Introduction
Colorectal cancer (CRC) is the third most common
cancer and accounts for ~9% of all cancer death, ranking the second
cause of cancer death worldwide (1). Over 50% of patients with CRC will
develop metastasis and the median survival time of metastatic CRC
(mCRC) is only 24 months, although various modalities of treatments
including chemotherapy with anti-EGFR (epidermal growth factor
receptor) targeted monoclonal antibodies have been applied
(2–4). Therefore, early diagnosis and
prediction of potential metastasis or recurrence show the greatest
potential for mCRC therapy. There are a few of tumor biomarkers
currently applied for prediction of CRC prognosis, such as gene
mutations status of Kras, Braf, PI3K, TP53, and microsatellite
instability (MSI) for selection of PD-1 inhibitor in the literature
(4–6). It is still pivotal to find additional
molecular biomarkers for clinical translation in CRC, especially
those that can predict potential tumor recurrence or
metastasis.
Except for the only 2% protein-coding genes, the
majority in human genome are non-coding genes. Non-coding RNAs were
previously regarded as ‘noise’ or ‘junk’ RNAs lacking
protein-coding potential (7).
However, in recent years due to the development of high-throughput
sequencing, accumulating evidence revealed that these neglected
non-coding RNAs are constitutively expressed and play critical
roles in cellular functions and human diseases, such as
multigenetic diseases and tumors (8,9).
Although small non-coding RNAs [<200 nucleotides (nt)], such as
microRNAs, have been the focus of research in RNA biology, in the
past few years, long non-coding RNAs (lncRNAs) were also shown to
play important roles in human disease and received most attention
(9–11). LncRNAs, defined as cellular RNAs
>200 nt in length, lack an open reading frame of an important
region (<100 amino acids) and thus code no proteins (8). Dozens of lncRNAs are reported to be
potential biomarkers for cancer diagnosis and prognosis prediction
in many solid tumors (8,9). Recently, LncRNA-H19, UCA1 and HOTAIR
were also reported strongly correlated with CRC metastasis and
prognosis (12–14). Therefore, lncRNAs are believed to
represent a new category of cancer biomarkers and potential tumor
therapeutic targets.
In this study, we report on the lncRNA-CTD903
(Ensemble version: ENST00000553153), a transcript of
lncRNA-CTD-2314B22.3 (also known as linc01296 in the gene set)
(15). It is located in chromosome
14q11.2. We have abbreviated the name of this transcript as
lncRNA-CTD903 based on the length of 903 bp.
We previously identified aberrant expression of
lncRNAs between CRC and matched paratumor normal tissues by a
microarray analysis (13).
Briefly, we selected six pairs of CRC tissues and corresponding
paratumor normal tissues and assayed the expression of lncRNAs
(30,586 items) and protein-coding mRNAs (26,109 items). Then we
found lncRNA-CTD903 expression was markedly upregulated (fold
change, 15.73, P<0.001) in CRC tissue. The fold change of
lncRNA-CTD903 expression ranked in the second position in thousands
of abnormally expressed lncRNAs, only secondary to known H19
according to the microarray data. Thus, we selected it for further
research. In this study, we confirm CTD903 is upregulated in CRC
tissues, compared to peritumor normal tissues. Importantly, CTD903
serves as an independent prediction factor of favorable prognosis
with a longer recurrence-free survival (RFS). Furthermore, cell
assays find overexpressed CTD903 could suppress cell invasion and
migration by inhibiting epithelial-mesenchymal transition (EMT).
Finally, our results indicate that CTD903 might repress
Wnt/β-catenin signaling and subsequently inhibit expression of the
transcription factors Twist and Snail to affect the EMT process.
These findings reveal that CTD903 may be a new biomarker for
prognosis and a potential target for treating mCRC.
Materials and methods
Patients and sample collection
The study was approved by Human Medical Ethics
Committee of Sun Yat-Sen University (SYSU). Fresh CRC tissues
(n=115) and paired paratumor normal mucosa samples (2 cm away from
the tumor border), and distant normal mucosa samples (≥5 cm away
from the tumor border) were collected from patients after surgery.
Informed consent was obtained from patients enrolled in this study.
Clinical tissue samples were all confirmed histopathologically and
stored in RNAlater solution (Invitrogen, USA) at −80°C until
extraction of total RNAs. Clinicopathological parameters were
collected from an online CRC database of this hospital, and we
confirmed the information by checking original medical records
manually. TNM stage was defined according to the 6th version of
American Joint Committee on Cancer (AJCC) staging Manual. Follow-up
was conducted according to the guideline of National Comprehensive
Cancer Network (NCCN). The recurrence-free survival (RFS) was
defined as the time interval from radical surgery to relapse,
metastasis or death of cancer.
Cell culture
Human CRC cell lines (RKO, SW480, SW620, Caco2,
HCT116, DLD1, and HT29) were all purchased in March 2013 from
Chinese Academy of Science, Shanghai, China. All cell lines were
routinely cultured in the DMEM or RPMI-1640 medium (Gibco, USA),
which were supplemented with 10% fetal bovine serum and 1%
penicillin/streptomycin (Gibco). All cell cultures were maintained
in an incubator at 37°C with a 5% CO2 humidity.
Quantitative real-time PCR analysis
Tissues were cut up into small pieces, and then
ground by a Tissue Lyser (Qiagen). Total RNAs were extracted using
TRIzol reagent (Invitrogen) according to the manufacturer's
instructions. Reverse transcription was conducted with Revertra Ace
aPCR RT master mix with gDNA remover (Toyobo, Japan). The
quantitative real-time polymerase chain reaction (qPCR) was
conducted by using SYBR Green mix (Applied Biosystems, USA)
according to the instructions. The conditions were as follows: 95°C
for 10 min; and 40 cycles (denaturation at 95°C for 15 sec,
annealing/extension temperature at 60°C for 1 min). All experiments
were performed in triplicates, including a negative control without
template. A melting curve was conducted to analyze the appropriate
amplification. Agarose gel (1%) was applied to validate the size of
PCR products. The oligo primers were used for lack of a poly(A)
tail of CTD903. The housekeeping gene glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was applied as the endogenous reference gene.
Relative gene expression was normalized to GAPDH using the
2−ΔΔCT method. The primers were as follows: CTD903
forward, 5′-TGGCAGTTTAAGAGTCTGGCA-3′; reverse,
5′-GAAGACTCGGGGATCAAGGT-3′. GAPDH forward,
5′-GACAGTCAGCCGCATCTTCTT-3′; reverse: 5′-AATCCGTTGACTCCGACCTTC-3′.
All qPCR assays were performed by an ABI 7500 system (Applied
Biosystems).
siRNA transfection
According to the manufacturer's instructions, the
optimum cell plating density was explored. Cells were plated in the
growth medium supplemented with 10% FBS without antibiotics in a
6-well plate, and then cultured for 24 h until 50–70% of confluent.
An appropriate density of 3×105 cells per well were
plated, then the volumes of siRNA were used for different cell
lines from 2 to 10 μl, to reach ≥70% of decreased expression
level after silencing. The optimum volumes of transfected siRNAs
for RKO, SW480, and SW620 were 2, 3 and 5 μl, respectively.
Moreover, an equivalent volume of scramble control was used. After
establishing the optimum volume, siRNA or scramble control was
mixed with Lipofectamine RNAiMAX reagent (Invitrogen) in reduced
serum medium Opti-MEM (Gibco), and then co-transfected with cells.
Forty-eight hours after the transfection, cells were harvested for
further studies. The siRNA oligonucleotides were designed and
synthesized by Ribobio (Guangzhou, China). The siRNA sequences of
lncRNA-CTD903 in this study were as follows: forward,
5′-GCUACCUUGUGGCAGUUUAdTdT-3′; reverse,
5′-UAAACUGCCACAAGGUAGCTdTd-3′.
Overexpression of lncRNA-CTD903
To obtain cell lines over-expressing lncRNA-CTD903,
the whole length of CTD903 was synthesized by Invitrogen Branch
Corp. (Guangzhou, China), and then was inserted into pcDNA3.1(+)
plasmid at the multiple cloning site using HindIII and
EcoRI restriction enzymes (New England Biolabs) in our
laboratory, and then confirmed by sequencing. HCT116 and DLD1, at
an appropriate density of 3×105 cells per well, were
transfected with 1 μg of plasmid pcDNA3.1-lncRNA-CTD903
(named pcDNA-CTD903 for short) or control pcDNA3.1(+) (named pcDNA
for short) using Lipofectamine 3000 (Invitrogen, USA) according to
the manufacturer's instructions. The CTD903 levels in overexpressed
cell lines were identified by qRT-PCR.
Cell invasion and migration assays
Cell invasion ability was assessed using the cell
culture insert (Corning, USA) according to the manufacturer's
protocols. An appropriate density of 40–60 thousand cells per well
for different cell lines were plated onto membrane (8.0-μm
pore size) coated with Matrigel and fibronectin (BD, USA) in the
upper chamber of the 24-well insert that contains serum-free
medium. The bottom chamber contained growth medium with 20% FBS.
Then cells were incubated at 37°C and 5% CO2 humidity
for 48 h, and then the bottom of the upper chamber insert was fixed
with 4% paraformaldehyde and stained with crystal violet. The cells
that remained on the inner membrane of the upper chamber were
removed by a cotton swab. The images of invaded cells were captured
on a 100X inverted microscope (five random fields for each well),
and then invaded cells were counted. Cell migration assay was
performed with the same method, but without Matrigel. The
experiments were repeated at least three times independently.
Cell proliferation, apoptosis and cell
cycle assays
Cell proliferation was assessed by cell counting
kit-8 (CCK-8) (Dojindo, Japan) and real-time cellular analysis
(RTCA) DP device (ACEA Biosciences, USA) at the same time. Briefly,
cells were plated in 96-well plate at an appropriate density of
6,000–8,000 cells per well for different cell lines, 10 μl
of CCK-8 was added to each well at the indicated time-points and
incubated for 2 h. Then the optical density was measured by a
multimode spectrum system (Thermo Scientific, USA) at 450 nm. The
methods of RTCA cell proliferation assays, apoptosis and cell cycle
assays by flow cytometric analysis were described in our previous
study (13).
Cell adhesion assays
Cells were plated onto a 96-well plate in triplicate
wells coated with fibronectin (BD) at an appropriate density of
10–20 thousand cells per well for different cell lines, and
incubated for 30 min with normal growth medium containing 10% FBS.
Then cells were washed with PBS two times to remove cells that did
not adhere. Adhering cells were fixed with 4% paraformaldehyde, and
the images of cells were captured under an inverted microscope
(five random ×100 fields), and the number of cells was calculated
by ImageJ software.
Western blot analysis
Cells were harvested and lysed by RIPA buffer
(Beyotime, China) with protease inhibitor (Thermo Scientific).
Lysates were then clarified by centrifugation and the
concentrations of extracted proteins were measured using BCA
Protein Assay kit (Beibo, China). Proteins were incubated with
SDS-PAGE loading buffer at 95°C for denaturation. Proteins (35
μg) were separated by using 5% stacking gel and 10% running
gel. The prestained protein ladder (Thermo Scientific) was loaded
in parallel as a reference of protein molecular weight. Then
proteins were transferred onto NC membranes (Millipore, USA). The
blots were blocked with 5% non-fat milk (BD) in TBS with Tween-20
(TBST). Then the membranes were incubated with primary antibody at
4°C overnight. After washing with TBST, secondary antibodies
HRP-labeled IgG (Abcam, USA) were incubated for 1 h, and then the
bound antibodies were detected using enhanced chemiluminescence
system (Odyssey, USA). Quantitative analysis was performed by
ImageJ software. Protein levels were all normalized to β-actin. The
primary antibodies used in this study were as follows: E-cadherin,
N-cadherin, Vimentin, ZEB1, Snail, and ZO-1 from an EMT kit (Cell
Signaling Technology, USA), β-catenin (BD), Twist 2 (Abcam),
β-actin (Proteintech Group, USA).
Statistical analysis
All of statistical analyses were performed by SPSS
version 17.0 software (Chicago, IL, USA). For the comparisons,
Student's t-tests or Chi-square test was performed appropriately.
Potential risk factors of RFS were evaluated by univariate
analysis. These risk factors with P<0.10 in univariate analysis
were included in the subsequent multivariate analysis using Cox
proportional hazards model. All P-values reported were two-sided
and the difference with a P<0.05 was considered to be
statistically significant.
Results
The novel lncRNA-CTD903 expression is
upregulated in CRC tissues
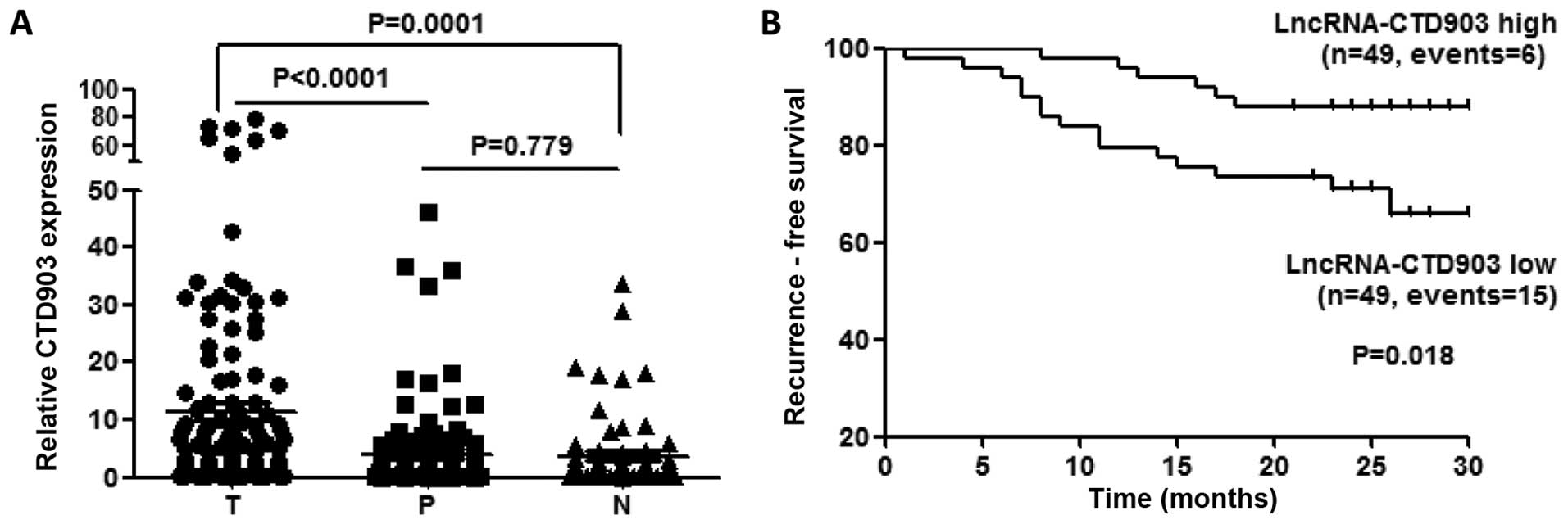
We analyzed CTD903 expression in 115 pairs of CRC
tissues (T) and corresponding paratumor tissues (P) using qRT-PCR
analysis. The distant normal intestine mucosas (N) from 66 cases of
these 115 patients were also assessed. We found that the CTD903
expression was upregulated more obviously in T than that in P
(P<0.0001) and N (P=0.0001), while no significant difference was
found between P and N (P=0.779) (Fig.
1A).
The correlations of CTD903 and
clinicopathological parameters and prognosis
We further examined the correlation of CTD903
expression and clinicopathological data in 115 CRC patients.
Patients were categorized as high or low CTD903 expression group
according to the median value. The higher CTD903 group showed more
percentages of colon cancers comparing with rectum cancers
(P=0.031), less mucinous tumors (P=0.030), and smaller tumor size
(P=0.041). No significant correlations between CTD903 expression
and other clinicopathological parameters were observed, such as
age, gender, differentiation, stages, lymphatic and distant
metastasis, venous and nerve invasion (Table I).
 | Table IThe correlations between
lncRNA-CTD903 and clinicopathological characteristics in 115
CRC. |
Table I
The correlations between
lncRNA-CTD903 and clinicopathological characteristics in 115
CRC.
| LncRNA-CTD903
expression |
|---|
|
|
|---|
|
Characteristics | High (n=55) | Low (n=60) | P-valuea |
|---|
| Age (years) | | | 0.424 |
| <60 | 27 | 25 | |
| ≥60 | 28 | 35 | |
| Gender | | | 0.945 |
| Male | 37 | 40 | |
| Female | 18 | 20 | |
| Location | | | 0.031 |
| Rectum | 11 | 23 | |
| Colon | 44 | 37 | |
| Colon cancer | | | 0.708 |
| Left-sided | 31 | 19 | |
| Non
left-sided | 13 | 18 | |
| Histologic
grade | | | 0.692b |
|
Well/moderately | 48 | 49 | |
| Poorly/others | 4 | 2 | |
| Mucinous
cancer | | | 0.030 |
| Yes | 4 | 13 | |
| No | 51 | 47 | |
| Tumor size | | | 0.041 |
| ≥5 cm | 18 | 30 | |
| <5 cm | 37 | 28 | |
| Depth of
invasion | | | 0.898 |
| T1/T2 | 6 | 7 | |
| T3/T4 | 49 | 53 | |
| Lymphatic node
stage | | | 0.708 |
| N0 | 35 | 33 | |
| N1 | 11 | 14 | |
| N2 | 9 | 12 | |
| Distant
metastasis | | | 0.503 |
| Present | 8 | 11 | |
| Absent | 47 | 46 | |
| AJCC stage | | | 0.379 |
| I/II | 32 | 30 | |
| III/IV | 23 | 30 | |
| Venous
invasion | | | 0.323 |
| Present | 8 | 13 | |
| Absent | 47 | 47 | |
| Nerve invasion | | | 0.923 |
| Present | 7 | 8 | |
| Absent | 48 | 52 | |
To explore whether CTD903 expression could influence
the clinical outcomes of CRC patients, we plotted ~2-year RFS
curves in two independent cohorts. In the total of 115 patients, 17
were excluded for RFS analysis, which included five lost to
follow-up, six died of non-cancerous causes, and six who had
pre-operative distant metastasis and did not undergo radical
resection of metastatic sites. Ninety-eight patients were enrolled
in final survival analysis, the results showed CTD903 high
expression group had a significantly longer RFS (P=0.018) than
CTD903 low expression (Fig. 1B),
using the median value of CTD903 expression as the cutoff similarly
to previous studies (11,16). Univariate analysis showed that
CTD903 expression (P=0.066), lymphatic metastasis (P<0.001),
distant metastasis (P<0.001), mucinous invasion (P=0.013),
venous invasion (P<0.001) and AJCC stages (P<0.001) were
prognostic indicators of CRC (Table
II). Furthermore, Cox's multivariate proportional hazards model
revealed that CTD903 low expression [HR: 3.430 (1.283–9.167),
P=0.014] and distant metastasis [HR: 3.808 (1.297–11.177), P=0.015]
were two independent prognostic factors associated with tumor
recurrence (Table III). Taken
together, these above data suggested the important role of CTD903
in CRC carcinogenesis and metastasis.
 | Table IIUnivariate analysis of
clinicopathological parameters for recurrence-free survival. |
Table II
Univariate analysis of
clinicopathological parameters for recurrence-free survival.
| Variable | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥60/<60
years) | 1.153 | 0.518–2.568 | 0.727 |
| Gender
(male/female) | 0.930 | 0.411–2.106 | 0.863 |
| Lymphatic
metastasis (positive/negative) | 7.058 | 2.928–17.014 | <0.001 |
| Differentiation
(well+moderate)/(poor+other) | 0.047 | 0.000–1666.333 | 0.561 |
| Distant metastasis
(yes/no) | 4.959 | 2.126–11.564 | <0.001 |
| T stages
(T3+T4)/(T1+T2) | 3.743 | 0.506–27.678 | 0.196 |
| Mucinous invasion
(yes/no) | 3.052 | 1.270–7.335 | 0.013 |
| Venous invasion
(yes/no) | 6.3111 | 2.742–14.524 | <0.001 |
| Nerve invasion
(yes/no) | 2.370 | 0.813–6.914 | 0.114 |
| AJCC stage
(III+IV)/(I+II) | 7.354 | 2.916–18.545 | <0.001 |
| Tumor size (≥5
cm/<5 cm) | 1.371 | 0.926–2.030 | 0.115 |
| CTD903 expression
(low/high) | 2.156 | 0.950–4.892 | 0.066 |
 | Table IIIMultivariate analysis
clinicopathological parameters for recurrence-free survival. |
Table III
Multivariate analysis
clinicopathological parameters for recurrence-free survival.
| Variable | Hazard ratio | 95% CI | P-value |
|---|
| Lymphatic
metastasis | 1.079 | 0.983–1.185 | 0.108 |
| Distant
metastasis | 3.808 | 1.297–11.177 | 0.015a |
| Mucinous
cancer | 2.251 | 0.796–6.368 | 0.126 |
| Venous
invasion | 2.052 | 0.718–5.863 | 0.179 |
| AJCC stage
(III+IV) | 2.670 | 0.811–8.794 | 0.106 |
| CTD903 low
expression | 3.430 | 1.283–9.167 | 0.014a |
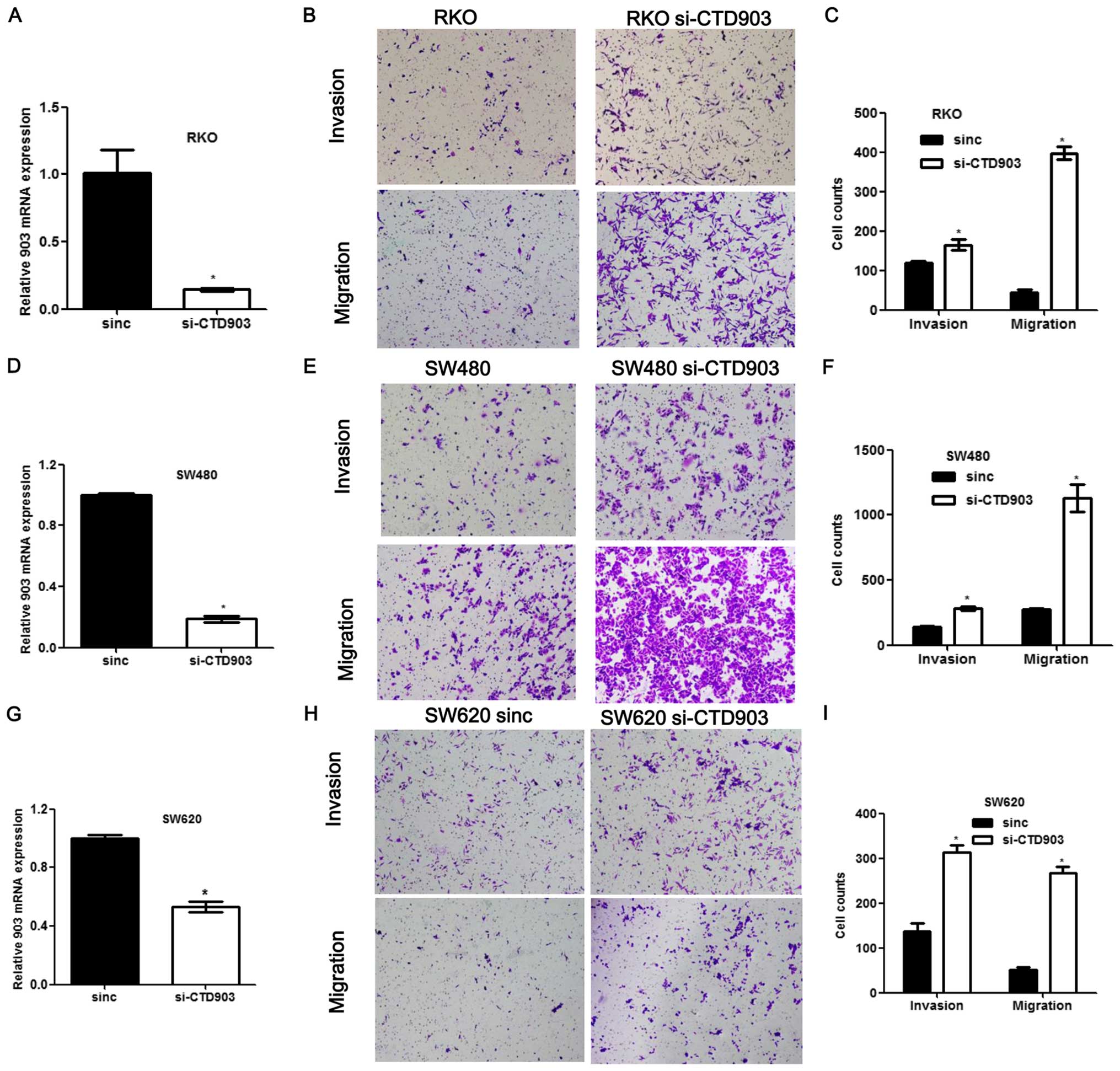
Reduced level of LncRNA CTD903 enhances
cell invasion/ migration and alters cell morphology
To evaluate the effects of CTD903 on cellular
behavior, we first examined CTD903 expression in a variety of CRC
cell lines, and then selected cell lines for subsequent
experiments. For silencing the expression of CTD903, cell lines
with relatively higher CTD903 expression including RKO and SW480
were chosen, along with SW620 derived from metastasis. For the
silencing assays, cells were treated with CTD903 siRNA or scramble
siRNA controls. CTD903 levels were significantly reduced in RKO,
SW480 and SW620 after siRNA treatments, compared to controls
(Fig. 2A, D and G). Transwell
assays showed that both cell invasion and migration were remarkably
increased in RKO, SW480 and SW620 after silencing CTD903, compared
to controls (Fig. 2B, C, E, F, H and
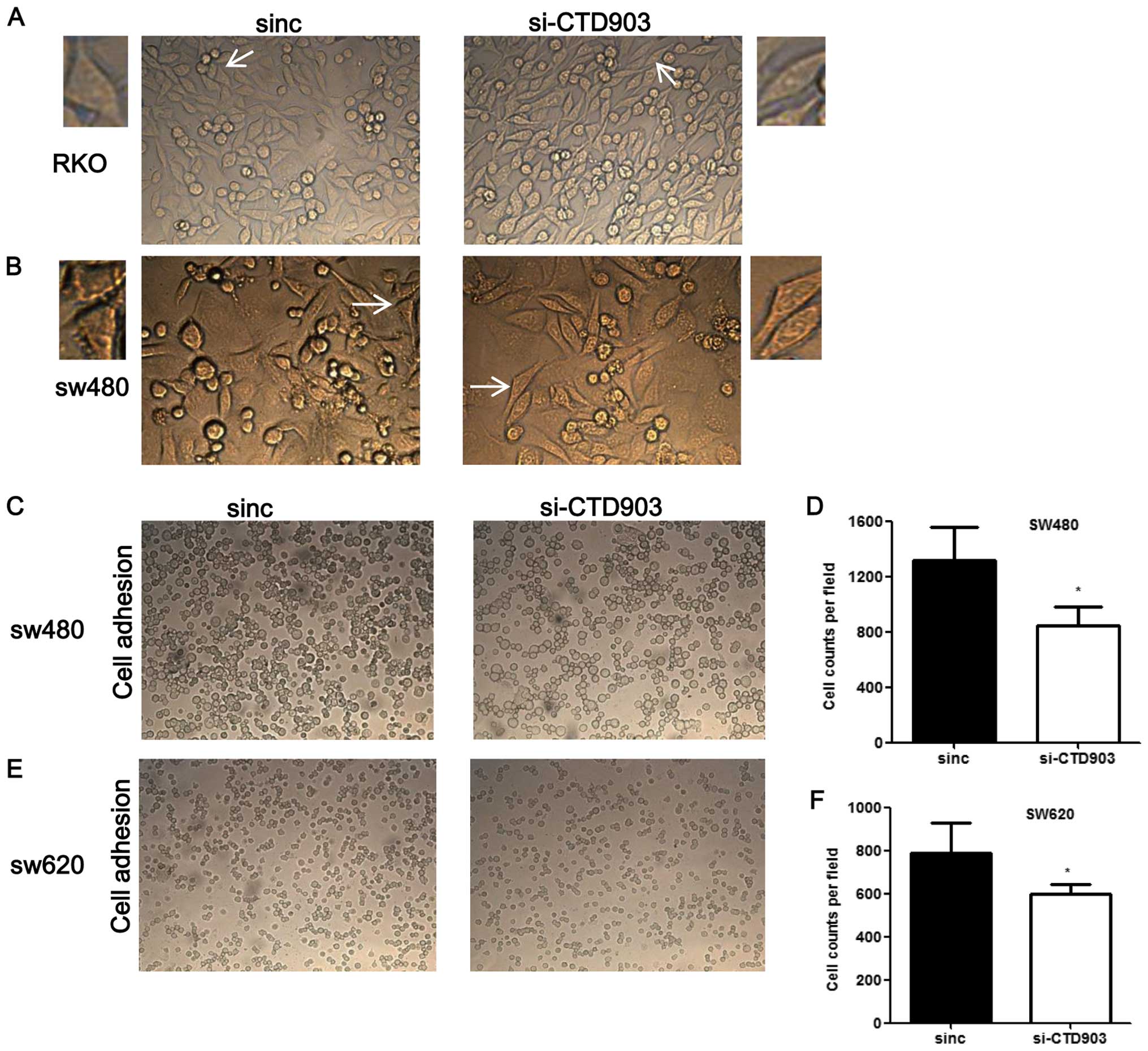
I). Furthermore, after siRNA transfection, cells exhibited more
elongated and spindle-like appearance in RKO and SW480, the typical
mesenchymal cell morphology, which indicated that CTD903 silencing
could induce EMT-like phenotypes (Fig.
3A and B). Reduced adhering cells were also observed by cell
adhesion assays after knockdown of CTD903 expression in SW480
(Fig. 3C and D) and SW620
(Fig. 3E and F). Cell
proliferation, apoptosis rate and cell cycle profile were also
analyzed, but no significant changes of these cell biological
functions were observed after silencing CTD903 expression (data not
shown). These results suggest reduced level of CTD903 would not
perturb the cell division per se.
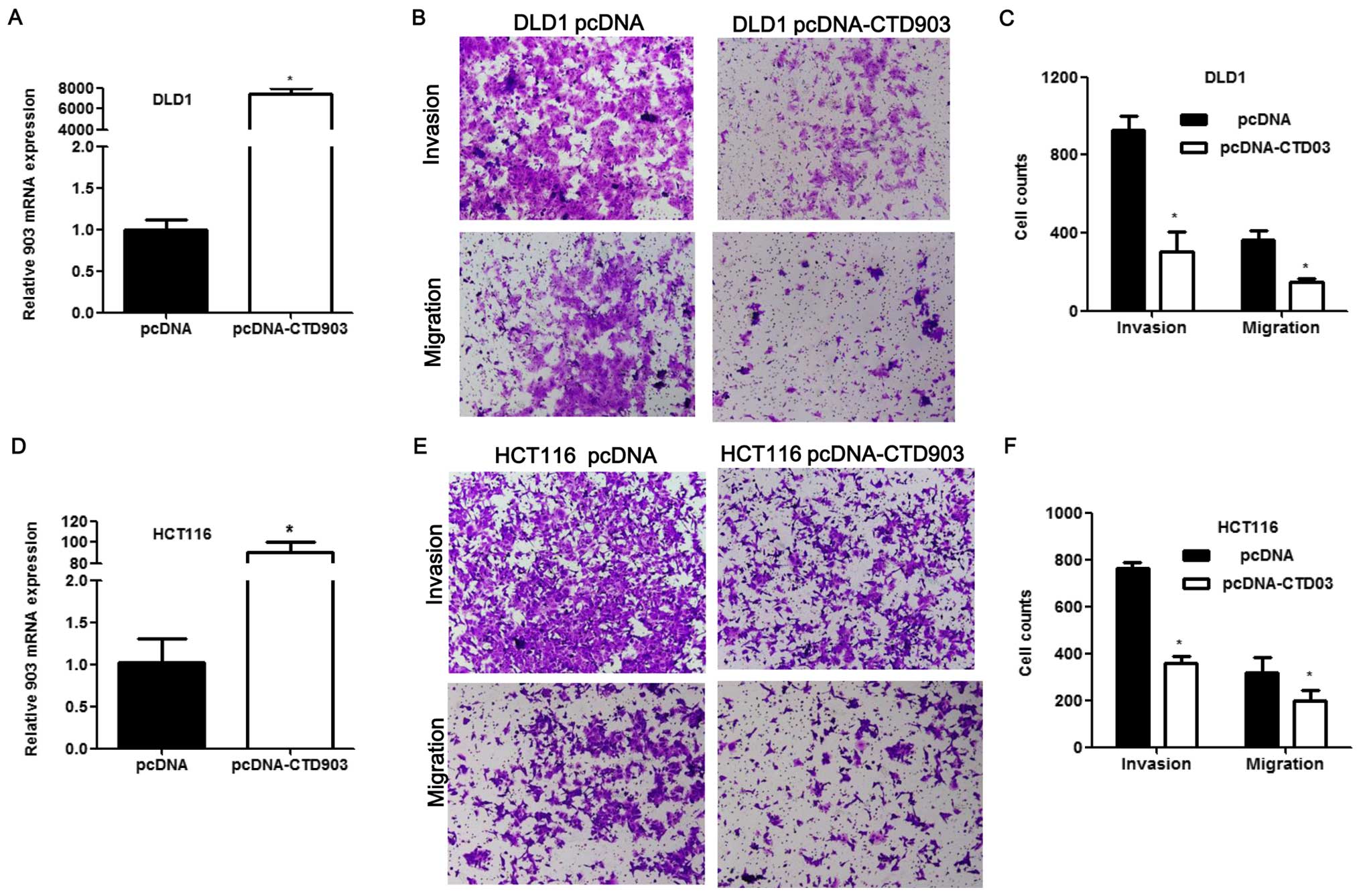
Overexpression of CTD903 impairs cell
invasion, migration and proliferation
To elucidate the effects of CTD903 over-expression
on cell biological functions, HCT116 and DLD1 with relatively low
CTD903 expression were transfected with pcDNA3.1-CTD903 or control
pcDNA3.1 plasmids. CTD903 levels were greatly elevated after
overexpression of CTD903 both in HCT116 and DLD1 (Fig. 4A and D). Transwell assays revealed
that cell invasion and migration were both impaired after
transfection in DLD1 (Fig. 4B and
C) and HCT116 (Fig. 4E and F).
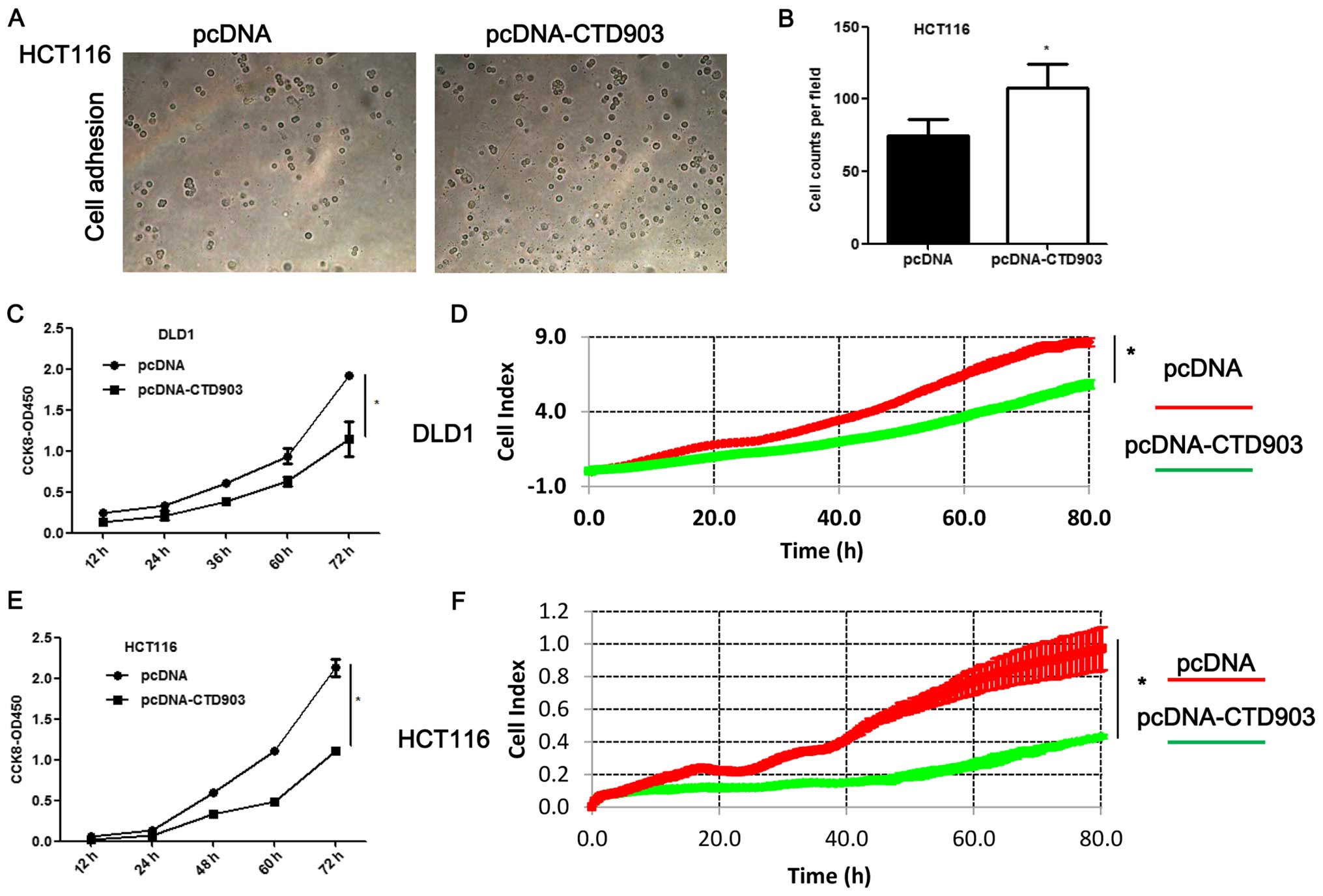
Cell adhesion was increased in HCT116 (Fig. 5A and B). Obvious decreased cell
proliferation rate was observed both in DLD1 (Fig. 5C and D) and HCT116 (Fig. 5E and F) after CTD903
overexpression. Taken together, these results showed CTD903 has the
ability to inhibit cell invasion and migration and therefore it is
reasoned to be a tumor suppressor gene, according to its silencing
and overexpression as indicated above.
CTD903 represses Wnt/β-catenin signaling
and inhibits EMT-associated transcription factors Twist and Snail
expression
To explore the effects of CTD903 on EMT-associated
proteins, the expression of epithelial markers were detected by
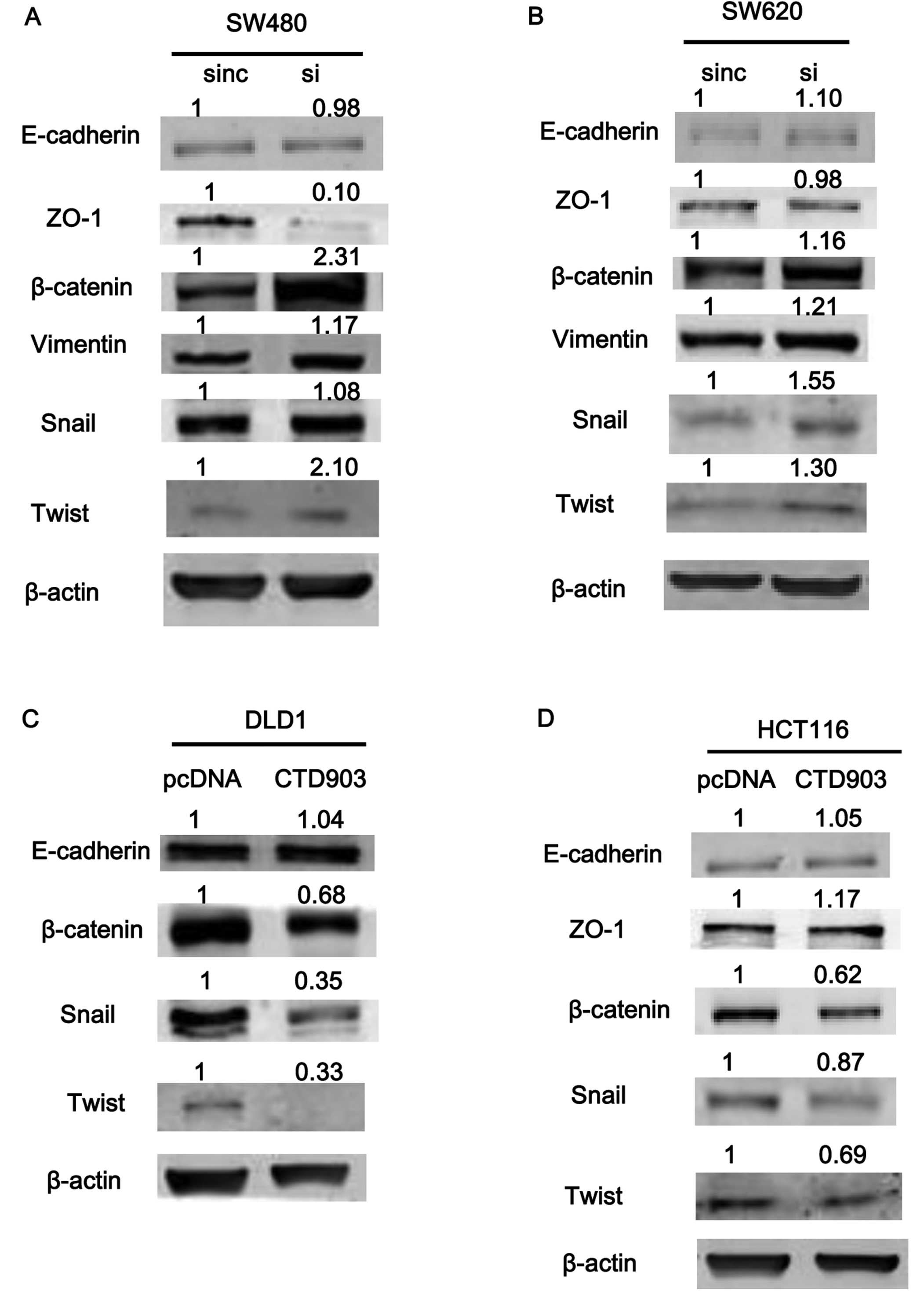
western blotting. After silencing CTD903 in SW480 and SW620, the
levels of β-catenin, vimentin, and snail increased significantly,
while ZO-1 protein level decreased (Fig. 6A and B). However, no significant
changes of E-cadherin (Fig. 6A and
B), N-cadherin and ZEB1 protein levels were observed (data not
shown). Overexpression of CTD903 in DLD1 and HCT116 cells,
decreased levels of β-catenin, Twist and Snail, along with slightly
increased ZO-1 expression (Fig. 6C and
D), and still no obvious changes of E-cadherin (Fig. 6C and D), N-cadherin and ZEB1
expression (data not shown) were detected. As for the inconsistent
changes between E-cadherin and β-catenin expression, previous
studies have explained that the reason is E-cadherin was not
essential for activating Wnt/β-catenin signaling to play roles in
EMT (17). We speculated that
knockdown of CTD903 can activate Wnt/β-catenin signaling
independently of cadherin expression.
Discussion
The human transcriptome is proved to be more complex
than known protein-coding genes, while ~90% of the genome is
non-coding transcripts (18).
Newly discovered lncRNAs have shown important roles in diverse
biological functions and tumors in recent years. The underling
mechanisms are mainly focused on regulating the processes of gene
expression, transcription and translation (8). LncRNA-H19 can increase cell invasion
and metastasis in bladder cancer through associating with EZH2 and
inhibiting E-cadherin (19), while
LncRNA-EBIC is found to promote tumor invasion by similar mechanism
in cervical cancer (18). Aberrant
expressed lncRNAs also unravelled their importance in CRC (20–25).
However, most of lncRNA studies focused on correlations of
clinicopathological characteristics and prognosis, the mechanisms
are rarely clarified, especially for mCRC.
To the best of our knowledge, we are the first to
report the function and mechanism of CTD903 in CRC. In this study,
CTD903 overexpression is found and is associated with smaller tumor
size, less mucinous adenocarcinomas, and favorable prognosis in
CRC, although prognosis did not reach statistical significance due
to the small sample size. These results reveal that tumors with
CTD903 high expression have less aggressive biological behavior
than low expression tumors. CTD903 acts as a tumor suppressor gene
and play protective roles in CRC. We also found the parameters,
including lymphatic and distant metastasis, mucinous and venous
invasion, and AJCC stages in univariate analysis, which are known
indicators of poor CRC outcome, and in turn reveal the reliable
results of our study. Furthermore, CTD903 is found to be an
independent prognostic factor for RFS. Thus, we believe CTD903
might be a potential novel biomarker for future clinical
translation.
Recently Qiu and Yan also reported that linc01296
was upregulated and can predict favorable prognosis in CRC by
conducting a meta-analysis of online databases in European
population (15). Their finding
was also consistent with our results. However, the function and
underlying mechanism of this lncRNA in tumors remain unknown.
Furthermore, as many as sixteen transcripts of linc01296 are
documented, but only three of them are overexpressed and others
were not aberrantly expressed in CRC according to our previous
microarray data, which indicate dramatic heterogeneity among these
transcripts (13). CTD903 is the
most overexpressed transcript of these three, and ranks in the
second position in thousands of aberrant expressed lncRNAs in CRC.
Therefore, CTD903 has shown biomarker potential and absorbed our
interest for further research.
EMT is characterized by the impairment of cell-cell
adhesion and can increase cell motility to promote cancer
progression and metastasis (26).
EMT is induced through activation of Wnt signaling (27). β-catenin is the key initial protein
in the Wnt signaling. After activation, β-catenin can translocate
from the cytoplasm to the nucleus, then regulate expression of
several transcription factors, and subsequently induce EMT
(17,28,29).
In accordance with previous studies, in this study, CTD903
overexpression can repress Wnt/β-catenin expression, and then
inhibits expressions of downstream mesenchymal marker Vimentin,
epithelial marker ZO-1 and the transcriptional factors Twist and
Snail. Finally, phenotypes of EMT and cell invasion and migration
are inhibited.
E-cadherin and N-cadherin are two key molecules for
EMT. Loss of E-cadherin expression and overexpression of N-cadherin
are usually observed when EMT occurs (30). However, E-cadherin is not
essentially correlated with activation of Wnt/β-catenin, because
other compartments such as APC or AXIN can also confer β-catenin
into the nucleus to induce EMT (17). In our study, no significant
expression changes of E-cadherin and N-cadherin were observed,
neither when EMT was induced. Thus, the results provide additional
evidence that activation of Wnt/β-catenin signaling can be
independent and not triggered by E-cadherin/N-cadherin. Further
research is needed to elucidate the underlying mechanism. No change
of ZEB1 expression was observed, which indicated that it may not be
involved in CTD903-mediated EMT.
Although most of the reported oncogenes are
overexpressed in cancer, small proportion of tumor suppressor genes
can also be upregulated and predict good prognosis, such as Rab27A
in CRC and FOXO3a in gastric cancer (31,32).
In this study, the level of CTD903 is upregulated significantly and
is also suggested to be a tumor suppressor gene. In addition,
overexpressed CTD903 level can decrease cell proliferation in
HCT116 and DLD1, but no increased cell proliferation was observed
when CTD903 was knocked down. The reason is perhaps for the limited
effects of CTD903 on cell proliferation, like others reported in
lncRNA-EBIC, and microRNA-10b that can remarkably affect cell
migration but have no effects on cell proliferation (18,30).
Cell cycle and apoptosis are not affected in knockdown of CTD903,
which may be explained by limited roles on cell cycle and
apoptosis, similarly to many other lncRNAs (18,33).
Thus, we focus on the invasion and migration to elucidate potential
mechanisms of tumor metastasis, which is currently a popular issue
(34,35) and the leading cause of
cancer-related death that remains not well answered. The most
important limitations of this study are lack of in vivo
experiments, which may provide more robust evidence for our
findings.
In conclusion, lncRNA-CTD903 acts as a tumor
suppressor gene in CRC, and can repress cell invasion and migration
by repressing Wnt/β-catenin signaling to inhibit EMT and CRC
metastasis. Our study enriches the underling molecular mechanisms
of carcinogenesis and metastasis, and provides a novel biomarker
and potential therapeutic target for CRC, which is promising for
precision medicine.
Acknowledgements
This study was supported by the following grants:
National Natural Scientific Foundation of China (nos. 81201581,
81372566 and 81573078), Guangdong Provincial Scientific Research
Foundation (no. S2013010013478), National Science and Technology
Support Project of Ministry of Science and Technology (no.
2014BAI09B06) and Young Teacher Training Program of Sun Yat-Sen
University (no. 12ykpy48). We thank Zhemiao Wang (Department of
Pathology) for tissue sample collection.
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
CRC
|
colorectal cancer
|
|
mCRC
|
metastatic colorectal cancer
|
|
EMT
|
epithelial-mesenchymal transition
|
|
RFS
|
recurrence-free survival
|
|
ZO-1
|
zonula occludens-1
|
|
EGFR
|
epidermal growth factor receptor
|
|
CTD903
|
CTD-2314B22.3-006, 903 bp
|
|
RTCA
|
real-time cellular analysis
|
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sastre J, Argiles G, Benavides M, Feliu J,
Garcia-Alfonso P, Garcia-Carbonero R, Grávalos C, Guillén-Ponce C,
Martínez-Villacampa M and Pericay C: Clinical management of
regorafenib in the treatment of patients with advanced colorectal
cancer. Clin Transl Oncol. 16:942–953. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ye LC, Liu TS, Ren L, Wei Y, Zhu DX, Zai
SY, Ye QH, Yu Y, Xu B, Qin XY, et al: Randomized controlled trial
of cetuximab plus chemotherapy for patients with KRAS wild-type
unresectable colorectal liver-limited metastases. J Clin Oncol.
31:1931–1938. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yuan ZX, Wang XY, Qin QY, Chen DF, Zhong
QH, Wang L and Wang JP: The prognostic role of BRAF mutation in
metastatic colorectal cancer receiving anti-EGFR monoclonal
antibodies: A meta-analysis. PLoS One. 8:e659952013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grady WM and Markowitz SD: The molecular
pathogenesis of colorectal cancer and its potential application to
colorectal cancer screening. Dig Dis Sci. 60:762–772. 2015.
View Article : Google Scholar
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long non-coding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C,
Zhou XY and Du X: Low expression of LOC285194 is associated with
poor prognosis in colorectal cancer. J Transl Med. 11:1222013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Wu Z, Fu X and Han W: Long
non-coding RNAs: Insights from biological features and functions to
diseases. Med Res Rev. 33:517–553. 2013. View Article : Google Scholar
|
|
11
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long non-coding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H19
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni B, Yu X, Guo X, Fan X, Yang Z, Wu P,
Yuan Z, Deng Y, Wang J, Chen D, et al: Increased urothelial cancer
associated 1 is associated with tumor proliferation and metastasis
and predicts poor prognosis in colorectal cancer. Int J Oncol.
47:1329–1338. 2015.PubMed/NCBI
|
|
14
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long non-coding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qiu JJ and Yan JB: Long non-coding RNA
LINC01296 is a potential prognostic biomarker in patients with
colorectal cancer. Tumour Biol. 36:7175–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W, et al: Oncofetal long
non-coding RNA PVT1 promotes proliferation and stem cell-like
property of hepatocellular carcinoma cells by stabilizing NOP2.
Hepatology. 60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ghahhari NM and Babashah S: Interplay
between microRNAs and WNT/β-catenin signalling pathway regulates
epithelial-mesenchymal transition in cancer. Eur J Cancer.
51:1638–1649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang
SB, Jin ZJ, Sun SH, Wang F and Li W: Long non-coding RNA-EBIC
promotes tumor cell invasion by binding to EZH2 and repressing
E-cadherin in cervical cancer. PLoS One. 9:e1003402014. View Article : Google Scholar
|
|
19
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long non-coding
RNA activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y,
Li R, Ying H, Wang F, Liu X, et al: Up-regulation of 91H promotes
tumor metastasis and predicts poor prognosis for patients with
colorectal cancer. PLoS One. 9:e1030222014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu
Y, Li X, Cai G and Cai S: Low expression of novel lncRNA
RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal
cancer. Med Oncol. 31:312014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yin D, He X, Zhang E, Kong R, De W and
Zhang Z: Long non-coding RNA GAS5 affects cell proliferation and
predicts a poor prognosis in patients with colorectal cancer. Med
Oncol. 31:2532014. View Article : Google Scholar
|
|
24
|
Kam Y, Rubinstein A, Naik S, Djavsarov I,
Halle D, Ariel I, Gure AO, Stojadinovic A, Pan H, Tsivin V, et al:
Detection of a long non-coding RNA (CCAT1) in living cells and
human adeno-carcinoma of colon tissues using FIT-PNA molecular
beacons. Cancer Lett. 352:90–96. 2014. View Article : Google Scholar
|
|
25
|
Yan B, Gu W, Yang Z, Gu Z, Yue X, Gu Q and
Liu L: Downregulation of a long non-coding RNA-ncRuPAR contributes
to tumor inhibition in colorectal cancer. Tumour Biol.
35:11329–11335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji Q, Liu X, Han Z, Zhou L, Sui H, Yan L,
Jiang H, Ren J, Cai J and Li Q: Resveratrol suppresses
epithelial-to-mesenchymal transition in colorectal cancer through
TGF-β1/Smads signaling pathway mediated Snail/E-cadherin
expression. BMC Cancer. 15:972015. View Article : Google Scholar
|
|
27
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar
|
|
28
|
Vincan E and Barker N: The upstream
components of the Wnt signalling pathway in the dynamic EMT and MET
associated with colorectal cancer progression. Clin Exp Metastasis.
25:657–663. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin JJ, Zhao TZ, Cai WK, Yang YX, Sun C,
Zhang Z, Xu YQ, Chang T and Li ZY: Inhibition of histamine receptor
3 suppresses glioblastoma tumor growth, invasion, and
epithelial-to-mesenchymal transition. Oncotarget. 6:17107–17120.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Sun J, Wang B, Ren JC, Su W and
Zhang T: MicroRNA-10b triggers the epithelial-mesenchymal
transition (EMT) of laryngeal carcinoma Hep-2 cells by directly
targeting the E-cadherin. Appl Biochem Biotechnol. 176:33–44. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi C, Yang X, Ni Y, Hou N, Xu L, Zhan F,
Zhu H, Xiong L and Chen P: High Rab27A expression indicates
favorable prognosis in CRC. Diagn Pathol. 10:682015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu S, Yu Y, Sun Y, Wang X, Luo R, Zhao N,
Zhang W, Li Q, Cui Y, Wang Y, et al: Activation of FOXO3a suggests
good prognosis of patients with radically resected gastric cancer.
Int J Clin Exp Pathol. 8:2963–2970. 2015.PubMed/NCBI
|
|
33
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long non-coding RNA (lncRNA) down-regulated
expression by HBx (Dreh) inhibits hepatocellular carcinoma
metastasis by targeting the intermediate filament protein vimentin.
Hepatology. 57:1882–1892. 2013. View Article : Google Scholar
|
|
34
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fang JH, Zhou HC, Zhang C, Shang LR, Zhang
L, Xu J, Zheng L, Yuan Y, Guo RP, Jia WH, et al: A novel vascular
pattern promotes metastasis of hepatocellular carcinoma in an
epithelial-mesenchymal transition-independent manner. Hepatology.
62:452–465. 2015. View Article : Google Scholar : PubMed/NCBI
|