Introduction
Osteosarcoma is a primary mesenchymal tumor, which
is considered an aggressive type of malignancy. Metastasis is an
important process in malignant tumor formation (1,2).
Conventional chemotherapy is not efficient in patients with
osteosarcoma, and ~80% of patients with distant metastasis develop
lung metastasis (3,4). Evidence has shown that 10-20% of
patients with osteosarcoma are at risk of developing pulmonary
metastasis, and ~40-50% of the patients develop pulmonary
heterochrony although without metastasis (5). By contrast, patients with
osteosarcoma with metastasis usually present with a lower 5-year
survival rate, compared with those patients without metastasis
(6). Conventional chemotherapy is
not efficient for treatment of the disease, and 30-50% of patients
eventually succumb to mortality as a result of the pulmonary
metastasis (7). As the occurrence
and development of the disease is closely associated with molecular
biology, investigating the molecular mechanism of osteosarcoma has
significant value for developments in early diagnosis and
treatment.
The underlying biological mechanism of osteosarcoma
tumors and metastasis has been widely investigated, and genes and
pathways involved in osteosarcoma have been identified. For
example, Notch proteins are important for normal bone development
and homeostasis, and also for angiogenesis (8,9).
Dysregulation of the Notch pathway is present in human diseases
associated with congenital skeletal abnormalities. The Notch
pathway genes have been found to be overexposed in osteosarcoma
cells, compared with normal bone cells, which include Hes related
family bHLH transcription factor with YRPW motif 1 (HEY1), Hes
family BHLH transcription factor 1 (HES1) and NOTCH2 (10-12).
Runt related transcription factor 2 (RUNX2) is an important
transcription factor involved in osteoblast differentiation
(13). Previous studies have
suggested that the Notch pathway transcription of Runx2 is
decreased via HES and HEY proteins. As a result, it is hypothesized
that the increase of immature osteoblasts may be one of the
potential factors in malignant transformation (14,15).
The Wnt pathway manages several processes, including cell growth,
normal bone development and carcinogenesis (16,17).
According to previous studies, the Wnt pathway was found to be
highly activated in human osteosarcoma, therefore, it may be also
potential pathway involved in osteosarcoma (18-20).
Mammalian target of rapamycin (mTOR), one of the phosphoinositide
3-kinase (PI3K) family members, is important in the regulation of
cell cycle, growth and development (21,22).
It was shown that, by suppressing the expression of mTOR or
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α,
growth arrest was induced in a murine osteosarcoma model (23). Based on the evidence so far,
molecular regulation appears to be important in the occurrence and
development of osteosarcoma. The investigation of genes which are
correlated with osteosarcoma are likely to be beneficial for the
diagnosis and treatment of the disease.
RNA sequencing (RNA-seq) is an advanced technique
based on the deep-sequencing approach. It provides a relatively
unbiased and more precise measurement for the quantitation of the
transcripts and their isoforms (24). As RNA-seq technology presents
apparent advantages against conventional hybridization-based
microarrays, it has been widely applied and utilized in the
screening the differentially expressed genes in multiple
experimental conditions. In the present study, candidate genes
involved in osteosarcoma were screened by analyzing the Gene
Expression Omnibus (GEO) dataset. A large number of differentially
expressed genes were screened based on the results of the
microarray (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21257).
The present study also verified the differentially expressed genes,
which have not been reported in osteosarcoma, in six cell lines
(hFOB, SW1353, SAOS-2, MG-63, 143B and U2-OS) by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis. The results indicated that the expression levels of Kelch
domain containing 1 (KLHDC1), TRIM2, Laccase domain-containing
protein 1 (LACC1), Mitochondrial transcription factor A (TFAM),
Laminin subunit β1 (LAMB1), Stanniocalcin 2 (STC2) and Vimentin
(VIM) in the osteosarcoma cell lines were significantly higher than
those in normal osteoblasts (data not shown). As the TRIM2, STC2
and VIM genes were significantly expressed in osteosarcoma cells,
they were selected for metastatic analysis and analysis of
prognosis. The prognosis and metastasis analyses indicated that
TRIM2 was important in the pathogenic process. Relevant cell assays
and RNA-seq were performed in order to verify the mechanism
underlying the effect of TRIM2.
Materials and methods
Prognosis and metastasis analysis
The expression profiles and clinical data of the
patients with osteosarcoma were downloaded from the NCBI GEO
database (accession no. GSE21257). The methods of pretreatment of
specimens, RNA isolation, and microarray assay setup were described
on the website. The expression values were used for statistical
analysis. The expression differences are presented via box-plots.
Differences in overall survival rates between 'high' and 'low'
expression groups were compared using Kaplan-Meier curves, with
P-values calculated via the log-rank test, using the Survival
package in R version 3.2.3 (https://www.r-project.org/).
Immunohistochemical staining
Fresh primary osteosarcoma samples and paired
adjacent non-tumor soft tissues from 53 patients undergoing
surgical resection at Zhuhai Hospital, Jinan University (Zhuhai,
China) between April 2010 and June 2011 were collected,
formalin-fixed and paraffin-embedded. The present study was
approved by the Institutional Research Ethics Committee of Jinan
University. Informed consent was provided by all participants prior
to the study. The clinicopathological information of the 53
patients were recorded and are presented in Table I.
 | Table IClinicopathological information of
the 53 patients with osteosarcoma. |
Table I
Clinicopathological information of
the 53 patients with osteosarcoma.
| Characteristic | n |
|---|
| Age (years) | |
| <20 | 42 |
| >20 | 11 |
| Sex | |
| Male | 34 |
| Female | 19 |
| Distant
metastasis | |
| Absent | 19 |
| Present | 34 |
| Huvos grade | |
| Unknown | 6 |
| 1 | 13 |
| 2 | 16 |
| 3 | 13 |
| 4 | 5 |
| Status | |
| Alive | 30 |
| Deceased | 23 |
The formalin-fixed and paraffin-embedded (FFPE)
tissue sections (5 μm) were subjected to antibody thermal
remediation, the activity of endogenous peroxidase was inhibited by
hydrogen peroxide and then closed with goat or mouse serum (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) following
dewaxing and hydration. The primary antibodies TRIM2 (cat. no.
ab3942; 1:1,000), E-cadherin (cat. no. ab76055; 1:1,000),
N-cadherin (cat. no. ab18203; 1:500) or Vimentin (cat. no. ab92547,
1:500) (all from Abcam, Cambridge, UK) diluted in 1X PBS solution
were used to cover the tissue sections and incubated overnight at
4°C. Anti-mouse immunoglobulin G (IgG) (cat. no. sc-516102;
1:2,000), anti-rabbit IgG (cat. no. sc-3753; 1:500) or anti-goat
IgG (cat. no. sc-2489; 1:500) (all from Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) secondary antibodies were used to bind the
specific primary antibodies at 37°C for 30 min, and DAB was applied
for coloration. The stained tissues were observed using IX71
inverted microscopy (Olympus Corporation, Tokyo, Japan). Six
samples for each group were used for the immunohistochemical
staining.
Cell culture
The hFOB, U2-OS, SW1353, MG63, 143B and SAOS-2 human
osteosarcoma cell lines provided by American Type Culture
Collection (Manassas, VA, USA) were used in the present study. The
cells were incubated in DMEM (Gibco; Thermo Fisher Scientific,
Inc.). The medium was supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and the cells were all maintained at 37°C
in 5% CO2. On reaching 70-80% confluence, the cells were
washed with PBS and detached with 0.25% trypsin/0.2% EDTA. Cell
morphology was viewed under a light microscope, and suspended to
the concentration of 1×106/ml.
Lentivirus production and
transfection
The night prior to transfection, 3×106
293T (CRL-3216™; American Type Culture Collection, Manassas, VA,
USA) cells were seeded into 10 cm dishes. Small interfering RNA
(siRNA) sequences targeted to TRIM2 were cloned into the PLKO.1
lentiviral vector (Dharmacon, Inc., Lafayette, CO, USA). The cells
were transfected with the TRIM2 siRNA-expressing PLKO.1 lentiviral
vectors and control empty vector along with packaging vectors
(psPAX2 and pMD2.G) using FuGENE-6 transfection reagent (Promega
Corporation, Madison, WI, USA). The lentivirus-containing
supernatants were harvested 72 h following transfection and were
filtered through 0.45-μm PVDF filters. The supernatant was
then concentrated by ultracentrifugation (at 100,000 x g for 2 h at
room temperature) in a Beckman Optima L-90K ultracentrifuge
(Beckman Coulter, Inc., Brea, CA, USA). The virus-containing pellet
was dissolved in DMEM, aliquoted and stored at −80°C. The sequences
of the siRNAs were as follows: siTRIM2-1 (human), forward,
5'-CCTGGAACG GTACAAGAAT-3' and reverse, 5'-ATTCTTGTACCGTTCCAGG-3';
siTRIM2-2 (human), forward, 5'-CCAGTGAAGGCACCAACAT-3' and reverse,
5'-ATGTTGGTGCCTTCACTGG-3'.
The U2OS and MG63 cells were infected by adding 1 ml
of concentrated virus supplemented with 2 μg/ml polybrene to
4×105 cells in 12-well plates. After 36 h at room
temperature, the viral supernatant was replaced with standard
growth medium, and transduction efficiency was monitored by GFP
expression 48 h following replacement of the virus-containing
medium with normal medium.
Cell viability assay
Cell viability was examined using the MTS included
in the CellTiter 96 Aqueous One Solution Cell Proliferation Assay
(Promega Corporation) according to the manufacturer's protocol. The
U2OS and MG63 cells were seeded in a 96-well plate at a cell
density of 200 cells per well. The cells were incubated with 15
μl of MTS reagent solution for 4 h and the produced formazan
was measured at 490 nm in a Sunrise Basic Tecan cell plate reader
with Magellan 6 software (both from Tecan Group, Ltd., Grödig,
Austria).
Transwell migration and invasion
assays
For the Transwell migration assays,
2.5-5×104 cells were plated in the top chamber with a
non-coated membrane (8-μm pore size; Corning Incorporated,
Corning, NY, USA). For the cell invasion assay, the Transwell
chambers were pretreated with Matrigel (EMD Millipore, Billerica,
MA, USA) and the cells were resuspended at a density of
2×105 cells/ml in serum-free medium at 48 h
post-transfection. In the two assays, the upper chamber was filled
with 300 μl cell suspensions and the lower well was filled
with complete medium. After 24 h, the cells in the upper chambers
were removed with a cotton swab, and the migrated or invasive cells
in the lower chambers were fixed with 4% formaldehyde and stained
with crystal violet. The cells were observed and counted by IX71
inverted microscopy (Olympus Corporation).
Cell apoptosis assay
The cells were seeded in 6-well plates and incubated
for 48 h. The cells were detached with trypsin and the apoptosis
assay was performed using an Annexin V assay kit according to the
manufacturer's protocol. The assay was performed with a BD
FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA). The apoptosis data were analyzed with FACSCanto II flow
cytometer software (BD Biosciences).
RNA-seq
Total RNA was extracted with TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. mRNA was isolated from the total RNA using
oligo(dT) magnetic beads, and fragmentation buffer was subsequently
added for the fragmentation of mRNA. The mRNA fragments were
subsequently used as templates for first-strand cDNA synthesis
using random primers. The second-strand cDNA was synthesized using
DNA polymerase I. Adaptors were ligated to the fragments and 200-bp
fragments were subsequently enriched by PCR amplification. The
quality of the library was confirmed using the Agilent 2100
Bioanalyzer. The sequencing was performed on the Hiseq2000
platform.
The primary raw reads were trimmed of adaptors and
filtered to remove low quality reads using SOAP nuke software
version 1.5.3 (http://soap.genomics.org.cn). The length of raw reads
used were 150-bp in pair end. The cleaned reads were aligned to the
hg19 reference genome using hierarchical indexing for spliced
alignment of transcripts. DEGseq2 were used to detect the
differentially expressed genes. The P-value threshold was
determined by the false discovery rate (FDR) using an FDR threshold
≤0.001. Gene Ontology (GO) functional enrichment and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis were
performed using the Blast2GO program version 5.1 (https://www.blast2go.com/).
RT-qPCR assay
Total RNA was prepared using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) following the manufacturer's
protocol. The genomic DNA-free RNA was then converted into cDNA
using M-MLV Reverse Transcriptase (Promega Corporation), according
to the manufacturer's protocol. PCR was carried out on the CFX96
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) in a 20
μl reaction mixture containing 10 μl
GoTaq® Green Master Mix (Promega Corporation), 1.5
μl cDNA, 0.5 μl each primer and 7.5 μl
nuclease-free water under the following conditions: Initial
denaturation at 95°C for 120 sec, followed by 40 cycles of
denaturation at 95 °C for 15 sec and extension at 60°C for 30 sec.
The primer sequences were as follows: Sirtuin 4 (SIRT4), forward,
5'-AAGCCTCCATTGGGTTATTTGT-3' and reverse,
5'-TGTAGTCTGGTATCCCCGATTC-3'; DNA damage-inducible transcript 3
(DDIT3), forward, 5'-GAACGGCTCAAGCAGGAAATC-3' and reverse,
5'-ATTCACCATTCGGTCAATCAGAG-3'; cAMP responsive element binding
protein 5 (CREB5), forward, 5'-AAAGACTGCCCAATAACAGCC-3' and
reverse, 5'-AAGCTGGGACAGGACTAGCA-3'; G protein-coupled receptor 65
(GPR65), forward, 5'-GAAATGGCAAATCAACCTCAAC-3' and reverse,
5'-TTCCTTGTTTTCCGTGGCTT-3'; frizzled class receptor 8 (FZD8),
forward, 5'-GGAGTGGGGTTACCTGTTGG-3' and reverse
5'-CTTGCGTGTCGTGGTTGAA-3'; SHC adaptor protein 2 (SHC2), forward,
5'-TACGTCGTGCGGTACATGG-3' and reverse, 5'-AAGCGAAGGTTGCTCTTGC-3';
ADP-ribosyltransferase 5 (ART5), forward, 5'-CAG
ATTTGGTAATGCCACCCTC-3' and reverse, 5'-CAAAGCCCAGAATGGAATAGC-3';
TRIM2, forward, 5'-AGGACAAAGACGGTGAGCTG-3' and reverse,
5'-CTCTTCACGCCTTCTGTGGT-3'; and GAPDH, forward,
5'-GAGTCAACGGATTTGGTCGT-3' and reverse, 5'-CCCAGTAGCAGTTCAGGTGG-3'.
GAPDH was used as the internal control. The relative expression
levels of genes were quantified using the 2−ΔΔCq method,
as described previously (25). At
least three biological repeats and three technical replicates were
performed.
Western blot analysis
Total protein from the osteosarcoma cells was lysed
and extracted using RIPA lysis buffer and quantified using a BCA
Protein Assay kit (both from Beyotime Institute of Biotechnology,
Haimen, China) according to the manufacturer's protocol. The
protein samples (40 μg) were separated by 10% SDS-PAGE and
transferred onto NC membranes (EMD Millipore). The membranes were
blocked in 5% non-fat milk and incubated with the following primary
antibodies: TRIM2 (cat. no. ab3942; 1:1,000), matrix
metal-loproteinase (MMP)9 (cat. no. ab38898; 1:1,000), MMP2 (cat.
no. ab37150; 1:500), N-cadherin (cat. no. ab18203; 1:500),
E-cadherin (cat. no. ab76055; 1:500), Snail (cat. no. ab180714;
1:1,000), Vimentin (cat. no. ab92547; 1:1,000), protein kinase B
(AKT; cat. no. ab8805; 1:500), phosphorylated-AKT (cat. no.
ab38449; 1:1,000), protein kinase A (PKA; cat. no. ab75991;
1:2,000), CREB (cat. no. ab31387; 1:1,000), phosphorylated-CREB
(cat. no. ab32096; 1:3,000) and anti-GAPDH (cat. no. ab9485;
1:2,000) (all from Abcam) at 4°C overnight. The membranes were then
incubated with secondary antibodies, anti-rabbit IgG (cat. no.
ab205718; 1:2,000), anti-goat IgG (cat. no. ab6566; 1:2,000) or
anti-mouse IgG (cat. no. ab6728; 1:2,000) (all from Abcam) for 2 h
at room temperature. The complexes were detected by ECL (Forevergen
Biosciences, Guangzhou, China). GAPDH was applied as the internal
standard. Six samples for each group were used for western blot
analysis.
In vivo nude mouse models
All animal experiments were approved by the
Institute Research Medical Ethics Committee of Sun Yat-Sen
University (Guangzhou, China). Male BALB/C nude mice (age, 5-6
weeks; weight, 18-22 g) were obtained from the Shanghai Laboratory
Animal Center (Shanghai, China). Animals were maintained in a
specific pathogen-free environment throughout the experiments under
the following conditions: Controlled humidity, 50±10%; temperature,
23±2°C; 12-h light/dark cycle and ad libitum access to food
and water. U2OS cells (2×106) in a volume of 100
μl were subcutaneously injected into the caudal veins of the
mice. Post-implantation, five of the mice were sacrificed and the
tumors were surgically dissected at 5 days. The weight and size of
the cancerous tissues were then examined.
Statistical analysis
Data are presented as the mean ± standard error of
the mean unless otherwise indicated. Significance was established
using SPSS PASW Statistics 18.0 (SPSS, Inc., Chicago, IL, USA)
software and GraphPad Prism 5 (GraphPad Software, Inc., La Jolla,
CA, USA) software. For data with a normal distribution, comparisons
were performed using independent t-tests, one-way analysis of
variance (ANOVA) and two-way ANOVA, with the LSD-t-test used for
the post hoc test. The non-parametric Mann-Whitney U test, K-S
test, Kruskal-Wallis test and Wilcoxon test were performed if the
data did not have a normal distribution, and the Nemenyi test was
used for multiple comparisons. P<0.05 was considered to indicate
a statistically significant difference. The significant level was
corrected if necessary.
Results
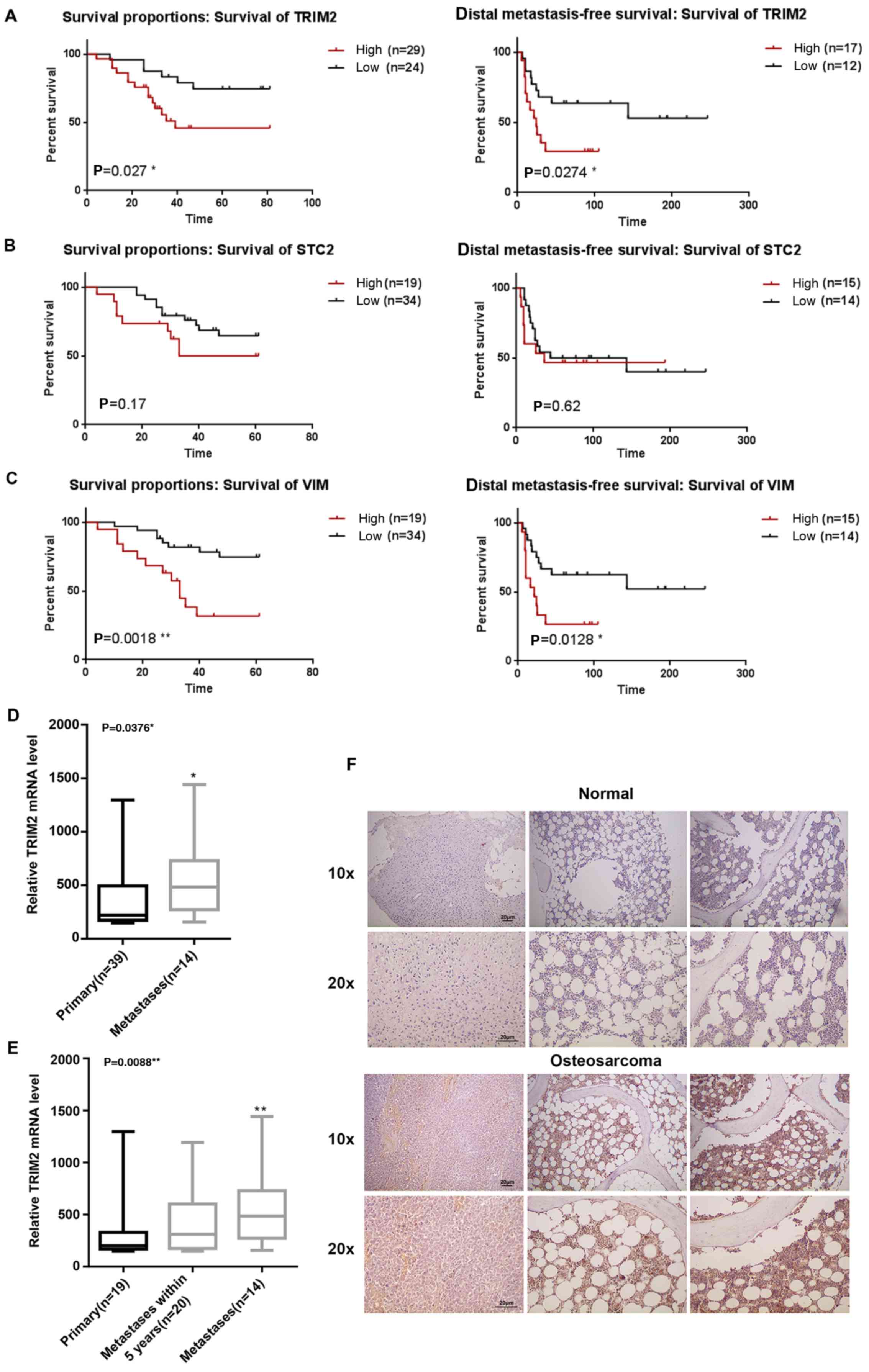
High expression of TRIM2 in osteosarcoma
tissue is correlated with lower survival rates and induction of
metastasis
The expression profiles were collected from the NCBI
GEO (GSE21257). By combining the expression profiles and the
clinical properties, the association between the expression levels
of TRIM2, STC2 and VIM and survival rate were analyzed. The results
showed that the survival difference was statistically significant
between the high and low expression level of TRIM2 and VIM.
Regardless of whether overall survival or distal metastasis-free
survival rate, a high expression level of TRIM2 and VIM led to a
reduced rates of survival and distal metastasis-free survival
(Fig. 1A–C). Higher expression
levels of TRIM2 and VIM reduced the survival percentage by 50% at
40 days. However, for STC2, although the survival percentage was
reduced when its expression level was high, the difference between
a low expression level of STC2 was not significant (P>0.05).
Therefore, the evidence confirmed that a high expression of TRIM2
was correlated with lower survival rate and distal metastasis-free
survival rate. By contrast, when the expression level of TRIM2 was
compared in primary tumor and metastatic tumor tissues, only the
metastatic tissues exhibited a significantly higher level of TRIM2,
compared with the primary tumor tissues or in tumor tissues with
metastasis within 5 years (Fig. 1D and
E). Therefore, actual osteosarcoma tumor tissues and normal
tissues were collected for the immunohistochemical staining to
determine the expression level of TRIM2. The TRIM2 molecules were
stained a brown color specifically. The results showed that the
level and density of the TRIM2 molecules were significantly higher
in the osteosarcoma tissues compared with the normal tissues
(Fig. 1F). According to the
preliminary analysis, it was concluded that TRIM2 in osteosarcoma
tissues performs specific functions that lead to lower survival
rate and induces metastasis. Therefore, the high expression level
of TRIM2 may also be an indicator of osteosarcoma.
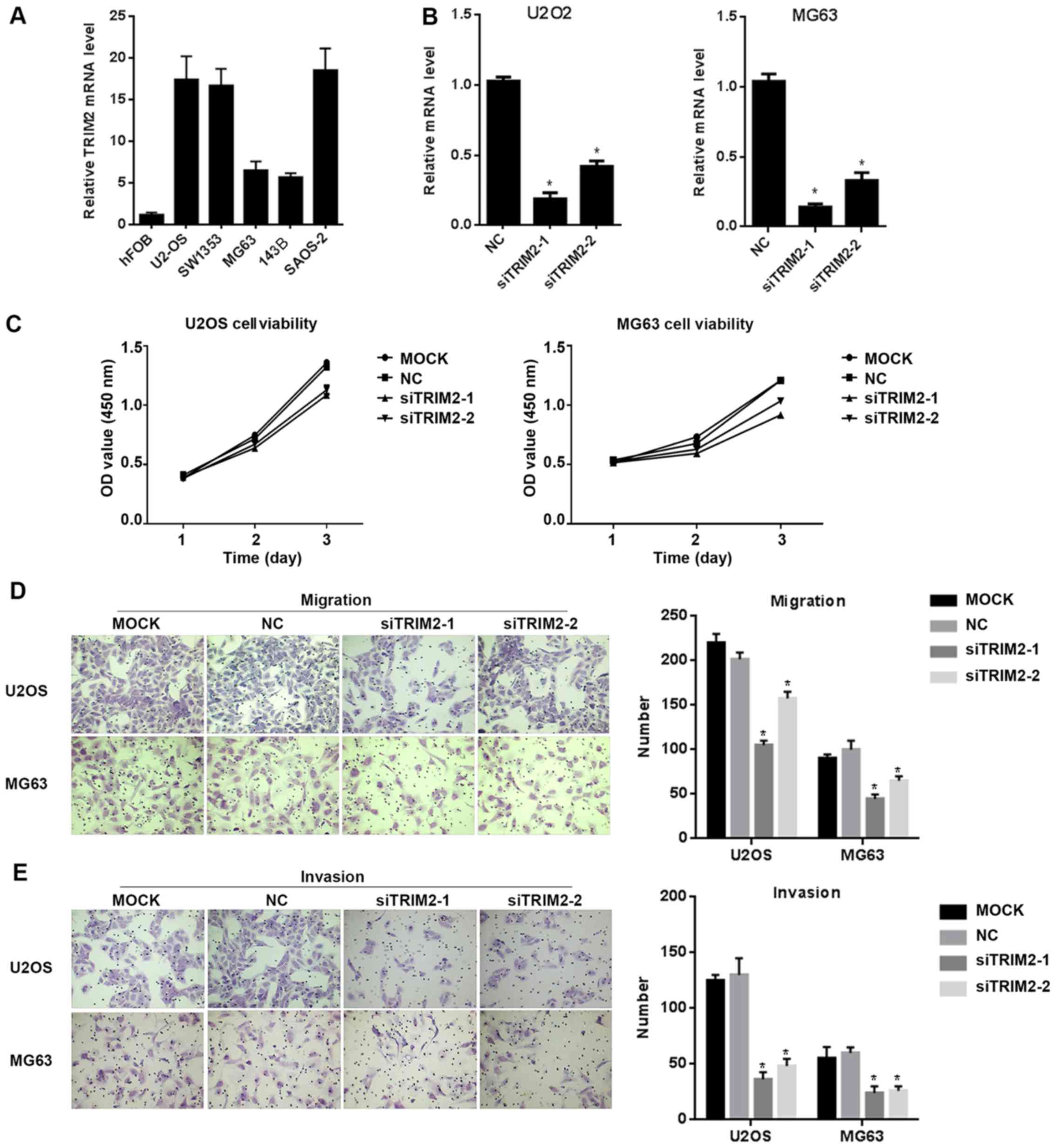
TRIM2 performs functions in regulating
cell proliferation, migration, invasion and apoptosis
In order to verify the functions of TRIM2, in
vitro cell assays were performed. The cell lines were examined
for the expression level of TRIM2 in order to identify the
appropriate line for the following assays. From the results, the
hFOB cell line exhibited the lowest expression signal of TRIM2
among the examined cell lines. In the remaining cell lines, U2OS,
SW1353 and SAOS2 cells exhibited relatively high expression levels
of TRIM2 among the six cell lines, whereas MG63 and 143B expressed
comparatively lower levels of TRIM2 (Fig. 2A). Therefore, U2OS and MG63 were
selected for the following assays, which exhibit high and low
expression level of TRIM2, respectively. siRNAs targeting TRIM2
were designed. The sequences of the siRNAs were synthesized and
integrated into the vector, and the vector was packed into the
lentivirus and transfected into U2OS and MG63 cells in order to
suppress the expression of TRIM2. The results revealed that
siTRIM2-1 and siTRIM2-2 successfully decreased the expression level
of TRIM2 (Fig. 2B). Among the two
siRNAs, siTRIM2-1 decreased the level to ~25% whereas siTRIM2-2
decreased the level to ~50%, compared with the control group, in
the U2OS and MG63 cell lines. Cell viability was then determined
via an MTS assay. The assay was performed every day for a total of
3 days. The results showed that, for the two cell lines, the
viability of the cells transfected with empty vector or without
treatment exhibited similar optical density (OD) values at 450 nm.
However, for those cells transfected with siTRIM2-1 and siTRIM2-2,
the cell viability decreased by ~25%, which was detected on day 3
(Fig. 2C). When the cell migration
and invasion capacity of those cells were determined, the migrated
cells and invasive cells in the control group and the mock group
were increased, with cells exhibiting in higher density in the
area; in the siTRIM2-1 and siTRIM2-2 treatment groups, fewer
migrated and invasive cells were observed than in the control group
and mock group (Fig. 2D and E).
This indicated that the migration and invasion capacities of the
cells were inhibited when the level of TRIM2 was decreased.
Subsequently, the protein from these cells was extracted in order
to determine the protein levels of TRIM2 via western blot analysis.
According to the results, the protein level of TRIM2 was decreased
in the MG63 and U2OS cell lines when transfected the TRIM2-targeted
siRNAs (Fig. 3). Therefore, the
protein levels of cell migration- and invasion-related molecules,
including N-Cadherin, E-Cadherin, Snail and Vimentin, were
examined. The results corresponded to those of the cell migration
and invasion assays: When the expression of TRIM2 was inhibited,
the levels of MMP9, MMP2, N-Cadherin, Snail and Vimentin were
accordingly decreased, whereas the protein level of E-Cadherin was
increased, indicating weak cell migration and invasion
capacities.
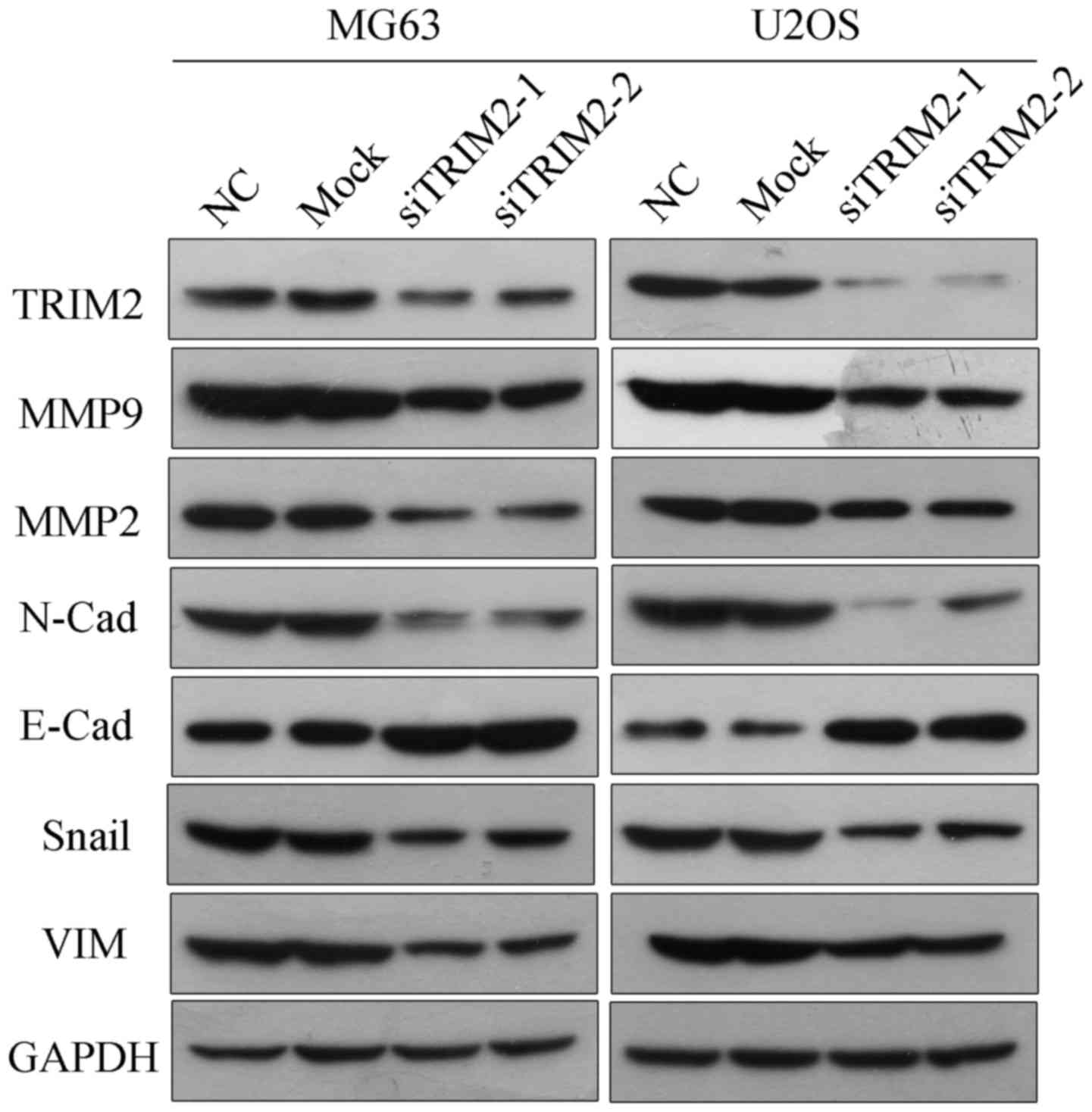 | Figure 3Protein levels of TRIM2 and
migration-related genes as detected via western blot analysis.
Protein levels of cell migration- and invasion-related molecules,
including MMP2, MMP9, N-Cadherin, E-Cadherin, Snail and VIM, were
determined by western blot analysis. GAPDH was applied as the
internal standard. Six samples were used for each group. TRIM2,
tripartite motif-containing protein 2; si, small interfering RNA;
NC, negative control; MMP, matrix metalloproteinase; VIM,
vimentin. |
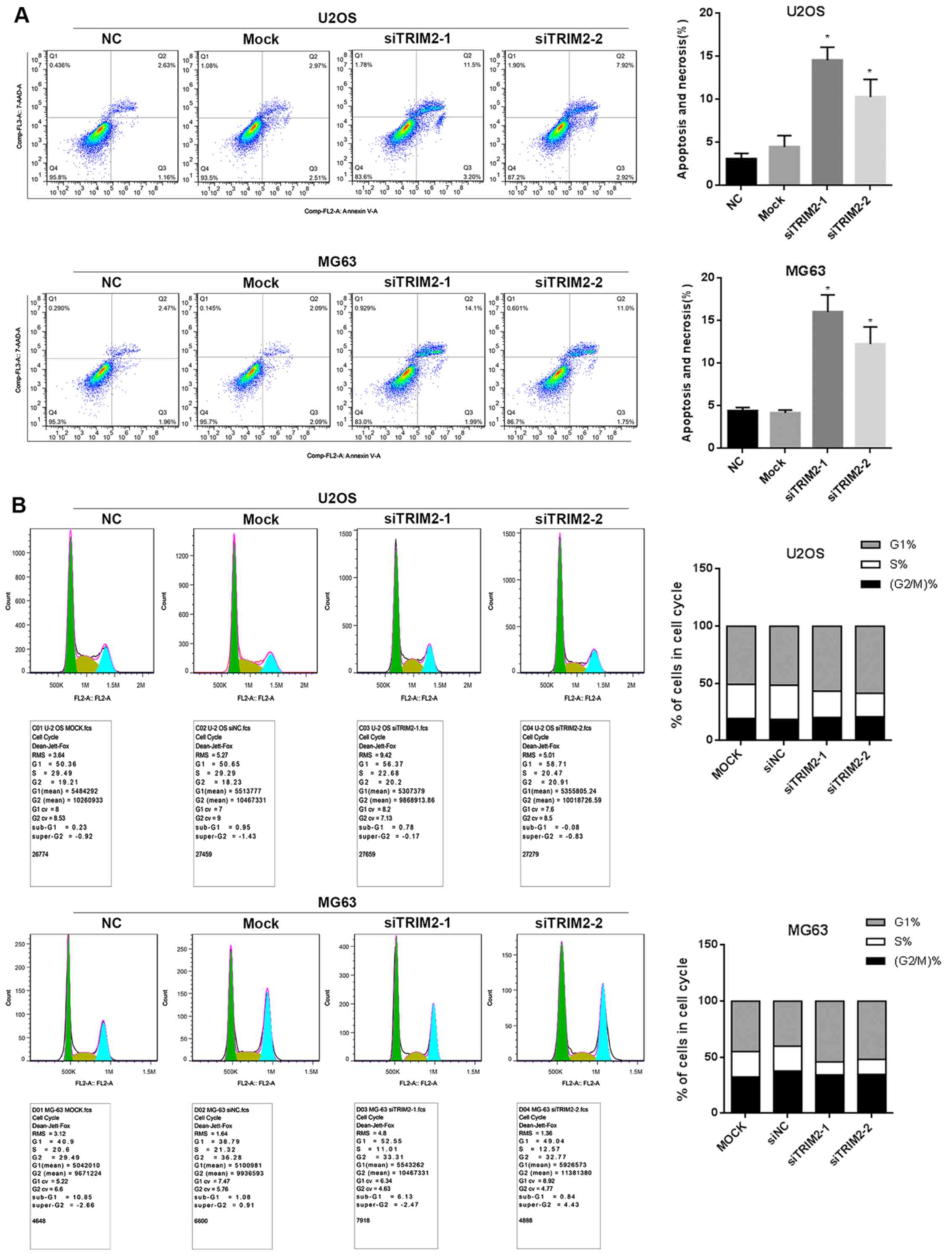
Flow cytometry was also performed to determine cell
apoptosis. This involved comparing the cells with and without
treatment of TRIM2-targeted siRNA transfection. As shown when
siTRIM2-1 and siTRIM2-2 inhibited the level of TRIM2 and thereby
led to lower cell viability, the cell apoptosis was increased when
TRIM2 was decreased in the results (Fig. 4A). The apoptotic percentage
increased ~5-fold in the siRNA-transfected cells compared with the
cells in the control group and mock group. In addition, the cell
cycle assay demonstrated that the proportion of MG63 and U2OS cells
in S phase was decreased, whereas the proportion in G1 phase was
increased post-transfection with siTRIM2-1 and siTRIM2-2 (Fig. 4B). These findings indicated that
most of the cells exhibited reduced viability due to the occurrence
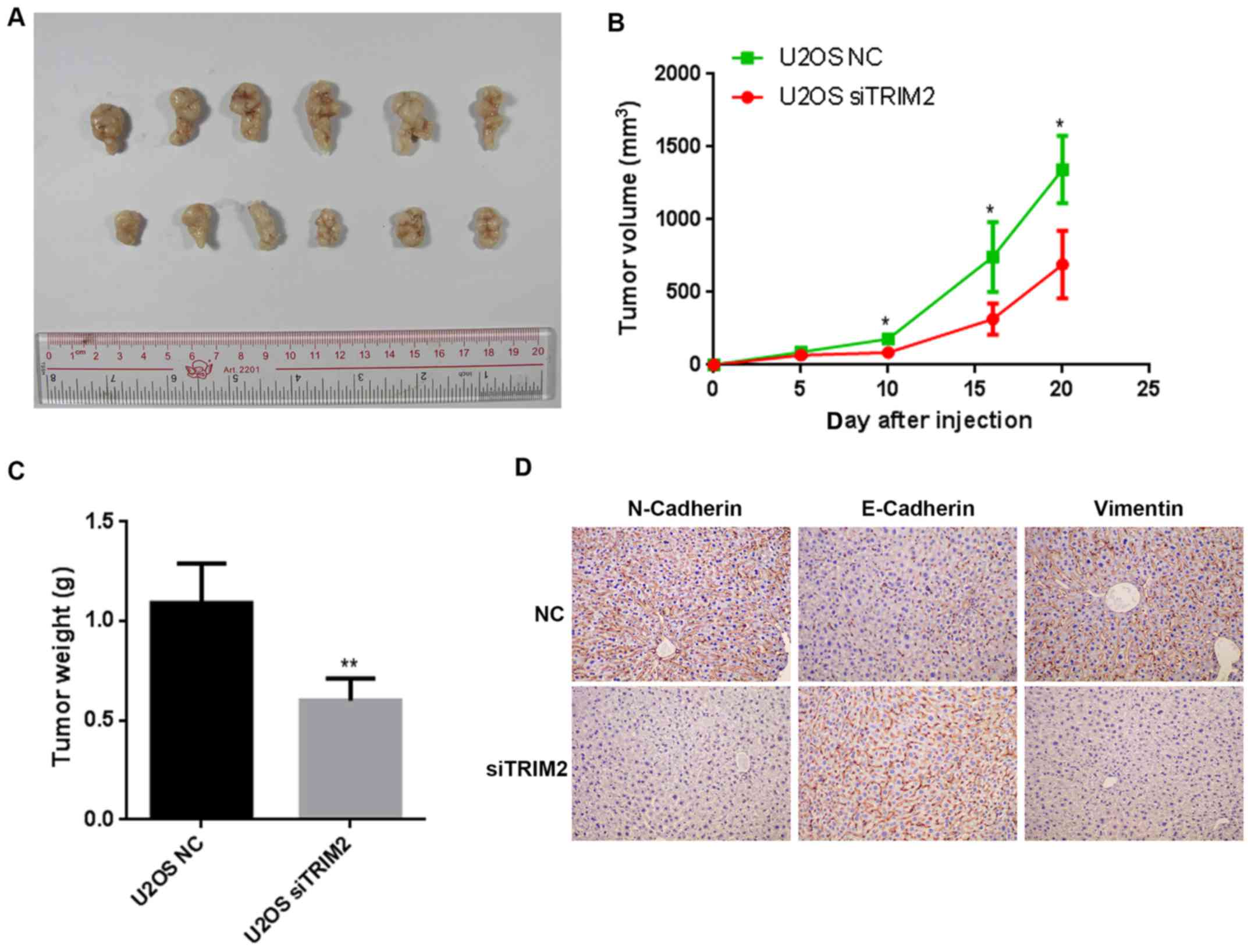
of cell apoptosis. Based on the evidence above, nude mice were used
to perform the tumor formation assay. When the cells were injected
into nude mice, the tumors formed gradually. The tumors were
collected and compared on day 20 (Fig.
5). The tumor was shown to be marginally smaller in the siTRIM2
group than that in negative control (NC) group. The volume of the
tumors were determined every 5 days for a total of 20 days. The
difference between the NC group and siTRIM2 group was significant
on day 10, and the size of the tumor in the NC group was ~2-fold
larger than that in the siTRIM2 on day 20. In terms of the weights
of the tumor tissues, the tumor weight in the NC group was 2-fold
higher than that in the siTRIM2 group, and the difference was
statistically significant. According to the immunohistochemical
staining of the metastasis-related proteins, the expression of
E-Cadherin was upregulated, whereas the expression of Vimentin and
N-Cadherin were downregulated expression when TRIM2 was inhibited
(Fig. 5D). These results suggested
that the tumor metastasis was suppressed when TRIM2 was at a lower
level compared with that in the control group. Therefore, according
to the results above, TRIM2 appeared to be an activator of cell
proliferation, cell migration and cell invasion, thus inducing
tumor metastasis.
Verifying the regulation network of
TRIM2
In order to further verify the regulation performed
by TRIM2 at the molecular level, transcriptome sequencing was
performed for the cells transfected with siTRIM2 and empty vector.
The criteria for the significant expression was considered as FDR
≤0.01 and log2Ratio ≥1. Under these criteria, the significantly
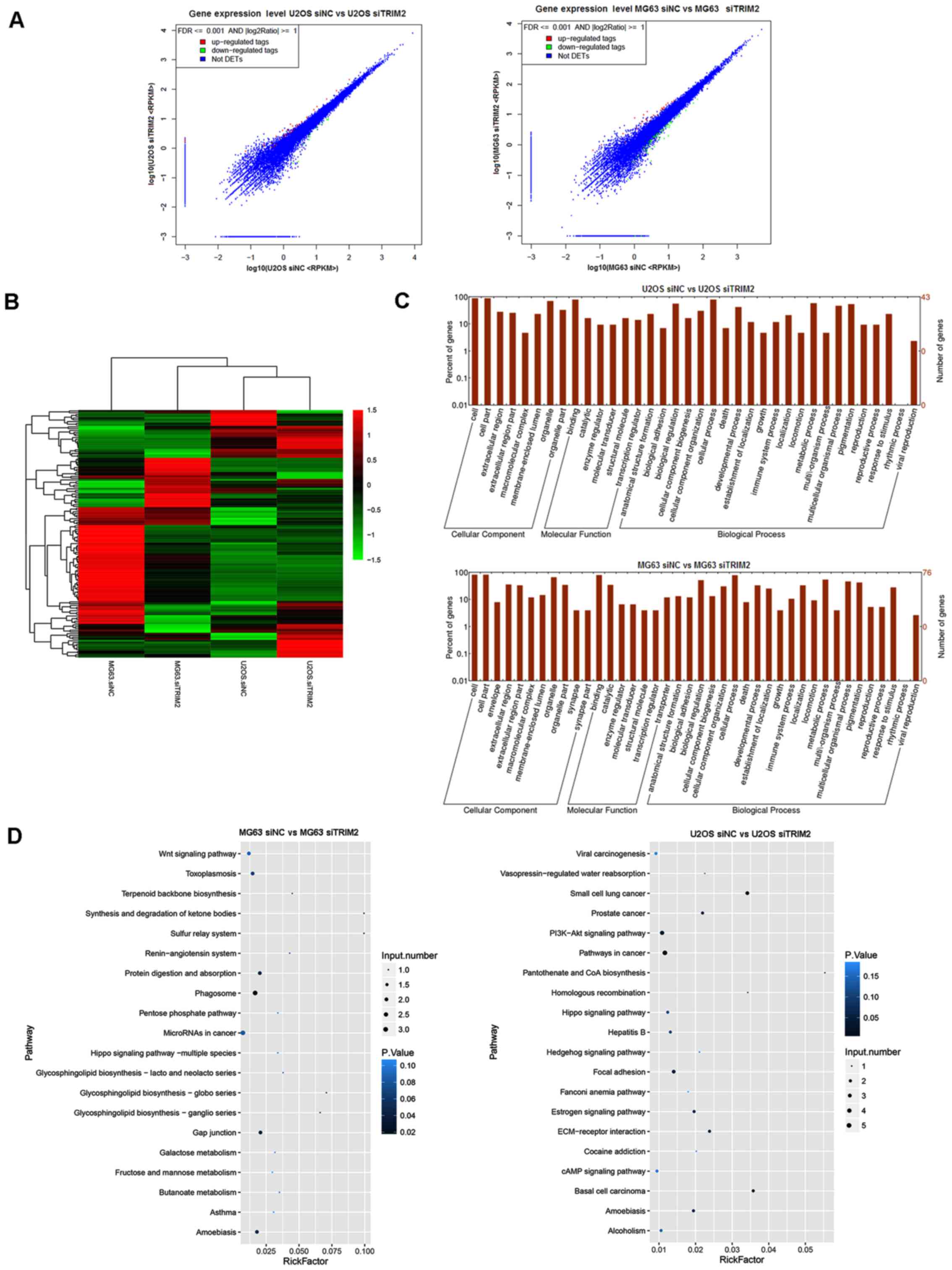
differential expressed genes were tagged in red and green (Fig. 6A). The hierarchical clustering for
the overview of the genes in the transcriptome sequencing is shown
in Fig. 6B. In the graph, the
genes with upregulated expression are tagged in red, and those with
downregulated expression are tagged in green. According to the
results, the effect of the inhibition of TRIM2 was different
between the MG63 and U2OS cells. For the MG63 cells, the
differentially expressed genes were predominantly upregulated,
whereas, the majority of the genes in the U2OS cells were
downregulated (Fig. 6B). The genes
were then subjected to GO and KEEG pathway analysis. As shown in
the plot graphs (Fig. 6C), the
genes involved in transcriptome analysis were involved in several
cellular components, molecular functions, and biological processes.
From the cellular component perspective, 'cell', 'cell part', and
'organelle' were the top three significantly enriched terms. From
the molecular function perspective, 'binding' was the top
significantly overrepresented term. From the perspective of
biological process, 'biological regulation', 'cellular process' and
'metabolic process' were the top three significantly enriched
terms. As shown in Fig. 6D, the
'Wnt signaling pathway' and 'MicroRNAs in cancer' were the top
enriched term in the MG63 cells, whereas 'Viral carcinogenesis',
'PI3K-Akt signaling pathway', 'Pathways in cancer', and 'cAMP
signaling pathway' were the most significantly enriched terms in
the U2OS cells.
According to the data from the cell assays, the
expression level of TRIM2 differed in the MG63 and U2OS cells, with
MG63 cells showing lower expression of TRIM2 and U2OS cells showing
higher expression of TRIM2. However, as the two cell lines showed
similar results in terms of lower cell viability and capacity to
migrate and invade when TRIM2 was inhibited, regulation was
expected to be common in these cell lines. Therefore, seven
significantly differentially expressed genes related to TRIM2 were
extracted from the expression profiles and further validated via
RT-qPCR analysis. In the U2OS cells, the TRIM2 gene was
significantly downregulated, whereas the remaining genes, with the
exception of FZD8 and SHC2, were significantly upregulated in the
two cell lines (Fig. 7). For FZD8
and SHC2, they were significantly increased in the MG63 cells but
did not in the U2OS cells. These two genes may not be closely
associated with TRIM2. As the consistently significantly expressed
genes are involved in the regulation of cell cycle, it was
hypothesized that TRIM2 performs its functions in osteosarcoma by
regulating these genes.
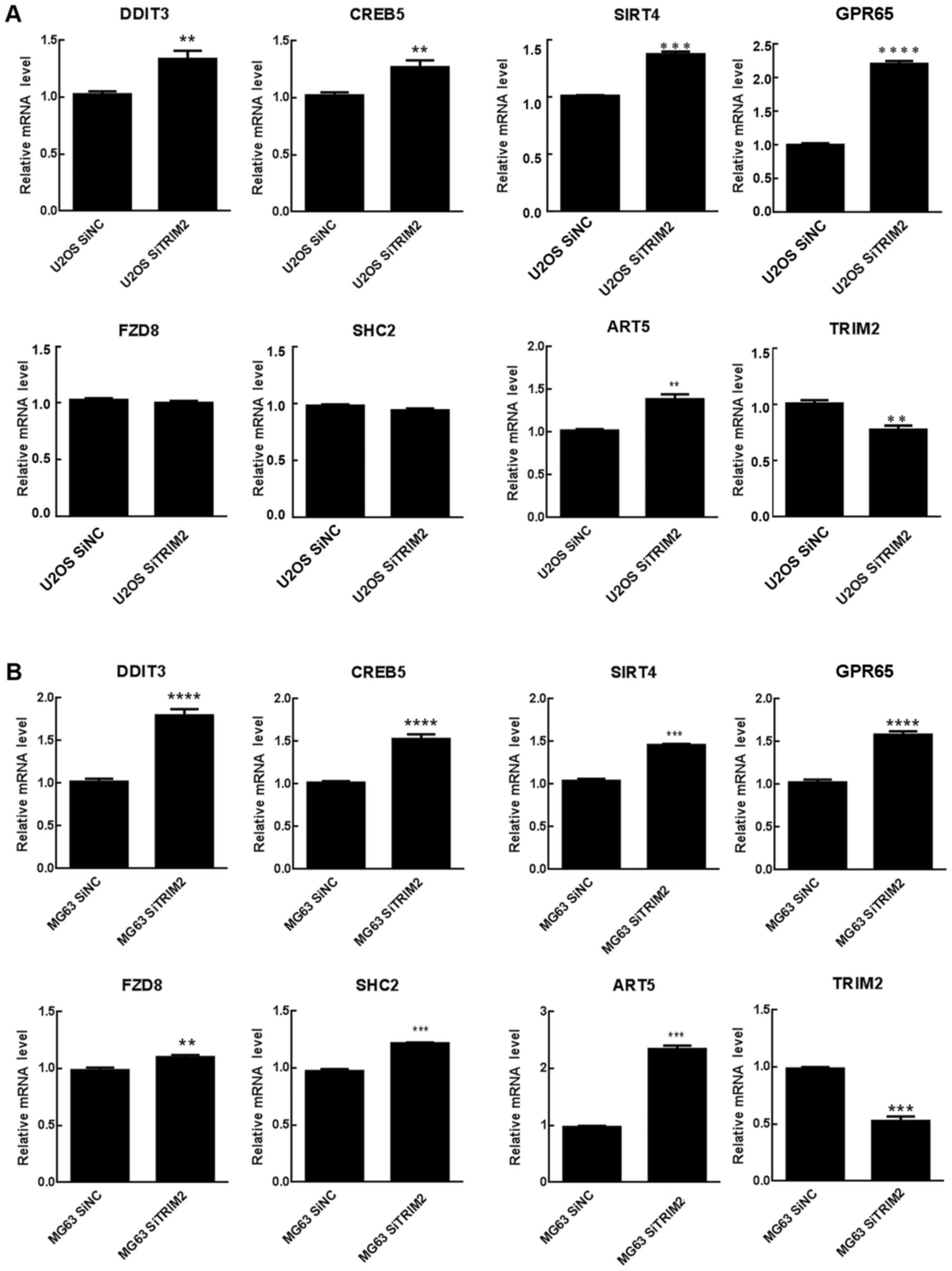 | Figure 7Genes involved in the regulation
network of TRIM2. Relative mRNA level of genes associated with
TRIM2 were detected in (A) U20S and (B) MG63 cells by reverse
transcription-quantitative polymerase chain reaction analysis.
**P<0.01, ***P<0.001 and
****P<0.0001. ART5, ADP-ribosyltransferase 5; CREB5,
cAMP responsive element binding protein 5; DDIT3, DNA
damage-inducible transcript 3; FZD8, frizzled class receptor 8;
GPR65, G protein-coupled receptor 65; NC, negative control; SHC2,
SHC adaptor protein 2; si, small interfering RNA; SIRT4, Sirtuin 4;
TRIM2, tripartite motif-containing protein 2. |
Western blot analysis was used to verify those genes
that may involved in the regulatory pathways indicated by the KEEG
analysis. The results, as shown in Fig. 8, indicated that, when TRIM2 was
inhibited in the U2OS and MG63 cell lines, the protein level of
p-AKT was decreased, whereas AKT was not changed. The protein
levels of PKA, CREB and phosphorylated CREB were not affected by
the inhibition of TRIM2. The results suggested that TRIM2 regulates
the development and metastasis of tumorous cells of osteosarcoma
via the PI3K/AKT pathway.
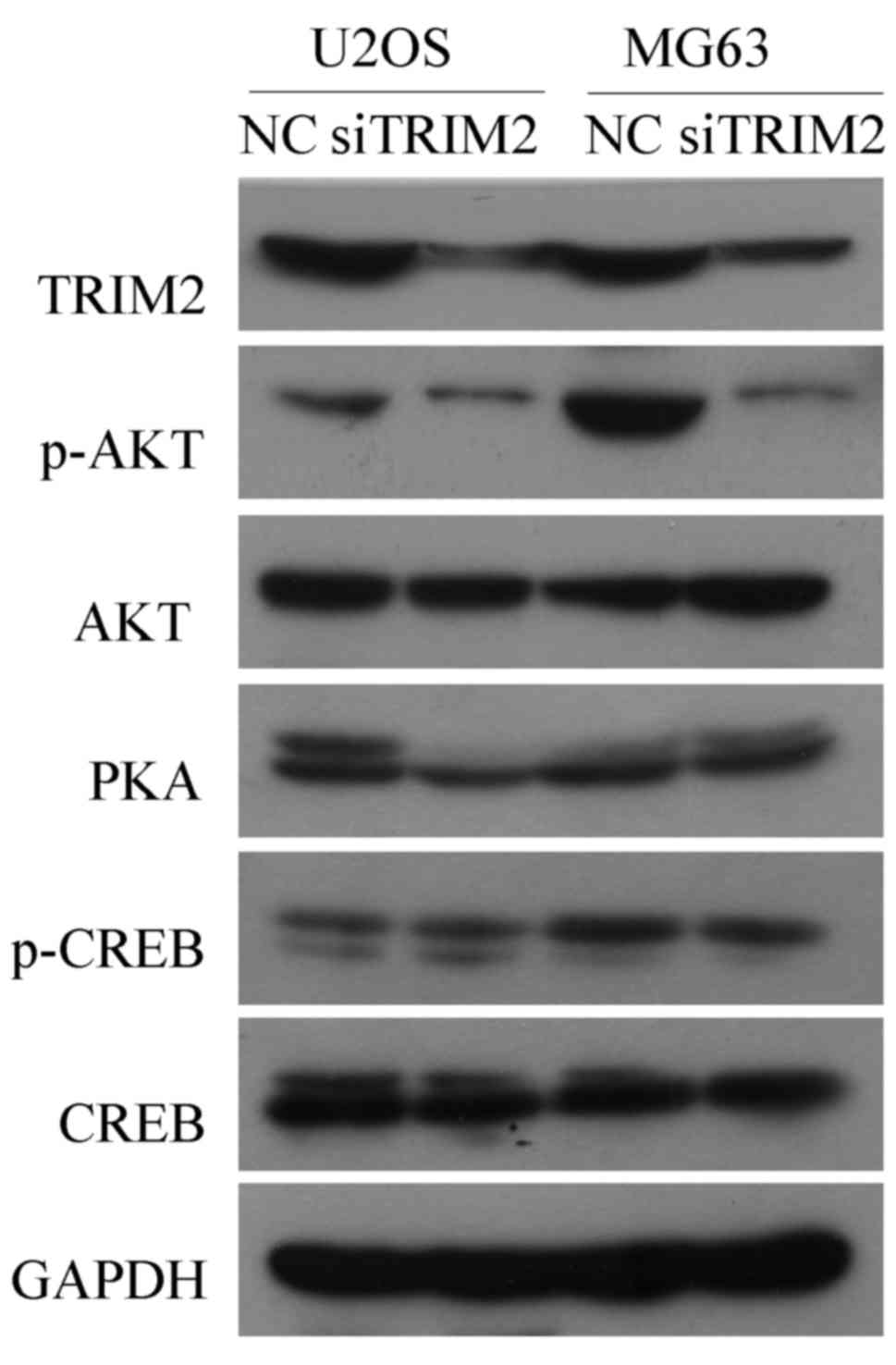 | Figure 8Western blot analysis to verify genes
that may be involved in the regulating pathways of TRIM2. Protein
levels of the genes that may be involved in the regulating pathways
of TRIM2, including p-AKT, AKT, PKA, p-CREB and CREB, were detected
by western blot analysis. GAPDH was applied as the internal
standard. Six samples were used for each group. AKT, protein kinase
B; CREB, cAMP response element binding protein; NC, negative
control; p-, phosphorylated; PKA, protein kinase A; si, small
interfering RNA; TRIM2, tripartite motif-containing protein 2. |
Discussion
TRIM2 was selected and its effects were verified
using TRIM2 siRNA in vitro and in vivo in the present
study; TRIM2 was shown to be correlated with tumorous cell
proliferation, invasion, migration and apoptosis. It may be a novel
therapeutic target for the diagnosis and treatment of osteosarcoma.
TRIM2 is a member of the TRIM-NHL protein family, which functions
as an E3 ubiquitin ligase. It can interact with B-cell lymphoma
2-interacting mediator (Bim) of cell death (26). TRIM2 usually binds to Bim when it
is phosphorylated by the p42/p44 mitogen-activated protein kinase
(MAPK) pathway. As TRIM2 was expressed at a higher level in
patients with osteosarcoma, the level of Bim is expected to be
accordingly decreased, thus leading to the enhanced cell
proliferation of the tumorous cells (26,27).
In previous studies, the association between TRIM2 and osteosarcoma
have not been reported. In studies focused on different types of
carcinoma, TRIM2 has been detected to be significantly
differentially expressed in ovarian cancer, breast cancer, cervical
carcinoma, buccal mucosa carcinoma and follicular carcinoma
(27-32). In a study of ovarian cancer, the
expression of TRIM2 was detected as upregulated, and negatively
regulated Bim (7). This finding is
consistent with the results of the present study. However, certain
studies have shown the opposite results, with the expression of
TRIM2 found to be downregulated (28,31,32).
The bias between different types of cancer suggests that TRIM2 is
an important gene involved in the pathogenic process of cancer,
although the underlying mechanism may be different and complex.
However, the dysfunction of TRIM2 may be one of the factors
involved in the occurrence and development of osteosarcoma to a
certain extent.
The epithelial-mesenchymal transition (EMT) is a
process whereby epithelial cells transition into cells of the
mesenchymal phenotype, and it has long been known to be involved in
cellular differentiation during development and tumor invasion. EMT
results in cells with migratory and invasive properties, the
mesenchymal state is associated with the capacity of cells to
migrate to distant organs and maintain stemness, enabling their
subsequent differentiation into multiple cell types during
development and the initiation of metastasis (33). EMT can be induced by various
factors. Transforming growth factor-β is a major inducer of EMT, it
downregulates genes expressed in epithelial cells, including
E-cadherin, and upregulates genes normally expressed in mesenchymal
cells, including N-cadherin and vimentin (34,35).
The degradation of extracellular matrix (ECM) through activating
MMPs is an essential step in tumor cell migration. The expression
of MMP-2 and MMP-9 is reported to be closely linked with tumor
progression and metastasis (36).
In the present study, the protein levels of the above-mentioned
genes, which are associated with cancer cell development and
metastasis, were examined in vitro and in vivo. The
results indicated that the inhibition of TRIM2 led to decreased
cell migration and invasion capacities, which confirmed that TRIM2
is important in regulating tumorous cell proliferation, migration
and invasion.
To investigate the underlying regulatory mechanism
of TRIM2, the gene expression was inhibited in expression and
screened by RNA-seq for candidate genes. There are few reports on
the association between TRIM2 and the candidate genes screened in
the present study. Among these genes, SIRT4 and CREB5 are reported
to be functionally correlated with tumor development. SIRT4, it is
a member of the sirtuin family of NAD+-dependent
enzymes, which have various roles in metabolism, stress response
and longevity (37,38). It has been reported that SIRT4
negatively regulates mitochondrial glutamine metabolism via the
inhibition of glutamate dehydrogenase and has tumor-suppressive
functions (39). The decreased
level of SIRT4 can lead to poor prognosis in breast cancer
(40), and the increased level of
SIRT4 contributed to tumorous cell proliferation when TRIM2 was
inhibited in the present study. It has been reported that CREB5 may
be key in the metastatic signal network of colorectal cancer. A
previous report confirmed that metastasis was promoted when CREB5
was expressed at a high level (41). In this study, CREB5 was increased
when TRIM2 was downregulated. Therefore, the increase of CREB5 may
enhance the metastatic process that leads to inactive cell
migration and invasion. However, migrated and invasive cells in the
present study were decreased in number when TRIM2 was suppressed.
This suggests other genes are likely to regulate the events, rather
than CREB5 alone.
For the remaining three genes, ART5, DDIT3 and
GPR65, their expression was upregulated when TRIM2 was inhibited.
Reports on the functions of these genes in tumor occurrence and
development are limited. In analyzing the gene functions
independently, ART5 can interact with TIM23. It has been reported
that the overexpression of ART5 suppresses TIM23, and defects in
cell growth were observed (42).
This also explains the decreased cell viability in the TRIM2
inhibition group in the present study. DDIT3 is able to function in
inducing DNA damage. The overexpression of DDIT3 can damage cell
migration and invasion according to a previous study (43,44).
Therefore, it may be an important gene contributing to the
decreased cell migration and invasion observed when TRIM2 was
suppressed. GPR65, is a gene involved in the p38 MAPK signaling
pathway, which is correlated with the cell cycle (45). However, its exact function,
particularly involving tumor development, remains to be fully
elucidated.
The results from KEGG analysis indicated that 'Wnt
signaling pathway' and 'MicroRNAs in cancer' were the top enriched
terms in MG63 cells, and 'Pathways in viral carcinogenesis',
'PI3K-Akt signaling pathway', 'Pathways in cancer', and 'cAMP
signaling pathway' were the most significantly enriched terms in
U2OS cells. The present study mainly focused on PI3K/AKT and cAMP
signaling pathways and examined whether TRIM2 was implicated in
these regulating pathways. A number of studies have shown that the
PI3K/AKT pathway is involved in osteosarcoma (46-48).
mTOR, one of the PI3K family members, is important in the
regulation of cell cycle, growth and development; the PI3K/AKT/mTOR
signaling pathway is reported to be involved in the proliferation,
migration and survival of osteosarcoma (49). The results of the present study
showed that, in the U2OS and MG63 cell lines, the expression of
TRIM2 was inhibited, the protein level of phosphorylated-AKT was
decreased, and the level of AKT was unchanged, which indicated that
the AKT signaling was implicated in the regulatory pathway of
TRIM2. A previous study demonstrated that the cAMP/PKA signaling
pathway was involved in osteosarcoma chemoresistance (50). Parathyroid hormone (PTH) stimulates
bone formation in animals and humans. Studies have indicated that
the cAMP/PKA pathway is involved in PTH signaling transduction,
which subsequently affects the development of osteosarcoma cells
(51,52). The present study failed to show
evidence that the cAMP/PKA pathway was implicated in the
development of osteosar-coma, which may due to the small sample
size, and further investigation is required to confirm the results.
Taken together, the results suggested that TRIM2 regulated the
development and metastasis of osteosarcoma tumor cells via the
PI3K/AKT pathway.
In conclusion, the present study determined the
functions of TRIM2 in osteosarcoma cell lines. TRIM2 is important
in regulating the tumorous cell proliferation, migration and
invasion. The SIRT4, ART5 and DDIT3 genes appear to be functionally
correlated with TRIM2, which regulates the pathogenic processes of
osteosarcoma, and the PI3K/AKT signaling pathway may be involved in
the regulatory role of TRIM2 in the development and metastasis of
osteosarcoma, however, comprehensive assays are required to
validate this. The importance of TRIM2 requires further validation
in clinical samples, and the comprehensive analysis of clinical
samples is likely to benefit the development of diagnosis and
treatment for osteosarcoma.
Acknowledgments
Not applicable.
Funding
This study was supported by the Science and
Technology Planning Project of Guangdong Province, China (grant
nos. 2016A020225001 and 2017A020213024), the Administration of
Traditional Chinese Medicine of Guangdong Province, China (grant
no. 20161233) and the Medical Scientific Research Foundation of
Guangdong Province, China (grant no. A2016233).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YQ and JY conceived and designed the study, and
critically revised the manuscript. JY performed the experiments,
analyzed the data and drafted the manuscript. FZ, SH and SW was
involved in study design, study implementation and manuscript
revision. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Research Ethics Committee of Jinan University. All animal
experiments were approved by The Institute Research Medical Ethics
Committee of Sun Yat-Sen University. Informed consent was provided
by all participants prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kashima T, Nakamura K, Kawaguchi J,
Takanashi M, Ishida T, Aburatani H, Kudo A, Fukayama M and
Grigoriadis AE: Overexpression of cadherins suppresses pulmonary
metastasis of osteosarcoma in vivo. Int J Cancer. 104:147–154.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bacci G, Rocca M, Salone M, Balladelli A,
Ferrari S, Palmerini E, Forni C and Briccoli A: High grade
osteosarcoma of the extremities with lung metastases at
presentation: Treatment with neoadjuvant chemotherapy and
simultaneous resection of primary and metastatic lesions. J Surg
Oncol. 98:415–420. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harting MT, Blakely ML, Jaffe N, Cox CS
Jr, Hayes-Jordan A, Benjamin RS, Raymond AK, Andrassy RJ and Lally
KP: Long-term survival after aggressive resection of pulmonary
metastases among children and adolescents with osteosarcoma. J
Pediatr Surg. 41:194–199. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsuchiya H, Kanazawa Y, Abdel-Wanis ME,
Asada N, Abe S, Isu K, Sugita T and Tomita K: Effect of timing of
pulmonary metastases identification on prognosis of patients with
osteosarcoma: The Japanese Musculoskeletal Oncology Group study. J
Clin Oncol. 20:3470–3477. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu PK, Chen WM, Chen CF, Lee OK, Haung CK
and Chen TH: Primary osteogenic sarcoma with pulmonary metastasis:
Clinical results and prognostic factors in 91 patients. Jpn J Clin
Oncol. 39:514–522. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsuchiya H, Tomita K, Mori Y, Asada N and
Yamamoto N: Marginal excision for osteosarcoma with caffeine
assisted chemotherapy. Clin Orthop Relat Res. 358:27–35. 1999.
View Article : Google Scholar
|
|
8
|
Zanotti S and Canalis E: Notch signaling
and the skeleton. Endocr Rev. 37:223–253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kofler NM, Shawber CJ, Kangsamaksin T,
Reed HO, Galatioto J and Kitajewski J: Notch signaling in
developmental and tumor angiogenesis. Genes Cancer. 2:1106–1116.
2011. View Article : Google Scholar
|
|
10
|
Engin F, Bertin T, Ma O, Jiang MM, Wang L,
Sutton RE, Donehower LA and Lee B: Notch signaling contributes to
the pathogenesis of human osteosarcomas. Hum Mol Genet.
18:1464–1470. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tanaka M, Setoguchi T, Hirotsu M, Gao H,
Sasaki H, Matsunoshita Y and Komiya S: Inhibition of Notch pathway
prevents osteosarcoma growth by cell cycle regulation. Br J Cancer.
100:1957–1965. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dailey DD, Anfinsen KP, Pfaff LE, Ehrhart
EJ, Charles JB, Bønsdorff TB, Thamm DH, Powers BE, Jonasdottir TJ
and Duval DL: HES1, a target of Notch signaling, is elevated in
canine osteosarcoma, but reduced in the most aggressive tumors. BMC
Vet Res. 9:1302013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Komori T: Regulation of osteoblast
differentiation by transcription factors. J Cell Biochem.
99:1233–1239. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hilton MJ, Tu X, Wu X, Bai S, Zhao H,
Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, et al:
Notch signaling maintains bone marrow mesenchymal progenitors by
suppressing osteoblast differentiation. Nat Med. 14:306–314. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McManus MM, Weiss KR and Hughes DP:
Understanding the role of Notch in osteosarcoma. Adv Exp Med Biol.
804:67–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krishnan V, Bryant HU and Macdougald OA:
Regulation of bone mass by Wnt signaling. J Clin Invest.
116:1202–1209. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cai Y, Cai T and Chen Y: Wnt pathway in
osteosarcoma, from oncogenic to therapeutic. J Cell Biochem 1.
15:625–631. 2014. View Article : Google Scholar
|
|
18
|
Hoang BH, Kubo T, Healey JH, Sowers R,
Mazza B, Yang R, Huvos AG, Meyers PA and Gorlick R: Expression of
LDL receptor-related protein 5 (LRP5) as a novel marker for disease
progression in high-grade osteosarcoma. Int J Cancer. 109:106–111.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma Y, Ren Y, Han EQ, Li H, Chen D, Jacobs
JJ, Gitelis S, O'Keefe RJ, Konttinen YT, Yin G, et al: Inhibition
of the Wnt-β-catenin and Notch signaling pathways sensitizes
osteosarcoma cells to chemotherapy. Biochem Biophys Res Commun.
431:274–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mödder UI, Oursler MJ, Khosla S and Monroe
DG: Wnt10b activates the Wnt, notch, and NFκB pathways in U2OS
osteosarcoma cells. J Cell Biochem. 112:1392–1402. 2011. View Article : Google Scholar
|
|
21
|
Laplante M and Sabatini DM: mTOR signaling
in growth control and disease. Cell. 149:274–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu K, Dai HB and Qiu ZL: mTOR signaling in
osteosarcoma: Oncogenesis and therapeutic aspects (Review). Oncol
Rep. 36:1219–1225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Perry JA, Kiezun A, Tonzi P, Van Allen EM,
Carter SL, Baca SC, Cowley GS, Bhatt AS, Rheinbay E, Pedamallu CS,
et al: Complementary genomic approaches highlight the PI3K/mTOR
pathway as a common vulnerability in osteosarcoma. Proc Natl Acad
Sci USA. 111:E5564–E5573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Thompson S, Pearson AN, Ashley MD, Jessick
V, Murphy BM, Gafken P, Henshall DC, Morris KT, Simon RP and Meller
R: Identification of a novel Bcl-2-interacting mediator of cell
death (Bim) E3 ligase, tripartite motif-containing protein 2
(TRIM2), and its role in rapid ischemic tolerance-induced
neuroprotection. J Biol Chem. 286:19331–19339. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen X, Dong C, Law PT, Chan MT, Su Z,
Wang S, Wu WK and Xu H: MicroRNA-145 targets TRIM2 and exerts
tumor-suppressing functions in epithelial ovarian cancer. Gynecol
Oncol. 139:513–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Williams MD, Zhang L, Elliott DD, Perrier
ND, Lozano G, Clayman GL and El-Naggar AK: Differential gene
expression profiling of aggressive and nonaggressive follicular
carcinomas. Hum Pathol. 42:1213–1220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Panaccione A, Guo Y, Yarbrough WG and
Ivanov SV: Expression profiling of clinical specimens supports the
existence of neural progenitor-like stem cells in basal breast
cancers. Clin Breast Cancer. 17:298–306.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ivanov SV, Panaccione A, Nonaka D, Prasad
ML, Boyd KL, Brown B, Guo Y, Sewell A and Yarbrough WG: Diagnostic
SOX10 gene signatures in salivary adenoid cystic and breast
basal-like carcinomas. Br J Cancer. 109:444–451. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang K, Zhang G, Mei J, Chen D and Wu M:
Screening and analysis of pathogenic genes during DMBA-induced
buccal mucosa carcinogenesis in golden hamsters. Oncol Rep.
23:1619–1624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Miyatake T, Ueda Y, Nakashima R, Yoshino
K, Kimura T, Murata T, Nomura T, Fujita M, Buzard GS and Enomoto T:
Down-regulation of insulin-like growth factor binding protein-5
(IGFBP-5): Novel marker for cervical carcinogenesis. Int J Cancer.
120:2068–2077. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Willis BC, Liebler JM, Luby-Phelps K,
Nicholson AG, Crandall ED, du Bois RM and Borok Z: Induction of
epithelial-mesenchymal transition in alveolar epithelial cells by
transforming growth factor-beta1: Potential role in idiopathic
pulmonary fibrosis. Am J Pathol. 166:1321–1332. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Willis BC and Borok Z: TGF-beta-induced
EMT: Mechanisms and implications for fibrotic lung disease. Am J
Physiol Lung Cell Mol Physiol. 293:L525–L534. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moses MA, Wiederschain D, Loughlin KR,
Zurakowski D, Lamb CC and Freeman MR: Increased incidence of matrix
metalloproteinases in urine of cancer patients. Cancer Res.
58:1395–1399. 1998.PubMed/NCBI
|
|
37
|
Finkel T, Deng CX and Mostoslavsky R:
Recent progress in the biology and physiology of sirtuins. Nature.
460:587–591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Haigis MC and Guarente LP: Mammalian
sirtuins–emerging roles in physiology, aging, and calorie
restriction. Genes Dev. 20:2913–2921. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi Q, Liu T, Zhang X, Geng J, He X, Nu M
and Pang D: Decreased sirtuin 4 expression is associated with poor
prognosis in patients with invasive breast cancer. Oncol Lett.
12:2606–2612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qi L and Ding Y: Involvement of the CREB5
regulatory network in colorectal cancer metastasis. Yi Chuan.
36:679–684. 2014.PubMed/NCBI
|
|
42
|
Harada Y, Tamura Y and Endo T:
Identification of yeast Art5 as a multicopy suppressor for the
mitochondrial translocator maintenance protein Tam41. Biochem
Biophys Res Commun. 392:228–233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shih YL, Chou HM, Chou HC, Lu HF, Chu YL,
Shang HS and Chung JG: Casticin impairs cell migration and invasion
of mouse melanoma B16F10 cells via PI3K/AKT and NF-κB signaling
pathways. Environ Toxicol. 32:2097–2112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang J, Wang L, Ho CT, Zhang K, Liu Q and
Zhao H: Garcinol from Garcinia indica downregulates cancer
stem-like cell biomarker ALDH1A1 in nonsmall cell lung cancer A549
cells through DDIT3 activation. J Agric Food Chem. 65:3675–3683.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Čokić VP, Smith RD, Biancotto A, Noguchi
CT, Puri RK and Schechter AN: Globin gene expression in correlation
with G protein-related genes during erythroid differentiation. BMC
Genomics. 14:1162013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lian Z, Han J, Huang L, Wei C, Fan Y, Xu
J, Zhou M, Feng H, Liu Q, Chen L, et al: A005, a novel inhibitor of
phosphatidylinositol 3-kinase/mammalian target of rapamycin,
prevents osteosarcoma-induced osteolysis. Carcinogenesis. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu G, Liu G, Yuan D, Dai J, Cui Y and Tang
X: Long non-coding RNA ANRIL is associated with a poor prognosis of
osteosarcoma and promotes tumorigenesis via PI3K/Akt pathway. J
Bone Oncol. 11:51–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen L, Pei H, Lu SJ, Liu ZJ, Yan L, Zhao
XM, Hu B and Lu HG: SPOP suppresses osteosarcoma invasion via
PI3K/AKT/NF-κB signaling pathway. Eur Rev Med Pharmacol Sci.
22:609–615. 2018.PubMed/NCBI
|
|
49
|
Hu B, Lv X, Gao F, Chen S, Wang S, Qing X,
Liu J, Wang B and Shao Z: Downregulation of DEPTOR inhibits the
proliferation, migration, and survival of osteosarcoma through
PI3K/Akt/mTOR pathway. OncoTargets Ther. 10:4379–4391. 2017.
View Article : Google Scholar
|
|
50
|
Pu Y, Yi Q, Zhao F, Wang H, Cai W and Cai
S: MiR-20a-5p represses multi-drug resistance in osteosarcoma by
targeting the KIF26B gene. Cancer Cell Int. 16:642016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fujimori A, Cheng SL, Avioli LV and
Civitelli R: Structure-function relationship of parathyroid
hormone: Activation of phospholipase-C, protein kinase-A and -C in
osteosarcoma cells. Endocrinology. 130:29–36. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Miles RR, Sluka JP, Halladay DL, Santerre
RF, Hale LV, Bloem L, Thirunavukkarasu K, Galvin RJ, Hock JM and
Onyia JE: ADAMTS-1: A cellular disintegrin and metalloprotease with
thrombospondin motifs is a target for parathyroid hormone in bone.
Endocrinology. 141:4533–4542. 2000. View Article : Google Scholar : PubMed/NCBI
|