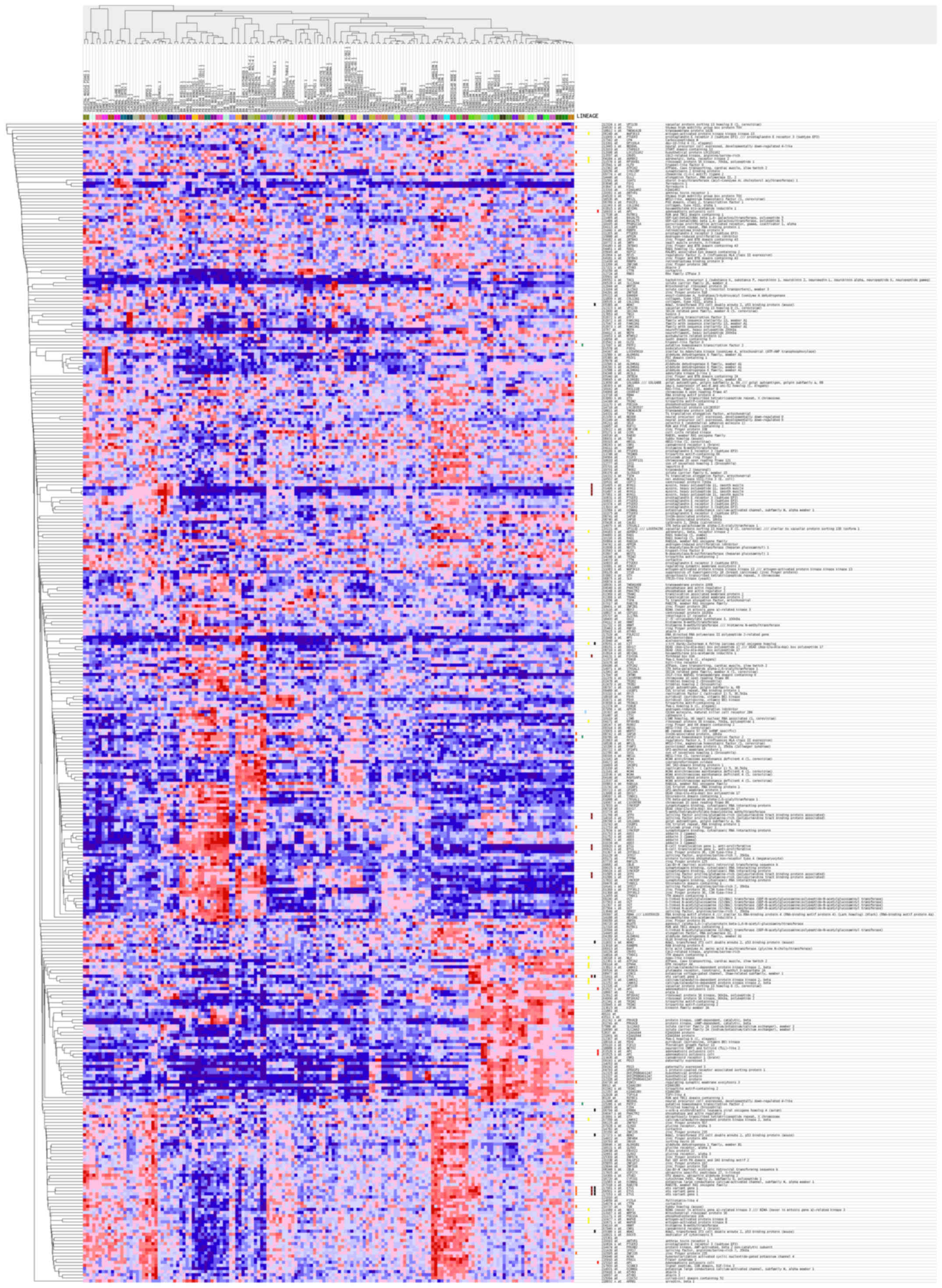
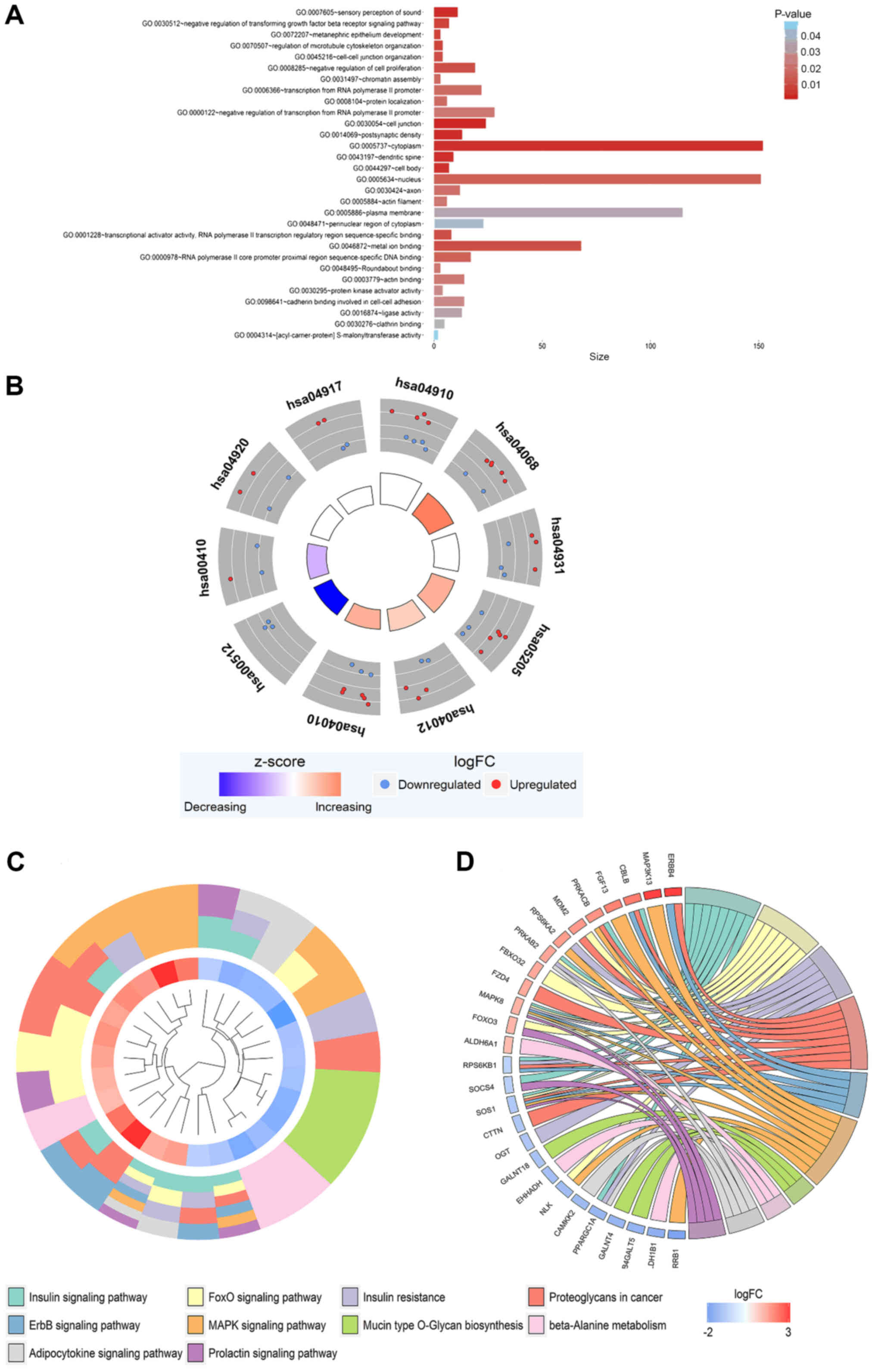
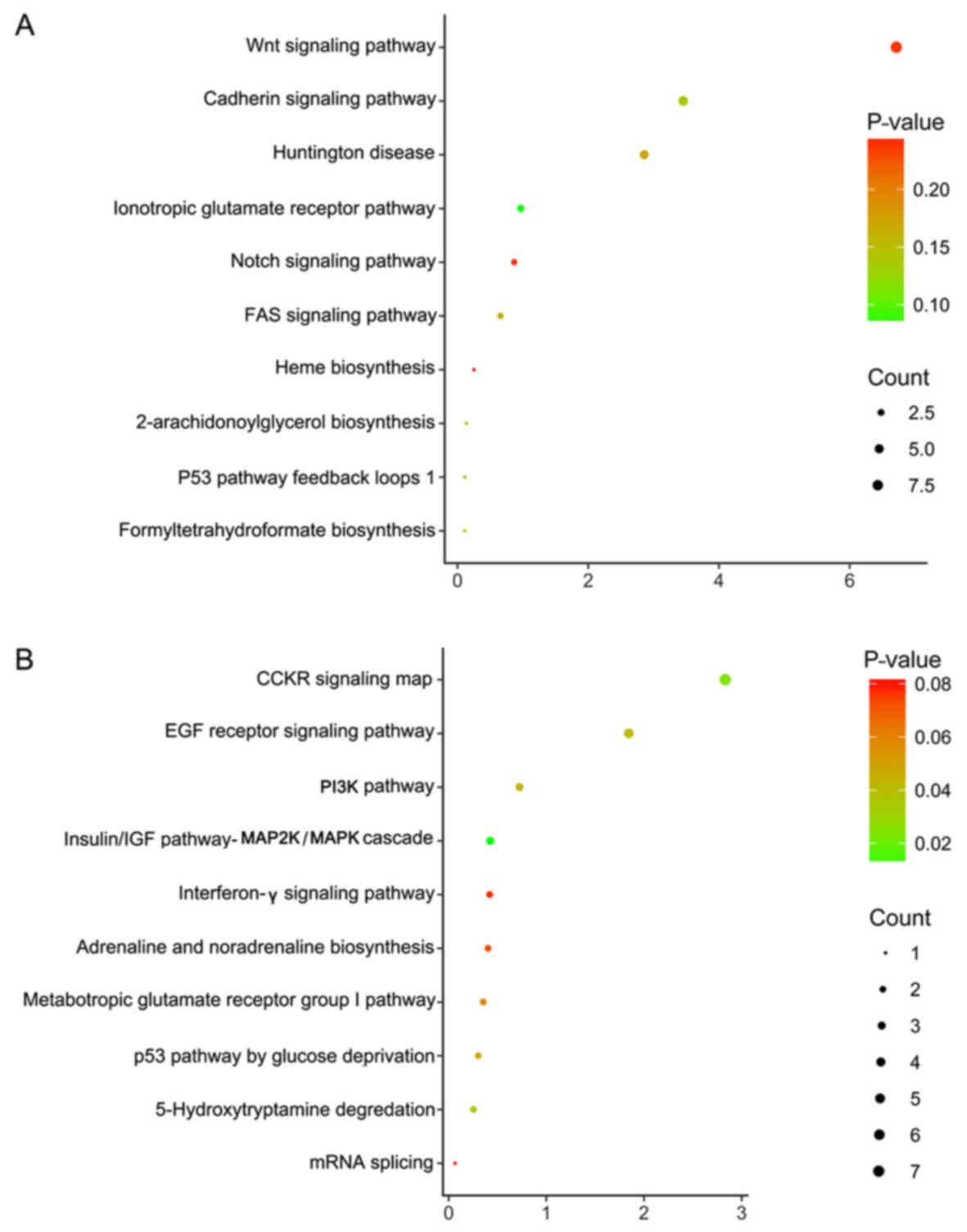
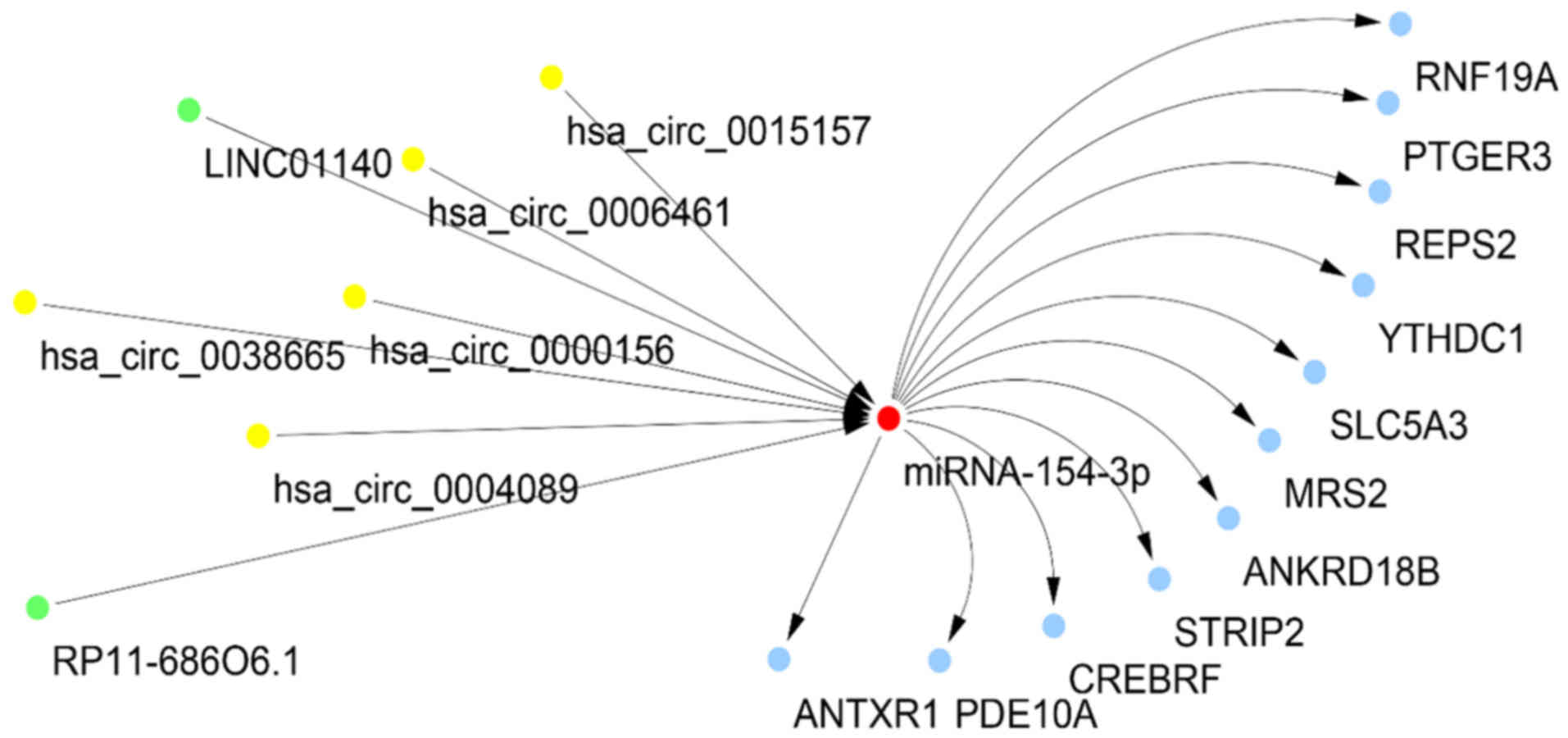
|
1
|
Ryan DP, Hong TS and Bardeesy N:
Pancreatic adenocarcinoma. N Engl J Med. 371:2140–2141. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Swords DS, Firpo MA, Scaife CL and
Mulvihill SJ: Biomarkers in pancreatic adenocarcinoma: Current
perspectives. OncoTargets Ther. 9:7459–7467. 2016. View Article : Google Scholar
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song HY, Wang Y, Lan H and Zhang YX:
Expression of Notch receptors and their ligands in pancreatic
ductal adenocarcinoma. Exp Ther Med. 16:53–60. 2018.PubMed/NCBI
|
|
6
|
Xu Q, Gao J and Li Z: Identification of a
novel alternative splicing transcript variant of the suppressor of
fused: Relationship with lymph node metastasis in pancreatic ductal
adenocarcinoma. Int J Oncol. 49:2611–2619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Q, Zhang L, Li X, Yan H, Yang L, Li Y,
Li T, Wang J and Cao B: The prognostic significance of human
epidermal growth factor receptor family protein expression in
operable pancreatic cancer: HER14 protein expression and prognosis
in pancreatic cancer. BMC Cancer. 16:9102016. View Article : Google Scholar
|
|
8
|
Li H, Hao X, Wang H, Liu Z, He Y, Pu M,
Zhang H, Yu H, Duan J and Qu S: Circular RNA Expression Profile of
Pancreatic Ductal Adenocarcinoma Revealed by Microarray. Cell
Physiol Biochem. 40:1334–1344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu L, Staley C, Kooby D, El-Rays B, Mao H
and Yang L: Current status of biomarker and targeted nanoparticle
development: The precision oncology approach for pancreatic cancer
therapy. Cancer Lett. 388:139–148. 2017. View Article : Google Scholar :
|
|
10
|
Kocaturk NM and Gozuacik D: Crosstalk
Between Mammalian Autophagy and the Ubiquitin-Proteasome System.
Front Cell Dev Biol. 6:1282018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong JC, Czito BG, Willett CG and Palta M:
A current perspective on stereotactic body radiation therapy for
pancreatic cancer. OncoTargets Ther. 9:6733–6739. 2016. View Article : Google Scholar
|
|
12
|
Marsoner K, Haybaeck J, Csengeri D, Waha
JE, Schagerl J, Langeder R, Mischinger HJ and Kornprat P:
Pancreatic resection for intraductal papillary mucinous neoplasm -
a thirteen-year single center experience. BMC Cancer. 16:8442016.
View Article : Google Scholar
|
|
13
|
Kang MJ, Jang JY and Kim SW: Surgical
resection of pancreatic head cancer: What is the optimal extent of
surgery. Cancer Lett. 382:259–265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ellerhoff TP, Berchtold S, Venturelli S,
Burkard M, Smirnow I, Wulff T and Lauer UM: Novel
epi-virotherapeutic treatment of pancreatic cancer combining the
oral histone deacetylase inhibitor resminostat with oncolytic
measles vaccine virus. Int J Oncol. 49:1931–1944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zuo C, Sheng X, Ma M, Xia M and Ouyang L:
ISG15 in the tumorigenesis and treatment of cancer: An emerging
role in malignancies of the digestive system. Oncotarget.
7:74393–74409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang JH, Jiang Y and Pillarisetty VG:
Role of immune cells in pancreatic cancer from bench to clinical
application: An updated review. Medicine (Baltimore). 95:e55412016.
View Article : Google Scholar
|
|
17
|
Kim KH and Lee MS: Autophagy--a key player
in cellular and body metabolism. Nat Rev Endocrinol. 10:322–337.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tao Z, Li T, Ma H, Yang Y, Zhang C, Hai L,
Liu P, Yuan F, Li J, Yi L, et al: Autophagy suppresses self-renewal
ability and tumorigenicity of glioma-initiating cells and promotes
Notch1 degradation. Cell Death Dis. 9:10632018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mowers EE, Sharifi MN and Macleod KF:
Autophagy in cancer metastasis. Oncogene. 36:1619–1630. 2017.
View Article : Google Scholar :
|
|
20
|
Tan YQ, Zhang J and Zhou G: Autophagy and
its implication in human oral diseases. Autophagy. 13:225–236.
2017. View Article : Google Scholar :
|
|
21
|
Green DR, Oguin TH and Martinez J: The
clearance of dying cells: Table for two. Cell Death Differ.
23:915–926. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sionov RV, Vlahopoulos SA and Granot Z:
Regulation of Bim in Health and Disease. Oncotarget. 6:23058–23134.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gafar AA, Draz HM, Goldberg AA, Bashandy
MA, Bakry S, Khalifa MA, AbuShair W, Titorenko VI and Sanderson JT:
Lithocholic acid induces endoplasmic reticulum stress, autophagy
and mitochondrial dysfunction in human prostate cancer cells.
PeerJ. 4:e24452016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jawhari S, Ratinaud MH and Verdier M:
Glioblastoma, hypoxia and autophagy: A survival-prone
‘ménage-à-trois’. Cell Death Dis. 7:e24342016. View Article : Google Scholar
|
|
25
|
Mao S and Zhang J: Role of autophagy in
chronic kidney diseases. Int J Clin Exp Med. 8:22022–22029.
2015.
|
|
26
|
Mo He Y, Luo Q, Qiao B, Xu Y, Zuo R, Deng
Z, Nong J, Peng X, He GW, et al: Induction of apoptosis and
autophagy via mitochondria- and PI3K/Akt/mTOR-mediated pathways by
E. adenophorum in hepatocytes of saanen goat. Oncotarget.
7:54537–54548. 2016.PubMed/NCBI
|
|
27
|
Goulielmaki M, Koustas E, Moysidou E,
Vlassi M, Sasazuki T, Shirasawa S, Zografos G, Oikonomou E and
Pintzas A: BRAF associated autophagy exploitation: BRAF and
autophagy inhibitors synergise to efficiently overcome resistance
of BRAF mutant colorectal cancer cells. Oncotarget. 7:9188–9221.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vera-Ramirez L, Vodnala SK, Nini R, Hunter
KW and Green JE: Autophagy promotes the survival of dormant breast
cancer cells and metastatic tumour recurrence. Nat Commun.
9:19442018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li SJ, Sun SJ, Gao J and Sun FB: Wogonin
induces Beclin-1/PI3K and reactive oxygen species-mediated
autophagy in human pancreatic cancer cells. Oncol Lett.
12:5059–5067. 2016. View Article : Google Scholar
|
|
30
|
Klein K, Werner K, Teske C, Schenk M,
Giese T, Weitz J and Welsch T: Role of TFEB-driven autophagy
regulation in pancreatic cancer treatment. Int J Oncol. 49:164–172.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kwon JJ, Willy JA, Quirin KA, Wek RC, Korc
M, Yin XM and Kota J: Novel role of miR-29a in pancreatic cancer
autophagy and its therapeutic potential. Oncotarget. 7:71635–71650.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ranjan A and Srivastava SK: Penfluridol
suppresses pancreatic tumor growth by autophagy-mediated apoptosis.
Sci Rep. 6:261652016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seo JW, Choi J, Lee SY, Sung S, Yoo HJ,
Kang MJ, Cheong H and Son J: Autophagy is required for PDAC
glutamine metabolism. Sci Rep. 6:375942016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan Y, Jiang K, Liu P, Zhang X, Dong X,
Gao J, Liu Q, Barr MP, Zhang Q, Hou X, et al: Bafilomycin A1
induces caspase-independent cell death in hepatocellular carcinoma
cells via targeting of autophagy and MAPK pathways. Sci Rep.
6:370522016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang S, Wang X, Contino G, Liesa M, Sahin
E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, et al:
Pancreatic cancers require autophagy for tumor growth. Genes Dev.
25:717–729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang A, Herter-Sprie G, Zhang H, Lin EY,
Biancur D, Wang X, Deng J, Hai J, Yang S, Wong KK, et al: Autophagy
sustains pancreatic cancer growth through both cell autonomous and
non-autonomous mechanisms. Cancer Discov. 8:276–287. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mukubou H, Tsujimura T, Sasaki R and Ku Y:
The role of autophagy in the treatment of pancreatic cancer with
gemcitabine and ionizing radiation. Int J Oncol. 37:821–828.
2010.PubMed/NCBI
|
|
38
|
Marinković M, Šprung M, Buljubašić M and
Novak I: Autophagy Modulation in Cancer: Current Knowledge on
Action and Therapy. Oxid Med Cell Longev. 2018.8023821:2018.
|
|
39
|
Islam MA, Reesor EK, Xu Y, Zope HR, Zetter
BR and Shi J: Biomaterials for mRNA delivery. Biomater Sci.
3:1519–1533. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu F, Gao S, Yang Y, Zhao X, Fan Y, Ma W,
Yang D, Yang A and Yu Y: Antitumor activity of curcumin by
modulation of apoptosis and autophagy in human lung cancer A549
cells through inhibiting PI3K/Akt/mTOR pathway. Oncol Rep.
39:1523–1531. 2018.PubMed/NCBI
|
|
41
|
Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu
DY, Tang CJ, De W and Yang F: STAT3-induced lncRNA HAGLROS
over-expression contributes to the malignant progression of gastric
cancer cells via mTOR signal-mediated inhibition of autophagy. Mol
Cancer. 17:62018. View Article : Google Scholar
|
|
42
|
Gong J, Muñoz AR, Chan D, Ghosh R and
Kumar AP: STAT3 down regulates LC3 to inhibit autophagy and
pancreatic cancer cell growth. Oncotarget. 5:2529–2541. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Raimondi M, Cesselli D, Di Loreto C, La
Marra F, Schneider C and Demarchi F: USP1 (ubiquitin specific
peptidase 1) targets ULK1 and regulates its cellular
compartmentalization and autophagy. Autophagy. Oct 18–2018.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pan Y, Li C, Chen J, Zhang K, Chu X, Wang
R and Chen L: The Emerging Roles of Long Noncoding RNA ROR
(lincRNA-ROR) and its Possible Mechanisms in Human Cancers. Cell
Physiol Biochem. 40:219–229. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bamodu OA, Huang WC, Lee WH, Wu A, Wang
LS, Hsiao M, Yeh CT and Chao TY: Aberrant KDM5B expression promotes
aggressive breast cancer through MALAT1 overexpression and
downregulation of hsa-miR-448. BM. Cancer. 16:1602016.
|
|
46
|
Zhang Y, He RQ, Dang YW, Zhang XL, Wang X,
Huang SN, Huang WT, Jiang MT, Gan XN, Xie Y, et al: Comprehensive
analysis of the long noncoding RNA HOXA11-AS gene interaction
regulatory network in NSCLC cells. Cancer Cell Int. 16:892016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Peng L, Yuan XQ and Li GC: The emerging
landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep.
33:2669–2674. 2015.Review. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Deng T, Yuan Y, Zhang C, Zhang C, Yao W,
Wang C, Liu R and Ba Y: Identification of Circulating MiR-25 as a
Potential Biomarker for Pancreatic Cancer Diagnosis. Cell Physiol
Biochem. 39:1716–1722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang YH, Fu J, Zhang ZJ, Ge CC and Yi Y:
LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability
and promotes apoptosis of colorectal cancer cells. Am J Transl Res.
8:5286–5297. 2016.
|
|
50
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li X, Yang L and Chen LL: The Biogenesis,
Functions, and Challenges of Circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T,
Wang X, Li Q, Tian L, Liu M, et al: Circular RNA: A novel biomarker
for progressive laryngeal cancer. Am J Transl Res. 8:932–939.
2016.PubMed/NCBI
|
|
53
|
Jin X, Feng CY, Xiang Z, Chen YP and Li
YM: CircRNA expression pattern and circRNA-miRNA-mRNA network in
the pathogenesis of nonalcoholic steatohepatitis. Oncotarget.
7:66455–66467. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J
and Ao Y: Circular RNA Related to the Chondrocyte ECM Regulates
MMP13 Expression by Functioning as a MiR-136 ‘Sponge’ in Human
Cartilage Degradation. Sci Rep. 6:225722016. View Article : Google Scholar
|
|
55
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ, et al: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
56
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Deng X, Feng N, Zheng M, Ye X, Lin H, Yu
X, Gan Z, Fang Z, Zhang H, Gao M, et al: PM2.5 exposure-induced
autophagy is mediated by lncRNA loc146880 which also promotes the
migration and invasion of lung cancer cells. Biochim Biophys Acta,
Gen Subj. 1861.112–125. 2017.
|
|
58
|
Li C, Zhao Z, Zhou Z and Liu R: Linc-ROR
confers gemcitabine resistance to pancreatic cancer cells via
inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis.
Cancer Chemother Pharmacol. 78:1199–1207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen ZH, Wang WT, Huang W, Fang K, Sun YM,
Liu SR, Luo XQ and Chen YQ: The lncRNA HOTAIRM1 regulates the
degradation of PML-RARA oncoprotein and myeloid cell
differentiation by enhancing the autophagy pathway. Cell Death
Differ. 24:212–224. 2017. View Article : Google Scholar :
|
|
60
|
Pawar K, Hanisch C, Palma Vera SE,
Einspanier R and Sharbati S: Down regulated lncRNA MEG3 eliminates
mycobacteria in macrophages via autophagy. Sci Rep. 6:194162016.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Vega-Rubín-de-Celis S, Zou Z, Fernández
AF, Ci B, Kim M, Xiao G, Xie Y and Levine B: Increased autophagy
blocks HER2-mediated breast tumorigenesis. Proc Natl Acad Sci USA.
115:4176–4181. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zarzynska JM: The importance of autophagy
regulation in breast cancer development and treatment. BioMed Res
Int. 2014.710345:2014.
|
|
63
|
Qin W, Li C, Zheng W, Guo Q, Zhang Y, Kang
M, Zhang B, Yang B, Li B, Yang H, et al: Inhibition of autophagy
promotes metastasis and glycolysis by inducing ROS in gastric
cancer cells. Oncotarget. 6:39839–39854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cai J, Li R, Xu X, Zhang L, Lian R, Fang
L, Huang Y, Feng X, Liu X, Li X, et al: CK1α suppresses lung tumour
growth by stabilizing PTEN and inducing autophagy. Nat Cell Biol.
20:465–478. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen PM, Gombart ZJ and Chen JW:
Chloroquine treatment of ARPE-19 cells leads to lysosome dilation
and intracellular lipid accumulation: possible implications of
lysosomal dysfunction in macular degeneration. Cell Biosci.
1:102011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li ML, Xu YZ, Lu WJ, Li YH, Tan SS, Lin
HJ, Wu TM, Li Y, Wang SY and Zhao YL: Chloroquine potentiates the
anticancer effect of sunitinib on renal cell carcinoma by
inhibiting autophagy and inducing apoptosis. Oncol Lett.
15:2839–2846. 2018.PubMed/NCBI
|
|
67
|
Maragkakis M, Vergoulis T, Alexiou P,
Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T
and Hatzigeorgiou AG: DIANA-microT Web server upgrade supports Fly
and Worm miRNA target prediction and bibliographic miRNA to disease
association. Nucleic Acids Res 39 (Web Server issue):. W145–W148.
2011. View Article : Google Scholar
|
|
68
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0:
Service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41(Web Server issue): W169–W173. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tsang JS, Ebert MS and van Oudenaarden A:
Genome-wide dissection of microRNA functions and cotargeting
networks using gene set signatures. Mol Cell. 38:140–153. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang X: Improving microRNA target
prediction by modeling with unambiguously identified
microRNA-target pairs from CLIP-ligation studies. Bioinformatics.
32:1316–1322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Vejnar CE, Blum M and Zdobnov EM: miRmap
web: Comprehensive microRNA target prediction online. Nucleic Acids
Res. 41(Web Server issue): W165–W168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC,
Hsu PW, Wong YH, Chen YH, Chen GH and Huang HD: miRNAMap 2.0:
Genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res.
36(Database): D165–D169. 2008. View Article : Google Scholar :
|
|
74
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M,
et al: Combinatorial microRNA target predictions. Nat Genet.
37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Michelson AM and Orkin SH:
Characterization of the homo-polymer tailing reaction catalyzed by
terminal deoxynucleotidyl transferase. Implications for the cloning
of cDNA. J Biol Chem. 257:14773–14782. 1982.PubMed/NCBI
|
|
80
|
Hoshino T and Inagaki F: A comparative
study of microbial diversity and community structure in marine
sediments using poly(A) tailing and reverse transcription-PCR.
Front Microbiol. 4:1602013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tajadini M, Panjehpour M and Javanmard SH:
Comparison of SYBR Green and TaqMan methods in quantitative
real-time polymerase chain reaction analysis of four adenosine
receptor subtypes. Adv Biomed Res. 3:852014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Mardis E and McCombie WR: Library
Quantification Using SYBR Green-Quantitative Polymerase Chain
Reaction (qPCR). Cold Spring Harb Protoc. 2017:pdb prot0947142017.
View Article : Google Scholar
|
|
83
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
84
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
85
|
Kang MJ, Jang JY, Chang YR, Kwon W, Jung W
and Kim SW: Revisiting the concept of lymph node metastases of
pancreatic head cancer: Number of metastatic lymph nodes and lymph
node ratio according to N stage. Ann Surg Oncol. 21:1545–1551.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Georgiadou D, Sergenta nis T N,
Sakellariou S, Vlachodimitropoulos D, Psaltopoulou T, Lazaris AC,
Gounaris A and Zografos GC: Prognostic role of sex steroid
receptors in pancreatic adenocarcinoma. Pathol Res Pract.
212:38–43. 2016. View Article : Google Scholar
|
|
87
|
Lovecek M, Skalicky P, Klos D, Bebarova L,
Neoral C, Ehrmann J, Zapletalova J, Svebisova H, Vrba R, Stasek M,
et al: Long-term survival after resections for pancreatic ductal
adeno-carcinoma. Single centre study. Biomed Pap Med Fac Univ
Palacky Olomouc Czech Repub. 160:280–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ramacciato G, Nigri G, Petrucciani N,
Pinna AD, Ravaioli M, Jovine E, Minni F, Grazi GL, Chirletti P,
Tisone G, et al: Pancreatectomy with Mesenteric and Portal Vein
Resection for Borderline Resectable Pancreatic Cancer: Multicenter
Study of 406 Patients. Ann Surg Oncol. 23:2028–2037. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li PF, Chen SC, Xia T, Jiang XM, Shao YF,
Xiao BX and Guo JM: Non-coding RNAs and gastric cancer. World J
Gastroenterol. 20:5411–5419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kishikawa T, Otsuka M, Ohno M, Yoshikawa
T, Takata A and Koike K: Circulating RNAs as new biomarkers for
detecting pancreatic cancer. World J Gastroenterol. 21:8527–8540.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Bolton EM, Tuzova AV, Walsh AL, Lynch T
and Perry AS: Noncoding RNAs in prostate cancer: the long and the
short of it. Clin Cancer Res. 20:35–43. 2014. View Article : Google Scholar
|
|
92
|
Zhao Z, Li S, Song E and Liu S: The roles
of ncRNAs and histone-modifiers in regulating breast cancer stem
cells. Protein Cell. 7:89–99. 2016. View Article : Google Scholar :
|
|
93
|
Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang
YF, Liu X, Che L, Ma CJ, Xie YK, et al: Landscape of RNAs in human
lumbar disc degeneration. Oncotarget. 7:63166–63176. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Xu CZ, Jiang C, Wu Q, Liu L, Yan X and Shi
R: A Feed-Forward Regulatory Loop between HuR and the Long
Noncoding RNA HOTAIR Promotes Head and Neck Squamous Cell Carcinoma
Progression and Metastasis. Cell Physiol Biochem. 40:1039–1051.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Dou C, Cao Z, Yang B, Ding N, Hou T, Luo
F, Kang F, Li J, Yang X, Jiang H, et al: Changing expression
profiles of lncRNAs, mRNAs, circRNAs and miRNAs during
osteoclastogenesis. Sci Rep. 6:214992016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200.
2016.PubMed/NCBI
|
|
97
|
Zhou X, Ye F, Yin C, Zhuang Y, Yue G and
Zhang G: The Interaction Between MiR-141 and lncRNA-H19 in
Regulating Cell Proliferation and Migration in Gastric Cancer. Cell
Physiol Biochem. 36:1440–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zeng JH, Xiong DD, Pang YY, Zhang Y, Tang
RX, Luo DZ and Chen G: Identification of molecular targets for
esophageal carcinoma diagnosis using miRNA-seq and RNA-seq data
from The Cancer Genome Atlas: A study of 187 cases. Oncotarget.
8:35681–35699. 2017.PubMed/NCBI
|
|
99
|
Massillo C, Dalton GN, Farré PL, De Luca P
and De Siervi A: Implications of microRNA dysregulation in the
development of prostate cancer. Reproduction. 154:R81–R97. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Murray MJ, Bell E, Raby KL, Rijlaarsdam
MA, Gillis AJ, Looijenga LH, Brown H, Destenaves B, Nicholson JC
and Coleman N: A pipeline to quantify serum and cerebrospinal fluid
microRNAs for diagnosis and detection of relapse in paediatric
malignant germ-cell tumours. Br J Cancer. 114:151–162. 2016.
View Article : Google Scholar :
|
|
101
|
Ouyang Q, Xu L, Cui H, Xu M and Yi L:
MicroRNAs and cell cycle of malignant glioma. Int J Neurosci.
126:1–9. 2016. View Article : Google Scholar
|
|
102
|
Del Vescovo V and Denti MA: microRNA and
Lung Cancer. Adv Exp Med Biol. 889:153–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang X, Tang W, Chen G, Ren F, Liang H,
Dang Y and Rong M: An Encapsulation of Gene Signatures for
Hepatocellular Carcinoma, MicroRNA-132 Predicted Target Genes and
the Corresponding Overlaps. PLoS One. 11:e01594982016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lan D, Zhang X, He R, Tang R, Li P, He Q
and Chen G: MiR-133a is downregulated in non-small cell lung
cancer: A study of clinical significance. Eur J Med Res. 20:502015.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Gan TQ, Tang RX, He RQ, Dang YW, Xie Y and
Chen G: Upregulated MiR-1269 in hepatocellular carcinoma and its
clinical significance. Int J Clin Exp Med. 8:714–721.
2015.PubMed/NCBI
|
|
107
|
Zhang X, Li P, Rong M, He R, Hou X, Xie Y
and Chen G: MicroRNA-141 is a biomarker for progression of squamous
cell carcinoma and adenocarcinoma of the lung: Clinical analysis of
125 patients. Tohoku J Exp Med. 235:161–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ren F, Ding H, Huang S, Wang H, Wu M, Luo
D, Dang Y, Yang L and Chen G: Expression and clinicopathological
significance of miR-193a-3p and its potential target astrocyte
elevated gene-1 in non-small lung cancer tissues. Cancer Cell Int.
15:802015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Liu Y, Ren F, Luo Y, Rong M, Chen G and
Dang Y: Down-Regulation of MiR-193a-3p Dictates Deterioration of
HCC: A Clinical Real-Time qRT-PCR Study. Med Sci Monit.
21:2352–2360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yang He R, Lin L, Chen X, Lin X, Wei X,
Liang F, Luo X, Wu Y, Gan YT, et al: MiR-30a-5p suppresses cell
growth and enhances apoptosis of hepatocellular carcinoma cells via
targeting AEG-1. Int J Clin Exp Pathol. 8:15632–15641. 2015.
|
|
111
|
Huang WT, Wang HL, Yang H, Ren FH, Luo YH,
Huang CQ, Liang YY, Liang HW, Chen G and Dang YW: Lower expressed
miR-198 and its potential targets in hepatocellular carcinoma: A
clinicopathological and in silico study. OncoTargets Ther.
9:5163–5180. 2016. View Article : Google Scholar
|
|
112
|
Zhang X, Tang W, Li R, He R, Gan T, Luo Y,
Chen G and Rong M: Downregulation of microRNA-132 indicates
progression in hepatocellular carcinoma. Exp Ther Med.
12:2095–2101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Frampton AE, Castellano L, Colombo T,
Giovannetti E, Krell J, Jacob J, Pellegrino L, Roca-Alonso L, Funel
N, Gall TM, et al: MicroRNAs cooperatively inhibit a network of
tumor suppressor genes to promote pancreatic tumor growth and
progression. Gastroenterology. 146:268-277e182014. View Article : Google Scholar
|
|
114
|
Frampton AE, Krell J, Jamieson NB, Gall
TM, Giovannetti E, Funel N, Mato Prado M, Krell D, Habib NA,
Castellano L, et al: microRNAs with prognostic significance in
pancreatic ductal adenocarcinoma: A meta-analysis. Eur J Cancer.
51:1389–1404. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Giovannetti E, Funel N, Peters GJ, Del
Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A,
Falcone A, et al: MicroRNA-21 in pancreatic cancer: Correlation
with clinical outcome and pharmacologic aspects underlying its role
in the modulation of gemcitabine activity. Cancer Res.
70:4528–4538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ouyang H, Gore J, Deitz S and Korc M:
microRNA-10b enhances pancreatic cancer cell invasion by
suppressing TIP30 expression and promoting EGF and TGF-β actions.
Oncogene. 33:4664–4674. 2014. View Article : Google Scholar
|
|
117
|
Nakata K, Ohuchida K, Mizumoto K,
Kayashima T, Ikenaga N, Sakai H, Lin C, Fujita H, Otsuka T, Aishima
S, et al: MicroRNA-10b is overexpressed in pancreatic cancer,
promotes its invasiveness, and correlates with a poor prognosis.
Surgery. 150:916–922. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Shao T, Wu A, Chen J, Chen H, Lu J, Bai J,
Li Y, Xu J and Li X: Identification of module biomarkers from the
dysregulated ceRNA-ceRNA interaction network in lung
adenocarcinoma. Mol Biosyst. 11:3048–3058. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z,
Shao T, Zhang J, Wang L and Li X: The mRNA related ceRNA-ceRNA
landscape and significance across 20 major cancer types. Nucleic
Acids Res. 43:8169–8182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Feng T, Shao F, Wu Q, Zhang X, Xu D, Qian
K, Xie Y, Wang S, Xu N, Wang Y, et al: miR-124 downregulation leads
to breast cancer progression via LncRNA-MALAT1 regulation and
CDK4/E2F1 signal activation. Oncotarget. 7:16205–16216.
2016.PubMed/NCBI
|
|
122
|
Jin H, Li Q, Cao F, Wang SN, Wang RT, Wang
Y, Tan QY, Li CR, Zou H, Wang D, et al: miR-124 Inhibits Lung
Tumorigenesis Induced by K-ras Mutation and NNK. Mol Ther Nucleic
Acids. 9:145–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Wang M, Meng B and Liu Y, Yu J, Chen Q and
Liu Y: MiR-124 Inhibits Growth and Enhances Radiation-Induced
Apoptosis in Non-Small Cell Lung Cancer by Inhibiting STAT3. Cell
Physiol Biochem. 44:2017–2028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhang X, Gao F, Zhou L, Wang H, Shi G and
Tan X: UCA1 Regulates the Growth and Metastasis of Pancreatic
Cancer by Sponging miR-135a. Oncol Res. 25:1529–1541. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Wang P, Zhang L, Chen Z and Meng Z:
MicroRNA targets autophagy in pancreatic cancer cells during cancer
therapy. Autophagy. 9:2171–2172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Ganley IG, Lam H, Wang J, Ding X, Chen S
and Jiang X: ULK1.ATG13.FIP200 complex mediates mTOR signaling and
is essential for autophagy. J Biol Chem. 284:12297–12305. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Hosokawa N, Hara T, Kaizuka T, Kishi C,
Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et
al: Nutrient-dependent mTORC1 association with the
ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell.
20:1981–1991. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Jung CH, Jun CB, Ro SH, Kim YM, Otto NM,
Cao J, Kundu M and Kim DH: ULK-Atg13-FIP200 complexes mediate mTOR
signaling to the autophagy machinery. Mol Biol Cell. 20:1992–2003.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Yan J, Kuroyanagi H, Tomemori T, Okazaki
N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, et
al: Mouse ULK2, a novel member of the UNC-51-like protein kinases:
Unique features of functional domains. Oncogene. 18:5850–5859.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Chan EY, Longatti A, McKnight NC and Tooze
SA: Kinase-inactivated ULK proteins inhibit autophagy via their
conserved C-terminal domains using an Atg13-independent mechanism.
Mol Cell Biol. 29:157–171. 2009. View Article : Google Scholar :
|
|
131
|
Kundu M, Lindsten T, Yang CY, Wu J, Zhao
F, Zhang J, Selak MA, Ney PA and Thompson CB: Ulk1 plays a critical
role in the autophagic clearance of mitochondria and ribosomes
during reticulocyte maturation. Blood. 112:1493–1502. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Mo J, Zhang D and Yang R: MicroRNA-195
regulates proliferation, migration, angiogenesis and autophagy of
endothelial progenitor cells by targeting GABARAPL1. Biosci Rep.
36:362016. View Article : Google Scholar
|
|
133
|
Zang W, Wang Y, Wang T, Du Y, Chen X, Li M
and Zhao G: miR-663 attenuates tumor growth and invasiveness by
targeting eEF1A2 in pancreatic cancer. Mol Cancer. 14:372015.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Huang W, Li J, Guo X, Zhao Y and Yuan X:
miR-663a inhibits hepatocellular carcinoma cell proliferation and
invasion by targeting HMGA2. Biomed Pharmacother. 81:431–438. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhang Y, Xu X, Zhang M, Wang X, Bai X, Li
H, Kan L, Zhou Y, Niu H and He P: MicroRNA-663a is downregulated in
non-small cell lung cancer and inhibits proliferation and invasion
by targeting JunD. BMC Cancer. 16:3152016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Kuroda K, Fukuda T, Krstic-Demonacos M,
Demonacos C, Okumura K, Isogai H, Hayashi M, Saito K and Isogai E:
miR-663a regulates growth of colon cancer cells, after
administration of antimicrobial peptides, by targeting CXCR4-p21
pathway. BMC Cancer. 17:332017. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Lin Y, Kuang W, Wu B, Xie C, Liu C and Tu
Z: IL-12 induces autophagy in human breast cancer cells through
AMPK and the PI3K/Akt pathway. Mol Med Rep. 16:4113–4118. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Wu L, Zhang Q, Mo W, Feng J, Li S, Li J,
Liu T, Xu S, Wang W, Lu X, et al: Quercetin prevents hepatic
fibrosis by inhibiting hepatic stellate cell activation and
reducing autophagy via the TGF-β1/Smads and PI3K/Akt pathways. Sci
Rep. 7:92892017. View Article : Google Scholar
|
|
139
|
Ren L, Han W, Yang H, Sun F, Xu S, Hu S,
Zhang M, He X, Hua J and Peng S: Autophagy stimulated proliferation
of porcine PSCs might be regulated by the canonical Wnt signaling
pathway. Biochem Biophys Res Commun. 479:537–543. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Lin R, Feng J, Dong S, Pan R, Zhuang H and
Ding Z: Regulation of autophagy of prostate cancer cells by
β-catenin signaling. Cell Physiol Biochem. 35:926–932. 2015.
View Article : Google Scholar
|
|
141
|
Zhao S, Li L, Wang S, Yu C, Xiao B, Lin L,
Cong W, Cheng J, Yang W, Sun W, et al: H2O2 treatment or serum
deprivation induces autophagy and apoptosis in naked mole-rat skin
fibroblasts by inhibiting the PI3K/Akt signaling pathway.
Oncotarget. 7:84839–84850. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Lu X, Lv S, Mi Y, Wang L and Wang G:
Neuroprotective effect of miR-665 against sevoflurane
anesthesia-induced cognitive dysfunction in rats through PI3K/Akt
signaling pathway by targeting insulin-like growth factor 2. Am J
Transl Res. 9:1344–1356. 2017.PubMed/NCBI
|
|
143
|
Su N, Wang P and Li Y: Role of
Wnt/β-catenin pathway in inducing autophagy and apoptosis in
multiple myeloma cells. Oncol Lett. 12:4623–4629. 2016. View Article : Google Scholar
|
|
144
|
Kai Y, Qiang C, Xinxin P, Miaomiao Z and
Kuailu L: Decreased miR-154 expression and its clinical
significance in human colorectal cancer. World J Surg Oncol.
13:1952015. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Xu H, Fei D, Zong S and Fan Z:
MicroRNA-154 inhibits growth and invasion of breast cancer cells
through targeting E2F5. Am J Transl Res. 8:2620–2630.
2016.PubMed/NCBI
|
|
146
|
Liu ZQ, Zhao S and Fu WQ: Insulin-like
growth factor 1 antagonizes lumbar disc degeneration through
enhanced autophagy. Am J Transl Res. 8:4346–4353. 2016.PubMed/NCBI
|
|
147
|
Liu Q, Guan JZ, Sun Y, Le Z, Zhang P, Yu D
and Liu Y: Insulin-like growth factor 1 receptor-mediated cell
survival in hypoxia depends on the promotion of autophagy via
suppression of the PI3K/Akt/mTOR signaling pathway. Mol Med Rep.
15:2136–2142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Ceperuelo-Mallafré V, Ejarque M, Serena C,
Duran X, Montori-Grau M, Rodríguez MA, Yanes O, Núñez-Roa C, Roche
K, Puthanveetil P, et al: Adipose tissue glycogen accumulation is
associated with obesity-linked inflammation in humans. Mol Metab.
5:5–18. 2015. View Article : Google Scholar
|
|
149
|
Ciarcia R, Damiano S, Montagnaro S,
Pagnini U, Ruocco A, Caparrotti G, d’ Angelo D, Boffo S, Morales F,
Rizzolio F, et al: Combined effects of PI3K and SRC kinase
inhibitors with imatinib on intracellular calcium levels,
autophagy, and apoptosis in CML-PBL cells. Cell Cycle.
12:2839–2848. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Hua R, Han S, Zhang N, Dai Q, Liu T and Li
J: cPKCγ-Modulated Sequential Reactivation of mTOR Inhibited
Autophagic Flux in Neurons Exposed to Oxygen Glucose
Deprivation/Reperfusion. Int J Mol Sci. 19:192018. View Article : Google Scholar
|
|
151
|
Shivaram S, Crist KA, Chaudhuri B, Mucci
SJ and Chaudhuri PK: Effect of CCK receptor antagonist on growth of
pancreatic adenocarcinoma. J Surg Res. 53:234–237. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Clerc P, Saillan-Barreau C, Desbois C,
Pradayrol L, Fourmy D and Dufresne M: Transgenic mice expressing
cholecystokinin 2 receptors in the pancreas. Pharmacol Toxicol.
91:321–326. 2002. View Article : Google Scholar
|
|
153
|
Shang C, Zhu W, Liu T, Wang W, Huang G,
Huang J, Zhao P, Zhao Y and Yao S: Characterization of long
non-coding RNA expression profiles in lymph node metastasis of
early-stage cervical cancer. Oncol Rep. 35:3185–3197. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Bronietzki AW, Schuster M and Schmitz I:
Autophagy in T-cell development, activation and differentiation.
Immunol Cell Biol. 93:25–34. 2015. View Article : Google Scholar
|
|
155
|
Martinez-Lopez N, Athonvarangkul D and
Singh R: Autophagy and aging. Adv Exp Med Biol. 847:73–87. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Tan Y, Gong Y, Dong M, Pei Z and Ren J:
Role of autophagy in inherited metabolic and endocrine myopathies.
Biochim Biophys Acta Mol Basis Dis. 1865.48–55. 2018.
|
|
157
|
Chu CT: Autophagy in Neurological
Diseases: An update. Neurobiol Dis. S0969–9961(18): 30728–9.
2018.
|
|
158
|
Yang A, Rajeshkumar NV, Wang X, Yabuuchi
S, Alexander BM, Chu GC, Von Hoff DD, Maitra A and Kimmelman AC:
Autophagy is critical for pancreatic tumor growth and progression
in tumors with p53 alterations. Cancer Discov. 4:905–913. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Zhan Y, Wang K, Li Q, Zou Y, Chen B, Gong
Q, Ho HI, Yin T, Zhang F, Lu Y, et al: The Novel Autophagy
Inhibitor Alpha-Hederin Promoted Paclitaxel Cytotoxicity by
Increasing Reactive Oxygen Species Accumulation in Non-Small Cell
Lung Cancer Cells. Int J Mol Sci. 19:192018. View Article : Google Scholar
|
|
160
|
Chen S, Wu J, Jiao K, Wu Q, Ma J, Chen D,
Kang J, Zhao G, Shi Y, Fan D, et al: MicroRNA-495-3p inhibits
multidrug resistance by modulating autophagy through GRP78/mTOR
axis in gastric cancer. Cell Death Dis. 9:10702018. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Verma SP and Das P: Monensin induces cell
death by autophagy and inhibits matrix metalloproteinase 7 (MMP7)
in UOK146 renal cell carcinoma cell line. In Vitro Cell Dev Biol
Anim. 54:736–742. 2018. View Article : Google Scholar : PubMed/NCBI
|