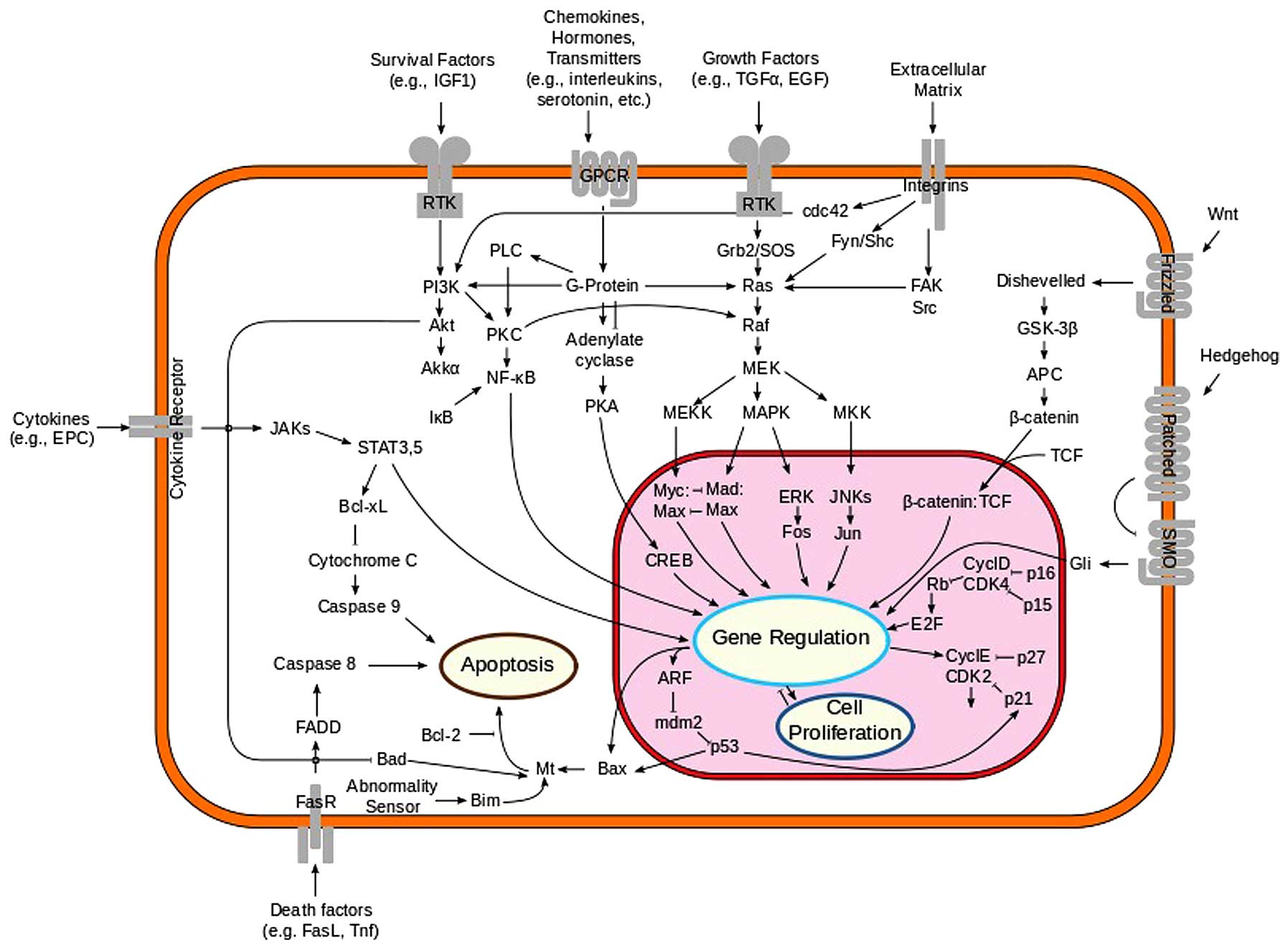
1. Introduction
Of the genes encoding the body plane/axis/design in
ontogenesis, and the stress-responses in adult life in the immense
numbers of life forms emerging in the Cambrian sea, nuclear factor
kappa B (NF-κB), STAT (signal transduction and transactivation),
and β-catenin-dependent transcriptional activators have gained
utmost and ever-lasting prominence. In the cnidarians, Toll-like
receptors activated NF-κB to function as a stress response inducing
cell survival pathway. The STATs are co-activators. In the advanced
mammalian vertebrate hosts (including Homo), the
β-catenin-based Wnt, and the NF-κB/Relp65 pathways have become the
principal inflammatory STAT-associated
proto-oncogenes/oncoproteins.
In the tunicate urochordate Ciona (C.
intestinalis) the Wnt/β-catenin pathway regulates body design,
first the trans-speciation of the swimming larva to the sessile
‘intestinal’ adult. In the decapitated hydra (H.
magnipapillata/viridis), the Wnt/β-catenin (with Notch and
PI3K) pathways regenerate a new head. In the other cnidarian,
Nematostella vectensis (Fig.
1) next to the Wnt pathway, the NF-κB/STAT pathway fulfills
essential physiological tasks. In the advanced multicellular
vertebrate hosts (including Homo), β-catenin has become one
of the most malignant oncoproteins, as it acts for example, in
colon carcinogenesis. In Wnt carcinogenesis, the cytoplasmic
‘β-catenin destroying complex’ fails to phosphorylate β-catenin,
thus, the complex does not mark it for ubiquitination. Intact
accumulating β-catenin is allowed to translocate from the cytoplasm
into the nucleus for the activation of oncoprotein promoter DNAs;
primarily that of tcf/TCF and lef/LEF (Fig. 2). Furthermore, the Wnt antagonist
Dickkopf tumor suppressor gene is forcefully eliminated. In the
human genome, the NF-κB/STAT pathway has assumed the role of a
proto-oncogene. When constitutively activated, it induces B cell
malignant lymphomas in the mediastinum and elsewhere. It is highly
active in the Reed-Sternberg cells of Hodgkin's lymphoma.
The rapidly expanding comb jelly ctenophores
(represented here by Mnemiopsis leidyi) (Fig. 3) operate a physiologically
defective cytoplasmic β-catenin destroying complex promoting
β-catenin's intranuclear transfer for activation of the
predecessors of those genes (tcf/lef), which have become
proto-oncogenes in the human genome. The ctenophore genomes do not
possess ancestral Dickkopf genes, which have become tumor
suppressors in the human genome. Furthermore, the ctenophore genome
operates a full set of sox/Sox genes/proteins, which sustain
pluripotent stem cells through the entire eukaryotic evolution, and
may convert into oncogenes in the human genome. The ctenophore
genome is a physiological oncogenome.
2. The cnidaria class anthozoa
Cell survival pathways
In 2006, it was surprising that the little
invertebrate basal animal, the burrowing sea anemone,
Nematostella vectensis (Fig.
1), carried an abundance of ‘human disease genes’ (1). The numbers and positions of the
introns in orthologous cnidarian and human genes reveal unusually
high concordance in 47 and 69%, respectively, surpassing those of
invertebrate insects and nematodes (2,3).
Most of these genes are stress responder metabolism regulators
(4,5). Prominent among them are oxidative
stress-activated receptors and others, the aryl hydrocarbon
receptor, AhR; and the hypoxia-inducible factor, HIF. Furthermore,
operational are the ligand-activated nuclear receptors, ancestral
predecessor of the hepatocyte nuclear factor; retinoic acid
receptor (RAR); signal transduction proteins; transcription factors
(including NF-κB, nuclear factor kappa B cell lymphoma); oxidizing,
reducing, conjugating enzymes; oxidative cytochrome P450 enzymes
(CYPs); and heat shock proteins (HSP), in several subfamilies. The
cyp gene progenitor of animals (not plants) created a tandem set of
duplicated genes, which utilized oxygen to modify substrate
structures. Plant cells have acquired cyp genes from marine animals
by horizontal transfer. Of the animal cyp gene clans,
Nematostella possesses up to 46. Cyp genes are absent in the
anaerobic Giardia, but are present in the aerobic
Saccharomyces. The sponge (Amphimedon) operates 35,
the amphioxus (Branchiostoma) has 236 cyp genes. Precursor
cyp genes existed interspersed with hox, wnt and arf (homeodomain
box; wingless; alternative reading frame GTPases) gene clusters
(6). Later in evolution, the P450
genes lying downstream from the AhRs, will assume prominent roles,
especially in hormone-dependent molecular carcinogenesis (breast
cancer) (vide infra).
There are active efflux pumps in cnidarians operated
by 64 ABC genes. The HSP (probably inherited from Archaeoglobus
fulgidus) have assumed the evolutionary role of chaperoning
oncoproteins in multicellular vertebrate hosts (including
Homo). The descendant enzymes of the original ATP-binding
cassettes (ABC) present in extant (and presumably ancient extinct)
zooxantellae, act as ATP-dependent efflux transporters in pumping
out molecular chemotherapeuticals from malignantly transformed
(cancer) cells in multicellular vertebrate mammalian (human) hosts
(as referenced in 7). Of
biotransformators, there are glutathione- and sulfotransferases,
but no acetyl transferases. Of antioxidant proteins, there are 6
superoxide dismutase (SOD) genes (4,5).
Virus-carrier algal symbionts
Some cnidarians and para-mecia accept the
intracellular invasion by chloroplast- and virus-carrier eukaryotic
chlorella green algae (Chlorella vulgaris; C.
variabilis) and live with them in symbiosis (8). In the case of the Florida strain of
Hydra virilis (in culture for over 20 years; fed with tiny
pieces of brine shrimp), and the chlorella green alga, the
symbiotic algal cells taken out from the hydra immediately
succumbed to the replication of the Hydra virilis dsDNA
chlorella virus 1, 2 and 3 (HVCV). The virus proved to be lytic to
the algal cells taken from the hydra, but not to the hydra cells.
Thin section of hydra cells viewed in transmission electron
microscopy failed to show viral particles. These original
observations remain cited in the literature of better technology;
recent metagenomic studies reveal the widely spread presence of
herpes-like viral agents in cnidarians, especially in
Porites corals, but also including Hydra and
Nematostella. Viral particles invade cells of the hosts and
the cells of their symbiotic zoochlorellae and zooxantellae
(9). The symbiotic life style and
the virology of zooxantella Symbiodinia algae have been
discussed (10). Proteobacteria
and Bacteriodetes and their phages colonize various species of
Hydra. Myoviridae and Podoviridae
Burkholderia; Syphoviridae Staphylococcus; and
Herpesviridae and Phycodnaviridae make up the
microbiome flora of the holobiont (11).
The genomes of Hydra magnipapillata and
Nematostella vectensis do not tolerate these symbiotic
relationships, thus, reject the potential symbionts (10,11).
Maltose-producing algae gained protection against cnidarian host
cell lysosomes. Intolerant (symbiosis-free) host cells mobilized
the ancient MAMP-PRR interaction (microbe-associated molecular
pattern; pattern recognition receptor) (10). In this interaction, host cells
release lectins that bind cell surface glycans (galactose; mannose)
of the invaders.
Proto-oncogene predecessor NF-κB
The activation of NF-κB is ancestrally TLR Toll-like
receptors-mediated (4). By its RHD
(reticuloendothelial viral antigen homology domain), the two
alleles Nv-NF-κB proteins bind their target-DNA. The consensus
sequence of the DNA recognition loop has either cysteine or serine
at position 6, and the serine variants are the more potent
activators. All other (including human) 440 aa NF-κB proteins have
cysteine in position six in the consensus (except for one of
Drosophila's four RHD proteins; there, the Relish RHD has
serine). The choanoflagellates (Monosiga) do not encode RHD
proteins, but the Amphimedon demosponge possesses one, thus,
predating the appearance of numerous RHDs after the triploblast
divergence. The human genome encodes twenty RHD proteins. Some
human RHD proteins may also undergo a Cys-to-Ser mutation (12,13).
Both in the cnidarian and in the human RHD, serine in position six
results in increased transactivational potency. It would be
interesting to know, if resting cells operate cysteins, vs.
constitutively activated cells (including the so-called malignantly
transformed cells), serins, in the DNA recognizing consensus loops
of their NF-κB RHDs. Ancestral ADAM (A disintegrin and
metallo-proteinase) exists in cnidarians; STATs, TNFα (tumor
necrosis factor), and its signaling proteins, SOD and
anti-apoptotic Bcl-proteins (B-cell lymphoma), are functional as
well. In the human genome encoding the mantle cell lymphoma, it
will be ADAM that activates the TNFα/NF-κB pathway (14) (vide infra). Promoters of
intranuclear translocation of NF-κB are interleukin IL-1,
Ca++-ionophore, PMA (phorbol myristate acetate) and
TNFα. NF-κB-inducing kinase (NIK) and the inhibitor of apoptosis
(IAP) are antagonistic to each other (15–17).
The cytoplasmic IκB kinase phosphorylates IκBα, thus initiating its
ubiquitination, that leads to the release of NF-κB. The dominant
form of the NF-κB family is its p50/p65 heterodimer, which is
acetylated. Deacetylase inhibitors (trichostatin) initiate and
potentiate the activation of NF-κB by TNFα, including its
intranuclear entry and attachment to its numerous pro-inflammatory
DNA targets. Among the NF-κB targets is the gene of its own
inhibitor IκBα. The cytoplasmic IκBα gene-product protein enters
the nucleus, binds the NF-κB proteins and leads them back
neutralized into the cytoplasm. Deacetylase inhibitors delay this
process and thus increase NF-κB activity. After its release from
its cytoplasmic inhibitor IκB, the intranuclear translocation and
mobility of the NF-κB/p50/p65 heterodimer is dependent on the
acetylation of five of its lysins; four of these residues contact
the targeted DNA domains. The transcriptional activity of the NF-κB
p65 subunit is further regulated by phosphorylation of its various
serine residues (18,19).
The Nematostella operates at least one STAT
pathway in co-existence with NF-κB (20,21).
The origins of the p53 pathway date back to the ancestors of
choanoflagellates and cnidarians (N. vectensis) (22,23).
However, the ancestral and extant choanoflagellate, cnidarian and
Spongiae genomes operate without an MDM (murine double minute)
pathway (24). The first signs of
MDM protein appear in the placozoan Trichoplax adhaerens and
in the sea squirt C. intestinalis (25). Since the placozoan T.
adhaerens possesses the anti-apoptotic MDM protein (25), presumably cnidarians (N.
vectensis) also have it, but no proof was found (23). Choanoflagellates (Monosiga)
and spongiae (Amphimedon) are devoid of mdm/MDM possessions
(24).
Cnidaria-related proto-oncogene
NF-κB/STAT in the human genome
NF-κB is TCR/TNFα- (T cell receptor), IL-1β-, or
mitochondrial stress-induced; calcineurin forces the release of
NF-κB from inhibitor IκB (26,27).
In the cellular transformation process,
loss-of-function-mutated iκbα/IκBα (inhibitory/inducing
kappaB kinase) permanently releases the NF-κB molecule. In
Reed-Sternberg (RS) cells of Hodgkin's lymphoma, among others, the
constitutively activated NF-κB gene/protein targets the promoter
DNAs of STAT5a, chemokine receptor CCR7, cyclin D2, and inhibitors
of apoptosis c-IAP2 and Bcl-xL (B cell lymphoma extra large).
Interleukins (IL-13 and IL-15), macrophage-derived chemokines, and
cytokines induced by NF-κB in the RS cells invoke the extraordinary
cellular infiltrates which surround the RS cells (28). In the human lymphoma cell line YT,
STAT3 and STAT5 are the promoters of the malignant transformation;
the tyrosine- and serine-phosphorylated STAT3 is constitutively
activated, but both STATs are anti-apoptotic. Neither the jak1, 2,
3/JAK (Janus kinase), nor IL-2 are drivers of STAT3, even though
STAT3 expresses domains for IL-2 binding. Antisense STAT3 and STAT5
blockade induced apoptosis. IL-2 could rescue the lymphoma cells
from apoptotic death consequential of STAT3 blockade, but not in
case of STAT5 blockade. STAT5 depletion resulted in reduced NF-κB
and TNFα activity. Helenalin, the inhibitor of DNA-binding by
NF-κB, induced lymphoma YT cell death. The YT lymphoma cells
resisted the blockade of the IL-2-induced pathways MAPK, PI3K, mTOR
(mitogen activated protein kinase; phosphatidyl inositol kinase 3;
mammalian target of rapamycin) (29). Common promoters of NF-κB-induced
diffuse large B-cell lymphomagenesis are the B cell activation
factor (BAF), constitutively expressed B-lymphocyte stimulator
(BLyS), autochthonous B-lymphocyte stimulator ligand, and the
NF-κB-inducing kinase, accompanied by the degradation of the
TNF-R-associated factor (30,31).
The EZH2-mediated (enhancer zeste homolog histone 3 lysine
methyltransferase, Drosophila), hotspot-mutated,
NF-κB-induced, mediastinal diffuse large B cell lymphomas are rare,
but highly malignant (32).
In human lymphoid cells undergoing malignant
transformation (RS cells of Hodgkin's disease, mediastinal diffuse
large B cell malignant lymphoma, certainly; mucosa-associated
lymphoid tissue MALT lymphoma, multiple myeloma, mycosis fungoides,
probably), the classical NF-κB pathway undergoes amplification, or
gain-of-function mutation, resulting in constitutive expression of
the molecule. NF-κB remains frequently co-activated with STATs. In
these entities, amplification of the NF-κB pathway is encoded by
the Rel locus at 2p14–15 (reticuloendotheliosis viral oncogene
homolog). TNFα phosphorylates serine Ser-471 in c-Rel for the
activation of NF-κB (33–36). The activation of NF-κB in T cells
is initiated by p65/RelA through triggering TCR/CD3 (cluster of
differentiation) and completed by TNFα, secreted either autocrine
or paracrine. TNFα induces the degradation of IκB kinase and
activates the wild-type c-Rel by phosphorylation of its Ser-471.
This stimulation of TNFα to activate NF-κB cannot occur in
Burkitt's lymphoma T cell lines (Jurkat) with mutated Ser-471
(S471N, asparagine replacing serine) (15). In diffuse large B-cell lymphoma,
NF-κB is constitutively activated by an NF-κB-inducing kinase; in
addition, BAF is also constitutively expressed (but no attention
was turned toward STAT) (37).
Furthermore, translocations and fusions of NIK
(NF-κB-inducing kinase) and NF-κB1, two genes and gene-product
proteins occur with the involvement of apoptosis inhibitor IAP
(another hallmark of malignant transformation) (16,17).
In evolutionarily more advanced cells (taken from mice or humans),
mitochondria-to-nucleus stress-signaling activates NF-κB/Rel.
Calcineurin-mediated inactivation by dephos-phorylation of IκBB
leads to the release of NF-κB (26,27).
However, the metazoan mitochondria had the time to develop genetic
variations after the divergence of advanced triploblastic metazoa
from the ancestral lineage of cnidarians (38,39).
In the cytoplasm, the IκB/NF-κB complex is heat-labile and can be
disrupted by HSP, thus, liberating NF-κB. In the nucleus, NF-κB can
target and induce the transcription of the IκB-genes (among
others). The newly synthesized IκB protein can bind intranuclear
DNA-bound NF-κB, and remove it from its bond. Exportin proteins
return NF-κB from nucleus to cytoplasm. This interplay was observed
in HIV-1-infected cells, and in HeLa cells (40). The Rel protein (reticuloendothelial
viral antigen) homology domain (RHD) of NvNF-κB is 50% identical
with the human p50/p52 Rel homology domain (RHD) proteins. Both the
cnidarian and the human NF-κB proteins interact with the Bcl-3 (B
cell lymphoma; cell cycle; anti-apoptosis) and c-Myc proteins
(12,13) (vide infra).
The ancient combined TNFα, STAT and NF-κB pathways
recur in the pathogenesis of human malignant lymphoma.
Phosphorylation of human cytoplasmic inhibitory IκB releases NF-κB
for its intranuclear transfer. In the nucleus, the NF-κB/RHD
complex activates directly or indirectly some of its 140 DNA
targets, including proinflammatory interleukins (IL-1, -2 and -6).
Point-mutated codon S525P serine to proline human rel/REL
oncogene/oncoprotein is active in human mediastinal B cell
lymphoma. The mutated REL protein transactivates the manganese SOD
promoter and renders transformed cells resistant to TNFα-induced
apoptosis (41).
The promoter of miR-155 expresses NF-κB-responsive
sites (42). Physiologically,
miR-155 positively regulates the immune response of CD8+
T cells (43). The promoter of the
human pro-inflammatory microRNA-155 cluster carries a NF-κB p50/p65
responsive site. The miR-155 promoter is a target gene for the
NF-κB protein. Activation of miR-155 (by NF-κB) is essential for
optimal CD8+ immune response (42,43).
In the human cutaneous T cell lymphoma mycosis fungoides, miR-155
suppresses STAT4 expression by targeting its mRNA for its
neutralization, and thus canceling the Th1 phenotype of the host;
in the opponent established STAT6/Th2 environment, tumor growth
prevailed. The Janus kinases JAK1 and 3 activate and sustain
different STAT3, 5 levels (44).
The miR-155 is a driver of the aggressive folliculotropic mycosis
fungoides (45).
NF-κB/STAT/PI3K proto-oncogenes in
virus-carrier human cells
In the human genome the NF-κB/STAT association is
prominently active in infectious processes, and in the malignant
transformation of selected cells. STAT4 and NF-κB (p65) were
expressed highly in tuberculoid leprosy, according to immune
reactions induced by interferon-γ (46). N. vectensis is not known to
possess IFNγ (3), but as a
predecessor of the system to come, it expresses alternatively
spliced mRNAs to bind tRNAs and GAIT element RNAs
(interferon-γ-activated inhibitor of translation) (47).
As a distant upward derivative of the viral-carrier
N. vectensis genome (vide supra), retrovirally and
herpesvirally infected human mesenchymal cells frequently express
NF-κB/STAT/PI3K (phosphatidyl inositol kinase) co-existing
pathways, often constitutively and in cells malignantly
transformed. In RS cells, STATs and NF-κB co-exist. STATs act in
antagonistic terms: STAT6 anti-, and STAT1 pro-apoptotically. RS
cells operate constitutively activated JAK/STAT pathways; the
DNA-binding STAT proteins entering from the cytoplasm to the
nucleus, activate their targets for the malignant transformation of
the cell (here, not by the stimulation of the cell cycle, since RS
cells are known for long periods of rest) (48,49).
It is the gene bcl-3 (B-cell lymphoma) at chromosome 19q13, which
encodes elements IκB of the NF-κB pathway. The IκB molecules form
complexes with the five NF-κB/Rel family member proteins in the
cytoplasm. Degraded IκB molecules release NF-κB proteins for their
translocation into the nucleus, in order to accomplish their
DNA-bindings. Aberrant Bcl-3 protein constitutively releases an
excess of NF-κB proteins for intranuclear transfer, where they bind
the promoters of their target genes, which encode chemokines,
cytokines and their receptors; anti-apoptotic molecules and various
transcription factors, including persistently activated STATs
(50). Viral DNA (herpesviral,
HHV4, HHV6 and HHV8) (51–55), and RNA (retroviral, HIV; HTLV-I)
(56) infections can force the
cytoplasmic IκB inhibitors for the release of NF-κB molecules. In
particular, open reading frame ORF-1 (referred to as
disease-related DR7) of the extant HHV6 encodes a p53-binding
protein, which renders it anti-apoptotic. In RS cells, DR7B is an
oncoprotein; it downregulates p53 and activates NF-κB (54). The herpesvirus-carrier N.
vectensis does not operate a pro-apoptotic p53, but has its
ancestral predecessors, p63/p73 (23). In the human genome, p53 eliminates
mutated cells by apoptosis. In the process of malignant
transformation, it is acting as a tumor suppressor gene, unless,
the cell eliminates the MDM-bound p53 protein by ubiquitination
(vide supra).
The genomes of herpesvirally infected human cells
(HHV4, EBV; HHV8, KSHV) readily respond with NF-κB/STAT activation
(51,52,55,56).
Human malignant cells transformed by circular dsDNA papillomaviral
genomes resist apoptosis, gain immortality (the HeLa cells), and
chemo-radiation resistance (58).
Could the DNA virom-carrier Hydra and Nematostella
cells (8) be driven by
constitutively activated NF-κB/STAT pathways, thus, resembling
cells in the process of malignant transformation higher up on the
evolutionary scale?
NF-κB/STAT-inhibitors
Dexamethasone, pyrrolidine dithio-carbamate,
BAY-11-7082; the sponge-derivative marine natural product
microsclerodermin inhibit or alter the excretions of inflammatory
chemokines (CXCL8/10) and interleukins (IL-1β, IL-6 and IL-8), but
have not been enlisted as effective anticancer agents (59–61).
Anti-NF-κB RNA-aptamers (specifically target-binding short ss
nucleotides) and siRNAs are powerful NF-κB inhibitors (62), not yet in clinical use. In
gram-negative septic shock, lipopolysaccharide endotoxins degrade
IκBβ and liberate NF-κB, which may induce an interleukin (IL-1β,
IL-6 and IL-12β) storm (63). In
gram-positive septic shock lipoteichoic acid LTA, activates
toll-like receptor TLR2, which induces the proto-oncogene cascade
of Raf/MEK/ERK/NF-κB. LTA promotes the transfer of NF-κB from
cytoplasm to nuclei; this is inhibited by propofol (64).
The sesquiterpene lactone parthenolide inhibits
NF-κB by alkylating p65 at Cys (65). Synthetic epoxyquinone A monomer is
a potent inhibitor of NF-κB resulting in the cessation of growth of
human lymphoma cells (66,67). The NF-κB inhibitor parthenolide
induced apoptotic death of p31 ras- and PI3K/Akt-driven malignantly
transformed cells (rat sarcoma oncogene; phosphatidylinositol-3
kinase; AK mouse thymic lymphoma retroviral oncogene). This effect
was mediated by downregulation of the inhibitor of apoptosis IAP2,
and degradation of polyadenosine-diphosphate-ribosyl-transferase
(PARP). In p210 BCR/ABL-transformed cells (breakpoint cluster
region; Abelson mouse retroviral oncogene), parthenolide treatment
failed to induce apoptotic deaths due to antagonism by STAT5 and
BCL-xL activation (B cell lymphoma extra large) (32). Under natural circumstances (in the
cell) siRNA and RNA aptamers (specific target-binding nucleotide or
peptide biomolecules) disable NF-κB mRNAs. In the RNA-induced
silencing complex (RISC), helicases unwind the dsRNA molecules,
thus, the antisense RNA strand can align with the receptive UTR
(untranslated) domain of the mRNA strand and neutralize it
(62). In contrast, O-linked
N-acetyglucosamine by glycolysation (oligosaccharide; N-terminal)
post-translationally activates the NF-κB molecule by acylating its
serine 350. Interacting are TCR and TNFα in encoding NF-κB target
gene IκBα. TCR and TNFα may activate NF-κB with or without
O-GlcNAcylation (OGT, O-linked GlcNAc transferase; N-glucoseamine
acetyl) (68). The Cys-to-Ser
mutated human NF-κB RHDs are not only gaining translational
activity, they become resistant to their natural inhibitors
sesquiterpenes and possibly to curcumins (vide supra). The
post-translationally functioning nucleo-cytoplasmic protein for
O-GlcNAcylation first appeared in prokaryota, and remained
functional in the low-branching Giardia and in the placozoan
Trichoplax adhaerens (69),
cultured and fed by red alga Rhodomonas salina. The ancient
system O-GlcNAcylation was discovered in the Drosophila
(70), but it must have been
present at the beginning of cellular life and remains active in
every cell. It is overexpressed in mitotic cells. Further
physiological blockade of NF-κB is brought about by the
p53-neutralizing protein, ubiquitin ligase MDM2 directed at p65Rel
(71).
Helicobacter pylori activates in gastric
epithelial cells the expression of IL-8, Jak1/STAT and NF-κB
(72). In the targeted cells
several ligands bind their receptors, as shown in Fig. 1 of the cited article (20). The physiological inhibitor of the
Jak/STAT pathways is SOCS (suppressor of cytokine signaling). The
ancestral Jak proteins appear in the choanoflagellates (C.
owczarzaki), and the STAT proteins in the cnidarians and
sponges (Nematostella vectensis; Amphimedon
queenslandica). The ankyrin repeats and the SH2 domains (Src
homologues, after Rous sarcoma virus) of the archetypical SOCS also
appear early in the cnidarians (N. vectensis) (20). Inhibition of the STAT3/5 pathways
in human breast cancer cells in vitro was accomplished
through siRNA-mediated knock-down of the gene SLC7A11, also known
ASCT (alanine, serine, cysteine transporter, xCT and solute carrier
family 7). It protected the cells from stress by reactive oxygen
species (73). Capsaicin was found
to inhibit disheveled in the Wnt pathway of human pancreatic
carcinoma cells. Consequentially, GSK (glycogen synthase kinase)
was not phosphorylated and the APC/Axin complex (adenomatosis
polyposis coli) was activated. Unphosphorylated cytoplasmic
β-catenin was degraded in the proteosome. Intranuclear β-catenin
target genes tcf/TCF, c-myc/Myc, cyclin D and STAT3 were not
activated; cytoplasmic STAT3 protein was not phosphorylated at
Tyr705. Capsaicin-treated pancreatic carcinoma cells undergo
caspase-3 cleavage and die in apoptosis. Antagonistically, IL-6
activated STAT3 (p-STAT-3 Tyr705), and increased intranuclear
β-catenin levels. Lithium chloride by inhibiting GSK, saves
β-catenin from phosphorylation and thus degradation; it is ready
for intranuclear transfer. Orally administered capsaicin reduced
the growth rate of orthotopic human pancreatic cancer xenografts in
athymic nude mice. These tumors showed low β-catenin, TCF and
p-STAT3 levels, and increased cleaved caspase-3 levels leading to
their apoptosis (74).
Autophagy
Under stress, Saccharomyceta and
Dictyostelia assume an autophagic stage of existence.
Autophagy has become an evolutionarily conserved process extended
to every eukaryotic species. Its potential reversal indicates that
autophagy is not a self-destructive process, but it is a
homeostatic phase in cellular life (75). Mitochondrial autophagy (mitophagy)
(76) indicates that the origin of
the process predated the appearance eukaryotes. The wild hydra may
live in a steady stage of autophagy and eventually recover. The
mTOR inhibitor rapamycin induces, wortmannin and bafilomycin
inhibit autophagy. In the hydra during starvation for weeks,
myoepithelial cells form autophagic vacuoles. Endodermal digestive
cells with knocked-out Kazal gene (serine peptidase inhibitor
Kazal, SPINK, vide infra) die, whereas ectodermal epithelial
cells survive in autophagy (77,78).
Break-down of the symbiotic relationship between host cnidarian and
symbiont dinoflagellate alga leads to autophagy and apoptotic death
of the host. Drastic environmental changes (temperature;
ultraviolet light; reactive oxygen species, ROS; loss of
antioxidant defenses; water pH changes; microbial invaders;
combination of unknown factors) induce the process. The process
could be induced by the TOR inhibitor rapamycin (target of
rapamycin). Hyperthermic-stressed (HTS) anemones released larger
number of symbionts than subjects kept on termed control
temperature (CT); however, exposure to rapamycin induced symbiont
release in CT anemones. It was either autophagy or apoptosis that
dominated, and possibly sphingosine kinase and PI3K signaling
regulated a see-saw relationship between the two outcomes (79). Alloreactivity after transitory
fusion of hydractinia (H. symbiolongicarpus) colonies with
minor incompatibilities is another cause of irreversible autophagy
terminating not in apoptosis, but in necrosis (observe electron
microscopic pictures in the cited article) (80).
The Wnt pathway remains dominant for the
regeneration of a new head in the decapitated hydra (81). The enzyme Kazal (vide supra)
in the hydra prevents autophagy and promotes self-preservation;
after amputation, it encodes de-differentiation and
transdifferentiation, blastemal transition, and activates the
ontogenic organizer pathways, for example those of the head
regeneration. View color pictures of Hydra proteomics
(metalloproteinases, thrombospondin, tubulin network, mitochondria,
and Kazal type 1 enzyme expression: its disappearance in autophagic
cells after its dsRNA knock-out (by feeding), and its reappearance
during regeneration. This study shows that the hydra Kazal(−)
phenotype mimics autophagic mammalian pancreatic cells with
non-functional trypsinogen-inhibitor SPINK1/3 (citing the
literature on the mouse experiments). The pancreas upregulates
SPINK3 expression in the gland regenerating after injury (82–84).
The ancient serine protease inhibitor (SPINK, Kazal type 1)
reappears in the fluid contents aspirated from human pancreatic
cysts; these aspirates are analyzed for carcinoembryonic antigens
CEA and CA 19-9, but it is the high SPINK content that indicates
the presence of malignant transformation (it is to be
differentiated from chronic pancreatitis by biopsy) (85). The Kazal type 1 serine protease
inhibitor SPINK1 gene is subject to point-mutations N34S and P55S
(asparagine, proline and serine) in patients with chronic
pancreatitis, but not in healthy controls (86). The SPINK1 N34S mutation is
considered to be a high risk factor for cancer, especially when
increased numbers of pancreatic stellate cells (PSC) co-exist; PSC
release proliferation and colony formation inducing molecular
mediators to the parenchymal or ductal cells (87).
3. The primordial cell survival pathways
become proto-oncogenes
Proto-oncogenes ascend to the human
genome
In the human genome, all these cnidarian genes and
gene-product proteins can function as proto-oncogenes,
oncoproteins.
SPARC
The primordial SPARC glycoproteins (secreted protein
acidic rich in cystein) forms the mesoglea in between the ectoderm
and endoderm of the diploblast cnidarians. In N. vectensis,
the Ca++-binding glutamic acid-rich acidic domain at its
N terminus is absent. Even in the absence of frank mesenchyme,
N. vectensis operates the primordial winged helix
transcription factor forkhead/and zink finger transcription factor
snail gene-product proteins. Snail formats the endodermal cells
from invaginating ectodermal cells (88). The N. vectensis larva
practices the evolutionarily conserved epithelial-to-mesenchymal
transition (89). SPARC proteins
in advanced multicellular vertebrate eukaryotes (including human
cells) are encoded by the genes snail and act as major contributors
to epithelial-to-mesenchymal transition of invasive cancer
cells-in-locomotion; co-activated are the ligands and receptors of
TGFβ and ERK kinase (extracellular signal-regulated kinase), while
miR-29b acts as an inhibitor of the process (90). Prominent examples are non-small
cell lung cancer and pancreatic and breast cancers expressing
melanoma antigen MAGE (91–93).
RUNX
The runt-related Runx transcriptional regulatory
factors bind the CAAT box (Runx/core binding factor CBFβ genes)
which appeared even before the cnidarians and sponges and continued
to evolve in triploblasts and bilateralia. In the figures of the
cited article (94), the ancestral
eukaryotic protists appearing first 987 mya are compared with the
cnidarians (Anthozoa and Hydrozoa) and sponges
(Porifera), appearing first 867 mya. The study presents the
genes as boxes connected with introns as lines and gives the
exon-products in amino acid numbers. These are 35–284 for the
Nematostella and 35–219 for the Hydra. Depictions of
the groucho-binding motif and the runt domain (RD) are given.
Nucleotides 56–434 are shown with digoxygenin labeling. DNA and
protein bindings are shown in three dimensions. The NvCBFβ protein
is in 59% identical and in 79% similar to the HsCBFβ protein. The
sponge AqCBFβ protein in 54% identical with the HsCBFβ, and in 47%
identical with the NcCBFβ proteins. As ‘packages of stem cells’,
cnidarians and sponges are able to regenerate from single cells of
adult hosts (the archaeocytes of the sponges), including the
regeneration of the nerve cells (94). (Further referenced in 7).
The ancestral Runx-CBFβ-DNA complex remained
evolutionarily conserved in advanced vertebrate hosts including
Homo. In the human genome, runx/RUNX may promote, induce, or
obliterate target genes as a multifunctional transcription factor.
The expression of FoxP3 in Treg cells is susceptible to suppression
by the Runxl/CBFβ transcription complex (95). In the human host, CBF-leukemias
arise due to inv(16) (pl3.1q22)
or t(8;21)(q22;q22) involving the NF-κB family kit (tyrosine kinase
receptor) gene (96). The Runxl
gene (of the Runx family Runx1, 2, 3, same as AML1, acute
myelogenous leukemia) forms fusion product with CBFβ. Further
fusion of the amino terminus of AML1 with MLL (mixed linkage
leukemias), carrier of histone methyltransferase activity at its
C-terminus, SET (suppressor of variegation, Su/var; enhancer of
zeste; trithorax, Drosophila) maintains lysine 4 (K)
trimethylation of histone 3 (H3K4me3) in the AML1 target gene PU1
(same as Spi, spleen focus-forming retrovirus proviral integration
oncogene). The ternary complex consisting of RUNX1/AML1, CBFβ and
MLL regulates hematopoiesis, but when mutated and constitutively
expressed, it induces leukemogenesis (AML or B-ALL) (97). The fusion oncoprotein EWS/FLI1
t(11;p22)(q24;q12) (Ewing's sarcoma; Fos-like immunoreactivity;
Finkel osteosarcoma retroviral oncogene; Friend leukemia insertion;
here: Friend leukemia insertion at 11q24.3) binds RUNX2 and
prevents its action. Runx3 protein promoted oncoprotein
EWS/FLI-stimulated target gene responses, and Runx2 and
transcriptional co-activator core binding factor-β (CBFβ) promote
invasiveness (98). Runx2 and bone
morphogenetic protein (BMP, member of TGFβ family) upregulated
snail-induced lung cancer cell migration and
epithelial-to-mesenchymal transition (99). The RUNX3 promoter in inv(16)(p13.1q22) acute myelogenous and
lymphocytic leukemias (AML; ALL) is hypermethylated with adverse
prognosis. In contrast, non-inv(16)(p13.1q22) and not hypermethylated
leukemic patients experienced relapse-free survival. Demethylator
decitabine restored RUNX3 mRNA and protein levels with expectation
of improved prognosis (100). In
the human embryo, the runx/Runx proteins work in hematopoiesis
(R1), osteogenesis (R2) and neurogenesis (R3). Gene/protein
runx2/RUNX2 is very active in embryonic life in regulating
endochondral ossification. Metastatic osteosarcoma with amplified
region 6p12-p27, the locus of runx2, remains a lethal tumor; this
locus operates two promoters, which constitutively release high
mRNA levels of the large DNA-binding protein RUNX2. Directly and by
cascading targeted major genes are IGF, BMP, PI3K/Akt, TCF/LEF,
FGF, iHh, Wnt (insulin-like growth factor; bone morphogenetic
protein; phosphatidyl inositol kinase 3; AK mouse thymus retroviral
oncogene; T-cell factor, lymphoid enhancer-binding factor,
fibroblast growth factor, Indian hedgehog, wingless/integrated).
Suppressed are CDKwaf/cip; and p53 by MDM (cycline-dependent
kinase; wild-type p53-activated fragment; cycline-dependent kinase
interacting protein; murine double minute protein) (101). Osteosarcoma tumors readily
outgrow standard chemotherapy of high dose methotrexate with
leucovorine rescue; doxorubicin and ifosfamide; cisplatin. A STAT3
molecular inhibitor (S31–201) inhibited growth, and induced
apoptosis of human osteosarcoma cell lines (102). The licensed multikinase inhibitor
sorafenib in reduced dosage in combination with cisplatin
suppressed phosphorylation of extracellular signal-regulated kinase
ERK, thus, inhibiting the growth and inducing apoptosis of human
osteosarcoma cell lines (103)
(see comments in Discussion). The fact that patients with chondro-
and osteosarcomas generate immune T- and NK cell-mediated attacks
on autologous osteosarcoma cells (104) raises high expectations that CAR
cell (chimeric antigen receptor), and checkpoint inhibitor
(PD-1/PD-1L) therapy might induce clinical remissions, especially
when the tumor cells express NY-ESO antigens (105).
Hedgehog
The first Hog domain proteins appeared in
dinoflagellate algae. The hedgehog (Hh), notch, sox and Wnt genomic
pathways operational in the cnidarians remained highly conserved by
the triploblastic bilaterians up to vertebrate mammalians
(including Homo). The Nematostella Hh orthologs are
among other cnidarian precursors (Wnt, TGFβ/BMP and FGF) of
bilaterian genomic pathways. Here, the Hh ligand-receptor patched
(Ptc) represses smoothened (Smo), the inhibitor of Gli (glioma).
Capturing the ligand by Ptc, releases Smo, that liberates Gli
(glioma-associated oncogene homolog 1). The Nematostella
genome contains six genes encodig Hh ligands and receptors, and
genes coordinating Wnt, Hh, TGFβ and FGF (106–108). The role of sensory cilia in Hh
signaling emerged in the sea urchin (Strongylocentrotus) and
was conserved up to vertebrates. The kinesin motor proteins
originated in the chlamydomonas (C. reinhardtii). The
choanoflagellates (Monosiga brevicollis) operate an
elementary Hh pathway in expressing receptor Ptc (109). Various co-expressions of these
genes guide the cell through its ontogenesis and final
differentiation. The enriched mammalian genomes operate three Hh
pathways (sonic, desert and indian), that evolved through gene
duplication. In chronic lymphocytic leukemia, the Hh pathway
enlists PI3K (110). In
hepatocellular carcinoma of whatever etiology (B&C viral;
alcoholic cirrhosis) the Hh pathway is united with the
Wnt/β-catenin (vide supra) and North (vide infra)
pathways with frequently silenced Dickkopf gene/protein (111).
Notch
The notch/Notch signaling pathway was functional in
the ancestral cnidarians. The delta and serrate/jagged
ligands-bound receptors, which are endocytosed and cleaved.
Intracellular protein cleavage results in the fragments NICD (Notch
intracellular domain). These are the DNA-binding proteins CSL (core
binding factor, CBF/suppressor of hairless, SuH/lymphocyte
activation gene, Lag2) directed at their targets. Involved are
Su(H), Hairy and Enhancer of Split (Espl; HES, Drosophila),
CBF/RBPJκ (recombination signal binding protein for IgκJ), gene
mastermind, mam/MAM and NF-κB. Most of the Notch proteins are
histone acetyl-transferases; most acetylated genes are silenced.
The Notch pathway is active in the embryos, larvae and planula of
the extant N. vectensis. The Notch pathway regulates
cell-to-cell communications, and the arising of neural precursors
from epithelial cells. Ancestral early multicellular eukaryotic
life forms (metazoa) utilized notch/Notch for nerve cell
differentiation. The process was duplicated in N. vectensis
by regulating the expression of gene Nvnotch. The Notch
cascade suppressed neural differentiation markers; Notch inhibited
the expression of differentiation inducer genes (among them some
Nvsox genes). In N. vectensis appear the first achaete scute
genes (chaete, bristle, Drosophila; plaque), which will be
prominent in the protochordate amphioxus, represented by
Branchiostoma floridae (112–114) (vide infra). In metazoa,
Notch will run the vertebrate segmentation clock and generate
secondary mesenchymal cells (115).
The human oncogenic dsDNA vir uses activate
notch/Notch. Human herpesvirus 8, Kaposi sarcoma herpesvirus (HHV8;
KSHV) uses the Rta protein (replication transcription activation)
as its lytic switch and RBPJκ (recombination signal-binding
protein-1 immunoglobulin kappa J region) is essential for the
transactivation. The protein-folding cellular peptyl-prolyl
cis/trans isomerase, ‘never in mitosis’ (Pin1) had
been conserved from the yeast to the human genome, where it is a
notch/Notch activator (and RBPJκ is a Notch effector), and as such
it is overexpressed in some tumors (Kaposi sarcoma;
adenocarcinomas). Co-opted by KSHV, Pin1 by direct binding,
enhances the viral Rta transactivational functioning and protects
Rta from auto-ubiquitylation, thus, contributing to Notch
activation (view Fig. 6 of the cited article) (116). In primary effusion lymphoma cells
KSHV and EBV commonly co-exist and activate the Notch signaling
pathway (117). In HPV-induced
squamous cell carcinomas activated Notch may be an aggravating or
an alleviating factor (118).
Observe: Kaposi sarcoma virologists use abbreviation vSOX for viral
shut-off exonuclease, that has no connotation none whatsoever with
eukaryotic stem cell proto-oncogene sox/Sox (vide
infra).
Gamma secretase and ADAM proteases (A disintegrin
and metalloprotease domain) cleave the Notch receptor; Delta and
Jagged ligands activate it so, that its intracellular domain
translocates into the nucleus. The ancient Notch pathway had been
evolutionarily conserved. A major Notch target gene is the RBP-Jκ
(vide supra). NF-κB activation is mediated by protein
interaction with IKKα (inhibitory kappa kinase). Notch negatively
interacting with tumor suppressor protein PTEN (phosphatase tensin
chromosome ten), liberates PI3K/Akt from suppression, and opening
up a pathway to mTOR activation. View Fig. 1 of the cited article (119). Hypoxia is a major activator of
proto-oncogene Notch in glioblastoma (vide infra).
Sox
N. vectensis utilizes the ortholog of the
coral Acrospora millepora's six sox/Sox pathways (sex
determination region Y, SRY box). These transcription factor genes
encode high mobility group proteins (HMGP) functioning in a wide
range of developmental processes, gastrulation; and laying down
ectoderm and endoderm. The sox genes co-exist in pluripotent stem
cells with stem cell genes kfl(Krüppel factor-like); oct (octamer),
c-myc (myelocytosis) and Nanog (rejuvenating celtic tribe). The
SoxB stem cells of N. vectensis are positioned
interstitially and encode three neural cell types: sensory neurons,
ganglion neurons and nematocytes. Sox and Fox (forkhead box) genes
cooperate in N. vectensis in germ layer specification and in
the regulation of gastrulation (120–123).
The stem cell promoter proto-oncogene sox/Sox
family members (SRY-related sex-determining factor on Y chromosome
high mobility group box) are ancient transcription factors which
exist in a family of over twenty proteins in the human
genome/proteome. The Sox proteins functionally sustain stem cell
pluripotentialities and act as generators of tissue anlagen in
embryonic life, and as proto-oncogenes/oncogenes (example Sox2/9)
or tumor suppressor genes (example Sox7/15), or alternatingly both
(example Sox2/4/9) in adult life. Some Sox proteins (Sox9/21)
activate the oncogenic pathways Wnt; Gli; Hh; Notch. Sox15/20
inhibit the Wnt pathway in pancreatic and other (testicular) cancer
cells by dimerising with other oncoproteins (124).
Wnt
Unicellular eukaryotes do not operate the Wnt
pathway. The Wnt pathway (wingless; Drosophila; integrated,
mouse) in the ontogenesis of Ciona intestinalis was recently
reviewed (125). Cnidarians and
ctenophores in their ontogenesis utilize the Wnt pathway. Wnt
ligands are secreted glycoproteins specific to their transmembrane
receptors frizzled that trigger the β-catenin cascade by first
liberating the β-catenin destroying unit from the control of
disheveled. The β-catenin destruction unit consisting of proteins
axin, glycogen synthase kinase (GSK) and an ancestral adenomatosis
polyposis coli gene product in the cytoplasm is able to
phosphorylate β-catenin, thus, marking it for ubiquitination. The
cell rests. However, if unphosphorylated β-catenin is allowed to
accumulate, it will translocate from the cytoplasm to the nucleus.
There, it will by domain-attachment activate its target genes. Of
numerous β-catenin target genes, genes tcfTCF/lefLEF are activated
first (T cell factor; lymphocyte enhancing factor). (Evolutionarily
reviewed and extensively referenced in ref. 126,127).
Hh/Wnt
The hedgehog pathway is active in several human
cancers and is potentially susceptible to small molecular
inhibitors. Vismodegib induces remissions in advanced basal cell
carcinoma due to inhibiting the Hh receptor smoothened (128). The antifungal itraconazole
inhibits chemotherapy-resistant pancreatic adenocarcinoma cells
(129). Natural products
(cyclopamine, curcumin, epigallocatechin-3-gallate, genistein,
norcantharidin, resveratrol and zerumbone) exert blockade in the
sonic Hh pathway (130). The
STAT/NF-κB pathway is suppressed by curcumin and epigallocatechin
gallate (131). The basic nature
of malignant transformation is to advance in several pathways,
thus, rendering monotherapy often ineffective. The Hh/Gli and the
Wnt/β-catenin pathways appear in combination (132). The sponge Amphimedon
releases the antifungal cyclic peptide microsclerodermin (61). NF-κB is constitutively activated in
pancreatic adenocarcinoma cells. Microsclerodermin blocked NF-κB
production in pancreatic adenocarcinoma cells and forced cell death
in the form of apoptosis (61). In
the Wnt pathway, β-catenin and DNA methyltransferase (DNMT)
co-localize in tumor cell nuclei. Lysine-specific demethylase
(LSD), the regulator of DNMT, becomes a component of the complex.
The DNMTs usually hypermethylate CpG islands and LSDs demethylate
histones. The complex represses tumor suppressor genes and activate
oncogenes (133).
CYPs
Cytochrome oxygenizers are expressed by the
ligand-activated AhRs. CYP1 family proteins are overexpresed in
ER+ human breast cancer cells. Considered to be
co-carcinogens (breast cancer) due to their positive involvement in
estrogen metabolism, production of procarcinogenic estrogen
metabolites and induction of nucleotide polymorphism (134,135).
ASC
Ernst Haeckel's works from 1876 are recently cited
for placing the amphioxus (represented by the Branchiostoma
floridae) (Fig. 4), at the
base of vertebrate evolution (136). The amphioxus without duplicating
its genome preserved the body image of its ancestors living 550
mya. It is not only being a cephalocordate that makes it unique. It
has developed a large repertoire of TLRs, retained a large supply
of transposable elements, and acquired the most ancient
transposable elements of the adaptive immune system of vertebrate
mammalians, one of the recombination activating genes, the RAG
transposon (reviewed in ref. 137). The achaete scute (ASC,
Drosophila) neurogenic pathway has taken its beginning in
the Nematostella and reached an advanced stage in the
amphioxus represented by the Branchiostoma floridae
(112,113). The human homolog of achaete scute
encodes esthesioneuroblastoma, the malignant tumor of the olfactory
ganglion, and converts adenocarcinoma cells into
radiochemotherapy-resistant neuroectodermal cancers (138,139). The neuroectodermal small cell
bronchogenic carcinoma was treated long ago with cyclophosphamide,
vincristine and doxorubicin. Short lasting remissions were induced
at the price of toxicity; all remissions were lost to relapses.
Immunostimulation with BCG (Bacille Calmette-Guréin) failed to
improve results (140). Cisplatin
improved remission rates, exerted toxicity, but failed to cure the
disease. Targeting the causative oncogene/oncoprotein hASH with
small molecular inhibitors and monoclonal antibodies raises the
possibility to cure (141).
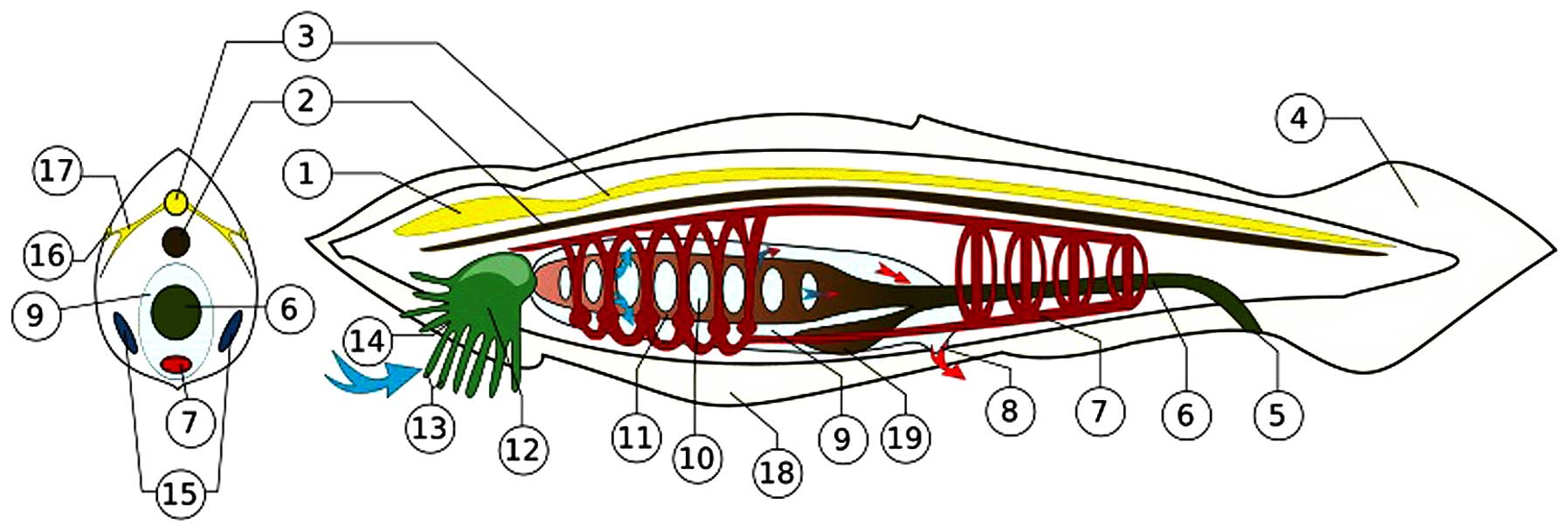 | Figure 4The amphioxus (represented by the
Branchiostoma floridae of the Everglades) operates a nervous
system consisting of peripheral sensory cells connected with the
notochord by axons and synapses. Gamma aminobutyric acid and
cholinergic molecular mediators circulate in the axons. The
primordial achaete scute (Ash) genes/proteins (with Notch) were
essential for the original encoding of the system. The ancestral
Ash genes appear first in N. vectensis. Conserved up to the
human genome, Ash gene homologs are proto-oncogenes for
esthesioneuroblastoma of the olfactory ganglion, small cell
undifferentiated carcinoma of the lung, and adenocarcinomas
transforming into neuroectodermal tumors (7,125).
The depiction shown in the Figure, is from the files of Wikimedia
Commons Information freely licensed media file repository,
description page three. Permission is granted to copy this document
under the terms of GNU Free Documentation License Version 1.2. 1,
brain-like blister; 2, notochord; 3, dorsal nerve cord; 4,
post-anal tail; 5, anus; 6, food canal; 7, blood system; 8,
abnominal porus; 9, overpharynx lacuna; 10, gill's slit; 11,
pharynx; 12, mouth lacuna; 13, mimosa; 14, mouth gap; 15, gonads
(ovary/testicles); 16, light sensor; 17, nerves; 18, abdominal ply;
19, hepatic caecum. |
MicroRNAs exert post-translational
control
The genomics and proteomics of the cnidaria cell
enable it to practice siRNA inhibition of the NF-κB mRNA, and
O-linked glycosylation of the NF-κB protein molecule (vide
supra), but all the target molecules thus diminished or
overexpressed have not as yet been reported or even identified. In
clinical medical oncology, naturally occurring and synthesized
molecular inhibitors are applied for treatment (and encounter
strong resistance by the NF-κB/STAT-transformed cancer cells)
(59,60).
The non-protein-coding RNAs (micro miRNAs; small
interfering siRNAs; piwi-interacting piRNAs) exist and function in
all eukaryotic cells primarily in post-translational downregulation
of mRNA expression. Drosha, Dicer and Ago-piwi (argonaute;
P-element-induced wimpy testis, Drosophila) RNAse, helicase
and RNA polymerase enzymes carry out the specific alignment and
degradation of the targeted mRNA molecules (142). Of the oldest miRs, only one, the
miR-100 possessed by the cnidarian, N. vectensis, was passed
through evolution to the human genome. Others, miR let-7 (the
ancestor of the let family miRs), appeared in the first true
bilaterians, the annelid Platynereis. Its antagonist Lin-28
expressed itself in the nematodes (Caenorhabditis) (143–146). All other genuine miRs the
cnidarians preserved for themselves, and the descendant organisms
generated their own new miRs. So much so, that the Hydra
does not have miR-100. The hydra PIWI protein/piRNA association
represses transposon expressions that might threaten genomic
integrity (147,148).
In the human genome, the major effect of miR-100 is
tumor growth suppression. In colon cancer, downregulation of
miR-100 results in accelerated tumor cell growth (149,150). The epithelial-to-mesenchymal
transition of tumor cells (breast cancer) is an adverse prognostic
event, except when it is induced by miR-100, in which case
cessation of tumor cell growth follows (151). Upregulated expression of miR-100
attenuates glioma cell growth due to decreased expression of ATM
(ataxia telangiectasia mutated) (152). In urothelial cancer of the
urinary bladder, miR-100 suppressed oncogenes mTOR and SMARCA
(mammalian target rapamycin; switch; sucrose non-fermentation;
matrix-associated actin-dependent regulator chromatin-A) (153). Pancreatic cancer cells suppress
miR-100; forced expression of miR-100 induced cessation of cancer
cell proliferation in vivo in xenografts; miR-100 expression
resulted in a blockade of fibroblast growth factor production
(captured by its receptor, FGFR3), the major driver of tumor cells
(154). In prostate cancer,
miR-100 directly targets for downregulation of AGO2c (argonaute)
and thus blocks downregulation of miR-34a and miR-125b; these
microRNAs suppress the expression of prostate cancer stem cells
(155). In HeLa cells, an
antagonism exists between the p53 and NF-κB proteins, in which the
former blocks the latter in its intranuclear binding to the
upstream sequence of the miR-100 gene (156). The presence of some of these
elements in N. vectensis (miR-100), allows the presumption,
that the entire human reaction originated in, and was inherited
from, the cnidarian.
The miR let-7 and Lin28 antagonist-collaborator did
not exist in the cnidarians; it was acquired and expanded to a
family in the first true bilaterians and their successors,
exhibiting itself in full in the nematode Caenorhabditis
elegans (146), from where it
continues its competition-collaboration in the human genome
(157). There, it determines if
chronic inflammatory processes will eventually terminate in
oncogenesis (158). The oncogenic
cascade is initiated by scr/SCR, Lin-28 and IL-6; let-7 is
depleted; STAT3 and NF-κB are co-activated and the process becomes
irreversible (159).
See images of cnidarian pyruvate dehydrogenase
kinase and hypoxia inducible factor in the internet (or in the
original literature). The presence of STAT/NF-κB and STK, the
src-type kinase, in the cnidarian cytoplasm (160) enables cells to live in the state
that is recognized in the mammalian host as a chronic inflammatory
process (the inflammasome) (158). Observe miRNAs regulating the
relationship between host (scleratinian corals) and the Red
Sea-isolate symbiont alga (Symbiodinium microadriaticum)
forming a holobiont (161) (viral
involvements received no comments in the study).
4. Ctenophores
Genomic proteomics of basal
metazoans
At the stage of baseline diploblastic metazoans,
beneath Cnidarians and Spongiae (162–164) lie the Placozoans, represented by
Trichoplax adhaerens (165,166), and the comb jelly Ctenophores,
here by Mnemiopsis leidyi and Pleurobrachia
pileus/bachei (167). These
ctenophores are best characterized by missing genes, which are
prominently present in other diploblastic metazoans. Ctenophores
are devoid of Toll-like receptors and MyD88 signals. Hedgehogs and
JAK/STATs are absent, but the numbers of homeodomain genes are in
excess of other diploblasts. Acetylcholine, adrenaline, dopamine,
histamine and serotonin are missing. Glutamate is a major signal
molecule; it induces action potentials and currents. G-protein
coupled neuropeptide receptors exist (without neurons). Ion
channels carry stimuli. DNA methylation and demethylation are
practiced. Argonaute units with Dicer are functional in the
cytoplasm (167). The
non-duplicated placozoan and cinadrian genomes carry the ancestral
P450 cyp/CYP sequences; so do the Spongiae (Amphimedon)
(168). The unicellular
choanoflagellates (Monosiga) do not show the presence of
Runx genes/proteins. Single Runt domain Runx gene-product proteins
are present in the placozoan Trichoplax, in the cnidarians and
Spongiae (Amphimedon). While the Amphimedon genome
encodes a Groucho protein, its Runx protein is devoid of the
Groucho recruitment motif amino acid sequence WRPY (tryptophan;
arginine; proline; tyrosine). Figs.
1 and 3 of the cited article
(169) show that all the other
listed organisms encoded this motif. The larva of the sea anemone
Edwardsiella lineata parasitizes the ctenophore M.
laidyi. The Edwardsiella NF-κB is ancestral to the split
and a further evolved NF-κB is operational in the non-parasitizing
N. vectensis (vide supra) (170).
The ctenophore Wnt pathway imitates
malignant transformation
Referenced in the Appendix of the volume in print
(7), the ctenophore genome is
shown encoding glycoprotein hormones (without glands), bone
morphogenetic protein (BMP, without bones), platelet-derived growth
factor (without platelets), nerve growth factor (without a nervous
system), and the Wnt activator and BMP/TGFβ-inhibitor norrin.
Norrin is the ligand of the leucine-rich repeat-containing G
protein-coupled receptor. The ctenophore Wnt pathway acts in an
upregulated state driven by ligand-bound Frizzled and lipoprotein
receptor-related protein receptors, and dishevelled, that can
inhibit by phosphorylation GSK-3 (glycogen synthase kinase). In the
cytoplasmic ‘β-catenin destructive complex’, the active GSK-3
phosphorylates cytoplasmic β-catenin, thus sending it to
ubiquitination. However, the inhibited GSK-3 allows excessive
cytoplasmic accumulation of non-phosphorylated β-catenin, which
translocates into the nucleus. There, it activates the promoters of
its target genes, primarily tcf; tcfTCF/lefLEF are T cell factor
and lymphocyte enhancing factor (without lymphocytes being in
existence). In mammalian cells, TCF/LEF have become
proto-oncogenes/oncoproteins. There, the target genes of the
TCF/LEF proteins are stimulatory to the cell cycle by PI3K/AKT and
inhibitory to apoptosis by survivin (171). The complete vertebrate mammalian
β-catenin destructive complex consists of GSK, axin and APC
(adenomatous polyposis coli) gene product proteins. The
ctenophore β-catenine destructive complex is deficient in that it
is devoid of axin expression, and its GSK may act downregulated.
The advanced mammalian Wnt pathway is antagonized by the tumor
suppressor proteins Dickkopf (Drosophila). In Wnt
carcinogenesis, the mammalian cell undergoing malignant
transformation eliminates the Dickkopf genes (which suffer
loss-of-function mutations). The ctenophores do not possess the
Dickkopf genes. Thus, the ctenophore Wnt pathway is similar to that
malignantly transformed vertebrate mammalian cells operate: the
β-catenin destructive complex is weak in antagonizing the
intranuclear transfer of β-catenin, thus, β-catenin readily
translocates. Furthermore, natural inhibitors of the process, the
Dickkopf proteins, are non-existent (172,173).
The sox/Sox transcription factors maintain stemness
of their host cells and operate by encoding high mobility group
proteins. In the so-called tumor stem cells, they are active in
association with other stem cell genes and proteins
(Krüppel-factor; c-Myc; Nanog; Oct4) (vide infra). In the
embryonic larval stage of ctenophores (Mnemiopsis;
Pleurobrachia), several members of five major Sox gene
groups perform essential regulatory functions for the directives of
orderly ontogenesis (174).
Extraordinary cell proliferations leading to deformities have not
been observed. If some functions are constitutively encoded due to
sox gene amplifications or gain-of-function point mutations, that
has not been as yet observed. However, in mammalian cells, the sox
genes (and several other stem cell genes) are known to convert into
proto-oncogenes (vide infra).
Wnt and Sox oncogenesis in human
cells
Within the Wnt/β-catenin oncogenic pathway in
hepatocellular carcinoma cells, the mRNA of the tumor suppressor
gene, Bednarek's WWOX (double tryptophan domain oxidoreductase), is
demolished in the AGO complex by miR-153, which by aligning
specifically to the WWOX mRNA 3′-untranslated region (UTR),
destroys it. The cell cycle inhibitory protein WWOX will not be
translated. The uninhibited intranuclearly transferred β-catenin
binds to its target DNA gene promoters of the cell cycle (primarily
to tcf/lef, vide supra) and constitutively activates them,
to produce their oncoproteins TCF/LEF and others (175). WWOX plasmid induced WWOX protein
expression in ovarian cancer stem cells. WWOX plasmid-treated
ovarian cancer stem cells reduced, or ceased, the expression of,
stem cell markers as follows: CD133, CD117, ATP-binding cassette G
member2 protein, Nanog, Oct4 (octamer-binding transcription factor
4) and breast cancer resistance proteins (for which specific
antibody is available at Chemicon, Billerica, CA, USA; cat. no.
#254515). A sign of differentiation induction was the increase of
E-cadherin presentation (taking control of β-catenin at the inner
cell membrane). The WWOX plasmid-treated cancer cells failed to
grow in xenografts and lost their chemotherapy resistance. The
DNA-binding WWOX protein of ancient derivation is naturally
released from the mitochondria and translocates into the nucleus,
where it interacts with TNF-related genes, oncogene Jun N-terminal
kinase (reticuloendothelial virus 17 oncogene; in Japanese 17 is
jun), being either inhibitory (to proto-oncogene jun) or
promotional (to pro-apoptotic TNF and p53) (176).
In advanced multicellular organisms (including
Homo), the cellular programing of twenty-some sox
gene-product SOX proteins regulate the development of hematopoietic
and central nervous systems in the embryo, and sustain pluripotency
in their resting stem cells. Expressing their faculties
constitutively, some sox/SOX proteins (human Sox2/9) transform
individual cells into immortal and independent life forms (cancer
cells), which overgrow and eventually kill their multicellular
host. Other sox/SOX proteins (human Sox7) undergo methylation
silencing it, so they may act as tumor suppressors. Such SOX
proteins bind oncoproteins β-catenin or TCF, thus neutralizing
them. If the sox/Sox protein resumes its cell polarity signaling,
that is physiological in embryonic cells, in mature cells of a
multicellular host, those cells will become metastasizing tumor
cells. Tumor cells with certain downregulated sox/SOX proteins gain
malignant potential, whereas tumor cells with overexpressed sox/SOX
undergo attenuation (124). It is
not known as yet which microRNAs influence translational expression
of sox/SOX15. Many tumor-promoting or inhibitory circulating
microRNAs are recognized in patients with cancer (177). Examples: miR-21 downregulating
tumor suppressor PTEN (phosphatase and tensin homolog) and
activating oncoprotein Akt (v-akt murine thymoma retroviral
oncogene); the differentiation-inducer tumor suppressor miR let-7
(letter, lethal) are downregulated in many cancers (lethal when
deleted); but SOX15 regulator miR has not been named as yet. In the
early multicellular bilobular life forms (cnidar-ians;
ctenophores), sox/Sox proteins serve in ontogenesis and in several
cell survival pathways of their hosts. It appears that the
ctenophore genome operates a weak β-catenin inhibitory system
without inhibitors. If sox/Sox proteins sustain undifferentiated
stem cells, the ctenophore genomes assume the imitation of an
oncogenome arising in selected individual cells of an advanced
multicellular host (including Homo). The sox/Sox2 stem cell
proto-oncogene promoted self-renewal, proliferation and
chemoresistance in head and neck squamous carcinoma cells (178). Chemoradiotherapy-resistant stem
cells arise in hypoxic regions of glioblastoma tumors. Hypoxia
inducible factors, vascular endothelial growth factor (VEGF) and
erythropoietin (EPO) accumulate in these regions. The tumor cells
assume stem cell characteristics and upregulate notch/Notch
proteins (179). Notch activation
in hypoxia may be a fundamental ancient reaction of the cells
living in anaerobic environment. When this fundamental faculty is
resumed in single cells of a multicellular host, it may
de-differentiate them to their ancestral pluripotentiality,
physicochemical resistance, and independence from the community
manifesting in incessant replicatory activity.
5. Discussion
Concerning the resistance of the Hydra and
the Nematostella to algal symbionts, what has not been
investigated and recognized is the ability of the invader
Symbiodinia cells to inactivate the NF-κB pathway of the
tolerant cnidarian host cell. One should conclude, if not proven
otherwise, that zoochlorellae and zooxantellae cannot overcome the
defensive NF-κB pathway in H. magnipapillata and N.
vectensis. Could these symbiosis-resistant cnidarian genomes
operate amplified or constitutively activated NF-κB/STAT pathways,
similarly to those of neoplastic cells, in order to achieve the
state of resistance? Affirmatively, run-away uncontrollable cell
replications tantamount to what will be referred to as malignant
transformation in the subjects occupying higher ranks on the
evolutionary scale do occur in the inhabitants of the lower
echelons (180). The Porifera
Acroporidae sponges are afflicted by the disease calicoblastic
neoplasm. The proliferating cells evict their zooxantellae
symbionts, and remain susceptible to apoptotic death induction. The
ancestral p53 gene protects the Nematostella (N.
vectensis) by inducing apoptotic death of its proliferating
germs cells (as referenced in 180).
Hundreds of million years of evolution enriched the
ancient genomes. While retaining their basic faculties, the
cnidarian NF-κB/STAT cell survival pathways phenomenally
complicated themselves during their trajectory of 600 million
years. As exercised in the human genome, the cell is rendered
immortal by mutations and recombinations. An example is the complex
encoding of a form of human acute myelogenous leukemia (181). The basic factor for
immunosurveillance raised from the avian genome (v-rel avian
reticuloendotheliosis viral oncogene homolog A) has become in the
human genome constitutively expressed dimers, which as the NF-κB
p50/p65 Rel proto-oncogene/oncoprotein family residing in
chromosome 11. The system is activated by RANK (receptor activator
of NF-κB) to enable it to act as the conductor of the cascading
orchestra consisting of members TNFα, TRAF (TNF-receptor associated
factor), STAT, IRF7 (interferon regulatory factor), TAK
(TGFβ-activated kinase), NIK (NF-κB-activated kinase), Bcl3 (B cell
leukemia) and proto-oncogenes fos/Fos, jun/Jun, src/Src. The T cell
leukemia virus (HTLV-1) Tax oncoprotein and the EBV and KSHV
equivalent FLIP oncoproteins (FLICE-like caspase inhibitory
protein; FADD-like IL-1 converting enzyme; Fas-associated death
domain; factor apoptosis stimulating) activate the NF-κB pathway.
At the 25th year after its discovery, NF-κB was recognized as a
basic immunoregulator and as an oncoprotein (182,183).
A complex network incorporating NF-κB drives a form
of acute human myeloid leukemia. Gain-of-function mutation of the
70 kb 21 exon c-kit gene at 4q11-q12 (originally c-kit → v-kit of
the Hardy-Zuckerman feline leukemia retrovirus) results in the
encoding of the 145 kDa KIT transmembrane protein receptor CD117,
whose ligand is the stem cell factor. The overexpressed KIT
activates Myc and RUNX1. KIT-activated MYC downregulates miR-29,
and may induce the Wnt pathway. Its specific siRNA, and
flavopiridol down-regulate KIT. Furthermore, the kit promoter
expresses binding sites for NF-κB and Sp1 (stimulating protein
encoded from 12p12-q13.1), and thus NF-κB/Sp1 induce KIT
overproduction. Sp1 inhibitor mithramycin, and NF-κB inhibitor Bay
11-7082 reduced KIT expression. Bortezomib induced Sp1
ubiquitination, and thus disrupted Sp1/NF-κB interaction.
Transactivation by the kit promoter Sp1/NF-κB unison was abrogated,
and thus KIT overproduction ceased. Bortezomib further inactivated
oncoproteins STAT3, AKT, and ERK by appropriate serine and tyrosine
phosphorylations. Codons 822- and 816-mutated KIT proteins are
inhibited by imatinib and dasatinib, respectively. Furthermore, Sp1
was found to repress miR-29 transcription and so acted as
antago-miR29, resulting in rising Sp1 levels; consequentially, KIT
was upregulated. In reverse, bortezomib, mithramycin, or Bay
11-7082 downregulated KIT and SP1 and increased miR-29 expression.
Sp1 and NF-κB have binding sites on the primary enhancer element of
the miR-29 transcript on chromosome 7, and thus reduce miR-29
expression. In reverse, siRNA-induced knock-down of Sp1 and NF-κB
(p65) resulted in miR-29 upregulation. Bortezomib blocked the
binding of Sp1/NF-κB to the miR-29 regulatory element on chromosome
7, and thus induced miR-29 production; it also reduced c-Myc
expression. Bortezomib downregulated Sp1/NF-κB, and upregulated
miR-29b by disrupting the Sp1/NF-κB/HDAC complex. The
hypophosphorylated KIT protein lost its leukemogenic potency.
Furthermore, Sp1/NF-κB and histone deacetylases (HDAC) interact.
Enrichment of HDACs and SP1/NF-κB on the miR-29 regulatory
sequences occurs in concert. In effect, HDAC inhibition by its
specific siRNA, or OSU-HDAC42 and MS275 resulted in upregulation of
miR-29 transcription, with concurrent drop of Sp1 and KIT levels.
Whereas HDAC expression resulted in downregulation of miR-29 and
overproduction of Sp1/NF-κB. Intact Sp1/NF-κB increased HDAC
binding to the miR-29 enhancer region, whereas decreased such
binding occurred when the Sp1/NF-κB physical interaction was
disrupted by the HDAC inhibitors. Upregulated miR-29 decreased the
binding of the Sp1/NF-κB complex to the kit gene promoter. The
leukemogenic oncogenes/oncoproteins could be so suppressed, NF-κB
by Bay 11-7082, Sp1 by mithramycin, and HDAC by its inhibitor
HDAC42; miR-29 acts by DNA methyltransferases; and bortezomib
disrupts the oncoprotein complex of Sp1/NF-κB (181).
When NF-κB is activated in melanoma, it suppresses
phosphatase tensin homologue PTEN, and thus liberates oncoprotein
PI3K, an activator of proto-oncogenes Akt and/or mTOR. NF-κB would
be inactivated by the SOCS (suppressor of cytokine signaling), were
it not constitutively expressed (184). The natural substance capsaicin is
an NF-κB inhibitor (185).
Several synthetic small molecular PI3K inhibitors are ready for
clinical trials (186). The FDA
approved the small molecular PI3K inhibitor idelalisib first for
lymphoma, lymphocytic leukemia, and as an investigational agent for
metastatic breast carcinoma (187). Molecular kinase inhibitory
therapy for the PI3K-driven rapidly replicating cancers (breast
carcinoma) is to follow. The complex oncogenome of the breast
cancer cell will not be switched off by single agent monotherapy
(view Fig. 2 in the cited article)
(188).
While the first cellular life forms succeeded in
populating the ancient Earth by using the ancestors of the extant
proto-oncogenes as cell survival pathways, the small molecular
inhibitors synthesized now against the human cancer oncoproteins
would probably render them defenseless first; but subsequent
mutations could rescue their new small subpopulations to begin
replicating again. Advanced protocols of combination chemotherapy
and targeted therapy readily induce remissions, that are lost to
relapses. Hundreds of million years of evolution superimposed an
adaptive immune system over the preserved innate immune system
(reviewed in refs. 90,137). Yet, infectious diseases are
eradicated not by natural herd immunity, but rather by external
interventions, such as preventive vaccinations. Is it possible that
malignantly transformed cell populations could be eradicated by the
immune system, even if the defensive cellular elements are the
malignant cells themselves? The treatment modalities
graft-versus-leukemia (189) and
HSCT (hematopoietic stem cell transplantations) induce remissions
in survivors of treatment mortality, especially when re-enforced
with NK cell, targeted chimeric antigen receptor- CAR-armed T cell,
PD-L1-directed (programmed death, ligand) monoclonal antibody, or
immunotoxin therapy (190). The
best examples are the adoptive immune T- and NK lymphocyte-mediated
protocols, the application of which eradicates malignant lymphomas
and lymphocytic leukemias (191).
In the leukemia-afflicted host, the native immune system does not
mobilize an effective reaction for the rejection of malignantly
proliferating cells. The elements for the rejection of the
malignant cell population are there, but they are not assembled and
are not self-applied. Properly assembled in the laboratory and
administered sequentially, host immune reactions are capable of
dissolving large masses of tumors (so much so that the patients are
to be protected from life-threatening lymphokine storms and tumor
lysis syndromes).
Predecessors of cnidarians and ctenophores survived
physicochemical insults in the ancient Earth in a stage of
autophagy. Malignantly transformed human cells (cancer cells)
resist the effects of chemical and radiological agents in the stage
of reversible autophagy followed by constitutively activated
proto-oncogenes for survival and recovery. These cells will resist
exposure to the same agents that induced the assumption of the
autophagic stage. The clinical oncologists diagnose
chemotherapy-resistant relapsed cancer. The practice of autophagy
by the malignantly transformed cells in advanced multicellular
hosts is a return to an atavistic physiological reaction widely
exercised by the ancestors of cellular life on Earth.
The intracellular environment with operational p53,
but absent MDM in the choanoflagellates, cnidarians and Spongiae
might have been chronically inflammatory, and thus hyperactive, but
not antiapoptotic. Their semblance to the malignantly transforming
cells higher up on the evolutionary scale is comparable to the Wnt
and NF-κB/STAT pathways, but not to the MDM anti-p53 pathway.
Environmentally stressed predecessors, the virus-resistant
cnidarians (Hydra magnipa-pillata; N. vectensis)
might have initiated for their survival the scenario that occurs in
advanced stages of the evolution, and manifests in cell survival by
the Wnt, and NF-κB/STAT co-activation, and the inhibition of
pro-apoptotic p53/p60/p70 by MDM generation. That would be
physiological metabolism in a stressed Ciona or amphioxus.
When activated in some selected individual cells of arthropod,
nematode, vertebrate piscine, reptilian, avian or mammalian
(including human) hosts, these reactions may be viewed as a
reversal to inherent ancient faculties conserved in their genomes
(7,125). The terminology for this condition
in vertebrate hosts is the malignant disease cancer.
What was not available to deal with for the
unicellular eukaryotic ancestors, is the multicellular host in
which somatic cells, germ cells and stem cells coexist. There, if
the malignant transformation was induced by an external pathogen
(an oncogenic virus), the malignant cell has to withstand an immune
attack by the host. Should the malignant transformation be induced
endogenously by a LINE transposon, as referenced in (7), the malignant cell induces no immune
reactions, or if it does, it is able to suppress them by the
mobilization of stromal cell-derived factor chemokine CXC and its
ligand CXCL12; Treg cells; myeloid-derived suppressor cells, and
dendritic cells remaining undifferentiated and tolerogenic.
Macrophages assuming M2 phenotypes, fibroblasts and endothelial
cells rise for direct defense and support of the transformed cell.
The transformed cell establishes cell-to-cell communications by
surface membrane-to-surface membrane contacts, or by the release of
microvesicle endosomes, all for its favor, within its
microenvironment with fibroblasts, vascular endothelial cells, and
M2 macrophages; microvesicles of tumor cell derivation de-activate
immune T cells or even NK cells (125). The tumor cell evokes host
tolerance by the production of IL-6, IL-10, IL-1β generating T17
lymphocytes, TGFβ, prostaglandins and other factors regularly
produced by syncytiotrophoblasts of the placenta, including IDO
(indoleamine 2,3-dioxygenase) (192,193). Thus, endogenously induced tumor
cells receive full support for their growth and spread from their
multicellular hosts. Human tumor cells grown in vitro in
cultures, or as xenografts in athymic mice, lose this support. When
tested for susceptibility to chemotherapeutical agents, or
oncolytic viruses, these cells are defenseless and display
deceivingly high sensitivity. No wonder, most of those treatment
regimens fail to induce complete remissions in the environment of
the natural hosts of the malignantly transformed cells in clinical
trials conducted for the cure of cancer.
6. Conclusion
The primordial cell survival pathways that were
assembled in the life forms in existence before and during the era
of the the Cambrian sea enabled the establishment and expansion of
eukaryotic cellular life on Earth. In extant descendants of all
eukaryotic cells continuing in unicellular, or arranging for
multicellular embodiments of life, the life survival pathways
rendered basic services for the individuals in their ontogenesis
and adult life, including the designs of body plans, generating
haploid germ cells and retaining stem cells for the construction of
organs, including a peripheral and a central nervous system, and
for the replacement of aging somatic cells. In these advanced
multicellular organisms, the cell survival pathways may revert to
their phylo- and ontogenetically ancestral structure and function
in a point-mutated constitutive manner, create new genes by
fusions, and keep the cell in an immature, undifferentiated,
individually independent and immortalized condition. Occurring in
advanced multicellular hosts, these structures are recognized as
proto-oncogenes and oncogenes and their gene products as
oncoproteins. In becoming parasites of their differentiated
multicellular hosts, these cells extract full support from their
host, while exacting mortal consumption. The clinics diagnose the
malady of cancer. The academic laboratories maintaining these
immortal cell populations in culture recognize a close to
indestructible ancient form of eukaryotic cellular life.
Note added in proof
An extraordinary observation concerns the evolution
of tumor necrosis factors (mentioned among Death Factors in
Fig. 2). The human TNF-α, as the
ligand of its receptors R1 and R2, activates different cytoplasmic
pathways. R1 releases NF-κB from its cytoplasmic anchor IκB, thus
allowing its transfer to the nucleus for the activation of its
numerous pro-inflammatory and proto-oncogenic target genes. This
receptor activates also cytoplasmic MAP3K (mitogene-activated
protein kinase). Further, commands emanating from R1 travel either
toward SODD (silencer of death domains), or to FADD (factor
apoptosis stimulator Fas-associated death domain), and to
activation of caspases 8/3. This latter effect culminating in
apoptosis induction was responsible for the unforgettable
sensational first introduction of the newly discovered ‘tumor
necrosis factor’ molecule in the mid-1970's (194). Pathways activated by R2 are those
of VEGF-R2 (vascular endothelial growth factor receptor) and PI3K
(phosphatidylinositol kinase) (195). The ancestor of the human system
was installed in the pre-Cambrian cnidarian corals Acropora, and
remains conserved throughout over 550 million years of evolution in
both the cnidarian and human genomes. The human TNF-α induces
apoptosis by myosin fragmentation in the extant cnidarian coral
cells, and vice versa: the cnidarian TNF equivalent remains an
apoptosis-inducer in human (WT-immortalized Jurkat T-lymphocytes)
cells, both operating through the FADD pathway (196). The derivation and the
evolutionary conservation of proto-oncogenes, from the ancestral
unicellular eukaryotes on, have been recognized early: yeasts and
amoebae (the Dictyostelium) operate the ancestral ras
proto-oncogenes (197).
References
|
1
|
Sullivan JC and Finnerty JR: A surprising
abundance of human disease genes in a simple ‘basal’ animal, the
starlet sea anemone (Nematostella vectensis). Genome. 50:689–692.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sullivan JC, Reitzel AM and Finnerty JR: A
high percentage of introns in human genes were present early in
animal evolution: Evidence from the basal metazoan Nematostella
vectensis. Genome Inform. 17:219–229. 2006.
|
|
3
|
Putnam NH, Srivastava M, Hellsten U, Dirks
B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov
VV, et al: Sea anemone genome reveals ancestral eumetazoan gene
repertoire and genomic organization. Science. 317:86–94. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reitzel AM, Sullivan JC, Traylor-Knowles N
and Finnerty JR: Genomic survey of candidate stress-response genes
in the estuarine anemone Nematostella vectensis. Biol Bull.
214:233–254. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldstone JV: Environmental sensing and
response genes in cnidaria: The chemical defensome in the sea
anemone Nematostella vectensis. Cell Biol Toxicol. 24:483–502.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nelson DR, Goldstone JV and Stegeman JJ:
The cytochrome P450 genesis locus: the origin and evolution of
animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci.
368:201204742013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sinkovics JG: RNA/DNA & Cancer.
Springer Verlag; 2015, (In print).
|
|
8
|
Blanc G, Duncan G, Agarkova I, Borodovsky
M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et
al: The Chlorella variabilis NC64A genome reveals adaptation to
photo-symbiosis, coevolution with viruses, and cryptic sex. Plant
Cell. 22:2943–2955. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vega Thurber RL, Barott KL, Hall D, Liu H,
Rodriguez-Mueller B, Desnues C, Edwards RA, Haynes M, Angly FE,
Wegley L, et al: Metagenomic analysis indicates that stressors
induce production of herpes-like viruses in the coral Porites
compressa. Proc Natl Acad Sci USA. 105:18413–18418. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Davy SK, Allemand D and Weis VM: Cell
biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol
Rev. 76:229–261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grasis JA, Lachnit T, Anton-Erxleben F,
Lim YW, Schmieder R, Fraune S, Franzenburg S, Insua S, Machado G,
Haynes M, et al: Species-specific viromes in the ancestral
holobiont Hydra. PLoS One. 9:e1099522014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sullivan JC, Wolenski FS, Reitzel AM,
French CE, Traylor-Knowles N, Gilmore TD and Finnerty JR: Two
alleles of NF-kappaB in the sea anemone Nematostella vectensis are
widely dispersed in nature and encode proteins with distinct
activities. PLoS One. 4:e73112009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wolenski FS, Garbati MR, Lubinski TJ,
Traylor-Knowles N, Dresselhaus E, Stefanik DJ, Goucher H, Finnerty
JR and Gilmore TD: Characterization of the core elements of the
NF-κB signaling pathway of the sea anemone Nematostella vectensis.
Mol Cell Biol. 31:1076–1087. 2011. View Article : Google Scholar :
|
|
14
|
Armanious H, Gelebart P, Anand M, Belch A
and Lai R: Constitutive activation of metalloproteinase ADAM10 in
mantle cell lymphoma promotes cell growth and activates the
TNFα/NF-κB pathway. Blood. 117:6237–6246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Martin AG and Fresno M: Tumor necrosis
factor-alpha activation of NF-kappa B requires the phosphorylation
of Ser-471 in the transactivation domain of c-Rel. J Biol Chem.
275:24383–24391. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Natoli G, Costanzo A, Moretti F, Fulco M,
Balsano C and Levrero M: Tumor necrosis factor (TNF) receptor 1
signaling downstream of TNF receptor-associated factor 2. Nuclear
factor kappaB (NFkappaB)-inducing kinase requirement for activation
of activating protein 1 and NFkappaB but not of c-Jun N-terminal
kinase/stress-activated protein kinase. J Biol Chem.
272:26079–26082. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee S, Challa-Malladi M, Bratton SB and
Wright CW: Nuclear factor-κB-inducing kinase (NIK) contains an
amino-terminal inhibitor of apoptosis (IAP)-binding motif (IBM)
that potentiates NIK degradation by cellular IAP1 (c-IAP1). J Biol
Chem. 289:30680–30689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schaaf MJM, Willetts L, Hayes BP, Maschera
B, Stylianou E and Farrow SN: The relationship between intranuclear
mobility of the NF-kappaB subunit p65 and its DNA binding affinity.
J Biol Chem. 281:22409–22420. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adam E, Quivy V, Bex F, Chariot A,
Collette Y, Vanhulle C, Schoonbroodt S, Goffin V, Nguyên TL-A,
Gloire G, et al: Potentiation of tumor necrosis factor-induced
NF-κB activation by deacetylase inhibitors is associated with a
delayed cytoplasmic reappearance of IκBα. Mol Cell Biol.
23:6200–6209. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liongue C and Ward AC: Evolution of the
JAK-STAT pathway. JAKSTAT. 2:e227562013.PubMed/NCBI
|
|
21
|
Horvath CM: STAT proteins and
transcriptional responses to extracellular signals. Trends Biochem
Sci. 25:496–502. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Belyi VA, Ak P, Markert E, Wang H, Hu W,
Puzio-Kuter A and Levine AJ: The origins and evolution of the p53
family of genes. Cold Spring Harb Perspect Biol. 2:a0011082010.
View Article : Google Scholar
|
|
23
|
Pankow S and Bamberger C: The p53 tumor
suppressor-like protein nvp63 mediates selective germ cell death in
the sea anemone Nematostella vectensis. PLoS One. 2:e7822007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Momand J, Villegas A and Belyi VA: The
evolution of MDM2 family genes. Gene. 486:23–30. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lane DP, Cheok CF, Brown C, Madhumalar A,
Ghadessy FJ and Verma C: Mdm2 and p53 are highly conserved from
placozoans to man. Cell Cycle. 9:540–547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Biswas G, Anandatheerthavarada HK, Zaidi M
and Avadhani NG: Mitochondria to nucleus stress signaling: A
distinctive mechanism of NFkappaB/Rel activation through
calcineurin-mediated inactivation of IkappaBbeta. J Cell Biol.
161:507–519. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Amuthan G, Biswas G, Zhang SY,
Klein-Szanto A, Vijayasarathy C and Avadhani NG:
Mitochondria-to-nucleus stress signaling induces phenotypic
changes, tumor progression and cell invasion. EMBO J. 20:1910–1920.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hinz M, Lemke P, Anagnostopoulos I, Hacker
C, Krappmann D, Mathas S, Dörken B, Zenke M, Stein H and
Scheidereit C: Nuclear factor kappaB-dependent gene expression
profiling of Hodgkin's disease tumor cells, pathogenetic
significance, and link to constitutive signal transducer and
activator of transcription 5a activity. J Exp Med. 196:605–617.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nagy ZS, Rui H, Stepkowski SM, Karras J
and Kirken RA: A preferential role for STAT5, not constitutively
active STAT3, in promoting survival of a human lymphoid tumor. J
Immunol. 177:5032–5040. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu L, Lin-Lee YC, Pham LV, Tamayo A,
Yoshimura L and Ford RJ: Constitutive NF-kappaB and NFAT activation
leads to stimulation of the BLyS survival pathway in aggressive
B-cell lymphomas. Blood. 107:4540–4548. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pham LV, Fu L, Tamayo AT, Bueso-Ramos C,
Drakos E, Vega F, Medeiros LJ and Ford RJ: Constitutive BR3
receptor signaling in diffuse, large B-cell lymphomas stabilizes
nuclear factor-κB-inducing kinase while activating both canonical
and alternative nuclear factor-κB pathways. Blood. 117:200–210.
2011. View Article : Google Scholar :
|
|
32
|
Gebauer N, Hardel TT, Gebauer J, Bernard
V, Merz H, Feller AC, Rades D, Biersack H, Lehnert H and Thorns C:
Activating mutations affecting the NF-kappa B pathway and
EZH2-mediated epigenetic regulation are rare events in primary
mediastinal large B-cell lymphoma. Anticancer Res. 34:5503–5507.
2014.PubMed/NCBI
|
|
33
|
Odqvist L, Montes-Moreno S,
Sánchez-Pacheco RE, Young KH, Martín-Sánchez E, Cereceda L,
Sánchez-Verde L, Pajares R, Mollejo M, Fresno MF, et al: NF-κB
expression is a feature of both activated B-cell-like and germinal
center B-cell-like subtypes of diffuse large B-cell lymphoma. Mod
Pathol. 27:1331–1337. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Demchenko YN and Kuehl WM: A critical role
for the NF-κB pathway in multiple myeloma. Oncotarget. 1:59–68.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Litvinov IV, Cordeiro B, Fredholm S, Ødum
N, Zargham H, Huang Y, Zhou Y, Pehr K, Kupper TS, Woetmann A, et
al: Analysis of STAT4 expression in cutaneous T-cell lymphoma
(CTCL) patients and patient-derived cell lines. Cell Cycle.
13:2975–2982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gelfanov VG, Burgess GS, Litz-Jackson S,
King AJ, Marshall MS, Nakasatri H and Boswell HS: Transformation of
interleukin-3-dependent cells without participation of
Stat5/bcl-xL: Cooperation leads to p65 nuclear factor κB-mediated
apoptosis involving c-IAP2. Blood. 98:2508–2517. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Feuerhake F, Kutok JL, Monti S, Chen W,
LaCasce AS, Cattoretti G, Kurtin P, Pinkus GS, de Leval L, Harris
NL, et al: NFkappaB activity, function, and target-gene signatures
in primary mediastinal large B-cell lymphoma and diffuse large
B-cell lymphoma subtypes. Blood. 106:1392–1399. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pont-Kingdon GA, Beagley CT, Okimoto R and
Wolstenholme DR: Mitochondrial DNA of the sea anemone, Metridium
senile (Cnidaria): Prokaryote-like genes for tRNA(f-Met) and
small-subunit ribosomal RNA, and standard genetic code
specificities for AGR and ATA codons. J Mol Evol. 39:387–399. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Beagley CT, Okimoto R and Wolstenholme DR:
The mitochondrial genome of the sea anemone Metridium senile
(Cnidaria): Introns, a paucity of tRNA genes, and a near-standard
genetic code. Genetics. 148:1091–1108. 1998.PubMed/NCBI
|
|
40
|
Kretz-Remy C, Munsch B and Arrigo AP:
NFkappa B-dependent transcriptional activation during heat shock
recovery. Thermolability of the NF-kappaB complex. J Biol Chem.
276:43723–43733. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Starczynowski DT, Trautmann H, Pott C,
Harder L, Arnold N, Africa JA, Leeman JR, Siebert R and Gilmore TD:
Mutation of an IKK phosphorylation site within the transactivation
domain of REL in two patients with B-cell lymphoma enhances REL's
in vitro transforming activity. Oncogene. 26:2685–2694. 2007.
View Article : Google Scholar
|
|
42
|
Thompson RC, Vardinogiannis I and Gilmore
TD: Identification of an NF-κB p50/p65-responsive site in the human
MIR155HG promoter. BMC Mol Biol. 14:242013. View Article : Google Scholar
|
|
43
|
Lind EF, Elford AR and Ohashi PS:
Micro-RNA 155 is required for optimal CD8+ T cell
responses to acute viral and intracellular bacterial challenges. J
Immunol. 190:1210–1216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Netchiporouk E, Litvinov IV, Moreau L,
Gilbert M, Sasseville D and Duvic M: Deregulation in STAT signaling
is important for cutaneous T-cell lymphoma (CTCL) pathogenesis and
cancer progression. Cell Cycle. 13:3331–3335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Marosvári D, Téglási V, Csala I,
Marschalkó M, Bödör C, Timár B, Csomor J, Hársing J and Reiniger L:
Altered microRNA expression in folliculotropic and transformed
mycosis fungoides. Pathol Oncol Res. 21:821–825. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Silva LM, Hirai KE, de Souza JR, Fuzii HT,
Dias LB Jr, Carneiro FR, de Souza Aarão TL and Quaresma JA:
Immunohistochemical analysis of the expression of cellular
transcription NFκB (p65). AP-1c-Fos and c-Jun and JAK/STAT in
leprosy. Human Pathol. 46:746–752. 2015. View Article : Google Scholar
|
|
47
|
Ray PS, Sullivan JC, Jia J, Francis J,
Finnerty JR and Fox PL: Evolution of function of a fused metazoan
tRNA synthetase. Mol Biol Evol. 28:437–447. 2011. View Article : Google Scholar
|
|
48
|
Sinkovics JG, Howe CD and Shullenberger
CC: Cellular activities in tissue culture of leukemic human bone
marrow. Blood. 24:389–401. 1964.PubMed/NCBI
|
|
49
|
Baus D, Nonnenmacher F, Jankowski S,
Döring C, Bräutigam C, Frank M, Hansmann ML and Pfitzner E: STAT6
and STAT1 are essential antagonistic regulators of cell survival in
classical Hodgkin lymphoma cell line. Leukemia. 23:1885–1893. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Canoz O, Rassidakis GZ, Admirand JH and
Medeiros LJ: Immunohistochemical detection of BCL-3 in lymphoid
neoplasms: A survey of 353 cases. Mod Pathol. 17:911–917. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cahir McFarland ED, Izumi KM and Mosialos
G: Epstein-Barr virus transformation: Involvement of latent
membrane protein 1-mediated activation of NF-kappaB. Oncogene.
18:6959–6964. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Espinoza JL, Takami A, Trung LQ, Kato S
and Nakao S: Resveratrol prevents EBV transformation and inhibits
the outgrowth of EBV immortalized human B cells. PLoS One.
7:e13062012. View Article : Google Scholar
|
|
53
|
Kashanchi F, Araujo J, Doniger J,
Muralidhar S, Hoch R, Khleif S, Mendelson E, Thompson J, Azumi N,
Brady JN, et al: Human herpesvirus 6 (HHV-6) ORF-1 transactivating
gene exhibits malignant transforming activity and its protein binds
to p53. Oncogene. 14:359–367. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lacroix A, Collot-Teixeira S, Mardivirin
L, Jaccard A, Petit B, Piguet C, Sturtz F, Preux P-M, Bordessoule D
and Ranger-Rogez S: Involvement of human herpesvirus-6 variant B in
classic Hodgkin's lymphoma via DR7 oncoprotein. Clin Cancer Res.
16:4711–4721. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sun Q, Matta H and Chaudhary PM: Kaposi's
sarcoma associated herpes virus-encoded viral FLICE inhibitory
protein activates transcription from HIV-1 Long Terminal Repeat via
the classical NF-kappaB pathway and functionally cooperates with
Tat. Retrovirology. 2:92005. View Article : Google Scholar
|
|
56
|
Hussain AR, Ahmed SO, Ahmed M, Khan OS, Al
Abdulmohsen S, Platanias LC, Al-Kuraya KS and Uddin S: Cross-talk
between NF-κB and the PI3-kinase/AKT pathway can be targeted in
primary effusion lymphoma (PEL) cell lines for efficient apoptosis.
PLoS One. 7:e399452012. View Article : Google Scholar
|
|
57
|
Sun SC and Ballard DW: Persistent
activation of NF-kappaB by the tax transforming protein of HTLV-1:
Hijacking cellular IkappaB kinases. Oncogene. 18:6948–6958. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shehata MF: Rel/Nuclear factor-kappa B
apoptosis pathways in human cervical cancer cells. Cancer Cell Int.
5:102005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Németh ZH, Deitch EA, Szabó C and Haskó G:
Pyrrolidine-dithiocarbamate inhibits NF-kappaB activation and IL-8
production in intestinal epithelial cells. Immunol Lett. 85:41–46.
2003. View Article : Google Scholar
|
|
60
|
Wang W, Nag SA and Zhang R: Targeting the
NF-κB signaling pathways for breast cancer prevention and therapy.
Curr Med Chem. 22:264–289. 2015. View Article : Google Scholar
|
|
61
|
Guzmán EA, Maers K, Roberts J,
Kemami-Wangun HV, Harmody D and Wright AE: The marine natural
product micro-sclerodermin A is a novel inhibitor of the nuclear
factor kappa B and induces apoptosis in pancreatic cancer cells.
Invest New Drugs. 33:86–94. 2015. View Article : Google Scholar
|
|
62
|
Chan R, Gilbert M, Thompson KM, Marsh HN,
Epstein DM and Pendergrast PS: Co-expression of anti-NFkappaB RNA
aptamers and siRNAs leads to maximal suppression of NFkappaB
activity in mammalian cells. Nucleic Acids Res. 34:e362006.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
McKenna S and Wright CJ: Inhibiting
IκBβ/NF-κB signaling attenuates the expression of select
pro-inflammatory genes. J Cell Sci. 128:2143–2155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chang HC, Lin KH, Tai YT, Chen JT and Chen
RM: Lipoteichoic acid-induced TNF-α and IL-6 gene expressions and
oxidative stress production in macrophages are suppressed by
ketamine through downregulating Toll-like receptor-mediated
activation of ERK1/2 and NF-κB. Shock. 33:486–492. 2010.
|
|
65
|
García-Piñeres AJ, Castro V, Mora G,
Schmidt TJ, Strunck E, Pahl HL and Merfort I: Cysteine 38 in
p65/NF-kappaB plays a crucial role in DNA binding inhibition by
sesquiterpene lactones. J Biol Chem. 276:39713–39720. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liang MC, Bardhan S, Porco JA Jr and
Gilmore TD: The synthetic epoxyquinoids jesterone dimer and
epoxyquinone A monomer induce apoptosis and inhibit REL (human
c-Rel) DNA binding in an IkappaBalpha-deficient diffuse large
B-cell lymphoma cell line. Cancer Lett. 241:69–78. 2006. View Article : Google Scholar
|
|
67
|
Liang MC, Bardhan S, Pace EA, Rosman D,
Beutler JA, Porco JA Jr and Gilmore TD: Inhibition of transcription
factor NF-kappaB signaling proteins IKKbeta and p65 through
specific cysteine residues by epoxyquinone A monomer: Correlation
with its anti-cancer cell growth activity. Biochem Pharmacol.
71:634–645. 2006. View Article : Google Scholar
|
|
68
|
Ramakrishnan P, Clark PM, Mason DE, Peters
EC, Hsieh-Wilson LC and Baltimore D: Activation of the
transcriptional function of the NF-κB protein c-Rel by O-GlcNAc
glycosylation. Sci Signal. 6:ra752013. View Article : Google Scholar
|
|
69
|
Selvan N, Maiappa D, van den Toom HWP,
Heck AJK, Ferenbach AT and van Aalten DMF: The early metazoan
Trichoplax adhaerens possesses a functional O-GlcNAc system. J Biol
Chem. 250:11969–11982. 2015. View Article : Google Scholar
|
|
70
|
Sümegi M, Hunyadi-Gulyás E, Medzihradszky
KF and Udvardy A: 26S proteasome subunits are O-linked
N-acetylglucosamine-modified in Drosophila melanogaster. Biochem
Biophys Res Commun. 312:1284–1289. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Heyne K, Winter C, Gerten F, Schmidt C and
Roemer K: A novel mechanism of crosstalk between p53 and NF-κB
pathways. Cell Cycle. 12:2479–2482. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cha B, Lim JW and Kim H: Jak1/Stat3 is an
upstream signaling of NF-κB activation in Helicobacter
pylori-induced IL-8 production in gastric epithelial AGS cells.
Yonsei Med J. 56:862–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Linher-Melville K, Haftchenary S, Gunning
P and Singh G: Signal transducer and activator of transcription 3
and 5 regulate system Xc- and redox balance in human breast cancer
cells. Mol Cell Biochem. 405:205–221. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Pramanik KC, Fofaria NM, Gupta P, Ranjan
A, Kim SH and Srivastava SK: Inhibition of β-catenin signaling
suppresses pancreatic tumor growth by disrupting nuclear
β-catenin/TCF-1 complex: Critical role of STAT-3. Oncotarget.
6:11561–11574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Martinou JC and Kroemer G: Autophagy:
Evolutionary and pathophysiological insights. Biochim Biophys Acta.
1793:1395–1396. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Katayama H, Kogure T, Mizushima N,
Yoshimori T and Miyawaki A: A sensitive and quantitative technique
for detecting autophagic events based on lysosomal delivery. Chem
Biol. 18:1042–1052. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Buzgariu W, Chera S and Galliot B: Methods
to investigate autophagy during starvation and regeneration in
hydra. Methods Enzymol. 451:409–437. 2008. View Article : Google Scholar
|
|
78
|
Chera S, Buzgariu W, Ghila L and Galliot
B: Autophagy in Hydra: A response to starvation and stress in early
animal evolution. Biochim Biophys Acta. 1793:1432–1443. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Dunn SR, Schnitzler CE and Weis VM:
Apoptosis and autophagy as mechanisms of dinoflagellate symbiont
release during cnidarian bleaching: Every which way you lose. Proc
Biol Sci. 274:3079–3085. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Buss LW, Anderson C, Westerman E,
Kritzberger C, Poudyal M, Moreno MA and Lakkis FG: Allorecognition
triggers autophagy and subsequent necrosis in the cnidarian
Hydractinia symbiolon-gicarpus. PLoS One. 7:e489142012. View Article : Google Scholar
|
|
81
|
Petersen HO, Höger SK, Looso M, Lengfeld
T, Kuhn A, Warnken U, Nishimiya-Fujisawa C, Schnölzer M, Krüger M,
Özbek S, et al: A comprehensive transcriptomic and proteomic
analysis of hydra head regeneration. Mol Biol Evol. April
2–2015.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Galliot B: Autophagy and
self-preservation: A step ahead from cell plasticity? Autophagy.
2:231–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Galliot B, Miljkovic-Licina M, Ghila L and
Chera S: RNAi gene silencing affects cell and developmental
plasticity in hydra. C R Biol. 330:491–497. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chera S, de Rosa R, Miljkovic-Licina M,
Dobretz K, Ghila L, Kaloulis K and Galliot B: Silencing of the
hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1
pancreatic phenotype. J Cell Sci. 119:846–857. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Räty S, Sand J, Laukkarinen J, Vasama K,
Bassi C, Salvia R and Nordback I: Cyst fluid SPINK1 may help to
differentiate benign and potentially malignant cystic pancreatic
lesions. Pancreatology. 13:530–533. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lempinen M, Paju A, Kemppainen E, Smura T,
Kylänpää ML, Nevanlinna H, Stenman J and Stenman UH: Mutations N34S
and P55S of the SPINK1 gene in patients with chronic pancreatitis
or pancreatic cancer and in healthy subjects: A report from
Finland. Scand J Gastroenterol. 40:225–230. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Shimosegawa T, Kume K and Satoh K: Chronic
pancreatitis and pancreatic cancer: Prediction and mechanism. Clin
Gastroenterol Hepatol. 7(Suppl 11): S23–S28. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Koehler A, Desser S, Chang B, MacDonald J,
Tepass U and Ringuette M: Molecular evolution of SPARC: Absence of
the acidic module and expression in the endoderm of the starlet sea
anemone, Nematostella vectensis. Dev Genes Evol. 219:509–521. 2009.
View Article : Google Scholar
|
|
89
|
Fritzenwanker JH, Saina M and Technau U:
Analysis of forkhead and snail expression reveals
epithelial-mesenchymal transitions during embryonic and larval
development of Nematostella vectensis. Dev Biol. 275:389–402. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Sinkovics JG: Horizontal gene transfers
and cell fusions in microbiology, immunology and oncology (Review).
Int J Oncol. 35:441–465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Grant JL, Fishbein MC, Hong LS, Krysan K,
Minna JD, Shay JW, Walser TC and Dubinett SM: A novel molecular
pathway for Snail-dependent, SPARC-mediated invasion in non-small
cell lung cancer pathogenesis. Cancer Prev Res (Phila). 7:150–160.
2014. View Article : Google Scholar
|
|
92
|
Kaleağasıoğlu F and Berger MR: SIBLINGs
and SPARC families: Their emerging roles in pancreatic cancer.
World J Gastroenterol. 20:14747–14759. 2014. View Article : Google Scholar
|
|
93
|
Yang F, Zhou X, Miao X, Zhang T, Hang X,
Tie R, Liu N, Tian F, Wang F and Yuan J: MAGEC2, an
epithelial-mesenchymal transition inducer, is associated with
breast cancer metastasis. Breast Cancer Res Treat. 145:23–32. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Sullivan JC, Sher D, Eisenstein M,
Shigesada K, Reitzel AM, Marlow H, Levanon D, Groner Y, Finnerty JR
and Gat U: The evolutionary origin of the Runx/CBFbeta
transcription factors--studies of the most basal metazoans. BMC
Evol Biol. 8:2282008. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Klunker S, Chong MMW, Mantel PY, Palomares
O, Bassin C, Ziegler M, Rückert B, Meiler F, Akdis M, Littman DR,
et al: Transcription factors RUNX1 and RUNX3 in the induction and
suppressive function of Foxp3+ inducible regulatory T
cells. J Exp Med. 206:2701–2715. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lück SC, Russ AC, Du J, Gaidzik V, Schlenk
RF, Pollack JR, Döhner K, Döhner H and Bullinger L: KIT mutations
confer a distinct gene expression signature in core binding factor
leukaemia. Br J Haematol. 148:925–937. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Huang G, Zhao X, Wang L, Elf S, Xu H, Zhao
X, Sashida G, Zhang Y, Liu Y, Lee J, et al: The ability of MLL to
bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is
impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood.
118:6544–6552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Bledsoe KL, McGee-Lawrence ME, Camilleri
ET, Wang X, Riester SM, van Wijnen AJ, Oliveira AM and Westendorf
JJ: RUNX3 facilitates growth of Ewing sarcoma cells. J Cell
Physiol. 229:2049–2056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hsu YL, Huang MS, Yang CJ, Hung JY, Wu LY
and Kuo PL: Lung tumor-associated osteoblast-derived bone
morphogenetic protein-2 increased epithelial-to-mesenchymal
transition of cancer by Runx2/Snail signaling pathway. J Biol Chem.
286:37335–37346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Estécio MR, Maddipoti S, Bueso-Ramos C,
DiNardo CD, Yang H, Wei Y, Kondo K, Fang Z, Stevenson W, Chang KS,
et al: RUNX3 promoter hypermethylation is frequent in leukaemia
cell lines and associated with acute myeloid leukaemia inv(16)
subtype. Br J Haematol. 169:344–351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Martin JW, Zielenska M, Stein GS, van
Wijnen AJ and Squire JA: The role of RUNX2 in osteosarcoma
oncogenesis. Sarcoma. 2011:2827452011. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Wang X, Goldstein D, Crowe PJ and Yang JL:
Impact of STAT3 inhibition on survival of osteosarcoma cell lines.
Anticancer Res. 34:6537–6545. 2014.PubMed/NCBI
|
|
103
|
Yang Q, Zhang S, Kang M, Dong R and Zhao
J: Synergistic growth inhibition by sorafenib and cisplatin in
human osteosar-coma cells. Oncol Rep. 33:2537–2544. 2015.PubMed/NCBI
|
|
104
|
Sinkovics JG: Cytolytic Immune
Lymphocytes. Schenk Buchverlag Passau; Germany; Dialog Campus,
Budapest: pp. 2802008
|
|
105
|
Burgess M and Tawbi H: Immunotherapeutic
approaches to sarcoma. Curr Treat Options Oncol. 16:3452015.
View Article : Google Scholar
|
|
106
|
Matus DQ, Magie CR, Pang K, Martindale MQ
and Thomsen GH: The Hedgehog gene family of the cnidarian,
Nematostella vectensis, and implications for understanding metazoan
Hedgehog pathway evolution. Dev Biol. 313:501–518. 2008. View Article : Google Scholar :
|
|
107
|
Bürglin TR: Evolution of hedgehog and
hedgehog-related genes, their origin from Hog proteins in ancestral
eukaryotes and discovery of a novel Hint motif. BMC Genomics.
9:1272008. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
McCabe JM and Leahy DJ: Smoothened goes
molecular: New pieces in the hedgehog signaling puzzle. J Biol
Chem. 290:3500–3507. 2015. View Article : Google Scholar
|
|
109
|
Warner JF, McCarthy AM, Morris RL and
McClay DR: Hedgehog signaling requires motile cilia in the sea
urchin. Mol Biol Evol. 31:18–22. 2014. View Article : Google Scholar :
|
|
110
|
Kern D, Regl G, Hofbauer SW, Altenhofer P,
Achatz G, Dlugosz A, Schnidar H, Greil R, Hartmann TN and Aberger
F: Hedgehog/GLI and PI3K signaling in the initiation and
maintenance of chronic lymphocytic leukemia. Oncogene. 2015,
doi.org/10.1038/onc.2014.450urisimpledoi.org/10.1038/onc.2014.450.
View Article : Google Scholar
|
|
111
|
Giakoustidis A, Giakoustidis D, Mudan S,
Sklavos A and Williams R: Molecular signalling in hepatocellular
carcinoma: Role of and crosstalk among WNT/β-catenin, Sonic
Hedgehog, Notch and Dickkopf-1. Can J Gastroenterol Hepatol.
29:212–211. 2015.
|
|
112
|
Layden MJ and Martindale MQ: Non-canonical
Notch signaling represents an ancestral mechanism to regulate
neural differentiation. Evodevo. 5:302014. View Article : Google Scholar
|
|
113
|
Layden MJ, Boekhout M and Martindale MQ:
Nematostella vectensis achaete-scute homolog NvashA regulates
embryonic ectodermal neurogenesis and represents an ancient
component of the metazoan neural specification pathway.
Development. 139:1013–1022. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Marlow H, Roettinger E, Boekhout M and
Martindale MQ: Functional roles of Notch signaling in the cnidarian
Nematostella vectensis. Dev Biol. 362:295–308. 2012. View Article : Google Scholar
|
|
115
|
Krejcí A and Bray S: Notch activation
stimulates transient and selective binding of Su(H)/CSL to target
enhancers. Genes Dev. 21:1322–1327. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Guito J and Lukac DM: KSHV reactivation
and novel implications of protein isomerization on lytic switch
control. Viruses. 7:72–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Spadavecchia S, Gonzalez-Lopez O, Carroll
KD, Palmeri D and Lukac DM: Convergence of Kaposi's
sarcoma-associated herpesvirus reactivation with Epstein-Barr virus
latency and cellular growth mediated by the notch signaling pathway
in coinfected cells. J Virol. 84:10488–10500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Rettig EM, Chung CH, Bishop JA, Howard JD,
Sharma R, Li RJ, Douville C, Karchin R, Izumchenko E, Sidransky D,
et al: Cleaved NOTCH1 expression pattern in head and neck squamous
cell carcinoma is associated with NOTCH1 mutation, HPV status, and
high risk features. Cancer Prev Res (Phila). 8:287–295. 2015.
View Article : Google Scholar
|
|
119
|
Ayaz F and Osborne BA: Non-canonical notch
signaling in cancer and immunity. Front Oncol. 4:3452014.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Jager M, Quéinnec E, Le Guyader H and
Manuel M: Multiple Sox genes are expressed in stem cells or in
differentiating neuro-sensory cells in the hydrozoan Clytia
hemisphaerica. Evodevo. 2:122011. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Shinzato C, Iguchi A, Hayward DC, Technau
U, Ball EE and Miller DJ: Sox genes in the coral Acropora
millepora: Divergent expression patterns reflect differences in
developmental mechanisms within the Anthozoa. BMC Evol Biol.
8:3112008. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Richards GS and Rentzsch F: Transgenic
analysis of a SoxB gene reveals neural progenitor cells in the
cnidarian Nematostella vectensis. Development. 141:4681–4689. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Magie CR, Pang K and Martindale MQ:
Genomic inventory and expression of Sox and Fox genes in the
cnidarian Nematostella vectensis. Dev Genes Evol. 215:618–630.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Thu KL, Becker-Santos DD, Radulovich N,
Pikor LA, Lam WL and Tsao MS: SOX15 and other SOX family members
are important mediators of tumorigenesis in multiple cancer types.
Oncoscience. 1:326–335. 2014.
|
|
125
|
Sinkovics JG: The cell survival pathways
of the primordial RNA-DNA complex remain conserved in the extant
genomes and may function as proto-oncogenes. Eur J Microbiol
Immunol (Bp). 5:25–43. 2015. View Article : Google Scholar
|
|
126
|
Ryan JF and Baxevanis AD: Hox, Wnt, and
the evolution of the primary body axis: Insights from the
early-divergent phyla. Biol Direct. 2:372007. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Holstein TW: The evolution of the Wnt
pathway. Cold Spring Harb Perspect Biol. 4:a0079222012. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Xin M: Hedgehog inhibitors: A patent
review (2013 - present). Expert Opin Ther Pat. 25:549–565. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lockhart NR, Waddell JA and Schrock NE:
Itraconazole therapy in a pancreatic adenocarcinoma patient: A case
report. J Oncol Pharm Pract. Feb 9–2015.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Huang YC, Chao KS, Liao HF and Chen YJ:
Targeting sonic hedgehog signaling by compounds and derivatives
from natural products. Evid Based Complement Alternat Med.
2013:7485872013. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Chung SS and Vadgama JV: Curcumin and
epigallocatechin gallate inhibit the cancer stem cell phenotype via
down-regulation of STAT3-NF-κB signaling. Anticancer Res. 35:39–46.
2015.PubMed/NCBI
|
|
132
|
Song L, Li ZY, Liu WP and Zhao MR:
Crosstalk between Wnt/β-catenin and Hedgehog/Gli signaling pathways
in colon cancer and implications for therapy. Cancer Biol Ther.
16:1–7. 2015. View Article : Google Scholar
|
|
133
|
[a] Song J, Du Z,
Ravasz M, Dong B, Wang Z and Ewing RM: A protein interaction
between beta-catenin and Dnmtl regulates Wnt signaling and DNA
methylation in colorectal cancer cells. Mol Cancer Res. 13:969–981.
2015. View Article : Google Scholar : PubMed/NCBI
[b] Jin L,
Hanigan CL, Wu Y, Wang W, Park BH, Woster PM and Casero RA: Loss of
LSD1 (lysine-specific demethylase 1) suppresses growth and alters
gene expression of human cancer cells in a p53- and DNMT1 (DNA
methyltransferase 1)-independent manner. Biochem J. 449:459–468.
2013. View Article : Google Scholar
|
|
134
|
Blackburn HL, Ellsworth DL, Shriver CD and
Ellsworth RE: Role of cytochrome P450 genes in breast cancer
etiology and treatment: Effects on estrogen biosynthesis,
metabolism, and response to endocrine therapy. Cancer Causes
Control. 26:319–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Go RE, Hwang KA and Choi KC: Cytochrome
P450 1 family and cancers. J Steroid Biochem Mol Biol. 147:24–30.
2015. View Article : Google Scholar
|
|
136
|
[a] Yue JX, Yu
JK, Putnam NH and Holland LZ: The tran-scriptome of an amphioxus,
Asymmetron lucayanum, from the Bahamas: A window into chordate
evolution. Genome Biol Evol. 6:2681–2696. 2014. View Article : Google Scholar : PubMed/NCBI
[b] Lu TM, Luo YJ
and Yu JK: BMP and Delta/Notch signaling control the development of
amphioxus epidermal sensory neurons: Insights into the evolution of
the peripheral sensory system. Development. 139:2020–2030. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Sinkovics JG: Horizontal gene transfers
with or without cell fusions in all categories of the living
matter. Adv Exp Med Biol. 714:5–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Mhawech P, Berczy M, Assaly M, Herrmann F,
Bouzourene H, Allal AS, Dulguerov P and Schwaller J: Human
achaete-scute homologue (hASH1) mRNA level as a diagnostic marker
to distinguish esthesioneuroblastoma from poorly differentiated
tumors arising in the sinonasal tract. Am J Clin Pathol.
122:100–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Carney ME, O'Reilly RC, Sholevar B,
Buiakova 01, Lowry LD, Keane WM, Margolis FL and Rothstein JL:
Expression of the human Achaete-scute 1 gene in olfactory
neuroblastoma. J Neurooncol. 26:35–43. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Holoye PY, Samuels ML, Smith T and
Sinkovics JG: Chemoimmunotherapy of small cell bronchogenic
carcinoma. Cancer. 42:34–40. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Augustyn A, Borromeo M, Wang T, Fujimoto
J, Shao C, Dospoy PD, Lee V, Tan C, Sullivan JP, Larsen JE, et al:
ASCL1 is a lineage oncogene providing therapeutic targets for
high-grade neuroendocrine lung cancers. Proc Natl Acad Sci USA.
111:14788–14793. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Shabalina SA and Koonin EV: Origins and
evolution of eukaryotic RNA interference. Trends Ecol Evol.
23:578–587. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Christodoulou F, Raible F, Tomer R,
Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P and
Arendt D: Ancient animal microRNAs and the evolution of tissue
identity. Nature. 463:1084–1088. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Grimson A, Srivastava M, Fahey B,
Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS and Bartel
DP: Early origins and evolution of microRNAs and Piwi-interacting
RNAs in animals. Nature. 455:1193–1197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Hertel J, Bartschat S, Wintsche A, Otto C
and Stadler PF; Students of the Bioinformatics Computer Lab.
Evolution of the let-7 microRNA family. RNA Biol. 9:231–241. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Ambros V: A hierarchy of regulatory genes
controls a larva-to-adult developmental switch in C. elegans. Cell.
57:49–57. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Krishna S, Nair A, Cheedipudi S, Poduval
D, Dhawan J, Palakodeti D and Ghanekar Y: Deep sequencing reveals
unique small RNA repertoire that is regulated during head
regeneration in Hydra magnipapillata. Nucleic Acids Res.
41:599–616. 2013. View Article : Google Scholar :
|
|
148
|
Juliano CE, Reich A, Liu N, Götzfried J,
Zhong M, Uman S, Reenan RA, Wessel GM, Steele RE and Lin H: PIWI
proteins and PIWI-interacting RNAs function in Hydra somatic stem
cells. Proc Natl Acad Sci USA. 111:337–342. 2014. View Article : Google Scholar :
|
|
149
|
Chen P, Xi Q, Wang Q and Wei P:
Downregulation of microRNA-100 correlates with tumor progression
and poor prognosis in colorectal cancer. Med Oncol. 31:2352014.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Peng H, Luo J, Hao H, Hu J, Xie SK, Ren D
and Rao B: MicroRNA-100 regulates SW620 colorectal cancer cell
proliferation and invasion by targeting RAP1B. Oncol Rep.
31:2055–2062. 2014.PubMed/NCBI
|
|
151
|
Chen D, Sun Y, Yuan Y, Han Z, Zhang P,
Zhang J, You MJ, Teruya-Feldstein J, Wang M, Gupta S, et al:
miR-100 induces epithelial-mesenchymal transition but suppresses
tumorigenesis, migration and invasion. PLoS Genet. 10:e10041772014.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Ng WL, Yan D, Zhang X, Mo YY and Wang Y:
Over-expression of miR-100 is responsible for the low-expression of
ATM in the human glioma cell line: M059J. DNA Repair (Amst).
9:1170–1175. 2010. View Article : Google Scholar
|
|
153
|
Morais DR, Reis ST, Viana N, Piantino CB,
Massoco C, Moura C, Dip N, Silva IA, Srougi M and Leite KRM: The
involvement of miR-100 in bladder urothelial carcinogenesis
changing the expression levels of mRNA and proteins of genes
related to cell proliferation, survival, apoptosis and chromosomal
stability. Cancer Cell Int. 14:1192014. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Li Z, Li X, Yu C, Wang M, Peng F, Xiao J,
Tian R, Jiang J and Sun C: MicroRNA-100 regulates pancreatic cancer
cells growth and sensitivity to chemotherapy through targeting
FGFR3. Tumour Biol. 35:11751–11759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Wang M, Ren D, Guo W, Wang Z, Huang S, Du
H, Song L and Peng X: Loss of miR-100 enhances migration, invasion,
epithelial-mesenchymal transition and stemness properties in
prostate cancer cells through targeting Argonaute 2. Int J Oncol.
45:362–372. 2014.PubMed/NCBI
|
|
156
|
Ghose J and Bhattacharyya NP:
Transcriptional regulation of microRNA-100, -146a, and -150 genes
by p53 and NFκB p65/RelA in mouse striatal STHdh(Q7)/Hdh(Q7) cells
and human cervical carcinoma HeLa cells. RNA Biol. 12:457–477.
2015. View Article : Google Scholar
|
|
157
|
Ma X, Li C, Sun L, Huang D, Li T, He X, Wu
G, Yang Z, Zhong X, Song L, et al: Lin28/let-7 axis regulates
aerobic glycolysis and cancer progression via PDK1. Nat Commun.
5:52122014. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Sinkovics JG: Molecular biology of
oncogenic inflammatory processes. I. Non-oncogenic and oncogenic
pathogens, intrinsic inflammatory reactions without pathogens, and
microRNA/DNA interactions (Review). Int J Oncol. 40:305–349.
2012.
|
|
159
|
Iliopoulos D, Hirsch HA and Struhl K: An
epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and
IL6 links inflammation to cell transformation. Cell. 139:693–706.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Bosch TC, Unger TF, Fisher DA and Steele
RE: Structure and expression of STK, a src-related gene in the
simple metazoan Hydra attenuata. Mol Cell Biol. 9:4141–4151.
1989.PubMed/NCBI
|
|
161
|
Baumgarten S, Bayer T, Aranda M, Liew YJ,
Carr A, Micklem G and Voolstra CR: Integrating microRNA and mRNA
expression profiling in Symbiodinium microadriaticum, a
dinoflagellate symbiont of reef-building corals. BMC Genomics.
14:7042013. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Ruiz-Ramos DV and Baums IB: Microsatellite
abundance across the Anthozoa and Hydrozoa in the phylum Cnidaria.
BMC Genomics. 15:9392014. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Srivastava M, Simakov O, Chapman J, Fahey
B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten
U, et al: The Amphimedon queenslandica genome and the evolution of
animal complexity. Nature. 466:720–726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Kerner P, Degnan SM, Marchand L, Degnan BM
and Vervoort M: Evolution of RNA-binding proteins in animals:
Insights from genome-wide analysis in the sponge Amphimedon
queenslandica. Mol Biol Evol. 28:2289–2303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Srivastava M, Begovic E, Chapman J, Putnam
NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter
ML, et al: The Trichoplax genome and the nature of placozoans.
Nature. 454:955–960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Schierwater B, de Jong D and Desalle R:
Placozoa and the evolution of Metazoa and intrasomatic cell
differentiation. Int J Biochem Cell Biol. 41:370–379. 2009.
View Article : Google Scholar
|
|
167
|
Moroz LL, Kocot KM, Citarella MR, Dosung
S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov
E, Buckley KM, et al: The ctenophore genome and the evolutionary
origins of neural systems. Nature. 510:109–114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Nelson DR, Goldstone JV and Stegeman JJ:
The cytochrome P-450 genesis locus: the origin and evolution of
animal cytochrome P450. Philos Trans R Soc Lond B Biol Sci.
368:2012.04742013.
|
|
169
|
Robertson AJ, Larroux C, Degnan BM and
Coffman JA: The evolution of Runx genes II. The C-terminal Groucho
recruitment motif is present in both eumetazoans and
homoscleromorphs but absent in a haplosclerid demosponge. BMC Res
Notes. 2:592009. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Stefanik DJ, Lubinski TJ, Granger BR, Byrd
AL, Reitzel AM, DeFilippo L, Lorenc A and Finnerty JR: Production
of a reference transcriptome and transcriptomic database
(EdwardsiellaBase) for the lined sea anemone, Edwardsiella lineata,
a parasitic cnidarian. BMC Genomics. 15:712014. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Fernández JG, Rodríguez DA, Valenzuela M,
Calderon C, Urzúa U, Munroe D, Rosas C, Lemus D, Díaz N, Wright MC,
et al: Survivin expression promotes VEGF-induced tumor angiogenesis
via PI3K/Akt enhanced β-catenin/Tcf-Lef dependent transcription.
Mol Cancer. 13:2092014. View Article : Google Scholar
|
|
172
|
Jager M, Dayraud C, Mialot A, Quéinnec E,
le Guyader H and Manuel M: Evidence for involvement of Wnt
signalling in body polarities, cell proliferation, and the
neuro-sensory system in an adult ctenophore. PLoS One.
8:e843632013. View Article : Google Scholar
|
|
173
|
Pang K, Ryan JF, Mullikin JC, Baxevanis AD
and Martindale MQ; NISC Comparative Sequencing Program. Genomic
insights into Wnt signaling in an early diverging metazoan, the
ctenophore Mnemiopsis leidyi. Evodevo. 1:102010. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Schnitzler CE, Simmons DK, Pang K,
Martindale MQ and Baxevanis AD: Expression of multiple Sox genes
through embryonic development in the ctenophore Mnemiopsis leidyi
is spatially restricted to zones of cell proliferation. Evodevo.
5:152014. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Hua HW, Jiang F, Huang Q, Liao Z and Ding
G: MicroRNA-153 promotes Wnt/β-catenin activation in hepatocellular
carcinoma through suppression of WWOX. Oncotarget. 6:3840–3847.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Yan HC, Xu J, Fang LS, Qiu YY, Lin XM,
Huang HX and Han QY: Ectopic expression of the WWOX gene suppresses
stemness of human ovarian cancer stem cells. Oncol Lett.
9:1614–1620. 2015.PubMed/NCBI
|
|
177
|
Ma R, Jiang T and Kang X: Circulating
microRNAs in cancer: Origin, function and application. J Exp Clin
Cancer Res. 31:382012. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Lee SH, Oh S-Y, Do SI, Lee HJ, Kang HJ,
Rho YS, Bae WJ and Lim YC: SOX2 regulates self-renewal and
tumorigenicity of stem-like cells of head and neck squamous cell
carcinoma. Br J Cancer. 111:2122–2130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Irshad K, Mohapatra SK, Srivastava C, Garg
H, Mishra S, Dikshit B, Sarkar C, Gupta D, Chandra PS,
Chattopadhyay P, et al: A combined gene signature of hypoxia and
notch pathway in human glioblastoma and its prognostic relevance.
PLoS One. 10:e01182012015. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Robert J: Comparative study of
tumorigenesis and tumor immunity in invertebrates and nonmammalian
vertebrates. Dev Comp Immunol. 34:915–925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Liu S, Wu LC, Pang J, Santhanam R, Schwind
S, Wu YZ, Hickey CJ, Yu J, Becker H, Maharry K, et al:
Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid
leukemia. Cancer Cell. 17:333–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Ghosh S and Hayden MS: Celebrating 25
years of NF-κB research. Immunol Rev. 246:5–13. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Xiao G and Fu J: NF-κB and cancer: A
paradigm of Yin-Yang. Am J Cancer Res. 1:192–221. 2011.
|
|
184
|
Ueda Y and Richmond A: NF-kappaB
activation in melanoma. Pigment Cell Res. 19:112–124. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Patel PS, Varney ML, Dave BJ and Singh RK:
Regulation of constitutive and induced NF-kappaB activation in
malignant melanoma cells by capsaicin modulates interleukin-8
production and cell proliferation. J Interferon Cytokine Res.
22:427–435. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Martini M, Ciraolo E, Gulluni F and Hirsch
E: Targeting PI3K in cancer: Any good news? Front Oncol. 3:1082013.
View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Editorial. FDA approves PI3K inhibitor,
idelalisib for treatment of relapsed CLL, follicular jymphoma and
small lymphocytic lymphoma. Science & Education on Oncology
PRO. http://oncologypro.esmo.org.
|
|
188
|
Mukohara T: PI3K mutations in breast
cancer: prognostic and therapeutic implications. Breast Cancer.
7:111–123. 2015.PubMed/NCBI
|
|
189
|
Sinkovics JG: Antileukemia and antitumor
effects of the graft-versus-host disease: A new immunovirological
approach. Acta Microbiol Immunol Hung. 57:253–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Geiger TL and Rubnitz JE: New approaches
for the immunotherapy of acute myeloid leukemia. Discov Med.
19:275–284. 2015.PubMed/NCBI
|
|
191
|
Magee MS and Snook AE: Challenges to
chimeric antigen receptor (CAR)-T cell therapy for cancer. Discov
Med. 18:265–271. 2014.PubMed/NCBI
|
|
192
|
Sinkovics JG and Horvath JC: Human natural
killer cells: A comprehensive review. Int J Oncol. 27:5–47.
2005.PubMed/NCBI
|
|
193
|
Bruchard M and Ghiringhelli F:
Microenvironment tumoral. Cellules régulatrices et cytokines
immunosuppressives. Med Sci. 30:429–435. 2014.
|
|
194
|
Old LJ: Tumor necrosis factor (TNF).
Science. 230:630–632. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Urschel K and Cicha I: TNF-α in the
cardiovascular system: from physiology to therapy. Internat J
Interferon Cytokine Med Res. 7:9–25. 2015.
|
|
196
|
Quistad SD, Stotland A, Barott KL,
Smurthwaite CA, Hilton BJ, Grasis JA, Wolkowicz R and Rohwer FL:
Evolution of TNF-induced apoptosis reveals 550 My functional
conservation. Proc Natl Acad Sci USA. 111:9567–9572. 2014.
View Article : Google Scholar
|
|
197
|
Spandidos DA and Lang JC: In vitro cell
transformation by ras oncogenes. Crit Rev Oncog. 1:195–209.
1989.PubMed/NCBI
|
Journal Information
Journal ID
(publisher-id): IJO
Title:
International Journal of Oncology
ISSN
(print): 1019-6439
ISSN
(electronic): 1791-2423
Publisher:
D.A. Spandidos
Article Information
Copyright © 2015, Spandidos
Publications
Copyright:
2015
License
(open-access, https://creativecommons.org/licenses/by-nc-nd/4.0):
This is an open access article
distributed under the terms of a Creative Commons Attribution
License
Publication date
(collection): October 2015
Volume:
47
Issue:
4
Page:
1229
Publisher
ID: ijo-47-04-1211
Addendum
Int J Oncol 19: 473–488, 2001; DOI:
10.3892/ijo.19.3.473
The authors would like to acknowledge that the
Coriphosphine O stain shown in the upper panel of Figure 8 was a
kind gift from Dr T.P. Loughran of the H.L. Moffitt Cancer Center,
Tampa, FL, USA.
Article Information (continued)
Categories:
Subject:
Articles
Process warnings
Warnings reported by the processor due to anomalous or
incomplete markup follow:
Elements are cross-referenced without labels.
Either the element should be provided a label, or their
cross-reference(s) should have literal text content.
{ label needed for
corresp[@id='c1-ijo-47-04-1211'] }
This display is generated from NLM/NCBI Journal Publishing 3.0
XML with jpub3-html.xsl. The XSLT engine is Saxonica