Introduction
Breast cancer is one of the most common female
maligant tumors and a leading cause of cancer mortality worldwide
each year (1). Genetic mutations
have been demonstrated to be causative of tumorigenesis in breast
cancer. At this point, it is essential to develop more effective
methods for early diagnosis and treatment.
MicroRNAs (miRNAs) are small, non-coding RNAs of
approximately 19–25 nucleotides acting as critical regulators of
gene expression (2,3). Mature miRNA plays its role by binding
to the 3′-untranslated regions (3′-UTRs) of certain mRNAs,
suppressing target gene expression (4,5). In
recent decades, miRNAs are confirmed to be involved in many
important physiological and pathological processes, such as cell
proliferation, differentiation, virus infection and tumorigenesis,
and are widely dyregulated in various cancers, suggesting that they
may function as either tumor-suppressor genes or oncogenes
(6,7). Alteration of miRNA expression has
emerged to be one of the key features in cancer-associated
dysfunction of gene regulatory networks, which can improve cancer
classification, diagnosis, and clinical prognostic information
(8). miRNA has become a hot spot
in breast cancer research whereby miRNAs are believed to have broad
prospects in terms of diagnosis and treatment of this disease.
The Hippo pathway, firstly discovered in
Drosophila melanogaster, is widely considered to be a
signaling pathway that is important in controlling organ size and
tumor progression by regulating cellular proliferation and
promoting apoptosis (9). In recent
years, accumulated evidence suggests that the Hippo signaling
pathway plays crucial roles in breast cancer. LATS2 (large tumor
suppressor kinase 2) is an AGC kinase of the NDR family of kinases.
It is a tumor suppressor of the LATS family, and plays a
significant role in centrosome duplication, maintenance of mitotic
fidelity, and genomic stability (10). LATS2 inhibits cell growth at the
G1/S transition by downregulating cyclin E/CDK2 kinase activity
(11). As an upstream regulator in
the Hippo pathway, LATS2 can regulate its downstream gene YAP
(Yes-associated protein). Specifically, phosphorylated and
activated LATS2 can phosphorylate transcription coactivators YAP,
leading to the YAP cytoplasm retention by 14-3-3 protein or
degradation (12,13). To date, several other miRNAs have
been proved to target LATS2 in different types of cancer. miR-181b
regulates ovarian cancer cell growth and invasion by targeting
LATS2 (14). miR-93 can promote
tumor angiogenesis and metastasis by suppressing LATS2 in human
breast cancer cells (15). miR-372
disrupts cell cycle in gastric cancer cells through direct
regulation of LATS2 (16). Recent
studies indicate that miR-135b is elevated in a variety of cancers
such as lung cancer, colorectal cancer and hepatocellular cancer,
and it is also confirmed to be implicated in cancer growth,
survival, motility, and invasiveness (17–19).
However, the specific expression features of miR-135b in breast
cancer remains undefined, and the potential role and its mechanism
of action are still unknown.
Our present study aimed to investigate the function
of miR-135b in breast cancer cells. Using qRT-PCR, both our breast
cancer tissue samples and cancer cell lines had higher miR-135b
expression levels, as predicted. Moreover, miR-135b serves as an
oncogene in breast cancer and is a vital regulator of cellular
proliferation, migration, invasion and cell cycle. LATS2 was found
to have binding sites for miR-135b in the 3′UTR region. We
demonstrated that LATS2 is a direct target of miR-135b. As the
downstream gene of LATS2, CDK2 and p-YAP was regulated by miR-135b
and LATS2 axis. These results suggest that miR-135b may act as a
biomarker in breast cancer and that downregulation of miR-135b is a
feasible therapeutic approach for breast cancer that merits further
evaluation.
Materials and methods
Specimens and cell lines
In our study, 16 pairs of breast cancer and adjacent
normal specimens were collected from the Department of Breast and
Thyroid Surgery of Shanghai Tenth People's Hospital, Shanghai,
China. The samples were immediately snap-frozen in liquid nitrogen.
Both tumor and normal tissues were histologically confirmed by more
than one experienced pathologist according to the World Health
Organization (WHO) using H&E (hematoxylin and eosin) staining,
and none of these patients had received any chemotherapy or
radiotherapy prior to surgery.
The human breast cancer cell lines MDA-MB-231,
MCF-7, MDA-MB-436, HCC1937 and non-malignant breast epithelial cell
line MCF-10A were purchased from Chinese Academy of Sciences in
Shanghai. The breast cancer cells were maintained in Dulbecco's
modified Eagle's medium (DMEM) (Gibco, Grand Island, NY, USA)
supplemented with 10% Fetal Bovine Serum (FBS) (Gibco), penicillin
(100 U/ml) and streptomycin (100 μg/ml) (Enpromise, Hangzhou,
China). MCF-10A cells were cultured in Mammary Epithelial Basal
Medium (MEBM) (Cambrex). All the cells were incubated at 37°C in a
humidified chamber supplemented with 5% CO2.
Transfection assay
MiR-135b mimics, inhibitor, LATS2-siRNA and their
negative control (NC) were chemosynthesized by Shanghai Genepharma
Co., Ltd. (Shanghai, China). Cells (1×106) were added
into each well of a 6-well plate and cultured with DMEM medium
without serum and antibiotics. When the confluency of breast cancer
cells reached 30–50%, miR-135b mimics, miR-135b inhibitor,
LATS2-siRNA and NC were transfected at working concentrations using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions. After 4–5 h of incubation, DMEM medium
was replaced by DMEM with 10% FBS, and all the cells were incubated
at 37°C in a CO2 incubator for 24 h prior to further
testing.
RNA extraction and quantitative
reverse-transcription PCR (qRT-PCR)
According to the manufacturer's protocol, total RNA
was extracted from the cells or tissues using TRIzol (Invitrogen).
For detection of miR-135b expression, primer design and qRT-PCR
were carried out according to the manufacturer's instructions. The
primers used were as followed: miR-135b forward,
5′-GCTTATGGCTTTTCATT CCT-3′, reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6
forward, 5′-CTCGCTTCGGCAGCACA-3′, reverse, 5′-AAC
GCTTCACGAATTTGCGT-3′. cDNA was generated by reverse transcription
using the PrimeScript™ RT-PCR kit in accordance with the
manufacturer's instructions (Takara, Tokyo, Japan). Real-time PCR
was performed on a 7900HT Fast RT-PCR instrument (Applied
Biosystems, Singapore). The amplification procedure was as follows:
5 min at 95°C, followed by 40 cycles at 95°C for 30 sec and 65°C
for 45 sec. The relative expression was evaluated following the
relative quantification equation, 2−ΔΔCT. Each sample
was tested in triplicate.
Quantitative detection of LATS2 was implemented
using the same strategy. The primers used were as followed: LATS2
forward, 5′-CTGGAATTCGAAGTGTGAGCAAGGTG ATG-3′, reverse,
5′-ACGACTAGTGACTTGAGTATGCC ACTCAC-3′; β-actin forward,
5′-CATGTACGTTGCTAT CCAGGC-3′, reverse, 5′-CTCCTTAATGTCACGCACGAT-3′.
The PCR parameters for relative quantification were as follows: 2
min at 95°C, followed by 40 cycles of 45 sec at 57°C and 45 sec at
72°C. The relative expression was evaluated following the relative
quantification equation, 2−ΔΔCT. Each sample was tested
in triplicate.
Cell proliferation assay (MTT assay)
Cell proliferation was detected using an MTT assay
kit (Sigma, Santa Clara, CA, USA) in accordance with the
manufacturer's instructions. In brief, the transfected cells
(2×103 cells/well) were seeded into 96-well culture
plates (BD Biosciences, Franklin Lakes, NJ, USA) and incubated
overnight at 37°C in 5% CO2. Cell proliferation was
assessed at 24, 48, 72 and 96 h following addition of 0.5 mg/ml MTT
(Sigma) solution. After a 4-h incubation, the medium was replaced
by 150 μl dimethylsulfoxide (DMSO; Sigma). After 10 min of
agitation (100 rpm), optical density at 490 nm was determined on a
microplate spectrophotometer. Each sample was tested with six
replicates. All experiments were performed in biological
triplicate.
Colony formation assay
Three hundred cells of each group were plated in a
6-well plate in complete medium 4 h after transfection. The plates
were shaken to disperse the cells equally. After 7–10 days, or when
the colonies were visible, the culture was terminated and the
plates were washed twice in phosphate buffered saline (PBS) after
removing the complete medium. Then the colonies were fixed in 95%
ethanol for 10 min, dried and stained with 0.1% crystal violet
solution for 10 min. Then, each plate was washed three times with
water, and the number of colonies was counted only if the well
contained >50 cells. The experiment was performed three
times.
Wound-healing assay
In the in vitro wound healing assay,
transfected cells were cultured in 6-well plates until the cell
confluence reached ~90%. Then the plates were washed in PBS after
making a scratch in each well using a sterile pipette tip.
Wound-healing was observed under a light microscope and images were
captured at the same view at 0, 12, 24 and 48 h after scratching to
observe the process of wound healing. The experiments were repeated
twice and representative photographs are shown.
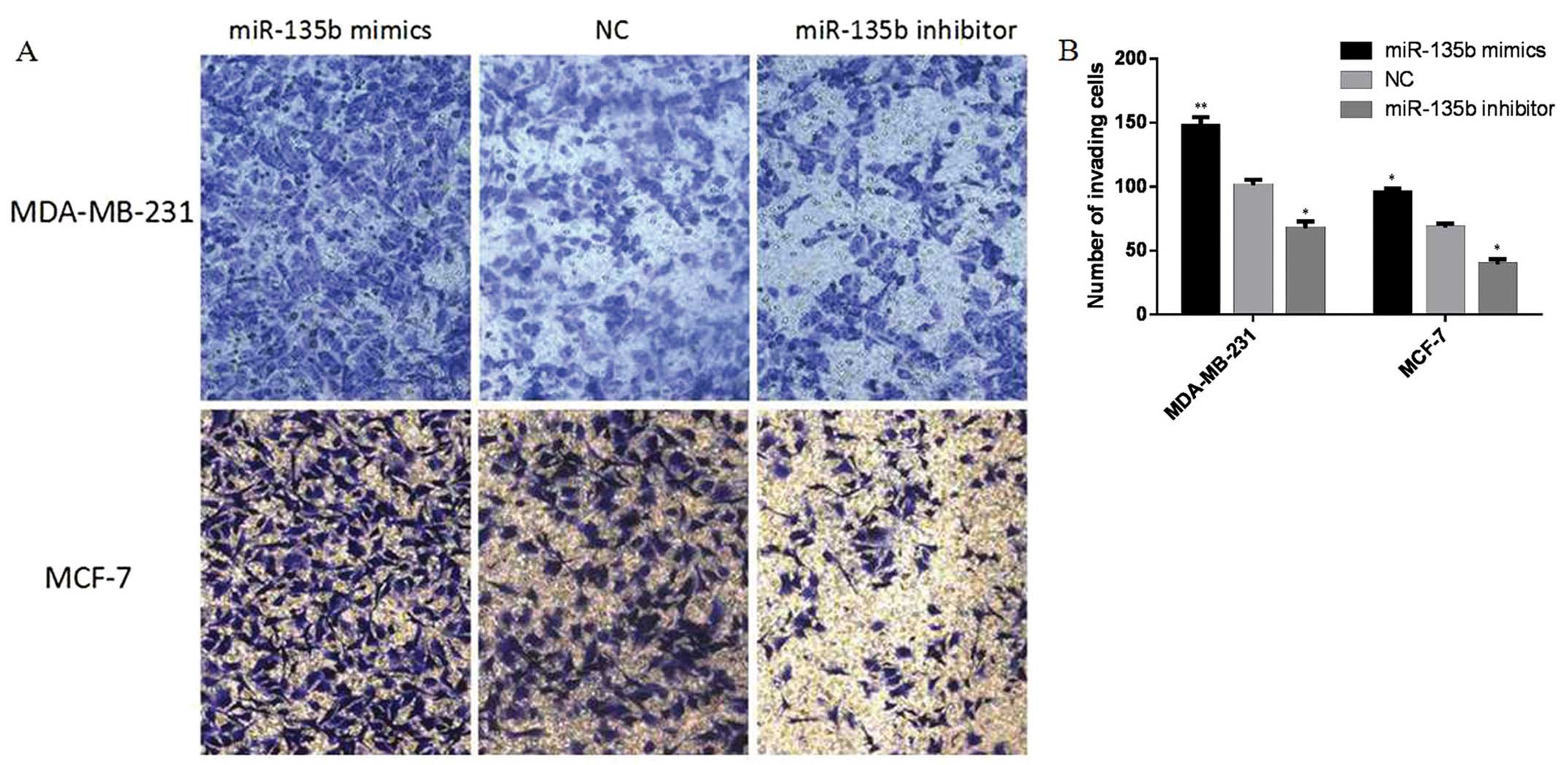
Transwell invasion assay
A Transwell invasion assay was performed by using
Chemicon Cell Invasion Assay kit (Chemicon, Temecula, CA, USA). The
miR-135b mimics or miR-135b inhibitor transfected cells
(5×104 cells/Transwell) were plated in the top chamber
of Transwells with a Matrigel (2 mg/ml)-coated membrane containing
8-mm diameter pores in 200 μl serum-free DMEM. The lower chambers
were filled with 500 μl of DMEM containing 10% FBS. After 48 h of
incubation, the membrane was stained with 0.1% crystal violet and
observed under a microscope after removing the Matrigel and cells
in the upper chambers. Five fields were randomly selected from each
membrane, and the number of cells penetrating the membrane was
counted at a magnification of ×200. The invasion ability was
described as the number of invading cells. Each experiment was
carried out in triplicate.
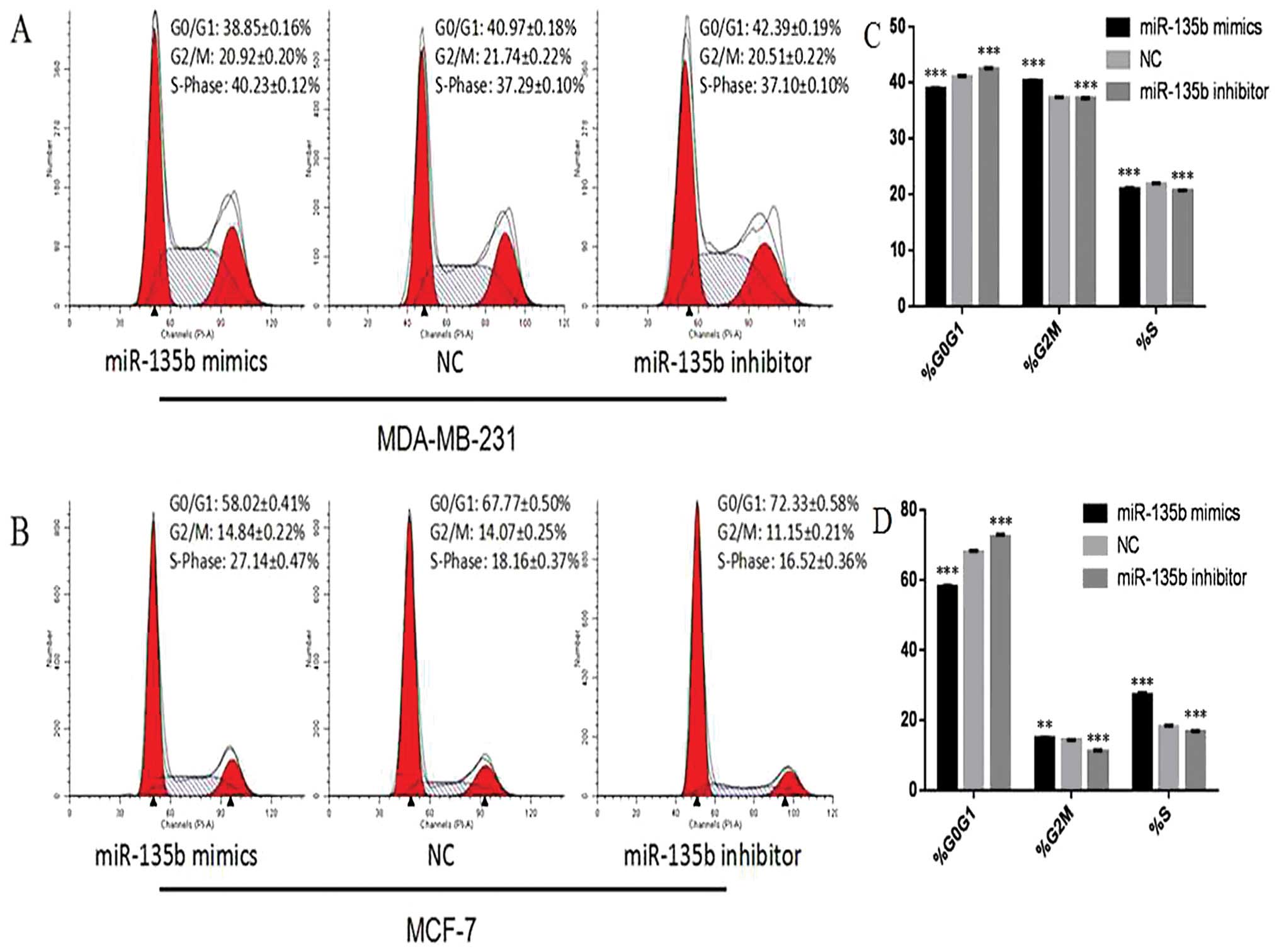
Cell cycle assay
At 24 h after transfection, cells were trypsinized
and centrifuged at 1000 rpm for 5 min, followed by two washes in
cold PBS. Then cells were fixed in 70% ice-cold ethanol at 4°C for
24 h. A total of 250 μl 0.05 g/l propidium iodide (PI) staining
solution was added into each sample and incubated for 30 min at
room temperature, and cell cycle distribution was analyzed using
flow cytometry (FACSCanto™ II, BD Biosciences, San Jose, CA,
USA).
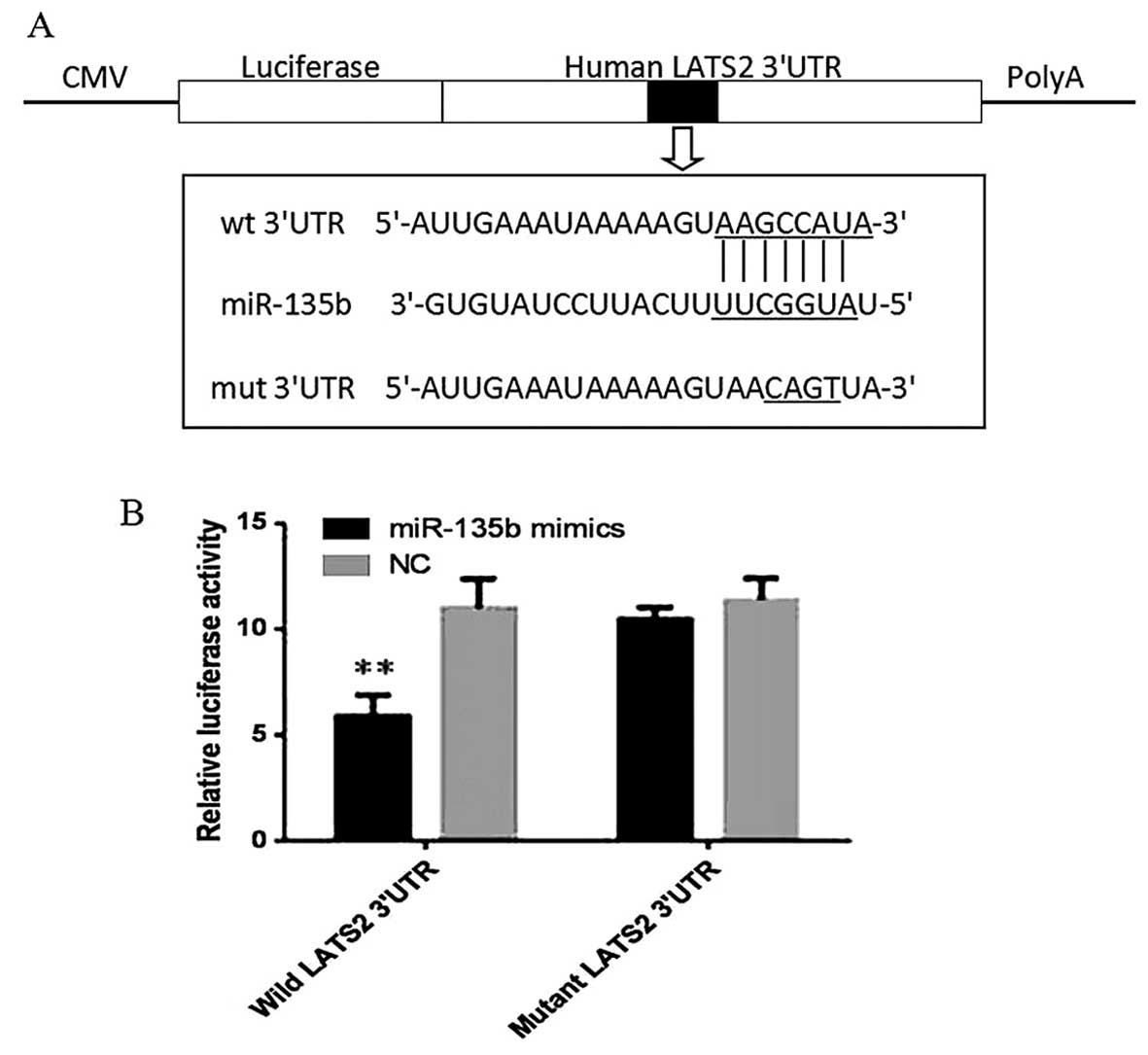
Dual-luciferase reporter assay
HEK293T cells were seeded in 12-well plates (BD
Biosciences) and cultured until the cells reached 80–90%
confluence. The 3′-UTR segments of the LATS2 mRNA sequence
containing the predicted miR-135b binding sites were amplified by
PCR using the PrimerStar kit (Takara). The corresponding mutant
constructs were created by mutating the seed regions of the
miR-135b-binding sites (5′-AAGCCAUA-3′ to 5′-AACAGTUA-3′). The
mutant constructs were generated by mutation. The forward primer
used in the reaction was 5′-CTGGAATTCGAAGTGTGAGC AAGGTGATG-3′; and
the reverse primer was 5′-ACGA CTAGTGACTTGAGTATGCCACTCAC-3′.
Fragments were subcloned into the XhoI site in the 3′-UTR of
Renilla luciferase of the psiCHECK-2 reporter vector. Cells were
transiently cotransfected with 0.2 μg psiCHECK-2/LATS2 3′-UTR or
psiCHECK-2/LATS2 3′-UTR mutant reporter plasmids and together with
100 nmol/l miR-135b or miR-NC using Lipofectamine™ 2000
(Invitrogen), according to the manufacturer's instructions. After
48 h, firefly and Renilla luciferase activities were measured by
using a Dual Luciferase Assay (Promega, Madison, WI, USA). Firefly
luciferase activity was normalized to Renilla, and the ratio of
firefly/renilla was recorded.
Western blotting
Cells were washed in ice-cold PBS and resuspended in
RIPA lysis buffer (100 μl/well, Beyotime). Then the cells were
collected and centrifuged for 30 min at 4°C (Eppendorf 5804R,
Eppendorf Biotech, Hamburg, Germany). Supernatants were collected
and the protein concentrations were quantified using a BCA protein
assay kit (Beyotime). Protein samples were denatured with 5X SDS
loading buffer (Beyotime) at 100°C for 10 min. Total protein was
separated by 8% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE, Beyotime) and transferred to a 0.45-μm
nitrocellulose membrane (Beyotime). The membrane was incubated at
4°C with primary antibodies against LATS2 (1:1,000; Bioworld
Technology; Nanjing, China), CDK2 (1:1,000; Bioworld Technology),
p-YAP (1:1,000; Cell Signaling Technology, USA) and β-actin
(1:1,000; Bioworld Technology). After washing with PBST (Shanghai
Engineering Co.), the membranes were incubated with secondary
antibodies for 60 min. Immunereactive protein bands were detected
with an Odyssey Scanning system.
Statistical analysis
Data are presented as the means ± standard deviation
(SD) from at least three independent experiments. The Student's
t-test was used to evaluate the differences between each group in
SPSS 20.0 software. Differences were considered significant for
P-values <0.05.
Results
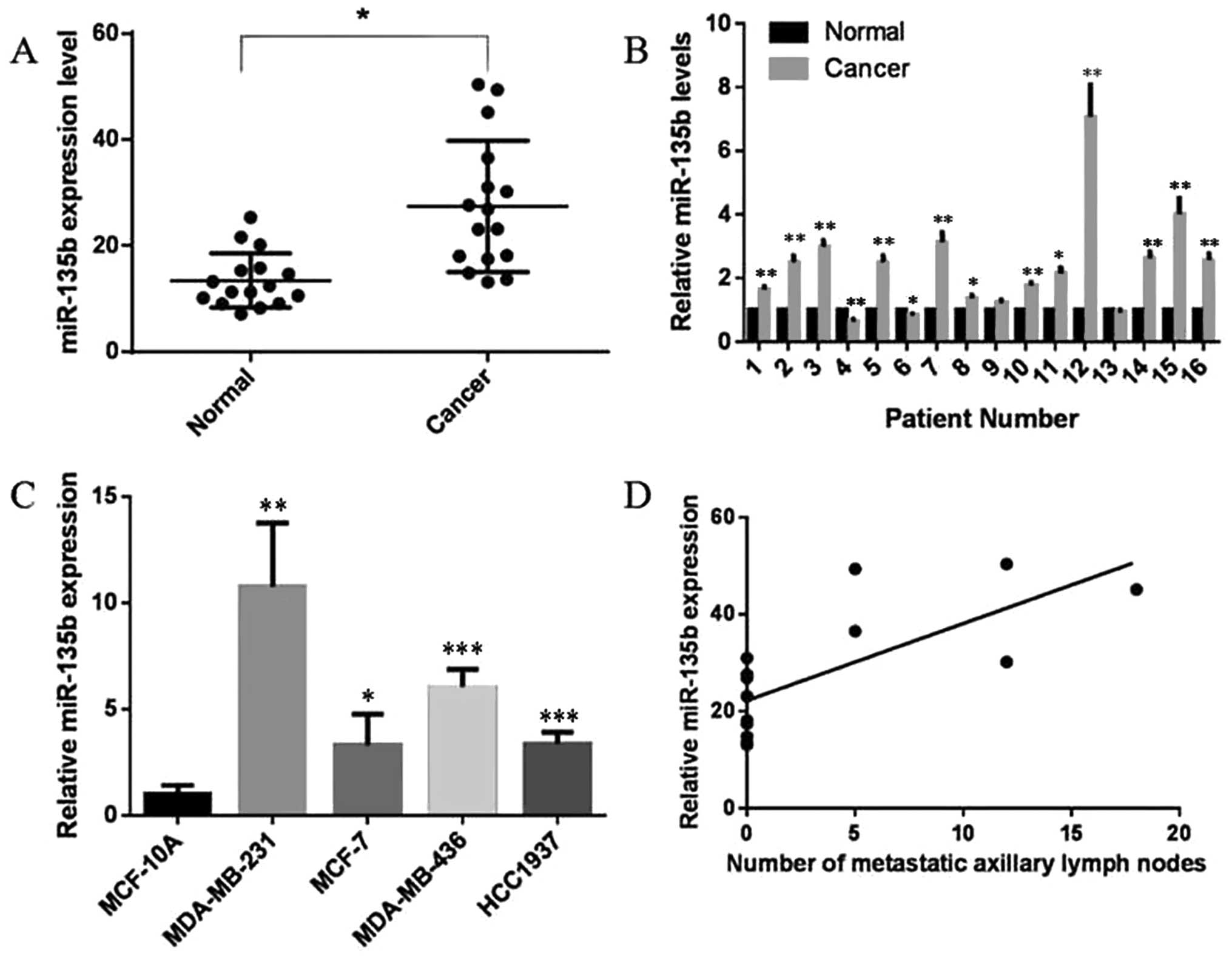
miR-135b is upregulated in both human
breast cancer specimens and breast cancer cell lines
Abnormal expression of miR-135b has been
demonstrated in a variety of cancers (20–27).
To explore the role of miR-135b in human breast cancer, we analyzed
16 pairs of breast cancer tissues and adjacent normal specimens in
this study. Total RNAs were isolated from excised tumor tissues and
benign tissues of patients with breast cancer. Analysis of miR-135b
by real-time PCR indicated that miR-135b levels were obviously
upregulated in breast cancer tissues compared with benign tissues
(P<0.05, Fig. 1A), and a
significant upregulation was found in 12 of 16 patients (P<0.05,
P<0.01, Fig. 1B). Moreover,
expression of miR-135b was also demontrated to be upregulated in
all four collected breast cancer cell lines compared to MCF-10A, a
non-malignant breast epithelial cell line (P<0.05, P<0.01,
P<0.001, Fig. 1C). These
results imply a potential role of miR-135b in breast cancer. In
this experiment, we also explored the relationship between the
number of metastatic axillary lymph nodes and the relative miR-135b
expression, noteworthy, a positive correlation was found between
the two variables (r=0.737, P<0.01, Fig. 1D).
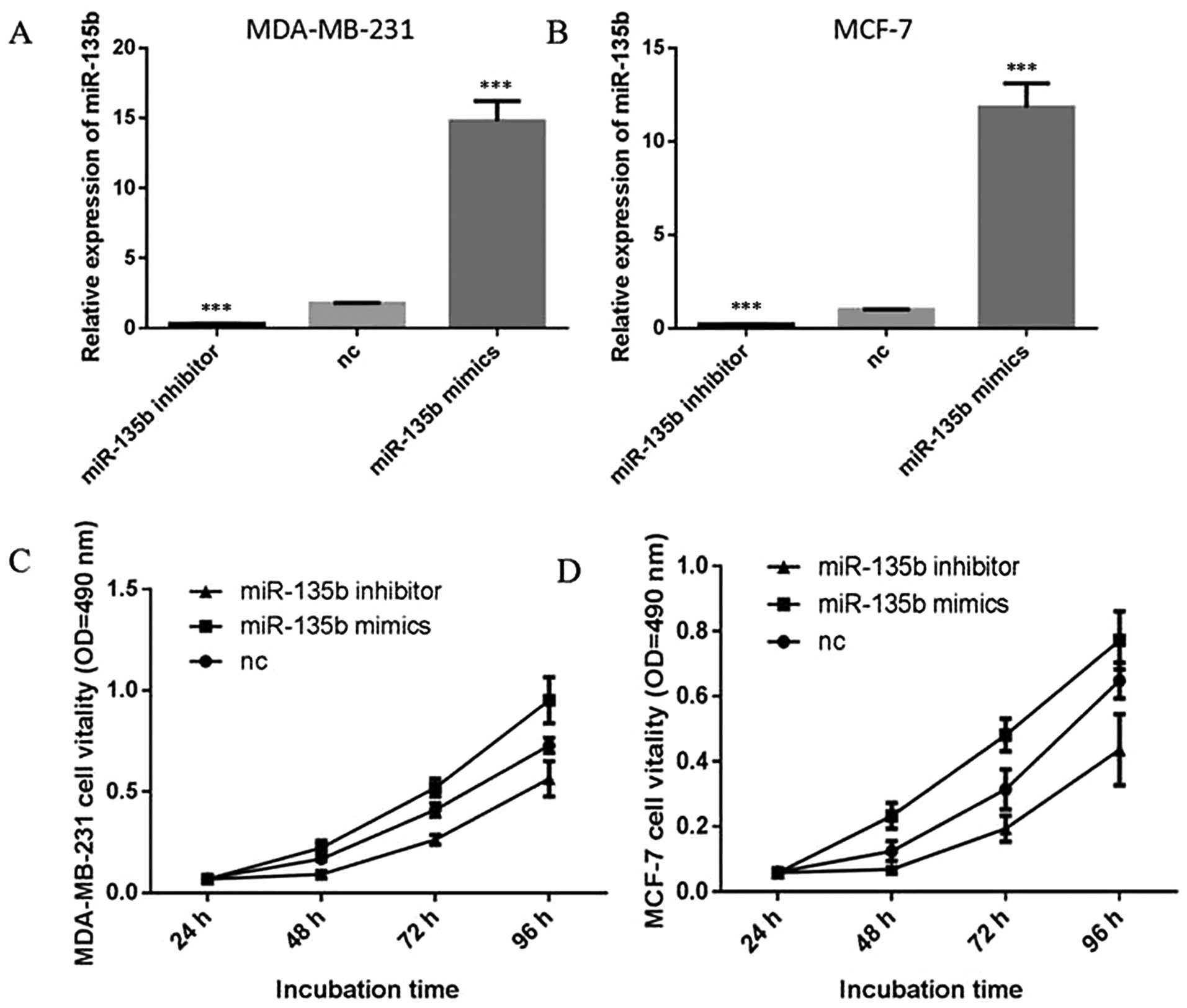
miR-135b promotes the proliferation of
breast cancer cells in vitro
To explore the potential role of miR-135b in human
breast cancer, we first detected the effect of miR-135b on
proliferation of breast cancer cells. Two of the most
representative breast cancer cell lines, MDA-MB-231 and MCF-7 cells
were selected in our following experiments. These two cells were
transiently transfected with miR-135b mimics, miR-135b inhibitor or
negative control (NC) and the expression levels of miR-135b were
detected by quantitative RT-PCR (qRT-PCR) (P<0.001, Fig. 2A and B). Cell proliferation assay
(MTT assay) indicated that overexpression of miR-135b in MDA-MB-231
and MCF-7 cells promoted an increment in cell proliferation
(P<0.05, Fig. 2C and D).
Inhibition of the endogenous miR-135b by miR-135b inhibitor led to
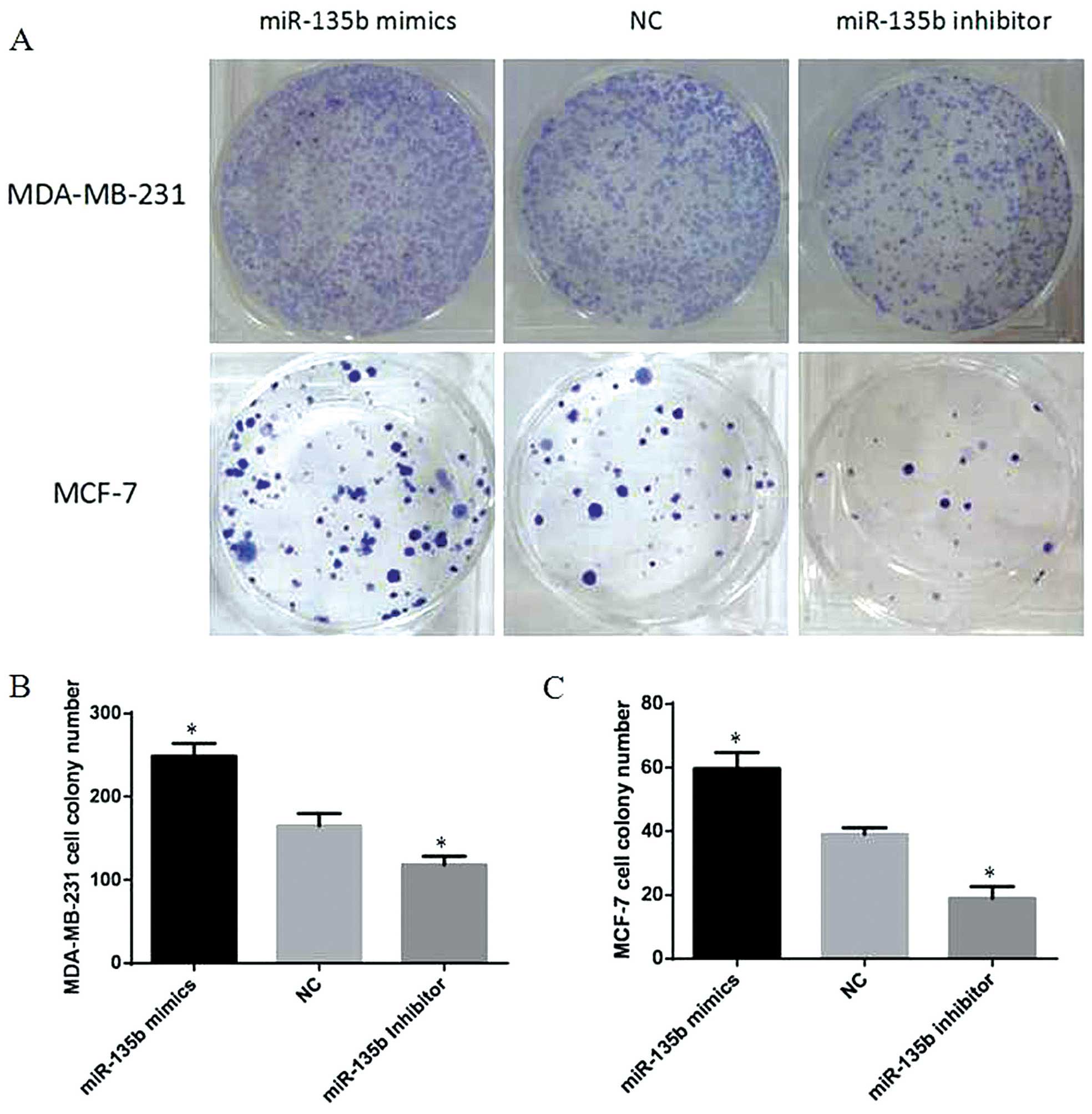
a significant reduction in cell proliferation (P<0.05). Colony
formation assays also showed much more colony formation in the
group transfected with miR-135b mimics compared with the NC group,
and miR-135b inhibitor group showed the opposite result (P<0.05,
Fig. 3). Our data indicated that
miR-135b can promote cell proliferation in breast cancer cells
in vitro.
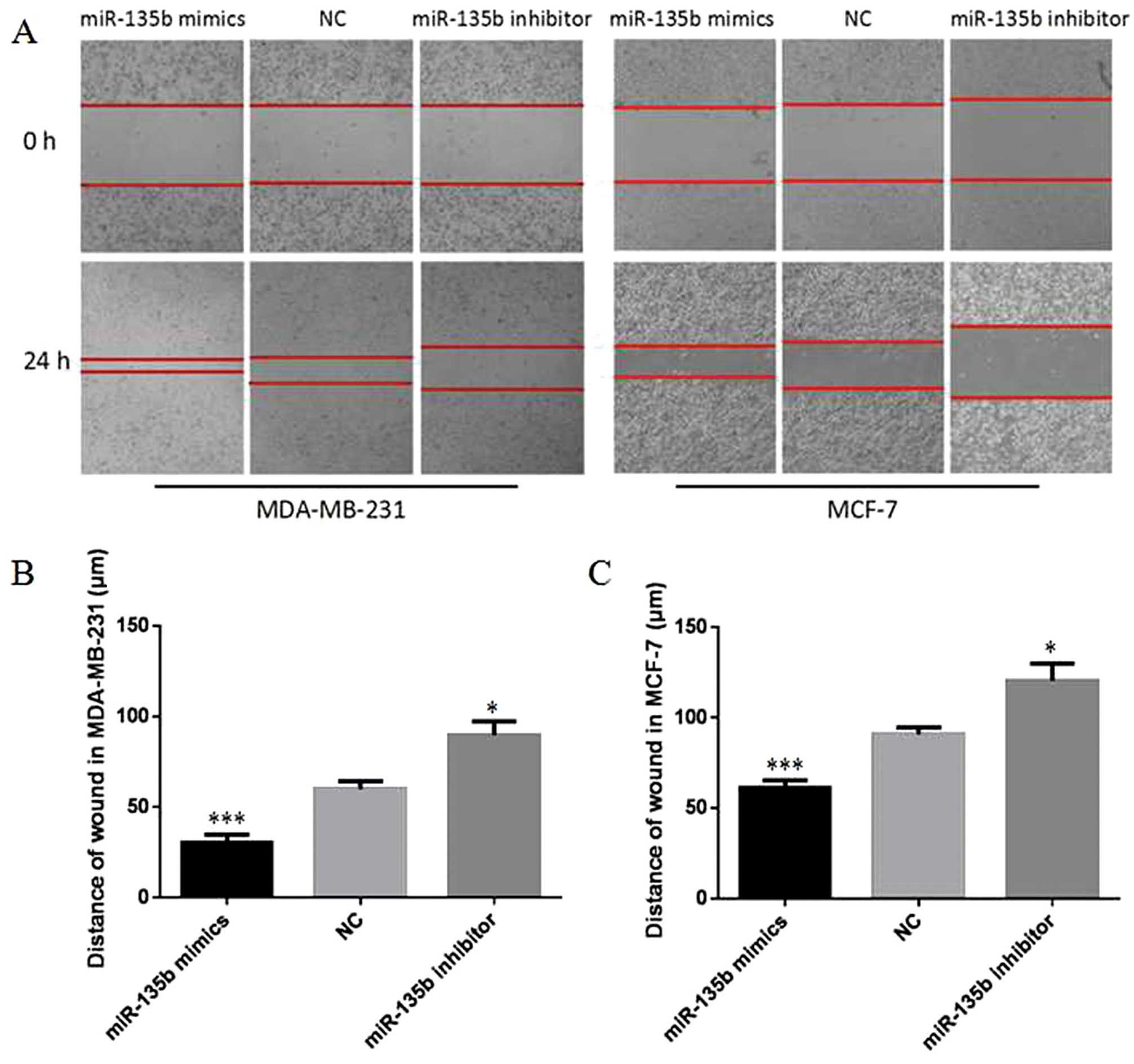
miR-135b accelerates cell migration and
invasion of breast cancer cells in vitro
To investigate how forced expression of miR-135b
affects cellular migration and invasion, wound healing assays and
Transwell assays were performed in MDA-MB-231 and MCF-7 cells.
Cells were transfected with miR-135b mimics, miR-135b inhibitor or
NC. As shown in Fig. 4, 24 h after
drawing the ‘scratch’ line in the monolayer MDA-MB-231 cells, the
miR-135b mimics group nearly filled the gap, the NC group still
showed a clear gap in the scratched region, and miR-135b inhibitor
group showed the opposite result (P<0.05, P<0.001). The
experiments carried out in MCF-7 cells also showed a similar trend
(P<0.05, P<0.001). The results indicate that overexpression
of miR-135b in MDA-MB-231 and MCF-7 cells accelerated cellular
migration. The Transwell invasion assay revealed that the number of
MDA-MB-231 and MCF-7 cells penetrating the membrane significantly
increased at 48 h after miR-135b mimic-transfection as compared to
the NC group and miR-135b inhibitor group (P<0.05, P<0.01,
Fig. 5). Together these results
showed that overexpression of miR-135b can accelerate cellular
migration and invasion in vitro.
MiR-135b regulates the cell cycle of
breast cancer cells
Twenty-four hours after the transfection with
miR-135b mimics, miR-135b inhibitor or NC in MCF-7 cells, flow
cytometry analysis indicated that the percentage of G0/G1 phase
cells (58.02±0.41%) dramatically decreased in the miR-135b mimics
group, when compared with that of the NC group (67.77±0.50%) and
miR-135b inhibitor group (72.33±0.58%) (P<0.05). At the same
time, the proportion of S-phase cells increased in the miR-135b
mimics group (27.14±0.47%) compared with that of the NC group
(18.16±0.37%) and miR-135b inhibitor group (16.52±0.36%)
(P<0.01, P<0.001). The percentage of G2/M phase cells was
also elevated in the miR-135b mimics group (14.84±0.22%) compared
with that of the NC group (14.07±0.25%) and miR-135b inhibitor
group (11.15±0.21%) (P<0.01, P<0.001). The experiments
carried out in MDA-MB-231 cells also showed a similar trend
(Fig. 6A and B). These findings
revealed that miR-135b can lead to the upregulation of S-phase and
G2/M phase cells.
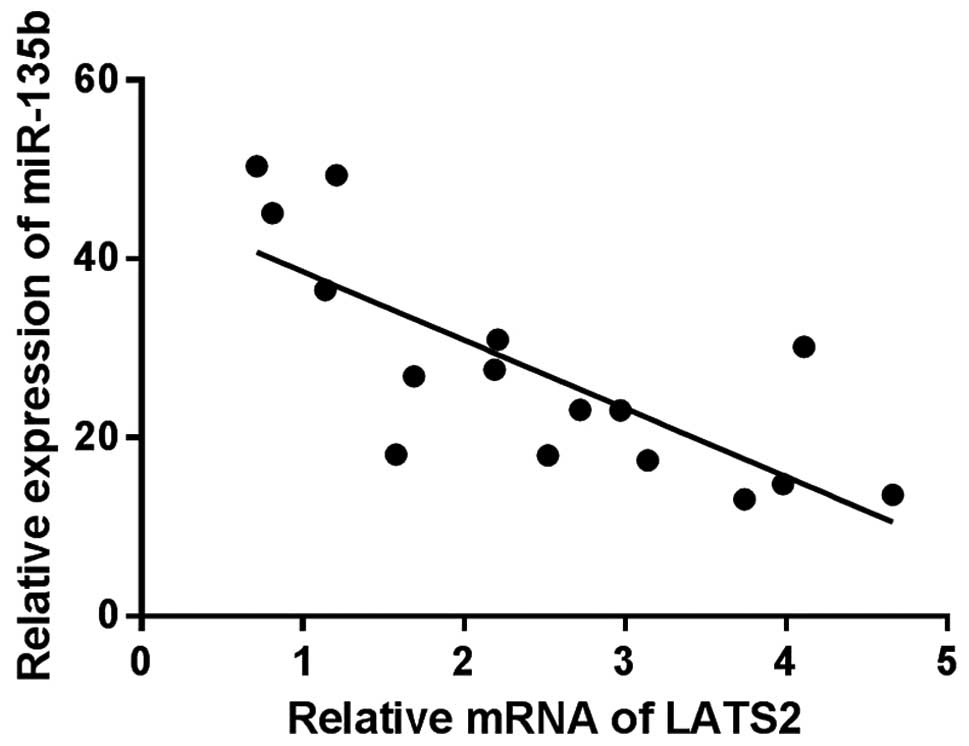
miR-135b regulates the expression of
LATS2 and its downstream gene in the Hippo pathway
To explore the action mechanism of miR-135b in
breast cancer, we screened the target genes of miR-135b using
Targetscan and microRNA.org (http://www.targetscan.org/ and http://www.microrna.org). LATS2, an upstream regulator
in the Hippo pathway, was identified as a candidate. Next, we
investigate the correlation between the expression of miR-135b and
LATS2 in clinical breast cancer specimens. Noteworthy, miR-135b
levels were found to be markedly inversely correlated with LATS2
expression (r= −0.765, P<0.01, Fig.
7), indicating miR-135b might target LATS2 mRNA in breast
cancer. To confirm that miR-135b can bind to the predicted site, we
performed a luciferase reporter assay in the 293T cell line. As
shown in Fig. 8, the luciferase
activity significantly decreased after co-transfection with
psi-CHECK-2/LATS2 3′-UTR and miR-135b mimics in comparison with
control cells, suggesting that miR-135b specifically binds to the
3′-UTR of LATS2 mRNA. The impact of miR-135b transfection on LATS2
protein expression in MDA-MB-231 and MCF-7 cell line was
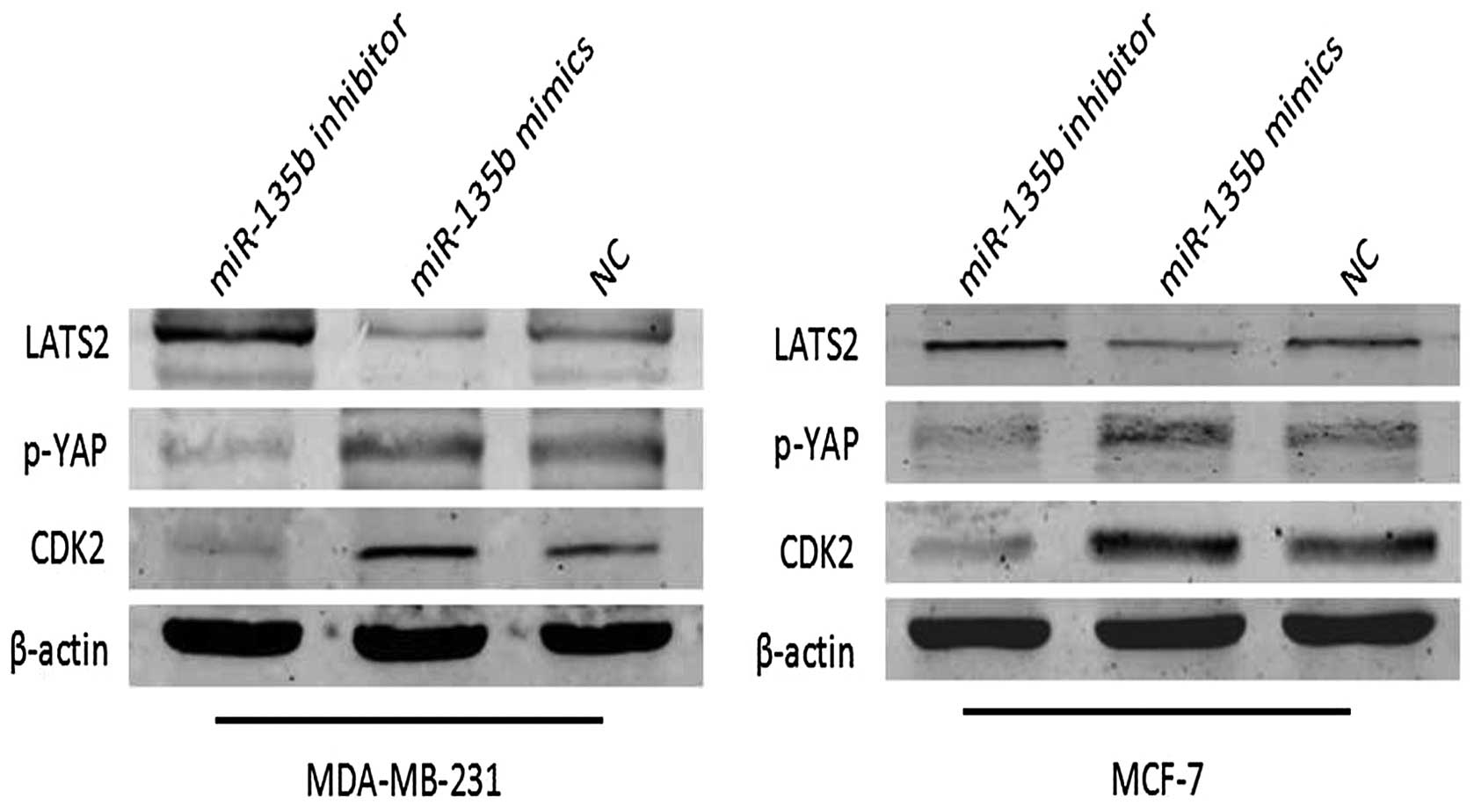
respectively assessed using western blot assays (Fig. 9). We observed that miR-135b was
capable of downregulating LATS2 in the cells at the protein level.
The opposite result was obtained when the cells were treated with
miR-135b inhibitor, indicating that miR-135b is able to attenuate
the expression of LATS2 in breast cancer cells. Further study
demonstrated that the downstream gene of LATS2 in the Hippo
pathway, such as CDK2 and p-YAP, can also be inhibited by miR-135b
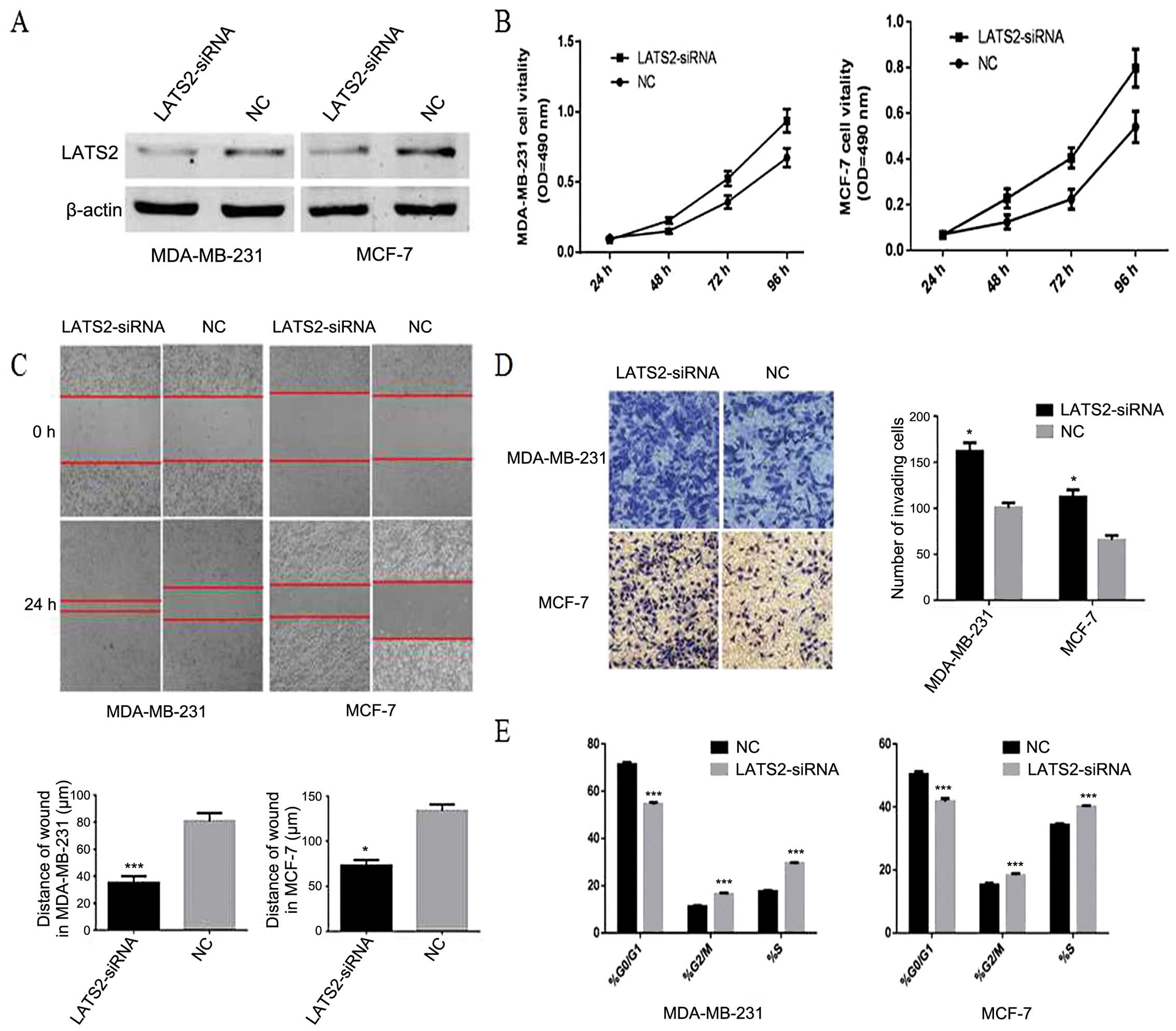
and LATS2 axis at the protein level (Fig. 9). Furthermore, silencing of LATS2
by LATS2-siRNA can promote proliferation, invasion, migration and
disrupte the cell cycle in MDA-MB-231 and MCF-7 breast cancer cells
(Fig. 10). Thus, our data
indicated that miR-135b can promote cell growth and disrupt the
cell cycle by regulating LATS2 and the Hippo pathway in breast
cancer cells.
Discussion
The miRNAs have been confirmed as important
regulators involved in different biological processes such as cell
proliferation, metastasis, differentiation, transcriptional
regulation and tumorigenesis (28). Dysregulation of miRNAs is connected
with initiation and progression of breast cancer, since they may
serve as oncogenes or tumor suppressors (29,30).
In the present study, we investigated the role of miR-135b in
breast cancer and speculate the Hippo pathway as one of its
possible mechanism.
The Hippo pathway was discovered 20 years ago in
Drosophila melanogaster (31), it is regulated by various upstream
signals, such as extracellular matrix (32), cell-cell contact (33), and cell stress (34). When the Hippo pathway is activated,
phosphorylated mammalian sterile 20-like kinase 1/2 (Mst1/2) forms
a complex by interacting with Sav1 (also known as WW45) (35). The Mst1/2 complex directly
phosphorylates the large tumor suppressor 1 and 2 (LATS1/2)
(36) and MOBKL1A/B (also known as
MOB1) that forms another kinase complex with LATS1/2 (37). Phosphorylated and activated LATS1/2
phosphorylates transcription coactivators YAP and TAZ at S127 and
S89, respectively (38–40), leading to the YAP/TAZ cytoplasm
retention by 14-3-3 or degradation (41). Unphosphorylated YAP and TAZ
translocate into the nucleus to interact with transcription
factors, such as TEAD1-4 (42),
Smads (43), and p73 (44). As a regulatory factor in the Hippo
pathway, LATS2 plays its tumor-suppressor role by downregulating
YAP, a gene promoting breast cancer cell proliferation (45). It is reported that LATS2 can also
inhibit cell growth at the G1/S transition by downregulating CDK2
kinase activity (11).
In our experiment, we first investigated the
expression levels of miR-135b in 16-paired clinical breast cancer
and adjacent normal specimens. Of note, we observed that the
expression levels of miR-135b were remarkably increased in 13 of 16
samples relative to paired non-tumor tissues. Similar findings were
reported in many different tumors, including hepatocellular
carcinoma (19), colorectal cancer
(21), osteosarcoma (26), as well as in glioblastoma
multiforme cells (25). Based on
this finding, we hypothesized that miR-135b might be a novel
tumor-promoter miRNA in breast cancer. Then we investigated the
specific role of miR-135b in two typical breast cancer cell lines,
MDA-MB-231 and MCF-7. Cells were transfected with miR-135b mimics,
miR-135b inhibitor or NC, respectively, to detect the gain-or-loss
of function effects on various aspects of breast cancer biology.
Our results showed that the exogenous overexpression of miR-135b
regulated by miR-135b mimics significantly promoted proliferation
and colony formation ability of breast cancer cells as measured by
MTT and colony formation assays.
Moreover, cell migration and invasion ability was
also significantly enhanced by overexpression of miR-135b. We found
that miR-135b inhibitor distinctly arrests cancer cells at the G1
phase when compared with the cell cycle of NC group. Moreover, we
explored the correlation between the expression levels of miR-135b
and LATS2 in clinical breast cancer specimens. As expected, we
observed a negative correlation between the two variables. We then
further investigated the effects of miR-135b on LATS2 expression in
breast cancer cells. Western blot assays demonstrated that miR-135b
was able to downregulate LATS2 protein and its direct target genes
(CDK2 and YAP) in the Hippo pathway. Luciferase reporter assay also
identified that miR-135b could directly bind to the 3′UTR of LATS2.
We concluded that knockdown of endogenous LATS2 can mimic the
result of miR-135b upregulation in breast cancer. Thus, our results
showed that miR-135b affected the Hippo pathway by downregulating
the levels of LATS2 in breast cancer cells.
In resent years, several studies on miR-135b and
cancer was accomplished. Lin et al identified that
expression of miR-135b, LZTS1, LATS2 and nuclear TAZ predicts poor
outcomes of non-small cell lung cancer (17). Li et al found that up
regulation of miR-135b level was found to be inversely correlated
with tumor capsule occurrence and serum hepatitis B virus E antigen
level (19). In this experiment,
we also tried to explore the relationship between clinical data and
miR-135b levels. The number of metastatic axillary lymph nodes was
found to have a positive correlation with the relative miR-135b
expression in our cases, which suggested that miR-135b could be a
reliable biological marker in diagnosis of breast cancer. Actually,
we also analyzed the probable connections of miR-135b levels with
age, tumor size, tumor stage, and molecular subtypes of breast
cancer, but no significant correlation was observed. Since this is
a pattern verified only in 16 clinical cases, the reliability is
relatively low and needs to be confirmed in a larger number of
samples.
Taken together, our findings demonstrated that
miR-135b is upregulated in breast cancer tissues and cell lines,
and is able to promote cellular proliferation, migration and
invasion as well as disrupt the cell cycle in vitro via
direct regulation of the expression of LATS2 and the Hippo pathway,
indicating that miR-135b can serve as a potential therapeutic
target for breast cancer.
Acknowledgements
This work was supported by grants from National
Natural Science Foundation of China (no. 82172240). We sincerely
thank all the teachers at the Central Laboratory of the Shanghai
Tenth People's Hospital for their help and support.
References
|
1
|
World Health Organization. WHO fact sheet
No. 297. World Health Organization; Geneva: 2006
|
|
2
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gregory RI, Chendrimada TP, Cooch N and
Shiekhattar R: Human RISC couples microRNA biogenesis and
post-transcriptional gene silencing. Cell. 123:631–640. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jay C, Nemunaitis J, Chen P, Fulgham P and
Tong AW: miRNA profiling for diagnosis and prognosis of human
cancer. DNA Cell Biol. 26:293–300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pan D: The hippo signaling pathway in
development and cancer. Dev Cell. 19:491–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yabuta N, Okada N, Ito A, Hosomi T,
Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, et
al: Lats2 is an essential mitotic regulator required for the
coordination of cell division. J Biol Chem. 282:19259–19271. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Pei J, Xia H, Ke H, Wang H and Tao
W: Lats2, a putative tumor suppressor, inhibits G1/S transition.
Oncogene. 22:4398–4405. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong J, Feldmann G, Huang J, Wu S, Zhang
N, Comerford SA, Gayyed MF, Anders RA, Maitra A and Pan D:
Elucidation of a universal size-control mechanism in Drosophila and
mammals. Cell. 130:1120–1133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oka T, Mazack V and Sudol M: Mst2 and Lats
kinases regulate apoptotic function of Yes kinase-associated
protein (YAP). J Biol Chem. 283:27534–27546. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xia Y and Gao Y: MicroRNA-181b promotes
ovarian cancer cell growth and invasion by targeting LATS2. Biochem
Biophys Res Commun. 447:446–451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang L, Du WW, Yang W, Rutnam ZJ, Peng C,
Li H, O'Malley YQ, Askeland RW, Sugg S, Liu M, et al: MiR-93
enhances angiogenesis and metastasis by targeting LATS2. Cell
Cycle. 11:4352–4365. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cho WJ, Shin JM, Kim JS, Lee MR, Hong KS,
Lee JH, Koo KH, Park JW and Kim KS: miR-372 regulates cell cycle
and apoptosis of ags human gastric cancer cell line through direct
regulation of LATS2. Mol Cells. 28:521–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin CW, Chang YL, Chang YC, Lin JC, Chen
CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, et al: MicroRNA-135b
promotes lung cancer metastasis by regulating multiple targets in
the Hippo pathway and LZTS1. Nat Commun. 4:18772013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He Y, Wang J, Wang J, Yung VY, Hsu E, Li
A, Kang Q, Ma J, Han Q, Jin P, et al: MicroRNA-135b regulates
apoptosis and chemoresistance in colorectal cancer by targeting
large tumor suppressor kinase 2. Am J Cancer Res. 5:1382–1395.
2015.PubMed/NCBI
|
|
19
|
Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao
F, Ding J, Liang L, Wang Q, Liu L, et al: MicroRNA-135b, a HSF1
target, promotes tumor invasion and metastasis by regulating RECK
and EVI5 in hepatocellular carcinoma. Oncotarget. 6:2421–2433.
2015. View Article : Google Scholar :
|
|
20
|
Zhang L, Sun ZJ, Bian Y and Kulkarni AB:
MicroRNA-135b acts as a tumor promoter by targeting the
hypoxia-inducible factor pathway in genetically defined mouse model
of head and neck squamous cell carcinoma. Cancer Lett. 331:230–238.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagel R, le Sage C, Diosdado B, van der
Waal M, Oude Vrielink JA, Bolijn A, Meijer GA and Agami R:
Regulation of the adenomatous polyposis coli gene by the miR-135
family in colorectal cancer. Cancer Res. 68:5795–5802. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valeri N, Braconi C, Gasparini P, Murgia
C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat
F, et al: MicroRNA-135b promotes cancer progression by acting as a
downstream effector of oncogenic pathways in colon cancer. Cancer
Cell. 25:469–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu W, Wang Z, Yang P, Yang J, Liang J,
Chen Y, Wang H, Wei G, Ye S and Zhou Y: MicroRNA-135b regulates
metastasis suppressor 1 expression and promotes migration and
invasion in colorectal cancer. Mol Cell Biochem. 388:249–259. 2014.
View Article : Google Scholar
|
|
24
|
He YQ, Sheng JQ, Ling XL, Fu L, Jin P, Yen
L and Rao J: Estradiol regulates miR-135b and mismatch repair gene
expressions via estrogen receptor-β in colorectal cells. Exp Mol
Med. 44:723–732. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao S, Yang Z, Lv R, Zhao J, Wu M, Liao Y
and Liu Q: miR-135b contributes to the radioresistance by targeting
GSK3β in human glioblastoma multiforme cells. PLoS One.
9:e1088102014. View Article : Google Scholar
|
|
26
|
Pei H, Jin Z, Chen S, Sun X, Yu J and Guo
W: MiR-135b promotes proliferation and invasion of osteosarcoma
cells via targeting FOXO1. Mol Cell Biochem. 400:245–252. 2015.
View Article : Google Scholar
|
|
27
|
Wu CW, Ng SC, Dong Y, Tian L, Ng SS, Leung
WW, Law WT, Yau TO, Chan FK, Sung JJ, et al: Identification of
microRNA-135b in stool as a potential noninvasive biomarker for
colorectal cancer and adenoma. Clin Cancer Res. 20:2994–3002. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo Q, Li X, Li J, Kong X, Zhang J, Chen
L, Huang Y and Fang L: MiR-15a is underexpressed and inhibits the
cell cycle by targeting CCNE1 in breast cancer. Int J Oncol.
43:1212–1218. 2013.PubMed/NCBI
|
|
30
|
Li J, Kong X, Zhang J, Luo Q, Li X and
Fang L: Correction: MiRNA-26b inhibits proliferation by targeting
PTGS2 in breast cancer. Cancer Cell Int. 13:172013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Justice RW, Zilian O, Woods DF, Noll M and
Bryant PJ: The Drosophila tumor suppressor gene warts encodes a
homolog of human myotonic dystrophy kinase and is required for the
control of cell shape and proliferation. Genes Dev. 9:534–546.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dupont S, Morsut L, Aragona M, Enzo E,
Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M,
Bicciato S, et al: Role of YAP/TAZ in mechanotransduction. Nature.
474:179–183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim
J, Xie J, Ikenoue T, Yu J, Li L, et al: Inactivation of YAP
oncoprotein by the Hippo pathway is involved in cell contact
inhibition and tissue growth control. Genes Dev. 21:2747–2761.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mohseni M, Sun J, Lau A, Curtis S,
Goldsmith J, Fox VL, Wei C, Frazier M, Samson O, Wong KK, et al: A
genetic screen identifies an LKB1-MARK signalling axis controlling
the Hippo-YAP pathway. Nat Cell Biol. 16:108–117. 2014. View Article : Google Scholar :
|
|
35
|
Callus BA, Verhagen AM and Vaux DL:
Association of mammalian sterile twenty kinases, Mst1 and Mst2,
with hSalvador via C-terminal coiled-coil domains, leads to its
stabilization and phosphorylation. FEBS J. 273:4264–4276. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chan EH, Nousiainen M, Chalamalasetty RB,
Schäfer A, Nigg EA and Silljé HH: The Ste20-like kinase Mst2
activates the human large tumor suppressor kinase Lats1. Oncogene.
24:2076–2086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Praskova M, Xia F and Avruch J:
MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell
proliferation. Curr Biol. 18:311–321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hao Y, Chun A, Cheung K, Rashidi B and
Yang X: Tumor suppressor LATS1 is a negative regulator of oncogene
YAP. J Biol Chem. 283:5496–5509. 2008. View Article : Google Scholar
|
|
39
|
Oka T, Mazack V and Sudol M: Mst2 and Lats
kinases regulate apoptotic function of Yes kinase-associated
protein (YAP). J Biol Chem. 283:27534–27546. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lei QY, Zhang H, Zhao B, Zha ZY, Bai F,
Pei XH, Zhao S, Xiong Y and Guan KL: TAZ promotes cell
proliferation and epithelial-mesenchymal transition and is
inhibited by the hippo pathway. Mol Cell Biol. 28:2426–2436. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kanai F, Marignani PA, Sarbassova D, Yagi
R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley
LC, et al: TAZ: A novel transcriptional co-activator regulated by
interactions with 14-3-3 and PDZ domain proteins. EMBO J.
19:6778–6791. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang H, Liu CY, Zha ZY, Zhao B, Yao J,
Zhao S, Xiong Y, Lei QY and Guan KL: TEAD transcription factors
mediate the function of TAZ in cell growth and
epithelial-mesenchymal transition. J Biol Chem. 284:13355–13362.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Varelas X, Sakuma R, Samavarchi-Tehrani P,
Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW and Wrana JL:
TAZ controls Smad nucleocytoplasmic shuttling and regulates human
embryonic stem-cell self-renewal. Nat Cell Biol. 10:837–848. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Strano S, Munarriz E, Rossi M, Castagnoli
L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G and Blandino G:
Physical interaction with Yes-associated protein enhances p73
transcriptional activity. J Biol Chem. 276:15164–15173. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Su L and Ou Q: Yes-associated
protein promotes tumour development in luminal epithelial derived
breast cancer. Eur J Cancer. 48:1227–1234. 2012. View Article : Google Scholar
|