Introduction
Gastric cancer (GC) is the third leading cause of
cancer-related mortality worldwide following lung and liver cancer
(1). Currently, the highest
incidence and mortality rates of GC are observed in Eastern Asia
and Latin America (2,3). The majority of GC-related deaths are
caused by cancer cell invasion, metastasis and tumor recurrence
(4). Thus, to gain a better
understanding of the pathogenesis of GC, the underlying mechanisms
of GC progression are worthy of further exploration.
The nuclear factor-κB (NF-κB) transcription factor
plays a pivotal role in tumor inflammation, cell differentiation,
proliferation and survival. In addition to its roles in immunity,
NF-κB activity is frequently detected in a number of cancer types
(5,6). During tumorigenesis and tumor
development, tumor cells have been reported to respond to chronic
inflammatory stimuli within the tumor microenvironment, which is
dependent on NF-κB activation (7,8).
Previous studies have demonstrated that NF-κB signaling is
constitutively activated in a variety of human cancer types and is
associated with tumor initiation, progression and metastasis
(9-11). The NF-κB complex refers to a group
of transcription factors (RelA, RelB, c-Rel, NF-κB1/p50 and
NF-κB2/p52), which form homo or heterodimers and can upregulate or
suppress the expression of a number of genes during cancer
progression (11,12). In response to a variety of stimuli,
the activation of NF-κB signaling promotes tumor cells to produce
numerous tumorigenic factors, including the angiogenesis regulator,
vascular endothelial growth factor (VEGF), matrix metalloproteinase
(MMP)-2, MMP-9, the chemokines, interleukin (IL)-1, IL-8, tumor
necrosis factor (TNF), IL-6 and monocyte chemoattractant protein-1
(MCP-1), and the anti-apoptotic factors, Bcl-xL and cellular
inhibitor of apoptosis proteins (cIAPs), which are essential to
tumor progression (12). Helbig
et al (13) found that
NF-κB promoted breast cancer cell invasiveness by increasing CXCR4
expression. Furthermore, the aberrant activation of NF-κB signaling
promotes lung tumorigenesis via the induction of
angiogenesis-related factors, such as VEGF and IL-8 (14). In addition to these findings,
accumulating evidence has indicated that the activation of NF-κB
signaling is essential for the bone metastasis of prostate cancers
(15,16). It has previously been demonstrated
that the NF-κB signaling system is also deregulated in GC (17). Further research has revealed that
RelA and NF-κB1/p50 are upregulated in GC and cancer cell lines and
that the expression of these proteins in GC tissue is strongly
associated with the abundance of other tumor- or
metastasis-promoting markers, including signal transducer and
activator of transcription (STAT)3, MMP-2 (18,19),
cyclooxygenase (COX)2 and VEGF (20,21).
In previous studies, the siRNA-mediated knockdown of RelA and
NF-κB1/p50 exerted an anti-tumor effect both in vitro and
in vivo (22,23). These results indicate that the
NF-κB signaling pathway may serve as a therapeutic target for the
treatment of GC. However, the underlying mechanisms of the
constitutive activation of NF-κB signaling in GC remain poorly
understood.
MicroRNAs (miRNAs or miRs), which are a series of
small non-coding RNAs composed of 18-24 nucleotides, function in
mRNA degradation and the post-transcriptional regulation of target
genes by specific binding to their 3'-untranslated region (3'-UTR)
(24,25). Abundant evidence has indicated that
the aberrant expression of miRNAs affects the capacity of cancer
cells to invade, migrate and metastasize (26,27).
Moreover, miRNAs have also been reported to serve as a modulator of
the NF-κB pathway. For example, miR-199a has been shown to activate
the NK-κB pathway and to be associated with the tumor inflammatory
microenvironment by regulating IKKβ (28). miR-146 also plays regulatory roles
in the NF-κB pathway, as it negatively regulates the protein levels
of IL-1 receptor-associated kinase 1 (IRAK1) and TNF
receptor-associated factor 6 (TRAF6) (29,30).
miRNA-301a, which is located on chromosome 17q22, has been shown to
be upregulated in a number of types of cancer, including
hepatocellular carcinoma, pancreatic cancer, small cell lung cancer
and breast cancer, which indicates a potential role for miRNA-301a
in cancer development (31-34).
In GC, Wang et al (35)
reported that the high expression of miRNA-301a was associated with
GC cell proliferation and invasion by targeting Runt-related
transcription factor 3 (RUNX3). In a previous study by the authors,
it was also found that the abnormal expression of miRNA-301a-3p in
GC was associated with progression and a poor prognosis (36). However, the underlying biological
processes and molecular mechanisms of action of miRNA-301a-3p in
GC, particularly as regards the regulation of the NK-κB pathway,
remain poorly understood.
In the present study, it was first found that the
upregulation of miRNA-301a-3p in GC was associated with tumor
progression and a worse prognosis. The function and molecular
mechanisms of miRNA-301a-3p were also investigated. An in
vitro assay indicated that the suppression of miRNA-301a-3p
attenuated cancer cell growth and migration, as well as tumor
progression. Additionally, the miRanda database was searched and it
was found that NF-κB repressing factor (NKRF) was a candidate
target gene of miRNA-301a-3p. A previous study indicated that NKRF
was involved in the negative regulation of NF-κB (37). These results demonstrated that the
upregulation of miRNA-301a-3p contributed to tumor progression in
GC by regulating NKRF expression, which led to the induction of
NF-κB activation and tumor growth. Therefore, this NF-κB activation
mechanism may be a target for therapeutic intervention in GC.
Materials and methods
Samples and patients
All fresh GC tissue and paired adjacent
non-cancerous samples were collected after obtaining written
informed consent from 30 patients who underwent resection for GC at
the Zhejiang Provincial People's Hospital from 2012 to 2013
(clinical characteristics are presented in Table SI). At the same time, a GC tissue
microarray (TMA) containing 120 GC tissue samples was collected at
the Zhejiang Provincial People's Hospital from 2012 to 2013 and
used for the in situ hybridization (ISH) detection of
miRNA-301a-3p (the characteristics of the cases are presented in
Table I). The histological tumor
type for all GC cases was diagnosed by 3 independent pathologists,
and all cases were classified according to the American Joint
Committee on Cancer (AJCC) classification (8th and 7th editions) of
GC tumors (38,39). The matched normal gastric
epithelial tissues, which were collected from an area at distance
of >5 cm from the tumors, were also verified at the same time.
None of the patients had received chemotherapy prior to surgery.
The study design and method were approved by the Ethics Committee
of Zhejiang Provincial People's Hospital. Upon admission, all
patients or their relatives provided informed consent within the
written treatment contract prior to their inclusion in the study.
All patients were followed-up for >5 years or until December,
2018. The survival time was calculated from the date of surgery to
the end of the follow-up period and/or the date of death. The age
of the patients with GC ranged from 29 to 82 years (median age,
61.22 years). The clinicopathological characteristics of the
patients with GC are summarized in Table I.
 | Table IAssociation between miRNA-301a
expression and clinicopathological factors. |
Table I
Association between miRNA-301a
expression and clinicopathological factors.
| Clinical
parameters | miR-301a expression
|
t/χ2 | P-value |
|---|
| Low | High |
|---|
| Age (years) | 60.02±12.97 | 61.96±10.97 | 2.266 | 0.135 |
| Sex | | | 1.984 | 0.159 |
| Male | 30 (65.2%) | 57 (77.0%) | | |
| Female | 16 (34.8%) | 17 (23.0%) | | |
| Location | | | 4.347 | 0.114 |
| Proximal | 7 (15.2%) | 12 (16.2%) | | |
| Middle | 24 (52.2%) | 25 (33.8%) | | |
| Distal | 15 (32.6%) | 37 (50.0%) | | |
| Tumor size | | | 4.971 | 0.026 |
| ≥5 cm | 19 (41.3%) | 46 (62.2%) | | |
| <5 cm | 27 (58.7%) | 28 (37.8%) | | |
| Histological
type | | | 0.509 | 0.917 |
| Papillary
adenocarcinoma | 1 (2.2%) | 2 (2.7%) | | |
| Tubular
adenocarcinoma | 36 (78.3%) | 56 (75.7%) | | |
| Mucinous
adenocarcinoma | 2 (4.3%) | 2 (2.7%) | | |
| Signet-ring cell
carcinoma | 7 (15.2%) | 14 (18.9%) | | |
| Lauren
classification | | | 6.398 | 0.011 |
| Diffuse type | 25 (54.3%) | 23 (31.1%) | | |
| Intestinal
type | 21 (45.7%) | 51 (68.9%) | | |
|
Differentiation | | | 5.530 | 0.063 |
| Well | 5 (10.9%) | 1 (1.4%) | | |
| Moderately | 10 (21.7%) | 20 (27.0%) | | |
| Poorly | 31 (67.4%) | 53 (71.6%) | | |
| Invasion depth (T
grade) | | | 31.685 | 6.10E-07 |
| T1 | 11 (23.9%) | 2 (2.7%) | | |
| T2 | 25 (54.3%) | 19 (25.7%) | | |
| T3 | 9 (19.6%) | 45 (60.8%) | | |
| T4 | 1 (3.2%) | 8 (10.8%) | | |
| Lymphatic
metastasis (N grade) | | | 17.880 | 4.66E-04 |
| N0 | 25 (54.3%) | 22 (29.7%) | | |
| N1 | 0 (0.0%) | 6 (8.1%) | | |
| N2 | 19 (41.3%) | 24 (32.4%) | | |
| N3 | 2 (4.3%) | 22 (29.7%) | | |
| Distant metastasis
(M grade) | | | 8.588 | 0.003 |
| M0 | 40 (87.0%) | 46 (62.2%) | | |
| M1 | 6 (13.0%) | 28 (37.8%) | | |
| TNM stage | | | 32.346 | 4.42E-07 |
| I | 21 (45.7%) | 5 (6.8%) | | |
| II | 17 (37.0%) | 24 (32.4%) | | |
| III | 2 (4.3%) | 17 (23.0%) | | |
| IV | 6 (13.0%) | 28 (37.8%) | | |
| TNM stage | | | 21.687 | 3.21E-06 |
| I + II | 38 (82.6%) | 29 (39.2%) | | |
| III + IV | 8 (17.4%) | 45 (60.8%) | | |
| Lymphatic
invasion | | | 3.487 | 0.062 |
| Yes | 11 (23.9%) | 30 (40.5%) | | |
| No | 35 (76.1%) | 44 (59.5%) | | |
| Vascular
invasion | | | 10.042 | 0.002 |
| No | 25 (54.3%) | 19 (25.7%) | | |
| Yes | 21 (45.7%) | 55 (74.3%) | | |
ISH detection of miRNA-301a-3p in a
GC
For the ISH detection of miRNA-301a-3p in GC
tissues, 5-µm-thick sections of the GC TMA were used. The sections
were deparaffinized, rehydrated and subjected to ISH signal
detection, as previously described (36). Briefly, a digoxin-labeled
LNA-miRNA-301a miRCURY probe (Qiagen, Inc.) was used to detect
miRNA-301a-3p expression in GC tissues. ISH was performed using a
Dig Labeled Probe Detection kit I (POD) (MK1003, Boster Biological
Technology) according to the manufacturer's instructions. For each
sample, the immunoreactivity levels of miRNA-301a-3p were estimated
under a light microscope by assessing the average signal intensity
(on a scale of 0-3). The proportion of cells that indicated
positive staining (0, <5%; 1, 5-25%; 2, 26-50%; 3, 51-75%; and
4, 76-100%) was independently estimated by 2 pathologists in the
absence of clinical information, as previously described (36). The intensity and percentage scores
were subsequently multiplied to obtain a composite score; a score
of 0 to 3 was defined as negative, while a score of 4 to 12 was
defined as positive.
Cells and cell culture
Human GC cell lines (AGS, MKN-45, HGC-27 and GES-1)
were purchased from the Cell Bank of Shanghai Institute of Cell
Biology and cultured in RPMI-1640 (HyClone; GE Healthcare Life
Sciences) supplemented with 10% fetal bovine serum (Sigma-Aldrich;
Merck KGaA) at 37°C in a humidified atmosphere containing 5%
CO2.
Transfection with miRNA inhibitor
The AGS and MKN-45 cells (1x105 cells per
well) were seeded in 6-well plates. After 24 h, the cells were
transfected with a micrOFF miRNA-301a inhibitor
(miR21111890035-1-5, RiboBio), micrON hsa-miR-301a-5p mimic
(miR10022696-1-5, RiboBio) and the corresponding negative controls
at a final concentration of 50 nM using Lipofectamine 3000
transfection reagent (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol, respectively. Following transfection
for 48 h, the cells were collected for use in further assays, such
as reverse transcription-quantitative polymerase chain reaction
(RT-qPCR), cell proliferation assay, invasion assay and western
blot analysis.
Inhibition of NF-κB activities by
dehydroxymethylepoxyquin- omicin (DHMEQ)
DHMEQ is a potent NF-κB inhibitor and it was
dissolved in dimethyl sulfoxide (DMSO) at 20 mg/ml and maintained
in aliquots at -30°C. Prior to use in the MKN-45 cell culture to
inhibit NF-κB activities, it was diluted with the medium to a final
concentration of 15 µg/ml and treated for 24 h to abrogate
constitutive NF-κB activity and downregulate NF-κB
transcription.
Immunofluorescence assay
MKN-45 cells were seeded onto glass coverslips in
12-well plates at 1x104 cells/well. After 24 h, the
cells were pre-treated with 15 µg/ml of DHMEQ for 16 h. The cells
were then washed twice with cold PBS (PBS) and fixed with
formaldehyde for 20 min, and then permeabilized with 0.1% Triton
X-100 in PBS for 20 min. After blocking with blocking buffer (1%
BSA in TBST) for 30 min, the cells were then incubated with NF-κB
p65 antibody (dilution, 1:500; cat. no. 8242, Cell Signaling
Technology, Inc.) overnight. After washing with TBST 3 times, the
cells were incubated the Cy3-labled secondary antibody (dilution,
1:5,000; cat. no. A0516, Beyotime Institute of Biotechnology, Inc.)
for 1 h at room temperature and counterstained with DAPI (cat. no.
C1005, Beyotime Institute of Biotechnology, Inc.) for 5 min at room
temperature. Fluorescence images were captured on a Leica SP2
confocal microscope (Leica Microsystems GmbH).
Luciferase reporter assays
The miRBase Targets (http://www.mirbase.org/), TargetScan Release 5.0
(http://www.targetscan.org/vert_72/)
and PicTar databases (https://pictar.mdc-berlin.de/) were searched and found
that NKRF may be a possible target of miRNA-301a-3p. The
pYr-mirTarget-NKRF-3'UTR-WT (site1, site2 and site3) and the
corresponding mutated luciferase vector, which contained the
putative binding site or mutated site of miRNA-301a-3p, were
purchased from Yinrun Biotechnology. 293 cells were purchased from
the Chinese Academy Of Sciences Cell Bank and seeded in 96-well
plates. The cells were then co-transfected with the wild-type (WT)
or mutated (Mut) reporter plasmids using Lipofectamine 3000 reagent
(Thermo Fisher Scientific, Inc.) along with the miRNA-301a-3p
inhibitor (100 nM). After 48 h, the luciferase activity was
assessed using the DualGlo Luciferase Assay System (Promega Corp.)
according to the manufacturer's instructions on a Sirius single
tube luminometer (Berthold Technologies, GmbH & Co. KG,). The
Firefly luciferase activity was normalized to Renilla
luciferase activity for each transfected well. All transfection
experiments were conducted in triplicate and were repeated
independently 3 times.
To further confirm NKRF as a target gene of
miRNA-301a-3p in GC cells, following the transfection of GC cells
(AGS and MKN-45) with the miRNA-301a-3p inhibitor and mimic, the
mRNA and protein expression levels of NKRF and downstream molecules
in NF-κB signaling were also assessed by RT-qPCR and western blot
analysis, respectively.
RNA isolation and RT-qPCR
Total RNA was isolated from the tissue samples and
GC cells according to the protocol of the RNAsimple Total RNA kit
[DP419; Tiangen Biotech (Beijing) Co., Ltd.]. Reverse transcription
was performed using a One-step PrimeScript miRNA cDNA synthesis kit
[D350A; Takara Biotechnology (Dalian) Co., Ltd]. and a PrimeScript™
RT reagent kit with gDNA Eraser [RR047A; Takara Biotechnology
(Dalian) Co., Ltd.]. To detect the expression levels of
miRNA-301a-3p, NKRF, NF-κB, VEGF, MMP-2 and MMP-9, RT-qPCR was
performed on an MX3000P system (Stratagene; Agilent Technologies,
Inc.) using gene-specific primers with a SYBR Premix ExTaq kit
[DRR081A; Takara Biotechnology (Dalian) Co., Ltd.]. All reactions
were performed in triplicate. U6 (RNU6B) and glyc-eraldehyde
3-phosphate dehydrogenase (GAPDH) were used as internal standards
for normalization of the gene expression levels. The primers of
candidate genes were selected from PrimerBank (https://pga.mgh.harvard.edu/primerbank/)
and are listed in Table II. The
qPCR reaction conditions were as follows: Initial denaturation (4
min at 95°C) and then 40 cycles of denaturation at 95°C for 20 sec,
annealing at 58°C for 20 sec, and extension at 72°C for 20 sec. The
relative expression levels were calculated using the
2-∆∆cq method (40).
 | Table IISequences of the primers used in the
present study. |
Table II
Sequences of the primers used in the
present study.
| Primer name | Sequence
(5'-3') | Annealing
temperature (°C) |
|---|
| miRNA-301a-3p |
CAGTGCAATAGTATTGTCAAAGC | 58 |
| U6B |
CGCTTCACGAATTTGCGTGTCAT | 58 |
| GAPDH-F |
ACAACTTTGGTATCGTGGAAGG | 50-60 |
| GAPDH-F |
GCCATCACGCCACAGTTTC | 50-60 |
| NKRF-F |
GTCAAAAACGCCACCTCTCAA | 55 |
| NKRF-R |
CTCGCATGGAATTTGGAACCG | 55 |
| NF-κB-F |
GGTGCGGCTCATGTTTACAG | 58 |
| NF-κB-R |
GATGGCGTCTGATACCACGG | 58 |
| VEGF-F |
AGGGCAGAATCATCACGAAGT | 58 |
| VEGF-R |
AGGGTCTCGATTGGATGGCA | 58 |
| MMP-9-F |
TGTACCGCTATGGTTACACTCG | 56 |
| MMP-9-R |
GGCAGGGACAGTTGCTTCT | 56 |
| MMP-2-F |
TACAGGATCATTGGCTACACACC | 58 |
| MMP-2-R |
GGTCACATCGCTCCAGACT | 58 |
| CXCL8-F |
ACTGAGAGTGATTGAGAGTGGAC | 55 |
| CXCL8-R |
AACCCTCTGCACCCAGTTTTC | 55 |
| PTGS2-F |
TAAGTGCGATTGTACCCGGAC | 56 |
| PTGS2-R |
TTTGTAGCCATAGTCAGCATTGT | 56 |
| STAT3-F |
ACCAGCAGTATAGCCGCTTC | 58 |
| STAT3-F |
GCCACAATCCGGGCAATCT | 58 |
| IL-6-R |
ACTCACCTCTTCAGAACGAATTG | 55 |
| IL-6-R |
CCATCTTTGGAAGGTTCAGGTTG | 55 |
| c-FLIP-F |
TGCTCTTTTTGTGCCGGGAT | 55 |
| c-FLIP-R |
CGACAGACAGCTTACCTCTTTC | 55 |
| BCL2L1-F |
GAGCTGGTGGTTGACTTTCTC | 58 |
| BCL2L1-R |
TCCATCTCCGATTCAGTCCCT | 58 |
| BIRC2-F |
GAATCTGGTTTCAGCTAGTCTGG | 58 |
| BIRC2-R |
GGTGGGAGATAATGAATGTGCAA | 58 |
| CCL2-F |
AGAATCACCAGCAGCAAGTGTCC | 56 |
| CCL2-R |
TCCTGAACCCACTTCTGCTTGG | 56 |
Cell proliferation assay
Cell proliferation was assessed using the Cell
Proliferation MTS Assay kit (G3580, Promega Corp.) following the
manufacturer's protocol. All cell lines (pre-transfected with the
miRNA-301a-3p inhibitor, mimic, and the corresponding negative
control cells) were seeded into 96-well plates with
3x103 cells in 200 µl culture medium per well. Following
attachment, 20 µl of MTS reagents was added to each well every 24
h. Following an additional 4-h incubation, the absorbance was
measured at 570 nm using a Tecan Infinite 200 microplate reader
(Tecan Group Ltd. Switzerland.)
Migration and invasion assays
The migration assay was performed using Transwell
plates containing membranes with 8 µm pores (3422; Corning Inc.).
Cell invasion assays were performed using invasion chambers
precoated with Matrigel (354480; BD Biosciences). The GC cells,
which were pre-transfected with the miRNA-301a-3p inhibitor or
mimic or 15 µg/ml DHMEQ, and the controls (2x105 for
invasion assays and 5x104 cells for migration assays)
were resuspended in serum-free medium and seeded into the upper
chamber. RPMI-1640 medium containing 20% FBS was added to the lower
chamber as a chemoattractant. After 24 or 48 h, the cells were
fixed and stained. Non-invading cells in the upper chambers were
removed with cotton swabs. The number of migrating or invading
cells that had attached to the lower surface was then counted in 5
random fields under an Olympus IX71 microscope (x200 magnification,
Olympus Corp.).
Western blot analysis
Briefly, protein was extracted from the cells using
radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime
Institute of Biotechnology). Sample protein concentrations were
quantitated using the BCA method and 30 µg total protein per lane
were separated by 10% SDS-PAGE and then transferred onto PVDF
membranes. The PVDF membranes were blocked with 5% BSA for 2 h and
then incubated with primary anti-human antibodies against NKRF
(ab168829, at 1:1,000 dilution; Abcam), MMP-9 (EM1801-22, at 1:500
dilution, HuaBio Inc.), MMP-2 (ER40806, at 1:1,000 dilution, HuaBio
Inc.), VEGF (ER30607, at 1:1,000 dilution, HuaBio Inc.), NF-κB-p65
(D14E12, #8242, at 1:2,000 dilution, Cell Signaling Technology,
Inc.) and GAPDH (M1310-2, at 1:3,000 dilution; HuaBio Inc.)
overnight at 4°C. The membranes were incubated with the
corresponding horseradish peroxidase (HRP)-labeled goat anti-rabbit
(A0208, at 1:10,000 dilution; Beyotime Institute of Biotechnology
Inc.) or anti-mouse IgG antibody (A0216, at 1:10,000 dilution;
Beyotime Institute of Biotechnology Inc.) for 1 h at room
temperature. After washing, the WB signal was detected using an
enhanced chemiluminescence system (Bio-Rad Laboratories, Inc.) and
the densitometric analysis of the images was performed using Image
Lab 6.0.1 software (Bio-Rad Laboratories, Inc.).
Statistical analyses
All statistical analyses were performed using
Statistical Package for the Social Sciences version 13.0 (SPSS
Inc.) and Prism software. Statistically significant differences in
miRNA-301a-3p expression between cancer tissues and normal tissues
were determined by a two-tailed paired Student's t-test. Data from
the comparisons of multiple groups from the cell migration and cell
invasion assays, luciferase assay, western blot analysis and
RT-qPCR are expressed as the means ± SE, and the significant
differences were determined by the one-way ANOVA S-N-K method and
Tukey's post hoc test. The association between miRNA-301a
expression and the patient clinicopathological characteristics was
determined using the Chi-square test. Survival curves were plotted
using the Kaplan-Meier method and were compared using the log-rank
test. The significance of various survival-related variables was
assessed by a Cox regression model in a multivariate analysis in
this GC cohort. A P-value <0.05 (P<0.05) was considered to
indicate a statistically significant difference.
Results
miRNA-301a-3p is upregulated in GC
tissues and GC cell lines
The results of RT-qPCR indicated that miRNA-301a-3p
expression in the GC tissues was significantly higher than that in
the matched normal tissues (0.001727±0.002114 vs. 0.000172±0.00012,
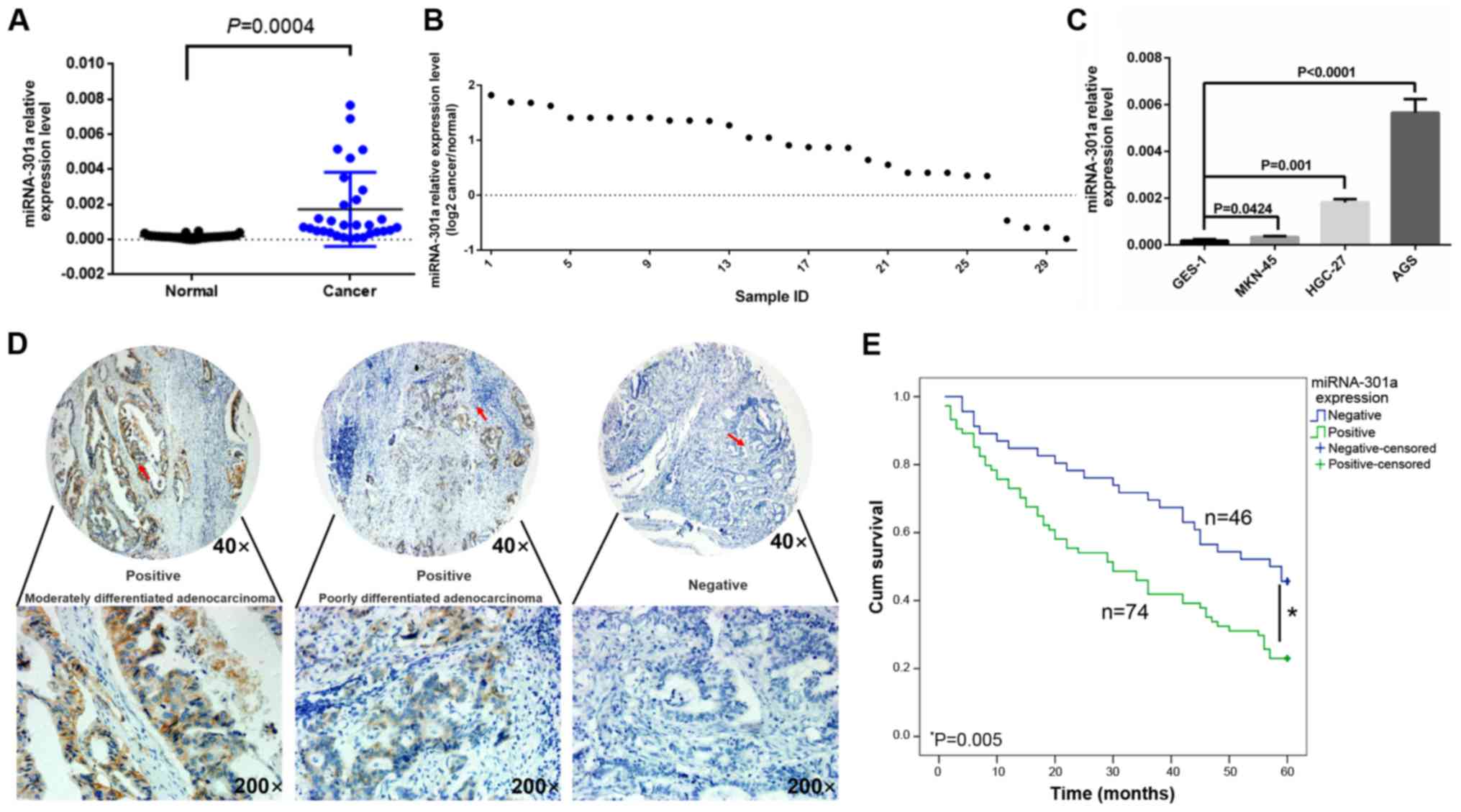
t=3.998, P<0.05) (Fig. 1A).
Among these 30 cases of GC, only 4 cases were found to have a lower
miRNA-301a-3p expression in the cancer tissues than in the adjacent
non-cancerous tissue (Fig. 1B).
Moreover, miRNA-301a-3p expression was increased in all 4 cancer
cell lines compared with the normal gastric epithelial cell line,
GES-1. Among these, the AGS cells exhibited the highest expression
level of miRNA-301a-3p and the MKN-45 cells the lowest (Fig. 1C). These 2 cell lines were
therefore selected for use in miRNA-301a-3p inhibition and mimic
in vitro experiments, respectively.
Clinical significance of miRNA-301a-3p in
GC
To further investigate the association between
miRNA-301a-3p expression and patient clinicopathological
characteristics, miRNA-301a-3p expression was validated
independently by ISH assay using a TMA with 120 clinical GC tissue
samples (Table I). Based on the
miRNA-301a-3p immunoreactive scores, high levels of miRNA-301a-3p
expression were detected in 74 (61.7%) of tumors, while low levels
of miRNA-301a-3p expression were detected in 46 (38.3%) of tumors
(Fig. 1D). miRNA-301a-3p
expression was found to be associated with tumor size, Lauren
classification, vascular invasion, invasion depth (T grade),
lymphatic metastasis (N grade) and distant metastasis (P<0.05,
Table I); however, it was not
associated with sex, tumor location, age, histological type,
lymphatic invasion and differentiation (P>0.05, Table I).
In patients with a tumor size ≥5 cm, the
miRNA-301a-3p positivity rate was higher than that in patients with
a tumor size <5 cm (70.8 vs. 50.9%, P<0.05). The
miRNA-301a-3p positivity rate in GC patients with the intestinal
type was higher than that in patients with the diffuse type (70.8
vs. 47.9%, P<0.05).
The miRNA-301a-3p positivity rate in patients with
lymph node metastasis (71.2%, or 52/73) was higher than that in
patients without lymph node metastasis (46.8%, or 22/47,
P<0.05). The miRNA-301a-3p positivity rate in patients with
distant metastasis (82.4%, or 28/34) was also higher than that in
patients without distant metastasis (53.5%, or 46/86; P<0.05).
In patients with GC, the miRNA-301a-3p positivity rate was
significantly increased from 19.2% (grade I) to 82.4% (grade IV,
P<0.05). Additionally, patients with vascular invasion also
exhibited a higher positivity rate than those without vascular
invasion (72.3%, 55/76 vs. 43.2%, 19/44, P<0.05, Table I).
High miRNA-301a-3p expression in GC is
associated with a poor prognosis
The association between the miRNA-301a-3p expression
level and the prognosis of patients with GC was also analyzed. In
this cohort of patients in the TMA (n=120), the 5-year survival
rate was 31.7%, and the mean survival time was 36.77±1.99 months.
By contrast, the mean survival time of miRNA-301a-3p-positive
patients was significantly shorter than that of
miRNA-301a-3p-negative patients (32.31±2.53 vs. 43.94±2.94 months
for miRNA-301a-3p-positive and -negative patients, respectively,
P<0.05). The 5-year survival rate of miRNA-301a-3p-positive
patients (23.0%) was also significantly lower than that of
miRNA-301a-3p-negative patients (45.7%, P<0.05, Fig. 1E). A Cox multivariate analysis
showed that Lauren classification, differentiation and distant
metastasis were independent prognostic factors, whereas
miRNA-301a-3p expression was not an independent prognostic factor
in this cohort of patients with GC (Table III).
 | Table IIIMultivariate analysis as determined
by Cox regression analysis in 120 patients with GC. |
Table III
Multivariate analysis as determined
by Cox regression analysis in 120 patients with GC.
| Clinicopathological
parameters | 95% CI
| HR
| P-value |
|---|
| Lower | Upper |
|---|
| Lauren
classification | 1.217 | 6.248 | 5.906 | 0.015 |
|
Differentiation | 0.347 | 0.986 | 4.045 | 0.044 |
| Distant
metastasis | 1.234 | 16.110 | 5.204 | 0.024 |
| miRNA-301a
expression | 0.400 | 1.440 | 0.713 | 0.399 |
miRNA-301a-3p expression affects GC cell
proliferation, invasion and migration
To evaluate the possible role of miRNA-301a-3p in
the proliferation and invasiveness of human GC cells, the AGS and
MKN-45 cells were selected for the transfection assay as these 2
cell lines were found to have the highest or lowest endogenous
miRNA-301a-3p expression out of all the cell lines tested (Fig. 1C).
The results of MTS assay revealed that when
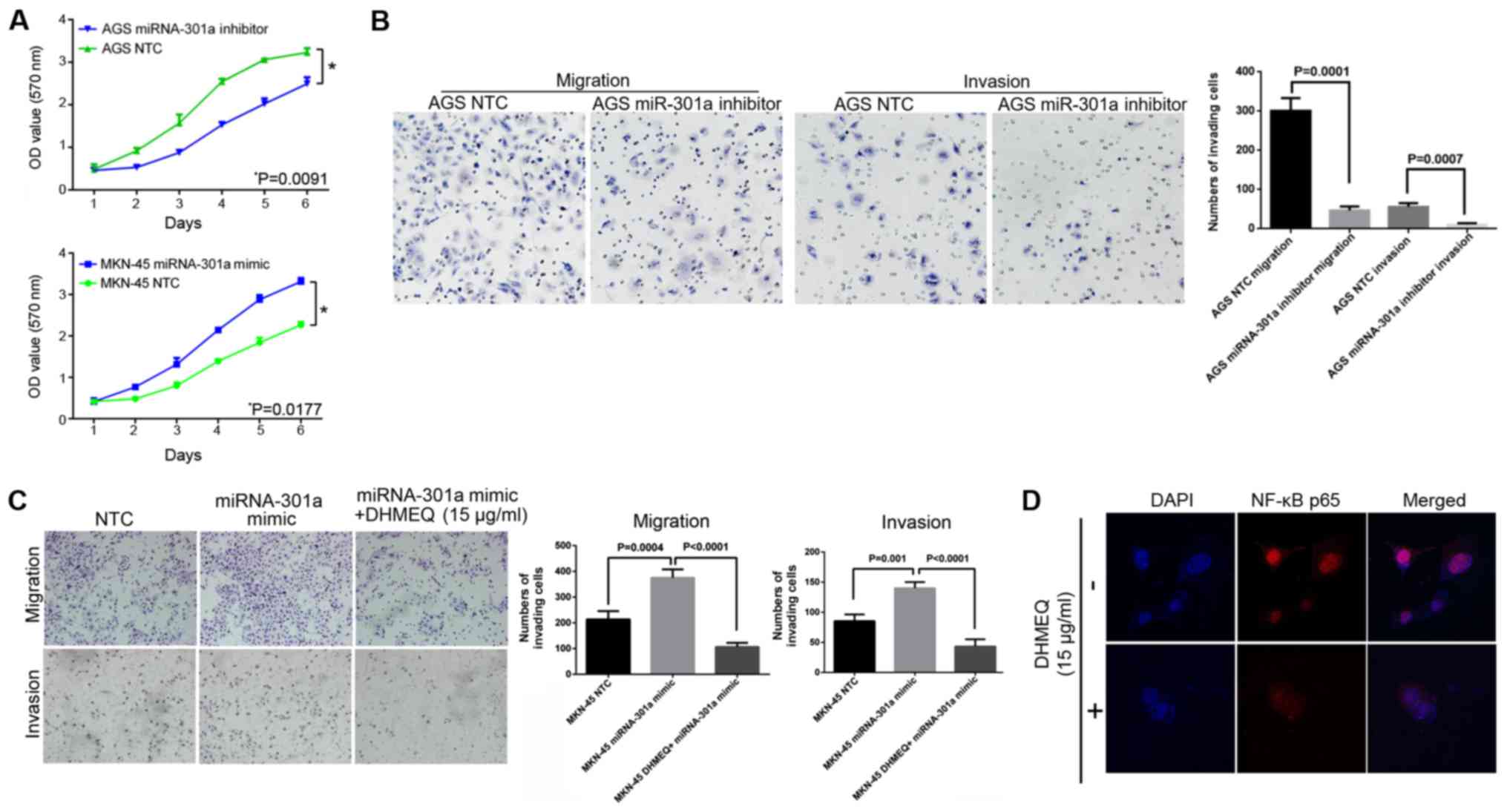
miRNA-301a-3p expression was inhibited in the AGS cells, they
exhibited a significantly slower proliferation rate compared with
the negative control cells (Fig.
2A, top panel; P<0.05). At the same time, as was expected,
the results of migration and invasion assays indicated that
following the inhibition of miRNA-301a-3p in the AGS cells, the
migratory and invasive abilities were significantly decreased
compared with the control cells (Fig.
2B; P<0.05). Furthermore, after increasing the miRNA-301a-3p
expression in the MKN-45 cells, the proliferative, migratory and
invasive abilities were significantly increased (Fig. 2A, bottom panel and C;
P<0.05).
miRNA-301a-3p targets NKRF and involves
NF-κB signaling
To investigate the possible mechanisms through which
miRNA-301a-3p affects GC cell invasiveness, the miRBase Targets,
TargetScan Release 5.0 and PicTar databases were searched and found
that NKRF may be a possible target of miRNA-301a-3p. To clarify
whether miRNA-301a-3p interacts directly with the 3'UTR region of
NKRF, a binding site investigation was performed and it was found
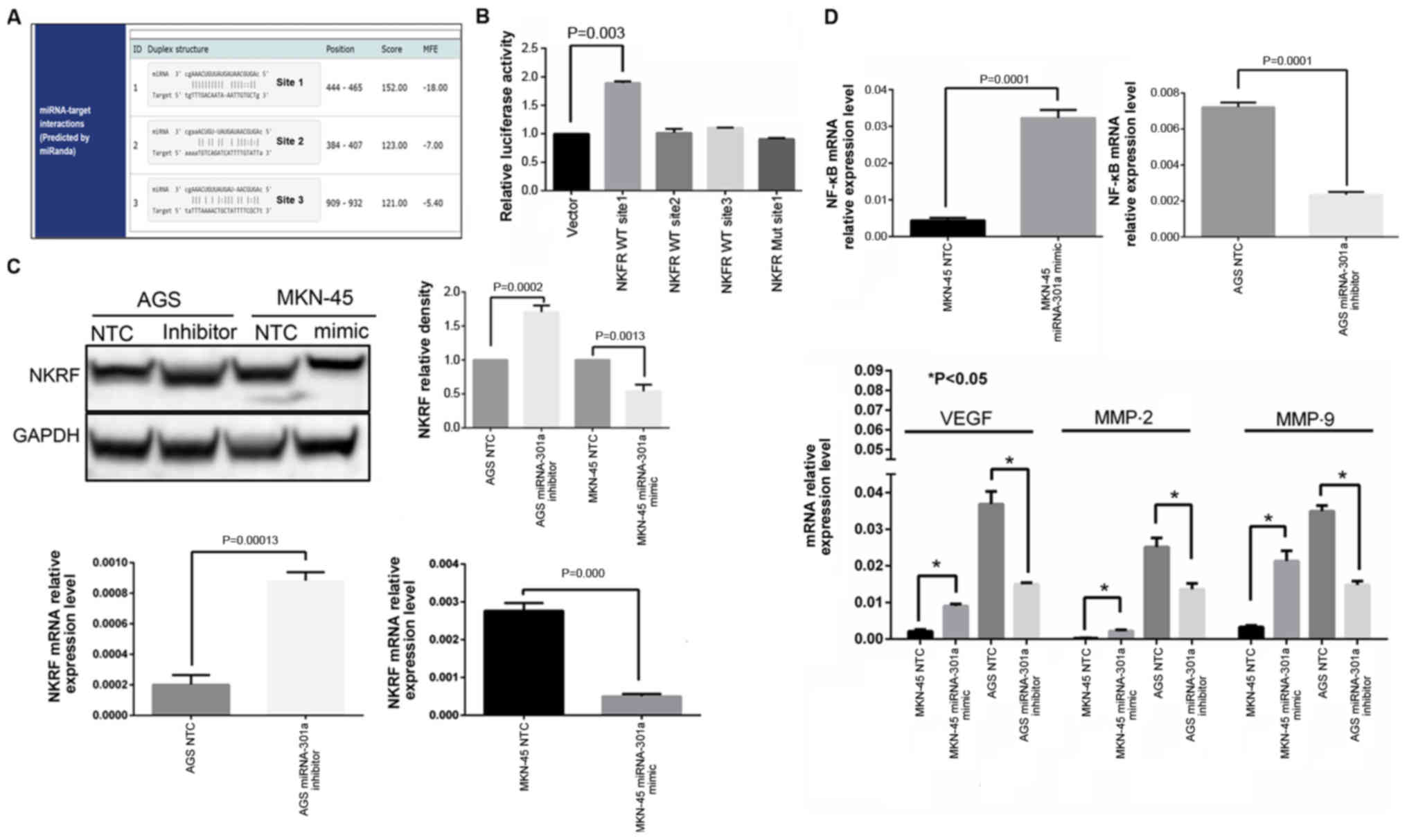
that the 3'UTR region of NKRF mRNA contained 3 miRNA-301a-3p
binding sites (Fig. 3A). Thus,
these 3 WT sequences of the human NKRF 3'UTR regions were inserted
into a luciferase reporter vector and then co-transfected into 293
cells with the miRNA-301a-3p inhibitor. The results of the
luciferase assay indicated that the relative luciferase activity
from NKRF WT site 1 group was significantly increased in the
presence of the miRNA-301a-3p inhibitor compared with the negative
control (P<0.05, Fig. 3B),
whereas other two WT sites (site 2 and site 3) of the NKRF 3'UTR
exhibited no significant changes in luciferase activity. These data
indicated that site 1 of the NKRF 3'UTR region may be a true
binding site of miRNA-301a-3p. To confirm this finding, the
corresponding mutated binding regions of site 1 of the NKRF 3'UTR
(NKRF Mut site 1) were cloned into the luciferase reporter vector,
which was subsequently co-trans-fected into the cells with the
miRNA-301a-3p inhibitor. Finally, unlike the NKRF WT site 1 group,
comparing with negative control group the miRNA-301a-3p inhibitor
did not alter the activity of luciferase of NKRF MUT site 1 group
(Fig. 3B). Thus, these results
indicated that miRNA-301a-3p specifically and directly targeted
NKRF by binding to the predicted site (site 1) of the NKRF 3'UTR
region.
Furthermore, the results of RT-qPCR and western blot
analysis also revealed that the NKRF expression levels were
inversely associated with miRNA-301a-3p expression in GC cells at
both the mRNA and protein level (Fig.
3C).
A previous study indicated that the NKRF protein
binds to NF-κB by a direct protein-protein interaction, and in a
cellular assay, NKRF inhibited NF-κB basal activity (37). Further research revealed that NF-κB
signaling was activated in GC and cancer cell lines and that its
expression in GC tissue was strongly associated with the abundance
of other tumor- or metastasis-promoting markers, including STAT3,
MMP-2, MMP-9, VEGF and others (21,22),
which are essential for GC progression (10). Therefore, these downstream
molecules of NF-κB signaling were also investigated in the present
study. From the RT-qPCR screening results and validation by western
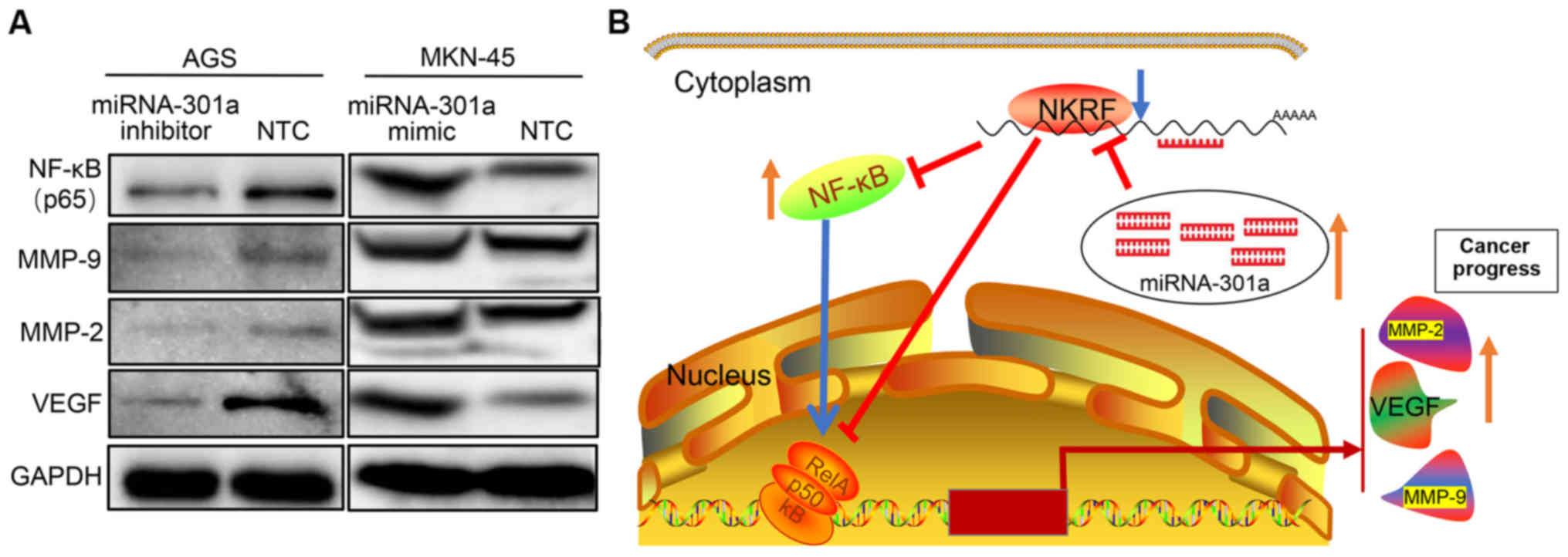
blot analysis, it was confirmed that NKRF expression was
significantly increased in the AGS cells transfected with the
miRNA-301a-3p inhibitor, while the expression of NF-κB and its
downstream effector molecules, such as MMP-2, MMP-9 and VEGF was
significantly decreased (Figs. 3C and
D, and 4A). By contrast, when
miRNA-301a-3p expression was increased in the MKN-45 cells, the
expression of NKRF was significantly decreased, whereas the
expression of NF-κB, MMP-2, MMP-9 and VEGF was significantly
increased (Figs. 3C and D, and
4A). However, no significant
changes were observed in the levels of other downstream molecules
of the NF-κB pathway, such as STAT3, IL-6, IL-8 (as known as
CXCL8), MCP-1 (as known as CCL2), COX2 (as known as PTGS2) and the
pro- and anti-apoptotic factors, cIAP1 (as known as BIRC2), c-FLIP
and BCL2L1 (as known as Bcl-xL), in GC cells in which miRNA-301a-3p
expression was downregulated (data not shown).
In order to verify whether the NF-κB pathway was
involved in the miRNA-301a-3p-induced invasiveness of GC, the
specific NF-κB inhibitor, DHMEQ, was used. It was found that, after
pre-blocking NF-κB activities in the MKN-45 cells, the promoting
effects of miRNA-301a-3p mimic on cell invasion and migration were
inhibited (Fig. 2C and D). Thus,
these data indicate that miRNA-301a-3p is an important regulator of
the candidate target gene NKRF, which participates in GC
progression by regulating the NF-κB signaling axis.
Discussion
A number of miRNAs have been reported to function as
oncogenes or tumor suppressors in cancer through their epigenetic
regulation of target gene expression; these miRNAs form a complex
regulatory network that influences cancer invasion, metastasis,
drug resistance, stemness, EMT and signaling pathways (41,42).
miRNA-301a has been reported to be upregulated in a number of types
of cancer, which indicates that miRNA-301a serves as an important
tumor gene in cancer development (43-45).
In the present study, miRNA-301a-3p expression was compared in 30
paired GC tissues and normal gastric tissues and the findings
provide important evidence to support the upregulation of
miRNA-301a-3p in GC tissue. Further analysis of clinical samples
using the ISH method demonstrated that a high level of
miRNA-301a-3p was associated with tumor size, differentiation,
invasion depth, lymphatic metastasis, TNM stage, vascular invasion
and poor prognosis. The in vitro Transwell and MTS assays
also indicated that decreased miRNA-301a-3p levels in GC cells
inhibited GC cell proliferative, invasive and migratory abilities.
These findings support the hypothesis that miRNA-301a-3p may play a
tumorigenic role in GC.
NF-κB comprises a family of transcription factors
that are involved in the regulation of a wide variety of biological
responses. NF-κB was first found to have multiple functions in the
regulation of immune responses and inflammation; however, in recent
years, increasing evidence has indicated that NF-κB may play a
major role in oncogenesis (12).
NF-κB and its downstream genes are involved in a number of
processes that participate in the development and progression of
cancer such as proliferation, migration and apoptosis (12). Aberrant or constitutive NF-κB
activation, which affects apoptosis, proliferation and
invasiveness, has been detected in several types of cancer
(12). In recent years, numerous
studies have focused on elucidating the functional consequences of
NF-κB activation, and miRNAs were found to be important regulators
of the NF-κB signaling axis (46,47).
In the present study, to explore the possibility that miRNA-301a-3p
is involved in NF-κB activation in GC, a strategy was formulated
with which to screen target genes of miRNA-301a-3p that could
modulate NF-κB signaling. It was found that NKRF was the most
potent target involved in NF-κB signaling regulation. NKRF is known
as a transcriptional silencer protein that binds negative
regulatory elements (NREs) specific for the suppression of
NF-κB/Rel basal activity as well as several NF-κB regulated
downstream genes (37). NKRF has
also been shown to interact with NF-κB/p65 through a minimal-core
sequence under basal or stimulated conditions (37,48).
Therefore, multiple external factors that induce or suppress NKRF
expression may contribute to NF-κB-driven transcriptional
regulation, with important implications in inflammation and cancer.
The luciferase assay in 293 cells demonstrated that miRNA-301a-3p
epigenetically targets and regulates NKRF by binding to its 3'-UTR
regions. This reverse targeted regulatory mechanism is supported by
the finding that downregulation of miRNA-301a-3p expression in GC
cells enhances NKRF gene expression at both the mRNA and protein
levels. At the same time, to inhibit the expression of
miRNA-301a-3p in GC cells, the NF-κB expression level was
downregulated, whereas the NF-κB expression level was upregulated
by miRNA-301a-3p mimic. Furthermore, the levels of a number of
downstream molecules of NF-κB signaling that are involved in cancer
invasion and progression, such as MMP-9, MMP-2 and VEGF, were also
decreased according to miRNA-301a-3p inhibition. Substantial
evidence has already validated that MMP-9, MMP-2 and VEGF are
highly expressed in GC and are important for GC invasion and
progression and are even associated with a poor prognosis (49). Thus, miRNA-301a-3p may promote GC
cell invasion by affecting the expression of these genes by NF-κB
signal regulation. Taken together, these results demonstrated that
miRNA-301a-3p may promote GC invasion and progression by targeting
NKRF expression and then involving the NF-κB signaling pathway.
However, in order to increase the opportunity to suppress GC
progression, additional research is required to determine the exact
mechanisms underlying miRNA-301a-3p-mediated functions in GC
cells.
In conclusion, the results of the present study
indicated that the dysregulation of miRNA-301a-3p may play a role
in the tumor progression and the prognosis of GC patients.
Furthermore, these data suggested that miRNA-301a-3p may modulate
GC cell invasion and progression by directly and negatively
regulating NKRF and then activating the NF-κB pathway. Therefore,
the restoration of miRNA-301a-3p expression or the targeted
inhibition of NF-κB activity may be a potential therapeutic
strategy for GC.
Supplementary Data
Funding
The present study was supported by grants from the
Medicine and Health Research Foundation of Zhejiang Province
(2013KYB022, 2014KYA160 and 2020357780), the National
Natural Science Foundation of China (81502090), the
Zhejiang Provincial Natural Science Foundation (LY14H160039,
LY18H160043, Y18H030007, Y18H160037 and 2013RCA002), the Zhejiang
Provincial Medical Science Research Foundation (2011RCA004) and The
Taizhou science and technology project (1802ky85).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
XH made a substantial contribution to the conception
and design of the study. XX, YX, WL, NN and XL participated in the
experimental design, conducted the experiment, and conducted data
analysis and interpretation. JM and JX performed the in situ
hybridization data analysis. XX, XH and XL wrote and modified the
manuscript. HT, YX and XH analyzed and interpret the data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All fresh GC tissue samples were collected with the
written informed consent of the 30 patients who underwent GC
resection. The study design and method were approved by the Ethics
Committee of Zhejiang Provincial People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Voutsadakis IA: Epithelial-Mesenchymal
Transition (EMT) and Regulation of EMT Factors by Steroid Nuclear
Receptors in Breast Cancer: A Review and in Silico Investigation. J
Clin Med. 5:52016. View Article : Google Scholar
|
|
2
|
Nie Y, Wu K, Yu J, Liang Q, Cai X, Shang
Y, Zhou J, Pan K, Sun L, Fang J, et al: A global burden of gastric
cancer: The major impact of China. Expert Rev Gastroenterol
Hepatol. 11:651–661. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Strong VE, Wu AW, Selby LV, Gonen M, Hsu
M, Song KY, Park CH, Coit DG, Ji JF and Brennan MF: Differences in
gastric cancer survival between the U.S. and China. J Surg Oncol.
112:31–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feng RM, Zong YN, Cao SM and Xu RH:
Current cancer situation in China: Good or bad news from the 2018
Global Cancer Statistics? Cancer Commun (Lond). 39:222019.
View Article : Google Scholar
|
|
5
|
Monisha J, Roy NK, Bordoloi D, Kumar A,
Golla R, Kotoky J, Padmavathi G and Kunnumakkara AB: Nuclear Factor
Kappa B: A Potential Target to Persecute Head and Neck Cancer. Curr
Drug Targets. 18:232–253. 2017. View Article : Google Scholar
|
|
6
|
Fouani L, Kovacevic Z and Richardson DR:
Targeting Oncogenic Nuclear Factor Kappa B Signaling with
Redox-Active Agents for Cancer Treatment. Antioxid Redox Signal.
30:1096–1123. 2019. View Article : Google Scholar
|
|
7
|
Wang Y, Ni H, Li H, Deng H, Xu LS, Xu S,
Zhen Y, Shen H, Pan H and Yao M: Nuclear factor kappa B regulated
monocyte chemoat-tractant protein-1/chemokine CC motif receptor-2
expressing in spinal cord contributes to the maintenance of
cancer-induced bone pain in rats. Mol Pain.
14:17448069187886812018. View Article : Google Scholar
|
|
8
|
Fujimura T, Kambayashi Y, Furudate S,
Kakizaki A, Hidaka T and Aiba S: Possible mechanisms of the
crosstalk between Langerhans cells and regulatory T cells in
extramammary Paget disease by receptor activator of nuclear factor
kappa B (RANK) ligand/RANK pathways. Br J Dermatol. 176:387–394.
2017. View Article : Google Scholar
|
|
9
|
Cao HJ, Fang Y, Zhang X, Chen WJ, Zhou WP,
Wang H, Wang LB and Wu JM: Tumor metastasis and the reciprocal
regulation of heparanase gene expression by nuclear factor kappa B
in human gastric carcinoma tissue. World J Gastroenterol.
11:903–907. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tomonaga M, Hashimoto N, Tokunaga F,
Onishi M, Myoui A, Yoshikawa H and Iwai K: Activation of nuclear
factor-kappa B by linear ubiquitin chain assembly complex
contributes to lung metastasis of osteosarcoma cells. Int J Oncol.
40:409–417. 2012.
|
|
11
|
Gambhir S, Vyas D, Hollis M, Aekka A and
Vyas A: Nuclear factor kappa B role in inflammation associated
gastrointestinal malignancies. World J Gastroenterol. 21:3174–3183.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dolcet X, Llobet D, Pallares J and
Matias-Guiu X: NF-kB in development and progression of human
cancer. Virchows Arch. 446:475–482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Helbig G, Christopherson KW II,
Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE and
Nakshatri H: NF-kappaB promotes breast cancer cell migration and
metastasis by inducing the expression of the chemokine receptor
CXCR4. J Biol Chem. 278:21631–21638. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai Z, Tchou-Wong KM and Rom WN: NF-kappaB
in lung tumorigenesis. Cancers (Basel). 3:4258–4268. 2011.
View Article : Google Scholar
|
|
15
|
Tong HB, Zou CL, Qin SY, Meng J, Keller
ET, Zhang J and Lu Y: Prostate cancer tends to metastasize in the
bone-mimicking microenvironment via activating NF-κB signaling. J
Biomed Res. 32:343–353. 2018.PubMed/NCBI
|
|
16
|
Khurana N and Sikka SC: Targeting
Crosstalk between Nrf-2, NF-κB and Androgen Receptor Signaling in
Prostate Cancer. Cancers (Basel). 10:102018. View Article : Google Scholar
|
|
17
|
Sokolova O and Naumann M: NF-κB Signaling
in Gastric Cancer. Toxins (Basel). 9:92017. View Article : Google Scholar
|
|
18
|
Jiang XJ, Lin J, Cai QH, Zhao JF and Zhang
HJ: CDH17 alters MMP-2 expression via canonical NF-κB signalling in
human gastric cancer. Gene. 682:92–100. 2019. View Article : Google Scholar
|
|
19
|
Xu GY and Tang XJ: Troxerutin (TXN)
potentiated 5-Fluorouracil (5-Fu) treatment of human gastric cancer
through suppressing STAT3/NF-κB and Bcl-2 signaling pathways.
Biomed Pharmacother. 92:95–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang F, Yao Y, Wu J, Liu Q, Zhang J, Pu
X, Zhang Q and Xia L: Curcumin inhibits gastric cancer-derived
mesenchymal stem cells mediated angiogenesis by regulating
NF-κB/VEGF signaling. Am J Transl Res. 9:5538–5547. 2017.
|
|
21
|
Li YS, Wu LP, Li KH, Liu YP, Xiang R,
Zhang SB, Zhu LY and Zhang LY: Involvement of nuclear factor κB
(NF-κB) in the downregulation of cyclooxygenase-2 (COX-2) by
genistein in gastric cancer cells. J Int Med Res. 39:2141–2150.
2011. View Article : Google Scholar
|
|
22
|
Chen W, Wang X, Bai L, Liang X, Zhuang J
and Lin Y: Blockage of NF-kappaB by IKKbeta- or RelA-siRNA rather
than the NF-kappaB super-suppressor IkappaBalpha mutant potentiates
adriamycin-induced cytotoxicity in lung cancer cells. J Cell
Biochem. 105:554–561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lun M, Zhang PL, Pellitteri PK, Law A,
Kennedy TL and Brown RE: Nuclear factor-kappaB pathway as a
therapeutic target in head and neck squamous cell carcinoma:
Pharmaceutical and molecular validation in human cell lines using
Velcade and siRNA/NF-kappaB. Ann Clin Lab Sci. 35:251–258.
2005.PubMed/NCBI
|
|
24
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, Maturation, Target Recognition and Regulatory
Functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
26
|
Xu X, Yang X, Xing C, Zhang S and Cao J:
miRNA: The nemesis of gastric cancer (Review). Oncol Lett.
6:631–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Singh R, Ramasubramanian B, Kanji S,
Chakraborty AR, Haque SJ and Chakravarti A: Circulating microRNAs
in cancer: Hope or hype? Cancer Lett. 381:113–121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen R, Alvero AB, Silasi DA, Kelly MG,
Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T and Mor G:
Regulation of IKKbeta by miR-199a affects NF-kappaB activity in
ovarian cancer cells. Oncogene. 27:4712–4723. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee HM, Kim TS and Jo EK: MiR-146 and
miR-125 in the regulation of innate immunity and inflammation. BMB
Rep. 49:311–318. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Öztemur Islakoğlu Y, Noyan S and Gür
Dedeoğlu B: hsa-miR-301a- and SOX10-dependent miRNA-TF-mRNA
regulatory circuits in breast cancer. Turk J Biol. 42:103–112.
2018. View Article : Google Scholar
|
|
32
|
Xia X, Zhang K, Cen G, Jiang T, Cao J,
Huang K, Huang C, Zhao Q and Qiu Z: MicroRNA-301a-3p promotes
pancreatic cancer progression via negative regulation of SMAD4.
Oncotarget. 6:21046–21063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou P, Jiang W, Wu L, Chang R, Wu K and
Wang Z: miR-301a is a candidate oncogene that targets the homeobox
gene Gax in human hepatocellular carcinoma. Dig Dis Sci.
57:1171–1180. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Z, Li Y and Zhang G: Downregulation of
microRNA-301a inhibited proliferation, migration and invasion of
non-small cell lung cancer by directly targeting DLC1. Oncol Lett.
14:6017–6023. 2017.PubMed/NCBI
|
|
35
|
Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu
Y, Gu Q, Zhu Z and Liu B: Overexpressed miR-301a promotes cell
proliferation and invasion by targeting RUNX3 in gastric cancer. J
Gastroenterol. 48:1023–1033. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu XD, He XJ, Tao HQ, Zhang W, Wang YY, Ye
ZY and Zhao ZS: Abnormal expression of miR-301a in gastric cancer
associated with progression and poor prognosis. J Surg Oncol.
108:197–202. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Coccia M, Rossi A, Riccio A, Trotta E and
Santoro MG: Human NF-κB repressing factor acts as a
stress-regulated switch for ribosomal RNA processing and nucleolar
homeostasis surveillance. Proc Natl Acad Sci USA. 114:pp.
1045–1050. 2017, View Article : Google Scholar
|
|
38
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The Eighth Edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
'personalized' approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 7:3077–3079. 2010.
View Article : Google Scholar
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Method Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
41
|
Zaravinos A: The Regulatory Role of
MicroRNAs in EMT and Cancer. J Oncol. 2015:8658162015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma F, Zhang J, Zhong L, Wang L, Liu Y,
Wang Y, Peng L and Guo B: Upregulated microRNA-301a in breast
cancer promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014. View Article : Google Scholar
|
|
44
|
Ni Z, Shang XF, Wang YF, Sun YJ and Fu DJ:
Upregulated microRNA-301a in osteosarcoma promotes tumor
progression by targeting CDC14A. Genet Mol Res. 15:152016.
View Article : Google Scholar
|
|
45
|
Liu L, Nie J, Chen L, Dong G, Du X, Wu X,
Tang Y and Han W: The oncogenic role of microRNA-130a/301a/454 in
human colorectal cancer via targeting Smad4 expression. PLoS One.
8:e555322013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bano N, Yadav M, Mohania D and Das BC: The
role of NF-κB and miRNA in oral cancer and cancer stem cells with
or without HPV16 infection. PLoS One. 13:e02055182018. View Article : Google Scholar
|
|
47
|
Alderton GK: Microrna: Micro loops in NF
κB signalling. Nat Rev Cancer. 11:62011. View Article : Google Scholar
|
|
48
|
Qiu W, He JH, Zuo H, Niu S, Li C, Zhang S,
Weng S, He J and Xu X: Identification, characterization, and
function analysis of the NF-κB repressing factor (NKRF) gene from
Litopenaeus vannamei. Dev Comp Immunol. 76:83–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zheng H, Takahashi H, Murai Y, Cui Z,
Nomoto K, Niwa H, Tsuneyama K and Takano Y: Expressions of MMP-2,
MMP-9 and VEGF are closely linked to growth, invasion, metastasis
and angiogenesis of gastric carcinoma. Anticancer Res. 26(5A): pp.
3579–3583. 2006, PubMed/NCBI
|