Introduction
Ovarian cancer is the eighth most frequent tumor
type in women worldwide, with 225,500 new cases and 140,200 deaths
each year. There is almost no difference in mortality between
developed and developing countries (1). Epithelial ovarian cancer comprises
the majority of malignant ovarian tumors. Borderline epithelial
tumors have been classified as a separate category, although their
origin is still unclear. Their clinicopathological features are
between the benign cystadenomas and malignant cystadenocarcinomas
(2). Borderline tumors and
low-grade serous carcinomas are associated with KRAS and
BRAF mutations. High-grade serous carcinomas are associated
with loss of BRCA1 or BRCA2, and with TP53
mutations, but no BRAF and rarely KRAS mutations.
This suggests that low-grade and high-grade serous carcinoma arise
via different genetic pathways (3). High BIRC5 gene expression
(Survivin) is often detected in various tumors, and is correlated
with poor outcome. Survivin protein expression correlates with
progression of epithelial ovarian carcinomas (4). Epithelial ovarian cancer shows a
diverse pattern of genomic rearrangements which lead to altered
gene expression and upregulation of cancer pathways, including
Hedgehog signaling (5).
Hedgehog signaling pathway is a developmental
pathway involved in formation of various tissues and organs. In
mammals, the signal transduction is triggered by binding of the
ligand Hedgehog (HH) to the transmembrane receptor Patched (PTCH).
This leads to internalization of PTCH and exposure of the protein
Smoothened (SMO) on the cell surface. SMO triggers a cytoplasmic
phosphorylation cascade leading to the release of transcription
factor GLI (GLI1-3) from Suppressor of Fused (SUFU) and
translocation of GLI to the nucleus, where it triggers
transcription of genes involved in cell proliferation. In the
absence of the HH signal, the cascade is inactive and GLI proteins
are retained in the cytoplasm and subsequently degraded in the
proteasome (6). GLI proteins are
regulated by SUFU, protein kinase A (PKA), glycogen synthase
kinase-3β (GSK3β) and casein kinase 1 (CK1). The role of GSK3β in
the Hedgehog signaling pathway is 2-fold: it phosphorylates GLI
proteins, tagging them for degradation and processing into
repressor forms, but it can also elicit a positive effect on the
signal transduction in ligand-stimulated cells, binding to SUFU and
enabling the release of GLI from the complex (7). Genes activated by the Hedgehog
pathway are involved in cell cycle regulation, proliferation,
adhesion, epithelial-mesenchymal transition, self-renewal and
pathway autoregulation. A recent study by Brun et al
implicated Survivin in Hedgehog-driven medulloblastoma (8), and a study by vlčková et al
demonstrated a direct link between Survivin expression and Hedgehog
signaling, where expression of Survivin was directly regulated by
GLI2 and GLI3 proteins (9).
Therefore, Survivin is considered a novel Hedgehog pathway target
in a large panel of tumor cell lines (9).
The Hedgehog signaling pathway is involved in
embryonic development and maturation of Drosophila ovaries.
In the adult fly it is involved in division of somatic stem cells
which give rise to follicle cells surrounding the gamete.
Inactivation of the pathway causes a reduction in follicle
production, while hyperactivation causes proliferation of somatic
cells between egg chambers (10).
In mice, activation of the pathway by Sonic Hedgehog (SHH) protein
increases growth and proliferation in cultured granulosa cells, and
cyclopamine was shown to prevent this induction. Russell et
al claim that many of the Hedgehog pathway genes are expressed
in immature and adult murine ovaries, granulosa cells and corpora
luteum: IHH, SHH, DHH, PTCH1, PTCH2 and SMO; while
GLI1 and HHIP are expressed in all ovarian tissues
(11). Another more recent study
by Huang et al indicates that Hedgehog signaling is inactive
in fetal, but active in the adult mouse ovary (12). A paracrine model of signaling was
suggested in the ovary: Hedgehog target genes GLI1 and
PTCH1 are primarily expressed in theca cells, while the
ligand is produced in the granulosa cells (13,14).
According to most authors Hedgehog pathway is
activated in ovarian carcinomas, where the protein or gene
expression is higher than in healthy tissue or benign tumors
(15–18). Other authors report Hedgehog
pathway protein expression in a subset of tumors, but claim it is
not a frequent event in ovarian cancer (19). The authors mostly focus on the four
most frequently examined components of the pathway: SHH, PTCH1, SMO
and GLI1. In healthy human ovaries, the Hedgehog pathway
genes/proteins are usually not expressed (15,17).
Expression of GLI1 and PTCH1 correlates with poor clinical outcome
of ovarian carcinoma patients (16,20).
Cell culture experiments on ovarian cancer cell lines show that
inhibition of Hedgehog signaling leads to inhibition of
proliferation, migration and invasion and to apoptosis (15–17,21,22).
It has also been demonstrated that inhibition of Hedgehog signaling
on the level of GLI1 inhibits the growth of ovarian cancer
xenografts (17,23,24),
and growth of ovarian cancer spheroids (25).
We and others have shown that loss of heterozygosity
(LOH) of the PTCH1 gene is a frequent event in various
malignant and benign tumors of the ovary (26–30).
Benign tumors, such as dermoids and fibromas, also show methylation
of the PTCH1 promoter (31), but this does not seem to be the
case for ovarian carcinoma (32).
In this study, we confirm that the Hedgehog
signaling pathway is active in ovarian tumors, and that GLI1
and SUFU are associated with tumor type and FIGO stage. It
seems that the activation occurs downstream of the membrane
components of the pathway, suggesting non-canonical activation. We
also demonstrate the activity in a primary tumor cell line
developed from the ovarian tumor sample, which seems to be a
superior model for study of the pathway compared to the established
lines.
Materials and methods
Sample collection
Twenty-three samples of ovarian tumors [16
carcinomas (CA), 7 atypical proliferative (borderline) tumors
(BDL)] and tissue from 9 healthy ovaries (OV) and 9 healthy
fallopian tubes (FT) excised for reasons other than malignant
transformation, were collected at the Department of Obstetrics and
Gynaecology, University Hospital Centre Zagreb, School of Medicine,
University of Zagreb. Of the 16 carcinoma samples, 3 were diagnosed
at stage I, 2 at stage II, 7 at stage III and 4 at stage IV
according to the International Federation of Gynecology and
Obstetrics (FIGO) classification. Of 16 carcinomas, 4 were grade 1
and 12 were grade 3. By type, 12 were serous adenocarcinoma and 4
were clear cell carcinoma. Of the 7 borderline tumors, 1 was
diagnosed at stage III and the remaining 6 were at stage I of FIGO
classification. All patients gave their informed consent before the
samples were taken, and samples were collected with the approval of
the hospital's ethics committee. All tissue samples taken during
surgery were immediately placed in a vial containing 1 ml RNALater
solution (invitrogen), kept at 4°C overnight, and RNA was extracted
using TRIzol reagent (Invitrogen) the following day. RNA
concentration was determined spectrophotometrically, and quality
was checked on 1% agarose gel.
Quantitative real-time PCR (qRT-PCR)
RNA was reverse- transcribed using TaqMan Reverse
Transcription Reagents (Applied Biosystems), and qRT-PCR was
performed on a CFX96 machine (Bio-Rad) using EVAGreen dye
(Bio-Rad). Primers and conditions used were described previously
(33,34). Expression levels for each gene were
first calculated as ΔCt relative to the housekeeping gene
RPLP0. Some samples showed no expression of a specific
target after 40 cycles of qRT-PCR, but for purposes of statistical
tests the Ct for these samples was set to 40 (35). Expression relative to the healthy
fallopian tube tissue (ΔΔCt) was then calculated using the
2−ΔΔCt formula.
Cell culture experiments
Primary culture was established from a high-grade
ovarian tumor sample (FIGO IIIC) as previously described (33) and maintained in DMEM+10%FBS, as was
the human ovarian adenocarcinoma cell line SKOV-3. Both cultures
were mycoplasma-free. Cells were treated with 7.5 µM
cyclopamine (Toronto Research Chemicals), 3 ng/µl SHH
protein (a kind gift from Professor A. Kenney, USA) for 96 h, 2.5
and 5 µM GANT-61 (Selleckchem, S8075) or 5 and 10 mM lithium
chloride (LiCl, Kemika) for 48 h. For MTT assay, primary cells or
SKOV-3 cells were plated in 96-well plates, and treated the
following day. Cell proliferation was measured after 24, 48 and 72
h.
For transfection experiments, SKOV-3 cells were
seeded in 6-well plates and transfected the following day with
pcDNA4nlSMtGLI1 plasmid (a kind gift from Professor F. Aberger,
Austria) using Lipofectamine reagent (Invitrogen). RNA was
extracted 24 h post-transfection and analyzed using qRT-PCR.
For silencing experiments, cells were seeded in
6-well plates and transfected with siPTCH1 (Ambion, Silencer Select
s11442) using siPORT reagent (Ambion) in DMEM without serum. After
24 h, the transfection was repeated, and 24 h later medium was
changed to DMEM+10% FBS. For negative control, cells were
transfected with negative control siRNA (Ambion, Silencer Select
Negative Control #1 siRNA) in the same way. Cells were left to
recover for additional 24 h and then RNA was extracted.
For migration analysis, cells were starved for 24 h
in media without serum, and plated in Transwell migration chambers
with or without cyclopamine (7.5 µM) or SHH protein (3
ng/µl). Serum-containing media (10% FBS) was used as
chemoattractant in the lower chamber. Cells were fixed and stained
with crystal violet 24 h (for the primary cells) or 4 h (for
SKOV-3) after plating, since SKOV-3 were too migratory after 24 h
and all plated cells migrated across the membrane. Minimum of 5
optical fields at ×400 magnification were counted for each
experiment.
Western blotting
Total proteins were extracted using standard methods
and 50 µg was loaded on polyacrylamide gel. Primary
antibodies were rabbit polyclonal anti-GLI3 (ProteinTech,
19949-1-AP, 1:600), rabbit polyclonal anti-GLI2 (Aviva Systems
Biology, ARP31885_T100, 1:1000), rabbit polyclonal anti-PTCH1
(ProteinTech, 17520-1-AP, 1:600), mouse monoclonal anti-GSK3β (Cell
Signaling Technology, #9832, 1:1000), rabbit monoclonal
anti-phospho-Ser9 GSK3β (Cell Signaling Technology, #5558, 1:1000)
and mouse monoclonal anti-β actin (ProteinTech, 60008-1-Ig,
1:2000). Secondary antibodies were HRP-conjugated anti-rabbit
(Santa Cruz Biotechnology, sc-2370, 1:2000) and anti-mouse (GE
Healthcare, NA931V, 1:2000), and the signal was detected using
SuperSignal West Pico and Femto (thermo fisher scientific).
Immunofluorescent staining
Cells were grown on cover slides in DMEM
complemented with 10% FBS and treated with 7.5 µM
cyclopamine for 24 h. Cells were paraformaldehyde-fixed,
permeabilized with methanol and heated in epitope-retrieval
solution at 85°C for 10 min. the primary antibodies diluted 1:100
were: rabbit polyclonal anti-GLI1 (Santa Cruz Biotechnology,
sc-20687), goat polyclonal anti- SUFU (Santa Cruz Biotechnology,
sc-10933), rabbit polyclonal anti-HH (Santa Cruz Biotechnology,
sc-9024) and goat polyclonal anti-PTCH1 (Santa Cruz Biotechnology,
sc-6147). Secondary antibodies, at 1:100 dilution, were mouse
anti-goat-FITC (Santa Cruz Biotechnology, sc-2356) and donkey
anti-rabbit-TR (Santa Cruz Biotechnology, sc-2784). Nuclei were
stained with DAPI. Negative control slides were treated in the same
way omitting the primary antibody. Co-localization coefficient was
determined using the ImageJ Software v.1.45e with the
Co-localization test plugin.
Statistical analysis
D'Agostino-Pearson test was used for testing the
normality of the data distribution. Since data did not show a
Gaussian distribution, non-parametric tests were used for
statistical analyses. Kruskal-Wallis test with a post- hoc test for
pairwise comparison of subgroups according to Conover were used to
determine the association between gene expression (Ct values) and
tumor type (ovarian carcinoma or borderline), FIGO stages (I-IV) or
grade. Correlations between gene expression levels were assessed by
Spearman's rank correlation coefficient ρ and ranked according to
Colton (36). Mann-Whitney test
was used for comparing co-localization coefficients, while
Kruskal-Wallis test was used to compare treated and non-treated
values for the migration assay. Two-tailed P-values <0.05 were
considered to indicate statistically significant differences.
Statistical analyses were performed using MedCalc for Windows
v.15.8 (MedCalc Software bvba).
Results
Gene expression of Hedgehog pathway genes (PTCH1,
SMO, GLI1, GLI2, GLI3, SHH, SUFU) and tumor survival marker
Survivin (BIRC5), was examined in tumor samples (CA or BDL),
healthy ovary (OV) and healthy fallopian tube (FT). Hedgehog
pathway genes were found expressed in the majority of ovarian tumor
samples: PTCH1 (23/23, 100%), SMO (23/23, 100%),
GLI1 (17/23, 73.9%), GLI2 (17/23, 73.9%), GLI3
(23/23, 100%), SHH (21/23, 91.3%) and SUFU (22/23,
95.7%). GLI1 was not detected in OV or FT, while all the
other genes were detected in both control tissues. BIRC5
expression was used as a tool for deciding on appropriate control
tissue, since it is differentially expressed in healthy tissue
compared to tumor tissue (4) and
its downregulation has potential application in ovarian tumor
therapy (37). BIRC5 was
upregulated in both tumor types (22/23 samples, 95.7%), as well as
in healthy ovaries, compared to the fallopian tube tissue where it
was not detected. Therefore we decided that fallopian tube
represents a better control tissue for ovarian carcinomas, which
also corresponds to recent literature data (38). GLI1 and SUFU were the
only differentially expressed genes. Expression of GLI1 was
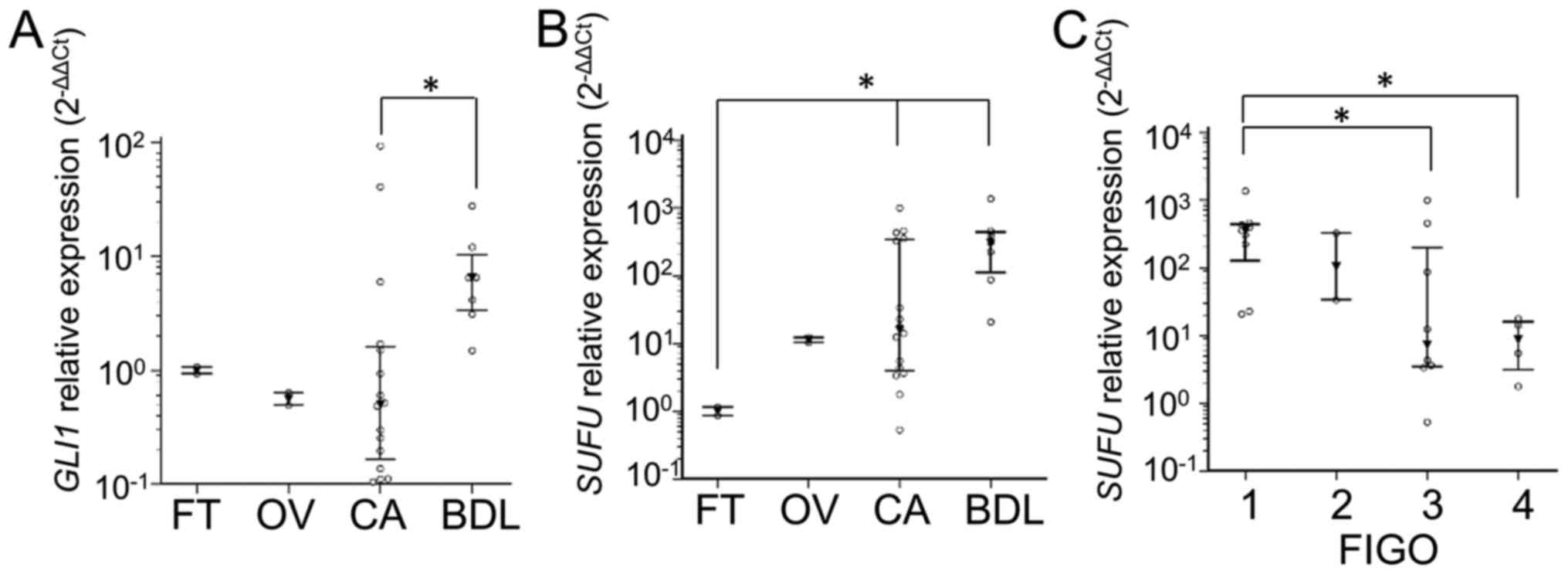
significantly higher in BDL compared to CA (P=0.042) (Fig. 1A). The same was observed for
SUFU (P= 0.025) (Fig. 1B).
When ovarian tumors were classified according to FIGO, we showed
that higher FIGO stages had lower expression of SUFU
(P=0.044) (Fig. 1C). Other tested
Hedgehog pathway genes were not associated with either tumor type
or FIGO, but similarly expressed between the categories. There was
no difference in gene expression levels in carcinoma samples
regarding the grade (data not shown).
Significant correlation was detected between the
tested genes in the dataset of ovarian carcinoma samples. Weak or
moderate correlation was detected between many components of the
Hedgehog signaling, with excellent correlation between PTCH1
and SUFU (ρ= 0.85, P<0.0001). Hedgehog signaling genes
were also positively correlated with BIRC5 expression
(GLI2, PTCH1 and SUFU). The correlation table also
confirms the negative correlation between FIGO status and
SUFU but also GLI1 and PTCH1 expression (for
details see Table I).
 | Table ICorrelation of gene expression and
FIGO stage in ovarian tumors. |
Table I
Correlation of gene expression and
FIGO stage in ovarian tumors.
| GLI2 | GLI3 | PTCH1 | SHH | SMO | SUFU | BIRC5 | FIGO | | Correlation
coefficienta |
|---|
| 0.30 | 0.26 | 0.45 | 0.61 | 0.71 | 0.39 | 0.06 | −0.42 | GLI1 | ϱ |
| 0.165 | 0.224 | 0.034 | 0.002 | 0.0002 | 0.069 | 0.795 | 0.047 | | P-value |
| 0.70 | 0.67 | 0.04 | 0.38 | 0.61 | 0.43 | −0.37 | GLI2 | ϱ |
| 0.0002 | 0.0005 | 0.851 | 0.073 | 0.002 | 0.040 | 0.081 | | P-value |
| | 0.53 | 0.04 | 0.37 | 0.42 | 0.29 | −0.20 | GLI3 | ϱ |
| | 0.010 | 0.869 | 0.084 | 0.044 | 0.187 | 0.364 | | P-value |
| | | −0.04 | 0.61 | 0.85 | 0.53 | −0.54 | PTCH1 | ϱ |
| | | 0.872 | 0.002 |
<0.0001 | 0.010 | 0.008 | | P-value |
| | | | 0.43 | 0.11 | −0.39 | −0.13 | SHH | ϱ |
| | | | 0.039 | 0.612 | 0.066 | 0.557 | | P-value |
| | | | | 0.62 | 0.22 | −0.33 | SMO | ϱ |
| | | | | 0.002 | 0.321 | 0.127 | | P-value |
| | | | | | 0.51 | −0.60 | SUFU | ϱ |
| | | | | | 0.012 | 0.002 | | P-value |
| | | | | | | −0.22 | BIRC5 | ϱ |
| | | | | | | 0.314 | | P-value |
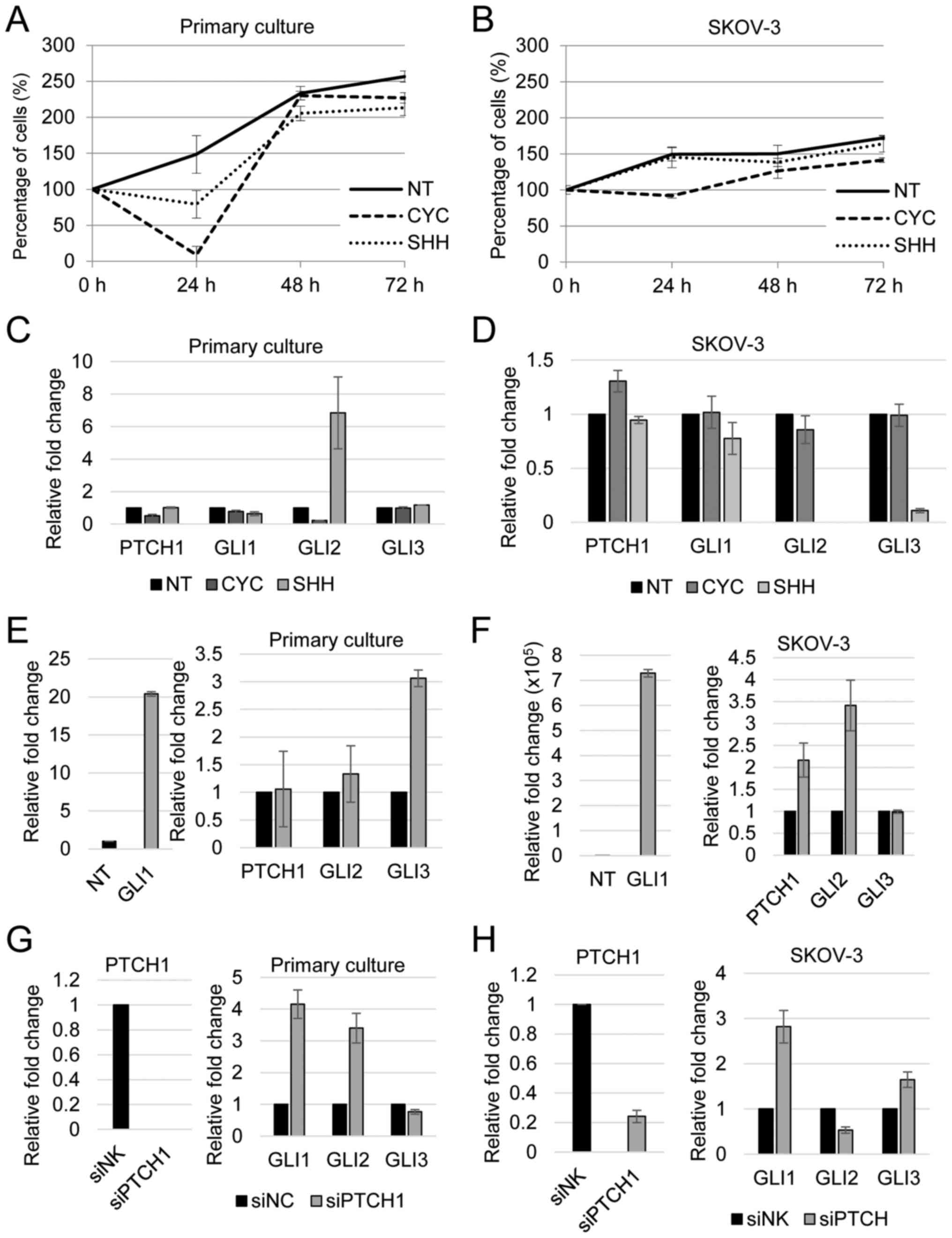
To determine if the pathway is fully functional in
ovarian tumors, we established a primary culture from one of the
tumor samples (FIGO IIIC). Cyclopamine shows an inhibitory effect
on primary cell growth 24 h after treatment, but cells quickly
recover and achieve the proliferation equal to non-treated cells
after 48–72 h (Fig. 2A). The
effect on primary culture is stronger than on the ovarian tumor
cell line SKOV-3, although the pattern is similar (Fig. 2B). SHH protein shows no additional
proliferative effect on either primary cells or the SKOV-3 cell
line (Fig. 2A and B). On gene
expression level, cyclopamine treatment of the primary culture
induces significant downregulation of Hedgehog pathway genes
PTCH1, GLI1 and GLI2, while GLI3 remains
mostly unchanged (Fig. 2C). The
gene expression level for the tested genes remains unchanged in
cylopamine-treated SKOV-3 cell line (Fig. 2D). SHH treatment of the primary
culture shows no change in gene expression of most pathway genes,
except an upregulation of GLI2 expression (Fig. 2C). The SKOV-3 cell line is also
mostly unresponsive to SHH stimulation, and even shows
downregulation of GLI2 and GLI3 genes (Fig. 2D).
To establish the effect of GLI1 upregulation, cells
were transiently transfected with GLI1. The primary culture
was difficult to transfect, and only a 20-fold increase in
GLI1 expression was achieved. This proved to be too weak to
demonstrate the effect of pathway activation, but a slight
upregulation was detected (Fig.
2E). Therefore we tested the effect of GLI1 transfection
on SKOV-3 cell line, which was easier to transfect. Apart from the
expected upregulation of GLI1, other Hedgehog signaling
pathway genes PTCH1 and GLI2, are also upregulated
after transfection with GLI1. GLI3 levels remain
unaltered, showing upregulation of Hedgehog pathway activity
(Fig. 2F).
PTCH protein is the negative regulator of Hedgehog
signaling. Therefore, PTCH1 silencing should increase
Hedgehog functional pathway activity in cells. In the primary cell
line, upon PTCH1 silencing GLI1 and GLI2 are
upregulated and GLI3 downregulated (Fig. 2G). For the SKOV-3 cell line,
GLI1 is also upregulated upon PTCH1 silencing, but
GLI2 is downregulated while GLI3 is upregulated
(Fig. 2H).
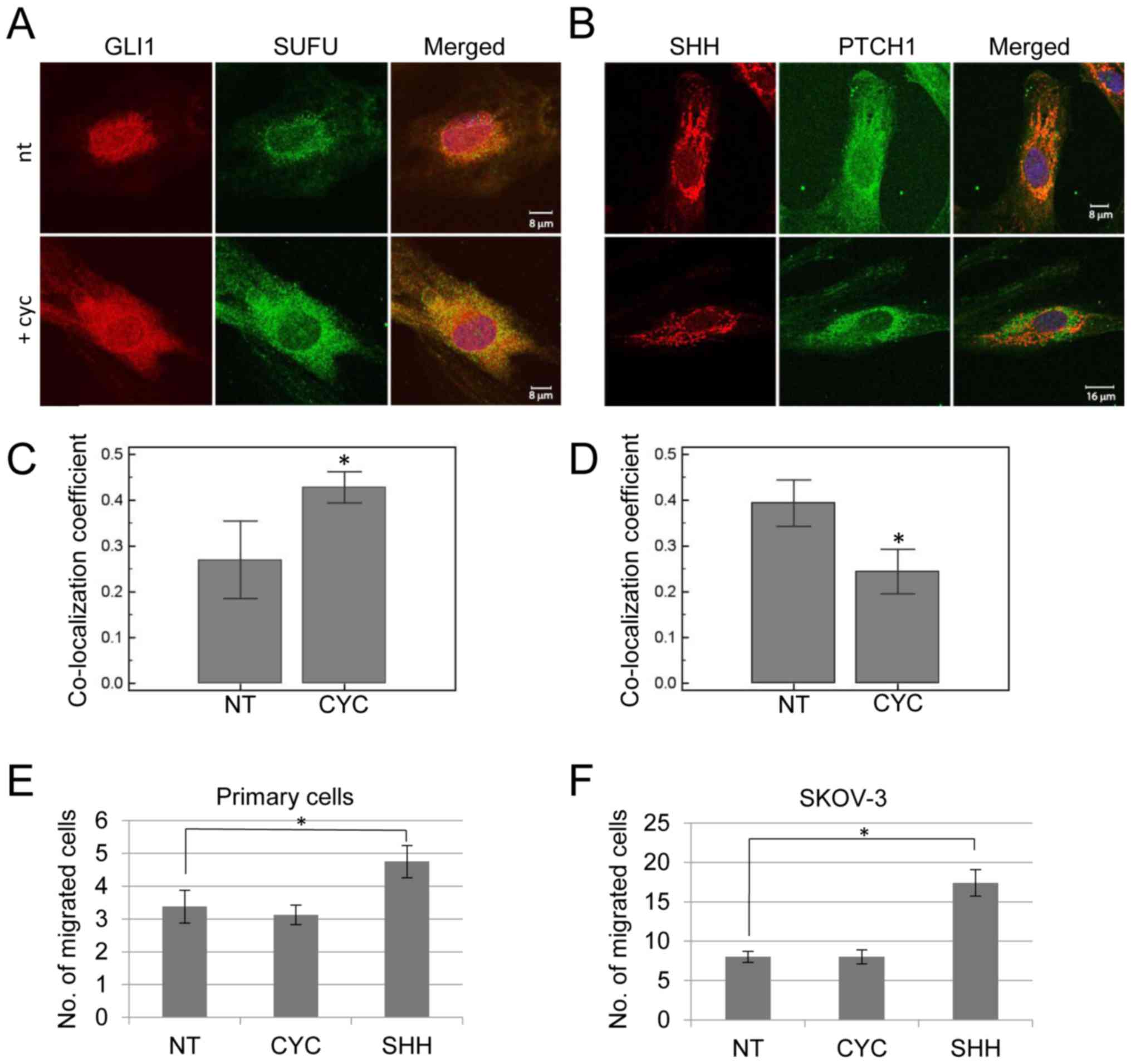
Immunofluorescent staining of the primary cells
shows nuclear localization of GLI1 in non-treated conditions, while
SUFU is located mostly at the nuclear periphery and the cytoplasm.
After treatment with cyclopamine GLI1 shows a tendency to move to
the nuclear periphery and cytoplasm and co-localizes with SUFU
(Fig. 3a), with significant
increase of the co-localization coefficient (P=0.0495) (Fig. 3C). In non- treated conditions SHH
shows cytoplasmic localization, while PTCH1 localizes to the
cytoplasm with slightly more intense staining in the region of SHH,
suggesting their interaction. After cyclopamine treatment PTCH1 is
located in different parts of the cytoplasm and ceases to
co-localize with SHH (P=0.021) (Fig.
3B and D).
Although SHH protein treatment shows no effect on
the level of gene expression of Hedgehog pathway genes, its effect
is noticeable in migration assays. SHH protein treatment results in
significant increase in cell migration in both the primary cells
and in the sKOv-3 cell line (P=0.045 and P=0.009, respectively),
while cyclopamine shows no effect on cell migration (Fig. 3E and F).
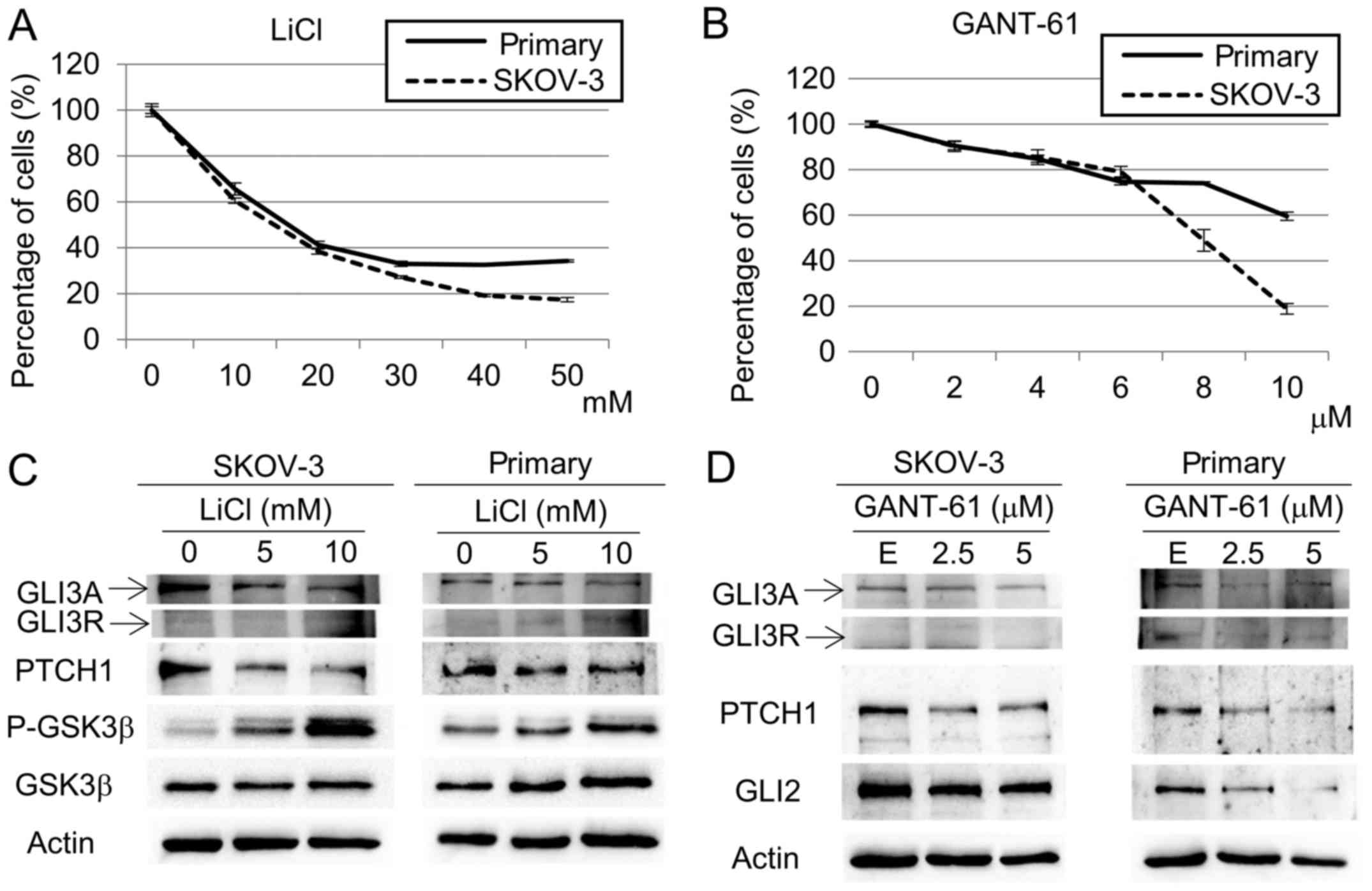
Since cyclopamine treatment affected both the
primary and SKOV-3 cell lines only in the first 24 h of treatment,
after which their proliferation rate returned to the level of
non-treated cells we decided to test for potential non-canonical
activation. We tested the responsiveness of cells to pathway
modulation down-stream of SMO. Lithium chloride is an inhibitor of
GSK3β (39), a protein kinase
involved in GLI protein processing into activator or repressor
forms, while GANT-61 is a specific GLI inhibitor. Both LiCl and
GANT-61 decrease the proliferation rates of the primary and SKOV-3
cell lines after 72 h (Fig. 4A and
B). On the protein level, it can be noted that LiCl increases
the Ser9 phosphorylated fraction of GSK3β in both cell lines as we
expected, while there is almost no change in total GSK3β levels in
either cell line (Fig. 4C). Our
laboratory has previously shown that dysregulated GSK3β can cause
upregulation of the Hedgehog pathway (40) and increasing the fraction of Ser9
phosphorylated GSK3β promotes GLI3 processing into the
transcriptional repressor form and subsequent downregulation of the
pathway. After treatment with LiCl a decrease in the levels of GLI3
activator form (GLI3A) can be noted in both cell lines. The GLI3
repressor form (GLI3R) was undetectable under these conditions in
the SKOV-3 cell line, but in the primary cell line an increase in
GLI3R levels can be observed after treatment with 10 mM LiCl
(Fig. 4C). Moreover, levels of
PTCH1, a marker of Hedgehog pathway activity, are decreased in both
cell lines after LiCl treatment, indicating signaling
downregulation (Fig. 4C). GLI
inhibition with GANT-61 causes a decrease in GLI2 protein levels in
both the primary and SKOV-3 cell line, as well as a decrease in the
levels of PTCH1 protein indicating pathway downregulation. A
decrease in GLI3A levels is detected in both cell lines, and a
decrease in GLI3R levels in the primary cell line (Fig. 4D). GLI1 is weakly expressed in both
cell lines and was undetectable on western blotting.
Discussion
Hedgehog pathway genes are expressed in the majority
of ovarian tumor samples, with GLI1 and SUFU
differing between tumor types, and SUFU between FIGO stages.
Majority of studies examining Hedgehog signaling used healthy
ovaries as control tissue, which makes sense when examining the
tumors immunohistochemically. However, ovarian carcinoma can arise
from either the fallopian tube or the ovary itself, with each
tissue giving rise to different tumor type (41). For this research we used both types
of controls: healthy ovary samples and healthy fallopian tube
samples. For our purposes FT controls seem a better control than
the OV samples, although the differences between the two control
groups were mostly not significant. The only difference between FT
and OV groups was observed for BIRC5 expression, where FT
has no expression of the gene, while OV expresses it. Since
Survivin is a protein associated with normal function of mitotic
spindle, and the ovarian tissue undergoes periodic cycles of cell
division, some expression of the protein is to be expected in
OV.
There is not much data on Hedgehog pathway
expression in the healthy human ovary. The groups that used OV as
control tissue for immunohistochemical staining report no or weak
signal of Hedgehog pathway proteins (15,17,20).
Studies on mouse ovaries show reactivation of the pathway in the
adult ovary (12). Since tissues
in the adult ovary go through periodical phases of proliferation,
it is not surprising to detect Hedgehog pathway activity. Still, we
did not detect the major effector of the pathway, GLI1, in
the OV or FT. Given that expression of PTCH1 and SMO
were detected, it is possible that in these tissues the Hedgehog
signaling is poised for reactivation, and requires a specific
stimulus to become active. The key proteins that seem to be
involved in pathway upregulation in clinical samples are GLI1 and
SUFU protein, suggesting non-canonical activation of the pathway in
ovarian tumors. The expression of pathway genes GLI2, PTCH1
and SUFU is correlated with BIRC5 expression in these
samples. This, taken together with the recent work by vlčkova et
al (9), suggests that Hedgehog
signaling regulates BIRC5 expression in ovarian tumors as
well. Our group has previously demonstrated a very similar
correlation of Hedgehog signaling with BIRC5 expression in
oropharyngeal squamous cell carcinoma (PTCH1 positively and
SHH negatively correlated with BIRC5 expression)
(34).
Vlčkova et al also demonstrated that the
major regulator of BIRC5 expression in their luciferase
model system is GLI2 protein, followed by GLI3 (9). In our samples we detect the
expression of all three GLI genes, but the only one that
seems to be associated with tumor progression is GLI1.
However, it must be taken into account that regulation of activity
of GLI proteins occurs on post-translational level, so gene
expression may not be the best choice for monitoring these effects.
Protein expression, on the other hand, is limited by quality of
antibodies currently on the market, and their inability to
distinguish between activator and repressor forms on tissue
sections.
Experiments on primary cells developed from ovarian
carcinoma tissue demonstrate that the Hedgehog signaling pathway
can be temporarily downregulated using cyclopamine, and cell
proliferation is delayed but not completely inhibited. This is
further supported by the retention of GLI1 and SUFU in the
cytoplasm and reduced PTCH1-SHH co-localization following
cyclopamine treatment. SKOV-3 cell line also shows a weak effect of
cyclopamine on cell proliferation (a slight delay), but no effect
on gene expression level. Exogenous SHH stimulation shows no direct
effect on gene expression of Hedgehog pathway genes, but it
positively affects the migratory potential of the cells. It is
possible that the SHH ligand binds to an alternative target and
stimulates cell migration, as we have shown recently on breast
cancer (42), and this effect
should be investigated further. Endogenous stimulation of the
pathway by GLI1 results in upregulation of PTCH1 and
GLI2/3. Silencing of PTCH1, a negative regulator of
the pathway, also upregulates signaling, especially GLI
transcription factors. These findings suggest that the Hedgehog
signaling in ovarian tumors is activated in a non- canonical
manner, at least partially bypassing the ligand-receptor signal and
instead activating the SUFU-GLI1 axis directly.
Inhibition of SMO by cyclopamine has a short-term
effect which blocks the canonical pathway, but the activity is
retained through the non-canonical signaling. Inhibitors targeting
GLI proteins or their regulators may be more relevant in this
context. One potential candidate compound is LiCl, which acts on
GSK3β, one of the regulators of GLI processing. It has been
demonstrated previously that GSK3β usually acts as a tumor
suppressor gene, but if it is overexpressed and dysregulated it can
have tumorigenic properties, for example in colon cancer (43,44).
This effect was also observed in ovarian cancer cells (45,46).
Herein, we observed that treatment with LiCl increases the Ser9
phosphorylated fraction of GSK3β in both cell lines. As a
consequence the levels of GLI3 protein decreased in both cell
lines. However, the effect of increased processing of GLI3 into
repressor form was only observed in the primary cell line.
Nevertheless, both cell lines showed downregulated Hedgehog
signaling, after LiCl treatment, indicated by decreased PTCH1
protein levels. The effect of GLI inhibition with GANT-61 was very
similar to the effect of LiCl. GANT-61 inhibits GLI1 and GLI2
proteins but according to literature does not affect GLI3 (47,48).
The proliferation rate of both cell lines decreased after treatment
with GANT-61, and the Hedgehog signaling pathway was downregulated.
GANT-61 significantly decreased Gli2 protein levels in both cell
lines, which in turn caused a decrease in PTCH1 protein levels.
These results indicate that the activity of Hedgehog signaling
pathway can be modulated through GSK3β inhibition in ovarian cancer
cells, as well as through direct GLI inhibition.
Downstream effectors play a role in regulation of
GLI proteins, and may be the key to successful Hedgehog pathway
modulation. For example, it has recently been shown that
diindolylmethane inhibits GLI1 and leads to anoikis of ovarian
cancer cells (24), and GLI
proteins are becoming preferred therapy targets since they can
inhibit both canonical and non- canonical Hedgehog signaling
(49). All these results signify
that the signaling pathway is fully functional in ovarian carcinoma
primary cells and could be targeted by compounds modifying Hedgehog
signaling. Finally, primary cell cultures seem to be a superior
model compared to the cell line SKOV-3. It has been shown that
ovarian cancer cell lines show great genomic instability, which
definitely affects their reliability in cell culture experiments
and explains the discrepancy in results obtained by different
research groups (50). The primary
culture more accurately reflects conditions in vivo, but has
a limited lifespan.
Acknowledgments
This work was funded by the Croatian Ministry of
Science, Education and Sports, grant no. 098-0982464-2461, the City
of Zagreb, URBROJ: 251-03-02-15-2, and the Croatian Society for
Gynecological Urology. The authors wish to thank all the patients
who participated in this study. We also thank Professor Fritz
Aberger for the GLI1 expression vector used in the study, Professor
Anna M. Kenney for the recombinant SHH protein, and Lucija Horvat,
BSc for help with confocal microscopy.
References
|
1
|
American Cancer Society: Global Cancer
Facts & Figures. 2nd Edition. American Cancer Society; Atlanta,
GA: 2011, https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/global-cancer-facts-and-figures/global-cancer-facts-and-figures-2nd-edition.pdf.
|
|
2
|
Hart WR: Borderline epithelial tumors of
the ovary. Mod Pathol. 18(Suppl 2): S33–S50. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bell DA: Origins and molecular pathology
of ovarian cancer. Mod Pathol. 18(Suppl 2): S19–S32. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takai N, Miyazaki T, Nishida M, Nasu K and
Miyakawa I: Expression of survivin is associated with malignant
potential in epithelial ovarian carcinoma. Int J Mol Med.
10:211–216. 2002.PubMed/NCBI
|
|
5
|
Hoogstraat M, de Pagter MS, Cirkel GA, van
Roosmalen MJ, Harkins TT, Duran K, Kreeftmeijer J, Renkens I,
Witteveen PO, Lee CC, et al: Genomic and transcriptomic plasticity
in treatment-naive ovarian cancer. Genome Res. 24:200–211. 2014.
View Article : Google Scholar :
|
|
6
|
Huntzicker EG, Estay IS, Zhen H, Lokteva
LA, Jackson PK and Oro AE: Dual degradation signals control Gli
protein stability and tumor formation. Genes Dev. 20:276–281. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takenaka K, Kise Y and Miki H: GSK3beta
positively regulates Hedgehog signaling through Sufu in mammalian
cells. Biochem Biophys Res Commun. 353:501–508. 2007. View Article : Google Scholar
|
|
8
|
Brun SN, Markant SL, Esparza LA, Garcia G,
Terry D, Huang JM, Pavlyukov MS, Li XN, Grant GA, Crawford JR, et
al: Survivin as a therapeutic target in Sonic hedgehog-driven
medulloblastoma. Oncogene. 34:3770–3779. 2015. View Article : Google Scholar :
|
|
9
|
Vlčková K, Ondrušová L, Vachtenheim J,
Réda J, Dundr P, Zadinová M, Žáková P and Poučková P: Survivin, a
novel target of the Hedgehog/GLI signaling pathway in human tumor
cells. Cell Death Dis. 7:e20482016. View Article : Google Scholar
|
|
10
|
Zhang Y and Kalderon D: Hedgehog acts as a
somatic stem cell factor in the Drosophila ovary. Nature.
410:599–604. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Russell MC, Cowan RG, Harman RM, Walker AL
and Quirk SM: The hedgehog signaling pathway in the mouse ovary.
Biol Reprod. 77:226–236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang C-CJ and Yao HH-C: Diverse functions
of Hedgehog signaling in formation and physiology of steroidogenic
organs. Mol Reprod Dev. 77:489–496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wijgerde M, Ooms M, Hoogerbrugge JW and
Grootegoed JA: Hedgehog signaling in mouse ovary: Indian hedgehog
and desert hedgehog from granulosa cells induce target gene
expression in developing theca cells. Endocrinology. 146:3558–3566.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Spicer LJ, Sudo S, Aad PY, Wang LS, Chun
SY, Ben-Shlomo I, Klein C and Hsueh AJ: The hedgehog-patched
signaling pathway and function in the mammalian ovary: A novel role
for hedgehog proteins in stimulating proliferation and
steroidogenesis of theca cells. Reproduction. 138:329–339. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Horiuchi A, Kikuchi N, Osada R,
Yoshida J, Shiozawa T and Konishi I: Hedgehog signal pathway is
activated in ovarian carcinomas, correlating with cell
proliferation: It's inhibition leads to growth suppression and
apoptosis. Cancer Sci. 98:68–76. 2007. View Article : Google Scholar
|
|
16
|
Liao X, Siu MKY, Au CWH, Wong ES, Chan HY,
Ip PP, Ngan HY and Cheung AN: Aberrant activation of hedgehog
signaling pathway in ovarian cancers: Effect on prognosis, cell
invasion and differentiation. Carcinogenesis. 30:131–140. 2009.
View Article : Google Scholar
|
|
17
|
Bhattacharya R, Kwon J, Ali B, Wang E,
Patra S, Shridhar V and Mukherjee P: Role of hedgehog signaling in
ovarian cancer. Clin Cancer Res. 14:7659–7666. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schmid S, Bieber M, Zhang F, Zhang M, He
B, Jablons D and Teng NNH: Wnt and hedgehog gene pathway expression
in serous ovarian cancer. Int J Gynecol Cancer. 21:975–980. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang L, He J, Huang S, Zhang X, Bian Y, He
N, Zhang H and Xie J: Activation of hedgehog signaling is not a
frequent event in ovarian cancers. Mol Cancer. 8:1122009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ciucci A, De Stefano I, Vellone VG, Lisi
L, Bottoni C, Scambia G, Zannoni GF and Gallo D: Expression of the
glioma-associated oncogene homolog 1 (gli1) in advanced serous
ovarian cancer is associated with unfavorable overall survival.
PLoS One. 8:e601452013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Q, Xu R, Zeng C, Lu Q, Huang D, Shi
C, Zhang W, Deng L, Yan R, Rao H, et al: Down-regulation of Gli
transcription factor leads to the inhibition of migration and
invasion of ovarian cancer cells via integrin β4-mediated FAK
signaling. PLoS One. 9:e883862014. View Article : Google Scholar
|
|
22
|
Steg AD, Burke MR, Amm HM, Katre AA,
Dobbin ZC, Jeong DH and Landen CN: Proteasome inhibition reverses
hedgehog inhibitor and taxane resistance in ovarian cancer.
Oncotarget. 5:7065–7080. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McCann CK, Growdon WB, Kulkarni-Datar K,
Curley MD, Friel AM, Proctor JL, Sheikh H, Deyneko I, Ferguson JA,
Vathipadiekal V, et al: Inhibition of Hedgehog signaling
antagonizes serous ovarian cancer growth in a primary xenograft
model. PLoS One. 6:e280772011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kandala PK and Srivastava SK:
Diindolylmethane-mediated Gli1 protein suppression induces anoikis
in ovarian cancer cells in vitro and blocks tumor formation ability
in vivo. J Biol Chem. 287:28745–28754. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ray A, Meng E, Reed E, Shevde LA and
Rocconi RP: Hedgehog signaling pathway regulates the growth of
ovarian cancer spheroid forming cells. Int J Oncol. 39:797–804.
2011.PubMed/NCBI
|
|
26
|
Byrom J, Mudaliar V, Redman CW, Jones P,
Strange RC and Hoban PR: Loss of heterozygosity at chromosome
9q22-31 is a frequent and early event in ovarian tumors. Int J
Oncol. 24:1271–1277. 2004.PubMed/NCBI
|
|
27
|
Tsuji T, Catasus L and Prat J: Is loss of
heterozygosity at 9q22.3 (PTCH gene) and 19p13.3 (STK11 gene)
involved in the pathogenesis of ovarian stromal tumors? Hum Pathol.
36:792–796. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Levanat S, Musani V, Komar A and Oreskovic
S: Role of the hedgehog/patched signaling pathway in oncogenesis: A
new polymorphism in the PTCH gene in ovarian fibroma. Ann NY Acad
Sci. 1030:134–143. 2004. View Article : Google Scholar
|
|
29
|
Levanat S, Kappler R, Hemmerlein B, Döring
P, Musani V, Komar A, Oreskovic S, Pavelic B and Hahn H: Analysis
of the PTCH1 signaling pathway in ovarian dermoids. Int J Mol Med.
14:793–799. 2004.PubMed/NCBI
|
|
30
|
Musani V, Sabol M, Car D, Ozretić P,
Kalafatić D, Maurac I, Orešković S and Levanat S: PTCH1 gene
polymorphisms in ovarian tumors: Potential protective role of
c.3944T allele. Gene. 517:55–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cretnik M, Musani V, Oreskovic S, Leovic D
and Levanat S: The Patched gene is epigenetically regulated in
ovarian dermoids and fibromas, but not in basocellular carcinomas.
Int J Mol Med. 19:875–883. 2007.PubMed/NCBI
|
|
32
|
Löf-Öhlin ZM, Levanat S, Sabol M, Sorbe B
and Nilsson TK: Promoter methylation in the PTCH gene in cervical
epithelial cancer and ovarian cancer tissue as studied by eight
novel Pyrosequencing® assays. Int J Oncol. 38:685–692.
2011.
|
|
33
|
Maurac I, Sabol M, Musani V, Car D,
Ozretic P, Kalafatic D, Oreskovic S, Babic D and Levanat S: A
low-grade ovarian carcinoma case with coincident LOH of PTCH1 and
BRCA1, and a mutation in BRCA1. Int J Gynecol Pathol. 31:264–271.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Leovic D, Sabol M, Ozretic P, Musani V,
Car D, Marjanovic K, Zubcic V, Sabol I, Sikora M, Grce M, et al:
Hh-Gli signaling pathway activity in oral and oropharyngeal
squamous cell carcinoma. Head Neck. 34:104–112. 2012. View Article : Google Scholar
|
|
35
|
McCall MN, McMurray HR, Land H and
Almudevar A: On non- detects in qPCR data. Bioinformatics.
30:2310–2316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Colton T: Statistics in Medicine. Little
Brown & Co; Boston: 1974
|
|
37
|
Xing J, Jia C-R, Wang Y, Guo J and Cai Y:
Effect of shRNA targeting survivin on ovarian cancer. J Cancer Res
Clin Oncol. 138:1221–1229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kurman RJ and Shih IeM: The origin and
pathogenesis of epithelial ovarian cancer: A proposed unifying
theory. Am J Surg Pathol. 34:433–443. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jope RS: Lithium and GSK-3: One inhibitor,
two inhibitory actions, multiple outcomes. Trends Pharmacol Sci.
24:441–443. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stecca B, Ruiz I and Altaba A:
Context-dependent regulation of the GLI code in cancer by HEDGEHOG
and non-HEDGEHOG signals. J Mol Cell Biol. 2:84–95. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Koshiyama M, Matsumura N and Konishi I:
Recent concepts of ovarian carcinogenesis: type I and type II.
Biomed Res Int. 2014:9342612014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sabol M, Trnski D, Uzarevic Z, Ozretic P,
Musani V, Rafaj M, Cindric M and Levanat S: Combination of
cyclopamine and tamoxifen promotes survival and migration of mcf-7
breast cancer cells - interaction of hedgehog-gli and estrogen
receptor signaling pathways. PLoS One. 9:e1145102014. View Article : Google Scholar
|
|
43
|
Shakoori A, Ougolkov A, Yu ZW, Zhang B,
Modarressi MH, Billadeau DD, Mai M, Takahashi Y and Minamoto T:
Deregulated GSK3β activity in colorectal cancer: Its association
with tumor cell survival and proliferation. Biochem Biophys Res
Commun. 334:1365–1373. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Trnski D, Sabol M, Gojević A, Martinić M,
Ozretić P, Musani V, Ramić S and Levanat S: GSK3β and Gli3 play a
role in activation of Hedgehog-Gli pathway in human colon cancer -
Targeting GSK3β downregulates the signaling pathway and reduces
cell proliferation. Biochim Biophys Acta. 1852:2574–2584. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rask K, Nilsson A, Brännström M, Carlsson
P, Hellberg P, Janson PO, Hedin L and Sundfeldt K: Wnt-signalling
pathway in ovarian epithelial tumours: Increased expression of
β-catenin and GSK3β. Br J Cancer. 89:1298–1304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hilliard TS, Gaisina IN, Muehlbauer AG,
Gaisin AM, Gallier F and Burdette JE: Glycogen synthase kinase 3β
inhibitors induce apoptosis in ovarian cancer cells and inhibit
in-vivo tumor growth. Anticancer Drugs. 22:978–985. 2011.PubMed/NCBI
|
|
47
|
Lauth M, Bergström A, Shimokawa T and
Toftgård R: Inhibition of GLI-mediated transcription and tumor cell
growth by small- molecule antagonists. Proc Natl Acad Sci USA.
104:8455–8460. 2007. View Article : Google Scholar
|
|
48
|
Tostar U, Toftgård R, Zaphiropoulos PG and
Shimokawa T: Reduction of human embryonal rhabdomyosarcoma tumor
growth by inhibition of the hedgehog signaling pathway. Genes
Cancer. 1:941–951. 2010. View Article : Google Scholar
|
|
49
|
Gonnissen A, Isebaert S and Haustermans K:
Targeting the Hedgehog signaling pathway in cancer: Beyond
Smoothened. Oncotarget. 6:13899–13913. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Elias KM, Emori MM, Papp E, MacDuffie E,
Konecny GE, Velculescu VE and Drapkin R: Beyond genomics: Critical
evaluation of cell line utility for ovarian cancer research.
Gynecol Oncol. 139:97–103. 2015. View Article : Google Scholar : PubMed/NCBI
|