Introduction
The incidence of papillary thyroid cancer (PTC) has
been rising dramatically for the past three decades (1,2). It
was the fifth most commonly diagnosed type of cancer among women in
United States in 2015 (3). The
prognosis of PTC patients is known to be excellent, with a 10-year
survival rate exceeding 90% (4,5).
Nevertheless, up to 10% of patients would eventually die of the
disease and an even greater proportion would face recurrence
(6,7). Therefore, avoiding recurrence and
early diagnosis of recurrent disease is of great importance in the
long term management of PTC. Efforts have been made in identifying
various clinicopathological as well as biological predictors for
PTC. Using these predictors, risk-group stratification or staging
systems can thus be established and be applied to differentiate PTC
prognosis. The ultimate purpose is to treat PTC patients
individually and hopefully, lower the recurrence rate of the
disease. Undisputedly, in order to reach this goal, a better
understanding of the PTC pathogenesis and progression is
needed.
Sirtuins (SIRTs) are a class of
NAD+-dependent protein deacetylases involved in stress
resistance and metabolic homeostasis. There are 7 members in the
family: SIRT1-SIRT7. Many of these proteins have been reported to
regulate important pathways and are responsible for vital
biological process (8–10). SIRT6, like the other members in the
family, also exhibits many pivotal functions and multiple enzymatic
activities (11–13). It has been reported to be in
connection with a number of human diseases, including cancer
(14–20). However, whether SIRT genes are
involved in PTC has not been elucidated yet.
This study aimed to investigate the roles of SIRTs
in PTC. We analyzed all SIRT family genes in 391 PTC cases from The
Cancer Genome Atlas (TCGA) cohort and an additional 45 PTC cases
from Microarray data. Tissues from 130 in-house surgically treated,
pathologically confirmed PTC patients were used for verification of
SIRT6 expression. We also generated SIRT6 silenced K1 and TPC-1
cells to explore the influence of SIRT6 on cancer cell
aggressiveness.
Materials and methods
Patients and tissue samples
This study acquired Institutional Review Board
approval from Fudan University Shanghai Cancer Center (Shanghai,
China). Written informed consent was obtained from all subjects.
The methods were carried out in accordance with the approved
guidelines.
Pathologically confirmed thyroid cancer tissues,
were obtained from 130 patients who received thyroidectomy between
2012 and 2015 at the Department of Head and Neck Surgery, Fudan
University Shanghai Cancer Center. For the TCGA cohort, SIRT gene
expression and clinical data of TCGA database are available from
the website of Cancer Genomics Browser of University of California
Santa Cruz (UCSC) (https://genome-cancer.ucsc.edu/). A total number of 7
members of the SIRT family are included in the database. Patients
who had PTC as the only malignancy or the first of multiple
malignancies and received surgical therapy were included. Other
inclusion criteria were: patients with no pre-treatment, with
sufficient description of primary tumors and intact recurrence-free
survival (RFS)/overall survival (OS) information. Follow-up was
completed on December 21, 2014. The patients were excluded if with:
insufficient data or unknown clinicopathologic profile,
undetermined histology, other types of thyroid cancer (follicular
thyroid cancer, medullary thyroid cancer and anaplastic thyroid
cancer), or secondary tumor. The following variables were used for
the analysis: gender, age at diagnosis, maximal tumor size,
bilateral lesions, multifocal lesions, and lymph node status.
Patients were staged using the latest 7th AJCC TNM staging system
according to the final pathology reports. The primary tumor and
nodal status were assessed in (or by) the final pathology.
Demographic and clinicopathological characteristics of enrolled
patients were obtained from electronic records and summarized in
Table I.
 | Table IClinical characteristics of patients
with papillary cancer in The Cancer Genome Atlas cohort. |
Table I
Clinical characteristics of patients
with papillary cancer in The Cancer Genome Atlas cohort.
| Variables | TCGA
|
|---|
| No. (%), total
n=391 |
|---|
| Gender | |
| Male | 108 (27.6) |
| Female | 283 (72.4) |
| Age, years | |
| Mean age | 47.0±15.8 |
| ≤45 | 192 (49.1) |
| >45 | 199 (50.9) |
| First degree
relative history | |
| No | 371 (94.9) |
| Yes | 20 (5.1) |
| Histological
type | |
|
Classical/usual | 275 (70.3) |
| Follicular | 85 (21.8) |
| Tall cell | 31 (7.9) |
| Primary tumor
size | |
| Mean size | 1.7±0.9 |
| <2 cm | 246 (62.9) |
| 2–4 cm | 136 (34.8) |
| >4 cm | 9 (2.3) |
| Primary tumor
focus | |
| Unifocal | 226 (57.8) |
| Multifocal | 165 (42.2) |
| Residual tumor | |
| R0 | 307 (78.5) |
| R1 | 44 (11.2) |
| R2 | 40 (10.3) |
| Hashimoto's
thyroiditis | |
| Negative | 331 (84.7) |
| Positive | 60 (15.3) |
| Pathologic T
stage | |
| T1 | 105 (26.9) |
| T2 | 139 (35.5) |
| T3 | 129 (33.0) |
| T4 | 18 (4.6) |
| Pathologic N
stage | |
| N0 | 217 (55.5) |
| N1 | 174 (44.5) |
| Pathologic M
stage | |
| M0 | 383 (98.0) |
| M1 | 8 (2.0) |
Selection of cut-off value
X-tile software (version 3.6.1, Yale University
School of Medicine, Newhaven, CT, USA) was used to divide the TCGA
cohort according to SIRT6 expression into 3 subsets: low, middle
and high. X-tile plots were generated for assessment of SIRT6
expression and RFS, optimization of cut-points and correction by
the use of minimum p statistics.
Microarray data analysis
Microarray data discussed in this study have been
deposited in the Gene Expression Omnibus (GEO) of U.S. National
Center for Biotechnology Information (NCBI; Bethesda, MD, USA;
http://www.ncbi.nlm.nih.gov/geo). The
results of the microarray experiment are accessible through GEO
series accession number GSE33630.
We hybridized 45 PTC tissues and the corresponding
para tumor tissues onto Affymetrix U133 Plus 2.0 arrays. Paired RNA
samples of PTCs and non-tumor thyroid tissues were obtained from
Ukraine via the Chernobyl Tissue Bank (www.chernobyltissuebank.com). Diagnoses were confirmed
by the members of the International Pathology Panel of the
Chernobyl Tissue Bank.
Immunohistochemistry (IHC) staining and
scoring
IHC staining was carried out according to the
manufacturer's instruction. Briefly, formalin-fixed and
paraffin-embedded tissue sections were deparaffinized in xylene and
hydrated with decreasing concentrations of ethanol (100, 95, 80 and
75%). The slices were then soaked in 10% BSA to inhibit endogenous
peroxidase activity and incubated with SIRT6 rabbit polyclonal
antibody (working dilution 1:50; Proteintech Group Inc., Chicago,
IL, USA) at 4°C overnight. A horseradish peroxidase-conjugated
rabbit secondary antibody was added for 60 min at room temperature;
then, 3,3′-diaminobenzidine (DAB) development (DAB Substrate
Chromogen System; Dako, Glostrup, Denmark) and hematoxylin staining
were performed according to standard protocols. Slides were fixed
and images were obtained through an Olympus IX71 inverted
microscope with a DP2-BSW Olympus image acquisition software
system. The sections were read separately by two experienced
pathologists blinded to the clinicopathologic data. The sections
were scored on the basis of the extent of staining (0, no staining;
1, ≤10%; 2, 10–50%; and 3, >50%) and the staining intensity (0,
negative; 1, weak; 2, moderate; and 3, strong). These scores were
multiplied to calculate an immunoreactivity score (IS) for each
case.
Cell lines and cell culture
Two human PTC cell lines (TPC-1 and K1) were used in
this study. TPC-1 cell line was kindly provided by Professor Haixia
Guan from China Medical University (Shenyang, China). K1 cell line
was purchased from University of Colorado Cancer Center Cell Bank.
All cells were cultured in RPMI-1640 medium supplemented with 10%
FBS (Invitrogen, Carlsbad, CA, USA) at 37°C with 5%
CO2.
shRNA plasmid and transfection
Two separate sequences (GCAGTCTTCCAGTGTGGTGTT and
GCTGGGTAC ATCGCTGCAGAT) of shRNA targeting SIRT6 gene were designed
and cloned into a pLKO.1 TRC (Addgene plasmid 10878) cloning
vector. Scramble RNA (Addgene plasmid 1864) was used as control
vector. Knockdown of SIRT6 in TPC-1 and K1 cells was accomplished
by lentiviral infection. All these plasmids were transfected into
PTC cells using Lipofectamine® 2000 under the
instruction of the product manual (Invitrogen). The knockout of
SIRT6 was confirmed by western blotting.
RNA extraction, reverse transcription and
quantitative PCR (qPCR)
Total RNA was extracted from 130 pairs of surgically
treated, pathologically diagnosed PTC tissues and cultured PTC
cells using TRIzol Reagent (Invitrogen). cDNA was then obtained
through reverse transcription using 1 µg of total RNA with a
PrimeScript™ RT reagent kit (Takara Bio, Inc., Otsu, Japan). The
SIRT6 expression status was assessed by Real-time PCR, which was
carried out in triplicate by a SYBR Premix Ex Taq™ kit (Takara Bio)
and ABI 7900HT Real-Time PCR system (Applied Biosystems Life
Technologies, Foster City, CA, USA). The SIRT6 primers used in qPCR
are as follows: Forward: 5′-ACTCGCCGATGAGGCCAGCAGGAA-3′; reverse:
5′-ATGCGGAGGTCAGCATGGCGGTCGT-3′. β-actin was used as an internal
control for mRNA assays. The comparative cycle threshold values
(2−ΔΔCt) were adopted to analyze the final results.
Colony formation assay
TPC-1 and K1 cells were digested and were seeded at
200 cells per well in a 6-well plate. After incubation for 5 days,
colonies were fixed in methanol and stained with 0.1% crystal
violet solution for 30 min. The cells were then washed three times
with PBS and clones were counted under an inverted microscope.
Clones were defined as colonies of ≥50 cells. Three different
fields were counted for each cell culture and the average was
calculated.
ATPLite luminescence assay
PTC cell proliferation was determined using the
ATPLite luminescent assay (PerkinElmer, Inc., Waltham, MA, USA)
according to the manufacturer's instructions. Cells were incubated
in 96-well plates for 120 h before testing.
Western blot analysis
Cell lysates were obtained from 1×106
cells with RIPA lysis buffer (containing 50 mM Tris-HCl, pH 7.4,
150 mM NaCl, 0.1% SDS, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, and 1
mM Protease Inhibitor Cocktail). Approximately 20 µg protein
was extracted from each sample and separated by 10% SDS-PAGE gels.
Proteins were then transferred to nitrocellulose membranes (0.45
mm; Solarbio, Beijing, China). After blocking the membrane with 5%
non-fat milk, it was immunoblotted with primary antibodies: SIRT6
rabbit polyclonal antibody (Proteintech Group Inc.) and GAPDH
(1:5,000; Abcam, Cambridge, MA, USA). The bands were visualized
with 1-step™ NBT/BCIP reagents (Thermo Fisher Scientific, Rockford,
IL, USA) and developed by Alpha Imager (Alpha Innotech, San
Leandro, CA, USA).
In vitro migration and invasion
assays
Cell migration and invasion were analyzed by a
transwell plate (24-well insert, 8 µm pore size; BD
Biosciences, Bedford, MA, USA). The filters (Corning Inc., Acton,
MA, USA) were coated with (invasion) or without (migration) 40
µl Matrigel (1:8 dilution; BD Biosciences). SIRT6-silenced
TPC-1 and SIRT6-silenced K1 cells were used in these assays. For
migration assays, 3×104 cells were suspended in 100
µl serum-free medium and seeded into the Matrigel-uncoated
upper chambers. A total of 600 µl of medium containing 10%
serum was added to the lower chamber as a chemoattractant. After
incubation at 37°C for 72 h, non-invading cells were wiped off the
upper surface with a cotton swab. The membranes were fixed with 4%
formaldehyde for 20 min and stained with 0.1% crystal violet at
room temperature. For invasion assays, 1×105 cells were
used with an incubation time of 48 h. The rest of the protocol was
identical to that described above. The cells were counted and
photographed under an inverted microscope over 10 different fields
of each triplicate filter.
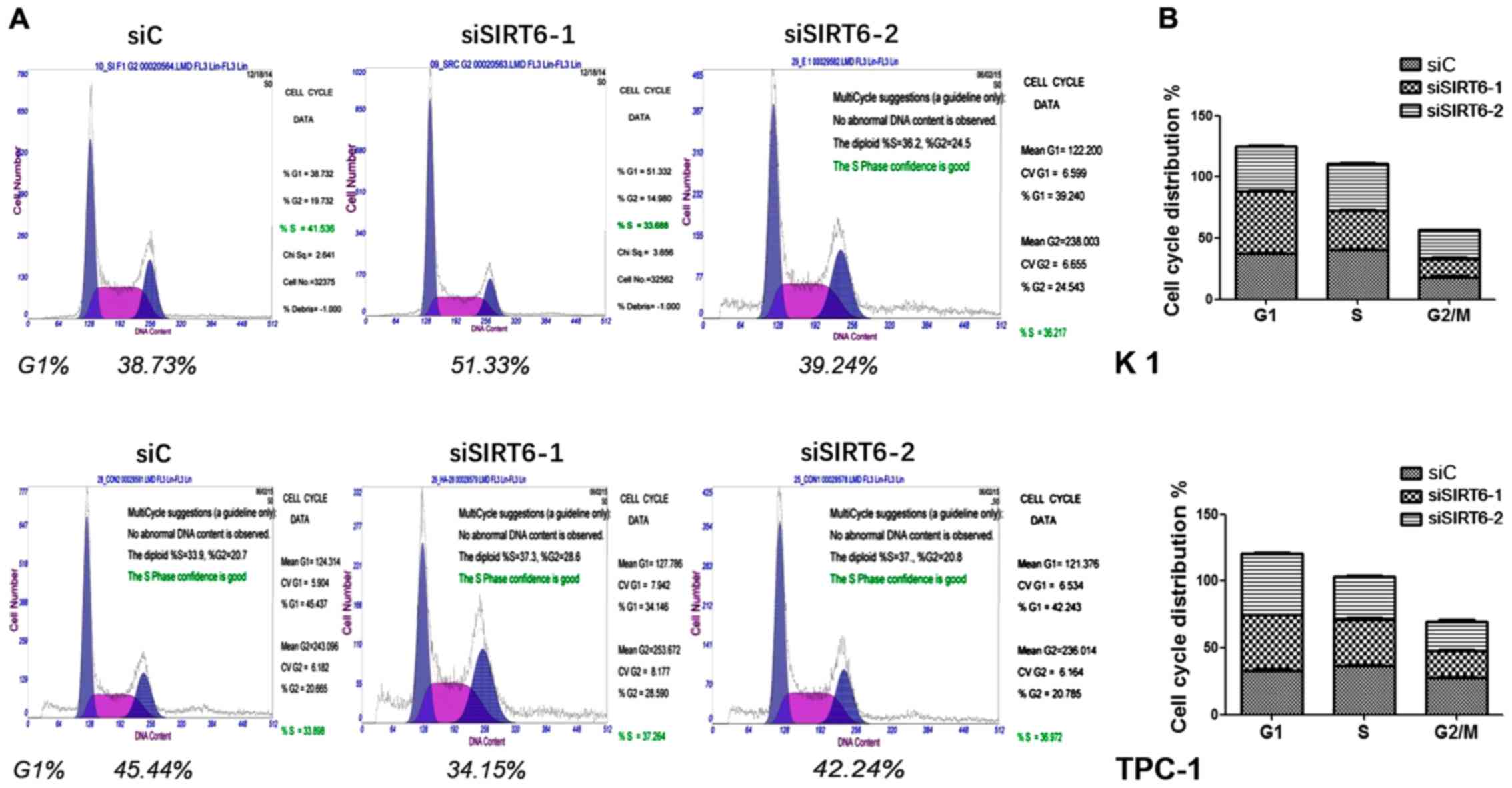
Cell cycle distribution
To evaluate the effect of silenced SIRT6 on PTC cell
cycle progression, cell cycle distribution was detected by flow
cytometry in TPC-1 and K1 cells after lentivirus transduction.
Briefly, 1.0×106 cells of each sample were harvested,
fixed in 70% ethanol, and stored at 4°C overnight. Cells were then
stained with NaCl/Pi staining solution (50 µg/ml PI and 100
µg/ml RNase A) for 1 h in the dark at room temperature
followed by flow cytometry (FACSCalibur; BD Biosciences, San Jose,
CA, USA) analysis. The fractions of the cells in G1, S, and
G2/phases were calculated with dedicated software (BD
Biosciences).
Statistical analysis
All statistical analysis was performed using SPSS
software (version 19.0, IBM Corp., Armonk, NY, USA). Independent
t-tests (for continuous variables) and Pearson's χ2
tests (for categorical variables) were used. Continuous variables
were categorized using certain cut-offs. The age of 45 was used as
the cut-off point to divide all patients into two groups: younger
patients (≤45 years old) and older patients (>45 years old). The
odds ratio (OR) for relationships between clinicopathological
factors, SIRT gene expression and regional node metastasis were
calculated using binary logistic regression. For the survival
analysis, patients who were alive and did not relapse were censored
at the date of their last follow-up visit. Neck or distant RFS was
defined as the time between the date of initial surgery and the
first adverse event or death. OS was defined as the time between
the date of initial surgery and death (all causes or
cancer-specific). Survival rates were estimated by the Kaplan-Meier
method. The hazard ratio (HR) for relationships between each
variable and recurrence were calculated using binary Cox regression
model. P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient characteristics and main outcomes
in follow-up (TCGA cohort)
We identified 391 eligible patients with PTC in TCGA
cohort. There were 108 (27.6%) males and 283 (72.4%) females. The
mean age was 47.0 years (range from 15 to 89 years). Of these
patients, 275 (70.3%) had been confirmed as classical/usual
papillary variant histologically, while follicular and tall cell
variants were found in 85 (21.8%) and 31 (7.9%) patients,
respectively. Combined with the size and extrathyroidal-extensional
status of primary tumor, 105 cases (26.9%) were classified as T1
stage, 139 (35.5%) were T2 stage, 129 (33.0%) were T3 stage, and
only 18 (1.6%) were T4 stage. According to the pathology, 174
(44.5%) patients had lymph node metastasis involving either the
central compartment (N1a) or both central and lateral compartment
(N1b). Using the latest 7th AJCC TNM staging system,
there were 220 patients (56.3%) with PTC stage I, 43 (11.0%) with
stage II, 86 (22.0%) with stage III, and 42 (10.7%) with stage IV.
The other clinicopathological characteristics, such as first degree
relative history, the primary tumor focus, the coexistent
Hashimoto's thyroiditis, and the surgical completeness (residual
tumor) of all patients are summarized in Table I.
The median follow-up duration was 574 days (range
1–4,780 months). During this time, 9 patients (2.3%) died from a
specific cancer cause. Disease recurrence occurred in 26 patients
(6.6%), including neck and distant recurrence. The OS rates were
92.0% at 1,854 days (approximately 5 years) and 88.0% at 2,973 days
(approximately 8 years) in this cohort. On the other hand, the RFS
rates were 86.1% at 1,785 days (approximately 5 years) and 83.9% at
2,093 days (approximately 6 years) in this cohort.
Influence of clinicopathological
characteristics and SIRT genes expression on lymph node metastasis
and RFS
The Pearson's χ2 tests was used to
analyze the influences of clinicopathologic variables and genes
expression on regional node metastasis. The results showed that the
risk of nodal metastasis was significantly increased by classical
papillary variant (compared to follicular or tall cell variant),
advanced T stage, and higher expression of SIRT6. Instead of
limiting the multivariate analysis to the significant terms from
the univariate analysis, we included all variables because these
factors had been previously demonstrated to be important in
predicting disease recurrence in adult PTC patients. The results
are presented in Table II. The
risk of nodal metastasis still increased with the independent
variables identified by univariate analysis. In particular, the
expressions of SIRT6 [OR=1.794, 95% confidence interval (CI):
1.256–1.920, p=0.012] was an independent biomarker predictor of
nodal metastasis (Table II).
 | Table IIUnivariate and multivariate logistic
regression analysis of SIRT gene expression and regional node
metastasis for patients with papillary thyroid cancer in the TCGA
cohort. |
Table II
Univariate and multivariate logistic
regression analysis of SIRT gene expression and regional node
metastasis for patients with papillary thyroid cancer in the TCGA
cohort.
| Variables | Univariate analysis
| Multivariate
analysis
|
|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Gender (female vs.
male) | 0.745
(0.517–1.381) | 0.402 | 0.664
(0.367–1.201) | 0.176 |
| Age (>45 vs.
≤45 years) | 0.703
(0.452–1.094) | 0.219 | 0.597
(0.350–1.018) | 0.058 |
| First degree
relative history | 0.651
(0.349–2.718) | 0.719 | 0.992
(0.311–3.165) | 0.989 |
| Histological
type | 0.510
(0.353–0.739) | 0.001 | 0.540
(0.416–0.984) | 0.042 |
| Primary tumor
focus | 1.226
(0.789–1.904) | 0.365 | 1.064
(0.634–1.786) | 0.814 |
| Hashimoto's
thyroiditis | 1.393
(0.747–2.525) | 0.208 | 2.172
(0.585–4.348) | 0.694 |
| Pathologic T
stage | 2.020
(1.340–3.650) | 0.001 | 1.777
(1.279–2.468) | 0.001 |
| Pathologic M
stage | 0.499
(0.051–2.207) | 0.324 | 0.398
(0.063–2.503) | 0.326 |
| SIRT1 | 0.731
(0.418–0.881) | 0.004 | 0.917
(0.686–1.507) | 0.065 |
| SIRT2 | 0.937
(0.713–1.232) | 0.642 | 0.854
(0.639–1.142) | 0.287 |
| SIRT3 | 0.714
(0.313–1.627) | 0.422 | 0.612
(0.252–1.485) | 0.277 |
| SIRT4 | 1.031
(0.501–2.122) | 0.933 | 0.995
(0.470–2.106) | 0.989 |
| SIRT5 | 1.019
(0.554–1.876) | 0.951 | 1.001
(0.513–1.952) | 0.999 |
| SIRT6 | 1.659
(1.120–2.812) | 0.007 | 1.794
(1.256–1.920) | 0.012 |
| SIRT7 | 0.976
(0.824–1.158) | 0.784 | 1.005
(0.837–1.208) | 0.956 |
The Kaplan-Meier method and log-rank test showed
that higher expression of SIRT6 was an independent predictor for
disease recurrence, while none of the clinicopathological
characteristics was significantly associated (Table III). However, multivariate
analysis after adjustment for all the potential prognostic factors
demonstrated that the expression of SIRT6 (HR=1.466, 95% CI:
1.195–1.913, p=0.086) lacked significance as an independent
predictor of RFS (Table
III).
 | Table IIIUnivariate and multivariate Cox
proportional hazards analysis of SIRT gene expression and
recurrence free survival for patients with papillary thyroid cancer
in the TCGA cohort. |
Table III
Univariate and multivariate Cox
proportional hazards analysis of SIRT gene expression and
recurrence free survival for patients with papillary thyroid cancer
in the TCGA cohort.
| Variables | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender (female vs.
male) | 0.427
(0.188–0.869) | 0.032 | 0.684
(0.190–2.457) | 0.760 |
| Age (>45 vs. ≤45
years) | 1.587
(0.669–4.693) | 0.304 | 1.271
(0.305–7.296) | 0.742 |
| First degree
relative history | 0.673
(0.083–5.439) | 0.710 | 0.770
(0.041–14.334) | 0.861 |
| Histological
type | 1.189
(0.607–2.292) | 0.666 | 1.095
(0.392–4.057) | 0.862 |
| Primary tumor
focus | 0.424
(0.194–1.037) | 0.068 | 0.303
(0.067–1.370) | 0.121 |
| Residual tumor | 1.086
(0.658–1.727) | 0.795 | 2.489
(0.760–3.917) | 0.246 |
| Hashimoto's
thyroiditis | 2.040
(0.624–4.747) | 0.177 | 1.340
(0.304–5.909) | 0.699 |
| Pathologic T
stage | 2.044
(0.652–3.671) | 0.858 | 0.912
(0.408–2.082) | 0.845 |
| Pathologic N
stage | 1.871
(0.801–4.368) | 0.148 | 3.348
(0.930–12.048) | 0.064 |
| Pathologic M
stage | 0.989
(0.120–8.132) | 0.992 | 0.108
(0.005–2.513) | 0.165 |
| SIRT1 | 0.361
(0.130–1.005) | 0.051 | 0.339
(0.114–1.006) | 0.051 |
| SIRT2 | 0.613
(0.295–1.274) | 0.190 | 0.596
(0.268–1.323) | 0.203 |
| SIRT3 | 1.935
(0.967–3.875) | 0.062 | 1.522
(0.658–3.519) | 0.326 |
| SIRT4 | 1.137
(0.484–2.674) | 0.768 | 1.881
(0.592–5.980) | 0.284 |
| SIRT5 | 1.988
(0.460–8.587) | 0.358 | 2.588
(0.480–13.961) | 0.269 |
| SIRT6 | 1.400
(1.183–2.873) | 0.021 | 1.466
(1.195–1.913) | 0.086 |
| SIRT7 | 1.689
(0.742–3.842) | 0.212 | 1.170
(0.414–3.305) | 0.767 |
To further explore the relationship between SIRT6
expression and RFS, we used X-tile software to calculate the cutoff
values on TCGA cohort. According to their SIRT6 expression, we
divided the cohort into three subpopulations: low, middle and high.
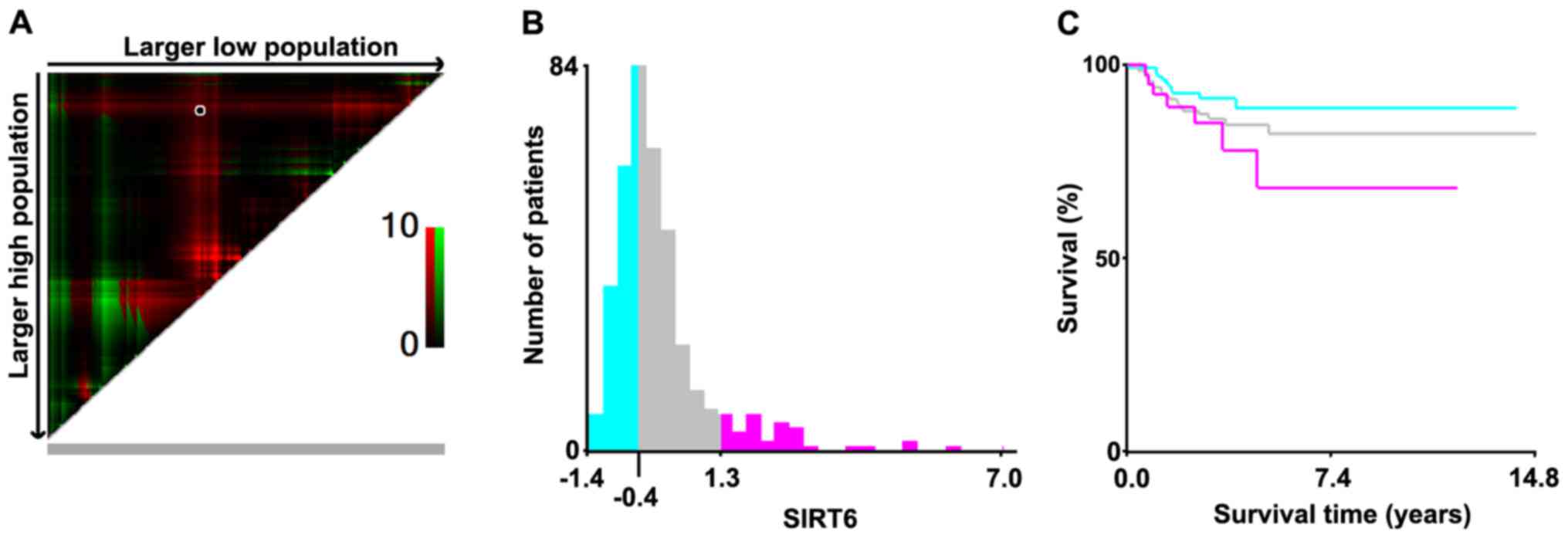
X-tile plots were generated accordingly and the maximum of
χ2 log-rank values was produced (Fig. 1). The low-middle cut-point was −0.4
and the middle-high cut-point was 1.3, individually. The three
subsets displayed a significant difference on Kaplan-Meier plot,
which showed that the patients with the highest expression of SIRT6
had the worst RFS and those who possessed lowest expression of the
gene had the best RFS.
SIRT6 expression is upregulated in PTC
tumor tissues
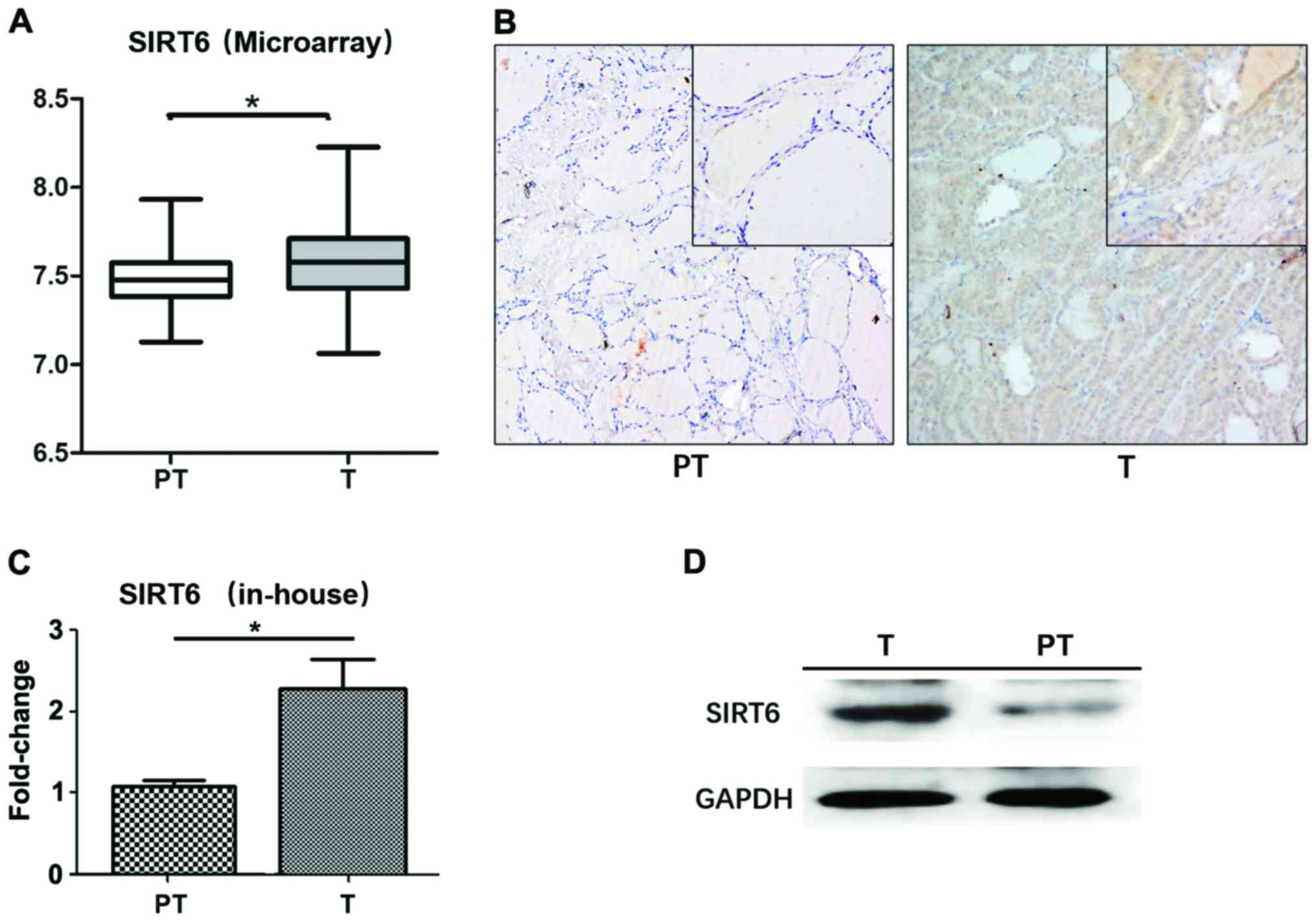
Microarray data (GSE33630) was used to evaluate
SIRT6 gene expression in PTC. A total number of 45 pairs of PTC
tissues and their corresponding para-tumor tissues were analyzed.
The expression of SIRT6 was significantly higher in tumor compared
to non-tumor tissues (p=0.029; Fig.
2A). IHC was also adapted to analyze the protein level of SIRT6
expression and subcellular localization. As is shown in Fig. 2B, SIRT6 was expressed in the nuclei
of cells and was higher in PTC tumor tissues.
Tissues from 130 patients who were surgically
treated and pathologically diagnosed as PTC were obtained to verify
SIRT6 expression. The upregulation of SIRT6 was observed in tumor
tissues at both mRNA (p=0.011; Fig.
2C) and protein levels (Fig.
2D).
Silencing SIRT6 weaken PTC cell
proliferation in vitro
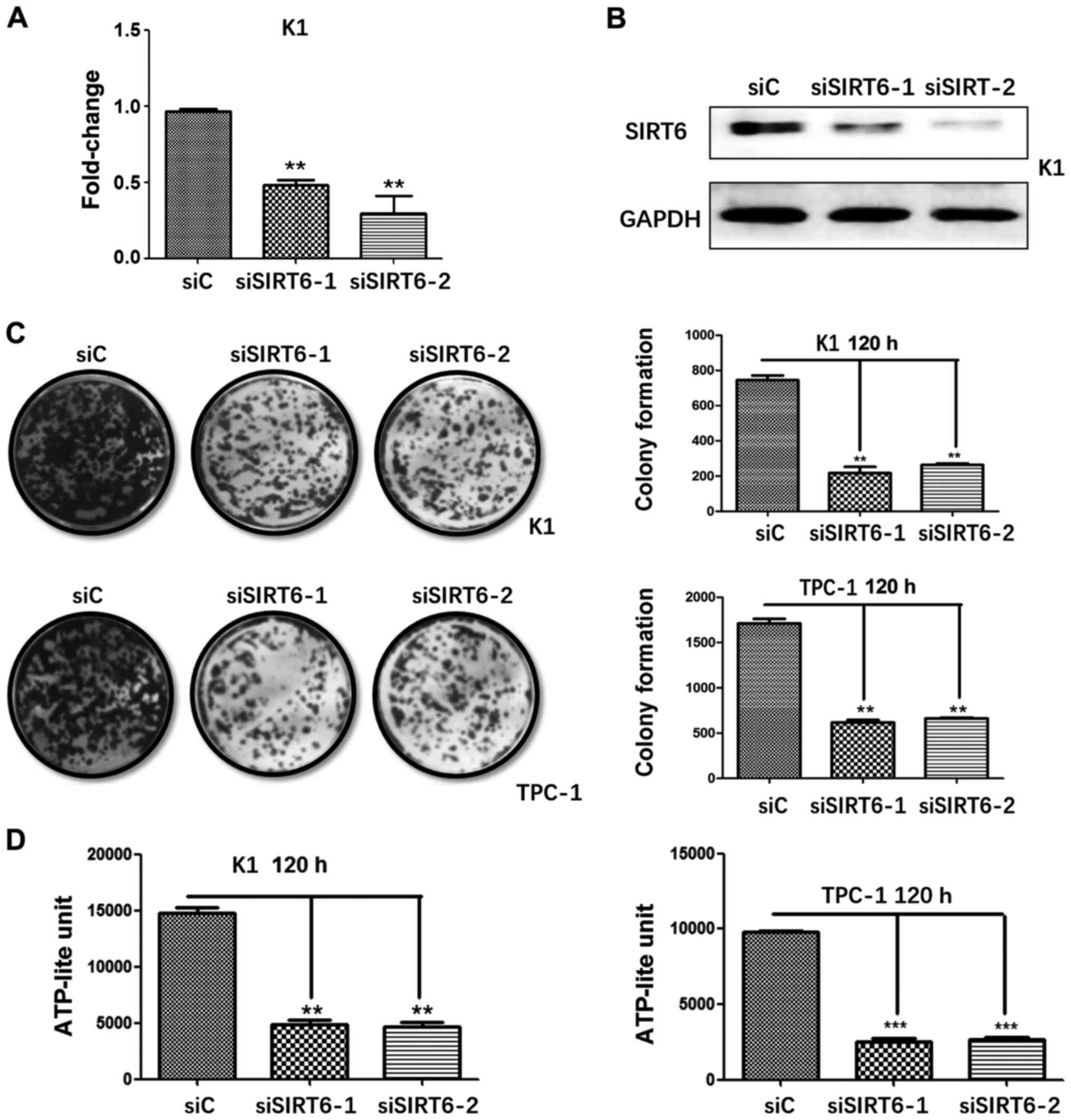
To investigate the role of SIRT6 on PTC cell
proliferation, we generated SIRT6-silenced TPC-1 and SIRT6-silenced
K1 cells. The two shRNAs against SIRT6 decreased gene expression in
PTC cells, which was confirmed by qRT-PCR (Fig. 3A) and western blotting (Fig. 3B). Colony formation assay showed
that silencing of SIRT6 resulted in significant decrease of cloning
capacity of PTC cells in vitro (Fig. 3C). Cell viability test was then
performed by using ATPlite luminescence assay, which displayed that
the ATP-lite unit of each experiment group (siSIRT6-1 and
siSIRT6-2) was lower than the control group in both cell lines
(Fig. 3D). Combined, these results
demonstrated that SIRT6 may promote the proliferation in TPC-1 and
K1 cells.
Silencing SIRT6 decreases PTC cell
migration and invasion in vitro
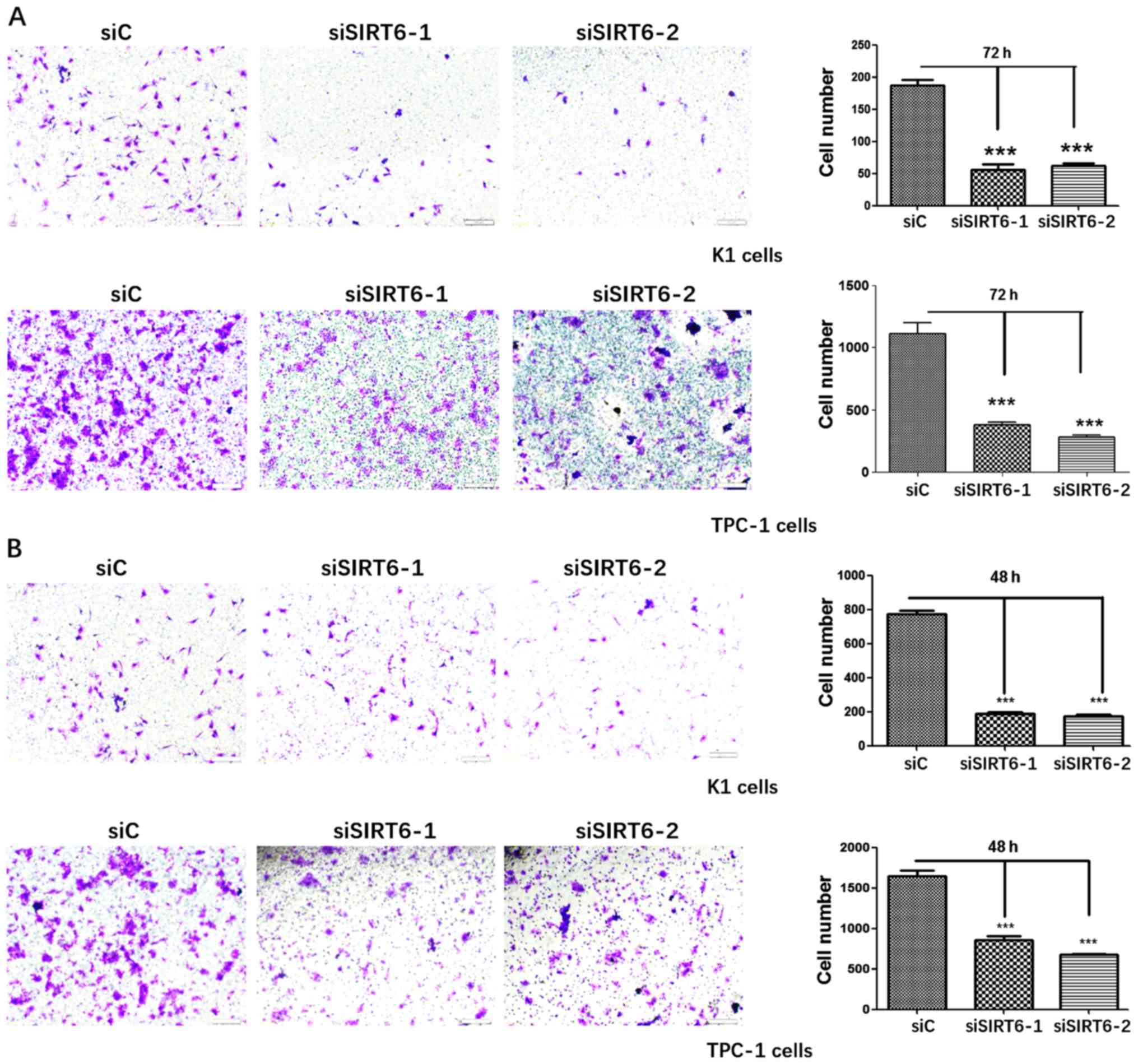
Transwell assays were performed on SIRT6-silenced
TPC-1 and SIRT6-silenced K1 cells to determine the influence of
silenced SIRT6 on PTC cell migration and invasion, respectively.
The migration assays showed a significant decrease in the number of
migrated cells in both cell lines (Fig. 4A). Similarly, the invasion assays
also revealed a huge decrease of cells invaded in K1 and TPC-1 cell
(Fig. 4B). These results pointed
out that SIRT6 stimulated cell migration and invasion in
vitro.
Influence of silencing SIRT6 gene on PTC
cell cycle distribution
To further investigate whether silenced SIRT6 alters
PTC cell cycle distribution, flow cytometry analysis was conducted
in TPC-1 and K1 cells. Results unveiled that SIRT6-1 silenced K1
cells possessed a significant increase in the proportion of cells
in G1 phase compared to the control group. However, there was only
slightly elevated number of cells in G1 phase of SIRT6-2 silenced
K1 cells. As for TPC-1 cells, the proportion of cells in G1 phase
decreased in both SIRT6-1 and SIRT6-2 silenced TPC-1 cells
(Fig. 5). The inconsistency in the
outcome indicated that SIRT6 had a rather complicated role in
regulating cell cycle distribution.
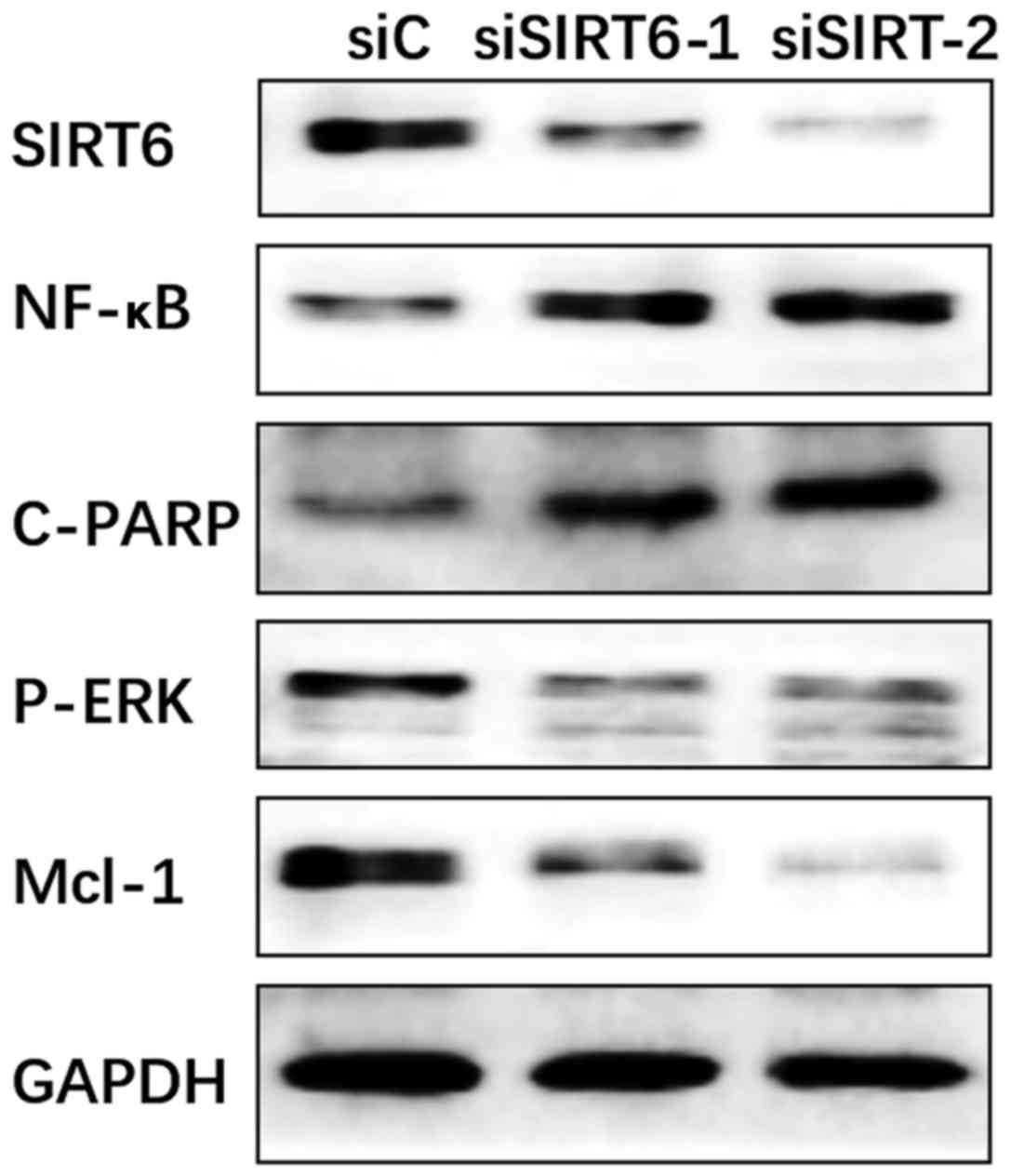
SIRT6 enhances PTC cell aggressiveness
via BRAF/ERK/Mcl-1 pathway
Western blot analysis was performed to determine if
SIRT6 is involved in BRAF/ERK/Mcl-1 signaling pathway. Apoptosis
related proteins including NF-κB and C-PARP were also tested. The
protein expression of SIRT6, NF-κB, C-PARP, phosphorylated ERK and
Mcl-1 were analyzed in control and SIRT6-silenced K1 cells,
separately. Silencing SIRT6 was accompanied by downregulation of
phosphorylated ERK and the apoptosis associated protein Mcl-1,
while the expression of NF-κB and C-PARP were enhanced (Fig. 6).
Discussion
SITR6 has been reported in connection with several
types of human cancers. Under most of the circumstances, it acts as
a tumor suppressor, such as in pancreatic cancer, breast cancer and
in hepatocellular carcinomas (14–18).
Regulation of aerobic glycolysis in cancer cells and
cancer-associated point mutations in SIRT6, may be the foundation
of its function as a tumor suppressor (21,22).
However recently, some researchers have found SIRT6 can be an
oncogene in lung cancer, prostate cancer and skin cancer where it
was overexpressed (19,20,23).
Thus, the function of SIRT6 in cancer is likely context and tissue
specific. The underlying mechanisms between SIRT6 and tumor
progression is unclear. To our best knowledge, this study is the
first to explore the involvement of SIRT6 gene in PTC.
We analyzed SIRTs expression and clinicopathological
parameters of the TCGA cohort. Among the 7 SIRTs, SIRT6 was an
independent biomarker for nodal metastasis (OR=1.794, 95% CI:
1.256–1.920, p=0.012) and related to poor RFS. The X-tile plots
further pointed out that when divided by SIRT6 expression volume,
PTC prognosis can differ significantly. Notably, the higher the
expression of SIRT6 in tumor was, the poorer their RFS would be. By
microarray analysis, we found SIRT6 was higher in PTC tumor tissues
and this result was also confirmed by IHC and the qRT-PCR of our
in-house cohort. After successful establishment of SIRT6 silenced
K1 and TPC-1 cell lines, we then further demonstrated that the
silencing of SIRT6 decreased PTC cell aggressiveness through
BRAF/ERK/Mcl-1 pathway in vitro. Taken together, these
results may indicate SIRT6 acts as an oncogene in PTC.
Our cell cycle distribution analysis received
inconsistent results from SIRT6 silenced K1 and TPC-1 cells. The
proportion of cell numbers in G1 phase of SIRT6 silenced K1 cells
was significantly increased compared to untreated K1 cells,
suggesting SIRT6 may be responsible for cell proliferation.
However, no significant increase of cells in G1 phase were observed
in the other three cell lines. The BRAFV600E
mutation may be responsible for the disagreement since K1, instead
of TPC-1 cell line is known to harbor BRAFV600E
mutation. Activating BRAF mutation is frequent, being the most
common oncogenic mutation in PTC (24). Also, the presence of the
BRAFV600E mutation was acknowledged to be
associated with poor prognosis among patients with PTC (6,25).
It can be inferred that SIRT6 plays a quite complicated role in
controlling PTC cell cycle distribution, which concerns BRAF.
We have demonstrated that silencing SIRT6 was
accompanied by a downregulation of Mcl-1 and phosphorylated ERK,
while NF-κB and C-PARP were elevated. On these grounds, we
speculate that SIRT6 is involved in the BRAF/ERK/Mcl-1 pathway.
BRAF promotes the stabilizing phosphorylation of the anti-apoptotic
protein Mcl-1, which was frequently reported in melanoma and
colorectal cancer (26,27). ERK is known to be a member of the
mitogen-activated protein kinase (MAPK) signaling pathway and is
markedly increased in PTC cells driven by oncogenic BRAF. The ERK
pathway suppresses cellular senescence by inhibiting the expression
of p16 and p21, thus it is crucial in sustaining cell proliferation
and activating tumorigenesis (28). ERK was also found to mediate Mcl-1
stabilization in colon cancer and hepatic cancers, which may be
another essential factor in cancer cell survival (29,30).
Activation of ERK and Mcl-1 stabilization is
BRAFV600E-dependent and related to the disruption
of S6K1-IRS-2/PI3K negative feedback loop (29). Bai et al presented that
SIRT6 overexpression increased ERK activation via ERK1/2/MMP9 axis
in non-small cell lung cancer (23). In hepatocellular carcinoma,
transforming growth factor-β1/H2O2/HOCl could upregulate SIRT6
expression by inducing the sustained activation of ERK (31). Anti-apoptosis related proteins such
as NF-κB and c-PAPP which is a fragment of caspase-3 cleaved PARP
that exists during apoptosis were analyzed. Previous studies also
showed that SIRT6 attenuates NF-κB signaling via H3K9 deacetylation
at chromatin (32). NF-κB levels
were significantly and negatively correlated with SIRT6 mRNA levels
in myocardial hypoxia/reoxygenation induced injury and other
diseases (12,33). Despite the above, SIRT6 was found
to directly inhibit the expression of a subset of NF-κB target
genes, especially those associated with senescence (32). This implies that SIRT6 have the
potential to promote tumor development considering its negative
effect on cellular senescence and that NF-κB is an important
anti-apoptotic factor. Combined, SIRT6 may be involved in the
BRAF/ERK/Mcl-1 axis, by which it promotes cell aggressiveness in
PTC.
In conclusion, our study demonstrated that SIRT6
upregulation was associated with poor RFS in PTC patients and may
promote PTC development and progression. Furthermore, we
demonstrated that SIRT6 promoted tumor progression through the
pathway. SIRT6 may serve as a potential therapeutic target in PTC
and its utility as a prognostic indicator warrants further
study.
Acknowledgments
This study was supported by funds from the National
Science Foundation of China (nos. 81572622 and 81272934 to
QHJ).
References
|
1
|
Davies L and Welch HG: Current thyroid
cancer trends in the United States. JAMA Otolaryngol Head Neck
Surg. 140:317–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Colonna M, Uhry Z, Guizard AV, Delafosse
P, Schvartz C, Belot A and Grosclaude P; FRANCIM network: Recent
trends in incidence, geographical distribution, and survival of
papillary thyroid cancer in France. Cancer Epidemiol. 39:511–518.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McLeod DS, Sawka AM and Cooper DS:
Controversies in primary treatment of low-risk papillary thyroid
cancer. Lancet. 381:1046–1057. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
La Vecchia C, Malvezzi M, Bosetti C,
Garavello W, Bertuccio P, Levi F and Negri E: Thyroid cancer
mortality and incidence: A global overview. Int J Cancer.
136:2187–2195. 2015. View Article : Google Scholar
|
|
6
|
Xing M, Alzahrani AS, Carson KA, Shong YK,
Kim TY, Viola D, Elisei R, Bendlová B, Yip L, Mian C, et al:
Association between BRAF V600E mutation and recurrence of papillary
thyroid cancer. J Clin Oncol. 33:42–50. 2015. View Article : Google Scholar :
|
|
7
|
Grogan RH, Kaplan SP, Cao H, Weiss RE,
Degroot LJ, Simon CA, Embia OM, Angelos P, Kaplan EL and Schechter
RB: A study of recurrence and death from papillary thyroid cancer
with 27 years of median follow-up. Surgery. 154:1436–1446;
discussion 1446–1447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartosch C, Monteiro-Reis S, Almeida-Rios
D, Vieira R, Castro A, Moutinho M, Rodrigues M, Graça I, Lopes JM
and Jerónimo C: Assessing sirtuin expression in endometrial
carcinoma and non-neoplastic endometrium. Oncotarget. 7:1144–1154.
2016.
|
|
9
|
Mouchiroud L, Houtkooper RH, Moullan N,
Katsyuba E, Ryu D, Cantó C, Mottis A, Jo YS, Viswanathan M,
Schoonjans K, et al: The NAD(+)/Sirtuin pathway modulates longevity
through activation of mitochondrial UPR and FOXO signaling. Cell.
154:430–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu
ML, Hsu CM and Yang MY: Altered expression of SIRT gene family in
head and neck squamous cell carcinoma. Tumour Biol. 34:1847–1854.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang X, Khan S, Jiang H, Antonyak MA,
Chen X, Spiegelman NA, Shrimp JH, Cerione RA and Lin H: Identifying
the functional contribution of the defatty-acylase activity of
SIRT6. Nat Chem Biol. 12:614–620. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toiber D, Erdel F, Bouazoune K, Silberman
DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor
B, Giacosa S, et al: SIRT6 recruits SNF2H to DNA break sites,
preventing genomic instability through chromatin remodeling. Mol
Cell. 51:454–468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang WW, Zeng Y, Wu B, Deiters A and Liu
WR: A chemical biology approach to reveal Sirt6-targeted Histone H3
sites in nucleosomes. ACS Chem Biol. 11:1973–1981. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kugel S, Sebastián C, Fitamant J, Ross KN,
Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al:
SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell.
165:1401–1415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marquardt JU, Fischer K, Baus K, Kashyap
A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, et al:
Sirtuin-6-dependent genetic and epigenetic alterations are
associated with poor clinical outcome in hepatocellular carcinoma
patients. Hepatology. 58:1054–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Elhanati S, Ben-Hamo R, Kanfi Y, Varvak A,
Glazz R, Lerrer B, Efroni S and Cohen HY: Reciprocal regulation
between SIRT6 and miR-122 controls liver metabolism and predicts
hepatocarcinoma prognosis. Cell Rep. 14:234–242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thirumurthi U, Shen J, Xia W, LaBaff AM,
Wei Y, Li CW, Chang WC, Chen CH, Lin HK, Yu D, et al: MDM2-mediated
degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis
and trastuzumab resistance in breast cancer. Sci Signal.
7:ra712014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Khongkow M, Olmos Y, Gong C, Gomes AR,
Monteiro LJ, Yagüe E, Cavaco TB, Khongkow P, Man EP, Laohasinnarong
S, et al: SIRT6 modulates paclitaxel and epirubicin resistance and
survival in breast cancer. Carcinogenesis. 34:1476–1486. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ming M, Han W, Zhao B, Sundaresan NR, Deng
CX, Gupta MP and He YY: SIRT6 promotes COX-2 expression and acts as
an oncogene in skin cancer. Cancer Res. 74:5925–5933. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie Q, Wong AS and Xia W: SIRT6 induces
EMT and promotes cancer cell invasion and migration in prostate
cancer. In: Proceedings of the 105th Annual Meeting of the American
Association for Cancer Research. Cancer Res. 74(Suppl 19): Abst
1151. 2014. View Article : Google Scholar
|
|
21
|
Kugel S, Feldman JL, Klein MA, Silberman
DM, Sebastián C, Mermel C, Dobersch S, Clark AR, Getz G, Denu JM,
et al: Identification of and molecular basis for SIRT6
loss-of-lunction point mutations in cancer. Cell Rep. 13:479–488.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sebastián C, Zwaans BM, Silberman DM,
Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber
D, et al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bai L, Lin G, Sun L, Liu Y, Huang X, Cao
C, Guo Y and Xie C: Upregulation of SIRT6 predicts poor prognosis
and promotes metastasis of non-small cell lung cancer via the
ERK1/2/MMP9 pathway. Oncotarget. 7:40377–40386. 2016.PubMed/NCBI
|
|
24
|
Liu X, Qu S, Liu R, Sheng C, Shi X, Zhu G,
Murugan AK, Guan H, Yu H, Wang Y, et al: TERT promoter mutations
and their association with BRAF V600E mutation and aggressive
clinicopathological characteristics of thyroid cancer. J Clin
Endocrinol Metab. 99:E1130–E1136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Howell GM, Nikiforova MN, Carty SE,
Armstrong MJ, Hodak SP, Stang MT, McCoy KL, Nikiforov YE and Yip L:
BRAF V600E mutation independently predicts central compartment
lymph node metastasis in patients with papillary thyroid cancer.
Ann Surg Oncol. 20:47–52. 2013. View Article : Google Scholar
|
|
26
|
Becker TM, Boyd SC, Mijatov B,
Gowrishankar K, Snoyman S, Pupo GM, Scolyer RA, Mann GJ, Kefford
RF, Zhang XD, et al: Mutant B-RAF-Mcl-1 survival signaling depends
on the STAT3 transcription factor. Oncogene. 33:1158–1166. 2014.
View Article : Google Scholar
|
|
27
|
Kawakami H, Huang S, Pal K, Dutta SK,
Mukhopadhyay D and Sinicrope FA: Mutant BRAF upregulates MCL-1 to
confer apoptosis resistance that is reversed by MCL-1 antagonism
and cobimetinib in colorectal cancer. Mol Cancer Ther.
15:3015–3027. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu D, Liu J, Lin B, Liu S, Hou R, Hao Y,
Liu Q, Zhang S and Iwamori M: Lewis y regulate cell cycle related
factors in ovarian carcinoma cell RMG-I in vitro via ERK and Akt
signaling pathways. Int J Mol Sci. 13:828–839. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He K, Chen D, Ruan H, Li X, Tong J, Xu X,
Zhang L and Yu J: BRAFV600E-dependent Mcl-1 stabilization leads to
everolimus resistance in colon cancer cells. Oncotarget.
7:47699–47710. 2016.PubMed/NCBI
|
|
30
|
Gao M, Kong Q, Hua H, Yin Y, Wang J, Luo T
and Jiang Y: AMPK-mediated up-regulation of mTORC2 and MCL-1
compromises the anti-cancer effects of aspirin. Oncotarget.
7:16349–16361. 2016.PubMed/NCBI
|
|
31
|
Feng XX, Luo J, Liu M, Yan W, Zhou ZZ, Xia
YJ, Tu W, Li PY, Feng ZH and Tian DA: Sirtuin 6 promotes
transforming growth factor-β1/H2O2/HOCl-mediated enhancement of
hepatocellular carcinoma cell tumorigenicity by suppressing
cellular senescence. Cancer Sci. 106:559–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kawahara TL, Michishita E, Adler AS,
Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang
HY, et al: SIRT6 links histone H3 lysine 9 deacetylation to
NF-kappaB-dependent gene expression and organismal life span. Cell.
136:62–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheng MY, Cheng YW, Yan J, Hu XQ, Zhang H,
Wang ZR, Yin Q and Cheng W: SIRT6 suppresses mitochondrial defects
and cell death via the NF-κB pathway in myocardial
hypoxia/reoxygenation induced injury. Am J Transl Res. 8:5005–5015.
2016.
|