Introduction
Colorectal carcinoma (CRC) is among the top three
malignancies with increasing rates of morbidity and mortality
worldwide (1). Despite significant
advances in cancer treatment, individuals with relapse or
metastases still exhibit poor prognosis (2). Therefore, the mechanisms of CRC
progression require further investigation.
RAB proteins regulate diverse pathways in
intracellular membrane trafficking and dynamics (3). Distinct RAB proteins exert different
effects. For example, RAB-1 and RAB-2 modulate the innate immunity,
neuron maturation and vesicle trafficking (4). Meanwhile, RAB14 is overexpressed in
multiple human malignancies, including ovarian (5), non-small cell lung (6), breast (7) and gastric (8,9)
cancer. RAB14 appears to act as an oncogene in human tumorigenesis
and metastasis. However, how RAB14 affects CRC tumorigenesis
remains unclear. Therefore, identifying and characterizing RAB14 is
essential for understanding its roles in the pathogenesis of
CRC.
MicroRNAs (miRNAs) constitute important mediators,
which regulate target genes via binding to the 3′-untranslated
region (UTR) of mRNAs (10).
Multiple miRNAs are aberrantly expressed in CRC, which promotes
malignant behavior (11,12). Previous findings by the authors
demonstrated that the overexpression of miR-194 or miR-217
significantly decreased the proliferation and invasion of CRC
cells. miR-199b might target Sirtuin 1 to suppress the metastasis
of CRC to the liver (13-15). Meanwhile, miR-490-3p promotes cell
proliferation and metastasis in liver cancer by targeting ERGIC and
Golgi 3 (16) while inhibiting
other malignant tumors, including gastric cancer (17,18),
ovarian cancer (19), breast
cancer (20) and osteosarcoma
(21). Although Xu et al
(22) and Zheng et al
(23) reported that miR-490-3p was
downregulated in colorectal carcinoma, the mechanism associated
with its role is not completely elucidated.
The present study aimed to assess whether RAB14 is
directly targeted by miR-490-3p, and evaluate the role of the
miR-490-3p/RAB14 pathway in the progression of CRC.
Materials and methods
Cells and human tissue specimens
Human colorectal cancer cells, SW480, SW620, RKO,
HT29, HCT116 and LoVo, were purchased from the American Type
Culture Collection (Manassas, VA, USA). NCM460 cells were from
INCELL Corporation LLC (San Antonio, TX, USA). HT29, LoVo, HCT116
and RKO cells were cultured in RPMI-1640. SW480 and SW620 were
cultured in Leibovitz's L-15 medium. All cells were cultured at
37°C in a humidified environment containing 5% CO2, and
the medium was supplemented with 10% fetal bovine serum (FBS,
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml
penicillin (Sigma-Aldrich; Merck KGaA), and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA).
A total of 90 patients with CRC were involved in the
present study. Accordingly, 90 human colorectal cancerous specimens
and the corresponding adjacent non-cancerous tissues were collected
during operations at Peking University People's Hospital during
January 2013 to December 2016 in subjects that underwent
coloproctectomy according to the National Comprehensive Cancer
Network guidelines for colon/rectal carcinoma (https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
or https://www.nccn.
org/professionals/physician_gls/pdf/rectal.pdf, version 1.
2013-2016). The samples were kept at −80°C until use. Written
informed consent was provided by all patients prior to sample
collection. The present study received approval from the Research
Ethics Committee of Peking University (Beijing, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The purification of total RNA and RT-qPCR were
performed with PrimeScript RT reagent kit and SYBR Green PCR Master
Mix, respectively (Takara Bio, Inc., Otsu, Japan) as described by
the manufacturer. The thermocycling conditions were 5 sec at 95°C
and 30 sec at 60°C. The data were analyzed using the
2−ΔΔCq method as previously described by Livak and
Schmittgen (24) and Wang et
al (15). GAPDH and U6 RNA
were employed as references for mRNAs and miRNAs, respectively. The
primers used were: miR-490-3p forward,
5′-CAACCTGGAGGACTCCATGCTG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′;
RAB14 forward, 5′-CGCTCGAGATGGCAACTGCACCATACAAC-3′ and reverse,
5′-CGGAATTCCTAGCAGCCACAGCCTTCTC-3′; GAPDH forward,
5′-CCCCGGTTTCTATAAATTGAGC-3′ and reverse,
5′-CACCTTCCCCATGGTGTCT-3′. The universal primer from Takara Bio,
Inc. was used as the reverse primer for miR-490-3p and U6.
Cell transfection or infection
The miRNAs and siRNAs were synthesized by Guangzhou
RiboBio Co., Ltd., (Guangzhou, China). RAB14 plasmids were
manufactured by Invitrogen (Thermo Fisher Scientific, Inc.).
Transfection was carried out with Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). RAB14 siRNA,
miR-490-3p mimics and respective negative controls were used at 50
nM for transient transfection. Lentiviral vectors (3E+8 TU/ml)
(Shanghai GeneChem Co., Ltd., Shanghai, China) expressing
miR-490-3p (LV-miR-490-3p) or the negative control sequence were
employed to infect CRC cells for in vivo experiments. The
oligonucleotide sequences for transfection were: miR-490-3p mimics,
5′-CAACCUGGAGGACUCCAUGCUG-3′; miR-NC, 5′-UUUGUACUACACAAAAGUACUG-3′
and RAB14 siRNA, 5′-TGCAAGGAATCTCACCAAT-3′.
Western blot assay
Cell lysis was performed with hypo-tonic lysis
buffer (Thermo Fisher Scientific, Inc.). Equal quantities of
protein (20 µg/lane) were resolved by 10% or 15% SDS-PAGE and
electro-transferred onto nitrocellulose membranes (Pall
Corporation, Pensacola, FL, USA). The detailed description of the
assay was recently reported (14).
Briefly, the membranes were blocked with 5% non-fat milk powder in
Tris-buffered saline containing 0.1% Tween-20 and probed with
primary antibodies overnight at 4°C. Following incubation with
secondary antibodies in room temperature, the bands were visualized
using enhanced chemiluminescence (Pierce; Thermo Fisher Scientific
Inc.) and imaged using a ChemiDoc™ XRSC system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The primary antibodies used
were: anti-GAPDH (1:1,000; cat. no. 2118; Cell Signaling
Technology, Inc., Danvers, MA, USA) and anti-RAB14 (1:500; cat. no.
ab28639; Abcam, Cambridge, UK). The secondary antibody used was
anti-rabbit IgG (1:5,000; cat. no. ab286397074; Cell Signaling
Technology, Inc.).
Cell proliferation and colony formation
assay
Cell proliferation was assessed using the Cell
Counting Kit-8 assay. After the cells (SW480 or SW620) were seeded
into 96-well plates at 2,000 cells/well, incubation was carried out
at 37°C for 48 h. Viable cells were quantified at various times at
450 nm on a microplate reader (Bio-Rad Laboratories, Inc.).
Sextuplicate assays were performed for three times.
To assess the ability of the cells to form colonies,
the cells were seeded into 6-well plates and cultured for two weeks
at 37°C following transfection. Next, the cells were washed with
PBS and fixed with 4% paraformaldehyde then subjected to staining
with 0.1% crystal violet for 20 min at room temperature. Duplicate
assays were carried out three times.
Cell cycle and apoptosis analysis
To assess cell cycle distribution, the cells (SW480
or SW620) were stained with the BD Cycletest™ plus DNA kit (BD
Biosciences, Franklin Lakes, NJ, USA) as suggested by the
manufacturer. To detect apoptosis, the transfected cells (72 h)
were incubated with the Alexa Fluor® 488 Annexin V/Dead
cell apoptosis kit (Invitrogen; Thermo Fisher Scientific, Inc.).
Apoptosis analysis was performed according to the manufacturer's
instructions. The temperature of incubation was different depending
on the steps undertaken. Data analysis was carried out with FlowJo
(version 7.0; Tree Star, Inc., Ashland, OR, USA) on a flow
cytometer (BD Biosciences).
Cell invasion assay
Cell invasion assay was performed using the
Transwell method. Cancer cells (SW480 or SW620) were added to the
upper chambers (24-well plate, pore size, 8 µm; Corning
Incorporated, Corning, NY, USA) with Matrigel pre-coated membranes.
Leibovitz's L-15 medium with 30% FBS was employed as a
chemoattractant in the lower chambers. The Transwell plate was
incubated for 2 days. The cells that passed through the membrane
were washed with PBS. The cells were fixed with 4%
paraformal-dehyde for 20 min and stained with 0.1% crystal violet
for 20 min at room temperature. Subsequently, the cells were
counted and imaged using an inverted microscope (magnification,
×200; Leica DM IL LED; Leica Microsystems GmbH, Wetzlar, Germany).
Duplicate assays were carried out three times.
Luciferase reporter assay
The miR-490-3p-binding site in RAB14 was predicted
using TargetScan 7.1 (http://www.targetscan.org/), microRNA (http://www.microrna.org/) and miRDB (http://www.mirdb.org/). SW480 cells in 24-well plates
were co-transfected with luciferase plasmids (wild-type or mutant,
500 ng/well) for RAB14 and miR-490-3p or NC mimics as directed by
the manufacturer (RiboBio Co., Ltd.). The samples were incubated
for 48 h at 37°C, and luciferase activities were assessed using the
Dual-Luciferase reporter assay system (Promega Corporation,
Madison, WI, USA). The data were reported as a ratio of Firefly to
Renilla luciferase activity.
Xenograft mice model
A total of 8 BALB/c-nude mice (female, 6-weeks-old;
Vital River Laboratories, Beijing, China) were randomly allocated
to two groups with 4 mice in each group to examine tumorigenicity.
In this assay, 200 µl cell suspensions (1×107
SW480 cells) were subcutaneously administered to the right flank of
each mouse. The volume of the tumors was assessed at 4-day
intervals using the formula: V = 0.5 × L. The animals were
scarified 40 days after cell inoculation and the tumors were
extracted for volume and weight measurements. All animal
experiments received approval from the Animal Research Committee of
the Peking University People's Hospital (Beijing, China). The mice
were handled following guidelines provided by the Institutional and
Animal Care and Use Committee.
Statistical analysis
The data are expressed as the mean ± standard
deviation and were assessed with SPSS (version 20.0; SPSS, Inc.,
Chicago, IL, USA). Student's t-test was employed for group
comparisons while LSD t-test was performed for multiple
comparisons. The associations of miR-490-3p levels with
clinicopathologic parameters in CRC were evaluated using the
Pearson χ2 test. The Kaplan-Meier method was employed to
assess overall survival with group differences determined by the
log-rank test. Cox regression analysis was used for multivariate
analysis. The association between the expression of miR-490-3p and
RAB14 was evaluated using the Spearman's correlation. P<0.05
indicated statistical signifi-cance.
Results
Low miR-490-3p levels indicate poor
prognosis in CRC
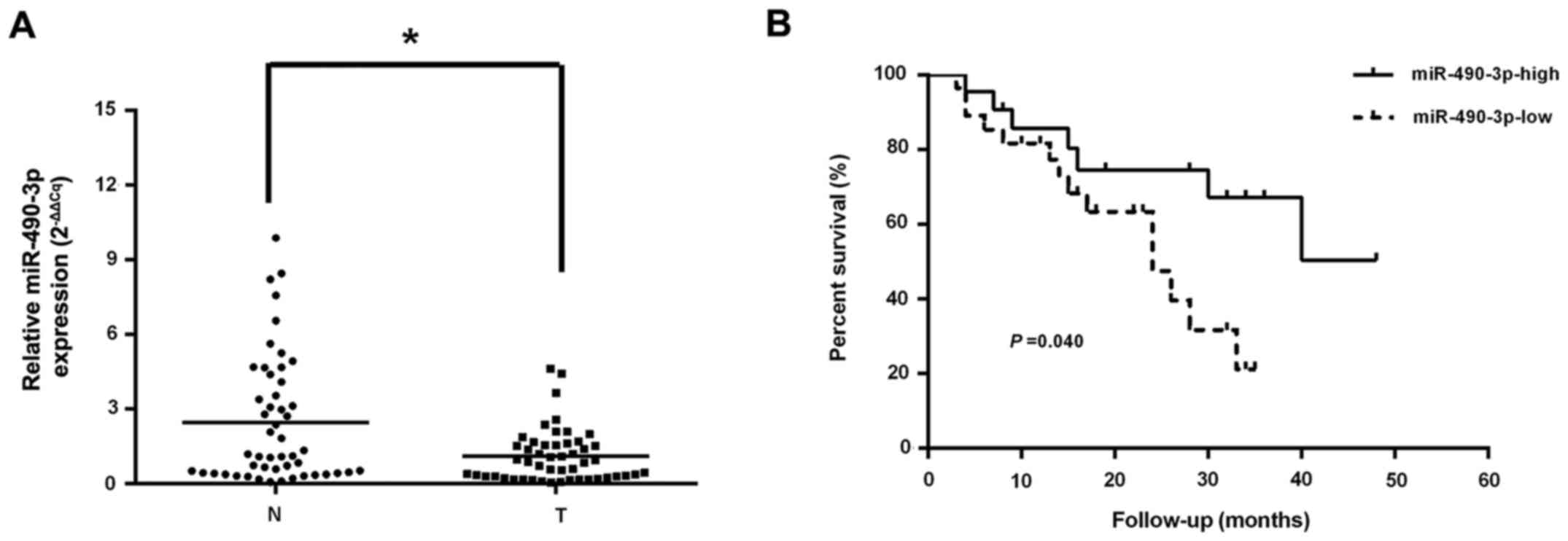
RT-qPCR was employed to assess the levels of
miR-490-3p, which were markedly reduced in CRC tissue specimens
compared with adjacent normal colorectal samples (Fig. 1A). In addition, decreased
miR-490-3p levels were highly correlated with late tumor-node
metastasis (TNM) stage and lymph node metastasis, but not age, sex,
tumor size, tumor differentiation and distant metastasis (Table I). Using the Kaplan-Meier method,
it was identified that CRC cases expressing low levels of
miR-490-3p had a reduced median survival compared with those
exhibiting elevated miR-490-3p expression (Fig. 1B). Cox's multivariate analysis
demonstrated that distant metastasis was closely associated with
overall survival in CRC, and it was an independent predictor of
survival (Table II).
 | Table IAssociation between miR-490-3p
expression and clinicopathologic characteristics in patients with
colorectal cancer. |
Table I
Association between miR-490-3p
expression and clinicopathologic characteristics in patients with
colorectal cancer.
| Parameters | miR-490-3p
expression
|
|---|
| High (n=22) | Low (n=28) | Total (n=50) | P-value |
|---|
| Age, years | | | | |
| ≤60 | 7 | 11 | 18 | 0.585 |
| >60 | 15 | 17 | 32 | |
| Sex | | | | |
| Female | 11 | 14 | 25 | 1.000 |
| Male | 11 | 14 | 25 | |
| Tumor size, cm | | | | |
| ≤2 | 9 | 11 | 20 | 0.907 |
| >2 | 13 | 17 | 30 | |
| Tumor
differentiation | | | | |
| Well/moderate | 12 | 14 | 26 | 0.750 |
| Poor | 10 | 14 | 24 | |
| TNM stage | | | | |
| I+II | 14 | 9 | 23 | 0.027a |
| III+IV | 8 | 19 | 27 | |
| Lymph node
metastasis | | | | |
| Positive | 7 | 18 | 25 | 0.023a |
| Negative | 15 | 10 | 25 | |
| Distant
metastasis | | | | |
| Positive | 5 | 12 | 17 | 0.136 |
| Negative | 17 | 16 | 33 | |
| Vascular
infiltration | | | | |
| Positive | 10 | 14 | 24 | 0.750 |
| Negative | 12 | 14 | 26 | |
 | Table IIMultivariate analysis of factors that
are associated with overall survival in patients with colorectal
cancer. |
Table II
Multivariate analysis of factors that
are associated with overall survival in patients with colorectal
cancer.
| Variables | Multivariate
analysis
|
|---|
| HR (95% CI) | P-value |
|---|
| Age | 1.021
(0.989-1.054) | 0.197 |
| Sex | 0.560
(0.165-1.900) | 0.352 |
| Tumor size | 0.956
(0.321-2.847) | 0.936 |
| Tumor
differentiation | 1.292
(0.467-3.572) | 0.622 |
| TNM stage | 9.829
(0.724-133.442) | 0.086 |
| Lymph node
metastasis | 0.257
(0.036-1.838) | 0.274 |
| Distant
metastasis | 6.538
(1.615-26.473) | 0.009a |
| miR-490-3p | 0.707
(0.205-2.439) | 0.583 |
| RAB14 | 2.019
(0.658-6.202) | 0.220 |
miRNA-490-3p suppresses the proliferation
of CRC cells in vitro and in vivo
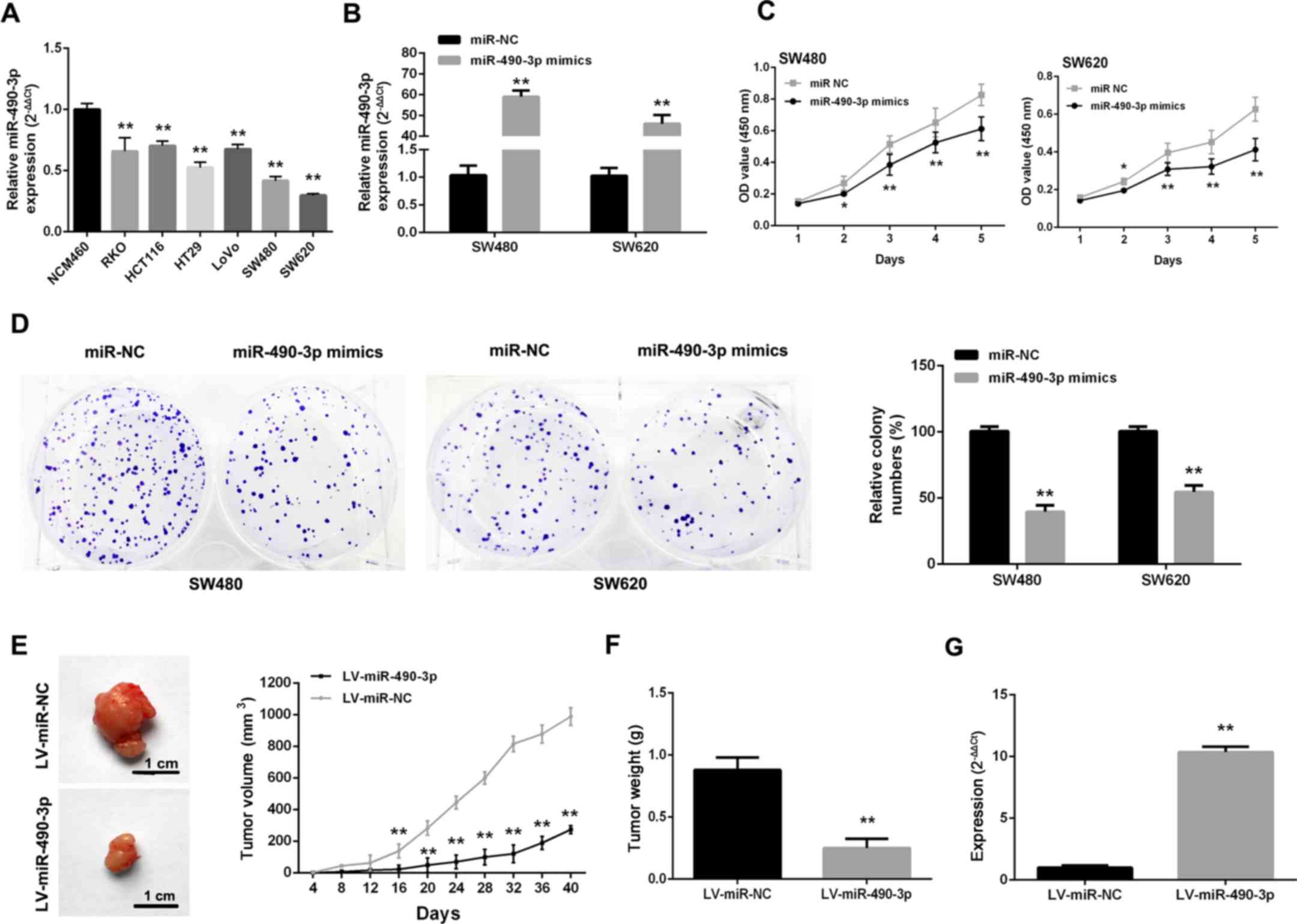
As CRC cells express low levels of miR-490-3p
(Fig. 2A), gain-of-function
studies of miR-490-3p were performed by transfecting miR-490-3p
mimics into cells (Fig. 2B). After
upregulating miR-490-3p, proliferative ability was markedly
repressed in SW480 and SW620 cells (Fig. 2C). Additionally, SW480 and SW620
cells transfected with miR-490-3p mimics formed markedly fewer
colonies compared with the NC controls (Fig. 2D). To further confirm these
findings, an in vivo study was carried out where SW480 cells
were infected with LV-miR-490-3p and LV-miR-NC. Tumors from the
LV-miR-490-3p-infected cells grew much more slowly compared with
the LV-miR-NC group (Fig. 2E and
F). In addition, miR-490-3p levels in the LV-miR-490-3p-
infected tumors were higher compared with the control values
(Fig. 2G).
miR-490-3p induces cancer cell cycle
arrest
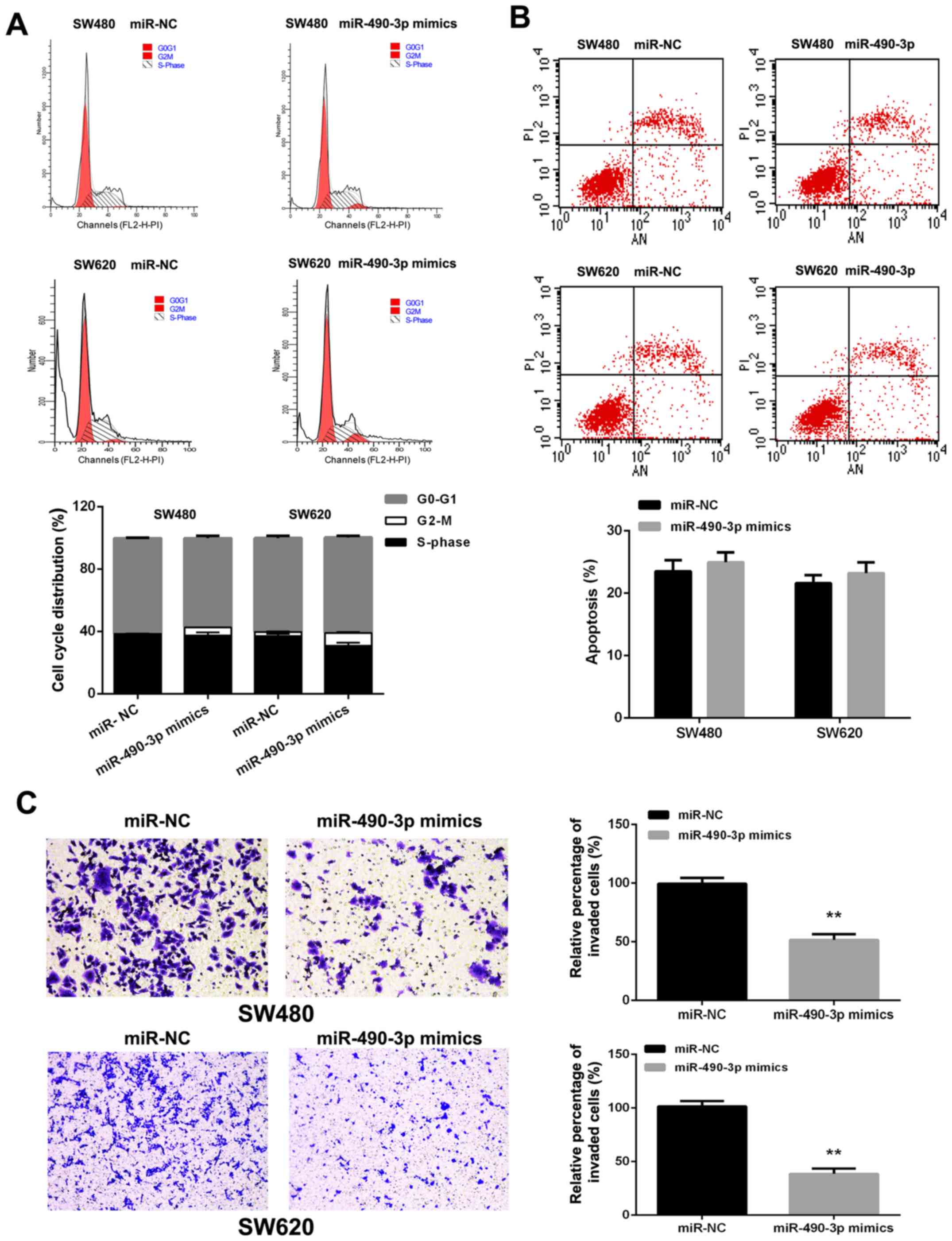
miR-490-3p overexpression resulted in the
accumulation of SW480 (1.235±0.078 vs. 5.175±0.106%, P<0.01) and
SW620 (2.655±0.332 vs. 8.110±0.665%, P<0.01) cells in the G2-M
phase (Fig. 3A). However,
miR-490-3p overexpression seemed to have no significant effects on
apoptosis in CRC cells (Fig.
3B).
miR-490-3p suppresses the invasive
ability of CRC cells
To assess the function of miR-490-3p in CRC
metastasis, miR-490-3p mimics were transfected into SW480 and SW620
cells. Transwell assays indicated that miR-490-3p overexpres-sion
decreased the invasive abilities of SW480 and SW620 cells (Fig. 3C).
miR-490-3p directly targets RAB14 by
interacting with its 3′UTR
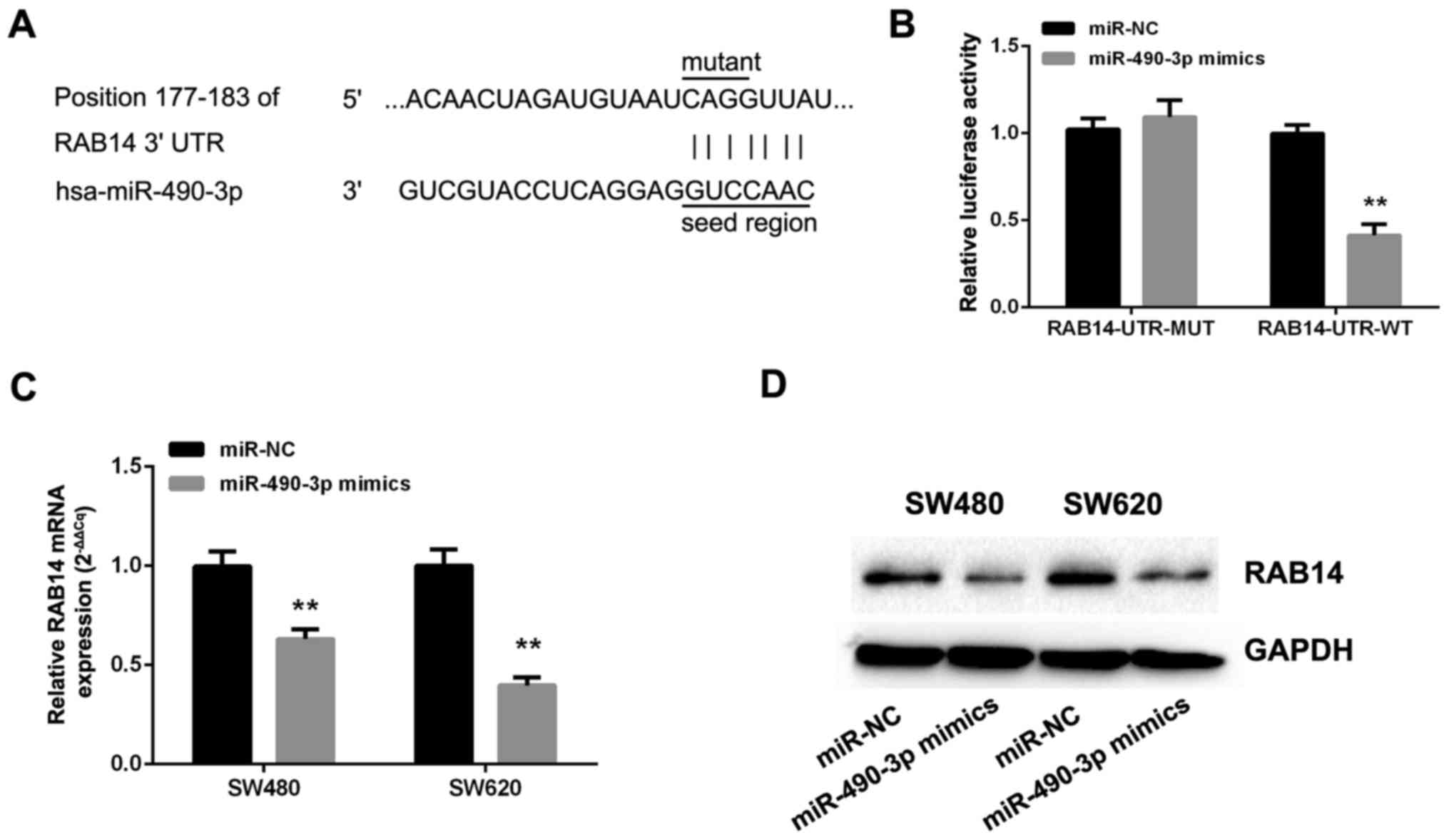
To explore the mechanisms by which miR-490-3p
suppresses the malignant behavior of CRC cells, TargetScan 7.1,
microRNA and miRDB were employed to identify potential miR-490-3p
targets. The databases revealed that the miRNA responsive element
within the 3′UTR of RAB14 is a putative miR-490-3p target (Fig. 4A). Therefore, dual-luciferase
reporters comprising the 3′-UTR of RAB14 with wild-type and mutated
seed sequence were cloned (Fig.
4A). Following the co-transfection of miR-490-3p or NC mimics
with RAB14-UTR-WT or RAB14-UTR-MUT plasmid into SW480 cells, the
relative luciferase activity in the wild-type group was markedly
reduced, while the mutant group exhibited no significant changes
(Fig. 4B). In addition, the
effects of miR-490-3p on endogenous RAB14 expression were assessed
by RT-qPCR and immunoblotting. Interestingly, the overexpression of
miR-490-3p markedly reduced the levels of RAB14 mRNA (Fig. 4C) and protein (Fig. 4D).
Association of miR-490-3p with RAB14
expression in colorectal tissue specimens
Next, the association of miR-490-3p with RAB14
expression was examined in CRC and normal colorectal tissue
specimens. RAB14 levels were markedly elevated in CRC tissues
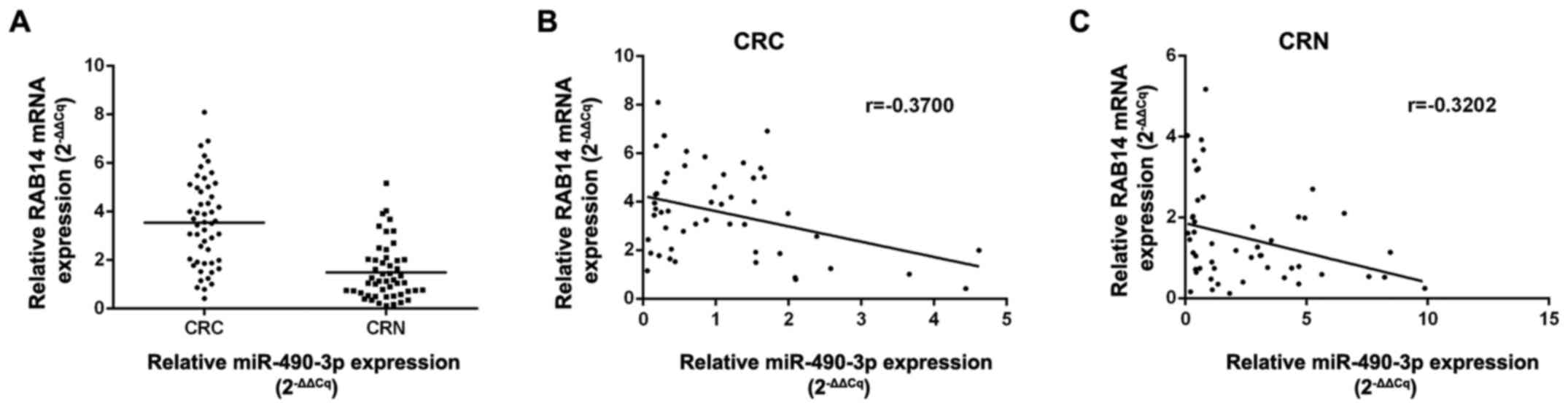
compared with non- cancerous specimens (P<0.01; Fig. 5A). Notably, an inverse correlation
was obtained between miR-490-3p and RAB14 levels in CRC (r=−0.3700,
P<0.05) and normal (r=−0.3202, P<0.01) tissues by Pearson's
correlation analysis (Fig. 5B and
C).
Upregulation of RAB14 counteracts the
inhibitory effects of miR-490-3p on malignant behavior in SW480
cells
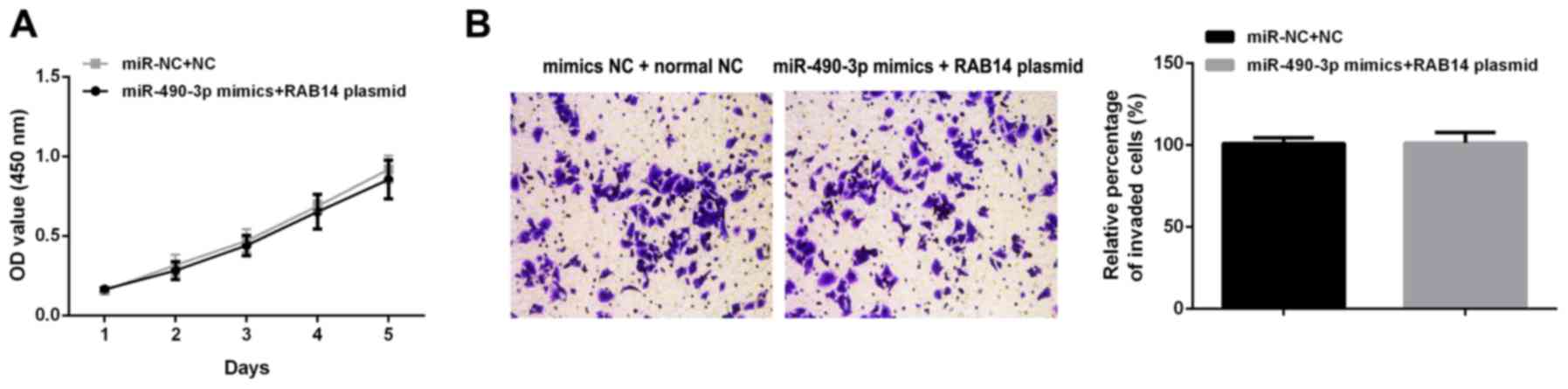
To confirm the role of RAB14 in miR-490-3p
associated anticancer process, rescue experiments were carried out
by upregulating RAB14 after transfecting miR-490-3p mimics. The
overexpression of miR-490-3p and RAB14 had no signifi-cant effects
on proliferation (Fig. 6A) and
invasion (Fig. 6B) in SW480
cells.
Discussion
The biological roles of many miRNAs have been
described in malignancy, revealing that they might function as
oncogenes or tumor suppressors (25,26).
The present study assessed miR-490-3p, which was reported to be
aberrantly expressed in various malignancies (17-20).
However, the roles of miR-490-3p in tumorigenesis and cell growth
are unclear.
In the present study, the clinicopathological
importance of miR-490-3p in CRC was first assessed. The mean
miR-490-3p expression in CRC samples were markedly reduced compared
with paired adjacent non-cancerous specimens. In addition,
miR-490-3p levels in patients with CRC were correlated with the
pathological stage and lymph node metastasis. Furthermore, low
miR-490-3p expression was associated with poor survival in CRC. As
the sample size in the present study was small, additional studies
with larger sample sizes are required to confirm the clinical and
prognostic values of miR-490-3p in CRC. Additionally, Cox's
multivariate analysis revealed distant metastasis as an independent
predictive factor of overall survival in CRC.
The overexpression of miR-490-3p markedly suppressed
the proliferation and colony formation of CRC cells, inducing cell
cycle arrest at the G2/M phase. Reportedly, miR-490-3p regulated
cyclin dependent kinase 1 that resulted in G2/M phase arrest in
ovarian cancer (19). Therefore in
colorectal cancer, miR-490-3p might exert in a similar mechanism.
However, in the present study, the upregulation of miR-490-3p had
no effects on cell apoptosis. This difference in results might be
because the effects of miRNA and its diverse downstream targets are
dependent on the type of cancer.
Zhang et al (16) revealed that miR-490-3p promoted the
growth and metastasis of hepatocellular carcinoma cells by
repressing cell apoptosis. By contrast, Shen et al (17) reported that miR-490-3p targeted
regulator of chromatin subfamily D member 1 (SMARCD1) to stimulate
apoptosis in gastric cancer. Moreover, the in vivo
experiments in the present study firstly confirmed miR-490-3p
upregulation markedly inhibited the growth of CRC xenograft tumors
in nude mice. These findings strongly indicated that miR-490-3p
inhibited tumorigenesis in CRC.
Metastasis results from a multistep-process with
cancer cells that respond to multiple intrinsic and extrinsic
stimuli, detach from the primary tumor, invade the contiguous
stroma, migrate to distant sites and colonize different organs
(27,28). Among distortions in the epigenome,
aberrant expression or function of miRNAs considerably contributes
to metastasis (29,30). An increasing number of studies
involving preclinical models of different human tumor types
indicated that specific miRNAs have functions in various steps of
CRC metastasis (31-33). Therefore, how miRNAs affect CRC
metastasis might provide a basis for developing new therapeutics
for CRC. As aforementioned, miR-490-3p was closely related to TNM
stage and lymph node metastasis. In addition, the over-expression
of miR-490-3p resulted in significant repression of invasive
ability in CRC cells. These findings suggested that restoring
miR-490-3p in CRC might represent a novel therapeutic approach for
CRC, particularly in metastatic cases.
The context-dependent roles of miRNAs in various
types of cancer are likely to depend on the signaling pathways
targeted. miR-490-3p was predicted to target numerous
cancer-associated genes, including SMARCD1 in gastric cancer
(17), TNKS2 in breast cancer
(20), HMGA2 in osteosarcoma
(21) and ABCC2 in ovarian cancer
(34).
Notably, RAB14 was also predicted to be a target of
miR-490-3p. RAB14 belongs to the RAB family of proteins. RAB14 is
involved in intracellular vesicle trafficking as well as regulating
signal transduction and recycling of diverse membrane receptors
(35). RAB14 has a role in the
progression of human cancer; however, its biological significance
in CRC is largely unknown. The present study assessed the
importance of RAB14 in individuals with CRC. Survival analysis
indicated no significant difference between the high and low RAB14
expression groups. However, overall survival was prolonged in CRC
cases with reduced RAB14 levels. Consistent with these findings,
the downregulation of RAB14 in cancer cells suppressed cell
proliferation and invasion as observed following the overexpression
of miR-490-3p. These results confirmed that RAB14 promotes the
progression of CRC.
Finally, whether RAB14 was directly targeted by
miR-490-3p was examined. The ectopic expression of miR-490-3p
induced marked RAB14 downregulation at the mRNA and protein levels.
In addition, in CRC tissues, a low expression level of miR-490-3p
was observed with a high expression level of RAB14. Conversely, a
high expression level of miR-490-3p was detected with decreased
RAB14 in colorectal normal tissues. These findings supported that
RAB14 was a direct target of miR-490-3p and that the downregulation
of RAB14 constitutes a mechanism by which miR-490-3p suppresses
tumor growth.
To further test this hypothesis, a rescue experiment
was carried out. The overexpression of RAB14 markedly reversed
miR-490-3p-induced inhibition of proliferation and invasion in
SW480 cells. However, due to distinct differences in body
structures and functions of cells between animals and humans, mouse
xenograft models in the present study work do not closely reflect
clinical CRC, notably concerning metastasis. Therefore, further
in vivo investigations are warranted to clarify the effects
of miR-490-3p on the metastasis of CRC.
Recently, Xu et al (22) and Zheng et al (23) reported that miR-490-3p inhibits the
metastatic and aggressive phenotype in colorectal cancer by
targeting transforming growth factor (TGF)-β and Wnt/β-catenin
signaling pathways. TGF-β, a cytokine with multiple biological
effects, was first reported as an inducer of epithelia-mesenchymal
transition (EMT) in normal mammary epithelial cells. Several
subsequent studies reported important roles of TGF-β-induced EMT in
tumor metastasis, including colorectal cancer (36). The canonical Wnt signaling pathway
is aberrantly activated in the majority of patients with CRC due to
mutations in adenomatous polyposis coli or β-catenin, and it was
demonstrated that the hyperactivation of Wnt signaling has an
essential role in the pathogenesis of CRC (37,38).
Recent studies have indicated that TGF-β and TGFβ-induced factor
homeobox 1 were able to activate the Wnt/β-catenin signaling
pathway in colorectal cancer (39), breast cancer (40) and systemic sclerosis (41). Hou et al (5) revealed that the Wnt inhibitor
abolished the effect of RAB14 on cell proliferation and Wnt target
genes in ovarian cancer, demonstrating that RAB14 regulated cell
aggressiveness via the Wnt/β-catenin pathway. Furthermore, RAB
family protein, RAB11, with similar structure to RAB14, was
reported to directly regulate the recycling of TGF-β receptors
irrespective of the activation state of the receptors (42). Therefore, based on these recent
studies, RAB14 might be an upstream key factor in regulating, or at
least, affecting the functions of TGF-β and the Wnt/β-catenin
signaling pathway. In the present study, miR-490-3p targeted RAB14
to affect the biological functions of colorectal cancer. RAB14
might be the main target of miR-490-3p in regulating the malignant
phenotype of colorectal cancer, which remains to be verified in
further investigations.
In summary, the present study indicated that human
CRC tissue samples and cells have decreased miR-490-3p expression
compared with colorectal normal tissues and cells respectively,
which was also reflected by a poor prognosis of patients with CRC.
Meanwhile, miR-490-3p suppressed tumorigenesis and aggressiveness
in CRC by directly targeting RAB14. These findings provide novel
insights into targeting miR-490-3p/RAB14 interactions as a
therapeutic option for CRC.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fakih MG: Metastatic colorectal cancer:
Current state and future directions. J Clin Oncol. 33:1809–1824.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stenmark H: Rab GTPases as coordinators of
vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reiner DJ and Lundquist EA: Small GTPases.
WormBook 1.67.2. 1–99. 2016.
|
|
5
|
Hou R, Jiang L, Yang Z, Wang S and Liu Q:
Rab14 is overexpressed in ovarian cancers and promotes ovarian
cancer proliferation through Wnt pathway. Tumour Biol.
37:16005–16013. 2016. View Article : Google Scholar
|
|
6
|
Sun J, Feng X, Gao S and Xiao Z:
microRNA-338-3p functions as a tumor suppressor in human
non-small-cell lung carcinoma and targets Ras-related protein 14.
Mol Med Rep. 11:1400–1406. 2015. View Article : Google Scholar
|
|
7
|
Yu J, Wang L, Yang H, Ding D, Zhang L,
Wang J, Chen Q, Zou Q, Jin Y and Liu X: Rab14 suppression mediated
by miR-320a inhibits cell proliferation, migration and invasion in
breast cancer. J Cancer. 7:2317–2326. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Liu H, Shao J and Xing G: miR-320a
serves as a negative regulator in the progression of gastric cancer
by targeting RAB14. Mol Med Rep. 16:2652–2658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo B, Wang W, Zhao Z, Li Q, Zhou K, Zhao
L, Wang L, Yang J and Huang C: Rab14 act as oncogene and induce
proliferation of gastric cancer cells via AKT signaling pathway.
PLoS One. 12:e01706202017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Valeri N, Croce CM and Fabbri M:
Pathogenetic and clinical relevance of microRNAs in colorectal
cancer. Cancer Genomics Proteomics. 6:195–204. 2009.PubMed/NCBI
|
|
12
|
Shen Z, Zhou R, Liu C, Wang Y, Zhan W,
Shao Z, Liu J, Zhang F, Xu L, Zhou X, et al: MicroRNA-105 is
involved in TNF-α-related tumor microenvironment enhanced
colorectal cancer progression. Cell Death Dis. 8:32132017.
View Article : Google Scholar
|
|
13
|
Shen ZL, Wang B, Jiang KW, Ye CX, Cheng C,
Yan YC, Zhang JZ, Yang Y, Gao ZD, Ye YJ, et al: Downregulation of
miR-199b is associated with distant metastasis in colorectal cancer
via activation of SIRT1 and inhibition of CREB/KISS1 signaling.
Oncotarget. 7:35092–35105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang B, Shen ZL, Gao ZD, Zhao G, Wang CY,
Yang Y, Zhang JZ, Yan YC, Shen C, Jiang KW, et al: miR-194,
commonly repressed in colorectal cancer, suppresses tumor growth by
regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle.
14:1046–1058. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang B, Shen ZL, Jiang KW, Zhao G, Wang
CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al: MicroRNA-217
functions as a prognosis predictor and inhibits colorectal cancer
cell proliferation and invasion via an AEG-1 dependent mechanism.
BMC Cancer. 15:4372015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang LY, Liu M, Li X and Tang H:
miR-490-3p modulates cell growth and epithelial to mesenchymal
transition of hepatocellular carcinoma cells by targeting
endoplasmic reticulum-Golgi intermediate compartment protein 3
(ERGIC3). J Biol Chem. 288:4035–4047. 2013. View Article : Google Scholar :
|
|
17
|
Shen J, Xiao Z, Wu WK, Wang MH, To KF,
Chen Y, Yang W, Li MS, Shin VY, Tong JH, et al: Epigenetic
silencing of miR-490-3p reactivates the chromatin remodeler SMARCD1
to promote Helicobacter pylori-induced gastric carcinogenesis.
Cancer Res. 75:754–765. 2015. View Article : Google Scholar
|
|
18
|
Qu M, Li L and Zheng WC: Reduced
miR-490-3p expression is associated with poor prognosis of
Helicobacter pylori induced gastric cancer. Eur Rev Med Pharmacol
Sci. 21:3384–3388. 2017.PubMed/NCBI
|
|
19
|
Chen S, Chen X, Xiu YL, Sun KX and Zhao Y:
MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial
carcinoma tumori-genesis and progression. Cancer Lett. 362:122–130.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jia Z, Liu Y, Gao Q, Han Y, Zhang G, Xu S,
Cheng K and Zou W: miR-490-3p inhibits the growth and invasiveness
in triple- negative breast cancer by repressing the expression of
TNKS2. Gene. 593:41–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu W, Xu G, Liu H and Li T:
MicroRNA-490-3p regulates cell proliferation and apoptosis by
targeting HMGA2 in osteo-sarcoma. FEBS Lett. 589B:3148–3153. 2015.
View Article : Google Scholar
|
|
22
|
Xu X, Chen R, Li Z, Huang N, Wu X, Li S,
Li Y and Wu S: MicroRNA-490-3p inhibits colorectal cancer
metastasis by targeting TGFβR1. BMC Cancer. 15:10232015. View Article : Google Scholar
|
|
23
|
Zheng K, Zhou X, Yu J, Li Q, Wang H, Li M,
Shao Z, Zhang F, Luo Y, Shen Z, et al: Epigenetic silencing of
miR-490-3p promotes development of an aggressive colorectal cancer
phenotype through activation of the Wnt/β-catenin signaling
pathway. Cancer Lett. 376:178–187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ: Schmittgen TD. Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Method Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Bracken CP, Scott HS and Goodall GJ: A
network-biology perspective of microRNA function and dysfunction in
cancer. Nat Rev Genet. 17:719–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvi-ronment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Profumo V and Gandellini P: MicroRNAs:
Cobblestones on the road to cancer metastasis. Crit Rev Oncog.
18:341–355. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen Z, Wang B, Luo J, Jiang K, Zhang H,
Mustonen H, Puolakkainen P, Zhu J, Ye Y and Wang S: Global-scale
profiling of differential expressed lysine acetylated proteins in
colorectal cancer tumors and paired liver metastases. J Proteomics.
142:24–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doldi V, Pennati M, Forte B, Gandellini P
and Zaffaroni N: Dissecting the role of microRNAs in prostate
cancer metastasis: Implications for the design of novel therapeutic
approaches. Cell Mol Life Sci. 73:2531–2542. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
White NM, Fatoohi E, Metias M, Jung K,
Stephan C and Yousef GM: Metastamirs: A stepping stone towards
improved cancer management. Nat Rev Clin Oncol. 8:75–84. 2011.
View Article : Google Scholar
|
|
31
|
Gandellini P, Doldi V and Zaffaroni N:
microRNAs as players and signals in the metastatic cascade:
Implications for the development of novel anti-metastatic
therapies. Semin Cancer Biol. 44:132–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka
H, Boland CR and Goel A: Circulating microRNA-203 predicts
prognosis and metastasis in human colorectal cancer. Gut.
66:654–665. 2017. View Article : Google Scholar
|
|
33
|
Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang
DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, et al: Identification
of microRNA-214 as a negative regulator of colorectal cancer liver
metastasis by way of regulation of fibroblast growth factor
receptor 1 expression. Hepatology. 60:598–609. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tian J, Xu YY, Li L and Hao Q: miR-490-3p
sensitizes ovarian cancer cells to cisplatin by directly targeting
ABCC2. Am J Transl Res. 9:1127–1138. 2017.PubMed/NCBI
|
|
35
|
Takai Y, Sasaki T and Matozaki T: Small
GTP-binding proteins. Physiol Rev. 81:153–208. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jung B, Staudacher JJ and Beauchamp D:
Transforming growth factor β superfamily signaling in development
of colorectal cancer. Gastroenterology. 152:36–52. 2017. View Article : Google Scholar
|
|
37
|
Morin PJ, Sparks AB, Korinek V, Barker N,
Clevers H, Vogelstein B and Kinzler KW: Activation of
beta-catenin-Tcf signaling in colon cancer by mutations in
beta-catenin or APC. Science. 275:1787–1790. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kinzler KW and Vogelstein B: Lessons from
hereditary colorectal cancer. Cell. 87:159–170. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang JL, Qi Z, Li YH, Zhao HM, Chen YG and
Fu W: TGFβ induced factor homeobox 1 promotes colorectal cancer
development through activating Wnt/β-catenin signaling. Oncotarget.
8:70214–70225. 2017.PubMed/NCBI
|
|
40
|
Ma F, Li W, Liu C, Li W, Yu H, Lei B, Ren
Y, Li Z, Pang D and Qian C: miR-23a promotes TGF-β1-induced EMT and
tumor metastasis in breast cancer cells by directly targeting CDH1
and activating Wnt/β-catenin signaling. Oncotarget. 8:69538–69550.
2017.PubMed/NCBI
|
|
41
|
Gillespie J, Ross RL, Corinaldesi C,
Esteves F, Derrett-Smith E, McDermott MF, Doody GM, Denton CP,
Emery P and Del Galdo F: Transforming growth factor β activation
primes canonical Wnt signaling through down-regulation of Axin-2.
Arthritis Rheumatol. 70:932–942. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mitchell H, Choudhury A, Pagano RE and
Leof EB: Ligand- dependent and -independent transforming growth
factor-beta receptor recycling regulated by clathrin-mediated
endocytosis and Rab11. Mol Biol Cell. 15:4166–4178. 2004.
View Article : Google Scholar : PubMed/NCBI
|