Introduction
Lung cancer is the leading cause of cancer-related
mortality (1). As the first line
of immune defense, neutrophils fight against infectious agents via
phagocytosis, degranulation (2)
and neutrophil extracellular traps (NETs) (3). NETs, were firstly identified as an
immune defense mechanism against bacteria (3), have been well documented in a diverse
range of diseases (4-6). In response to various stimuli,
neutrophils release extracellular chromatins coupled with granular
and selected cytoplasmic proteins. In patients with lung cancer,
elevated number of circulating neutrophils are a potential
biomarker of poor prognosis (7).
In systemic sepsis, intravascular neutrophils produce NETs, which
sequester the circulating lung tumor cells (8). Although infection in patients with
lung cancer is concomitant (9),
systemic sepsis is not inevitable. In aseptic inflammation, whether
neutrophils that have infiltrated the parenchyma of patients with
lung cancer can form NETs, is unknown.
In the central dogma of biology, RNA function is
cell autonomous (10). With the
aid of ribosomal RNA and transfer RNA, messenger RNA transcribed
from the genome is translated into protein in the cell. Previous
studies have demonstrated the transfer of macromolecular RNA
between mammalian cells (11,12)
may trigger a broad range of physiologic and pathologic processes.
Compared with non-tumor cells, tumor cells secrete higher levels of
exRNAs (13). Therefore, in the
sera of patients with lung cancer the concentration of
extracellular RNA (exRNA) s is significantly elevated (14). The profile of microRNAs (miRNAs),
the major population of exRNAs, is considered as a diagnostic
marker and therapeutic candidate for lung cancer (15-17).
Besides miRNAs, extracellular mRNAs may also be functional. For
example, Gag-encoding mRNA has the potential to promote the
secretion of tumor necrosis factor-α (TNF-α) and activate dendritic
cells (18). In the myocardial
ischaemia reperfusion injury, RNase1 has been demonstrated to be
protective via degrading mRNAs (19). RNase1/exRNA balance is also linked
with tumor invasion (13).
However, the cross-talk between lung cancer exRNAs and neutrophils
still remains unknown.
The aim of the present study was to investigate the
roles of exRNA in the formation of NETs in a mouse model of lung
cancer and in patients with lung cancer. Furthermore, the
contribution of NETs to the activation and damage of epithelial
cells was investigated. Collectively, the present findings
indicated that the cross-talk between exRNAs from lung cancer cells
and NETs may contribute to the oncogenesis of lung cancer which may
shed light on a new strategy for treating lung cancer.
Materials and methods
Animals
A total of 80 wild-type female C57BL/6 mice, aged
6-8 weeks old, weighing 25-33 g were purchased from the College of
Veterinary Medicine, Yangzhou University (Yangzhou, China) and bred
in the Animal Laboratory of Nanjing Medical University (Nanjing,
China), under standard laboratory conditions (12:12 h light: dark
cycle, relative humidity 60±5%, temperature 25±2°C) in individually
ventilated cages with free access to water or food. All animal
procedures were approved by the Institutional Animal Care Committee
of Nanjing Medical University.
Cell culture
The murine lung cancer cell line Lewis lung
carcinoma (LLC) was acquired from the Cell Bank of Shanghai
Institutes for Biological Sciences (Shanghai, China). And the
catalog number for LLC was TCM 7 (www.cellbank.org.cn/detail_1.asp?id=78&serial=TCM%207).
The Murine Lung Epithelial-12 (MLE-12) cell line was obtained from
the American Type Culture Collection. The LLC and MLE-12 cells were
maintained in high-glucose Dulbecco's modified Eagle's medium
(DMEM; HyClone; GE Healthcare Life Sciences) with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a 5%
CO2 humidified atmosphere at 37°C.
Protocol of harvesting exRNAs
exRNAs were prepared as previously described by
Laurent and Alexander (20).
Briefly, LLC or MLE-12 cells were seeded in 25-cm2 cell
culture flasks. Once the cells had covered 75% of the flask, the
culture medium was removed and the flask was washed gently with
pre-warmed PBS. The PBS was then discarded and 2 ml low-glucose
DMEM (HyClone; GE Healthcare Life Sciences) without FBS was added
to the flask for 48 h at 37°C. After centrifugation at 2,000 × g
for 10 min at 4°C, the supernatant (cell culture supernatant; CCS)
was collected and stored at -80°C. A total of 20 and 50% CCS in the
culture medium significantly increased IL-1β transcription (data
not shown). Therefore, 50% CCS was selected for the subsequent
experiments. To validate whether the CCS contained exRNAs, it was
treated with RNase1 (0.5 µg/µl; cat. no. R4875;
Sigma-Aldrich; Merck KGaA) and then subjected to electrophoresis on
a 3% agarose gel. The results were analyzed in a Tanon system
(Tanon 4600SF; Tanon Science and Technology Co., Ltd.) at 310 nm
UV.
The RNA in the LLC cells prior to and following
starvation was also directly stained with SYTO®
RNASelect™ Green Fluorescent Cell Stain (cat. no. S32703; Molecular
Probes; Thermo Fisher Scientific, Inc.). Briefly, the LLC cells
were seeded onto cell slides and starved for 24 h at 37°C.
Subsequently, the cells on the slides were washed gently with
pre-warmed PBS and stained with RNA-selective dye (same as above;
500 nM) and DAPI for 20 min at room temperature. The stained cells
were observed under a confocal microscope (magnification,
×200).
Activation of MLE-12 cells with
exRNAs
To determine whether the CCS could activate MLE-12
cells (cultured for 12 h at 1×106/ml cells), different
concentrations of the CCS (10, 20 and 50%) were added to the
culture medium of the MLE-12 cells for 12 h at 37°C. The MLE-12
cells were washed in PBS for 5 times and then collected for
quantitative PCR or western blot analyses. To validate the roles of
exRNAs in the activation of MLE-12 cells, CCS with or without
RNase1 (0.5 µg/µl) was added to the cell culture.
Induction of NETs
Bone marrow cells were collected from the two hind
leg femurs of 8-10 week-old mice (n=15), were suspended in PBS, and
centrifuged at 450 × g for 10 min at 4°C. The red blood cells were
lysed with ACK Lysing Buffer (cat. no. A1049201; Thermo Fisher
Scientific, Inc.). Briefly, prepare a lysing solution by adding 0.5
ml of lysing buffer to 4.5 ml of sterile water and lyse the
remaining red blood cells with 5 ml of lysing solution. Lysing
solution was transferred into centrifuged tube contains pellets on
ice in the dark for 10 min (21).
The cells were then subjected to a discontinuous 72-64%
Percoll® (cat. no. P4937; Sigma-Aldrich; Merck KGaA)
density gradient centrifugation at 450 × g for 30 min at 4°C. The
neutrophils were collected at the 72-64% Percoll®
interface and washed three times with PBS at 4°C. The neutrophils
(5×106 cells) were seeded in 10-cm culture plates that
were pre-treated with attachment factor protein (cat. no. S006100;
Gibco; Thermo Fisher Scientific, Inc.). To mimic the roles of
exRNAs, the cells were stimulated with 10 µg/ml poly(I:C;
InvivoGen) at 37°C for 4 h (21).
The supernatant was removed slowly and the plate was gently washed
with pre-warmed PBS. The plate was then flushed with 1 ml PBS at
4°C for 5 times to separate the NETs from the bottom of the plate.
After centrifugation at 450 × g for 10 min at 4°C, the supernatant
containing the NETs was transferred to 1.5 ml Eppendorf tubes. The
supernatant containing the NETs was further centrifuged at 18,000 ×
g for 10 min at 4°C. The sediment (NETs) with 100 µl residue
was collected and stored at -80°C.
NET induction by activated MLE-12
cells
MLE-12 cells were seeded on the sterile slides of a
6-well plate. When the cells covered 75% of the plates, CCS was
added to replenish the medium and activate the MLE-12 cells. RNase1
(0.5 µg/μl) or interleukin (IL)-1β inhibitor AS101 (2.5
µg/ml; cat. no. S8301; Selleck Chemicals) was added to the
CCS to block the potential roles of exRNAs or IL-1β. After
incubation for 12 h at 37°C, the MLE-12 cells were gently washed
for 5 times with pre-warmed PBS. Neutrophils were added to each
well for the induction of NETs. After 4 h, the supernatant was
collected for quantitative analysis. The cell slides were gently
washed in 4°C PBS, fixed in 4°C acetone for 10 min and stained with
DAPI at room temperature for half an hour in the dark.
MLE-12 cells treated with NETs
MLE-12 cells were seeded in a 6-well plate. The
confluent MLE-12 cells were treated with NETs of 10, 20 and 50%
concentration for 4 h at 37°C. After the incubation, the MLE-12
cells were washed with PBS for 5 times. A number of the MLE-12
cells were harvested for western blot analysis of the tight
junction protein claudin-5 and the apoptotic protein caspase 3. The
remaining MLE-12 cells were seeded in a glass-bottomed dish. The
NET-treated MLE-12 cells were stained with the dye propidium iodide
(PI; P1304MP; Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature in the dark for 10 min in order to identify the cell
death. exRNAs from NET-damaged MLE-12 cells were quantified using
the Quant-iT™ RiboGreen™ RNA Assay Kit (cat. no. R11490; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The confluent living MLE-12 cells in the glass-bottomed dish were
stained with 1 µm CellTrace™ Far Red DDAO-SE (C34553;
Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature in
the dark for 15 min. The poly(I:C)-induced NETs were stained with 1
µm SYTOX Green (cat. no. s7020; Invitrogen; Thermo Fisher
Scientific, Inc.) at room temperature in the dark for 20 min. The
interactions between the MLE-12 cells and NETs were directly
observed under a confocal microscope (magnification, × 200).
Reverse transcription-quantitative
PCR
The total RNA was extracted from the MLE-12 cells
using the Takara universal total RNA extraction kit (Takara
Biotechnology Co., Ltd.) and cDNA was synthesized using PrimeScript
RT Master Mix (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. Quantitative PCR was performed using
SYBR-Green Universal PCR Master mix (Takara Biotechnology Co.,
Ltd.). The RNA expression was quantified using a StepOnePlus
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) under the following conditions: Denaturation for 30 sec at
95°C, followed by 40 cycles of denaturation for 3 sec at 95°C, and
extension for 30 sec at 60°C. Melting curve analysis was performed
at the end to validate the specificity of the expected PCR product.
The relative expression levels of each mRNA were calculated using
standard curve method (22). Three
independent samples were prepared for each assay, and each
experiment was performed at least three times. The primer sequences
were designed using PrimerBank (pga.mgh.harvard. edu/primerbank), a
public resource for PCR primers, and were as follows: IL-1β
forward, 5′-AGCTCTCCACCTCAATGGA-3′ and reverse,
5′-TTGCTTGGGATCCACACTCT-3′; IL-6 forward,
5′-GACTGATGCTGGTGACAACC-3′ and reverse, 5′-AGACAGGTCTGTTGGGAGTG-3′;
TNF-α forward, 5′-GGTGAGGCAGCAAGAGATTG-3′ and reverse, 5′-GAG
CAGCAGGTTTCAGGATG-3′; vascular cell adhesion molecule (VCAM)-1
forward, 5′-TTGGGAGCTGAACACTTTTCC CAG-3′ and reverse,
5′-TGTGGTGCTGCAAGTCAGGAGC-3′; and GAPDH forward
5′-AACTTTGGCATTGTGGAAGG-3′ and reverse,
5′-GGATGCAGGGATGATGTTCT-3′.
Western blotting
Total protein from the MLE-12 cells was isolated
using radioimmunoprecipitation lysis buffer with protease and
phosphatase inhibitor cocktail (Beyotime Institute of
Biotechnology). The proteins were separated in a 10 or 15%
SDS-polyacrylamide gel and transferred to a polyvinylidene fluoride
membrane. The membranes were blocked for 1 h with 5% bovine serum
albumin (A1933; Sigma-Aldrich; Merck KGaA) in PBS and then were
incubated with anti-VCAM1 (cat. no. ab134047), anti-claudin 5 (cat.
no. ab15106) (both from Abcam) and anti-caspase-3 (cat. no. 9665)
at a 1:1,000 dilution or with anti-β-actin (cat. no. 4970) (both
from Cell Signaling Technology, Inc.) at a 1:2,000 dilution
overnight at 4°C. After washing for 5 times with PBS with Tween-20,
the membranes were incubated with a horseradish-conjugated
anti-rabbit IgG antibody (65-6120; Invitrogen; Thermo Fisher
Scientific, Inc.) at a 1:2,000 dilution for 1 h at room
temperature. The blots were washed a further 5 times with PBS and
then incubated with enhanced chemiluminescence substrate (cat. no.
35055; Thermo Fisher Scientific, Inc.) with gentle agitation for 1
min at room temperature. The individual target proteins were
visualized and recorded using a G:BOX instrument (GENESys V1.3.5.0;
Syngene).
ELISA
The concentration of IL-1β in the cell culture
supernatant was determined via ELISA (cat. no. BMS6002;
eBioscience; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
LLC cell-recruited and -activated
neutrophils in vivo
Mice were anesthetized with a mixture of ketamine
(100 mg/kg) and xylazine (10 mg/kg) via intraperitoneal injection.
After exposure of the trachea, 1×106 LLC cells were
injected into the lungs through a trimmed sterile 31-gauge needle
inserted into the tracheal lumen. After 4 h, the bronchoalveolar
lavage fluid (BALF) and lung tissues were collected from each
mouse. The bronchial and alveolar spaces were washed three times
with 1 ml PBS. The BALF from the two lungs per mouse was pooled and
centrifuged at 1,000 × g for 5 min at 4°C. A total of 400 µl
supernatant was transferred to a new Eppendorf tube and the
quantity of NETs was tested. The rest of the fluid was re-suspended
for flow cytometry. After the BALF was obtained, PBS was pumped
into the right ventricle to clear blood in the pulmonary
vasculature. The lung tissues from one mouse were divided into
three parts. The upper right lung lobe was removed and fixed in 10%
neutral-buffered formalin for histopathology imaging. The left lung
lobe was removed and immobilized with OCT (Sakura Finetek Europe
B.V.) at −80°C for ≥24 h for fluorescence microscopy. The lower
right lung lobe was pulverized in 70-µm cell strainers and
washed with 1.0 ml PBS. ACK lysis buffer (A1049201; Gibco; Thermo
Fisher Scientific, Inc.) was added to the suspension to lyse the
erythrocytes. After the lysis of the erythrocytes, the cell
suspension was centrifuged 700 × g for 10 min at room temperature.
After the centrifugation, cell pellets rom the lung homogenates
were re-suspended in PBS and stained for flow cytometry.
Flow cytometry
The leukocytes from the BALF or pulverized lung were
labeled with fluorescent antibodies in order to quantify the
neutrophils. Briefly, the cells were first incubated with
anti-CD16/32 (cat. no. 14-0161-82; Invitrogen; Thermo Fisher
Scientific, Inc.; 1:60 in PBS) to reduce the non-specific binding.
Subsequently, anti-CD45 conjugated with FITC (cat. no. 11-0451-82;
Invitrogen; Thermo Fisher Scientific, Inc.; 1:40 in PBS),
anti-mouse Ly-6G conjugated with PE (cat. no. 12-5931-82;
Invitrogen; Thermo Fisher Scientific, Inc.; 1:240 in PBS) and
anti-CD11b conjugated with allophycocyanin (cat. no. 17-0112-82;
eBioscience; Thermo Fisher Scientific, Inc.; 1:120 in PBS) or
isotype controls were added at 37°C for 30 min, in the dark. The
cells were centrifuged and re-suspended in 500 µl PBS for
flow cytometric analysis in the BD FACSCalibur (Becton-Dickinson
and Company). All of the FACS data were analyzed with FlowJo v10
(FlowJo LLC).
Fluorescence microscopy
The lung samples from the mice were immobilized with
OCT (Sakura Finetek Europe B.V.) at −80°C for ≥24 h. The frozen
samples were cut on a cryostat microtome, and the 7-µm
sections were placed on polylysine-coated glass slides. The tissue
sections were fixed in 4°C acetone and rehydrated in PBS. The
slides were gently washed in PBS for three times and then blocked
with 5% goat serum (cat. no. 16210-064; Gibco; Thermo Fisher
Scientific, Inc.; 1:500 in PBS) at 37°C to reduce non-specific
binding. After 30 min, the sections were washed with PBS and
stained with Histone H3 (citrulline R2+R8+R17) antibody (cat. no.
ab5103; Abcam; 1:300 diluted) (23) overnight at 4°C in the dark.
Subsequently, the sections were gently washed in PBS and then
stained with the fluorescent-conjugated secondary antibody [Goat
anti-Rabbit IgG (H+L) Alexa Fluor® 555 conjugate, cat.
no. A-21428; Invitrogen; Thermo Fisher Scientific, Inc.] at 37°C
for 60 min in the dark. After the unbound fluorescent antibody was
removed, the sections were incubated with SYTOX Green diluted in
PBS (1:2,000) at 37°C for 15 min. The sections were washed again,
and then observed and recorded using confocal microscopy
(magnification, ×100; CarlZeiss LSM710; Zeiss AG).
Histopathology imaging
The lungs were fixed in 10% neutral-buffered
formalin for 24 h in room temperature, and then were dehydrated and
embedded in paraffin. The fixed embedded tissues were cut into
7-µm sections on a Leica RM2165 rotary microtome (Leica
Microsystems GmbH) and stained with hematoxylin and eosin for 5 min
at room temperature. The histological analyses were performed by
two independent pathologists blinded to the treatment groups.
Clinical samples
A total of 2 ml blood was collected from the
antecubital veins of patients with or without lung cancer who had
undergone pulmonary surgery under general anesthesia from January
to March 2017 in the First Affiliated Hospital of Nanjing Medical
University. The inclusion criteria were that patients should not
have pulmonary infection. Approximately 0.1 ml sputum was also
collected from the patients via endotracheal intubation. The sputum
was treated with 1.0 ml 0.1% DTT for 15 min at room temperature,
re-suspended and filtrated with a 40-µm cell strainer, and
centrifuged at 1,000 × g for 5 min at 4°C. The supernatant was
analyzed for the quantity of NET elastase according to the
manufacturer's protocol. Briefly, the lung tissues with or without
lung cancer were fixed in OCT (Sakura Finetek Europe B.V.) at −80°C
for ≥24 h. Fluorescence microscopy was conducted as described for
the mouse samples. All of the human experiments were approved by
the Institutional Human Ethics Committee of the First Affiliated
Hospital of Nanjing Medical University (Nanjing, China; approval
no. 2017-SR-243) with written informed consent from all subjects
and it was conducted in accordance with the Declaration of Helsinki
ethical guidelines.
NET quantification
NETs were quantified using a NETs assay kit (cat.
no. 601010; Cayman Chemical Company) according to the
manufacturer's protocol (24).
Briefly, the free elastase was washed away followed the generation
of NETs induced by LLC cell culture supernatant or activated MLE-12
cells. The samples were further digested with S7 nuclease
(10107921001; Sigma-Aldrich; Merck KGaA). The supernatant
containing neutrophil elastase was added to the selective substrate
and quantified at 405 nm. The concentration of NETs in each sample
was calculated according to the NET standards provided by the kit.
A total of 1 ml blood sample from mice was collected before mice
eutanasia. In the BALF and serum of mice, sputum and serum of
patients, free elastase was quantified based on the standard.
Statistical analysis
All statistical analyses were conducted using
GraphPad Prism 7 (GraphPad Software, Inc.) and SPSS version 12.0
(SPSS, Inc.). Statistically significant differences were determined
using Student's t-test for two groups or analysis of variance for
more than two groups, followed by the Dunnett comparison. All data
are expressed as the mean ± standard error of the mean. P<0.05
was considered to indicate a statistically significant
difference.
Results
exRNAs from lung cancer cells activate
MLE-12 cells
RNA-selective dye was used to stain intracellular
RNAs in the LLC cells. Without FBS and enough glucose, the starving
LLC cells underwent nuclear condensation and enhanced distribution
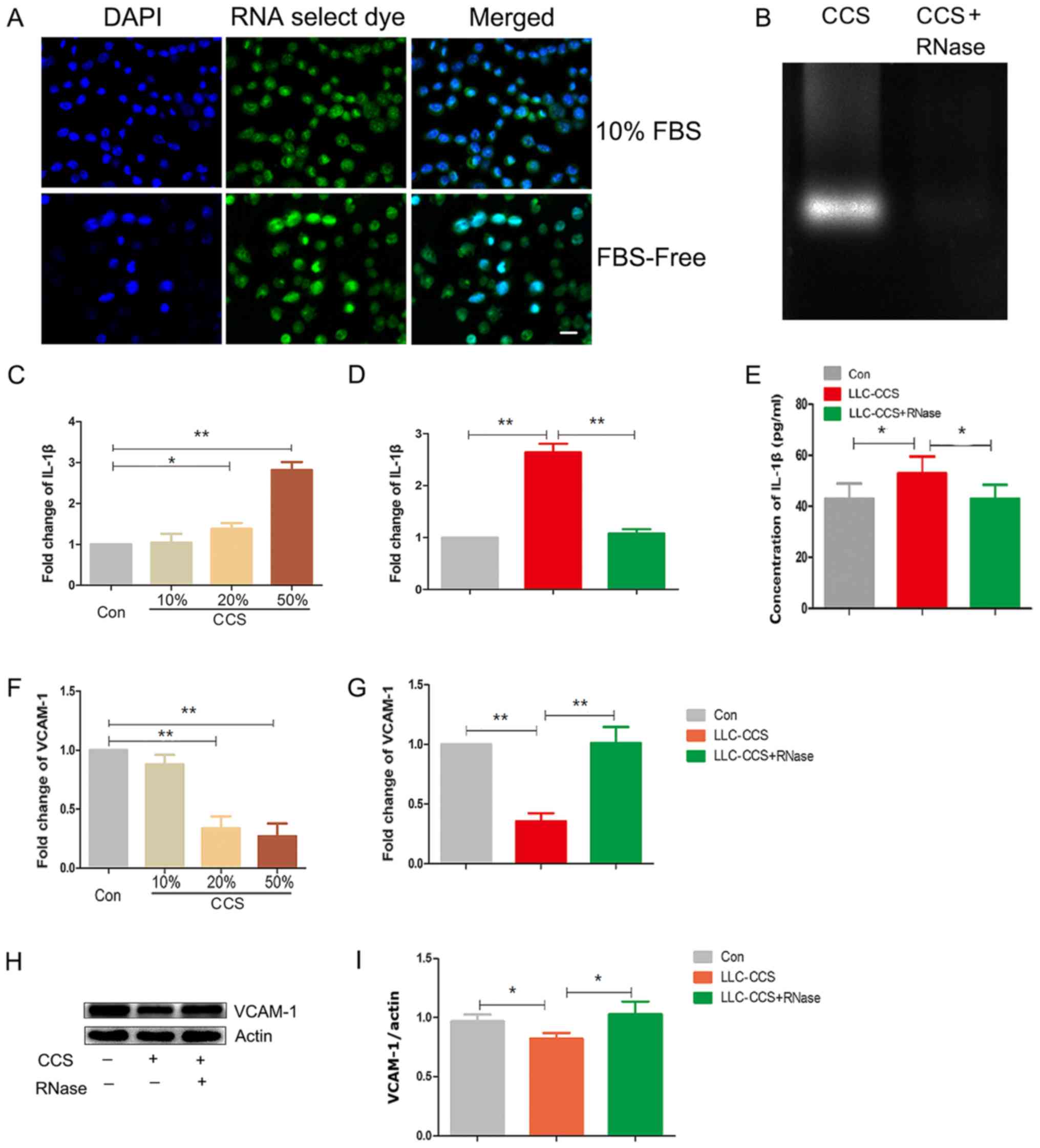
of RNAs in the nucleus (Fig. 1A).
In the CCS from starving LLC cells, RNase1 digested nucleic acids
in the agarose gel, suggesting that RNase1-sensitive exRNAs existed
in the CCS from starving LLC cells (Fig. 1B). The CCS from the starving LLC
cells increased the levels of the inflammatory cytokine IL-1β in
the MLE-12 cells (Fig. 1C).
Notably, RNase1 pretreatment abolished the effects of CCS on IL-1β
in the MLE-12 cells (Fig. 1D and
E). Conversely, the levels of the adhesion molecule VCAM-1 were
significantly reduced upon CCS treatment (Fig. 1F). In accordance, RNase1
pretreatment rescued VCAM-1 transcription (Fig. 1G) and protein expression (Fig. 1H and I). In summary, exRNAs from
starving lung cancer cells upregulated IL-1β and reduced VCAM-1 in
MLE-12 cells, implying that exRNAs from lung cancer cells may
activate MLE-12 cells.
Activated MLE-12 cells promote NETs
Neutrophil infiltration occurs frequently in lung
cancer (25). To observe whether
activated MLE-12 cells can provoke NETs, neutrophils were seeded on
a MLE-12 cell monolayer that was treated with or without CCS. As
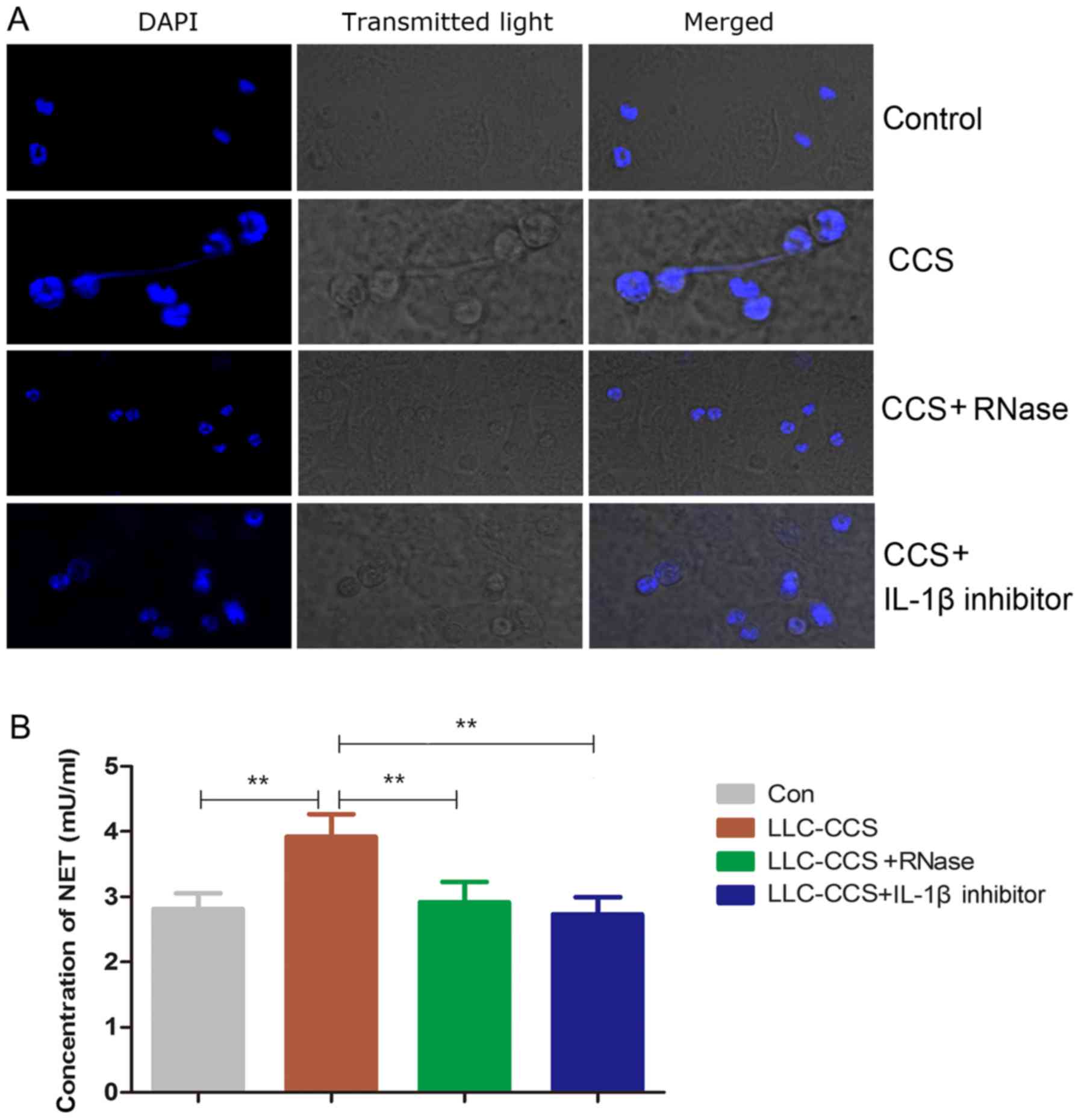
shown in Fig. 2A, the CCS-treated
MLE-12 cells induced NETs. RNase1 treatment abolished the formation
of NETs, suggesting that exRNAs may be essential in the development
of NETs. IL-1β is a potent inducer of NETs (26). In the present study, IL-1β
inhibitor blocked the NETs induced by activated MLE-12 cells. In
the formation of NETs, DNA from neutrophils is associated with
elastase (27). Therefore,
NET-specific elastase DNA was quantified in Fig. 2B. Compared to the medium control,
the CCS-treated MLE-12 cells had significantly increased levels of
elastase DNA. As expected, RNase1 or IL-1β downregulated the NETs,
suggesting that promotion of NETs by CCS-activated MLE-12 cells is
at least partially dependent on exRNAs in the CCS and IL-1β
released from the MLE-12 cells.
NETs damage MLE-12 cells
Activated MLE-12 cells initiate the formation of
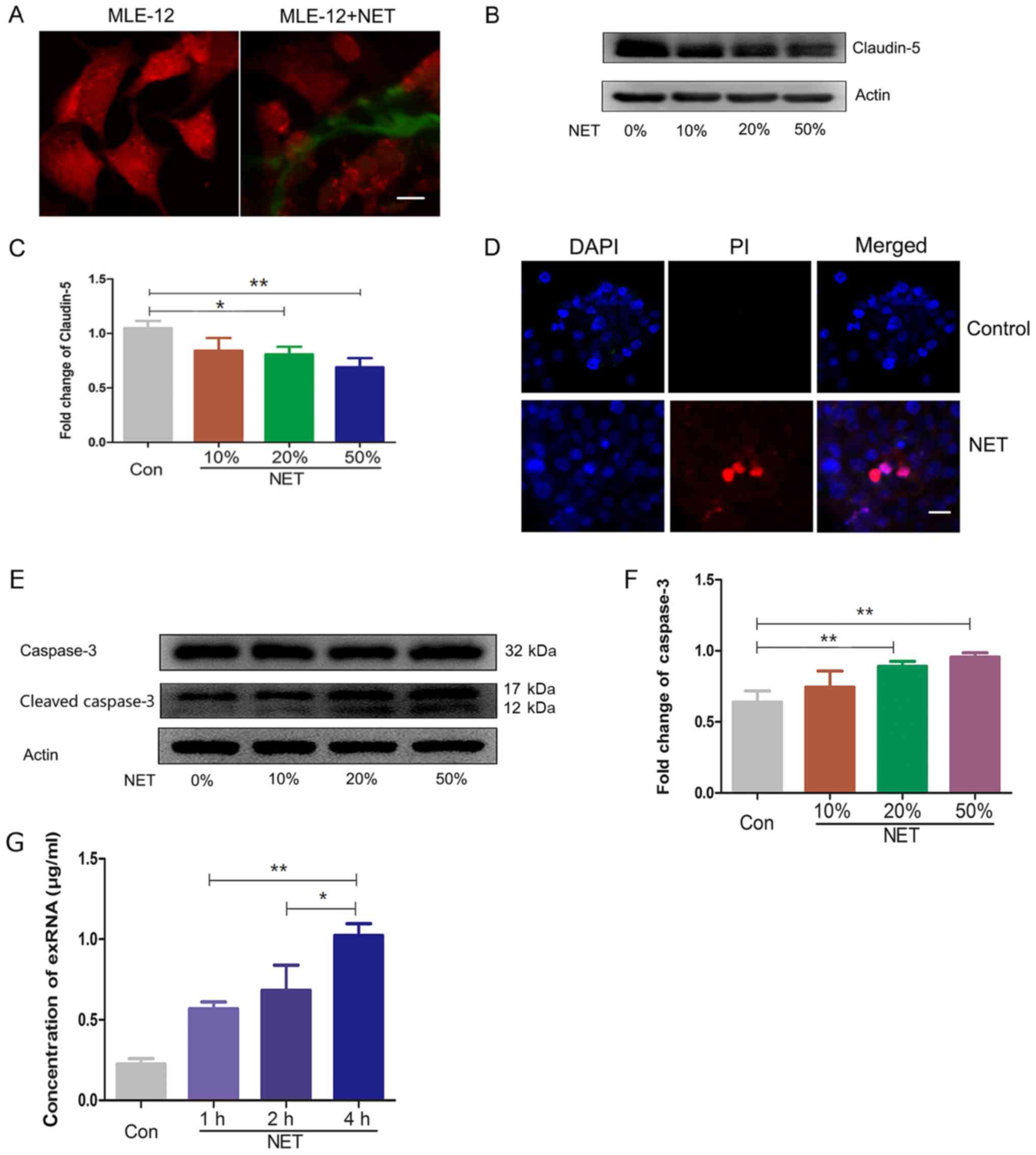
NETs and their cross-talk was directly observed in Fig. 3A. The integrity of the epithelium
is closely associated with junction proteins. Claudins are
considered as gatekeepers of lung epithelial function (28,29).
In the present study, the levels of tight junction claudin-5 in the
MLE-12 cells were significantly reduced upon treatment with NETs
(Fig. 3B and C), suggesting that
NETs may damage MLE-12 cells. Furthermore, the NETs promoted the
death of MLE-12 cells, as indicated by the PI-positive cells
(Fig. 3D). The master regulatory
factor caspase-3 was significantly activated upon treatment with
the NETs (Fig. 3E and F).
Therefore, the death of MLE-12 cells may be mediated by caspase-3
activation. As the dead LLC cells produced exRNAs, the death of the
MLE-12 cells also released exRNAs into the supernatant (Fig. 3G). Collectively, NETs damage MLE-12
cells and promote the secretion of exRNAs.
Self-activation of MLE-12 cells via
exRNAs
To address whether exRNAs from dead MLE-12 cells can
activate healthy MLE-12 cells, MLE-12 cells were stimulated with
CCS from dead lung cancer cells or MLE-12 cells. With the increased
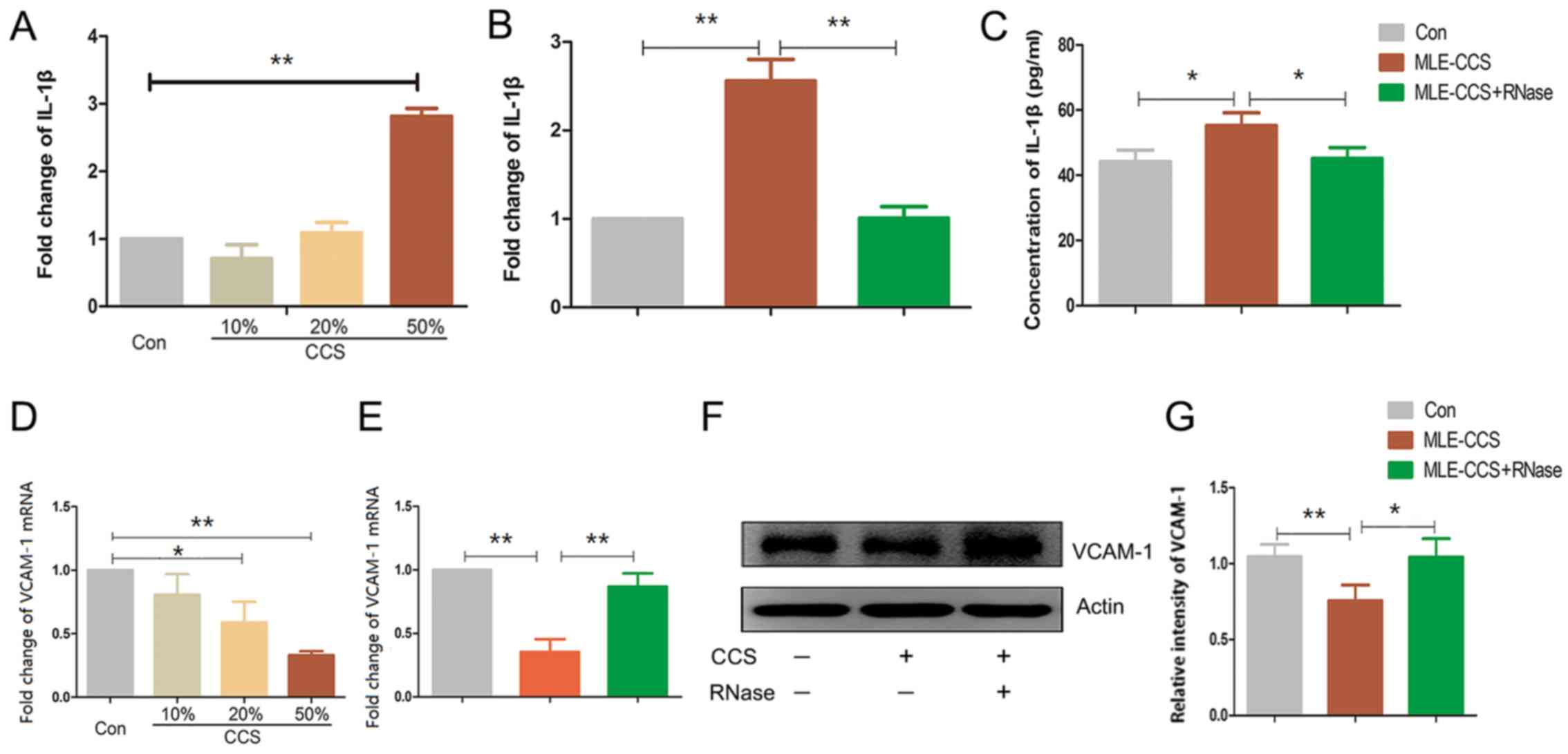
concentration of CCS in the culture medium, the levels of the
pro-inflammatory cytokine IL-1β were progressively increased
(Fig. 4A). RNase1 significantly
reduced the transcription and translation of IL-1β in the MLE-12
cells treated with CCS from the epithelial cells (Fig. 4B and C). As observed in the LLC
cell CCS, the MLE-12 cell CCS downregulated the cell adhesion
molecule VCAM-1 at mRNA and protein expression levels, which was
partially attenuated by RNase1 (Fig.
4D-G). Therefore, exRNAs from MLE-12 cells activate epithelial
cells.
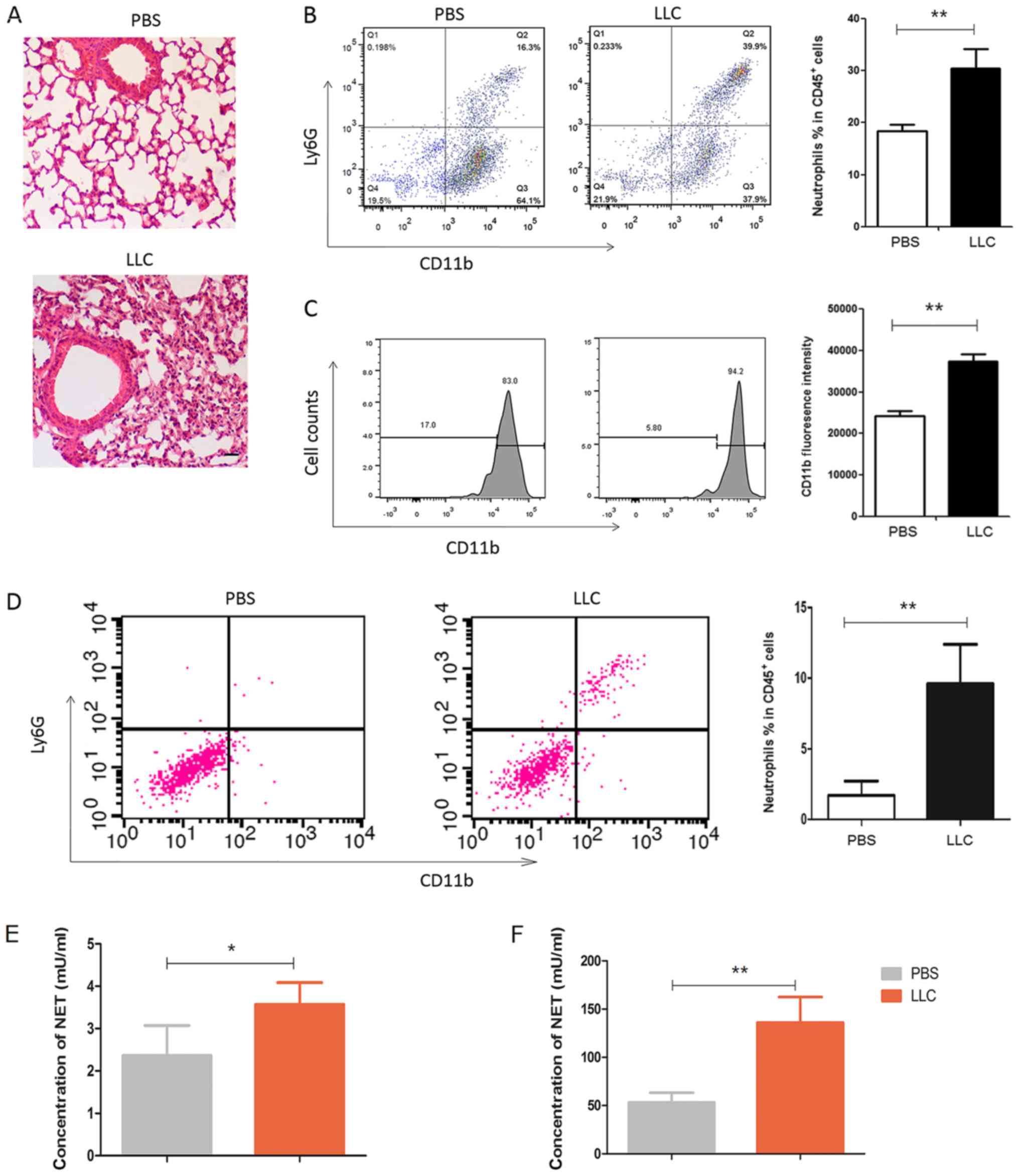
Lung cancer cells recruit and activate
neutrophils in vivo
In the murine model of lung cancer, LLC cells were
intratracheally instilled into the lung. As shown in Fig. 5A, the instillation of the LLC cells
significantly exacerbated the inflammation in the lung.
Neutrophils, which were
CD45+CD11b+Ly6G+, were recruited
into the lung parenchyma (Fig.
5B). Notably, CD11b expression on the neutrophils was
significantly enhanced in the mice that received LLC cells
(Fig. 5C), suggesting that LLC
cells activate neutrophils in vivo. In line with this
observation, the levels of neutrophils in the BALF were also
significantly increased (Fig. 5D).
The levels of elastase were significantly augmented in the BALF
(Fig. 5E) and sera (Fig. 5F) from the LLC-treated mice,
suggesting that the lung cancer cells induced neutrophil activation
and potential NET formation in vivo.
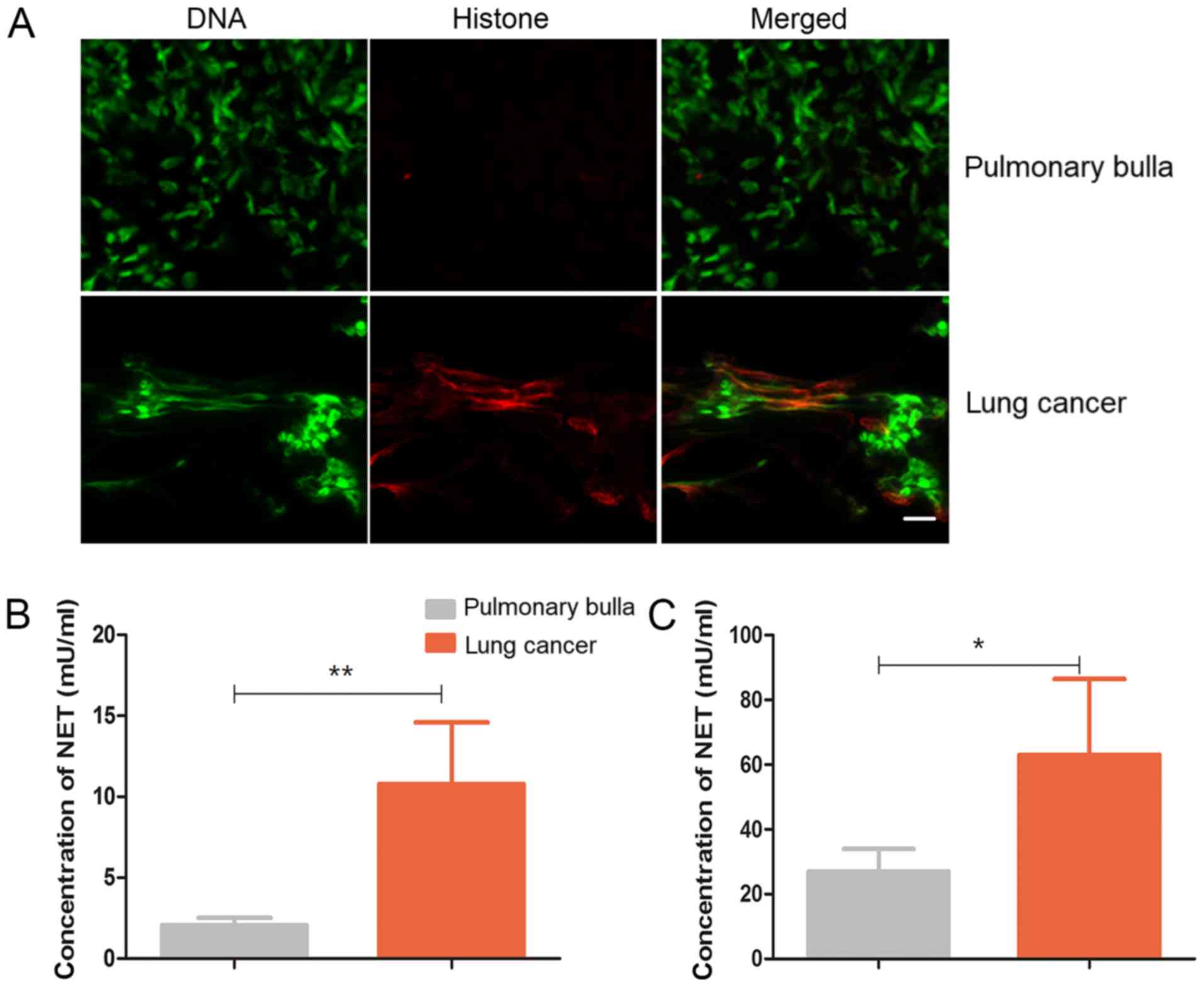
NETs in patients with lung cancer
The aforementioned results demonstrated that exRNAs
from lung cancer cells provoke NETs in vitro and in the
mice. In the clinical samples, the formation of NETs was observed
in the lung tissues from the patients with lung cancer but not in
those from the patients with pulmonary bulla (Fig. 6A). In line with the
immunofluorescence observation, the levels of the NET hallmark
elastase were significantly increased in either the sputum or the
peripheral blood (Fig. 6B and C),
suggesting that lung cancer may be accompanied with NETs.
Discussion
The present study showed that the exRNAs released
from lung cancer cells indirectly promoted the formation of NETs
via activating epithelial cells. Administration of RNase1
significantly blocked the roles of exRNAs in the NETs induction and
epithelial cells activation. Outside of the cell, miRNAs containing
~22 nucleotides are stable. In contrast to miRNAs, mRNAs and long
non-coding RNAs (lncRNAs) in the extracellular medium are
relatively sensitive to RNase1 (30). Similarly, it was postulated that
lung cancer cells may release RNase1-sensitive mRNAs and/or lncRNAs
into the extracellular space.
Lung cancer cells exRNAs evoke the secretion of
IL-1β from bronchia epithelial cells. Epithelial cells express Toll
like receptor3, Retinoic acid-inducible gene-1 and Melanoma
differentiation-associated protein-5, which may be responsible for
exRNA recognition and activation of the signaling pathway to
epithelial cells (31). VCAM-1 is
an inducible adhesion molecule expressed by respiratory endothelial
and epithelial cells (32). In
endothelial cells, IL-1β increases the levels of VCAM-1 expression
(33,34). However, in respiratory epithelial
cells, VCAM-1 expression is not affected by IL-1β (32). Instead, exRNAs in the lung cancer
cell CCS upregulate IL-1β and reduce VCAM-1 expression on
epithelial cells. VCAM-1 may mediate the leukocyte infiltration
across respiratory epithelial cells (32). As an adhesion molecule, VCAM-1 is
bound with integrin α4β1 mediating leukocyte transmigration. The
tight junction protein JAM may also interact with integrin α4β1
(35), indicating that VCAM-1
contributes to epithelial integrity. Soluble VCAM-1 impairs the
integrity of the blood-brain barrier (BBB) via α4β1 (36). In the present study, intratracheal
instillation of LLC cells recruited neutrophils into the lung
parenchyma and BALF, suggesting that leukocyte infiltration was
enhanced. Due to lack of special marker for LLC, lung cancer cells
were not directly detected in the pulmonary parenchyma.
Collectively, it was postulated that CCS exRNAs damage epithelial
cells, resulting in reduced integrity and increased leukocyte
infiltration, but this needs to be verified.
It was previously reported that activated
endothelial cells induce NETs, which is partially dependent on IL-8
(37). IL-1β is also a potential
inducer of NETs (26). In the
present study, exRNA-treated MLE-12 cells promoted the formation of
NETs, which was closely associated with exRNAs and IL-1β. NETs not
only kill pathogens but can also cause tissue injury (27). As NETs damage endothelial cells
(38), in the present study NETs
directly reduced the expression of claudin-5 in the epithelial
cells. In claudin-5-deficient mice, BBB integrity against small
molecules is severely compromised (39). In respiratory epithelial cells,
increased claudin-5 expression reduces alveolar epithelial barrier
function (29). Therefore, the
downregulation of claudin-5 in the MLE-12 cells by NETs in the
present study is arguable and requires further research. In the
present study, NETs induced the death of MLE-12 cells, which may be
associated with caspase-3. Furthermore, NETs triggered the
secretion of exRNAs from the starving MLE-12 cells. As observed in
the lung cancer cell exRNAs, the MLE-12 cell exRNAs also affected
IL-1β and VCAM-1 in epithelial cells. Thus, there may be positive
feedback in the reaction cascade as follows: i) exRNAs from damaged
lung tumor cells activate epithelial cells; ii) activated
epithelial cells promote NETs; ii) NETs cause the secretion of
exRNAs from necrotic epithelial cells; and iv) exRNAs from the
necrotic epithelial cells activate the neighboring healthy
epithelial cells. As we demonstrated that poly I:C induced NETs in
the lung (21) and other organs
(40,41), the double RNA analogy poly I:C
could directly induce the formation of NETs in vitro (data
unpublished). Indeed, poly I:C induced NETs were used in this study
to explore the interactions between NETs and epithelial cells.
Therefore, we could not preclude the possibility that exRNAs from
cancer cells may directly trigger NETs formation.
It has been widely recognized that NETs facilitate
tumor progression and metastasis (42). In the present study, NETs were
recorded in the patients with lung cancer, not only in the lung
tissues but also in the peripheral blood and sputum. The
danger-associated molecular pattern protein high mobility group box
1 (HMGB1) can induce NET formation (43). HMGB1 serves essential roles in lung
cancer tumorigenesis and metastasis (44). In the consideration that cell
culture supernatant may contain exosomes, cytokines and other
biological components, the possibility that all of these factors,
including exRNAs and HMGB1, may be jointly involved with NETs
formation and tumor progression, cannot be excluded.
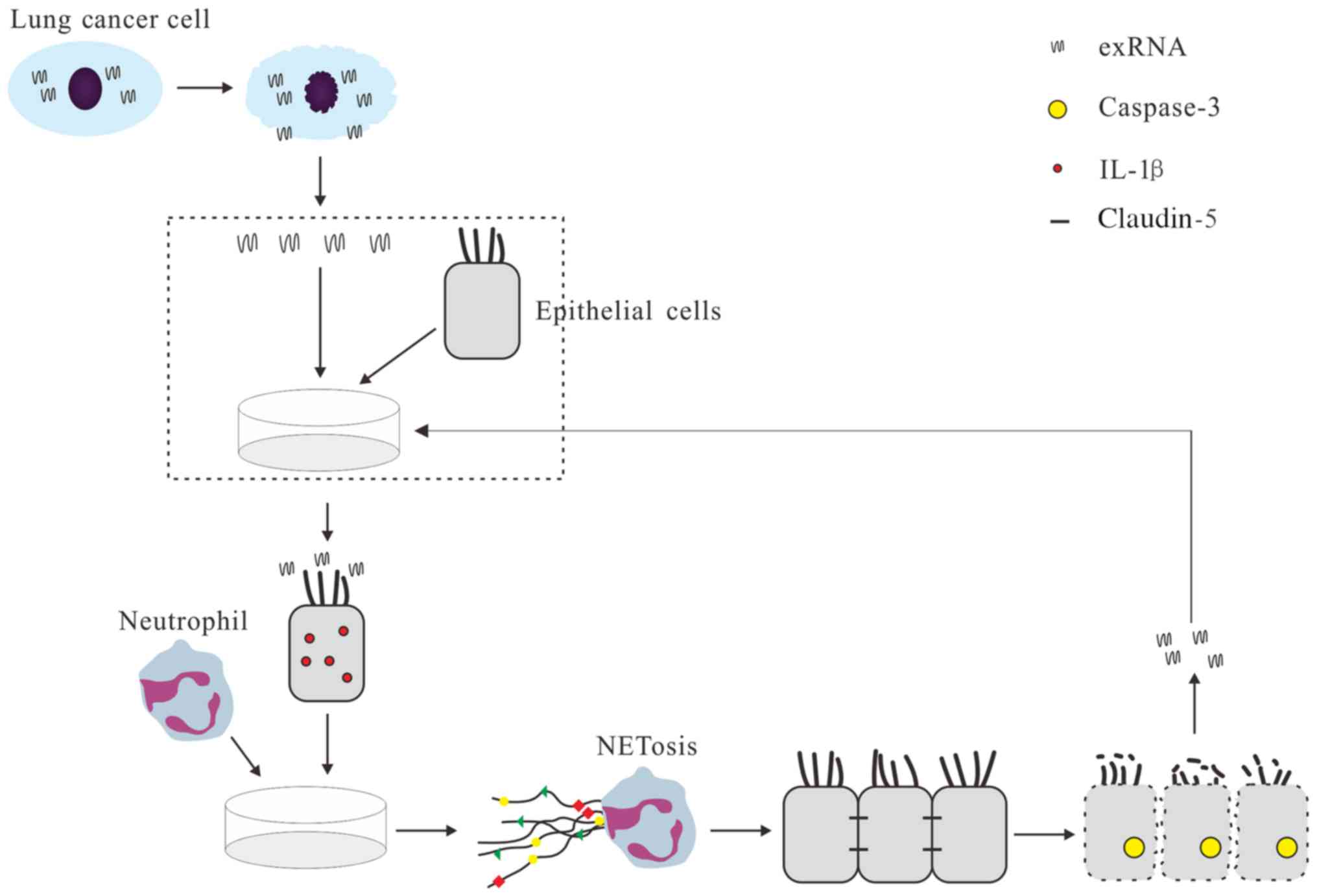
In summary, the results of the present study
demonstrated that activated epithelial cells induce NETs via exRNAs
from lung cancer cells (Fig. 7),
adding the recognition of novel roles of exRNAs for cancer
development (42). RNase1 and
IL-1β inhibitor may be potential tools to block the formation of
NETs induced by exRNAs and activated epithelial cells. Further
studies on the cross-talk between exRNAs and NETs in lung cancer
and other types of cancer are required.
Funding
The present study was supported by National Natural
Science Foundation of China (grant no. 81671563).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
YC and MZ conceived and designed the study. YL, YY,
TG and JZ conducted the experiments. FH, NH, BY, and MZ analyzed
the results. MZ wrote the paper. All the authors reviewed and
approved the manuscript.
Clinics approval and consent to
participate
The present study was carried out in accordance with
the recommendations of 'IACUC of Nanjing Medical University' with
written informed consent from all subjects. All subjects gave
written informed consent in accordance with the Declaration of
Helsinki. The protocol was approved by the 'IACUC of Nanjing
Medical University'.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The present study was supported by National Natural
Science Foundation of China (grant no. 81671563), Natural Science
Foundation of Jiangsu Province (grant no. BK2015155) and Nanjing
Medical University key project (grant no. 2014NJMUZD010).
References
|
1
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar
|
|
2
|
Wright HL, Moots RJ, Bucknall RC and
Edwards SW: Neutrophil function in inflammation and inflammatory
diseases. Rheumatology (Oxford). 49:1618–1631. 2010. View Article : Google Scholar
|
|
3
|
Brinkmann V, Reichard U, Goosmann C,
Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y and Zychlinsky A:
Neutrophil extracellular traps kill bacteria. Science.
303:1532–1535. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Erpenbeck L and Schön MP: Neutrophil
extracellular traps: Protagonists of cancer progression? Oncogene.
36:2483–2490. 2017. View Article : Google Scholar
|
|
5
|
Barnado A, Crofford LJ and Oates JC: At
the Bedside: Neutrophil extracellular traps (NETs) as targets for
biomarkers and therapies in autoimmune diseases. J Leukoc Biol.
99:265–278. 2016. View Article : Google Scholar
|
|
6
|
Döring Y, Soehnlein O and Weber C:
Neutrophil extracellular traps in atherosclerosis and
atherothrombosis. Circ Res. 120:736–743. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bar-Ad V, Palmer J, Li L, Lai Y, Lu B,
Myers RE, Ye Z, Axelrod R, Johnson JM, Werner-Wasik M, et al:
Neutrophil to lymphocyte ratio associated with prognosis of lung
cancer. Clin Transl Oncol. 19:711–717. 2017. View Article : Google Scholar
|
|
8
|
Cools-Lartigue J, Spicer J, McDonald B,
Gowing S, Chow S, Giannias B, Bourdeau F, Kubes P and Ferri L:
Neutrophil extracellular traps sequester circulating tumor cells
and promote metastasis. J Clin Invest. 123:674842013. View Article : Google Scholar
|
|
9
|
Akinosoglou KS, Karkoulias K and Marangos
M: Infectious complications in patients with lung cancer. Eur Rev
Med Pharmacol Sci. 17:8–18. 2013.PubMed/NCBI
|
|
10
|
Patton JG, Franklin JL, Weaver AM, Vickers
K, Zhang B, Coffey RJ, Ansel KM, Blelloch R, Goga A, Huang B, et
al: Biogenesis, delivery, and function of extracellular RNA. J
Extracell Vesicles. 4:274942015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kolodny GM: Evidence for transfer of
macromolecular RNA between mammalian cells in culture. Exp Cell
Res. 65:313–324. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kolodny GM: Cell to cell transfer of RNA
into transformed cells. J Cell Physiol. 79:147–150. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fischer S, Gesierich S, Griemert B,
Schänzer A, Acker T, Augustin HG, Olsson AK and Preissner KT:
Extracellular RNA liberates tumor necrosis factor-α to promote
tumor cell trafficking and progression. Cancer Res. 73:5080–5089.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rabinowits G, Gerçel-Taylor C, Day JM,
Taylor DD and Kloecker GH: Exosomal microRNA: A diagnostic marker
for lung cancer. Clin Lung Cancer. 10:42–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Munagala R, Aqil F and Gupta RC: Exosomal
miRNAs as biomarkers of recurrent lung cancer. Tumour Biol.
37:10703–10714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou Q, Huang SX, Zhang F, Li SJ, Liu C,
Xi YY, Wang L, Wang X, He QQ, Sun CC, et al: MicroRNAs: A novel
potential biomarker for diagnosis and therapy in patients with
non-small cell lung cancer. Cell Prolif. 50:e123942017. View Article : Google Scholar
|
|
17
|
Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z,
Xiang Y, Wu N, Wu L, Bai L, et al: Circulating exosomal microRNAs
as prognostic biomarkers for non-small-cell lung cancer.
Oncotarget. 8:13048–13058. 2017.PubMed/NCBI
|
|
18
|
Ni H, Capodici J, Cannon G, Communi D,
Boeynaems JM, Karikó K and Weissman D: Extracellular mRNA induces
dendritic cell activation by stimulating tumor necrosis
factor-alpha secretion and signaling through a nucleotide receptor.
J Biol Chem. 277:12689–12696. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cabrera-Fuentes HA, Ruiz-Meana M,
Simsekyilmaz S, Kostin S, Inserte J, Saffarzadeh M, Galuska SP,
Vijayan V, Barba I, Barreto G, et al: RNase1 prevents the damaging
interplay between extracellular RNA and tumour necrosis factor-α in
cardiac ischaemia/reperfusion injury. Thromb Haemost.
112:1110–1119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laurent LC and Alexander RP: Cell Culture
Supernatant Collection. PROTOCOL (Version 1) Protocol Exchange.
2015, https://dio.org/10.1038/protex.2015.107.
Accessed December 21, 2015.
|
|
21
|
Gan T, Yang Y, Hu F, Chen X, Zhou J, Li Y,
Xu Y, Wang H, Chen Y and Zhang M: TLR3 regulated poly I:C-induced
neutrophil extracellular traps and acute lung injury partly through
p38 MAP kinase. Front Microbiol. 9:31742018. View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Hirose T, Hamaguchi S, Matsumoto N,
Irisawa T, Seki M, Tasaki O, Hosotsubo H, Yamamoto N, Yamamoto K,
Akeda Y, et al: Presence of neutrophil extracellular traps and
citrullinated histone H3 in the bloodstream of critically ill
patients. PLoS One. 9:e1117552014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yizengaw E, Getahun M, Tajebe F, Cruz
Cervera E, Adem E, Mesfin G, Hailu A, Van der Auwera G, Yardley V,
Lemma M, et al: Visceral leishmaniasis patients display altered
composition and maturity of neutrophils as well as impaired
neutrophil effector functions. Front Immunol. 7:5172016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stankovic B, Bjørhovde HAK, Skarshaug R,
Aamodt H, Frafjord A, Müller E, Hammarström C, Beraki K, Bækkevold
ES, Woldbæk PR, et al: Immune cell composition in human non-small
cell lung cancer. Front Immunol. 9:31012019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Keshari RS, Jyoti A, Dubey M, Kothari N,
Kohli M, Bogra J, Barthwal MK and Dikshit M: Cytokines induced
neutrophil extracellular traps formation: Implication for the
inflammatory disease condition. PLoS One. 7:e481112012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaplan MJ and Radic M: Neutrophil
extracellular traps: Double-edged swords of innate immunity. J
Immunol. 189:2689–2695. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Soini Y: Claudins in lung diseases. Respir
Res. 12:702011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schlingmann B, Molina SA and Koval M:
Claudins: Gatekeepers of lung epithelial function. Semin Cell Dev
Biol. 42:47–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsui NB, Ng EK and Lo YM: Stability of
endogenous and added RNA in blood specimens, serum, and plasma.
Clin Chem. 48:1647–1653. 2002.PubMed/NCBI
|
|
31
|
Slater L, Bartlett NW, Haas JJ, Zhu J,
Message SD, Walton RP, Sykes A, Dahdaleh S, Clarke DL, Belvisi MG,
et al: Co-ordinated role of TLR3, RIG-I and MDA5 in the innate
response to rhinovirus in bronchial epithelium. PLoS Pathog.
6:e10011782010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Papi A and Johnston SL: Respiratory
epithelial cell expression of vascular cell adhesion molecule-1 and
its up-regulation by rhinovirus infection via NF-kappaB and GATA
transcription factors. J Biol Chem. 274:30041–30051. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marui N, Offermann MK, Swerlick R, Kunsch
C, Rosen CA, Ahmad M, Alexander RW and Medford RM: Vascular cell
adhesion molecule-1 (VCAM-1) gene transcription and expression are
regulated through an antioxidant-sensitive mechanism in human
vascular endothelial cells. J Clin Invest. 92:1866–1874. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Skerrett SJ, Liggitt HD, Hajjar AM, Ernst
RK, Miller SI and Wilson CB: Respiratory epithelial cells regulate
lung inflammation in response to inhaled endotoxin. Am J Physiol
Lung Cell Mol Physiol. 287:L143–L152. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ebnet K, Suzuki A, Ohno S and Vestweber D:
Junctional adhesion molecules (JAMs): More molecules with dual
functions? J Cell Sci. 117:19–29. 2004. View Article : Google Scholar
|
|
36
|
Haarmann A, Nowak E, Deiss A, van der Pol
S, Monoranu CM, Kooij G, Müller N, van der Valk P, Stoll G, de
Vries HE, et al: Soluble VCAM-1 impairs human brain endothelial
barrier integrity via integrin α-4-transduced outside-in
signalling. Acta Neuropathol. 129:639–652. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gupta AK, Joshi MB, Philippova M, Erne P,
Hasler P, Hahn S and Resink TJ: Activated endothelial cells induce
neutrophil extracellular traps and are susceptible to
NETosis-mediated cell death. FEBS Lett. 584:3193–3197. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Villanueva E, Yalavarthi S, Berthier CC,
Hodgin JB, Khandpur R, Lin AM, Rubin CJ, Zhao W, Olsen SH, Klinker
M, et al: Netting neutrophils induce endothelial damage, infiltrate
tissues, and expose immunostimulatory molecules in systemic lupus
erythematosus. J Immunol. 187:538–552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H,
Hashimoto N, Furuse M and Tsukita S: Size-selective loosening of
the blood-brain barrier in claudin-5-deficient mice. J Cell Biol.
161:653–660. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jenne CN, Wong CH, Zemp FJ, McDonald B,
Rahman MM, Forsyth PA, McFadden G and Kubes P: Neutrophils
recruited to sites of infection protect from virus challenge by
releasing neutrophil extracellular traps. Cell Host Microbe.
13:169–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Arai Y, Yamashita K, Kuriyama K, Shiokawa
M, Kodama Y, Sakurai T, Mizugishi K, Uchida K, Kadowaki N,
Takaori-Kondo A, et al: Plasmacytoid dendritic cell activation and
IFN-α production are prominent features of murine autoimmune
pancreatitis and human IgG4-related autoimmune pancreatitis. J
Immunol. 195:3033–3044. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Redzic JS, Balaj L, van der Vos KE and
Breakefield XO: Extracellular RNA mediates and marks cancer
progression. Semin Cancer Biol. 28:14–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tadie JM, Bae HB, Jiang S, Park DW, Bell
CP, Yang H, Pittet JF, Tracey K, Thannickal VJ, Abraham E, et al:
HMGB1 promotes neutrophil extracellular trap formation through
interactions with Toll-like receptor 4. Am J Physiol Lung Cell Mol
Physiol. 304:L342–L349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu J, Luo J, Li Y, Jia M, Wang Y, Huang Y
and Ke S: HMGB1 induces human non-small cell lung cancer cell
motility by activating integrin αvβ3/FAK through TLR4/NF-κB
signaling pathway. Biochem Biophys Res Commun. 480:522–527. 2016.
View Article : Google Scholar : PubMed/NCBI
|