Introduction
Lung cancer is the most common cause of
cancer-associated mortality worldwide and >80% of lung cancer
cases are non-small cell lung cancer (NSCLC) (1,2).
Despite significant improvement in NSCLC treatment over previous
decades, the 5-year survival rate remains <20% (3). Chemotherapy is an important component
of the current first-line treatment for patients with NSCLC.
However, chemoresistance remains a major obstacle to the clinical
application of chemotherapeutic drugs. Therefore, investigating the
mechanisms underlying chemoresistance may uncover novel promising
molecules that can be exploited as therapeutic targets.
Long non-coding RNAs (lncRNAs) are a heterogeneous
class of non-coding RNA with a minimum length of 200 nucleotides
and limited protein-coding potential (4,5).
There is increasing evidence that a number of lncRNAs are vital in
tumor development and progression (6-9).
Previous studies have implicated lncRNAs in NSCLC chemotherapy
resistance, cell proliferation and metastasis, although their exact
roles in the pathophysiology of NSCLC remain to be fully elucidated
as their biological and molecular functions are complex (10-12).
A previous study showed that epidermal growth factor receptor
antisense RNA 1 (EGFR-AS1) promotes hepatocellular carcinoma cell
proliferation and invasion by promoting cell cycle progression
(13). EGFR-AS1 also mediates EGFR
addiction and modulates treatment response in squamous cell
carcinoma (14). However, the
expression and functions of EGFR-AS1 in NSCLC remain to be fully
elucidated.
In the present study, it was first identified that
the overex-pression of EGFR-AS1 was associated with a poor
prognosis in patients with NSCLC. Furthermore, it was found that
the increased expression of EGFR-AS1 induced proliferation and
chemoresistance through an EGFR-AS1/microRNA (miR)-223/insulin-like
growth factor 1 receptor (IGF1R) signaling pathway in NSCLC cells.
The data also suggested that plasma EGFR-AS1 may be a promising
biomarker for predicting chemoresistance in patients with
NSCLC.
Materials and methods
Cell lines
The human NLCLC cell lines, A549, NCI-H460,
NCI-H1299 and NCI-H358, were purchased from the American Type
Culture Collection (Manassas, VA, USA), and the HCC827, NCI-H292,
and NCI-H838 cells were purchased from the Chinese Academy of
Sciences (Shanghai, China). All cells were cultured in Dulbecco’s
modified Eagle’s medium (DMEM; HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco, Carlsbad, CA, USA) and maintained in a humidified
incubator at 37°C with 5% CO2.
Clinical samples
Serum, tumor tissues and corresponding adjacent
normal tissues for reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) analysis were randomly collected from 78
patients with NSCLC who underwent curative resection between
January, 2008 and December, 2010 at Jiangxi Chest Hospital
(Nanchang, China) and The First Affiliated Hospital of Nanchang
University (Nanchang, China). In addition, 22 healthy serum
controls were randomly collected from Jiangxi Chest Hospital and
The First Affiliated Hospital of Nanchang University. Retrospective
data were also obtained from 46 patients with advanced recurrent
NSCLC receiving gemcitabine and cisplatin (GP) who had undergone
NSCLC resection 2-60 months prior to the GP therapy; patient
demographics and overall survival (OS) were recorded. Written
informed consent was obtained from all patients and healthy donors,
and the study was approved by the institutional Ethics Review
Committee of Jiangxi Chest Hospital and The First Affiliated
Hospital of Nanchang University.
Immunohistochemistry
The immunohistochemistry procedure for IGF1R was
performed according to a previously described protocol (15). The percentage of IGF1R-positive
cells was scored in five groups: 0 (0%), 1 (1 to ≤25%), 2 (25 to
≤50%), 3 (50 to ≤75%) and 4 (>75%). Groups 0, 1, and 2 groups
were defined as low expression, whereas groups 3 and 4 were defined
as high expression.
Western blot analysis
Western blot analysis was performed as described in
a previous study (16). Cell
lysates were collected and centrifuged for 15 min at 13,200 × g,
4°C. The supernatant was transferred to a clean tube and proteins
concentrations were then quantified using the BCA kit (Pierce,
Rockford, IL, USA). Proteins were then separated on 10% SDS-PAGE
gels and transferred onto nitrocellulose membranes. The membranes
were then blocked with 5% skim milk for 2 h at room temperature and
incubated overnight at 4°C with primary antibodies. Primary
antibodies against IGF1R (ab39675; Abcam, Cambridge, UK), AKT
(ab179463; Abcam), p-AKT (ab176657; Abcam) and tubulin (ab6160;
Abcam) were used. Immune complexes were then detected by incubating
the nitrocellulose membranes with HRP-conjugated goat
anti-mouse/rabbit antibody (ab6789/ab6721; Abcam) for 2 h at room
temperature, followed by exposure of the membrane to enhanced
chemiluminescence reagents (Pierce).
RNA isolation and RT-qPCR analysis
Total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) from
serum, cell lines, frozen tumor tissues and corresponding adjacent
normal tissues according to the manufacturer’s protocol. To detect
the levels of lncRNA and mRNA, 1 µg sample of total RNA was used as
a template for single strand cDNA synthesis performed using random
primers and Primescript reverse transcriptase kit (Takara Bio,
Inc., Otsu, Japan) according to the manufacturer’s instructions.
GAPDH served as the internal control. To detect the levels of
miR-223, we used a MicroRNA Assays kit (Applied Biosystems, Foster
City, CA, USA). miR-223 data was normalized to endogenous U6 small
RNA. The 2-∆∆Cq method was used for calculating the
relative expression levels as reference (17). The primer sequence: EGFR-AS1,
forward, 5′-TCTGT CAGCT CC TTG CACCT C-3′ and reverse, 5′-TGCTC
AGTGT GGTCT GATGT C-3′; GAPDH, forward, 5′-TGTTC GTCAT GGGTG
TGAAC-3′ and reverse, 5′-ATGGC ATGGA CTGTG GTCAT-3′; miR-223,
forward, 5′-AGCTG GTGTT GTGAA TCAGG CCG-3′ and reverse, 5′-TGGTG
TCGTG GAGTC G-3′; U6 forward, 5′-CTCGC TTCGG CAGCA CA-3′ and
reverse, 5′-AACGC TTCAC GAATT TGCGT-3′. RT-qPCR analysis was
performed using a SYBR-Green PCR kit (Takara Bio, Inc.) according
to the manufacturer’s protocol. RT-qPCR was performed as follows:
pre-denaturation at 95°C for 5 min, followed by 40 cycles of
denaturation at 95°C for 10 sec and elongation at 60°C for 30
sec.
Cell proliferation and cisplatin
sensitivity assays
To assess cell proliferation, the cells were seeded
into a 96-well plate at a density of 500 cells per well. At various
time-points (24, 48, 72, 96 and 120 h), CCK-8 solution (Dojindo
Molecular Technologies, Inc., Tokyo, Japan) was added to each well,
and each plate was incubated for 2 h at 37°C with 5%
CO2. The optical density was measured at 450 nm. For the
cisplatin sensitivity assay, the cells were seeded in 96-well
plates at a density of 2,000 cells per well. After 24 h, the cells
were cultured in complete medium containing the indicated
concentrations (0, 2, 4, 6, 8 and 10 µM) of cisplatin or
gemcitabine (0, 10, 20, 30, 40 and 50 µM) (both from Qilu
Pharmaceutical Co., Ltd., Jinan, China) for 72 h at 37°C with 5%
CO2, and the sensitivity of the cells to cisplatin was
measured using a CCK-8 assay.
Luciferase reporter assay
To construct the reporter vector, EGFR-AS1
[wild-type (wt) or miR-223 target mutant (mu), respectively]
amplified and fused to a into the luciferase reporter pGL3
Dual-Luciferase miRNA Target Expression Vector (Invitrogen; Thermo
Fisher Scientific, Inc.). Mutant reporter vectors were performed as
instructions using the QuickMutation kit (Beyotime, Shanghai,
China). The NSCLC NCI-H358 cells were seeded into 24-well plates
and transfected with the different reporter plasmid including
pGL3-wtEGFR-AS1 or pGL3-muEGFR-AS1, together with miR-223 mimics or
negative control using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Forty-eight hours after the transfection,
luciferase activity was then measured using a Dual Luciferase
Reporter Assay system (Promega Corp., Madison, WI, USA) according
to the manufacturer’s protocol.
Cell transfection
The full-length of EGFR-AS1 sequence was transfected
into Trans-OETM plasmid (GenePharma, Shanghai, China) and the empty
Trans-OE™ vector was used as a Mock. The specific short-hairpin RNA
(shRNA) against human EGFR-AS1 was cloned into pENTRTM/U6 plasmid
(GenePharma). A non-targeting shRNA (shNC; GenePharma) was used as
a negative control. Cells (5×104 cells/well) were seeded
into 6-well plates and were transfected with above plasmid using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer’s instructions. The target sequences
for EGFR-AS1 shRNAs was 5′-GACGC AUGAA UGCGA UCUU-3′.
Bioinformatics analysis
The prediction programs (TargetScan and miRGen) were
used to identify potential binding sites with miRNA in the EGFR-AS1
and the target association was found between the EGFR-AS1 and
miR-223 among the results.
Statistical analysis
Experimental data in the present study are presented
as the mean ± standard deviation. Statistical analyses were
performed using the SPSS software package (version 16.0; SPSS Inc.,
Chicago, IL, USA) with Student’s t-test or one-way analysis of
variance. Multiple groups were compared by the one-way ANOVA and
post hoc Dunnett’s test, and data among three groups were compared
by the Kruskal-Wallis test followed by a Mann-Whitney U post hoc
test with Bonferroni’s correction. A χ2 test was used to
analyze the association between the expression of EGFR-AS1 and
clinicopathological parameters. Survival curves were analyzed using
the Kaplan-Meier method and assessed using a log-rank test. The Cox
proportional hazard regression model was applied to identify
independent prognostic factors. The association between the
expression of EGFR-AS1 and mRNA expression of IGF1R in NSCLC
tissues was analyzed using Pearson’s correlation. P<0.05 was
considered to indicate a statistically significant difference.
Results
EGFR-AS1 is upregulated in human NSCLC
tissues and is positively associated with a poor prognosis
The level of EGFR-AS1 was detected in 78 paired
NSCLC tissues and adjacent normal tissues using RT-qPCR analysis.
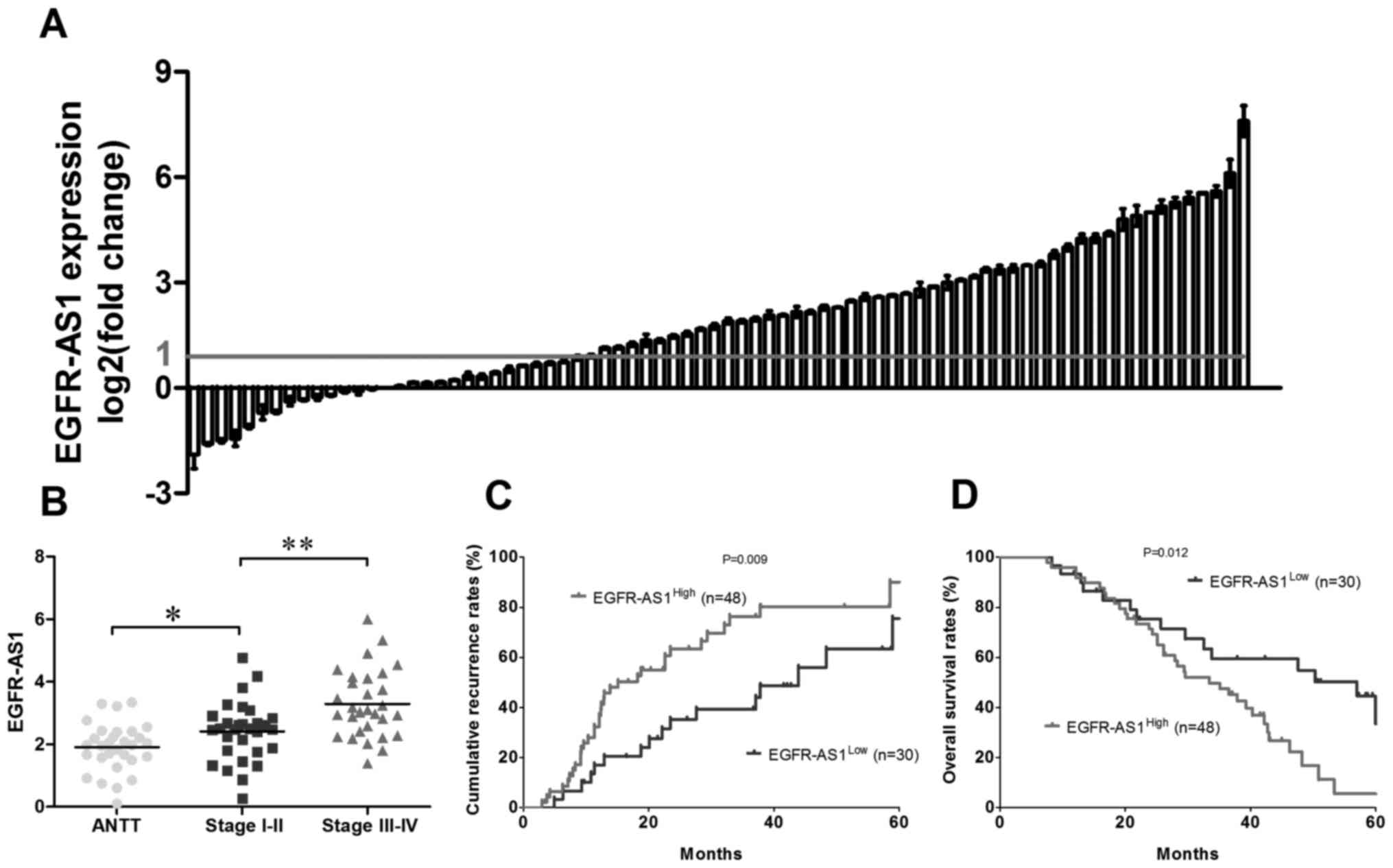
As shown in Fig. 1A, the
expression of EGFR-AS1 was significantly increased in 61.5% (48/78)
of the tumor tissues compared with the adjacent normal tissues.
Furthermore, the expression level of EGFR-AS1 was positively
associated with the clinical stage in patients with NSCLC (Fig. 1B). To further investigate the role
of EGFR-AS1 in NSCLC, the association between EGFR-AS1 and the
clinicopathologic characteristics of patients with NSCLC was
analyzed. As shown in Table I, the
results indicated that the expression of EGFR-AS1 was positively
associated with tumor size (P=0.004) and clinical stage (P=0.016).
For further analysis, the patients were dichotomized into
EGFR-AS1High (n=48) or EGFR-AS1Low (n=30)
groups. Statistically, there was a positive association between the
expression level of EGFR-AS1 and the cumulative recurrence rate
(P=0.009; Fig. 1C) and, to a
lesser extent, the OS rate (P=0.012; Fig. 1D). Multivariate analysis revealed
that the expression of EGFR-AS1 in tumors was an independent
prognostic predictor for cumulative recurrence and OS in patients
with NSCLC (Tables II and
III).
 | Table IAssociation between EGFR-AS1 and
clinicopatho-logical characteristics in 78 patients with non-small
cell lung cancer. |
Table I
Association between EGFR-AS1 and
clinicopatho-logical characteristics in 78 patients with non-small
cell lung cancer.
| Variable | EGFR-AS1
|
|---|
| | |
|---|
| Low | High | P-value |
|---|
| (n=30) | (n=48) | |
| Age (years) | | | |
| ≤50 | 17 | 22 | 0.485 |
| >50 | 13 | 26 | |
| Sex | | | |
| Female | 12 | 14 | 0.337 |
| Male | 18 | 34 | |
| Smoking | | | |
| Yes | 14 | 27 | 0.487 |
| No | 16 | 21 | |
| Tumor size
(diameter, cm) | | | |
| ≤3 | 21 | 13 | 0.004 |
| >3 | 9 | 35 | |
| Histological
differentiation | | | |
| Low and
moderate | 15 | 29 | 0.482 |
| High | 15 | 19 | |
| TNM stage | | | |
| I/II | 17 | 13 | 0.016 |
| III/IV | 13 | 35 | |
| Lymph node
metastasis | | | |
| Yes | 16 | 21 | 0.177 |
| No | 14 | 27 | |
 | Table IIUnivariate and multivariate analyses
of factors associated with cumulative recurrence. |
Table II
Univariate and multivariate analyses
of factors associated with cumulative recurrence.
| Factor | Cumulative
recurrence
|
|---|
| Univariate
P-value | Multivariate
|
|---|
| HR | 95% CI | P-value |
|---|
| Age (≤50, vs.
>50 years) | 0.736 | | | NA |
| Sex (female, vs.
male) | 0.544 | | | NA |
| Smoking (yes, vs.
no) | 0.213 | | | NA |
| Tumor size
(diameter; >3 vs. ≤3 cm) | 0.147 | | | NA |
| Histological
differentiation (low and moderate, vs. high) | 0.068 | | | NA |
| TNM stage (I/II,
vs. III/IV) | 0.026 | 1.746 | 1.327-2.468 | NS |
| Lymph node
metastasis (yes, vs. no) | 0.074 | | | NA |
| EGFR-AS1 expression
(high, vs. low) | 0.008 | 1.542 | 1.071-1.927 | 0.027 |
 | Table IIIUnivariate and multivariate analyses
of factors associated with OS rate. |
Table III
Univariate and multivariate analyses
of factors associated with OS rate.
| Factor | OS rate
|
|---|
| Univariate
P-value | Multivariate
|
|---|
| HR | 95% CI | P-value |
|---|
| Age (≤50, vs.
>50 years) | 0.319 | | | NA |
| Sex (female, vs.
male) | 0.525 | | | NA |
| Smoking (yes, vs.
no) | 0.713 | | | NA |
| Tumor size
(diameter; >3 vs. ≤3 cm) | 0.014 | 1.732 | 1.517-3.242 | NS |
| Histological
differentiation (low and moderate, vs. high) | 0.079 | | | NA |
| TNM stage (I/II,
vs. III/IV) | 0.029 | 1.922 | 1.055-2.913 | NS |
| Lymph node
metastasis (yes, vs. no) | 0.014 | 1.375 | 1.071-2.415 | NS |
| EGFR-AS1 expression
(high, vs. low) | 0.008 | 1.218 | 0.922-2.073 | 0.031 |
Expression of EGFR-AS1 is inversely
associated with the sensitivity of NSCLC to cisplatin and
gemcitabine-based therapy
Subsequently, the present study analyzed
retrospective data from 46 patients with advanced recurrent NSCLC
receiving gemcitabine and cisplatin (GP) who had undergone NSCLC
resection 2-60 months prior to the GP therapy; patient demographics
were recorded (Table IV). The
expression levels of EGFR-AS1 were then measured using RT-qPCR
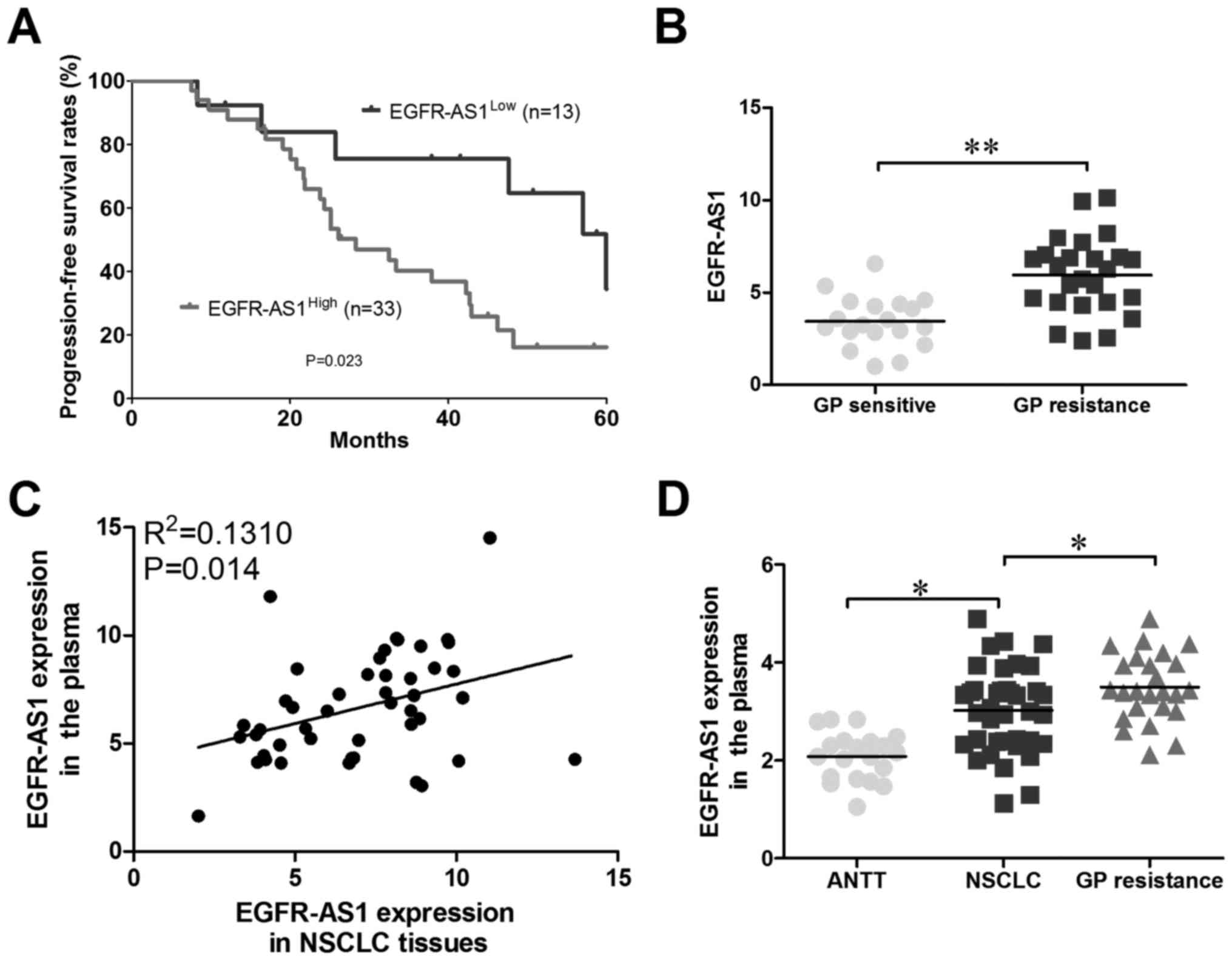
analysis, and Kaplan-Meier survival analysis revealed that the OS
for the EGFR-AS1high group was lower than that for the
EGFR-AS1Low group (Fig. 2A
and B); therefore, it was hypothesized that high expression
levels of EGFR-AS1 lead to NSCLC GP resistance. To examine whether
EGFR-AS1 was present in patient plasma, RNA was extracted from the
plasma of 46 patients with NSCLC. As shown in Fig. 2C, EGFR-AS1 was detectable in the
plasma and positively correlated with that in NSCLC tissues.
Compared with healthy donors, the plasma levels of EGFR-AS1 were
increased in patients with NSCLC, particularly in GP-resistant
patient plasma (Fig. 2D).
Together, these results suggested that plasma EGFR-AS1 may be a
promising non-invasive biomarker for predicting the response to GP
therapy in patients with NSCLC.
 | Table IVDemographic and baseline
characteristics of the patients (gemcitabine and cisplatin-treated
population). |
Table IV
Demographic and baseline
characteristics of the patients (gemcitabine and cisplatin-treated
population).
| Variable |
EGFR-AS1Low (n=13) |
EGFR-AS1High (n=33) |
|---|
| Age (years) | 49.13±5.19 | 52.44 ± 7.71 |
| Sex | | |
| Male | 8 | 20 |
| Female | 5 | 13 |
| Smoking | | |
| Yes | 7 | 22 |
| No | 6 | 11 |
| Tumor size
(diameter, cm) | | |
| >3 | 7 | 16 |
| ≤3 | 6 | 17 |
| Histological
differentiation | | |
| Low and
moderate | 11 | 23 |
| High | 2 | 10 |
| TNM stage | | |
| I/II | 8 | 18 |
| III/IV | 5 | 15 |
| Lymph node
metastasis | | |
| Yes | 9 | 24 |
| No | 4 | 9 |
Enforced expression of EGFR-AS1 promotes
proliferation and chemoresistance in lung cancer cells
To further determine the biological roles of
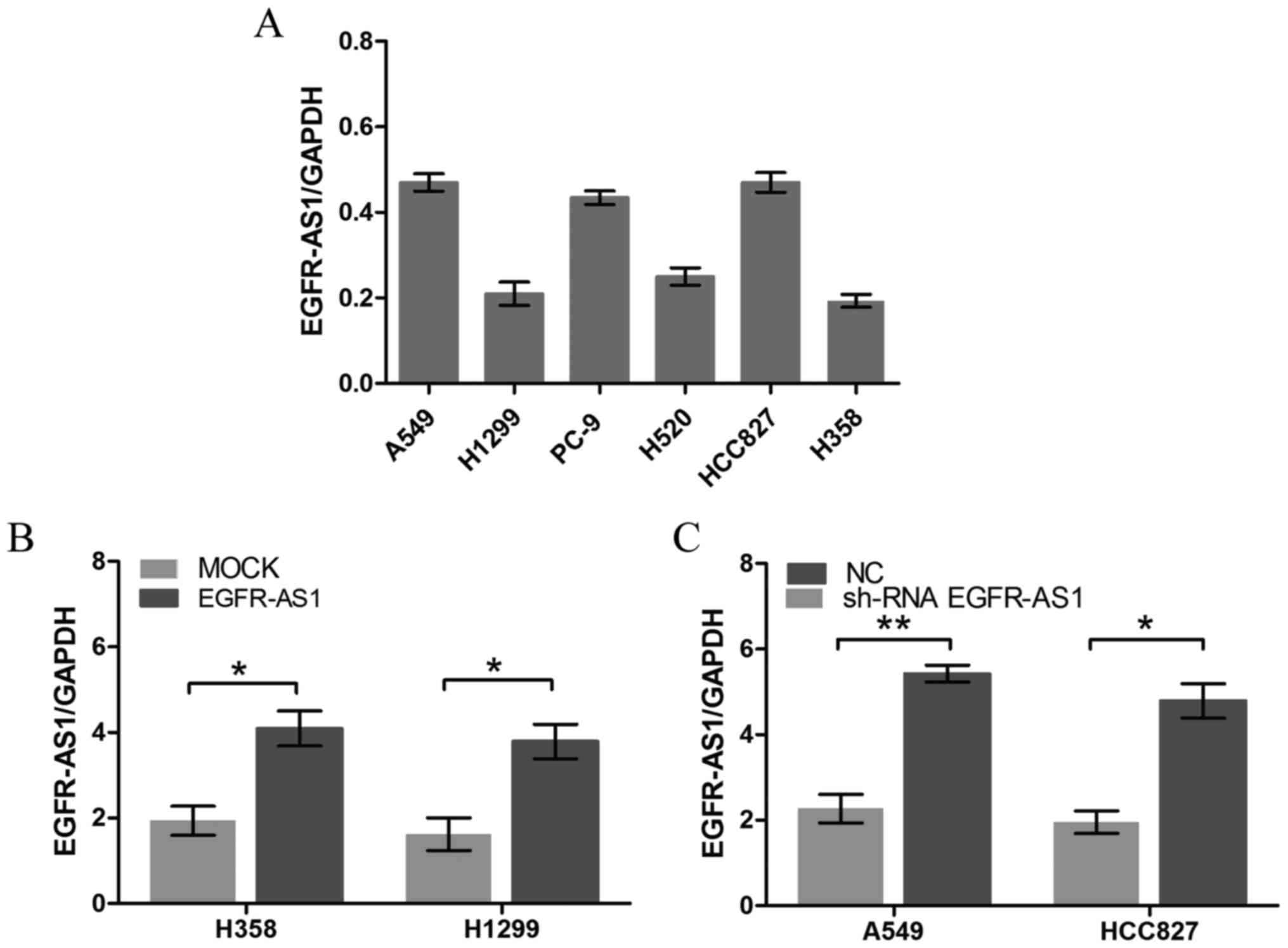
EGFR-AS1 in NSCLC, RT-qPCR analysis was performed to detect the
relative expression levels of EGFR-AS1 in different NSCLC cell
lines. The A549 and HCC827 cells expressed relatively high levels
of EGFR-AS1, whereas the NCI-H1299 and NCI-H358 cells expressed
relatively low levels of EGFR-AS1 (Fig. 3A). NSCLC cell lines with stably
enforced or reduced expression of EGFR-AS1 were established
(Fig. 3B and C). A cell
proliferation assay revealed that the enforced expression of
EGFR-AS1 significantly increased cell proliferation, whereas the
reduced expression of EGFR-AS1 inhibited cell proliferation
(Fig. 4A-D). Furthermore, the
enforced expression of EGFR-AS1 significantly promoted NSCLC cell
resistance to gemcitabine and cisplatin, whereas reduced EGFR-AS1
increased the sensitivity of NSCLC cells to gemcitabine and
cisplatin (Fig. 4E-H).
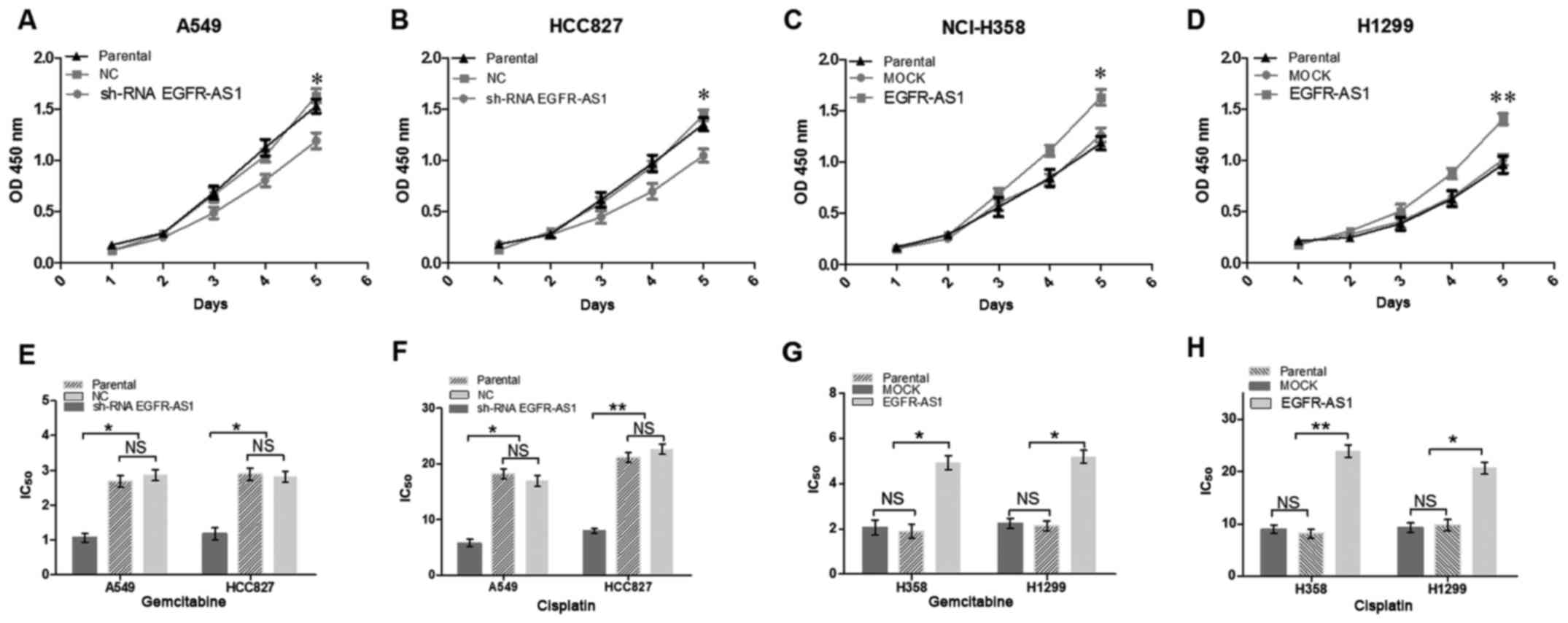 | Figure 4EGFR-AS1 promotes proliferation and
chemoresistance of NSCLC cells in vitro. NSCLC cell lines
infected with EGFR-AS1 shRNA, EGFR-AS1 or control expression
lentivirus were used. Growth of (A) A549, (B) HCC827, (C) NCI-H358,
and (D) H1299 NSCLC cells was examined with CCK-8 assays. Knockdown
of the expression of EGFR-AS1 in (E and F) A549 and HCC827 cells
increased their sensitivity to gemcitabine and cisplatin. However,
the enforced expression of EGFR-AS1 in (G and H) NCI-H358 and H1299
NSCLC cells reduced their sensitivity to gemcitabine and cisplatin.
Data are presented as the mean ± standard deviation (n=3).
*P<0.05; **P<0.01. NSCLC, non-small
cell lung cancer; EGFR-AS1, epidermal growth factor receptor
antisense RNA 1; shRNA, short hairpin RNA; NC, negative control;
NS, not significant. |
EGFR-AS1 functions as a competitive
endogenous RNA (ceRNA) for miR-223 to facilitate the expression of
IGF1R
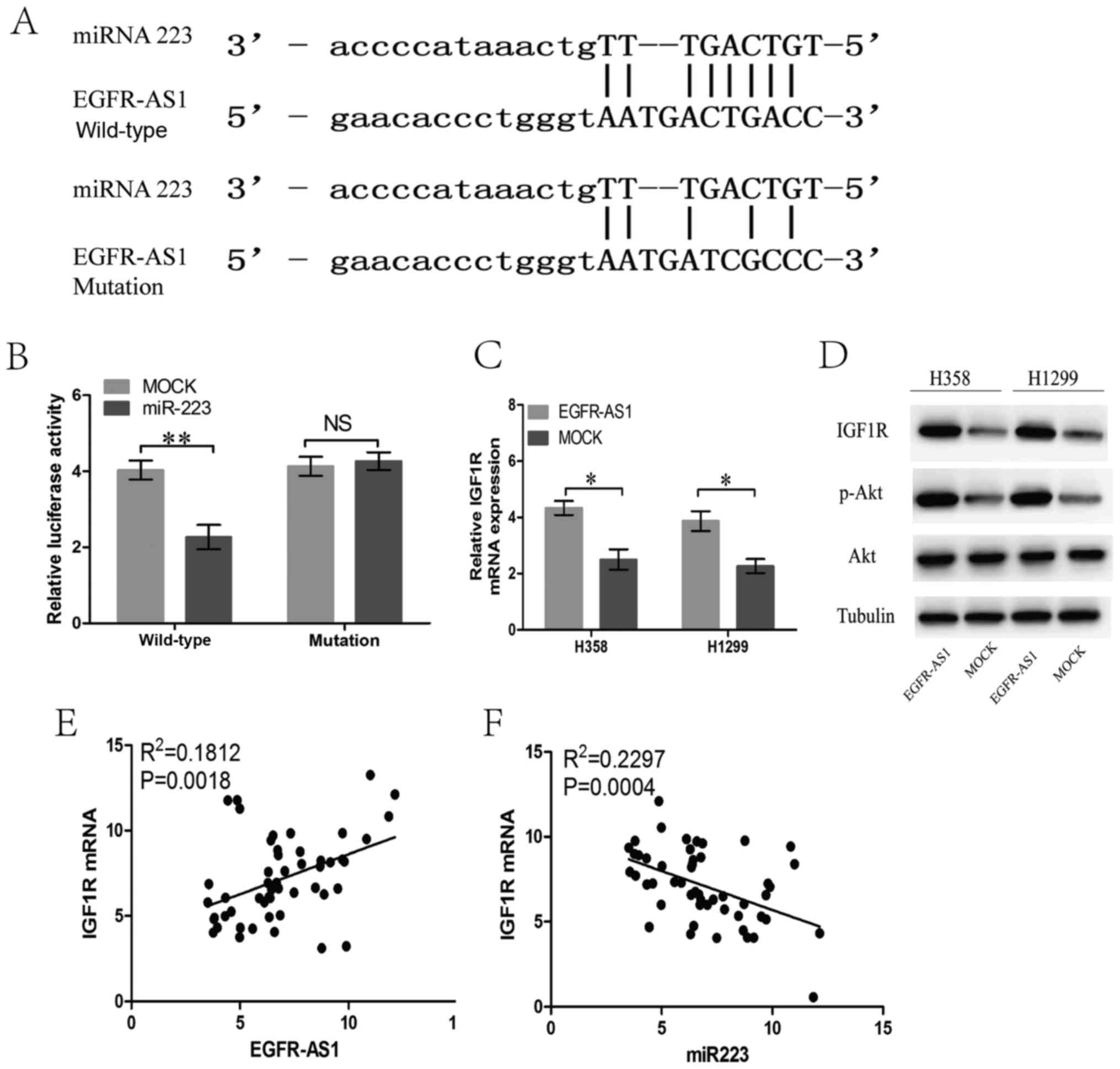
The bioinformatics analysis revealed that the
EGFR-AS1 transcript contains a potential binding site for miR-223
(Fig. 5A), which is downregulated
in several types of cancer, suggesting that EGFR-AS1 may act as a
ceRNA in NSCLC cells. To confirm whether EGFR-AS1 is a ceRNA of
miR-223 in NSCLC, luciferase reporter assays were performed
following the transfection of NSCLC NCI-H358 cells containing a
synthetic miR-223 mimic with the wild-type or mutated miR-223
target sequence of EGFR-AS1. The results showed that miR-223
reduced the luciferase activity in cells trans-fected with
wild-type EGFR-AS1 but had no effect on the luciferase activity in
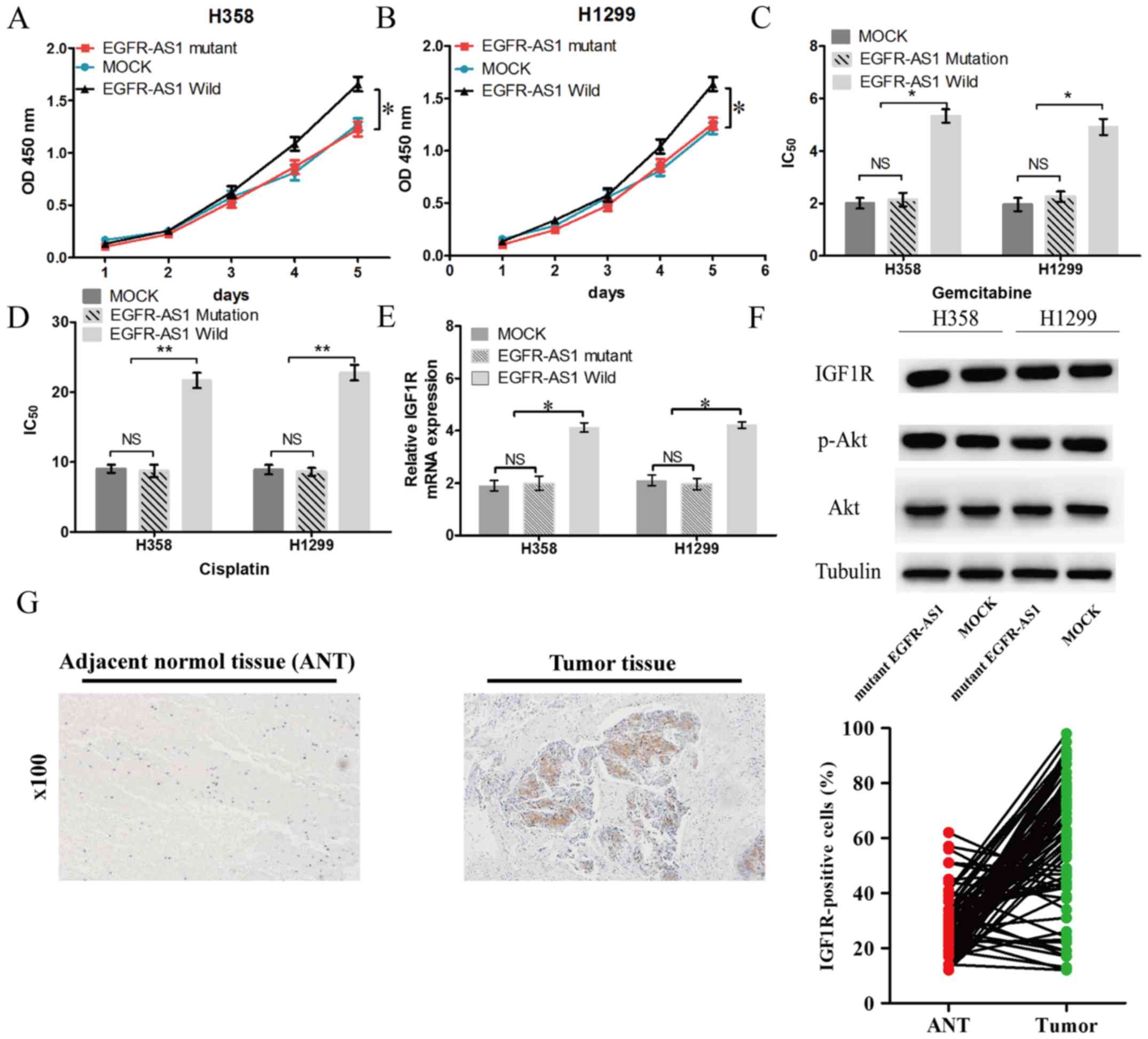
cells transfected with mutant EGFR-AS1 (Fig. 5B). In previous studies, IGF1R was
verified as a target gene of miR-223 in NSCLC cells (18,19).
In the present study, EGFR-AS1 elevated the mRNA and protein
expression of IGF1R, leading to the upregulation of the activity of
AKT (Fig. 5C and D). Furthermore,
the mRNA expression of IGF1R in human NSCLC tissues was positively
correlated with the level of EGFR-AS1 (Fig. 5E). The expression of miR-223 in
human NSCLC tissues was negatively correlated with the level of
IGF1R (Fig. 5F).
EGFR-AS1 mediates NSCLC proliferation and
resistance to cisplatin and gemcitabine through a conserved miR-223
target sequence
To further determine the biological roles of
EGFR-AS1 in NSCLC, NCI-H1299 and NCI-H358 cell lines with stable
forced expression of the miR-223 target sequence EGFR-AS1 mutant
were established. A cell proliferation assay revealed that forced
miR-223 target sequence mutant EGFR-AS1 expression neither
increased cell proliferation nor promoted NSCLC cell resistance to
cisplatin and gemcitabine (Fig.
6A-D). Additionally, mutant EGFR-AS1 did not elevate the mRNA
or protein expression of IGF1R in NSCLC cells (Fig. 6E and F). Subsequently, the
expression of IGF1R was investigated in NSCLC tissues. It was found
that the expression of IGF1R was significantly increased in 66.7%
(52/78) of tumor tissues compared with adjacent normal tissues
(Fig. 6G).
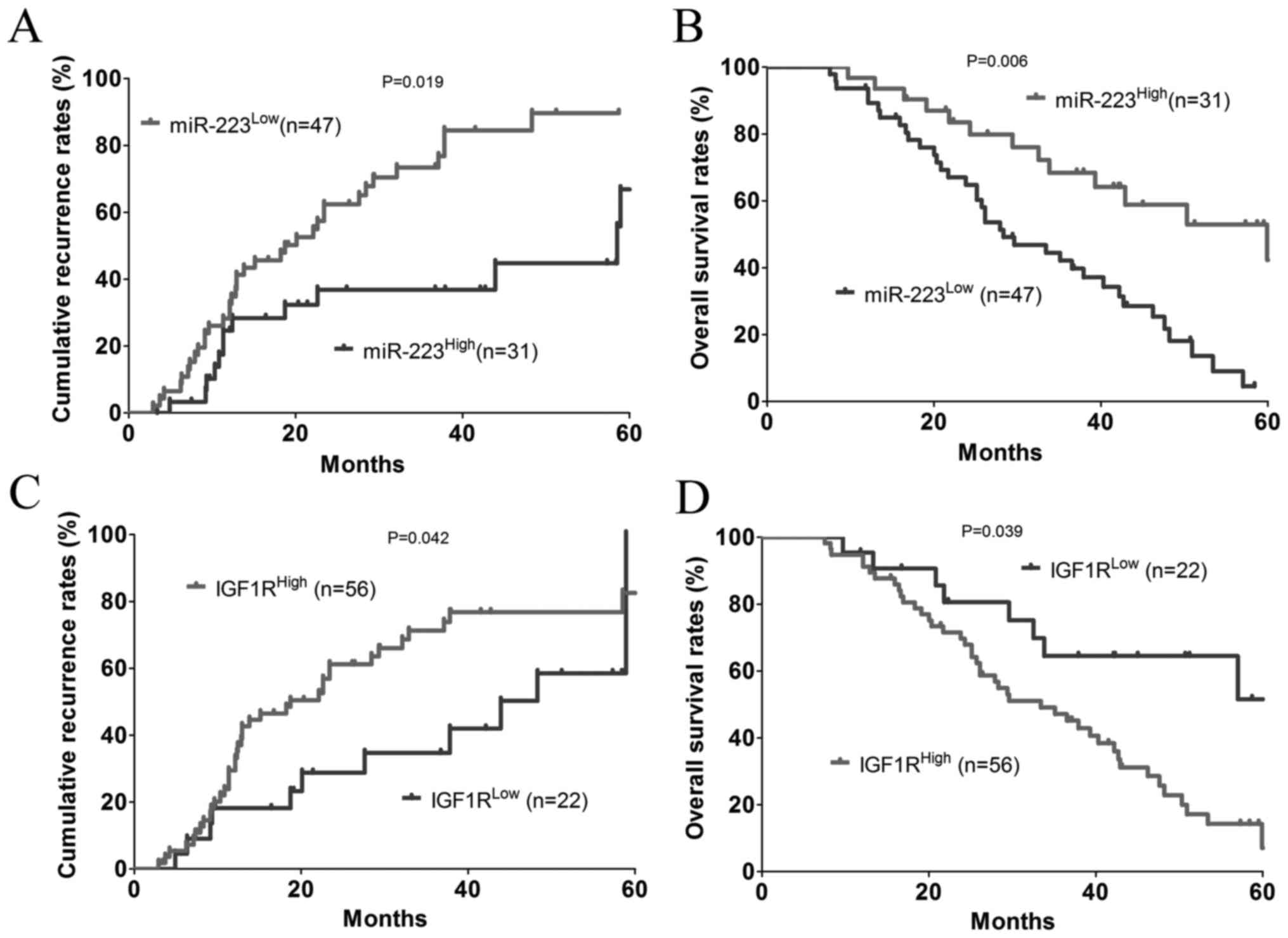
Expression of miR-223 and IGF1R are
associated with a prognosis of patients with CRC
The patients were dichotomized into a
miR-223High (n=31) or a miR-223Low (n=47)
group. The results showed that there was a significant negative
association between the expression of miR-223 and cumulative
recurrence rate (P=0.019; Fig. 7A)
and, to a greater extent, the OS rate (P=0.006; Fig. 7B). In addition, the patients were
dichotomized into a IGF1RHigh (n=56) or
IGF1RLow (n=22) group. Statistically, there was a
significant positive association between the expression of IGF1R
and the cumulative recurrence rate (P=0.039; Fig. 7C) and, to a lesser extent, the OS
rate (P=0.042; Fig. 7D).
Discussion
lncRNAs are a novel class of potential therapeutic
targets for several complex diseases, including cancer. In the
present study, the lncRNA EGFR-AS1 was identified in NSCLC and it
was found to be significantly upregulated in NSCLC tissues and
patient plasma using a RT-qPCR assay, indicating the potential
oncogenic function of EGFR-AS1 in NSCLC. EGFR-AS1 was found to be a
ceRNA of miR-223, and there was an interactive inhibition between
them. EGFR-AS1 acts as an oncogene to promote tumor cell
proliferation and chemo-resistance in NSCLC, and this activity can
be attributed to the direct inhibition of miR-223 and subsequent
activation of the IGF1R/AKT/PI3K signaling pathway. In the present
study, it was found that the expression of EGFR-AS1 was
significantly higher in larger tumors and at advanced stages of
tumor development. The study also revealed a positive association
between the expression levels of EGFR-AS1 and poor NSCLC prognosis
or chemotherapy resistance.
Previous studies have verified that lncRNAs are
dysregulated in several types of human cancer, including NSCLC
(6-8). Understanding the molecular mechanism
of lncRNAs in human cancer may reveal novel potential therapeutic
targets for the treatment of human NSCLC. EGFR-AS1 is an lncRNA
that was initially characterized in epithelial ovarian cancer
(20). The forced expression of
EGFR-AS1 promoted cell proliferation in vitro, whereas the
knockdown of EGFR-AS1 inhibited cell proliferation via the
regulation of cell cycle progression (21). However, the molecular mechanism by
which EGFR-AS1 exerts its oncogenic functions requires further
investigation. Mechanistically, the present study confirmed the
direct binding sequence of the predicted miR-223 binding site on
EGFR-AS1 via bioinformatics analysis and luciferase reporter
assays.
Previous studies have shown that miR-223 is an
important tumor-suppressive miRNA, and was significantly decreased
in the serum of patients with osteosarcoma and osteosarcoma cancer
cells compared with healthy controls (22). Furthermore, the downregulation of
miR-223 can induce activation of the IGF1R/AKT/PI3K signaling
pathway in NSCLC cells and is responsible for the resistance of
NSCLC cells to erlotinib, suggesting that miR-223 is a potential
molecule for overcoming EGFR-tyrosine kinase inhibitor resistance
(18,19).
To investigate whether the EGFR-AS1-induced
inhibition of miR-223 results in derepression of its target mRNA
and promotes cell proliferation and chemoresistance in NSCLC, the
present study focused on the miR-223 target gene IGF1R for further
investigations. IGF1R is a transmembrane glycoprotein that is
important in a number of biological functions (23). Previous studies have demonstrated
that IGF1R is vital in cancer development and progression (24-26).
IGF1R can be used as a novel marker for targeted therapy for
several types of tumor, including glioma, colorectal cancer and
breast cancer (27-29). Several previous studies have
revealed a significant association between the expression level of
IGF1R and NSCLC carcinogenicity, which supports the critical role
of IGF1R in tumor epithelial-mesenchymal transition and
chemotherapy resistance (30,31).
In the present study, the novel EGFR-AS1/miR-223/IGF1R/AKT/PI3K
signaling pathway regulatory network was further examined, in which
EGFR-AS1 was found to act as a ceRNA to repress the biological
function of miR-223, resulting in increased expression of IGF1R and
activation of the IGF1R/AKT/PI3K signaling pathway in NSCLC.
Therefore, these findings provide novel insight into the
pathogenesis of NSCLC and provide a promising target for the
treatment of NSCLC.
Collectively, the findings obtained in the present
study indicate that EGFR-AS1 is an oncogenic lncRNA that promotes
NSCLC cell proliferation and chemotherapy resistance through the
miR-223/IGF1R/AKT axis. EGFR-AS1 is an independent prognostic
factor in NSCLC, and current evidence suggests that plasma EGFR-AS1
may be a promising biomarker for predicting chemoresistance in
patients with NSCLC.
Funding
No funding was received.
Availability of data and materials
The datasets generated during the study are
available from the corresponding author on reasonable request.
Authors’ contributions
SGX and SBT conceived and designed the experiments.
YHX and JRT performed the experiments. TTZ analyzed the data. SGX
and SBT wrote and modified the manuscript. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and healthy donors, and the study was approved by the
institutional Ethics Review Committee of Jiangxi Chest Hospital and
The First Affiliated Hospital of Nanchang University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
NSCLC
|
non-small cell lung cancer
|
|
EGFR-AS1
|
epidermal growth factor receptor
antisense RNA 1
|
|
PRC2
|
proteasome component 2
|
|
LSD1
|
lysine demethylase 1A
|
|
DNMT1
|
DNA methyltransferase 1
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
ceRNA
|
competitive endogenous RNA
|
|
IGF1R
|
insulin-like growth factor 1
receptor
|
|
PI3K
|
phosphatidylinositol 3-kinase
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buyukcelik A, Yalcin B and Utkan G:
Multidisciplinary management of lung cancer. N Engl J Med.
350:2008–2010; author reply 2008–2010 2004. PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guttman M, Russell P, Ingolia NT, Weissman
JS and Lander ES: Ribosome profiling provides evidence that large
noncoding RNAs do not encode proteins. Cell. 154:240–251. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu Q, Deng F, Qin Y, Zhao Z, Wu Z, Xing Z,
Ji A and Wang QJ: Long non-coding RNA regulation of
epithelial-mesenchymal transition in cancer metastasis. Cell Death
Dis. 7:e22542016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li W, Sun M, Zang C, Ma P, He J, Zhang M,
Huang Z, Ding Y and Shu Y: Upregulated long non-coding RNA
AGAP2-AS1 represses LATS2 and KLF2 expression through interacting
with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death
Dis. 7:e22252016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun X, Du P, Yuan W, Du Z, Yu M, Yu X and
Hu T: Long non-coding RNA HOTAIR regulates cyclin J via inhibition
of microRNA-205 expression in bladder cancer. Cell Death Dis.
6:e19072015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng N, Cai W, Ren S, Li X, Wang Q, Pan
H, Zhao M, Li J, Zhang Y, Zhao C, et al: Long non-coding RNA UCA1
induces non-T790M acquired resistance to EGFR-TKIs by activating
the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer.
Oncotarget. 6:23582–23593. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang NS, Chi YY, Xue JY, Liu MY, Huang S,
Mo M, Zhou SL and Wu J: Long non-coding RNA metastasis associated
in lung adenocarcinoma transcript 1 (MALAT1) interacts with
estrogen receptor and predicted poor survival in breast cancer.
Oncotarget. 7:37957–37965. 2016.PubMed/NCBI
|
|
13
|
Qi HL, Li CS, Qian CW, Xiao YS, Yuan YF,
Liu QY and Liu ZS: The long noncoding RNA, EGFR-AS1, a target of
GHR, increases the expression of EGFR in hepatocellular carcinoma.
Tumour Biol. 37:1079–1089. 2016. View Article : Google Scholar
|
|
14
|
Tan DSW, Chong FT, Leong HS, Toh SY, Lau
DP, Kwang XL, Zhang X, Sundaram GM, Tan GS, Chang MM, et al: Long
noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor
addiction and modulates treatment response in squamous cell
carcinoma. Nat Med. 23:1167–1175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kang Q, Cai JB, Dong RZ, Liu LX, Zhang C,
Zhang PF, Zou H, Xie N, Zhang L, Zhang XY, et al: Mortalin promotes
cell proliferation and epithelial mesenchymal transition of
intrahepatic cholangiocarcinoma cells in vitro. J Clin Pathol.
70:677–683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang PF, Li KS, Shen YH, Gao PT, Dong ZR,
Cai JB, Zhang C, Huang XY, Tian MX, Hu ZQ, et al: Galectin-1
induces hepato-cellular carcinoma EMT and sorafenib resistance by
activating FAK/PI3K/AKT signaling. Cell Death Dis. 7:e22012016.
View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Zhao FY, Han J, Chen XW, Wang J, Wang XD,
Sun JG and Chen ZT: miR-223 enhances the sensitivity of non-small
cell lung cancer cells to erlotinib by targeting the insulin-like
growth factor-1 receptor. Int J Mol Med. 38:183–191. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han J, Zhao F, Zhang J, Zhu H, Ma H, Li X,
Peng L, Sun J and Chen Z: miR-223 reverses the resistance of
EGFR-TKIs through IGF1R/PI3K/Akt signaling pathway. Int J Oncol.
48:1855–1867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Richards EJ, Permuth-Wey J, Li Y, Chen YA,
Coppola D, Reid BM, Lin HY, Teer JK, Berchuck A, Birrer MJ, et al:
A functional variant in HOXA11-AS, a novel long non-coding RNA,
inhibits the oncogenic phenotype of epithelial ovarian cancer.
Oncotarget. 6:34745–34757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Q, Zhang J, Liu Y, Zhang W, Zhou J,
Duan R, Pu P, Kang C and Han L: A novel cell cycle-associated
lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA
transcript and is a biomarker of progression in glioma. Cancer
Lett. 373:251–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dong J, Liu Y, Liao W, Liu R, Shi P and
Wang L: miRNA-223 is a potential diagnostic and prognostic marker
for osteosarcoma. J Bone Oncol. 5:74–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Werner H and Sarfstein R: Transcriptional
and epigenetic control of IGF1R gene expression: Implications in
metabolism and cancer. Growth Horm IGF Res. 24:112–118. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ball MW, Bezerra SM, Chaux A, Faraj SF,
Gonzalez-Roibon N, Munari E, Sharma R, Bivalacqua TJ, Netto GJ and
Burnett AL: Overexpression of insulin-like growth factor-1 receptor
is associated wth penile cancer progression. Urology. 92:51–56.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park E, Park SY, Kim H, Sun PL, Jin Y, Cho
SK, Kim K, Lee CT and Chung JH: Membranous insulin-like growth
factor-1 receptor (IGF1R) expression is predictive of poor
prognosis in patients with epidermal growth factor receptor
(EGFR)-mutant lung adenocarcinoma. J Pathol Transl Med. 49:382–388.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rota LM, Albanito L, Shin ME, Goyeneche
CL, Shushanov S, Gallagher EJ, LeRoith D, Lazzarino DA and Wood TL:
IGF1R inhibition in mammary epithelia promotes canonical Wnt
signaling and Wnt1-driven tumors. Cancer Res. 74:5668–5679. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Q, Zhang J, Cui Q, Li X, Gao G, Wang
Y, Xu Y and Gao X: GSK1904529A, an insulin-like growth factor-1
receptor inhibitor, inhibits glioma tumor growth, induces apoptosis
and inhibits migration. Mol Med Rep. 12:3381–3385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Puzanov I, Lindsay CR, Goff L, Sosman J,
Gilbert J, Berlin J, Poondru S, Simantov R, Gedrich R, Stephens A,
et al: A phase I study of continuous oral dosing of OSI-906, a dual
inhibitor of insulin-like growth factor-1 and insulin receptors, in
patients with advanced solid tumors. Clin Cancer Res. 21:701–711.
2015. View Article : Google Scholar
|
|
29
|
Di Cosimo S, Sathyanarayanan S, Bendell
JC, Cervantes A, Stein MN, Braña I, Roda D, Haines BB, Zhang T,
Winter CG, et al: Combination of the mTOR inhibitor ridaforolimus
and the anti-IGF1R monoclonal antibody dalotuzumab: Preclinical
characterization and phase I clinical trial. Clin Cancer Res.
21:49–59. 2015. View Article : Google Scholar
|
|
30
|
Nurwidya F, Takahashi F, Kobayashi I,
Murakami A, Kato M, Minakata K, Nara T, Hashimoto M, Yagishita S,
Baskoro H, et al: Treatment with insulin-like growth factor 1
receptor inhibitor reverses hypoxia-induced epithelial-mesenchymal
transition in non-small cell lung cancer. Biochem Biophys Res
Commun. 455:332–338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park JH, Choi YJ, Kim SY, Lee JE, Sung KJ,
Park S, Kim WS, Song JS, Choi CM, Sung YH, et al: Activation of the
IGF1R pathway potentially mediates acquired resistance to
mutant-selective 3rd-generation EGF receptor tyrosine kinase
inhibitors in advanced non-small cell lung cancer. Oncotarget.
7:22005–22015. 2016.PubMed/NCBI
|