Introduction
Breast cancer is one of the most common female
cancers in the world. The American Cancer Society provides an
overview of female breast cancer statistics in the United States,
~252,710 new cases of invasive breast cancer and 40,610 breast
cancer deaths are expected to occur among US women in 2017
(1). Breast cancer is also the
highest incidence of female malignant tumor in China, accounting
for ~30% (2). The treatments for
breast cancer include surgical resection, radiotherapy,
endocrinotherapy and chemotherapy. Chemotherapy is one of the
standard therapies which have been shown to inhibit tumor growth
and prolong survival in patients (3). Paclitaxel has been recognized as the
first-line therapy in breast cancer treatment. However, its
efficacy is often limited by the development of drug resistance
(4,5). Therefore, identifying the underlying
mechanism is responsible for regulating chemotherapy resistance for
improving breast cancer treatment.
MicroRNAs (miRNAs) are small non-coding RNAs of
~19–25 nucleotides in length, which functions in
post-transcriptional regulation of gene expression by binding to
the 3′-untranslated region (3′-UTR) (6). Several studies have demonstrated that
miRNA misregulation can increase chemo-resistance in cancer if
specific proteins are affected (7–9),
which indicated that miRNAs might play important roles in drug
resistance. For instance, miR-205 enhanced chemosensitivity of
breast cancer cells to TAC (docetaxol, doxorubicin plus
cyclophosphamide) chemotheraphy by suppressing both VEGFA and FGFZ,
leading to evasion of apoptosis (10). A study showed that miR-221/222
confers tamoxifen resistance in breast cancer by targeting
P27kip1, a downstream modulator of PI3K/Akt pathway
(11). Raza et al reported
that miR-644a/CTBP1/P53 axis could suppress drug resistance by
simultaneous inhibition of cell survival and epithelial-mesenchymal
transition (EMT) in breast cancer (12). Another study revealed that
miR-125a-3p function as a tumor suppressor potentiates docetaxel
sensitivity by regulating the BRCA1 signaling (13). Moreover, miR-452 could regulate the
expression of insulin-like growth factor-1 receptor (IGF-1R), and
mediated the change of adriamycin (ADR) resistance of breast cancer
MCF-7 cells (14). Furthermore,
miR-3646 is an important regulator responsible to Doc-resistant
phenotype of breast cancer cells, and manipulates GSK-3β-dependent
activation of β-catenin signaling pathway (15). Over the past few years, numerous
miRNAs have been reported to be involved in breast cancer
tumorigenesis, such as miR-17-59 (16), miR-25 (17), miR-222 (18), miR-375 (19), miR-200c (20), miR-29a (21), miR-145 (22), miR-489 (23,24).
Given the major roles of miRNAs in regulating protein expression in
general, it is reasonable to infer that targeting miRNAs could be a
promising approach to overcome drug resistance.
A growing body of data indicates that miR-335 plays
an essential role in drug resistance (25–27).
Kim et al found that miR-335/SIAH2/HDAC3 axis regulates the
response to anticancer drugs (25). Another study revealed that the
expression of miR-335 was downregulated in all the ovarian cancer
resistant cells, suggesting a direct involvement in the development
of chemoresistance (26).
Interestingly, Tang et al found that miR-335 could regulate
WW domain binding protein 5 induces multidrug resistance of small
cell lung cancer through the Hippo pathway (27). Although these studies have shown
the potential role of miR-335 in chemoresistance in human cancer
cells, the function of miR-335 in breast cancer remains poorly
understood.
Here, we report that miR-335 was downregulated in
paclitaxel-resistant (PR) human breast cancer cells and describe
the mechanism of PR mediated by miR-335. Our previous study showed
that the 21-residue N-terminal of vMIP-II (NT21MP), is a potent
antagonist of SDF-1α and CXCR4 (28), could inhibit breast cancer
progression and metastasis in vivo (29). In this study, we provide
experimental evidence that the use of NT21MP and overexpression of
miR-335 suppressed the cells migration and invasion, increased the
PR cell apoptosis and arrested cells into
G0/G1 phase. Furthermore, SETD8 was validated
as a potential target of miR-335, miR-335/SETD8 promoted PR by
regulating Wnt/β-catenin signaling pathway activities. Our data
provide new insights that miR-335 is potentially a clinically
significant breast cancer biomarker.
Materials and methods
Cell culture
Human breast cancer cell lines SKBR-3, MCF-7,
paclitaxel-resistant cells (SKBR-3/PR and MCF-7/PR) were cultured
at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium
(DMEM; Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal
bovine serum (FBS). MCF-7/PR and SKBR-3/PR cells were maintained in
culture medium with 10 and 25 µg/ml paclitaxel (30).
Reagents and antibodies
NT21MP was designed by our laboratory and
synthesized by GL Biochem Ltd. (Shanghai, China). The amino acid
sequence information of the NT21MP is
H-D-leu-D-Gly-D-Ala-D-Ser-D-Trp-Dhis-D-Arg-D-Pro-D-Asp-D-Lys-Cys-Cys-Leu-Gly-Tyr-GlnLys-Arg-Pro-Leu-Pro-OH.
Annexin V and Dead Cell and Cell Cycle Detection kit were purchased
from Beyotime (Shanghai, China). Superscript First-Strand Synthesis
system was purchased from Thermo Scientific (Waltham, MA, USA).
SYBR GreenER™ qPCR Super Mix was from Life Technologies (Carlsbad,
CA, USA). Tumor Invasion assay kit was from BD Biosciences
(Bedford, MA, USA). Primary antibodies against SETD8, β-catenin,
c-Myc and cyclin D1 and β-actin were from Santa Cruz Biotechnology
(Santa Cruz, CA, USA).
miRNA real-time RT-PCR
The miRNA RT-PCR was used to detect the alterations
of miR-335 expression in breast cancer cells. Briefly, 10 ng of
total RNA was reverse transcribed into cDNA using TaqMan miRNA
hsa-miR-335-specifc primers (Applied Biosystems). Then real-time
PCR was performed using a TaqMan MicroRNA Reverse Transcription kit
(Applied Biosystems). RNA U6 was carried out as endogenous control
in each sample. The relative expression was analyzed using the
comparative Ct method.
Wound healing assay
The PR cells were seeded in 6-well plate until the
cells reached 80–90% confluency. The confluent monolayers were
scratched with a 10-µl pipette tip to generate the wound.
Then cells were washed twice with PBS and further cultured for 24 h
to allow wound healing at 37°C with 5% CO2. The
photographic images were taken at 0 and 24 h. At least three
independent experiments were performed in each cell line.
Western blot analysis
Cells were harvested and lysed with RIPA buffer. The
protein concentrations were measured using Bio-Rad protein assay
kit (Bio-Rad Laboratories, CA, USA). The proteins were resolved
through 10% SDS-polyacrylamide gel electrophoresis and then
electrotransferred to PVDF membranes. These membranes were
immunoblotted with indicated antibodies for western blotting as
described previously (30).
Quantification of protein bands was performed using the ImageJ
software.
Transwell invasion assays
The invasive activity of cells was measured using
Transwell inserts precoated with Matrigel as described earlier
(17). Briefly, cells were added
to the 24-well upper chamber of the inserts. Cell culture medium
with 10% FBS was added to the lower chamber. After incubation for
20 h at 37°C in 5% CO2. Then cells on the upper side of
the Transwell were removed, the invading cells on the underside
were fixed with 4% paraformaldehyde, and stained with Giemsa
solution. The stained invaded cells were photographed under a
microscope in five randomly-selected fields.
Apoptosis assay and cell cycle assay
Cells were collected by trypsinization, flow
cytometry analysis was performed using an Annexin V and Dead Cell
kit and Cell Cycle Detection kit according to the manufacturer's
instructions. All experiments were performed in triplicate.
Luciferase reporter assay
The miR-335 response element (wild-type or mutated)
in the 3′-UTR of SETD8 was cloned into pMIR-Report (Ambion) plasmid
downstream of luciferase reporter gene with firefly luciferase.
Cells seeded into 24-well plates were co-transfected with miR-335
mimics (miR-335 inhibitor) and luciferase reporter constructs
containing WT or MT SETD8 3′-UTR. After 48 h of transfection, the
luciferase activities were measured according to the manufacturer's
protocols (Promega). Each experiment was repeated in
triplicate.
qRT-PCR assay for gene expression
Total RNA was isolated with TRIzol (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocols.
Real-time quantitative PCR was performed using an IQ5 Multicolor
Detection system (Bio-Rad). The SYBR green RT-PCR assay was
described previously (31). The
primers used in PCR reactions are: SETD8 forward, 5′-ACT TAC GGA
TTT CTA CCC TGT C-3′; reverse, 5′-CGA TGA GGT CAA TCT TCA TTC C-3′.
GAPDH forward, 5′-CAG CCT CAA GAT CAT CAG CA-3′; reverse, 5′-TGT
GGT CAT GAG TCC TTC CA-3′. β-catenin forward, 5′-GGC TAC TGT TGG
ATT GAT TCG AA-3′; reverse, 5′-GCT GGG TAT CCT GAT GTG CAC-3′.
c-Myc forward, 5′-GCG ACT CTG AGG AGG AAC A-3′; reverse, 5′-TGA GGA
CCA GTG GGC TGT-3′. Cyclin D1 forward, 5′-CCC TCC GTA TCT TAC TTC
AA-3′; reverse, 5′-GAT GGT CTG CTT GTT CTC AT-3′.
Transfection
Cells were seeded in 6-well plates and transfected
with SETD8 siRNA, or control siRNA using Lipofectamine 2000 as
described before (32). The
sequences used for SETD8 siRNA are as follows: SETD8 siRNA1,
forward oligo, 5′-CAG GAA GAG AAC UCA GUU ATT-3′; reverse oligo,
5′-UAA CUG AGU UCU CUU CCU GTT-3′. SETD8 siRNA2, forward oligo,
5′-GCA ACA GAA UCG CAA ACU UTT-3′; reverse oligo, 5′-AAG UUU GCG
AUU CUG UUG CTT-3′. SETD8 siRNA3, forward oligo, 5′-CCU AGG AAG ACU
GAU CAA UTT-3′; reverse oligo, 5′-AUU GAU CAG UCU UCC UAG GTT-3′.
The non-targeting control siRNA, forward oligo, 5′-UUC UCC GAA CGU
GUC ACG UTT-3′; reverse oligo, 5′-ACG UGA CAC GUU CGG AGA ATT-3′.
After the transfection, the cells were used for further analysis as
described under the results section.
Statistical analysis
Data were expressed as mean ± SEM from at least
three independent experiments. All statistical analyses were done
using GraphPad Prism 4.0 (GraphPad software, La Jolla, CA, USA).
Student's t-test was used to compare different groups. P<0.05
was considered statistically significant.
Results
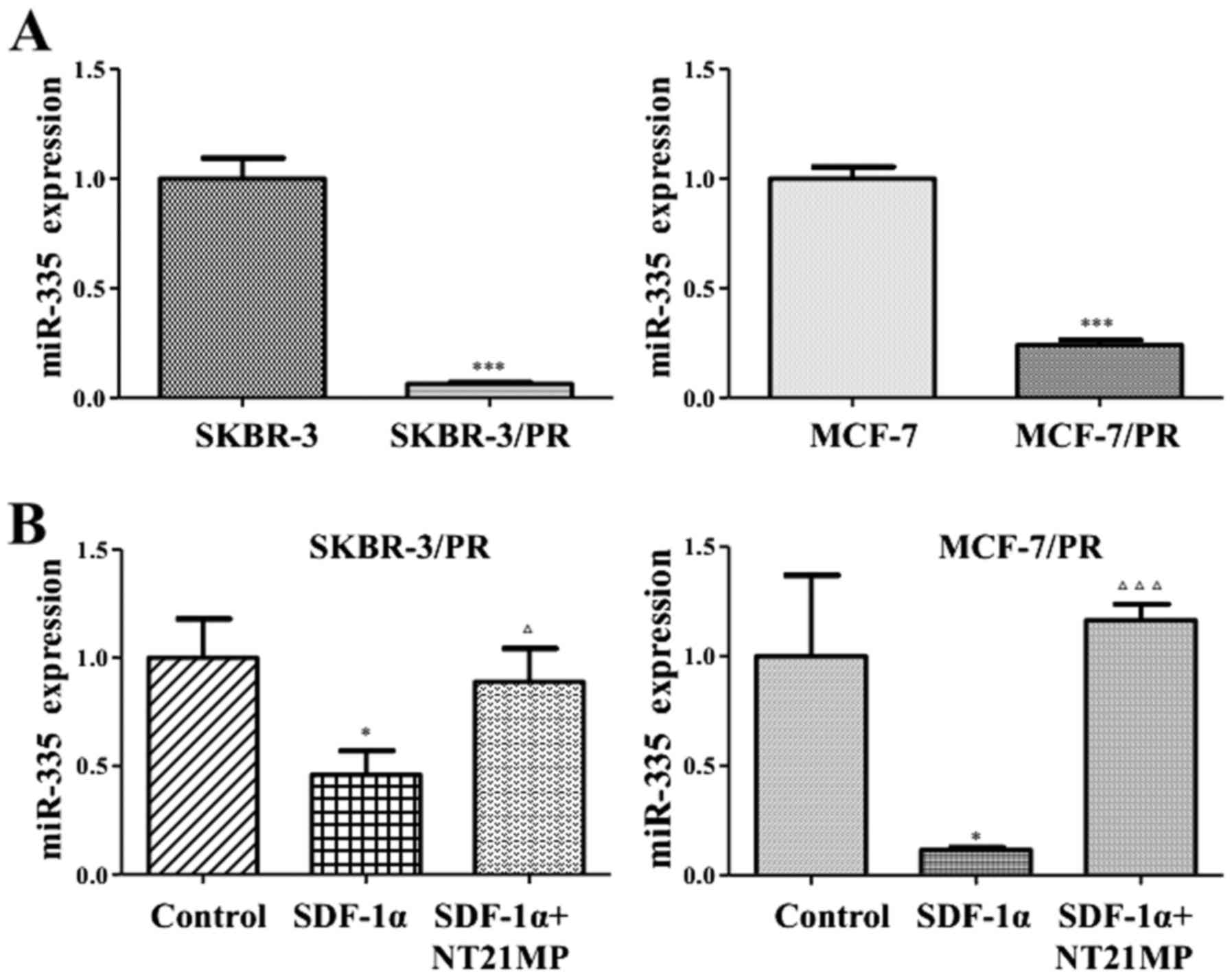
Downregulation of miR-335 in PR
cells
Multiple miRNAs were upregulated and some miRNAs
were downregulated in PR cells by miRNA microarray (data not
shown). Our previous study on miRNA microarray analysis revealed
that miR-335 was significantly downregulated in PR cells compared
with parental cells. Heidary et al confirmed that miR-335
critically served as anti-oncogene in breast cancer (33). qRT-PCR was performed to validate
that miR-335 was significantly reduced in PR cells, and miR-335
might play a pivotal role in both SKBR-3/PR and MCF-7/PR cells
(Fig. 1A).
NT21MP inhibits SDF-1α-induced decrease
of miR-335 expression in PR cells
Our previous studies have shown that SDF-1α could
promote breast cancer cellular proliferation, metastasis combined
with CXCR4. SKBR-3/PR and MCF-7/PR cell treatment with SDF-1α (0.1
µg/ml) lead to the decrease of miR-335 expression, while
NT21MP (1 µg/ml) could inhibit the effect of SDF-1α and
upregulation of the expression of miR-335 (Fig. 1B).
NT21MP and overexpression of miR-335
inhibit biological activity in PR cells
To further confirm the effect of NT21MP and
overexpression of miR-335 in regulation of cell motility, we
performed the migration and invasion assays in SKBR-3/PR and
MCF-7/PR cells treated with NT21MP and miR-335 mimics. Our wound
healing assay showed that NT21MP or miR-335 mimics inhibited the
cell migration respectively, and the combine use of NT21MP and
miR-335 mimics made a more obvious suppression (Fig. 2A and B). In line with these
findings, our invasion assay results revealed that NT21MP and
miR-335 mimics suppressed cell invasion in PR cells (Fig. 2C and D). Additionally, we performed
cell apoptosis and cell cycle experiments to observe the function
of NT21MP and miR-335. Results showed that both miR-335 mimics and
NT21MP increased the number of cell apoptosis (Fig. 2E and F) and arrested cells in
G0/G1 phase (Fig. 2G and Tables I and II) in both SKBR-3/PR and MCF-7/PR breast
cancer cells compared with the control.
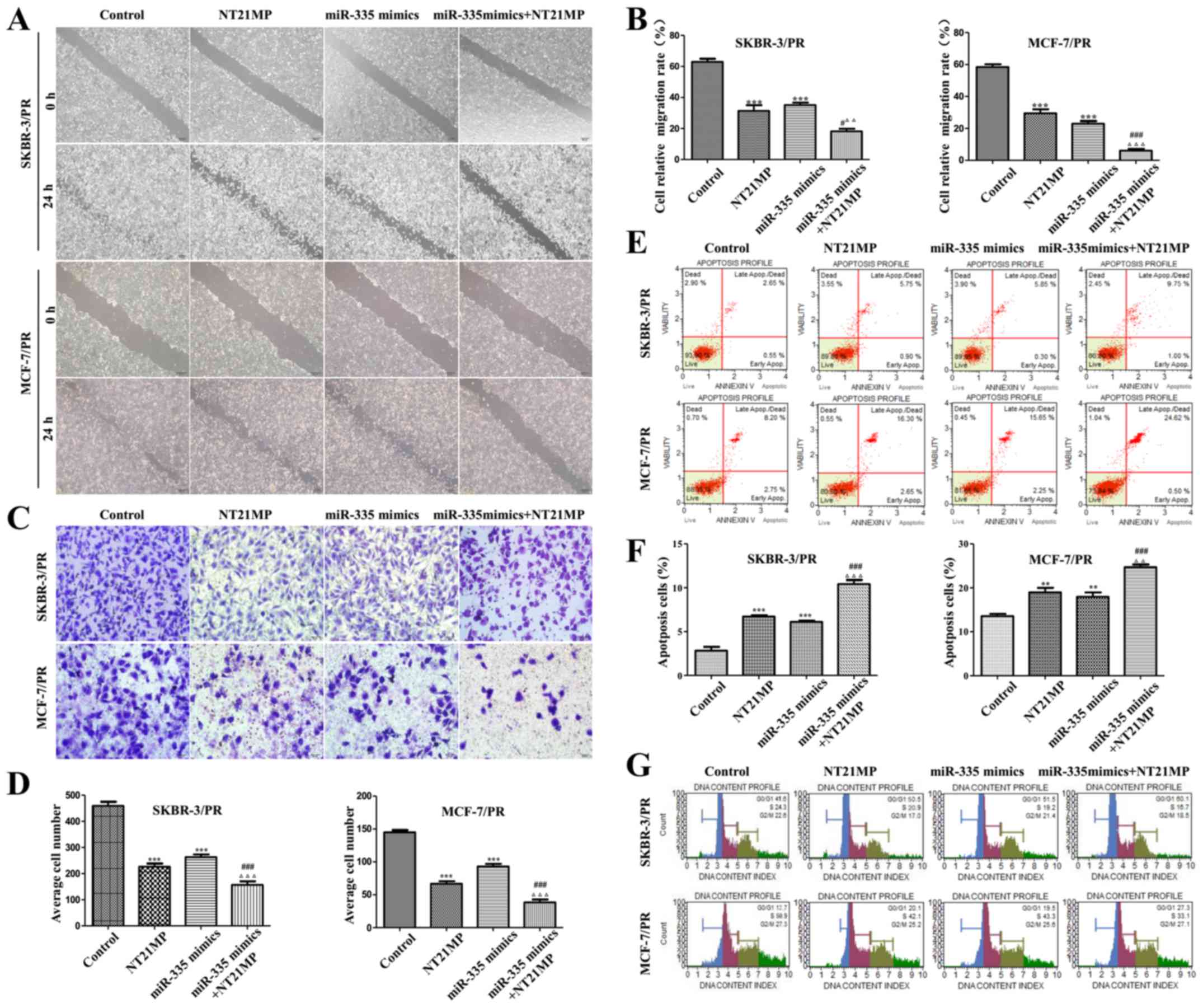 | Figure 2NT21MP and overexpression of miR-335
inhibit biological activity in PR cells. (A) Images of wound assay
of the SKBR-3/PR and MCF-7/PR cells treated with control, NT21MP,
miR-335 mimics or both. (B) Quantitative results are illustrated
for (A). (C) Transwell assays were performed in SKBR-3/PR and
MCF-7/PR cells after treatment with NT21MP and miR-335 mimics. (D)
Quantitative results are illustrated for (B). (E and G) Evaluation
of the effect of miR-335 mimics and NT21MP on cell apoptosis and
cell cycle in SKBR-3/PR and MCF-7/PR cells using Muse Cell
Analyzer. (F) Quantitative results are illustrated for (E).
Compared with control group, *P<0.05,
**P<0.01, ***P<0.001, compared with
NT21MP group, #P<0.05, ##P<0.01,
###P<0.001, compared with miR-335 mimics group,
ΔP<0.05, ΔΔP<0.01,
ΔΔΔP<0.001. |
 | Table ICell cycle was arrested in
G0/G1 phase treated with NT21MP and miR-335
mimics in SKBR-3/PR cells. |
Table I
Cell cycle was arrested in
G0/G1 phase treated with NT21MP and miR-335
mimics in SKBR-3/PR cells.
| Groups |
G0/G1 phase (%) | S phase (%) |
|---|
| Control | 41.5±0.43 | 24.3±0.33 |
| NT21MP | 50.5±0.56a | 20.9±0.31 |
| miR-335 mimics | 51.5±0.49a | 19.2±0.20a |
| miR-335
mimics+NT21MP | 60.1±0.98a,b | 16.7±0.25a,b |
 | Table IICell cycle was arrested in
G0/G1 phase treated with NT21MP and miR-335
mimics in MCF-7/PR cells. |
Table II
Cell cycle was arrested in
G0/G1 phase treated with NT21MP and miR-335
mimics in MCF-7/PR cells.
| Groups |
G0/G1 phase (%) | S phase (%) |
|---|
| Control | 12.7±0.24 | 50.9±1.04 |
| NT21MP | 20.1±0.33a | 40.2±0.75a |
| miR-335 mimics | 19.3±0.19a | 43.3±0.69a |
| miR-335
mimics+NT21MP | 27.3±0.20a,b | 33.1±0.70a,b |
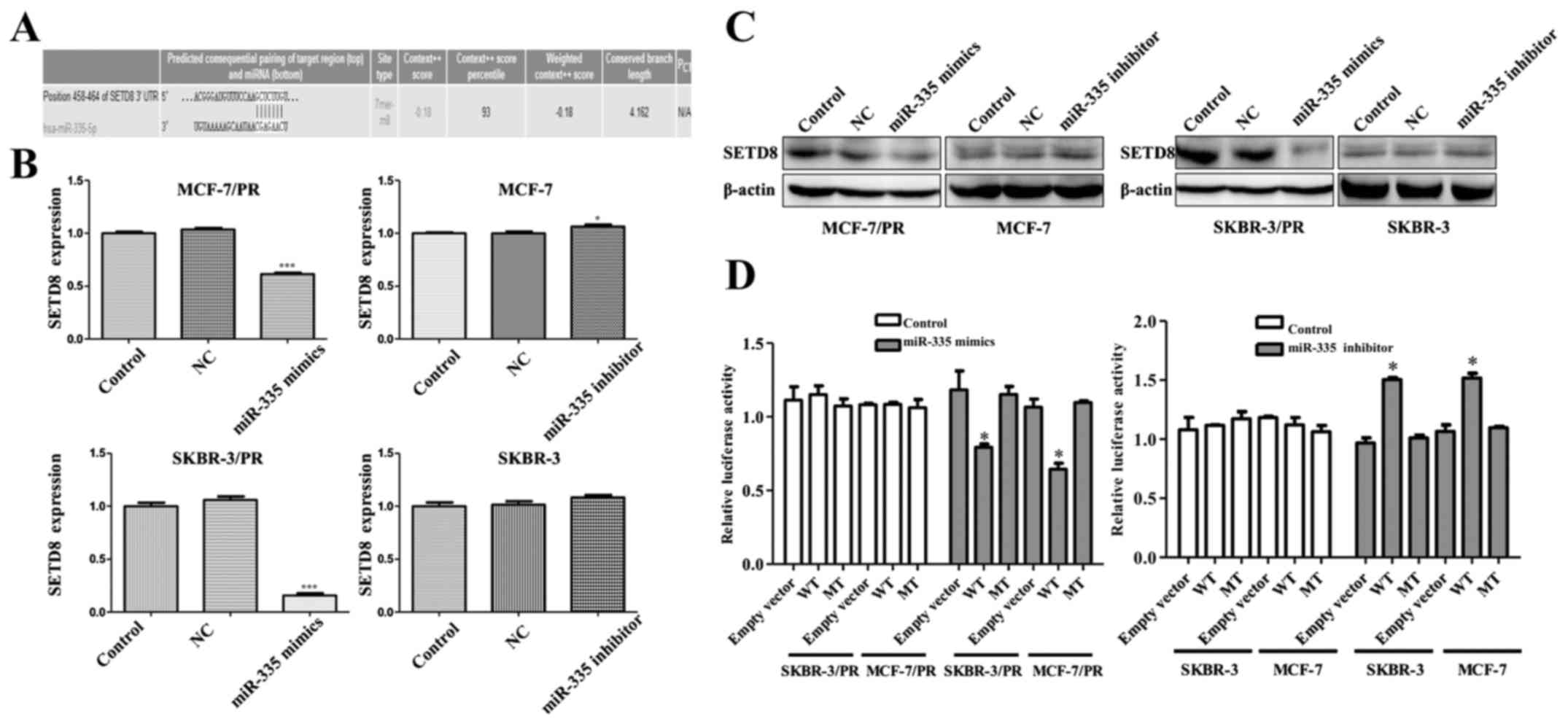
miR-335 specifically suppresses SETD8
expression in PR cells
To explore the molecular mechanism by which miR-335
contributes to breast cancer progression. we measured the putative
miR-335 target genes, three computational algorithms including
TargetScan, miRanda and PicTar were used in combination to search
for potential targets of miR-335. Among these genes, SETD8 was
identified as a potential target based on a predicted binding site
of miR-335 at its 3′-UTR (Fig.
3A). To ascertain whether miR-335 regulates SETD8, we
investigated the effects of miR-335 on SETD8 expression in
SKBR-3/PR and MCF-7/PR cells. qRT-PCR demonstrated that miR-335
mimics treatment led to a downregulation of SETD8 in SKBR-3/PR and
MCF-7/PR cells, whereas miR-335 inhibitor treatment induced an
upregulation in SKBR-3 and MCF-7 cells (Fig. 3B). Western blot analysis results
showed that SETD8 expression was suppressed by transfection of the
miR-335 mimics in SKBR-3/PR and MCF-7/PR cells and enhanced by
transfection of the miR-335 inhibitor in MCF-7 and SKBR-3 cells
(Fig. 3C). To confirm that miR-335
directly targets the presumed binding sites in the SETD8 3′-UTR and
negatively regulates SETD8 expression, we constructed luciferase
reporter plasmid with the wild-type and mutant SETD8 3′-UTR region.
Results showed a significant decrease in luciferase activity with
SETD8 3′-UTR wild-type, but not SETD8 3′-UTR mutation in SKBR-3/PR
and MCF-7/PR cells. Consistently, miR-335 inhibitor treatment
remarkably increased luciferase activity with wild-type SETD8 in
MCF-7 and SKBR-3 cells (Fig. 3D).
Collectively, these results indicated that SETD8 was a target gene
of miR-335 and might contribute to PR in breast cancer.
NT21MP and overexpression of miR-335
regulate the downstream genes of Wnt/β-catenin signaling
pathway
In order to elucidate the mechanism of NT21MP and
miR-335 mimics in regulation of the biological activity of PR
cells, SETD8 as a target gene of miR-335 was measured. As miR-335
mimics, SETD8 was downregulated after treatment with NT21MP, and it
is obviously decreased in combined use of NT21MP and miR-335 mimics
(Fig. 4). We then examined the
protein and gene expression level of β-catenin by western blot
analysis and real-time PCR, respectively. As expected, we observed
a reduction of β-catenin in the treatment of NT21MP and miR-335
mimics (Fig. 4). Consistently the
expression of the downstream genes of Wnt/β-catenin signaling c-Myc
and cyclin D1 were also decreased in both PR cell lines. These
findings provided the evidences that NT21MP and miR-335 could
regulate Wnt/β-catenin signaling.
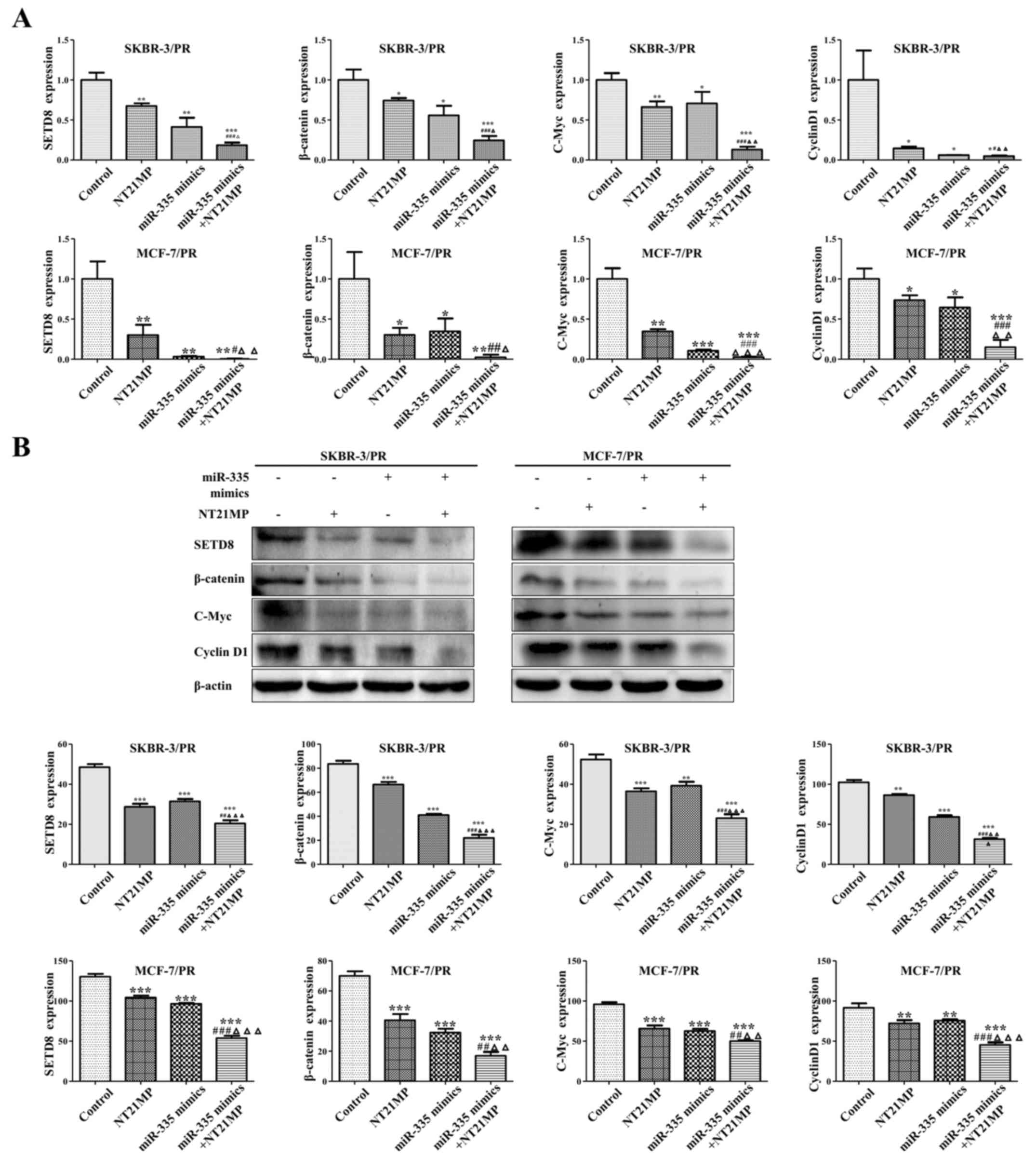 | Figure 4NT21MP and overexpression of miR-335
by regulation SETD8 and Wnt/β-catenin signaling pathway. (A)
Effects of NT21MP and miR-335 mimics on SETD8 and the downstream
genes of Wnt/β-catenin signaling pathway using qRT-PCR. (B) Western
blot analysis was performed to measure the effects of miR-335
mimics and NT21MP on SETD8 and the downstream genes of
Wnt/β-catenin signaling pathway. Compared with control group,
*P<0.05, **P<0.01,
***P<0.001, compared with NT21MP group,
#P<0.05, ##P<0.01,
###P<0.001, compared with miR-335 mimics gourp,
ΔP<0.05, ΔΔP<0.01,
ΔΔΔP<0.001. |
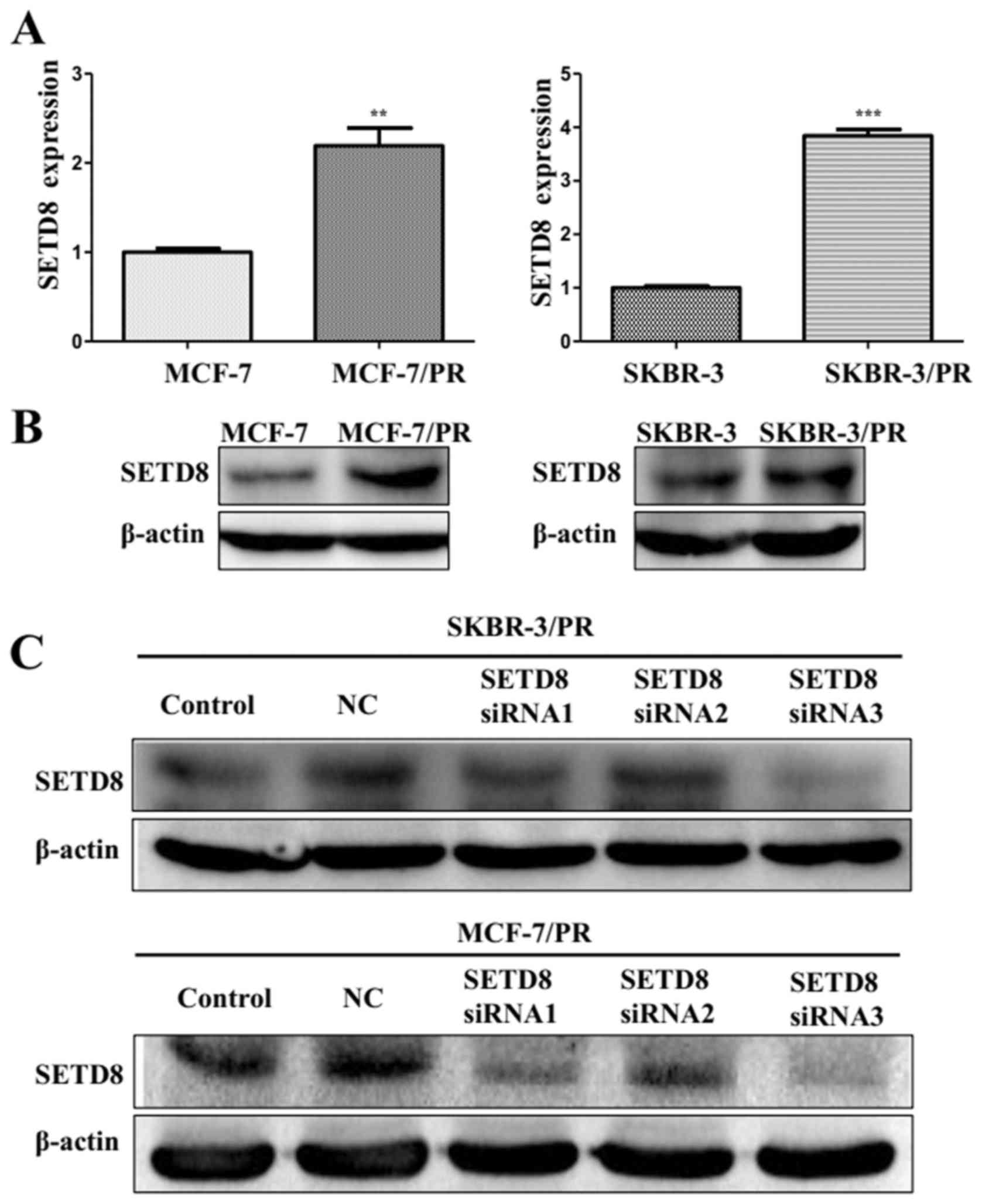
NT21MP and si-SETD8 affected PR cells
partially through SETD8 mediated-Wnt/β-catenin signaling
To further confirm the function of SETD8 in PR
cells, we performed experiment to confirmed that high expression of
SETD8 in SKBR-3/PR and MCF-7/PR cells (Fig. 5A and B), which is consistent with
low expression of miR-335 in PR cells. To determine whether SETD8
plays a key role in Wnt/β-catenin signaling pathway, we depleted
the SETD8 using its specific siRNAs in SKBR-3/PR and MCF-7/PR
cells. We found significant decrease of SETD8 with its siRNAs by
western blot analysis (Fig. 5C).
Then, we used SETD8 siRNA3 for the following experiments. To
further illuminate the function of NT21MP and silence of SETD8
reverse drug resistance through Wnt/β-catenin signaling pathway, we
detected the expression of β-catenin. Our qRT-PCR results showed
that depletion of SETD8 and NT21MP could decreased the expression
of β-catenin, more remarkable reduction was seen with combination
of both. As the target genes of Wnt/β-catenin signaling pathway,
there is also a corresponding reduction of c-Myc and cyclin D1
(Fig. 6A). Western blot analysis
showed similar changes in the protein levels (Fig. 6B). Taken together, these results
indicated that NT21MP and SETD8 plays a critical role in regulating
PR via suppressing Wnt/β-catenin signaling pathway in breast cancer
cells.
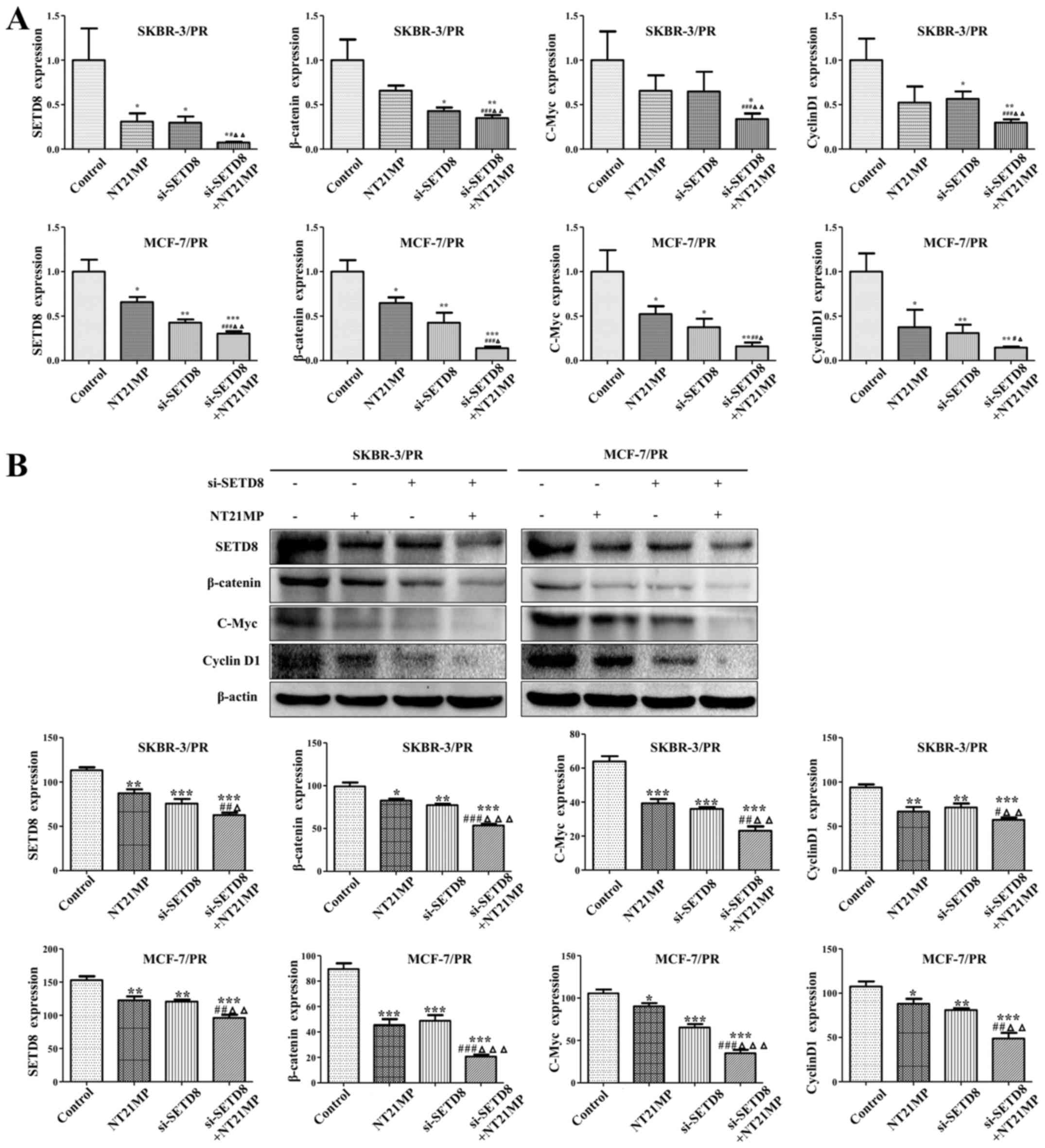 | Figure 6NT21MP and si-SETD8 affect PR cells
partially through SETD8 mediated-Wnt/β-catenin signaling. (A)
Effects of si-SETD8 and NT21MP on the downstream genes of
Wnt/β-catenin signaling pathway using qRT-PCR. (B) SKBR-3/PR and
MCF-7/PR cells transfected with si-SETD8 and NT21MP were used for
assessing the expression of Wnt/β-catenin markers by western blot
analysis. Compared with control group, *P<0.05,
**P<0.01, ***P<0.001, compared with
NT21MP group, #P<0.05,
##P<0.01,###P<0.001, compared with
si-SETD8 groups, ΔP<0.05,
ΔΔP<0.01,ΔΔΔP<0.001. |
NT21MP and depletion of SETD8 inhibit
biological activity in PR cells
To explore whether NT21MP and si-SETD8 are involved
in PR-mediated the variation of cell biological activity. we
investigated whether reduction of expression SETD8 could mimic the
growth of miR-335. Our wound healing assay showed that NT21MP and
SETD8 siRNA transfection could retarded cell motility in PR cells
(Fig. 7A and B). Moreover, in line
with this findings, our invasion assay results revealed NT21MP and
depletion of SETD8 suppressed the cell invasion in SKBR-3/PR and
MCF-7/PR cells (Fig. 7C and D),
the proportion of apoptotic cells increased (Fig. 7E and F), cell cycle were arrested
in G0/G1 phase (Fig. 7G and Tables III and IV), these affects are more significant
after combination use of NT21MP and si-SETD8 than used alone. These
results suggested that NT21MP and si-SETD8 to a certain extent,
reversed the resistance of PR cells in breast cancer.
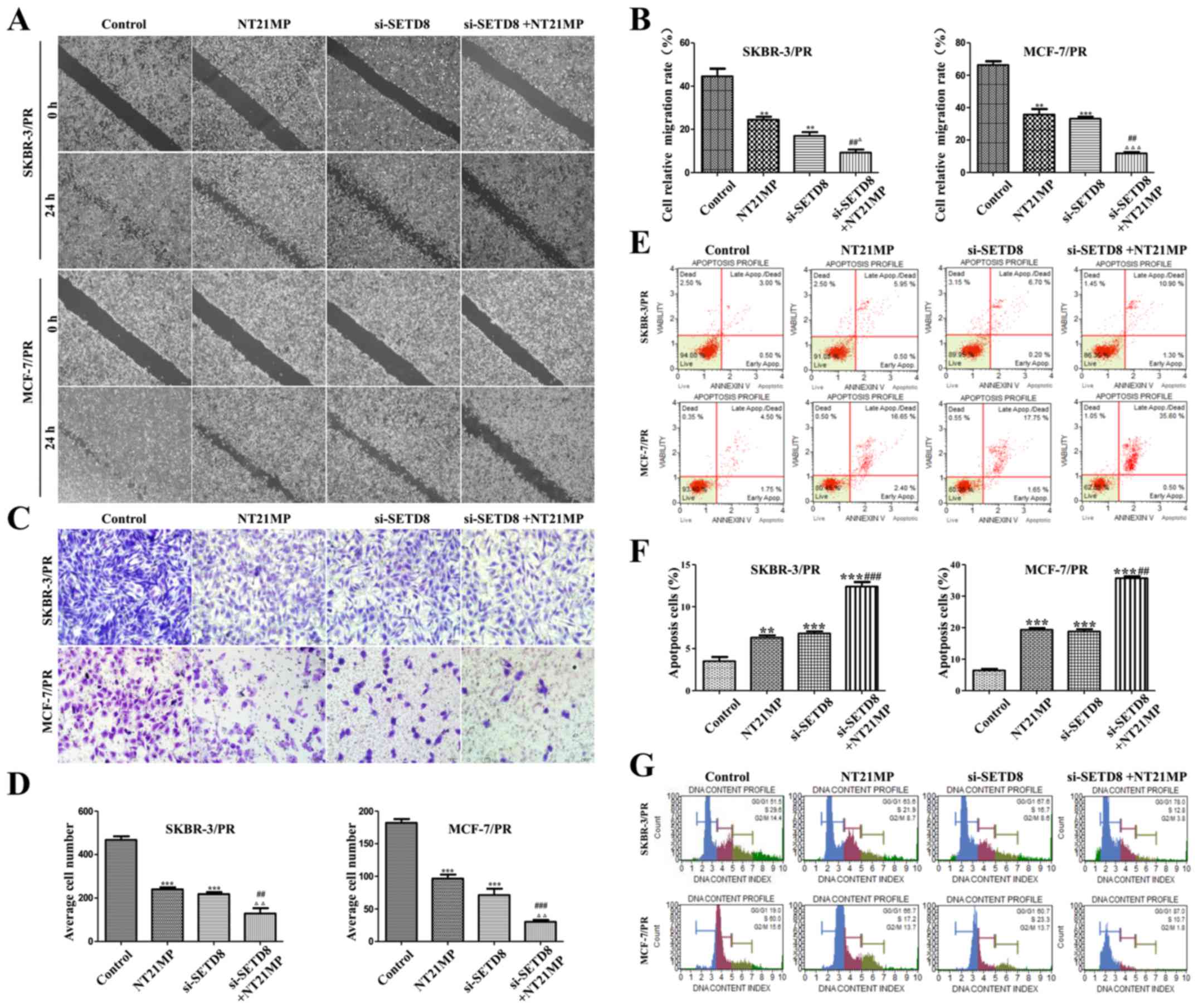 | Figure 7Dealing with NT21MP or low-expression
of SETD8 inhibits cell biological activity in PR cells. (A) NT21MP
and si-SETD8 inhibited the migration of PR cells. (B) Quantitative
results are illustrated (A). (C) Transwell assay was performed in
SKBR-3/PR and MCF-7/PR cells after treatment with NT21MP and
si-SETD8. (D) Quantitative results are illustrated (C). (E) NT21MP
and si-SETD8 induced the apoptosis of SKBR-3/PR and MCF-7/PR cells.
(F) Quantitative results are illustrated (E). (G) Effects of
si-SETD8 and NT21MP on the cell cycle of SKBR-3/PR and MCF-7/PR
cells. Compared with control group, *P<0.05,
**P<0.01, ***P<0.001, compared with
NT21MP group, #P<0.05, ##P<0.01,
###P<0.001, compared with si-SETD8 group,
ΔP<0.05, ΔΔP<0.01,
ΔΔΔP<0.001. |
 | Table IIICell cycle was arrested in
G0/G1 phase treated with NT21MP and si-SETD8
in SKBR-3/PR cells. |
Table III
Cell cycle was arrested in
G0/G1 phase treated with NT21MP and si-SETD8
in SKBR-3/PR cells.
| Groups |
G0/G1 phase (%) | S phase (%) |
|---|
| Control | 51.5±1.20 | 29.6±0.64 |
| NT21MP | 63.6±0.83a | 21.9±0.49a |
| si-SETD8 | 67.6±1.11a | 16.7±0.57a |
| si-SETD+NT21MP | 78.0±0.97a,b | 12.8±0.55a,b |
 | Table IVCell cycle was arrested in
G0/G1 phase treated with NT21MP and si-SETD8
in MCF-7/PR cells. |
Table IV
Cell cycle was arrested in
G0/G1 phase treated with NT21MP and si-SETD8
in MCF-7/PR cells.
| Groups |
G0/G1 phase (%) | S phase (%) |
|---|
| Control | 19.0±0.31 | 60.0±0.55 |
| NT21MP | 66.7±1.20a | 17.2±0.43a |
| si-SETD8 | 60.7±1.11a | 23.3±0.59a |
|
si-SETD8+NT21MP | 87.0±1.25a,b | 10.7±0.50a,b |
Synergistic effect of combined miR-335
and SETD8 siRNA
We explored whether combination use of si-SETD8 and
overexpression of miR-335 had a synergistic or additive effect. As
shown in Fig. 8, western blot
analysis was performed to detect the expression of SETD8 and its
downstream target genes, combination use of miR-335 mimics and
si-SETD8 significantly enhanced the function of individual
treatment group in both SKBR-3/PR and MCF-7/PR cells. To further
detect the correlations and to determine the functional
consequences of this combination, cell migration and invasion
analysis were performed after treatment with miR-335 mimics and
si-SETD8. Results showed that combination of both enhanced the
inhibition compared with individual treatment group in both PR
cells (Fig. 9A–D). Furthermore,
combination of both treatments also showed a synergistic effect in
cell apoptosis and accumulated cell cycle into
G0/G1 phase (Fig. 9 and Tables V and VI).
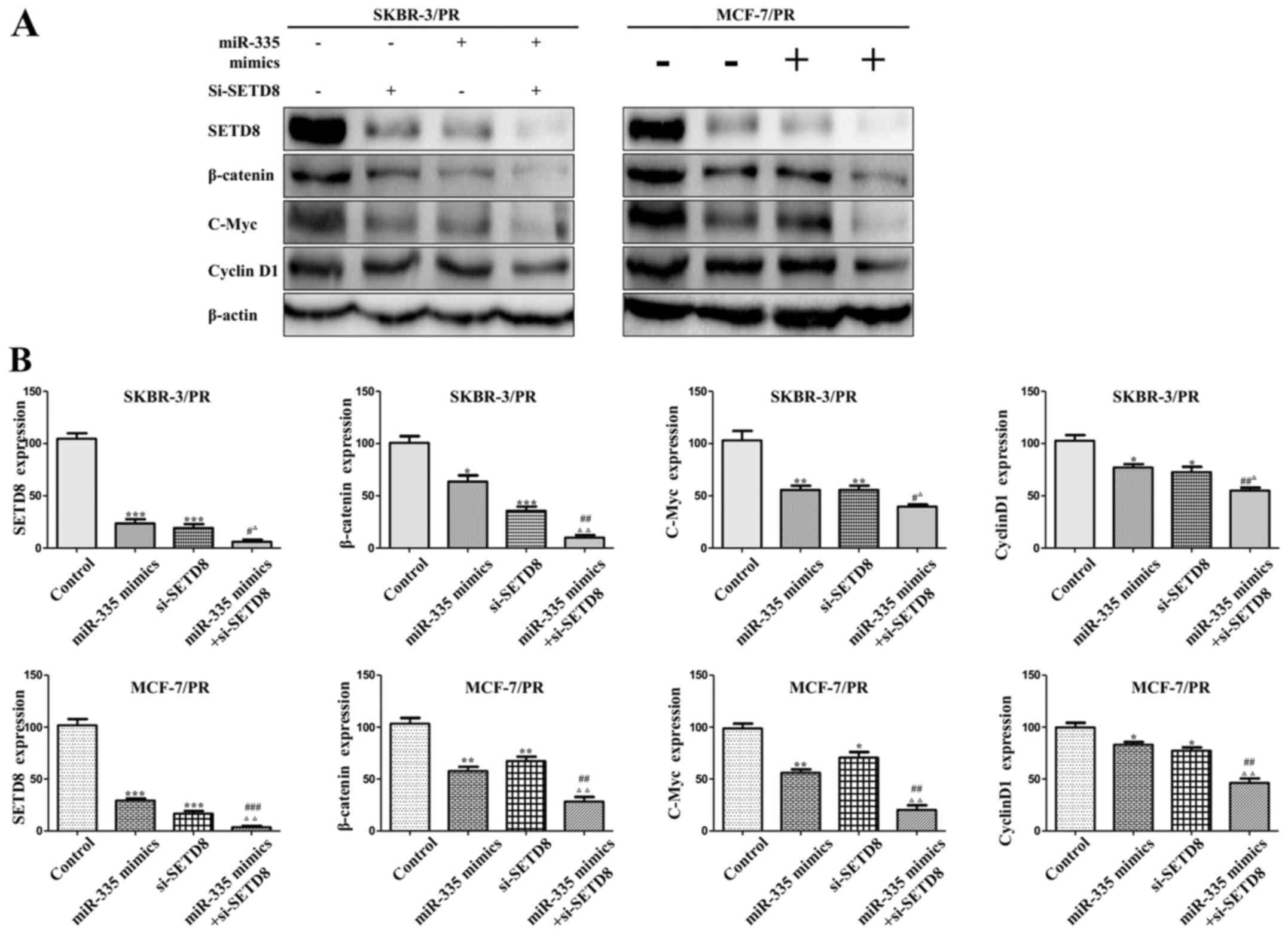 | Figure 8Combination of miR-335 mimics and
si-SETD8 treatment shows enhanced effect of SETD8 inhibition. (A)
Western blot analysis was performed in SKBR-3/PR and MCF-7/PR cells
treated with miR-335 mimics and si-SETD8. (B) Quantitative results
are illustrated (A). Compared with control group,
*P<0.05, **P<0.01,
***P<0.001, compared with miR-335 mimics group,
#P<0.05, ##P<0.01,
###P<0.001, compared with si-SETD8 group,
ΔP<0.05, ΔΔP<0.01,
ΔP<0.001. |
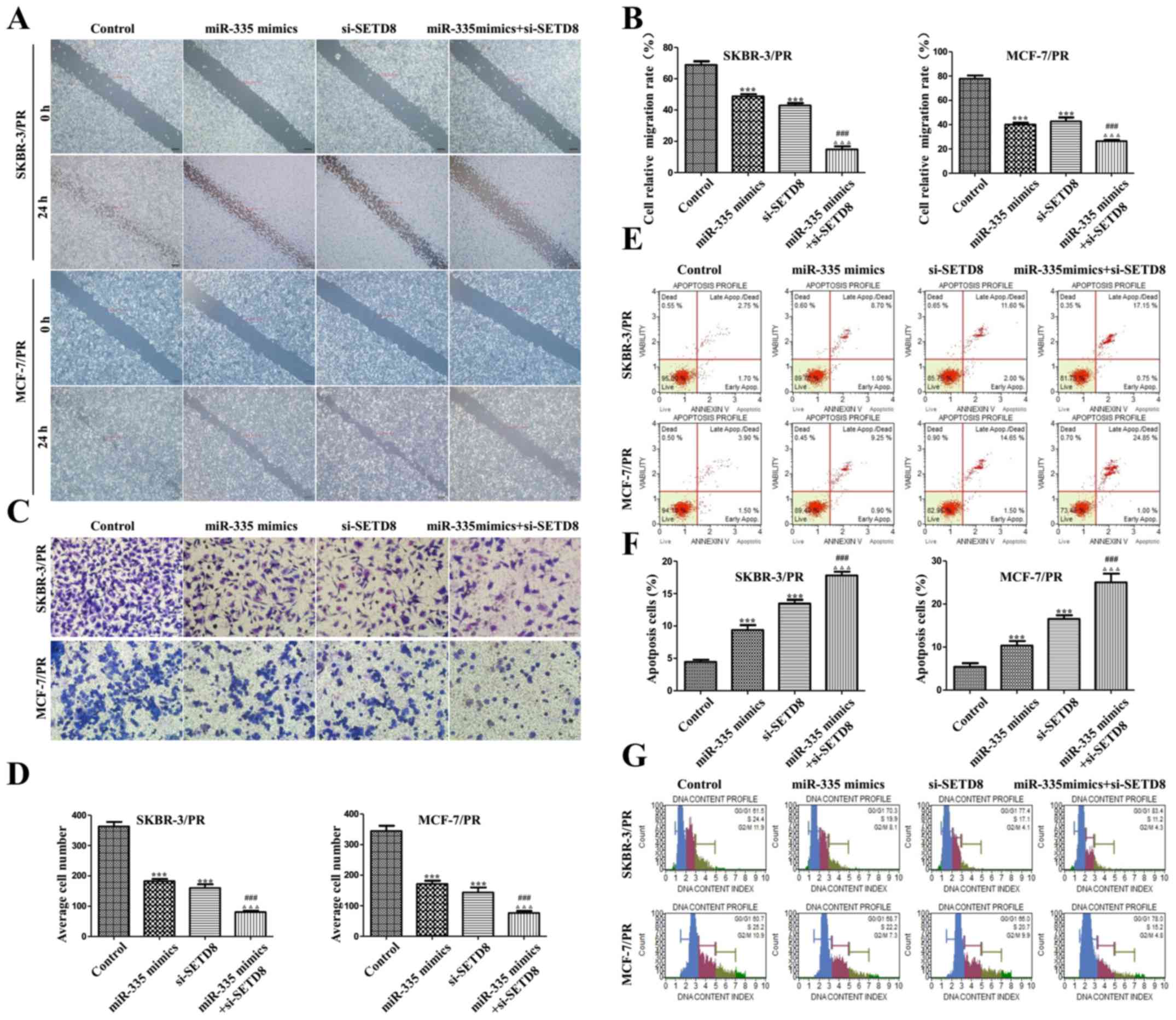 | Figure 9Combination of miR-335 mimics and
si-SETD8 treatment inhibits cell biological activity in PR cells.
(A) miR-335 mimics and si-SETD8 inhibited the migration of PR
cells. (B) Quantitative results are illustrated (A). (C) Transwell
assay were performed in SKBR-3/PR and MCF-7/PR cells after
treatment with miR-335 mimics and si-SETD8. (D) Quantitative
results are illustrated (C). (E) miR-335 mimics and si-SETD8
induced the apoptosis of SKBR-3/PR and MCF-7/PR cells. (F)
Quantitative results are illustrated (E). (G) Effects of si-SETD8
and miR-335 mimics on the cell cycle of SKBR-3/PR and MCF-7/PR
cells. Compared with control group, *P<0.05,
**P<0.01, ***P<0.001, compared with
NT21MP group, #P<0.05, ##P<0.01,
###P<0.001, compared with si-SETD8 group,
ΔP<0.05, ΔΔP<0.01,
ΔΔΔP<0.001. |
 | Table VCell cycle was arrested in
G0/G1 phase treated with miR-335 mimics and
si-SETD8 in SKBR-3/PR cells. |
Table V
Cell cycle was arrested in
G0/G1 phase treated with miR-335 mimics and
si-SETD8 in SKBR-3/PR cells.
| Groups |
G0/G1 phase (%) | S phase (%) |
|---|
| Control | 61.5±0.26 | 24.4±0.41 |
| miR-335 mimics | 70.3±0.43a | 19.9±0.81a |
| si-SETD8 | 77.4±0.28a | 17.1±0.14a |
| miR-335
mimics+si-SETD8 | 83.4±0.74a,b | 11.2±0.63a,b |
 | Table VICell cycle was arrested in
G0/G1 phase treated with miR-335 mimics and
si-SETD8 in MCF-7/PR cells. |
Table VI
Cell cycle was arrested in
G0/G1 phase treated with miR-335 mimics and
si-SETD8 in MCF-7/PR cells.
| Groups |
G0/G1 phase (%) | S phase (%) |
|---|
| Control | 60.7±0.19 | 25.2±0.85 |
| miR-335 mimics | 68.7±0.51a | 22.2±0.56a |
| si-SETD8 | 66.0±0.63a | 20.7±0.66a |
| miR-335
mimics+si-SETD8 | 78.0±0.41a,b | 15.2±0.24a,b |
Discussion
A number of studies have demonstrated that miRNAs
are involved in the occurrence and development of breast cancer.
Experimental evidence has demonstrated that dysregulation of
miR-335 play an important role in breast cancer progression and
metastasis (34). For instance,
miR-335 inhibits proliferation, cell cycle progression, colony
formation and invasion via targeting PAX6 in breast cancer cells
(35). A study showed that miR-335
inhibits migration of breast cancer cells through targeting
oncoprotein c-met (36). Some
studies have demonstrated that miR-335 is a suppressor of breast
cancer metastasis, loss of miR-335 leads to the activation of SOX4
and TNC (encoding tenascin C), which are responsible for the
acquisition of metastatic properties (37,38),
additionally, miR-335 is directly associated with relapse.
Interestingly, Png et al identified miR-335 as a robust
inhibitor of breast cancer reinitiation (39), and miR-335 acts as a metastasis
suppressor, as a decrease in its expression could be connected to
the ability to form breast cancer metastasis in mice (40). In this study, we compared the
expression of two paclitaxel sensitive and resistant cell line
pairs and identified miR-335 as a miRNA that can modulate the
sensitivity to paclitaxel in breast cancer cells.
Nowadays, studies increasingly focus on the
regulation function of individual miRNAs on cancer cells, but only
for a very small fraction, their targets are experimentally
affirmed. In our study, we explored the molecular mechanism
underlying the function of miR-335 and found that SETD8 is one of
the target gene. SETD8 (also known as SET8, PR-SET7 or KMT5A),
first characterized in 2002, is the only known PKMT (protein lysine
methyltransferase) that catalyzes the monomethylation of histone H4
lysine 20 (H4K20) (41–43). Several studies have demonstrated
that SETD8 is involved in cell cycle regulation, invasion cell
maturation and survival (44,45),
SETD8 also plays an important role in the progress of human cancer.
For instance, Zhang et al found that miR-127-3p inhibits
proliferation and invasion by targeting SETD8 in human osteosarcoma
cells (46). Moreover, high
expression of SETD8 was associated with a shorter survival time in
gastric cancer patients, and SETD8 was found to be an independent
predictor of gastric cancer outcome (47). Additionally, modulation of SETD8 is
associated with breast cancer (48–50).
In line with these observations, our data showed that SETD8 was
also necessary for the migration and invasion activity and PR of
breast cancer cells, co-treatment with miR-335 mimics and si-SETD8
significantly enhanced the effect compared with individual
treatment group, which is mediated by Wnt/β-catenin signaling.
Recent studies revealed that Wnt signaling plays a
critical role in a wide range of biological and pathophysiological
processes (51,52). Accumulating evidence has suggested
that SETD8 was critically involved in Wnt/β-catenin signaling.
Knockdown of SETD8 reduces H4K20mel, and consequently, several of
the tested Wnt target genes show reduced activation (53) and it was plausible to test whether
SETD8 is recruited through interaction with β-catenin. Li et
al reported that SETD8 is a Wnt signaling mediator and is
recruited by LEF1/TCF4 to regulate the transcription of
Wnt-activated genes (54).
Additionally, reduced PPARγ-setd8-H4K20mel would be associated with
reduced Wnt signaling genes β-catenin, and Wnt target gene Axin2
(55). In line with the role of
miRNAs in regulating Wnt/β-catenin, we found that upregulation
miR-335 could decrease β-catenin expression in PR cells. In
particular, knockdown of SETD8 by siRNA displays a phenomenon with
the effect of miR-335 in PR cells.
As β-catenin is a critical component of the
well-studied Wnt/β-catenin signaling pathway, when Wnt/β-catenin
signaling is activated, β-catenin degradation is inhibited. Cyclin
D1 is a cycle related gene which can promote the cell cycle from
G1-S phase transition period at the downstream of
Wnt/β-catenin signaling pathway (56). Boonmuen et al also
demonstrated that Wnt/β-catenin signaling target genes include
c-Myc, cyclin D1 and Axin2 in MCF-7 and MDA-MB-231 cells (57). Thus, our findings suggested that
SETD8 as a target gene of miR-335, may represent a promising
therapeutic target of PR breast cancer cells through Wnt signaling
and its downstream genes c-Myc and cyclin D1. However, the
mechanism by which SETD8 regulates Wnt/β-catenin signaling activity
remains to be investigated.
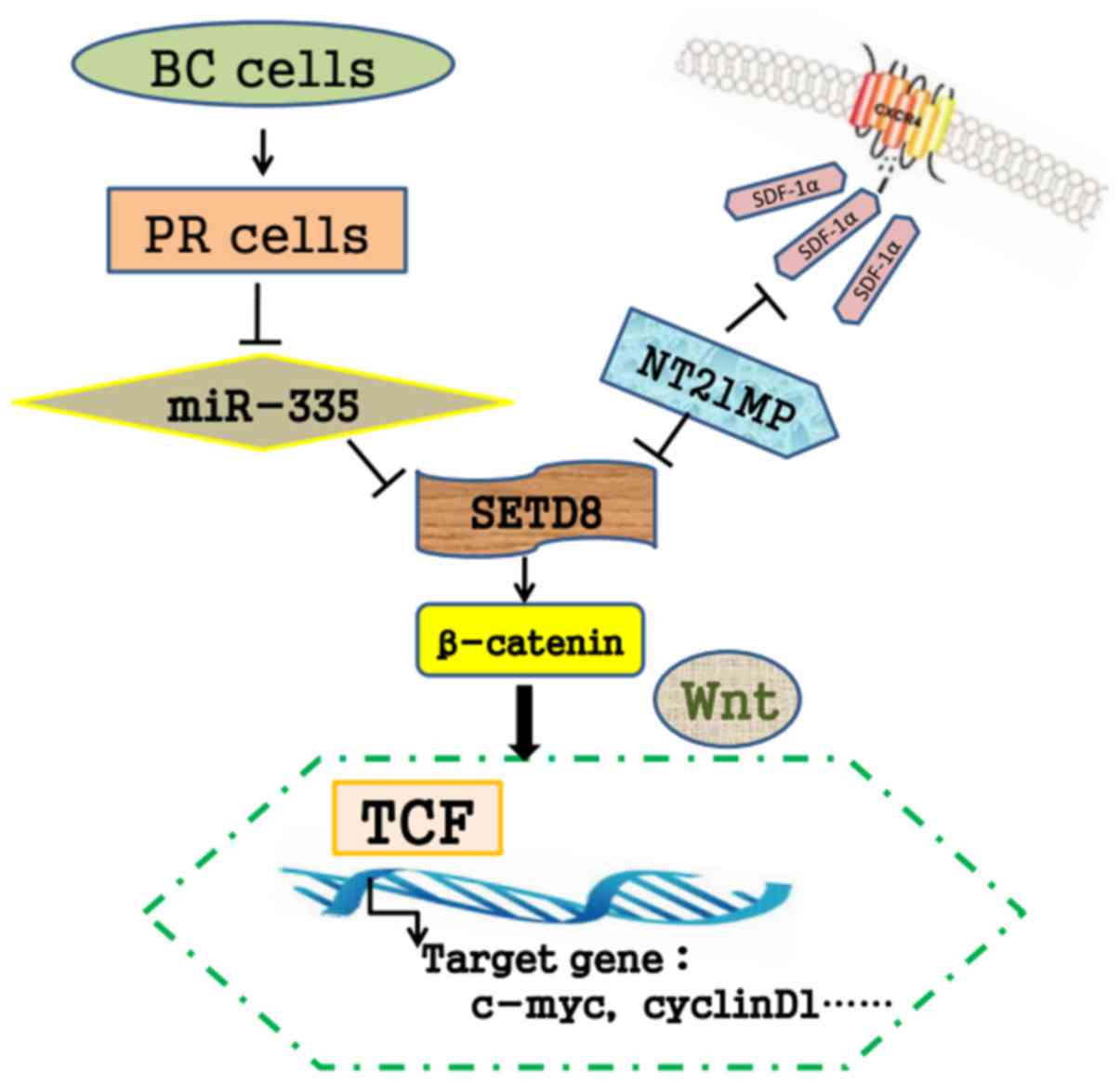
We previously synthesized NT21MP and demonstrated
that NT21MP could inhibit cellular proliferation, promote apoptosis
and inhibit the progression of breast cancer in vivo
(29). As shown in Fig. 10, our results showed that SDF-1α
acting at CXCR4 induced the downregulation of miR-335, NT21MP as a
potent antagonist could attenuates these changes. The combined use
of NT21MP and overexpression of miR-335 or silence of SETD8
contributes to the reduction of PR breast cancer cell migration and
invasion, influencing the proportion of apoptotic cells and the
cell cycle.
To this end, our data suggested that NT21MP is a
potent antagonist, and miR-335 level might serve as a potential
biomarker of PR breast cancer and that miR-335 overexpression might
aid in overcoming breast cancer cell drug resistance. Our findings
revealed that miR-335 might function as an important miRNAs in
paclitaxel resistance by regulating SETD8 and could serve as a
promising candidate for therapeutic intervention.
In conclusion, we have identified that miR-335 can
regulate breast cancer paclitaxel-resistance through its direct
target gene SETD8 and Wnt/β-catenin signaling pathway. Both miR-335
and SETD8 may be used to predict the prognosis of patients with
drug resistance of breast cancer. NT21MP, as a small molecules
compound may represent new therapeutic strategy of breast cancer.
However, more investigation needs to be carried out to illustrate
how SETD8 regulates Wnt signaling, the potential therapeutic role
of miRNAs and NT21MP remains a major clinical challenge.
Acknowledgments
This study was supported by funding from the Major
Program of Anhui Educational Committee (nos. KJ2015ZD29 and
KJ2016SD37), the Natural Science Foundation of Anhui (no.
1508085MH159), the Key Program of college discipline (major)
top-notch talent academic subsidy of Anhui (no. gxbjZD2016069), the
grant from a Key Program of Anhui Educational Committee
(KJ2016A474), the Program for science research of Bengbu Medical
College (no. BYKY14147ZD), the Program for graduates research of
Bengbu Medical College (no. Byycx1615) and the Bengbu municipal
scientific research Key projects (no. 20150309).
References
|
1
|
Rebecca L, Siegel MPH, Kimberly D and
Miller MPH: Issue Information. CA Cancer J Clin. 67:1–85. 2017.
View Article : Google Scholar
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zardavas D, Baselga J and Piccart M:
Emerging targeted agents in metastatic breast cancer. Nat Rev Clin
Oncol. 10:191–210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang X, Jin L, Cao P, Cao K, Huang C, Luo
Y, Ma J, Shen S, Tan M, Li X, et al: MicroRNA-16 sensitizes breast
cancer cells to paclitaxel through suppression of IKBKB expression.
Oncotarget. 7:23668–23683. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Armat M, Oghabi Bakhshaiesh T, Sabzichi M,
Shanehbandi D, Sharifi S, Molavi O, Mohammadian J, Saeid Hejazi M
and Samadi N: The role of Six1 signaling in paclitaxel-dependent
apoptosis in MCF-7 cell line. Bosn J Basic Med Sci. 16:28–34.
2016.PubMed/NCBI
|
|
6
|
Mirnezami AH, Pickard K, Zhang L, Primrose
JN and Packhamå G: MicroRNAs: Key players in carcinogenesis and
novel therapeutic targets. Eur J Surg Oncol. 35:339–347. 2009.
View Article : Google Scholar
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Verghese ET, Hanby AM, Speirs V and Hughes
TA: Small is beautiful: microRNAs and breast cancer-where are we
now? J Pathol. 215:214–221. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu Y, Qiu Y, Yagüe E, Ji W, Liu J and
Zhang J: miRNA-205 targets VEGFA and FGF2 and regulates resistance
to chemotherapeutics in breast cancer. Cell Death Dis. 7:e22912016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu J, Zhou Q and Tan S: Targeting miRNAs
associated with surface expression of death receptors to modulate
TRAIL resistance in breast cancer. Cancer Lett. 383:154–160. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Raza U, Saatci Ö, Uhlmann S, Ansari SA,
Eyüpoğlu E, Yurdusev E, Mutlu M, Ersan PG, Altundağ MK, Zhang JD,
et al: The miR-644a/CTBP1/p53 axis suppresses drug resistance by
simultaneous inhibition of cell survival and epithelial-mesenchymal
transition in breast cancer. Oncotarget. 7:49859–49877. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu X, Lv YG, Yan CY, Yi J and Ling R:
Enforced expression of hsa-miR-125a-3p in breast cancer cells
potentiates docetaxel sensitivity via modulation of BRCA1
signaling. Biochem Biophys Res Commun. 479:893–900. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu Q, Gong JP, Li J, Zhong SL, Chen WX,
Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH, et al: Down-regulation of
miRNA-452 is associated with adriamycin-resistance in breast cancer
cells. Asian Pac J Cancer Prev. 15:5137–5142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Zhong S, Xu Y, Yu D, Ma T, Chen
L, Zhao Y, Chen X, Yang S, Wu Y, et al: MicroRNA-3646 contributes
to docetaxel resistance in human breast cancer cells by
GSK-3β/β-catenin signaling pathway. PLoS One. 11:e01531942016.
View Article : Google Scholar
|
|
16
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17–5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie
T, Zhang J, Peng C, Lin Y and Chen J: MicroRNA-25 regulates
chemoresistance-associated autophagy in breast cancer cells, a
process modulated by the natural autophagy inducer
isoliquiritigenin. Oncotarget. 5:7013–7026. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen H, Wang D, Li L, Yang S, Chen X, Zhou
S, Zhong S, Zhao J and Tang J: MiR-222 promotes drug-resistance of
breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1
pathway. Gene. 596:110–118. 2017. View Article : Google Scholar
|
|
19
|
Ward A, Balwierz A, Zhang JD, Küblbeck M,
Pawitan Y, Hielscher T, Wiemann S and Sahin Ö: Re-expression of
microRNA-375 reverses both tamoxifen resistance and accompanying
EMT-like properties in breast cancer. Oncogene. 32:1173–1182. 2013.
View Article : Google Scholar
|
|
20
|
Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang
B, Min L and Liu W: miRNA-200c increases the sensitivity of breast
cancer cells to doxorubicin through the suppression of
E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 7:1579–1584.
2013.PubMed/NCBI
|
|
21
|
Shen H, Li L, Yang S, Wang D, Zhong S,
Zhao J and Tang J: MicroRNA-29a contributes to drug-resistance of
breast cancer cells to adriamycin through PTEN/AKT/GSK3β signaling
pathway. Gene. 593:84–90. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao M, Miao L, Liu M, Li C, Yu C, Yan H,
Yin Y, Wang Y, Qi X and Ren J: miR-145 sensitizes breast cancer to
doxorubicin by targeting multidrug resistance-associated protein-1.
Oncotarget. 7:59714–59726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang L, He D, Yang D, Chen Z, Pan Q, Mao
A, Cai Y, Li X, Xing H, Shi M, et al: MiR-489 regulates
chemoresistance in breast cancer via epithelial mesenchymal
transition pathway. FEBS Lett. 588:2009–2015. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Wang YW, Xing AY, Xiang S, Shi DB,
Liu L, Li YX and Gao P: Suppression of SPIN1-mediated PI3K-Akt
pathway by miR-489 increases chemosensitivity in breast cancer. J
Pathol. 239:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim Y, Kim H, Park D and Jeoung D: miR-335
targets SIAH2 and confers sensitivity to anti-cancer drugs by
increasing the expression of HDAC3. Mol Cells. 38:562–572. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang R, Lei Y, Hu B, Yang J, Fang S, Wang
Q, Li M and Guo L: WW domain binding protein 5 induces multidrug
resistance of small cell lung cancer under the regulation of
miR-335 through the Hippo pathway. Br J Cancer. 115:243–251. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang QL, Li CH, Ding YX, Chen CJ, Zhang J
and Wang H: Inhibitory effect of polypeptide to inhibit CXCR4 on
metastasis of breast cancer cell line. CTM. 20:89–92. 2008.
|
|
29
|
Yang QL, Ding YX, Chen CJ, Tang J, Zhang J
and Yang ZF: Suppression of murine breast cancer metastasis by
selective inhibition of CXCR4 by synthetic polypeptide derived from
viral macrophage inflammatory protein II. Chin Sci Bull.
55:2152–2159. 2010. View Article : Google Scholar
|
|
30
|
Yang Q, Huang J, Wu Q, Cai Y, Zhu L, Lu X,
Chen S, Chen C and Wang Z: Acquisition of epithelial-mesenchymal
transition is associated with Skp2 expression in
paclitaxel-resistant breast cancer cells. Br J Cancer.
110:1958–1967. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen S, Zhu L, Huang J, Cai Y, Lu X, Yang
Q, Wu Q, Chen C and Wang Z: Arsenic trioxide targets miR-125b in
glioma cells. Curr Pharm Des. 20:5354–5361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu Q, Wang R, Yang Q, Hou X, Chen S, Hou
Y, Chen C, Yang Y, Miele L, Sarkar FH, et al: Chemoresistance to
gemcitabine in hepatoma cells induces epithelial-mesenchymal
transition and involves activation of PDGF-D pathway. Oncotarget.
4:1999–2009. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Heidary MF, Mahmoodzadeh Hosseini H,
Mehdizadeh Aghdam E, Nourani MR, Ranjbar R, Mirnejad R and Imani
Fooladi AA: Overexpression of metastatic related microRNAs, mir-335
and mir-10b, by Staphylococcal enterotoxin B in the metastatic
breast cancer cell line. Adv Pharm Bull. 5:255–259. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang R, Dick M, Marme F, Schneeweiss A,
Langheinz A, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, Varon
R, et al: Genetic variants within miR-126 and miR-335 are not
associated with breast cancer risk. Breast Cancer Res Treat.
127:549–554. 2011. View Article : Google Scholar
|
|
35
|
Meng Y, Zou Q, Liu T, Cai X, Huang Y and
Pan J: microRNA-335 inhibits proliferation, cell-cycle progression,
colony formation, and invasion via targeting PAX6 in breast cancer
cells. Mol Med Rep. 11:379–385. 2015.
|
|
36
|
Gao Y, Zeng F, Wu JY, Li HY, Fan JJ, Mai
L, Zhang J, Ma DM, Li Y and Song FZ: MiR-335 inhibits migration of
breast cancer cells through targeting oncoprotein c-Met. Tumour
Biol. 36:2875–2883. 2015. View Article : Google Scholar
|
|
37
|
Negrini M and Calin GA: Breast cancer
metastasis: A microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Png KJ, Yoshida M, Zhang XH, Shu W, Lee H,
Rimner A, Chan TA, Comen E, Andrade VP, Kim SW, et al: MicroRNA-335
inhibits tumor reinitiation and is silenced through genetic and
epigenetic mechanisms in human breast cancer. Genes Dev.
25:226–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Erturk E, Cecener G, Egeli U, Tunca B,
Tezcan G, Gokgoz S, Tolunay S and Tasdelen I: Expression status of
let-7a and miR-335 among breast tumors in patients with and without
germ-line BRCA mutations. Mol Cell Biochem. 395:77–88. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nishioka K, Rice JC, Sarma K,
Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P,
Tempst P, Steward R, et al: PR-Set7 is a nucleosome-specific
methyltransferase that modifies lysine 20 of histone H4 and is
associated with silent chromatin. Mol Cell. 9:1201–1213. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fang J, Feng Q, Ketel CS, Wang H, Cao R,
Xia L, Erdjument-Bromage H, Tempst P, Simon JA and Zhang Y:
Purification and functional characterization of SET8, a nucleosomal
histone H4-lysine 20-specific methyltransferase. Curr Biol.
12:1086–1099. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Beck DB, Oda H, Shen SS and Reinberg D:
PR-Set7 and H4K20me1: At the crossroads of genome integrity, cell
cycle, chromosome condensation, and transcription. Genes Dev.
26:325–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
DeVilbiss AW, Sanalkumar R, Hall BD,
Katsumura KR, de Andrade IF and Bresnick EH: Epigenetic
determinants of erythropoiesis: Role of the histone
methyltransferase SetD8 in promoting erythroid cell maturation and
survival. Mol Cell Biol. 35:2073–2087. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Malik J, Getman M and Steiner LA: Histone
methyltransferase Setd8 represses Gata2 expression and regulates
erythroid maturation. Mol Cell Biol. 35:2059–2072. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang J, Hou W, Chai M, Zhao H, Jia J, Sun
X, Zhao B and Wang R: MicroRNA-127-3p inhibits proliferation and
invasion by targeting SETD8 in human osteosarcoma cells. Biochem
Biophys Res Commun. 469:1006–1011. 2016. View Article : Google Scholar
|
|
47
|
Shi XL, Guo ZJ, Wang XL, Liu XL and Shi
GF: SET8 expression is associated with overall survival in gastric
cancer. Genet Mol Res. 14:15609–15615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Song F, Zheng H, Liu B, Wei S, Dai H,
Zhang L, Calin GA, Hao X, Wei Q, Zhang W, et al: An miR-502-binding
site single-nucleotide polymorphism in the 3′-untranslated region
of the SET8 gene is associated with early age of breast cancer
onset. Clin Cancer Res. 15:6292–6300. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu N, Huangyang P, Yang X, Han X, Yan R,
Jia H, Shang Y and Sun L: microRNA-7 suppresses the invasive
potential of breast cancer cells and sensitizes cells to DNA
damages by targeting histone methyltransferase SET8. J Biol Chem.
288:19633–19642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang F, Sun L, Li Q, Han X, Lei L, Zhang H
and Shang Y: SET8 promotes epithelial-mesenchymal transition and
confers TWIST dual transcriptional activities. EMBO J. 31:110–123.
2012. View Article : Google Scholar
|
|
51
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schotta G: H4K20 monomethylation faces the
WNT. Proc Natl Acad Sci USA. 108:3097–3098. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Z, Nie F, Wang S and Li L: Histone H4
Lys 20 monomethylation by histone methylase SET8 mediates Wnt
target gene activation. Proc Natl Acad Sci USA. 108:3116–3123.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ke X, Xing B, Yu B, Yu X, Majnik A, Cohen
S, Lane R and Joss-Moore L: IUGR disrupts the
PPARγ-Setd8-H4K20me(1) and Wnt signaling pathways in the juvenile
rat hippocampus. Int J Dev Neurosci. 38:59–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tamamori-Adachi M, Ito H,
Sumrejkanchanakij P, Adachi S, Hiroe M, Shimizu M, Kawauchi J,
Sunamori M, Marumo F, Kitajima S, et al: Critical role of cyclin D1
nuclear import in cardiomyocyte proliferation. Circ Res.
92:e12–e19. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Boonmuen N, Thongon N, Chairoungdua A,
Suksen K, Pompimon W, Tuchinda P, Reutrakul V and Piyachaturawat P:
5-Acetyl goniothalamin suppresses proliferation of breast cancer
cells via Wnt/β-catenin signaling. Eur J Pharmacol. 791:455–464.
2016. View Article : Google Scholar : PubMed/NCBI
|