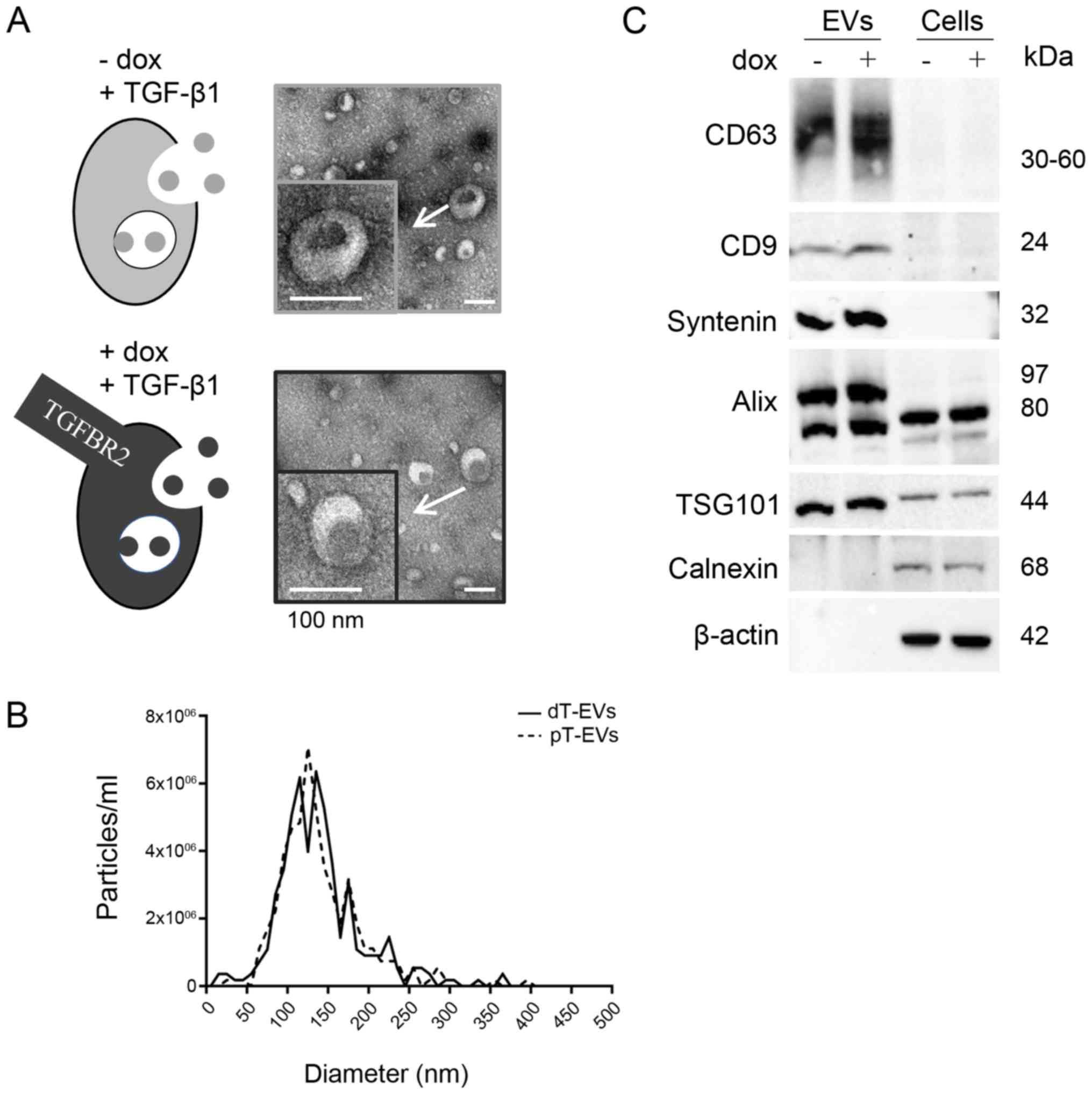
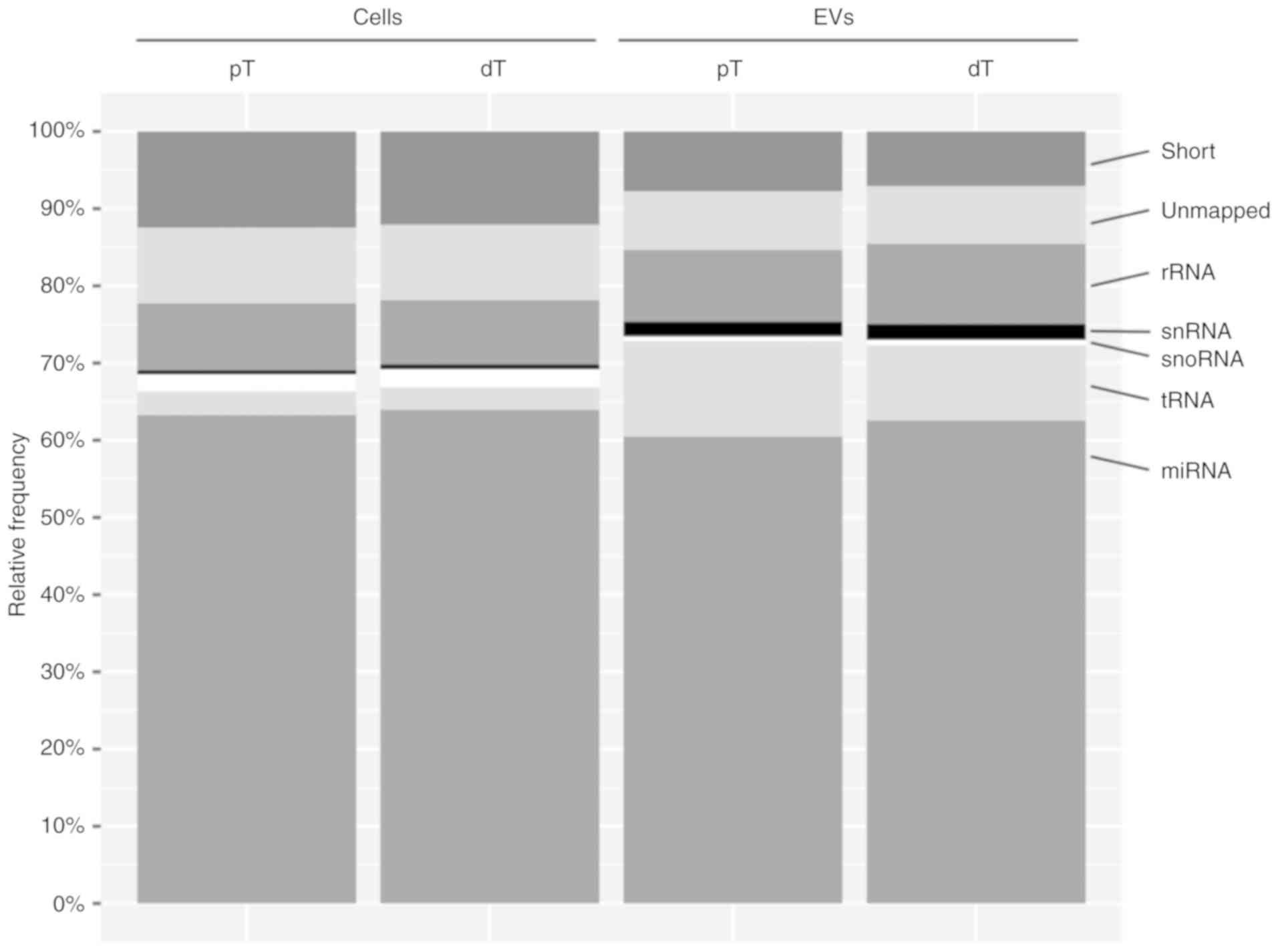
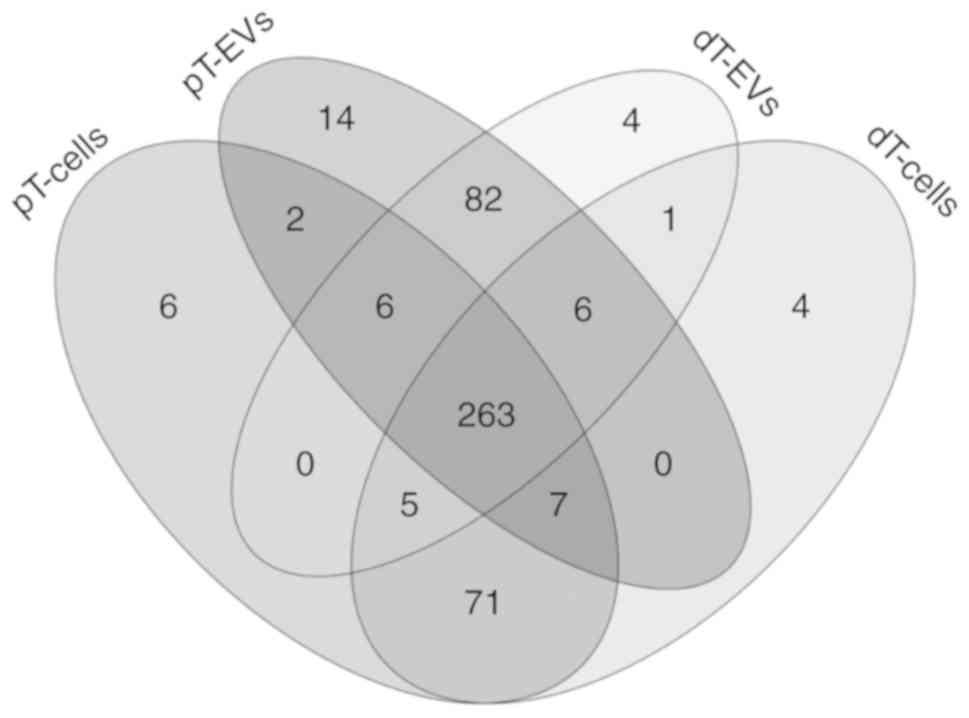
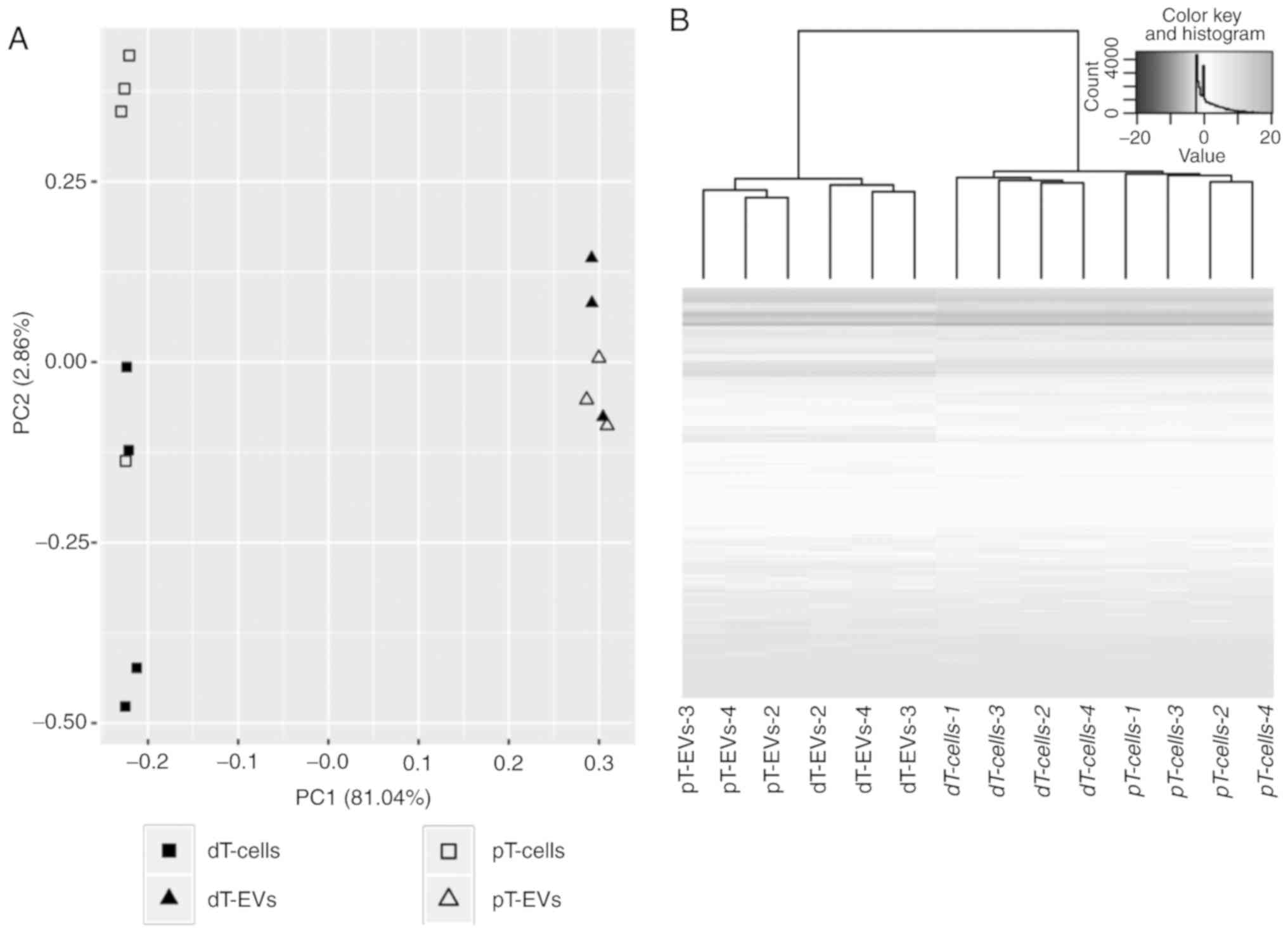
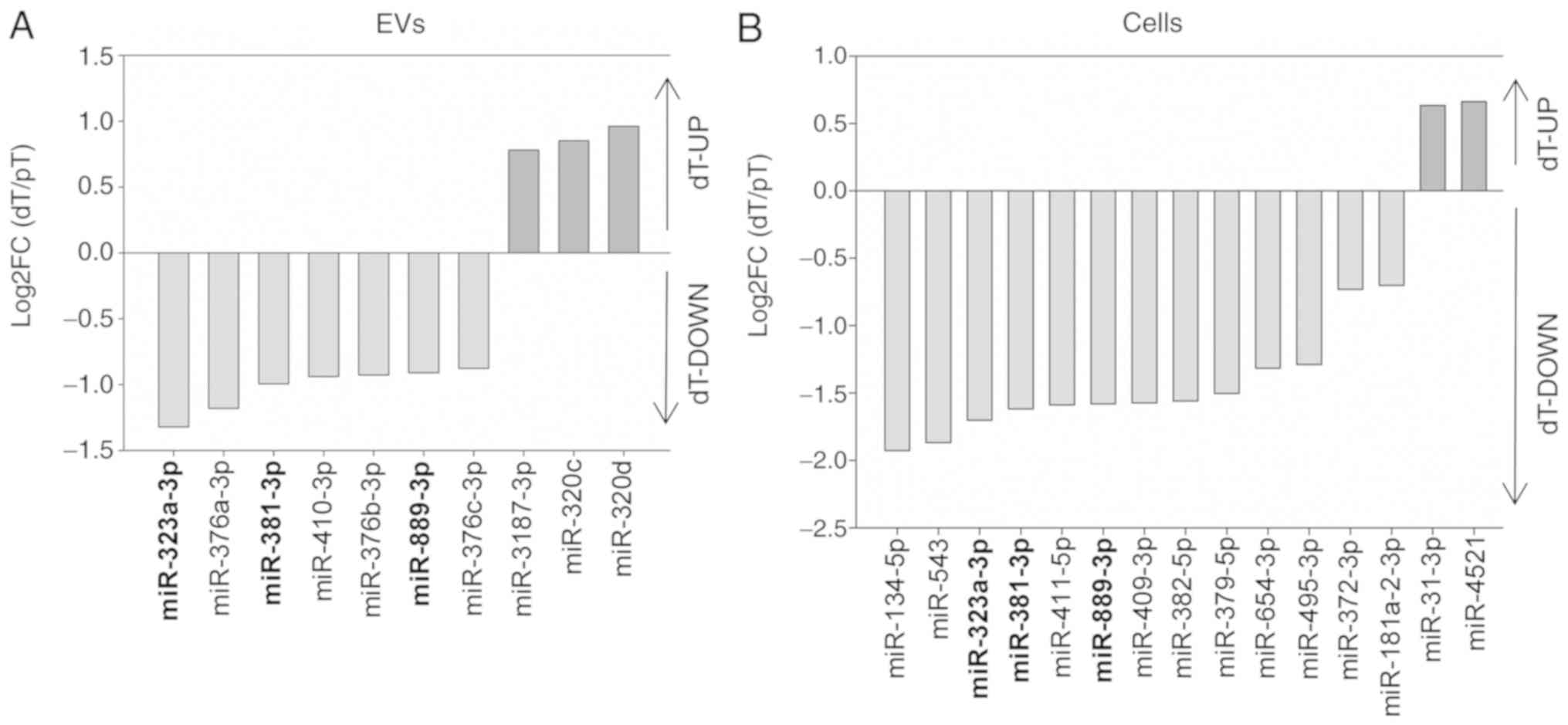
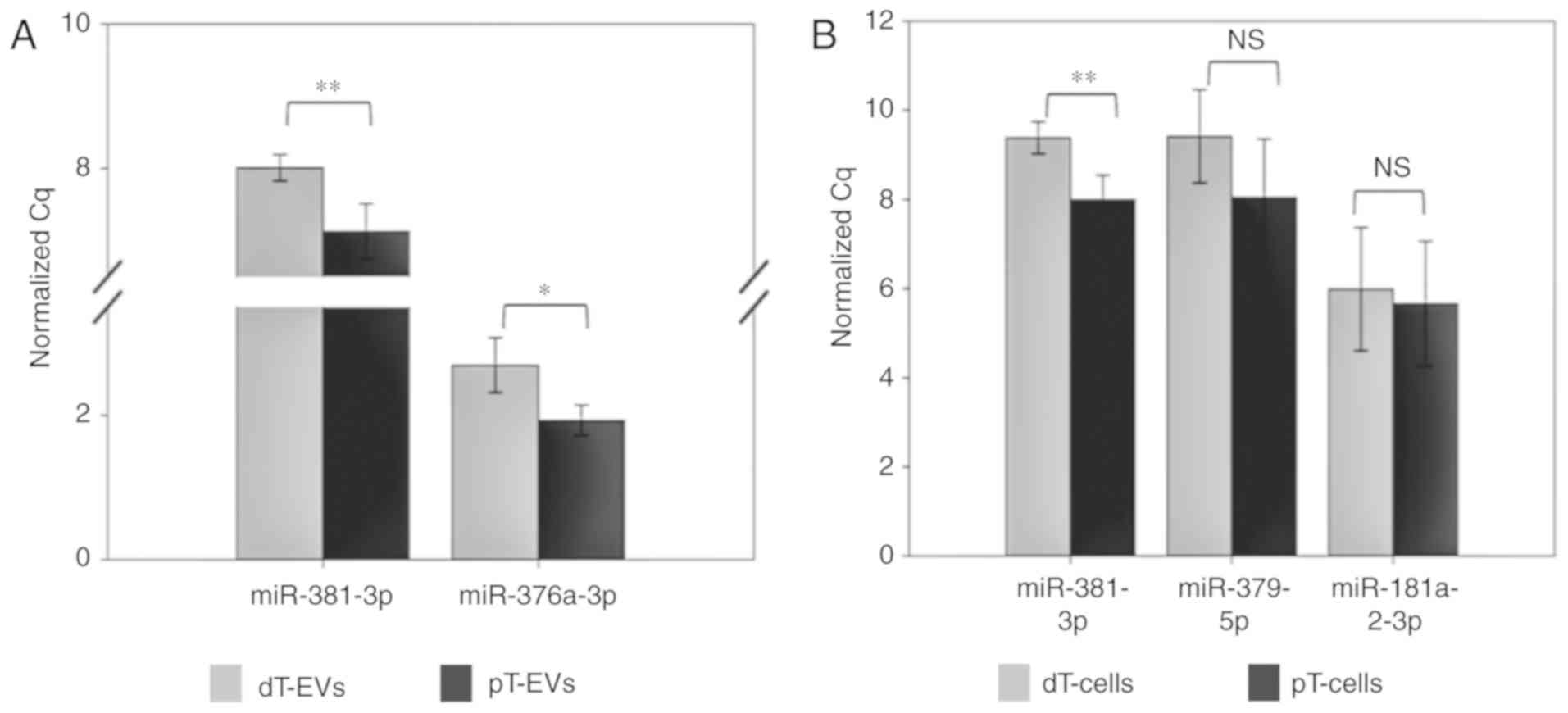
|
1
|
Boland CR and Goel A: Microsatellite
instability in colorectal cancer. Gastroenterology.
138:2073–2087.e3. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Woerner SM, Benner A, Sutter C, Sutter C,
Schiller M, Yuan YP, Keller G, Bork P, Doeberitz Mv and Gebert JF:
Pathogenesis of DNA repair-deficient cancers: A statistical
meta-analysis of putative Real Common Target genes. Oncogene.
22:222622352003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Markowitz S, Wang J, Myeroff L, Parsons R,
Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein
B, et al: Inactivation of the type II TGF beta receptor in colon
cancer cells with microsatellite instability. Science.
268:1336–1338. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Massagué J: TGFβ signalling in context.
Nat Rev Mol Cell Biol. 13:616–630. 2012. View Article : Google Scholar
|
|
5
|
Butz H, Rácz K, Hunyady L and Patócs A:
Crosstalk between TGF β signaling and the microRNA machinery.
Trends Pharmacol Sci. 33:382–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar :
|
|
8
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mamma lian mRNAs are conserved targets of microRNAs.
Genome Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
9
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cantini L, Isella C, Petti C, Picco G,
Chiola S, Ficarra E, Caselle M and Medico E: MicroRNA mRNA
interactions under lying colorectal cancer molecular subtypes. Nat
Commun. 6:88782015. View Article : Google Scholar
|
|
11
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bellingham SA, Shambrook M and Hill AF:
Quantitative Analysis of Exosomal miRNA via qPCR and Digital PCR.
Methods Mol Biol. 1545:55–70. 2017. View Article : Google Scholar
|
|
13
|
Mathieu M, Martin-Jaular L, Lavieu G and
Théry C: Specificities of secretion and uptake of exosomes and
other extracellular vesi cles for cell to cell communication. Nat
Cell Biol. 21:9–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fricke F, Lee J, Michalak M, Warnken U,
Hausser I, Suarez Carmona M, Halama N, Schnölzer M, Kopitz J and
Gebert J: TGFBR2 dependent alterations of exosomal cargo and
functions in DNA mismatch repair-deficient HCT116 colorectal cancer
cells. J Cell Commun Signal. 15:142017. View Article : Google Scholar
|
|
15
|
Lee J, Ballikaya S, Schönig K, Ball CR,
Glimm H, Kopitz J and Gebert J: Transforming growth factor beta
receptor 2 (TGFBR2) changes sialylation in the microsatellite
unstable (MSI) Colorectal cancer cell line HCT116. PLoS One.
8:e570742013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Welman A, Barraclough J and Dive C:
Generation of cells expressing improved doxycycline regulated
reverse transcrip tional transactivator rtTA2S-M2. Nat Protoc.
1:803–811. 2006. View Article : Google Scholar
|
|
17
|
Andrews S: FastQC: A quality control tool
for high throughput sequence data. Bioinformatics Babraham.
http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Accessed 18 Dec, 2018.
|
|
18
|
Kong Y: Btrim: A fast, lightweight adapter
and quality trim ming program for next-generation sequencing
technologies. Genomics. 98:152–153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
The RNAcentral Consortium; Petrov AI, Kay
SJE, Kalvari I, Howe KL, Gray KA, Bruford EA, Kersey PJ, Cochrane
G, Finn RD, et al: A comprehensive database of non coding RNA
sequences. Nucleic Acids Res. 45:D28–D134. 2017.
|
|
20
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:68–73. 2014. View Article : Google Scholar
|
|
21
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10:R252009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Benjamini Y and Hochberg Y: Controlling
the False discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
1995.
|
|
24
|
Fan Y, Siklenka K, Arora SK, Ribeiro P,
Kimmins S and Xia J: miRNet dissecting miRNA-target interactions
and functional associations through network based visual analysis.
Nucleic Acids Res. 44:W135–W141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fabregat A, Sidiropoulos K, Garapati P,
Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F,
McKay S, et al: The Reactome pathway Knowledgebase. Nucleic Acids
Res. 44:D481–D487. 2016. View Article : Google Scholar :
|
|
26
|
Pfaffl MW: A new mathematical model for
relative quantification in real time RT PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expres sion data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Clin Cancer Res. 64:5245–5250. 2004.
|
|
29
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar
|
|
30
|
Théry C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin Smith GK, et al: Minimal information for studies of extra
cellular vesicles 2018 (MISEV2018): A position statement of the
International Society for Extracellular Vesicles and update of the
MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar
|
|
31
|
Ewing B, Hillier L, Wendl MC and Green P:
Base calling of automated sequencer traces using phred. I Accuracy
assessment. Genome Res. 8:175–185. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Patsos G, André S, Roeckel N, Gromes R,
Gebert J, Kopitz J and Gabius HJ: Compensation of loss of protein
function in microsat ellite-unstable colon cancer cells (HCT116): A
gene dependent effect on the cell surface glycan profile.
Glycobiology. 19:726–734. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee J, Warnken U, Schnölzer M, Gebert J
and Kopitz J: A new method for detection of tumor driver dependent
changes of protein sialylation in a colon cancer cell line reveals
nectin 3 as TGFBR2 target. Protein Sci. 24:1686–1694. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee J, Katzenmaier EM, Kopitz J and Gebert
J: Reconstitution of TGFBR2 in HCT116 colorectal cancer cells
causes increased LFNG expression and enhanced
N-acetyl-d-glucosamine incorporation into Notch1. Cell Signal.
28:1105–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee J, Fricke F, Warnken U, Schnölzer M,
Kopitz J and Gebert J: Reconstitution of TGFBR2 mediated signaling
causes upregulation of GDF-15 in HCT116 colorectal cancer cells.
PLoS One. 10:e01315062015. View Article : Google Scholar
|
|
36
|
He X, Wei Y, Wang Y, Liu L, Wang W and Li
N: MiR-381 func tions as a tumor suppressor in colorectal cancer by
targeting Twist1. Onco Targets Ther. 9:1231–1239. 2016.
|
|
37
|
Cha DJ, Franklin JL, Dou Y, Liu Q,
Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N,
Levy S, et al: KRAS-dependent sorting of miRNA to exosomes. Elife.
4:e071972015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tang H, Wang Z, Liu Q, Liu X, Wu M and Li
G: Disturbing miR-182 and -381 inhibits BRD7 transcription and
glioma growth by directly targeting LRRC4. PLoS One. 9:e841462014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang M, Huang S and Long D: MiR-381
inhibits migration and invasion in human gastric carcinoma through
downregulatedting SOX4. Oncol Lett. 14:3760–3766. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liang Y, Zhao Q, Fan L, Zhang Z, Tan B,
Liu Y and Li Y: Down-regulation of MicroRNA-381 promotes cell
proliferation and invasion in colon cancer through up-regulation of
LRH 1. Biomed Pharmacother. 75:137–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu WW, Chen PC, Chen JM, Wu YM, Liu PY, Lu
CH, Lin YF, Tang CH and Chao CC: Periostin promotes epithelial
mesen chymal transition via the MAPK/miR-381 axis in lung cancer.
Oncotarget. 8:62248–62260. 2017.PubMed/NCBI
|
|
42
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heldin CH, Vanlandewijck M and Moustakas
A: Regulation of EMT by TGFβ in cancer. FEBS Lett. 586:1959–1970.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Buckowitz A, Knaebel HP, Benner A, Bläker
H, Gebert J, Kienle P, von Knebel Doeberitz M and Kloor M:
Microsatellite instability in colorectal cancer is associated with
local lympho cyte infiltration and low frequency of distant
metastases. Br J Cancer. 92:1746–1753. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nishimura J, Handa R, Yamamoto H, Tanaka
F, Shibata K, Mimori K, Takemasa I, Mizushima T, Ikeda M, Sekimoto
M, et al: microRNA-181a is associated with poor prognosis of
colorectal cancer. Oncol Rep. 28:2221–2226. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang
Z, Xi J, Yan L and Gu J: MicroRNA-181a promotes tumor growth and
liver metastasis in colorectal cancer by targeting the tumor
suppressor WIF 1. Mol Cancer. 13:862014. View Article : Google Scholar
|
|
47
|
Taylor MA, Sossey Alaoui K, Thompson CL,
Danielpour D and Schiemann WP: TGF-β upregulates miR-181a
expression to promote breast cancer metastasis. J Clin Invest.
123:150–163. 2013. View Article : Google Scholar
|
|
48
|
Ji LJ, Su J, Xu AL, Pang B and Huang QM:
MiR-134-5p attenu ates neuropathic pain progression through
targeting Twist1. J Cell Biochem. Sep 6–2018. View Article : Google Scholar : Epub ahead of
print.
|
|
49
|
El Daly SM, Abba ML, Patil N and Allgayer
H: miRs-134 and-370 function as tumor suppressors in colorectal
cancer by inde pendently suppressing EGFR and PI3K signalling. Sci
Rep. 6:247202016. View Article : Google Scholar
|
|
50
|
Oh BY, Kim SY, Lee YS, Hong HK, Kim TW,
Kim SH, Lee WY and Cho YB: Twist1 induced epithelial -mesenchymal
transition according to microsatellite instability status in colon
cancer cells. Oncotarget. 7:57066–57076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu F, Cao QH, Lu DJ, Luo B, Lu XF, Luo RC
and Wang XG: TMEM16A overexpression contributes to tumor invasion
and poor prognosis of human gastric cancer through TGF β signaling.
Oncotarget. 6:11585–11599. 2015.PubMed/NCBI
|
|
52
|
Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W
and Wang L: MicroRNA 381 inhibits the metastasis of gastric cancer
by targeting TMEM16A expression. J Exp Clin Cancer Res. 36:292017.
View Article : Google Scholar
|
|
53
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar
|
|
54
|
Liu L, Yao J, Li Z, Zu G, Feng D, Li Y,
Qasim W, Zhang S, Li T, Zeng H and Tian X: miR-381-3p knockdown
improves intestinal epithelial proliferation and barrier function
after intestinal isch emia/reperfusion injury by targeting nurr1.
Cell Death Dis. 9:4112018. View Article : Google Scholar
|
|
55
|
Tang YT, Huang YY, Li JH, Qin SH, Xu Y, An
TX, Liu CC, Wang Q and Zheng L: Alterations in exosomal miRNA
profile upon epithelial mesenchymal transition in human lung cancer
cell lines. BMC Genomics. 19:8022018. View Article : Google Scholar
|
|
56
|
Yuan T, Huang X, Woodcock M, Du M, Dittmar
R, Wang Y, Tsai S, Kohli M, Boardman L, Patel T and Wang L: Plasma
extracellular RNA profiles in healthy and cancer patients. Sci Rep.
6:194132016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang J, Yan F, Zhao Q, Zhan F, Wang R,
Wang L, Zhang Y and Huang X: Circulating exosomal miR 125a 3p as a
novel biomarker for early-stage colon cancer. Sci Rep. 7:41502017.
View Article : Google Scholar
|
|
58
|
Slattery ML, Trivellas A, Pellatt AJ,
Mullany LE, Stevens JR, Wolff RK and Herrick JS: Genetic variants
in the TGFβ-signaling pathway influence expression of miRNAs in
colon and rectal normal mucosa and tumor tissue. Oncotarget.
8:16765–16783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fang Y, Xiang J, Chen Z, Gu X, Li Z, Tang
F and Zhou Z: miRNA expression profile of colon cancer stem cells
compared to non-stem cells using the SW1116 cell line. Oncol Rep.
28:2115–2124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mishra L, Derynck R and Mishra B:
Transforming growth factor-beta signaling in stem cells and cancer.
Science. 310:68–71. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Valmiki S, Ahuja V and Paul J: MicroRNA
exhibit altered expression in the inflamed colonic mucosa of
ulcerative colitis patients. World J Gastroenterol. 23:5324–5332.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fang K, Law IKM, Padua D, Sideri A, Huang
V, Kevil CG, Iliopoulos D and Pothoulakis C: MicroRNA-31-3p is
involved in substance P (SP)-associated inflammation in human
colonic epithelial cells and experimental colitis. Am J Pathol.
188:586–599. 2018. View Article : Google Scholar :
|
|
63
|
Olaru AV, Selaru FM, Mori Y, Vazquez C,
David S, Paun B, Cheng Y, Jin Z, Yang J, Agarwal R, et al: Dynamic
changes in the expression of MicroRNA-31 during inflammatory bowel
disease-associated neoplastic transformation. Inflamm Bowel Dis.
17:221–231. 2011. View Article : Google Scholar
|
|
64
|
Li T, Luo W, Liu K, Lv X and Xi T: miR 31
promotes prolifera tion of colon cancer cells by targeting E2F2.
Biotechnol Lett. 37(523): 5322015. View Article : Google Scholar
|
|
65
|
Nosho K, Igarashi H, Nojima M, Ito M,
Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, et
al: Association of microRNA-31 with BRAF mutation, colorectal
cancer survival and serrated pathway. Carcinogenesis. 35:776–783.
2014. View Article : Google Scholar
|
|
66
|
Laurent Puig P, Grisoni ML, Heinemann V,
Liebaert F, Neureiter D, Jung A, Montestruc F, Gaston Mathe Y,
Thiébaut R and Stintzing S: Validation of miR 31 3p expression to
predict cetuximab efficacy when used as first-line treatment in RAS
Wild-type metastatic colorectal cancer. Clin Cancer Res.
25:134–141. 2019. View Article : Google Scholar
|
|
67
|
Harrison PW, Alako B, Amid C, Cerdeño
Tárraga A, Cleland I, Holt S, Hussein A, Jayathilaka S, Kay S,
Keane T, et al: The European nucleotide archive in 2018. Nucleic
Acids Res. 47:D84–D88. 2019. View Article : Google Scholar :
|
|
68
|
EV-TRACK Consortium; Van Deun J, Mestdagh
P, Agostinis P, Akay Ö, Anand S, Anckaert J, Martinez ZA, Baetens
T, Beghein E, et al: EV TRACK: Transparent reporting and
centralizing knowledge in extracellular vesicle research. Nat
Methods. 14:228–232. 2017. View Article : Google Scholar : PubMed/NCBI
|