Introduction
Glioma is the most common and life-threatening type
of brain tumor (1). Even following
surgery, radiation and chemotherapeutic treatments, the majority of
patients with glioma succumb to mortality within 2 years of
diagnosis (2,3). How to improve the efficacy of
clinical diagnosis and treatment for glioma has become the focus of
investigations on glioma.
Traditional two-dimensional (2D) cell culture
systems are often used to assess the sensitivity of tumor cells to
radiotherapy and chemotherapy, and guide the clinical treatments.
However, 2D cultured cells perform poorly and are not suitable for
investigating solid tumors (4,5), as
they do not accurately reproduce tissue architecture or have
interactions between cells and their microenvironment. This leads
to deviations in drug sensitivities between in vitro tests
and in vivo clinical evaluations. Therefore, a novel
research model is crucial for the development of effective
anti-glioma therapeutics.
Three-dimensional (3D) cell culture systems,
including sphere (6,7) and material culture (8–12)
have been applied for several type of tumor, as they better
simulate the native tumor microenvironment and provide more
accurate drug efficacy analysis. The biomaterials used to establish
3D culture system include poly (lactic-co-glycolic) acid, chitosan,
alginate, Matrigel and collagen. Among these, collagen is an ideal
biomaterial for 3D scaffolds, as it is the main component of the
extracellular matrix (ECM) in connective tissues, and has low
antigenicity. The commonly applied biomaterials in studies of
glioma are Matrigel and hydrogel, and their application is mainly
focused on detection of the sensitivities of co-cultured tumor
cells to radiation and drugs (13–25).
There have been few reports on collagen scaffold culture in glioma,
and its effects on whole gene expression profiles and the functions
of glioma cells remain to be fully elucidated.
In the present study, glioma cells (U87, U251 and
HS683) were cultured in 3D collagen scaffolds with different
pore-diameters, and the cell morphology, gene expression profiles,
biological functions and associated signaling pathways of the 3D
cultured cells were compared with those of 2D monolayer cultured
cells.
Materials and methods
Preparation of 3D collagen scaffolds
The collagen scaffolds were prepared as previously
described (26). According to the
pore diameter, they were subdivided into scaffold A (diameter,
30–50 µm) and scaffold B (diameter, 70–100 µm)
types.
Cell culture
The 2D culture was performed as follows: Three
glioma cell lines (U87, U251 and HS683 cells) were purchased from
Xiangya Central Laboratory (Xingya, China). The U87 and HS683 cells
were grown and maintained in Dulbecco's modified Eagle's medium
(DMEM; Sigma-Aldrich; EMD Millipore, Billerica, MA, USA), and the
U251 cells were in RPMI-1640 medium (Sigma-Aldrich; EMD Millipore).
Both media were supplemented with 10% fetal bovine serum
(Biological Industries, Kibbutz Beit Haemek, Israel), 100 U/ml
penicillin and 100 mg/ml streptomycin (termed complete medium). All
the cells were cultured in cell culture flasks at 37°C with 5%
CO2.
The 3D-culture was performed as follows: Following
immersion in homologous cell culture mediums for 24 h at 37°C, the
collagen scaffolds were loaded with cell suspensions
(1×105 cells in 20 µl medium per scaffold) and
maintained at 37°C for 4 h; every scaffold with seeded cells was
then transferred to one well of a 12-well cell culture plate
containing 2 ml complete medium, which was replaced every 2 days.
The process of harvesting cells from the 3D collagen scaffold was
performed mainly through trypsin digestion. In brief, every
scaffold with cells was washed with phosphate buffer solution (PBS)
three times, and then submerged in 0.25% trypsin (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA USA) at 37°C for 10
min. During the digestion, the scaffold was blown using pipette
tips 2–3 times. The digestion was terminated by the complete medium
which contains the fetal bovine serum. The whole process was
repeated once to harvest as many cells as possible. The
twice-digested fluid was collected, and the supernatant was
discarded following centrifugation (300 × g, 5 min, room
temperature). The resulting pure cells were used for the subsequent
experiments.
Cell morphology analysis
Cell morphology was observed via FDA (Sigma-Aldrich;
EMD Millipore) staining and hematoxylin and eosin (H&E;
Sigma-Aldrich; EMD Millipore). For the FDA staining, the scaffolds
with cells cultured for 1, 5 and 10 days were washed with PBS three
times, then submerged in FDA solution (1%FDA in PBS) for 1 min, and
washed twice with PBS. The stained scaffolds were observed under
the fluorescent inverted phase contrast microscope (Nikon Imaging
Japan Inc., Tokyo, Japan; cat. no. Elipse E2000-S). For the H&E
staining, the three glioma cell lines growing on glass coverslips
were examined. Scaffolds on day 10 were fixed in 4%
paraformaldehyde, embedded in paraffin, cut into 5-µm
sections and stained with H&E.
Cell proliferation assay
The three glioma cell lines were seeded at a density
of 1×105 cells/scaffold or 3×104 cells/well
in a 6-well plate, respectively. Following culture for 1, 5 and 10
days, every scaffold or well (n=3) was digested with 0.25% trypsin,
following which the cell numbers were counted. Cell count was
determined as a relative value, and the number of seeded cells was
set as 1.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total mRNA was isolated from the 2D and 3D (on day
10) cultured cells using TRIzol™ reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), following the manufacturer's protocol. The
RT-qPCR analysis was performed as previously described (27). Reverse transcription (RT) was
carried out with 2 µg total RNA per 20 µl reaction
using the 5X All-In-One MasterMix (ABM, Richmond, BC, Canada). qPCR
was performed with the CFX96 Real-Time PCR detection system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA; cat. no. 185-5195)
using AceQ® qPCR SYBR® Green Master mix
(Vazyme, Piscataway, NJ, USA). The final volume of the reaction mix
was 25 µl, consisting of AceQ qPCR SYBR-Green Master mix
(2X) 10 µl, 0.2 µM of each specific forward and
reverse primer, the resulting cDNA 1 µl and sterile purified
water. Amplifications were done under standard conditions (5 min at
95°C followed by 40 cycles of 10 sec at 95°C and 30 sec at 60°C).
Sequence-specific primers were quoted from an official website
'PrimerBank' (http://pga.mgh.harvard.edu/primerbank/) for the
indicated genes (Tables I and
II). All reactions were performed
in triplicate, and relative changes in transcript level normalized
by β-actin mRNA were calculated by the ΔΔCt method (28).
 | Table IPrimer sequences used for reverse
transcription-quantitative polymerase chain reaction amplification
of genes related to stemness and cell cycle. |
Table I
Primer sequences used for reverse
transcription-quantitative polymerase chain reaction amplification
of genes related to stemness and cell cycle.
| Gene | Primer sequence
(5′-3′) |
|---|
| CD133 | F:
ATTGACTTCTTGGTGCTGTTGA |
| R:
GATGGAGTTACGCAGGTTTCTC |
| Nestin | F:
CTTGCCTGCTACCCTTGAGAC |
| R:
GTTTCCTCCCACCCTGTGT |
| Oct4 | F:
TATTCAGCCAAACGACCATCT |
| R:
TCAGCTTCCTCCACCCACTT |
| Sox2 | F:
TGTCAAGGCAGAGAAGAGAGTG |
| R:
GCCGCCGATGATTGTTATTAT |
| Nanog | F:
CCCCAGCCTTTACTCTTCCTA |
| R:
CCAGGTTGAATTGTTCCAGGTC |
| c-Myc | F:
GGCTCCTGGCAAAAGGTCA |
| R:
CTGCGTAGTTGTGCTGATGT |
| MSI1 | F:
CCAACCGGCACCGAGGGTTC |
| R:
GCTGAGCCCGTTGGCGACAT |
| MSI2 | F:
ACGACTCCCAGCACGACC |
| R:
GCCAGCTCAGTCCACCGATA |
| BMI-1 | F:
CGTGTATTGTTCGTTACCTGGA |
| R:
TTCAGTAGTGGTCTGGTCTTGT |
| CCNA | F:
TGGAAAGCAAACAGTAAACAGCC |
| R:
GGGCATCTTCACGCTCTATTT |
| CCNB | F:
AATAAGGCGAAGATCAACATGGC |
| R:
TTTGTTACCAATGTCCCCAAGAG |
| CCND | F:
CAATGACCCCGCACGATTTC |
| R:
CATGGAGGGCGGATTGGAA |
| CCNE | F:
GCCAGCCTTGGGACAATAATG |
| R:
CTTGCACGTTGAGTTTGGGT |
| p21 | F:
TGTCCGTCAGAACCCATGC |
| R:
AAAGTCGAAGTTCCATCGCTC |
| p27 | F:
ATCACAAACCCCTAGAGGGCA |
| R:
GGGTCTGTAGTAGAACTCGGG |
| β-actin | F:
CCTGTACGCCAACACAGTGC |
| R:
ATACTCCTGCTTGCTGATCC |
 | Table IIPrimer sequences used for reverse
transcription-quantitative polymerase chain reaction amplification
of genes related to epithelial-mesenchymal transition, migration,
invasion and glioma malignancy. |
Table II
Primer sequences used for reverse
transcription-quantitative polymerase chain reaction amplification
of genes related to epithelial-mesenchymal transition, migration,
invasion and glioma malignancy.
| Gene | Primer sequence
(5′-3′) |
|---|
|
N-cadherin | F:
AGCCAACCTTAACTGAGGAGT |
| R:
GGCAAGTTGATTGGAGGGATG |
|
Vimentin | F:
AGTCCACTGAGTACCGGAGAC |
| R:
CATTTCACGCATCTGGCGTTC |
| MMP1 | F:
CTCTGGAGTAATGTCACACCTCT |
| R:
TGTTGGTCCACCTTTCATCTTC |
| MMP2 | F:
GATACCCCTTTGACGGTAAGGA |
| R:
CCTTCTCCCAAGGTCCATAGC |
| MMP3 | F:
CTGGACTCCGACACTCTGGA |
| R:
CAGGAAAGGTTCTGAAGTGACC |
| MMP7 | F:
GAGTGAGCTACAGTGGGAACA |
| R:
CTATGACGCGGGAGTTTAACAT |
| GFAP | F:
AGGTCCATGTGGAGCTTGAC |
| R:
GCCATTGCCTCATACTGCGT |
| EGFR | F:
AGGCACGAGTAACAAGCTCAC |
| R:
ATGAGGACATAACCAGCCACC |
| Ki67 | F:
GCCTGCTCGACCCTACAGA |
| R:
GCTTGTCAACTGCGGTTGC |
Western blot analysis
Western blot analysis was performed in accordance
with the previously described method (29). In brief, on day 10, the cells
cultured in 2D or 3D environments were lysed in RIPA (Beyotime
Institute of Biotechnology, Haimen, China) for 30 min and total
proteins were obtained. Following high-speed centrifugation (12,000
× g, 30 min, 4°C), the proteins were denatured, and the proteins
(40 µg) were loaded and separated by 6–12% SDS-PAGE gels.
The samples were transferred onto a PVDF membrane (EMD Millipore)
followed by blocking with 5% milk in TBST and then immunoblotting
with target primary antibodies and anti-β-actin antibody overnight
at 4°C, respectively. Finally, the Gel Imaging system (Bio-Rad
Laboratories, Inc.; cat. no. Universal Hood II, Chemi, XR+, XRS+)
was used to visualize the protein bands following incubation with
corresponding peroxidase-conjugated anti-IgG antibody (1:40,000,
A0545 or A9044, Sigma-Aldrich; EMD Millipore) for 1 h at room
temperature. The visualisation reagent was Luminata™ Crescendo
Western HRP Substrate (EMD Millipore). The primary antibodies used
in the present study included the following: Anti-CD133 (1:500,
18470-1-AP), anti-Nestin (1:500, 19483-1-AP), anti-octamer-binding
transcription factor 4 (Oct4) (1:500, 11263-1-AP), anti-SRY-Box 2
(Sox2) (1:500, 11064-1-AP), anti-Nanog (1:500, 14295-1-AP),
anti-c-Myc (1:500, 10828-1-AP), anti-Musashi RNA binding protein
(MSI)1 (1:500, 27185-1-AP), anti-MSI2 (1:500, 10770-1-AP),
anti-cyclin (CCN)A1 (1:500, D151775), anti-CCNB1 (1:500,
55004-1-AP), anti-CCND1 (1:500, 60186-1-Ig), anti-CCNE1 (1:500,
11554-1-AP), anti-p21 (1:500, 10355-1-AP), anti-p27 (1:500,
26714-1-AP), anti-N-cadherin (1:400, BA0673), anti-vimentin (1:400,
BM0135), anti-matrix metal-loproteinase (MMP)1 (1:400, BM4305),
anti-MMP2 (1:400, BM4075), anti-MMP3 (1:300, BM4074), anti-MMP7
(1:300, PB0070), anti-glial fibrillary acidic protein (GFAP)
(1:500, 23935-1-AP), anti-epidermal growth factor receptor (EGFR)
(1:500, 22542-1-AP), anti-Ki67 (1:500, BS1454), anti-p53 (1:500,
10442-1-AP), anti-programmed death-ligand 1 (PDL1) (1:500,
17952-1-AP), and anti-Livin (1:500, 27543-1-AP), the Apoptosis
Antibody Sampler kit [including caspase (Cas)3, 7 and 9, PARP]
(1:1,000, #9915), Notch Isoform Antibody Sampler kit (1:1,000,
#3640), anti-Wnt3a (1:400, BA2628-2), anti-Wnt5a (1:400, BA2839),
anti-SHH (1:500, 20697-1-AP) and anti-β-actin (1:3,000, A5441).
With the exception of anti-caspase 9, anti-vimentin, anti-p53,
CCND1 and anti-β-actin, which were mouse monoclonal antibodies, the
primary antibodies mentioned above were rabbit poly-clonal
antibodies. The Apoptosis Antibody Sampler kit and the Notch
Isoform Antibody Sampler kit were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA), and anti-β-actin primary
antibody was from Sigma-Aldrich; EMD Millipore. The anti-Ki67
primary antibody was from Bioworld Technology, Inc. (St. Louis
Park, MN, USA), and the anti-CCNA1 primary antibody was from BBI
Life Sciences Corp. (Shanghai, China). The anti-N-cadherin,
anti-vimentin, anti-MMP1, anti-MMP2, anti-MMP3, anti-MMP7, Wnt3a
and Wnt5a primary antibodies were purchased from Boster Biological
Technology, Ltd. (Wuhan, Hubei, China), and the remainder of the
primary antibodies were from ProteinTech Group, Inc. (Chicago, IL,
USA).
Colony formation assay
For the colony formation assays, glioma cells from
the different culture models were plated at 1,000 cells/well in
different complete medium in 6-well plates, and were allowed to
form colonies for 10 days. The colonies were fixed with 4%
paraformaldehyde for 15 min and stained with 0.4% crystal violet
for 30 min. Colonies containing >50 cells were counted manually
using the inverted phase contrast microscope mentioned above.
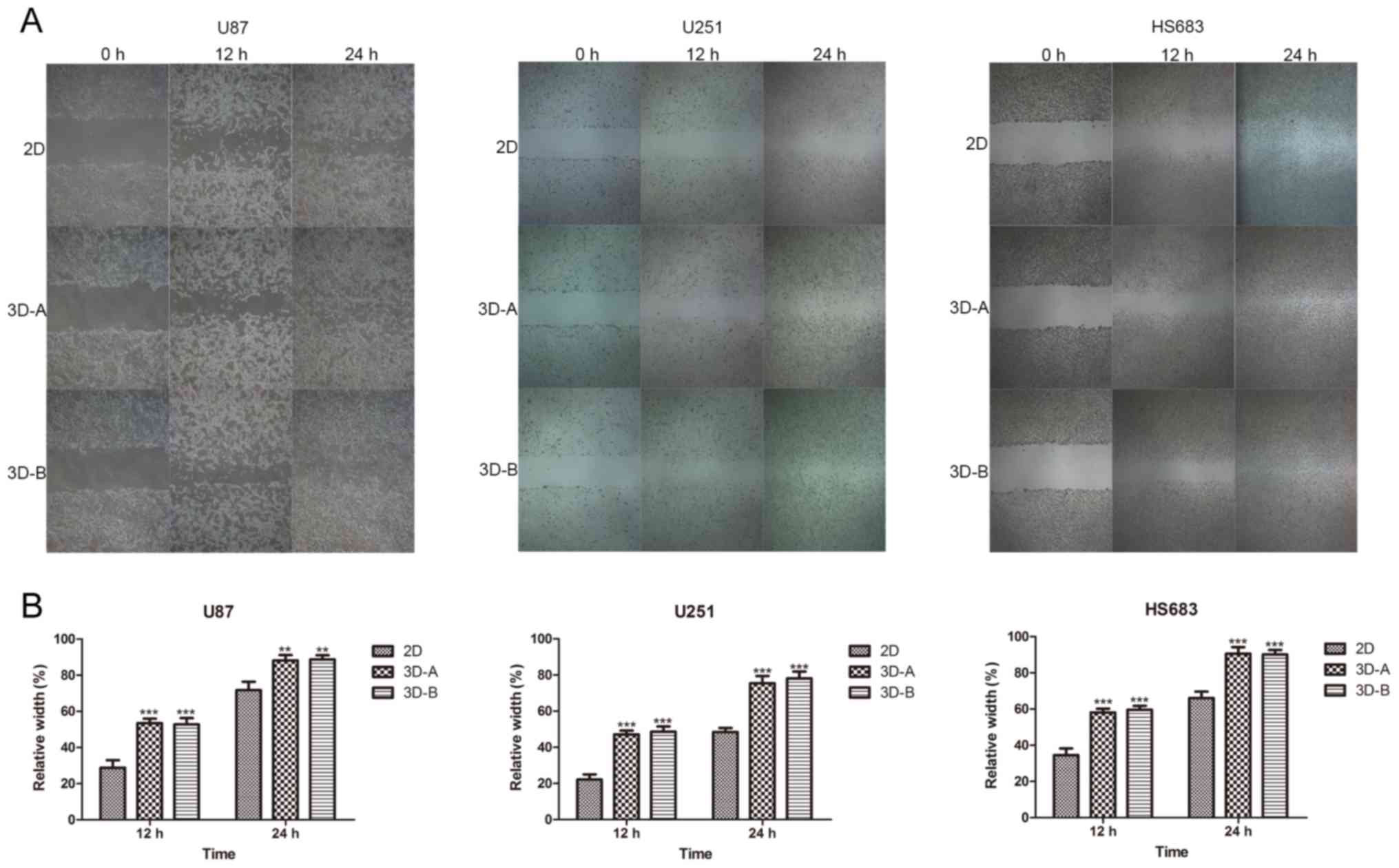
Wound-healing assay
A wound-healing assay was performed according to a
previously described protocol (30). The glioma cells from the different
culture models were plated in 6-well plates. On reaching 95%
confluence, the cell monolayers were wounded with a P-200 pipette
tip, and the wounded monolayers were gently washed three times with
PBS; medium containing 2% FBS was then added for further
incubation. Images were captured at 0, 12 and 24 h, and the
distances between the two wound edges were scaled for three
positions at different time-points. The distances at 0, 12 and 24 h
were counted as d0, d1 and d2, respectively. Relative width = (d1
or d2 − d0) / d0.
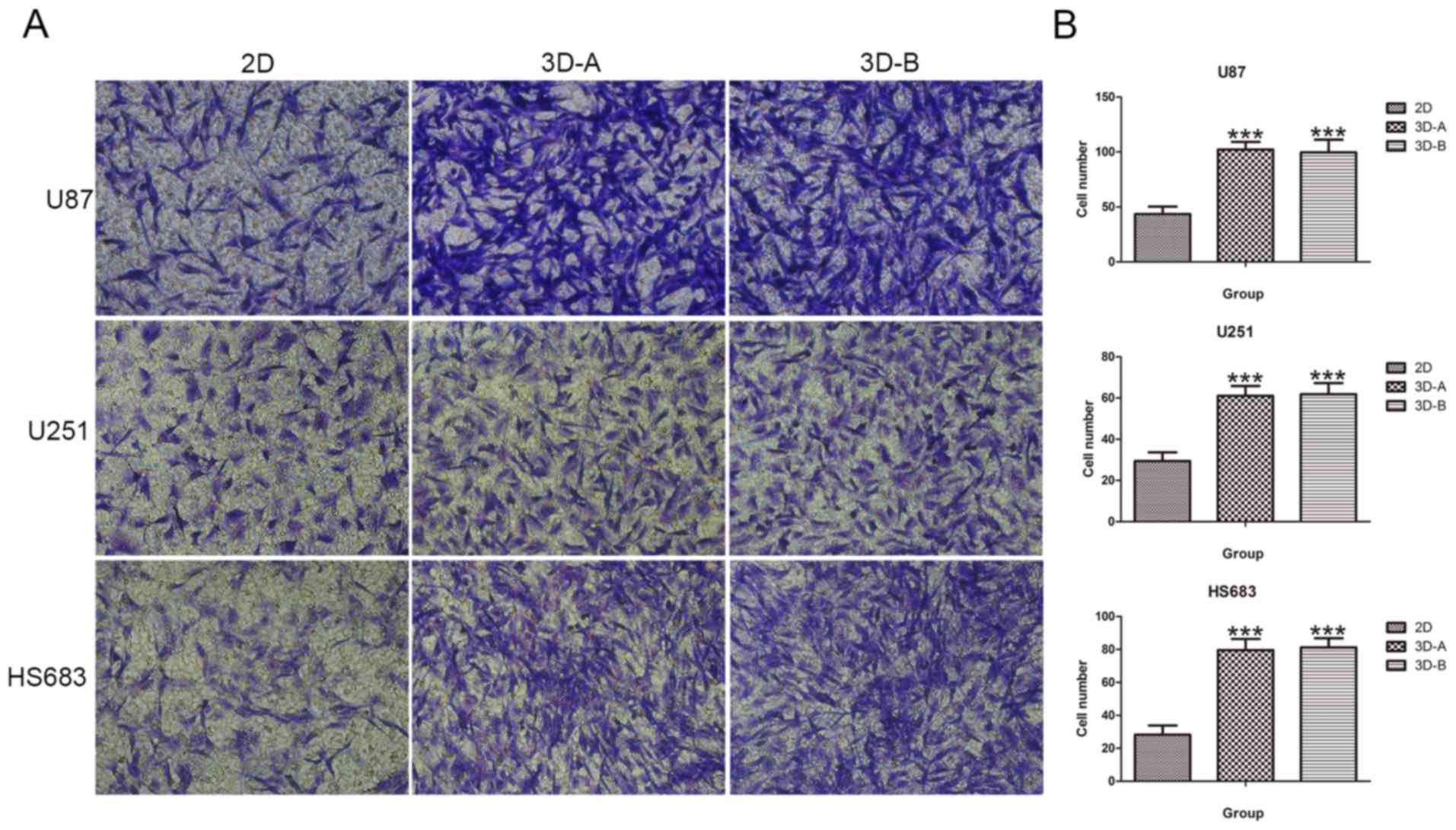
Transwell invasion assay
To examine the invasive capacity of the glioma
cells, Transwell invasion assays were performed using 24-well MILLI
cell hanging cell culture inserts (8 mm PET; EMD Millipore) coated
with Matrigel matrix gel (BD Biosciences, Franklin Lakes, NJ, USA)
according to the manufacturer's protocol. Cells from the different
culture models were suspended in serum-free medium and
5×104 cells were added into the upper chamber. Following
incubation for 48 h, the cells on the underside of the membrane
were fixed with 4% paraformaldehyde and stained with 0.1% crystal
violet solution. The cells attached to the lower surface were
counted under a microscope (Nikon, Tokyo, Japan) at ×400
magnification in six randomly selected fields.
Statistical analysis
Data are presented as the mean ± standard deviation
of at least three independent experiments. Statistical significance
was determined using one-way analysis of variance (ANOVA) with
Tukey's multiple comparisons test performed in IBM SPSS Statistics
24.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
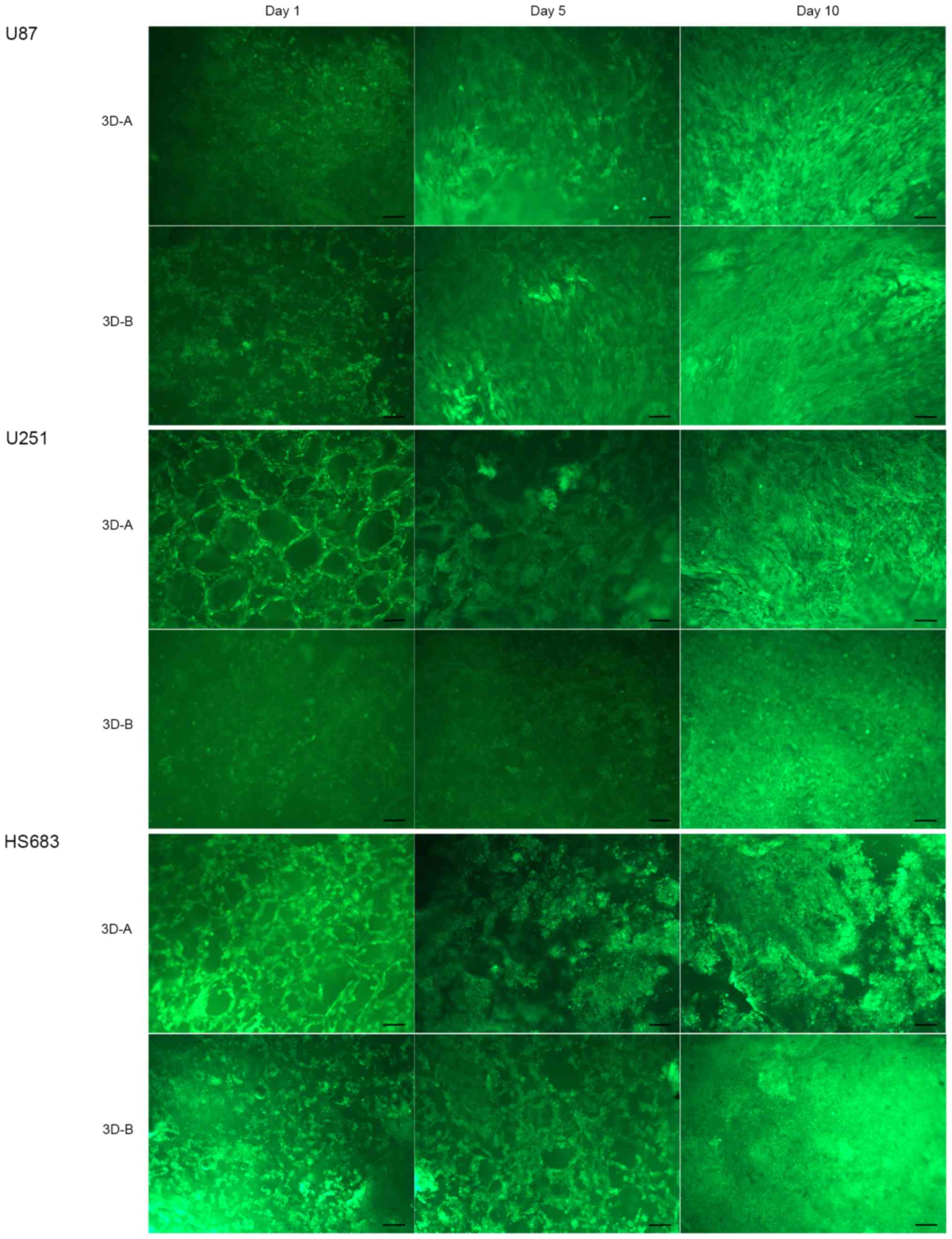
Cell morphology and proliferation
Firstly, the present study examined the morphology
of the three glioma cell lines (U87-MG, U251 and HS683) in the
different collagen scaffolds by FDA staining on days 1, 5 and 10.
As shown in Fig. 1, all three cell
lines adhered to the scaffolds and grew along the skeleton. With
the increase of culture duration, the numbers of cells also
increased. These cells appeared stereoscopic and formed a
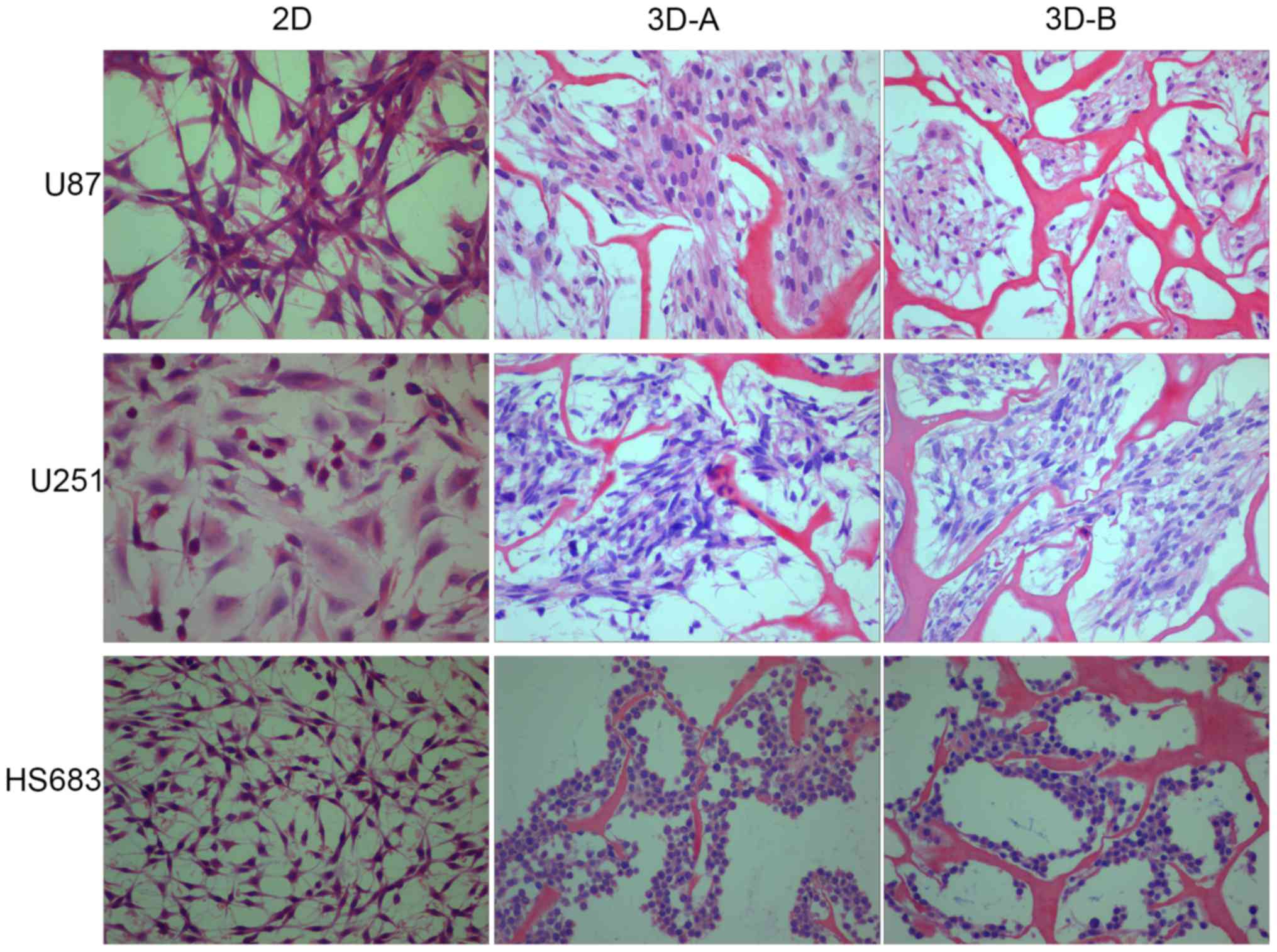
multilayer structure. The shape of the 3D-cultured cells on day 10
was compared with those on the 2D culture plates by H&E
staining. As shown in Fig. 2, the
glioma cells in 2D culture were fusiform or polygonal, flat and
epithelioid or fibroblast-like, whereas those in the 3D scaffolds
grew as small, round or ovoid conglomerate cells. The latter also
exhibited trachychromatic and heteromorphous nuclei. Among the
three cell lines, the morphological change of the U87 cells was the
most marked. The U87 and U251 cells gathered to form masses, and
more HS683 cells grew along the skeleton. Between scaffold A and
scaffold B, cells in the latter exhibited increased variance.
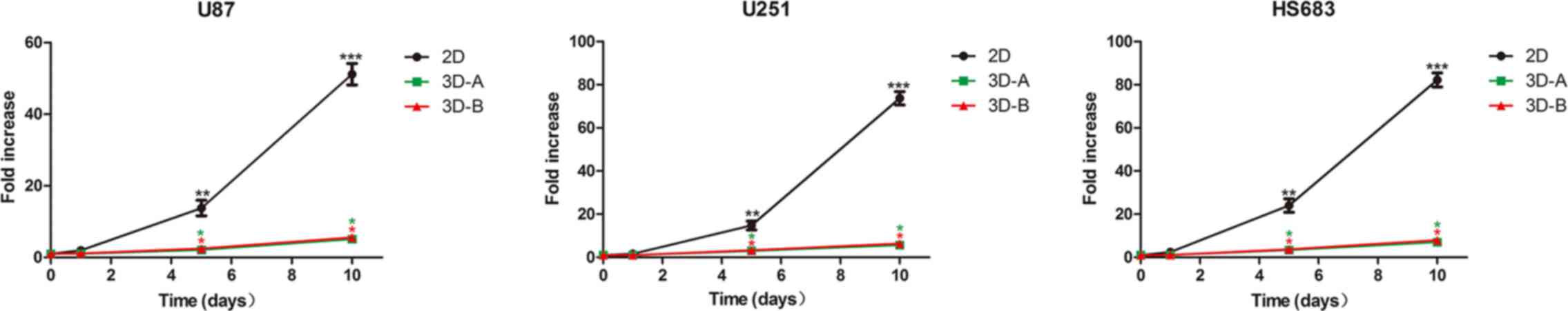
The present study then examined the number of cells
in the three glioma cell lines cultured under 2D and 3D conditions
on days 1, 5 and 10 by cell number counting. For all three glioma
cell lines, similar proliferation curves were observed between the
3D-cultured cells and 2D cells (Fig.
3), which showed a lag phase of 1–5 days and an exponential
increase between 5 and 10 days. However, the cells grew more slowly
in 3D scaffolds than in 2D monolayer cultures. Statistically
significant differences were observed on days 5 and 10 of culture
(P<0.05). However, there was no statistically significant
difference between the scaffold A group and scaffold B group in any
of the cell lines.
Changes in gene expression profiles in 3D
scaffold-cultured cells
The present study compared the differences in gene
expression profiles between the 3D cultured cells and 2D monolayer
cells using RT-qPCR and western blot analyses. The examined genes
were related to stemness, cell cycle, epithelial-mesenchymal
transition (EMT), migration, invasion and glioma malignancy.
The stemness-related genes, including CD133,
Nestin, Oct4, Sox2, c-Myc, Nanog, MSI1, MSI2 and BMI-1,
were examined. As shown in Fig.
4A, the majority of these genes were upregulated to different
degrees in the glioma cells cultured in 3D collagen scaffolds,
compared with those cultured on 2D plates, following culture for 10
days. The RT-qPCR analysis showed that CD133, Oct4, Sox2 and
Nanog were markedly upregulated in all three of the cell
lines, indicating these four genes were important in the glioma
cell lines. Other genes were also upregulated in each of the cell
lines. In the U87 cells, Nestin was upregulated; in U251
cells, MSI1, MSI2 and BMI-1 were upregulated; in
HS683 cells, c-Myc and BMI-1 were upregulated. These
changes of stemness markers were in accordance with the results of
the morphological analysis. The western blot experiments (Fig. 4B) indicated that CD133, Nestin,
Oct4, Sox2, Nanog and MSI2 were upregulated in all three cell
lines, and the expression of MSI1 and c-Myc was increased in the
HS683 cells. These results were consistent with the RT-qPCR data.
Statistically significant differences were observed between the 3D
cells and 2D cells for each of the glioma cell lines.
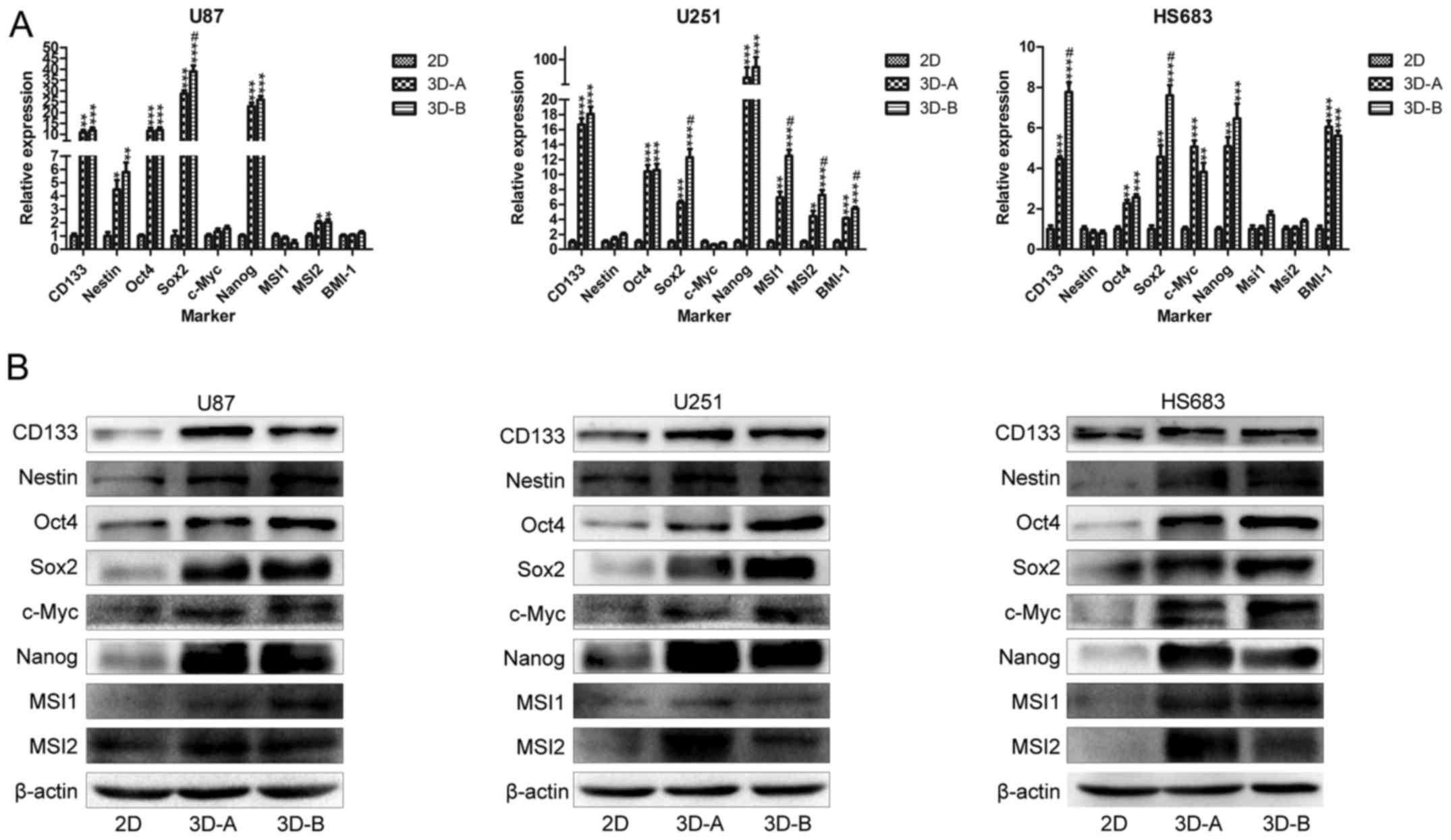 | Figure 4Expression of stemness-related genes.
(A) mRNA expression levels of stem cell genes CD133, Nestin,
Oct4, Sox2, c-Myc, Nanog, MSI1, MSI2 and BMI-1,
determined by reverse transcription-quantitative polymerase chain
reaction analysis. *P<0.05, **P<0.01
and ***P<0.001, compared with 2D groups; #P<0.05,
compared with 3D-A groups. (B) Protein expression levels of the
above stem cell genes, determined by western blot analysis. The
majority of the genes were upregulated in all the three cell lines.
3D, three-dimensional; 2D, two-dimensional; Oct4,
octamer-binding transcription factor 4; Sox2, SRY-Box 2;
MSI, Musashi RNA binding protein. |
Subsequently, the present study analyzed the
expression of cell cycle-related genes in the 2D and 3D cultured
cells. The RT-qPCR results (Fig.
5A) indicated that the genes in all three of the glioma cell
lines under 3D conditions shared similar changing trends, compared
with those in the corresponding 2D cultured cells, which included
significantly upregulated p21 and p27, but no changes
in CCNA, CCNB, CCND or CCNE. Statistically
significant differences were observed between the 3D cells and 2D
cells for all three glioma cell lines. The western blot data
(Fig. 5B) showed a degree of
variance, compared with the RT-qPCR data. Compared with the 2D
groups, the cells in 3D scaffolds exhibited upregulated p21 and
p27, and increased levels of CCNA1, CCNB1, CCND1 and CCNE1. The
differences between the RT-qPCR and western blot data suggested
that the effect of the culture surroundings on cell cycle proteins
may be predominantly at the post-transcriptional level. Although
the cyclins (CCNA1, CCNB1, CCND1 and CCNE1) and cyclin-dependent
kinase inhibitors (p21 and p27) were upregulated in the 3D culture
systems, their comprehensive effect was to suppress the
proliferation of glioma cells, indicating that the effect of the
latter was more marked.
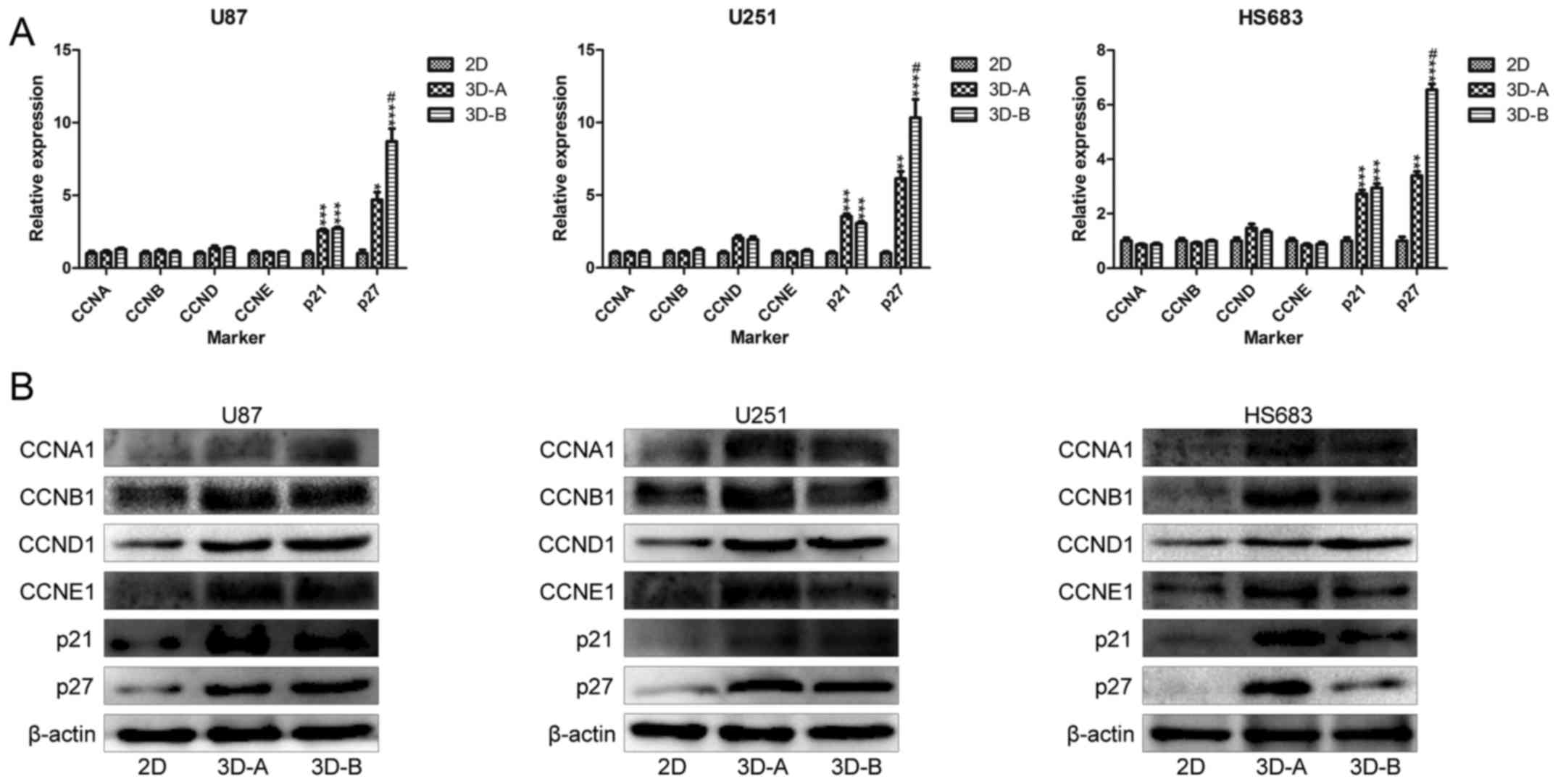 | Figure 5Expression of cell cycle-related
genes. (A) mRNA expression levels of cell cycle-related genes
CCNA, CCNB, CCND, CCNE, p21 and p27, determined by
RT-qPCR analysis. *P<0.05, **P<0.01 and
***P<0.001, compared with 2D groups;
#P<0.05, compared with 3D-A groups. (B) Protein
expression levels of CCNA1, CCNB1, CCND1, CCNE1, p21 and p27,
determined by western blot analysis. Results of RT-qPCR showed
upregulated mRNA levels of p21 and p27, and western
blot data showed higher expression of all proteins in the 3D-A
group. RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; 3D, three-dimensional; 2D, two-dimensional; CCN,
cyclin. |
The present study also observed the expression of
genes related to EMT (N-cadherin and vimentin) and
invasion (MMP1, MMP2, MMP3 and MMP7). The data are
shown in Fig. 6A for RT-qPCR
analysis and Fig. 6B for western
blot analysis. The data obtained via RT-qPCR and western blot
analyses exhibited increases in the expression of these genes to
differing degrees in the 3D collagen scaffolds, compared with those
in the cells cultured on 2D plates.
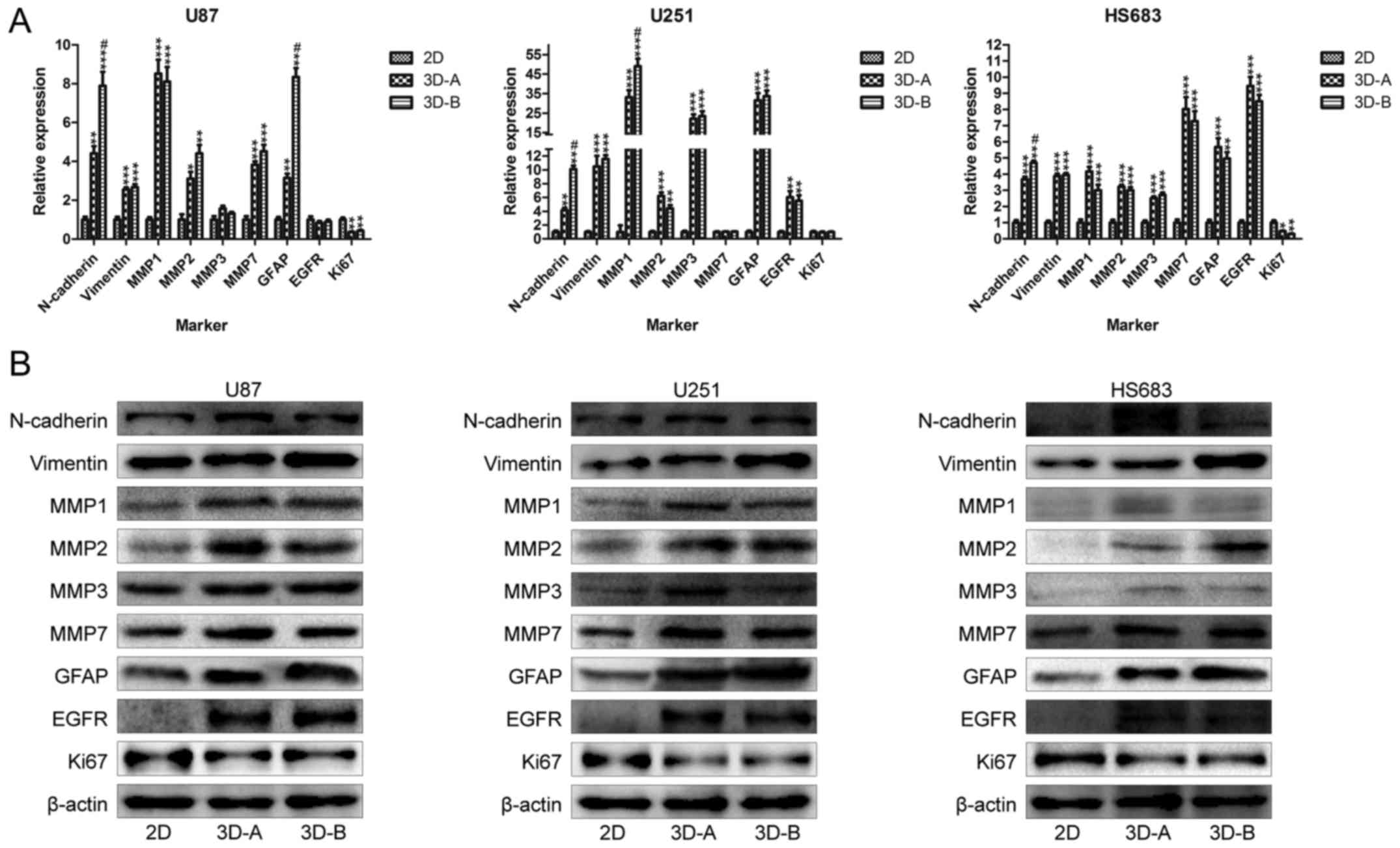 | Figure 6Expression of genes related to
epithelial-mesenchymal transition, migration, invasion and glioma
malignancy. (A) mRNA expression levels of genes, determined by
reverse transcription-quantitative polymerase chain reaction
analysis. *P<0.05, **P<0.01 and
***P<0.001, compared with 2D groups;
#P<0.05, compared with 3D-A groups. (B) Protein
expression levels of the above genes, determined by western blot
analysis. These genes included N-cadherin, vimentin, MMP1, MMP2,
MMP3, MMP7, GFAP, EGFR and Ki67. The majority of the
genes were upregulated in all the three cell lines cultured in the
3D system. 3D, three-dimensional; 2D, two-dimensional; MMP,
matrix metalloproteinase; GFAP, glial fibrillary acidic
protein; EGFR, epidermal growth factor receptor. |
Finally, glioma malignancy-related markers,
including GFAP, EGFR and Ki67, were detected. The
RT-qPCR results showed that GFAP and EGFR were
upregulated and Ki67 was downregulated in glioma cells
cultured in the 3D system, compared with those cultured in the 2D
system. The western blot analysis revealed similar trends (Fig. 6A and B). These changes were
concordant among the three cell lines. The upregulation of
GFAP and EGFR indicated that the 3D collagen culture
enhanced the malignancy of the glioma cells. As a tumor
proliferation marker, the downregulation of Ki67 indicated
the suppression of cell growth, which was consistent with the
results of the cell counting and cell cycle protein assays. For the
expression of all the above genes, statistically significant
differences were observed between the 3D and 2D groups for each of
the glioma cell lines.
Notably, in addition to the comparison between the
3D scaffold and the 2D plate groups, the expression differences of
the above genes were also examined between the A-type scaffold and
B-type scaffold in the three glioma cells. As indicated by the
results of the RT-qPCR analysis (Figs.
2A, 3A and 4A), common differentially expressed genes
of the three cell lines were Sox2 and p27. In the U87
and U251 cells, N-cadherin was the shared differential gene.
GFAP was the specific differential gene for U87 cells, and
MSI1 and MMP1 were uniquely differentially expressed
in the U251 cells. These differential genes were upregulated in
B-type scaffolds, compared with the A-type scaffolds. The results
of the western blot analysis (Figs.
2B, 3B and 4B) showed that Oct4, Sox2, Nanog, MSI2,
CCNB1, CCNE1, vimentin and GFAP were the common differential
proteins to all the three cell lines. Among these proteins, the
expression levels of Sox2, Oct4, vimentin and GFAP were higher in
the B-type scaffold groups, and those of Nanog, MSI2, CCNB1 and
CCNE1 were higher in the A-type scaffold groups. The differences
between the A-type scaffold and B-type scaffold groups were
significant. Compared with the results of the RT-qPCR analysis, the
data from the western blot analysis showed additional differential
genes and the trends were not completely the same. These data
suggested that the scaffold aperture affected the gene expression
of glioma cells, and that the effects were exerted mainly at the
protein level rather than at the mRNA level.
Changes in the biological functions of 3D
system-cultured cells
Considering the variance of gene expression
profiles, the present study aimed to determine whether these
changes affected the relevant biological functions of glioma cells.
Therefore, the colony forming ability, migratory behavior and
invasive ability were compared between the 3D-scaffold cultured
cells and 2D cells on plates. The analyses performed included a
colony formation assay, wound-healing assay and Transwell invasion
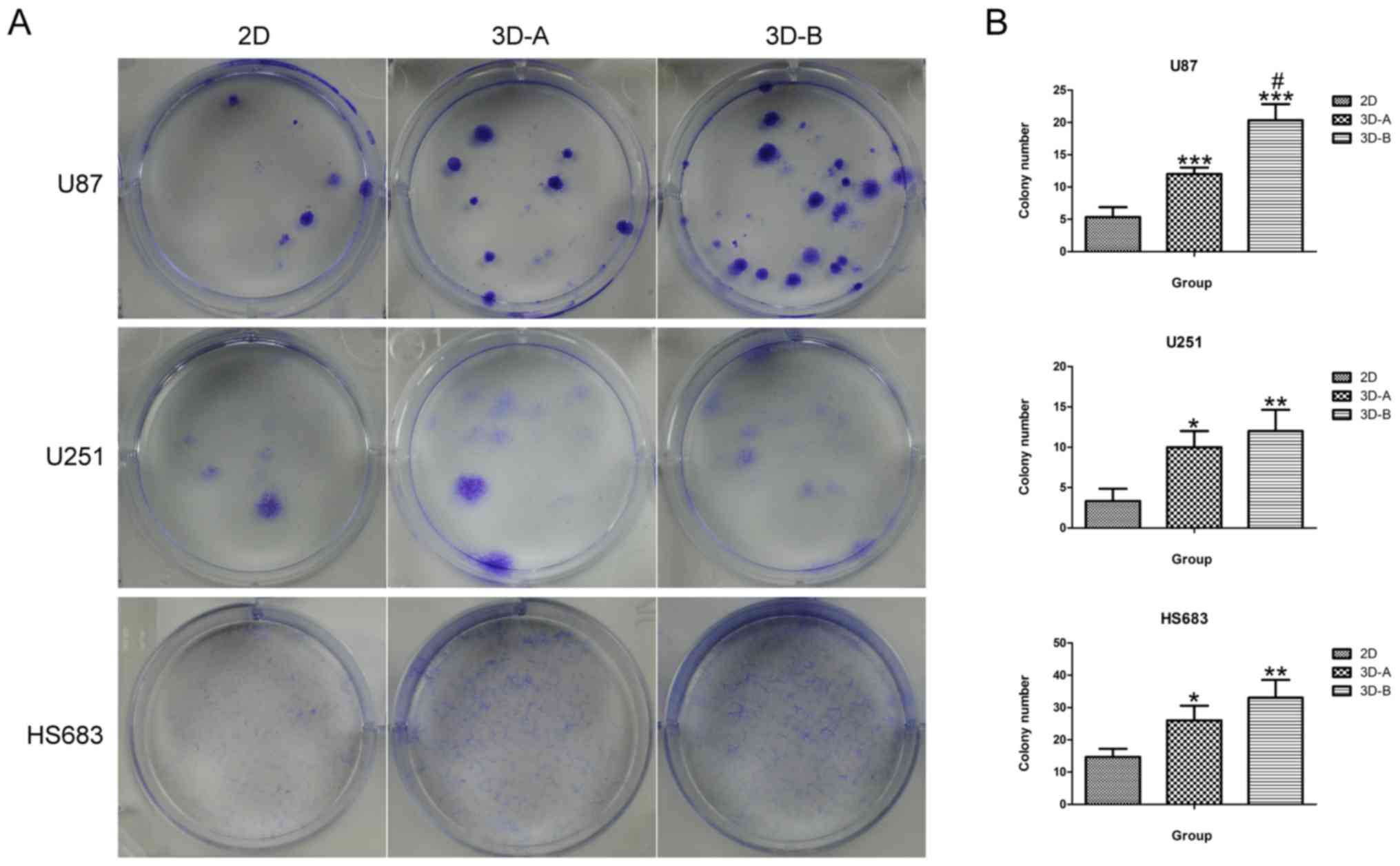
assay. As shown in Fig. 7A and B,
the cells cultured in the 3D collagen scaffolds and on the 2D
plates were able to form colonies, however, more colonies were
formed in all three cell lines when cultured under 3D conditions.
The differences between the 2D cells and 3D cells were significant.
Among the three cell lines, the U87 cells exhibited the most marked
colony formation ability, which was consistent with the results of
stemness-related gene expression. As shown in Fig. 8A and B, the 3D collagen scaffold
culture environment enhanced the migration ability of the glioma
cells, compared with the 2D plate culture environment for all three
cell lines.
Similarly, the results of Transwell invasion assay
confirmed the effects of 3D culture methods on glioma cells. As
shown in Fig. 9A and B, a higher
number of 3D-cultured glioma cells passed through the Matrigel
matrix and appeared on the underside of the membranes.
Statistically significant differences were found between the 3D
cells and 2D cells for all three glioma cell lines
(P<0.001).
In addition to the comparison between 3D and 2D
cells, the differences in biological function between the A-type
scaffold and B-type scaffold in glioma cells were examined. With
the exception of the colony formation assay for the U87 groups, no
significant differences were found in any of the functional
analyses for the cell lines, although differential genes existed
between the A-type scaffold and B-type scaffold. These data
indicated that the aperture size of the collagen-scaffold had no
clear effect on the biological functions of the glioma cells.
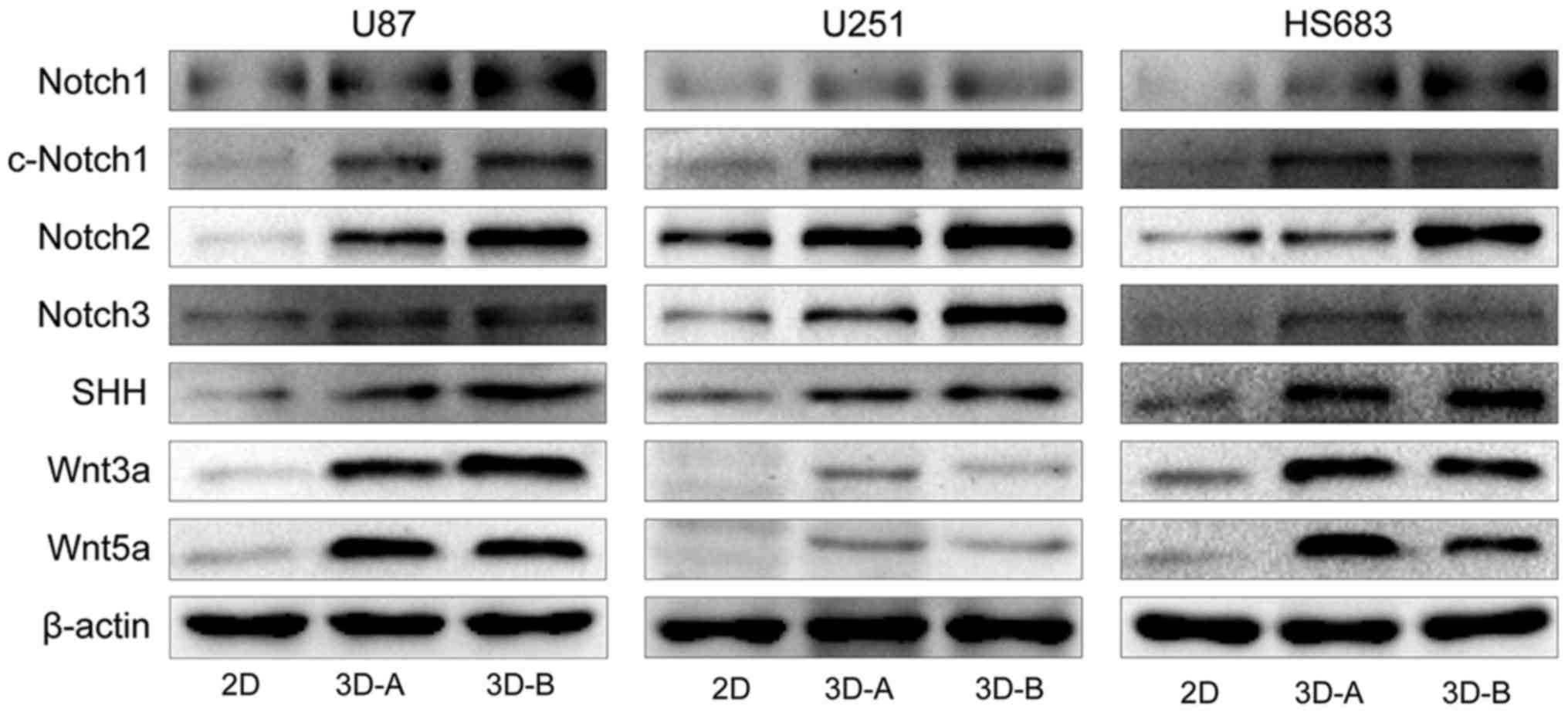
Changes in associated signaling pathways
in 3D system-cultured cells
To examine the molecular mechanisms underlying the
changes in gene expression and biological functions, the present
study detected typical signaling pathways using western blot
analysis, including the apoptotic, Wnt, SHH and Notch pathways. As
shown in Fig. 10A and B, compared
with the 2D-cultured cells, pro-apoptotic factors, including
caspases, poly (ADP-ribose) polymerase (PARP) and p53, were
downregulated and anti-apoptotic factors (PDL-1 and Livin) were
upregulated in cells cultured in 3D scaffolds for all three cell
lines. These results suggested that the 3D culture environment
inhibited the apoptosis of glioma cells. The Wnt pathway, SHH
pathway and Notch pathway are three representative signal
transduction pathways, which are involved in regulating multiple
functions of cells and affecting the occurrence and development of
glioma. Therefore, the present study also detected key proteins in
these pathways. As shown in Fig.
11, Notch1, 2 and 3, Wnt3a, Wnt5a and SHH were all expressed at
high levels in the three types of 3D-cultured cells, compared with
those in the 2D-cultured cells, suggesting that the 3D collagen
scaffold culture affected several important signaling pathways,
followed by changes in gene expression and biological
functions.
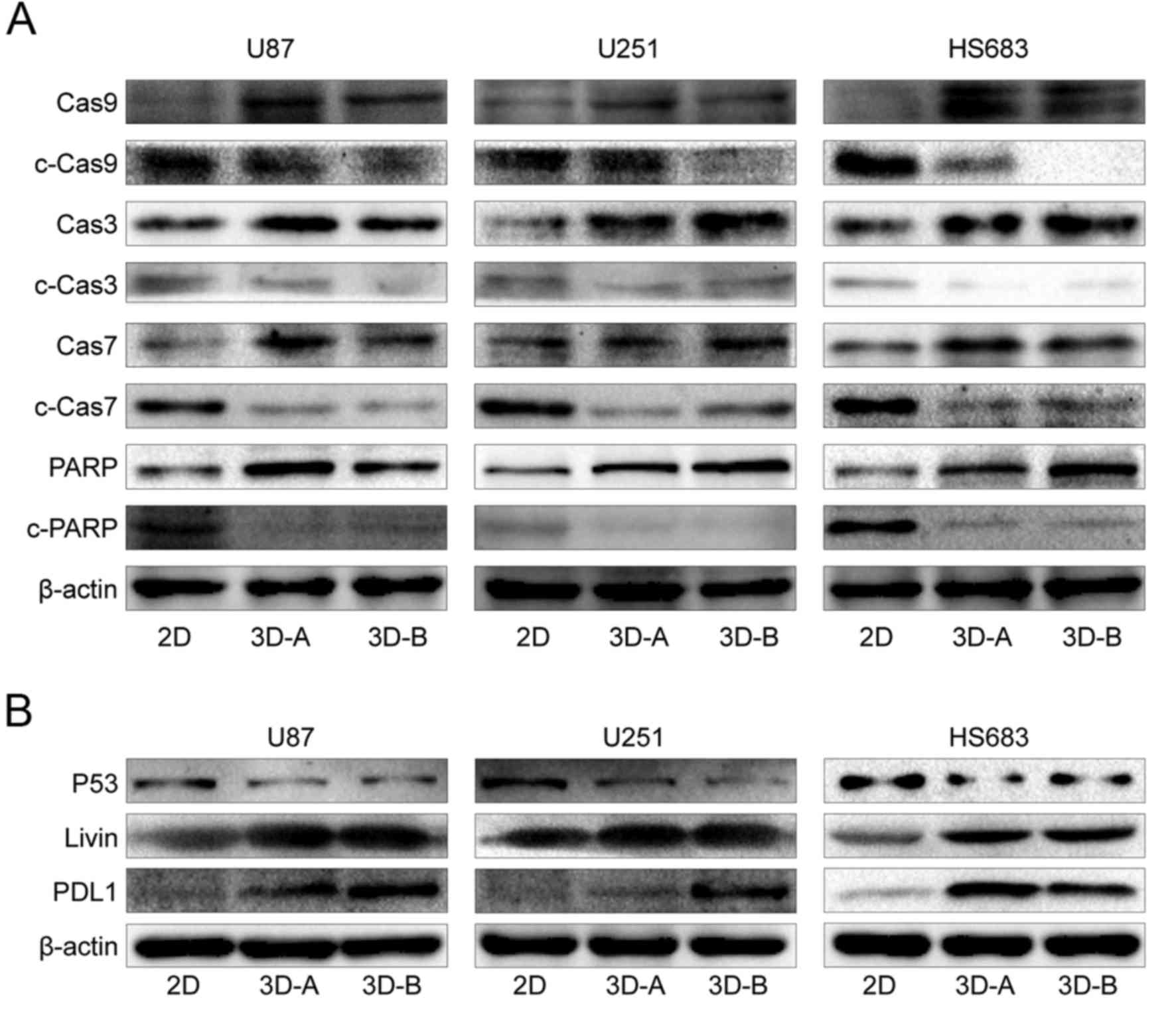 | Figure 10Expression of apoptosis-related
genes, detected by western blot analysis. (A) Apoptosis pathway
regulated by caspases. (B) Other apoptosis-related genes, including
p53, Livin and PDL-1. Compared with 2D cells, pro-apoptotic factors
(caspases, PARP and p53) were downregulated and anti-apoptotic
factors (PDL-1 and Livin) were upregulated in all three cell lines
cultured in 3D scaffolds. 3D, three-dimensional; 2D,
two-dimensional; Cas, caspase; PARP, poly (ADP-ribose) polymerase;
PDL1, programmed death-ligand 1. |
The signaling differences between the A- and B-type
scaffolds for all three glioma cell lines were also examined. Among
apoptotic-related factors, PDL-1 was the only differential gene. It
was upregulated in the B scaffold group for the U87 and U251 cells,
and in the A scaffold group for the HS683 cells (Fig. 10B). For the Wnt, SHH and Notch
pathways (Fig. 11), Notch2 was
the common differentially expressed gene and was expressed at a
high level in the B group of all three glioma cell lines. For the
U87 cells, the majority of these multifunctional signaling proteins
were enhanced in group B; for the U251 cells, c-Notch1 and Notch3
were increased in group B; for HS683 cells, although Notch1 and SHH
were upregulated in group B, c-Notch1, Wnt3a and Wnt5a were higher
in group A.
Discussion
Due to better simulating the microenvironment where
in vivo cells grow, 3D culture has attracted increasing
attention, and has been used in several studies of malignant
tumors, including squamous cell carcinoma, pancreatic cancer and
oral cancer (8,11,12).
3D culture has also been used in glioma, the most common and
life-threatening type of adult brain tumor. In previous studies,
gels, including Matrigel or hydrogel, have been commonly used in 3D
glioma culture systems (13–25).
Although these systems exhibit good biocompatibility, the
experimental steps of these systems are cumbersome, and the cells
planted in these biomaterials show limited digestion and recycling.
Therefore, 3D collagen scaffolds have become a focus of attention,
not only due to their biological compatibility as a main component
of the ECM, but also for the convenience of use. Previous studies
have shown that rat neural stem cells, mouse embryonic stem cells
and human mesenchymal stem cells grow well in this collagen
scaffold, and that the stemness and self-renewal properties are
maintained (31–33). However, there have been few reports
on the effects of the collagen scaffold on tumor cells. Therefore,
this collagen scaffold was used in the present study to observe the
effect of 3D culture on glioma cells. Cell morphology and
proliferation analyses indicated that the three glioma cells
examined exhibited suitable biocompatibility with these 3D collagen
scaffolds. The cells planted in collagen scaffolds formed clusters,
exhibited a small and ovoid appearance, and had heteromorphic and
deeply stained nuclei, indicating that co-culture with the
scaffolds promoted stem cell-like changes of the glioma cells,
which was coincident with the data from the previous studies
mentioned.
As the effects of the collagen scaffold culture on
the gene expression profile and associated functions of glioma
cells remained to be fully elucidated, these alterations were
systematically observed in the cells following planting in 3D
collagen scaffolds. Compared with the 2D groups, the expression of
stemness-related genes was increased in the 3D groups, consisted
with the results of morphology analysis. Jiguet et al and Lv
et al reported similar results (13,17).
The present study also surveyed other important genes involved in
cell cycle, EMT, invasion and glioma malignancy. Cell cycle-related
genes were upregulated to differing degrees in the 3D-cultured
cells, and the comprehensive efficiency inhibited cell
proliferation. The expression of the remainder of the genes also
increased to differing degrees. Considering the changes in gene
expression, the present study aimed to determine whether these
genetic variations cause corresponding changes in biological
function. The data from colony formation, wound-healing and
Transwell invasion assays showed that 3D collagen culture enhanced
the colony-forming, migration and invasion abilities of the glioma
cells. These results suggested that the 3D collagen culture
patterns increased the malignancy of the in vitro cultured
glioma cells, and are thus closer to the environments surrounding
glioma cells in vivo.
Finally, the present study examined the signaling
pathways involved in these changes in gene expression and
biological functions via western blot analysis. A number of vital
pathways, including the apoptotic pathway, Wnt pathway, SHH pathway
and Notch pathway, were examined. Apoptosis mediated by caspases
influences cellular growth, differentiation and programmed death.
The Notch, SHH and Wnt pathways are involved in regulating multiple
functions of cells and affecting the occurrence and development of
glioma (34–38). The results showed that the
apoptotic pathway was inhibited, and the Notch, SHH and Wnt
pathways were activated in the 3D culture groups for all three
glioma cell lines. The data from these analyses indicated that the
3D collagen scaffold culture influenced crucial cellular signaling
pathways, followed by changes in gene expression and biological
functions.
In addition to the comparisons between 3D scaffold
and 2D plate cultures, the present study compared the differences
in the above-mentioned indicators between the A-type scaffold and
B-type scaffold in the three glioma cell lines. Cell morphology and
proliferation analysis showed no notable difference between the
two. However, differences in gene expression were found. The
differential genes included Oct4, Sox2, vimentin, GFAP, Nanog,
MSI2, CCNB1 and CCNE1. The expression levels of the
first four of these genes were higher in the B scaffold group,
whereas those of Nanog, MSI2, CCNB1 and CCNE1 were
higher in the A scaffold group, suggesting that the large aperture
collagen scaffold facilitated the expression of stemness-related
and EMT genes, but that the small aperture had a more marked effect
on the expression of cell cycle-related proteins. However, these
differences in gene expression did not cause changes in biological
functions, including clone formation, migration and invasion.
Finally, the disparities in signaling pathways between the A and
B-type scaffold groups were examined. Notch2 was upregulated in the
B-type group for all three glioma cell lines, indicating that it
was closely associated with the pore diameter of the scaffolds. For
the U87 and U251 cells, the levels of the majority of these
foregoing signaling proteins were increased in the B-type group. In
the HS683 cells, the expression levels of certain genes, including
Notch1 and SHH, were higher in the B-type group, whereas others,
including c-North1, Wnt3a and Wnt5a, were higher in the A-type
group. These differences among the cell lines may be due to the
degree of malignancy of the cells. The highly malignant U87 and
U251 glioma cells in the collagen scaffolds grow in clumps more
readily, owing to their adhesion and proliferation abilities,
therefore, the large aperture may be more appropriate for these
cells and induce increased activity in the signaling pathways. As a
less malignant glioma cell, HS683 cells in collagen scaffolds grow
preferentially along the skeleton rather than in clusters, with
lower adhesion and proliferation abilities; therefore the advantage
of the large aperture in activating the signaling pathways was less
apparent. The results of the H&E staining were in accordance
with these hypotheses.
Notably, we used the controversial U87 MG ATCC
(American Type Culture Collection, Manassas, VA, USA) cell line in
the present study according to the STR profile test performed by us
(data not shown). In the past, the U87 cell line from ATCC was
widely applied in studies on glioma as a glioblastoma cell line.
However, Allen et al reported that this cell line from ATCC
was not the original glioblastoma cell line established in 1968 at
the University of Uppsala, and it was most probably also a
glioblastoma cell line, but whose origin was unknown (39). As we were concerned that the
misidentification of the U87 MG ATCC cell line might affect the
outcomes of the present study, in this study, we observed the cell
morphology and gene expression profile of the U87 MG ATCC cells and
the results revealed that these cells exhibited the characteristics
of glioblastoma. Furthermore, following implantation in 3D collagen
scaffolds, the U87 MG ATCC cell groups exhibited similar changes as
the U251 cell groups (another high-grade glioma cell line) and
exhibited a greater malignancy than the low-grade glioma cell line
(HS683), not only from the gene expression analysis, but also from
the corresponding biological function analysis. These data
indicated that this misidentification may not affect the outcomes
of the present study. In conclusion, the present study found that
3D collagen scaffolds had good biocompatibility with glioma cells,
and enhanced the malignancy of the glioma cells by affecting gene
expression and biological functions. The increase in the degree of
malignancy was regulated by several signal transduction pathways,
including the apoptotic, Wnt, SHH and Notch pathways. The in
vitro glioma culture models based on 3D collagen scaffolds may
better reflect the characteristics of in vivo tumor growth
and have widespread application potential as platforms for
screening novel anti-glioma therapeutics.
Acknowledgments
The authors gratefully acknowledge the cooperation
of all participating institutes for experimental technical
support.
Funding
The present study was supported by the National Key
Research and Development Program of China (grant no.
2016YFC1101502), the National Natural Science Foundation of China
(grant nos. 81472355, 81773179 and 81272972), the Strategic
Priority Research Program of the Chinese Academy of Sciences (grant
no. XDA01030000), the Hunan Provincial Science and Technology
Department (grant nos. 2014FJ6006 and 2016JC2049) and the Open-End
Fund for the Valuable and Precision Instruments of Central South
University (grant no. SUZC201634 and CSUZC201638).
Availability of data and materials
All the data supporting the conclusions of this
article are included in the article.
Authors' contributions
WJ performed the major experiments and wrote the
manuscript. CR, XJ and JD contributed equally to the conception and
design of the study proposal. JD prepared the collagen scaffolds.
WL and CR revised the manuscript. WL, LW, BZ, WH and WJ directed
the writing and layout of the manuscript. SL and XL contributed to
data analysis. XZ and DC reviewed the manuscript and provided
suggestions. HZ, XL, MZ and DX provided experimental technical
support.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Ostrom QT, Gittleman H, Liao P, Rouse C,
Chen Y, Dowling J, Wolinsky Y, Kruchko C and Barnholtz-Sloan J:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2007-2011. Neuro Oncol.
16(Suppl 4): iv1–iv63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hong J, Peng Y, Liao Y, Jiang W, Wei R,
Huo L, Han Z, Duan C and Zhong M: Nimotuzumab prolongs survival in
patients with malignant gliomas: A phase I/II clinical study of
concomitant radiochemotherapy with or without nimotuzumab. Exp Ther
Med. 4:151–157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the Central Nervous System: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Birgersdotter A, Sandberg R and Ernberg I:
Gene expression perturbation in vitro - a growing case for
three-dimensional (3D) culture systems. Semin Cancer Biol.
15:405–412. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fallica B, Makin G and Zaman MH:
Bioengineering approaches to study multidrug resistance in tumor
cells. Integr Biol. 3:529–539. 2011. View Article : Google Scholar
|
|
6
|
Shannon S, Vaca C, Jia D, Entersz I,
Schaer A, Carcione J, Weaver M, Avidar Y, Pettit R, Nair M, et al:
Dexamethasone-mediated activation of fibronectin matrix assembly
reduces dispersal of primary human glioblastoma cells. PLoS One.
10:e01359512015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Le HT, Nguyen HT, Min HY, Hyun SY, Kwon S,
Lee Y, Le THV, Lee J, Park JH and Lee HY: Panaxynol, a natural
Hsp90 inhibitor, effectively targets both lung cancer stem and
non-stem cells. Cancer Lett. 412:297–307. 2018. View Article : Google Scholar
|
|
8
|
Jiang YJ, Lee CL, Wang Q, Zhou ZW, Yang F,
Jin C and Fu DL: Establishment of an orthotopic pancreatic cancer
mouse model: Cells suspended and injected in Matrigel. World J
Gastroenterol. 20:9476–9485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang Z, Han B, Li H, Li X, Yang Y and Liu
W: Preparation and anti-tumor metastasis of carboxymethyl chitosan.
Carbohydr Polym. 125:53–60. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Shi L, Tu Q, Wang H, Zhang H, Wang
P, Zhang L, Huang Z, Zhao F, Luan H, et al: Treating cutaneous
squamous cell carcinoma using 5-aminolevulinic acid
polylactic-co-glycolic acid nanoparticle-mediated photodynamic
therapy in a mouse model. Int J Nanomedicine. 10:347–355.
2015.PubMed/NCBI
|
|
11
|
Yu L, Ni C, Grist SM, Bayly C and Cheung
KC: Alginate core-shell beads for simplified three-dimensional
tumor spheroid culture and drug screening. Biomed Microdevices.
17:332015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sakuma K, Tanaka A and Mataga I: Collagen
gel droplet-embedded culture drug sensitivity testing in squamous
cell carcinoma cell lines derived from human oral cancers: Optimal
contact concentrations of cisplatin and fluorouracil. Oncol Lett.
12:4643–4650. 2016. View Article : Google Scholar
|
|
13
|
Jiguet Jiglaire C, Baeza-Kallee N,
Denicolaï E, Barets D, Metellus P, Padovani L, Chinot O,
Figarella-Branger D and Fernandez C: Ex vivo cultures of
glioblastoma in three-dimensional hydrogel maintain the original
tumor growth behavior and are suitable for preclinical drug and
radiation sensitivity screening. Exp Cell Res. 321:99–108. 2014.
View Article : Google Scholar
|
|
14
|
Bayat N, Ebrahimi-Barough S,
Norouzi-Javidan A, Saberi H, Tajerian R, Ardakan MMM, Shirian S, Ai
A and Ai J: Apoptotic effect of atorvastatin in glioblastoma
spheroids tumor cultured in fibrin gel. Biomed Pharmacother.
84:1959–1966. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan Y, Nguyen DT, Akay Y, Xu F and Akay M:
Engineering a brain cancer chip for high-throughput drug screening.
Sci Rep. 6:250622016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heffernan JM, Overstreet DJ, Srinivasan S,
Le LD, Vernon BL and Sirianni RW: Temperature responsive hydrogels
enable transient three-dimensional tumor cultures via rapid cell
recovery. J Biomed Mater Res A. 104:17–25. 2016. View Article : Google Scholar
|
|
17
|
Lv D, Yu SC, Ping YF, Wu H, Zhao X, Zhang
H, Cui Y, Chen B, Zhang X, Dai J, et al: A three-dimensional
collagen scaffold cell culture system for screening anti-glioma
therapeutics. Oncotarget. 7:56904–56914. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang K, Kievit FM, Erickson AE, Silber JR,
Ellenbogen RG and Zhang M: Culture on 3D chitosan-hyaluronic acid
scaffolds enhances stem cell marker expression and drug resistance
in human glioblastoma cancer stem cells. Adv Healthc Mater.
5:3173–3181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gonçalves DPN, Rodriguez RD, Kurth T, Bray
LJ, Binner M, Jungnickel C, Gür FN, Poser SW, Schmidt TL, Zahn DRT,
et al: Enhanced targeting of invasive glioblastoma cells by
peptide-functionalized gold nanorods in hydrogel-based 3D cultures.
Acta Biomater. 58:12–25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gomez-Roman N, Stevenson K, Gilmour L,
Hamilton G and Chalmers AJ: A novel 3D human glioblastoma cell
culture system for modeling drug and radiation responses. Neuro
Oncol. 19:229–241. 2017.
|
|
21
|
Heffernan JM, McNamara JB, Borwege S,
Vernon BL, Sanai N, Mehta S and Sirianni RW:
PNIPAAm-co-Jeffamine®(PNJ) scaffolds as in vitro models
for niche enrichment of glioblastoma stem-like cells. Biomaterials.
143:149–158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Narayan RS, Fedrigo CA, Brands E, Dik R,
Stalpers LJ, Baumert BG, Slotman BJ, Westerman BA, Peters GJ and
Sminia P: The allosteric AKT inhibitor MK2206 shows a synergistic
interaction with chemotherapy and radiotherapy in glioblastoma
spheroid cultures. BMC Cancer. 17:2042017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pedron S, Hanselman JS, Schroeder MA,
Sarkaria JN and Harley BAC: Extracellular hyaluronic acid
influences the rfficacy of EGFR tyrosine kinase inhibitors in a
biomaterial model of glioblastoma. Adv Healthc Mater.
6:17005292017. View Article : Google Scholar
|
|
24
|
Schiariti MP, Restelli F, Ferroli P,
Benetti A, Berenzi A, Ferri A, Ceserani V, Ciusani E, Cadei M,
Finocchiaro G, et al: Fibronectin-adherent peripheral blood derived
mononuclear cells as Paclitaxel carriers for glioblastoma
treatment: An in vitro study. Cytotherapy. 19:721–734. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Tong X, Jiang X and Yang F: Effect
of matrix metalloproteinase-mediated matrix degradation on
glioblastoma cell behavior in 3D PEG-based hydrogels. J Biomed
Mater Res A. 105:770–778. 2017. View Article : Google Scholar :
|
|
26
|
Chen L, Xiao Z, Meng Y, Zhao Y, Han J, Su
G, Chen B and Dai J: The enhancement of cancer stem cell properties
of MCF-7 cells in 3D collagen scaffolds for modeling of cancer and
anti-cancer drugs. Biomaterials. 33:1437–1444. 2012. View Article : Google Scholar
|
|
27
|
Liu H, Ren C, Zhu B, Wang L, Liu W, Shi J,
Lin J, Xia X, Zeng F, Chen J, et al: High-efficient transfection of
human embryonic stem cells by single-cell plating and starvation.
Stem Cells Dev. 25:477–491. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
29
|
Huang W, Liu J, Feng X, Chen H, Zeng L,
Huang G, Liu W, Wang L, Jia W, Chen J, et al: DLC-1 induces
mitochondrial apoptosis and epithelial mesenchymal transition
arrest in naso-pharyngeal carcinoma by targeting EGFR/Akt/NF-κB
pathway. Med Oncol. 32:1152015. View Article : Google Scholar
|
|
30
|
Feng X, Li C, Liu W, Chen H, Zhou W, Wang
L, Zhu B, Yao K, Jiang X and Ren C: DLC-1, a candidate tumor
suppressor gene, inhibits the proliferation, migration and
tumorigenicity of human nasopharyngeal carcinoma cells. Int J
Oncol. 42:1973–1984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui Y, Xiao Z, Chen T, Wei J, Chen L, Liu
L, Chen B, Wang X, Li X and Dai J: The miR-7 identified from
collagen biomaterial-based three-dimensional cultured cells
regulates neural stem cell differentiation. Stem Cells Dev.
23:393–405. 2014. View Article : Google Scholar :
|
|
32
|
Du M, Liang H, Mou C, Li X, Sun J, Zhuang
Y, Xiao Z, Chen B and Dai J: Regulation of human mesenchymal stem
cells differentiation into chondrocytes in extracellular
matrix-based hydrogel scaffolds. Colloids Surf B Biointerfaces.
114:316–323. 2014. View Article : Google Scholar
|
|
33
|
Wei J, Han J, Zhao Y, Cui Y, Wang B, Xiao
Z, Chen B and Dai J: The importance of three-dimensional scaffold
structure on stemness maintenance of mouse embryonic stem cells.
Biomaterials. 35:7724–7733. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Filbin MG, Dabral SK, Pazyra-Murphy MF,
Ramkissoon S, Kung AL, Pak E, Chung J, Theisen MA, Sun Y,
Franchetti Y, et al: Coordinate activation of Shh and PI3K
signaling in PTEN-deficient glioblastoma: New therapeutic
opportunities. Nat Med. 19:1518–1523. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Borcherding N, Kusner D, Kolb R, Xie Q, Li
W, Yuan F, Velez G, Askeland R, Weigel RJ and Zhang W: Paracrine
WNT5A signaling inhibits expansion of tumor-initiating cells.
Cancer Res. 75:1972–1982. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tian J, He H and Lei G: Wnt/β-catenin
pathway in bone cancers. Tumour Biol. 35:9439–9445. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zong D, Ouyang R, Li J, Chen Y and Chen P:
Notch signaling in lung diseases: Focus on Notch1 and Notch3. Ther
Adv Respir Dis. 10:468–484. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu L, Yang ZL, Wang C, Miao X, Liu Z, Li
D, Zou Q, Li J, Liang L, Zeng G, et al: The Expression of Notch 1
and Notch 3 in gallbladder cancer and their clinicopathological
significance. Pathol Oncol Res. 22:483–492. 2016. View Article : Google Scholar
|
|
39
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016. View Article : Google Scholar : PubMed/NCBI
|