Introduction
Globally, gastric cancer (GC) is the fifth most
common malignant tumor and the third leading cause of
cancer-related mortality (1). The
5-year survival rate of GC remains low (25%) (2,3).
Although great progress has been made in the availability of
treatment techniques for GC, the prognosis for GC patients remains
poor due to the biological properties of tumor cells, including a
high propensity for invasiveness and metastasis, rapid
proliferation, and anti-apoptotic behavior (4). Therefore, studying the pathogenesis
of GC may improve the diagnosis, treatment and prognosis of GC.
Recently, the migration, invasion and other biological functions of
GC have attracted considerable attention, and various proteins and
signaling pathways associated with the invasiveness and metastasis
of GC in patients have been found (5-7).
However, many of the molecular mechanisms of GC metastasis remain
unknown.
Stathmin (STMN) is a microtubule-regulating protein
that has a critical role in the aggregation and depolymerization of
mitotic spindles. In previous studies, STMN overexpression has been
demonstrated in a variety of tumors and to contribute to the
occurrence and development of gastrointestinal malignancies
(8-11). In addition, previous studies have
confirmed that STMN is associated with cell differentiation,
proliferation and prognosis in tumors, particularly in correlation
with migration and invasion of GC cells (12-14).
These findings indicate that STMN could be a potential molecular
marker and target in GC gene therapy. However, few studies have
focused on the mechanism by which STMN promotes migration and
invasion in stomach neoplasm.
In the present study, the isobaric tags for relative
and absolute quantitation (iTRAQ) methodology was used to identify
the differentially expressed proteins (DEPs) in GC cells with or
without silencing of the STMN gene. Following verification and
further analysis by reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and western blotting, the DEPs clusterin,
cystatin C, and matrix metalloproteinases (MMPs) were focused on,
which have recently been reported to be closely associated with
metastasis and invasiveness in malignant tumors. Additionally, the
biological processes of STMN-silenced GC cells were studied.
Elucidating the underlying molecular mechanisms in STMN-induced GC
metastasis may provide a theoretical basis for gene therapy in GC
patients, and a novel pathway was identified that may be
responsible for the promotional effect of STMN in GC
metastases.
Materials and methods
Reagents
Eight-plex iTRAQ kits were purchased from Applied
Biosystems; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). All
electrophoresis reagents used in this study were acquired from
Bio-Rad Laboratories, Inc. (Hercules, CA, USA). CytoSelect™ 24-well
Cell Migration and Invasion assay kits (8 µm, colorimetric
format) were purchased from Cell Biolabs, Inc. (San Diego, CA,
USA). Opti-MEM was purchased from Gibco; Thermo Fisher Scientific,
Inc. Lipofectamine 2000, STMN-specific small interfering (si) RNA
oligonucleotides (HSS180637 and HSS142799) and a negative control
(12935-400) were acquired from Invitrogen; Thermo Fisher
Scientific, Inc. Monoclonal antibodies against STMN (TA325913S) and
apolipoprotein E (APOE; TA805358) were obtained from OriGene
Technologies, Inc. (Rockville, MD, USA). Antibodies against LI
Cadherin (CDH17; ab109220), clusterin (sCLU; ab92548), cystatin-C
(CST3; ab133495), cathepsin D (CTSD; ab134169), fascin (FSCN;
ab126772), heat shock protein 90 (HSP90; ab13492), MMP9 (ab38898),
MMP1 (ab137332), signal transducer and activation of transcription
(STAT3; ab68153), phosphorylated (p)-STAT3 (ab76315), protein
kinase B (AKT; ab8805), p-AKT (ab38449), cyclin-dependent kinase 1
(CDK1; ab18) and superoxide dismutase 1 (SOD1; ab13498) were
acquired from Abcam (Cambridge, MA, USA).
Patients and cell lines
The tumor specimens used in this study were 40
samples obtained from 40 GC patients (11 female and 29 male
patients; mean age, 59 years) who underwent curative resection at
Infectious Disease Department of the Second Affiliated Hospital of
Chongqing Medical University between September 2015 and October
2016 (Chongqing, China; Table I).
All the studied cases met the following conditions: i) Patients
with resectable GC; ii) diagnosed via at least two types of
examination; iii) clinical staging was made by at least two types
of imaging examination; iv) patients without preoperative
chemotherapy orradiotherapy; and v) patients without a history of a
drug-related allergy. Among the clinical patients, the majority of
GC patients had neoplasm of intermediate differentiation (stage III
or IV). Non-cancerous tissues were obtained from the distal edge of
the resection, ≥10 cm from the GC tissues. All methods used in the
present study were approved by the Ethics Committee of Chongqing
Medical University (Chongqing, China), and all patients provided
written informed consent prior to participation. The human GC cell
line (AGS) was purchased from the American Type Culture Collection
(Manassas, VA, USA), grown in Dulbecco's modified Eagle's medium
(DMEM; GE Healthcare Life Sciences, Logan, UT, USA) medium
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and penicillin, and incubated in an atmosphere of
5.0% CO2 at 37°C.
 | Table IClinical and pathological data of 40
gastric cancer patients. |
Table I
Clinical and pathological data of 40
gastric cancer patients.
| Sample no. | Sex | Age (years) | Tumor position | Pathology | Grade | Stage | TNM | Type |
|---|
| 1 | F | 78 | Stomach | Adenocarcinoma | 1 | II | T3N0M0 | Malignant |
| 2 | M | 67 | Stomach | Adenocarcinoma | 1 | IB | T2N0M1 | Malignant |
| 3 | F | 59 | Stomach | Adenocarcinoma | 1 | II | T3N0M0 | Malignant |
| 4 | M | 50 | Stomach | Adenocarcinoma | 1 | II | T3N0M0 | Malignant |
| 5 | F | 51 | Stomach | Adenocarcinoma | 1 | II | T3N0M0 | Malignant |
| 6 | F | 68 | Stomach | Adenocarcinoma | 2 | Ib | T2N0M0 | Malignant |
| 7 | M | 48 | Stomach | Adenocarcinoma | 1 | Ib | T2N0M0 | Malignant |
| 8 | M | 56 | Stomach | Adenocarcinoma | 2 | IB | T2bN0M0 | Malignant |
| 9 | M | 57 | Stomach | Adenocarcinoma | 2 | IB | T2bN0M0 | Malignant |
| 10 | M | 74 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 11 | M | 52 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 12 | M | 68 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 13 | M | 51 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 14 | F | 55 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 15 | M | 53 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 16 | M | 54 | Stomach | Adenocarcinoma | 2 | Ib | T2N0M0 | Malignant |
| 17 | M | 69 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 18 | M | 72 | Stomach | Adenocarcinoma | 3 | II | T3N0M0 | Malignant |
| 19 | M | 56 | Stomach | Adenocarcinoma | 3 | II | T3N0M0 | Malignant |
| 20 | F | 53 | Stomach | Adenocarcinoma | 3 | II | T3N0M0 | Malignant |
| 21 | M | 61 | Stomach | Adenocarcinoma | 2 | IIIA | T3N1M0 | Malignant |
| 22 | M | 49 | Stomach | Adenocarcinoma | 2 | II | T3N0M0 | Malignant |
| 23 | M | 53 | Stomach | Adenocarcinoma | 3 | II | T3N0M0 | Malignant |
| 24 | M | 54 | Stomach | Adenocarcinoma | 3 | II | T3N0M0 | Malignant |
| 25 | M | 50 | Stomach | Adenocarcinoma | 3 | II | T3N0M0 | Malignant |
| 26 | F | 63 | Stomach | Adenocarcinoma | 3 | II | T3N0M0 | Malignant |
| 27 | F | 68 | Stomach | Adenocarcinoma | 3 | I | T2N0M0 | Malignant |
| 28 | M | 71 | Stomach | Adenocarcinoma | 3 | IIIA | T3N1M0 | Malignant |
| 29 | M | 61 | Stomach | Adenocarcinoma | 3 | Ib | T2N0M0 | Malignant |
| 30 | M | 47 | Stomach | Adenocarcinoma | 3 | II | T2N1M0 | Malignant |
| 31 | M | 58 | Stomach | Adenocarcinoma | 3 | II | T3N0M1 | Malignant |
| 32 | F | 63 | Stomach | Adenocarcinoma | 3 | Ib | T2N0M0 | Malignant |
| 33 | M | 66 | Stomach | Adenocarcinoma | 3 | Ib | T2N0M0 | Malignant |
| 34 | M | 52 | Stomach | Adenocarcinoma | 3 | Ib | T2N0M0 | Malignant |
| 35 | F | 54 | Stomach | Adenocarcinoma | 3 | IIIB | T4N1M0 | Malignant |
| 36 | F | 51 | Stomach | Adenocarcinoma | 3 | IIIa | T3N1M0 | Malignant |
| 37 | M | 64 | Stomach | Carcinoma | - | II | T3N0M0 | Malignant |
| 38 | M | 73 | Stomach | Adenocarcinoma | 3 | Ib | T2N0M0 | Malignant |
| 39 | M | 62 | Stomach | Carcinoid | - | IIIa | T3N1M0 | Carcinoid |
| 40 | M | 50 | Stomach | Adenocarcinoma | - | IIIa | T4N0M0 | Malignant |
Immunohistochemistry (IHC) and tissue
microarrays
Tissue microarrays, containing 40 GC tissues and 40
adjacent non-cancerous gastric tissues (ST801a; Alenabio, Xian,
China) were procured from US Biomax, Inc. (Rockville, MD, USA). The
tissue samples were fixed (24 h, 4°C) with 1% formalin, embedded in
paraffin and cut into sections (thickness, 5 µm). Following
dewaxing with xylene, the sections were rehydrated using a
descending alcohol series (100, 95 and 70%). The samples were
washed thrice with PBS (5 min/wash) and subjected to heat-induced
(100°C) antigen retrieval in a 0.01 M sodium citrate buffer for 5
min. Endogenous peroxidase activity was quenched with 3%
H2O2 (10 min). The sections were blocked with
5% bovine serum albumin (Beijing Solarbio Science & Technology
Co., Ltd., Beijing, China) for 0.5 h and incubated with primary
antibodies against STMN (1:100) overnight at 4°C. Detection was
performed on an Envision/Horseradish Peroxidase system (K4006;
DakoCytomation, Glostrup, Denmark), and all slides were
counterstained at room temperature with Gill's hematoxylin for 1
min, dehydrated and mounted for light microscope analysis
(magnification, ×20).
The IHC score was used to evaluate the different
expression of stathmin in GC and non-cancerous tissues. The score
was calculated as the product of the percentage of positive cells
and the intensity of staining. Staining intensity scoring criteria
were as follows: No staining scored 0, yellow staining scored 1,
yellow-brown staining scored 2, and brown staining scored 3.
Scoring for the proportion of positive cells in the scale was
performed as follows: The number of positive cells <10% scored
0, 10-40% scored 1, 40-70% scored 2 and >70% scored 3.
STMN siRNA transfection
AGS cells were transfected with 50 nM of either
STMN-specific siRNA (Invitrogen; Thermo Fisher Scientific, Inc.), a
negative control siRNA (12935-400) or blank control using
Lipofectamine 2000 (Life Technologies; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol, and opti-MEM
(Gibco). Following transfection, the cells were cultured (37°C) in
high-glucose DMEM supplemented with 10% FBS. Following 48 h of
incubation, follow-up experiments were performed, siRNA sequences
used were as follows: STMN1 siRNA, 5′-AGCCCUCGGUCAAAAGA AU-3′;
STMN2 siRNA, 5′-CAACAUCUAUACUUACGAU-3′; STMN3 siRNA,
5′-CUGUCUAGAUGCAACUUUU-3′ and negative control siRNA,
5′-UUCUCCGAACGUGUCACGU-3′.
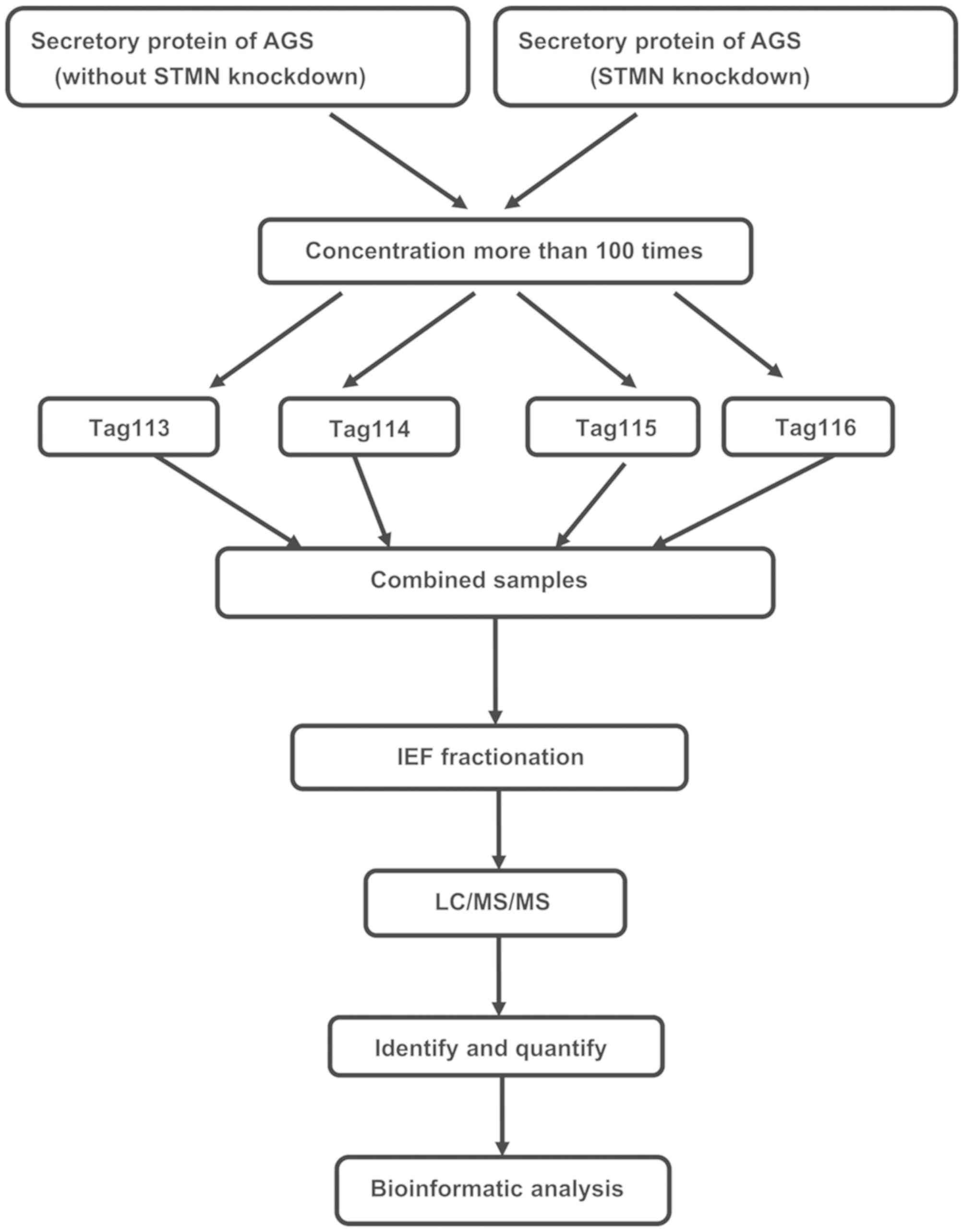
Protein collection and iTRAQ
labeling
Secretory proteins of AGS were collected, filtered
with a needle filter, and concentrated by centrifugation (3,000 ×
g; 4°C; 45 min) with an Amicon® centrifugal filter (EMD
Millipore, Billerica, MA, USA). Protein concentration was
determined using a 2-D Quant kit (GE Healthcare, Chicago, IL, USA),
according to the manufacturer's instructions. A total of 100
µg each protein sample was precipitated, denatured, cysteine
blocked (60°C; 1 h; Sigma-Aldrich; Merck KGaA), and digested with
sequencing-grade modified trypsin. Following the use of iTRAQ
Reagent-8PLEX mixed with the corresponding protein samples, the
pooled proteins from the STMN-silenced AGS cells were labeled with
tags 114/116, and the pooled proteins from AGS cells without STMN
knockdown were labeled with tags 113/115. All labels were from the
iTRAQ Reagent-8PLEX Multiplex kit (Sigma-Aldrich; Merck KGaA) and
the manufacturer's protocol was followed. The labeled samples were
pooled prior to analysis.
Fractionation of peptides
The pooled iTRAQ-labeled samples were dissolved in
300 µl 1% Pharmalyte (GE Healthcare) and 8 M urea, and
applied to IPG gel strips (pH 3-10; GE Healthcare) at 30 V for 14 h
(20°C). The peptides were electrofocused successively at 500 V for
1 h, 1,000 V for 1 h, 3,000 V for 1 h and 8,000 V for 8.5 h for a
total of 68 kV/h on an IPGphor system (GE Healthcare). The strips
were removed and sliced into 36 sections of 5-mm thickness.
Peptides were extracted from the gel using a 0.1% formic acid and
2% acetonitrile solution. The fractions were purified and
concentrated on a C18 Discovery DSC-18 SPE column (Sigma-Aldrich;
Merck KGaA). The purified fractions were lyophilized and stored at
−20°C prior to mass spectrometric analysis.
Mass spectrometry and database
search
Mass spectrometry was performed using a Qstar Elite
mass spectrometer coupled to a Dionex Ultimate 3000 liquid
chromatography system (Dionex Corporation, Sunnyvale, CA, USA). The
purified, labeled peptides were reconstituted in a solution
containing 2% acetonitrile solution and 0.1% formic acid and
injected into the mass spectrometer. A gradient series for each
analysis was loaded on a C18-PepMap column (2 µm x 1,000
µm x 50 mm; Dionex Corporation) at a flow rate of 0.3
μl/min. The two most abundantly charged ions above 20 counts were
selected for mass spectrometry. Dynamic exclusion criteria was set
to 30 sec with a ±50 kDa mass tolerance. Protein identification and
quantification was performed using ProteinPilot v.2.0 (Thermo
Fisher Scientific, Inc.). Mass spectrometry data were processed by
searching the UniprotKB database (https://www.uniprot.org/help/uniprotkb/). Methane
thiosulfate was set as a fixed cysteine modification. Protein
identification was based on selection thresholds of
ProtScore>1.3 or ProtScore<0.77, and false discovery rate
P-values <0.05. The PANTHER Classification System (www.pantherdb.org) was utilized to group the DEPs
according to their biological processes and molecular
functions.
RNA extraction and RT-qPCR
Total RNA of AGS was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. First-strand cDNA was synthesized from 2
µg total RNA using a Reverse Transcription kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
In order to determine the relative level of cDNA in the reverse
transcribed samples, a SYBR Fast qPCR kit (Kapa Biosystems; Roche
Diagnostics, Basel, Switzerland) was used to perform qPCR using the
following primers: STMN, forward 5′-AGAACCGAGAGGCACAAATGGC-3′ and
reverse 5′-TCTCGTCAGCAGGGTCTTTGGA-3′; actin, forward
5′-CACCATTGGCAATGAGCGGTTC-3′ and reverse
5′-AGGTCTTTGCGGATGTCCACGT-3′.
The Kapa SYBR Fast qPCR kit was also used to perform
qPCR with primers (Invitrogen; Thermo Fisher Scientific, Inc.) for
APOE (NM_000041), forward 5′-GGGTCGCTTTTGGGATTACCTG-3′ and reverse
5′-CAACTCCTTCATGGTCTCGTCC-3′; HSP90 (NM_005348), forward
5′-TCTGCCTCTGGTGATGAGATGG-3′ and reverse
5′-CGTTCCACAAAGGCTGAGTTAGC-3′; FSCN1 (NM_003088), forward
5′-GACACCAAAAAGTGTGCCTTCCG-3′ and reverse
5′-CAAACTTGCCATTGGACGCCCT-3′; CA LM2 (NM_001743), forward
5′-AGTGCTGCAGAACTTCGCCA TG-3′ and reverse
5′-CAAGGTCTTCACTTTGCTGTCATC-3′; CTSD (NM_001909), forward
5′-GCAAACTGCTGGACATCGCTTG-3′ and reverse
5′-GCCATAGTGGATGTCAAACGAGG-3′; CST3 (NM_000099), forward
5′-CCTTCCATGACCAGCCACATCT-3′ and reverse
5′-AGGCGTCCTGACAGGTGGATTT-3′; CDH17 (NM_004063), forward
5′-GGCAATGTGACTGCCAAGGATC-3′ and reverse
5′-GCTTCTCTGTCCAATGGAGCCA-3′; MMP1 (NM_002421), forward
5′-ATGAAGCAGCCCAGATGTGGAG-3′ and reverse
5′-TGGTCCACATCTGCTCTTGGCA-3′; CLU (NM_001831), forward
5′-TGCGGATGAAGGACCAGT GTGA-3′ and reverse
5′-TTTCCTGGTCAACCTCTCAGCG-3′; MAPK1 (NM_002745), forward
5′-ACACCAACCTCTCGTACATCGG-3′ and reverse
5′-TGGCAGTAGGTCTGGTGCTCAA-3′; LGALS1 (NM_002305), forward
5′-AGCAGCGGGAGGCTGTCTTTC-3′ and reverse
5′-ATCCATCTGGCAGCTTGACGGT-3′; PARK7 (NM_007262), forward
5′-GTCCTACTGCTCTGTTGGCTCA-3′ and reverse
5′-CCACACGATTCTCAGAGTAGGTG-3′; BSG (NM_001728), forward
5′-GGCTGTGAAGTCGTCAGAACAC-3′ and reverse
5′-ACCTGCTCTCGGAGCCGTTCA-3′; CD9 (NM_001769), forward
5′-TCGCCATTGAAATAGCTGCGGC-3′ and reverse
5′-CGCATAGTGGATGGCTTTCAGC-3′; SDCBP2 (NM_080489), forward
5′-AGTCCAGGCAACAGCCATT TCC-3′ and reverse
5′-AAGCAGGCTCTCCTGGACTTCT-3′; CST4 (NM_001899), forward
5′-TGTGCCTTCCATGAACAGCCAG-3′ and reverse
5′-CCTAGGCTTCTTGACACCTGGA-3′; PFN1 (NM_005022), forward
5′-CATCGTGGGCTACAAGGACTCG-3′ and reverse
5′-CCAAGTGTCAGCCCATTCACGT-3′; CACYBP (NM_014412), forward
5′-CTGCTGTGGTTGCTCCCATTAC-3′ and reverse
5′-CACCTGCACATTCTCAGTGGGA-3′; GSTM5 (NM_000851), forward
5′-CACATGGAGCTGGTCAGACTGT-3′ and reverse 5′-
CTTGTCTCCTGCAAACCATGGC-3′; COT L1 (NM_021149), forward
5′-AAATATGACGGCTCCACCATC GT-3′ and reverse
5′-TGGACCTCTTGCTCATGGCATC-3′; COL4A2 (NM_001846), forward
5′-GGATAACAGGCGTGACTGGAGT-3′ and reverse
5′-CTTTGCCACCAGGCAGTCCAAT-3′; and CTSH (NM_004390), forward
5′-TACCTTCGAGGTACTGGTCCCT-3′ and reverse
5′-GGTGGAGAAAGTCCAGCAACTG-3′. qPCR reaction was performed according
to the instructions included in the Kapa SYBR Fast qPCR kit. Data
were normalized to actin levels in all samples, and qPCR was
carried out at 94°C for 60 sec, 37°C for 60 sec, and at 72°C for
120 sec, for 25-30 cycles in total. Quantification of gene
expression was calculated using the 2-ΔΔCq method
(15). RT-PCR analyses were
conducted in triplicate.
Western blot analysis
GC cells were lysed with lysis buffer containing 1
mM EDTA (pH 8.0), 0.5% IGEPAL, 50 mM, pH 7.5 Tris-HCl, 50 mM sodium
fluoride, 150 mM NaCl, 1 mM sodium orthovanadate, 0.5% Triton X-100
and protease inhibitors. The concentration of extracted proteins
was determined via an Enhanced BCA Protein Assay kit (Beyotime
Institute of Biotechnology, Haimen, China). Approximately 30
µg protein specimens were separated by 10% SDS-PAGE and
transferred to polyvinylidene difluoride membranes. The membranes
were blocked with 5% skimmed milk in TBS-Tween-20 buffer (TBST; pH
7.6; 0.5% Tween-20) for 1 h at room temperature. The membranes were
incubated with monoclonal antibodies against STMN (1:300), sCLU
(1:2,000), CST3 (1:4,000), CTSD (1:2,000), MMP9 (1:1,000), MMP1
(1:1,000), STAT3 (1:2,000), p-STAT3 (1:2,000), AKT (1:500), p-AKT
(1:500), CDK1 (1:500), SOD1 (1:1,000), FSCN (1:1,000), CDH17
(1:1,000), APOE (1:2,000) and Hsp90 (1:500), overnight at 4°C. The
membranes were washed thrice with TBST and incubated with a
horseradish peroxidase-conjugated goat anti-mouse (sc-2039)
immunoglobulin G (IgG) or goat anti-rabbit (sc-2040) IgG antibody
(both 1:5,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for
1 h at room temperature. The membranes were washed thrice with TBST
and visualized with an ECL detection system (Bio-Rad Laboratories,
Inc.). Western blot analyses were performed in triplicate using
Image Lab software (Bio-Rad Laboratories, Inc).
Transwell and wound-healing assays
Wound-healing, and cell migration and invasion
assays were conducted 2 days following transfection. The
wound-healing assay was performed in 6-well plates with 60% cells.
When the cultured cells reached 100% confluence, a sterile p200
pipette tip was used to incise a wound in the cell monolayer and
the debris was removed by gently washing with PBS. Images of the
scratches were captured at 0 and 24 h under (37°C) a phase contrast
microscope (magnification, ×20). The capacity of cell migration was
determined by the extent of gap closure.
The Transwell migration and invasion assays were
performed using a 24-well Cell Migration and Invasion Assay kit
(Cell Biolabs, Inc., San Diego, CA, USA). In brief, following
transfection with STMN or control siRNA, AGS cells starved for 24 h
(37°C), and harvested and resuspended in serum-free media,
according to the manufacturer's protocol. The lower chambers were
filled with 500 µl media (RPMI-1640 plus 10% FBS), and
~3×105 cells/300 µl media were loaded into the
upper chamber, with or without Matrigel. The migrating/invading
cells on the bottom of the filters were stained, fixed and
extracted, according to the manufacturer's instructions, and the
optical density was measured at 560 nm at 12 or 24 h following
seeding. The determination of STMN downregulation was made via
western blotting analysis, as detailed above. All experiments were
performed in triplicate.
Cell proliferation assay and flow
cytometry
Cell proliferation was analyzed using an MTT assay.
Briefly, AGS cells were seeded in 96-well plates at a density of
1.5×103 cells/well. Cells were transfected with STMN
siRNA or control siRNA, as detailed above, and cultured in DMEM
media supplemented with 10% FBS for 0, 24, 48, 72 and 96 h at 37°C.
The cells were then incubated with 20 µl MTT (Sigma-Aldrich;
Merck KGaA) at 37°C for 4 h. The MTT substrate was dissolved in 200
µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) for 5 min.
Absorbance at 570 nm was then measured. All experiments were
performed in triplicate.
Cell apoptosis and cell cycle tests were analyzed by
flow cytometry (FACSCanto II; Becton, Dickinson and Company,
Franklin, Lakes, NJ, USA) after AGS cell were stained with an
Annexin V-FLUOS Staining kit (Sigma-Aldrich; Merck KGaA) and
propidium iodide and treated with STMN-specific siRNA and negative
control siRNA. All assays were performed independently at least
three times.
Statistical analysis
All experiments were performed at least in
triplicate. Continuous variables are presented as the mean ±
standard deviation. All statistical analyses were performed using
SPSS 20.0 software (IBM Corp., Armonk, NY, USA). Differences
between two groups were analyzed by paired or unpaired Student's
t-tests. One-way analysis of variance was used to compare multiple
groups, and when appropriate, it was followed by Fisher's least
significant difference post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Differential expression of STMN in
tissues
IHC was performed on tissue microarrays of GC and
non-cancerous tissues to identify the differential expression of
STMN. All 40 stomach neoplasm samples exhibited strong staining,
whereas the 40 non-cancerous samples exhibited no staining. The
results revealed that STMN was expressed at greater levels in GC
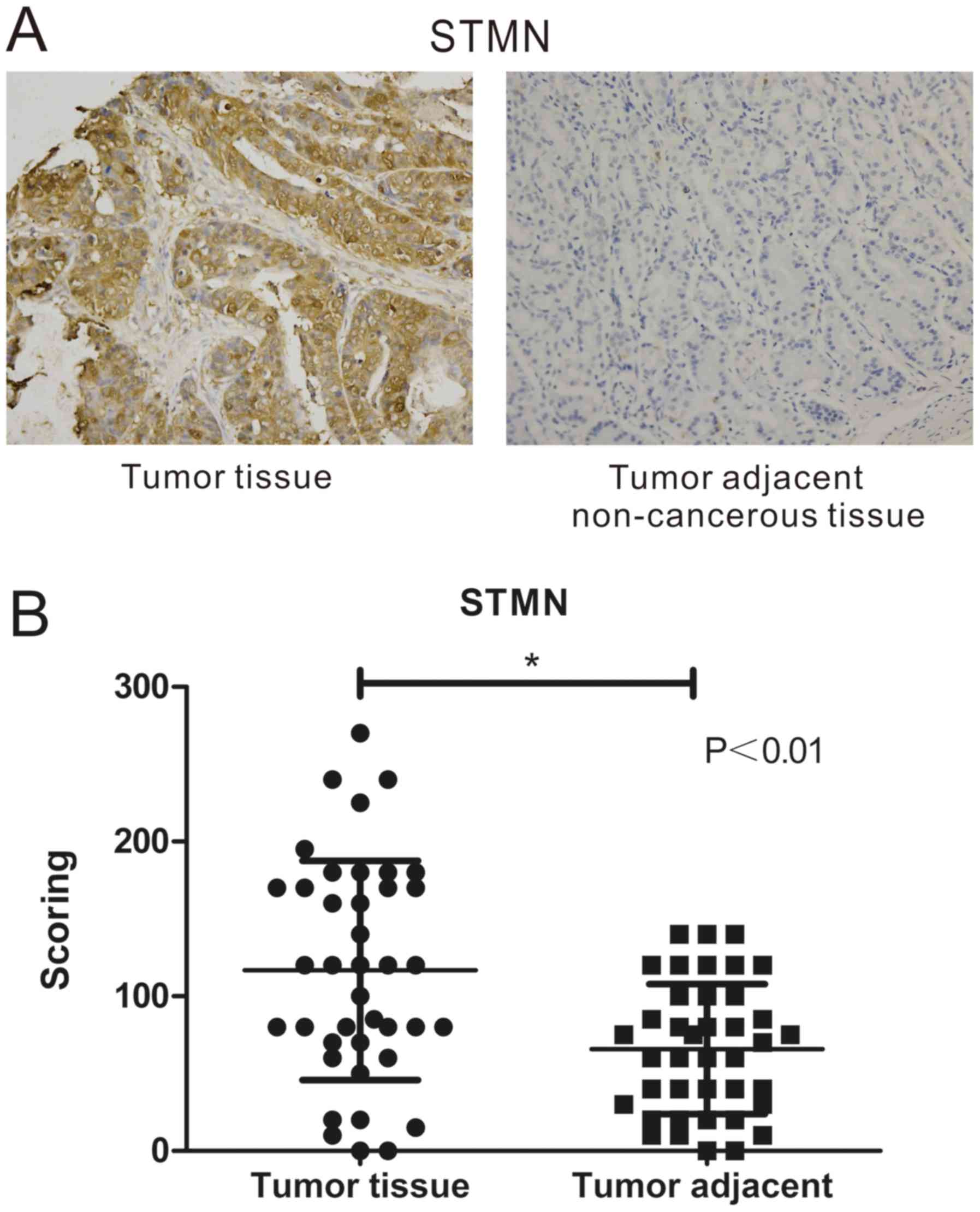
samples than in non-cancerous samples (Fig. 1A). The IHC score was used to
evaluate the different expression of STMN in GC and non-cancerous
tissues. IHC score values of STMN were significantly higher in the
GC tissue group than in the adjacent normal tissues group (Fig. 1B), Furthermore, paired Student's
t-test analysis denoted that the difference in STMN expression
between the stomach neoplasm tissues and the adjacent normal
tissues was statistically significant (P<0.001).
Effect of STMN on cell biological
processes in GC
As upregulation of STMN in GC samples was
demonstrated in the IHC results, the association of STMN with
biological processes was validated. RNA interference was used to
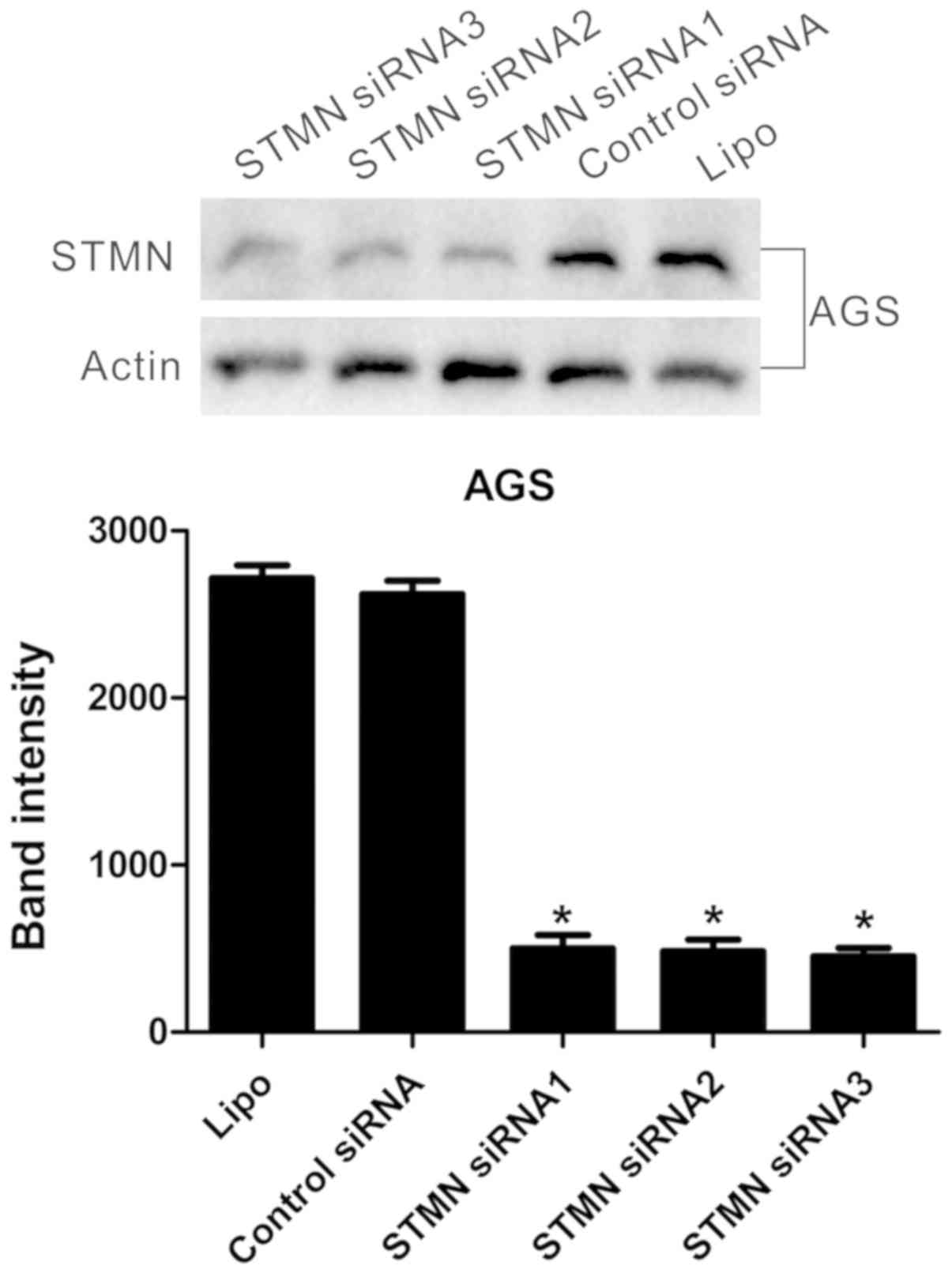
inhibit STMN expression in AGS GC cells. The cells were transfected
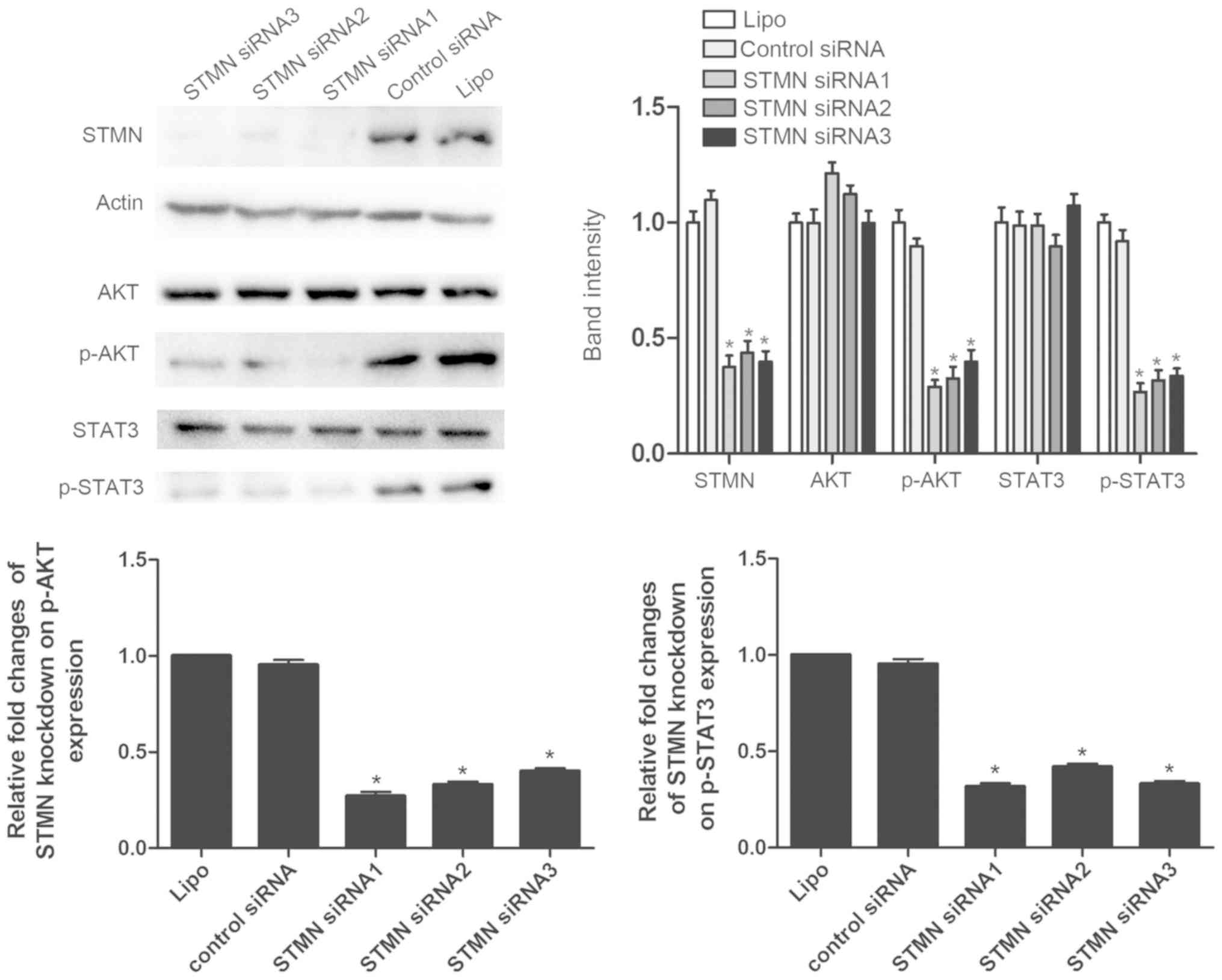
with three STMN-specific siRNA sequences and control siRNA. Western
blot analysis demonstrated that the STMN-specific siRNA
significantly downregulated the expression of STMN in the AGS GC
cell line (P<0.001; Fig.
2).
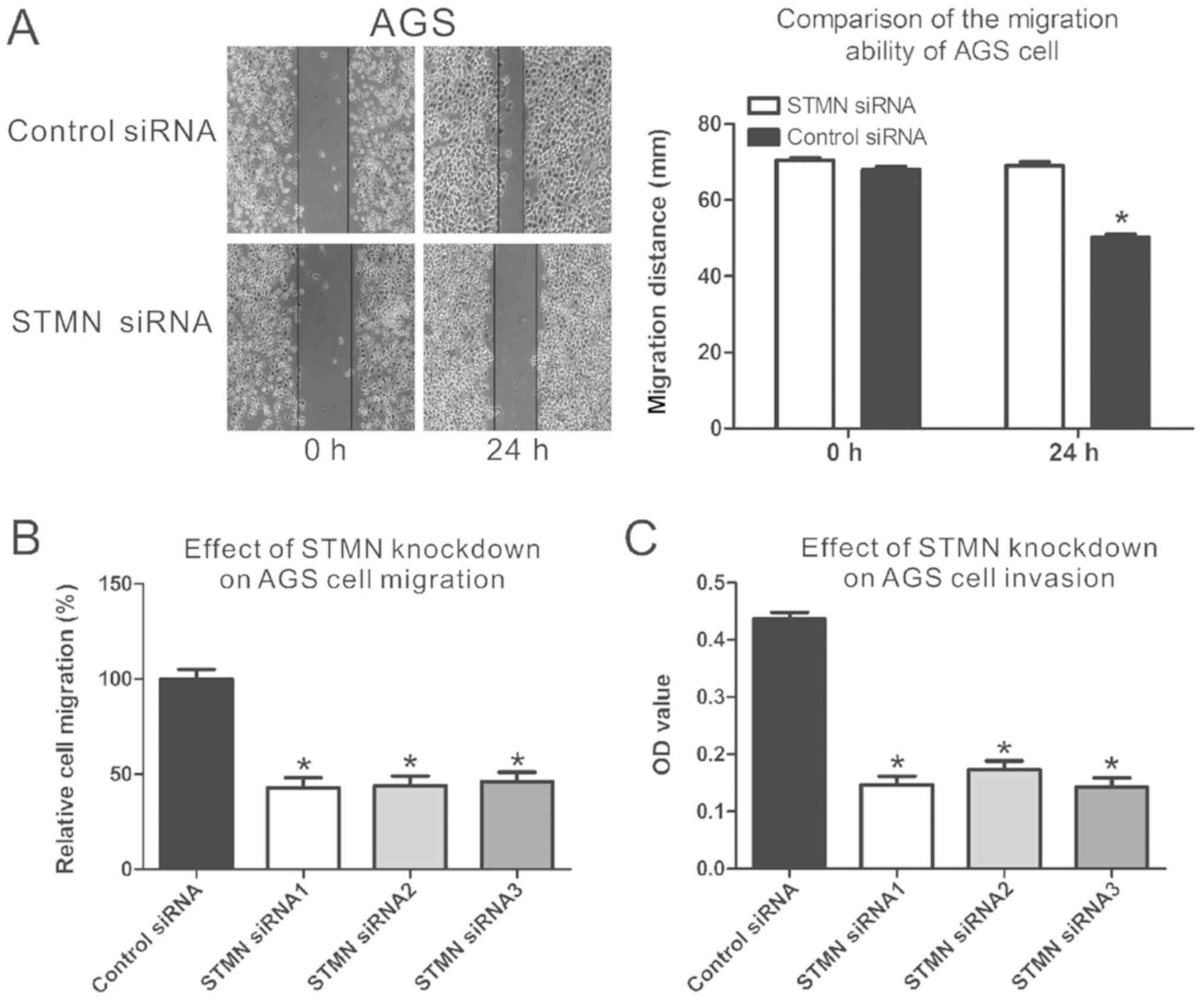
The results of the wound-healing assay demonstrated
that siRNA-mediated silencing of STMN significantly decreased the
ability of transfected cells to close scratch wounds in the AGS
cells (P<0.05; Fig. 3A). The
migration assay demonstrated that downregulated expression of STMN
weakened the migration ability of AGS cells by 45-48%, compared
with the control group (P<0.05; Fig. 3B). The invasion assay confirmed
that downregulation of STMN expression inhibited the invasive
capacity of AGS cells by 60%, in comparison with control siRNA
(P<0.05; Fig. 3C). These
results indicated that STMN has a crucial role in GC metastasis and
invasiveness.
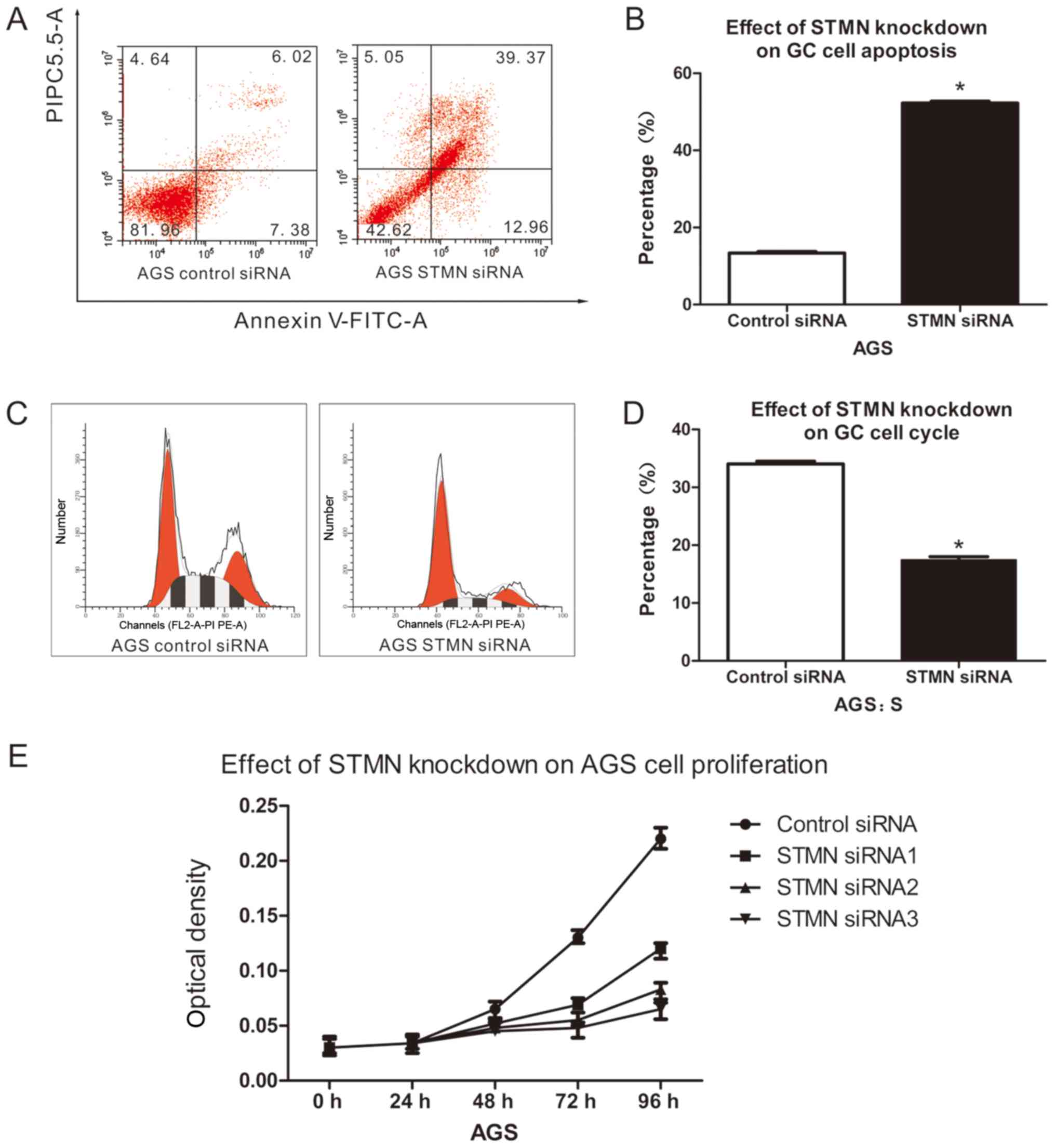
In order to determine whether the anti-apoptotic,
proliferation and cell cycle progression properties of GC were due
to upregulation of STMN, these capacities were assayed in cell
lines with and without STMN knockdown. STMN silencing resulted in a
×3.9 increase in apoptosis in AGS cells, compared with controls
(P<0.05; Fig. 4A and B).
Knockdown of STMN also increased S-phase cell cycle arrest by 50%
in the AGS cell line, compared with the siRNA control (P<0.05;
Fig. 4C and D). The MTT assay
demonstrated that proliferation of STMN-silenced AGS cells was
decreased compared with the control cells (P<0.05; Fig. 4E). Together, these results
indicated that STMN promotes invasion and metastasis, proliferation
and anti-apoptotic behavior in stomach neoplasm.
Analysis of iTRAQ data of DEPs
In order to investigate the mechanism by which STMN
influences biological functions in GC cells, the iTRAQ proteomics
approach was employed to discern differentially expressed proteins.
Fig. 5 presents the flow chart of
the iTRAQ proteomics methodology. iTRAQ-based MS was performed on
proteins isolated from AGS cells following siRNA-mediated STMN
knockdown and from un-silenced control cells. In order to improve
the reliability and enhance the range of protein identification,
specimens were iTRAQ-labeled in duplicate. The ratios of 114:113
and 116:115 identified the differential protein expression in AGS
cells. A ProteinPilot-based database search was performed, with
thresholds set to achieve 95% confidence at a 5% false discovery
rate, and hundreds of proteins were identified. Conforming to
commonly accepted iTRAQ-based MS conventions (16), proteins with ratios of <0.77
(1/1.3) or >1.3 (1.3/1) were classified as downregulated or
upregulated, respectively. The technical variation of data from
duplicate experiments was <30%. A total of 96 proteins were
identified with a confidence of 95%, of which 45 were significantly
upregulated and 51 downregulated. Table II presents a list of 31 proteins
representative of the total identified proteins.
 | Table IIDifferentially expressed proteins in
supernatant of AGS cells: STMN knockdown vs. control. |
Table II
Differentially expressed proteins in
supernatant of AGS cells: STMN knockdown vs. control.
| Accession | Gene symbol | Protein name | Peptides (95%) | AGS STMN, knockdown
114:control 113 | P-value |
|---|
|
sp|Q99497|PARK7_HUMAN | PARK7 | Protein DJ-1 | 9 | 0.73154 | 0.0104 |
|
tr|Q54A51|Q54A51_HUMAN | BSG | Basigin | 5 | 0.729301 | 0.0104 |
|
tr|G8JLH6|G8JLH6_HUMAN | CD9 | Tetraspanin | 3 | 0.514838 | 0.0052 |
|
sp|Q9H190|SDCB2_HUMAN | SDCBP2 | Syntenin-2 | 3 | 0.71116 | 0.0028 |
|
sp|P01036|CYTS_HUMAN | CST4 | Cystatin-S | 25 | 0.287784 | 0.0004 |
|
sp|P02649|APOE_HUMAN | APOE | Apolipoprotein
E | 19 | 0.488746 | 0.0001 |
|
sp|P07339|CATD_HUMAN | CTSD | Cathepsin D | 39 | 0.629353 | 0.0006 |
|
sp|P01034|CYTC_HUMAN | CST3 | Cystatin-C | 30 | 0.457414 | 0.0020 |
|
sp|Q12864|CAD17_HUMAN | CDH17 | Cadherin-17 | 22 | 0.499042 | 0.0069 |
|
tr|A8K690|A8K690_HUMAN | HSP90 | Hsp90 | 18 | 0.739335 | 0.0407 |
|
sp|P10909|CLUS_HUMAN | CLU | Clusterin | 19 | 0.594053 | 0.0002 |
|
sp|Q14764|MVP_HUMAN | MVP | Major vault
protein | 18 | 0.706437 | 0.0005 |
|
sp|P08572|CO4A2_HUMAN | COL4A2 | Collagen α-2(IV)
chain | 17 | 1.038458 | 0.0007 |
|
sp|P09382|LEG1_HUMAN | LGALS1 | Galectin-1 | 15 | 0.687828 | 0.0039 |
|
tr|Q6IBC3|Q6IBC3_HUMAN | CTSH | CTSH protein | 14 | 0.874563 | 0.0387 |
|
tr|Q53G96|Q53G96_HUMAN | MMP1 | Matrix
metalloproteinase-1 preproprotein | 10 | 0.662887 | 0.0049 |
|
sp|P14780|MMP9_HUMAN | MMP9 | Matrix
metalloproteinase-9 | 3 | 0.57889 | 0.0030 |
|
tr|J3QR44|J3QR44_HUMAN | CDK1 | Cyclin-dependent
kinase 1 | 3 | 0.577492 | 0.0076 |
|
sp|Q9Y3F4|STRAP_HUMAN | AKT | Serine-threonine
kinasereceptor-associated protein | 7 | 0.621259 | 0.0092 |
|
tr|B5BTZ6|B5BTZ6_HUMAN | STAT3 | Signal transducer
and activator of transcription | 9 | 0.71708 | 0.0261 |
|
sp|Q13347|EIF3I_HUMAN | EIF3I | Eukaryotic
translation initiation factor 3 subunit I | 3 | 0.807284 | 0.0469 |
|
tr|B3KQF4|B3KQF4_HUMAN |
Metalloproteinaseinhibitor 1 | Metalloproteinase
inhibitor 1 | 6 | 0.536465 | 0.0046 |
|
sp|P02647|APOA1_HUMAN | APOA1 | Apolipoprotein
A-I | 6 | 0.668486 | 0.0140 |
|
tr|B5BU83|B5BU83_HUMAN | STMN1 | Stathmin | 4 | 0.461021 | 0.0020 |
|
tr|B3KTA3|B3KTA3_HUMAN | Fascin | Fascin | 16 | 1.283847 | 0.0150 |
|
tr|H0Y7A7|H0Y7A7_HUMAN | CALM2 | Calmodulin | 35 | 1.389 | 0.0040 |
|
sp|P07737|PROF1_HUMAN | PFN1 | Profilin-1 | 35 | 1.266123 | 0.0080 |
|
tr|Q6NVY0|Q6NVY0_HUMAN | CACYBP | Calcyclin binding
protein | 10 | 1.202297 | 0.0260 |
|
sp|P46439|GSTM5_HUMAN | GSTM5 | Glutathione
S-transferase µ5 | 5 | 1.757358 | 0.0005 |
|
sp|Q14019|COTL1_HUMAN | COTL1 | Coactosin-like
protein | 6 | 1.835 | 0.0014 |
|
sp|P00441|SODC_HUMAN | SOD1 | Superoxide
dismutase | 9 | 1.305398 | 0.0023 |
Cellular and molecular functional
annotation of the DEPs
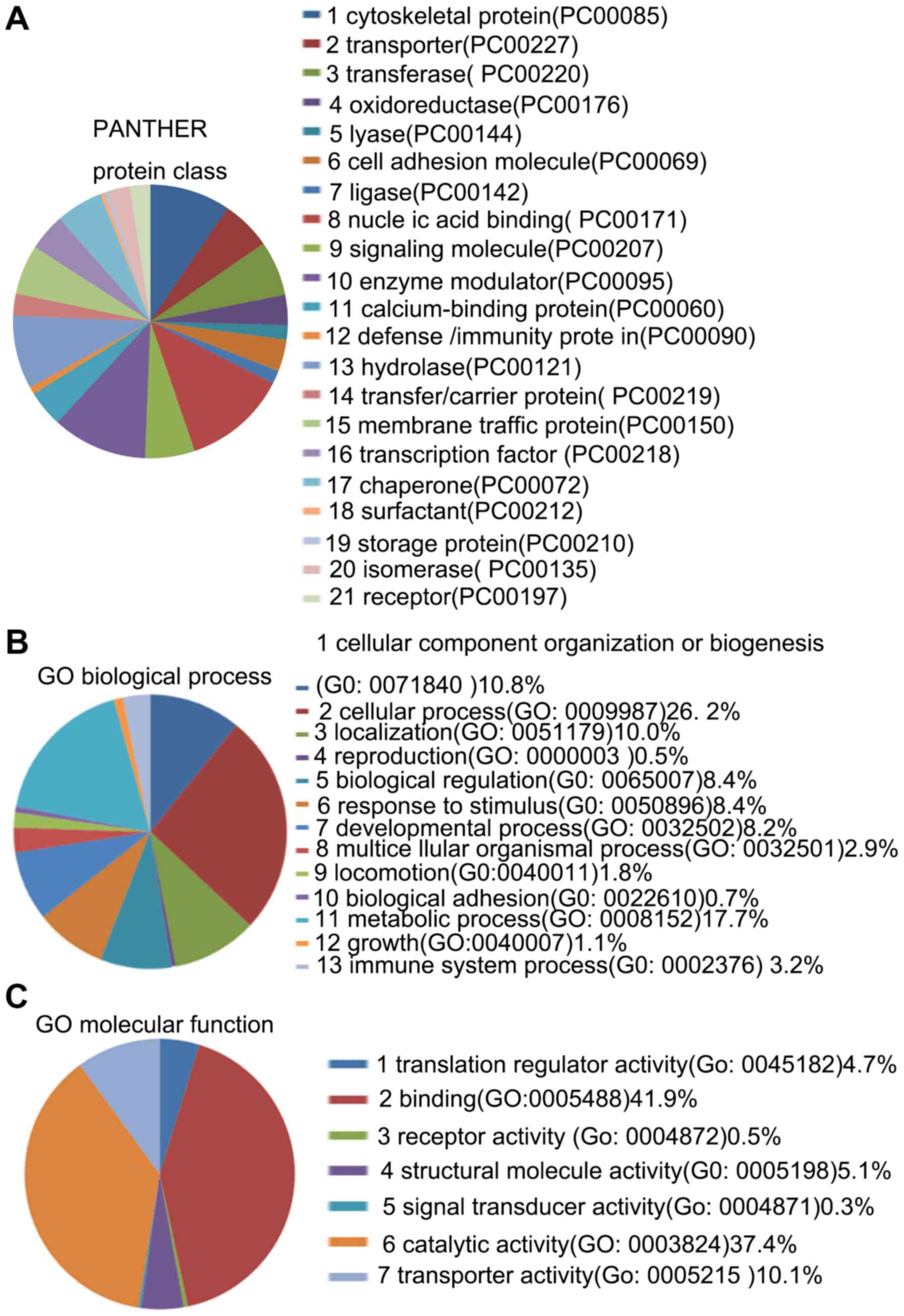
The 96 DEPs belonged to 21 protein classes, 13
biological processes and 7 molecular functions (Fig. 6). The results demonstrated that
'catalytic activity' and 'binding' were the most common molecular
functions and that 'metabolic process' was the most common
biological process.
Validation of differentially expressed
proteins identified by iTRAQ
RT-qPCR and western blot analyses were performed to
validate the differentially expressed proteins identified by iTRAQ.
RT-PCR determined the mRNA expression levels of PARK7, BSG, CD9,
SDCBP2, CST4, APOE, HSP90, FSCN1, CALM2, PFN1, CACYBP, GSTM5,
COTL1, CTSD, CST3, CDH17, LGALS1, sCLU, COL4A2, CTSH, MAPK1 and
MMP1. Actin was used as the control. The mRNA levels were
consistent with the results obtained via iTRAQ. Following knockdown
of STMN, the mRNA expression levels of BSG, FSCN, PFN1, CACYBP,
GSTM5, COTL1, COL4A2, CTSH and MAPK were increased, whereas CTSD,
CST3, CDH17, LGALS1, sCLU, MMP1, PARK7, CD9, SDCBP2, CST4, APOE,
HSP90 and CALM2 were decreased (P<0.05; Fig. 7A). Western blot analysis was
utilized to quantify the expression levels of the identified
proteins that were identified by iTRAQ and RT-qPCR. The levels of
the extracellular sCLU, CST3, CTSD, MMP1, MMP9 and the
intracellular CDK1, CTSD, HSP90 and SOD1 corresponded with the
results in the above iTRAQ and RT-qPCR analysis (P<0.05;
Fig. 7B). The expression levels of
CALM2, APOE and LGALS1 were too low to be detected by western blot
analysis and the differential expression of FSCN, CTSH and MAPK
were deemed to be without clinical significance.
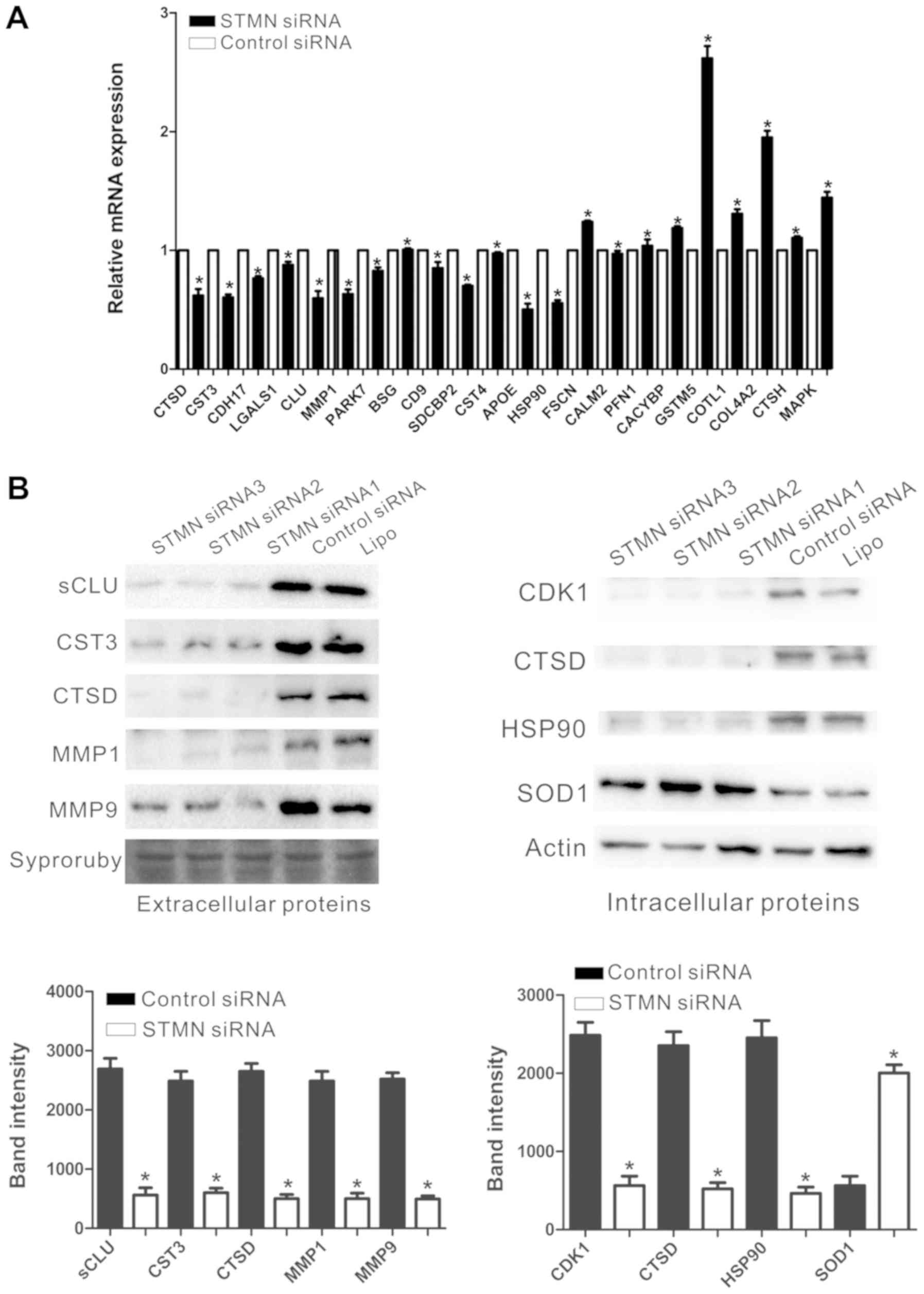 | Figure 7Validation of DEPs. (A) Reverse
transcription-quantitative polymerase chain reaction detected the
relative mRNA expression levels of a number of DEPs, as normalized
to actin. (B) A representative western blot analysis for sCLU,
CST3, CTSD, MMP1, MMP9, CDK1, HSP90 and SOD1 expression in the
experimental and control groups. *P<0.05 vs. control.
DEP, differentially expressed protein; sCLU, secretory clusterin;
CST3, cystatin-C; CTSD, cathepsin D; MMP, matrix metalloproteinase;
CDK, cyclin-dependent kinase; HSP90, heat shock protein 90; SOD,
superoxide dismutase. |
STMN promotes the development of GC via
different pathways
As has been confirmed by extensive studies,
activated AKT and STAT3 signaling is closely associated with the
biological functions of many tumors, also associated with GC
(17-26). Western blot analyses demonstrated
that the activity of AKT and STAT3 signaling was decreased
following inhibition of STMN expression (P<0.05; Fig. 8). These results revealed that STMN
promotes the development of GC via different pathways.
Discussion
GC is the third most common cause of cancer-related
mortality worldwide, and has attracted much attention due to its
relatively high rates of incidence and mortality (3). Although surgical resection and
chemotherapy are the mainstream methods of treating this
malignancy, patients suffering from advanced stage GC have poor
prognoses and high mortality rates. The vast majority of patients
already have metastasis by the time a diagnosis is made (27). Previous studies have confirmed that
the 5-year survival rate of tumors is substantially reduced for
patients with metastases (2,28).
Metastasis and recurrence are major obstacles to the improvement of
survival rates and quality of life in GC patients. Therefore,
investigations into the molecular mechanisms involved in GC
progression are necessary and may provide insights leading to
improved diagnosis and therapeutic approaches.
STMN is a microtubule regulating protein that has a
critical role in the aggregation and depolymerization of mitotic
spindles. The critical functions of STMN in cancer cells have been
investigated and it is known to participate in regulating many
cellular functions of gastrointestinal malignancies (14). STMN is frequently overexpressed in
many human cancers including lung (29), bladder (30), endometrial (31) and oral cancer (32).
Furthermore, STMN expression has been demonstrated
to be significantly associated with tumor cell biological
functions. For instance, STMN is associated with the proliferation,
differentiation and metastasis of cancers (33). High levels of STMN expression are
required for maintaining high proliferation rates in tumor cells
(34). A previous study also
concluded that high STMN expression was significantly associated
with tumor invasion and TNM clinical classification in esophageal
carcinoma (9). STMN has been
demonstrated to be an oncogene in many kinds of tumors, which
promotes proliferation, invasion and metastasis in a variety of
tumors (35,36). In the present study, the results of
the comparison of STMN expression between GC tissues and adjacent
non-cancerous tissues were in accordance with previous research. It
was demonstrated that STMN could significantly promote tumor cell
migration, invasion and proliferation and had an anti-apoptotic
effect in GC cells. These effects may be because the
microtubule-destabilizing activity of STMN interferes with
microtubule dynamics. The findings suggested that STMN may be a
pivotal factor contributing to the formation and progression of GC
and that STMN could be considered a valuable prognostic indicator
and therapeutic target in patients with GC.
Using the iTRAQ proteomics methodology, 96 DEPs were
identified in GC samples, the majority of which were involved in
metabolic and cellular processes. A number of them, including
HSP90, CTSD, CST3, sCLU, MMP1, SOD1, CDK1 and MMP9 were confirmed
using RT-qPCR and western blot analyses. Based on protein classes,
biological processes and molecular functions, the DEPs were further
classified into different types and several vital proteins that are
responsible for the apparent differences following silencing STMN
were evaluated.
CLU is a highly conserved glycoprotein with
ubiquitous tissue distribution. It appears to have two main
isoforms with vastly different functions: Secretory CLU (sCLU) and
intracellular CLU (nCLU). sCLU represents the major product of the
CLU gene (37,38). Recently, a number of studies have
demonstrated that expression of sCLU is significantly higher in
many types of cancer, compared with adjacent non-cancerous tissue
(37-40). Overexpression of sCLU has been
identified to be associated with tumor invasion, metastasis,
anti-apoptotsis, cell proliferation and survival in neoplasm of the
human bladder and in renal, liver, prostate, breast, lung and
gastrointestinal malignant tumors (37,41).
For example, overexpression of clusterin correlates with tumor
progression and metastasis in GC (40). The present results revealed that
inhibition of STMN significantly downregulated sCLU protein
expression in GC. These findings suggest that STMN promotes GC cell
invasion, metastasis, anti-apoptotsis, cell proliferation, survival
ability by regulating the expression of sCLU.
Signaling pathways involved in interactions with
sCLU were further investigated. A number of previous studies have
verified that numerous intracellular and extracellular proteins
promote tumor cell biological functions through activation of the
AKT pathway via upregulation of sCLU. A recent study reported that
CLU can promote HCC metastasis through AKT-MMP13 signaling
(42), and several studies have
mentioned that CDK1, SOD1 and CTSD regulate tumor cell biological
functions by influencing AKT signaling (43-47).
In the present study, it was demonstrated that the expression of
sCLU, CDK1, SOD1 and CTSD in the STMN-silenced group was
downregulated, compared with the control group. The AKT signaling
pathway is believed to act downstream of sCLU. For example, the
sCLU-AKT signaling pathway is responsible for cisplatin resistance
in human lung cancer (48).
Insulin-like growth factor-1 activates the P13K/AKT signaling
pathway via upregulation of sCLU in lung carcinoma (49). Clusterin facilitates metastasis by
eukaryotic translation initiation factor 3 subunit I
(EIF3I)/AKT/MMP13 signaling in hepatocellular carcinoma (42). Previous studies confirmed that
abnormal activation of the AKT signaling pathway was common in GC,
was associated with tumorigenesis of GC, and promoted tumor
migration and tumor aggressiveness in GC cells (17-21).
According to the western blot analysis in the present study, the
results demonstrated that the expression of sCLU and the
phosphorylation level of the AKT protein in the AGS cells were
markedly inhibited following knockdown of STMN.
Our results demonstrated that, following STMN
silencing, the expression levels of MMP9 and MMP1 were
downregu-lated and sCLU expression decreased. MMPs are produced by
various types of cancer cells. It has been reported that sCLU
induces matrix metalloproteinase-9 expression via phosphoinositide
3-kinase/AKT/nuclear factor-κB pathways in monocytes/macrophages
(50). This suggests that STMN
activates the AKT-MMP9 signaling pathway to promote GC cell
invasion, metastasis, anti-apoptotsis, cell proliferation and
survival via upregulation of sCLU.
A previous study on hepatocellular carcinoma
confirmed that overexpression of sCLU was accompanied by an
upregulation in levels of the EIF3I protein and that sCLU may
protect EIF3I from degradation. The sCLU-EIF3I complex may function
as a cooperative unit in cancer cells (42). Wang et al (42) also reported that EIF3I could form a
complex with AKT and lead to a constitutive activation of AKT
signaling. AKT phosphorylation was significantly inhibited when
EIF3I was silenced. These results suggest that sCLU forms a complex
with EIF3I and prevents its degradation, leading to upregulation of
AKT activity. Furthermore, according to iTRAQ results, the
expression of EIF3I was obviously decreased in GC when STMN was
silenced. In agreement with these analyses, the present findings
reveal a potential mechanism in GC wherein STMN significantly
regulates sCLU protein expression, and sCLU forms a complex with
EIF3I to activate the AKT signaling pathway. This, in turn,
promotes expression of MMP9 and leads to facilitated metastasis,
invasion, anti-apoptosis and cell proliferation of GC cells.
Conversely, the results of ITRAQ and western blot
analyses demonstrated that STMN-knockdown attenuated STAT3
activation in GC cells. Previous studies have demonstrated that the
STAT3 signaling pathway acts downstream of STMN and that activated
STAT3 upregulates MMP expression (51-54).
STAT3, which is considered a point of convergence for numerous
oncogenic signaling pathways, is constitutively activated in tumor
cells and is known to promote cell proliferation and angiogenesis
and serve a role in tumor avoidance of immune responses. Therefore,
the proliferative and anti-apoptotic effects of STMN in GC are
possibly associated with STAT3 signaling activation and concomitant
expression of MMPs.
The present results also revealed that inhibition of
STMN was accompanied by an abnormal regulation in the levels of
CST3, CDK1, CTSD, SOD1 and HSP90 proteins. Previous studies
reported the aberrant expression of these proteins in GC and that a
number of them are associated with AKT signaling pathway
activation, thus contributing to cell proliferation and metastasis
(55-62). As such, the present findings
suggested that STMN may promote the biological functions in GC by
combined interactions with these intracellular and extracellular
proteins.
In summary, the present study revealed several
mechanisms by which STMN regulates GC tumor cells. Primarily, STMN
significantly regulates sCLU protein expression and sCLU
facilitates metastasis, invasion, anti-apoptosis and cell
proliferation of GC cells via the EIF3I/AKT/MMP9 signaling pathway,
Secondly, STMN is possibly associated with STAT3 signaling
activation and overexpression of MMPs to influence the occurrence
and development of GC. These results indicate that targeting STMN
may be a rational strategy for suppressing the progression of GC.
The present results suggest a potential molecular pathway of STMN
mediating its influence on GC. Additional studies to verify the
pathway identified in the present study and to demonstrate the
conclusion in animal models are necessary to further elucidate the
molecular mechanisms underlying the effect of STMN in GC.
Furthermore, a drawback of the present study is that, generally, at
least two cell lines should be used in a well-structured study,
whereas only one GC cell line was used in the present study.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81171560, 30930082,
81171561, 30972584 and 81372399), the National Science and
Technology Major Project of China (grant nos. 2008ZX10002-006,
2012ZX1002007001, 2011ZX09302005, 2012ZX09303001-001 and
2012ZX10002003) and the Capital's Funds for Health Improvement and
Research (grant no. 2016-1-2112).
Availability of data and materials
Datasets included in the present study can be
obtained from the corresponding author.
Authors' contributions
FS and XZ performed all experiments. HT, SS and JH
performed a number of experiments. HR, HH and SP gave some critical
suggestions to this project. FS and XZ analyzed data and wrote the
manuscript. JW and YY were involved in the conception, design and
final approval of manuscript. YY obtained financial support, and
final approval of manuscript.
Ethics approval and consent to
participate
All methods used in the present study were approved
by the Ethics Committee of Chongqing Medical University (Chongqing,
China), and all patients provided written informed consent prior to
participation.
Patient consent for publication
All patients provided written informed consent prior
to participation.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Takeno S, Noguchi T, Kikuchi R, Sato T,
Uchida Y and Yokoyama S: Analysis of the survival period in
resectable stage IV gastric cancer. Ann Surg Oncol. 8:215–221.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bouriez D, Giraud J, Gronnier C and Varon
C: Efficiency of All-Trans Retinoic Acid on Gastric Cancer: A
Narrative Literature Review. Int J Mol Sci. 19:192018. View Article : Google Scholar
|
|
4
|
Li X, Liu Y, Cao B, Liu B, Bai T, Li X,
Mei L and Che X: Metastatic lymph node ratio and prognosis of
gastric cancer at different pT stages. Hepatogastroenterology.
62:507–511. 2015.PubMed/NCBI
|
|
5
|
Wang Q, Chen Q, Zhu L, Chen M, Xu W,
Panday S, Wang Z, Li A, Røe OD, Chen R, et al: JWA regulates
TRAIL-induced apoptosis via MARCH8-mediated DR4 ubiquitination in
cisplatin-resistant gastric cancer cells. Oncogenesis. 6:e3532017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu LP, Sheng XP, Shuai TK, Zhao YX, Li B
and Li YM: Helicobacter pylori promotes invasion and metastasis of
gastric cancer by enhancing heparanase expression. World J
Gastroenterol. 24:4565–4577. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie Y, Jin P, Sun X, Jiao T, Zhang Y, Li Y
and Sun M: SIX1 is upregulated in gastric cancer and regulates
proliferation and invasion by targeting the ERK pathway and
promoting epithelial-mesenchymal transition. Cell Biochem Funct.
Oct 31–2018, Epub ahead of print. View
Article : Google Scholar
|
|
8
|
Song Y, Mu L, Han X, Liu X and Fu S: siRNA
targeting stathmin inhibits invasion and enhances chemotherapy
sensitivity of stem cells derived from glioma cell lines. Acta
Biochim Biophys Sin (Shanghai). 46:1034–1040. 2014. View Article : Google Scholar
|
|
9
|
Wang F, Xuan XY, Yang X, Cao L, Pang LN,
Zhou R, Fan QX and Wang LX: Stathmin is a marker of progression and
poor prognosis in esophageal carcinoma. Asian Pac J Cancer Prev.
15:3613–3618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X, Liu H, Liang J, Yin B, Xiao J, Li
J, Feng D and Li Y: Stathmin is a potential molecular marker and
target for the treatment of gastric cancer. Int J Clin Exp Med.
8:6502–6509. 2015.PubMed/NCBI
|
|
11
|
Lu Y, Liu C, Cheng H, Xu Y, Jiang J, Xu J,
Long J, Liu L and Yu X: Stathmin, interacting with Nf-κB, promotes
tumor growth and predicts poor prognosis of pancreatic cancer. Curr
Mol Med. 14:328–339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ke B, Wu LL, Liu N, Zhang RP, Wang CL and
Liang H: Overexpression of stathmin 1 is associated with poor
prognosis of patients with gastric cancer. Tumour Biol.
34:3137–3145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Byrne FL, Yang L, Phillips PA, Hansford
LM, Fletcher JI, Ormandy CJ, McCarroll JA and Kavallaris M:
RNAi-mediated stathmin suppression reduces lung metastasis in an
orthotopic neuroblastoma mouse model. Oncogene. 33:882–890. 2014.
View Article : Google Scholar
|
|
14
|
Lu Y, Liu C, Xu YF, Cheng H, Shi S, Wu CT
and Yu XJ: Stathmin destabilizing microtubule dynamics promotes
malignant potential in cancer cells by epithelial-mesenchymal
transition. Hepatobiliary Pancreat Dis Int. 13:386–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Moriyama T, Ito A, Omote S, Miura Y and
Tsumoto H: Heat Resistant Characteristics of Major Royal Jelly
Protein 1 (MRJP1) Oligomer. PLoS One. 10:e01191692015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang ZQ, Cai Q, Hu L, He CY, Li JF, Quan
ZW, Liu BY, Li C and Zhu ZG: Long noncoding RNA UCA1 induced by SP1
promotes cell proliferation via recruiting EZH2 and activating AKT
pathway in gastric cancer. Cell Death Dis. 8:e28392017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pan T, Chen W, Yuan X, Shen J, Qin C and
Wang L: miR-944 inhibits metastasis of gastric cancer by preventing
the epithelial-mesenchymal transition via MACC1/Met/AKT signaling.
FEBS Open Bio. 7:905–914. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim JW, Lee HS, Nam KH, Ahn S, Kim JW, Ahn
SH, Park DJ, Kim HH and Lee KW: PIK3CA mutations are associated
with increased tumor aggressiveness and Akt activation in gastric
cancer. Oncotarget. 8:90948–90958. 2017.PubMed/NCBI
|
|
20
|
Wang C, Jiang J, Ji J, Cai Q, Chen X, Yu
Y, Zhu Z and Zhang J: PKM2 promotes cell migration and inhibits
autophagy by mediating PI3K/AKT activation and contributes to the
malignant development of gastric cancer. Sci Rep. 7:28862017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song Y, Li ZX, Liu X, Wang R, Li LW and
Zhang Q: The Wnt/β-catenin and PI3K/Akt signaling pathways promote
EMT in gastric cancer by epigenetic regulation via H3 lysine 27
acetylation. Tumour Biol. 39:10104283177126172017. View Article : Google Scholar
|
|
22
|
Zhang X, Tang J, Zhi X, Xie K, Wang W, Li
Z, Zhu Y, Yang L, Xu H and Xu Z: Correction: miR-874 functions as a
tumor suppressor by inhibiting angiogenesis through STAT3/VEGF-A
pathway in gastric cancer. Oncotarget. 8:295352017.PubMed/NCBI
|
|
23
|
You W, Tang Q, Zhang C, Wu J, Gu C, Wu Z
and Li X: IL-26 promotes the proliferation and survival of human
gastric cancer cells by regulating the balance of STAT1 and STAT3
activation. PLoS One. 8:e635882013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng L, Chen J, Zhou Z and He Z:
Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer
cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol.
39:10104283177053352017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu YY, Guo M, Yang LQ, Zhou F, Yu C, Wang
A, Pang TH, Wu HY, Zou XP, Zhang WJ, et al: Regulation of CD44v6
expression in gastric carcinoma by the IL-6/STAT3 signaling pathway
and its clinical significance. Oncotarget. 8:45848–45861.
2017.PubMed/NCBI
|
|
26
|
Merchant JL: What lurks beneath: IL-11,
via Stat3, promotes inflammation-associated gastric tumorigenesis.
J Clin Invest. 118:1628–1631. 2008.PubMed/NCBI
|
|
27
|
Zgodziński W, Grywalska E, Surdacka A,
Zinkiewicz K, Majewski M, Szczepanek D, Wallner G and Roliński J:
Surface CD200 and CD200R antigens on lymphocytes in advanced
gastric cancer: A new potential target for immunotherapy. Arch Med
Sci. 14:1271–1280. 2018. View Article : Google Scholar
|
|
28
|
Jin X, Zhu Z and Shi Y: Metastasis
mechanism and gene/protein expression in gastric cancer with
distant organs metastasis. Bull Cancer. Oct 8–2014, Epub ahead of
print. PubMed/NCBI
|
|
29
|
Han ZX, Wang HM, Jiang G, Du XP, Gao XY
and Pei DS: Overcoming paclitaxel resistance in lung cancer cells
via dual inhibition of stathmin and Bcl-2. Cancer Biother
Radiopharm. 28:398–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wosnitzer MS, Domingo-Domenech J,
Castillo-Martin M, Ritch C, Mansukhani M, Petrylack DP, Benson MC,
McKiernan JM and Cordon-Cardo C: Predictive value of microtubule
associated proteins tau and stathmin in patients with nonmuscle
invasive bladder cancer receiving adjuvant intravesical taxane
therapy. J Urol. 186:2094–2100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Werner HM, Trovik J, Halle MK, Wik E,
Akslen LA, Birkeland E, Bredholt T, Tangen IL, Krakstad C and
Salvesen HB: Stathmin protein level, a potential predictive marker
for taxane treatment response in endometrial cancer. PLoS One.
9:e901412014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kouzu Y, Uzawa K, Koike H, Saito K,
Nakashima D, Higo M, Endo Y, Kasamatsu A, Shiiba M, Bukawa H, et
al: Overexpression of stathmin in oral squamous-cell carcinoma:
Correlation with tumour progression and poor prognosis. Br J
Cancer. 94:717–723. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miceli C, Tejada A, Castaneda A and Mistry
SJ: Cell cycle inhibition therapy that targets stathmin in in vitro
and in vivo models of breast cancer. Cancer Gene Ther. 20:298–307.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yuan SF, Chen WJ, Zhu LJ, Zheng WE, Chen H
and Xiong JP: Effects of monoclonal antibodies against human
stathmin combined with paclitaxel on proliferation of the QG-56
human lung carcinoma cell line. Asian Pac J Cancer Prev.
13:2967–2971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
D'Andrea S, Berton S, Segatto I, Fabris L,
Canzonieri V, Colombatti A, Vecchione A, Belletti B and Baldassarre
G: Stathmin is dispensable for tumor onset in mice. PLoS One.
7:e455612012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ying L, Su D, Zhu J, Ma S, Katsaros D and
Yu H: Genotyping of stathmin and its association with clinical
factors and survival in patients with ovarian cancer. Oncol Lett.
5:1315–1320. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pucci S, Bonanno E, Sesti F, Mazzarelli P,
Mauriello A, Ricci F, Zoccai GB, Rulli F, Galatà G and Spagnoli LG:
Clusterin in stool: A new biomarker for colon cancer screening? Am
J Gastroenterol. 104:2807–2815. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Flanagan L, Whyte L, Chatterjee N and
Tenniswood M: Effects of clusterin over-expression on metastatic
progression and therapy in breast cancer. BMC Cancer. 10:1072010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rizzi F and Bettuzzi S: The clusterin
paradigm in prostate and breast carcinogenesis. Endocr Relat
Cancer. 17:R1–R17. 2010. View Article : Google Scholar
|
|
40
|
Bi J, Guo AL, Lai YR, Li B, Zhong JM, Wu
HQ, Xie Z, He YL, Lv ZL, Lau SH, et al: Overexpression of clusterin
correlates with tumor progression, metastasis in gastric cancer: A
study on tissue microarrays. Neoplasma. 57:191–197. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xiu P, Xu Z, Liu F, Li Z, Li T, Zou F, Sun
X and Li J: Downregulating sCLU enhances the sensitivity of
hepatocellular carcinoma cells to gemcitabine by activating the
intrinsic apoptosis pathway. Dig Dis Sci. 59:1798–1809. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang C, Jin G, Jin H, Wang N, Luo Q, Zhang
Y, Gao D, Jiang K, Gu D, Shen Q, et al: Clusterin facilitates
metastasis by EIF3I/Akt/MMP13 signaling in hepatocellular
carcinoma. Oncotarget. 6:2903–2916. 2015.PubMed/NCBI
|
|
43
|
Gong F, Peng X, Sang Y, Qiu M, Luo C, He
Z, Zhao X and Tong A: Dichloroacetate induces protective autophagy
in LoVo cells: Involvement of cathepsin D/thioredoxin-like protein
1 and Akt-mTOR-mediated signaling. Cell Death Dis. 4:e9132013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang WW, Tsai SC, Peng SF, Lin MW, Chiang
JH, Chiu YJ, Fushiya S, Tseng MT and Yang JS: Kaempferol induces
autophagy through AMPK and AKT signaling molecules and causes G2/M
arrest via downregulation of CDK1/cyclin B in SK-HEP-1 human
hepatic cancer cells. Int J Oncol. 42:2069–2077. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Y, Huang W, Ran Y, Xiong Y, Zhong Z,
Fan X, Wang Z and Ye Q: miR-582-5p inhibits proliferation of
hepatocellular carcinoma by targeting CDK1 and AKT3. Tumour Biol.
36:8309–8316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tibes R, McDonagh KT, Lekakis L,
Bogenberger JM, Kim S, Frazer N, Mohrland S, Bassett D, Garcia R,
Schroeder K, et al: Phase I study of the novel Cdc2/CDK1 and AKT
inhibitor terameprocol in patients with advanced leukemias. Invest
New Drugs. 33:389–396. 2015. View Article : Google Scholar
|
|
47
|
Li B, Xu W, Luo C, Gozal D and Liu R:
VEGF-induced activation of the PI3-K/Akt pathway reduces mutant
SOD1-mediated motor neuron cell death. Brain Res Mol Brain Res.
111:155–164. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang B, Zhang K, Liu Z, Hao F, Wang M, Li
X, Yin Z and Liang H: Secreted clusterin gene silencing enhances
chemo-sensitivity of a549 cells to cisplatin through AKT and ERK1/2
pathways in vitro. Cell Physiol Biochem. 33:1162–1175. 2014.
View Article : Google Scholar
|
|
49
|
Ma X and Bai Y: IGF-1 activates the
P13K/AKT signaling pathway via upregulation of secretory clusterin.
Mol Med Rep. 6:1433–1437. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shim YJ, Kang BH, Jeon HS, Park IS, Lee
KU, Lee IK, Park GH, Lee KM, Schedin P and Min BH: Clusterin
induces matrix metal-loproteinase-9 expression via ERK1/2 and
PI3K/Akt/NF-κB pathways in monocytes/macrophages. J Leukoc Biol.
90:761–769. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yadav P, Selvaraj BT, Bender FL, Behringer
M, Moradi M, Sivadasan R, Dombert B, Blum R, Asan E, Sauer M, et
al: Neurofilament depletion improves microtubule dynamics via
modulation of Stat3/stathmin signaling. Acta Neuropathol.
132:93–110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Morris EJ, Kawamura E, Gillespie JA, Balgi
A, Kannan N, Muller WJ, Roberge M and Dedhar S: Stat3 regulates
centrosome clustering in cancer cells via Stathmin/PLK1. Nat
Commun. 8:152892017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ng DC, Lin BH, Lim CP, Huang G, Zhang T,
Poli V and Cao X: Stat3 regulates microtubules by antagonizing the
depolymerization activity of stathmin. J Cell Biol. 172:245–257.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Verma NK, Dourlat J, Davies AM, Long A,
Liu WQ, Garbay C, Kelleher D and Volkov Y: STAT3-stathmin
interactions control microtubule dynamics in migrating T-cells. J
Biol Chem. 284:12349–12362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ebrahimpour S and Saadat I: Association of
CAT C-262T and SOD1 A251G single nucleotide polymorphisms
susceptible to gastric cancer. Mol Biol Res Commun. 3:223–229.
2014.PubMed/NCBI
|
|
56
|
Kantsaliev AL, Kozyreva EA, Kushlinskiĭ
NE, Rottenberg VI, Klimenkov AA and Vasil'ev AV: Cathepsin D
activity in gastric cancer. Vopr Onkol. 40:40–46. 1994.In
Russian.
|
|
57
|
Saku T, Sakai H, Tsuda N, Okabe H, Kato Y
and Yamamoto K: Cathepsins D and E in normal, metaplastic,
dysplastic, and carcinomatous gastric tissue: An
immunohistochemical study. Gut. 31:1250–1255. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gao SY, Li J, Qu XY, Zhu N and Ji YB:
Downregulation of Cdk1 and cyclinB1 expression contributes to
oridonin-induced cell cycle arrest at G2/M phase and growth
inhibition in SGC-7901 gastric cancer cells. Asian Pac J Cancer
Prev. 15:6437–6441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zeng Q, Zhao Y, Yang Y, Chen XX, Wang G,
Zhang P, Cui Y, Su S and Li K: Expression of Cystatin C in human
stomach neoplasms. Mol Med Rep. 3:607–611. 2010.
|
|
60
|
Allgayer H, Babic R, Grützner KU, Beyer
BC, Tarabichi A, Wilhelm Schildberg F and Heiss MM: An
immunohistochemical assessment of cathepsin D in gastric carcinoma:
Its impact on clinical prognosis. Cancer. 80:179–187. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lee MH, Cho Y, Kim DH, Woo HJ, Yang JY,
Kwon HJ, Yeon MJ, Park M, Kim SH, Moon C, et al: Menadione induces
G2/M arrest in gastric cancer cells by down-regulation of CDC25C
and proteasome mediated degradation of CDK1 and cyclin B1. Am J
Transl Res. 8:5246–5255. 2016.
|
|
62
|
Liu H, Lu J, Hua Y, Zhang P, Liang Z, Ruan
L, Lian C, Shi H, Chen K and Tu Z: Targeting heat-shock protein 90
with ganetespib for molecularly targeted therapy of gastric cancer.
Cell Death Dis. 6:pp. e15952015, View Article : Google Scholar : PubMed/NCBI
|