Introduction
Severe acute respiratory syndrome (SARS)
coronavirus-2 (SARS-CoV2) is the cause of COVID-19, which was first
identified at the end of 2019 and has evolved into a pandemic
during the following months (1).
Based on increasing data, older age and male sex predispose to
severe COVID-19, whilst a number of underlying diseases/conditions
are also directly related with significantly higher risk for
adverse clinical outcomes from COVID-19 (1). The latter include diabetes,
hypertension, obesity, immunosuppression, asthma, and chronic
obstructive pulmonary disease (1).
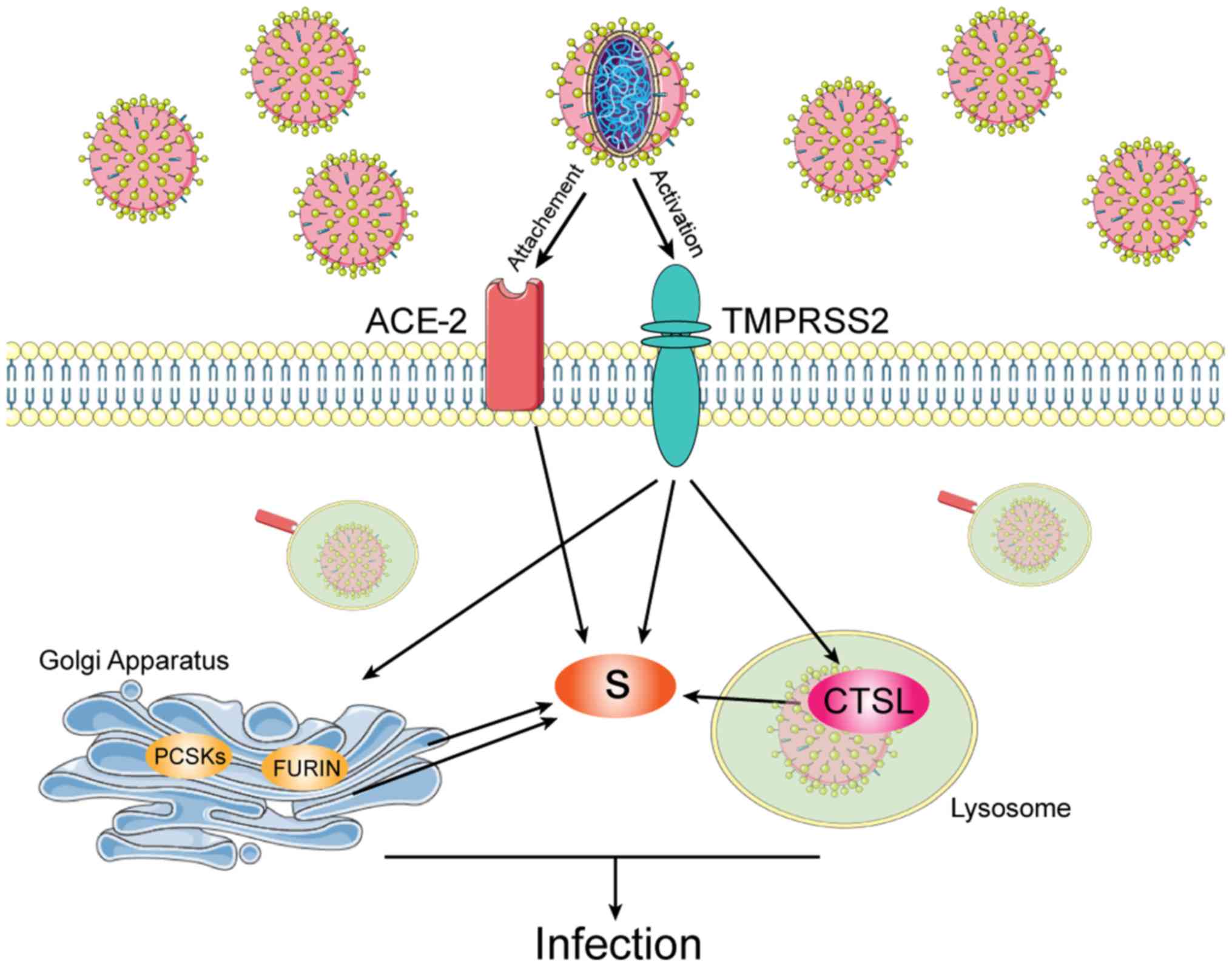
Entry of SARS-CoV2 into its host cells is
facilitated by its spike proteins which bind to the
angiotensin-converting enzyme 2 (ACE-2) (2). Moreover, the spike viral proteins are
primed by the transmembrane protease serine 2 (TMPRSS2) (2). As such, SARS-CoV2 infection of host
cells is facilitated by the cleavage of the spike proteins by
TMPRSS2 and host cell proteases, such as cathepsin L (CTSL)
(Fig. 1) (3).
Cancer has been identified as a risk factor for
severe COVID-19, as many patients can be immunocompromised
(4). In a very recent robust
systematic analysis, Chai et al curated a pan-cancer
analysis of ACE-2 detailing the expression and mutations across a
wide spectrum of tumors (5).
Following the same approach with this elegant study, here we
expanded on these excellent observations by curating data for the
other two mediators of SARS-CoV-2 infection, namely CTSL and
TMPRSS2.
Materials and methods
Bioinformatic analysis
TMPRSS2 and CTSL were validated in The Cancer Genome
Atlas (TCGA), GEPIA (http://gepia.cancer-pku.cn/), UALCAN (http://ualcan.path.uab.edu/cgi-bin/ualcan-res.pl).
The pan-cancer cohort of TCGA was downloaded through cBioPortal
(https://www.cbioportal.org/). The
datasets used for the two genes were: ACC, adrenocortical
carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast
invasive carcinoma; CESC, cervical squamous cell carcinoma and
endocervical adenocarcinoma; CHOL, cholangio carcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme;
HNSC, head and neck squamous cell carcinoma; KICH, kidney
chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG,
brain lower grade glioma; LIHC, liver hepato-cellular carcinoma;
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma;
MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD,
pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD,
stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA,
thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial
carcinoma; UCS, uterine carcinosarcoma and UVM, uveal melanoma.
Results and Discussion
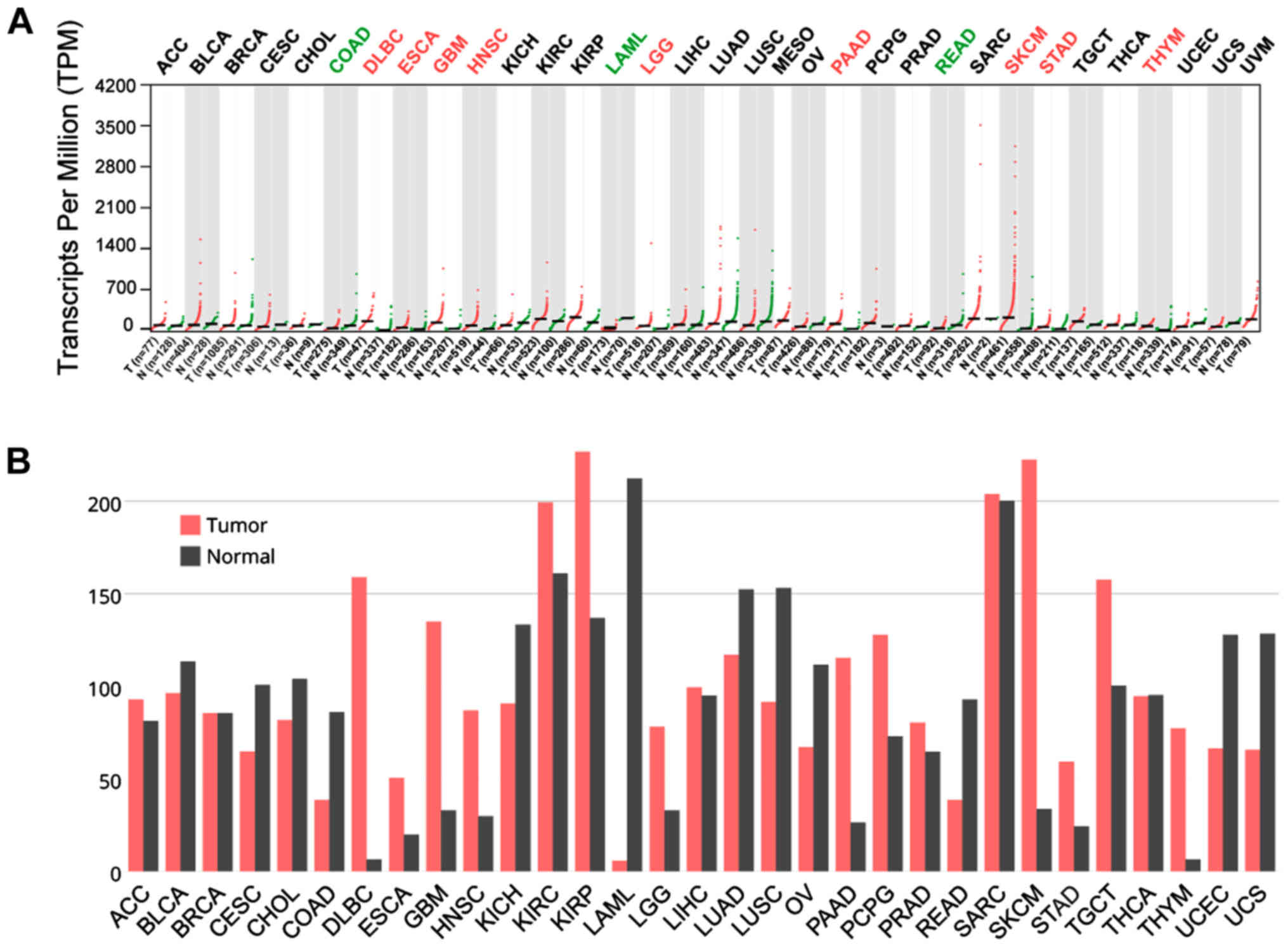
In TCGA datasets for all cancers, CTSL was
upregulated in DLBC, ESCA, GBM, HNSC, LGG, PADD, SKCM, STAD and
THYM, while it has lower expression than normal in COAD, LAML and
READ (Fig. 2A and B). On the other
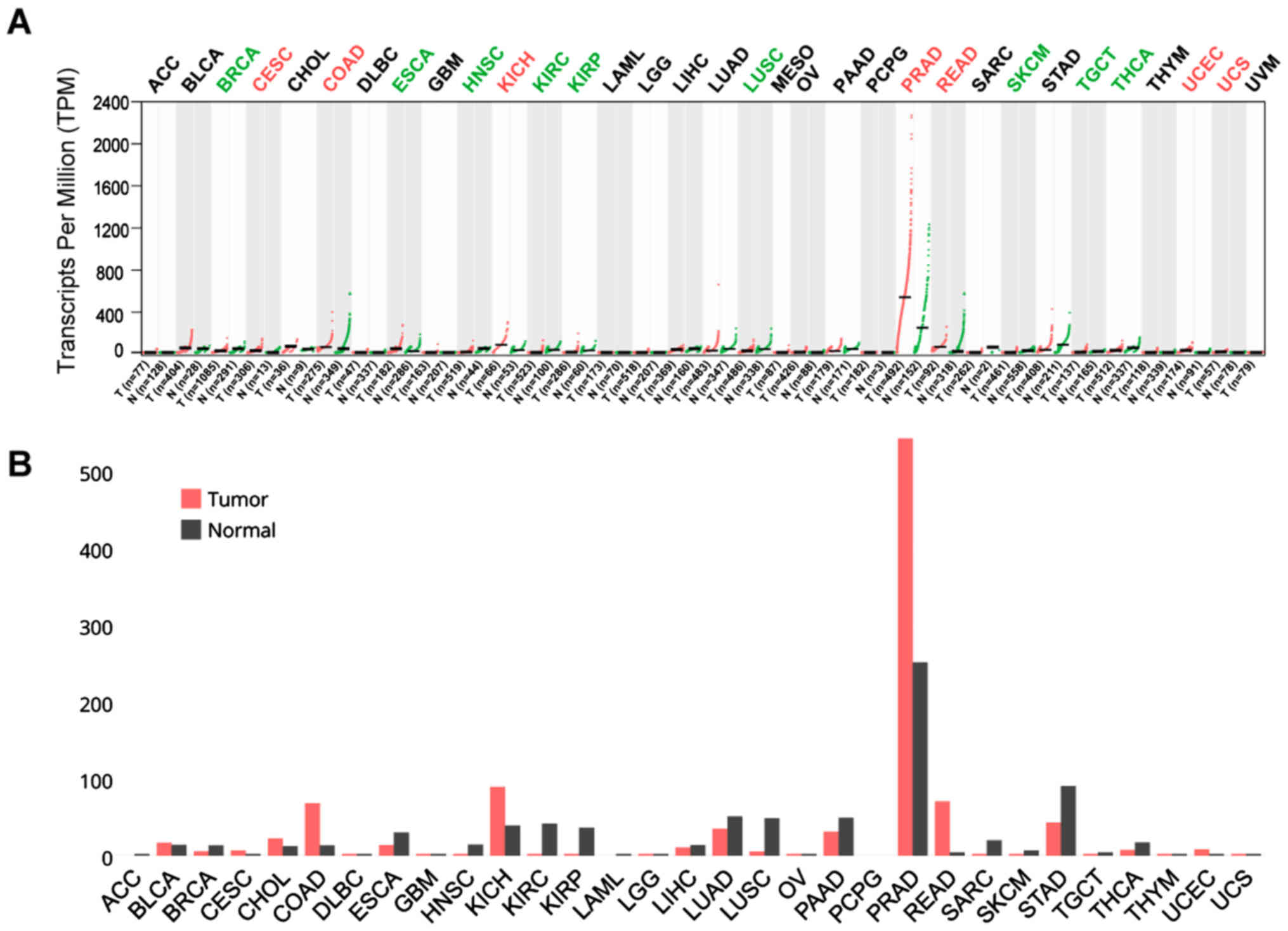
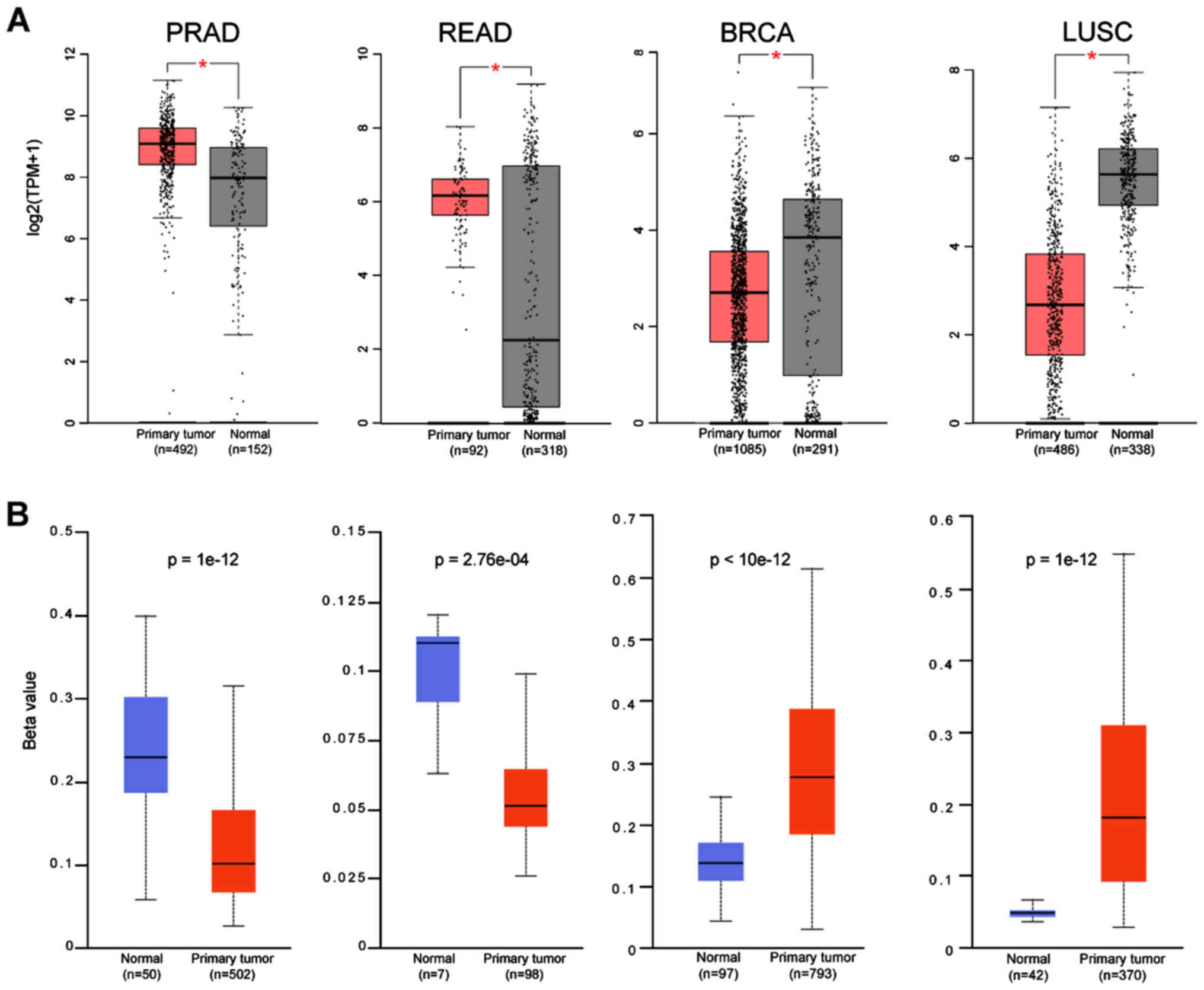
hand, TMPRSS2 was upregulated in CESC, COAD, KICH, PRAD, READ, UCEC
and UCS, with PRAD and READ exhibiting the highest expression of
all cancers (Fig. 3A and B).
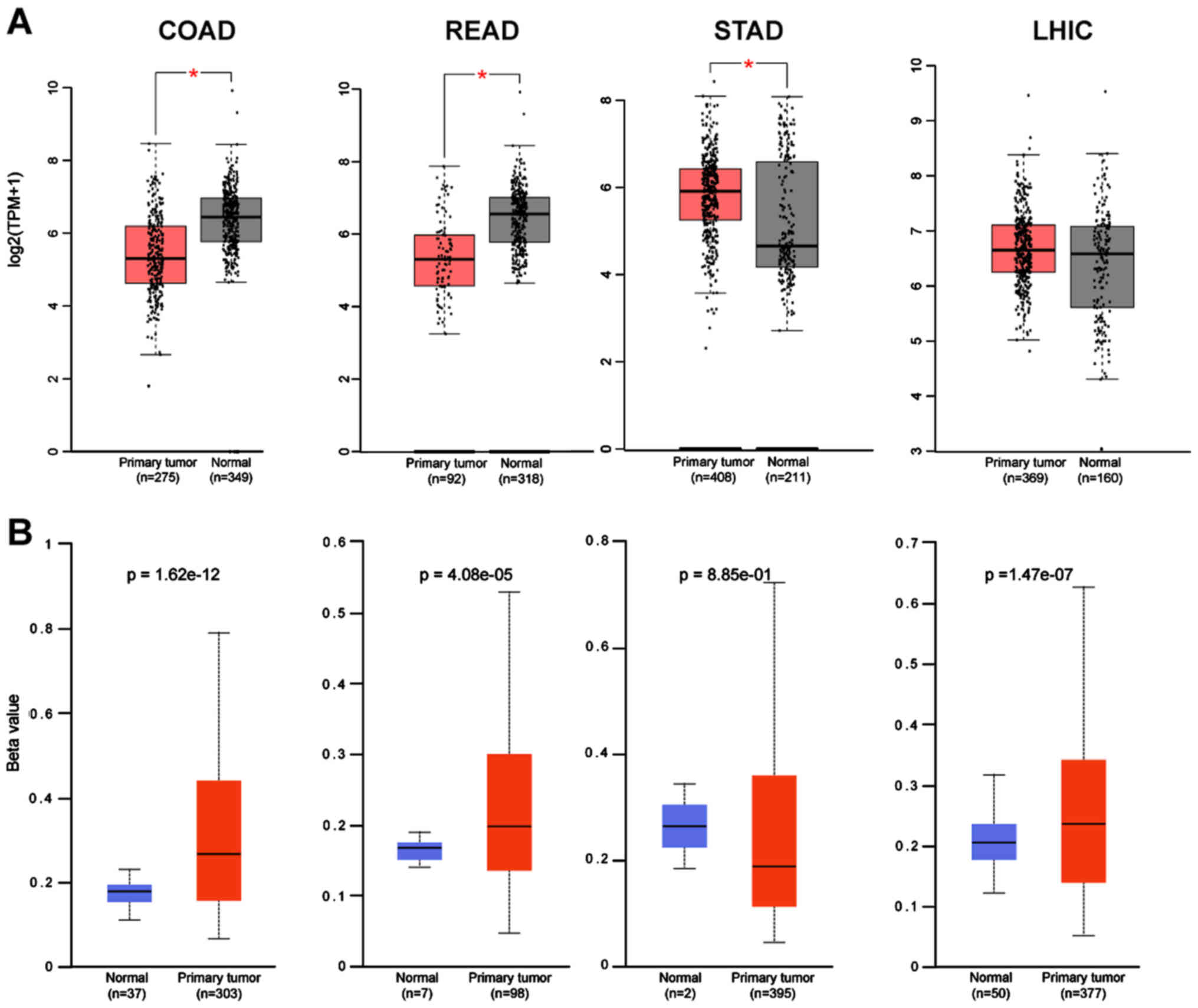
Subsequently, the expression of these two genes from
GEPIA were correlated with the methylation status using the Ualcan
database. For most of the cancers there was a strong correlation of
the gene expression with its promoter methylation status in
agreement with the findings for ACE-2 (5). CTSL in COAD and READ has a higher
beta value; hence, lower expression of the gene, while in STAD, the
lower methylation of the promoter, leads to higher gene expression
(Fig. 4A and B). Five cancers were
found to have lower methylation than normal for the CTSL, and seven
cancers for the TMPRSS2. In LUSC and BRCA, the very high
methylation rate of the TMPRSS2 gene leads to a very low expression
level (Fig. 5A and B), while the
very low methylation rate in PRAD and READ lead to very high
numbers of gene transcripts.
Moreover, using the Human Protein Atlas analysis, of
the consensus transcript expression levels that combines the Human
Protein Atlas (HPA), Genotype-Tissue Expression; (GTEx) and the
Functional Annotation of Mammalian Genomes 5 (FANTOM5) datasets,
the expression of these two genes in organ/tissue samples were
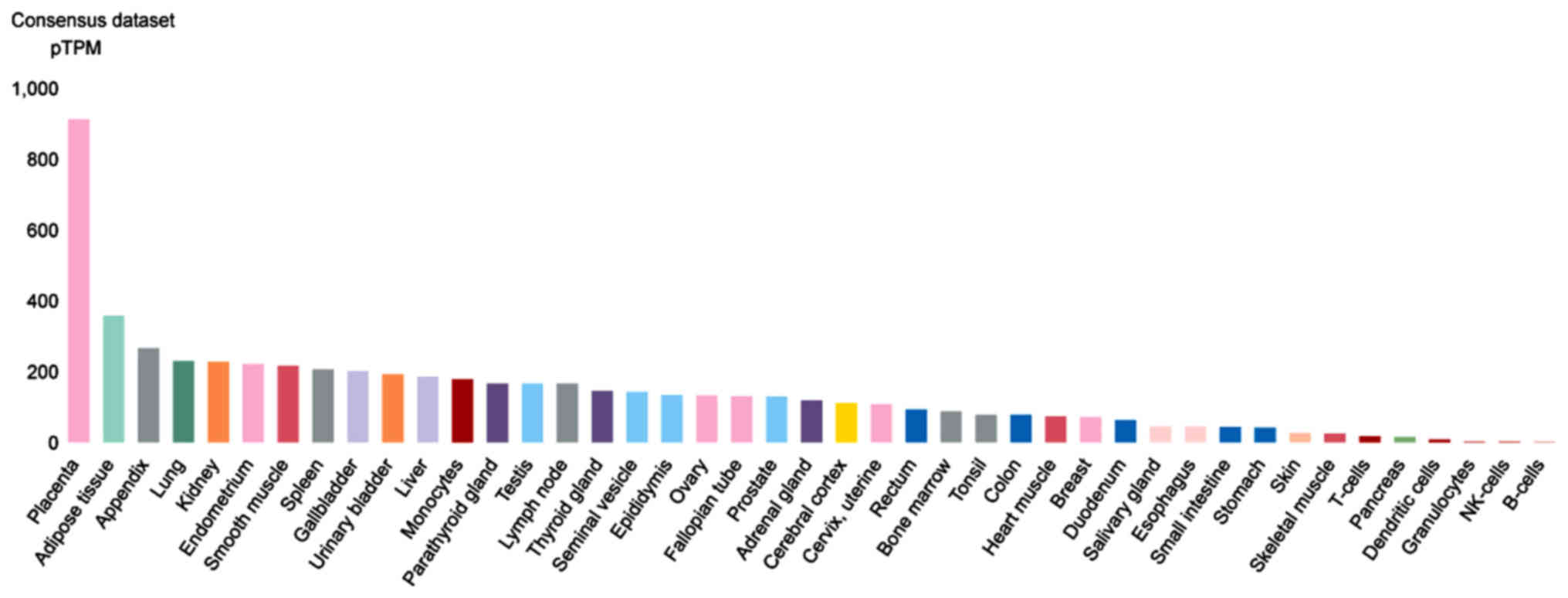
examined. CTSL is highly expressed in the placenta, adipose,
appendix and lung tissue (Fig. 6).
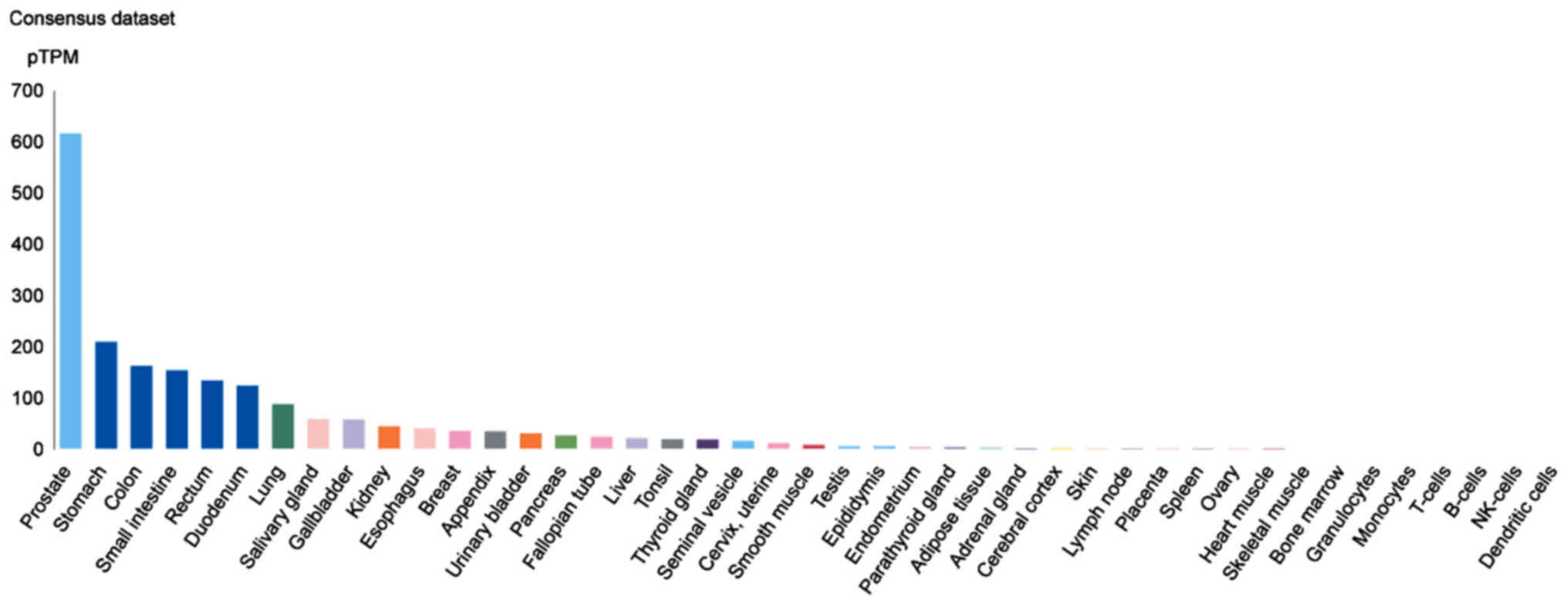
TMPRSS2 is highly expressed in prostate [indeed, TMPRSS2 and PCA
are utilized as biomarkers for prostate cancer (6)], stomach, colon and small intestine
tissues (Fig. 7).
Furthermore, using the cBioportal pan-cancer panel,
the region and the types of mutations were identified which these
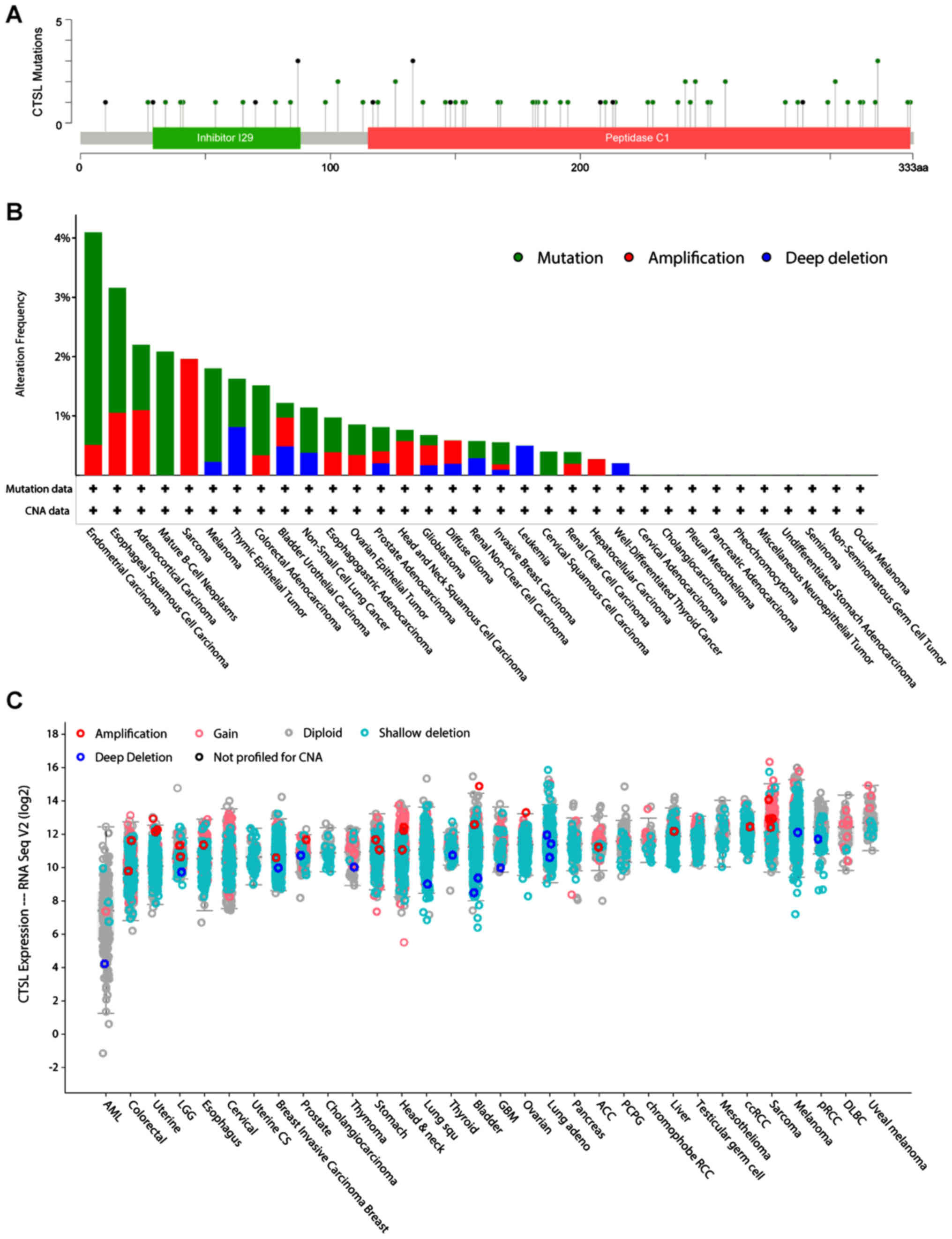
two genes have in all the examined cancer types (Figs. 8A and 9A). Most of the CTSL mutations are lying
on the peptidase region and are mostly found in CESC, ESCA, Mature
B-cell Neoplasms, Melanoma and COAD (Fig. 8B). Of note, in most of the cancers
the majority of the patients had deletions and partly some gains
and amplifications (Fig. 8C).
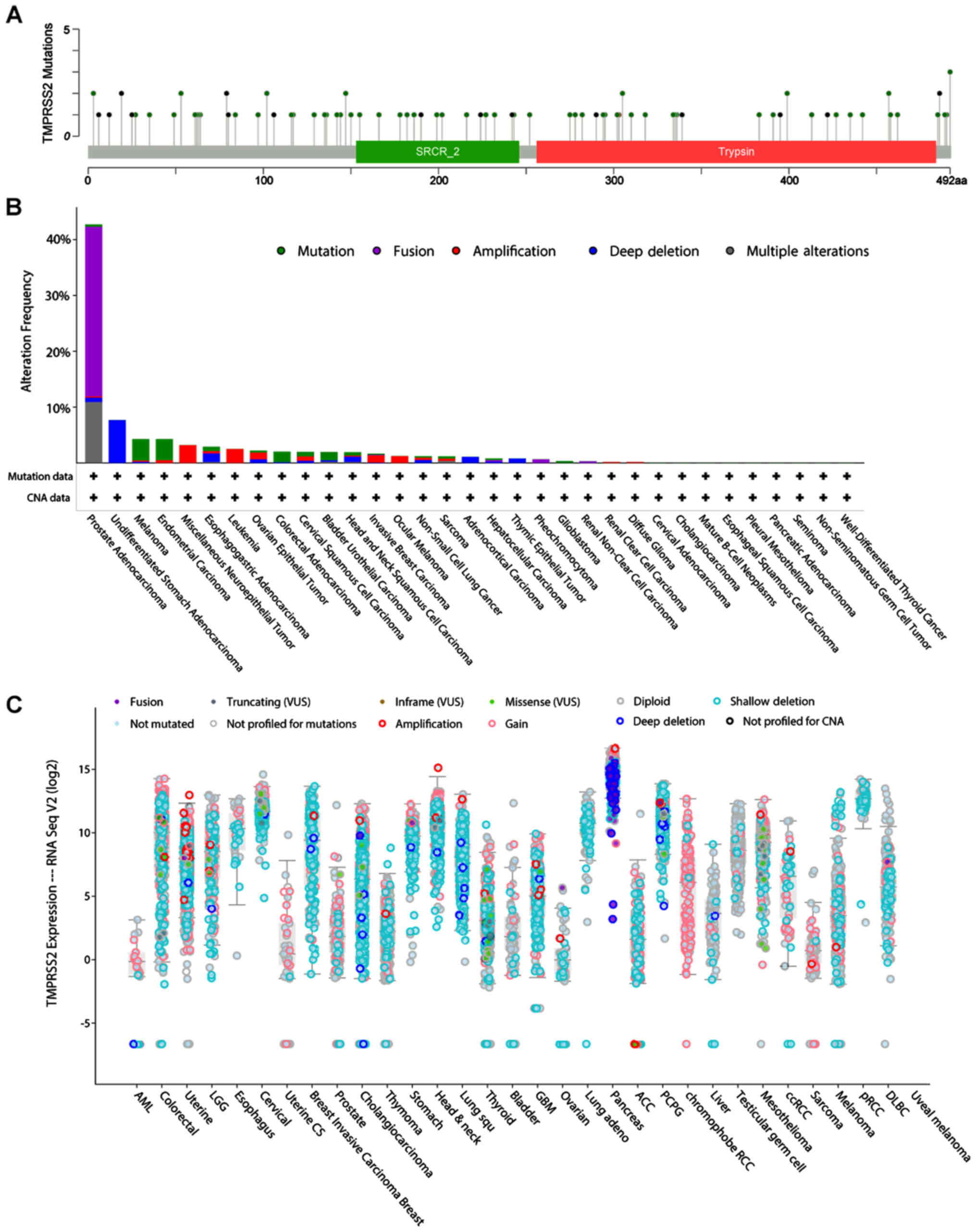
TMPRSS2 mutations were lying across the whole gene region and
mostly consist of gene fusions (TMPRSS2-ERG) in prostate
adenocarcinoma (Fig. 9B and
C).
Finally, examination of the overall survival (OS) of
all cancers, showed that CTSL low expression in KIRC (P=0.0001) has
poor prediction for the patients, while low expression of CTSL in
LUSC (P=0.0077) had better prognosis. High expression of TMPRSS2 in
BRCA, SARC and UM had poor prediction for the patients, while gene
expression was not statistically relevant for the patients of the
other types of cancers (Figs.
S1-S4).
Of note, neither of the two proteins were
differentially regulated in LUAD; a comorbidity of severe COVID-19
contrary to ACE-2 (7). TMPRSS2 is
significantly upregulated in prostate cancer, where it harbors gene
fusion events, as documented in this study. While preparing this
report, another interesting study was published which corroborated
the expression of TMPRSS2 in the prostate. In this study, the
authors have put forward the question of whether the TMPRSS2
increased expression in prostate is involved with the documented
sexual dimorphism of COVID-19 (8).
This is an exciting hypothesis that needs to be studied further
given that the clinical data indicate that male sex is among the
risk factors for adverse COVID-19 related outcomes.
In our analysis we also demonstrate that the
pancreas is riddled with deep deletions for TMPRSS2 where ACE-2 is
co-expressed. Interestingly, Liu et al (9) have recently shown that ACE-2
expression in the pancreas may lead to pancreatic damage following
SARS-CoV-2 infection. For CTSL, higher expression was noted in the
human placenta where ACE-2 is also co-expressed (10). This finding warrants further
investigation into the role of these viral entry mediators in
placentation, vertical transmission of SARS-CoV-2 to the fetus, as
well as their potential involvement in maternal and neonatal
complications (11). These
hypotheses remain to be investigated in large clinical studies.
Supplementary Data
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during this study are
included in this published article (and its supplementary
information files).
Authors' contributions
PK generated the data, and produced the figures; VA
and KC contributed to critical revision of the article; HSR, DAS
contributed to the writing of the manuscript and final edits; IK
and EK contributed equally to the conception of the work and data
analysis and interpretation. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Yuki K, Fujiogi M and Koutsogiannaki S:
COVID-19 patho-physiology: A review. Clin Immunol. 215:1084272020.
View Article : Google Scholar
|
|
2
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H,
Nitsche A, et al: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280.e8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smieszek SP, Przychodzen BP and
Polymeropoulos MH: Amantadine disrupts lysosomal gene expression: A
hypothesis for COVID19 treatment. Int J Antimicrob Agents. Apr
30–2020.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sidaway P: COVID-19 and cancer: What we
know so far. Nat Rev Clin Oncol. Apr 7–2020.Epub ahead of print.
View Article : Google Scholar
|
|
5
|
Chai P, Yu J, Ge S, Jia R and Fan X:
Genetic alteration, RNA expression, and DNA methylation profiling
of coronavirus disease 2019 (COVID-19) receptor ACE2 in
malignancies: A pan-cancer analysis. J Hematol Oncol. 13:432020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Filella X and Foj L: Prostate Cancer
Detection and Prognosis: From Prostate Specific Antigen (PSA) to
Exosomal Biomarkers. Int J Mol Sci. 17:17842016. View Article : Google Scholar :
|
|
7
|
Kong Q, Xiang Z, Wu Y, Gu Y, Guo J and
Geng F: Analysis of the susceptibility of lung cancer patients to
SARS-CoV-2 infection. Mol Cancer. 19:802020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stopsack KH, Mucci LA, Antonarakis ES,
Nelson PS and Kantoff PW: TMPRSS2 and COVID-19: Serendipity or
opportunity for intervention? Cancer Discov. Apr 10–2020.Epub ahead
of print. View Article : Google Scholar
|
|
9
|
Liu F, Long X, Zhang B, Zhang W, Chen X
and Zhang Z: ACE2 expression in pancreas may cause pancreatic
damage after SARS-CoV-2 infection. Clin Gastroenterol Hepatol. Apr
22–2020.Epub ahead of print. View Article : Google Scholar :
|
|
10
|
Li M, Chen L, Zhang J, Xiong C and Li X:
The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface
and fetal organs by single-cell transcriptome study. PLoS One.
15:e02302952020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stumpfe FM, Titzmann A, Schneider MO,
Stelzl P, Kehl S, Fasching PA, Beckmann MW and Ensser A: SARS-CoV-2
infection in pregnancy - a review of the current literature and
possible impact on maternal and neonatal outcome. Geburtshilfe
Frauenheilkd. 80:380–390. 2020.In German. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBio-Portal. Sci Signal. 6:l12013. View Article : Google Scholar
|