTA, followed by FPLC.
SA electrophoretic mobility shift
assay
We used 3 EP tubes (1.5 ml) to conduct this assay.
Two tubes were filled with either moderate biotinylated protein or
SA, and equal amounts of both biotinylated protein and SA were
added to the third tube. PBS was added to the tubes containing only
protein or SA to obtain equal reaction volumes in all tubes.
Subsequently, the tubes were incubated on ice for 0.5–1 h after
mixing. The shift results were assessed using 12% SDS-PAGE.
Bead-protein ligation reactions and
detection of ligation efficiency
Appropriate amounts of Dynabeads M-280 Streptavidin
were washed with PBST (0.05% Tween-20 in PBS) and PBS, and
resuspended in 500 μl PBS. Excessive and equimolar
h4-1BBL-BirA-tag-biotin and hIL-21-BirA-tag-biotin proteins were
mixed with the prepared beads. The ligation reaction system was
performed on a rotary mixer at 4°C for at least 6 h. Mouse anti-His
IgG-PE was added to a sample of the ligation reaction to detect the
ligation efficiency of hIL-21-BirA-tag-biotin and the beads, while
rabbit anti-His IgG-FITC was added to detect the ligation
efficiency of h4-1BBL-BirA-tag-biotin and the beads, and mouse
anti-His IgG-PE and rabbit anti-His IgG-FITC were consecutively
used to detect the ligation efficiency of the two proteins and the
beads. The same batch beads were used to repeat the experiments
described above, but phosphate-buffered saline (PBS) was
substituted for mouse anti-His IgG-PE, 4-1BB-His-tag protein and
rabbit anti-His IgG-FITC as important controls to exclude the
influence of non-specific ligation.
NK cell expansion
PBMCs were isolated from human concentrated
leukocyte blood, resuspended in X-VIVO™ 15 medium supplemented with
10% human AB serum, 10 mmol/l HEPES, 1 mmol/l sodium pyruvate, 2
mmol/l L-glutamine, 1% MEM NEAA, 1% penicillin-streptomycin and 100
IU/ml IL-2, seeded at a density of 1×106 cells/ml onto
24-well plates at 1 ml/well, and subsequently incubated at 37°C in
5% CO2. The following experimental groups were
designated: two proteins (4-1BBL-IL-21-beads), single fixed and
single soluble proteins (4-1BBL-beads+sIL-21, s4-1BBL+IL-21-beads),
the mixture of the two soluble proteins (s4-1BBL+sIL-21), and the
non-irritant control group. Each group was prepared with two
parallel wells. On the first day, each group was co-incubated with
PBMCs at 3:1. Repeated stimulation was performed weekly for 3
weeks. We should appropriately enlarge the culture system to ensure
the viable cell density of 0.5–2×106/ml during this
stimulus.
Cell counting and flow cytometric
analysis
After stimulation, we collected the cells of each
group, separated the beads from the culture system with magnetic
separation rack, and stained the cells with an LDS/PI mixture for
cell counting using Guava flow cytometry. Approximately
5×105 cells were washed twice with PBS, followed by
incubation with the appropriate antibodies (FITC-conjugated
anti-human CD3 or APC-conjugated anti-human CD56) in a 100-μl
reaction volume for 30 min at 4°C. Subsequently, the cells were
washed twice as described above and resuspended in 300 μl of PBS.
The data were acquired using a FACSCalibur flow cytometer (BD
Biosciences) and analyzed using FlowJo software (Ashland, OR,
USA).
In vitro killing experiment
K562-luc cells cultured in the basic RPMI-1640
medium supplemented with 10% FBS, 1% penicillin-streptomycin were
used as the target cells. The target cells were diluted to
1×105 cells/ml in basic medium and plated onto 96-well
plates at 100 μl/well. Subsequently, effector cells were added to
the 96-well plates and incubated with the target cells for 4 h in
5% CO2 at 37°C at ratios of 8:1, 4:1, 2:1, and 1:1, with
4 parallel experiment groups for each effector/target (E/T) ratio.
Subsequently, we added luciferin and measured the total
fluorescence in each well. The percent cytotoxicity was calculated
using the following formula: cytotoxicity (%) = (control group
fluorescence value-experimental group fluorescence value)/control
group fluorescence value × 100%.
Results
The expression of BirA-tagged 4-1BBL and
IL-21 recombinant proteins
4-1BBL (4-1BB ligand or CD137 ligand) is a member of
the tumor necrosis factor (TNF) ligand family. This 254-amino acid
protein contains a 28-amino acid cytoplasmic domain, a 21-amino
acid transmembrane domain, and a 205-amino acid extracellular
domain (31). In the present
study, the extracellular domain was cloned and the recombinant
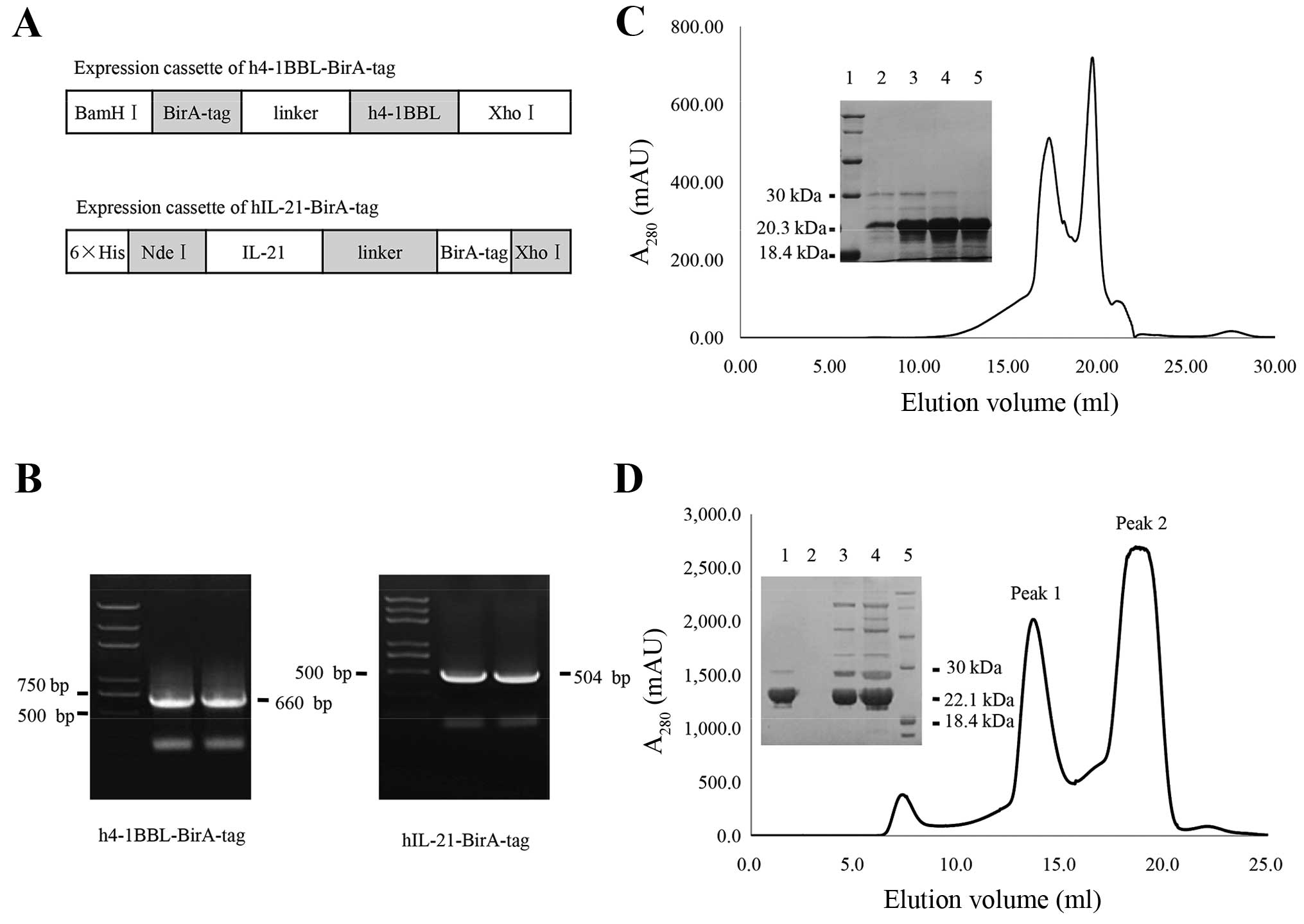
expression cassette is shown in Fig.
1A. Because 4-1BBL is a type II transmembrane protein (31), we generated an N-terminal
BirA-tagged fusion protein to ensure that the protein functions
normally, and the theoretical molecular weight of the recombinant
protein is ~22.1 kDa. However, for human IL-21, a secreted protein
belonging to the IL-15/IL-2 family, there is no significant
functional difference between the N- and C-terminal fusion proteins
(32). Therefore, we generated a
C-terminal BirA-tagged fusion for this protein (Fig. 1A), the theoretical molecular weight
of which is ~20.3 kDa. These constructs were confirmed through gel
electrophoresis of the PCR products for targeted cassettes
(Fig. 1B).
The constructs were expressed in E. coli
strain BL21 (DE3), and subsequently the h4-1BBL-BirA-tag, which
forms a dimer in solution, was cut using PSP, and the recombinant
proteins were purified using size-exclusion chromatography
(Fig. 1D). The hIL-21-BirA-tag
recombinant protein was obtained through denaturation and refolding
(33,34) and subsequently purified using
size-exclusion chromatography (Fig.
1C).
Biotinylation efficiency of 4-1BBL and
IL-21
Following biotinylation, h4-1BBL-BirA-tag-biotin
protein was purified using FPLC (Fig.
1D). SDS-PAGE was used to identify the purified target protein,
and the results showed that peak 1 was h4-1BBL-BirA-tag-biotin
protein (Fig. 1D). However, for
hIL-21-BirA-tag-biotin protein, the target protein could not be
purified from the other proteins in the first experiment (data not
shown). His-tag in hIL-21-BirA-tag protein was used to purify this
protein using Ni-NTA beads, followed by further purification
through FPLC (data not shown). Next, we used the SA electrophoretic
mobility shift assay (35) to
measure the biotinylation efficiency of these two proteins. The
data revealed that these two types of protein were nearly 100%
shifted with SA after conjugation, suggesting that both proteins
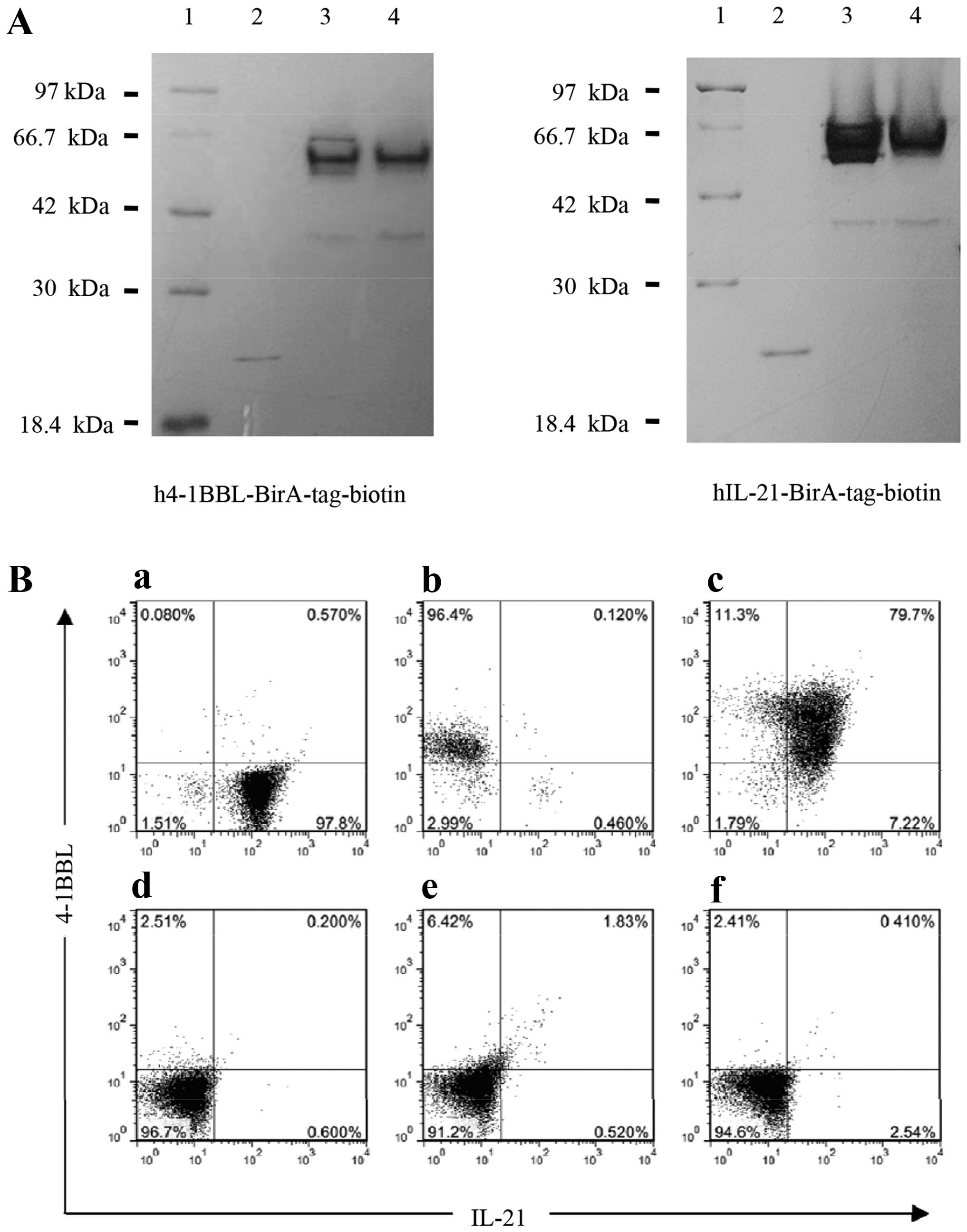
were markedly and efficiently biotinylated (Fig. 2A). To ensure that the stimulation
experiment could be effectively conducted, the bead-protein
ligation efficiency was also examined. The FACS analysis
demonstrated that each of two proteins could efficiently ligate
with the beads, with an efficiency of 96.4% for
h4-1BBL-BirA-tag-beads (Fig. 2B-b)
and 97.8% for hIL-21-BirA-tag-beads (Fig. 2B-a). Moreover, these proteins could
efficiently be simultaneously bound to the beads, with ~80%
efficiency (Fig. 2B-c).
Immobilized 4-1BBL and IL-21 induced the
expansion of NK cells in vitro
After stimulation for 3 weeks, the expanded cells
were harvested, purified from the culture system with magnetic
separation rack and stained with LDS/PI to evaluate cell viability
using flow cytometry. Appropriate amounts of the cells from each
group were sampled to detect the expression of the NK cell surface
molecular markers CD3 and CD56 via flow cytometry and to determine
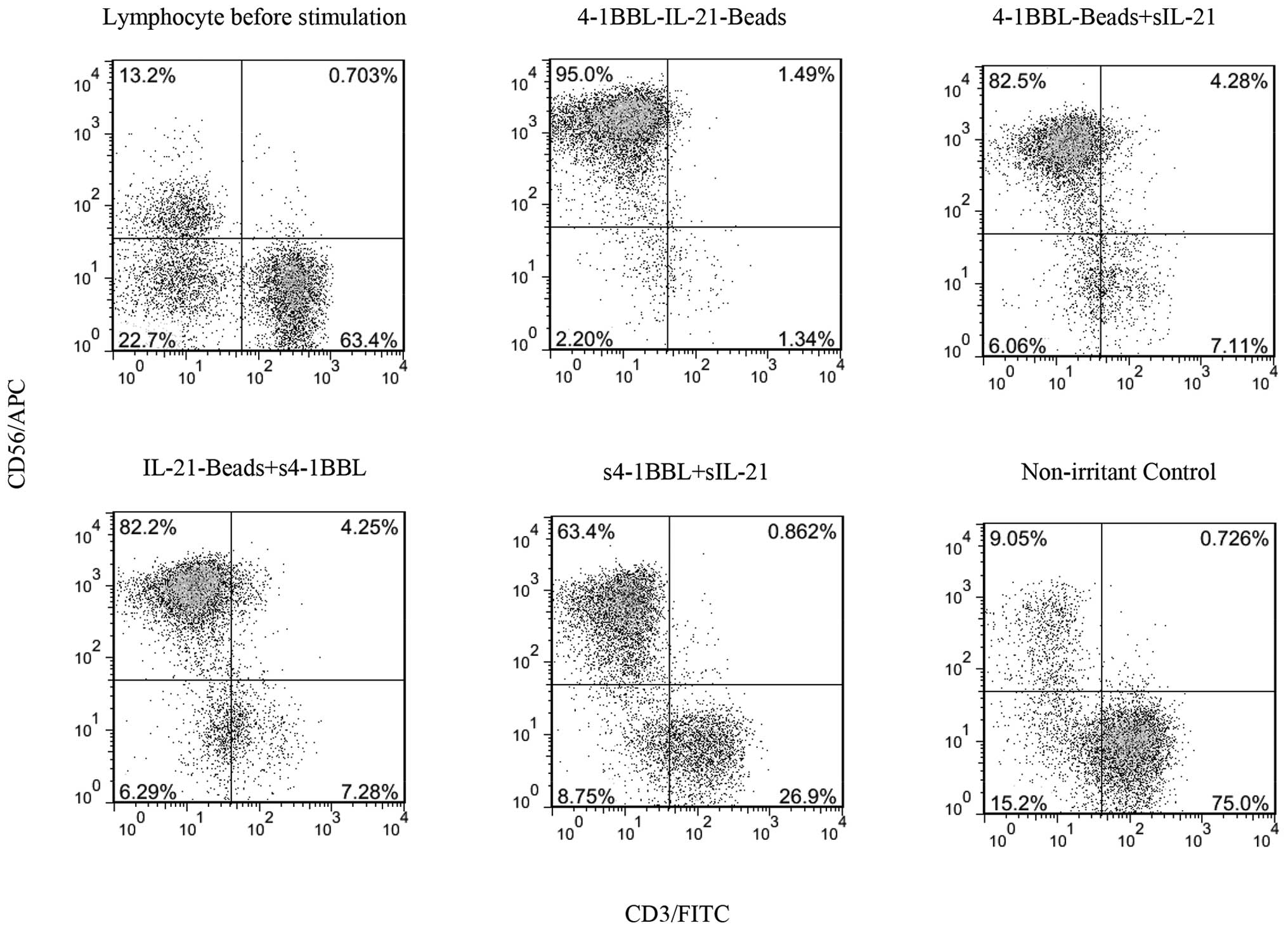
the NK cell purity in each group. The lymphocyte profile before
stimulation is shown in Fig. 3,
indicating that the sample was obtained from a normal source, with
10–15% NK cells, consistent with previous studies, and this
information was used to calculate the fold-expansion of each subset
in each group of cells. Compared with the NK purity before
stimulation in the non-irritant control group, the NK purity in
each of the stimulation groups was markedly increased, particularly
in the 4-1BBL-IL-21-beads stimulation group, obtaining 95% NK
purity, which was the highest level detected among all groups
(Fig. 3). This result suggests
that 4-1BBL-IL-21-beads could elicit high-purity NK cells.
Furthermore, according to the statistical and calculated results
shown in Table II, the
4-1BBL-IL-21-bead-stimulation group achieved the highest
fold-expansion of NK cells, with ~140-fold higher expansion than
the other groups. Moreover, the NKT cells in each group showed a
small increase in the ratio of lymphocytes, but a high proportion
of T cells was no longer observed (Fig. 3), potentially reflecting the fact
that 4-1BBL-IL-21-beads were beneficial for the expansion of NK
cells but not T cells. Thus, 4-1BBL-IL-21-beads induced significant
NK cell expansion.
 | Table IIThe expansion of NK cells induced
through 4-1BBL and IL-21 immobilized beads. |
Table II
The expansion of NK cells induced
through 4-1BBL and IL-21 immobilized beads.
| After stimulation
for 3 weeks | Before
stimulation | 4-1BBL-
IL-21-beads | 4-1BBL-beads | IL-21-beads
+s4-1BBL | sIL-21
+s4-1BBL | Non-irritant
control |
|---|
| The fold expansion
of total PBMC numbers | -/- | 19.477 | 1.975 | 1.983 | 3.175 | 2.579 |
| NK cell |
| Purity (%) | 13.2 | 95.00 | 82.50 | 82.20 | 63.40 | 9.05 |
| Folda | -/- | 140.17 | 12.34 | 12.35 | 15.24 | 1.77 |
| NKT cell |
| Purity (%) | 0.703 | 1.49 | 4.28 | 4.25 | 0.862 | 0.726 |
| Fold | -/- | 41.28 | 12.02 | 11.98 | 3.89 | 2.66 |
| T cell |
| Purity (%) | 63.4 | 1.34 | 7.11 | 7.28 | 26.9 | 75.0 |
| Fold | -/- | 0.41 | 0.22 | 0.22 | 1.34 | 3.05 |
Expanded NK cells were effectively
activated through immobilized 4-1BBL and IL-21
Luciferase quantitative assay, a simple and
sensitive method, has been increasingly and widely used to measure
the cytolytic effects of various effector cells (36–38).
In the present study, we used this method to test the cytotoxicity
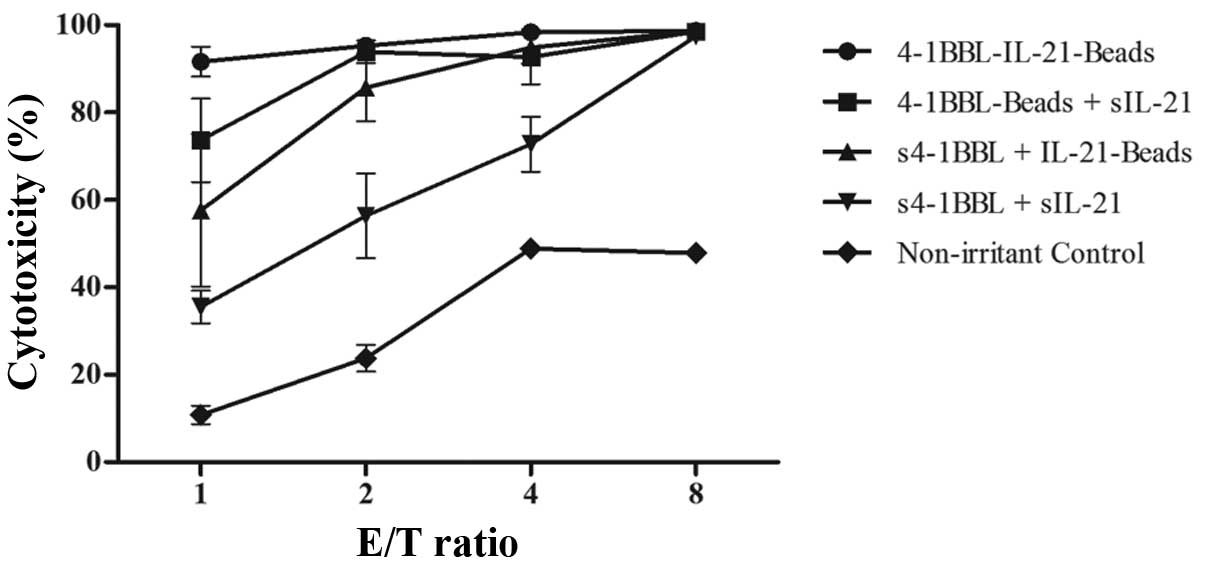
of stimulated cells against K562-luc cells. The results showed that
compared with the control group and other stimulation groups,
stimulation with 4-1BBL-IL-21-beads achieved the best cytotoxicity
against K562-luc cells at each E/T ratio, and this cell group
exhibited high cytotoxicity even at an E/T ratio of 1:1, with
nearly 100% cytotoxicity at an E/T ratio of 8:1 (Fig. 4). In contrast, the cells under
single fixed and single soluble cytokine stimulation or mixed
soluble cytokine stimulation showed higher killing activity than
the cells in the control group, and cytotoxicity increased with
increasing E/T ratio. Moreover, the control group cells also
exhibited killing activity in relation to the small number of NK
cells in the control group in the last graph of Fig. 3. Briefly, the cells stimulated with
4-1BBL-IL-21-beads showed the highest cytotoxicity against the
K562-luc cells, indicating that NK cells could be highly activated
using 4-1BBL-IL-21-beads.
Discussion
The use of adoptive NK cell transfer to treat
malignant tumors has gained increasing attention in the field of
cell-based therapy and has made rapid progress in research,
demonstrating promising clinically curative effects. However, it is
difficult to obtain large-scale clinical-grade NK cells from
donors; therefore, to overcome this limitation, many researchers
have developed methods to amplify NK cells ex vivo (4). In the present study, we used solid
phase cytokines of recombinant human IL-21 and 4-1BBL to
proliferate the NK cells in PBMCs obtained from healthy donors. We
not only achieved considerable NK cell expansion with high purity
and cytotoxicity but also realized the effective segregation of the
irritants and the expanded NK cells, an outstanding point and
innovation compared with previously published methods.
In recent years, a large number of expanded NK cells
have been acquired from the blood of healthy individuals, showing
190-fold NK expansions induced through anti-CD3 and IL-2 after 21
days, but with low NK cell purity of only 55% (16). In addition, 1,000-fold NK
expansions have also been obtained using the K562 cell line
expressing 4-1BBL on the surface and weekly stimulation for 21
days. However, the initiating cells were negatively selected NK
cells from PBMCs, and the feeder cell line could not be
efficaciously removed from the final cultures (39). Moreover, the number of NK cells
obtained from the blood of patients with myeloma showed on average
1,600-fold expansion after 20 days, but the cytotoxicity of these
cells against K562 cells was nearly 60% at a 10:1 E/T ratio and
<10% at a 1:1 E/T ratio (40).
In addition, we selected a three-week stimulation
cycle based on previous studies using similar cytokines to
stimulate NK cells (14,15,30,39),
and the cytotoxicity of the expanded NK cells was reported to peak
after 3–5 weeks, followed by a decline, although with continuously
growing numbers of NK cells (30).
Thus, the differences in experimental conditions between the
present study and previous reports suggest that it is worthwhile to
examine the expansion and killing activity of NK cell through
prolonged stimulation and increasing stimulation times.
Based on previous studies, we proposed that
expanding NK cells with cultures containing feeder cells is more
effective compared with cultures containing cytokines alone
(4). For example, K562 cells with
membrane-bound IL-15 and 4-1BBL were used as feeder cells to expand
NK cells and generate large numbers of highly cytotoxic NK cells
(41). Although the safeguards,
such as using cultures of irradiated K562 cells and monitoring the
cell growth and DNA synthesis rate, were provided, the inability to
deplete the feeder cell line provided difficulties for clinical
treatment. To overcome the problem of T and NKT cell contamination
and overgrowth, the initial NK cells was purified from PBMCs
(13,42,43),
but such purification procedure was time-consuming, arduous and
costly. In this study NK cells in PBMCs without purification were
used which certainly is a significant advantage.
Finally, an assessment of the anti-tumor activity of
expanded NK cells in vivo is needed to meet clinical
requirements although high purity and substantial fold-expansion of
NK cells was achieved, and potent cytotoxicity was verified in
vitro in the present study.
Acknowledgements
This study was supported by a grant from the Basic
Research Program of China (973 Program, no. 2013CB531502), the
Ministry of Science and Technology of China (S&T major Program,
no. 2012ZX1004701-001-002), and the National Nature Science
Foundation of China (nos. 31370889, 31400754 and 31170829).
References
|
1
|
Nagler A, Lanier LL, Cwirla S and Phillips
JH: Comparative studies of human FcRIII-positive and negative
natural killer cells. J Immunol. 143:3183–3191. 1989.PubMed/NCBI
|
|
2
|
Lanier LL, Le AM, Civin CI, Loken MR and
Phillips JH: The relationship of CD16 (Leu-11) and Leu-19 (NKH-1)
antigen expression on human peripheral blood NK cells and cytotoxic
T lymphocytes. J Immunol. 136:4480–4486. 1986.PubMed/NCBI
|
|
3
|
Davies JO, Stringaris K, Barrett AJ and
Rezvani K: Opportunities and limitations of natural killer cells as
adoptive therapy for malignant disease. Cytotherapy. 16:1453–1466.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Childs RW and Berg M: Bringing natural
killer cells to the clinic: Ex vivo manipulation. Hematology (Am
Soc Hematol Educ Program). 2013:234–246. 2013. View Article : Google Scholar
|
|
5
|
Vivier E, Raulet DH, Moretta A, Caligiuri
MA, Zitvogel L, Lanier LL, Yokoyama WM and Ugolini S: Innate or
adaptive immunity? The example of natural killer cells. Science.
331:44–49. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ljunggren H-G and Malmberg K-J: Prospects
for the use of NK cells in immunotherapy of human cancer. Nat Rev
Immunol. 7:329–339. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Srivastava S, Lundqvist A and Childs RW:
Natural killer cell immunotherapy for cancer: A new hope.
Cytotherapy. 10:775–783. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miller JS, Soignier Y,
Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, McKenna
D, Le C, Defor TE, Burns LJ, et al: Successful adoptive transfer
and in vivo expansion of human haploidentical NK cells in patients
with cancer. Blood. 105:3051–3057. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Klingemann H and Boissel L: Targeted
cellular therapy with natural killer cells. Horm Metab Res.
40:122–125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sutlu T, Stellan B, Gilljam M, Quezada HC,
Nahi H, Gahrton G and Alici E: Clinical-grade, large-scale,
feeder-free expansion of highly active human natural killer cells
for adoptive immunotherapy using an automated bioreactor.
Cytotherapy. 12:1044–1055. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Herberman RB: Cancer immunotherapy with
natural killer cells. Seminars in Oncology. Elsevier; pp. 27–30.
2002, View Article : Google Scholar
|
|
12
|
Klingemann H-G and Martinson J: Ex vivo
expansion of natural killer cells for clinical applications.
Cytotherapy. 6:15–22. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Berg M, Lundqvist A, McCoy P Jr, Samsel L,
Fan Y, Tawab A and Childs R: Clinical-grade ex vivo-expanded human
natural killer cells up-regulate activating receptors and death
receptor ligands and have enhanced cytolytic activity against tumor
cells. Cytotherapy. 11:341–355. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Imai C, Iwamoto S and Campana D: Genetic
modification of primary natural killer cells overcomes inhibitory
signals and induces specific killing of leukemic cells. Blood.
106:376–383. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Denman CJ, Senyukov VV, Somanchi SS,
Phatarpekar PV, Kopp LM, Johnson JL, Singh H, Hurton L, Maiti SN,
Huls MH, et al: Membrane-bound IL-21 promotes sustained ex vivo
proliferation of human natural killer cells. PLoS One.
7:e302642012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carlens S, Gilljam M, Chambers BJ, Aschan
J, Guven H, Ljunggren HG, Christensson B and Dilber MS: A new
method for in vitro expansion of cytotoxic human
CD3-CD56+ natural killer cells. Hum Immunol.
62:1092–1098. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smith CA, Farrah T and Goodwin RG: The TNF
receptor superfamily of cellular and viral proteins: Activation,
costimulation, and death. Cell. 76:959–962. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
DeBenedette MA, Shahinian A, Mak TW and
Watts TH: Costimulation of CD28− T lymphocytes by 4-1BB
ligand. J Immunol. 158:551–559. 1997.PubMed/NCBI
|
|
19
|
Hurtado JC, Kim SH, Pollok KE, Lee ZH and
Kwon BS: Potential role of 4-1BB in T cell activation. Comparison
with the costimulatory molecule CD28. J Immunol. 155:3360–3367.
1995.PubMed/NCBI
|
|
20
|
Vinay DS and Kwon BS: 4-1BB signaling
beyond T cells. Cell Mol Immunol. 8:281–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Melero I, Johnston JV, Shufford WW,
Mittler RS and Chen L: NK1.1 cells express 4-1BB (CDw137)
costimulatory molecule and are required for tumor immunity elicited
by anti-4-1BB monoclonal antibodies. Cell Immunol. 190:167–172.
1998. View Article : Google Scholar
|
|
22
|
Wu C, Guo H, Wang Y, Gao Y, Zhu Z and Du
Z: Extracellular domain of human 4-1BBL enhanced the function of
cytotoxic T-lymphocyte induced by dendritic cell. Cell Immunol.
271:118–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo H, Jiang W, Liu W, Gao Y, Yang M, Zhou
Y, Wang J, Qi J, Cheng X, Zhu Z, et al: Extracellular domain of
4-1BBL enhanced the antitumoral efficacy of peripheral blood
lymphocytes mediated by anti-CD3 × anti-Pgp bispecific diabody
against human multidrug-resistant leukemia. Cell Immunol.
251:102–108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rabu C, Quéméner A, Jacques Y,
Echasserieau K, Vusio P and Lang F: Production of recombinant human
trimeric CD137L (4-1BBL). Cross-linking is essential to its T cell
co-stimulation activity. J Biol Chem. 280:41472–41481. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parrish-Novak J, Dillon SR, Nelson A,
Hammond A, Sprecher C, Gross JA, Johnston J, Madden K, Xu W, West
J, et al: Interleukin 21 and its receptor are involved in NK cell
expansion and regulation of lymphocyte function. Nature. 408:57–63.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parrish-Novak J, Foster DC, Holly RD and
Clegg CH: Interleukin-21 and the IL-21 receptor: Novel effectors of
NK and T cell responses. J Leukoc Biol. 72:856–863. 2002.PubMed/NCBI
|
|
27
|
Zeng R, Spolski R, Casas E, Zhu W, Levy DE
and Leonard WJ: The molecular basis of IL-21-mediated
proliferation. Blood. 109:4135–4142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burgess SJ, Marusina AI, Pathmanathan I,
Borrego F and Coligan JE: IL-21 down-regulates NKG2D/DAP10
expression on human NK and CD8+ T cells. J Immunol.
176:1490–1497. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mehta DS, Wurster AL and Grusby MJ:
Biology of IL-21 and the IL-21 receptor. Immunol Rev. 202:84–95.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Lee DA, Wang Y, Wang L, Yao Y, Lin
Z, Cheng J and Zhu S: Membrane-bound interleukin-21 and CD137
ligand induce functional human natural killer cells from peripheral
blood mononuclear cells through STAT-3 activation. Clin Exp
Immunol. 172:104–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goodwin RG, Din WS, Davis-Smith T,
Anderson DM, Gimpel SD, Sato TA, Maliszewski CR, Brannan CI,
Copeland NG, Jenkins NA, et al: Molecular cloning of a ligand for
the inducible T cell gene 4-1BB: A member of an emerging family of
cytokines with homology to tumor necrosis factor. Eur J Immunol.
23:2631–2641. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fa P, Zhang Z, Li J, Hu Z and Gao J:
Expression, purification and bioactivity evaluation of
streptavidin-tagged human interleukin-21 fusion protein. Nan Fang
Yi Ke Da Xue Xue Bao. 30:1240–1243. 12492010.(In Chinese).
|
|
33
|
Lee CM, McGuire H, Basten A, King C and
Christ D: Expression, purification and characterization of
recombinant interleukin-21. J Immunol Methods. 362:185–189. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Asano R, Kudo T, Makabe K, Tsumoto K and
Kumagai I: Antitumor activity of interleukin-21 prepared by novel
refolding procedure from inclusion bodies expressed in Escherichia
coli. FEBS Lett. 528:70–76. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Garboczi DN, Utz U, Ghosh P, Seth A, Kim
J, VanTienhoven EA, Biddison WE and Wiley DC: Assembly, specific
binding, and crystallization of a human TCR-alphabeta with an
antigenic Tax peptide from human T lymphotropic virus type 1 and
the class I MHC molecule HLA-A2. J Immunol. 157:5403–5410.
1996.PubMed/NCBI
|
|
36
|
Fu X, Tao L, Rivera A, Williamson S, Song
XT, Ahmed N and Zhang X: A simple and sensitive method for
measuring tumor-specific T cell cytotoxicity. PLoS One.
5:e118672010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brown CE, Wright CL, Naranjo A, Vishwanath
RP, Chang WC, Olivares S, Wagner JR, Bruins L, Raubitschek A,
Cooper LJ, et al: Biophotonic cytotoxicity assay for
high-throughput screening of cytolytic killing. J Immunol Methods.
297:39–52. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma J, Han H, Liu D, Li W, Feng H, Xue X,
Wu X, Niu G, Zhang G, Zhao Y, et al: HER2 as a promising target for
cytotoxicity T cells in human melanoma therapy. PLoS One.
8:e732612013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Cui Y, Voong N, et al: Activating
signals dominate inhibitory signals in CD137L/IL-15 activated
natural killer cells. J Immunother. 34:187–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Alici E, Sutlu T, Björkstrand B, Gilljam
M, Stellan B, Nahi H, Quezada HC, Gahrton G, Ljunggren HG and
Dilber MS: Autologous antitumor activity by NK cells expanded from
myeloma patients using GMP-compliant components. Blood.
111:3155–3162. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fujisaki H, Kakuda H, Shimasaki N, Imai C,
Ma J, Lockey T, Eldridge P, Leung WH and Campana D: Expansion of
highly cytotoxic human natural killer cells for cancer cell
therapy. Cancer Res. 69:4010–4017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lapteva N, Durett AG, Sun J, Rollins LA,
Huye LL, Fang J, Dandekar V, Mei Z, Jackson K, Vera J, et al:
Large-scale ex vivo expansion and characterization of natural
killer cells for clinical applications. Cytotherapy. 14:1131–1143.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luhm J, Brand J-M, Koritke P, Höppner M,
Kirchner H and Frohn C: Large-scale generation of natural killer
lymphocytes for clinical application. J Hematother Stem Cell Res.
11:651–657. 2002. View Article : Google Scholar : PubMed/NCBI
|