Introduction
Hepatocellular carcinoma (HCC) remains a serious
health burden worldwide (1). In
China, >466,100 new cases and 422,100 HCC-associated deaths were
reported in 2015 (2). Clinical
experience indicates that patients affected by tumors of a large
size usually have a poor prognosis (3). This may be ascribed to two main
reasons: The loss of surgical opportunity due to the large tumor
size, and fatal complications following a massive hepatectomy.
Thus, the control of tumor growth is a critical strategy for
improving the prognosis of patients with HCC.
Long non-coding RNAs (lncRNAs) are a class of
abundant and largely uncharacterized non-protein-coding RNAs that
are >200 nucleotides in length (4). lncRNAs have been demonstrated to
regulate cell proliferation in HCC (5). lncRNA TUG1 has been shown to promote
HCC growth in vitro and in vivo by epigenetically
suppressing Kruppel-like factor 2 (KLF2) transcription (6). lncRNA HOTAIR has been shown to
increase cell viability by enhancing glucose transporter isoform 1
(GLUT1)-mediated glycolysis (7).
lncRNA KCNQ1 opposite strand/antisense transcript 1 (known as
KCNQ1OT1) was initially identified as an aberrantly expressed
lncRNA in Beckwith-Wiedemann syndrome, which is an overgrowth
disorder usually presenting in the embryonic stages (8). Recently, KCNQ1OT1 was found to be
required for the proliferation of cancer cells. In breast cancer, a
high KCNQ1OT1 expression has been shown to be associated with a
large tumor size (9). Yoshizawa
et al (10) found that
pyrrole-imidazole polyamide bound to the CCAAT boxes of the
KCNQ1OT1 promoter region to silence its expression. Moreover,
KCNQ1OT1 silencing by this drug induced the death of Wilms'
tumor-derived G401 cells (10).
These results indicate that KCNQ1OT1 may be a novel therapeutic
target for patients with high intra-tumoral levels of KCNQ1OT1.
In the present study, we confirmed the cell
growth-related lncRNA KCNQ1OT1 to be critical for HCC cell growth
in vitro and in vivo. The overexpression of KCNQ1OT1
was found to be associated with a large tumor size and a poor
prognosis of patients with HCC. Additionally, KCNQ1OT1 was found to
function as a competing endogenous RNA sponge for microRNA-504
(miR-504). Thus, cyclin-dependent kinase 16 (CDK16), a downstream
target of miR-504, was harbored by KCNQ1OT1 to facilitate its tumor
growth-promoting effects.
Materials and methods
Clinical samples and cell lines
A total of 50 pairs of liquid nitrogen-stored HCC
tissues and matched tumor-adjacent tissues were collected from 30
male patients and 20 female patients at the Department of
Hepatobiliary Surgery, First Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China) between January 2011 and January 2013.
Written informed consent was obtained from all patients enrolled in
this study. The use of clinical samples was approved by the Ethics
Committee of First Affiliated Hospital of Xi'an Jiaotong
University. In total, 5 HCC cell lines (Huh-7, SMMC-7721, Hep3B,
MHCC97-H and MHCC97-L), the human immortalized normal hepatocyte
cell line, LO2, and 293T cells were obtained from the Shanghai
Institute of Biochemistry and Cell Biology, Chinese Academy of
Sciences (Shanghai, China), and maintained in our laboratory. The
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
containing 10% fetal bovine serum (FBS) (both from Gibco, Carlsbad,
CA, USA) and 1% V/V penicillin/streptomycin (MilliporeSigma,
Bedford, MA, USA) in a humidified atmosphere containing 5%
CO2 at 37°C.
Cancer Genome Atlas dataset
Gene Expression Omnibus (GEO) data for 95 HCC
samples and 39 normal liver samples were collected from the dataset
entitled Gene Expression Profiles of Human Hepatocellular Carcinoma
(GSE45436), and pre-analyzed using R2: Genomics Analysis and
Visualization Platform (http://r2.amc.nl).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from tissues and cells was isolated using
the miRNeasy Mini kit (Qiagen, Germantown, MD, USA). The KCNQ1OT1
levels, miR-504 levels and CDK16 mRNA levels were measured by
RT-PqCR using a SuperScript III Reverse Transcriptase kit
(Invitrogen, Carlsbad, CA, USA) and an iTaq Universal SYBR-Green
Supermix kit (Bio-Rad Laboratories, Hercules, CA, USA). The
Bulge-Loop miR-504 qPCR Primer Set was purchased from RiboBio Co.,
Ltd. (Guangzhou, China). Other primer sequences were as follows:
KCNQ1OT1 forward, 5′-CTTTGCAGCAACCTCCTTGT-3′ and reverse,
5′-TGGGGTGAGGGATCTGAA-3′; CDK16 forward, 5′-CCGTCGTGTCAGCCTATCT-3′
and reverse, 5′-CTTCTCCGTGTGGATAATGTCA-3′; and GAPDH forward,
5′-CCAGGGCTGCTTTTAACTCT-3′ and reverse, 5′-GGACTCCACGACGTACTCA-3′.
An ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City,
CA, USA) was used to perform the qPCR assay. The thermocycling
conditions were as follows: A holding stage at 50°C for 2 min, and
95°C for 10 min; PCR stage (40 cycles) at 95°C for 15 sec, and 60°C
for 1 min. The 2−ΔΔCq method (11) was used to calculate the relative
gene expression normalized by GAPDH.
Cell transduction and transfection
KCNQ1OT1-specific shRNA (sh-KCNQ1OT1) and negative
control shRNA (sh-Ctrl) were designed and synthesized by
GeneCopoeia (Guangzhou, China), and cloned into the psi-LVRH1GP
lentiviral vectors. Recombinant lentiviruses were transduced into
the Huh-7 and SMMC-7721 cells. miR-504 mimics (miR-504), negative
control mimics (miR-Ctrl), miR-504 inhibitor (anti-miR-504) and a
negative control inhibitor (anti-miR-Ctrl) were purchased from
RiboBio Co., Ltd. The vectors, mimics and respective inhibitors
were transfected into the cells using Lipofectamine 3000 reagent
(Invitrogen/Thermo Fisher Scientific, Inc., Waltham, MA, USA), as
described in a previous study of ours (12).
Cell counting kit-8 (CCK-8) assay
The cancer cells were seeded into a 96-well plate at
2,000/well in quintuplicate, and cell viability was determined by
CCK-8 assay (E606335; Sangon, Shanghai, China) at 0, 24, 48 and 72
h. The absorbance of the samples was measured using a model 550
microplate reader (Bio-Rad Laboratories) at a wavelength of 450
nm.
Colony formation assay
A total of 400 cells were seeded into a 6-well plate
and maintained in DMEM containing 10% FBS for 2 weeks. Cell
colonies were fixed with 20% methanol and stained with 0.1% crystal
violet at room temperature for 15 min. The colonies were counted
using a ELIspot Bioreader 5000 (BIO-SYS, Karben, Germany).
Flow cytometry
Apoptotic cells were stained at the time point of 72
h of transfection with the Annexin V-PE/7-AAD apoptosis detection
kit (KGA-1017; KeyGEN, Nangjing, China). The cells
(2×105) were suspended with 50 µl binding buffer,
and received the following treatments: 5 µl/sample 7-AAD
with 15 min incubation; 1 µl/sample Annexin V-PE and 450
µl/sample binding buffer with 15 min of incubation.
Positively stained cells were detected using a BD FACSCanto II Flow
Cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Apo-ONE® homogeneous
caspase-3/7 assay
A total of 1×104 cells (100 µl)
were seeded into a 96-well plate. This was followed by the thawing
and mixing of the Caspase Substrate and Apo-ONE®
Caspase-3/7 Buffer to produce the Apo-ONE® Caspase-3/7
Reagent according to the manufacturer's instructions (Promega,
Madison, WI, USA) and then by the addition of 100 µl reagent
to each well. The wells were then incubated at room temperature for
4 h. The fluorescence of each well at 499 nm was measured using a
model 550 microplate reader (Bio-Rad Laboratories).
In vivo tumor growth assays
The in vivo experiments were performed after
obtaining ethics approval from the Biomedical Ethics Committee of
Xi'an Jiaotong University Health Science Center. A total of 12
BALB/cA male nude mice (6 weeks old, weighing 16–18 g) were
obtained from the Laboratory Animal Center of Xi'an Jiaotong
University Health Science Center, kept in sterilized cages in an
appropriate environment (25°C, 45% humidity, 12:12 light:dark
cycles), and fed a regular chow diet with water available ad
libitum. The mice were randomly divided into 4 groups as
follows: The Huh-7 sh-control group, Huh-7 sh-KCNQ1OT1 group,
SMMC-7721 sh-control group and the SMMC-7721 sh-KCNQ1OT1 group.
Modified Huh-7 and SMMC-7721 cells were subcutaneously injected
into the outside of the right hind limb of the nude mice to
establish tumor xenograft models. The tumor diameter was measured
using a vernier caliper every 7 days to monitor the tumor growth
from the time of the injection. The tumor volume was calculated as
follows: (length × width2)/2. All mice were kept under
the aforementioned conditions. After 4 weeks, the mice were
sacrificed by CO2 euthanasia (the flow rate of
CO2 was 20% displacement/min). The final weights of the
mice at the time of sacrifice were 20-23 g. Each mouse only had a
single tumor nodule. The maximum diameter of a single tumor
measured was 14.9 mm, and the largest single tumor volume was
1,251.31 mm3. Paraformaldehyde-fixed paraffin-embedded
mouse subcutaneous tumor tissue sections were created as previously
described (12).
Immunohistochemistry
Specific Ki-67 rabbit anti-human primary antibodies
(#9027) were purchased from Cell Signaling Technology (Danvers, MA,
USA) and diluted 1:100 with PBS to label the antigens at 4°C
overnight. Biotinylated goat anti-rabbit secondary antibodies were
used to label the combined primary antibodies. Complexes were
detected using HRP-streptavidin conjugates and visualized with DAB.
In total, 20 high power (×400) fields were randomly selected to
calculate the total cell numbers and Ki-67-positive cell numbers.
The percentage of Ki-67-positive cells was identified as the Ki-67
index.
Western blot analysis
The cells were lysed with RIPA buffer (HEART
Biotech, Xi'an, China) and quantified using a BCA protein assay kit
II (#5000002; Bio-Rad Laboratories). Protein samples (40 µg)
were separated by 10% SDS-PAGE and transferred onto nitrocellulose
membranes (Invitrogen/Thermo Fisher Scientific, Inc.) using a
Bio-Rad tank blotting system (Bio-Rad Laboratories). The membranes
were incubated with the appropriate primary antibodies at a 1:1,000
dilution overnight at 4°C. Horseradish peroxidase-conjugated
secondary antibodies at a 1:2,000 dilution were used to incubate
the membranes for 1 h at room temperature, after washing them with
TBST 3 times for 10 min. The targeting proteins on the membrane
were visualized with ECL reagents (Millipore, Plano, TX, USA). The
primary antibodies used in this study were purchased from Cell
Signaling Technology, and included those against glycogen synthase
kinase 3β (GSK3β) (#5676), GSK3βser9 (#9322), β-catenin (#8480),
Bcl-2 (#2872) and GAPDH (#5174). The secondary antibodies used,
included the Amersham ECL Donkey Anti-Mouse IgG Horseradish
Peroxidase-Linked Species-Specific Whole Antibody (NA931; GE
Healthcare Life Sciences, Pittsburgh, PA, USA). The relative band
density was determined using ImageJ software (NIH, Bethesda, ML,
USA).
Luciferase reporter assay
StarBase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php) software
was used to predict the target mRNA of miR-504. The wild-type (Wt)
fragments from KCNQ1OT1 containing the predicted miR-504 binding
sites and the corresponding mutant-type (Mt) fragments were cloned
into the pEZX-MT06 vectors (RiboBio Co., Ltd.), respectively.
Similarly, three online MicroRNA targets prediction tools,
TargetScan (http://www.targetscan.org/), PicTar (http://www.pictar.org/) and MiRanda (http://www.microrna.org/), were used to located the
binding sites between miR-1271 and PTP4A1 3′-UTR region. The Wt
fragment from the CDK16 3′-UTR region and the corresponding Mt
fragment were cloned into the pEZX-MT06 vectors. The vectors and
miR-504 mimics were then co-transfected into 293T cells using
Lipofectamine 3000 reagent, and the Luc-Pair Duo-Luciferase assay
kit 2.0 (GeneCopoeia) was used to determine the relative Rluc/Luc
ratio.
Statistical analysis
Continuous data are presented as the means ± SD.
Statistical analysis was performed using SPSS version 21.0 (SPSS
Inc., Chicago, IL, USA) and GraphPad Prism 5 (GraphPad Software,
Inc., La Jolla, CA, USA). Correlations between KCNQ1OT1 and
clinical characteristics were analyzed using the Pearson's
Chi-squared test. The differences between 2 groups were analyzed
using a Student's t-test. ANOVA was used to compare the data from
multiple groups. The Bonferroni method was used as the post hoc
test. Survival analysis was performed using a Kaplan-Meier curve.
The hazard ratio and confidence interval were determined using the
log-rank (Mantel-Cox) test. The respective correlations between
KCNQ1OT1 and miR-504, and CDK16 mRNA levels were analyzed by
Spearman's correlation analysis. A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of KCNQ1OT1 is increased in
HCC tissues and predicts a worse prognosis
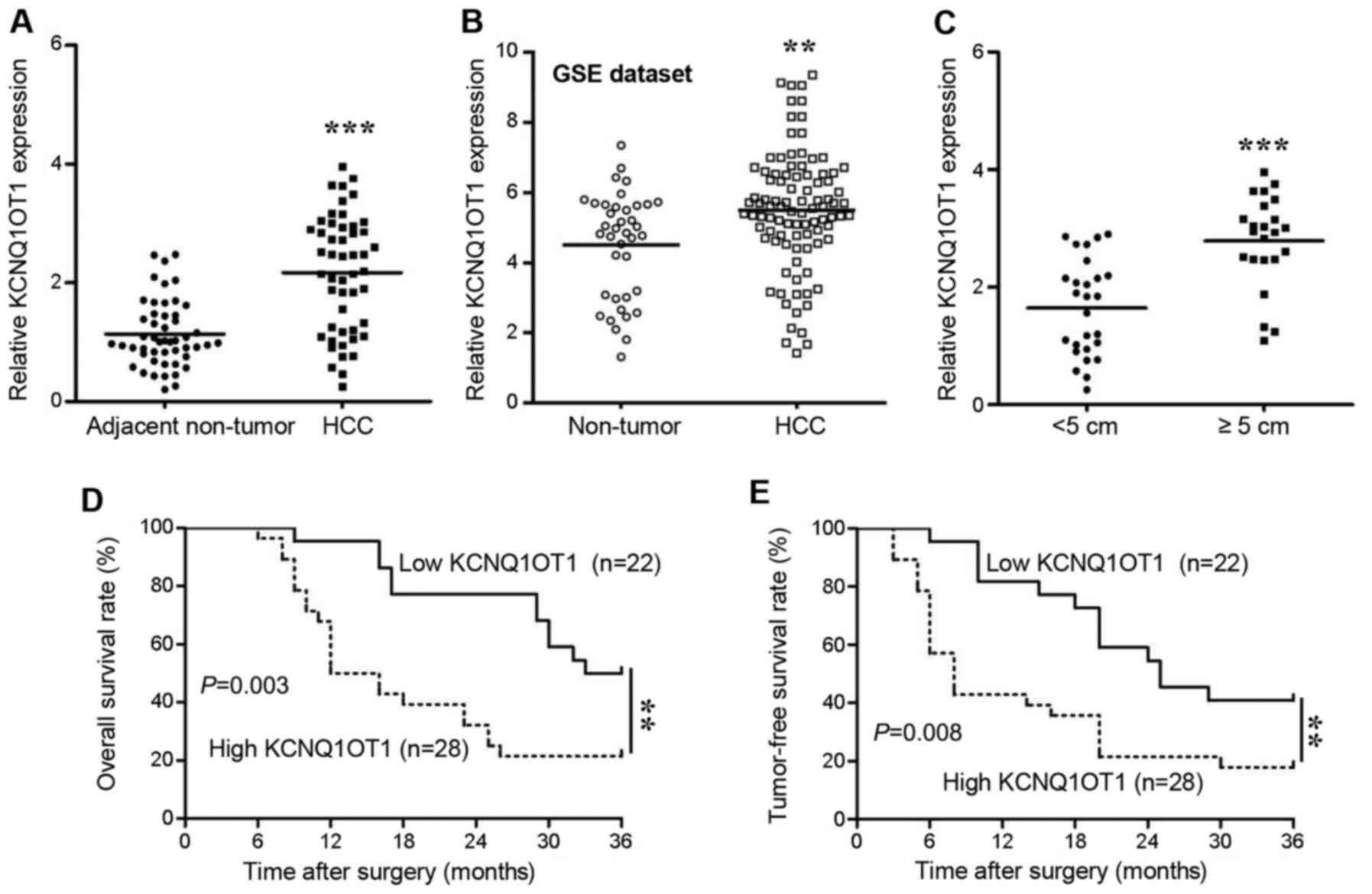
The expression of KCNQ1OT1 in human tissues was
screened using RT-qPCR. We found that KCNQ1OT1 expression was
significantly increased in the HCC tissues compared with in the
adjacent non-tumor tissues (Fig.
1A, P<0.001). To confirm the results obtained by our PCR
analysis, we further analyzed KCNQ1OT1 expression data available in
the GSE dataset (GSE45436). As expected, KCNQ1OT1 expression was
also upregulated in those 95 HCC tissues (Fig. 1B, P=0.002). Additionally, in
comparison to the HCC tissues obtained from small tumors (<5 cm
in diameter), the KCNQ1OT1 levels were significantly higher in the
tissues obtained from large tumors (≥5 cm in diameter) (Fig. 1C, P<0.001). This result
indicated that KCNQ1OT1 may be associated with tumor growth in
HCC.
Therefore, in order to further determine the
clinical and prognostic significance of KNQ1OT1 in HCC, we selected
the mean value of KCNQ1OT1 (2.112) as a cut-off value to divide the
50 patients with HCC into a low KCNQ1OT1 expression group (n=22,
<2.112) and a high KCNQ1OT1 expression group (n=28, ≥2.112). The
correlations between KCNQ1OT1 expression and the clinical
characteristics were analyzed using the Chi-squared test (Table I).
High expression of KCNQ1OT1 was confirmed to be associated with
liver cirrhosis (P=0.027), a large tumor size (>5 cm,
P<0.001) and an advanced TNM stage (stages III and IV, P=0.002).
Moreover, patients with HCC and a high KCNQ1OT1 expression
experienced a shorter overall survival (Fig. 1D; hazard ratio, 2.962; 95%
confidence interval, 1.440-6.091; P=0.003) and tumor-free survival
(Fig. 1E; hazard ratio, 2.588; 95%
confidence interval, 1.282-5.221; P=0.008) than patients with a low
KCNQ1OT1 expression. These findings thus suggest that KCNQ1OT1 has
the potential to predict the outcomes of patients with HCC.
KCNQ1OT1 enhances HCC proliferation and
tumorige- nicity in vitro and in vivo
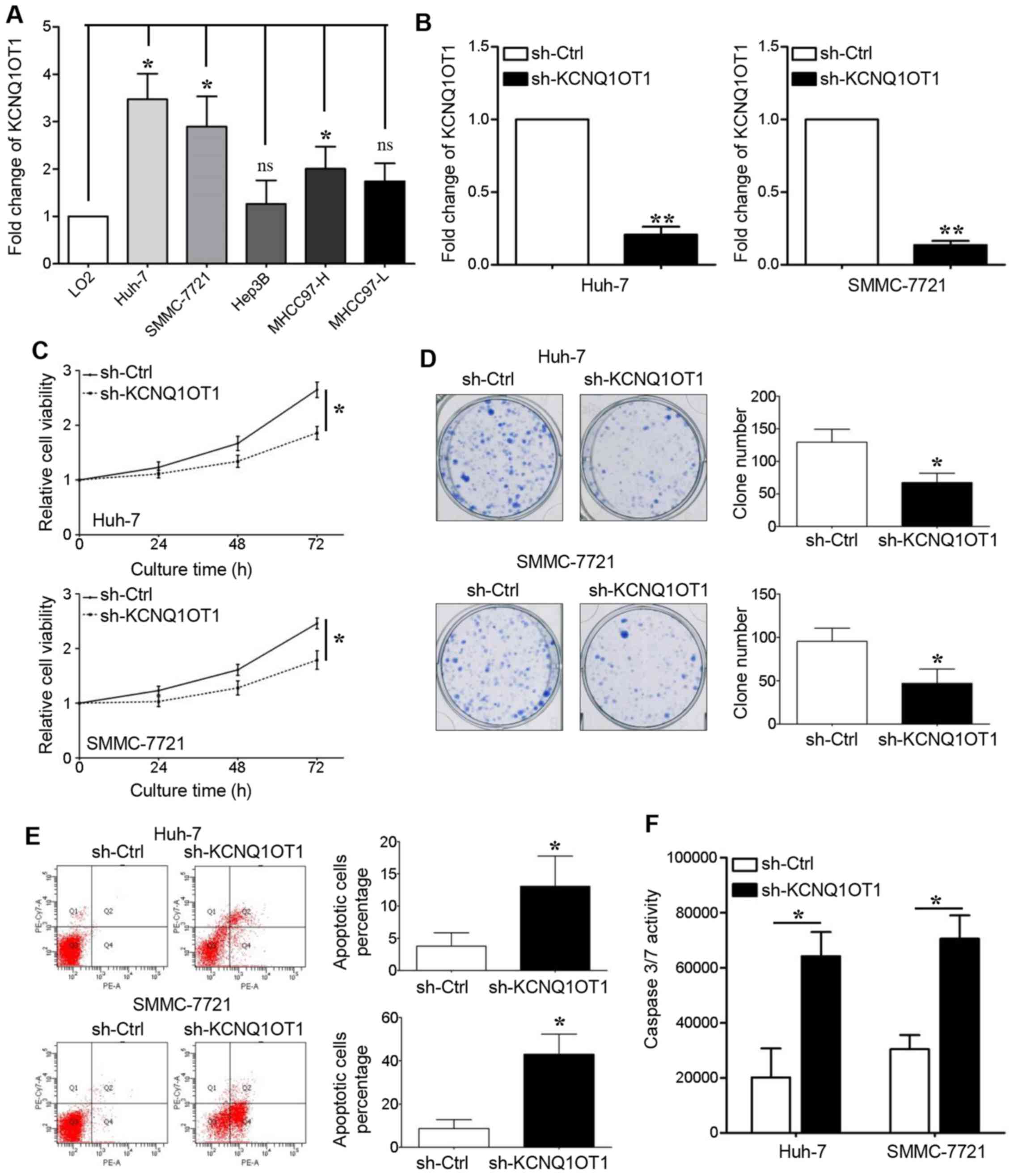
Next, in vitro cell proliferation and
apoptosis experiments were carried out to determine the role of
KCNQ1OT1 in tumor growth. We first detected KCNQ1OT1 expression in
LO2 cells and in 5 HCC cell lines, and found that KCNQ1OT1
expression was upregulated in all the HCC cell lines, apart from
the Hep3B and MHCC97-L cell lines (Fig. 2A, P<0.05, respectively). Two HCC
cell lines with high KCNQ1OT1 expression levels, namely Huh-7 and
SMMC-7721, were transduced with a KCNQ1OT1-specific shRNA
lentivirus. The PCR results revealed that KCNQ1OT1 expression was
silenced in these two cell lines following transduction (Fig. 2B, P<0.01, respectively).
KCNQ1OT1 knockdown significantly suppressed the
viability of the Huh-7 and SMMC-7721 cells (Fig. 2C, P<0.05, respectively). The
capacity of the Huh-7 and SMMC-7721 cells for clone formation was
also inhibited by KCNQ1OT1 knockdown (Fig. 2D, P<0.05, respectively). The
effect of KCNQ1OT1 on cell apoptosis was also examined in this
study. We found that KCNQ1OT1 silencing increased the percentage of
apoptotic cells (Fig. 2E,
P<0.05, respectively), as well as caspase-3/7 activity (Fig. 2F, P<0.05, respectively) in the
Huh-7 and SMMC-7721 cells.
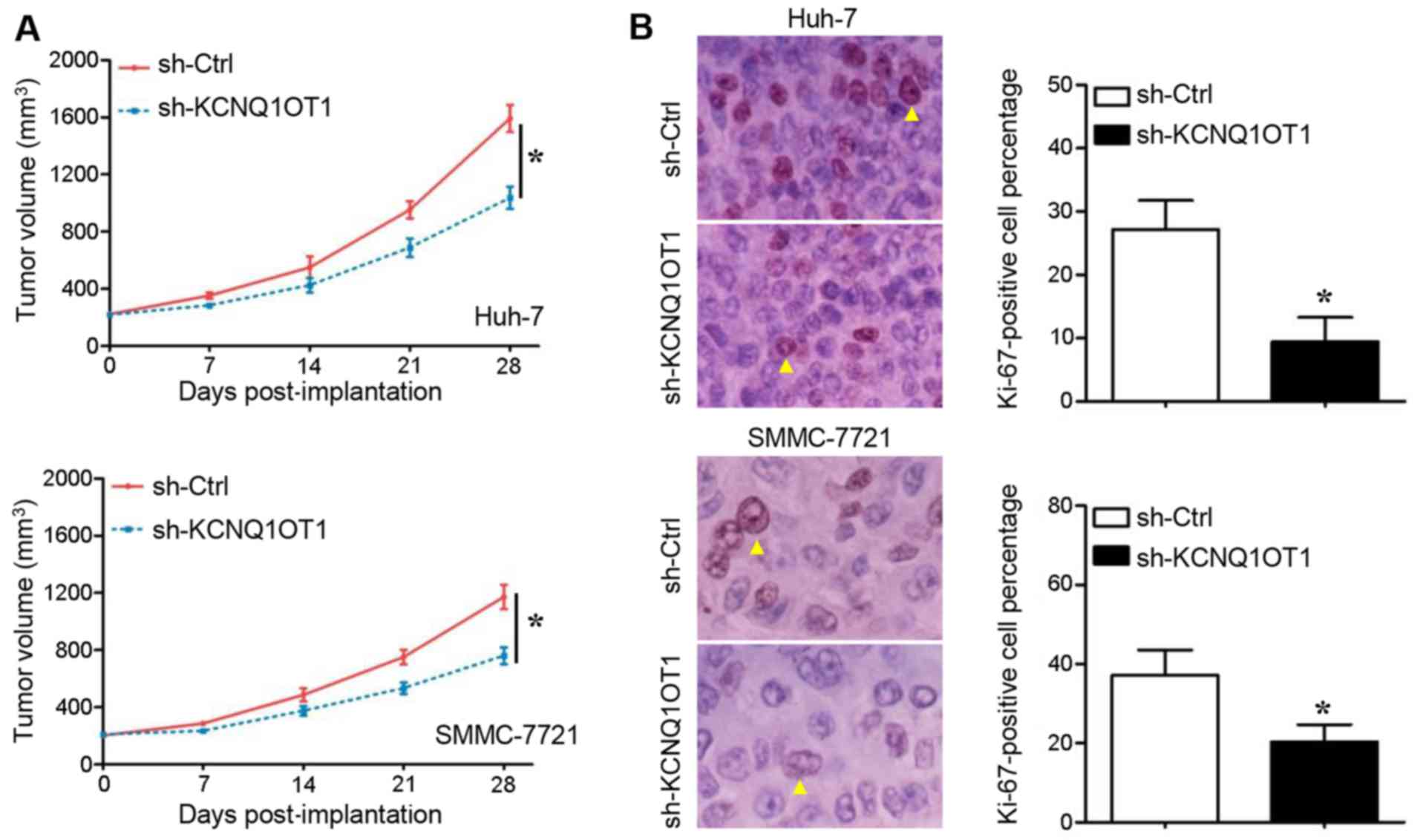
The cells in which KCNQ1OT1 was stably knocked down
were implanted into nude mice to examine the growth of HCC cells
in vivo. Compared with the mice in the control group, the
tumors in mice injected with KCNQ1OT1 shRNA-transduced Huh-7
(Fig. 3A, upper panel) and
SMMC-7721 (Fig. 3A, lower panel)
cells grew at a slower rate during the 4-week observation period
(P<0.05, respectively). Subsequently, a classical proliferation
marker, Ki-67, was stained in order to measure the cell
proliferation in the tumors xenografts. As was expected, the
Ki-67-positive percentage of Huh-7 (Fig. 3B, upper panel) and SMMC-7721
(Fig. 3B, lower panel) cells in
the tumor tissues derived from cells in which KCNQ1OT1 was knocked
down was significantly lower than that in the tumor tissues derived
from the negative controls (P<0.05, respectively). These results
thus suggest that KCNQ1OT1 promotes HCC growth by promoting cell
proliferation and inhibiting apoptosis.
KCNQ1OT1 drives tumor growth in HCC by
sponging miR-504
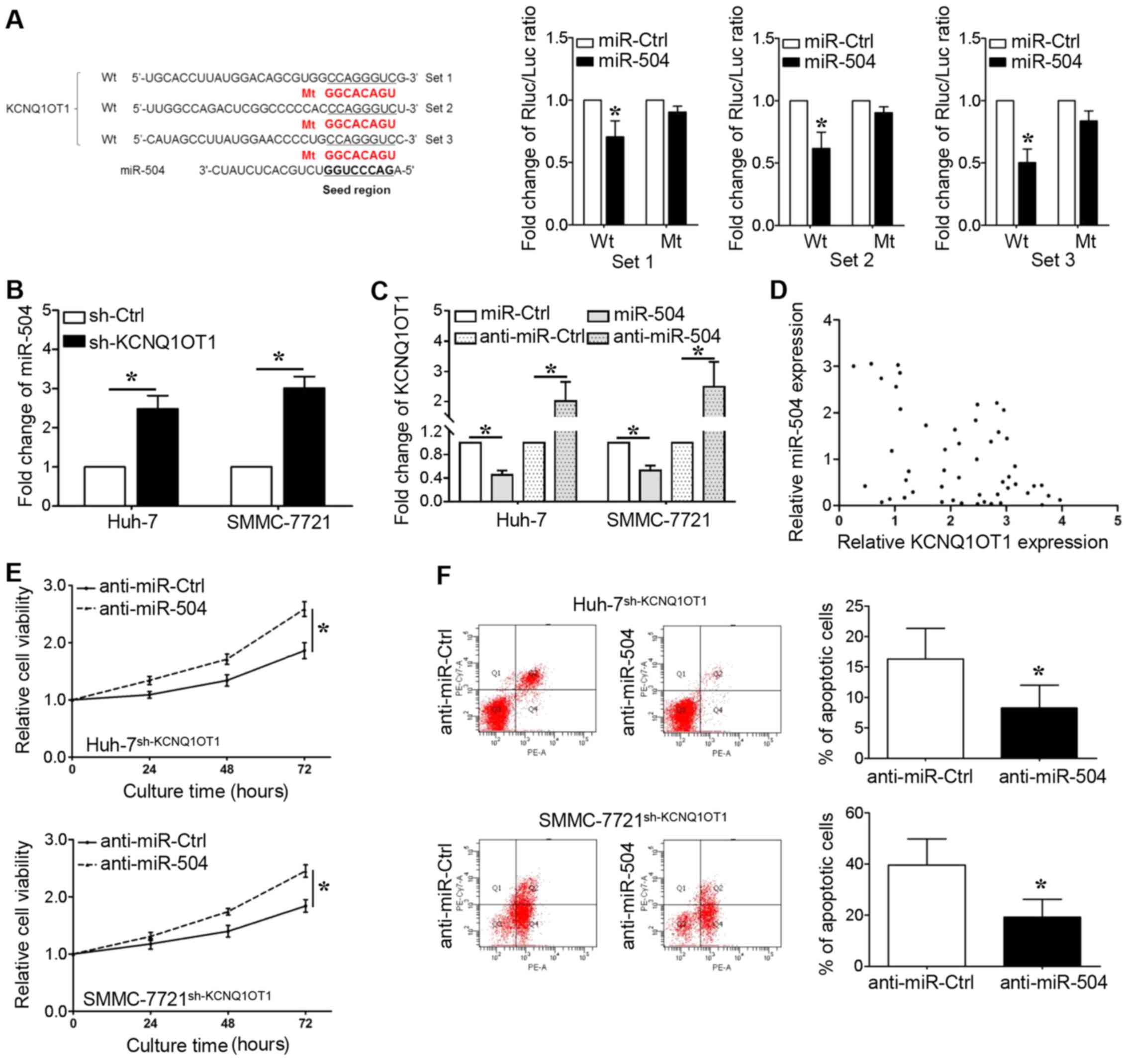
lncRNAs can bind to miRNAs and function as sponges
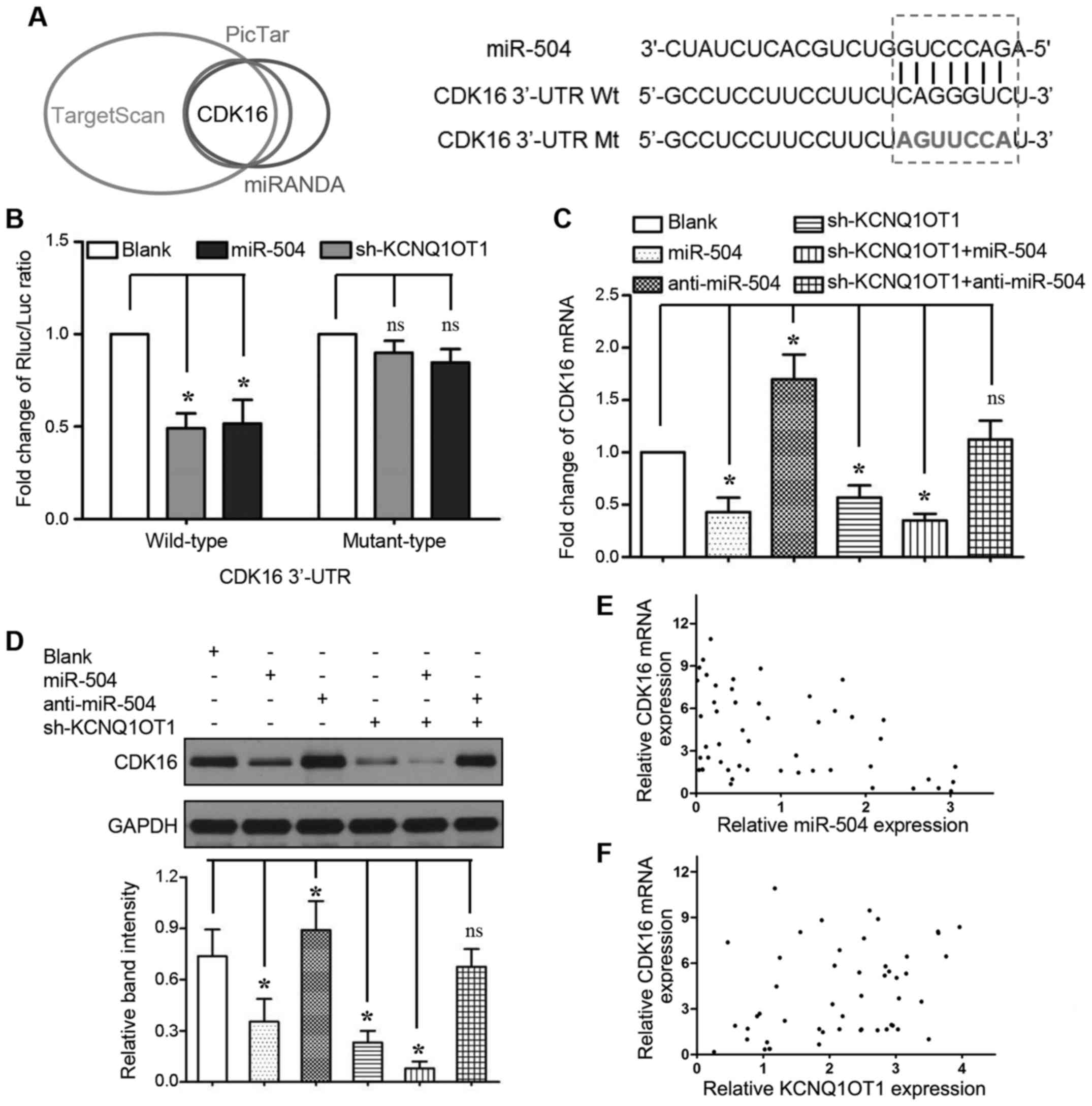
to regulate the expression of certain miRNAs. As shown in Fig. 4A, we identified that the seed
region of miR-504 (2–8 bp from 5′) has putative binding sites for
KCNQ1OT1. We cloned these sequences into reporter vectors; the
reporter vector and miR-504 were co-transfected into 293T cells.
The luciferase activities were significantly inhibited by the
miR-504 mimics (P<0.05, respectively). The expression of miR-504
was upregulated when KCNQ1OT1 was knocked down in the Huh-7 and
SMMC-7721 cells (Fig. 4B,
P<0.05, respectively). We also found that the overexpression of
miR-504 decreased KCNQ1OT1 expression, whereas miR-504 knockdown
increased KCNQ1OT1 expression (Fig.
4C, P<0.05, respectively). Furthermore, a significant
inverse correlation was demonstrated between the KCNQ1OT1 and
miR-504 levels (Fig. 4D, r=-0.337;
95% confidence interval, -0.5680 to -0.05574; P=0.017). In
addition, the miR-504 inhibitor was transfected into the Huh-7 and
SMMC-7721 cells in which KCNQ1OT1 was knocked down to further
examine whether KCNQ1OT1-induced HCC cell proliferation could be
ascribed to miR-504 inhibition. Even though KCNQ1OT1 knockdown
inhibited the viability and promoted the apoptosis of Huh-7 and
SMMC-7721 cells, these effects were markedly abrogated by miR-504
inhibition (Fig. 4E and F,
P<0.05, respectively).
CDK16 is a direct target of miR-504 in
HCC
To explore the underlying mechanisms of the
KCNQ1OT1/miR-504 axis in tumor growth regulation, we used the
TargetScan (http://www.targetscan.org/), PicTar (http://www.pictar.org/) and MiRanda (http://www.microrna.org/) algorithms to predict the
putative targets and binding sites of miR-504. In silico
analyses revealed that the CDK16 3′-UTR contains a corresponding
sequence to the miR-504 seed region (Fig. 5A). Indeed, relative luciferase
activity was significantly inhibited only when we co-transfected
the wild-type CDK16 3′-UTR reporter vector with miR-504 mimics
(Fig. 5B, 2nd bar from the left;
P<0.05) or KCNQ1OT1 shRNA (Fig.
5B, 3rd bar from the left; P<0.05). Moreover, both miR-504
overexpression and KCNQ1OT1 knockdown markedly inhibited CDK16 mRNA
(Fig. 5C, 2nd and 4th bars from
the left; P<0.05, respectively) and protein (Fig. 5D, lanes 2 and 4; P<0.05,
respectively) expression. To decrease CDK16 expression, miR-504
overexpression and KCNQ1OT1 knockdown functioned synergistically
(Fig. 5C, 5th bar from the left;
and Fig. 5D, lane 5; P<0.05,
respectively). By contrast, the inhibition of miR-504 expression
increased CDK16 mRNA (Fig. 5C, 3rd
bar from the left; P<0.05) and protein (Fig. 5D lane 3; P<0.05) expression, and
abrogated the effects of KCNQ1OT1 knockdown on CDK16 expression
(Fig. 5C, 6th bar from the left;
and Fig. 5D, lane 6). A
significant inverse correlation was verified by Spearman's
correlation analysis between the miR-504 levels and the mRNA
expression levels of CDK16 (Fig.
5E, r=-0.406; 95% confidence interval, -0.6201 to -0.1354; P=
0.003). Importantly, we also found that KNCQ1OT1 expression
positively correlated with CDK16 mRNA expression (Fig. 5F, r=0.352; 95% confidence interval,
0.073 to 0.580; P=0.012). These results strongly indicate that
CDK16 is a downstream target of miR-504 mediated by KCNQ1OT1 in
HCC.
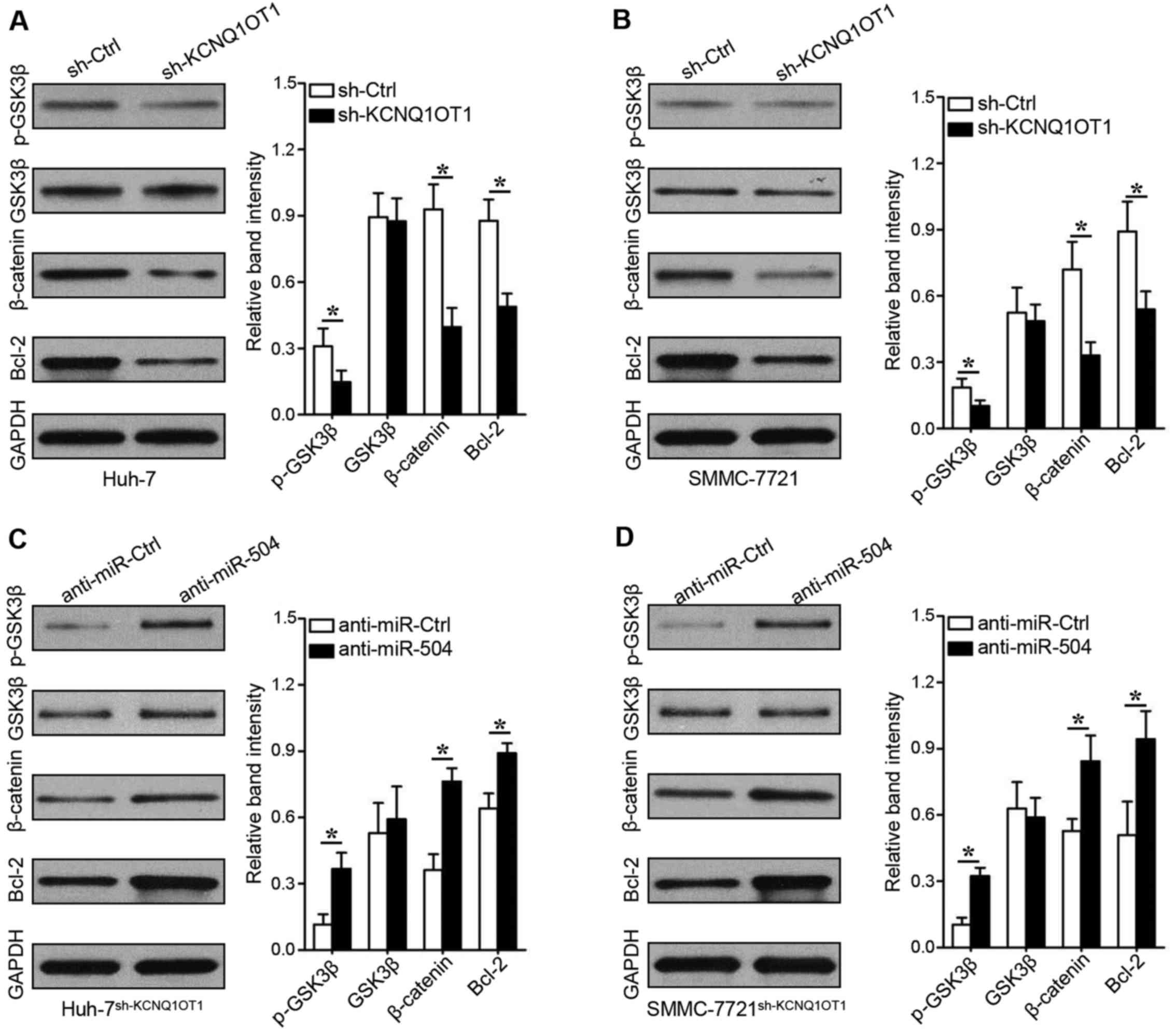
KCNQ1OT1 promotes GSK3β phosphorylation
to decrease β-catenin-mediated Bcl-2 expression
To explore the underlying mechanisms of
KCNQ1OT1/miR-504/CDK16, the GSK3β-β-catenin signaling pathway,
which plays an important role in HCC progression, particularly in
proliferation disorders (13), was
evaluated in this study. KCNQ1OT1 knockdown decreased GSK3β
phosphorylation and β-catenin expression in the Huh-7 (Fig. 6A, rows 1 and 3; P<0.05) and
SMMC-7721 (Fig. 6B, rows 1 and 3;
P<0.05) cells. We found that the expression of Bcl-2 was also
inhibited when KCNQ1OT1 was silenced in the Huh-7 (Fig. 6A, row 4, P<0.05) and SMMC-7721
(Fig. 6B, row 4, P<0.05) cells.
Importantly, the downregulation of miR-504 increased GSK3β
phosphorylation, β-catenin expression and Bcl-2 expression in the
KCNQ1OT1 shRNA-transduced Huh-7 (Fig.
6C, P<0.05) and SMMC-7721 (Fig.
6D, P<0.05) cells. These data thus suggest that KCNQ1OT1
mediates HCC proliferation via the miR-504/CDK16/GSK3β signaling
pathway.
Discussion
lncRNAs have critical biological functions, and
KCNQ1OT1 is one of the few well-characterized lncRNAs. In the
current study, we investigated the expression, clinical
significance and biological functions of KCNQ1OT1 in human HCC.
Several new discoveries were made. First, KCNQ1OT1 was found to be
upregulated in human HCC tissues. This was consistent with findings
that KCNQ1OT1 was overexpressed in a HCC cohort from the GSE
dataset. Second, high levels KCNQ1OT1 were found to correlate with
liver cirrhosis, an advanced TNM stage and a large tumor size.
Tissues from larger-sized tumors (>5 cm in diameter) also had
high KCNQ1OT1 expression levels. Third, patients with a high
intra-tumoral KCNQ1OT1 expression experienced worse 3-year overall
survival and tumor-free survival times. This evidence strongly
suggests that KCNQ1OT1 is a prognostic biomarker, and that it
functions to promote tumor growth. With this antecedent, we
subsequently found that the down-regulation of KCNQ1OT1 inhibited
cell proliferation and induced apoptosis in vitro, and
attenuated subcutaneous tumor growth in vivo.
Consistent with our findings, Ren et al
identified a high KCNQ1OT1 expression in lung adenocarcinoma
tissues (14). KCNQ1OT1 knockdown
suppressed the proliferation of and sensitized A549 cells to
paclitaxel treatment. KCNQ1OT1 silencing also exerted tumor
suppressive functions in glioma cells by acting as a sponge for
miR-370 (15). The ability to
function as a competing endogenous RNA (ceRNA) in order to sponge
miRNAs is commonly found in lncRNA networks (16). For example, lncRNA-CCAT1 has been
reported to sponge let-7 in HCC (17), miR-410 in glioma (18) and miR-181a in nasopharyngeal cancer
(19). In this study, we found
that KCNQ1OT1 had three fragments containing miR-504 seed region
sequences. Upregulated miR-504 was found in cells in which KCNQ1OT1
was knocked down. The inhibition of miR-504 abrogated the changes
in cell proliferation and apoptosis induced by KCNQ1OT1 knockdown.
In the 50 pairs of HCC tissues, an inverse correlation was also
confirmed between KCNQ1OT1 and miR-504 expression. We then used
different algorithms to assume that CDK16 is a putative target of
miR-504. The in vitro results from the dual-luciferase
reporter gene system, RT-qPCR and western blot analyses verified
our hypothesis. Moreover, the down-regulation of KCNQ1OT1 inhibited
CDK16 expression in HCC cells. Clinical analysis also confirmed the
inverse correlation between KCNQ1OT1 and CDK16. Clearly, KCNQ1OT1
modulates CDK16 expression by sponging miR-504 in order to regulate
cell proliferation in human HCC.
The underlying mechanisms through which KCNQ1OT1
increases HCC proliferation were also explored. CDKs are a family
of serine/threonine-protein kinases. CDK16 is a recently
recognized, atypical member of the CDK family (20). The activation of CDK16 usually
occurs upon binding to membrane-associated cyclin Y (CCNY)
(21). CDK16 has been reported to
exert its functions by phosphorylating Akt at Ser473 (22), KAP0 at Ser83 (23) and caspase-8 at Ser387 (24). In this study, we found that
KCNQ1OT1 silencing downregulated the phosphorylation of GSK3β at
Ser9. Ser9 functions as an inhibitory phosphorylating position
(25), which indicates that GSK3β
phosphorylation was decreased by KCNQ1OT1 knockdown. Therefore, we
detected the downregulation of β-catenin in the cells in which
KCNQ1OT1 was knocked down. The expression of Bcl-2, an
anti-apoptotic gene that can be regulated by β-catenin (26,27),
was also suppressed by transfection with KCNQ1OT1 shRNA. Notably,
all these changes were attenuated by miR-504 inhibition. Our
results suggested that the role of KCNQ1OT1 in the regulation of
the GSK3β/β-catenin/Bcl-2 signaling pathway depends on the
inhibition of miR-504. Recently, Sunamura et al reported
that β-catenin promoted KCNQ1OT1 transcription by directly binding
to the KCNQ1OT1 promoter (28),
which may explain why miR-504 overexpression decreased KCNQ1OT1
expression in Huh-7 and SMMC-7721 cells.
In conclusion, our data highlight the critical role
played by KCNQ1OT1 in the proliferation and apoptosis of HCC cells.
Our findings also indicate that lncRNA KCNQ1OT1 is a potential
biomarker for HCC post-surgical surveillance, in addition to a
molecular therapeutic target.
Acknowledgments
Not applicable.
Notes
[1]
Funding
This study was supported by a grant from the
National Natural Scientific Foundation of China (no. 81472247).
[2] Availability
of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
[3] Authors'
contributions
CLi designed the research and wrote the manuscript;
RM, JZ and KQ participated in the research work; and CLiu designed
the research.
[4] Ethics
approval and consent to participate
The use of clinical samples was approved by the
Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong
University. Written informed consent was obtained from all patients
enrolled in this study. The in vivo experiments were
performed after obtaining ethics approval from the Biomedical
Ethics Committee of Xi'an Jiaotong University Health Science
Center.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Sim HW, Knox J and Dawson LA: An update on
randomized clinical trials in hepatocellular carcinoma. Surg Oncol
Clin N Am. 26:647–666. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kalogeraki A, Papadakis GZ, Tamiolakis D,
Karvela-Kalogeraki I, Karvelas-Kalogerakis M, Segredakis J and
Moustou E: Fine Needle Aspiration Biopsy (FNAB) in the Diagnosis of
Hepatocellular Carcinoma: A Review. Rom J Intern Med. 53:209–217.
2015.PubMed/NCBI
|
|
4
|
Chi HC, Tsai CY, Tsai MM, Yeh CT and Lin
KH: Roles of long noncoding RNAs in recurrence and metastasis of
radiotherapy-resistant cancer stem cells. Int J Mol Sci. 18:182017.
View Article : Google Scholar
|
|
5
|
Klingenberg M, Matsuda A, Diederichs S and
Patel T: Non-coding RNA in hepatocellular carcinoma: Mechanisms,
biomarkers and therapeutic targets. J Hepatol. 67:603–618. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma
P and Shu YQ: Long non-coding RNA TUG1 is up-regulated in
hepatocellular carcinoma and promotes cell growth and apoptosis by
epigenetically silencing of KLF2. Mol Cancer. 14:1652015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong
Z, Hua X, Su D, Sun H, Li H, et al: Promotion of glycolysis by
HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep.
38:1902–1908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inoue T, Nakamura A, Matsubara K, Nyuzuki
H, Nagasaki K, Oka A, Fukami M and Kagami M: Continuous
hypomethylation of the KCNQ1OT1:TSS-DMR in monochorionic twins
discordant for Beckwith-Wiedemann syndrome. Am J Med Genet A.
173:2847–2850. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Z, Weaver DL, Olsen D, deKay J, Peng
Z, Ashikaga T and Evans MF: Long non-coding RNA chromogenic in situ
hybridisation signal pattern correlation with breast tumour
pathology. J Clin Pathol. 69:76–81. 2016. View Article : Google Scholar
|
|
10
|
Yoshizawa S, Fujiwara K, Sugito K, Uekusa
S, Kawashima H, Hoshi R, Watanabe Y, Hirano T, Furuya T, Masuko T,
et al: Pyrrole-imidazole polyamide-mediated silencing of KCNQ1OT1
expression induces cell death in Wilms' tumor cells. Int J Oncol.
47:115–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
12
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695. 2017.
|
|
13
|
Liu L, Zhang S, Hu L, Liu L, Guo W and
Zhang J: HMGA1 participates in MHCC97H cell proliferation and
invasion through the ILK/Akt/GSK3β signaling pathway. Mol Med Rep.
16:9287–9294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren K, Xu R, Huang J, Zhao J and Shi W:
Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance
to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol.
80:243–250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gong W, Zheng J, Liu X, Liu Y, Guo J, Gao
Y, Tao W, Chen J, Li Z, Ma J, et al: Knockdown of long non-coding
RNA KCNQ1OT1 restrained glioma cells' malignancy by activating
miR-370/CCNE2 axis. Front Cell Neurosci. 11:842017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu J, Li M, Zhong W and Hu C: Competing
endogenous RNA in cancer: A new pattern of gene expression
regulation. Int J Clin Exp Med. 8:17110–17116. 2015.
|
|
17
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
noncoding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang ZH, Guo XQ, Zhang QS, Zhang JL, Duan
YL, Li GF and Zheng DL: Long non-coding RNA CCAT1 promotes glioma
cell proliferation via inhibiting microRNA-410. Biochem Biophys Res
Commun. 480:715–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Q, Zhang W and Hao S: LncRNA CCAT1
modulates the sensitivity of paclitaxel in nasopharynx cancers
cells via miR-181a/CPEB2 axis. Cell Cycle. 16:795–801. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mikolcevic P, Rainer J and Geley S: Orphan
kinases turn eccentric: A new class of cyclin Y-activated,
membrane-targeted CDKs. Cell Cycle. 11:3758–3768. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shehata SN, Deak M, Morrice NA, Ohta E,
Hunter RW, Kalscheuer VM and Sakamoto K: Cyclin Y phosphorylation-
and 14-3-3-binding-dependent activation of PCTAIRE-1/CDK16. Biochem
J. 469:409–420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ćwiek P, Leni Z, Salm F, Dimitrova V,
Styp-Rekowska B, Chiriano G, Carroll M, Höland K, Djonov V,
Scapozza L, et al: RNA interference screening identifies a novel
role for PCTK1/CDK16 in medulloblastoma with c-Myc amplification.
Oncotarget. 6:116–129. 2015. View Article : Google Scholar :
|
|
23
|
Iwano S, Satou A, Matsumura S, Sugiyama N,
Ishihama Y and Toyoshima F: PCTK1 regulates integrin-dependent
spindle orientation via protein kinase A regulatory subunit KAP0
and myosin X. Mol Cell Biol. 35:1197–1208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yanagi T, Shi R, Aza-Blanc P, Reed JC and
Matsuzawa S: PCTAIRE1-knockdown sensitizes cancer cells to TNF
family cytokines. PLoS One. 10:e01194042015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang L, Zhang C, Su L and Song Z: GSK3β
attenuates TGF-β1 induced epithelial-mesenchymal transition and
metabolic alterations in ARPE-19 cells. Biochem Biophys Res Commun.
486:744–751. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ye X, Lin J, Lin Z, Xue A, Li L, Zhao Z,
Liu L, Shen Y and Cong B: Axin1 up-regulated 1 accelerates
stress-induced cardiomyocytes apoptosis through activating
Wnt/β-catenin signaling. Exp Cell Res. 359:441–448. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Zhou J, Xu YJ and Hu HB: Long
non-coding RNA LINC00968 acts as oncogene in NSCLC by activating
the Wnt signaling pathway. J Cell Physiol. 233:3397–3406. 2018.
View Article : Google Scholar
|
|
28
|
Sunamura N, Ohira T, Kataoka M, Inaoka D,
Tanabe H, Nakayama Y, Oshimura M and Kugoh H: Regulation of
functional KCNQ1OT1 lncRNA by β-catenin. Sci Rep. 6:206902016.
View Article : Google Scholar
|