Introduction
Chondrosarcoma is cancer of the cartilage that
eventually metastasize. The disease can affect multiple organs,
such as long bones, spine, pelvis, larynx and head. Conventional
therapies are not effective in this disease treatment and there is
urgency in seeking new approaches (1,2). The
signaling events resulting in mesenchymal cell transformation to
sarcoma have yet to be fully elucidated. Proline rich polypeptide
1, (PRP-1), also known as (galarmin) is produced by the brain
neurosecretory cells and comprised of 15 amino acids (3), and is a mTOR kinase (mTORC1)
inhibitor in chondrosarcoma, which causes 80–90% inhibition of
chondrosarcoma cell growth, halting G1/S phase cell cycle
progression in chondrosarcoma (4,5) and
other mesenchymal tumors (6). The
ability of PRP-1 to upregulate tumor suppressor miRNAs and
downregulate onco-miRNAs in human chondrosarcoma JJ012 cell line
was demonstrated (7). The
upregulation of most tumor suppressors in chondrosarcoma (8) including inflammation related TET1/2
and SOCS3 is one of the unique PRP-1 properties, however, it
depends on which molecular pathway these tumor suppressors are part
of (9). PRP-1 epigenetically
downregulates embryonic stem cell marker miR-302c in human
chondrosarcoma and its targets Nanog, c-Myc and Bmi1 (10). To understand better the mechanism
of PRP-1 action and its potential as therapeutic agent in the
future, it is very important to identify the receptor it binds to.
In the present study, we present evidence that PRP-1 exerts its
effect via interacting with toll like receptor family TLR1/2, TLR6
and mucin MUC5B. Innate immunity toll-like receptors (TLRs), or
pattern recognition receptors are sensitive both to endogenous and
exogenous ligands (11,12) and can be found both inside the
cells and at the cell surface. Intracellular TLRs start their
journey from the endoplasmic reticulum (ER) through the Golgi and
eventually to endolysosomes (13).
TLRs play active roles in carcinogenesis and tumor progression or
its inhibition (14,15) where the activation of TLR
signalling could regulate antitumor immunity of the host (16). The term alarmins is often used when
referring to endogenous TLR ligands. The innate immune system can
be activated by recognizing pathogen associated molecular patterns
(PAMPs). The injured cells in their turn have ability to release
danger-associated molecular patterns (DAMPs) and contribute to the
activation of innate immune system. Thus, immune system is involved
not only in fighting the infection by mobilizing the immunologic
arsenal, but also in the process of tissue repair. Hence, the term
non-infectious inflammation response, whenever TLR signaling is
mediated by endogenous ligands, which secure autoimmune disease and
tumorigenesis in addition to tissue repair and injury (17,18).
TLRs1 (cluster of differentiation 281), 2, 4, 5 and 6 are expressed
on the cell surface, whereas TLRs3, 7, 8 and 9 are intracellular
nucleic acid receptors. The ligand for TLR10 remains to be found
(19). The antitumorigenic role of
TLR2 is recognized, its deficiency led to early intestinal tumor
formation (20). Most of
endogenous TLR ligands are agonists of TLR4 and TLR2 (21). There is a reported link berween TLR
signaling andmucins (MUCs) leading to effective pathogen
elimination (22-24). Mucins are glycosylated large
extracellular proteins that are found not only in mucous cells but
also in connective tissue and goblet cells. Mucin expression
glycosylation alterations can lead to the development of cancer and
cellular transformation (25–31).
Apomucin with the attached O-linked oligosaccharides is the protein
backbone for mucin. There are 'secreted (gel-forming and
non-gel-forming)' and 'membrane-bound' mucins, with transmemebrane
domain (32). The goblet cells
from the epithelium and mucous cells from submucosal glands
generate secreted mucins. Secreted mucins on the chromosome 11p15
include MUC2, MUC5AC, MUC5B, MUC6 and MUC19. Some of the mucins can
manifest themselves as tumor suppressors, for example MUC4
(33,34). MUC5B expression has protumorigenic
(28,35) or antitumorigenic consequence for
the cell growth (36,37) and was linked both to decreased
survival or better prognosis in cancer patients correspondingly,
depending on the disease and organ specificity. MUC5B was shown to
have very beneficial effects in human airway defense (38). The epigenetic mechanism,
hypermethylation of MUC5B promoter was attributed to the silencing
of its tumor suppressor activity (39). Both overexpression and
downregulation of mucins in different organs can contribute to
cancer pathology and inflammation (26,40).
Materials and methods
PRP-1 initial isolation and chemical
synthesis
Initially, PRP-1 was isolated from the
neurosecretory granules of bovine neurohypophysis by the method
described (3,41) followed by its chemical synthesis
(42).
PRP-1 antiserum affinity chromatography
purification
Antiserum for PRP1 was generated (43), then affinity chromatography
purified, AminoLink Plus Immobilization kit instructions (44894;
Thermo Fisher Scientific, Waltham, MA, USA) were followed for
protein sample desalting with Zeba Spin columns (89891; Thermo
Fisher Scientific).
Tissue culture
The human JJ012 chondrosarcoma cell line was
received from Dr Joel Block's Laboratory (Rush University, Chicago
IL, USA). JJ012 chondrosarcoma cells were cultured as previously
described (8). The medium
composition: Dulbecco's modified Eagle's medium (DMEM),
supplemented with F12, 10% fetal bovine serum (FBS), 25
μg/ml ascorbic acid, 100 ng/ml insulin, 100 nM
hydrocortisone and 1% penicillin/streptomycin.
Brief immunocytochemistry protocol
Adherent cells were grown directly on coverslips
with 5×105 cells/coverslip in 6-well clusters, where
they were cultured overnight at 37°C in an incubator. Twenty-four
hours later the medium was removed and samples were fixed in 1 ml
of 4% formaldehyde solution, (F8775; Sigma-Aldrich St. Louis, MO,
USA) in phosphate-buffered saline (PBS), pH 7.4 1X Gibco,
(10010-023) PBS for 15 min in the incubator. Samples were washed
with PBS twice, then were permeabilized with PBS/Triton X-100
(T9284; Sigma-Aldrich), 1% for 5 min at room temperature. Detergent
was removed and non-specific sites were blocked in PBS containing
2% bovine serum albumin (BSA, A2153; Sigma-Aldrich) at room
temperature for 30 min. Samples were further incubated overnight in
cold room along with all primary antibodies for the experiment,
followed by two consecutive washes the next morning and incubation
in BSA solution with secondary antibodies at room temperature for 2
h along with Zenon complex and two washes with PBS, for 10 min
each. Second fixation step with formaldehyde for 15 min at room
temperature in the dark was performed, followed by two washing
steps.
Zenon complex formation
PRP-1 serum antibody and Zenon rabbit IgG, Alexa
Fluor 488 (Z-25302; Molecular Probes, Eugene, OR, USA) were mixed
according to the manual and the procedures. The mixture was
incubated for 10 min at room temperature with labeling reagent A,
then another 10 min incubation with the blocking reagent B and 1 ml
of the resulting mixture was applied to each well. Cells were
stained with 3 μM of 4′,6-diamino-2-phenylindole
dihydrochloride (DAPI, D1306; Thermo Fisher Scientific) for nuclear
staining or 10 min at room temperature, the washed with PBS twice.
The samples on coverslips were mounted in Antifade mounting medium,
followed by microscopy. ProLong Gold Antifade reagent (P10144; Life
Technologies) was applied as a liquid mountant directly to
fluorescently labeled cells on microscope slides. The reagent
contains chemicals to protect fluorescent dyes from fading during
fluorescence microscopy.
Antibodies used for
immunocytochemistry
For plasma membrane staining wheat germ agglutinin
Alexa Fluor 594 conjugate was used (W11262; Thermo Fisher
Scientific); TLR1 rabbit antibody (ab180798; Abcam); goat
anti-rabbit H&L (DyLight 550) (ab96884; Abcam); mouse
anti-MUC5B, Abcam (ab77995); goat anti-mouse IgG secondary antibody
Alexa Fluor 647 (A2124; Life Technologies).
Imaging
Image acquisition was performed by the Analytical
Imaging Core Facility at DRI/SCCC, University of Miami (FL,
USA).
Zeiss 200M, ApoTome fluorescent microscope, DAPI 49,
GFP 38HE, Cy3 43, Cy5 50 filter cubes, heated stage, Orca II ERG
Hamamatsu b/w 14 bit camera and AxioVision acquisition software
were used. The coverslips were placed in regular 35-mm Petri dishes
and the cells grown on them, covered with medium. Once the cells
were grown, the coverslips were taken out, the cells were fixed,
stained and mounted on the glass slides. For imaging controls
secondary antibodies were used without the primaries.
Human MUC5B ELISA and electrophoresis and
western blotting
MUC5B protein was measured with human mucin -5
subtype (MUC5B) ELISA kit (MyBioSource, San Diego, CA, USA) (MBS
704534-48T).
The cells were trypsinized once they reached
confluency and then seeded in 6-well clusters at a concentration of
1×106 cells/ml. PRP-1 was added only to the experimental
samples but not to controls. The overnight incubation in 5%
CO2 incubator at 37°C was followed by cell wash with
ice-cold PBS with added protease inhibitor. The cell lysis buffer
(C2978; Sigma-Aldrich) was supplemented with the protease inhibitor
in a 1:100 ratio. The cells were collected with a scraper and
centrifuged at 15,000 × g at 4°C. The supernatant was collected and
the protein concentration was measured. The pellets were frozen at
−80°C until loading on the gel (20 μg/lane). Polyacrylamide
gel electrophoresis and western blotting reagents were supplied by
Lonza, Inc. (Allendale, NJ, USA), and all the related procedures
followed the company's protocol. The catalog numbers for the
reagents and the suppliers are listed below for convenience,
although they were reported in our previous communication (8). PAGEr™ Gold Precast Gels (59502; 10%
Tris-Glycine; Lonza); ECL reagent (RPN2109; GE Healthcare, Little
Chalfont, UK); Western Blocker solution (W0138; Sigma-Aldrich);
ProSieve QuadColor Protein marker (4.6–300 kDa, 00193837; Lonza);
20X Reducing Agent for ProSieve ProTrack Dual Color Loading buffer
(00193861; Lonza); ProTrack Loading buffer (00193861; Lonza);
ProSieve ProTrack Dual Color Loading buffer EX running buffer
(00200307; Lonza); ProSieve EX Western Blot Transfer buffer
(00200309; Lonza); Immobilon®-P PVDF Membranes (P4188;
Sigma-Aldrich).
Immunoblot antibodies
Rabbit polyclonal anti-TLR6 (ab37072), MW 92 kDa
(Abcam); rabbit anti-TLR1 cell (2209), MW 86 kDa (Cell Signaling
Technology, Danvers, MA, USA); rabbit anti-TLR1 (ab68158), MW 90
kDa (Abcam); mouse anti-TLR2 [TL2.1], (ab9100), MW 90 kDa (Abcam);
Mouse anti-TLR3 (TLR3.7) (sc-32232), MW 104 kDa (Santa Cruz
Biotechnology, Santa Cruz, CA, USA); mouse anti-TLR4 (25) (sc-293072), MW 95-120 kDa (Santa
Cruz Biotechnology); mouse anti-TLR5 (19D759.2), (sc-57461), MW
110-120 kDa (Santa Cruz Biotechnology); rabbit anti-TLR7, (5632),
MW 140 kDa (Cell Signaling Technology); mouse TLR 8 (9A6),
(sc-135584), MW 119.8 kDa (Santa Cruz Biotechnology); rabbit
anti-TLR9, (5845), MW 130 kDa (Cell Signaling Technology); mouse
TLR10 (2A11), sc-293300, MW 90 kDa (Santa Cruz Biotechnology);
mouse anti-tubulin, (T5168; Sigma-Aldrich); rabbit
anti-TRIF/TICAM1, NBP2-31189, MW 75 kDa (Novus Biologicals,
Littleton, CO, USA); mouse anti-TICAM2, MW 21 kDa (Santa Cruz
Biotechnology); rabbit anti-TRAF6 (3566R-100), MW 54 kDa
(BioVision, Inc., Milpitas, CA, USA); goat anti-rabbit IgG,
HRP-linked (7074; Cell Signaling Technology); anti-mouse IgG,
HRP-linked (7076; Cell Signaling Technology).
Lead Hunter discovery services
(DiscoveRx)
Nuclear Hormone Receptor Assays:
PathHunter® NHR Protein Interaction (Pro) and Nuclear
Translocation (NT) assays monitor the activation of a nuclear
hormone receptor in a homogeneous, non-imaging assay format using a
technology developed by DiscoveRx called enzyme fragment
complementation (EFC). The company described NHR Pro assay detects
of protein-protein interactions between an actvated NHR protein and
a nuclear fusion protein containing steroid receptor co-activator
peptide (SRCP). When bound by ligand, the NHR will migrate to the
nucleus and recruit the SRCP domain, whereby complementation
occurs, generating a unit of active β-galactosidase (β-gal) and
production of chemiluminescent signal.
Arrestin pathway
The PathHunter® β-arrestin assay based on
activation of a GPCR using a method developed by DiscoveRx called
enzyme fragment complementation (EFC) with β-galactosidase (β-gal)
as the functional reporter (44).
In brief, according to the manufacturer's protocol: the enzyme is
split into two inactive complementary portions (EA for enzyme
acceptor and ED for enzyme donor) expressed as fusion proteins in
the cell. EA is fused to β-arrestin and ED is fused to the GPCR of
interest. When the GPCR is activated and β-arrestin is recruited to
the receptor, ED and EA complementation occurs, restoring β-gal
activity which is measured using chemiluminescent
PathHunter® detection reagents.
Data analysis
The GPCR max panel % agonist was calculated as 100%
(mean of test samples - mean of vehicle control)/mean Max control
ligand - mean of vehicle control). For antagonist mode assays,
percentage inhibition was calculated using the following formula: %
Inhibition =100% × (1 – (mean RLU of test sample − mean RLU of
vehicle control)/(mean RLU of EC80 control − mean RLU of vehicle
control). For the orphan max panel, % agonist activity was
calculated as 100% × (mean of test sample − mean of vehicle
control)/mean of vehicle control.
gpcrMAX and NHR - Agonist mode
calculation
To determine if a compound is potentially acting as
an agonist to activate the receptor and induce arrestin recruitment
the following factors should be considered: Is the % activity
>30%? If so, is the compound mean RLU >Baseline RLU + 3 ×
Baseline SD.
gpcrMAX and NHR - Antagonist mode
Inhibition of GPCR activation by a compound acting
as an antagonist of ligand binding results in a decrease in
β-arrestin recruitment to the target GPCR. The NHR panel measures
agonist interactions during a 6-h period and antagonist are
preincubated for 1 h prior to agonist challenge. To determine if a
compound is potentially acting as an antagonist to inhibit receptor
activation the following factors should be considered: Is the %
inhibition >35%, if so, is the compound mean RLU <EC80 RLU −
3 × EC80 SD.
orphanMAX - Agonist mode
Activation of Orphan GPCR by a compound acting as an
agonist will result in an increase in β-arrestin recruitment to the
target orphan GPCR. To determine if a compound is potentially
acting as an agonist to activate an orphan receptor and induce
arrestin recruitment the following factor should be considered: Is
the % activity >50%. If so, is the compound mean RLU
>Baseline RLU + 3 × Baseline SD.
TriCEPS technology
This technology from Dualsystems Biotech AG (Zurich,
Switzerland) was implemented to detect PRP-1 receptor or
interacting partners. Specific cell surface protein receptors are
involved in the drug, peptide ligand mediated physiological
responses. The TriCEPS method is based on the Ligand-based receptor
capture (LRC) technology where special reagent can be coupled to a
ligand of interest, which allows to capture the ligand when bound
to corresponding receptors. One can picture TriCEPS with three
arms: one that binds to amino group containing ligands, a second
for the ligand-based capture of glycosylated and a third one with
biotin tag for purifying receptor peptides to be analyzed by
quantitative mass spectrometry (MS). Specific receptors for the
ligand of interest are identified through quantitative comparison
of the identified peptides with a sample generated by a control
probe with known (e.g., insulin) receptor.
TriCEPS protocol: Ligand coupling
This procedure implemented processing of ligand and
the identification of receptor candidates (3 ligand and 3 control
samples, 300 μg control ligand or 300 μg ligand of
interest to 120 μl of 25 mM HEPES pH 8.2, 1.5 μl (150
μg) TriCEPS v.3 was added to both reactions and mixed
immediately by pipetting up and down using a 200 μl pipette,
then incubated at 20°C under gentle agitation (350 rpm) in a
ThermoMixer for 90 min. Cell preparation and oxidation
1.2×108 cells were utilized for the experiment in
triplicates. Cells were centrifuged at 300 × g for 5 min at 4°C,
then were resuspended in 49 ml LRC buffer. The oxidation agent 1 ml
(75 mM sodium metaperiodate) were added to a final concentration of
1.5 mM metaperiodate, followed by incubation for 15 min at 4°C. The
mild oxidant sodium metaperiodate generates aldehydes from
carbohydrates that link to the proteins of cell surface. When the
ligand binds to the receptor, the hydrazine group formed a bond
with the aldehyde for protein labeling.
Mass spectrometry
The LRC-TriCEPS samples were analyzed on a Thermo
LTQ Orbitrap XL spectrometer. The samples were processed in data
dependent acquisition mode in a 90-min gradient with 10 cm C18
packed column. The statistical ANOVA model was applied to the six
remaining samples in the CaptiRec dataset. With models of Gaussian
distribution the system tests each protein for differential
abundance in all pairwise comparisons of ligand and control samples
and calculates P-values. P-values are undergoing multiple
comparisons to control the experiment-wide false discovery rate
(FDR). Then, this adjusted P-value from each individual protein is
plotted against the magnitude of the fold enrichment between the
two experimental conditions. The receptor candidate space is
defined based on the criteria where the area in the volcano plot
that is limited by an enrichment factor of ≥4-fold and an
FDR-adjusted P≤0.01.
RT2 qPCR primer assays
These custom designed assays by Qiagen (Valencia,
CA, USA) served as sensitive gene expression profiling tool for
real-time PCR analyses. Assay utilized RT2 SYBR-Green qPCR Master
Mixes. Mature RNA isolated using RNA extraction according to the
manufacturer's instructions. RNA quality was determined using a
spectrophotometer and was reverse transcribed using a cDNA
conversion. The cDNA in combination with RT2 SYBR-Green qPCR Master
Mix (cat. no. 330529) was used with RT2 qPCR assays. Ct values were
uploaded on web portal at http://www.qiagen.com/geneglobe. Samples were assigned
to control and test groups. Ct values were normalized based on a
manual selection of reference genes. The data analysis web portal
calculates fold change/regulation using ∆∆Ct method, in which ∆Ct
is calculated between gene of interest (GOI) and an average of
housekeeping genes (HKG) followed by ∆∆Ct calculations
[∆Ct(experiment) − ∆Ct(control)]. Fold change is then calculated
using the 2−∆∆Ct formula.
Results
PRP-1 receptors are not G protein
coupled, neither nuclear nor orphan receptors
The search for PRP-1 binding partners started with
DiscoverX platform of G protein coupled receptors, (GPCR) in
agonist, (Table I) and antagonist
modes (Table II). There was no
indication that PRP-1 was G protein coupled, neither that this
peptide was agonist for orphan receptors (Table III) or agonist/antagonist for
nuclear receptor in nhrMax panels (Table IV). MUC5B was identified as PRP-1
receptor binding partner in human chondrosarcoma JJ012 cell line
using Ligand-receptor capture technology. We proceeded further in
the attempt to identify binding partners for PRP-1 using TriCEPS
Ligand-receptor capture (LRC) technology from Dualsystems Biotech
AG (45). LRC was used to identify
novel ligand-receptor interactions. After the TriCEPS coupled
ligand (PRP-1) bound to its targets in or at the cell membrane the
second arm of TriCEPS coupled to the glycans of that target
receptor. The third arm of TriCEPS was used to isolate the proteins
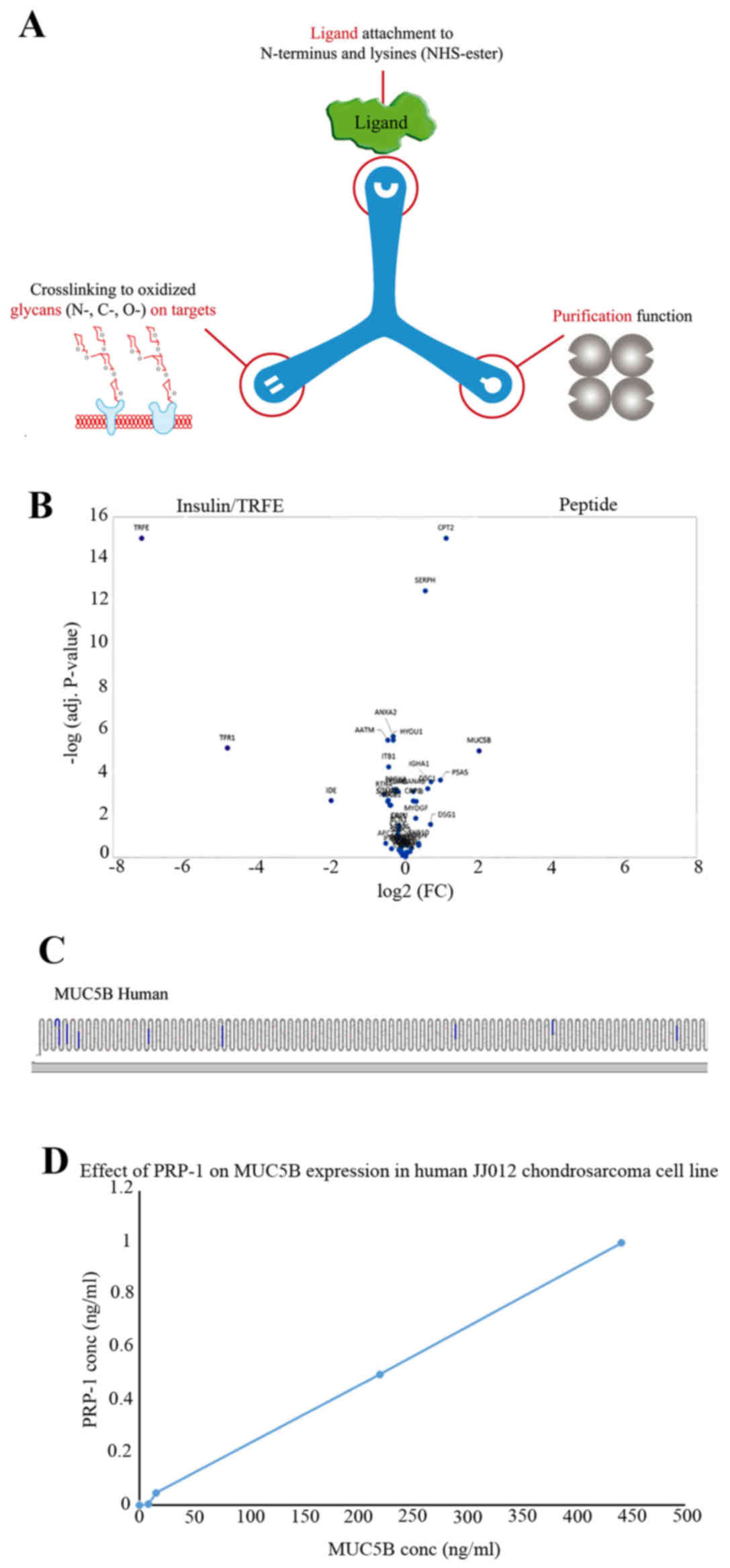
that are bound to TriCEPS (Fig.
1A). In the next step the isolated proteins were subjected to a
trypsin digest. The resulting peptides of the digest were
identified and quantified using liquid chromatography, tandem mass
spectrometry (LC-MS/MS) (45,46).
Then, the quantified peptides from the control reaction
(transferrin as ligand) were compared to the ligand of interest
(PRP-1) reaction (labelled in the volcano plot as peptide). The
proteins that were 4-fold enriched in one of the treatments
compared to the other treatments were considered as the binding
partners of the ligand used. When the cells were treated with
TriCEPS coupled transferrin, the transferrin receptor protein
(TFR1) was enriched (left side of the volcano plot), whereas in the
ligand of interest treated samples the MUC5B was enriched. Thus,
MUC5B was identified as receptor for PRP-1 (Fig. 1B). The data of the experiment was
presented in biological triplicates. The P-value obtained for every
protein was plotted against the log2 of the magnitude of the fold
enrichment. The space for positive control receptors and
high-confidence receptor candidates was designated and visualized
based on (fold-change >4) significant enrichment (adjusted
P<0.01). True positive receptor candidates that contain only few
tryptic peptides can be enriched substantially but will rarely get
adjusted P<0.01. For the final selection of receptor candidates
for follow-up investigations, all proteins in the receptor space
should be viewed based on the following parameters: proteins that
were increased >4 times (log2=2 on the x-axis) with adjusted
P<0.01 [−log (0.01)=2] were considered to be in the receptor
space (white). The protter figure (Fig. 1C) usually displays peptides
belonging to the ligand's receptor as being enriched (shown in
highlighted stripes) compared with the control sample (no ligand).
Protter is an interactive tool for protein data analysis (47). The experimental results with MUC5B
ELISA confirmed the binding, PRP-1 at 1 μg/ml detected MUC5B
presence at 440 ng/ml in the cell lysates of human JJ012
chondrosarcoma cells (Fig. 1D).
Toll like receptors TLR1/2 and TLR6 were identified by western blot
as binding interaction partners with PRP-1 in human chondrosarcoma
JJ012 cell line lysates. Polyacrylamide gel electrophoresis,
immunoblot results indicated that PRP-1 caused strong upregulation
of TLR1 and TLR2 in comparison to untreated control. TLR3
expression was present but weak and data is not shown. TLR4 and
TLR5 were expressed, but PRP-1 did not have any effect (Fig. 2). TLR6 protein expression was also
increased in dose-dependent manner in PRP-1 treated samples. TLR7
was not expressed at all in the cell line, whereas TLR8, 9 and 10
were expressed but there was no indication of PRP-1 effect
(Fig. 2). Thus, TLR1/2 and TLR6
were identified as interacting binding partners for PRP-1. Fig. 2 depicts the PRP-1 action on the
protein expression of the adaptors TICAM1 (TRIF) and TICAM2 (TRAM).
PRP-1 upregulated in dose-dependent manner the adaptor TICAM2 but
not TICAM1. Toll like receptors TLR1/2 and TLR6 and MUC5B were
detected with PRP-1 in the nucleus of human chondrosarcoma cells by
immunocytochemistry. Since the experiments with immunoblot and
TriCEPS indicated that cell surface receptors TLR1/2, TLR6 and gel
forming mucin MUC5B were the binding partners for PRP-1, the
immunocytochemistry experiments followed next; not only to prove
PRP-1 endogenous presence in chondrosarcoma, but also its cellular
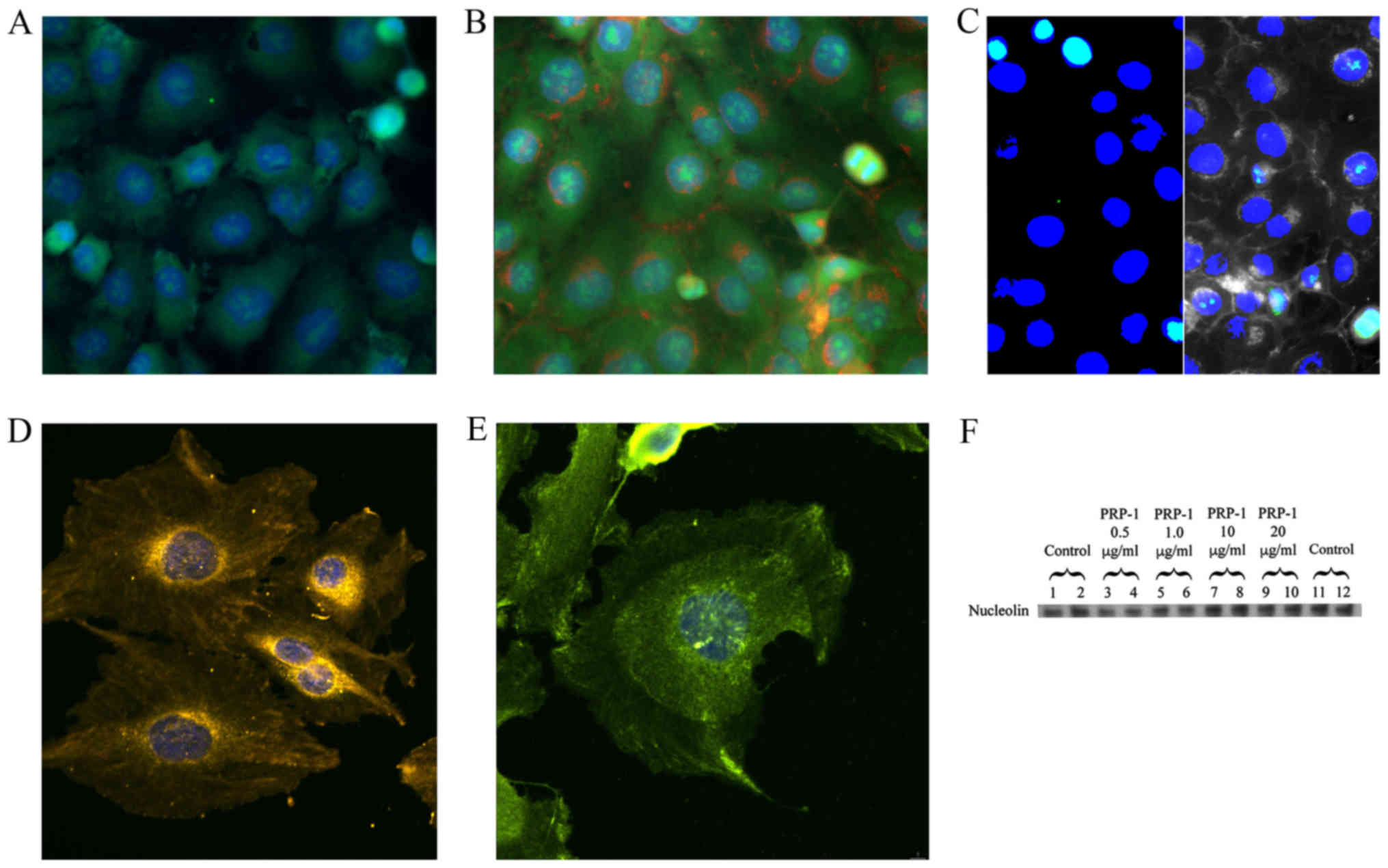
colocalization with the binding partners. The images are displayed
in Fig. 3. PRP-1 antibody,
isolated from rabbit serum and affinity chromatography purified was
labeled with Zenon Alexa Fluor 488 IgG complex (green) and
manifested its presence in the nucleus (labeled with DAPI in blue)
of the chondrosarcoma cells (Fig.
3A). The green speckles and dots can be seen both inside and
outside the nucleus. In the separate experiment without PRP-1 we
have demonstrated that MUC5B is present in the nucleus of these
cells as well (Fig. 3B). The
plasma membrane is seen in red and MUC5B, which was labeled with
DyLight 488 is green. The composite image (Fig. 3C) demonstrates nuclear localization
of both MUC5B (left panel) and PRP-1 (right panel) in aqua green
color on the background of blue nucleus. Fig. 3D illustrates the presence of TLR1
receptor, (labeled in yellow with H&L DyLight 550) in the
nucleus and around it. Fig. 3E
depicts colocalization experiment of TLR6 and PRP-1 and whereas it
was problematic to show TLR1 and PRP-1 colocalization in the
previous figure due to spectral overlaps, here TLR6 receptor
nuclear and cytoplasmic localization is demonstrated with PRP-1,
which was stained with Zenon Alexa Fluor 488 IgG (green). H&L
DyLight 550 was used as a secondary antibody (yellow) for TLR6. The
immunoblot experiment with the nucleolin antibody, marker for
nucleoli indicated that PRP-1 was not located in the nucleoli, as
no changes in nucleolin protein expression was observed on PRP-1
treatment (Fig. 3F).
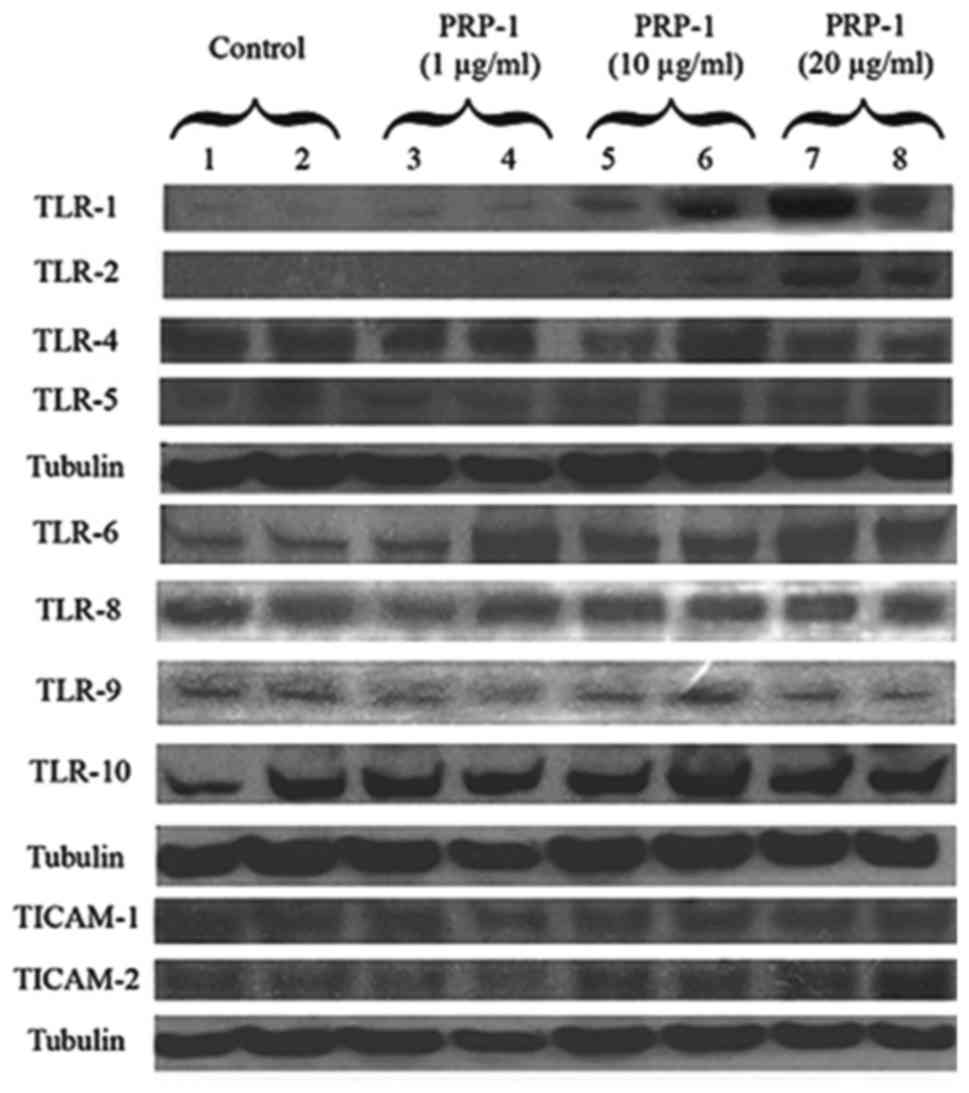 | Figure 2PRP-1 effect on TLR receptors and
adaptor proteins in human chon-drosarcoma JJ012 cell line. PRP-1
upregulated protein expression of TLR1 and TLR2 in dose-dependent
manner in human JJ012 chondrosarcoma cell lysates. TLR3 expression
was very weak and the data is not shown. TLR4, TLR5 proteins were
expressed but PRP-1 did not have any effect. Tubulin was used as
loading control. The bands were detected for TLR1, TLR2, TLR4 and
TLR5 and tubulin at 120, 119, 130, 90 and 50 kDa, correspondingly.
PRP-1 upregulated protein expression of TLR6 in dose-dependent
manner in human JJ012 chondrosarcoma cell lysates. TLR7 was not
expressed in this cell line at all. TLR8, 9 and 10 were expressed
but no effect of PRP-1 was observed. Tubulin was used as
housekeeping control. The bands were detected for TLR6, TLR8, TLR9,
TLR10 and tubulin at 100, 86, 100, 120 and 50 kDa, correspondingly.
PRP-1 upregulated TICAM2 (TRAM) adaptor protein in dose-response
manner but did not have any effect on TICAM1 (TRIF) adaptor protein
in human JJ012 chondrosarcoma cell line. Tubulin was used as
loading control. The bands were detected for TICAM1, TICAM2 and
tubulin. |
 | Table IPRP-1 effect on GPCR receptors
(agonist mode). |
Table I
PRP-1 effect on GPCR receptors
(agonist mode).
| GPCR ID | Assay mode | Conc
(μM) | Mean RLU | % Activity |
|---|
| ADCYAP1R1 | Agonist | 6 | 271200 | 1 |
| ADORA3 | Agonist | 6 | 213700 | 1 |
| ADRA1B | Agonist | 6 | 372900 | 1 |
| ADRA2A | Agonist | 6 | 312500 | 0 |
| ADRA2B | Agonist | 6 | 315400 | 4 |
| ADRA2C | Agonist | 6 | 296300 | 0 |
| ADRB1 | Agonist | 6 | 184300 | 1 |
| ADRB2 | Agonist | 6 | 18800 | 3 |
| AGTR1 | Agonist | 6 | 424900 | 2 |
| AGTRL1 | Agonist | 6 | 429300 | 1 |
| AVPR1A | Agonist | 6 | 23600 | 0 |
| AVPR1B | Agonist | 6 | 35200 | 0 |
| AVPR2 | Agonist | 6 | 822800 | 0 |
| BDKRB1 | Agonist | 6 | 30100 | 1 |
| BDKRB2 | Agonist | 6 | 663600 | 0 |
| BRS3 | Agonist | 6 | 209300 | 0 |
| C3AR1 | Agonist | 6 | 55900 | 0 |
| C5AR1 | Agonist | 6 | 119400 | 0 |
| C5L2 | Agonist | 6 | 164800 | 0 |
| CALCR | Agonist | 6 | 42500 | 1 |
| CALCRL-RAMP1 | Agonist | 6 | 92800 | 0 |
| CALCRL-RAMP2 | Agonist | 6 | 219200 | 1 |
| CALCRL-RAMP3 | Agonist | 6 | 425200 | 0 |
| CALCR-RAMP2 | Agonist | 6 | 139500 | 2 |
| CALCR-RAMP3 | Agonist | 6 | 28600 | 7 |
| CCKAR | Agonist | 6 | 44600 | 0 |
| CCKBR | Agonist | 6 | 894800 | 0 |
| CCR10 | Agonist | 6 | 93400 | 0 |
| CCR1 | Agonist | 6 | 540700 | 7 |
| CCR2 | Agonist | 6 | 67000 | 0 |
| CCR3 | Agonist | 6 | 272100 | 2 |
| CCR4 | Agonist | 6 | 180300 | 0 |
| CCR5 | Agonist | 6 | 89800 | 0 |
| CCR6 | Agonist | 6 | 141000 | 0 |
| CCR7 | Agonist | 6 | 766200 | 1 |
| CCR8 | Agonist | 6 | 35900 | 0 |
| CCR9 | Agonist | 6 | 119300 | 1 |
| CHRM1 | Agonist | 6 | 1181100 | 1 |
| CHRM2 | Agonist | 6 | 54600 | 1 |
| CHRM3 | Agonist | 6 | 166300 | 2 |
| CHRM4 | Agonist | 6 | 787900 | 16 |
| CHRM5 | Agonist | 6 | 2995100 | 5 |
| CMKLR1 | Agonist | 6 | 81900 | 0 |
| CNR1 | Agonist | 6 | 80000 | 0 |
| CNR2 | Agonist | 6 | 315400 | −2 |
| CRHR1 | Agonist | 6 | 361400 | 1 |
| CRHR2 | Agonist | 6 | 161100 | 0 |
| CRTH2 | Agonist | 6 | 172600 | 0 |
| CX3CR1 | Agonist | 6 | 342700 | 1 |
| CXCR1 | Agonist | 6 | 219900 | 0 |
| CXCR2 | Agonist | 6 | 165200 | 1 |
| CXCR3 | Agonist | 6 | 387900 | 1 |
| CXCR4 | Agonist | 6 | 72500 | 2 |
| CXCR5 | Agonist | 6 | 230900 | 1 |
| CXCR6 | Agonist | 6 | 27700 | 2 |
| CXCR7 | Agonist | 6 | 194500 | 0 |
| DRD1 | Agonist | 6 | 73000 | 0 |
| DRD2L | Agonist | 6 | 83500 | 0 |
| DRD2S | Agonist | 6 | 247200 | 0 |
| DRD3 | Agonist | 6 | 414700 | 2 |
| DRD4 | Agonist | 6 | 22800 | 3 |
| DRD5 | Agonist | 6 | 21000 | 1 |
| EBI2 | Agonist | 6 | 150100 | 0 |
| EDG1 | Agonist | 6 | 165500 | 0 |
| EDG3 | Agonist | 6 | 877900 | 0 |
| EDG4 | Agonist | 6 | 234300 | 4 |
| EDG5 | Agonist | 6 | 174300 | 1 |
| EDG6 | Agonist | 6 | 574100 | −2 |
| EDG7 | Agonist | 6 | 154400 | 0 |
| EDNRA | Agonist | 6 | 38500 | 0 |
| EDNRB | Agonist | 6 | 68900 | 0 |
| F2R | Agonist | 6 | 470700 | −2 |
| F2RL1 | Agonist | 6 | 566200 | 0 |
| F2RL3 | Agonist | 6 | 909900 | −1 |
| FFAR1 | Agonist | 6 | 560800 | 3 |
| FPR1 | Agonist | 6 | 1133900 | 4 |
| FPRL1 | Agonist | 6 | 63900 | 0 |
| FSHR | Agonist | 6 | 197900 | −2 |
| GALR1 | Agonist | 6 | 263400 | 1 |
| GALR2 | Agonist | 6 | 307700 | 1 |
| GCGR | Agonist | 6 | 295600 | 0 |
| GHSR | Agonist | 6 | 524600 | 2 |
| GIPR | Agonist | 6 | 17500 | −1 |
| GLP1R | Agonist | 6 | 123500 | 0 |
| GLP2R | Agonist | 6 | 101400 | 1 |
| GPR1 | Agonist | 6 | 58400 | 0 |
| GPR103 | Agonist | 6 | 45100 | 2 |
| GPR109A | Agonist | 6 | 458200 | 4 |
| GPR109B | Agonist | 6 | 410400 | 1 |
| GPR119 | Agonist | 6 | 289300 | 3 |
| GPR120 | Agonist | 6 | 27800 | 1 |
| GPR35 | Agonist | 6 | 287100 | 1 |
| GPR92 | Agonist | 6 | 257600 | 1 |
| GRPR | Agonist | 6 | 39000 | 0 |
| HCRTR1 | Agonist | 6 | 45600 | 0 |
| HCRTR2 | Agonist | 6 | 69900 | 0 |
| HRH1 | Agonist | 6 | 345600 | 1 |
| HRH2 | Agonist | 6 | 91300 | 1 |
| HRH3 | Agonist | 6 | 47100 | 2 |
| HRH4 | Agonist | 6 | 910300 | 4 |
| HTR1A | Agonist | 6 | 879000 | 0 |
| HTR1B | Agonist | 6 | 1214900 | −4 |
| HTR1E | Agonist | 6 | 2963300 | 3 |
| HTR1F | Agonist | 6 | 345900 | 2 |
| HTR2A | Agonist | 6 | 455100 | 1 |
| HTR2C | Agonist | 6 | 921600 | 1 |
| HTR5A | Agonist | 6 | 990000 | 0 |
| KISS1R | Agonist | 6 | 45800 | 1 |
| LHCGR | Agonist | 6 | 24700 | 0 |
| LTB4R | Agonist | 6 | 187300 | 1 |
| MC1R | Agonist | 6 | 17000 | −2 |
| MC3R | Agonist | 6 | 25100 | 3 |
| MC4R | Agonist | 6 | 25900 | −1 |
| MC5R | Agonist | 6 | 54900 | 2 |
| MCHR1 | Agonist | 6 | 140300 | 1 |
| MCHR2 | Agonist | 6 | 62800 | 1 |
| MLNR | Agonist | 6 | 227800 | 1 |
| MRGPRX1 | Agonist | 6 | 869600 | 1 |
| MRGPRX2 | Agonist | 6 | 355800 | 0 |
| MTNR1A | Agonist | 6 | 79100 | 2 |
| NMBR | Agonist | 6 | 62800 | 0 |
| NMU1R | Agonist | 6 | 98100 | 1 |
| NPBWR1 | Agonist | 6 | 69600 | 3 |
| NPBWR2 | Agonist | 6 | 169400 | 1 |
| NPFFR1 | Agonist | 6 | 131100 | 3 |
| NPSR1B | Agonist | 6 | 89900 | 2 |
| NPY1R | Agonist | 6 | 106100 | 0 |
| NPY2R | Agonist | 6 | 362000 | 0 |
| NTSR1 | Agonist | 6 | 313400 | 0 |
| OPRD1 | Agonist | 6 | 91200 | 0 |
| OPRK1 | Agonist | 6 | 36400 | 0 |
| OPRL1 | Agonist | 6 | 229600 | 0 |
| OPRM1 | Agonist | 6 | 137000 | 1 |
| OXER1 | Agonist | 6 | 86900 | −2 |
| OXTR | Agonist | 6 | 25800 | 0 |
| P2RY1 | Agonist | 6 | 123800 | −1 |
| P2RY11 | Agonist | 6 | 68900 | 1 |
| P2RY12 | Agonist | 6 | 250500 | 1 |
| P2RY2 | Agonist | 6 | 384500 | 2 |
| P2RY4 | Agonist | 6 | 397000 | −1 |
| P2RY6 | Agonist | 6 | 324200 | 0 |
| PPYR1 | Agonist | 6 | 34900 | 0 |
| PRLHR | Agonist | 6 | 35900 | 4 |
| PROKR1 | Agonist | 6 | 49600 | 2 |
| PROKR2 | Agonist | 6 | 15700 | 0 |
| PTAFR | Agonist | 6 | 480900 | 1 |
| PTGER2 | Agonist | 6 | 30800 | 4 |
| PTGER3 | Agonist | 6 | 246700 | 1 |
| PTGER4 | Agonist | 6 | 95700 | 1 |
| PTGFR | Agonist | 6 | 16400 | 0 |
| PTGIR | Agonist | 6 | 135100 | 1 |
| PTHR1 | Agonist | 6 | 129100 | 1 |
| PTHR2 | Agonist | 6 | 123100 | 0 |
| RXFP3 | Agonist | 6 | 149000 | 6 |
| SCTR | Agonist | 6 | 502600 | 1 |
| SSTR1 | Agonist | 6 | 15000 | −3 |
| SSTR2 | Agonist | 6 | 11800 | 0 |
| SSTR3 | Agonist | 6 | 82000 | 1 |
| SSTR5 | Agonist | 6 | 176900 | 1 |
| TACR1 | Agonist | 6 | 702100 | 1 |
| TACR2 | Agonist | 6 | 415100 | 1 |
| TACR3 | Agonist | 6 | 145800 | 0 |
| TBXA2R | Agonist | 6 | 203300 | 1 |
| TRHR | Agonist | 6 | 27600 | 1 |
| TSHR(L) | Agonist | 6 | 9600 | 4 |
| UTR2 | Agonist | 6 | 31200 | 3 |
| VIPR1 | Agonist | 6 | 399800 | 0 |
| VIPR2 | Agonist | 6 | 342300 | 1 |
 | Table IIPRP-1 effect on GPCR receptors
(antagonist mode). |
Table II
PRP-1 effect on GPCR receptors
(antagonist mode).
| GPCR ID | Assay mode | Conc
(μM) | Mean RLU | % Inhibition |
|---|
| ADCYAP1R1 | Antagonist | 6 | 1676100 | −6 |
| ADORA3 | Antagonist | 6 | 891300 | −3 |
| ADRA1B | Antagonist | 6 | 2076200 | 0 |
| ADRA2A | Antagonist | 6 | 1075700 | 0 |
| ADRA2B | Antagonist | 6 | 856100 | 2 |
| ADRA2C | Antagonist | 6 | 1511200 | −4 |
| ADRB1 | Antagonist | 6 | 674600 | −5 |
| ADRB2 | Antagonist | 6 | 179900 | −3 |
| AGTR1 | Antagonist | 6 | 2568500 | 0 |
| AGTRL1 | Antagonist | 6 | 2263600 | 0 |
| AVPR1A | Antagonist | 6 | 789300 | −1 |
| AVPR1B | Antagonist | 6 | 258900 | −2 |
| AVPR2 | Antagonist | 6 | 3487000 | 0 |
| BDKRB1 | Antagonist | 6 | 225900 | −8 |
| BDKRB2 | Antagonist | 6 | 4293900 | 5 |
| BRS3 | Antagonist | 6 | 1248800 | 4 |
| C3AR1 | Antagonist | 6 | 1933800 | −2 |
| C5AR1 | Antagonist | 6 | 2029300 | 3 |
| C5L2 | Antagonist | 6 | 547000 | −5 |
| CALCR | Antagonist | 6 | 358200 | 1 |
| CALCRL-RAMP1 | Antagonist | 6 | 1395000 | 4 |
| CALCRL-RAMP2 | Antagonist | 6 | 892100 | −1 |
| CALCRL-RAMP3 | Antagonist | 6 | 2293600 | 1 |
| CALCR-RAMP2 | Antagonist | 6 | 698200 | 0 |
| CALCR-RAMP3 | Antagonist | 6 | 59000 | −8 |
| CCKAR | Antagonist | 6 | 1490300 | −3 |
| CCKBR | Antagonist | 6 | 3697600 | −3 |
| CCR10 | Antagonist | 6 | 1295800 | −3 |
| CCR1 | Antagonist | 6 | 1214200 | −3 |
| CCR2 | Antagonist | 6 | 1380800 | −3 |
| CCR3 | Antagonist | 6 | 1136200 | −3 |
| CCR4 | Antagonist | 6 | 2075100 | 2 |
| CCR5 | Antagonist | 6 | 2359300 | −2 |
| CCR6 | Antagonist | 6 | 1487700 | 2 |
| CCR7 | Antagonist | 6 | 3413600 | −2 |
| CCR8 | Antagonist | 6 | 1376000 | −4 |
| CCR9 | Antagonist | 6 | 1538300 | −6 |
| CHRM1 | Antagonist | 6 | 2892100 | −12 |
| CHRM2 | Antagonist | 6 | 540500 | −6 |
| CHRM3 | Antagonist | 6 | 1187400 | −6 |
| CHRM4 | Antagonist | 6 | 1390200 | −18 |
| CHRM5 | Antagonist | 6 | 4585200 | −4 |
| CMKLR1 | Antagonist | 6 | 3258400 | −2 |
| CNR1 | Antagonist | 6 | 377300 | 2 |
| CNR2 | Antagonist | 6 | 580500 | 5 |
| CRHR1 | Antagonist | 6 | 4103800 | 1 |
| CRHR2 | Antagonist | 6 | 2968600 | −2 |
| CRTH2 | Antagonist | 6 | 1149200 | −8 |
| CX3CR1 | Antagonist | 6 | 3207000 | 2 |
| CXCR1 | Antagonist | 6 | 3818300 | −1 |
| CXCR2 | Antagonist | 6 | 565700 | −3 |
| CXCR3 | Antagonist | 6 | 1392600 | −1 |
| CXCR4 | Antagonist | 6 | 139700 | 2 |
| CXCR5 | Antagonist | 6 | 1098600 | −8 |
| CXCR6 | Antagonist | 6 | 101400 | −6 |
| CXCR7 | Antagonist | 6 | 2856200 | −2 |
| DRD1 | Antagonist | 6 | 696300 | −5 |
| DRD2L | Antagonist | 6 | 393800 | 2 |
| DRD2S | Antagonist | 6 | 1242000 | 6 |
| DRD3 | Antagonist | 6 | 1263000 | −15 |
| DRD4 | Antagonist | 6 | 65500 | −2 |
| DRD5 | Antagonist | 6 | 149600 | −8 |
| EBI2 | Antagonist | 6 | 2425000 | −1 |
| EDG1 | Antagonist | 6 | 936700 | 0 |
| EDG3 | Antagonist | 6 | 4548500 | 1 |
| EDG4 | Antagonist | 6 | 657500 | 5 |
| EDG5 | Antagonist | 6 | 2180100 | −8 |
| EDG6 | Antagonist | 6 | 1169200 | 4 |
| EDG7 | Antagonist | 6 | 1432500 | −3 |
| EDNRA | Antagonist | 6 | 953300 | 0 |
| EDNRB | Antagonist | 6 | 1248000 | −1 |
| F2R | Antagonist | 6 | 1612200 | −16 |
| F2RL1 | Antagonist | 6 | 3638100 | 3 |
| F2RL3 | Antagonist | 6 | 3027600 | −1 |
| FFAR1 | Antagonist | 6 | 1070500 | 0 |
| FPR1 | Antagonist | 6 | 3077200 | −4 |
| FPRL1 | Antagonist | 6 | 2991600 | −2 |
| FSHR | Antagonist | 6 | 621700 | −2 |
| GALR1 | Antagonist | 6 | 1759100 | −4 |
| GALR2 | Antagonist | 6 | 1686800 | −11 |
| GCGR | Antagonist | 6 | 3115100 | −4 |
| GHSR | Antagonist | 6 | 2068100 | −6 |
| GIPR | Antagonist | 6 | 79600 | −24 |
| GLP1R | Antagonist | 6 | 1983100 | −9 |
| GLP2R | Antagonist | 6 | 727800 | −11 |
| GPR1 | Antagonist | 6 | 1076400 | −5 |
| GPR103 | Antagonist | 6 | 103800 | 6 |
| GPR109A | Antagonist | 6 | 1141200 | −4 |
| GPR109B | Antagonist | 6 | 2871700 | −6 |
| GPR119 | Antagonist | 6 | 521500 | −1 |
| GPR120 | Antagonist | 6 | 130400 | −6 |
| GPR35 | Antagonist | 6 | 910400 | −6 |
| GPR92 | Antagonist | 6 | 854500 | 12 |
| GRPR | Antagonist | 6 | 1570000 | −7 |
| HCRTR1 | Antagonist | 6 | 3242300 | 0 |
| HCRTR2 | Antagonist | 6 | 2611400 | −1 |
| HRH1 | Antagonist | 6 | 1912200 | 0 |
| HRH2 | Antagonist | 6 | 347800 | 1 |
| HRH3 | Antagonist | 6 | 180200 | −6 |
| HRH4 | Antagonist | 6 | 2264500 | −9 |
| HTR1A | Antagonist | 6 | 2576600 | −5 |
| HTR1B | Antagonist | 6 | 2334400 | 4 |
| HTR1E | Antagonist | 6 | 5487000 | 5 |
| HTR1F | Antagonist | 6 | 982500 | 2 |
| HTR2A | Antagonist | 6 | 3103200 | −5 |
| HTR2C | Antagonist | 6 | 4188400 | −3 |
| HTR5A | Antagonist | 6 | 4536500 | 0 |
| KISS1R | Antagonist | 6 | 270900 | 3 |
| LHCGR | Antagonist | 6 | 144000 | −15 |
| LTB4R | Antagonist | 6 | 1844800 | 0 |
| MC1R | Antagonist | 6 | 69300 | −2 |
| MC3R | Antagonist | 6 | 153200 | −6 |
| MC4R | Antagonist | 6 | 128200 | −6 |
| MC5R | Antagonist | 6 | 179900 | −5 |
| MCHR1 | Antagonist | 6 | 1023000 | −4 |
| MCHR2 | Antagonist | 6 | 530900 | −3 |
| MLNR | Antagonist | 6 | 2032800 | −7 |
| MRGPRX1 | Antagonist | 6 | 3890200 | −2 |
| MRGPRX2 | Antagonist | 6 | 1945500 | −8 |
| MTNR1A | Antagonist | 6 | 241200 | −9 |
| NMBR | Antagonist | 6 | 720100 | −5 |
| NMU1R | Antagonist | 6 | 985900 | −3 |
| NPBWR1 | Antagonist | 6 | 188200 | 0 |
| NPBWR2 | Antagonist | 6 | 1070300 | −3 |
| NPFFR1 | Antagonist | 6 | 290800 | −3 |
| NPSR1B | Antagonist | 6 | 699900 | 0 |
| NPY1R | Antagonist | 6 | 973600 | 4 |
| NPY2R | Antagonist | 6 | 3477300 | −2 |
| NTSR1 | Antagonist | 6 | 2192400 | −1 |
| OPRD1 | Antagonist | 6 | 795800 | −2 |
| OPRK1 | Antagonist | 6 | 215800 | −12 |
| OPRL1 | Antagonist | 6 | 1062000 | −7 |
| OPRM1 | Antagonist | 6 | 2828200 | −2 |
| OXER1 | Antagonist | 6 | 246400 | −16 |
| OXTR | Antagonist | 6 | 540000 | 1 |
| P2RY1 | Antagonist | 6 | 530600 | 1 |
| P2RY11 | Antagonist | 6 | 484300 | −12 |
| P2RY12 | Antagonist | 6 | 699600 | 8 |
| P2RY2 | Antagonist | 6 | 1093000 | −4 |
| P2RY4 | Antagonist | 6 | 1323200 | 0 |
| P2RY6 | Antagonist | 6 | 1825600 | 3 |
| PPYR1 | Antagonist | 6 | 276600 | −4 |
| PRLHR | Antagonist | 6 | 122100 | −3 |
| PROKR1 | Antagonist | 6 | 499800 | −2 |
| PROKR2 | Antagonist | 6 | 111500 | 8 |
| PTAFR | Antagonist | 6 | 3532600 | −8 |
| PTGER2 | Antagonist | 6 | 72100 | −11 |
| PTGER3 | Antagonist | 6 | 967100 | −2 |
| PTGER4 | Antagonist | 6 | 852400 | 4 |
| PTGFR | Antagonist | 6 | 453000 | 1 |
| PTGIR | Antagonist | 6 | 380600 | −2 |
| PTHR1 | Antagonist | 6 | 2941200 | −2 |
| PTHR2 | Antagonist | 6 | 2882300 | −1 |
| RXFP3 | Antagonist | 6 | 330200 | −3 |
| SCTR | Antagonist | 6 | 3424900 | −2 |
| SSTR1 | Antagonist | 6 | 38300 | −14 |
| SSTR2 | Antagonist | 6 | 797000 | −11 |
| SSTR3 | Antagonist | 6 | 771000 | −5 |
| SSTR5 | Antagonist | 6 | 1327400 | −11 |
| TACR1 | Antagonist | 6 | 4862800 | −2 |
| TACR2 | Antagonist | 6 | 2331100 | 0 |
| TACR3 | Antagonist | 6 | 2745900 | −1 |
| TBXA2R | Antagonist | 6 | 1032900 | −10 |
| TRHR | Antagonist | 6 | 301600 | −5 |
| TSHR(L) | Antagonist | 6 | 83000 | −4 |
| UTR2 | Antagonist | 6 | 156000 | −7 |
| VIPR1 | Antagonist | 6 | 3344300 | −9 |
| VIPR2 | Antagonist | 6 | 3604300 | −3 |
 | Table IIIPRP-1 effect on orphan receptors
(agonist mode). |
Table III
PRP-1 effect on orphan receptors
(agonist mode).
| GPCR ID | Assay mode | Conc
(μM) | Mean RLU | % Inhibition |
|---|
| ADCYAP1R1 | Antagonist | 6 | 1676100 | −6 |
| ADORA3 | Antagonist | 6 | 891300 | −3 |
| ADRA1B | Antagonist | 6 | 2076200 | 0 |
| ADRA2A | Antagonist | 6 | 1075700 | 0 |
| ADRA2B | Antagonist | 6 | 856100 | 2 |
| ADRA2C | Antagonist | 6 | 1511200 | −4 |
| ADRB1 | Antagonist | 6 | 674600 | −5 |
| ADRB2 | Antagonist | 6 | 179900 | −3 |
| AGTR1 | Antagonist | 6 | 2568500 | 0 |
| AGTRL1 | Antagonist | 6 | 2263600 | 0 |
| AVPR1A | Antagonist | 6 | 789300 | −1 |
| AVPR1B | Antagonist | 6 | 258900 | −2 |
| AVPR2 | Antagonist | 6 | 3487000 | 0 |
| BDKRB1 | Antagonist | 6 | 225900 | −8 |
| BDKRB2 | Antagonist | 6 | 4293900 | 5 |
| BRS3 | Antagonist | 6 | 1248800 | 4 |
| C3AR1 | Antagonist | 6 | 1933800 | −2 |
| C5AR1 | Antagonist | 6 | 2029300 | 3 |
| C5L2 | Antagonist | 6 | 547000 | −5 |
| CALCR | Antagonist | 6 | 358200 | 1 |
| CALCRL-RAMP1 | Antagonist | 6 | 1395000 | 4 |
| CALCRL-RAMP2 | Antagonist | 6 | 892100 | −1 |
| CALCRL-RAMP3 | Antagonist | 6 | 2293600 | 1 |
| CALCR-RAMP2 | Antagonist | 6 | 698200 | 0 |
| CALCR-RAMP3 | Antagonist | 6 | 59000 | −8 |
| CCKAR | Antagonist | 6 | 1490300 | −3 |
| CCKBR | Antagonist | 6 | 3697600 | −3 |
| CCR10 | Antagonist | 6 | 1295800 | −3 |
| CCR1 | Antagonist | 6 | 1214200 | −3 |
| CCR2 | Antagonist | 6 | 1380800 | −3 |
| CCR3 | Antagonist | 6 | 1136200 | −3 |
| CCR4 | Antagonist | 6 | 2075100 | 2 |
| CCR5 | Antagonist | 6 | 2359300 | −2 |
| CCR6 | Antagonist | 6 | 1487700 | 2 |
| CCR7 | Antagonist | 6 | 3413600 | −2 |
| CCR8 | Antagonist | 6 | 1376000 | −4 |
| CCR9 | Antagonist | 6 | 1538300 | −6 |
| CHRM1 | Antagonist | 6 | 2892100 | −12 |
| CHRM2 | Antagonist | 6 | 540500 | −6 |
| CHRM3 | Antagonist | 6 | 1187400 | −6 |
| CHRM4 | Antagonist | 6 | 1390200 | −18 |
| CHRM5 | Antagonist | 6 | 4585200 | −4 |
| CMKLR1 | Antagonist | 6 | 3258400 | −2 |
| CNR1 | Antagonist | 6 | 377300 | 2 |
| CNR2 | Antagonist | 6 | 580500 | 5 |
| CRHR1 | Antagonist | 6 | 4103800 | 1 |
| CRHR2 | Antagonist | 6 | 2968600 | −2 |
| CRTH2 | Antagonist | 6 | 1149200 | −8 |
| CX3CR1 | Antagonist | 6 | 3207000 | 2 |
| CXCR1 | Antagonist | 6 | 3818300 | −1 |
| CXCR2 | Antagonist | 6 | 565700 | −3 |
| CXCR3 | Antagonist | 6 | 1392600 | −1 |
| CXCR4 | Antagonist | 6 | 139700 | 2 |
| CXCR5 | Antagonist | 6 | 1098600 | −8 |
| CXCR6 | Antagonist | 6 | 101400 | −6 |
| CXCR7 | Antagonist | 6 | 2856200 | −2 |
| DRD1 | Antagonist | 6 | 696300 | −5 |
| DRD2L | Antagonist | 6 | 393800 | 2 |
| DRD2S | Antagonist | 6 | 1242000 | 6 |
| DRD3 | Antagonist | 6 | 1263000 | −15 |
| DRD4 | Antagonist | 6 | 65500 | −2 |
| DRD5 | Antagonist | 6 | 149600 | −8 |
| EBI2 | Antagonist | 6 | 2425000 | −1 |
| EDG1 | Antagonist | 6 | 936700 | 0 |
| EDG3 | Antagonist | 6 | 4548500 | 1 |
| EDG4 | Antagonist | 6 | 657500 | 5 |
| EDG5 | Antagonist | 6 | 2180100 | −8 |
| EDG6 | Antagonist | 6 | 1169200 | 4 |
| EDG7 | Antagonist | 6 | 1432500 | −3 |
| EDNRA | Antagonist | 6 | 953300 | 0 |
| EDNRB | Antagonist | 6 | 1248000 | −1 |
| F2R | Antagonist | 6 | 1612200 | −16 |
| F2RL1 | Antagonist | 6 | 3638100 | 3 |
| F2RL3 | Antagonist | 6 | 3027600 | −1 |
| FFAR1 | Antagonist | 6 | 1070500 | 0 |
| FPR1 | Antagonist | 6 | 3077200 | −4 |
| FPRL1 | Antagonist | 6 | 2991600 | −2 |
| FSHR | Antagonist | 6 | 621700 | −2 |
| GALR1 | Antagonist | 6 | 1759100 | −4 |
| GALR2 | Antagonist | 6 | 1686800 | −11 |
| GCGR | Antagonist | 6 | 3115100 | −4 |
| GHSR | Antagonist | 6 | 2068100 | −6 |
| GIPR | Antagonist | 6 | 79600 | −24 |
| GLP1R | Antagonist | 6 | 1983100 | −9 |
| GLP2R | Antagonist | 6 | 727800 | −11 |
| GPR1 | Antagonist | 6 | 1076400 | −5 |
| GPR103 | Antagonist | 6 | 103800 | 6 |
| GPR109A | Antagonist | 6 | 1141200 | −4 |
| GPR109B | Antagonist | 6 | 2871700 | −6 |
| GPR119 | Antagonist | 6 | 521500 | −1 |
| GPR120 | Antagonist | 6 | 130400 | −6 |
| GPR35 | Antagonist | 6 | 910400 | −6 |
| GPR92 | Antagonist | 6 | 854500 | 12 |
| GRPR | Antagonist | 6 | 1570000 | −7 |
| HCRTR1 | Antagonist | 6 | 3242300 | 0 |
| HCRTR2 | Antagonist | 6 | 2611400 | −1 |
| HRH1 | Antagonist | 6 | 1912200 | 0 |
| HRH2 | Antagonist | 6 | 347800 | 1 |
| HRH3 | Antagonist | 6 | 180200 | −6 |
| HRH4 | Antagonist | 6 | 2264500 | −9 |
| HTR1A | Antagonist | 6 | 2576600 | −5 |
| HTR1B | Antagonist | 6 | 2334400 | 4 |
| HTR1E | Antagonist | 6 | 5487000 | 5 |
| HTR1F | Antagonist | 6 | 982500 | 2 |
| HTR2A | Antagonist | 6 | 3103200 | −5 |
| HTR2C | Antagonist | 6 | 4188400 | −3 |
| HTR5A | Antagonist | 6 | 4536500 | 0 |
| KISS1R | Antagonist | 6 | 270900 | 3 |
| LHCGR | Antagonist | 6 | 144000 | −15 |
| LTB4R | Antagonist | 6 | 1844800 | 0 |
| MC1R | Antagonist | 6 | 69300 | −2 |
| MC3R | Antagonist | 6 | 153200 | −6 |
| MC4R | Antagonist | 6 | 128200 | −6 |
| MC5R | Antagonist | 6 | 179900 | −5 |
| MCHR1 | Antagonist | 6 | 1023000 | −4 |
| MCHR2 | Antagonist | 6 | 530900 | −3 |
| MLNR | Antagonist | 6 | 2032800 | −7 |
| MRGPRX1 | Antagonist | 6 | 3890200 | −2 |
| MRGPRX2 | Antagonist | 6 | 1945500 | −8 |
| MTNR1A | Antagonist | 6 | 241200 | −9 |
| NMBR | Antagonist | 6 | 720100 | −5 |
| NMU1R | Antagonist | 6 | 985900 | −3 |
| NPBWR1 | Antagonist | 6 | 188200 | 0 |
| NPBWR2 | Antagonist | 6 | 1070300 | −3 |
| NPFFR1 | Antagonist | 6 | 290800 | −3 |
| NPSR1B | Antagonist | 6 | 699900 | 0 |
| NPY1R | Antagonist | 6 | 973600 | 4 |
| NPY2R | Antagonist | 6 | 3477300 | −2 |
| NTSR1 | Antagonist | 6 | 2192400 | −1 |
| OPRD1 | Antagonist | 6 | 795800 | −2 |
| OPRK1 | Antagonist | 6 | 215800 | −12 |
| OPRL1 | Antagonist | 6 | 1062000 | −7 |
| OPRM1 | Antagonist | 6 | 2828200 | −2 |
| OXER1 | Antagonist | 6 | 246400 | −16 |
| OXTR | Antagonist | 6 | 540000 | 1 |
| P2RY1 | Antagonist | 6 | 530600 | 1 |
| P2RY11 | Antagonist | 6 | 484300 | −12 |
| P2RY12 | Antagonist | 6 | 699600 | 8 |
| P2RY2 | Antagonist | 6 | 1093000 | −4 |
| P2RY4 | Antagonist | 6 | 1323200 | 0 |
| P2RY6 | Antagonist | 6 | 1825600 | 3 |
| PPYR1 | Antagonist | 6 | 276600 | −4 |
| PRLHR | Antagonist | 6 | 122100 | −3 |
| PROKR1 | Antagonist | 6 | 499800 | −2 |
| PROKR2 | Antagonist | 6 | 111500 | 8 |
| PTAFR | Antagonist | 6 | 3532600 | −8 |
| PTGER2 | Antagonist | 6 | 72100 | −11 |
| PTGER3 | Antagonist | 6 | 967100 | −2 |
| PTGER4 | Antagonist | 6 | 852400 | 4 |
| PTGFR | Antagonist | 6 | 453000 | 1 |
| PTGIR | Antagonist | 6 | 380600 | −2 |
| PTHR1 | Antagonist | 6 | 2941200 | −2 |
| PTHR2 | Antagonist | 6 | 2882300 | −1 |
| RXFP3 | Antagonist | 6 | 330200 | −3 |
| SCTR | Antagonist | 6 | 3424900 | −2 |
| SSTR1 | Antagonist | 6 | 38300 | −14 |
| SSTR2 | Antagonist | 6 | 797000 | −11 |
| SSTR3 | Antagonist | 6 | 771000 | −5 |
| SSTR5 | Antagonist | 6 | 1327400 | −11 |
| TACR1 | Antagonist | 6 | 4862800 | −2 |
| TACR2 | Antagonist | 6 | 2331100 | 0 |
| TACR3 | Antagonist | 6 | 2745900 | −1 |
| TBXA2R | Antagonist | 6 | 1032900 | −10 |
| TRHR | Antagonist | 6 | 301600 | −5 |
| TSHR(L) | Antagonist | 6 | 83000 | −4 |
| UTR2 | Antagonist | 6 | 156000 | −7 |
| VIPR1 | Antagonist | 6 | 3344300 | −9 |
| VIPR2 | Antagonist | 6 | 3604300 | −3 |
| ADCYAP1R1 | Agonist | 6 | 271200 | 1 |
| ADORA3 | Agonist | 6 | 213700 | 1 |
| ADRA1B | Agonist | 6 | 372900 | 1 |
| ADRA2A | Agonist | 6 | 312500 | 0 |
| ADRA2B | Agonist | 6 | 315400 | 4 |
| ADRA2C | Agonist | 6 | 296300 | 0 |
| ADRB1 | Agonist | 6 | 184300 | 1 |
| ADRB2 | Agonist | 6 | 18800 | 3 |
| AGTR1 | Agonist | 6 | 424900 | 2 |
| AGTRL1 | Agonist | 6 | 429300 | 1 |
| AVPR1A | Agonist | 6 | 23600 | 0 |
| AVPR1B | Agonist | 6 | 35200 | 0 |
| AVPR2 | Agonist | 6 | 822800 | 0 |
| BDKRB1 | Agonist | 6 | 30100 | 1 |
| BDKRB2 | Agonist | 6 | 663600 | 0 |
| BRS3 | Agonist | 6 | 209300 | 0 |
| C3AR1 | Agonist | 6 | 55900 | 0 |
| C5AR1 | Agonist | 6 | 119400 | 0 |
| C5L2 | Agonist | 6 | 164800 | 0 |
| CALCR | Agonist | 6 | 42500 | 1 |
| CALCRL-RAMP1 | Agonist | 6 | 92800 | 0 |
| CALCRL-RAMP2 | Agonist | 6 | 219200 | 1 |
| CALCRL-RAMP3 | Agonist | 6 | 425200 | 0 |
| CALCR-RAMP2 | Agonist | 6 | 139500 | 2 |
| CALCR-RAMP3 | Agonist | 6 | 28600 | 7 |
| CCKAR | Agonist | 6 | 44600 | 0 |
| CCKBR | Agonist | 6 | 894800 | 0 |
| CCR10 | Agonist | 6 | 93400 | 0 |
| CCR1 | Agonist | 6 | 540700 | 7 |
| CCR2 | Agonist | 6 | 67000 | 0 |
| CCR3 | Agonist | 6 | 272100 | 2 |
| CCR4 | Agonist | 6 | 180300 | 0 |
| CCR5 | Agonist | 6 | 89800 | 0 |
| CCR6 | Agonist | 6 | 141000 | 0 |
| CCR7 | Agonist | 6 | 766200 | 1 |
| CCR8 | Agonist | 6 | 35900 | 0 |
| CCR9 | Agonist | 6 | 119300 | 1 |
| CHRM1 | Agonist | 6 | 1181100 | 1 |
| CHRM2 | Agonist | 6 | 54600 | 1 |
| CHRM3 | Agonist | 6 | 166300 | 2 |
| CHRM4 | Agonist | 6 | 787900 | 16 |
| CHRM5 | Agonist | 6 | 2995100 | 5 |
| CMKLR1 | Agonist | 6 | 81900 | 0 |
| CNR1 | Agonist | 6 | 80000 | 0 |
| CNR2 | Agonist | 6 | 315400 | −2 |
| CRHR1 | Agonist | 6 | 361400 | 1 |
| CRHR2 | Agonist | 6 | 161100 | 0 |
| CRTH2 | Agonist | 6 | 172600 | 0 |
| CX3CR1 | Agonist | 6 | 342700 | 1 |
| CXCR1 | Agonist | 6 | 219900 | 0 |
| CXCR2 | Agonist | 6 | 165200 | 1 |
| CXCR3 | Agonist | 6 | 387900 | 1 |
| CXCR4 | Agonist | 6 | 72500 | 2 |
| CXCR5 | Agonist | 6 | 230900 | 1 |
| CXCR6 | Agonist | 6 | 27700 | 2 |
| CXCR7 | Agonist | 6 | 194500 | 0 |
| DRD1 | Agonist | 6 | 73000 | 0 |
| DRD2L | Agonist | 6 | 83500 | 0 |
| DRD2S | Agonist | 6 | 247200 | 0 |
| DRD3 | Agonist | 6 | 414700 | 2 |
| DRD4 | Agonist | 6 | 22800 | 3 |
| DRD5 | Agonist | 6 | 21000 | 1 |
| EBI2 | Agonist | 6 | 150100 | 0 |
| EDG1 | Agonist | 6 | 165500 | 0 |
| EDG3 | Agonist | 6 | 877900 | 0 |
| EDG4 | Agonist | 6 | 234300 | 4 |
| EDG5 | Agonist | 6 | 174300 | 1 |
| EDG6 | Agonist | 6 | 574100 | −2 |
| EDG7 | Agonist | 6 | 154400 | 0 |
| EDNRA | Agonist | 6 | 38500 | 0 |
| EDNRB | Agonist | 6 | 68900 | 0 |
| F2R | Agonist | 6 | 470700 | −2 |
| F2RL1 | Agonist | 6 | 566200 | 0 |
| F2RL3 | Agonist | 6 | 909900 | −1 |
| FFAR1 | Agonist | 6 | 560800 | 3 |
| FPR1 | Agonist | 6 | 1133900 | 4 |
| FPRL1 | Agonist | 6 | 63900 | 0 |
| FSHR | Agonist | 6 | 197900 | −2 |
| GALR1 | Agonist | 6 | 263400 | 1 |
| GALR2 | Agonist | 6 | 307700 | 1 |
| GCGR | Agonist | 6 | 295600 | 0 |
| GHSR | Agonist | 6 | 524600 | 2 |
| GIPR | Agonist | 6 | 17500 | −1 |
| GLP1R | Agonist | 6 | 123500 | 0 |
 | Table IVPRP-1 activity with nhrMAX panel. |
Table IV
PRP-1 activity with nhrMAX panel.
| Assay format | Assay target | Conc
(μM) | Value 1 | Value 2 | Average value | SD | % Efficacy |
|---|
| Agonist | AR | 6 | 3600 | 3200 | 3400 | 282.84 | −0.3 |
| Antagonist | AR | 6 | 17200 | 17400 | 17300 | 141.42 | 4.3 |
| Agonist | ERalpha | 6 | 50400 | 50000 | 50200 | 282.84 | 0.6 |
| Antagonist | ERalpha | 6 | 293600 | 288200 | 290900 | 3818.4 | −2.8 |
| Antagonist | ERRalpha | 6 | 65800 | 61200 | 63500 | 3252.7 | −4.7 |
| Inverse
agonist | ERRalpha | 6 | 134600 | 120000 | 127300 | 10324 | −7.2 |
| Agonist | FXR | 6 | 4000 | 6200 | 5100 | 1555.6 | 0.9 |
| Antagonist | FXR | 6 | 95400 | 103400 | 99400 | 5656.9 | −4.5 |
| Agonist | GR | 6 | 15200 | 16800 | 16000 | 1131.4 | 0.3 |
| Antagonist | GR | 6 | 999800 | 1086200 | 1043000 | 61094 | −9.7 |
| Agonist | LXRalpha | 6 | 232800 | 216800 | 224800 | 11314 | −0.6 |
| Antagonist | LXRalpha | 6 | 1660000 | 1807600 | 1733800 | 104370 | −10.7 |
| Agonist | LXRbeta | 6 | 311400 | 330200 | 320800 | 13294 | 1.6 |
| Antagonist | LXRbeta | 6 | 1208200 | 1379200 | 1293700 | 120920 | −4 |
| Agonist | MR | 6 | 16200 | 18000 | 17100 | 1272.8 | 0.3 |
| Antagonist | MR | 6 | 295400 | 303800 | 299600 | 5939.7 | −0.7 |
| Agonist | PPARalpha | 6 | 14600 | 15000 | 14800 | 282.84 | 0.9 |
| Antagonist | PPARalpha | 6 | 108400 | 105000 | 106700 | 2404.2 | −3.3 |
| Agonist | PPARdelta | 6 | 1077800 | 1247000 | 1162400 | 119640 | 0.9 |
| Antagonist | PPARdelta | 6 | 3209000 | 2884800 | 3046900 | 229240 | 2.2 |
| Agonist | PPARgamma | 6 | 4400 | 5800 | 5100 | 989.95 | 0.3 |
| Antagonist | PPARgamma | 6 | 29600 | 28400 | 29000 | 848.53 | 6.5 |
| Agonist | PRalpha | 6 | 25800 | 22800 | 24300 | 2121.3 | −1.6 |
| Antagonist | PRalpha | 6 | 185400 | 190400 | 187900 | 3535.5 | −3.1 |
| Agonist | PRbeta | 6 | 3200 | 2400 | 2800 | 565.69 | 1.4 |
| Antagonist | PRbeta | 6 | 28600 | 33600 | 31100 | 3535.5 | −0.6 |
| Agonist | RARalpha | 6 | 29400 | 44800 | 37100 | 10889 | −3.3 |
| Antagonist | RARalpha | 6 | 104000 | 106800 | 105400 | 1979.9 | 14 |
| Agonist | RARbeta | 6 | 263600 | 243600 | 253600 | 14142 | −3.9 |
| Antagonist | RARbeta | 6 | 475000 | 572200 | 523600 | 68731 | −0.5 |
| Agonist | RXRalpha | 6 | 226000 | 230000 | 228000 | 2828.4 | −3.8 |
| Antagonist | RXRalpha | 6 | 851800 | 821400 | 836600 | 21496 | 0 |
| Agonist | RXRgamma | 6 | 367800 | 354200 | 361000 | 9616.7 | −2.7 |
| Antagonist | RXRgamma | 6 | 1266600 | 1310200 | 1288400 | 30830 | −1.2 |
| Agonist | THRalpha | 6 | 43400 | 34600 | 39000 | 6222.5 | −0.2 |
| Antagonist | THRalpha | 6 | 383000 | 415800 | 399400 | 23193 | −4.8 |
| Agonist | THRbeta | 6 | 732400 | 819600 | 776000 | 61660 | 9.3 |
| Antagonist | THRbeta | 6 | 1274000 | 1246600 | 1260300 | 19375 | −5.2 |
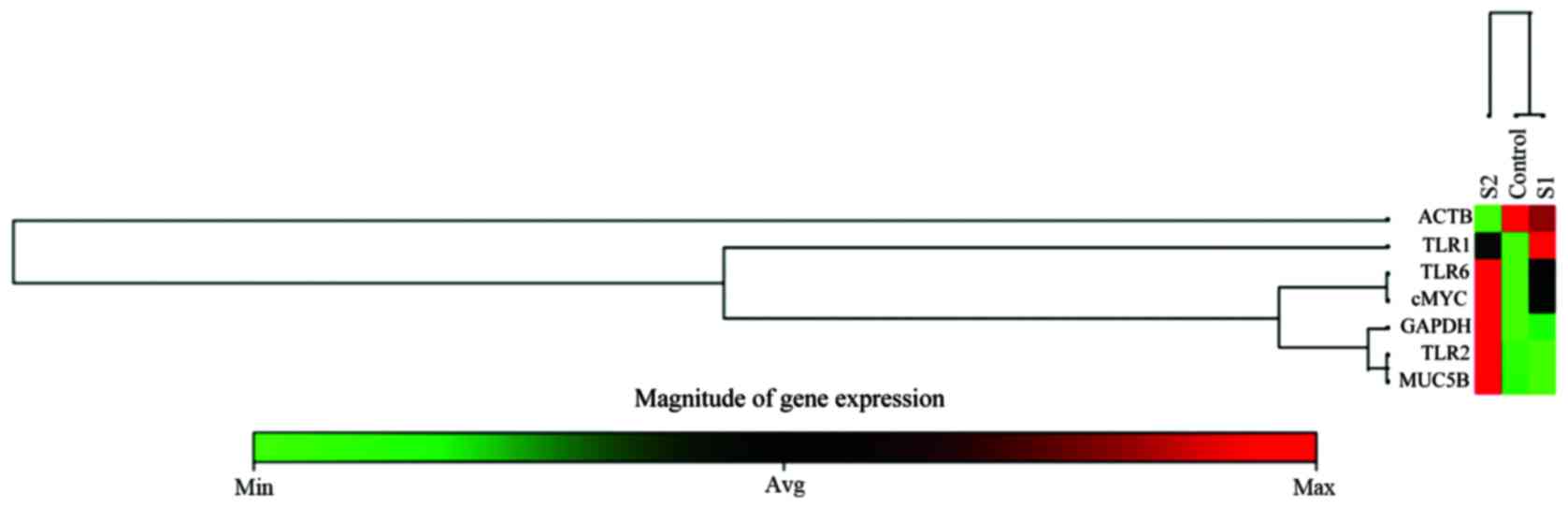
RT2 qPCR primer assays show the effect of
PRP-1 on gene expression of TLR receptors and MUC5B
RT2 qPCR custom designed primer assays were
performed by Qiagen to understand the effect of PRP-1 on gene
expression of TLR receptors and MUC5B. Mature RNA was isolated
using RNA extraction according to the manufacturer's instructions.
RNA was subjected to spectrophotometrical quality control and then
reverse transcribed to cDNA. RT2 SYBR-Green qPCR Master Mix was
used with RT2 qPCR assays. In this study, 7 genes (TLR1, TLR2,
TLR6, MUC5B, c-Myc and 2 housekeeping genes GAPDH and ACTB) were
profiled on three samples with technical triplicates. c-Myc was
included, as we wanted to confirm its drastic downregulation after
PRP-1 treatment in luciferase assay (4) and western blot experiments, reported
earlier (10). The heat map,
clustergram of average Ct values across the gene of each sample,
with the magnitude of gene expression scale below, is presented in
Fig. 4. As evident from the
figure, there is dose-dependent effect of PRP-1 on the expression
of the above mentioned genes, except the control housekeeping
genes. The TLR1 receptor was well expressed in the cells treated
with 10 μg/ml, whereas no expression was detected at 1
μg/ml peptide treatment. TLR2, TLR6, MUC5B demonstrated high
expression with 1 μg/ml treatment when compared to
nontreated control. c-Myc expression went down drastically when
treated with 10 μg/ml PRP-1. The data analysis web portal
calculated fold change/regulation using ∆∆Ct method, in which ∆Ct
is calculated between gene of interest (GOI) and an average of
housekeeping genes (HKG) followed by ∆∆Ct calculations (∆Ct
(experiment) − ∆Ct (control). Fold change is then calculated using
the 2−∆∆Ct formula. The system detected only
statistically significant upregulation (P<0.0001) of TLR2 for
the samples treated with 1 μg/ml PRP-1, with 5.26-fold
upregulation when calculated with the ∆∆Ct.
Discussion
Metastatic chondrosarcoma is fatal because of
metastatic spread and absence of the effective therapies.
Therefore, search for new approaches is of the utmost importance.
PRP-1, inhibits chondrosarcoma cell growth by >80% (4,6) it
halts cell cycle progression in G1/S transition (6). This mTORC1 inhibitor, cytostatic
peptide is potent upregulator of tumor suppressors and inhibitor of
oncoproteins and embryonic stem cell markers (7–9). We
have demonstrated also that intracellular expression of PRP-1 is
associated with the early stages of lymphocyte activation by
phytohemagglutinin, (PHA) (Fig.
5). However, the interacting partners or receptors for this
important peptide has not been identified. Using triCEPS (ligand
based receptor capture technology), we were able to identify MUC5B,
one of the members of mucin family as the receptor for PRP-1.
Notably, proline rich proteins in saliva, different from the
neuropeptide PRP-1, were found in reported literature to interact
with mucins (48). Immunoblot
results of this study indicated that TLR1, TLR2 (which are usually
dimerized) and TLR6 are binding interaction partners for PRP-1, as
their expression increased in dose-response manner upon PRP-1
treatment. Indeed, the link between TLR1/2 and TLR6 was documented
in the literature. TLR1 and TLR6 was shown to pair with TLR2 and
that interaction was needed for pattern recognition of pathogens
(49,50). TLR7 was not expressed in human
JJ012 cell line at all, but all the other TLR groups were present.
No changes in the expression of TLR10 were observed with PRP-1,
although sometimes it was reported that in certain cases TLR10 is
able to homodimerize or heterodimerize with TLR1 and TLR2, but its
ligand remains unknown (19). We
have demonstrated that PRP-1 upregulates the expression of adaptor
protein TICAM2 (TRAM) but not of TICAM1 (TRIF). Most TLRs share a
common signaling pathway in which myeloid differentiation factor 88
(MyD88) plays a central role (51). It is known also that TLR2 can be
TRAM dependent in addition to the MyD88-dependent pathway with
certain MyD88 independent exceptions (51). TLR2 is also internalized following
ligand binding, but in this case, MyD88-dependent signaling
continues from an intracellular location away from the plasma
membrane and stimulates type I IFN production through an as yet
unknown mechanism (52). Due to
the importance of TLR signaling in tumorigenesis, TLR agonists have
potential for antitumor therapy (53–57).
Both TLR and mucins have important role in host defense mechanism,
however, the link between two of them in non-infectious conditions
and cancer pathology deserves attention. Understanding connecting
crosstalk between two of them will open new avenues for therapeutic
intervention. Cancer cells might use the TLR signaling pathways
much in the same way to upregulate the expression of MUCs which in
turn may also regulate TLR signaling (22). Mucins are a class of major
differentially expressed proteins between normal and cancer cells,
which makes them a potential target for anticancer therapies. As a
class of glycoproteins, MUCs are recognized as potential markers of
disease progression or inhibition (58) and are currently investigated as
therapeutic targets for cancer (59). Our experimental results indicated
the nuclear localization of both MUC5B and TLR1/2. Indeed the
evidence of their nuclear translocation was reported in the
literature (24,60). Due to the importance of TLR
signaling in tumorigenesis, TLR agonists have potential for
antitumor therapy (22,54–58).
The fact that PRP-1 has receptors of innate immunity explains
observed antibacterial properties of PRP (61). The biochemical evidence for the
direct interaction of TLRs or MUC5B with endogenous stimulators is
limited. There is no doubt that it is of great significance to
identify those ligands and elucidate their biological functions,
especially if upon their binding with the ligand, the
antiproliferative effect in tumor is manifested. The western blot
analysis and immunocytochemistry data indicated upregulated protein
expression of TLR1, TLR6, MUC5B after the treatment with PRP-1 in
JJ012 human chondrosarcoma cell line, the custom designed RT2 qPCR
primer assays proved that PRP-1 indeed has effect on expression
levels of its interacting partner genes as well. However, depending
on the method it showed some dose response differences. For
example, in case of TLR2 the protein expression upregulation with 1
and 10 μg/ml PRP-1 was observed in dose-response manner in
western blot experiments. However, the heat map and qRT-PCR ∆Ct
calculations proved that the highest upregulation of TLR2
expression is taking place at 1 μg/ml (>5-fold
upregulation). On the gene expression level, the qRT-PCR did not
report significant fold change in ∆Ct for TLR1 or TLR6, whereas in
western blot experiments we saw obvious upregulation of protein
expression for these respective receptors after the dose response
treatment with PRP-1. MUC5B on the heat map demonstrated the most
upregulation after the treatment with 1 μg/ml, which
coincided with MUC5B ELISA results, though TriCEPS technology
detected MUC5B as binding partner with 10 μg/ml PRP-1
treatment. c-Myc results demonstrated downregulation of its gene
expression in dose-response manner with PRP-1, being downregulated
very strongly at 10 μg/ml, which is concordant with our
previous results (10). Despite
these differences, it is important to mention that most of
inhibitory responses caused by PRP-1 treatment on cell growth of
tumor cell lines or upregulation of tumor suppressors and
down-regulation of oncoproteins, were maximally observed when
treated with 1–20 μg/ml PRP-1 range. PRP-1 is a compound
naturally produced in the body and the fact it was detected in the
nucleus of chondrosarcoma cells and that it upregulated TLR1/2
dimer and TLR6 possibly indicates PRP-1 as an endogenous
ligand.
The ability of TLRs to recognize endogenous
stimulators appears to be essential to their function in regulating
noninfectious (sterile) inflammation. TLR-induced innate immune
responses regulate non-infectious sterile inflammation and
subsequently, adaptive immune response. The endogenous TLR ligands
and their receptors can be localized in different cellular
compartments and cannot interact physiologically. However, when the
tissue is injured, the passive release of endogenous ligand or its
active transport utilizing non-conventional lysosomal route. In the
present study, we were able to identify the pattern recognition
receptors of adaptive immunity TLR1, TLR2 and TLR6, and secreted
mucin MUC5B as binding partners for cytostatic PRP-1 peptide. The
mentioned results allow to understand the immunomodulatory,
antibacterial effect of PRP-1, reported by our group before
(3,42,61).
From oncologic standpoint it is important information that immune
receptors play antitumorigenic role when bound to PRP-1 ligand.
Acknowledgments
The present study was supported in part by a gift
from the Ratcliffe Foundation to Miami Center of Orthopedic
Research and Education. We would like to thank Qiagen Inc., Service
Core for the qRT-PCR experiments, the Analytical Imaging Core
Facility at DRI/SCCC, University of Miami, which provided
immunocytochemistry imaging service. Our thanks to Ms. Maria
Boulina for the help and guidance with the imaging. We would like
to thanks Dr Paul Helbling and Dr Florian Marty from Dual Systems
Biotech (Zurich), Switzerland, for their productive collaboration
and triCEPS methodology experiments.
References
|
1
|
Ozaki T, Hillmann A, Lindner N, Blasius S
and Winkelmann W: Metastasis of chondrosarcoma. J Cancer Res Clin
Oncol. 122:625–628. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mirra J: Bone Tumors: Clinical,
radiologic, and pathologic correlations. Lea and Febiger;
Philadelphia, PA: 1989
|
|
3
|
Galoyan A: Neurochemistry of brain
neuroendocrine immune system: Signal molecules. Neurochem Res.
25:1343–1355. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Galoian K, Temple TH and Galoyan A:
Cytostatic effect of the hypothalamic cytokine PRP-1 is mediated by
mTOR and cMyc inhibition in high grade chondrosarcoma. Neurochem
Res. 36:812–818. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galoian K, Temple HT and Galoyan A: mTORC1
inhibition and ECM-cell adhesion-independent drug resistance via
PI3K-AKT and PI3K-RAS-MAPK feedback loops. Tumour Biol. 33:885–890.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Galoian KA, Temple TH and Galoyan A:
Cytostatic effect of novel mTOR inhibitor, PRP-1 (galarmin) in MDA
231 (ER-) breast carcinoma cell line. PRP-1 inhibits mesenchymal
tumors. Tumour Biol. 32:745–751. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Galoian KA, Guettouche T, Issac B, Qureshi
A and Temple HT: Regulation of onco and tumor suppressor MiRNAs by
mTORC1 inhibitor PRP-1 in human chondrosarcoma. Tumour Biol.
35:2335–2341. 2014. View Article : Google Scholar
|
|
8
|
Galoian K, Qureshi A, Wideroff G and
Temple HT: Restoration of desmosomal junction protein expression
and inhibition of H3K9-specific histone demethylase activity by
cytostatic proline-rich polypeptide-1 leads to suppression of
tumorigenic potential in human chondrosarcoma cells. Mol Clin
Oncol. 3:171–178. 2015. View Article : Google Scholar
|
|
9
|
Galoian K, Luo S, Qureshi A, Patel P,
Price R, Morse AS, Chailyan G, Abrahamyan S and Temple HT: Effect
of cytostatic proline rich polypeptide-1 on tumor suppressors of
inflammation pathway signaling in chondrosarcoma. Mol Clin Oncol.
5:618–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Galoian K, Qureshi A, D'Ippolito G,
Schiller PC, Molinari M, Johnstone AL, Brothers SP, Paz AC and
Temple HT: Epigenetic regulation of embryonic stem cell marker
miR302C in human chondrosarcoma as determinant of antiproliferative
activity of proline-rich polypeptide 1. Int J Oncol. 47:465–472.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu L, Wang L and Chen S: Exogenous or
endogenous Toll-like receptor ligands: Which is the MVP in
tumorigenesis? Cell Mol Life Sci. 69:935–949. 2012. View Article : Google Scholar
|
|
12
|
Rakoff-Nahoum S and Medzhitov R: Toll-like
receptors and cancer. Nat Rev Cancer. 9:57–63. 2009. View Article : Google Scholar
|
|
13
|
Joshi S, Kumar S, Choudhury A, Ponnusamy
MP and Batra SK: Altered Mucins (MUC) trafficking in benign and
malignant conditions. Oncotarget. 5:7272–7284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blasius AL and Beutler B: Intracellular
toll-like receptors. Immunity. 32:305–315. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang B, Zhao J, Unkeless JC, Feng ZH and
Xiong H: TLR signaling by tumor and immune cells: A double-edged
sword. Oncogene. 27:218–224. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Apetoh L, Ghiringhelli F, Tesniere A,
Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E,
Saulnier P, et al: Toll-like receptor 4-dependent contribution of
the immune system to anticancer chemotherapy and radiotherapy. Nat
Med. 13:1050–1059. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Medzhitov R: Origin and physiological
roles of inflammation. Nature. 454:428–435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seong SY and Matzinger P: Hydrophobicity:
An ancient damage-associated molecular pattern that initiates
innate immune responses. Nat Rev Immunol. 4:469–478. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hasan U, Chaffois C, Gaillard C, Saulnier
V, Merck E, Tancredi S, Guiet C, Brière F, Vlach J, Lebecque S, et
al: Human TLR10 is a functional receptor, expressed by B cells and
plasmacytoid dendritic cells, which activates gene transcription
through MyD88. J Immunol. 174:2942–2950. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lowe EL, Crother TR, Rabizadeh S, Hu B,
Wang H, Chen S, Shimada K, Wong MH, Michelsen KS and Arditi M:
Toll-like receptor 2 signaling protects mice from tumor development
in a mouse model of colitis-induced cancer. PLoS One. 5:e130272010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu L, Wang L and Chen S: Endogenous
toll-like receptor ligands and their biological significance. J
Cell Mol Med. 14:2592–2603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tarang S, Kumar S and Batra SK: Mucins and
toll-like receptors: Kith and kin in infection and cancer. Cancer
Lett. 321:110–119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanzler H, Barrat FJ, Hessel EM and
Coffman RL: Therapeutic targeting of innate immunity with Toll-like
receptor agonists and antagonists. Nat Med. 13:552–559. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lakshminarayanan V, Thompson P, Wolfert
MA, Buskas T, Bradley JM, Pathangey LB, Madsen CS, Cohen PA,
Gendler SJ and Boons GJ: Immune recognition of tumor-associated
mucin MUC1 is achieved by a fully synthetic aberrantly glycosylated
MUC1 tripartite vaccine. Proc Natl Acad Sci USA. 109:261–266. 2012.
View Article : Google Scholar :
|
|
25
|
Hollingsworth MA and Swanson BJ: Mucins in
cancer: Protection and control of the cell surface. Nat Rev Cancer.
4:45–60. 2004. View Article : Google Scholar
|
|
26
|
Remmers N, Anderson JM, Linde EM, DiMaio
DJ, Lazenby AJ, Wandall HH, Mandel U, Clausen H, Yu F and
Hollingsworth MA: Aberrant expression of mucin core proteins and
o-linked glycans associated with progression of pancreatic cancer.
Clin Cancer Res. 19:1981–1993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sóñora C, Mazal D, Berois N, Buisine MP,
Ubillos L, Varangot M, Barrios E, Carzoglio J, Aubert JP and
Osinaga E: Immunohistochemical analysis of MUC5B apomucin
expression in breast cancer and non-malignant breast tissues. J
Histochem Cytochem. 54:289–299. 2006. View Article : Google Scholar
|
|
28
|
Kim YS, Gum J Jr and Brockhausen I: Mucin
glycoproteins in neoplasia. Glycoconj J. 13:693–707. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Turner MS, McKolanis JR, Ramanathan RK,
Whitcomb DC and Finn OJ: Mucins in gastrointestinal cancers. Cancer
Chemother Biol Response Modif. 21:259–274. 2003. View Article : Google Scholar
|
|
30
|
Berois N, Varangot M, Sóñora C,
Zarantonelli L, Pressa C, Laviña R, Rodríguez JL, Delgado F,
Porchet N, Aubert JP, et al: Detection of bone marrow-disseminated
breast cancer cells using an RT-PCR assay of MUC5B mRNA. Int J
Cancer. 103:550–555. 2003. View Article : Google Scholar
|
|
31
|
Moniaux N, Andrianifahanana M, Brand RE
and Batra SK: Multiple roles of mucins in pancreatic cancer, a
lethal and challenging malignancy. Br J Cancer. 91:1633–1638. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Andrianifahanana M, Moniaux N and Batra
SK: Regulation of mucin expression: Mechanistic aspects and
implications for cancer and inflammatory diseases. Biochim Biophys
Acta. 1765:189–222. 2006.PubMed/NCBI
|
|
33
|
Kufe DW: Mucins in cancer: Function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Velcich A, Yang W, Heyer J, Fragale A,
Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K and
Augenlicht L: Colorectal cancer in mice genetically deficient in
the mucin Muc2. Science. 295:1726–1729. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Van Seuningen I, Perrais M, Pigny P,
Porchet N and Aubert JP: Sequence of the 5′-flanking region and
promoter activity of the human mucin gene MUC5B in different
phenotypes of colon cancer cells. Biochem J. 348:675–686. 2000.
View Article : Google Scholar
|
|
36
|
Aziz MA, AlOtaibi M, AlAbdulrahman A,
AlDrees M and AlAbdulkarim I: Mucin family genes are downregulated
in colorectal cancer patients. J Carcinogene Mutagene.
S10:009:2014.
|
|
37
|
Wakata K, Tsuchiya T, Tomoshige K, Takagi
K, Yamasaki N, Matsumoto K, Miyazaki T, Nanashima A, Whitsett JA,
Maeda Y, et al: A favourable prognostic marker for EGFR mutant
non-small cell lung cancer: Immunohistochemical analysis of MUC5B.
BMJ Open. 5:e0083662015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Roy MG, Livraghi-Butrico A, Fletcher AA,
McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK,
Song AS, Petrova YM, et al: Muc5b is required for airway defence.
Nature. 505:412–416. 2014. View Article : Google Scholar
|
|
39
|
Vincent A, Perrais M, Desseyn JL, Aubert
JP, Pigny P and Van Seuningen I: Epigenetic regulation (DNA
methylation, histone modifications) of the 11p15 mucin genes (MUC2,
MUC5AC, MUC5B, MUC6) in epithelial cancer cells. Oncogene.
26:6566–6576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Macha MA, Krishn SR, Jahan R, Banerjee K,
Batra SK and Jain M: Emerging potential of natural products for
targeting mucins for therapy against inflammation and cancer.
Cancer Treat Rev. 41:277–288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Markossian KA, Gurvits BY and Galoyan AA:
Isolation and identification of novel peptides from secretory
granules of neuro-hypophysis. Neurochem Res. 16:221999.
|
|
42
|
Galoyan AA: Brain neurosecretory
cytokines: immune response and neuronal survival. Kluwer Academic
Plenum Publishers; New York: 2004, https://doi.org/10.1007/978-1-4419-8893-5.
View Article : Google Scholar
|
|
43
|
Abrahamyan SS, Davtyan TK, Khachatryan AR,
Tumasyan NV, Sahakyan IK, Harutyunyan HA, Chailyan SG and Galoyan
AA: Quantification of the hypothalamic proline rich polypeptide-1
in rat blood serum. Neurochem J. 8:38–43. 2014. View Article : Google Scholar
|
|
44
|
Yan YX, Boldt-Houle DM, Tillotson BP, Gee
MA, D'Eon BJ, Chang XJ, Olesen CE and Palmer MA: Cell-based
high-throughput screening assay system for monitoring G
protein-coupled receptor activation using beta-galactosidase enzyme
complementation technology. J Biomol Screen. 7:451–459. 2002.
View Article : Google Scholar
|
|
45
|
Frei AP, Moest H, Novy K and Wollscheid B:
Ligand-based receptor identification on living cells and tissues
using TRICEPS. Nat Protoc. 8:1321–1336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Slavoff SA and Saghatelian A: Discovering
ligand-receptor interactions. Nat Biotechnol. 30:959–961. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Omasits U, Ahrens CH, Müller S and
Wollscheid B: Protter: Interactive protein feature visualization
and integration with experimental proteomic data. Bioinformatics.
30:884–886. 2014. View Article : Google Scholar
|
|
48
|
Senapati S, Das S and Batra SK:
Mucin-interacting proteins: From function to therapeutics. Trends
Biochem Sci. 35:236–245. 2010. View Article : Google Scholar
|
|
49
|
Ozinsky A, Underhill DM, Fontenot JD,
Hajjar AM, Smith KD, Wilson CB, Schroeder L and Aderem A: The
repertoire for pattern recognition of pathogens by the innate
immune system is defined by cooperation between toll-like
receptors. Proc Natl Acad Sci USA. 97:13766–13771. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Janssens S and Beyaert R: Role of
Toll-like receptors in pathogen recognition. Clin Microbiol Rev.
16:637–646. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nilsen N, Nonstad U, Khan N, Knetter CF,
Akira S, Sundan A, Espevik T and Lien E: Lipopolysaccharide and
double-stranded RNA up-regulate toll-like receptor 2 independently
of myeloid differentiation factor 88. J Biol Chem. 279:39727–39735.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Seibert SA, Mex P, Köhler A, Kaufmann SH
and Mittrücker HW: TLR2-, TLR4- and Myd88-independent acquired
humoral and cellular immunity against Salmonella enterica serovar
Typhimurium. Immunol Lett. 127:126–134. 2010. View Article : Google Scholar
|
|
53
|
Jeung HC, Moon YW, Rha SY, Yoo NC, Roh JK,
Noh SH, Min JS, Kim BS and Chung HC: Phase III trial of adjuvant
5-fluorouracil and adriamycin versus 5-fluorouracil, adriamycin,
and polyad-enylic-polyuridylic acid (poly A:U) for locally advanced
gastric cancer after curative surgery: final results of 15-year
follow-up. Ann Oncol. 19:520–526. 2008. View Article : Google Scholar
|
|
54
|
Smits EL, Ponsaerts P, Berneman ZN and Van
Tendeloo VF: The use of TLR7 and TLR8 ligands for the enhancement
of cancer immunotherapy. Oncologist. 13:859–875. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Leonard JP, Link BK, Emmanouilides C,
Gregory SA, Weisdorf D, Andrey J, Hainsworth J, Sparano JA, Tsai
DE, Horning S, et al: Phase I trial of toll-like receptor 9 agonist
PF-3512676 with and following rituximab in patients with recurrent
indolent and aggressive non Hodgkin's lymphoma. Clin Cancer Res.
13:6168–6174. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mikulandra M, Pavelic J and Glavan TM:
Recent findings on the application of Toll- like receptors agonists
in cancer therapy. Curr Med Chem. 24:2011–2032. 2017. View Article : Google Scholar
|
|
57
|
Kaczanowska S, Joseph AM and Davila E: TLR
agonists: Our best frenemy in cancer immunotherapy. J Leukoc Biol.
93:847–863. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rachagani S, Torres MP, Moniaux N and
Batra SK: Current status of mucins in the diagnosis and therapy of
cancer. Biofactors. 35:509–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Planque N: Nuclear trafficking of secreted
factors and cell-surface receptors: New pathways to regulate cell
proliferation and differentiation, and involvement in cancers. Cell
Commun Signal. 4:72006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huhta H, Helminen O, Lehenkari PP, Saarnio
J, Karttunen TJ and Kauppila JH: Toll-like receptors 1, 2, 4 and 6
in esophageal epithelium, Barrett's esophagus, dysplasia and
adenocarcinoma. Oncotarget. 7:23658–23667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Galoyan AA: Brain immune system signal
molecules in protection from aerobic and anaerobic infections.
Advances in Neurobiology. 6. Springer; 2012, View Article : Google Scholar
|