Prostate cancer (PC) is the most commonly diagnosed
cancer affecting males in developed countries and a major cause of
cancer-related mortality among males (1). The disease is highly heterogeneous
and progresses with a large degree of disparity. PC evolves from
high-grade prostatic intra-epithelial neoplasia (HGPIN) to local
carcinoma; some local tumors will develop into metastatic disease
with bone as the preferential site (2). Primary tumors are managed through
watchful waiting (active surveillance) and curative therapies:
Radical prostatectomy (RP) or radiation therapy (RT) (3-6). The
disease may relapse in the form of biochemical recurrence (BCR)
with elevations in serum prostate-specific antigen (PSA) levels of
>0.2 ng/ml following RP and >2 ng/ml above the nadir
following RT (7). Approximately
30% (20-40%) of patients following RP (8-10)
and 30-50% of males treated with RT will experience BCR (11,12)
within 10 years posy-therapy. BCR represents a major progression
and is associated with a significantly increased risk of PC
metastasis; 24-34% of patients with BCR will develop metastasis
(13,14). The standard treatment for
metastatic PC remains androgen deprivation therapy (ADT); however
it is largely a palliative care as metastatic castration-resistant
PCs (mCRPCs) commonly develop (15). Although multiple treatment options
are currently available for mCRPCs, these therapies only marginally
prolong the median overall survival (OS) and resistance develops
rapidly. This is the major challenge with therapies targeting
mCRPCs using docetaxel (16,17)
or the second generation anti-androgens (abiraterone and
enzalutamide) approved by the FDA in 2011 and 2012 (18,19).
Collectively, with this knowledge of PC development and the current
limitations in treating metastasis, the most beneficial management
of prostate cancer is through the accurate stratification of
patients with PC with a low risk of BCR progression from those with
a high risk. This capacity of BCR risk stratification is of
particular relevance to patients with low- and intermediate-risk
PCs; low-risk and intermediate-risk PCs are defined by the European
Association of Urology (EAU)-European Society for Radiotherapy and
Oncology (ESTRO)-International Society of Geriatric Oncology (SIOG)
as PSA <10 ng/ml, Gleason score (GS) <7, cT1-2a, and
local-ized (low risk) and PSA levels of 10-20 ng/ml or GS 7 or cT2c
and localized (intermediate risk) (3).
The current stratification of the risk of BCR in
clinical practice remains poor; improvement in this capacity
remains a major focus of the research community. Attributing to
this massive effort and the involvement of complex networks
affecting BCR progression, there are enriched data for BCR risk
classification for localized tumors following primary curative
treatments, particularly RP. The risk stratification is based on
two general aspects of PC: Clinical characteristics and molecular
properties or biomarkers. The latter includes alterations in gene
expression at both the gene and protein level. Due to the
overwhelming amount (search for 'prostate cancer AND biomarkers AND
biochemical recurrence' in PubMed resulted in 2,500 articles) and
the heterogeneity of the data, in this review, we focus on
RNA-based biomarkers, which can be effective in nature. We also
briefly discuss other types of BCR biomarkers to make this review
comprehensive.
The clinical and tumor characteristics have long
been investigated for the estimation of the risk of BCR. By using
pre-treatment PSA, the GS, clinical T stage, the percentage of
biopsy cores positive for cancer, and age in 1,493 patients treated
with RP between 1992 and 2001, the University of California, San
Francisco Cancer of the Prostate Risk Assessment (UCSF-CAPRA or
CAPRA) was developed in 2005 to appraise the BCR risk; this is a
score system with scale of 0-10 and higher scores represent a
higher risk of BCR (20). Up to
2017, CAPRA has been validated on BCR risk stratification following
RP and RT by 12 investigations carried out in the USA, Germany,
Japan, Australia, Korea and Canada; these studies involved a total
of 17,457 patients and demonstrated that CAPRA classifies the risk
of BCR with a concordance index (c-index) ranging from 0.67 to 0.81
(20). The status of CAPRA has
recently been updated by Brajtbord et al (21); the modified version, CAPRA-S, was
subsequently developed by the same group in 2011 and independently
validated (21,22). Prior to CAPRA, the D'Amico
classification of the risk of BCR was generated by D'Amico et
al in 1998 (23). The CAPRA
score system seems superior to the D'Amico classification (21).
While approximately 30% of males undergoing RP will
experience BCR within 10 years (8-10),
two-thirds of these recurrences occur during the first 2 years
(24-26). Early recurrence is associated with
a higher risk of metastasis (27,28).
To assess early BCR, the Walz nomogram was constructed in 2009
(29), which has recently been
updated with 13,797 patients who had undergone radical
prostatectomy from Hamburg (2005-2016) and validated using 5,952
males treated with RP in Vienna (30). The validation using the Vienna
dataset revealed the best estimation of BCR risk by the updated
nomogram in comparison to the Walz nomogram, MSKCC nomogram, and
CAPRA-S (30). The nomogram
estimates BCR risk at 12 and 24 months post-RP based on PSA, GS, pT
stage, surgical margin status and lymph node status (30).
MYC plays multiple roles in tumorigenesis, which
includes the regulation of cancer metabolism (36,37).
It is upregulated in PC (38) and
contributes to PC progression in part via telomerase overexpression
and the loss of PTEN (39,40). While increases in MYC protein
expression are associated with higher a GS and T-stage, an
association between MYC and BCR remains unclear (33).
The overexpression of ERG in PC results from the
fusion of the androgen target gene transmembrane serine protease 2
(TMPRSS2) with ERG (TMPRSS2-ERG) (41). The ERG protein can be detected in
PC by immunohistochemistry (IHC) (42). In a systemic review, the
overexpression of the ERG protein was shown to be modestly
associated with BCR with P-values of 0.04, 0.006, or 0.002
(33).
In a study of 52 males with PC, an association of
p53 expression with BCR was demonstrated (P=00097) (43), which was corroborated by another
small cohort involving 86 patients with PC (P<0.01) (44). Collectively, IHC-detected p53
protein expression is associated with BCR (33). In a systemic review published in
2018 on the IHC-based detection of BCR (33), the loss of PTEN was found to be
associated with BCR in 8 investigations.
Nonetheless, while IHC-detected protein expression
can display significant associations with BCR, the associations are
modest in most cases and their applications in clinical practice
are limited. This is likely attributed to the limited number of
proteins that can be simultaneously detected by IHC; the
examination of the expression status of a panel of proteins or
signatures consisting of multiple factors is critical to
effectively stratify the risk of BCR.
While the impact of genomic alterations on PC
progression will not be covered in this review, it is important to
summarize the recent developments related to the impact of germline
mutations on PC progression. A family history is a well-recognized
risk factor of PC (45);
nonetheless, hereditary PCs, which constitute approximately 9% of
all PCs, do not differ from spontaneous PCs based on the 2016
EAU-ESTRO-SIOG guidelines (3).
Thus, it was generally accepted that germline mutations do not
promote PC progression and are thus without prognostic value. The
exception was first observed with BRCA2 germ-line mutations that
increase the incidence of PC along with the risk of PC progression
(46,47); these mutations drive the evolvement
of PC by causing genomic instability (48). In line with this concept, germline
mutations in other factors regulating the DNA damage response (DDR)
also increase the risk of PC progression, including ATM, CHEK2,
BRCA1, RAD51D and PALB2 (49). The
observation that BRCA1/2 germline mutations are associated with the
risk of PC and PC progression provides additional support for the
similarities between PC and breast cancer. This is consistent with
a recent study demonstrating that PCs can be grouped into
PAM50-based luminal A and luminal B subtypes (50), the well-known subtypes of estrogen
receptor-positive breast cancer (51).
It will thus be of interest to investigate the
contributions of mutations in BRCA2, ATM, CHEK2, BRCA1, RAD51D and
PALB2 in a variety of combinations in the assessment of the risk of
BCR. Of note, genomic alterations in 9 DDR pathways involving 17
gene sets are able to classify the risk of BCR [population size,
n=545; hazard ratio (HR), 1.89; 95% confidence interval (95% CI),
1.44-2.48; P=5.01e-6] (52).
In light of the important function of long
non-coding RNAs (lncRNAs) in preventing miRNA-mediated mRNA
degradation via competing or sponging, we also discuss the
association of lncRNAs with BCR.
An increase in platelet-derived growth factor
receptor (PDGFR)-β expression in the stroma significantly enhances
BCR (Table II) (87). An elevated stromal PDGFR-β
expression has been shown to be associated with a poor prognosis in
both breast and prostate cancer (88).
The downregulation of metallothionein 1E (MT1E) is a
risk factor for BCR in association with promoter methylation
(89). MT1E belongs to the
metallothionein (MT) family consisting of cysteine-rich small
proteins that regulate metal homeostasis (90). In addition to PC, MT1E is also
downregulated in endometrial carcinoma (91), intrahepatic cholangiocarcinoma
(92), melanoma (93), non-small cell lung cancer (94), papillary thyroid carcinoma
(95) and renal cell carcinoma
(96); in the majority of these
cancer types, the reductions are associated with hypermethylation
(90). However, the upregulation
of MT1E has been reported in estrogen receptor-negative breast
cancer (97) and it also
facilitates glioma progression (98,99).
An elevation in neuropilin-1 (NRP1) mRNA expression
is associated with BCR following RT (Table II) (102). This transmembrane glycoprotein
can activate PDGFR-β (103) and
contributes to the stemness of breast CSCs via the activation of
Wnt signaling (104). NRP1 has
been reported to be upregulated in PC (105) and may contribute to BCR in part
through the regulation of endothelial cell functions (106).
Increases in sterile alpha motif domain containing 5
(SAMD5) mRNA expression display biomarker values in predicting BCR
(Table II) (107). SAMD5 facilitates small cell lung
cancer cell proliferation (108),
is upregulated in cholangiocarcinoma (109) and is associated with the response
to chemotherapy in rectal cancer (110). SAMD5 facilitates the Eph receptor
tyrosine kinase signaling (111),
suggesting a mechanism mediating SAMD5 oncogenic potential and its
association with BCR.
Consistent with SMAD4 as a tumor suppressor in the
inhibition of PTEN inactivation-induced PC progression (112), a reduction in SMAD4 mRNA
expression enhances the risk of BCR (113).
The downregulation of pleomorphic adenoma gene
like-2 (PLAGL2) mRNA expression is a risk factor of BCR (114). PLAGL2 is a transcription factor
that has been shown to activate Wnt/β-catenin signaling through
unidentified mechanisms in colorectal cancer (115) and gliomas (116). PLAGL2 also contributes to
hematopoietic tumorigenesis (117,118); however, its involvement in PC has
not yet been fully investigated.
In an analysis of 7,826 prospectively collected RP
tissues and 1,567 retrospectively obtained samples, while PD-L1 did
not exhibit prognostic values, an increase in PD-L2 expression was
associated with a decrease in BCR-free survival (Table II), distant-free metastasis
survival (HR, 1.25; 95% CI, 1.05-1.49; P= 0.01) and PC-specific
survival (HR, 1.45; 95% CI, 1.13-1.86; P=0.003) (119). These observations are in line
with the actions of the immune checkpoint in the downregulation of
immunoresponses to cancers. Nonetheless, these associations are not
particularly robust.
RNase khas been shown to be downregulated in PC
(n=111) in comparison to benign prostatic hyperplasia (BPH); the
downregulation was associated with BCR (Table II) (120). The contributions of RNase k to
tumorigenesis in general remain unclear (121).
An upregulation of glioma tumor suppressor candidate
region gene 1 (GLTSCR1) in PC vs. normal prostate tissues has been
reported; the upregulation is a risk factor of BCR (122). Evidence suggests an oncogenic
role of GLTSCR1 in oligodendrogliomas (123). Although the functionality of
GLTSCR1 in tumorigenesis remains unclear, recent evidence indicates
its role in chromatin remodeling (124), implying GLTSCR1 may contribute to
BCR progression via epigenetic regulations.
Butyrylcholinesterase (BChE) was recently reported
to display a biphasic alteration in PCs in both the MSKCC (n=140)
and TCGA (n=245) databases; elevations in BChE mRNA expression have
been shown to be associated with BCR in both cohorts (P=0.008 for
MSKCC and P=0.04 for TCGA) (Table
II) (125). BChE has been
shown to hydrolyze butyrylcholine (126), succinylcholine (127) and ghrelin (the hunger hormone)
(128-131), and thus may play a role in PC
metabolism.
Collectively, the above individual mRNAs stratify
BCR risk through different pathways, including the Wnt pathway,
growth factor receptor-mediated cell proliferation, androgen
signaling, cytokines, immune checkpoints, RNA metabolism and others
(Table II). While this is in
accordance with the complex nature of BCR progression, it also
reveals the challenge of using individual mRNA to effectively
predict BCR risk and the calls for developing multigene sets or
signatures for assessing BCR development.
To enhance the accuracy of predicting BCR risk,
there have been numerous efforts made towards the construction of
multigene panels; the rapid accumulation of cancer genomic data
owing to technology advances in DNA sequencing [next generation
sequencing (NGS)] greatly facilitates this exploration. Among these
multigene panels, only three are commercially available to assist
patient management. The 22-gene Decipher is intended to predict
metastasis following RP (132-134); both the 17-gene Oncotype DX
[Genomic Prostate Score (GPS)] and the 31-gene Prolaris [Cell Cycle
Progression (CCP)] stratify patients at risk of PC recurrence at
the time of diagnosis (135-139) and following RP (140,141). Herein, we briefly review Oncotype
DX GPS and CCP and discuss other multi-gene panels regarding their
potentials and limitations.
Oncotype DX Prostate Cancer Assay was developed by
Genomic Health Inc. as an assay in the Oncotype DX assays for
multiple cancer types. Oncotype DX GPS is a RT-PCR assay on 12
cancer-related and 5 reference genes (ARF1, ATP5E,
CLTC, GSP1 and PGK1) using biopsy tissues
(135); the 12 genes function in
4 aspects of PC tumorigenesis, including a stromal process
(BGN, COL1A1 and SFRP4), cellular organization
pathway (FLNC, GSN, TPM2 and GSTM2),
androgen signaling (FAM13C, KLK2, AZGP1 and
SRD5A2) and cell proliferation regulation (TPX2)
(135). They were selected from
732 candidate genes, which were narrowed down from an initial set
of 1,082 nominating candidates, through a variety of processes
involving multiple data-mining models (136). PGS in the scale of 0-100 can be
calculated based on the normalized expressions of 12 cancer-related
genes with increased scores indicating elevations in BCR risk
(136). In patients with low-risk
(GS 6) or intermediate-risk (GS 3+4) PC, GPS predicts BCR (n=382;
HR, 2.73; 95% CI, 1.84-3.96; P<0.001) (140). In a recent validation study, GPS
classified PCs at risk of BCR (n=259; HR, 2.5; 95% CI, 1.28-3.03;
P=0.002) (142). Furthermore, in
a late multiple institutional investigation involving 1,200 males
with very low-, low- and intermediate-risk PCs, GPS predicted
adverse pathological features of PC (143). Although GPS has been
independently validated for the better management of patients with
low- and intermediate-risk PC, the system could be improved. For
instance, GPS does not significantly predict BCR in patients who
are <56 years old (n=100) (140); the cellular organization group
score, 3 of 4 component genes of this group, and the proliferation
group score do not individually predict BCR risk (140), which reduces the biomarker value
of GPS. Although the 12 cancer-related genes were selected via a
thorough and complex process from 732 candidates (136), it is of concern whether too many
manipulations may not produce the best model.
Genes regulating CCP possess prognostic potential in
assessing cancer progression (144). Of note, a panel of 31 CCP genes
has been selected from 126 cell cycle progression genes, which
together with 15 housekeeping genes form the Prolaris (CCP)
multigene panel (Myriad Genetics Int.) (137). Prolaris is a RT-PCR based assay
on formalin-fixed paraffin-embedded tumor tissues and provides risk
assessment of BCR progression (137). The risk stratification has been
validated (Table III) (141,145-147). Evidence also indicates its
utilization in the risk stratification of PC fatality (n=349; HR,
2.02; 95% CI, 1.62-2.53; P<1e-9) (148). However, variations in the
effectiveness of BCR risk stratification of some studies were
apparent; for instance, in the study involving 236 patients
(Table III), HR was modest and
the lower HR in the 95% CI range was marginal (Table III). Additionally, it remains
uncertain whether the Prolaris CCP test will have an impact on PC
death and is unlikely to facilitate treatment decision; the cost of
test is also high (149).
Nonetheless, both Oncotype DX GPS and Prolaris CCP are commercially
available to assess BCR risk.
Even with the construction of Oncotype DX GPS and
Prolaris CCP multigene panels, there is clearly a need to improve
the assessment of BCR. To fulfill this need, there are numerous
additional multigene sets reported (Table IV), including a 6-differentially
expressed gene (DEG) panel (150), an 8-gene panel with its risk
scores predicting BCR at P=5e-7 (151), and a 10-gene panel HDDA10
(152) (Table IV).
Hypoxia is well known to promote PC progression via
multiple pathways, including inflammation and notch signaling
(153,154). To examine the prognostic values
of hypoxia-induced events in PC progression, Yang et al
derived a 28-gene hypoxia-related prognostic signature from 848
differentially expressed genes that were identified in human PC
cell lines cultured under hypoxic and normoxic conditions (155). The signature modestly predicts
BCR in RP patients receiving post-operative radiotherapy (155) (Table IV).
Instead of focusing on a particular pathway, a
15-gene signature has recently been formulated from the MUC1
network (SigMuc1NW) (156); the
signature was validated in the MSKCC dataset. SigMuc1NW stratifies
the BCR risk in the MSKCC dataset at P-value 3.11e-15 (156). MUC1 is the most intensively
investigated tumor-associated antigen (157-159) and is an attractive target for
developing immunotherapies for multiple tumor types (160). MUC1 upregulation is weakly
associated with BCR occurrence and PC mortality (161,162). The biomarker potential of MUC1
alterations in the classification of BCR risk was significantly
enhanced in a 9-gene genomic signature (163). The 15-gene SigMuc1NW was derived
using the 9-gene signature-associated DEGs (156). SigMuc1NW is an independent risk
factor of BCR (HR, 2.44; 95% CI, 1.53-3.87; P=1.62e-4) after
adjusting for age at diagnosis, GS, surgical margin and tumor stage
(156). Among its 15 component
genes, 8 (SLCO2A1, SUPV3L1, TATDN2, MGAT4B, VAV2, SLC25A33, ASNS
and OIP5) individually predict BCR after adjusting the clinical
features (156). Another
attractive feature of SigMuc1NW lies in its novelty; among the 15
component genes, 11 have not been reported in PC particularly
and/or tumorigenesis in general (156).
The inclusion of Opa interacting protein 5 (OIP5)
in SigMuc1NW is intriguing; it is a cancer-testis antigen and thus
a tumor-associated antigen (TAA) detected in other cancer types
(164). OIP5 is likely a novel
PC-associated TAA. More appealingly, recent developments revealed
an essential role of OIP5 in chromosome segregation during cell
cycle progression. OIP5 is also known as Miss18β, that plays a
critical role in centromere formation during the G1 phase (165,166). In accordance with this knowledge,
OIP5 is an independent risk factor for BCR (HR, 1.94; 95% CI,
1.20-3.12; P=0.00638) after adjusting for age at diagnosis, GS,
surgical margin and tumor stage (156); OIP5 promotes bladder cancer
metastasis and chemoresistance (167), glioblastoma metastasis (168), it displays a biomarker potential
in clear cell renal cell carcinoma (169), and it is upregulated in
colorectal and breast cancer (170,171).
In line with the concept of the involvement of
multiple pathways in BCR progression and the robustness of
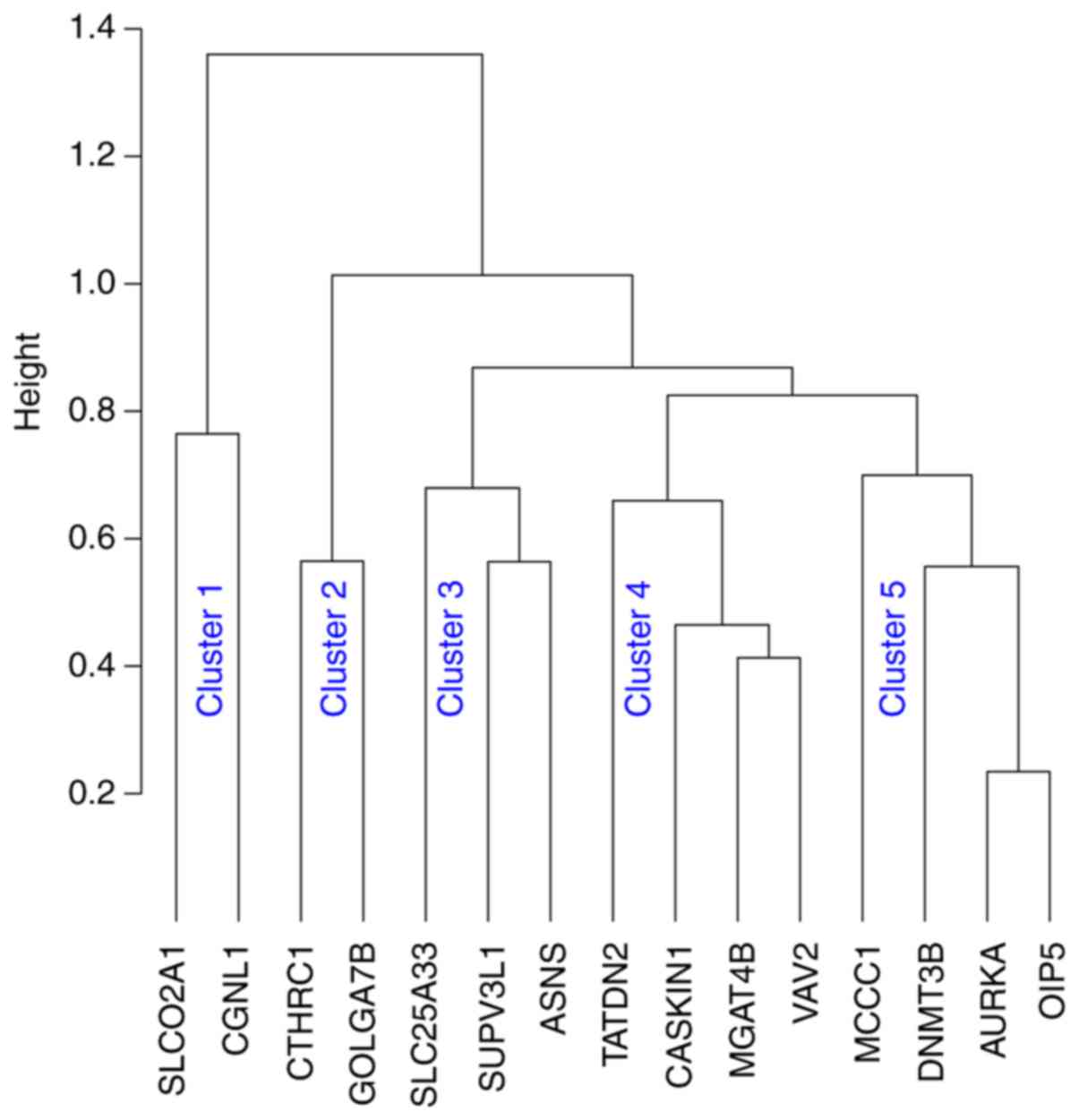
SigMuc1NW in the classification of BCR risk (Table IV) (156), our recent analysis revealed the
signature's 15 component genes (Table
IV) being grouped into 5 clusters using Kendall, Spearman's and
Pearson correlation (Fig. 3).
Collectively, evidence supports SigMuc1NW as a novel and robust
multigene signature. Nonetheless, its biomarker value has not been
independently tested.
While the mechanisms underlying the lncRNA-mediated
regulation of gene expression remain incompletely understood, they
are likely regulated through complex actions at the genome
(chromatin remodeling), mRNA and protein levels (172). Of these, its function as miRNA
sponges is emerging as a prevalent mechanism (172,173). In this regard, this section
reviews the current evidence for lncRNAs as classifiers of BCR
risk. For a comprehensive review, we first searched PubMed for
'lncRNA' AND 'prostate cancer' AND 'biochemical recurrence', and
retrieved 15 articles. With exclusion of one non-accessible
publication and three articles in which the association of lncRNAs
with BCR was not clear, 11 manuscripts are included (179) and Tables V and VI.
Elevations in the levels of lncRNA LOC400891 have
been observed in tumors vs. prostate tissue (181). The upregulation increases BCR
risk in patients (Table V); its
overexpression and knockdown accordingly enhance and inhibit PC
cell proliferation in vitro. There is evidence to indicate a
role of LOC400891 in the activation of the PI3K pathway (181). Nonetheless, the involvement of
LOC400891 in PC and other cancer types has yet to be further
investigated.
Second chromosome locus associated with prostate-1
(SChLAP1; LINC00913) is upregulated in PC and promotes tumor
invasion and metastasis (195).
In a multicenter study involving 937 patients, SChLAP1
overexpression was associated with lethal PC (196). Of note, elevations in SChLAP1
expressoin have been shown to predict PSA relapse (Table V) (197), an event which has also been
observed by others (179), and PC
metastasis (198). While SChLAP1
has been reported to prevent the association of the SWI/SNF complex
with chromatin and thereby inhibiting the complex-associated tumor
suppression in PC (195), late
development revealed a SWI/SNF-independent action of SChLAP1 in PC
tumorigenesis (199); the
mechanisms through which SChLAP1 affects PC require further
investigation.
The lncRNA urothelial carcinoma-associated 1 (UCA1)
marginally predicts the risk of BCR (200). The prediction is consistent with
the associations of UCA1 with reductions in the 5-year disease-free
survival in PC (n=130; HR, 2.88; 95% CI, 1.36-6.21; P= 0.007)
(200) and in overall survival
(n=40, P<0.001) (201).
Additionally, the upregulation of UCA1 has also been shown to be a
risk factor for the progression of ovarian cancer (202), gastric cancer (203), melanoma (204), pancreatic cancer (205), glioma (206) and others (207). Mechanistically, UCA1 facilitates
PC at least in part through upregulations of ATF2 and CXCR4 by
sponging miR-204 (208,209). Intriguingly, UCA1 sequesters
miR-204, leading to EMT in glioma, TGF-β signaling in oral cancer
and Sox4 actions in esophageal cancer (207); UCA1 also sponges other miRNAs in
promoting tumorigenesis in other cancer types (207). In this regard, the association of
UCA1 with BCR could be strengthened by consideration of
UCA1-regulated oncogenic factors.
The downregulation of the lncRNA prostate
cancer-associated transcript 7 (PCAT7) is an independent factor
predicting BCR (Table V) (210), consistent with its reductions
following advance in GS and its downregulations independently
predicting metastasis (210).
Similar clinical associations were also confirmed by a multicenter
study, in which PCAT14 was found to be an independent risk factor
of metastasis (n=910; HR, 0.56, 95% CI, 0.41-0.71; P=1.09e-6),
prostate cancer-specific survival (HR, 0.53; 95% CI. 0.39-0.72;
P=6.54e-5) and overall survival (HR, 0.67; 95% CI, 0.54-0.83;
P=0.00019) (211). Apart from
these two investigations, the involvement of PCAT14 in PC and other
cancer types has not yet been thoroughly examined; the potential
mechanisms of PCAT14 downregulation and its impact on PC
progression have yet to be reported. Nonetheless, it appears that
PCAT14 affects tumorigenesis in a complex manner; in HCC, PCAT14 is
upregulated and promotes HCC cell proliferation and invasion
(212).
Multi-lncRNA panels have been constructed to
stratify the risk of BCR, including a 4-lncRNA (213), 5-lncRNA (214), 7-lncRNA (215) and 8-lncRNA panels (Table VI) (216). All these studies were
bioinformatics analyses of the TCGA dataset using different modules
and sub-datasets. Differentially expressed lncRNAs (DE-lncRNAs) in
the setting of PCs vs. prostate tissues were derived, followed by
selection for their associations with BCR using either univariate
Cox analysis (213,215,216) or the LASSO (least absolute
shrinkage and selection operator) Cox regression (214); DE-lncRNAs with significant
associations with BCR constituted the individual lncRNA panels
(Table VI). Risk scores of these
panels were used to stratify the risk of BCR; the scores were
calculated based on the following formula: Risk scores=sum
(coefi x DE-lncRNAi), where
DE-lncRNAi is the ith DE-lncRNA expression
(i=1, … n) and coefi is the Cox coefficient of
DE-lncRNAi (213-216).
PSA relapse offers the early identification of
patients with failure following initial curative therapies with RP
and RT. While BCR precedes clinical disease recurrence, the
management of males with PSA relapse needs to consider multiple
factors including tumor recurrence (234,235). The nature of BCR is heterogeneous
with local and distant recurrence (236). Additionally, not all patients
with BCR will progress to lethal disease (13). In addition to these variations are
the improvements in risk stratification of BCR and metastasis as
well as advances in salvage treatment. The heterogeneity of BCR
along with the aforementioned advances complicates the management
of patients with BCR. This topic has been recently discussed by
several recent reviews (236-238). We also highlight the recent
advances and suggest improvement on management of these patients in
the context of BCR risk stratification using RNA-based
biomarkers.
Recent developments have improved the diagnosis of
clinical recurrence following BCR using the prostate-specific
membrane antigen (PSMA)-based positron emission tomography (PET)
imaging in comparison to conventional imaging modalities: Computed
tomography (CT), magnetic resonance imaging (MRI) and bone scan
(239,240). PMSA (glutamate carboxypeptidase
II) is an enzyme encoded by the folate hydrolase 1 (FOLH1)
gene (https://en.wikipedia.org/wiki/Glutamate_carboxypep-tidase_II)
(241). It is mainly expressed in
the prostate with weaker expressions detected in the brain,
salivary gland and small intestine (242). PSMA expression is markedly
upregulated in PC and the level of overexpression is associated
with PC progression, including castration-resistant prostate
adenocarcinoma (242-245). Nonetheless, its expression is
suppressed in neuroendocrine prostate cancer (NEPC) (246), which will produce false
negativities. False positivity is also a concern (246). Nonetheless, PSMA-PET has higher
sensitivities in detecting recurrent sites at BCR in comparison to
other imaging modalities (247).
In a recent single-arm clinical trial on patients with BCR (n=635)
to assess the accuracy of 68Ga-PSMA-11 PET in detecting
recurrent PCs, the overall detection rate was 75% (475/635) and the
PET-positive rates in different PSA groups were 38% for <0.5
ng/ml, 57% for 0.5−<1.0 ng/ml, 84% for 1.0−<2.0 ng/ml, 86%
for 2.0−<5.0 ng/ml, and 97% for ≥5.0 ng/ml respectively
(248). In a recent diagnostic
study of 100 patients with BCR using 18F-PSMA-1007
PET/CT, the PET-positive rate was 86, 89, 100 and 100% for patients
with PSA levels ≤0.5, 0.51-1.0, 1.0-2.0, and ≥2.0 ng/ml,
respectively (249).
Clinical recurrence in the setting of BCR can also
be at distant sites or metastasis. The diagnosis of metastasis can
be facilitated using the Decipher test (GenomeDx Bioscience), a
22-gene genomic classifier (GC). This is an RNA-based gene panel
consisting of coding and non-coding transcripts that function in
multiple pathways including cell proliferation, adhesion, immune
response, cell cycle progression and others (132). The Decipher GC predicts
metastasis in patients following RP (132-134). In a recent multicenter study on
561 males with adverse pathological features, GC independently
stratified the risk of prostate cancer-specific mortality (PCSM)
following RP (250). The
prediction was improved by combining GS with CAPRA-S (251) a classifier of BCR risk following
PR (21,22). In this regard, it would be expected
that combination of GS with those RNA-based biomarkers discussed
herein may strengthen the accuracy in predicting PCSM in the
setting of RP; this will facilitate management of patients with BCR
with respect to decision making on salvage treatment selection.
Other biomarkers could also be considered. RP
produces excellent outcomes in patients with localized low- and
intermediate-risk PCs. However, the biochemical relapse rates for
high-risk localized disease [PSA>20 ng/ml, GS>7, or cT2c
(3)] can increase to 50-80%
(252). Males with high-risk
tumors can be managed with adjuvant therapy following RP; in a
small group of patients (n=127) treated with adjuvant hormone
therapy, high level of PDL1 expression is an independent risk
factor of BCR (253). The PDL1
expression status could facilitate the diagnosis of BCR following
RP.
Treatment selection for patients with BCR depends
on the site of recurrence and the extent of progression; this
information will be derived using imaging and other assessment
including biomarker-based (such as GS) risk evaluation and PSA
changes (236). Life expectancy,
quality of life (QOL) and the time span of approximately 8 years
for metastatic progression from BCR (7,13)
are among the factors that affect treatment decision making
(237,254).
Salvage radiotherapy (SRT) to the prostate bed is
commonly used in patients with BCR following RP; it controls
biochemical failure in approximately 50% cases, reduces distant
metastasis and improves PCSM (236,255,256). The PSA status can guide local
salvage treatment. EAU-ESTRO-SIOG recommends surveillance and
delayed SRT in males exhibiting an increase in PSA with a favorable
prognostic setting [≤pT3a; time to BCR, >3 years; PSA doubling
time (DT), >12 months; and GS ≤7], and beginning SRT at PSA
<0.5 ng/ml (7). On the other
hand, the National Comprehensive Cancer Network (NCCN) recommends
the initiation of SRT with confirmed increasing PSA levels, and
many favor SRT at PSA 0.2 ng/ml (238). For patients with BCR following
RT, salvage RP is an option with confirmed local recurrence
according to EAU-ESTRO-SIOG guidelines (7). Similarly, the prostate cancer
guidelines from the European Association of Nuclear Medicine
(EAU-EANM)-European Society of Urogenital Radiology
(ESTRO-ESUR)-SIOG classify males with BCR into a low-risk [PSA-DT
>1 year and pathological GS (pGS) <8 or International Society
of Urological Pathology (ISUP) grade <4] and high-risk group
(PSA-DT ≤1 year, pGS 8-10 or ISUP grade 4-5) for biological
recurrence following RP or a low-risk [IBF (interval from primary
therapy to biochemical failure) >18 months and biopsy GC (bGS)
<8 or ISUP grade <4] and high-risk (IBF ≤18 months and)
groups (pGS 8-10 or ISUP grade 4-5) (254). The stratification was recently
validated based on the 5-year risk of developing metastasis and
PCSM in a large cohort of patients with BCR (n=1,040) (257). The guidelines call for the
surveillance for males with BCR in the low-risk group and salvage
ADT should not be given to these patients (254). It appears that SRT plus hormone
therapy (bicalutamide) improved the outcome (258,259). The risk of metastasis following
SRT in patients with BCR can be stratified using Decipher GC
(260). It is thus possible to
assign patients with BCR following RP with combination therapy of
SRT and ADT based on GC scores. Following this logic, whether
incorporating BCR risk stratification with GS will enhance the
decision making warrants further investigations in the future.
BCR precedes clinical disease recurrence and is
significantly associated with increases in metastasis development
and CRPC (13,14,261), conditions to which our knowledge
and ability to intervene remain poor. While more than half of
patients with high-risk PCs will experience BCR following RP
(252), the curative therapy
yields good results in males with low- and intermediate-risk
tumors. Accurately predicting the risk of BCR is thus highly
relevant in the management of these patients. In view of the
metastasis progression following BCR, the stratification of the
risk of BCR also contributes to the management of males with PSA
relapse (please see section above entitled 'Salvage therapies
following BCR'). Collectively, the effective evaluation of the risk
of BCR is an essential aspect of patient management. With this
recognition, a major research focus has been searching for
biomarkers to robustly assess BCR risk, which is evident by 2,502
articles listed under 'prostate cancer' AND 'biomarker' AND
'biochemical recurrence' by PubMed. However, none of these had
succeeded in progressing to routine clinical application (262); this clearly outlines the
challenges in the identification of effective biomarkers.
While individual biomarkers, regardless of whether
they are clinical feature-, DNA-, RNA-, and protein-based, may
display a significant association with BCR, it is unlikely that
they can effectively stratify BCR risk individually. BCR is
regulated by complex mechanisms, which is likely an attribute to
the lack of overlapping genes between two commercially available
multigene panels, Oncotype DX GPS and Prolaris, despite both
assessing the risk of BCR (135,137). It is thus conceivable that
multigene panels will certainly enhance the effectiveness of BCR
biomarkers. In this regard, it will be intriguing to systemically
analyze Oncotype DX, Prolaris and other RNA-based biomarkers along
with clinical feature-based (PSA, GS, stage, surgical margin
status, lymph node status and others) BCR risk classifiers
(CAPRA-S, Walz nomogram, and others) for the stratification of the
risk of BCR. This may produce a much more robust system, covering
essential pathways leading to BCR, in predicting the risk of BCR,
which will greatly improve patient management with prostate
cancer.
Another avenue worthy of exploration for the
improvement of the stratification of the risk of BCR is the process
of DNA damage response (DDR). Genomic instability is a hallmark of
cancer and the driving force of cancer progression (263); genomic stability is maintained
through DDR by coordinating checkpoint activation and DNA lesion
repairs (264-266). It is surprising that factors in
DDR regulation have not been intensively investigated for their
biomarker potential.
The same situation applies to stromal factors.
While a variety of tumor properties have been examined for
prognostic purposes, the stromal contributions and the
communications between he stroma and tumor have not been actively
determined for biomarker purposes. A potential mechanism causing
stromal alterations is through PC-associated metabolic
reprogramming, which results in the accumulation of metabolic
intermediates (267); these
materials affect gene expression via epigenetic alterations
(268). Metabolic reprogramming
is a well-established mechanism supporting not only tumorigenesis,
but also cancer progression (36,267,268). In this regard, PC-associated
metabolic alterations will have a prognostic potential which has
been recently reviewed by Lucarelli et al (267). It is of interest that PCs can be
grouped into two metabolic profiles:
Phopho-AKThigh/MYClow or
phopho-AKTlow/MYChigh with the former and
latter affecting the glucose-related processes and lipid
metabolism, respectively (269).
Nonetheless, the prognostic potential of PC-associated metabolic
alterations remains complex. For instance, the AKT- and MYC-related
metabolic signatures are not associated with GS and pathological
stage (269); of note, neither
MYC overexpression nor AKT phosphorylation displays a strong
prognostic potential in PC (267,270,271). While increases in body mass index
(BMI) and obesity are associated with PC-related mortality
(272), there is also evidence to
support the reverse association (273). A similar situation also applies
to the association between cholesterol and PC progression. A
meta-analysis of 27 clinical studies up to 2012 with a pooled
population of 1.8 million males revealed a 7% reduction in PC cases
and a 20% decrease in PC progression in statin users (274). Statins were reported to reduce
BCR following RT (275) and RP
(276). However, other studies
observed no clinical benefits in males with PC who were statin
users (277,278) and reported statins having no
impact on BCR following RP (279). Clearly, the prognostic values of
metabolic alterations in PC warrant further investigations.
The plasticity of cancer, including PC, presents a
major challenge not only in cancer therapy, but also in assessing
the risk of cancer progression. Cancer plasticity is regulated by
complex mechanisms, including those functioning in CSCs and DDR
(280,281). It is noteworthy that BMI1, a
well-established factor in maintaining CSC (282), also compromises genomic
instability via attenuating ATM and ATR functions (264,283-285). In this regard, DDR regulations
and stroma-cancer cell communications, both of which contribute to
cancer plasticity, should be actively brought into the picture of
BCR risk assessment; with these components incorporated, the
ability to accurately classify BCR risk will likely be
significantly improved.
PC is associated with high levels of intratumoral
and intertumoral heterogeneity (286). This aspect has not been given
sufficient consideration and should be pursued in PC biomarker
development. Collaborative efforts involving multiple institutes in
sharing materials and expertise will certainly be helpful to
achieve this goal.
Not applicable.
DT was supported by grants from the Cancer Research
Society, Canadian Cancer Society (grant no. 319412), CIHR and by
funds from the Urological Cancer Center for Research and Innovation
(UCCRI). PM was supported by CIHR. YG was supported by Studentship
provided by Ontario Graduate Scholarships and Research Institute of
St. Joe's Hamilton.
Not applicable.
XL, AK, HX, PM and DT were involved in the
conception of the study. XL, YG and MJC were involved in the
literature search. XL, PM and DT were involved in the writing and
preparation of the original draft of the manuscript. All authors
were involved in the writing and reviewing of the article. YG, PM
and DT were involved in the writing and editing of the article. DT
supervised the study. All authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heidenreich A, Bastian PJ, Bellmunt J,
Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T,
Zattoni F, et al: EAU guidelines on prostate cancer. Part II:
Treatment of advanced, relapsing, and castration-resistant prostate
cancer. Eur Urol. 65:467–479. 2014. View Article : Google Scholar
|
|
3
|
Mottet N, Bellmunt J, Bolla M, Briers E,
Cumberbatch MG, De Santis M, Fossati N, Gross T, Henry AM, Joniau
S, et al: EAU-ESTRO-SIOG Guidelines on prostate cancer. Part 1:
Screening, diagnosis, and local treatment with curative intent. Eur
Urol. 71:618–629. 2017. View Article : Google Scholar
|
|
4
|
Bill-Axelson A, Holmberg L, Garmo H, Rider
JR, Taari K, Busch C, Nordling S, Häggman M, Andersson SO,
Spångberg A, et al: Radical prostatectomy or watchful waiting in
early prostate cancer. N Engl J Med. 370:932–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayes JH, Ollendorf DA, Pearson SD, Barry
MJ, Kantoff PW, Lee PA and McMahon PM: Observation versus initial
treatment for men with localized, low-risk prostate cancer: A
cost-effectiveness analysis. Ann Intern Med. 158:853–860. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Godtman RA, Holmberg E, Khatami A, Stranne
J and Hugosson J: Outcome following active surveillance of men with
screen-detected prostate cancer. Results from the Göteborg
randomised population-based prostate cancer screening trial. Eur
Urol. 63:101–107. 2013. View Article : Google Scholar
|
|
7
|
Cornford P, Bellmunt J, Bolla M, Briers E,
De Santis M, Gross T, Henry AM, Joniau S, Lam TB, Mason MD, et al:
EAU-ESTRO-SIOG Guidelines on prostate cancer. Part II: Treatment of
relapsing, metastatic, and castration-resistant prostate cancer.
Eur Urol. 71:630–642. 2017. View Article : Google Scholar
|
|
8
|
Zaorsky NG, Raj GV, Trabulsi EJ, Lin J and
Den RB: The dilemma of a rising prostate-specific antigen level
after local therapy: What are our options? Semin Oncol. 40:322–336.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roehl KA, Han M, Ramos CG, Antenor JA and
Catalona WJ: Cancer progression and survival rates following
anatomical radical retropubic prostatectomy in 3,478 consecutive
patients: Long-term results. J Urol. 172:910–914. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Freedland SJ, Humphreys EB, Mangold LA,
Eisenberger M, Dorey FJ, Walsh PC and Partin AW: Risk of prostate
cancer-specific mortality following biochemical recurrence after
radical prostatectomy. JAMA. 294:433–439. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kupelian PA, Mahadevan A, Reddy CA,
Reuther AM and Klein EA: Use of different definitions of
biochemical failure after external beam radiotherapy changes
conclusions about relative treatment efficacy for localized
prostate cancer. Urology. 68:593–598. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Artibani W, Porcaro AB, De Marco V,
Cerruto MA and Siracusano S: Management of biochemical recurrence
after primary curative treatment for prostate cancer: A review.
Urol Int. 100:251–262. 2018. View Article : Google Scholar
|
|
13
|
Pound CR, Partin AW, Eisenberger MA, Chan
DW, Pearson JD and Walsh PC: Natural history of progression after
PSA elevation following radical prostatectomy. JAMA. 281:1591–1597.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boorjian SA, Thompson RH, Tollefson MK,
Rangel LJ, Bergstralh EJ, Blute ML and Karnes RJ: Long-term risk of
clinical progression after biochemical recurrence following radical
prostatectomy: The impact of time from surgery to recurrence. Eur
Urol. 59:893–899. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heinlein CA and Chang C: Androgen receptor
in prostate cancer. Endoc Rev. 25:276–308. 2004. View Article : Google Scholar
|
|
16
|
Tannock IF, de Wit R, Berry WR, Horti J,
Pluzanska A, Chi KN, Oudard S, Théodore C, James ND, Turesson I, et
al: Docetaxel plus prednisone or mitoxantrone plus prednisone for
advanced prostate cancer. N Engl J Med. 351:1502–1512. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Berthold DR, Pond GR, Soban F, de Wit R,
Eisenberger M and Tannock IF: Docetaxel plus prednisone or
mitoxantrone plus prednisone for advanced prostate cancer: Updated
survival in the TAX 327 study. J Clin Oncol. 26:242–245. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
de Bono JS, Logothetis CJ, Molina A,
Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB Jr, Saad F,
et al: Abiraterone and increased survival in metastatic prostate
cancer. N Engl J Med. 364:1995–2005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Scher HI, Fizazi K, Saad F, Taplin ME,
Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, et
al: Increased survival with enzalutamide in prostate cancer after
chemotherapy. N Engl J Med. 367:1187–1197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cooperberg MR, Pasta DJ, Elkin EP, Litwin
MS, Latini DM, Du Chane J and Carroll PR: The University of
California, San Francisco Cancer of the Prostate Risk Assessment
score: A straightforward and reliable preoperative predictor of
disease recurrence after radical prostatectomy. J Urol.
173:1938–1942. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brajtbord JS, Leapman MS and Cooperberg
MR: The CAPRA Score at 10 Years: Contemporary perspectives and
analysis of supporting studies. Eur Urol. 71:705–709. 2017.
View Article : Google Scholar
|
|
22
|
Cooperberg MR, Hilton JF and Carroll PR:
The CAPRA-S score: A straightforward tool for improved prediction
of outcomes after radical prostatectomy. Cancer. 117:5039–5046.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
D'Amico AV, Whittington R, Malkowicz SB,
Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA,
Kaplan I, Beard CJ and Wein A: Biochemical outcome after radical
prosta-tectomy, external beam radiation therapy, or interstitial
radiation therapy for clinically localized prostate cancer. JAMA.
280:969–974. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dillioglugil O, Leibman BD, Kattan MW,
Seale-Hawkins C, Wheeler TM and Scardino PT: Hazard rates for
progression after radical prostatectomy for clinically localized
prostate cancer. Urology. 50:93–99. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han M, Partin AW, Zahurak M, Piantadosi S,
Epstein JI and Walsh PC: Biochemical (prostate specific antigen)
recurrence probability following radical prostatectomy for
clinically localized prostate cancer. J Urol. 169:517–523. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Simmons MN, Stephenson AJ and Klein EA:
Natural history of biochemical recurrence after radical
prostatectomy: Risk assessment for secondary therapy. Eur Urol.
51:1175–1184. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lange PH, Ercole CJ, Lightner DJ, Fraley
EE and Vessella R: The value of serum prostate specific antigen
determinations before and after radical prostatectomy. J Urol.
141:873–879. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim DK, Koo KC, Lee KS, Hah YS, Rha KH,
Hong SJ and Chung BH: Time to disease recurrence is a predictor of
metastasis and mortality in patients with High-risk prostate cancer
who achieved undetectable prostate-specific antigen following
Robot-assisted radical prostatectomy. J Korean Med Sci.
33:e2852018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Walz J, Chun FK, Klein EA, Reuther A, Saad
F, Graefen M, Huland H and Karakiewicz PI: Nomogram predicting the
probability of early recurrence after radical prostatectomy for
prostate cancer. J Urol. 181:601–608. 2009. View Article : Google Scholar
|
|
30
|
Pompe RS, Bandini M, Preisser F, Marchioni
M, Zaffuto E, Tian Z, Salomon G, Schlomm T, Huland H, Graefen M, et
al: Contemporary approach to predict early biochemical recurrence
after radical prostatectomy: Update of the Walz nomogram. Prostate
Cancer Prostatic Dis. 21:386–393. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ross AE, D'Amico AV and Freedland SJ:
Which, when and why? Rational use of tissue-based molecular testing
in local-ized prostate cancer. Prostate Cancer Prostatic Dis.
19:1–6. 2016. View Article : Google Scholar
|
|
33
|
Carneiro A, Barbosa ARG, Takemura LS,
Kayano PP, Moran NKS, Chen CK, Wroclawski ML, Lemos GC, da Cunha
IW, Obara MT, et al: The role of immunohistochemical analysis as a
tool for the diagnosis, prognostic evaluation and treatment of
prostate cancer: A systematic review of the literature. Front
Oncol. 8:3772018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iatropoulos MJ and Williams GM:
Proliferation markers. Exp Toxicol Pathol. 48:175–181. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fantony JJ, Howard LE, Csizmadi I,
Armstrong AJ, Lark AL, Galet C, Aronson WJ and Freedland SJ: Is
Ki67 prognostic for aggressive prostate cancer? A multicenter
real-world study. Biomark Med. 12:727–736. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wong N, Ojo D, Yan J and Tang D: PKM2
contributes to cancer metabolism. Cancer Lett. 356:184–191. 2015.
View Article : Google Scholar
|
|
37
|
Carabet LA, Rennie PS and Cherkasov A:
Therapeutic inhibition of myc in cancer. Structural bases and
computer-aided drug discovery approaches. Int J Mol Sci. 20:pii:
E120. 2018. View Article : Google Scholar
|
|
38
|
Gurel B, Iwata T, Koh CM, Jenkins RB, Lan
F, Van Dang C, Hicks JL, Morgan J, Cornish TC, Sutcliffe S, et al:
Nuclear MYC protein overexpression is an early alteration in human
prostate carcinogenesis. Mod Pathol. 21:1156–1167. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Baena-Del Valle JA, Zheng Q, Esopi DM,
Rubenstein M, Hubbard GK, Moncaliano MC, Hruszkewycz A, Vaghasia A,
Yegnasubramanian S, Wheelan SJ, et al: MYC drives overex-pression
of telomerase RNA (hTR/TERC) in prostate cancer. J Pathol.
244:11–24. 2018. View Article : Google Scholar
|
|
40
|
Hubbard GK, Mutton LN, Khalili M, McMullin
RP, Hicks JL, Bianchi-Frias D, Horn LA, Kulac I, Moubarek MS,
Nelson PS, et al: Combined MYC activation and pten loss are
sufficient to create genomic instability and lethal metastatic
prostate cancer. Cancer Res. 76:283–292. 2016. View Article : Google Scholar :
|
|
41
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park K, Tomlins SA, Mudaliar KM, Chiu YL,
Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald
T, et al: Antibody-based detection of ERG rearrangement-positive
prostate cancer. Neoplasia. 12:590–598. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Inoue T, Segawa T, Shiraishi T, Yoshida T,
Toda Y, Yamada T, Kinukawa N, Kinoshita H, Kamoto T and Ogawa O:
Androgen receptor, Ki67, and p53 expression in radical
prostatectomy specimens predict treatment failure in Japanese
population. Urology. 66:332–337. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Osman I, Drobnjak M, Fazzari M, Ferrara J,
Scher HI and Cordon-Cardo C: Inactivation of the p53 pathway in
prostate cancer: Impact on tumor progression. Clin Cancer.
5:2082–2088. 1999.
|
|
45
|
Hemminki K: Familial risk and familial
survival in prostate cancer. World J Urol. 30:143–148. 2012.
View Article : Google Scholar
|
|
46
|
Castro E, Goh C, Olmos D, Saunders E,
Leongamornlert D, Tymrakiewicz M, Mahmud N, Dadaev T, Govindasami
K, Guy M, et al: Germline BRCA mutations are associated with higher
risk of nodal involvement, distant metastasis, and poor survival
outcomes in prostate cancer. J Clin Oncol. 31:1748–1757. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Castro E, Goh C, Leongamornlert D,
Saunders E, Tymrakiewicz M, Dadaev T, Govindasami K, Guy M, Ellis
S, Frost D, et al: Effect of BRCA mutations on metastatic relapse
and Cause-specific survival after radical treatment for localised
prostate cancer. Eur Urol. 68:186–193. 2015. View Article : Google Scholar
|
|
48
|
Taylor RA, Fraser M, Livingstone J,
Espiritu SM, Thorne H, Huang V, Lo W, Shiah YJ, Yamaguchi TN,
Sliwinski A, et al: Germline BRCA2 mutations drive prostate cancers
with distinct evolutionary trajectories. Nat Commun. 8:136712017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pritchard CC, Mateo J, Walsh MF, De Sarkar
N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R,
et al: Inherited DNA-repair gene mutations in men with metastatic
prostate cancer. N Engl J Med. 375:443–453. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cooperberg MR, Erho N, Chan JM, Feng FY,
Fishbane N, Zhao SG, Simko JP, Cowan JE, Lehrer J, Alshalalfa M, et
al: The diverse genomic landscape of clinically Low-risk prostate
cancer. Eur Urol. 74:444–452. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Evans JR, Zhao SG, Chang SL, Tomlins SA,
Erho N, Sboner A, Schiewer MJ, Spratt DE, Kothari V, Klein EA, et
al: Patient-level DNA damage and repair pathway profiles and
prognosis after prostatectomy for high-risk prostate cancer. JAMA
Oncol. 2:471–480. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Magi-Galluzzi C, Tsusuki T, Elson P,
Simmerman K, LaFargue C, Esgueva R, Klein E, Rubin MA and Zhou M:
TMPRSS2-ERG gene fusion prevalence and class are significantly
different in prostate cancer of Caucasian, African-American and
Japanese patients. Prostate. 71:489–497. 2011. View Article : Google Scholar
|
|
54
|
Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X,
Cui Z, Zhang J, Yi K, Xu W, et al: RNA-seq analysis of prostate
cancer in the Chinese population identifies recurrent gene fusions,
cancer-associated long noncoding RNAs and aberrant alternative
splicings. Cell Res. 22:806–821. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Song C and Chen H: Predictive significance
of TMRPSS2-ERG fusion in prostate cancer: A meta-analysis. Cancer
Cell Int. 18:1772018. View Article : Google Scholar :
|
|
56
|
Shamseer L, Moher D, Clarke M, Ghersi D,
Liberati A, Petticrew M, Shekelle P and Stewart LA; PRISMA-P Group:
Preferred reporting items for systematic review and meta-analysis
protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ.
350:g76472015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Moher D, Shamseer L, Clarke M, Ghersi D,
Liberati A, Petticrew M, Shekelle P and Stewart LA; PRISMA-P Group:
Preferred reporting items for systematic review and meta-analysis
protocols (PRISMA-P) 2015 statement. Syst Rev. 4:12015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nam RK, Benatar T, Wallis CJ, Amemiya Y,
Yang W, Garbens A, Naeim M, Sherman C, Sugar L and Seth A: MiR-301a
regulates E-cadherin expression and is predictive of prostate
cancer recurrence. Prostate. 76:869–884. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li T, Li RS, Li YH, Zhong S, Chen YY,
Zhang CM, Hu MM and Shen ZJ: miR-21 as an independent biochemical
recurrence predictor and potential therapeutic target for prostate
cancer. J Urol. 187:1466–1472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sun X, Yang Z, Zhang Y, He J, Wang F, Su
P, Han J, Song Z and Fei Y: Prognostic implications of tissue and
serum levels of microRNA-128 in human prostate cancer. Int J Clin
Exp Pathol. 8:8394–8401. 2015.PubMed/NCBI
|
|
61
|
Aakula A, Kohonen P, Leivonen SK, Mäkelä
R, Hintsanen P, Mpindi JP, Martens-Uzunova E, Aittokallio T,
Jenster G, Perälä M, et al: Systematic identification of MicroRNAs
that impact on proliferation of prostate cancer cells and display
changed expression in tumor tissue. Eur Urol. 69:1120–1128. 2016.
View Article : Google Scholar
|
|
62
|
Ling XH, Han ZD, Xia D, He HC, Jiang FN,
Lin ZY, Fu X, Deng YH, Dai QS, Cai C, et al: MicroRNA-30c serves as
an independent biochemical recurrence predictor and potential tumor
suppressor for prostate cancer. Mol Biol Rep. 41:2779–2788. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Avgeris M, Stravodimos K, Fragoulis EG and
Scorilas A: The loss of the tumour-suppressor miR-145 results in
the shorter disease-free survival of prostate cancer patients. Br J
Cancer. 108:2573–2581. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Tao Z, Xu S, Ruan H, Wang T, Song W, Qian
L and Chen K: MiR-195/-16 family enhances radiotherapy via T cell
activation in the tumor microenvironment by blocking the PD-L1
immune checkpoint. Cell Physiol Biochem. 48:801–814. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mei W, Lin X, Kapoor A, Gu Y, Zhao K and
Tang D: The contributions of prostate cancer stem cells in prostate
cancer initiation and metastasis. Cancers (Basel). 11:pii: E434.
2019. View Article : Google Scholar
|
|
67
|
Yang Y, Guo JX and Shao ZQ: miR-21 targets
and inhibits tumor suppressor gene PTEN to promote prostate cancer
cell proliferation and invasion: An experimental study. Asian Pac J
Trop Med. 10:87–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhao Y, Yin Z, Li H, Fan J, Yang S, Chen C
and Wang DW: MiR-30c protects diabetic nephropathy by suppressing
epithelial-to-mesenchymal transition in db/db mice. Aging Cell.
16:387–400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhu C, Hou X, Zhu J, Jiang C and Wei W:
Expression of miR-30c and miR-29b in prostate cancer and its
diagnostic significance. Oncol Lett. 16:3140–3144. 2018.PubMed/NCBI
|
|
70
|
Sachdeva M, Liu Q, Cao J, Lu Z and Mo YY:
Negative regulation of miR-145 by C/EBP-β through the Akt pathway
in cancer cells. Nucleic Acids Res. 40:6683–6692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ozen M, Creighton CJ, Ozdemir M and
Ittmann M: Widespread deregulation of microRNA expression in human
prostate cancer. Oncogene. 27:1788–1793. 2008. View Article : Google Scholar
|
|
72
|
Wach S, Nolte E, Szczyrba J, Stöhr R,
Hartmann A, Ørntoft T, Dyrskjøt L, Eltze E, Wieland W and Keck B:
MicroRNA profiles of prostate carcinoma detected by multiplatform
microRNA screening. Int J Cancer. 130:611–621. 2012. View Article : Google Scholar
|
|
73
|
Liu B, Li J and Cairns MJ: Identifying
miRNAs, targets and functions. Brief Bioinform. 15:1–19. 2014.
View Article : Google Scholar :
|
|
74
|
Chen CS, Huang CY, Huang SP, Lin VC, Yu
CC, Chang TY and Bao BY: Genetic interaction analysis of TCF7L2 for
biochemical recurrence after radical prostatectomy in localized
prostate cancer. Int J Med Sci. 12:243–247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Malhotra S, Lapointe J, Salari K, Higgins
JP, Ferrari M, Montgomery K, van de Rijn M, Brooks JD and Pollack
JR: A tri-marker proliferation index predicts biochemical
recurrence after surgery for prostate cancer. PLoS One. 6. pp.
e202932011, View Article : Google Scholar
|
|
76
|
Mo RJ, Han ZD, Liang YK, Ye JH, Wu SL, Lin
SX, Zhang YQ, Song SD, Jiang FN, Zhong WD and Wu CL: Expression of
PD-L1 in tumor-associated nerves correlates with reduced
CD8+ tumor-associated lymphocytes and poor prognosis in
prostate cancer. Int J Cancer. 144:3099–3110. 2019. View Article : Google Scholar
|
|
77
|
Gevensleben H, Dietrich D, Golletz C,
Steiner S, Jung M, Thiesler T, Majores M, Stein J, Uhl B, Müller S,
et al: The immune checkpoint regulator PD-L1 is highly expressed in
aggressive primary prostate cancer. Clin Cancer Res. 22:1969–1977.
2016. View Article : Google Scholar
|
|
78
|
Luo JH, Yu YP, Cieply K, Lin F, Deflavia
P, Dhir R, Finkelstein S, Michalopoulos G and Becich M: Gene
expression analysis of prostate cancers. Mol Carcinog. 33:25–35.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sandsmark E, Andersen MK, Bofin AM,
Bertilsson H, Drabløs F, Bathen TF, Rye MB and Tessem MB: SFRP4
gene expression is increased in aggressive prostate cancer. Sci
Rep. 7:142762017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Mortensen MM, Høyer S, Lynnerup AS,
Ørntoft TF, Sørensen KD, Borre M and Dyrskjøt L: Expression
profiling of prostate cancer tissue delineates genes associated
with recurrence after prostatectomy. Sci Rep. 5:160182015.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Mazzoni SM and Fearon ER: AXIN1 and AXIN2
variants in gastrointestinal cancers. Cancer Lett. 355:1–8. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li J, Hu SB, Wang LY, Zhang X, Zhou X,
Yang B, Li JH, Xiong J, Liu N, Li Y, et al: Autophagy-dependent
generation of Axin2+ cancer stem-like cells promotes
hepatocarcinogenesis in liver cirrhosis. Oncogene. 36:6725–6737.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Martins-Neves SR, Corver WE,
Paiva-Oliveira DI, van den Akker BE, Briaire-de-Bruijn IH, Bovée
JV, Gomes CM and Cleton-Jansen AM: Osteosarcoma stem cells have
active Wnt/β-catenin and overexpress SOX2 and KLF4. J Cell Physiol.
231:876–886. 2016. View Article : Google Scholar
|
|
84
|
Lim X, Tan SH, Yu KL, Lim SB and Nusse R:
Axin2 marks quiescent hair follicle bulge stem cells that are
maintained by autocrine Wnt/β-catenin signaling. Proc Natl Acad Sci
USA. 113:E1498–E1505. 2016. View Article : Google Scholar
|
|
85
|
Ma C, Liu C, Huang P, Kaku H, Chen J, Guo
K, Ueki H, Sakai A, Nasu Y, Kumon H, et al: Significant association
between the Axin2 rs2240308 single nucleotide polymorphism and the
incidence of prostate cancer. Oncol Lett. 8:789–794. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hu BR, Fairey AS, Madhav A, Yang D, Li M,
Groshen S, Stephens C, Kim PH, Virk N, Wang L, et al: AXIN2
expression predicts prostate cancer recurrence and regulates
invasion and tumor growth. Prostate. 76:597–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Nordby Y, Richardsen E, Rakaee M, Ness N,
Donnem T, Patel HR, Busund LT, Bremnes RM and Andersen S: High
expression of PDGFR-β in prostate cancer stroma is independently
associated with clinical and biochemical prostate cancer
recurrence. Sci Rep. 7:433782017. View Article : Google Scholar
|
|
88
|
Paulsson J, Ehnman M and Östman A: PDGF
receptors in tumor biology: Prognostic and predictive potential.
Future Oncol. 10:1695–1708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Demidenko R, Daniunaite K, Bakavicius A,
Sabaliauskaite R, Skeberdyte A, Petroska D, Laurinavicius A,
Jankevicius F, Lazutka JR and Jarmalaite S: Decreased expression of
MT1E is a potential biomarker of prostate cancer progression.
Oncotarget. 8:61709–61718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Si M and Lang J: The roles of
metallothioneins in carcinogenesis. J Hematol Oncol. 11:1072018.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tse KY, Liu VW, Chan DW, Chiu PM, Tam KF,
Chan KK, Liao XY, Cheung AN and Ngan HY: Epigenetic alteration of
the metallothionein 1E gene in human endometrial carcinomas. Tumour
Biol. 30:93–99. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Subrungruanga I, Thawornkunob C,
Chawalitchewinkoon-Petmitrc P, Pairojkul C, Wongkham S and Petmitrb
S: Gene expression profiling of intrahepatic cholangiocarcinoma.
Asian Pac J Cancer Prev. 14:557–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Faller WJ, Rafferty M, Hegarty S, Gremel
G, Ryan D, Fraga MF, Esteller M, Dervan PA and Gallagher WM:
Metallothionein 1E is methylated in malignant melanoma and
increases sensitivity to cisplatin-induced apoptosis. Melanoma Res.
20:392–400. 2010.PubMed/NCBI
|
|
94
|
Wer ynska B, Pula B, Muszczynska-Bernhard
B, Gomulkiewicz A, Piotrowska A, Prus R, Podhorska-Okolow M,
Jankowska R and Dziegiel P: Metallothionein 1F and 2A
over-expression predicts poor outcome of non-small cell lung cancer
patients. Exp Mol Pathol. 94:301–308. 2013. View Article : Google Scholar
|
|
95
|
Ferrario C, Lavagni P, Gariboldi M,
Miranda C, Losa M, Cleris L, Formelli F, Pilotti S, Pierotti MA and
Greco A: Metallothionein 1G acts as an oncosupressor in papillary
thyroid carcinoma. Lab Invest. 88:474–481. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Takahashi M, Rhodes DR, Furge KA, Kanayama
H, Kagawa S, Haab BB and Teh BT: Gene expression profiling of clear
cell renal cell carcinoma: Gene identification and prognostic
classification. Proc Natl Acad Sci USA. 98:9754–9759. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Jin R, Bay BH, Chow VT, Tan PH and Lin VC:
Metallothionein 1E mRNA is highly expressed in oestrogen
receptor-negative human invasive ductal breast cancer. Br J Cancer.
83:319–323. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hur H, Ryu HH, Li CH, Kim IY, Jang WY and
Jung S: Metallothinein 1E enhances glioma invasion through
modulation matrix metalloproteinases-2 and 9 in U87MG mouse brain
tumor model. J Korean Neurosurg Soc. 59:551–558. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ryu HH, Jung S, Jung TY, Moon KS, Kim IY,
Jeong YI, Jin SG, Pei J, Wen M and Jang WY: Role of metallothionein
1E in the migration and invasion of human glioma cell lines. Int J
Oncol. 41:1305–1313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Mavridis K, Stravodimos K and Scorilas A:
Quantified KLK15 gene expression levels discriminate prostate
cancer from benign tumors and constitute a novel independent
predictor of disease progression. Prostate. 73:1191–1201. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Obiezu CV and Diamandis EP: Human tissue
kallikrein gene family: Applications in cancer. Cancer Lett.
224:1–22. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Tse BWC, Volpert M, Ratther E, Stylianou
N, Nouri M, McGowan K, Lehman ML, McPherson SJ, Roshan-Moniri M,
Butler MS, et al: Neuropilin-1 is upregulated in the adaptive
response of prostate tumors to androgen-targeted therapies and is
prognostic of metastatic progression and patient mortality.
Oncogene. 36:3417–3427. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Muhl L, Folestad EB, Gladh H, Wang Y,
Moessinger C, Jakobsson L and Eriksson U: Neuropilin 1 binds PDGF-D
and is a co-receptor in PDGF-D-PDGFRβ signaling. J Cell Sci.
130:1365–1378. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang L, Wang H, Li C, Zhao Y, Wu L, Du X
and Han Z: VEGF-A/Neuropilin 1 pathway confers cancer stemness via
activating Wnt/β-catenin axis in breast cancer cells. Cell Physiol
Biochem. 44:1251–1262. 2017. View Article : Google Scholar
|
|
105
|
Latil A, Bièche I, Pesche S, Valéri A,
Fournier G, Cussenot O and Lidereau R: VEGF overexpression in
clinically localized prostate tumors and neuropilin-1
overexpression in metastatic forms. Int J Cancer. 89:167–171. 2000.
View Article : Google Scholar
|
|
106
|
Talagas M, Uguen A, Garlantezec R,
Fournier G, Doucet L, Gobin E, Marcorelles P, Volant A and DE
Braekeleer M: VEGFR1 and NRP1 endothelial expressions predict
distant relapse after radical prostatectomy in clinically localized
prostate cancer. Anticancer Res. 33:2065–2075. 2013.PubMed/NCBI
|
|
107
|
Li F, Xu Y and Liu RL: SAMD5 mRNA was
overexpressed in prostate cancer and can predict biochemical
recurrence after radical prostatectomy. Int Urol Nephrol.
51:443–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Matsuo T, Dat le T, Komatsu M, Yoshimaru
T, Daizumoto K, Sone S, Nishioka Y and Katagiri T: Early growth
response 4 is involved in cell proliferation of small cell lung
cancer through transcriptional activation of its downstream genes.
PLoS One. 9:e1136062014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yagai T, Matsui S, Harada K, Inagaki FF,
Saijou E, Miura Y, Nakanuma Y, Miyajima A and Tanaka M: Expression
and localization of sterile alpha motif domain containing 5 is
associated with cell type and malignancy of biliary tree. PLoS One.
12:e01753552017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Watanabe T, Kobunai T, Akiyoshi T, Matsuda
K, Ishihara S and Nozawa K: Prediction of response to preoperative
chemo-radiotherapy in rectal cancer by using reverse transcriptase
polymerase chain reaction analysis of four genes. Dis Colon Rectum.
57:23–31. 2014. View Article : Google Scholar
|
|
111
|
Wang Y, Shang Y, Li J, Chen W, Li G, Wan
J, Liu W and Zhang M: Specific Eph receptor-cytoplasmic effector
signaling mediated by SAM-SAM domain interactions. Elife. 7:pii:
e35677. 2018.
|
|
112
|
Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang
J, Perry SR, Labrot ES, Wu X, Lis R, et al: SMAD4-dependent barrier
constrains prostate cancer growth and metastatic progression.
Nature. 470:269–273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhang DT, Shi JG, Liu Y and Jiang HM: The
prognostic value of Smad4 mRNA in patients with prostate cancer.
Tumour Biol. 35:3333–3337. 2014. View Article : Google Scholar
|
|
114
|
Guo J, Wang M, Wang Z and Liu X:
Overexpression of pleomor-phic adenoma gene-like 2 is a novel poor
prognostic marker of prostate cancer. PLoS One. 11:e01586672016.
View Article : Google Scholar
|
|
115
|
Li N, Li D, Du Y, Su C, Yang C, Lin C, Li
X and Hu G: Overexpressed PLAGL2 transcriptionally activates Wnt6
and promotes cancer development in colorectal cancer. Oncol Rep.
41:875–884. 2019.
|
|
116
|
Zheng H, Ying H, Wiedemeyer R, Yan H,
Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al:
PLAGL2 regulates Wnt signaling to impede differentiation in neural
stem cells and gliomas. Cancer Cell. 17:497–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Landrette SF, Kuo YH, Hensen K, Barjesteh
van Waalwijk van Doorn-Khosrovani S, Perrat PN, Van de Ven WJ,
Delwel R and Castilla LH: Plag1 and Plagl2 are oncogenes that
induce acute myeloid leukemia in cooperation with Cbfb-MYH11.
Blood. 105:2900–2907. 2005. View Article : Google Scholar
|
|
118
|
Landrette SF, Madera D, He F and Castilla
LH: The transcription factor PlagL2 activates Mpl transcription and
signaling in hematopoietic progenitor and leukemia cells. Leukemia.
25:655–662. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhao SG, Lehrer J, Chang SL, Das R, Erho
N, Liu Y, Sjöström M, Den RB, Freedland SJ, Klein EA, et al: The
immune landscape of prostate cancer and nomination of PD-L2 as a
potential therapeutic target. J Natl Cancer Inst. 111:301–310.
2019. View Article : Google Scholar
|
|
120
|
Kladi-Skandali A, Mavridis K, Scorilas A
and Sideris DC: Expressional profiling and clinical relevance of
RNase kappa in prostate cancer: A novel indicator of favorable
progression-free survival. J Cancer Res Clin Oncol. 144:2049–2057.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Gkratsou AS, Fragoulis EG and Sideris DC:
Effect of cytostatic drugs on the mRNA expression levels of
ribonuclease κ in breast and ovarian cancer cells. Anticancer
Agents Med Chem. 14:400–408. 2014. View Article : Google Scholar
|
|
122
|
Ma X, Du T, Zhu D, Chen X, Lai Y, Wu W,
Wang Q, Lin C, Li Z, Liu L and Huang H: High levels of glioma tumor
suppressor candidate region gene 1 predicts a poor prognosis for
prostate cancer. Oncol Lett. 16:6749–6755. 2018.PubMed/NCBI
|
|
123
|
Yang P, Kollmeyer TM, Buckner K, Bamlet W,
Ballman KV and Jenkins RB: Polymorphisms in GLTSCR1 and ERCC2 are
associated with the development of oligodendrogliomas. Cancer.
103:2363–2372. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Alpsoy A and Dykhuizen EC: Glioma tumor
suppressor candidate region gene 1 (GLTSCR1) and its paralog
GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol
Chem. 293:3892–3903. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Gu Y, Chow MJ, Kapoor A, Mei W, Jiang Y,
Yan J, De Melo J, Seliman M, Yang H, Cutz JC, et al: Biphasic
alteration of butyrylcholinesterase (BChE) during prostate cancer
development. Transl Oncol. 11:1012–1022. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Chatonnet A and Lockridge O: Comparison of
butyrylcholines-terase and acetylcholinesterase. Biochem J.
260:625–634. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Evans FT, Gray PW, Lehmann H and Silk E:
Sensitivity to succinylcholine in relation to serum-cholinesterase.
Lancet. 1:1229–1230. 1952. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
De Vriese C, Gregoire F, Lema-Kisoka R,
Waelbroeck M, Robberecht P and Delporte C: Ghrelin degradation by
serum and tissue homogenates: Identification of the cleavage sites.
Endocrinology. 145:4997–5005. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Brimijoin S, Gao Y, Geng L and Chen VP:
Treating cocaine addiction, obesity, and emotional disorders by
viral gene transfer of butyrylcholinesterase. Front Pharmacol.
9:1122018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Schopfer LM, Lockridge O and Brimijoin S:
Pure human butyrylcholinesterase hydrolyzes octanoyl ghrelin to
desacyl ghrelin. Gen Comp Endocrinol. 224:61–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Brimijoin S, Chen VP, Pang YP, Geng L and
Gao Y: Physiological roles for butyrylcholinesterase: A
BChE-ghrelin axis. Chem Biol Interact. 259:271–275. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Erho N, Crisan A, Vergara IA, Mitra AP,
Ghadessi M, Buerki C, Bergstralh EJ, Kollmeyer T, Fink S, Haddad Z,
et al: Discovery and validation of a prostate cancer genomic
classifier that predicts early metastasis following radical
prostatectomy. PLoS One. 8:e668552013. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Karnes RJ, Bergstralh EJ, Davicioni E,
Ghadessi M, Buerki C, Mitra AP, Crisan A, Erho N, Vergara IA, Lam
LL, et al: Validation of a genomic classifier that predicts
metastasis following radical prostatectomy in an at risk patient
population. J Urol. 190:2047–2053. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Klein EA, Haddad Z, Yousefi K, Lam LL,
Wang Q, Choeurng V, Palmer-Aronsten B, Buerki C, Davicioni E, Li J,
et al: Decipher genomic classifier measured on prostate biopsy
predicts metastasis risk. Urology. 90:148–152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Knezevic D, Goddard AD, Natraj N,
Cherbavaz DB, Clark-Langone KM, Snable J, Watson D, Falzarano SM,
Magi-Galluzzi C, Klein EA and Quale C: Analytical validation of the
Oncotype DX prostate cancer assay-a clinical RT-PCR assay optimized
for prostate needle biopsies. BMC Genomics. 14:6902013. View Article : Google Scholar
|
|
136
|
Klein EA, Cooperberg MR, Magi-Galluzzi C,
Simko JP, Falzarano SM, Maddala T, Chan JM, Li J, Cowan JE, Tsiatis
AC, et al: A 17-gene assay to predict prostate cancer
aggressiveness in the context of Gleason grade heterogeneity, tumor
multifocality, and biopsy undersampling. Eur Urol. 66:550–560.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Cuzick J, Swanson GP, Fisher G, Brothman
AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E,
Foster CS, et al: Prognostic value of an RNA expression signature
derived from cell cycle proliferation genes in patients with
prostate cancer: A retrospective study. Lancet Oncol. 12:245–255.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Oderda M, Cozzi G, Daniele L, Sapino A,
Munegato S, Renne G, De Cobelli O and Gontero P: Cell-cycle
Progression-score might improve the current risk assessment in
newly diagnosed prostate cancer patients. Urology. 102:73–78. 2017.
View Article : Google Scholar
|
|
139
|
Albala D, Kemeter MJ, Febbo PG, Lu R, John
V, Stoy D, Denes B, McCall M, Shindel AW and Dubeck F: Health
economic impact and prospective clinical utility of oncotype DX®
genomic prostate score. Rev Urol. 18:123–132. 2016.
|
|
140
|
Cullen J, Rosner IL, Brand TC, Zhang N,
Tsiatis AC, Moncur J, Ali A, Chen Y, Knezevic D, Maddala T, et al:
A Biopsy-based 17-gene genomic prostate score predicts recurrence
after radical prostatectomy and adverse surgical pathology in a
racially diverse population of men with clinically low- and
intermediate-risk prostate cancer. Eur Urol. 68:123–131. 2015.
View Article : Google Scholar
|
|
141
|
Cooperberg MR, Simko JP, Cowan JE, Reid
JE, Djalilvand A, Bhatnagar S, Gutin A, Lanchbury JS, Swanson GP,
Stone S and Carroll PR: Validation of a cell-cycle progression gene
panel to improve risk stratification in a contemporary
prostatectomy cohort. J Clin Oncol. 31:1428–1434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Van Den Eeden SK, Lu R, Zhang N,
Quesenberry CP Jr, Shan J, Han JS, Tsiatis AC, Leimpeter AD,
Lawrence HJ, Febbo PG and Presti JC: A Biopsy-based 17-gene genomic
prostate score as a predictor of metastases and prostate cancer
death in surgically treated men with clinically localized disease.
Eur Urol. 73:129–138. 2018. View Article : Google Scholar
|
|
143
|
Eggener S, Karsh LI, Richardson T, Shindel
AW, Lu R, Rosenberg S, Goldfischer E, Korman H, Bennett J, Newmark
J and Denes BS: A 17-gene panel for prediction of adverse prostate
cancer pathologic features: Prospective clinical validation and
utility. Urology. 126:76–82. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Mosley JD and Keri RA: Cell cycle
correlated genes dictate the prognostic power of breast cancer gene
lists. BMC Med Genomics. 1:112008. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Freedland SJ, Gerber L, Reid J, Welbourn
W, Tikishvili E, Park J, Younus A, Gutin A, Sangale Z, Lanchbury
JS, et al: Prognostic utility of cell cycle progression score in
men with prostate cancer after primary external beam radiation
therapy. Int J Radiat Oncol Biol Phys. 86:848–853. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Bishoff JT, Freedland SJ, Gerber L,
Tennstedt P, Reid J, Welbourn W, Graefen M, Sangale Z, Tikishvili
E, Park J, et al: Prognostic utility of the cell cycle progression
score generated from biopsy in men treated with prostatectomy. J
Urol. 192:409–414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Tosoian JJ, Chappidi MR, Bishoff JT,
Freedland SJ, Reid J, Brawer M, Stone S, Schlomm T and Ross AE:
Prognostic utility of biopsy-derived cell cycle progression score
in patients with National Comprehensive Cancer Network low-risk
prostate cancer undergoing radical prostatectomy: Implications for
treatment guidance. BJU Int. 120:808–814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Cuzick J, Berney DM, Fisher G, Mesher D,
Møller H, Reid JE, Perry M, Park J, Younus A, Gutin A, et al:
Prognostic value of a cell cycle progression signature for prostate
cancer death in a conservatively managed needle biopsy cohort. Br J
Cancer. 106:1095–1099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Health Quality Ontario: Prolaris cell
cycle progression test for localized prostate cancer: A health
technology assessment. Ont Health Technol Assess Ser. 17:1–75.
2017.
|
|
150
|
Li F, Ji JP, Xu Y and Liu RL:
Identification a novel set of 6 differential expressed genes in
prostate cancer that can potentially predict biochemical recurrence
after curative surgery. Clin Transl Oncol. 21:1067–1075. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Chu J, Li N and Gai W: Identification of
genes that predict the biochemical recurrence of prostate cancer.
Oncol Lett. 16:3447–3452. 2018.PubMed/NCBI
|
|
152
|
Abou-Ouf H, Alshalalfa M, Takhar M, Erho
N, Donnelly B, Davicioni E, Karnes RJ and Bismar TA: Validation of
a 10-gene molecular signature for predicting biochemical recurrence
and clinical metastasis in localized prostate cancer. J Cancer Res
Clin Oncol. 144:883–891. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Russo MA, Ravenna L, Pellegrini L,
Petrangeli E, Salvatori L, Magrone T, Fini M and Tafani M: Hypoxia
and inflammation in prostate cancer progression. Cross-talk with
androgen and estrogen receptors and cancer stem cells. Endocr Metab
Immune Disord Drug Targets. 16:235–248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Marignol L, Rivera-Figueroa K, Lynch T and
Hollywood D: Hypoxia, notch signalling, and prostate cancer. Nat
Rev Urol. 10:405–413. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Yang L, Roberts D, Takhar M, Erho N, Bibby
BAS, Thiruthaneeswaran N, Bhandari V, Cheng WC, Haider S, McCorry
AMB, et al: Development and validation of a 28-gene Hypoxia-related
prognostic signature for localized prostate cancer. EBioMedicine.
31:182–189. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Jiang Y, Mei W, Gu Y, Lin X, He L, Zeng H,
Wei F, Wan X, Yang H, Major P and Tang D: Construction of a set of
novel and robust gene expression signatures predicting prostate
cancer recurrence. Mol Oncol. 12:1559–1578. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Apostolopoulos V, Stojanovska L and
Gargosky SE: MUC1 (CD227): A multi-tasked molecule. Cell Mol Life
Sci. 72:4475–4500. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Kufe DW: Mucins in cancer: Function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Nath S and Mukherjee P: MUC1: A
multifaceted oncoprotein with a key role in cancer progression.
Trends Mol Med. 20:332–342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Wurz GT, Kao CJ, Wolf M and DeGregorio MW:
Tecemotide: An antigen-specific cancer immunotherapy. Hum Vaccin
Immunother. 10:3383–3393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Eminaga O, Wei W, Hawley SJ, Auman H,
Newcomb LF, Simko J, Hurtado-Coll A, Troyer DA, Carroll PR, Carroll
PR, et al: MUC1 expression by immunohistochemistry is associated
with adverse pathologic features in prostate cancer: A
Multi-institutional Study. PLoS One. 11:e01652362016. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Wong N, Major P, Kapoor A, Wei F, Yan J,
Aziz T, Zheng M, Jayasekera D, Cutz JC, Chow MJ and Tang D:
Amplification of MUC1 in prostate cancer metastasis and CRPC
development. Oncotarget. 7:83115–83133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Lin X, Gu Y, Kapoor A, Wei F, Aziz T, Ojo
D, Jiang Y, Bonert M, Shayegan B, Yang H, et al: Overexpression of
MUC1 and genomic alterations in its network associate with prostate
cancer progression. Neoplasia. 19:857–867. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Tarnowski M, Czerewaty M, Deskur A,
Safranow K, Marlicz W, Urasińska E, Ratajczak MZ and Starzyńska T:
Expression of cancer testis antigens in colorectal cancer: New
prognostic and therapeutic implications. Dis Markers.
2016:19875052016. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Stellfox ME, Nardi IK, Knippler CM and
Foltz DR: Differential binding partners of the Mis18a/β YIPPEE
domains regulate Mis18 complex recruitment to centromeres. Cell
Rep. 15:2127–2135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Nardi IK, Zasadzińska E, Stellfox ME,
Knippler CM and Foltz DR: Licensing of centromeric chromatin
assembly through the Mis18a-Mis18β heterotetramer. Mol Cell.
61:774–787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Wang D, Chen Z, Lin F, Wang Z, Gao Q, Xie
H, Xiao H, Zhou Y, Zhang F, Ma Y, et al: OIP 5 promotes growth,
metastasis and chemoresistance to cisplatin in bladder cancer
cells. J Cancer. 9:4684–4695. 2018. View Article : Google Scholar :
|
|
168
|
He J, Zhao Y, Zhao E, Wang X, Dong Z, Chen
Y, Yang L and Cui H: Cancer-testis specific gene OIP5: A downstream
gene of E2F1 that promotes tumorigenesis and metastasis in
glioblastoma by stabilizing E2F1 signaling. Neuro Oncol.
20:1173–1184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Gong M, Xu Y, Dong W, Guo G, Ni W, Wang Y,
Wang Y and An R: Expression of Opa interacting protein 5 (OIP5) is
associated with tumor stage and prognosis of clear cell renal cell
carcinoma. Acta Histochem. 115:810–815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Chun HK, Chung KS, Kim HC, Kang JE, Kang
MA, Kim JT, Choi EH, Jung KE, Kim MH, Song EY, et al: OIP5 is a
highly expressed potential therapeutic target for colorectal and
gastric cancers. BMB Rep. 43:349–354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Mobasheri MB, Shirkoohi R and Modarressi
MH: Cancer/Testis OIP5 and TAF7L Genes are Up-regulated in breast
cancer. Asian Pac J Cancer Prev. 16:4623–4628. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
López-Urrutia E, Bustamante Montes LP,
Ladrón de Guevara Cervantes D, Pèrez-Plasencia C and Campos-Parra
AD: Crosstalk between long Non-coding RNAs, Micro-RNAs and mRNAs:
Deciphering molecular mechanisms of master regulators in cancer.
Front Oncol. 9:6692019. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Ramnarine VR, Kobelev M, Gibb EA, Nouri M,
Lin D, Wang Y, Buttyan R, Davicioni E, Zoubeidi A and Collins CC:
The evolution of long noncoding RNA acceptance in prostate cancer
initiation and progression, and its clinical utility in disease
management. Eur Urol. Aug 22–2019.Epub ahead of print. View Article : Google Scholar
|
|
175
|
Bussemakers MJ, van Bokhoven A, Verhaegh
GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N and Isaacs
WB: DD3: A new prostate-specific gene, highly overexpressed in
prostate cancer. Cancer Res. 59:5975–5979. 1999.PubMed/NCBI
|
|
176
|
Crawford ED, Rove KO, Trabulsi EJ, Qian J,
Drewnowska KP, Kaminetsky JC, Huisman TK, Bilowus ML, Freedman SJ,
Glover WL Jr and Bostwick DG: Diagnostic performance of PCA3 to
detect prostate cancer in men with increased prostate specific
antigen: A prospective study of 1,962 cases. J Urol. 188:1726–1731.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Deras IL, Aubin SM, Blase A, Day JR, Koo
S, Partin AW, Ellis WJ, Marks LS, Fradet Y, Rittenhouse H and
Groskopf J: PCA3: A molecular urine assay for predicting prostate
biopsy outcome. J Urol. 179:1587–1592. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Gittelman MC, Hertzman B, Bailen J,
Williams T, Koziol I, Henderson RJ, Efros M, Bidair M and Ward JF:
PCA3 molecular urine test as a predictor of repeat prostate biopsy
outcome in men with previous negative biopsies: A prospecAive
multicenter clinical study. J Urol. 190:64–69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Ma W, Chen X, Ding L, Ma J, Jing W, Lan T,
Sattar H, Wei Y, Zhou F and Yuan Y: The prognostic value of long
noncoding RNAs in prostate cancer: A systematic review and
meta-analysis. Oncotarget. 8:57755–57765. 2017.PubMed/NCBI
|
|
180
|
Wu XL, Zhang JW, Li BS, Peng SS and Yuan
YQ: The prognostic value of abnormally expressed lncRNAs in
prostatic carcinoma: A systematic review and meta-analysis.
Medicine (Baltimore). 96:e92792017. View Article : Google Scholar
|
|
181
|
Wang J, Cheng G, Li X, Pan Y, Qin C, Yang
H, Hua L and Wang Z: Overexpression of long non-coding RNA
LOC400891 promotes tumor progression and poor prognosis in prostate
cancer. Tumour Biol. 37:9603–9613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Xu S, Yi XM, Tang CP, Ge JP, Zhang ZY and
Zhou WQ: Long non-coding RNA ATB promotes growth and
epithelial-mesen-chymal transition and predicts poor prognosis in
human prostate carcinoma. Oncol Rep. 36:10–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocel-lular carcinoma. Cancer Cell. 25:666–681. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Xiao H, Zhang F, Zou Y, Li J, Liu Y and
Huang W: The function and mechanism of long Non-coding RNA-ATB in
cancers. Front Physiol. 9:3212018. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Wu J, Cheng G, Zhang C, Zheng Y, Xu H,
Yang H and Hua L: Long noncoding RNA LINC01296 is associated with
poor prognosis in prostate cancer and promotes cancer-cell
proliferation and metastasis. Onco Targets Ther. 10:1843–1852.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Qiu JJ and Yan JB: Long non-coding RNA
LINC01296 is a potential prognostic biomarker in patients with
colorectal cancer. Tumour Biol. 36:7175–7183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Seitz AK, Christensen LL, Christensen E,
Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM,
Palmfeldt J, Thomson M, et al: Profiling of long non-coding RNAs
identifies LINC00958 and LINC01296 as candidate oncogenes in
bladder cancer. Sci Rep. 7:3952017. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Wang K, Zhang M, Wang C and Ning X: Long
Noncoding RNA LINC01296 Harbors miR-21a to regulate colon carcinoma
proliferation and invasion. Oncol Res. 27:541–549. 2019. View Article : Google Scholar
|
|
189
|
Qin QH, Yin ZQ, Li Y, Wang BG and Zhang
MF: Long inter-genic noncoding RNA 01296 aggravates gastric cancer
cells progress through miR-122/MMP-9. Biomed Pharmacother.
97:450–457. 2018. View Article : Google Scholar
|
|
190
|
Zhang D, Li H, Xie J, Jiang D, Cao L, Yang
X, Xue P and Jiang X: Long noncoding RNA LINC01296 promotes tumor
growth and progression by sponging miR-5095 in human
cholangiocarcinoma. Int J Oncol. 52:1777–1786. 2018.PubMed/NCBI
|
|
191
|
Jiang M, Xiao Y, Liu D, Luo N, Gao Q and
Guan Y: Overexpression of long noncoding RNA LINC01296 indicates an
unfavorable prognosis and promotes tumorigenesis in breast cancer.
Gene. 675:217–224. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Hu X, Duan L, Liu H and Zhang L: Long
noncoding RNA LINC01296 induces non-small cell lung cancer growth
and progression through sponging miR-5095. Am J Transl Res.
11:895–903. 2019.PubMed/NCBI
|
|
193
|
Feng W, Zhai C, Shi W, Zhang Q, Yan X,
Wang J, Wang Q and Li M: Clinicopathological and prognostic value
of LINC01296 in cancers: A meta-analysis. Artif Cells Nanomed
Biotechnol. 47:3315–3321. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Wan Y, Li M and Huang P: LINC01296
promotes proliferation, migration, and invasion of HCC cells by
targeting miR-122-5P and modulating EMT activity. Onco Targets
Ther. 12:2193–2203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Prensner JR, Iyer MK, Sahu A, Asangani IA,
Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al:
The long noncoding RNA SChLAP1 promotes aggressive prostate cancer
and antagonizes the SWI/SNF complex. Nat Genet. 45:1392–1398. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Mehra R, Udager AM, Ahearn TU, Cao X, Feng
FY, Loda M, Petimar JS, Kantoff P, Mucci LA and Chinnaiyan AM:
Overexpression of the long Non-coding RNA SChLAP1 independently
predicts lethal prostate cancer. Eur Urol. 70:549–552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Prensner JR, Zhao S, Erho N, Schipper M,
Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A,
Siddiqui J, et al: RNA biomarkers associated with metastatic
progression in prostate cancer: A multi-institutional
high-throughput analysis of SChLAP1. Lancet Oncol. 15:1469–1480.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Mehra R, Shi Y, Udager AM, Prensner JR,
Sahu A, Iyer MK, Siddiqui J, Cao X, Wei J, Jiang H, et al: A novel
RNA in situ hybridization assay for the long noncoding RNA SChLAP1
predicts poor clinical outcome after radical prostatectomy in
clinically localized prostate cancer. Neoplasia. 16:1121–1127.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Raab JR, Smith KN, Spear CC, Manner CJ,
Calabrese JM and Magnuson T: SWI/SNF remains localized to chromatin
in the presence of SCHLAP1. Nat Genet. 51:26–29. 2019. View Article : Google Scholar :
|
|
200
|
Fotouhi Ghiam A, Taeb S, Huang X, Huang V,
Ray J, Scarcello S, Hoey C, Jahangiri S, Fokas E, Loblaw A, et al:
Long non-coding RNA urothelial carcinoma associated 1 (UCA1)
mediates radiation response in prostate cancer. Oncotarget.
8:4668–4689. 2017.
|
|
201
|
Na XY, Liu ZY, Ren PP, Yu R and Shang XS:
Long non-coding RNA UCA1 contributes to the progression of prostate
cancer and regulates proliferation through KLF4-KRT6/13 signaling
pathway. Int J Clin Exp Med. 8:12609–12616. 2015.PubMed/NCBI
|
|
202
|
Wang F, Zhou J, Xie X, Hu J, Chen L, Hu Q,
Guo H and Yu C: Involvement of SRPK1 in cisplatin resistance
related to long non-coding RNA UCA1 in human ovarian cancer cells.
Neoplasma. 62:432–438. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C,
Ye H, Zhou B, Chen JJ and Chen P: Aberrant expression of UCA1 in
gastric cancer and its clinical significance. Clin Transl Oncol.
17:640–646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
Malat-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Chen P, Wan D, Zheng D, Zheng Q, Wu F and
Zhi Q: Long non-coding RNA UCA1 promotes the tumorigenesis in
pancreatic cancer. Biomed Pharmacother. 83:1220–1226. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Zhao W, Sun C and Cui Z: A long noncoding
RNA UCA1 promotes proliferation and predicts poor prognosis in
glioma. Clin Transl Oncol. 19:735–741. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Xuan W, Yu H, Zhang X and Song D:
Crosstalk between the lncRNA UCA1 and microRNAs in cancer. FEBS
Lett. 593:1901–1914. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Zhang S, Dong X, Ji T, Chen G and Shan L:
Long non-coding RNA UCA1 promotes cell progression by acting as a
competing endogenous RNA of ATF2 in prostate cancer. Am J Transl
Res. 9:366–375. 2017.PubMed/NCBI
|
|
209
|
He C, Lu X, Yang F, Qin L, Guo Z, Sun Y
and Wu J: LncRNA UCA1 acts as a sponge of miR-204 to up-regulate
CXCR4 expression and promote prostate cancer progression. Biosci
Rep. 39:pii: BSR20181465. 2019. View Article : Google Scholar
|
|
210
|
Shukla S, Zhang X, Niknafs YS, Xiao L,
Mehra R, Cieślik M, Ross A, Schaeffer E, Malik B, Guo S, et al:
Identification and validation of PCAT14 as prognostic biomarker in
prostate cancer. Neoplasia. 18:489–499. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
White NM, Zhao SG, Zhang J, Rozycki EB,
Dang HX, McFadden SD, Eteleeb AM, Alshalalfa M, Vergara IA, Erho N,
et al: Multi-institutional analysis shows that low PCAT-14
expression associates with poor outcomes in prostate cancer. Eur
Urol. 71:257–266. 2017. View Article : Google Scholar
|
|
212
|
Wang Y, Hu Y, Wu G, Yang Y, Tang Y, Zhang
W, Wang K, Liu Y, Wang X and Li T: Long noncoding RNA PCAT-14
induces proliferation and invasion by hepatocellular carcinoma
cells by inducing methylation of miR-372. Oncotarget.
8:34429–34441. 2017.PubMed/NCBI
|
|
213
|
Huang TB, Dong CP, Zhou GC, Lu SM, Luan Y,
Gu X, Liu L and Ding XF: A potential panel of four-long noncoding
RNA signature in prostate cancer predicts biochemical
recurrence-free survival and disease-free survival. Int Urol
Nephrol. 49:825–835. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Shao N, Tang H, Qu Y, Wan F and Ye D:
Development and validation of lncRNAs-based nomogram for prediction
of biochemical recurrence in prostate cancer by bioinformatics
analysis. J Cancer. 10:2927–2934. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Shao N, Zhu Y, Wan FN and Ye DW:
Identification of seven long noncoding RNAs signature for
prediction of biochemical recurrence in prostate cancer. Asian J
Androl. Mar 5–2019.Epub ahead of print. PubMed/NCBI
|
|
216
|
Xu J, Lan Y, Yu F, Zhu S, Ran J, Zhu J,
Zhang H, Li L, Cheng S, Xiao Y and Li X: Transcriptome analysis
reveals a long non-coding RNA signature to improve biochemical
recurrence prediction in prostate cancer. Oncotarget.
9:24936–24949. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Diao P, Song Y, Ge H, Wu Y, Li J, Zhang W,
Wang Y and Cheng J: Identification of 4-lncRNA prognostic signature
in head and neck squamous cell carcinoma. J Cell Biochem.
120:10010–10020. 2019. View Article : Google Scholar
|
|
218
|
Zhang J, Yin M, Peng G and Zhao Y: CRNDE:
An important oncogenic long non-coding RNA in human cancers. Cell
Prolif. 51:e124402018. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Li J, Zhang Z, Xiong L, Guo C, Jiang T,
Zeng L, Li G and Wang J: SNHG1 lncRNA negatively regulates
miR-199a-3p to enhance CDK7 expression and promote cell
proliferation in prostate cancer. Biochem Biophys Res Commun.
487:146–152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Thin KZ, Tu JC and Raveendran S: Long
non-coding SNHG1 in cancer. Clin Chim Acta. 494:38–47. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Sun M, Geng D, Li S, Chen Z and Zhao W:
LncRNA PART1 modulates toll-like receptor pathways to influence
cell proliferation and apoptosis in prostate cancer cells. Biol
Chem. 399:387–395. 2018. View Article : Google Scholar
|
|
222
|
Zhu D, Yu Y, Wang W, Wu K, Liu D, Yang Y,
Zhang C, Qi Y and Zhao S: Long noncoding RNA PART1 promotes
progression of non-small cell lung cancer cells via JAK-STAT
signaling pathway. Cancer Med. Aug 22–2019.Epub ahead of print.
View Article : Google Scholar
|
|
223
|
Hu X, Feng H, Huang H, Gu W, Fang Q, Xie
Y, Qin C and Hu X: Downregulated long noncoding RNA PART1 inhibits
proliferation and promotes apoptosis in bladder cancer. Technol
Cancer Res Treat. 18:15330338198466382019. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Zhihua Z, Weiwei W, Lihua N, Jianying Z
and Jiang G: p53-induced long non-coding RNA PGM5-AS1 inhibits the
progression of esophageal squamous cell carcinoma through
regulating miR-466/PTEN axis. IUBMB Life. 71:1492–1502. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Liu Q, Wu Y, Xiao J and Zou J: Long
Non-coding RNA prostate cancer-associated Transcript 7 (PCAT7)
induces poor prognosis and promotes tumorigenesis by inhibiting
mir-134-5p in Non-small-cell lung (NSCLC). Med Sci Monit.
23:6089–6098. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Yuan Q, Chu H, Ge Y, Ma G, Du M, Wang M,
Zhang Z and Zhang W: LncRNA PCAT1 and its genetic variant rs1902432
are associated with prostate cancer risk. J Cancer. 9:1414–1420.
2018. View Article : Google Scholar :
|
|
227
|
Shang Z, Yu J, Sun L, Tian J, Zhu S, Zhang
B, Dong Q, Jiang N, Flores-Morales A, Chang C and Niu Y: LncRNA
PCAT1 activates AKT and NF-kappaB signaling in castration-resistant
prostate cancer by regulating the PHLPP/FKBP51/IKKa complex.
Nucleic Acids Res. 47:4211–4225. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Han Y, Rand KA, Hazelett DJ, Ingles SA,
Kittles RA, Strom SS, Rybicki BA, Nemesure B, Isaacs WB, Stanford
JL, et al: Prostate cancer susceptibility in men of african
ancestry at 8q24. J Natl Cancer Inst. 108:2016. View Article : Google Scholar
|
|
229
|
Huang L, Wang Y, Chen J, Wang Y, Zhao Y,
Wang Y, Ma Y, Chen X, Liu W, Li Z, et al: Long noncoding RNA PCAT1,
a novel serum-based biomarker, enhances cell growth by sponging
miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis.
10:5132019. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Yang ML, Huang Z, Wu LN, Wu R, Ding HX and
Wang BG: lncRNA-PCAT1 rs2632159 polymorphism could be a biomarker
for colorectal cancer susceptibility. Biosci Rep. 39:pii:
BSR20190708. 2019. View Article : Google Scholar
|
|
231
|
Zhao X, Fan Y, Lu C, Li H, Zhou N, Sun G
and Fan H: PCAT1 is a poor prognostic factor in endometrial
carcinoma and associated with cancer cell proliferation, migration
and invasion. Bosn J Basic Med Sci. 19:274–281. 2019.PubMed/NCBI
|
|
232
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Brockdorff N, Ashworth A, Kay GF, McCabe
VM, Norris DP, Cooper PJ, Swift S and Rastan S: The product of the
mouse Xist gene is a 15 kb inactive X-specific transcript
containing no conserved ORF and located in the nucleus. Cell.
71:515–526. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Ward JF and Moul JW: Biochemical
recurrence after definitive prostate cancer therapy. Part I:
Defining and localizing biochemical recurrence of prostate cancer.
Curr Opin Urol. 15:181–186. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Ward JF and Moul JW: Biochemical
recurrence after definitive prostate cancer therapy. Part II:
Treatment strategies for biochemical recurrence of prostate cancer.
Curr Opin Urol. 15:187–195. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
McCormick BZ, Mahmoud AM, Williams SB and
Davis JW: Biochemical recurrence after radical prostatectomy:
Current status of its use as a treatment endpoint and early
management strategies. Indian J Urol. 35:6–17. 2019.PubMed/NCBI
|
|
237
|
Spratt DE, McHugh DJ, Morris MJ and
Morgans AK: Management of biochemically recurrent prostate cancer:
Ensuring the right treatment of the right patient at the right
time. Am Soc Clin Oncol Educ Book. 38:355–362. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Gillessen S, Attard G, Beer TM, Beltran H,
Bossi A, Bristow R, Carver B, Castellano D, Chung BH, Clarke N, et
al: Management of patients with advanced prostate cancer: The
report of the advanced prostate cancer consensus conference APCCC
2017. Eur Urol. 73:178–211. 2018. View Article : Google Scholar
|
|
239
|
Rahbar K, Afshar-Oromieh A, Jadvar H and
Ahmadzadehfar H: PSMA theranostics: Current status and future
directions. Mol Imaging. 17:15360121187760682018. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Wester HJ and Schottelius M: PSMA-Targeted
radiopharmaceu-ticals for imaging and therapy. Semin Nucl Med.
49:302–312. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
O'Keefe DS, Bacich DJ, Huang SS and Heston
WDW: A perspective on the evolving story of PSMA biology,
PSMA-based imaging, and endoradiotherapeutic strategies. J Nucl
Med. 59:1007–1013. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Israeli RS, Powell CT, Corr JG, Fair WR
and Heston WD: Expression of the prostate-specific membrane
antigen. Cancer Res. 54:1807–1811. 1994.PubMed/NCBI
|
|
243
|
Sweat SD, Pacelli A, Murphy GP and
Bostwick DG: Prostate-specific membrane antigen expression is
greatest in prostate adenocarcinoma and lymph node metastases.
Urology. 52:637–640. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Wright GL Jr, Grob BM, Haley C, Grossman
K, Newhall K, Petrylak D, Troyer J, Konchuba A, Schellhammer PF and
Moriarty R: Upregulation of prostate-specific membrane antigen
after androgen-deprivation therapy. Urology. 48:326–334. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Marchal C, Redondo M, Padilla M, Caballero
J, Rodrigo I, García J, Quian J and Boswick DG: Expression of
prostate specific membrane antigen (PSMA) in prostatic
adenocarcinoma and prostatic intraepithelial neoplasia. Histol
Histopathol. 19:715–718. 2004.PubMed/NCBI
|
|
246
|
Sheikhbahaei S, Werner RA, Solnes LB,
Pienta KJ, Pomper MG, Gorin MA and Rowe SP: Prostate-specific
membrane antigen (PSMA)-targeted PET imaging of prostate cancer: An
update on important pitfalls. Semin Nucl Med. 49:255–270. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Bouchelouche K and Sathekge MM: Letter
from the editors. Semin Nucl Med. 49:245–246. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Fendler WP, Calais J, Eiber M, Flavell RR,
Mishoe A, Feng FY, Nguyen HG, Reiter RE, Rettig MB, Okamoto S, et
al: Assessment of 68Ga-PSMA-11 PET accuracy in localizing recurrent
prostate cancer: A prospective Single-Arm clinical trial. JAMA
Oncol. 5:856–863. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Rahbar K, Afshar-Oromieh A, Seifert R,
Wagner S, Schäfers M, Bögemann M and Weckesser M: Diagnostic
performance of 18F-PSMA-1007 PET/CT in patients with
biochemical recurrent prostate cancer. Eur J Nucl Med Mol Imaging.
45:2055–2061. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Karnes RJ, Choeurng V, Ross AE, Schaeffer
EM, Klein EA, Freedland SJ, Erho N, Yousefi K, Takhar M, Davicioni
E, et al: Validation of a genomic risk classifier to predict
prostate cancer-specific mortality in men with adverse pathologic
features. Eur Urol. 73:168–175. 2018. View Article : Google Scholar
|
|
251
|
Cooperberg MR, Davicioni E, Crisan A,
Jenkins RB, Ghadessi M and Karnes RJ: Combined value of validated
clinical and genomic risk stratification tools for predicting
prostate cancer mortality in a high-risk prostatectomy cohort. Eur
Urol. 67:326–333. 2015. View Article : Google Scholar :
|
|
252
|
Wiegel T, Bartkowiak D, Bottke D, Bronner
C, Steiner U, Siegmann A, Golz R, Störkel S, Willich N, Semjonow A,
et al: Adjuvant radiotherapy versus wait-and-see after radical
prosta-tectomy: 10-year follow-up of the ARO 96-02/AUO AP 09/95
trial. Eur Urol. 66:243–250. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Li H, Wang Z, Zhang Y, Sun G, Ding B, Yan
L, Liu H, Guan W, Hu Z, Wang S, et al: The immune checkpoint
regulator PDL1 is an independent prognostic biomarker for
biochemical recurrence in prostate cancer patients following
adjuvant hormonal therapy. J Cancer. 10:3102–3111. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
254
|
Van den Broeck T, van den Bergh RCN,
Briers E, Cornford P, Cumberbatch M, Tilki D, De Santis M, Fanti S,
Fossati N, Gillessen S, et al: Biochemical recurrence in prostate
cancer: The European association of urology prostate cancer
guidelines panel recommendations. Eur Urol Focus. Jun 24–2019.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Tendulkar RD, Agrawal S, Gao T, Efstathiou
JA, Pisansky TM, Michalski JM, Koontz BF, Hamstra DA, Feng FY,
Liauw SL, et al: Contemporary update of a Multi-institutional
predictive nomogram for salvage radiotherapy after radical
prostatectomy. J Clin Oncol. 34:3648–3654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Isharwal S and Stephenson AJ:
Post-prostatectomy radiation therapy for locally recurrent prostate
cancer. Expert Rev Anticancer Ther. 17:1003–1012. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Tilki D, Preisser F, Graefen M, Huland H
and Pompe RS: External validation of the European association of
urology biochemical recurrence risk groups to predict metastasis
and mortality after radical prostatectomy in a european cohort. Eur
Urol. 75:896–900. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Spratt DE, Dess RT, Zumsteg ZS, Lin DW,
Tran PT, Morgan TM, Antonarakis ES, Nguyen PL, Ryan CJ, Sandler HM,
et al: A systematic review and framework for the use of hormone
therapy with salvage radiation therapy for recurrent prostate
cancer. Eur Urol. 73:156–165. 2018. View Article : Google Scholar
|
|
259
|
Shipley WU, Seiferheld W, Lukka HR, Major
PP, Heney NM, Grignon DJ, Sartor O, Patel MP, Bahary JP, Zietman
AL, et al: Radiation with or without antiandrogen therapy in
recurrent prostate cancer. N Engl J Med. 376:417–428. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
260
|
Freedland SJ, Choeurng V, Howard L, De
Hoedt A, du Plessis M, Yousefi K, Lam LL, Buerki C, Ra S, Robbins
B, et al: Utilization of a genomic classifier for prediction of
metastasis following salvage radiation therapy after radical
prostatectomy. Eur Urol. 70:588–596. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
261
|
Semenas J, Allegrucci C, Boorjian SA,
Mongan NP and Persson JL: Overcoming drug resistance and treating
advanced prostate cancer. Curr Drug Targets. 13:1308–1323. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Cucchiara V, Cooperberg MR, Dall'Era M,
Lin DW, Montorsi F, Schalken JA and Evans CP: Genomic markers in
prostate cancer decision making. Eur Urol. 73:572–582. 2018.
View Article : Google Scholar
|
|
263
|
Pikor L, Thu K, Vucic E and Lam W: The
detection and implication of genome instability in cancer. Cancer
Metastasis Rev. 32:341–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Lin X, Ojo D, Wei F, Wong N, Gu Y and Tang
D: A novel aspect of tumorigenesis-BMI1 functions in regulating DNA
damage response. Biomolecules. 5:3396–3415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Lin X, Yan J and Tang D: ERK kinases
modulate the activation of PI3 kinase related kinases (PIKKs) in
DNA damage response. Histol Histopathol. 28:1547–1554.
2013.PubMed/NCBI
|
|
266
|
Wei F, Yan J and Tang D: Extracellular
signal-regulated kinases modulate DNA damage response-a
contributing factor to using MEK inhibitors in cancer therapy. Curr
Med Chem. 18:5476–5482. 2011. View Article : Google Scholar
|
|
267
|
Lucarelli G, Loizzo D, Ferro M, Rutigliano
M, Vartolomei MD, Cantiello F, Buonerba C, Di Lorenzo G,
Terracciano D, De Cobelli O, et al: Metabolomic profiling for the
identification of novel diagnostic markers and therapeutic targets
in prostate cancer: An update. Expert Rev Mol Diagn. 19:377–387.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Williams D and Fingleton B: Non-canonical
roles for metabolic enzymes and intermediates in malignant
progression and metastasis. Clin Exp Metastasis. 36:211–224. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Priolo C, Pyne S, Rose J, Regan ER, Zadra
G, Photopoulos C, Cacciatore S, Schultz D, Scaglia N, McDunn J, et
al: AKT1 and MYC induce distinctive metabolic fingerprints in human
prostate cancer. Cancer Res. 74:7198–7204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
270
|
Pettersson A, Gerke T, Penney KL, Lis RT,
Stack EC, Pértega-Gomes N, Zadra G, Tyekucheva S, Giovannucci EL,
Mucci LA and Loda M: MYC overexpression at the protein and mRNA
level and cancer outcomes among men treated with radical
prostatectomy for prostate cancer. Cancer Epidemiol Biomarkers
Prev. 27:201–207. 2018. View Article : Google Scholar :
|
|
271
|
Hammarsten P, Cipriano M, Josefsson A,
Stattin P, Egevad L, Granfors T and Fowler CJ: Phospho-Akt
immunoreactivity in prostate cancer: Relationship to disease
severity and outcome, Ki67 and phosphorylated EGFR expression. PLoS
One. 7:e479942012. View Article : Google Scholar : PubMed/NCBI
|
|
272
|
Flegal KM, Kit BK, Orpana H and Graubard
BI: Association of all-cause mortality with overweight and obesity
using standard body mass index categories: A systematic review and
meta-analysis. JAMA. 309:71–82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
273
|
Schiffmann J, Karakiewicz PI, Rink M,
Manka L, Salomon G, Tilki D, Budäus L, Pompe R, Leyh-Bannurah SR,
Haese A, et al: Obesity paradox in prostate cancer: Increased body
mass index was associated with decreased risk of metastases after
surgery in 13,667 patients. World J Urol. 36:1067–1072. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
274
|
Bansal D, Undela K, D'Cruz S and Schifano
F: Statin use and risk of prostate cancer: A meta-analysis of
observational studies. PLoS One. 7:e466912012. View Article : Google Scholar : PubMed/NCBI
|
|
275
|
Kollmeier MA, Katz MS, Mak K, Yamada Y,
Feder DJ, Zhang Z, Jia X, Shi W and Zelefsky MJ: Improved
biochemical outcomes with statin use in patients with high-risk
localized prostate cancer treated with radiotherapy. Int J Radiat
Oncol Biol Phys. 79:713–718. 2011. View Article : Google Scholar
|
|
276
|
Song C, Park S, Park J, Shim M, Kim A,
Jeong IG, Hong JH, Kim CS and Ahn H: Statin use after radical
prostatectomy reduces biochemical recurrence in men with prostate
cancer. Prostate. 75:211–217. 2015. View Article : Google Scholar
|
|
277
|
Freedland SJ, Hamilton RJ, Gerber L, Banez
LL, Moreira DM, Andriole GL and Rittmaster RS: Statin use and risk
of prostate cancer and high-grade prostate cancer: Results from the
REDUCE study. Prostate Cancer Prostatic Dis. 16:254–259. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
278
|
Nordström T, Clements M, Karlsson R,
Adolfsson J and Grönberg H: The risk of prostate cancer for men on
aspirin, statin or antidiabetic medications. Eur J Cancer.
51:725–733. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
279
|
Rieken M, Kluth LA, Xylinas E, Seitz C,
Fajkovic H, Karakiewicz PI, Lotan Y, Briganti A, Loidl W, Faison T,
et al: Impact of statin use on biochemical recurrence in patients
treated with radical prosta-tectomy. Prostate Cancer Prostatic Dis.
16:367–371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Pazhanisamy SK: Stem cells, DNA damage,
ageing and cancer. Hematol Oncol Stem Cell Ther. 2:375–384. 2009.
View Article : Google Scholar
|
|
281
|
Vitale I, Manic G, De Maria R, Kroemer G
and Galluzzi L: DNA damage in stem cells. Mol Cell. 66:306–319.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
282
|
Siddique HR and Saleem M: Role of BMI1, a
stem cell factor, in cancer recurrence and chemoresistance:
Preclinical and clinical evidences. Stem Cells. 30:372–378. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Lin X, Gu Y and Tang D: BMI1, ATM and DDR.
Oncoscience. 2:665–666. 2015.PubMed/NCBI
|
|
284
|
Lin X, Wei F, Whyte P and Tang D: BMI1
reduces ATR activation and signalling caused by hydroxyurea.
Oncotarget. 8:89707–89721. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
285
|
Wei F, Ojo D, Lin X, Wong N, He L, Yan J,
Xu S, Major P and Tang D: BMI1 attenuates etoposide-induced G2/M
checkpoints via reducing ATM activation. Oncogene. 34:3063–3075.
2015. View Article : Google Scholar
|
|
286
|
Wei L, Wang J, Lampert E, Schlanger S,
DePriest AD, Hu Q, Gomez EC, Murakam M, Glenn ST, Conroy J, et al:
Intratumoral and intertumoral genomic heterogeneity of multifocal
localized prostate cancer impacts molecular classifications and
genomic prognosticators. Eur Urol. 71:183–192. 2017. View Article : Google Scholar
|