Introduction
Colorectal cancer (CRC), one of the most common
types of cancer, is associated with a high mortality rate worldwide
(1). While the diagnosis of and
treatment strategies for CRC have improved, the overall survival of
patients with CRC remains unsatisfactory (2). Current CRC treatment includes
neoadjuvant radiotherapy and chemotherapy (3), surgical therapy (4,5) and
immunization therapy (6,7). In the majority of cases, however, CRC
is diagnosed at an advanced pathological stage before any symptoms
appear, resulting in poor survival rates. Recent studies have
indicated that long non-coding RNAs (lncRNAs) are critical factors
in regulating the development, differentiation and apoptosis
(8-10) of cancer cells. lncRNAs are also
associated with tumor progression and metastasis (11). In CRC specifically, lncRNA could
inhibit tumor growth by deactivating Akt signaling (12). The aberrant expression of lncRNAs
is associated with a poor survival (13), lymph node metastasis (12) and the modulation of
pithelial-mesenchymal transition (EMT) in CRC (14). Therefore, lncRNAs may function as
novel therapeutic targets and molecular markers for the diagnosis
of CRC. Several studies have identified molecular markers affecting
CRC dynamics (15-17). For instance, miRNA-126 expression
has been shown to exert a critical effect on CRC pathogenesis
(18). However, further
clarifications of the systemic correlation between miRNAs and
lncRNAs are required.
The lncRNA-miRNA complex forms a regulatory network
in non-small cell lung cancer A549 cells (19), indicating that lncRNAs are involved
in controlling tumor progression by interacting with miRNAs.
Additional molecules, such as the orphan nuclear receptors, Nur77,
RARγ and WNT, signal the inhibitors, OVOL2, Myb-like SWIRM and MPN
domains 1 (MYSM1), which influence the invasion, metastasis and
progression of CRC (14,20-22).
Some bioactive enzymes, such as serum β-glucuronidase, are also
considered potential markers of CRC (23). Neuropilin-1 (NRP1) is a
transmembrane glycoprotein that can function as an oncogene by
participating in the development and progression of various types
of cancer (24,25), including CRC (26). Numerous biomarkers have been
identified in CRC cells; however, there is still a need for further
clarification of the mechanisms driving the regulation between
biomarkers and tumor progression and metastasis.
In this study, we found that lncRNA 00152 (lnc00152)
was overexpressed in CRC tumors and cancer cells when compared to
adjacent non-tumor tissues. lnc00152 is an important factor
affecting cell proliferation and invasion in vitro and tumor
growth in vivo. The regulation of lnc00152 altered NRP1
expression and affected EMT by modulating N-cadherin and E-cadherin
expression. Finally, we found that the effects of lnc00152 were
regulated by miRNA-206. Our findings indicate that lnc00152 may
function as a therapeutic target against CRC.
Materials and methods
Ethics statements
The use of human samples was approved by the Ethics
Committee Guangzhou First People's Hospital and all patients
involved in the study provided written informed consent. The use of
animals in this study was approved by the Ethics Committee of the
Laboratory Animal Center of South China University of
Technology.
Patients and tissue samples
This study was conducted using a total of 80
paraffin-embedded CRC samples and 40 adjacent non-tumor tissue
samples. All the samples were histopathologically and clinically
diagnosed at Guangzhou First People's Hospital from 2010 to 2016.
None of the recruited participants had received adjunctive
treatment prior to surgery. The histological characterization and
clinicopathological staging of the samples were determined
according to the Union for International Cancer Control (UICC). The
clinicopathological information was available for all samples
(Table I). The CRC specimens and
the matched adjacent non-cancerous tissues were frozen and stored
in liquid nitrogen until further use. The overall survival (OS) was
defined as the period between diagnosis and either death or the
last follow-up. Follow-up ranged from 0 to 78 months. The
Kaplan-Meier overall survival curve was obtained according to
lnc00152 expression and the cumulative survival ratio was
calculated at different time-points during follow-up.
 | Table IDetailed clinical information of the
patients with colorectal cancer. |
Table I
Detailed clinical information of the
patients with colorectal cancer.
| Clinical
characteristics | Patients
|
|---|
| No. of
patients | lncRNA 00152
expression | P-value |
|---|
| Age (years) | | | |
| ≥60 | 60 | 288.76±150.94 | 0.968 |
| <60 | 20 | 321.38±129.96 | |
| Sex | | | |
| Male | 43 | 295.75±152.82 | 0.563 |
| Female | 37 | 298.27±139.44 | |
| Pathological
grade | | | |
| I–II | 37 | 222.71±83.22 | 0.008 |
| III–IV | 43 | 360.77±158.31 | |
| Tumor invasion | | | |
|
T1–T2 | 19 | 253.38±111.75 | 0.383 |
|
T3–T4 | 61 | 316.08±150.66 | |
| Lymph-node
metastasis | | | |
| N0 | 39 | 227.72±84.06 | 0.004 |
|
N1–N2 | 41 | 362.73±161.88 | |
| Distant
metastasis | | | |
| M0 | 60 | 279.70±144.43 | 0.429 |
| M1 | 20 | 348.55±141.26 | |
| Vascular
invasion | | | |
| No | 65 | 290.74±155.12 | 0.142 |
| Yes | 15 | 323.67±95.55 | |
Cell culture
We obtained the CRC cell lines LoVo, SW480 and SW620
from The Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China). The cells were grown at 37°C with 5%
CO2 in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA)
supplemented with 10% fatal bovine serum (FBS) (Invitrogen), 100
U/ml penicillin and 100 µg/ml streptomycin (Invitrogen).
RNA extraction and reverse transcription
(real-time) quantitative PCR (RT-qPCR)
RT-qPCR was used to determine the expression of
lnc00152. We extracted total RNA from the tissues and cultured
cells using TRIzol reagent (Invitrogen/Life Technologies) according
to the manufacturer's instructions. Subsequently, 2 µg of
the extracted total RNA was subjected to reverse transcription (RT)
using a reverse transcription kit (Cat. no. 639505; Takara, Tokyo,
Japan). In order to perform real-time (quantitative) PCR, we used
the One Step SYBR-Green® (Tokyo, Japan) PrimeScript™
RT-PCR kit (Cat. no. RR064A; Takara). For real-time fluorescence
detection we used the ABI7300 real-time PCR thermal cycle
instrument (Applied Biosystems, Foster City, CA, USA) according to
the manufacturer's instructions. The 20 µl total reaction
mixture contained 20 ng cDNA, 10 µl of 2X SYBR-Green master
mix, 200 nM of forward and reverse primers and PCR-grade water. The
PCR cycling conditions were set to 95°C for 3 min, followed by 35
cycles of 95°C for 30 sec, and 55°C for 30 sec. A melting curve
analysis verified the specificity of each primer after PCR to
ensure amplification specificity. Relative gene expression was
calculated using the 2−ΔΔCq method (27), and we normalized the results to the
expression of 18s rRNA. For miRNA-206 expression detection, we
validated human U6 as the normalizer. The sequences of the primers
used for PCR are presented in Table
II.
 | Table IISequences of primers used in
RT-qPCR. |
Table II
Sequences of primers used in
RT-qPCR.
| Gene name | Primer sequence
(5′-3′) |
|---|
| lnc00152 | F:
CCACCAGCCTCTCCTTGAAT |
| R:
GGCTGAGTCGTGATTTTCGGT |
| E-cadherin | F:
TGCCGCCATCGCTTACACCATC |
| R:
GGTCAGCAGCTTGAACCACCAG |
| N-cadherin | F:
CCCACAGCTCCACCATATGACTC |
| R:
CCTGCTCACCACCACTACTTGAG |
| Human U6 | F:
CTCGCTTCGGCAGCACA |
| R:
AACGCTTCACGAATTTGCGT |
| NRP1 | F:
GAGGCATGAAGGCAGACAGAG |
| R:
GAGGCATGAAGGCAGACAGAG |
| 18s rRNA | F:
CCTGGATACCGCAGCTAGGA |
| R:
GCGGCGCAATACGAATGCCCC |
| miRNA-206 | F:
ACACTCCAGCTGGGTGGAATGTAAGGAAGTGTG |
| R:
GCAGGGTCCGAGGTATTCG |
Transfection with small interfering RNA
(siRNA) targeting lnc00152
We selected the site in the β-catenin mRNA sequence
as a siRNA target. Three targeted siRNAs against lnc00152
(lnc00152-siRNAs) were created and the sequences were as follows:
siRNA1, GGGAAATAAATGACTGGAT; siRNA2, GGAGATGAAACAGGAAGCT; and
siRNA3, GGGAATGG AGGGAAATAAA. All the siRNAs, as well as the
negative control siRNA were synthesized and purified by RiboBio
Biotech Corp. (Guangzhou, China). The siRNA plasmids were
transfected into the SW620 cells with Lipofectamine 2000
(Invitrogen) at a working concentration of 200 nM and incubated for
24 h. After the incubation period, the cells were harvested for use
in RT-qPCR analysis.
lnc00152 overexpression, plasmid
construction and transfection
The full-length lnc00152 was synthesized by Shanghai
Sangon Biotech Corp. (Shanghai, China) and cloned into pLV4
plasmids (Promega Corp., Madison, WI, USA). Empty pLV4 plasmids,
without the insertion, served as the negative control. Cell
transfection was conducted with Lipofectamine 2000 reagent
according to the manufacturer's instructions. Further analyses were
conducted at 24 h following transfection.
Cell Counting kit-8 (CCK-8) assay
A Cell Counting kit-8 assay ('CCK-8', KeyGen
Biotech, Nanjing, China) was used to evaluate cell viability at 12,
24, 36, 48, 60 and 72 h of culture. Briefly, both the control and
infected cells were seeded at a density of 2×103
cells/well into a 96-well plate, to which 10 µl of the 10%
CCK-8 solution was subsequently added. The plate was incubated at
37°C for 4 h in a 5% CO2 incubator. A microplate reader
(Spectramax M5; Molecular Devices Corp., San Jose, CA, USA) was
used to measure the product absorbance at 490 nM, and the results
are reported as follows: (OD value of sample − OD value of
blank)/(OD value of blank). Data were collected from 3 independent
experiments.
Transwell invasion assay
A Transwell invasion assay was used to evaluate the
invasive potential of the transfected cells. We used Matrigel
invasion chambers (Corning Inc., Corning, NY, USA) with 8-µm
pores (BD Biosciences, San Jose, CA, USA). In brief, a
concentration of 2×104 cells/100 µl was
re-suspended in DMEM without fetal bovine serum (FBS) then
subsequently seeded on top of a Matrigel-coated Transwell. A total
of 600 µl of DMEM containing 10% FBS was added to the lower
chambers. Following a 24-h incubation period, the filters
separating the upper and lower chamber were washed twice with PBS,
fixed by methanol and stained with crystal violet at room
temperature for 10 min (Beijing Solarbio Science & Technology
Co. Ltd., Beijing, China). We then counted the number of stained
cells under a light microscope (Nikon Eclipse E200; Nikon, Tokyo,
Japan; magnification, ×100). The area of each membrane was, on
average, 5 visual fields. This experiment was repeated
independently, in triplicate.
Establishment of tumor xenograft
models
We purchased 12 athymic nude mice (6 males and 6
females; weighing 18–20 g, 4 weeks old) from Beijing Slac
Laboratory Animal Co. Ltd. (Beijing, China), which we housed in
high-efficiency particulate air-filtered cages in a pathogen-free
facility. The housing environment was maintained at 25±2°C, 45–55%
humidity, and a standard 12-h dark/12-h light cycle, and we fed
mice an autoclaved diet with free access to water. We cleaned and
sterilized the inoculation area (right upper limb) with ethanol and
iodine solutions before subcutaneously injecting the SW620 cells
(1×106/ml) transfected with pLV-NC or pLV-lnc00152 into
the mice (n=6 mice per group). Tumor volumes (0.5 × length ×
width2) were measured on days 10, 14, 18, 22 and 26
following implantation. Of note, the largest tumor diameter of a
single tumor in our study was 18 mm and the largest volume was
570.317 mm3. No mouse developed multiple tumors. After 4
weeks, the mice were sacrificed by CO2 inhalation (20% of the
chamber volume was displace per minute by the flow of
CO2). Tumor tissues were excised, then fixed in a 4%
paraformaldehyde solution for further analysis. All the animal
experiments were performed in the Animal Laboratory Center of
Guangzhou First Hospital.
Western blot analysis
For protein extraction, the cultured cells were
lysed in a cell lysis buffer containing 140 mM NaCl, 10 mM
Tris-HCl, 1% Triton X-100, 1 mM EDTA and 1X protease inhibitor. The
tumor tissues were homogenized in lysis buffer (pH 7.5) containing
300 mM NaCl, 50 mM Tris-HCl, 0.5% Triton X-100 and 1X protease
inhibitor, and then incubated at 4°C for 30 min. The cell and
tissue lysates were centrifuged at 3,000 × g at 4°C for 15 min. We
used the BCA method to determine the protein concentration (Sangon,
Shanghai, China). Subsequently, 10 µg aliquots of the cell
and tissue lysates were loaded onto each lane of a 10%
polyacrylamide gel, then blotted onto polyvinylidene difluoride
(PVDF) membranes. After blocking with a PBST containing 5% non-fat
dry milk, the membranes were incubated with antibodies against NRP1
(1:500; Cat. no. ab81321; Abcam, Cambridge, MA, USA), N-cadherin
(1:500; Cat. no. ab18203; Abcam), E-cadherin (1:500; Cat. no.
cst9961, 1:1,000; Cell Signaling Technology, Danvers, MA, USA) and
GAPDH (1:1,000; Cat. no. 8245; Abcam). The membranes were then
washed with TBST 3 times, then incubated them with
peroxidase-linked anti rabbit IgG secondary antibody (Cat. no.
A16096; Thermo Fisher Scientific, Waltham, MA, USA) for 1 h at room
temperature. These proteins were visualized by using an ECL western
blotting detection kit (Amersham Biosciences, Piscataway, NJ, USA).
This experiment was repeated in triplicate independently.
Visualization of proteins and band intensity was determined using
ImageJ software version 14.8 (NIH, MD, USA). The gray level was
analyzed using Quantity One software (Bio-Rad, Hercules, CA,
USA).
Hematoxylin and eosin (H&E)
staining
The tumor xenograft sections were deparaffinized in
xylene and dehydrated in alcohol. The sections were then stained in
Harris' hematoxylin solution for 8 min before bluing in 0.2%
ammonia water or saturated lithium carbonate solution for 1 min.
After rinsing in 95% alcohol, the sections were counterstained in
eosin-phloxine solution for 1 min. Finally, we mounted sections
with a xylene-based medium and visualized the slides with a Nikon
ECLIPSE 90i (Nikon; magnification, x100). Two pathologists
evaluated the images in a blinded manner.
Immunohistochemistry
The tumor xenograft sections were washed in PBS and
blocked for 60 min in 0.3% Triton X-100 and PBS with 5% bovine
serum albumin, before being incubated overnight at 4°C with
anti-E-cadherin (1:50; Cat. no. 9961) or anti-N-cadherin (1:50;
Cat. no. 4061) antibodies (Cell Signaling Technology). We then
applied HRP-conjugated secondary antibody (1:2,000; Cat. no. 7074;
Abcam, Cambridge, UK) to the slides before a 1 h room temperature
incubation. To develop color on the slide, we added
diaminobenzidine (DAB)/H2O2 before the slides
were visualized with a Nikon ECLIPSE 90i microscope (Nikon;
magnification, x100). As mentioned above, two pathologists
evaluated the images in a blinded manner.
Site-directed mutagenesis and
dual-luciferase reporter assay
We used a SQE-PCR to perform site mutation on
lnc00152 and then ligated the lnc00152 insert into a psiCHEK™-2
vector (Promega Corp.). Three fragments were cloned by PCR using
the following primers, including Psi-152F, CCGCTC GAGCATCAT
TGGGAATGGAGGGAAAT; Psi-152R, ATTTGCGGCCGCTT CTGTTTTCTTTAGTTTTGCTT;
Mut-152F1, GTTTCAAAT TGGAGCCTTCGACAAGCGGTGCCTGAGC; Mut-152R1,
CACCGCTTGTCGAAGGCTCCAATTTGAAACTTAAAA AGC; Mut-152F2,
GCCTCCATCCGAGCCTT CACCTCCGTC TGCATCCCTCG; and Mut-152R2,
AGACGGAGGTGAAGG CTCGGATGGAGGCTGGCAAGTTTC. Human 293T cells (Cell
Bank of Type Culture Collection of Chinese Academy of Sciences,
Shanghai, China) were co-transfected with 150 ng of miRNA-206
mimics, miRNA NC mimics, miRNA-206 inhibitors and miRNA-NC
inhibitor (RiboBio). Subsequently, 50 ng of wild-type lnc00152 or
lnc00152 mutant fluorescent vector were co-transfected with the
psiCHEK™-2 vector into the human 293T cells using Lipofectamine
2000 (Invitrogen). We used miRNA-NC as a negative control and
repeated the transfections in triplicate. Luciferase assay was
conducted at 48 h following transfection, and we normalized the
relative luciferase activity using the luciferase assay kit
(Promega Corp.) at 48 h after transfection. Finally, to elucidate
the interaction between lnc00152 and miRNA-206, we expressed miRNA
inhibitor and mimics into the 293T cells and subsequently applied
qPCR analysis for miRNA-206 and lnc00152 expression at 48 h
following transfection.
Statistical analysis
Summarized data are presented as the means ± SEM. We
performed all statistical analyses using SPSS19.0 software (SPSS
Inc., Chicago, IL, USA). A Chi-square test was used to analyze the
association between lnc00152 expression and the patient
clinicopathological characteristics. Survival curves were plotted
by the Kaplan-Meier method and compared using the log-rank test.
Comparisons between 2 groups for statistical significance were
carried out with two-tailed paired Student's t-tests. For analyses
involving multiple sample groups, statistical significance was
determined using one-way ANOVA followed by Tukey's test for
multiple comparisons. Statistical significance was set at a P-value
<0.05.
Results
High expression of lnc00152 is associated
with low survival rates of patients with CRC
In order to determine the potential role of lnc00152
in CRC, we first compared lnc00152 expression levels between CRC
tissues and adjacent non-tumor tissues. The lnc00152 expression
level was significantly higher in the CRC tissues compared with the
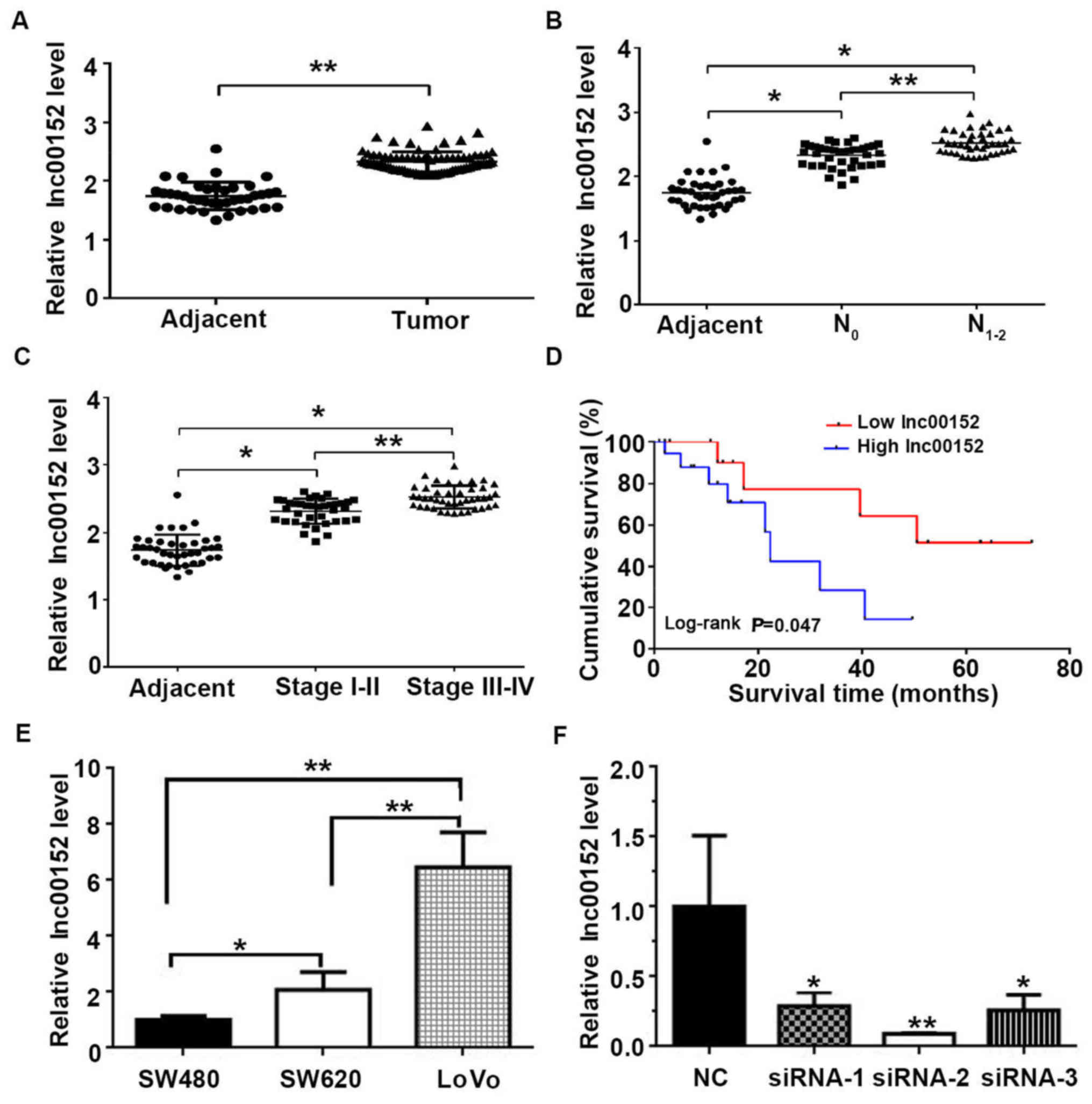
adjacent non-tumor tissues (Fig.
1A, P<0.01). In terms of lymph-node metastasis, the lnc00152
expression level was higher in the N1–2 grade tumors
than in the N0 grade tumors (Fig. 1B, P<0.01). Furthermore, both the
N1–2 and N0 grade tumors exhibited
significantly higher levels of lnc00152 than the adjacent non-tumor
tissues (Fig. 1B, P<0.05).
Similarly, the lnc00152 level was associated with the pathological
grade of colorectal tumors. The malignancy of CRC was associated
with the level of lnc00152 expression, with significantly higher
levels observed in stage III–IV than in stage I–II tumors (Fig. 1C, P<0.01). Furthermore, the
lnc00152 levels were associated with the overall survival, with
significantly shorter survival times observed in patients with CRC
with higher lnc00152 levels (Fig.
1D and Table I, log-rank test,
P=0.047). Neither age nor sex were significantly associated with
lnc00152 expression (P=0.968 and P=0.563, respectively); however,
lnc00152 expression was significantly associated with the
pathological grade (P=0.008) and lymph-node metastasis (P=0.004).
The human CRC cells (SW480, SW620 and LoVo) also expressed
lnc00152, with the highest expression observed in the LoVo cells
(Fig. 1E). These results suggest
that lnc00152 overexpression is an indicator of CRC in human
tissues and cells, and it may be useful in the prognosis of
CRC.
lnc00152 enhances the proliferative and
invasive ability of the CRC cells in vitro
To elucidate the mechanisms through which lnc00152
regulates CRC growth, we controlled the expression level of
lnc00152 in the CRC cancer cell line, SW620. lnc00152 expression
was significantly decreased in the siRNA-transfected cells, with an
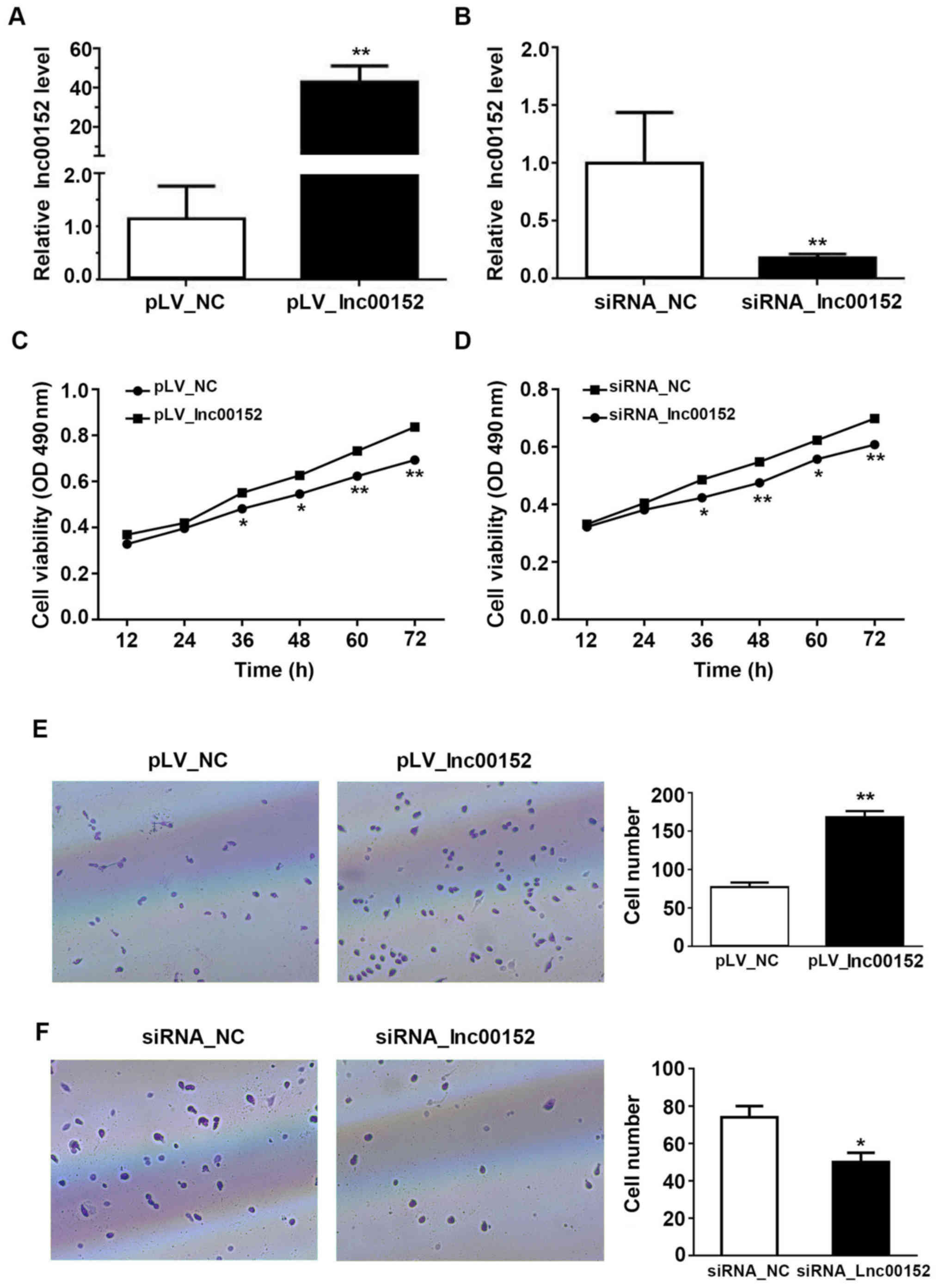
optimal silencing effect observed with siRNA-2 (P<0.01, Figs. 1F and 2B). Additionally, we established that the
expression of linc00152 in the SW620 cells was significantly higher
in the cells transfected with the lnc00152 overexpression vector
compared with the pLV-NC-transfected cells (Fig. 2A, P<0.01). The overexpression of
lnc00152 enhanced the viability of the SW620 cells at 36, 48, 60
and 72 h following transfection (Fig.
2C). The silencing of lnc00152, however, significantly
decreased cellular proliferation (Fig.
2D) at 36 (P<0.05), 48 (P<0.01), 60 (P<0.05), and 72 h
(P<0.01) following transfection, but not at 12 or 24 h. From the
Transwell assay evaluating the cell invasive potential, we found
that the overexpression of ln00152 enhanced the invasive ability of
the SW620 cells (Fig. 2E,
P<0.01). Conversely, lnc00152 silencing decreased the number of
invading cells relative to those transfected with the control siRNA
(siRNA_NC) (Fig. 2F, P<0.05).
Consequently, our findings indicate that lnc00152 enhances the
proliferative and invasive ability of CRC cells.
lnc00152 overexpression enhances
colorectal tumor growth in vivo
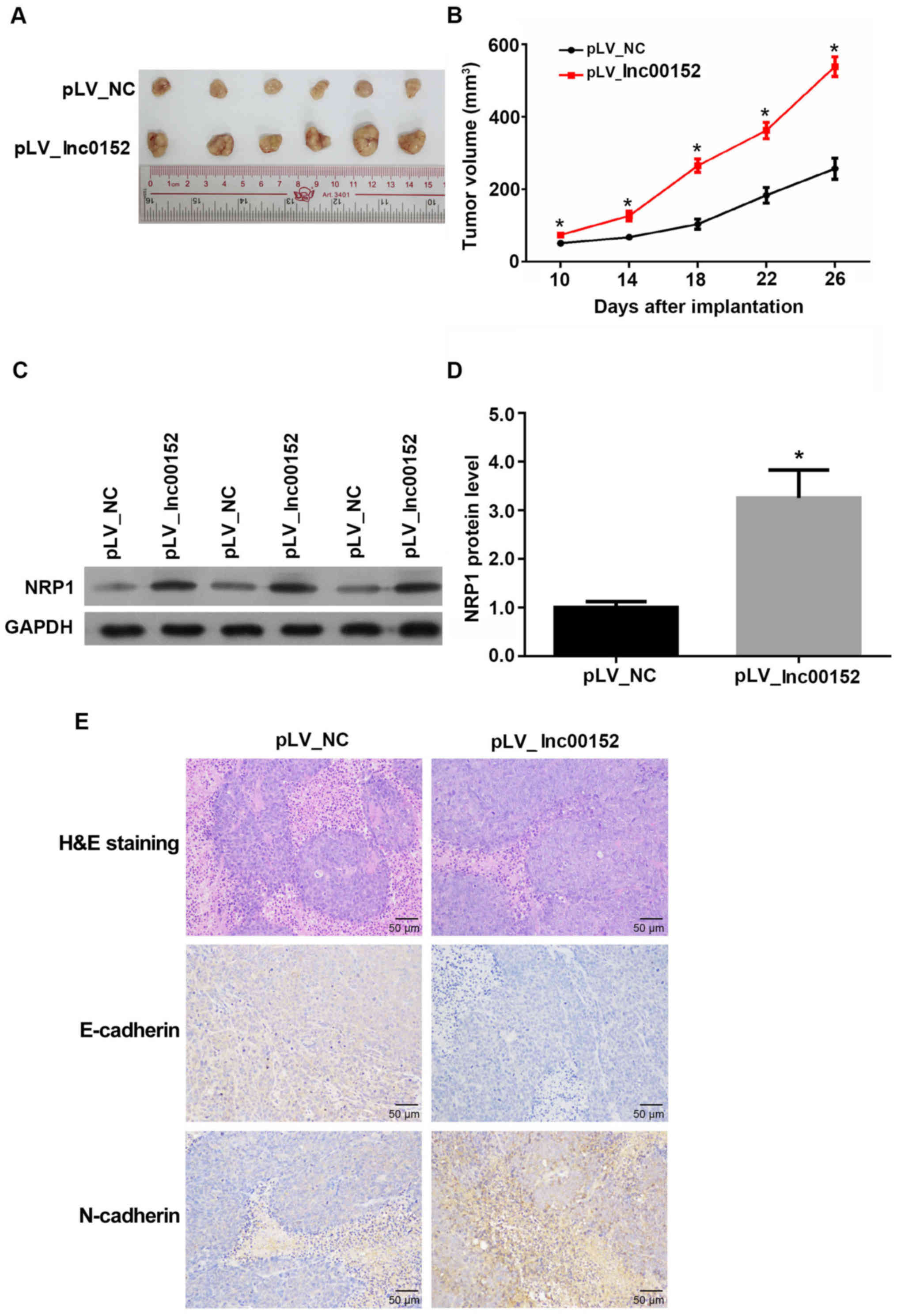
To determine whether lnc00152regulates tumor growth
in vivo, we xenografted lnc00152 stably expressing SW620
cells into nude mice. The volumes of tumors derived SW620 cells
overexpressing lnc00152 were significantly greater than those of
tumors derived from the pLV-NC-transfected control cells from days
10 to 26 following implantation (Fig.
3A and B, P<0.05). Additionally, the protein levels of NRP1
were increased in tumors derived from the lnc00152-overexpressing
cells (Fig. 3C and D, P<0.05).
Furthermore, lnc00152 overexpression also promoted EMT in the
SW620-derived tumors by decreasing the level of E-cadherin, while
increasing N-cadherin expression (Fig.
3E). H&E staining also confirmed the cellular alteration in
lnc00152-overexpressing colorectal tumors, which exhibited typical
morphological characteristics of more malignant tumors (large cell
morphology, large cell nucleus and malformed nuclei). Collectively,
these data indicate that lnc00152 overexpression promotes tumor
growth and EMT in vivo.
miRNA-206 is a target gene of
lnc00152
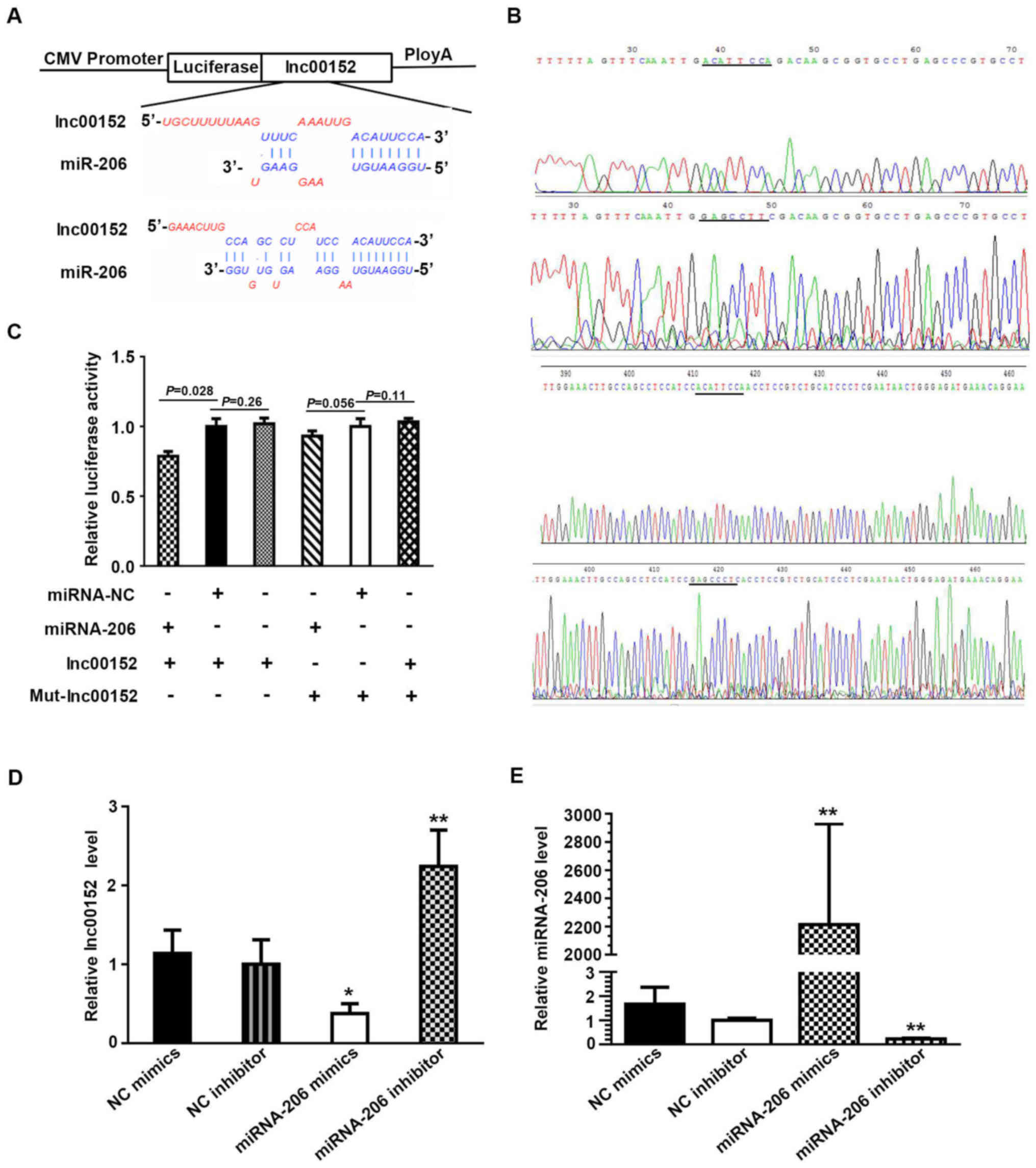
A dual luciferase assay verified the target gene of
miRNA-206 as lnc00152. Plasmid construction and sequence data and
the sequence of mutant lnc00152 are described in Fig. 4A and B. When the SW620 cells were
transfected with the lnc00152 overexpression vector together with
miRNA-NC, the relative luciferase activity did not differ
significantly from that in the cells transfected with the lnc00152
overexpression vector only (Fig.
4C, P=0.26>0.1). By contrast, the relative luciferase
activity decreased when the cells were transfected with the
lnc00152 overexpression vector and the miRNA-206 mimic (Fig. 4C, P=0.028<0.05), demonstrating a
direct interaction between lnc00152 and miRNA-206. Co-transfection
with mutated lnc00152 (Mut-Lnc00152) and miRNA-206 did not result
in reduced relative luciferase activity (Fig. 4C, P>0.05), indicating that
mutated lnc00152 disrupted the interaction between lnc00152 and
miRNA-206. In addition, the lnc00152 level was lower in the cells
transfected with miRNA-206 mimics compared to those transfected
with NC mimics (Fig. 4D,
P<0.05), while the lnc00152 level increased significantly
following incubation with the miRNA-206 inhibitor compared to the
NC inhibitor (Fig. 4D, P<0.01).
In the presence of miRNA-206 mimics, the miRNA-206 level increased
significantly (Fig. 4E,
P<0.01), while it decreased significantly following incubation
with miRNA-206 inhibitor (Fig. 4E,
P<0.01). This displayed the specific efficiency of miRNA-206
mimics and inhibitor; collectively, these data indicate that
miRNA-206 can directly bind to lnc00152 through miRNA recognition
sites.
lnc00152 regulates the miRNA-206 target
gene, NRP1
Yin et al (28) previously demonstrated that
miRNA-206 significantly inhibited the luciferase activity of the
RLuc-NRP1 3′-UTR reporter, confirming that NRP1 is a target gene of
miR-206. Thus, one of our aims in this study was to clarify whether
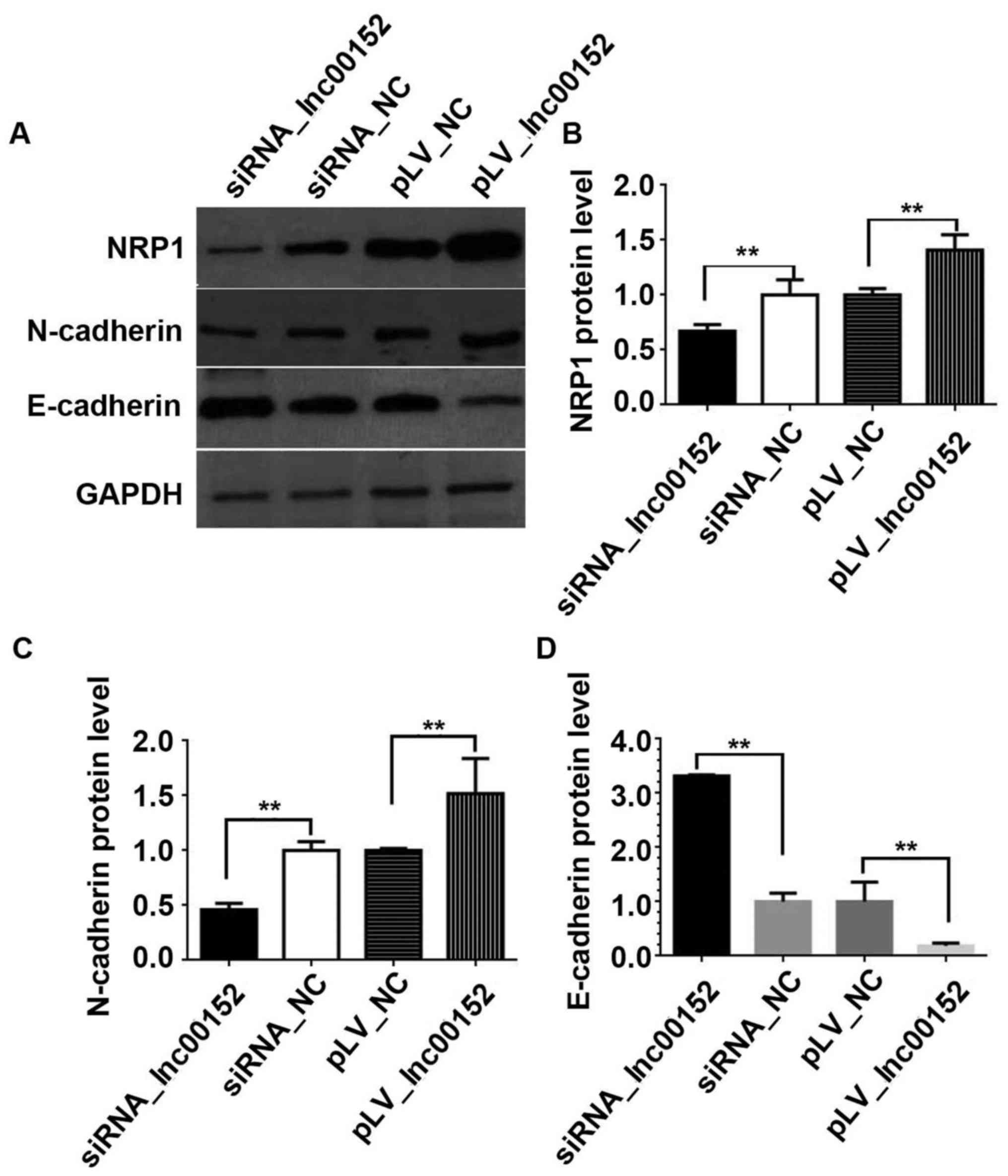
lnc00152 regulates NRP1 in CRC cells. Our results indicated that
lnc00152 silencing downregulated NRP1 expression, while lnc00152
overexpression elevated NRP1 expression (Fig. 5A and B, P<0.01). Additionally,
we validated the effects of lnc00152 expression on EMT in CRC
cells. lnc00152 silencing downregulated N-cadherin expression,
whereas it upregulated E-cadherin expression (Fig. 5A–D, P<0.01). By contrast,
lnc00152 overexpression increased N-cadherin expression, while it
decreased E-cadherin expression (Fig.
5A–D, P<0.01). Therefore, lnc00152 regulates NRP1 expression
and additionally affects EMT in CRC cells.
The mechanisms through which lnc00152
regulates NRP1 in colorectal cancer
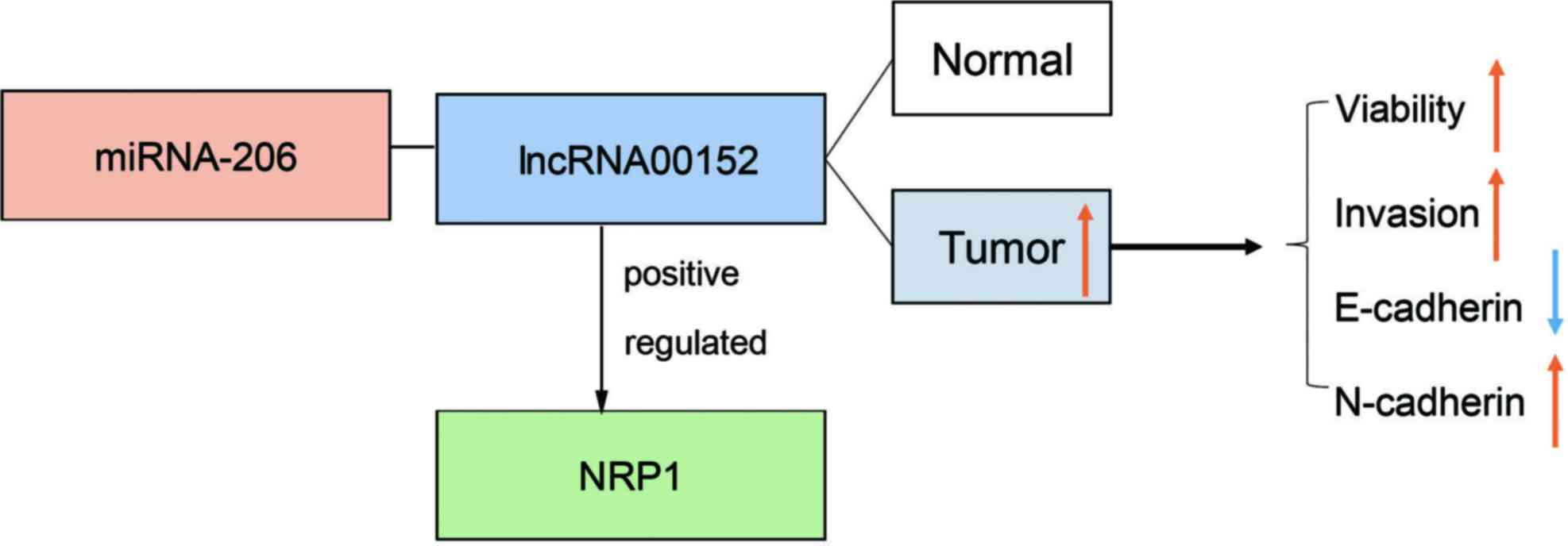
From the above-mentioned results, we drew a
schematic model to demonstrate the possible mechanisms through
which lnc00152 regulates NRP1 (Fig.
6). Compared with the adjacent normal tissues, lnc00152 was
highly expressed in CRC tumor tissues and promoted CRC cell
viability and invasion by competitively binding miRNA-206,
upregulating NRP1, and subsequently upregulating N-cadherin, while
downregulating E-cadherin.
Discussion
In this study, we found that lnc00152 was expressed
in colorectal tumor tissues and the CRC cell lines, SW480, LoVo and
SW620. The regulation of lnc00152 also affected CRC cell viability,
invasion and EMT in vitro, as well as tumor growth in
vivo. This process was associated with NRP1, a miRNA-206 target
gene, and miRNA-206 was associated with lnc00152 expression.
lncRNAs play critical roles in cancer development
and progression (29). Recent
studies have indicated that lncRNAs can regulate both the migration
and invasiveness of CRC. For example, the downregulation of lncRNA
H19 has been shown to inhibit CRC cell migration and metastasis
(30). This is consistent with our
findings in that the silencing of lnc00152 decreased the
proliferative and invasive ability of CRC cells. lncRNAs have also
been reported to suppress EMT in CRC (11). EMT is a hallmark in tumor
progression (31). Our findings
further validated this observation by showing that the
downregulation of lnc00152 reduced N-cadherin and increased
E-cadherin expression, while lnc00152 overexpression had the
opposite effect. This is consistent with the role of other lncRNAs
in the CRC cell lines LoVo and HCT116 (11).
Furthermore, we found that overexpression of
lnc00152 may increase the malignant potential of CRC cells. This
has also been observed in a previous study which showed that the
upregulation of lncRNA BANCR was associated with metastasis and the
poor survival of patients with CRC (12). In this study, patients with a
higher level of lnc00152 had a shorter survival time, displaying an
association between the lnc00152 level and CRC prognosis. This
finding, too, is supported by the findings of other studies that
have demonstrated an unfavorable prognosis of patients with CRC
with a lncRNA expression (32-34).
In our tumor xenograft experiment, lnc00152 overexpression promoted
tumor growth, which demonstrates that lnc00152 plays an essential
role in regulating the malignancy of CRC. Importantly, as lnc00152
silencing appears to have an anti-tumor effect, it may function as
a therapeutic target in the treatment of CRC, and may thus aid in
the treatment and prognosis of CRC.
Some miRNAs have been observed to be dysregulated in
CRC tissues (35). The function of
miRNAs in CRC involves controlling EMT (36) and affecting cell proliferation,
migration and invasion (37),
similar to the function of lncRNAs in CRC. In ovarian cancer, the
miRNA-lncRNA signature is important in patient survival (38). The miRNA-lncRNA complex is also
involved in the transformation process of gastric cancer initiation
to malignancy (39). Accordingly,
lncRNA may interact with miRNAs to regulate colorectal tumor
malignancy. Previous studies have reported that lnc00152 is
implicated in CRC by regulating cell cycle, apoptosis, cell
motility and EMT (40,41). In addition, lnc00152 has been
demonstrated to promote oncogenesis via several mechanisms,
including serving as a sponge for miR-4767 and miR-205 (42,43)
and a partner for EZH2 (41). In
this study, we observed the interaction of lnc00152 with miRNA-206
with a dual luciferase assay. This observation and other reports of
miR-206 inhibiting tumor invasion and migration in CRC (44), highlight an important association
between miRNA-206 and lnc00152 in CRC.
Our experiments have additionally located the
mutation site on lnc00152 that interacts with miRNA-206. NRP1 is
the target gene of miRNA-206 (28), and our data demonstrated that
regulating the lnc00152 levels altered NRP1 expression. NRP1 is a
co-receptor for vascular endothelial growth factor (VEGF), and the
blockaded of NRP1 suppresses tumor growth by inhibiting
angiogenesis or by directly inhibiting tumor cell proliferation
(45). In CRC, NRP1 is associated
with liver metastasis (46), and,
importantly, EMT in CRC is dependent on NRP1 (26). Future studies, therefore, are
warranted in order to focus on the molecular mechanisms that
underlie the synergic effects of miRNA-206 and lnc00152 on CRC
progression and on the signaling pathway of the NRP1-associated
modulation of CRC metastasis.
In conclusion, the results of the present study
identify lnc00152 as a competing endogenous RNA that positively
regulates NRP1 expression by sponging miRNA-206 (Fig. 6). Given its critical role in CRC,
it may be an effective therapeutic target in CRC prognosis and
treatment.
Acknowledgments
Not applicable.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanthan R, Senger JL and Kanthan SC:
Molecular events in primary and metastatic colorectal carcinoma: A
review. Pathol Res Int. 2012:5974972012. View Article : Google Scholar
|
|
3
|
Guan X, Jiang Z, Ma T, Liu Z, Hu H, Zhao
Z, Song D, Chen Y, Wang G and Wang X: Radiotherapy dose led to a
substantial prolongation of survival in patients with locally
advanced rectosigmoid junction cancer: A large population based
study. Oncotarget. 7:28408–28419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu DJ, Chen XW, OuYang MZ and Lu Y: Three
surgical planes identified in laparoscopic complete mesocolic
excision for right-sided colon cancer. World J Surg Oncol.
14:72016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang YW, Huang LY, Song CL, Zhuo CH, Shi
DB, Cai GX, Xu Y, Cai SJ and Li XX: Laparoscopic vs open
abdominoperineal resection in the multimodality management of low
rectal cancers. World J Gastroenterol. 21:10174–10183. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin T, Song C, Chuo DY, Zhang H and Zhao
J: Clinical effects of autologous dendritic cells combined with
cytokine-induced killer cells followed by chemotherapy in treating
patients with advanced colorectal cancer: A prospective study.
Tumour Biol. 37:4367–4372. 2016. View Article : Google Scholar
|
|
7
|
Du XH, Liu HL, Li L, Xia SY, Ning N, Zou
ZY, Teng D, Xiao CH, Li R and Xu YX: Clinical significance of
immunotherapy with combined three kinds of cells for operable
colorectal cancer. Tumour Biol. 36:5679–5685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen LL and Zhao JC: Functional analysis
of long noncoding RNAs in development and disease. Adv Exp Med
Biol. 825:129–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu W, Yuan B, Flygare J and Lodish HF:
Long noncoding RNA-mediated anti-apoptotic activity in murine
erythroid terminal differentiation. Genes Dev. 25:2573–2578. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rossi MN and Antonangeli F: LncRNAs: New
players in apoptosis control. Int J Cell Biol. 2014:4738572014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang M, Wu WB, Wang ZW and Wang XH:
lncRNA NEAT1 is closely related with progression of breast cancer
via promoting proliferation and EMT. Eur Rev Med Pharmacol Sci.
21:1020–1026. 2017.PubMed/NCBI
|
|
12
|
Shen X, Bai Y, Luo B and Zhou X:
Upregulation of lncRNA BANCR associated with the lymph node
metastasis and poor prognosis in colorectal cancer. Biol Res.
50:322017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou HB, Li Q, Liu M, Cao YQ and Xu JY:
Increased expression of long non-coding RNA SBDSP1 correlates with
poor survival in colorectal cancer. Eur Rev Med Pharmacol Sci.
21:3837–3841. 2017.PubMed/NCBI
|
|
14
|
Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng
ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, et al: Long noncoding
RNA XIST expedites metastasis and modulates epithelial-mesenchymal
transition in colorectal cancer. Cell Death Dis. 8:e30112017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu K, Zhao Z, Ma J, Chen J, Peng J, Yang S
and He Y: Deregulation of miR-193b affects the growth of colon
cancer cells via transforming growth factor-β and regulation of the
SMAD3 pathway. Oncol Lett. 13:2557–2562. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tong F, Ying Y, Pan H, Zhao W, Li H and
Zhan X: MicroRNA-466 (miR-466) functions as a tumor suppressor and
prognostic factor in colorectal cancer (CRC). Bosn J Basic Med Sci.
Jan 17–2018.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ebrahimi F, Gopalan V, Wahab R, Lu CT,
Smith RA and Lam AK: Deregulation of miR-126 expression in
colorectal cancer pathogenesis and its clinical significance. Exp
Cell Res. 339:333–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li DY, Chen WJ, Luo L, Wang YK, Shang J,
Zhang Y, Chen G and Li SK: Prospective lncRNA-miRNA-mRNA regulatory
network of long non-coding RNA LINC00968 in non-small cell lung
cancer A549 cells: A miRNA microarray and bioinformatics
investigation. Int J Mol Med. 40:1895–1906. 2017.PubMed/NCBI
|
|
20
|
Guo PD, Lu XX, Gan WJ, Li XM, He XS, Zhang
S, Ji QH, Zhou F, Cao Y, Wang JR, et al: RARγ downregulation
contributes to colorectal tumorigenesis and metastasis by
derepressing the Hippo-Yap pathway. Cancer Res. 76:3813–3825. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang JR, Gan WJ, Li XM, Zhao YY, Li Y, Lu
XX, Li JM and Wu H: Orphan nuclear receptor Nur77 promotes
colorectal cancer invasion and metastasis by regulating MMP-9 and
E-cadherin. Carcinogenesis. 35:2474–2484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ye GD, Sun GB, Jiao P, Chen C, Liu QF,
Huang XL, Zhang R, Cai WY, Li SN, Wu JF, et al: OVOL2, an inhibitor
of WNT signaling, reduces invasive activities of human and mouse
cancer cells and is down-regulated in human colorectal tumors.
Gastroenterology. 150:659–671.e16. 2016. View Article : Google Scholar
|
|
23
|
Waszkiewicz N, Szajda SD,
Konarzewska-Duchnowska E, Zalewska-Szajda B, Gałązkowski R, Sawko
A, Nammous H, Buko V, Szulc A, Zwierz K, et al: Serum
β-glucuronidase as a potential colon cancer marker: A preliminary
study. Postepy Hig Med Dosw. 69:436–439. 2015. View Article : Google Scholar
|
|
24
|
Miyauchi JT, Chen D, Choi M, Nissen JC,
Shroyer KR, Djordevic S, Zachary IC, Selwood D and Tsirka SE:
Ablation of Neuropilin 1 from glioma-associated microglia and
macrophages slows tumor progression. Oncotarget. 7:9801–9814. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X, Fan S, Pan X, Xiaokaiti Y, Duan J,
Shi Y, Pan Y, Tie L, Wang X, Li Y, et al: Nordihydroguaiaretic acid
impairs prostate cancer cell migration and tumor metastasis by
suppressing neuropilin 1. Oncotarget. 7:86225–86238. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tomida C, Yamagishi N, Nagano H, Uchida T,
Ohno A, Hirasaka K, Nikawa T and Teshima-Kondo S: Antiangiogenic
agent sunitinib induces epithelial to mesenchymal transition and
accelerates motility of colorectal cancer cells. J Med Invest.
64:250–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Yin K, Yin W, Wang Y, Zhou L, Liu Y, Yang
G, Wang J and Lu J: MiR-206 suppresses epithelial mesenchymal
transition by targeting TGF-β signaling in estrogen receptor
positive breast cancer cells. Oncotarget. 7:24537–24548.
2016.PubMed/NCBI
|
|
29
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liang W, Zou Y, Qin F, Chen J, Xu J, Huang
S, Chen J and Dai S: sTLR4/MD-2 complex inhibits colorectal cancer
migration and invasiveness in vitro and in vivo by lncRNA H19
down-regulation. Acta Biochim Biophys Sin (Shanghai). 49:1035–1041.
2017. View Article : Google Scholar
|
|
31
|
Cao H, Xu E, Liu H, Wan L and Lai M:
Epithelial-mesenchymal transition in colorectal cancer metastasis:
A system review. Pathol Res Pract. 211:557–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen SW, Zhu J, Ma J, Zhang JL, Zuo S,
Chen GW, Wang X, Pan YS, Liu YC and Wang PY: Overexpression of long
non-coding RNA H19 is associated with unfavorable prognosis in
patients with colorectal cancer and increased proliferation and
migration in colon cancer cells. Oncol Lett. 14:2446–2452. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao X, Wen J, Gao P and Zhang G and Zhang
G: Overexpression of the long non-coding RNA, linc-UBC1, is
associated with poor prognosis and facilitates cell proliferation,
migration, and invasion in colorectal cancer. OncoTargets Ther.
10:1017–1026. 2017. View Article : Google Scholar
|
|
34
|
Lu M, Liu Z, Li B, Wang G, Li D and Zhu Y:
The high expression of long non-coding RNA PANDAR indicates a poor
prognosis for colorectal cancer and promotes metastasis by EMT
pathway. J Cancer Res Clin Oncol. 143:71–81. 2017. View Article : Google Scholar
|
|
35
|
Chen X, Shi K, Wang Y, Song M, Zhou W, Tu
H and Lin Z: Clinical value of integrated-signature miRNAs in
colorectal cancer: miRNA expression profiling analysis and
experimental validation. Oncotarget. 6:37544–37556. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jaca A, Govender P, Locketz M and Naidoo
R: The role of miRNA-21 and epithelial mesenchymal transition (EMT)
process in colorectal cancer. J Clin Pathol. 70:331–356. 2017.
View Article : Google Scholar
|
|
37
|
Deng B, Wang B, Fang J, Zhu X, Cao Z, Lin
Q, Zhou L and Sun X: MiRNA-203 suppresses cell proliferation,
migration and invasion in colorectal cancer via targeting of
EIF5A2. Sci Rep. 6:283012016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo L, Peng Y, Meng Y, Liu Y, Yang S, Jin
H and Li Q: Expression profiles analysis reveals an integrated
miRNA-lncRNA signature to predict survival in ovarian cancer
patients with wild-type BRCA1/2. Oncotarget. 8:68483–68492.
2017.PubMed/NCBI
|
|
39
|
Mao Y, Liu R, Zhou H, Yin S, Zhao Q, Ding
X and Wang H: Transcriptome analysis of miRNA-lncRNA-mRNA
interactions in the malignant transformation process of gastric
cancer initiation. Cancer Gene Ther. 24:267–275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui
P, Zhang Y and Huang G: Long non-coding RNA Linc00152 is involved
in cell cycle arrest, apoptosis, epithelial to mesenchymal
transition, cell migration and invasion in gastric cancer. Cell
Cycle. 14:3112–3123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen WM, Huang MD, Sun DP, Kong R, Xu TP,
Xia R, Zhang EB and Shu YQ: Long intergenic non-coding RNA 00152
promotes tumor cell cycle progression by binding to EZH2 and
repressing p15 and p21 in gastric cancer. Oncotarget. 7:9773–9787.
2016.PubMed/NCBI
|
|
42
|
Wang Y, Liu J, Bai H, Dang Y, Lv P and Wu
S: Long intergenic non-coding RNA 00152 promotes renal cell
carcinoma progression by epigenetically suppressing P16 and
negatively regulates miR-205. Am J Cancer Res. 7:312–322.
2017.PubMed/NCBI
|
|
43
|
Teng W, Qiu C, He Z, Wang G, Xue Y and Hui
X: Linc00152 suppresses apoptosis and promotes migration by
sponging miR-4767 in vascular endothelial cells. Oncotarget.
8:85014–85023. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sun P, Sun D, Wang X, Liu T, Ma Z and Duan
L: miR-206 is an independent prognostic factor and inhibits tumor
invasion and migration in colorectal cancer. Cancer Biomark.
15:391–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jubb AM, Strickland LA, Liu SD, Mak J,
Schmidt M and Koeppen H: Neuropilin-1 expression in cancer and
development. J Pathol. 226:50–60. 2012. View Article : Google Scholar
|
|
46
|
Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P,
Zhang Q, Dong L, Liu Y and Dong J: microRNA-320a inhibits tumor
invasion by targeting neuropilin 1 and is associated with liver
metastasis in colorectal cancer. Oncol Rep. 27:685–694. 2012.
|