Introduction
Retinoblastoma (RB) is the most common type of
primary intra-ocular cancer, which mainly occurs in children <5
years (1). It has been reported
that RB comprises ~11% of cancers that appear in the first year of
life, with 95% diagnosed before 5 years of age (2). RB can be inherited in an autosomal
dominant fashion caused by inactivation of the RB1 gene (3). RB presents with numerous symptoms,
including abnormal appearance of the pupil, leukocoria, red and
irritated eyes, deterioration of vision, faltering growth or
delayed development (4). In
addition, RB tumors exhibit perivascular sleeves composed of viable
neoplastic cells and necrotic tissues, thus indicating that RB is
closely associated with blood vessels (5). According to a previous study,
microRNAs (miRNAs/miRs) serve a pivotal role in the development of
RB; therefore, further studies are required to investigate the role
of miRNAs in RB (6).
miRNAs are a class of small non-coding RNAs that
have vital roles in the regulation of target genes (7). miRNAs are involved in numerous
physiological and pathological processes, including tumor
development and exacerbation; aberrant miRNA expression has been
observed in various types of cancer, including RB (6,8,9).
miR-182 is a member of the miR-183 cluster, which is located in the
human chromosome 7q32 region (10). Notably, miR-182 overexpression has
been detected in numerous solitary malignancies, including ovarian
cancer (11). Cell adhesion
molecule 2 (CADM2), which is a member of the CADM family, is an
immunoglobulin-like cell adhesion molecule (12). A previous study revealed that
down-regulation of epithelial cell adhesion molecule (a member of
the CADM family) serves an essential role in the development of RB
(13). The catalytic subunit of
phosphatidylinositol-3-OH kinase (PI3K) can produce the
phosphoinositide phosphates PIP2 and PIP3, which can activate
several serine/threonine kinases, including protein kinase B (AKT)
(14). AKT inhibits cell apoptosis
by phosphorylating and inactivating the proapoptotic protein B-cell
lymphoma 2 antagonist of cell death (15). The PI3K/AKT pathway is involved in
cell growth, proliferation, survival and tumorigenesis (16), and according to a previous study,
the development of RB is associated with activation of the PI3K/AKT
pathway (17). However, there is
currently limited data regarding the role of miR-182 in RB and its
involvement with CADM2 and the PI3K/AKT pathway. Therefore, the
present study aimed to clarify how miR-182 regulates RB, and its
effects on the PI3K/AKT pathway and CADM2.
Materials and methods
Ethics statement
Written informed consent was obtained from patients
and their families prior to the study. The experiment was approved
by the Ethics Committee of Luoyang Central Hospital (Luoyang,
China). The animal studies conducted in the present study were
approved by the Animal Care Committee at Luoyang Central
Hospital.
Clinical sample collection
A total of 22 RB samples were collected from
patients with RB who underwent an ophthalmectomy at Luoyang Central
Hospital between December 2012 and December 2016. Each sample was
cut into 4-mm serial sections, and hematoxylin and eosin staining
was used to verify the diagnosis. The patients enrolled consisted
of 13 males and 9 females, aged between 3 months and 5 years, with
no family history of RB (patients with a family history of RB were
excluded). Of the patients, 17 had mono-ocular RB and 5 had
binocular RB. Following surgery, fresh tumor tissues were
immediately removed under aseptic conditions, fixed in formalin,
automatically dehydrated, infiltrated with wax, embedded and
sectioned. A total of 34 normal retinal tissue sections were
collected from the patients who underwent eye enucleation following
trauma.
Cell culture
Y79 and WERI-Rb-1 human RB cells were obtained from
the cell repository of Shanghai Institute of Biochemistry and Cell
Biology, Shanghai Institutes for Biological Sciences, Chinese
Academy of Sciences (Shanghai, China). Y79 cells in a cryogenic
vial were immediately placed in a water bath at a constant
temperature of 37°C, and the cryogenic vial was stirred to unfreeze
and recover the cells as quickly as possible. The thawed Y79 cells
were extracted with a pipette on a sterilized bench and placed in a
centrifuge tube. Serum-free RPMI-1640 medium (cat. no. PM150110;
Wuhan ProCell Life Science & Technology Co., Ltd., Wuhan,
China) was added to the centrifuge tube, in order to resuspend Y79
cells. The Y79 cells were then centrifuged at 376 × g for 5 min at
4°C, after which, the supernatant was removed and the precipitation
was washed 1-2 times. Subsequently, culture medium containing 15%
fetal bovine serum (FBS, cat. no. 10099-141; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was added and the cells were
incubated at 37°C in an atmosphere containing 5% CO2.
WERI-Rb-1 cells were maintained in Dulbecco’s modified Eagle’s
medium (DMEM) containing 10% FBS, penicillin (100 µg/ml) and
streptomycin (100 µg/ml, Thermo Fisher Scientific, Inc.), at
37°C in an atmosphere containing 5% CO2, and were
conventionally passaged (18).
Cells in the logarithmic growth phase were used for each
experiment.
Dual luciferase reporter assay
A biology prediction website (microRNA.org) was used to analyze the target genes of
miR-182 and to validate whether CADM2 is a target gene of miR-182.
After searching the human target gene sequences in GenBank
(National Institutes of Health, Bethesda, MA, USA; https://www.ncbi.nlm.nih.gov/genbank/),
according to the results of the prediction analysis, the 3′
untranslated region (UTR) sequence of CADM2 was designed, and the
one-step site-directed mutagenesis method (19) was applied to construct plasmid
vectors containing CADM2-3′UTR wild-type (pCADM2-Wt) and
CADM2-3′UTR mutant (pCADM2-Mut) sequences. Briefly, a
double-stranded DNA fragment containing the target sequence of the
wild/mutant 3′ UTR was cloned into the XhoI/NotI site
of the psiCHECK2 luciferase plasmid (cat. no. AY535006; Promega
Corporation, Madison, WI, USA). Subsequently, miR-182 mimic
negative control (NC) and miR-182 mimics (both from Shanghai
GenePharma Co., Ltd, Shanghai, China), at a concentration of 40 nM,
were separately co-transfected with pCADM2-Wt or pCADM2-Mut vectors
at a concentration of 50 nM into Y79 and WERI-Rb-1
(2.5×105 cells/ml) at 37°C for 24 h, according to the
instructions of the Lipofectamine® 2000 kit (cat. no.
11668-027; Invitrogen; Thermo Fisher Scientific, Inc.). The cells
were thus assigned into the following four groups: miR-182 mimic +
pCADM2-Wt, miR-182 mimic + pCADM2-Mut, miR-182 mimic NC + pCADM2-Wt
and miR-182 mimic NC + pCADM2-Mut. The cells were incubated for 6 h
at 37°C in an incubator containing 5% CO2, after which
the medium was removed and replaced with fresh medium. Following a
48-h incubation, the cells were lysed; to each well, 100 µl
passive lysis buffer (cat. no. 99912; Shanghai Open Biotechnology
Co., Ltd., Shanghai, China) was added and the cells were agitated
at low speed for 15 min and stored at 4°C. A dual luciferase
reporter assay was subsequently performed (cat. no. E1910; Inner
Mongolia Hengsheng Biotechnological Co., Ltd., Inner Mongolia,
China) according to the manufacturer’s protocol. Luciferase assay
reagent II (100 µl) was added to 1.5 ml-Eppendorf tubes, and
a luminometer (TD20/20; Turner Designs, San Jose, CA, USA) was used
to visualize the tubes for 2 and 10 sec before the addition of cell
lysate. Subsequently, cell lysate (20 µl) was added to the
Eppendorf tubes and completely mixed before being placed on the
luminometer to measure the activity of firefly luciferase (FLUC).
Subsequently, 100 µl 1X Stop&Glo was added and
completely mixed before being placed on the luminometer to measure
the activity of Renilla luciferase (RLUC). The RLUC/FLUC
ratio was calculated as the relative fluorescence activity, and
applied to determine the target site of miRNA.
Cell transfection and grouping
Y79 and WERI-Rb-1 cells in logarithmic growth phase
were inoculated in a 6-well culture plate at a concentration of
2×105 cells/well. Following adhesion of Y79 and
WERI-Rb-1 cells, and once confluence reached 30-50%, transfection
was performed using the Lipofectamine® 2000 kit (cat.
no. 11668-027; Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer’s protocol. A total of 100 µl
of each sequence was cloned into pcDNA3.1 plasmids (cat. no.
V79020; Thermo Fisher Scientific, Inc.), which were then diluted in
250 µl serum-free RPMI-1640 medium (the sequences were added
to the cells at a final concentration of 50 nM), gently mixed and
incubated at room temperature for 5 min. In addition, 5 µl
Lipofectamine® 2000 was diluted with 250 µl
serum-free RPMI-1640 medium, gently mixed and incubated at room
temperature for 5 min. The two mixtures were then mixed together,
incubated at room temperature for 20 min and added to the cell
culture wells. The cells (2.5×105 cells/ml) were
transfected for 48 h. After transfection, the Y79 and WERI-Rb-1
cells were incubated for 4-6 h at 37°C in an incubator containing
5% CO2 with saturated humidity, and the culture medium
containing the transfection solution was discarded and replaced
with RPMI-1640 containing FBS. The Y79 and WERI-Rb-1 cells were
then incubated for 24-48 h for subsequent experiments. The Y79 and
WERI-Rb-1 cells were assigned into the following seven groups:
Blank group (without transfection), miR-182 mimic group
(transfected with miR-182 mimic plasmid for 24 h; transfection
concentration, 50 nmol/l), mimic NC group (transfected with miR-182
mimic NC plasmid for 24 h; transfection concentration, 50 nmol/l),
miR-182 inhibitor group (transfected with miR-182 inhibitor plasmid
for 24 h; transfection concentration, 50 nmol/l), inhibitor NC
group (transfected with miR-182 inhibitor NC plasmid for 24 h;
transfection concentration, 50 nmol/l), siCADM2 group [transfected
with CADM2 small interfering (si)RNA plasmid, 50 nmol/l] and
miR-182 inhibitor + siCADM2 group (co-transfected with miR-182
inhibitor and CADM2 siRNA plasmid). The pcDNA3.1 plasmid was used
as a vector. The sequences used for transfection were purchased
from Shanghai GenePharma Co., Ltd. (Table I).
 | Table ISequences used for transfection. |
Table I
Sequences used for transfection.
| Plasmid | Sequence |
|---|
| miR-182 mimic | F:
5′-UUUGGCAAUGGUAGAACUCACACU-3′ |
| R:
5′-AGUGUGAGUUCUACCAUUGCCAAA-3′ |
| miR-182 mimic
NC | F:
5′-CCUGGUAAUGGUAGAAUCUACACU-3′ |
| R:
5′-AGUGUGAGUUCUACCAUUGCCAAA-3′ |
| miR-182
inhibitor |
5′-AGUGUGAGUUCUACCAUUGCCAAA-3′ |
| miR-182 inhibitor
NC |
5′-UAAUUCAAAAGACUAAAGGAAUCA-3′ |
| CADM2 siRNA |
5′-TACGTCCAAGGTCGGGCAGGAAGA-3′ |
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Tissues (300 mg) or cells was collected and washed
three times with PBS, after which total RNA was conventionally
extracted using the TRIzol® one-step extraction method
(cat. no. 15596-018; Invitrogen; Thermo Fisher Scientific, Inc.).
The quality of total RNA was assessed and RNA concentration was
adjusted. For the detection of U6 and miR-182, miRNA were reverse
transcribed using stem-loop-mediated RT primers, which were
synthesized by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai,
China). For the detection of genes, RNA was reverse transcribed
into cDNA using the RevertAid First Strand cDNA Synthesis kit
(Thermo Fisher Scientific, Inc.), according to the manufacturer’s
protocol. Briefly, RNA was gently agitated, centrifuged and
incubated at 42°C for 60 min, after which the samples were placed
in a 70°C water bath for 5 min to terminate the RT reaction. The
obtained cDNA was stored at -80°C. The mRNA expression levels of
CADM2, PI3K, AKT, vascular endothelial growth factor (VEGF), Cyclin
E, cyclin-dependent kinase (CDK)2, CDK4, Vimentin, matrix
metalloproteinase (MMP)-9, snail family transcriptional repressor 1
(Snail) and GAPDH were determined based on the extracted RNA.
Random hexamers were used as reverse primers. RT-qPCR was conducted
using the TaqMan probe method (TaqMan-MGB; Shanghai Jianglin
Biological Technology Co., Ltd., Shanghai, China), and the primer
sequences are shown in Table II.
The reaction conditions were as follows: Pre-denaturation at 95°C
for 30 sec, followed by a total of 40 cycles of denaturation at
95°C for 10 sec, annealing at 60°C for 20 sec and extension at 70°C
for 10 sec, and a final extension step at 70°C for 10 min. The
reaction system contained 12.5 µl Premix Ex Taq or
SYBR-Green Mix (cat. no. RR820A; Beijing Think Far Technology Co.,
Ltd., Beijing, China), 1 µl forward primer, 1 µl
reverse primer and 1-4 µl DNA template; ddH2O was
used to make up the final volume of 25 µl. The measurements
were performed using RT-qPCR apparatus (Bio-Rad iQ5; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). U6 was used as the internal
reference gene for miR-182, whereas GAPDH was applied as the
internal reference for the other genes. The 2-ΔΔCq
method (20,21) was employed to calculate the
relative expression levels of miR-182, CADM2, PI3K, AKT, VEGF,
Cyclin E, Vimentin, MMP-9 and Snail. The formula was as follows:
ΔΔCq = ΔCqRB group - ΔCqnormal group, ΔCq =
Cqtarget gene - CqU6/GAPDH. The experiment
was repeated independently three times.
 | Table IIReverse transcription-quantitative
polymerase chain reaction primer sequences. |
Table II
Reverse transcription-quantitative
polymerase chain reaction primer sequences.
| Gene | Primer
sequence |
|---|
| miR-182 | F:
5′-TGCGGTTTGGCAATGGTAGAAC-3′ |
| R:
5′-CCAGTGCAGGGTCCGAGGT-3′ |
| U6 | F:
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ |
| R:
5′-CCAGTGCAGGGTCCGAGGT-3′ |
| CADM2 | F:
5′-AAACCGGTCGCCACCATGATTTG |
| GAAACGCAGC-3′ |
| R:
5′-AAATCGATTAAATGAAATACTCTTTTTTCTC-3′ |
| PI3K | F:
5′-TTTCTCATGGCTGTCCTTCAG-3′ |
| R:
5′-CAGGAGAATCTAACGGATGC-3′ |
| AKT | F:
5′-CATCACATCTGGTTTCCTTGG-3′ |
| R:
5′-AACTGGAAATGTAATTTTGGG-3′ |
| VEGF | F:
5′-CAGCCCCAGCTACCACCTC-3′ |
| R:
5′-TCTTCCTCCTCCCCCTCCTC-3′ |
| CDK2 | F:
5′-CCTTGTTTGTCCCTTCTAC-3′ |
| R:
5′-CAAATCCACCCACTATGA-3′ |
| CDK4 | F:
5′-CATGTAGACCAGGACCTAAGC-3′ |
| R:
5′-AACTGGCGCATCAGATCCTAG-3′ |
| MMP-9 | F:
5′-CACTGTCCACCCCTCAGAGC-3′ |
| R:
5′-GCCACTTGTCGGCGATAAGG-3′ |
| Snail | F:
5′-CTCCTCTACTTCAGCCTCTT-3′ |
| R:
5′-CTTCATCAACGTCCTGTGGG-3′ |
| Vimentin | F:
5′-AAAGTGTGGCTGCCAAGAAC-3′ |
| R:
5′-AGCCTCAGAGAGGTCAGCAA-3′ |
| Cyclin E | F:
5′-GTTCCATTTGCAAAGGCTGT-3′ |
| R:
5′-CAGGTGGTTATCCCGAAAGA-3′ |
| GAPDH | F:
5′-TGGTGAAGGTCGGTGTGAAC-3′ |
| R:
5′-GCTCCTGGAAGATGGTGATGG-3′ |
Western blot analysis
The cells or tissues were collected, and 200
µl pre-cooled radioimmunoprecipitation assay buffer (cat.
no. R0020; Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) and 1 mmol/l phenylmethylsulfonyl fluoride were
added for cell lysis with gentle agitation. The cells were well
mixed using a pipette, and were disrupted for 30 min on ice. The
protein lysates were transferred to a new centrifuge tube and spun
at 2,414 × g for 5 min at 4°C. The proteins in the supernatant were
extracted and protein concentration was measured using the
Bicinchoninic Acid Protein Quantification kit (Wuhan Boster
Biological Technology, Ltd., Wuhan, China), according to the
manufacturer’s protocol. Extracted proteins (30 µg/well)
were subsequently added to sample buffer and boiled at 95°C for 10
min. The proteins were separated by 10% PAGE, after which they were
transferred onto polyvinylidene difluoride membranes (cat. no.
P2438; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) using
semi-dry electro-transfer, and blocked with 5% milk at room
temperature for 1 h. Subsequently, the membranes were incubated
with the following primary antibodies overnight at 4°C:
Rabbit-anti-CADM2 (1:1,000, cat. no. ab64873), rabbit-anti-PI3K
(1:500, cat. no. ab191606), rabbit-anti-AKT (1:500, cat. no.
ab8805), rabbit-anti-VEGF (1:5,000, cat. no. ab53465),
rabbit-anti-Vimentin (1:1,000, cat. no. ab45939), rabbit-anti-MMP-9
(1:1,000, cat. no. ab73734), rabbit-anti-Snail (1:333, cat. no.
ab82846), rabbit-anti-CDK2 (1:1,000, cat. no. ab32147),
rabbit-anti-CDK4 (1:1,000, cat. no. ab108357), rabbit-anti-Cyclin E
(1:1,000, cat. no. ab88259) and rabbit-anti-GAPDH (1:2,500, cat.
no. ab9485), which was used as a control (all from Abcam,
Cambridge, MA, USA). The membranes were washed three times with
Tris-buffered saline with 0.05% Tween-20 (5 min/wash), and a goat
anti-rabbit secondary antibody (1:2,000, cat. no. ab6721; Abcam)
was added and incubated for 1 h at room temperature. The membranes
were then washed a further three times (5 min/wash).
Chemiluminescent reagents (Shanghai Yanhui Biological Technology
Co., Ltd., Shanghai, China) were used for visualization, and GAPDH
served as an internal reference. Bio-Rad Gel Doc EZ imager (Bio-Rad
Laboratories, Inc.) was applied to develop the image, and ImageJ
software v1.8.0 (National Institutes of Health) was employed to
analyze the gray values of the target bands; the relative protein
expression levels were calculated using the ratio of the gray
values of the target band and the internal reference band. The
experiment was repeated independently three times.
MTT assay
After 12 h transfection, Y79 and WERI-Rb-1 cells
were washed with PBS twice, detached using 0.25% trypsin and a
single cell suspension was generated. The cells were counted and
then inoculated into a 96-well plate at a density of
3-6×103 cells/well. The volume in each well was 200
µl, including the blank well, with 6 parallel wells in each
plate. The cells were cultured in an incubator with constant
humidity at 37°C and 5% CO2; the 96-well plate was
removed from the incubator at a fixed time each day. After 24 and
48 h, 20 µl 5 mg/ml MTT solution (cat. no. A2776-1g;
Shanghai Shifeng Biological Technology Co., Ltd., Shanghai, China)
was added to each well. Following a 4-h incubation at 37°C, the
medium was discarded, 150 µl dimethyl sulfoxide was added to
each well and the plates were incubated at room temperature with
gently agitation for 10 min. The optical density (OD) was measured
at 570 nm using an enzyme-linked immunometric meter at 24 and 48 h.
Subsequently, the cell viability curve was generated with time as
the x-axis label and OD value as the y-axis label; cell viability
was compared between the experimental groups (the miR-182 mimic,
mimic NC, miR-182 inhibitor, inhibitor NC, siCADM2 and miR-182
inhibitor + siCADM2 groups) and the blank wells. The experiment was
repeated independently three times.
Transwell assay
Matrigel (cat. no. 40111ES08; Shanghai Yesen
Biological Technology Co., Ltd., Shanghai, China) was dissolved at
4°C overnight and diluted with DMEM at a ratio of 1:3. A total of
30 µl diluted Matrigel was added to the apical chamber of
each Transwell system in three separate additions (15, 7.5 and 7.5
µl) with a 10-min interval between each. The Matrigel was
evenly laid and covered all of the micropores on the bottom of the
apical chamber. After a 48-h transfection, Y79 and WERI-Rb-1 cells
from each group were collected, and a cell suspension at a final
concentration of 1×106 cells/ml was prepared, which was
inoculated into the apical chamber of the Transwell chamber. DMEM
containing 10% FBS was added into the basolateral chamber, and the
cells incubated for 48 h at 37°C and 5% CO2. Swabs were
used to gently remove non-penetrating cells in the apical chamber.
The membrane was then removed, fixed with 95% ethanol for 15-20
min, immersed in water and then stained with 1 mg/l methyl violet
for 10 min at 37°C. The membrane was immersed in water and observed
under high magnification with an inverted microscope. A total of
five high-power fields of view were selected from each sample to
count the cells and calculate the mean values. The number of cells
that invaded through the Matrigel in each group was used as an
index to evaluate their invasive ability. The experiment was
repeated independently three times.
Xenograft tumor model in nude mice
A total of 70 BALB/c nude mice (cat. no. J004;
Nanjing Junke Biological Engineering Co., Ltd., Nanjing, China;
age, 5 weeks; weight, 18-22 g) were housed under
specific-pathogen-free conditions: temperature, 18-22°C; humidity,
50-60%; ammonia concentration, ≤20 ppm; and ventilation rate, 10-20
times/h. The mice were given free access to sterilized food/water
and were maintained under a 10-h light/14-h dark cycle. The nude
mice were randomly allocated into the following seven groups: Blank
group (mouse model of RB was established using untransfected Y79
and WERI-Rb-1 cells), miR-182 mimic group (mice were injected with
Y79 and WERI-Rb-1 cells transfected with miR-182 mimic sequence),
mimic NC group (mice were injected with Y79 and WERI-Rb-1 cells
transfected with miR-182 mimic NC sequence), miR-182 inhibitor
group (mice were injected with Y79 and WERI-Rb-1 cells transfected
with miR-182 inhibitor sequence), inhibitor NC group (mice were
injected with Y79 and WERI-Rb-1 cells transfected with miR-182
inhibitor NC sequence), siCADM2 (mice were injected with Y79 and
WERI-Rb-1 cells transfected with CADM2 siRNA sequence) and miR-182
inhibitor + siCADM2 group (mice were injected with Y79 and
WERI-Rb-1 cells transfected with miR-182 inhibitor and CADM2 siRNA
sequences) (n=10 mice/group; male and female, 1:1). After 24 h
transfection, cells were treated with 0.25% trypsin and a cell
suspension was generated, with the cell density adjusted to
1×105 cells/ml. Local skin disinfection was performed
and 0.5 ml cell suspension was subcutaneously injected into the
right foreleg of each mouse. During injections, the needle was
cautiously inserted under the skin and slowly extracted. Following
extraction of the needle, slight pressure was placed onto the
injection site for 30 sec, in order to prevent leakage of the cell
suspension. The general condition of the nude mice and the local
condition of the inoculation site were observed. All nude mice were
sacrificed under deep anesthesia with pentobarbital sodium (100
mg/kg) 5 weeks post-inoculation, and gross tumor size was observed
and tumor weight was measured.
Immunohistochemistry
Tumor specimens were fixed with neutral-buffered
formalin, dehydrated, embedded in paraffin, serially sectioned at 5
µm and dehydrated. The sections were then treated with 3%
hydrogen peroxide at room temperature for 10 min and underwent
heated antigen retrieval with sodium citrate antigen repair
solution (cat. no. IH0305; Beijing Leagene Biotech Co., Ltd.,
Beijing, China) in a water bath. The subsequent experiment was not
conducted until the buffer was heated to boiling. The sections were
sealed with normal nonim-mune mouse serum (cat. no. GD-MY595J;
Shanghai Guduo Biotechnology Co., Ltd., Shanghai, China) at room
temperature for 3 h. A cluster of differentiation (CD) 34 rabbit
polyclonal primary antibody (1:200, cat. no. ab185732; Abcam) was
added to the sections and incubated at 4°C overnight, followed by
incubation with a biotin-labeled goat anti-rabbit immunoglobulin G
secondary antibody (1:2,000, cat. no. ab6720; Abcam) at 37°C for 20
min. DAB (BIOSS, Beijing, China) was applied for development. The
sections were counterstained with hematoxylin, dehydrated, cleared,
mounted and observed under a confocal microscope. According to CD34
expression, the number of microvessels in each of the 10 fields of
view was counted, and the mean microvessel density (MVD) was
calculated.
Statistical analysis
The experiments were repeated three times. SPSS
software version 21.0 (IBM Corp., Armonk, NY, USA) was used for
data analysis. Measurement data are presented as the means ±
standard deviation. Student’s t-test was applied for comparisons
between two groups. The Kolmogorov-Smirnov method was used to test
normality of data, and one-way analysis of variance and Tukey’s
post hoc test were used for comparisons among multiple groups with
normal distribution. The non-parametric Kruskal-Wallis method was
used to analyze data with skewed distribution, and Dunn’s multiple
comparison post hoc test was performed. P<0.05 was considered to
indicate a statistically significant difference.
Results
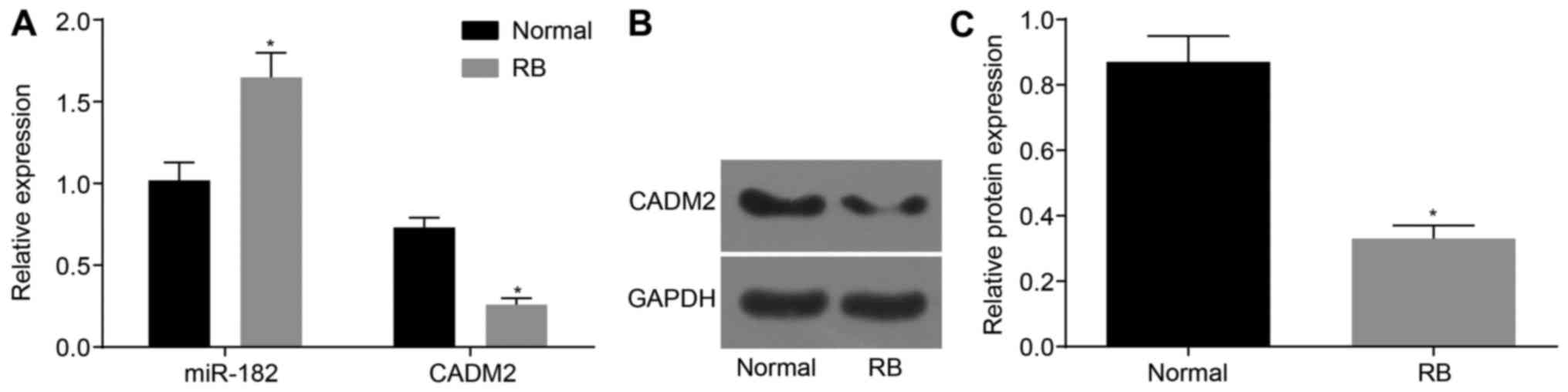
miR-182 is upregulated and CADM2 is
downregulated in RB retinal tissue
The expression levels of CADM2 mRNA and protein and
miR-182 were measured by RT-qPCR and western blot analysis. As
shown in Fig. 1A, miR-182
expression was increased in RB retinal tissues compared with in
normal retinal tissues (P<0.05). In addition, in RB retinal
tissues, the mRNA and protein expression levels of CADM2 were
decreased compared with in normal retinal tissues (P<0.05;
Fig. 1B and C). These results
indicated that the expression levels of miR-182 were upregulated,
whereas CADM2 was downregulated in RB retinal tissue.
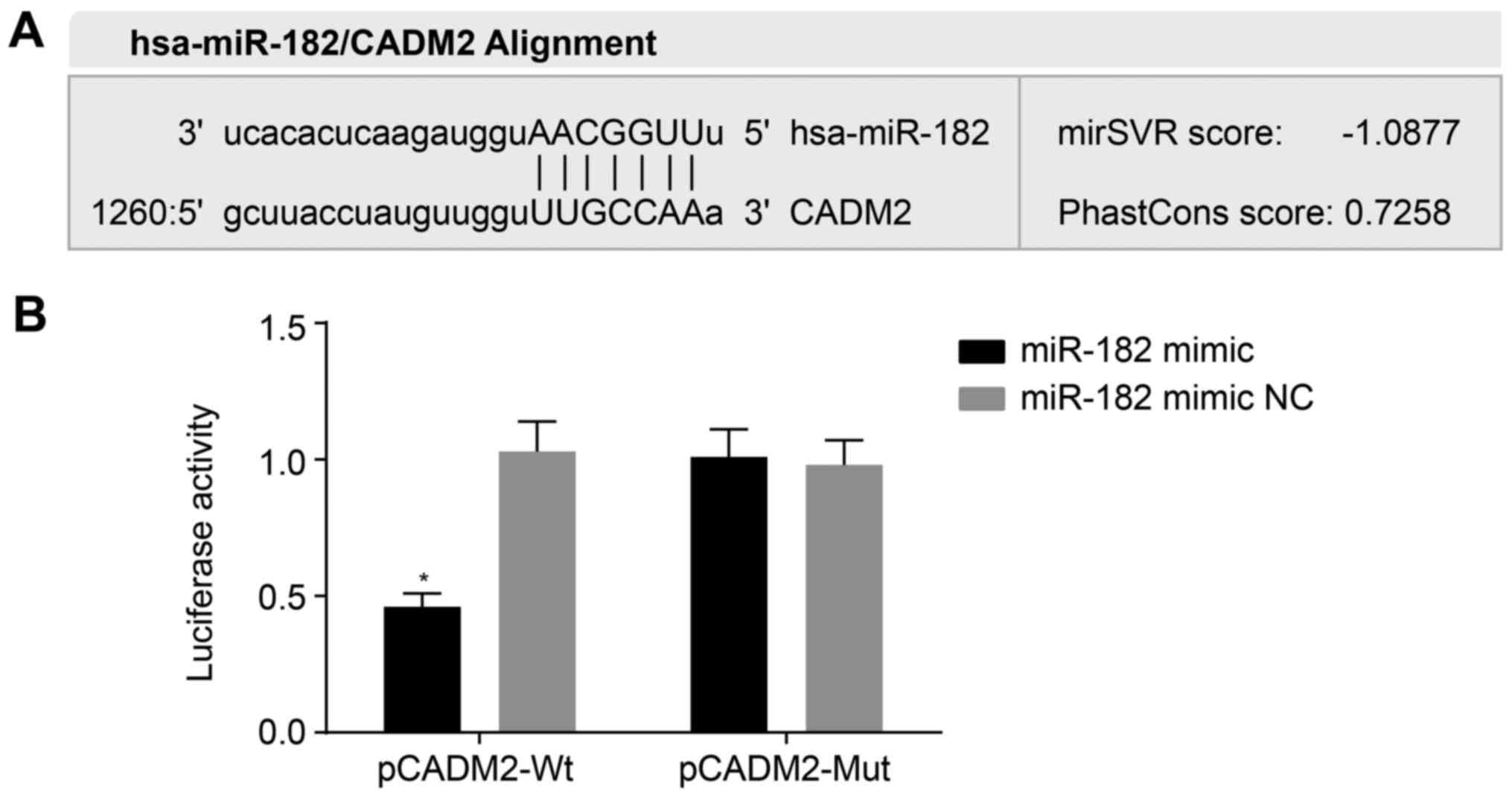
miR-182 binds to the 3′UTR of CADM2
The relationship between miR-182 and CADM2 was
predicted by microRNA.org and confirmed using the
dual luciferase reporter assay. microRNA.org
revealed that miR-182 targeted CADM2 (Fig. 2A). The results of a dual luciferase
reporter assay (Fig. 2B)
demonstrated that the luciferase signal was decreased by ~53% in
the miR-182 mimic + pCADM2-Wt group compared with the miR-182 mimic
NC + pCADM2-Wt group (P<0.05), whereas there were no differences
in the luciferase signal between the miR-182 mimic + pCADM2-Mut and
miR-182 mimic NC + pCADM2-Wt or miR-182 mimic NC + pCADM2-Mut
groups (P>0.05). These results indicated that miR-182 may target
and negatively regulate CADM2 expression.
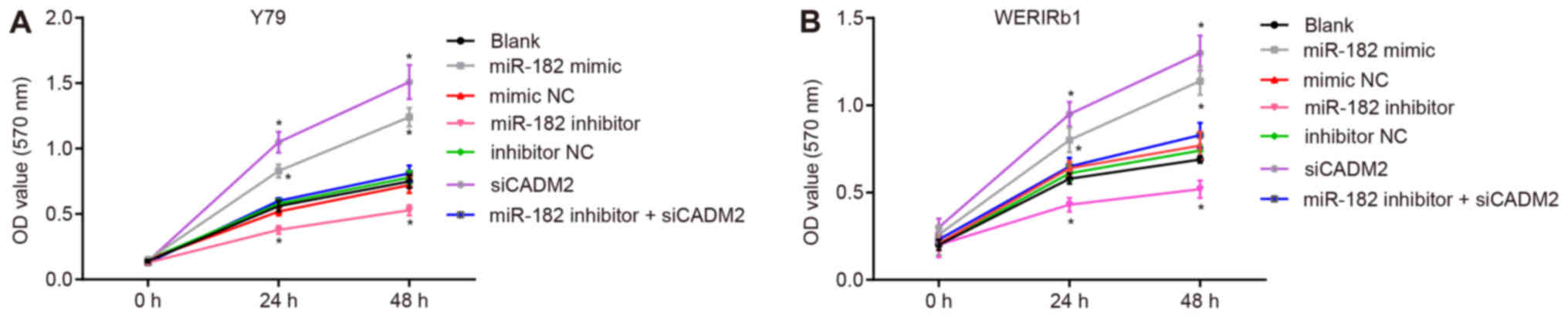
miR-182 overexpression or CADM2 silencing
promotes Y79 and WERI-Rb-1 cell viability
Cell viability was measured using the MTT assay
(Fig. 3). In human RB Y79 and
WERI-Rb-1 cells 24 and 48 h post-transfection, compared with the
blank group, the miR-182 mimic and siCADM2 groups exhibited
significantly elevated cell viability (P<0.05), whereas cell
viability was reduced in the miR-182 inhibitor group (P<0.05).
There was no difference in cell viability among the blank, mimic
NC, inhibitor NC and miR-182 inhibitor + siCADM2 groups
(P>0.05). These findings suggested that cell viability may be
promoted by miR-182 overexpression or CADM2 silencing.
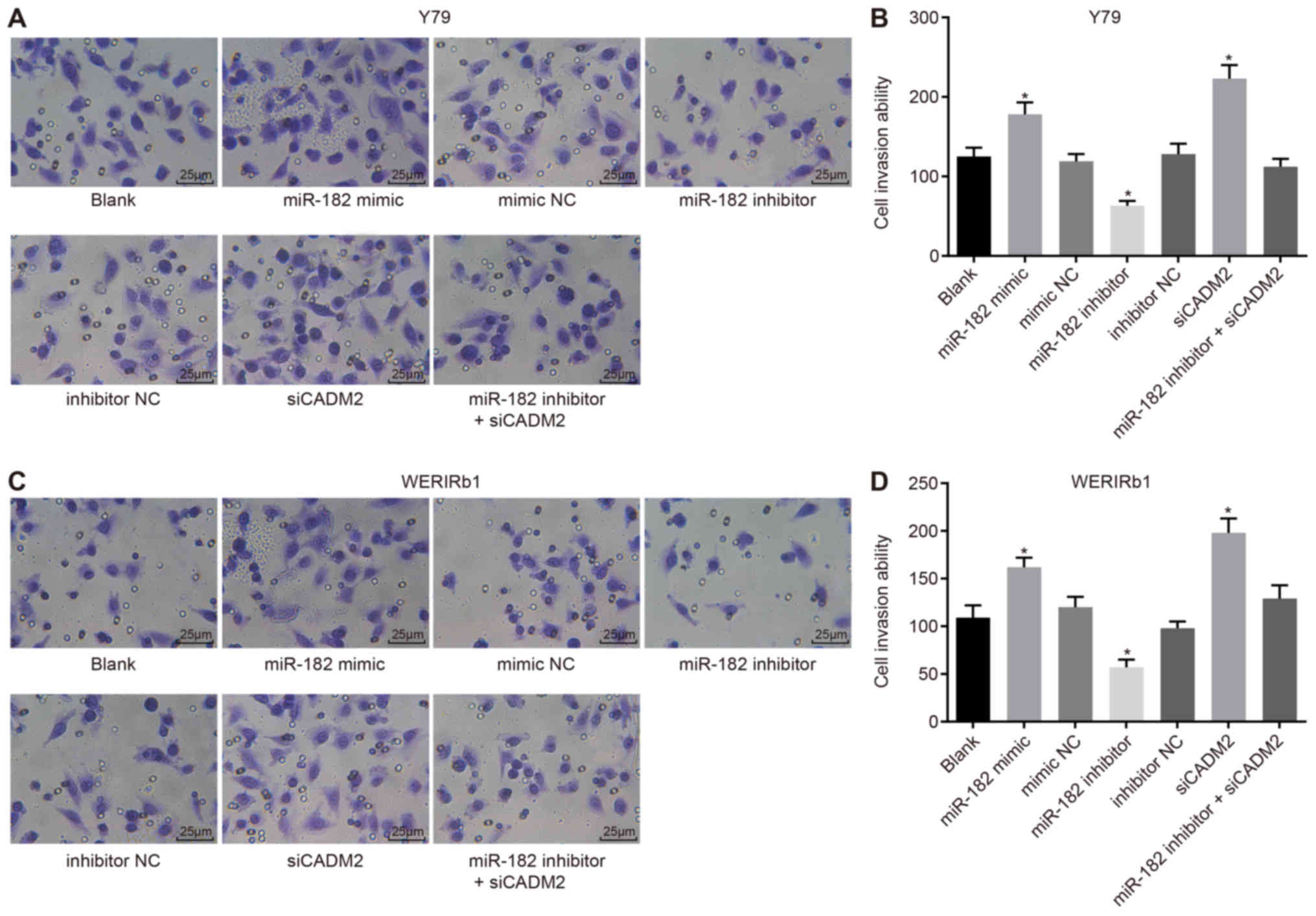
miR-182 overexpression or CADM2 silencing
promotes Y79 and WERI-Rb-1 cell invasion
In order to assess the invasive ability of RB
retinal cells, a Transwell assay was conducted. As shown in
Fig. 4, following 48-h culture of
Y79 and WERI-Rb-1 cells, compared with the blank group, the miR-182
mimic and siCADM2 groups exhibited a significantly elevated number
of invasive cells, whereas the number of invasive cells was reduced
in the miR-182 inhibitor group (P<0.05). There was no difference
in the number of invasive cells among the blank, mimic NC,
inhibitor NC and miR-182 inhibitor + siCADM2 groups (P>0.05).
These findings indicated that cell invasion may be promoted by
miR-182 overexpression or CADM2 silencing.
miR-182 overexpression or CADM2 silencing
increases the mRNA expression levels of PI3K, AKT, VEGF, Cyclin E,
CDK2, CDK4, Vimentin, MMP-9 and Snail
RT-qPCR was used to measure miR-182 expression and
the mRNA expression levels of CADM2, PI3K/AKT pathway-associated
genes (PI3K and AKT), an angiogenesis-associated gene (VEGF),
proliferation-associated genes (Cyclin E, CDK2 and CDK4) and
invasion-associated genes (Vimentin, MMP-9 and Snail) in Y79 and
WERI-Rb-1 cells post-transfection. As shown in Fig. 5, compared with the blank group, in
the miR-182 mimic group, miR-182 expression was significantly
elevated, whereas in the miR-182 inhibitor and miR-182 inhibitor +
siCADM2 groups, miR-182 expression was reduced (P<0.05). There
was no difference in miR-182 expression among the mimic NC,
inhibitor NC and siCADM2 groups (P>0.05). The mRNA expression
levels of CADM2 were significantly decreased in the miR-182 mimic
and siCADM2 groups, whereas the mRNA expression levels of PI3K,
AKT, VEGF, Cyclin E, CDK2, CDK4, Vimentin, MMP-9 and Snail were
increased (P<0.05). In the miR-182 inhibitor group, an increase
in CADM2 mRNA expression was observed, whereas PI3K, AKT, VEGF,
Cyclin E, CDK2, CDK4, Vimentin, MMP-9 and Snail were decreased
(P<0.05). There was no difference in the expression of the
aforementioned genes among the blank, mimic NC, inhibitor NC and
miR-182 inhibitor + siCADM2 groups (P>0.05). These findings
revealed that the PI3K/AKT pathway may be activated in response to
miR-182 overexpression or CADM2 silencing, which may contribute to
enhancement of angiogenesis, viability and invasion.
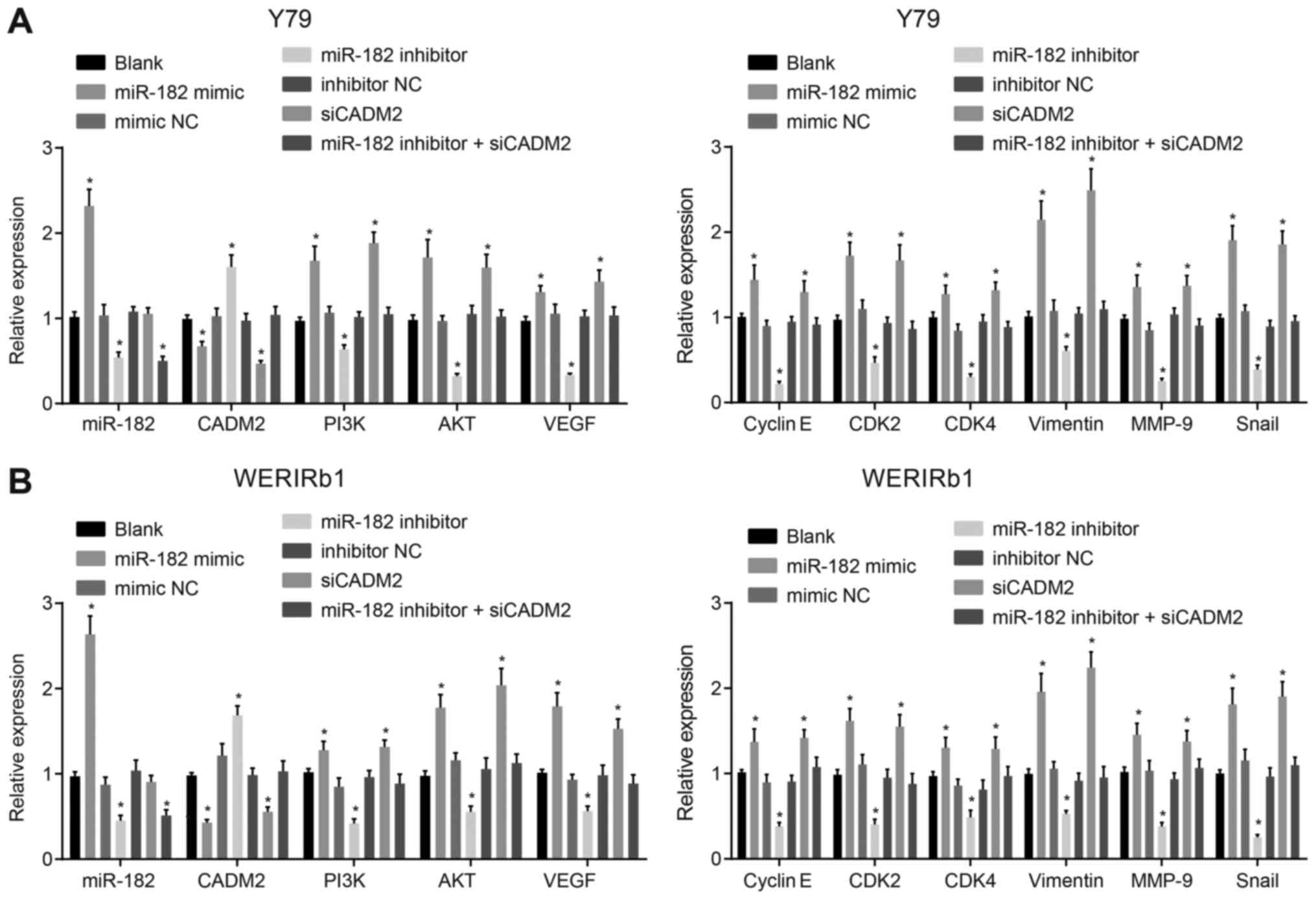 | Figure 5mRNA expression levels of PI3K/AKT
pathway-, angiogenesis-, proliferation- and invasion-associated
genes are increased by miR-182 overexpres-sion or CADM2 silencing,
as determined by reverse transcription-quantitative polymerase
chain reaction. (A) mRNA expression levels of PI3K, AKT, VEGF,
Cyclin E, CDK2, CDK4, Vimentin, MMP-9 and Snail were increased in
Y79 cells in response to miR-182 overexpression or CADM2 silencing.
(B) mRNA expression of PI3K, AKT, VEGF, Cyclin E, CDK2, CDK4,
Vimentin, MMP-9 and Snail was increased in WERI-Rb-1 cells in
response to miR-182 overexpres-sion or CADM2 silencing.
*P<0.05 compared with the blank group. The
measurement data are presented as the means ± standard deviation,
and were analyzed by one-way analysis of variance and Tukey’s post
hoc test. The experiment was repeated independently three times.
AKT, protein kinase B; miR-182, microRNA-182; CADM2, cell adhesion
molecule 2; CDK, cyclin-dependent kinase; MMP-9, matrix
metalloproteinase-9; NC, negative control; PI3K, phosphoinositide
3-OH kinase; siCADM2, CADM2 small interfering RNA; Snail, snail
family transcriptional repressor 1; VEGF, vascular endothelial
growth factor. |
miR-182 overexpression or CADM2 silencing
increases the protein expression levels of PI3K, AKT, VEGF, Cyclin
E, CDK2, CDK4, Vimentin, MMP-9 and Snail
Western blot analysis was conducted on Y79 and
WERI-Rb-1 cells to determine the protein expression levels of PI3K,
AKT, VEGF, Cyclin E, CDK2, CDK4, Vimentin, MMP-9 and Snail. As
shown in Fig. 6, compared with the
blank group, the miR-182 mimic and siCADM2 groups exhibited
significantly reduced expression of CADM2, and elevated PI3K, AKT,
VEGF, Cyclin E, CDK2, CDK4, Vimentin, MMP-9 and Snail expression
(P<0.05). An opposite trend was detected in the miR-182
inhibitor group, which exhibited upregulation of CADM2 protein
expression, and decreased expression of PI3K, AKT, VEGF, Cyclin E,
CDK2, CDK4, Vimentin, MMP-9 and Snail (P<0.05). There was no
difference in the protein expression levels of PI3K and AKT among
the blank, mimic NC, inhibitor NC and miR-182 inhibitor + siCADM2
groups (P>0.05). These findings revealed that the PI3K/AKT
pathway may be activated by miR-182 overexpression or CADM2
silencing, which may contribute to enhancement of angiogenesis,
viability and invasion.
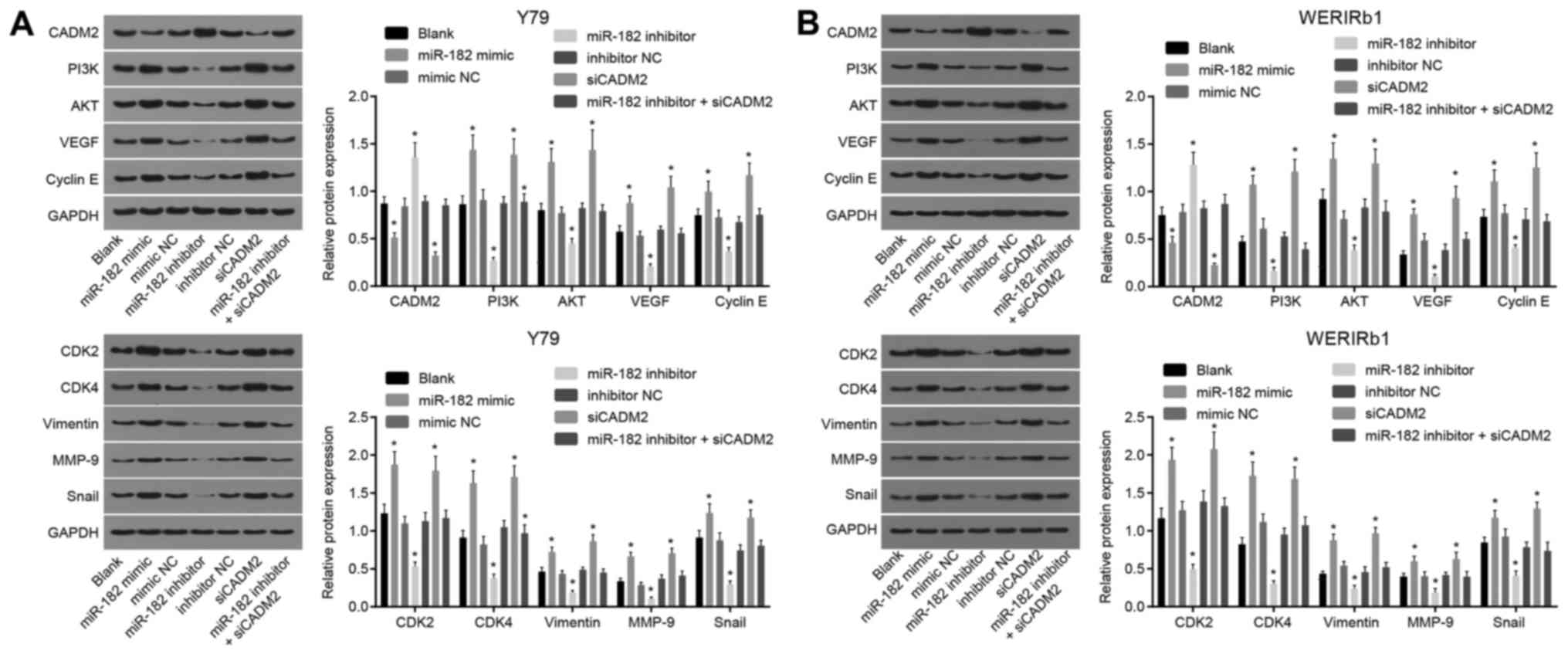 | Figure 6Expression levels of PI3K/AKT
pathway-, angiogenesis-, viability- and invasion-associated
proteins are increased by miR-182 overexpression or CADM2
silencing, as determined by western blot analysis. (A) Protein
expression levels of PI3K, AKT, VEGF, Cyclin E, CDK2, CDK4,
Vimentin, MMP-9 and Snail were increased in Y79 cells by miR-182
overexpression or CADM2 silencing. (B) Protein expression levels of
PI3K, AKT, VEGF, Cyclin E, CDK2, CDK4, Vimentin, MMP-9 and Snail
were increased in WERI-Rb-1 cells by miR-182 overexpression or
CADM2 silencing. *P<0.05, compared with the blank
group. The measurement data are presented as the means ± standard
deviation, and were analyzed by one-way analysis of variance and
Tukey’s post hoc test. The experiment was repeated independently
three times. AKT, protein kinase B; miR-182, microRNA-182; CADM2,
cell adhesion molecule 2; CDK, cyclin-dependent kinase; MMP-9,
matrix metalloproteinase-9; NC, negative control; PI3K,
phosphoinositide 3-OH kinase; siCADM2, CADM2 small interfering RNA;
Snail, snail family transcriptional repressor 1; VEGF, vascular
endothelial growth factor. |
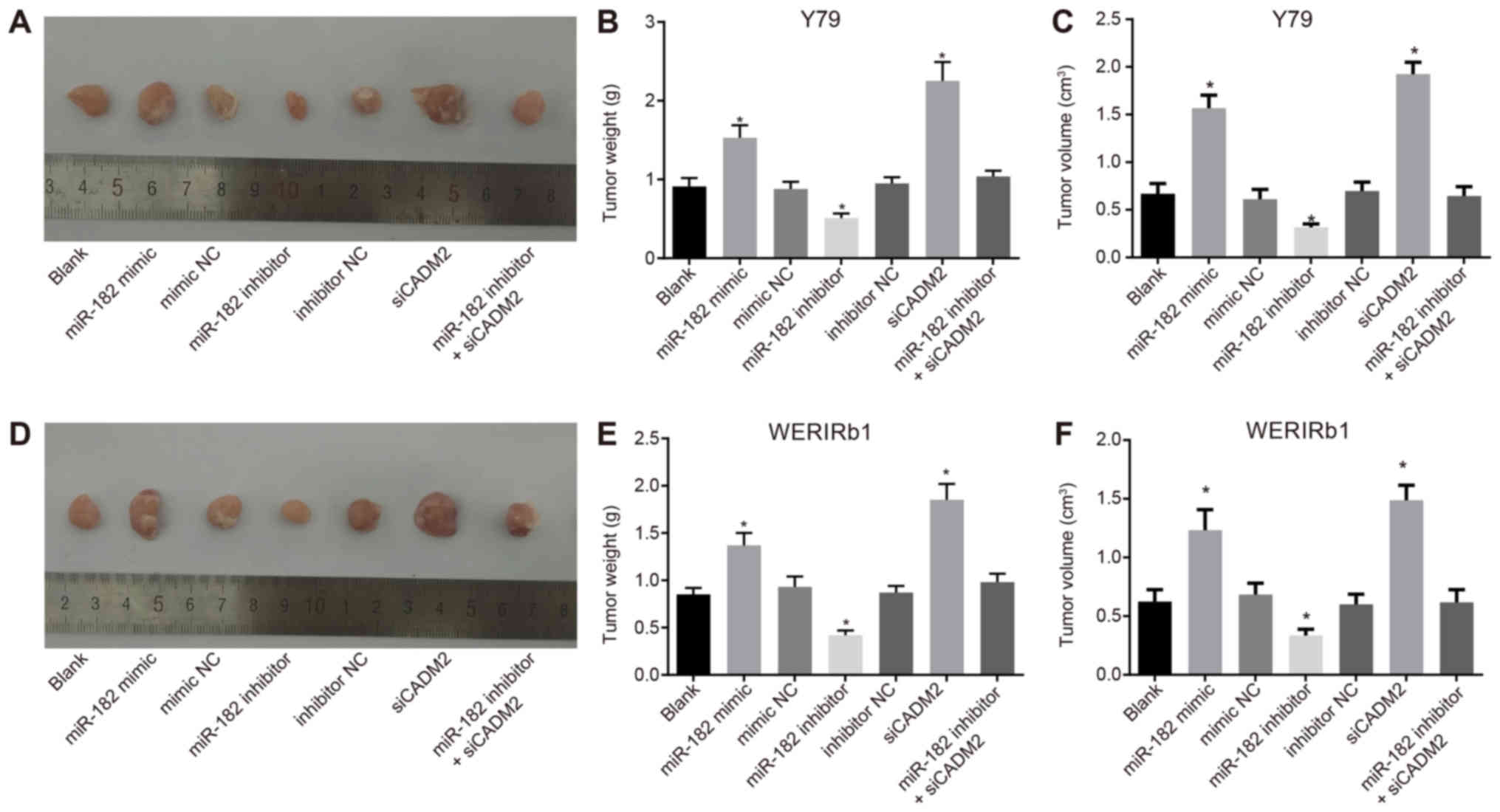
miR-182 overexpression or CADM2 silencing
promotes tumor growth in nude mice inoculated with Y79 and
WERI-Rb-1 cells post-transfection
A xenograft tumor model was generated in nude mice
to analyze whether miR-182 could affect the growth of RB by
regulating CADM2 in vivo. Following a tumor incubation
period of ~7 days, the inoculation site of all nude mice possessed
tumors visible to the naked eye; at the inoculation site,
subcutaneous nodules were oval at the beginning and gradually
became irregular with a lobulated concavo-convex surface. The tumor
formation rate was 100%. After 5 weeks, the nude mice were
sacrificed once ulceration was observed on the tumor surface. The
tumor was removed and weighed, and the tumor volume was calculated.
Compared with the blank group, the miR-182 mimic and siCADM2 groups
exhibited significantly elevated tumor weight and volume, whereas
the miR-182 inhibitor group exhibited reduced tumor weight and
volume (P<0.05). There was no difference in tumor weight and
volume among the blank, mimic NC, inhibitor NC and miR-182
inhibitor + siCADM2 groups (P>0.05) (Fig. 7). This observation revealed that
xenograft tumor growth in nude mice could be promoted by elevating
miR-182 expression.
miR-182 overexpression or CADM2 silencing
increases MVD in nude mice inoculated with Y79 and WERI-Rb-1 cells
post-transfection
Vascular endothelial cells were labeled with
anti-CD34 monoclonal antibodies, and the number of microvessels was
detected using immunohistochemistry. CD34 was localized in the
cytoplasm of vascular endothelial cells and was presented as pale
brown granules. The results demonstrated that, compared with the
blank group, the miR-182 mimic and siCADM2 groups exhibited
significantly elevated MVD, whereas the miR-182 inhibitor group had
reduced MVD (P<0.05; Fig. 8).
There was no difference in MVD among the blank, mimic NC, inhibitor
NC and miR-182 inhibitor + siCADM2 groups (P>0.05). These
results indicated that MVD may be promoted by miR-182
overexpression or CADM2 silencing.
Discussion
A previous study reported that RB has a high
incidence in developing countries (22); in addition, several miRNAs are
differentially expressed in RB (23). However, it is unclear how miR-182
regulates RB. The present study investigated the effects of miR-182
on RB. The findings provided evidence to suggest that inhibiting
miR-182 may activate CADM2 and suppress the PI3K/AKT pathway,
thereby suppressing cell viability, invasion and angiogenesis in
RB.
In the present study, miR-182 was significantly
increased and CADM2 was decreased in RB retinal tissues. It has
been reported that miR-182 is an oncogene (11); miR-182 is upregulated in numerous
types of cancer, including non-small cell lung cancer (24). In addition, a previous study
provided evidence to suggest that miR-182 is associated with
angiogenesis (25). Du et
al (26) observed more
capillary-like structures in a miR-182 mimic group compared with in
a normal group, as determined using an in vitro capillary
tube formation assay. Notably, a previous study demonstrated that
miR-182 is overexpressed in primary open-angle glaucoma, which is
characterized by retinal ganglion cell death (27). CADM2, also known as Nectin-like 3,
SynCAM2 and immunoglobulin superfamily, member 4D, is downregulated
in numerous types of cancer, including ovarian cancer, prostate
cancer, lymphoma and melanoma (28). Consistent with the present results,
Yang et al (29) detected
low expression of CADM2 in hepatocellular carcinoma. The present
study indicated that RB may be associated with miR-182
overexpression and low CADM2 expression. CADM2 is one of the target
genes of miR-182. RT-qPCR and western blot analysis revealed that
Y79 cells transfected with miR-182 inhibitor possessed
significantly decreased expression of PI3K/AKT pathway-associated
genes (PI3K and AKT) and the angiogenesis-associated gene VEGF, as
well as decreased MVD, but increased CADM2 expression. The majority
of cancer types exhibit activation of the PI3K/AKT pathway,
including RB (30). A previous
study confirmed that miR-182 activates the PI3K/AKT pathway
(31), whereas CADM3 inhibits
activation of the PI3K/AKT signaling pathway, and CADM3 can bind to
CADM4 (32). Non-overlapping
expression of CADM2 and CADM3 has been detected in the developing
retina, and binding of CADM2 and CADM3 to CADM1 and CADM4 in
vitro may indicate the functional role of CADMs during nervous
system development (33). VEGF is
an angiogenic growth factor that is involved in the pathogenesis of
retinal diseases, such as diabetic retinopathy (34). A previous study revealed that VEGF
overexpression is associated with RB (35). Furthermore, another study
identified that miR-182 overexpression is associated with VEGF-A
production and angiogenesis (34).
Liu et al (31) suggested
that inhibition of miR-182 decreases the level of
phosphorylated-AKT. MVD is used to measure angiogenesis, and it has
been reported that enhanced angiogenesis appears in
miR-182-overexpressing cells (36,37).
In addition, it has been revealed that miR-182 targets CADMs
(38). These findings indicated
that suppression of miR-182 may activate CADM2 and inhibit the
activation of PI3K/AKT, thus inhibiting angiogenesis in RB.
The present study revealed that miR-182 inhibition
reduced cell viability and invasion by activating CADM2 and
suppressing the PI3K/AKT signaling pathway, in addition to reducing
the expression of proliferation-associated genes (Cyclin E, CDK2
and CDK4) and invasion-associated genes (Vimentin, MMP-9 and
Snail). It is well known that miR-182 acts as an oncomiR, which
promotes cell proliferation (24).
miR-182 overexpression serves a pivotal role in enhancing cell
invasion via inhibition of various targets, such as Forkhead box
O3, programmed cell death 4 and microphthalmia-associated
transcription factor (39). A
recent study revealed that CADM2 overexpression suppresses cancer
cell proliferation and invasion (28). The PI3K/AKT signaling pathway is a
survival pathway essential in apoptosis and cell proliferation
(17); it has also been reported
that the PI3K/AKT signaling pathway contributes to the
proliferation of airway smooth muscle cells (40). In addition, activation of the
PI3K/AKT signaling pathway serves an important role in prostate
cancer cell invasion (41). A
previous study focused on miR-135b, miR-182 and the PI3K/AKT
signaling pathway in colorectal cancer (CRC), identifying that
overexpressed miR-182 promotes the chemoresistance of CRC by
targeting ST6 N-acetylgalactosaminide α-2,6-sialyltransferase 2 via
the PI3K/AKT signaling pathway (42). Furthermore, the ability of miR-182
to activate the PI3K/AKT signaling pathway has previously been
confirmed (31). These findings
indicated that inhibition of miR-182 suppressed cell viability and
invasion via the activation of CADM2 and by suppressing the
PI3K/AKT signaling pathway. Notably, the present findings suggested
that cells transfected with miR-182 inhibitor + siCADM2 exhibited
similar cell viability and invasive abilities as untreated cells.
These findings indicated that CADM2 knockdown may reverse the
suppressive effects of miR-182 inhibition on cell viability and
invasion.
In conclusion, the present study revealed that
downregulation of miR-182 may suppress cell viability, invasion and
angiogenesis in RB by upregulating CADM2 and inhibiting the
PI3K/AKT signaling pathway. Notably, this study indicated that
miR-182 and CADM2 are promising therapeutic targets for RB;
however, further studies are required to develop the practical
clinical application of these therapeutic targets in the treatment
of RB.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors’ contributions
YXH, XGN and GDL designed the study. DSF and LLS
were involved in data collection. YXH, XGN and XLZ analyzed the
data, and reviewed the results and discussion. GDL and DSF
collected and provided the samples for the study, prepared the
manuscript and revised it. All authors had final approval of the
submitted and published versions.
Ethics approval and consent to
participate
Written informed consent was obtained from patients
and their families prior to the study. The experiment was approved
by the Ethics Committee of Luoyang Central Hospital (Luoyang,
China). The animal studies conducted in the present study were
approved by the Animal Care Committee at Luoyang Central
Hospital.
Patient consent for publication
Written informed consent was obtained from patients
and their families prior to the study.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Carvalho IN, Reis AH, Cabello PH and
Vargas FR: Polymorphisms of CDKN1A gene and risk of retinoblastoma.
Carcinogenesis. 34:2774–2777. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wong JR, Tucker MA, Kleinerman RA and
Devesa SS: Retinoblastoma incidence patterns in the US
Surveillance, Epidemiology, and End Results program. JAMA
Ophthalmol. 132:478–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reis AH, Vargas FR and Lemos B: More
epigenetic hits than meets the eye: microRNAs and genes associated
with the tumorigenesis of retinoblastoma. Front Genet. 3:2842012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yun J, Li Y, Xu CT and Pan BR:
Epidemiology and Rb1 gene of retinoblastoma. Int J Ophthalmol.
4:103–109. 2011.PubMed/NCBI
|
|
5
|
Song HB, Park KD and Kim JH, Kim DH, Yu YS
and Kim JH: Tissue factor regulates tumor angiogenesis of
retinoblastoma via the extracellular signal-regulated kinase
pathway. Oncol Rep. 28:2057–2062. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Wang X, Li Z, Liu H and Teng Y:
MicroRNA-183 suppresses retinoblastoma cell growth, invasion and
migration by targeting LRP6. FEBS J. 281:1355–1365. 2014.
View Article : Google Scholar
|
|
7
|
Xu X, Ge S, Jia R, Zhou Y, Song X, Zhang H
and Fan X: Hypoxia-induced miR-181b enhances angiogenesis of
retinoblastoma cells by targeting PDCD10 and GATA6. Oncol Rep.
33:2789–2796. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu SS, Wang YS, Sun YF, Miao LX, Wang J,
Li YS, Liu HY and Liu QL: Plasma microRNA-320, microRNA-let-7e and
microRNA-21 as novel potential biomarkers for the detection of
retinoblastoma. Biomed Rep. 2:424–428. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang L, Mao P, Song L, Wu J, Huang J, Lin
C, Yuan J, Qu L, Cheng SY and Li J: miR-182 as a prognostic marker
for glioma progression and patient survival. Am J Pathol.
177:29–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiang CH, Hou MF and Hung WC:
Up-regulation of miR-182 by β-catenin in breast cancer increases
tumorigenicity and invasiveness by targeting the matrix
metalloproteinase inhibitor RECK. Biochim Biophys Acta.
1830:3067–3076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Z, Liu J, Segura MF, Shao C, Lee P,
Gong Y, Hernando E and Wei JJ: MiR-182 overexpression in
tumourigenesis of high-grade serous ovarian carcinoma. J Pathol.
228:204–215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang M, Xia Z, Wang Y, Huang L, Ma Q,
Chen X, Wang H, Lu B and Guo Y: Generation of a monoclonal antibody
specific to a new candidate tumor suppressor, cell adhesion
molecule 2. Tumour Biol. 35:7415–7422. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kandalam MM, Beta M, Maheswari UK,
Swaminathan S and Krishnakumar S: Oncogenic microRNA 17-92 cluster
is regulated by epithelial cell adhesion molecule and could be a
potential therapeutic target in retinoblastoma. Mol Vis.
18:2279–2287. 2012.PubMed/NCBI
|
|
14
|
Brunet A, Datta SR and Greenberg ME:
Transcription-dependent and -independent control of neuronal
survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol.
11:297–305. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
De Luca A, Maiello MR, D’Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: Role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16(Suppl 2):
S17–S27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu D, Hou P, Liu Z, Wu G and Xing M:
Genetic alterations in the phosphoinositide 3-kinase/Akt signaling
pathway confer sensitivity of thyroid cancer cells to therapeutic
targeting of Akt and mammalian target of rapamycin. Cancer Res.
69:7311–7319. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cohen Y, Merhavi-Shoham E, Avraham-Lubin
BC, Savetsky M, Frenkel S, Pe’er J and Goldenberg-Cohen N: PI3K/Akt
pathway mutations in retinoblastoma. Invest Ophthalmol Vis Sci.
50:5054–5056. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang B, Shen J and Wang J: UNBS5162
inhibits proliferation of human retinoblastoma cells by promoting
cell apoptosis. OncoTargets Ther. 10:5303–5309. 2017. View Article : Google Scholar
|
|
19
|
Qi D and Scholthof KB: A one-step
PCR-based method for rapid and efficient site-directed fragment
deletion, insertion, and substitution mutagenesis. J Virol Methods.
149:85–90. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ayuk SM, Abrahamse H and Houreld NN: The
role of photo-biomodulation on gene expression of cell adhesion
molecules in diabetic wounded fibroblasts in vitro. J Photochem
Photobiol B. 161:368–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Moreno F, Sinaki B, Fandiño A, Dussel V,
Orellana L and Chantada G: A population-based study of
retinoblastoma incidence and survival in Argentine children.
Pediatr Blood Cancer. 61:1610–1615. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Beta M, Khetan V, Chatterjee N,
Suganeswari G, Rishi P, Biswas J and Krishnakumar S: EpCAM
knockdown alters microRNA expression in retinoblastoma--functional
implication of EpCAM regulated miRNA in tumor progression. PLoS
One. 9:e1148002014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ning FL, Wang F, Li ML, Yu ZS, Hao YZ and
Chen SS: MicroRNA-182 modulates chemosensitivity of human non-small
cell lung cancer to cisplatin by targeting PDCD4. Diagn Pathol.
9:1432014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li N, Hwangbo C, Jaba IM, Zhang J,
Papangeli I, Han J, Mikush N, Larrivée B, Eichmann A, Chun HJ, et
al: miR-182 modulates myocardial hypertrophic response induced by
angiogenesis in heart. Sci Rep. 6:212282016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du C, Weng X, Hu W, Lv Z, Xiao H, Ding C,
Gyabaah OA, Xie H, Zhou L, Wu J, et al: Hypoxia-inducible MiR-182
promotes angiogenesis by targeting RASA1 in hepatocellular
carcinoma. J Exp Clin Cancer Res. 34:672015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Bailey JC, Helwa I, Dismuke WM, Cai
J, Drewry M, Brilliant MH, Budenz DL, Christen WG, Chasman DI, et
al: A common variant in MIR182 is associated with primary
open-angle glaucoma in the NEIGHBORHOOD consortium. Invest
Ophthalmol Vis Sci. 57:3974–3981. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He W, Li X, Xu S, Ai J, Gong Y, Gregg JL,
Guan R, Qiu W, Xin D, Gingrich JR, et al: Aberrant methylation and
loss of CADM2 tumor suppressor expression is associated with human
renal cell carcinoma tumor progression. Biochem Biophys Res Commun.
435:526–532. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang S, Yan HL, Tao QF, Yuan SX, Tang GN,
Yang Y, Wang LL, Zhang YL, Sun SH and Zhou WP: Low CADM2 expression
predicts high recurrence risk of hepatocellular carcinoma patients
after hepatectomy. J Cancer Res Clin Oncol. 140:109–116. 2014.
View Article : Google Scholar
|
|
30
|
Mao Y, Xi L, Li Q, Cai Z, Lai Y, Zhang X
and Yu C: Regulation of cell apoptosis and proliferation in
pancreatic cancer through PI3K/Akt pathway via Polo-like kinase 1.
Oncol Rep. 36:49–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y
and Jia L: Upregulation of microRNA-135b and microRNA-182 promotes
chemoresistance of colorectal cancer by targeting ST6GALNAC2 via
PI3K/AKT pathway. Mol Carcinog. 56:2669–2680. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen MS, Kim H, Jagot-Lacoussiere L and
Maurel P: Cadm3 (Necl-1) interferes with the activation of the PI3
kinase/Akt signaling cascade and inhibits Schwann cell myelination
in vitro. Glia. 64:2247–2262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim KH, Lee KY, Lee JB, Yang KS, Hwang J,
Je BK and Park HJ: Radiologic factors related to double-bar
insertion in minimal invasive repair of pectus excavatum. World J
Pediatr. 11:148–153. 2015. View Article : Google Scholar
|
|
34
|
Akiyama H, Tanaka T, Maeno T, Kanai H,
Kimura Y, Kishi S and Kurabayashi M: Induction of VEGF gene
expression by retinoic acid through Sp1-binding sites in
retinoblastoma Y79 cells. Invest Ophthalmol Vis Sci. 43:1367–1374.
2002.PubMed/NCBI
|
|
35
|
Areán C, Orellana ME, Abourbih D, Abreu C,
Pifano I and Burnier MN Jr: Expression of vascular endothelial
growth factor in retinoblastoma. Arch Ophthalmol. 128:223–229.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chiang CH, Chu PY, Hou MF and Hung WC:
MiR-182 promotes proliferation and invasion and elevates the
HIF-1α-VEGF-A axis in breast cancer cells by targeting FBXW7. Am J
Cancer Res. 6:1785–1798. 2016.
|
|
37
|
Foote RL, Weidner N, Harris J, Hammond E,
Lewis JE, Vuong T, Ang KK and Fu KK: Evaluation of tumor
angiogenesis measured with microvessel density (MVD) as a
prognostic indicator in nasopharyngeal carcinoma: Results of RTOG
9505. Int J Radiat Oncol Biol Phys. 61:745–753. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding
F and Gu X: miR-182 inhibits Schwann cell proliferation and
migration by targeting FGF9 and NTM, respectively at an early stage
following sciatic nerve injury. Nucleic Acids Res. 40:10356–10365.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Spitschak A, Meier C, Kowtharapu B,
Engelmann D and Pützer BM: MiR-182 promotes cancer invasion by
linking RET oncogene activated NF-κB to loss of the HES1/Notch1
regulatory circuit. Mol Cancer. 16:242017. View Article : Google Scholar
|
|
40
|
Liu Y, Yang K, Sun X, Fang P, Shi H, Xu J,
Xie M and Li M: MiR-138 suppresses airway smooth muscle cell
proliferation through the PI3K/AKT signaling pathway by targeting
PDK1. Exp Lung Res. 41:363–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shukla S, Maclennan GT, Hartman DJ, Fu P,
Resnick MI and Gupta S: Activation of PI3K-Akt signaling pathway
promotes prostate cancer cell invasion. Int J Cancer.
121:1424–1432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jia L, Luo S, Ren X, Li Y, Hu J, Liu B,
Zhao L, Shan Y and Zhou H: miR-182 and miR-135b mediate the
tumorigenesis and invasiveness of colorectal cancer cells via
targeting ST6GALNAC2 and PI3K/AKT pathway. Dig Dis Sci.
62:3447–3459. 2017. View Article : Google Scholar : PubMed/NCBI
|