Introduction
Upregulation of the Hedgehog (HH/GLI) signaling
pathway is responsible for the formation and progression of a
number of human cancers through the aberrant activation of
transcription factors GLI (1–3).
Autocrine and paracrine ligand sonic Hedgehog (Shh) binds to
Patched (PTCH) receptor, thereby relieving Patched repressive
activity on the 7-transmembrane protein Smoothened (SMO), which in
turn causes activation of the downstream effectors, zinc finger
containing GLI transcription factors (4–7).
Aberrant HH/GLI pathway activity has been initially identified in
basal cell carcinomas and medulloblastomas (8) and later found to be deregulated in
many common human cancers such as lung tumors, pancreas,
colorectal, ovarian and prostate carcinomas, glioblastomas as well
as melanomas (5,7,9).
Inhibitors of SMO vismodegib and cyclopamine have been used in many
clinical trials. However, as the HH pathway can be upregulated
non-canonically by direct activation of GLI factors by several
signaling pathways (10–12), using the inhibitor of GLI activity
GANT61 can overcome the possible ineffectiveness of upstream SMO
inhibitors. GANT61 prevents the binding of GLIs to DNA (13) while fully preserving their
expression.
GLI2 has been shown to control the invasiveness and
metastatic potential and to contribute to the
epithelial-to-mesenchymal transition in melanoma (14). Melanomas express MITF
(microphthalmia-associated transcription factor), a crucial factor
in the pigment cell transcriptional circuitry, activating a large
number of genes with various functions (15–17).
GLI2 expression was reported to be inversely correlated with MITF
expression. Thus, high GLI2 and low MITF levels characterize the
invasive cell phenotype in melanoma (18,19).
High GLI2 expression in melanoma was achieved through the HH and
TGF-β/SMAD pathways (20,21). HH/GLI1 signaling has an essential
role in controlling self-renewal and tumor initiation of melanoma
and GANT61 has been reported to reduce the number of
melanomaspheres formed from cells with features of tumor initiating
stem cells (22). However, despite
the importance of HH/GLI pathway and GLI2 in the melanoma
development, the data that would determine the effect of GANT61 on
melanoma cell cultures in vitro is lacking. The treatment of
melanoma by inhibitors against BRAF(V600E) or MAPK pathway were
initially promising, but resistence appeared almost invariably
after months through multiple mechanisms (23,24).
Other treatment approaches are therefore needed in melanoma.
In the present study, we undertook treatment of 9
melanoma cell lines with GANT61, a downstream Hedgehog/GLI pathway
inhibitor, and performed a combined incubation of GANT61 with
obatoclax, a BCL2 family inhibitor. We identify that melanoma cells
are efficiently eliminated by GANT61 through apoptosis. GANT61 with
obatoclax reveal synthetic lethality in cell lines in vitro
as assessed by colony formation assays. Although the results
require the in vivo verification, this targeted combined
therapy is promising to be beneficial for melanoma patients
irrespective of the BRAF or NRAS mutational status.
Materials and methods
Promoters and reporter assays
The ΔNGLI2 expression vector was previously
described (25). The MITF
promoter-reporter construct has been described (26). Professor Fritz Aberger (University
of Salzburg) provided the Patched promoter plasmid and Professor
Rune Toftgard (Karolinska Institutet) the 12xGLI reporter plasmid.
For reporter assays, melanoma cells were seeded in 12-well plates
and transfected at 70–80% confluency in fresh medium with LipoJet
(SignaGen Laboratories, Rockville, MD, USA) according to the
instructions of the manufacturer. Dual-luciferase assay kit
(Promega, Madison, WI, USA) was used to determine promoter-reporter
activity.
Cell lines
Melanoma cell lines MeWo, SK-MEL-3, SK-MEL-5,
SK-MEL-28, WM35, WM1552C and SW13 line (human cervical carcinoma)
were purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Beu and Hbl cells were previously described
(25). 501mel cell line was
obtained from Dr Ruth Halaban (Yale University) and maintained in
RPMI-1640 medium. Hbl and SW13 were grown in Dulbecco’s modified
Eagle’s medium (DMEM) and all other cell lines were cultivated in
EMEM medium. All types of media (Sigma-Aldrich, St. Louis, MI, USA)
were supplemented with 10% fetal calf serum (FCS; Life
Technologies, Carlsbad, CA, USA), L-glutamine and antibiotics
(Sigma-Aldrich).
Cell viability assay
Cells (50,000/well) were seeded in 12-well plates
(Nunc, Roskilde, Denmark) and treated the next day with 25 μM
GANT61 (Selleckchem, Munich, Germany) or DMSO (Sigma-Aldrich)
(control) for 72 h. Cultivation medium was removed and cell
viability was determined with MTT cell viability assay kit
(Sigma-Aldrich). Experiments were performed in duplicate and data
are expressed as a mean of duplicate measurements (mean ± SD) in
percentage of control-treated cells (100%). Two independent
experiments were carried out with similar results and one
experiment is shown.
Cell proliferation assay
To perform colony outgrowth assay, subconfluent
cells were trypsinised and seeded in 12-well plates (day 0). The
next day, cell lines were treated with inhibitors at concentrations
of 20 μM GANT61 or 100 nM obatoclax (Selleckchem), or their
combination for the indicated time intervals (9 days maximum). The
plates were fixed in 3% paraformaldehyde solution in PBS and
stained with 1% crystal violet. Two experiments were performed in
duplicate. Results of both experiments were similar. The density of
the remaining stained cells was quantified with the ImageJ software
and the results of one experiment are shown.
Soft agar assay
Assay was performed in 60-mm dishes containing 0.6%
lower layer of Noble agar (Difco, Radnor, PA, USA). Cells
(5×104) were seeded in the upper agar (0.4% agar in
cultivation medium containing 15% FCS), overlayed with the
cultivation medium with 15% FCS and DMSO or inhibitors. The media
with inhibitors or control vehicle were refreshed twice a week.
After 21 days colonies were stained with p-indonitrotetrazolium
violet (Sigma-Aldrich), counted, photo-documented and quantified by
the ImageJ software.
Detection of apoptotic cells
DNA fragmentation as a hallmark of advanced
apoptosis was detected by flow cytometric and microscopic analyses.
SK-MEL-3 and SK-MEL-5 cells were treated with 20 μM GANT61 or DMSO
(control) for 48 h and then trypsinized, combined with detached
cells, washed with PBS, incubated in the DNA extraction buffer (192
ml of 0.2 M Na2HPO4 with 8 ml of 0.1 M citric
acid, pH 7.8) for 10 min at room temperature, washed again with
cold PBS, resuspended in PBS containing 5% BSA (Merck, Darmstadt,
Germany) and stained with propidium iodide (Sigma-Aldrich) (20 μl
of 0.5 mg/ml solution per 1 ml of cell suspension). RNAse
(Sigma-Aldrich) was added simultaneously (2 μl of the 20 mg/ml
stock per 1 ml of cell suspension). Cell histograms were acquired
on a FACSCanto flow cytometer (Becton-Dickinson, Franklin Lakes,
NJ, USA) with the FACSDiva VI acquisition and analysis software. To
obtain microscopic images of apoptotic nuclei, cells were grown in
2-well chambers (NUNC) and treated with GANT61 as above. The
remaining attached cells were then fixed with 3% paraformaldehyde
in PBS. Slides were coverslipped with Vectashield mounting medium
with DAPI (Vector Laboratories, Burlingame, CA, USA) to visualize
the nuclei. Images were acquired on an Olympus BX61 microscope
(Olympus, Tokyo, Japan).
Western blot analysis
Cell extracts for immunoblotting analysis were
prepared by lysis the cells in RIPA buffer (1% NP-40, 150 mM NaCl,
5 mM EDTA, 0.5% sodium deoxycholate, 50 mM Tris-HCl pH 7.5, 0.1%
SDS) with added protease and phosphatase inhibitors: 1 μg/ml of
leupeptin, aprotinin and pepstatin, and cOmplete and PhosStop
(Roche Diagnostics, Mannheim, Germany) as recommended by the
manufacturer. Protein extracts (30 μg) in sample loading buffer (50
mM Tris-HCl pH 6.8; 2% sodium dodecyl sulphate; 100 mM
dithiotreitol; 10% glycerol; 0.1% bromophenol blue) were heated to
98°C for 2 min and proteins were separated on 10% polyacrylamide
gels. After transfer onto the PVDF membrane (Millipore, Billerica,
MA, USA), the membranes were blocked in 5% Blotto (Santa Cruz
Biotechnology, Dallas, TX, USA) in PBS containing 0.1% Tween-20
(Sigma-Aldrich) at room temperature for 1 h. The incubation with
the primary antibodies against GLI2 (GTX46056), purchased from
GeneTex (Irvine, CA, USA), MITF (Lab Vision, Fremont, CA, USA), or
β-actin (AC-74) from Sigma-Aldrich was conducted for several hours
at room temperature, diluted 1:1,000 in the blocking solution.
After washing in PBS-0.1% Tween, 1 h incubation in the HRP-labelled
secondary antibody (Cell Signaling Technology, Danvers, MA, USA)
and washing, signals were detected by the Pierce ECL
chemiluminiscent detection reagent (Thermo Fisher Scientific,
Waltham, MA, USA). In western blot experiments determining the
effect of GANT61 on MITF protein levels, GANT61 was left on cells
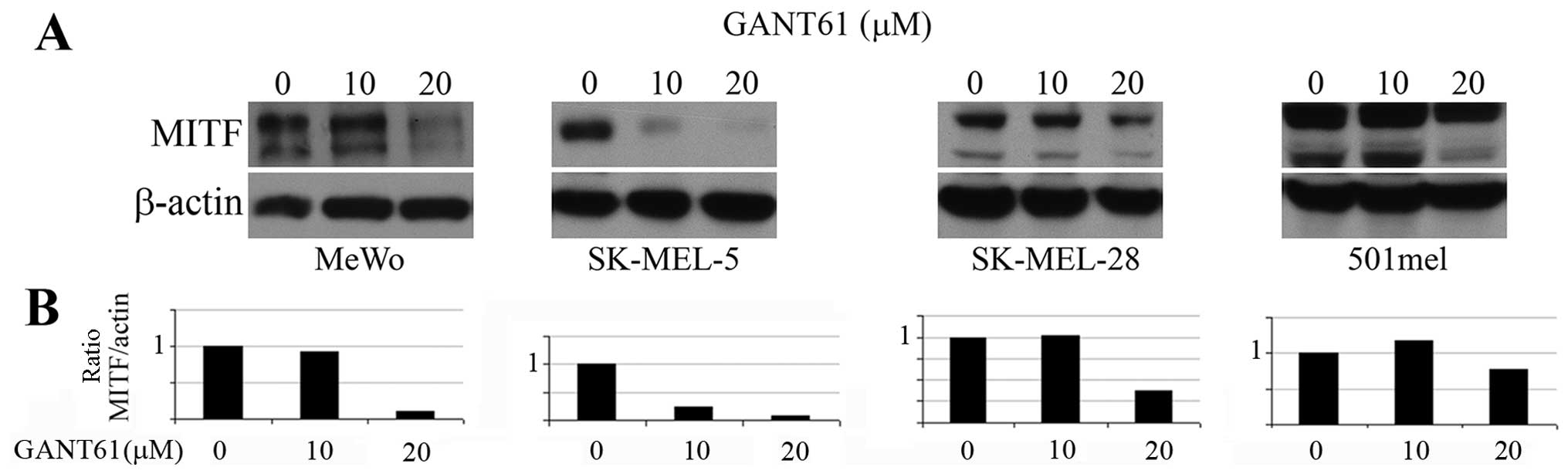
for 20 h and the cells were harvested (Fig. 2).
Statistical analysis
Statistical comparison was carried out using the
unpaired Student’s t-test. Significance was set at P<0.05.
Standard error (SE) is shown for triplicate or duplicate samples in
promoter-reporter or cell viability assays, respectively. The
combination index (CI) was calculated using CalcuSyn software. GLI2
and MITF proteins levels were corrected after densitometry (using
AIDA image analyzer software) to western blots with the β-actin
(control) expression (Figs. 1B and
2B).
Results
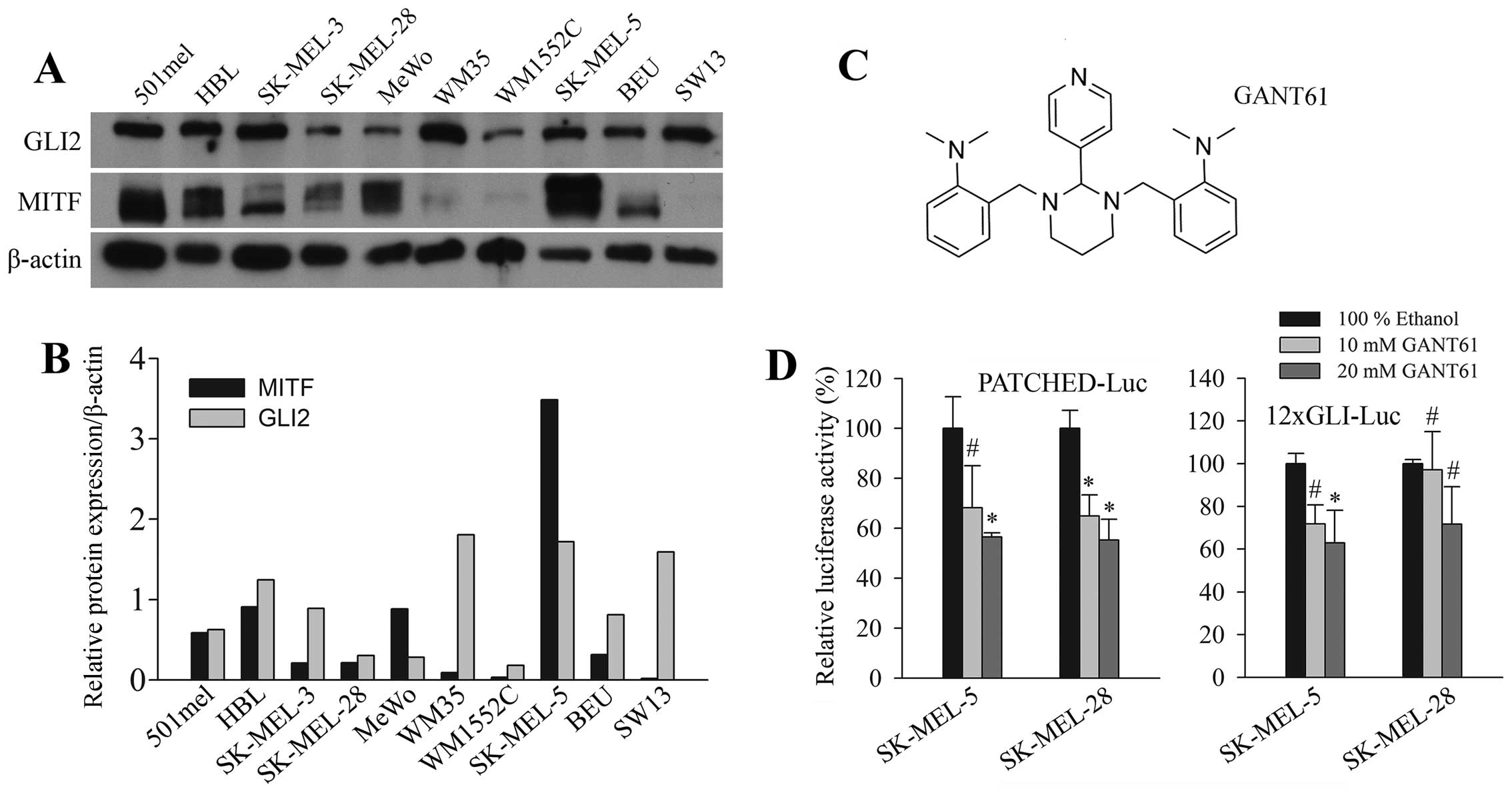
Expression of GLI2 in melanoma cell
lines
GLI2 was shown to be an essential protein for
maintaining the pro-oncogenic phenotype in melanoma (14). GLI2 expression was determined in
melanoma cell lines and control non-melanoma cells SW13 (adrenal
gland carcinoma) by western blotting. The BRAF and NRAS mutational
status of melanoma cell lines used is provided in Table I (27–32).
GLI2 was present in all cells examined (Fig. 1A). Expression was noted also in the
501mel cells, which were previously reported to be GLI2 negative
(18). In our recent study, these
cells were also clearly GLI2 positive for both protein and mRNA, as
detected by real-time PCR (25)
and RT-PCR (data not shown). While the use of a different antibody
might explain the difference in the western blot result, we can not
clarify the discrepancy in the result of RNA level in 501mel cells
at present. The western blot band density was quantitated and
corrected to β-actin levels (Fig.
1B). In 6 of 9 cell lines, similar trend of inverse correlation
between GLI2 and MITF was observed, as that previously reported
(18). We observed this trend in
different cell lines (with the exception of 501mel) than those
employed by others (18). Since we
have detected GLI2, a protein implicated in melanoma invasion and
metastasis, in all cell lines in our panel, they are predicted to
be good models for exploring the effect of GANT61. The chemical
structure of GANT61 is shown in Fig.
1C.
 | Table IMutational status of BRAF and NRAS in
melanoma cell lines. |
Table I
Mutational status of BRAF and NRAS in
melanoma cell lines.
GANT61 inhibits GLI2-dependent promoter
reporters
To test whether GANT61 inhibits GLI-dependent
transcription, we examined the activity of known HH/GLI responsive
promoter reporters, the 12xGLI arteficial super-promoter and the
Patched promoter. After stimulation of these promoters by
cotransfecting ΔNGLI2, the most effective GLI construct which was
also the most effective stimulant for the newly discovered GLI2
target survivin promoter (25),
increasing concentrations of GANT61 were added for 20 h. The
activity of both promoters was evidently decreased in two melanoma
cell lines tested (Fig. 1D),
albeit most results at l0 μM concentration and one result at 20 μM
of GANT61 (the 12xGLI promoter in SK-MEL-28 cells) were not
statistically significant (due to high SE values). Considering this
observation, together with the finding of the uniform presence of
GLI2 levels in melanoma cells (Fig.
1A), we hypothesized that GANT61 might possibly affect the
growth of melanoma cells.
GANT61 decreases the MITF expression,
cell viability and outgrowth of melanoma cells in soft agar
Since GLI2 expression was reported to be inversely
correlated with the MITF expression (18) and GLI2 was described to repress
MITF transcription (19), we
expected that GANT61, a GLI1/2 inhibitor, would increase the MITF
levels in melanoma cells. However, we observed no effect in MITF
protein levels on western blots after treatment of cells with 10 μM
GANT61 (Fig. 2A) in 3 melanoma
cell lines and a massive inhibition of MITF level in SK-MEL-5 cells
even at 10 μM (see Discussion) (Fig.
2A). The 20 μM concentration inhibited MITF levels strongly in
3 cell lines and only negligibly in 501mel cells. These results are
clearly evident after quantitation and correction of the MITF
levels to β-actin levels (Fig.
2B).
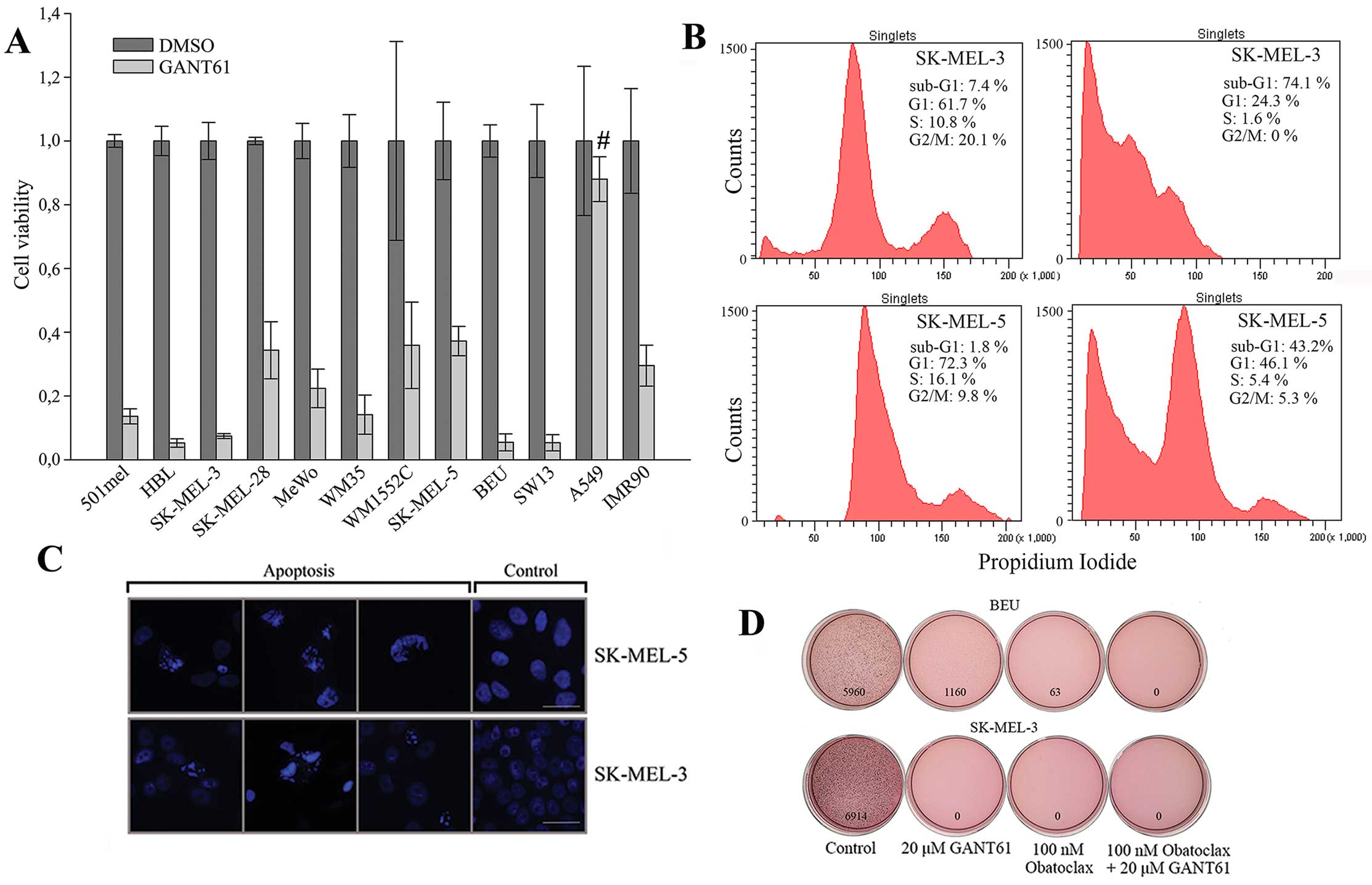
We next addressed whether GANT61 has effects on
cellular phenotype of melanoma cells. The viability of our panel of
9 melanoma cell lines and 3 non-melanoma cell lines was drastically
diminished after 72-h incubation with 25 μM GANT61. Only 5–35% of
viable cells remained. Only one of controls (A549 cells, lung
carcinoma) was resistant as viability decreased by <10%
(Fig. 3A). The soft agar growth of
the two most sensitive lines based on the viability assay, SK-MEL-3
and Beu, was examined. Whereas colony formation of SK-MEL-3 cells
was completely abrogated by 20 μM GANT61 after three weeks, Beu
cells formed only small number of colonies (related to control)
after quantitation of the dish images (Fig. 3D). We tested also 100 nM obatoclax
and its combination with GANT61 for clonogenic growth. Obatoclax
completely abrogated soft agar colony outgrowth in SK-MEL-3 cells
and it was more effective than GANT61 in Beu cells. Combined
treatment completely prevented the colony formation in both cell
lines (Fig. 3D), suggesting a
synergistic effect in eradication of melanoma cells.
GANT61 induces massive apoptosis in
melanoma cells
It is well established that GANT61 causes apoptosis
in cancer cells (5,23–35).
The possible accompanying caspase and PARP cleavage can be
dependent on the cell type. To demonstrate whether GANT61
eliminates viability of melanoma cells through apoptosis, flow
cytometric analysis was performed after 2 days of cultivation in at
concentration of 20 μM. Massive sub-G1 phase indicating the DNA
fragmentation was observed in SK-MEL-3 and SK-MEL-5 cells (Fig. 3B). Together with examining the
remaining attached cells, we analysed also the detached cells,
possibly explaining the appearance of such profound apoptosis. The
high sensitivity of SK-MEL-3 correlated with the colony formation
assays in which these cells appeared to be the most susceptible to
GANT61 treatment (Fig. 4). To
further substantiate that apoptosis was the mechanism of killing
the melanoma cells, the same GANT61 treatment as used for flow
cytometry was applied to cell monolayers and the remaining attached
cells were mounted in DAPI-containing medium. Nuclei fragmentation
and chromatin condensation indicating apoptosis were clearly seen,
whereas normal nuclei were observed in control cells (Fig. 3C). Taken together these findings
indicate that GANT61 induces powerful apoptosis in melanoma
cells.
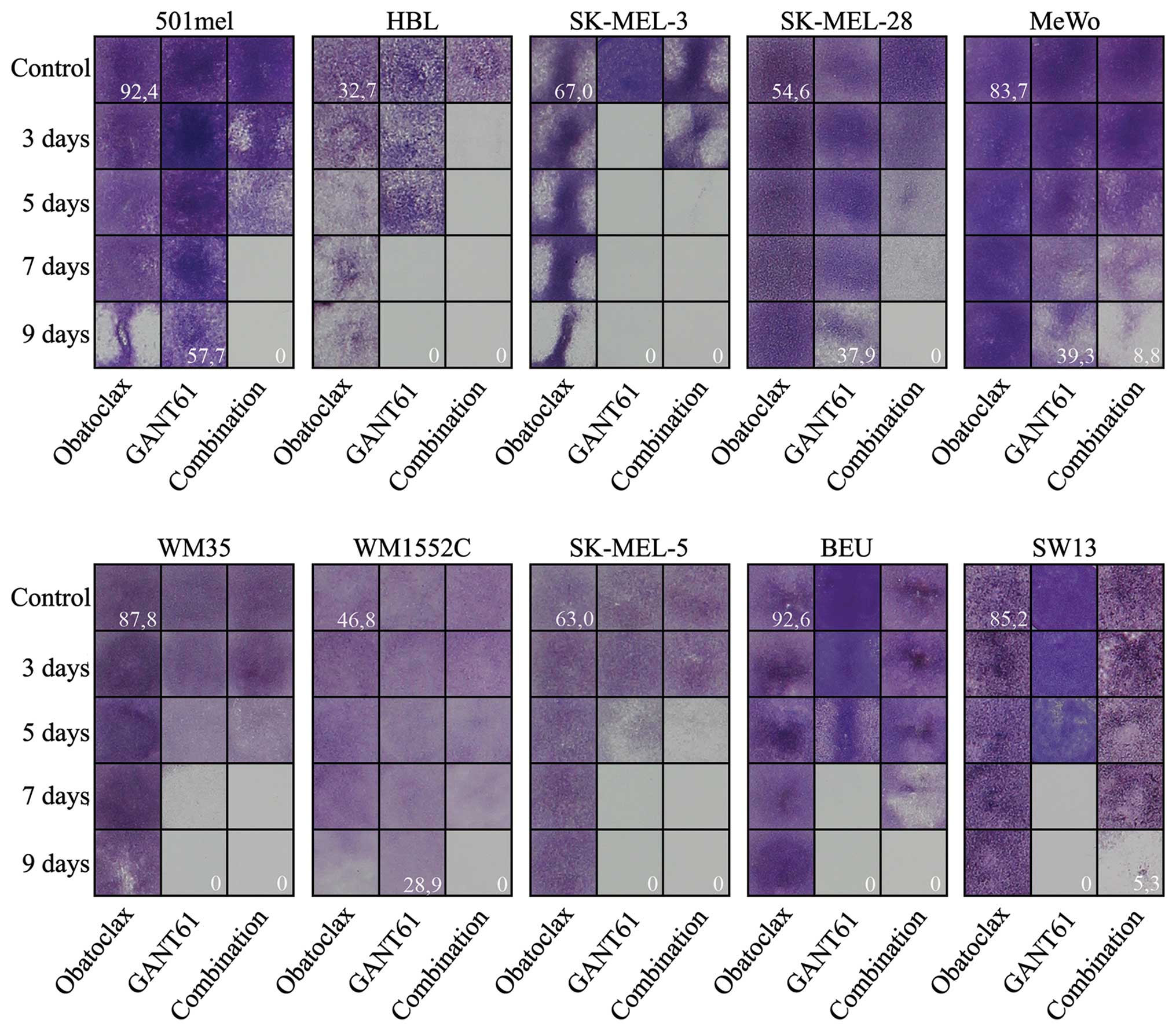
Obatoclax in synergy with GANT61
efficiently eradicate melanoma cells in vitro
Having shown that GANT61 can effectively abolish the
survival of melanoma cells, we decided to investigate the possible
synergistic effect of GANT61 when used with inhibitors related to
melanoma treatment. Firstly, we studied whether selumetinib, a MEK
inhibitor, and AZD5363, an AKT kinase inhibitor, could improve the
efficacy of 20 μM GANT61 in melanoma cells. We used 300 nM
concentration of both drugs, which is sufficiently high
concentration considering that the IC50 values for both
selumetinib and AZD5363 targets (MEK and ERK1/1 or AKT,
respectively) are below 20 nM (www.selleckchem.com). The experiments were designed
similarly as shown in Fig. 4.
However, the combined incubation of cell lines in the panel did not
lead to improvement of the use of GANT61 alone (data not shown).
Expectedly, selumetinib or AZD5363 alone (at 300 nM concentration)
did not reveal any considerable effect on any cell line even after
9 days of treatment (results not shown). We therefore used
obatoclax, an inhibitor of the BCL2 family of anti-apoptotic
proteins. Although it showed no effect on any cell line when tested
alone (at 100 nM concentration) (Fig.
4), obatoclax accelerated the effect of GANT61 treatment in 6
of 9 melanoma cell lines (501mel, Hbl, SK-MEL-28, MeWo, WM1552C and
SK-MEL-5), whereas no additional effect was observed in the
remaining 3 cell lines (SK-MEL-3, WM35 and Beu). In SK-MEL-3 (the
most GANT61 sensitive melanoma cells), Beu melanoma cells and SW13
non-melanoma control cells, the cell death appeared even earlier
when GANT61 was applied alone (Fig.
4). Combination index (CI) was calculated (Table II) revealing the ‘antagonistic’
effect in the Beu and SK-MEL-3 cell line, evidently due to the
earlier effect of GANT61 alone than the combined treatment.
Additive CI was calculated for WM35 cell line (Table II). Nonetheless, at the end of the
experiment (day 9), all 9 melanoma cell lines revealed the best
effect of GANT61 + obatoclax combination (the bottom right field in
each block; Fig. 4).
 | Table IICombination index (CI) of the
analyzed melanoma cell lines.a |
Table II
Combination index (CI) of the
analyzed melanoma cell lines.a
| Cell line | Combination index
(CI) |
|---|
| 501mel | 0.091 |
| HBL | 0.14 |
| SK-MEL-3 | 1.798b |
| SK-MEL-28 | 0.121 |
| MeWo | 0.414 |
| WM35 | 0.95 |
| WM1552C | 0.812 |
| SK-MEL-5 | 0.216 |
| BEU | 1.88b |
| SW13
(non-melanoma) | 10.949 |
Thus, GANT61 efficiently eliminated melanoma cells
in the clonogenic growth assay and exhibited synthetically lethal
effect with obatoclax in most melanoma cell lines.
Discussion
Malignant melanoma continues to increase in
incidence worldwide. The treatment of advanced melanoma with low
molecular weight inhibitors directed to mutated BRAF or MAPK
inhibitors results in acquired resistance. It has been found that
some kinase inhibitors induce a specific secretome increasing the
tumor outgrowth and metastasis, supporting growth of cell clones
with drug-resistance, actually promoting tumor progression after
treatment (24). Although the
combination of drugs improved the therapy, it remains questionable
if MAPK signaling pathway, despite its deregulation virtually in
all melanomas, is an ideal target for the therapy. More mechanisms
are involved in the acquired resistance to the MAPK signaling in
melanoma (23,36–39).
Therefore, alternative approaches should also be considered.
GLI2 transcription factor has been recognized as
important pro-invasive factor through its contribution to
maintaining the cancer stem cell subpopulations and progression to
metastasis in many cancers (21,40–43).
Many pro-oncogenic targets for GLI2 has been identified. Recently,
we found that survivin is also a GLI2 target in about half of a
large panel of human tumor cell lines, with several tumor lines
being absolutely reliant on GLI2 for survivin epression (25). In keeping with this, both survivin
expression and Hedgehog signaling are active during embryonic
development and in tumor cells, while normal adult
non-proliferating cells are silent or display very low activities
of both HH/GLI and survivin expression. Thus, HH/GLI can widely
contribute to the antiapoptotic activity through GLI2-directed
survivin expression in cancer cells. GANT61 is a powerful and
specific inhibitor of GLI1/2 factors activity. It also progresses
into the clinic as a promising anticancer drug for many types of
tumors. We therefore reasoned that it could constitute a possible
effective agent in melanoma. We have shown here that it causes
apoptosis in melanoma cells and inhibits their survival as assessed
by colony formation assay. As the emerging data from both
preclinical and clinical studies suggest that resistance is likely
to occur following the monotherapy, we have successfully treated
melanoma cells with GANT61 in combination with the BCL2 inhibitor
obatoclax. Deregulated expression of BCL2 family proteins is known
to play a central role in the resistance of melanoma to apoptosis
(39).
In the present study, obatoclax accelerated the
eradication of tumor cells in most melanoma lines (6 of 9), whereas
we have not observed its toxicity against any melanoma cell line
when used alone at 100 nM concentration. In contrast, we found that
either MEK inhibitor selumetinib or AKT inhibitor AZD-5363 did not
synergize with GANT61 in eradication of melanoma cells. As
obatoclax is effective towards the whole BLC2 family, the proposed
enhancement of apoptosis induced by obatoclax may improve the
results obtained with GANT61 and prevent the development of
resistance in patients. To conclude, the presented combined
treatment could be the basis for further research based on melanoma
cell elimination through apoptosis and independently of BRAF or
NRAS mutations and the activity of MAPK signaling.
Concomitantly, we estimated here the level of a
pivotal transcription factor and melanoma oncogene MITF after
GANT61 treatment. GLI2 has been demonstrated previously to repress
transcription of MITF (19) and
reciprocal expression of GLI2 and MITF is involved in the
‘phenotype switching’ model where high GLI2 functions to promote
melanoma cell phenotypic plasticity and invasive behavior (18). Surprisingly, we found that the
blockade of GLI2 activity by the addition of GANT61 does not
increase the MITF protein, but rather has no effect or causes a
decrease of its level. We therefore hypothesize that GLI2 may not
be the MITF transcriptional repressor in every cell context, or
GLI2 activity can eventually block the proteasomal degradation of
the MITF protein by a yet unknown mechanism, similarly as observed
for survivin in some tumor cell lines (25). However, although it is difficult to
interpret these data at present, the findings together argue that
in a specific cell context (SK-MEL-5 cells), GLIs are important to
maintain the MITF cellular level. We speculate that the MITF level
might not be a causative factor in the melanoma phenotype
switching, but rather is a consequence that mirrors the phenotype
changes governed by other factors. GLI2, on the other hand,
activate genes that confer the invasive phenotype to melanoma cells
and together with other factors such as SOX2, ZEB1, TWIST or SNAIL1
(44–46) could contribute to stem cell-like
and metastasis prone properties of small subpopulations of cells in
melanoma.
Acknowledgements
We thank Professor Fritz Aberger (University of
Salzburg) for providing the Patched promoter plasmid and Professor
Rune Toftgard (Karolinska Institutet) for the 12xGLI reporter
plasmid. The present study was supported by IGA, Ministry of Health
of the Czech Republic (grant NT/14005-3) and by the Institutional
research project PRVOUK-P25/LF1/2 from the Charles University,
Prague, Czech Republic.
References
|
1
|
Marini KD, Payne BJ, Watkins DN and
Martelotto LG: Mechanisms of Hedgehog signalling in cancer. Growth
Factors. 29:221–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Robbins DJ, Fei DL and Riobo NA: The
Hedgehog signal transduction network. Sci Signal. 5:re62012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Varjosalo M and Taipale J: Hedgehog:
Functions and mechanisms. Genes Dev. 22:2454–2472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katoh Y and Katoh M: Hedgehog target
genes: Mechanisms of carcinogenesis induced by aberrant hedgehog
signaling activation. Curr Mol Med. 9:873–886. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gonnissen A, Isebaert S and Haustermans K:
Targeting the Hedgehog signaling pathway in cancer: Beyond
Smoothened. Oncotarget. 6:13899–13913. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McMillan R and Matsui W: Molecular
pathways: The hedgehog signaling pathway in cancer. Clin Cancer
Res. 18:4883–4888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li H, Li J and Feng L: Hedgehog signaling
pathway as a therapeutic target for ovarian cancer. Cancer
Epidemiol. 40:152–157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Atwood SX, Chang AL and Oro AE: Hedgehog
pathway inhibition and the race against tumor evolution. J Cell
Biol. 199:193–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stecca B, Mas C, Clement V, Zbinden M,
Correa R, Piguet V, Beermann F, Ruiz I and Altaba A: Melanomas
require HEDGEHOG-GLI signaling regulated by interactions between
GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA.
104:5895–5900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lauth M and Toftgård R: Non-canonical
activation of GLI transcription factors: Implications for targeted
anti-cancer therapy. Cell Cycle. 6:2458–2463. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jenkins D: Hedgehog signalling: Emerging
evidence for non-canonical pathways. Cell Signal. 21:1023–1034.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shevde LA and Samant RS: Nonclassical
hedgehog-GLI signaling and its clinical implications. Int J Cancer.
135:1–6. 2014. View Article : Google Scholar
|
|
13
|
Agyeman A, Jha BK, Mazumdar T and Houghton
JA: Mode and specificity of binding of the small molecule GANT61 to
GLI determines inhibition of GLI-DNA binding. Oncotarget.
5:4492–4503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alexaki VI, Javelaud D, Van Kempen LC,
Mohammad KS, Dennler S, Luciani F, Hoek KS, Juàrez P, Goydos JS,
Fournier PJ, et al: GLI2-mediated melanoma invasion and metastasis.
J Natl Cancer Inst. 102:1148–1159. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Steingrímsson E, Copeland NG and Jenkins
NA: Melanocytes and the microphthalmia transcription factor
network. Annu Rev Genet. 38:365–411. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hoek KS, Schlegel NC, Eichhoff OM, Widmer
DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir
K, Schepsky A, Dummer R, et al: Novel MITF targets identified using
a two-step DNA microarray strategy. Pigment Cell Melanoma Res.
21:665–676. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vachtenheim J and Borovanský J:
‘Transcription physiology’ of pigment formation in melanocytes:
Central role of MITF. Exp Dermatol. 19:617–627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Javelaud D, Alexaki VI, Pierrat MJ, Hoek
KS, Dennler S, Van Kempen L, Bertolotto C, Ballotti R, Saule S,
Delmas V, et al: GLI2 and M-MITF transcription factors control
exclusive gene expression programs and inversely regulate invasion
in human melanoma cells. Pigment Cell Melanoma Res. 24:932–943.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pierrat MJ, Marsaud V, Mauviel A and
Javelaud D: Expression of microphthalmia-associated transcription
factor (MITF), which is critical for melanoma progression, is
inhibited by both transcription factor GLI2 and transforming growth
factor-β. J Biol Chem. 287:17996–18004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dennler S, André J, Alexaki I, Li A,
Magnaldo T, ten Dijke P, Wang XJ, Verrecchia F and Mauviel A:
Induction of sonic hedgehog mediators by transforming growth
factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression
in vitro and in vivo. Cancer Res. 67:6981–6986. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Javelaud D, Alexaki VI, Dennler S,
Mohammad KS, Guise TA and Mauviel A: TGF-β/SMAD/GLI2 signaling axis
in cancer progression and metastasis. Cancer Res. 71:5606–5610.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Santini R, Vinci MC, Pandolfi S,
Penachioni JY, Montagnani V, Olivito B, Gattai R, Pimpinelli N,
Gerlini G, Borgognoni L, et al: Hedgehog-GLI signaling drives
self-renewal and tumorigenicity of human melanoma-initiating cells.
Stem Cells. 30:1808–1818. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Davies MA and Kopetz S: Overcoming
resistance to MAPK pathway inhibitors. J Natl Cancer Inst.
105:9–10. 2013. View Article : Google Scholar
|
|
24
|
Obenauf AC, Zou Y, Ji AL, Vanharanta S,
Shu W, Shi H, Kong X, Bosenberg MC, Wiesner T, Rosen N, et al:
Therapy-induced tumour secretomes promote resistance and tumour
progression. Nature. 520:368–372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vlčková K, Ondrušová L, Vachtenheim J,
Réda J, Dundr P, Zadinová M, Žáková P and Poučková P: Survivin, a
novel target of the Hedgehog/GLI signaling pathway in human tumor
cells. Cell Death Dis. 7:e20482016. View Article : Google Scholar
|
|
26
|
Vachtenheim J, Sestáková B and Tuhácková
Z: Inhibition of MITF transcriptional activity independent of
targeting p300/CBP coactivators. Pigment Cell Res. 20:41–51. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Packer LM, East P, Reis-Filho JS and
Marais R: Identification of direct transcriptional targets of
(V600E)BRAF/MEK signalling in melanoma. Pigment Cell Melanoma Res.
22:785–798. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Herraiz C, Journé F, Ghanem G,
Jiménez-Cervantes C and García-Borrón JC: Functional status and
relationships of melanocortin 1 receptor signaling to the cAMP and
extracellular signal-regulated protein kinases 1 and 2 pathways in
human melanoma cells. Int J Biochem Cell Biol. 44:2244–2252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hao H, Muniz-Medina VM, Mehta H, Thomas
NE, Khazak V, Der CJ and Shields JM: Context-dependent roles of
mutant B-Raf signaling in melanoma and colorectal carcinoma cell
growth. Mol Cancer Ther. 6:2220–2229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smalley KS, Lioni M, Dalla Palma M, Xiao
M, Desai B, Egyhazi S, Hansson J, Wu H, King AJ, Van Belle P, et
al: Increased cyclin D1 expression can mediate BRAF inhibitor
resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther.
7:2876–2883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Singh S, Davis R, Alamanda V, Pireddu R,
Pernazza D, Sebti S, Lawrence N and Chellappan S: Rb-Raf-1
interaction disruptor RRD-251 induces apoptosis in metastatic
melanoma cells and synergizes with dacarbazine. Mol Cancer Ther.
9:3330–3341. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Domenzain-Reyna C, Hernández D,
Miquel-Serra L, Docampo MJ, Badenas C, Fabra A and Bassols A:
Structure and regulation of the versican promoter: The versican
promoter is regulated by AP-1 and TCF transcription factors in
invasive human melanoma cells. J Biol Chem. 284:12306–12317. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Desch P, Asslaber D, Kern D, Schnidar H,
Mangelberger D, Alinger B, Stoecher M, Hofbauer SW, Neureiter D,
Tinhofer I, et al: Inhibition of GLI, but not Smoothened, induces
apoptosis in chronic lymphocytic leukemia cells. Oncogene.
29:4885–4895. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pan D, Li Y, Li Z, Wang Y, Wang P and
Liang Y: Gli inhibitor GANT61 causes apoptosis in myeloid leukemia
cells and acts in synergy with rapamycin. Leuk Res. 36:742–748.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Graab U, Hahn H and Fulda S:
Identification of a novel synthetic lethality of combined
inhibition of hedgehog and PI3K signaling in rhabdomyosarcoma.
Oncotarget. 6:8722–8735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Johannessen CM, Boehm JS, Kim SY, Thomas
SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP,
Barretina J, et al: COT drives resistance to RAF inhibition through
MAP kinase pathway reactivation. Nature. 468:968–972. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nazarian R, Shi H, Wang Q, Kong X, Koya
RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al: Melanomas
acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS
upregulation. Nature. 468:973–977. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Poulikakos PI, Persaud Y, Janakiraman M,
Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al:
RAF inhibitor resistance is mediated by dimerization of aberrantly
spliced BRAF(V600E). Nature. 480:387–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Haq R, Yokoyama S, Hawryluk EB, Jönsson
GB, Frederick DT, McHenry K, Porter D, Tran TN, Love KT, Langer R,
et al: BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene
that confers resistance to BRAF inhibition. Proc Natl Acad Sci USA.
110:4321–4326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang DW, Li HY, Lau WY, Cao LQ, Li Y,
Jiang XF, Yang XW and Xue P: Gli2 silencing enhances TRAIL-induced
apoptosis and reduces tumor growth in human hepatoma cells in vivo.
Cancer Biol Ther. 15:1667–1676. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kumar K, Raza SS, Knab LM, Chow CR, Kwok
B, Bentrem DJ, Popovic R, Ebine K, Licht JD and Munshi HG:
GLI2-dependent c-MYC upregulation mediates resistance of pancreatic
cancer cells to the BET bromodomain inhibitor JQ1. Sci Rep.
5:94892015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nagao-Kitamoto H, Nagata M, Nagano S,
Kitamoto S, Ishidou Y, Yamamoto T, Nakamura S, Tsuru A, Abematsu M,
Fujimoto Y, et al: GLI2 is a novel therapeutic target for
metastasis of osteosarcoma. Int J Cancer. 136:1276–1284. 2015.
View Article : Google Scholar
|
|
43
|
Soengas MS and Lowe SW: Apoptosis and
melanoma chemoresistance. Oncogene. 22:3138–3151. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Caramel J, Papadogeorgakis E, Hill L,
Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J,
Hutchinson P, Tse G, et al: A switch in the expression of embryonic
EMT-inducers drives the development of malignant melanoma. Cancer
Cell. 24:466–480. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vandamme N and Berx G: Melanoma cells
revive an embryonic transcriptional network to dictate phenotypic
heterogeneity. Front Oncol. 4:3522014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Denecker G, Vandamme N, Akay O, Koludrovic
D, Taminau J, Lemeire K, Gheldof A, De Craene B, Van Gele M,
Brochez L, et al: Identification of a ZEB2-MITF-ZEB1
transcriptional network that controls melanogenesis and melanoma
progression. Cell Death Differ. 21:1250–1261. 2014. View Article : Google Scholar : PubMed/NCBI
|