The development of molecular characterization of
tumors two decades ago led to a revolution in cancer research and
therapeutic approaches. In the case of breast cancer, 'molecular
portraits' provided by the quantitative analysis of gene expression
patterns (1) have helped define
different intrinsic subtypes while multiparameter molecular tests
such as the PAM-50 provide useful prognostic information (2). However, despite the usefulness of
mRNA-based classifications, integrated proteogenomic analyses of
breast or colon tumors have revealed that protein abundance cannot
be reliably predicted from DNA- or RNA-level measurements as they
are only modestly correlated. In particular, proteomics identify
colorectal cancer subtypes similar to those detectable by
transcriptome profiles, but also reveal features not detectable in
transcript profiles, probably as a result of post-transcriptional
regulation (3). Further evidence
of this unique potential of proteomics-based subtyping was recently
provided in the study by Johansson et al who identified
protein products mapping to non-coding genomic regions, potentially
leading to a new class of tumor-specific immunotherapeutic targets
(4).
High-throughput proteomics is still an
underdeveloped field compared to transcriptomics (4) and its contribution to oncology has
probably not yet been fully realized. However, in two decades,
quantitative proteomics has rapidly evolved both technologically
(5) and strategically, allowing
researchers to explore the complexity of protein interaction
networks in a wide variety of situations, but also to formulate new
hypotheses to be further functionally tested (6). To identify tumor biomarkers to assist
in individualizing treatments for certain types of cancer, many
complementary technologies have also been developed (7). As an example, for the majority of the
common causes of cancer-related mortality worldwide, such as lung
cancer, these technologies have led to the identification of
predictive biomarkers of drug resistance, candidate biomarkers for
diagnosis and prognostic biomarkers (8). To deal with the limited success of
'targeted therapies', quantitative proteomics, together with other
major technological and conceptual developments, has reinforced the
search for characteristic features of the adhesive-migratory
phenotype of malignant cells (9).
Some other examples of important contributions include the study of
RNA-protein complexes (10), stem
cell plasticity (11), chromatin
remodeling (12) and more
recently, the regulation of mitochondrial function and dynamics
(13), shed microvesicles biology
(14) or mechanisms of
radioresistance (15).
It was recently demonstrated that proteomes and
transcrip-tomes were better associated in highly proliferative
tumors than in lowly proliferative tumors (4). Herein, data on cancer invasiveness
are reviewed with the aim of highlighting key findings within the
huge amount of information available in the literature. The present
review focuses in particular on a list of 76 proteins of interest,
selected after crossing with SWATH-MS data collected by our team on
experimental models and human tumor samples.
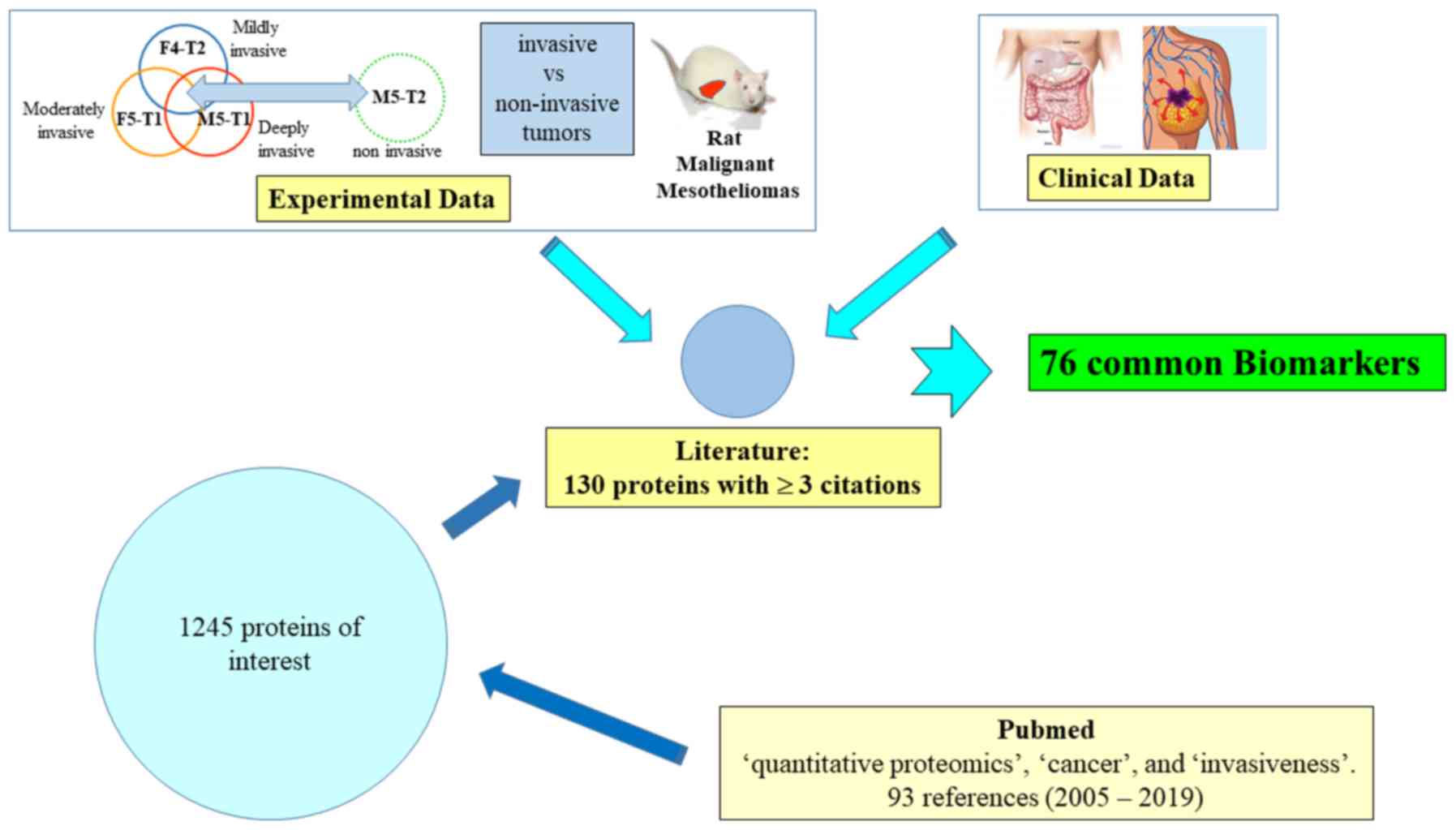
The procedure used to identify the list of proteins
of interest is summarized in Fig.
1. First, an initial search on the PubMed database was
performed on March 7, 2019, with the keywords 'quantitative
proteomics', 'cancer' and 'invasiveness'. In total, 93 studies with
full text in the English language were analyzed, published between
2005 and 2019. A file of 1,245 proteins mentioned in comparative
analyses between tumor cell lines of different invasiveness, tumor
cell lines versus normal cell lines, tumors versus normal tissues,
invasive or not, from 42 relevant articles was established. The
number of citations of each protein in these articles was then
recorded, and the 130 potential candidates for which quantitative
changes were documented at least in three different studies, were
listed for further examination.
Subsequently, this list of 130 candidates was
crossed with experimental data collected on rat malignant
mesotheliomas (MMs) differing by their invasiveness (21), and then with clinical data from
cohorts of patients with colon adenocarcinoma (22) or breast cancer (23). As a result of technological
improvements that occurred between these two studies, the number of
specific biomarkers identified in breast cancer was higher than
that in colon cancer. Although our team is primarily focused on
breast cancer, the authors wished to extend the present review to
other types of malignant tumors originating in two different
tissues in order to validate the most robust and generalizable
biomarkers.
This led to a final list of 76 upregulated or
downregulated proteins common to the three sources of data, which
represent potential tumor invasive biomarkers. The present review
is based on i) articles selected from the procedure illustrated in
Fig. 1; and ii) the screening of
literature for each individual protein listed in Table I with the following keywords 'name
of the protein' and 'cancer', without or with 'invasiveness'.
Finally, some articles combining two or more of these protein names
with these keywords were also analyzed.
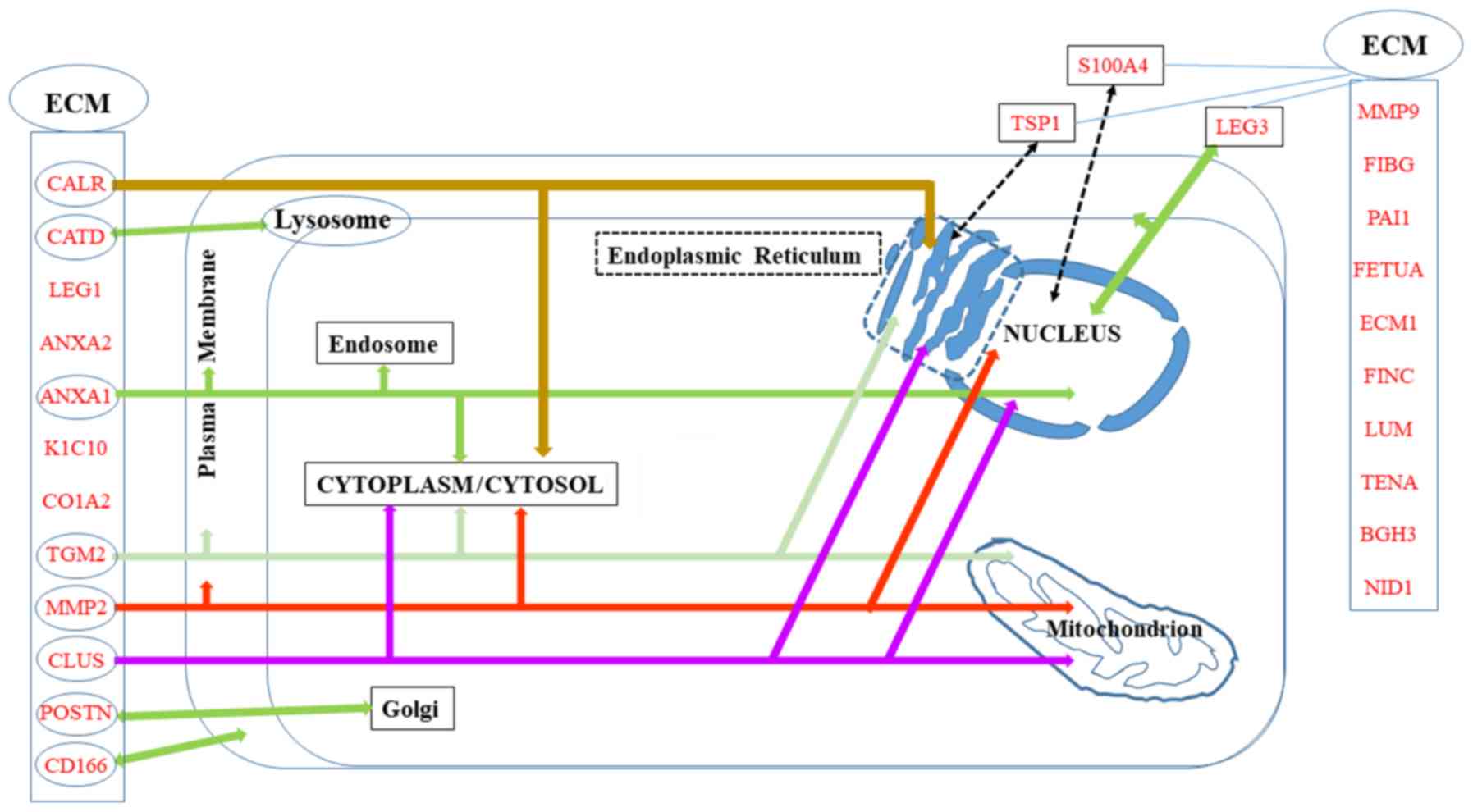
A number of the proteins in the list of potential
biomarkers of invasiveness are localized in the ECM, either
specifically or in parallel to other intracellular localizations,
which are summarized in Fig.
2.
Communications between stromal cells and cancer
cells represent a key parameter of invasiveness. The S100 family of
proteins in particular is involved in the formation and maintenance
of a pro-inflammatory environment. Among the 21 members of this
family present in humans, all regulated by Ca2+ binding,
11 are described in the literature in relation to the subject of
the present review (S100A4, S100A6, S100A7, S100A8, S100A9,
S100A10, S100A11, S100A13, S100A14, S100A16 and S100P). S100A4 and
S100A6 are the most extensively documented; however, S100A8, which
forms heterodimers with S100A9 [the overexpression of which is very
useful for the identification of circulating tumor cells (24)], is also involved in the regulation
of inflammatory responses and pre-metastatic niche formation,
together with S100A4 (25,26). The heterodimeric protein, named
calprotectin, is released by activated granulocytes, and functions
in a cytokine-like manner by triggering signaling pathways involved
in inflammatory processes. A recent review revealed the complex
functionality of this molecule, emphasizing the fact that its
function depends on its concentration and location inside or
outside the cell (27). Of note,
S100A9 and S100A4 exhibit common molecular interactions, including
heterodimerization which require levels of Zn2+ that are
found in the extracellular space, but not intracellularly (28).
The contribution of S100A4, also known as
metastasin, to tumor progression and metastasis in various tumors
has been well documented over the past two decades, as recently
reviewed (29). In normal and
benign lesions, S100A4 is restricted to a few stromal fibroblasts
and inflammatory cells (30). By
contrast, both tumor cells and stromal cells secrete the protein in
malignant tumors (25,30). Moreover, in the context of a
pre-metastatic niche (31), the
production of S100A4 serves as a link between inflammation and
tumor metastasis and is indicative of a poor prognosis (32). S100A4 is also involved in the
epithelial-mesenchymal transition (EMT) and is particularly highly
expressed in the peripheral leading edge of breast cancer (26) and non-small cell lung cancer
(33). S100A4 is related to tumor
characteristics associated with a poor prognosis. The combined gain
of S100A4 and the loss of membrane E-cadherin in cervical cancer
tend to confirm its link with an unfavorable prognosis (34), in good agreement with previous
studies by the authros on experimental malignant mesothelioma
(21,35).
S100A6, also known as calcyclin, is overexpressed in
the majority of cancers, and its involvement in tumor cell motility
has been widely documented (36).
In metastatic tumor tissues, S100A6 expression has also been found
to be higher than that in non-metastatic tissues (37). However, the molecular mechanisms
underpinning the ability of this protein to regulate cell motility
are not yet completely understood, as the down- or upregulation of
S100A6 expression has been shown to lead to increased or decreased
migration, respectively, in the particular case of osteosarcoma
(38). Intracellularly and in
vitro, its role in cytoskeletal reorganization and EMT has been
widely investigated, although to date, several questions remain
unanswered as regards its function/regulation in vivo, such
as the confirmation of direct interaction with the cyto-skeleton
and the determination of the primary cause(s) of its increased
expression (36). Finally, S100A6
can be secreted or released by some cell types and the observation
that serum levels are elevated in patients with gastric cancer,
which is associated with lymph node metastasis (39), raises the question of the
extracellular effects of this protein (36).
Although functionally unrelated, these two families
of proteins for which extracellular localization has been well
documented, both share evidence of secretion via direct
translocation and using a vesicle-based pathway, instead of the
conventional endoplasmic reticulum/Golgi network in the context of
cancer (40).
Although Annexins A1, A2 and A6 are all present in
endosomal compartments, the extracellular functions of Annexins A1
and A2 differ in several respects: Annexin A1 is involved in the
regulation of inflammation, apoptosis, leukocyte trafficking and
inhibition of neutrophil and monocyte extravasation (51), while Annexin A2 is a co-receptor
for tissue plasminogen and participates in neoangiogenesis
(41). Early studies have
demonstrated that Annexin A1 regulates leukocyte migratory events
through its function as an agonist of n-formylpeptide receptors
(FPRs), initiating a cascade of signaling events (52). Further investigations have
indicated that the binding of Annexin A1 may lead to both pro- and
anti-inflammatory effects depending on the type of ligand, its
expression being either increased or decreased in different types
of cancers(53). In the particular
case of mammary tumorigenesis, studies have revealed that the
expression and functional roles of Annexin A1 are controversial
(54,55), suggesting an association which may
be much more complex than initially thought (56). This is probably due to its diverse
actions on the many cell types of the tumor and also to the great
complexity of the network of ANXA1-regulated proteins (54). The contribution of hypoxia to the
combined upregulation of this protein and S100A4 has recently been
described (57). In addition, a
link between MMP9 and these two proteins has been reported
(58), mirroring previous findings
(59).
Annexin A2 is a multifunctional protein found at
various cellular locations. In addition to being present in soluble
form in the cytoplasm or associated with the actin cytoskeleton,
and both the intra- and extracellular sides of the plasma membrane,
this crucial protein is subjected to complex regulation via ligand
binding and post-translational modifications (60). Although it is overexpressed in
numerous types of cancer, both its upregulation and downregulation
have been suggested as prognostic biomarkers (61). The overexpression of Annexin A2 in
glioblastoma is associated with tumor aggressiveness and patient
survival, and with a mesenchymal phenotype (62). These observations are coherent with
the involvement of EMT in ovarian and colorectal cancer
inva-siveness (63,64). The role of EMT in pancreatic cancer
has also been reported to depend on the combined expression of
tenascin C and Annexin A2 (65).
Other studies in the same field have described links between
Annexin A2 expression and S100A4 or S100A6 as prognostic biomarkers
for invasive types of urothelial carcinoma (66) or gastric cancer (67). For invasive breast cancers
characterized by increased glycolysis and carbonyl stress, the
production of advanced glycation end products (AGEs) and
AGE-modified proteins leads to alterations, affecting several
proteins in parallel to the increase in Annexin A2, including
fibrinogen gamma chain and prohibitin present in the list of
biomarkers in the present review (68). Notably, a proteomic analysis of
pleural effusion from lung adenocarcinoma patients identified seven
proteins not previously reported in plasma, including Annexin A2,
as well as another protein from our list, BGH3 (69). Finally, an investigation of the
interaction of ovarian cancer and peritoneal cells identified
several ECM proteins including Annexin A2, and highlighted links
with BGH3, PAI1, fibronectin and periostin (70).
Since the pioneering work dedicated to the
comparative expression of galectins 1 and 3 in advanced cancers
(71), revealing in particular
their contribution to the stimulation of glioblastoma cell
migration (72), interest in these
proteins has continued to grow over the past two decades.
Galectin 1 belongs to the first group of this
family, characterized by the presence of one conserved
carbohydrate-recognition domain (CRD). It represents an interesting
target for the development of cancer therapies (73). Previous studies have confirmed the
pioneering discovery that galectin 1 overexpression is involved in
cancer invasiveness. By 2016, the role of galectin 1 in cancer
progression had been clearly demonstrated, although the mechanisms
underlying its different actions were not well understood (74). To overcome this gap in knowledge,
numerous studies have been conducted over the past few years. For
example, Bhat et al described the role of nuclear
localization, suggesting that differential glycosylation at the
level of tissue microanatomy regulates this parameter in breast
carcinoma (75). Shen et al
demonstrated the mechanisms through which galectin 1 mediates the
activity of matrix metalloproteinase (MMP)9 through the
Ras-Rac1-MEKK4-JNK-AP1 signaling pathway in urinary bladder
urothelial carcinoma cell invasion (76). In addition, the mechanisms through
which galectin 1 induces EMT have been investigated in detail,
including its secretion by cancer-associated fibroblasts and its
binding to integrin β1 (ITGB1) (77) or the non-canonical hedgehog pathway
(78) in gastric cancer, and the
activation of an αvβ3-integrin/FAK/PI3K/AKT signaling pathway in
hepatocellular carcinoma (79).
Finally, additional findings by Qian et al revealed that
galectin 1 induces the secretion of stromal cell-derived factor-1,
thus modulating stromal pancreatic stellate cells in the context of
pancreatic cancer metastasis (80), and plays an important role in
immune escape of gingival squamous cell carcinoma through induction
of T cell apoptosis (81).
However, galectin 3 has incited twice as much
interest as galectin 1 in the context of cancer invasiveness, and a
seven-fold increase was observed in the number of PubMed references
on galectin 3 between the years 1999 and 2014. The wealth of data
collected during this period has led to several reviews of
excellent quality. Galectin 3 expression is closely involved in
tumor cell transformation, migration, invasion and metastasis in a
wide variety of cancers (82). The
value of this protein as a prognostic biomarker for gastric cancer
was confirmed in 2018 by the observation that its overexpression is
associated with shorter overall survival in all patients (83). Galectin 3 orchestrates different
cell events in the tumor microenvironment, suppressing immune
surveillance by killing T cells and interfering with NK cell
function (84). Upon secretion,
galectin 3 can oligomerize, playing a homeostatic role in tumors by
favoring either the exit of tumor cells from a stressed environment
or the entry of endothelial and immune cells into the tumor
organoid (85). Finally, two other
biomarkers involved in cell-matrix interactions and present in the
list of invasive biomarkers in the present review, fibronectin and
S100A4, were found together with galectin 1 and galectin 3, in a
list of 61 statistically significant differentially expressed genes
associated with ERBB2 overexpression in breast cancers, and
their clinical relevance was demonstrated at the protein level
(86).
Another type of secreted protein affecting both
cancer cells and stromal cells is cathepsin D (CATD), a lysosomal
cysteine and aspartic proteinase, which is generally overexpressed
in aggressive cancers and is associated with a poor prognosis
(87). In the case of breast
cancer, Derocq et al demonstrated that CATD secreted in the
extracellular environment triggers fibroblast outgrowth by binding
to the β-chain of the LDL receptor-related protein-1 (LRP1), a
scavenger receptor mediating the endocy-tosis of various
extracellular ligands and involved in signal transduction and gene
transcription (88). The multiple
roles of CATD show increasing promise for the development of novel
anticancer agents (89).
The ubiquitous TGM2 belongs to a family of enzymes
that catalyze the formation of covalent bonds between a free
ε-amino group of a lysine and the γ-carboxyl group of a gluta-mine.
Although localized in different cellular compartments, it can also
be exported from the cell, then interacting with and/or
cross-linking numerous components of the ECM where together with
nuclear factor (NF)-κB it influences cellular sensitivity to
genotoxic agents and activates EMT (90). TGM2 is overexpressed in various
types of cancer, where it remodels and stabilizes the ECM in
association with MMP2 and MMP9, and high levels of TGM2 are
associated with lower survival rates (91).
Among the 28 types of collagens that represent the
most abundant proteins of the extracellular matrix, two are present
in our list of invasive biomarkers, both of which assemble into
higher-order supra-molecular structures such as fibrils (92). Almost ten years ago, Garamszegi
et al reported that collagen type I, among other ECM
molecules, induces Smad2 activation in human breast cancer cells,
suggesting that cell-matrix communication was more complex than
previously thought (93). The
interest for this molecule has grown during over the past decade
through the discovery that this major fibrillary component of the
stroma induces the dedifferentiation of epithelial cells and the
disruption of the E-cadherin adhesion complex (94). In parallel, the overexpression of
this type of collagen has been shown to be associated with tumor
development in gastric cancer (95) and medulloblastoma (96). Subsequently, a detailed proteomic
analysis of breast tissue collagens has confirmed that fibrillary
collagens, in particular, are increased in tumor tissue compared to
matched normal tissue, in agreement with findings by other authors
(97). A link with the presence of
bone marrow-derived fibrocyte-like cells, defined as α-1 type I
collagen-positive cells producing FGF2, has also been reported in
human malignant mesothelioma (98). More recently, Rong et al
demonstrated that the collagen type I alpha 2 chain (COL1A2) gene
modulated cell motility through interaction with the cytoskeleton
(99).
Cancer cells share with immune cells the ability to
penetrate the dense network of the BM. To cross this barrier, they
secrete different categories of proteases, in particular MMPs (also
known as matrixins) (92), among
which MMP2 and MMP9 (gelatinases a and b, respectively) expression
has been shown to be associated with invasiveness (100). MMP9 has the particularity of
degrading both type I and type IV collagens, and for this reason it
facilitates invasion across the BM (101) in association with Annexin A1
(59), resulting in a poor
prognosis (102). The association
of both MMP9 and MMP2 with tumor invasiveness was further confirmed
for a number of cancer types (103). A link with S100A4 was soon
established in the context of breast cancer invasiveness (104). These investigations demonstrated
that exposure to interleukin (IL)-1β induced EMT in MCF-7 cells,
which could then respond to chemokine CXCL12 (101), ultimately leading to the
expression of S100A4 and increased secretion of MMP2 and MMP9
(105). Matsuura et al
contributed further insight into the mechanistic process by
demonstrating that S100A4 binds to Smad3, an important mediator of
transforming growth factor (TGF)-β signaling, in a
Ca2+-dependent manner, which ultimately induces MMP9
expression (106).
Fibronectin is a major stromal protein associated
with tumors. It is a part of the four matrix components that are
common to breast cancer progression and mammary gland involution
(107). Its contribution to the
mechanisms through which a tumor cell undergoes metastasis to a
predetermined location was initially documented by Kaplan et
al in 2005, who demonstrated that bone marrow-derived
hematopoietic progenitors that express the vascular endothelial
growth factor receptor 1 (VEGFR1) form cellular clusters that
upregulate fibronectin, providing a niche for incoming tumor cells
(108). Other factors have been
identified more recently, including a platelet ADP receptor that
recruits VEGFR1+ cells in the lung, fibroblasts that
secrete both fibronectin and S100A4, or exosomes produced by
pancreatic ductal adenocarcinoma cells and taken up by Kupffer
cells, leading to upregulation of fibronectin production by hepatic
stellate cells (31). Another
advancement in the understanding of the metastatic process was the
discovery that several ECM proteins, including fibro-nectin are
processed via the plasminogen-plasmin pathway as a result of
interactions between ovarian cancer cells and peritoneal cells
(70). Interactions between
fibronectin and integrins, in particular α5β1, could provide
interesting prospects for the treatment of stroma-rich tumors.
Studies on the parameters that regulate such interactions have
highlighted the modulating role played by tenascin C (109). Finally, fibro-nectin plays other
important roles, revealed by silencing its gene which leads to the
inhibition of cell proliferation and the promotion of cell
senescence and apoptosis via the regulation of the PI3K/AKT
signaling pathway (110).
Proteoglycans, which consist of a core protein and
glycosaminoglycan side-chains, are a large family of proteins
involved in interactions of cells with the ECM. Two of these, ECM1
and LUM, appear in the list of invasive biomarkers in the present
review.
Extracellular matrix protein 1 (ECM1) is
significantly elevated in a number of epithelial tumors, giving
rise to metastases, and its high expression around blood vessels
has been suggested to play a role in angiogenesis (111). Interest for this protein grew
significantly after a 2008 study classifying tumors on the basis of
the expression of ECM components in relation to different clinical
outcomes (112). Subsequently,
Lal et al established this protein as a novel prognostic
biomarker for poor long-term survival in 134 women diagnosed with
invasive breast cancer (113),
and the link with EMT was documented by two independent studies on
breast cancer and hepatocellular carcinoma (114,115). The silencing of ECM1 in two
triple-negative breast cancer cell lines has revealed that it
regulates actin cytoskeletal architecture and decreases the
expression of the prometastatic protein S100A4, suggesting that it
is the primary effector of the changes observed in morphology,
migration, invasion and adhesion (116). Finally, ECM1 facilitates the
expression of genes associated with EMT by activating the
ITGB4/FAK/glycogen synthase kinase 3β signaling pathway (117).
In addition to the galectins discussed above, two
other proteins belonging to this category are present in the list
of invasive biomarkers in the present review, periostin and
thrombospondin-1 (TSP1), both of which have been reported to
represent secreted matrix molecules decorating ECM fibers in the
process of tissue-specific restricted guidance of cancer invasion
(125).
Periostin (also known as POST) interacts with
multiple cell-surface receptors, particularly integrins, promoting
cancer cell survival, EMT, invasion and metastasis (126). In a large cohort of 300 patients
with breast cancer, Kim et al demonstrated that a high
epithelial periostin expression was more frequently observed in the
distant metastatic relapse-positive group, compared with the
negative group and was associated with a reduced overall survival
(127). In relation to its
ability to induce EMT, Mino et al reported that periostin
expression was markedly higher in an undifferentiated intrahepatic
cholangiocarcinoma cell line compared with a moderately
differentiated one (128).
Similar contradictory findings were initially
reported for TSP1 in relation to cancer progression, as this
protein presents both stimulatory and inhibitory effects (129). An immunohisto-chemistry study of
80 cases of intraductal papillary-mucinous neoplasms of the
pancreas revealed an association between TSP1 and tumor
invasiveness: Patients in the strongly positive group exhibited a
significantly poorer prognosis compared with the negative group
(130). Of note, Firlej et
al described the capacity of TSP1 to increase both hypoxia and
cell migration which appear to be linked (131). In another type of cancer for
which hypoxia is a hallmark, the increased aggressiveness of
pancreatic ductal adenocarcinoma is dependent on the formation of
Annexin A6/LDL receptor-related protein 1/TSP1 complexes by
cancer-associated fibroblasts (CAFs) and their uptake by tumor
cells (49). Platelet-secreted
TSP1 contributes to colon cancer invasiveness by promoting the
signal regulation of MMP-9 via the p38MAPK pathway (132). Finally, Joshi et al
investigated the mechanisms through which bone marrow-derived
mesenchymal stromal cells and prostate cancer cells interact,
demonstrating that the bioactive principle responsible for this
chemotaxis in co-culture was present in a high-molecular weight
fraction containing TSP1, and the formation of complexes of this
protein with fragments of fibronectin function as matrikines
(133).
BGH3 (TGFBI, also known as βig-h3) is involved in
cell-collagen interactions by binding to types I, II and IV
collagen. It has been described as a promoter or a suppressor of
cancer growth as its effect is apparently highly cell
type-dependent. BGH3 is upregulated in a number of tumor types, and
Lebdai et al demonstrated that this overexpression in
aggressive clear cell renal cell carcinoma was associated with the
stage, size, grade and necrosis (SSIGN) score, as well as with
outcomes (134). In melanoma,
Nummela et al also reported that BGH3 impaired the adhesion
of melanoma cells to collagen type I, fibronectin and laminin, thus
confirming its role as an important regulator of invasive growth
(135). Additionally, Klamer
et al revealed that the upregulation of BGH3 in
hematopoietic stem and progenitor cells may help loosen the
adhesive contacts with the bone marrow niche from which they
originate, thus making them susceptible for polarization and
subsequent egress (136).
Finally, quantitative changes in BGH3 were also related to the
parallel evolution of other ECM proteins associated with
inva-siveness, such as fibronectin, periostin and Annexin A2
(70), enzymes involved in redox
regulation of the cell, such as peroxiredoxins (137), or S100A4 in the context of EMT
(138).
CD166 [also known as activated leukocyte cell
adhesion molecule (ALCAM)], is a member of the immunoglobulin
superfamily. It was first described as a novel actor in invasive
growth and control of matrix metalloproteinase activity (139). A first review of its interest in
cancer highlighted the existence of a marked heterogeneity of
expression in different tumors, with two additional levels of
complexity due to the fact that its expression is dependent on the
stage of tumor development and on RNA and protein levels in breast
cancer tissues (140). Weidle
et al subsequently confirmed and detailed the
context-dependent prognostic impact of CD166 expression in cancers
(141). The recent finding by von
Lersner et al that CD166 promotes malignant behavior through
the regulation of its availability (dynamic turnover of the protein
at the cell surface) rather than its specific activity, provides a
key explanation of its heterogeneous expression within malignant
diseases (142).
Basement membranes (BMs) consist mainly of collagen
type IV, laminins and glycoproteins, including nidogens and heparin
sulfate proteoglycans (143). The
main component of nidogens, NID1, binds to laminin and acts as a
bridge to the collagen network to complete the core basement
membrane scaffold (144).
However, in studies on the matrisome, which includes not only the
structural components of the BM, but also proteins that it
interacts with, or modifies, the ECM, NID1was additionally found to
be expressed by the stroma (145). Among the numerous studies
documenting the implication of NID1 in cancers, Zhou et al
demonstrated that the expression of NID1 in ovarian cancer cells
revealed an EMT phenotype characterized by the enhancement of
mobility, invasiveness and cisplatin resistance (146), while Pedrola et al
observed a significant increase in NID1 expression in the invasion
front of endometrial tumors compared to their paired superficial
zone (147).
PAI1 is a glycoprotein synthesized by various normal
cells and a large number of different tumor cells. It belongs to
the serine protease inhibitor super family (SERPIN), which probably
represents the most important component of the plasminogen
activator system (148). A high
tumor level of this protein has been associated with a poor patient
prognosis (149). This
association was confirmed in renal cell carcinoma (150) and breast cancer; higher
concentrations of PAI1 have been shown to be associated with an
aggressive phenotype and a poor prognosis, with a positive
correlation has been found with MMP9 (151). Rhone et al further
analyzed the clinicopathological determinants of patients with
invasive breast cancer and found significantly higher PAI1
concentrations in patients with ductal carcinoma compared to those
with lobular carcinoma, suggesting that a high PAI1 expression
predisposes to a pro-coagulant environment expressed by
simultaneous activation of coagulation and fibrinolysis suppression
(152). Finally, PAI1 is now
considered to be a prognostic factor, particularly in breast
cancer, and Li et al recently reviewed the numerous tumor
promoting factors involved in the modulation of PAI1 activity
(153).
Alpha-2-HS-glycoprotein [also known as fetuin-A
(FETUA)] is a serum glycoprotein involved in the adhesion of tumor
cells, functioning as a chemoattractant in breast cancer
progression (154), and
interacting synergistically with CXCL12 at low concentrations
(155). FETUA is endocytosed by
tumor cells, enhancing the secretion of exosomes to the
extracellular milieu that ultimately promotes cell spreading and
adhesion, a process that also requires Annexin A2 and A6 (44,46,50,156). Combined with ECM1, the diagnostic
potential of this protein has recently been confirmed in another
cancer type, non-small cell lung cancer (157).
Interest for this protein in cancer emerged through
the discovery of its presence in a list of seven molecules
associated with the formation of AGEs in tumors exhibiting
increased glycolysis and carbonyl stress (68). Together with other downstream
thrombin procoagulant targets, fibrinogen has long been
investigated as a promoter of tumor cell metastatic potential,
which was confirmed through the experimental demonstration that
tumor growth was markedly impeded in fibrinogen-deficient mice
(158). Additional evidence was
provided by Honda et al, who found a protein complex
containing FGG in plasma from patients with advanced ovarian cancer
(159), and by the report of its
association with gastric cancer (160). Finally, in a study investigating
the clinicopathological significance of this protein in the process
of migration and invasion of hepatocellular carcinoma cells, a
higher FGG expression was significantly associated with a higher
recurrence rate and a shorter survival through EMT signaling by
regulating the expression levels of Slug and zinc finger
E-box-binding homeobox 1 (ZEB1) (161).
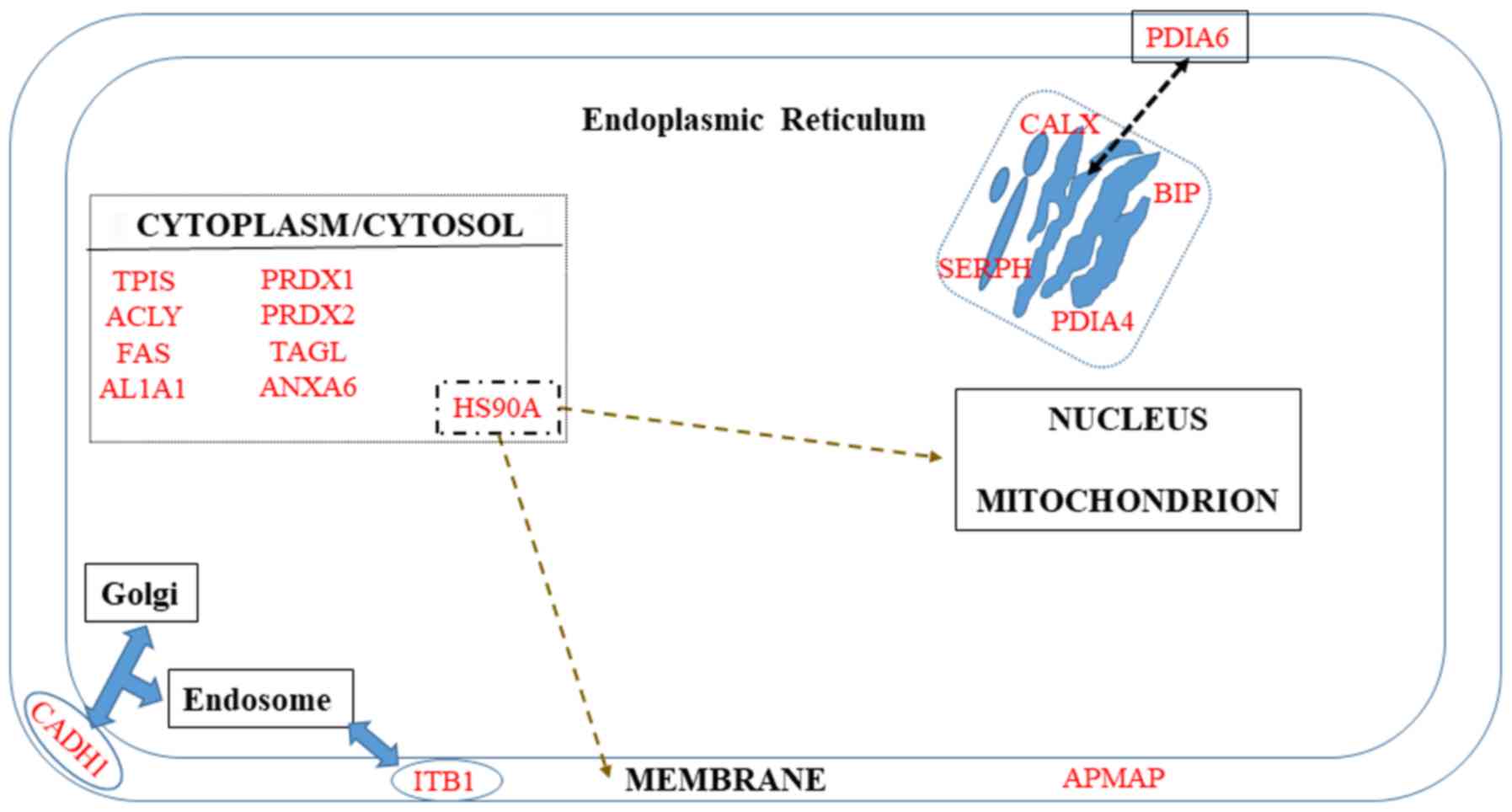
A number of proteins are known to be localized in
the membrane but are not restricted to it (Fig. 3). The first category includes ITB1
and APMAP. The second category, which corresponds to proteins
additionally localized in Golgi and endosomes (CADH1), ER (PDIA6),
or cytoplasm (HS90A), will be discussed in the third paragraph of
this chapter.
More than 10 molecules are known to bind to the
cytoplasmic tail of this protein, some of them acting as binding
platforms for cytoskeletal and signaling molecules; however, the
mechanisms through which integrin signaling is induced in the
intracellular space have not yet been elucidated (162). To date, the associations between
ITGB1 and the clinical features of patients with cancer are still
unclear and a number of studies have reported contradictory
conclusions depending on the type of cancer (163,164).
The keystone position of S100A4 in the acquisition
of invasiveness by cancer cells can be illustrated by its inverse
correlation with E-cadherin expression in different invasive
phenotypes of oral squamous cell carcinoma (169). In melanoma cells, this inhibition
of E-cadherin expression, which leads to EMT and increased
invasiveness, has been shown to be associated with the upregulation
of Annexin A1, which can be reversed by the use of small
interfering RNAs (170). Overall,
E-cadherin functional loss has been associated with a poor
prognosis and survival in various types of cancer (171). Notably, Yu et al recently
reported an inverse correlation between E-cadherin and
peroxiredoxin 1 expression, and demonstrated that these two
combined parameters were associated with EMT, poor differentiation,
deeper invasion and an advanced TNM stage of gastric cancer
(172).
Potential biomarkers of invasiveness present in the
cytosol/cytoplasm are summarized in Fig. 3.
Glycolysis dysregulation is a main hallmark of
cancer cells and has been the subject of extensive investigations
(173) since the research by
Warburg a century ago (174).
These studies have emphasized the key role of enzymes in the
adaptation to hypoxia, including triose phosphate isomerase (TPI)
which catalyzes the interconversion of dihydroxyacetone phosphate
and glyceraldehyde-3-phosphate, playing an important role in the
development of many types of cancers (175). Apart from its metabolic function,
Lincet and Icard demonstrated that this enzyme also participated in
cell cycle activation (176),
while its transcriptional regulation involves microRNAs
(miRNAs/miRs)-22/28 (177).
A growing interest in fatty acid synthase (FAS)
emerged in oncology in the mid-2000s, when this lipogenic enzyme
was found to confer growth and survival advantages to cancer cells
rather than functioning as an anabolic energy-storage pathway
(178). FAS upregulation has been
shown to be associated with a poor prognosis in a variety of
cancers, although the underlying mechanisms are yet not completely
understood. Its localization in the nucleus in a subset of prostate
cancer cells has been found to be associated with the Gleason grade
(179). Subsequently, Wang et
al revealed that the knockdown of this enzyme in human
colorectal cancer cell lines attenuated the activation of the Wnt
signaling pathway and metastasis; a positive correlation was
observed in patients between FAS expression and Wnt signal
biomarker gene expression (180).
These investigations opened up interesting therapeutic prospects
against prostate (181) and
breast cancer cells (182).
Another key enzyme involved in the biosynthesis of
fatty acids is ATP-citrate lyase (ACLY), which is upregulated or
activated in several types of cancer (183), particularly in gastric cancer,
where its regulation involves miR-133b (184). These observations have led to the
development of ACLY inhibitors attracting interest as promising
anticancer agents (185), with
citrate levels monitored as an indicator of cancer aggressiveness
and/or as a biomarker for response to therapy (186).
A number of cancer cells are characterized by an
increase in reactive oxygen species (ROS) and a dysregulation of
enzymes involved in the redox-regulating proteins, in particular
peroxiredoxins, which catalyze the peroxide reduction of
H2O2, organic hydroperoxides and
peroxynitrite (187). The
over-simplification of the role of 'antioxidants', which until now
was attributed to these enzymes has recently been questioned
(188), as the genetic disruption
of their expression in mice has been shown to lead to an increased
incidence of neoplasia, consistent with a probable role in the
protection of genomic integrity (189). Moreover, although their
overexpression has been mostly reported in various malignant
tumors, the suppression of 2-Cys peroxiredoxins has also been found
in certain metastatic cancers (190), raising questions as to their
complex functions related to tumor cell invasiveness, with
important implications for therapies (191). In the particular case of
peroxiredoxin 1, a major member of the family present mainly in the
cytosol, the recent study by Kim et al demonstrated that
this protein has RNA-binding properties, binding to a specific
subset of small nucleolar RNAs (snoRNAs) and regulating these
molecules at the post-transcriptional level (192).
PKM, which catalyzes the final step in glycolysis,
consists of four isoforms in mammals, two of which, PKM1 and PKM2,
are encoded by the PKM gene through alternative splicing of
mutually exclusive exons (193).
Although PKM2. but not PKM1 was initially considered to favor
cancer cell proliferation, the exclusive role of PKM2 in
tumorigenesis has recently been challenged. In the particular case
of liver tumorigenesis, various PKM1/PKM2 ratios and pyruvate
kinase activities can sustain the glucose catabolism required for
the process (194). Moreover,
although PKM2 has been suggested to be the predominant isoform in
cancer cells, providing a basis for the development of novel
therapeutic strategies (195),
mass spectrometry-based proteomic analyses have demonstrated that
PKM2 can be detected in both cancer and normal cells (196). Nevertheless, deeper
investigations evaluating the mechanisms through which the two
isoforms regulate the invasiveness of pancreatic ductal
adenocarcinoma have revealed that both regulate cell migration and
invasion in vitro, but only PKM2 overexpression the promotes
metastasis of cancer cells in vivo (197).
This superfamily consists of 19 proteins displaying
mainly catalytic functions involved in detoxification, and their
role in cancer has been widely emphasized over the past decade
(198), particularly in relation
to stem cells and resistance to chemotherapy (199). The overexpression of the first
member of this family, ALDH1A1, also known as retinal dehydrogenase
1, is generally associated with poor outcomes (200-202).
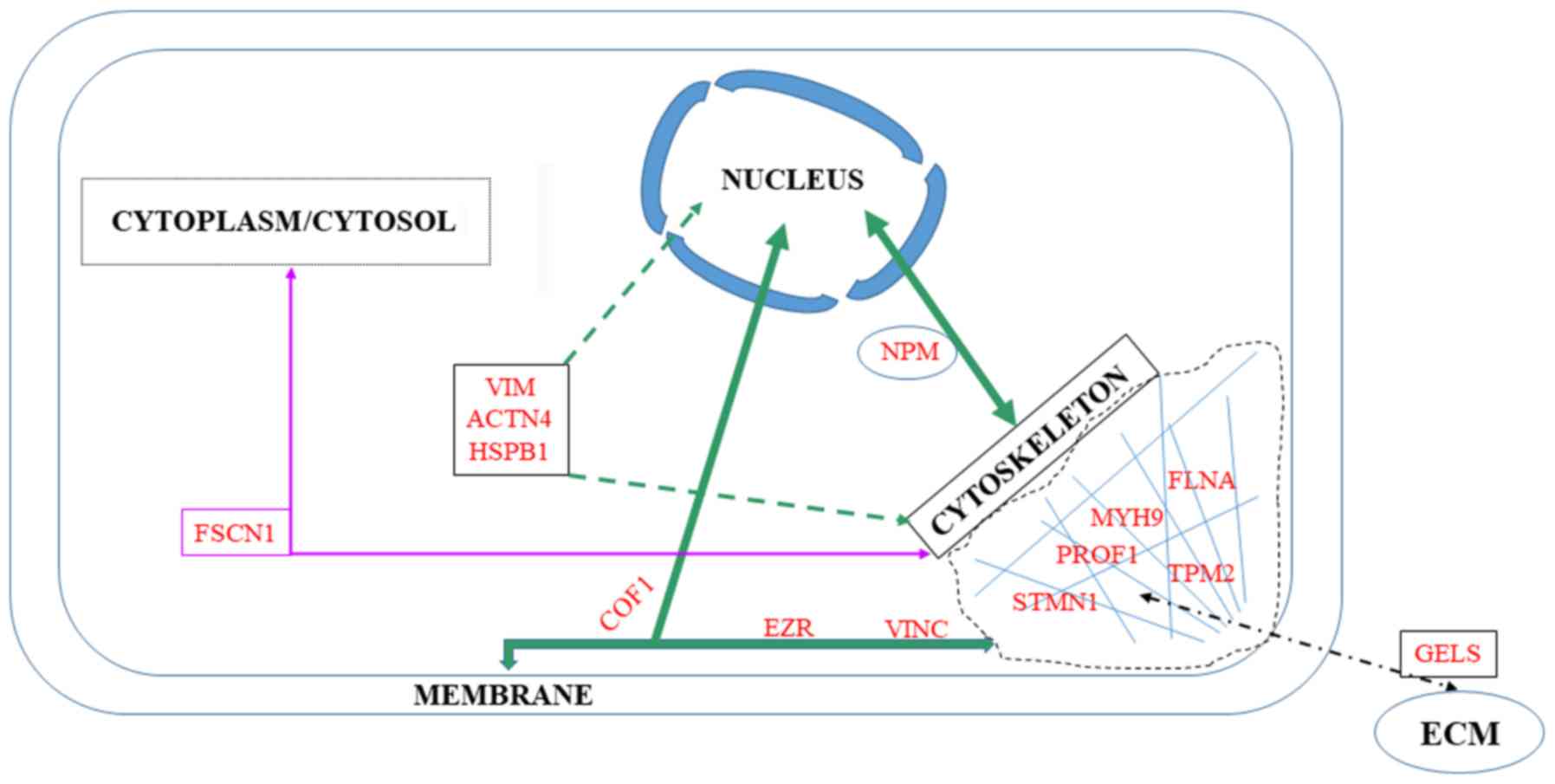
Among the 76 upregulated or downregulated proteins
in the list of potential tumor invasive biomarkers in the present
review, seven belong to the cytoskeleton, while another seven
interact with the membrane or the nucleus (Fig. 4).
Transgelin is a protein which affects the dynamics
of the actin cytoskeleton through the stabilization of actin
filaments. This biomarker is involved in a number of cancer-related
processes and was initially described as a tumor suppressor. The
transgelin level has been shown to be decreased in tumor cells
relative to cells of healthy tissues. However, it can be highly
expressed by the reactive tumor stroma and its re-expression in
tumor cells in the more advanced stages of cancer can support
migration and metastasis (203).
In support of this observation, transgelin positivity has been
associated with more aggressive tumors, a high Ki-67 index and low
estrogen and progesterone receptor expression levels in of breast
cancer (204).
Fascin is another protein regulating cytoskeletal
structures which coordinates motility and invasion and promotes
filopodia formation in carcinoma cells. Although absent from most
normal epithelia, its expression has been associated with
metastasis in colorectal and gastric cancers (205).
Stathmin is a major microtubule-destabilizing
protein which promotes microtubule depolymerization and mediates
the effects of p27Kip1, an inhibitor of cyclin-dependent
kinase complexes (206). This
action is obtained either through the sequestration of free tubulin
dimers or directly by the induction of the microtubule-catastrophe.
Thus, stathmin is an important target of the main regulator of the
M phase and offers interesting prospects for anti-metastatic
therapies (207). The value of
stathmin as a prognostic biomarker has been confirmed by the
observation that its overexpression is associated with tumor cell
differentiation, lymph node invasion and a high TNM stage (208).
Profilin-1 (PROF1) is an actin-monomer binding
protein ubiquitously expressed in all cell types. It regulates
actin dynamics and cell motility and plays an important role in the
migration of cancer cells (209).
In addition to its sequestering function on actin monomers, PROF1
promotes the assembly of globular-actin monomers (G-actin) into
filamentous-actin (F-actin) (210), interacts with certain membrane
lipids, and is also involved in regulating the expression of
several cancer stem cell genes (211). PROF1 has been shown to be
downregulated in different types of cancer, including breast,
pancreatic and hepatocellular carcinoma, and it is associated with
aggressive clinicopathological characteristics and a poor prognosis
(212).
Filamin A (FLNA) was the first actin filament
cross-linking protein identified in non-muscle cells and the
contribution of its structure and functions to cell migration and
adhesion has already been reviewed (213). Differences in subcellular
localizations and effects on cancer development have led to the
conclusions that an association exists between high cytoplasmic
levels and invasive cancers, whereas the localization of an active
form to the nucleus and its interaction with transcription factors
is linked to a decrease in invasiveness (214). Consequently, as a promising
prospect, drugs that can transpose FLNA from the cytoplasm to
nucleus are currently under development (215).
Myosin 9 has recently attracted the attention of
oncology researchers, as it has been found that this protein,
classified as a cytokine involved in cytoskeletal reorganization
and coded by a suppressor gene, plays an important role in the
formation of cellular pseudopodia and is closely related to the
progression and a poor prognosis of the majority of solid tumors
(216).
Finally, the tropomyosin isoform Tpm2.1 is
considered to be another tumor suppressor, regulating the
sensitivity to apoptosis beyond anoikis (217). Indeed, both the mRNA expression
and protein levels of this molecule have been shown to be
significantly decreased in colorectal cancer compared with paired
adjacent normal tissue (218). In
breast cancer, the downregulated expression of this protein is due
to its promoter methylation, induced by hypoxia, leading to cell
invasiveness, poor prognosis and chemoresistance (219). In another independent study, Shin
et al found that the loss of Tpm2.1 increased the efficiency
of migration of breast cancer cells out of the spheroids on
different coated extracellular matrices (220). Finally, Mitchell et al
completed these investigations by demonstrating the mechanisms
through which the loss of this high molecular weight tropomyosin
induced glioblastoma cell spreading and elongation in soft 3D
hydrogels, recapitulating the biomechanical architecture of the
brain (221).
According to Uniprot, among the cytoskeletal
proteins in the list of potential biomarkers of invasiveness in the
present review, three co-localize in the membrane: Cofilin-1, ezrin
and vinculin.
Cofilin-1 belongs to the actin-depolymerizing
family of proteins, which is essential for the dynamic changes in
the actin cytoskeleton associated with the reorganization of
cellular shape during the acquisition of invasiveness (222). A number of investigations have
led to the conclusion that cofilin-1 expression increases in
relation to cell cycle progression, migration, intravasation and
the invasion of cancer cells. Gasparski et al revealed how
the maturation of invadopodia, these actin-rich structures present
in invasive cancer cells which degrade the surrounding ECM to
facilitate invasion, was related to the downregulation of integrin
β3 expression, leading to an increase in cofilin activity (223). However, biphasic effects between
the cofilin level and locomotory rate have also been observed,
suggesting that the process will proceed via a complex dose- and
time-dependent manner, a partially documented complex mechanism
that still needs to be clarified (224).
Ezrin, which belongs to the ERM family, interacts
with membrane proteins by organizing
membrane-cytoskel-eton-associated complexes, thus creating
specialized membrane domains, and also promotes tumor metastasis
(225). The role of ezrin in the
mechanism of the activation of the Wnt-β-catenin signaling pathway
in the context of colorectal cancer has been well documented
(226). However, although ezrin
is clearly associated with a poor prognosis and metastasis in
different cancer types, Cihan pointed out that contradictory
results remain as regards the association between ezrin expression
and clinicopathological features or prognostic parameters,
suggesting that this field of research requires further
investigations before evaluating ezrin-based therapies (227).
The third protein of interest, vinculin, couples
the ECM to the acto-myosin cytoskeleton via β-integrins and
paxillin, and thus acts as a mechano-coupling and
mechano-regulating protein (228). As an orchestrator of mechanical
signaling events, its deregulation has important consequences on
cell adhesion, contractility, motility and growth, all of which are
crucial in the process of cancer metastasis (229). A positive association between
estrogen receptor alpha and vinculin expression has also been
demonstrated in breast cancer cells (230).
According to Uniprot, among cytoskeletal proteins
in the list of potential biomarkers of invasiveness in the present
review, four of these interact with the nucleus: Nucleophosmin
(NPM), heat shock protein β1 (HspB1), actinin-4 (ACTN4) and
vimentin.
NPM, an ubiquitous phosphoprotein belonging to the
nucleoplasmin family of chaperones, is mainly localized in the
nucleolus, a proportion of which continuously shuttles between the
nucleus and the cytoplasm (231).
Andersen et al initially identified this protein in a
mass-spectrometry-based proteomic analysis of human centrosomes in
the interphase of the cell cycle (232). NPM is involved in numerous
pathways, including mRNA transport, chromatin remodeling and genome
stability; however, Box et al demonstrated that its
multifunctional role within the cell also included DNA repair
pathways and the regulation of apoptosis in cancers (233). Of note, Werner et al
reported the discovery of a translocation of the anaplastic
lymphoma kinase gene (ALK) with the promoter region and a
proximal domain of the NPM gene (NPM1) on chromosome 5q35,
yielding a chimeric protein that modulates numerous genes involved
in the evasion of the antitumor immune response, protection from
hypoxia, angiogenesis, DNA repair, and cell migration and
invasiveness (234).
HspB1 (also known as Hsp-27) is a chaperone that
regulates a number of fundamental cellular processes, and whose
structural organization presents dynamic and complex rearrangements
in response to changes in the cellular environment. Its
sophisticated anti-apoptotic role acts both upstream and downstream
of the mitochondria, the former effect occurring by alterations in
F-actin or nucleus architecture integrity (235). Xie et al recently
discovered a novel interaction between HspB1 and ezrin: The
knockdown of HspB1 resulted in a decreased phosphorylation at ezrin
Thr567, thus markedly suppressing the ability of ezrin to bind to
the actin cytoskeleton, leading to the migration of esophageal
squamous cell carcinoma cells (236).
ACTN4 is a non-muscle isoform of α-actinin
initially found concentrated in the cytoplasm of breast cancer
cells migrating and located at the edge of cell clusters (239). Subsequently, Hayashida et
al revealed that the β-catenin and actinin-4 complex was highly
concentrated in actin-rich protrusions at the peripheries of cell
clusters, and that their colocalization in the nucleus, repressing
E-cadherin expression, induced cancer invasion (238). Thomas and Robinson also
demonstrated that although the two α-actinin isoforms 1 and 4 share
regulatory mechanisms, actinin-4 exhibits a unique mechanosensory
regulation, which warrants further, more detailed investigation
(239). In parallel, Yamaguchi
et al observed that the overexpression of actinin-4, but not
that of actinin-1, significantly promoted the formation of
invadopodia by carcinoma cells (240).
Actin remodeling in cancer cells may be the result
of the inactivation of several important actin-binding proteins
such as gelsolin (243). Interest
for this protein has increased as it is not only found in the
cytoplasm, but also in the extracellular environment, providing
future prospects for studies on its prognostic potential (244).
The endoplasmic reticulum (ER) is directly
concerned with four potential biomarkers of invasiveness in the
list in the present review, calnexin (CALX), BIP, serpin H1 and
PDIA4 (Fig. 3), but also with four
additional proteins, CALR, CLUS, TGM2 and CATD, which can be found
in multiple cell compartments (Fig.
2).
Calnexin is a resident chaperone of the ER which
was initially identified as an important protein involved in the
reduction or suppression of antigen presentation by the major
histocompatibility complex (MHC) on tumor cells (245). In a very interesting study aimed
at identifying proteins preferentially expressed in a poor
prognosis group of patients with lung adenocarcinoma relative to a
good prognosis group (exhibiting no recurrence), Okayama et
al reported that calnexin was preferentially expressed in the
former, and this result was further confirmed in a cell-culture
model (246). Ryan et al
subsequently confirmed the prognostic significance of CALX in
colorectal cancer (247).
Together with calreticulin, calnexin serves as a molecular
chaperone, which prevents the aggregation and export of
incompletely folded proteins from the ER, a mechanism involved in
the metastatic progression of tumors (248). As CALX can escape from the ER and
be transported to the plasma membrane or released outside the cell,
its impact on the human immune system was recently investigated by
Chen et al, who reported that its upregulation was
associated with the inhibition of T-cell infiltration in tumor
tissues (249).
The binding immunoglobulin protein (BIP) is an
ER-lumenal polypeptide chain binding protein, which belongs to the
heat shock protein 70 family and interacts with numerous partners.
Evidence of its role in various types of cancer began to emerge a
decade ago (250). BIP is an
essential factor of the translocation machinery for protein import
into the ER; it regulates Ca2+ homeostasis in the ER,
facilitates ER-associated protein degradation, and can initiate the
unfolded protein response, inducing autophagy and crosstalking with
the apoptosis machinery to assist in the cell survival decision
(251). Among the numerous
contributions of BIP to cancers found in the literature, Herroon
et al interestingly found a link between hemeoxygenase (HO),
an inducible enzyme involved in the resistance of cells against
oxidative stress, whose overexpression is associated with
aggressiveness, and the upregulation of BIP (252). The impact BIP on the success of
anticancer therapies has been described by Chen et al, who
reported that the inhibition of the hexosamine biosynthesis pathway
resulted in the downregulation of BIP, which exacerbated
cisplatin-induced non-small-cell lung cancer apoptosis (253).
Serpin H1 (Hsp47) specifically binds to procollagen
as a resident protein of the ER, and dissociates from it in the
cis-Golgi to allow fibril formation (254). A recent review of studies on this
protein in the context of cancer has revealed that it plays a role
in numerous steps of collagen synthesis, promoting tumor
angiogenesis, growth, migration and metastatic capacity (255).
Protein disulfide isomerases (PDIs), which correct
the arrangement of disulfide bonds in the ER through reductase,
oxidase and isomerase functions, are implicated in the development
of certain types of cancer, leading to the development of PDI
inhibitors as potential novel anticancer therapies (256). Among the 21 members of this
family documented so far in mammals, four of these, including PDIA6
and PDIA4, are upregulated in a variety of tumor cells. A
mechanistic study demonstrated the mechanisms through which PDIA4
negatively regulates tumor cell death by inhibiting degradation and
the activation of procaspases 3 and 7 via their mutual interaction
(257).
Calreticulin (CALR), belonging to the
damage-associated molecular patterns (DAMPs), is involved in the
immunogenicity of cell death, but also represents a major predictor
of a better prognosis in various types of cancer (258). However, it also plays an
additional role as a pro-tumorigenic multifunctional ER protein
with variable distribution as it has been shown to promote the
progression of pancreatic cancer cells via the
integrin/EGFR-ERK/MAPK pathway (259). The increased complexity of the
functions of this pleiotropic protein was recently illustrated by
the discovery that CALR functions outside the ER where its
translocation to the cell membrane serves as an 'eat me' signal,
promoting a silenced immune response (efferocytosis), while its
effects on cytokine production are dependent on its conformation
(260).
Clusterin is a highly glycosylated protein
initially described as a cytoprotective chaperone-like molecule
controlling cell-cell and cell-matrix interactions including
adhesion (138). Subsequently, a
link between this protein and TSP1 was established in the
regulation of MMP9 when tumors cells interact with platelets in
colonic cancer invasion (132).
Clusterin has been shown to facilitate metastasis in
hepato-cellular carcinoma through the formation of complexes with
eukaryotic translation initiation factor 3 subunit (EIF3I) and the
activation of the Akt pathway, promoting the expression of MMP13
(261). Additionally, Shapiro
et al reported that CLUS was overexpressed in metastatic
human colorectal cancer cells in association with the presence of
stem cells (262), while Liu
et al reported that the overexpression of clusterin promoted
the invasiveness of clear cell renal carcinoma cells, an effect
mediated by S100A4 (263).
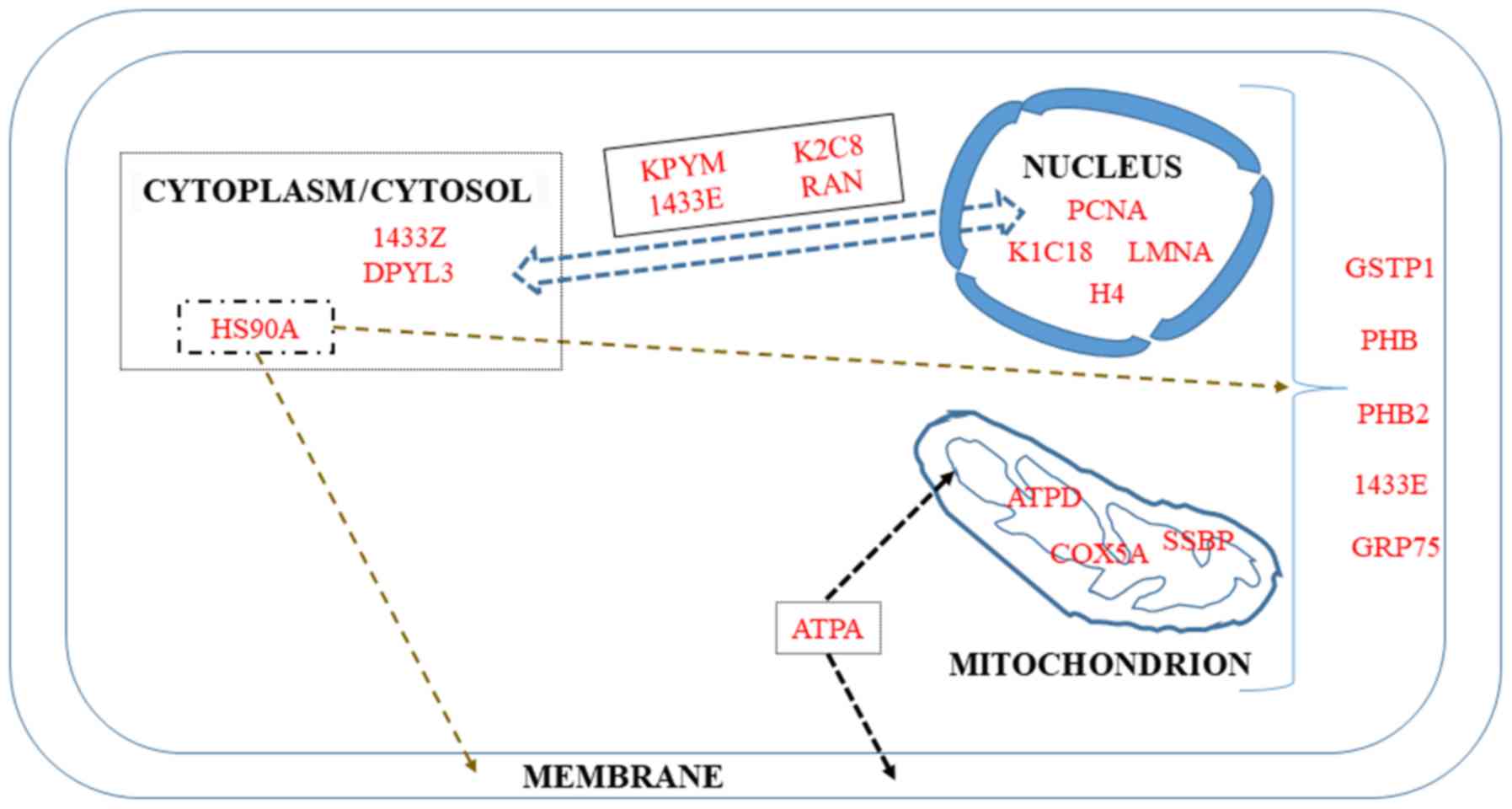
Four proteins in the list of biomarkers of
invasiveness in the present review are described as macromolecules
localized both in the mitochondria and nucleus, while five are
restricted to the mitochondria and another five are restricted to
the nucleus (Fig. 5).
The first protein in this category is the π isoform
of glutathione-S-transferase P1 (GSTP1). It belongs to a subgroup
of the GST family initially involved in cellular protection against
free radical and carcinogenic compounds (264). However, other functions have been
discovered for this protein, including the maintenance of cellular
redox homeostasis, the downregulation of which has been associated
with a poor prognosis (265).
14-3-3 proteins (their names are derived from the
elution profile on HPLC) are recognition structures at or near DNA
replication. They are cell cycle-regulated, being maximal at the
G1-S phase and minimal at the G0-G1 phase (266). This family consists mainly of
seven isoforms present in mammals whose dysregulated expression
contributes to tumorigenesis in different types of cancer. In
particular, the downregulation of the ε isoform (1433E) has been
associated with lung (267) and
gastric tumorigenesis (268).
Although an increase or decrease may occur according to cancer
type, leading to some confusion, high levels of the ε isoform have
been shown to predict a poor two-year overall survival of a group
of chemotherapy-resistant compared with chemotherapy-sensitive
patients (269).
Prohibitins are important intercellular
communicators between the nucleus and mitochondria, and their many
functions are highly dependent on their localization (270). Both prohibitins localize to the
inner membrane, functioning as mitochondrial chaperones, although
they are also present in the plasma membrane and nucleus where they
act in membrane signaling and independently as transcriptional
repressors of target genes, respectively (271-273). As regards the implication of PHB2
in transcription, Zhou et al recently described its
substantial localization in the nucleolus, where it maintains
nucleolar morphology, while promoting tumor proliferation and
probably repressing differentiation in rhabdomyosarcoma cells
(273). Mechanistically, PHB2 has
also been reported to promote prostate cancer cells by inhibiting
the expression of AKT serine/threonine kinase 2 (274). Finally, Yan et al reported
that PHB2 also mediates mitophagy (275).
GRP75 (also known as Hsp70 or mortalin) plays a
major role in the import and refolding of mitochondrial proteins,
representing a potential serum biomarker of high prognostic value
for patients with colorectal cancer (276). Additionally, Cruz et al
demonstrated that this protein is a candidate biomarker of
drug-resistant disease in ovarian cancer cell lines and tissues
(277), while Niu et al
described its involvement in modulating oncogenic Dbl-driven
endocytosis (278).
Among the 16 different subunits composing the ATP
synthase, two were initially reported to be overexpressed in
cancers and to be associated with histological grade (279), the α-subunit, a major component
of the catalytic F1 head, and the d-subunit, a major
component of the F0 membranous domain (280).
Single-stranded DNA binding protein (SSBP) is
another specific mitochondrial protein that maintains the
structural stability of the mitochondrial genome by binding to
single-stranded mtDNA. This protein regulates mitochondrial
function and metabolism, and its level correlates with cancer cell
aggressiveness. Thus, novel treatment strategies aimed at its
downregulation have been proposed to increase the accumulation of
ROS, decrease key glycolytic enzymes and finally, enhance the
radiosensitivity of lung cancer cells (283). The role of this protein in the
regulation of the base excision repair pathway (284) and in the protection against DNA
damage events (285) has recently
been investigated.
In addition to Ki-67, proliferating cell nuclear
antigen (PCNA) has been used in immunohistochemistry experiments,
demonstrating that these two proteins are associated with each
other, and with tumor grade and stage (286). Its value as an independent
predictor of histological grade, recurrence rate and prognosis were
subsequently confirmed in a number of studies on gastric cancer
(287), hepatocellular carcinoma
(288) and non-small cell lung
cancer (289), highlighting in
particular its interest, in association with p53, for the
characterization of the invasive front of carcinomas (290).
The development of chromatin immunoprecipitation
(ChIP) has been crucial to the study of protein-DNA interactions,
leading in particular to the elucidation of the role of the A
family of type V intermediate filaments (LMNA) (291) in maintaining the positional
stability of DNA repair foci in mammalian nuclei (292). As the principal component of the
lamina, the meshwork of proteins at the nucleoplasmic side of the
inner nuclear membrane, lamins provide mechanical steadiness to the
cell nucleus by protecting it from mechanical forces (293). Kim et al described the
mechansims through which lamin A/C mediates the formation of the
perinuclear apical actin cables to protect the nuclear structural
integrity (294). In complement
to that study, Taheri et al also demonstrated the role of
lamin A in determining the viscoelasticity of the chromatin network
(295). Notably, Zuo et al
recently reported that differences in lamin A/C expression patterns
between high and low Gleason scores in prostate cancer tissues was
not associated with LMNA mutations, but rather with EMT or
MET processes (296).
The past few years have seen a growing interest in
the study of keratin-cancer associations, with some important
recent reviews on this subject (297,298). In particular, certain researchers
have focused on cytokeratins 8 (K2C8) and 18 (K1C18), which allow
enrichment in circulating tumor cells (299). The fact that K2C8 constitutes an
important part of the cytoskeleton and is involved in the
migration, invasion and metastasis of small-cell lung carcinoma
cells led Erlandsson et al to develop a novel treatment
protocol for this type of cancer based on the use of an
anti-keratin 8 antibody (300). A
high expression of K1, K8 and K18 has been associated with a poor
survival and a higher risk of recurrence (301), suggesting that these keratins
function as sensors of changing epithelia (297). Finally, the K8/K18 pair has been
demonstrated to modulate α6β4 integrin-mediated signaling with an
impact on cancer progression (302).
Perturbations of chromatin remodeling complexes
have been well-documented in malignant progression, in particular
when EMT is involved (303). In
these studies, H4 can undergo post-translational modifications, as
it belongs to the four types of core histones forming the octamer
units of nucleosome core particles (304). Together with H3, H4 is also
subject to a wide variation in the abundance of acetylation of its
lysine residues during the reprogramming of somatic cells
(fibroblasts) into induced pluripotent stem cells (305).
The following two proteins have been suggested to
be localized in the cytoplasm, although to date, there is no
further documentation in the literature.
Another isoform of the 14-3-3 family of proteins
mentioned above, 1433Z, which binds to several different enzymes
and may reduce apoptosis, has been associated with induction of
tumorigenesis in mice (306).
Notably, the proteomic characterization of the tumor-promoting
rearrangements of the lungs in a model of metastatic breast cancer
in the mouse revealed that 1433Z was included in a small list of
proteins differentially expressed at a stage corresponding to
secretion of tumor-derived factors (307).
Dihydropyrimidinase-like 3 (DPYL3), which interacts
witih ezrin, was first identified as a biomarker differentially
expressed in several pancreatic ductal adenocarcinoma cell lines
originating from liver metastasis in contrast to others originating
from lymph node metastasis and the primary tumor (308). Of note, recent findings by
Matsunuma et al revealed that the dysregulation of this
protein was specific to a subset of triple-negative breast cancers
characterized by low expression of claudins and E-cadherin and high
levels of mesenchymal biomarkers (309), while Yang et al
demonstrated that its inhibition promoted the metastasis of lung
cancer (310).
The stringent methodology used in the present
review to select biomarkers of interest led to the exclusion of a
number of important candidates from the list. To extend the
approach of the present review to additional molecules reported in
the recent literature and which are involved in the cancer
metastatic process, below, three examples are provided that may
offer new perspectives.
A first example is represented by S100B, another
member of the S100 family discussed above in the section entitled
'S100 proteins'. This protein has been a subject of cancer research
since 1983 and since then, 377 references have been published, with
a continuous increased observed from 1995. The reason for this
interest is based on the observation that S100B improves the early
diagnosis, staging and prognosis of malignant melanoma (311,312), and that its high expression
promotes self-renewal and tumorigenicity in ovarian cancer stem
cells (313). Moreover, the serum
S100B level has been shown to be independently associated with a
poor outcome of patients with metastatic breast cancer (314), while treatment with S100B
inhibitors blocks glioma growth through the alteration of the
polarization and trafficking of tumor-associated myeloid-derived
cells (315). Notably, although
S100B was not detected in the data described in the present review,
a recent study based on an integrative analysis of transcriptomic
and proteomic data of MCF7 cells submitted to acid adaptation
reported high expression levels of both S100B and S100A6 in the
course of EMT process affecting breast cancer cells (316). Additionally, proteomics and
microarray data from breast cancer patients have revealed a shorter
long-term survival in two subsets of patients with a combined high
expression of S100B, kallikrein and S100A7 or S100A14 - S100A16
(317).
The third example is represented by the bZIP
transcription factor, nuclear factor erythroid 2-related factor 2
(NRF2), initially highlighted in studies on chemopreventive agents
during investigations of the benefit conferred by consumption of
fruits and vegetables on the reduced incidence of cancer (325). Interest for this protein in
cancerology grew considerably, beginning in 2004, and then
exponentially since 2010, reaching as many as 555 references in
2019. Recent reviews have highlighted the importance of its
modulation for cancer chemoprevention and therapy (326), and the crucial role of the
disruption of KEAP1 binding to NRF2 (327). To date, the NRF2 pathway
represents a driver of cancer progression, metastasis and
resistance to therapy (328). To
provide mechanical insight into the implications of this pathway in
lung cancer metastasis, Lignitto et al dissected the
molecular events regulating the simultaneous loss of KEAP1 and
stabilization of the transcriptional regulator Bach1 (329). These authors demonstrated that
free heme promoted the physical interaction between Bach1 and
Fbxo22, a substrate receptor of the CRL1 complex, by inducing Ho1,
a heme-catabolizing enzyme (329). Subsequently, Wiel et al
revealed that the reduction of free heme by long-term
supplementation with antioxidants also stimulated metastasis by
stabilizing Bach1, leading to increased glycolysis rates (330). Finally, an interesting point with
regard to NRF2 is the increased abundance in CD44 antigen found in
a most aggressive model of rat MM (M5-T1), compared to the two less
invasive ones (F4-T2 and F5-T1) (21). This suggests a link with HMGA1
mentioned above (320), and
illustrates the importance of the CD44-NRF2 axis described in
breast cancer stem cell-like cells (331).
The aim of the present review was to establish a
list of potential biomarkers of cancer invasiveness at the
crossroads between literature data and experimental and clinical
data, which may provide the groundwork for both basic science and
translational studies. Although a number of other proteins remain
outside the focus of this review, the main point is that this list
represents quantitative changes which are common to different
cancer types and locations. In the field of basic science, a first
question concerns a number of biomarkers that are increased or
decreased in most/all situations and which could therefore
represent an important tool with which to understand the biological
system considered as a whole (at the cancer cell or tumor scale).
This approach may assist in the detection of defects within the
network, the stoichiometry of components and their connectivity.
Another question is why some of these biomarkers exhibit various
quantitative patterns of change (increase or decrease) according to
the different situations considered. On the translational side, the
use of combined biomarkers could contribute to the diagnosis and
prognosis of certain types of aggressive cancers, for example
malignant mesothelioma, for which improvements are urgently
required. This tool could also help to define and evaluate more
accurate therapeutic strategies. Finally, a question which is
beyond the scope of this review, is how it would be possible to
prioritize these different biomarkers in a given context. This will
certainly offer interesting prospects in this fascinating field of
research.
The present study was conducted with the support of
the French National Health and Medical Research Institute (INSERM),
the Ligue contre le Cancer (Ligue inter-régionale du Grand Ouest,
comités 16, 29, 44, 72), and a grant from the 'Comité Féminin 49
Octobre Rose'.
Not applicable.
DLP wrote the manuscript on the basis of
experimental and clinical data obtained with the support of CG, AB
and OC, who were also involved in drafting the review. All authors
read and approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Coates AS, Winer EP, Goldhirsch A, Gelber
RD, Gnant M, Piccart-Gebhart M, Thürlimann B, Senn H-J, André F,
Baselga J, et al: Panel Members: Tailoring therapies - improving
the management of early breast cancer: St Gallen International
Expert Consensus on the Primary Therapy of Early Breast Cancer
2015. Ann Oncol. 26:1533–1546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi
Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al: NCI CPTAC:
Proteogenomic characterization of human colon and rectal cancer.
Nature. 513:382–387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johansson HJ, Socciarelli F, Vacanti NM,
Haugen MH, Zhu Y, Siavelis I, Fernandez-Woodbridge A, Aure MR,
Sennblad B, Vesterlund M, et al: Consortia Oslo Breast Cancer
Research Consortium (OSBREAC): Breast cancer quantitative proteome
and proteogenomic landscape. Nat Commun. 10:16002019. View Article : Google Scholar
|
|
5
|
Mann M: Quantitative proteomics? Nat
Biotechnol. 17:954–955. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Monti C, Zilocchi M, Colugnat I and
Alberio T: Proteomics turns functional. J Proteomics. 198:36–44.
2019. View Article : Google Scholar
|
|
7
|
Simpson RJ and Dorow DS: Cancer
proteomics: From signaling networks to tumor markers. Trends
Biotechnol. 19(Suppl): S40–S48. 2001. View Article : Google Scholar
|
|
8
|
Cheung CHY and Juan HF: Quantitative
proteomics in lung cancer. J Biomed Sci. 24:372017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Geiger T and Geiger B: Towards elucidation
of functional molecular signatures of the adhesive-migratory
phenotype of malignant cells. Semin Cancer Biol. 20:146–152. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jazurek M, Ciesiolka A, Starega-Roslan J,
Bilinska K and Krzyzosiak WJ: Identifying proteins that bind to
specific RNAs - focus on simple repeat expansion diseases. Nucleic
Acids Res. 44:9050–9070. 2016.PubMed/NCBI
|
|
11
|
Abazova N and Krijgsveld J: Advances in
stem cell proteomics. Curr Opin Genet Dev. 46:149–155. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eubanks CG, Dayebgadoh G, Liu X and
Washburn MP: Unravelling the biology of chromatin in health and
cancer using proteomic approaches. Expert Rev Proteomics.
14:905–915. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gómez-Serrano M, Camafeita E, Loureiro M
and Peral B: Mitoproteomics: Tackling mitochondrial dysfunction in
human disease. Oxid Med Cell Longev. 2018:14359342018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Suwakulsiri W, Rai A, Chen M, Greening DW
and Simpson RJ: Proteomic profiling reveals key cancer progression
modulators in shed microvesicles released from isogenic human
primary and metastatic colorectal cancer cell lines. Biochim
Biophys Acta Proteins Proteom. 1867:1401712019. View Article : Google Scholar
|
|
15
|
Li Z, Li N, Shen L and Fu J: Quantitative
proteomic analysis identifies MAPK15 as a potential regulator of
radioresistance in nasopharyngeal carcinoma cells. Front Oncol.
8:5482018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gillet L, Navarro P, Tate S, Röst H,
Selevsek N, Reiter L, Bonner R and Aebersold R: Targeted data
extraction of the MS/MS spectra generated by data-independent
acquisition: a new concept for consistent and accurate proteome
analysis. Mol Cell Proteomics. 11:pp. O111.0167172012, View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ludwig C, Gillet L, Rosenberger G, Amon S,
Collins BC and Aebersold R: Data-independent acquisition-based
SWATH-MS for quantitative proteomics: A tutorial. Mol Syst Biol.
14:pp. e81262018, View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krisp C and Molloy MP: SWATH mass
spectrometry for proteomics of non-depleted plasma. Methods Mol
Biol. 1619:373–383. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jamwal R, Barlock BJ, Adusumalli S,
Ogasawara K, Simons BL and Akhlaghi F: Multiplex and label-free
relative quantification approach for studying protein abundance of
drug metabolizing enzymes in human liver microsomes using SWATH-MS.
J Proteome Res. 16:4134–4143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Skotland T, Berge V, Sandvig K and
Llorente A: Exosomal proteins as prostate cancer biomarkers in
urine: From mass spectrometry discovery to immunoassay-based
validation. Eur J Pharm Sci. 98:80–85. 2017. View Article : Google Scholar
|
|
21
|
Nader JS, Abadie J, Deshayes S, Boissard
A, Blandin S, Blanquart C, Boisgerault N, Coqueret O, Guette C,
Grégoire M, et al: Characterization of increasing stages of
invasiveness identifies stromal/cancer cell crosstalk in rat models
of mesothelioma. Oncotarget. 9:16311–16329. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Besson D, Pavageau A-H, Valo I, Bourreau
A, Bélanger A, Eymerit-Morin C, Moulière A, Chassevent A,
Boisdron-Celle M, Morel A, et al: A quantitative proteomic approach
of the different stages of colorectal cancer establishes OLFM4 as a
new nonmetastatic tumor marker. Mol Cell Proteomics. 10:0097122011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Valo I, Raro P, Boissard A, Maarouf A,
Jézéquel P, Verriele V, Campone M, Coqueret O and Guette C: OLFM4
expression in ductal carcinoma in situ and in invasive breast
cancer cohorts by a SWATH-based proteomic approach. Proteomics.
19:pp. e18004462019, View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Powell AA, Talasaz AH, Zhang H, Coram MA,
Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et
al: Single cell profiling of circulating tumor cells:
Transcriptional heterogeneity and diversity from breast cancer cell
lines. PLoS One. 7:e337882012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lukanidin E and Sleeman JP: Building the
niche: The role of the S100 proteins in metastatic growth. Semin
Cancer Biol. 22:216–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bresnick AR, Weber DJ and Zimmer DB: S100
proteins in cancer. Nat Rev Cancer. 15:96–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shabani F, Farasat A, Mahdavi M and Gheibi
N: Calprotectin (S100A8/S100A9): A key protein between inflammation
and cancer. Inflamm Res. 67:801–812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Björk P, Källberg E, Wellmar U, Riva M,
Olsson A, He Z, Törngren M, Liberg D, Ivars F and Leanderson T:
Common interactions between S100A4 and S100A9 defined by a novel
chemical probe. PLoS One. 8:pp. e630122013, View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fei F, Qu J, Zhang M, Li Y and Zhang S:
S100A4 in cancer progression and metastasis: A systematic review.
Oncotarget. 8:73219–73239. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshimura H, Otsuka A, Michishita M,
Yamamoto M, Ashizawa M, Zushi M, Moriya M, Azakami D, Ochiai K,
Matsuda Y, et al: Expression and roles of S100A4 in anaplastic
cells of canine mammary carcinomas. Vet Pathol. 56:389–398. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y and Cao X: Characteristics and
significance of the pre-metastatic niche. Cancer Cell. 30:668–681.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hansen MT, Forst B, Cremers N, Quagliata
L, Ambartsumian N, Grum-Schwensen B, Klingelhöfer J, Abdul-Al A,
Herrmann P, Osterland M, et al: A link between inflammation and
metastasis: Serum amyloid A1 and A3 induce metastasis, and are
targets of metastasis-inducing S100A4. Oncogene. 34:424–435. 2015.
View Article : Google Scholar
|
|
33
|
Mahmood MQ, Ward C, Muller HK, Sohal SS
and Walters EH: Epithelial mesenchymal transition (EMT) and
non-small cell lung cancer (NSCLC): A mutual association with
airway disease. Med Oncol. 34:452017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu M, Liu J, Yang B, Gao X, Gao LL, Kong
QY, Zhang P and Li H: Inversed expression patterns of S100A4 and
E-cadherin in cervical cancers: Implication in
epithelial-mesenchymal transition. Anat Rec (Hoboken). 300:pp.
2184–2191. 2017, View Article : Google Scholar
|
|
35
|
Roulois D, Deshayes S, Guilly MN, Nader
JS, Liddell C, Robard M, Hulin P, Ouacher A, Le Martelot V,
Fonteneau JF, et al: Characterization of preneoplastic and
neoplastic rat mesothelial cell lines: The involvement of TETs,
DNMTs, and 5-hydroxy-methylcytosine. Oncotarget. 7:34664–34687.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Donato R, Sorci G and Giambanco I: S100A6
protein: Functional roles. Cell Mol Life Sci. 74:2749–2760. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lyu X, Li H, Ma X, Li X, Gao Y, Ni D, Shen
D, Gu L, Wang B, Zhang Y, et al: High-level S100A6 promotes
metastasis and predicts the outcome of T1-T2stage in clear cell
renal cell carcinoma. Cell Biochem Biophys. 71:279–290. 2015.
View Article : Google Scholar
|
|
38
|
Luo X, Sharff KA, Chen J, He T-C and Luu
HH: S100A6 expression and function in human osteosarcoma. Clin
Orthop Relat Res. 466:2060–2070. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Zhang K, Jiang X and Zhang J:
S100A6 as a potential serum prognostic biomarker and therapeutic
target in gastric cancer. Dig Dis Sci. 59:2136–2144. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Popa SJ, Stewart SE and Moreau K:
Unconventional secretion of annexins and galectins. Semin Cell Dev
Biol. 83:42–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gerke V, Creutz CE and Moss SE: Annexins:
Linking Ca2+ signalling to membrane dynamics. Nat Rev
Mol Cell Biol. 6:449–461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qi H, Liu S, Guo C, Wang J, Greenaway FT
and Sun M-Z: Role of annexin A6 in cancer (Review). Oncol Lett.
10:1947–1952. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Grewal T, Hoque M, Conway JRW, Reverter M,
Wahba M, Beevi SS, Timpson P, Enrich C and Rentero C: Annexin A6-A
multifunctional scaffold in cell motility. Cell Adhes Migr.
11:288–304. 2017. View Article : Google Scholar
|
|
44
|
Sakwe AM, Koumangoye R, Guillory B and
Ochieng J: Annexin A6 contributes to the invasiveness of breast
carcinoma cells by influencing the organization and localization of
functional focal adhesions. Exp Cell Res. 317:823–837. 2011.
View Article : Google Scholar :
|
|
45
|
Keklikoglou I, Cianciaruso C, Güç E,
Squadrito ML, Spring LM, Tazzyman S, Lambein L, Poissonnier A,
Ferraro GB, Baer C, et al: Chemotherapy elicits pro-metastatic
extracellular vesicles in breast cancer models. Nat Cell Biol.
21:190–202. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Koumangoye RB, Nangami GN, Thompson PD,
Agboto VK, Ochieng J and Sakwe AM: Reduced annexin A6 expression
promotes the degradation of activated epidermal growth factor
receptor and sensitizes invasive breast cancer cells to
EGFR-targeted tyrosine kinase inhibitors. Mol Cancer. 12:1672013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
García-Melero A, Reverter M, Hoque M,
Meneses-Salas E, Koese M, Conway JRW, Johnsen CH, Alvarez-Guaita A,
Morales-Paytuvi F, Elmaghrabi YA, et al: Annexin A6 and late
endosomal cholesterol modulate integrin recycling and cell
migration. J Biol Chem. 291:1320–1335. 2016. View Article : Google Scholar :
|
|
48
|
Widatalla SE, Korolkova OY, Whalen DS,
Goodwin JS, Williams KP, Ochieng J and Sakwe AM: Lapatinib-induced
annexin A6 upregulation as an adaptive response of triple-negative
breast cancer cells to EGFR tyrosine kinase inhibitors.
Carcinogenesis. 40:998–1009. 2019. View Article : Google Scholar :
|
|
49
|
Leca J, Martinez S, Lac S, Nigri J, Secq
V, Rubis M, Bressy C, Sergé A, Lavaut M-N, Dusetti N, et al:
Cancer-associated fibroblast-derived annexin A6+
extracellular vesicles support pancreatic cancer aggressiveness. J
Clin Invest. 126:4140–4156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Whalen DS, Widatalla SE, Korolkova OY,
Nangami GS, Beasley HK, Williams SD, Virgous C, Lehmann BD, Ochieng
J and Sakwe AM: Implication of calcium activated RasGRF2 in Annexin
A6-mediated breast tumor cell growth and motility. Oncotarget.
10:133–151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sheikh MH and Solito E: Annexin A1:
Uncovering the many talents of an old protein. Int J Mol Sci.
19:10452018. View Article : Google Scholar :
|
|
52
|
Babbin BA, Lee WY, Parkos CA, Winfree LM,
Akyildiz A, Perretti M and Nusrat A: Annexin I regulates SKCO-15
cell invasion by signaling through formyl peptide receptors. J Biol
Chem. 281:19588–19599. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Guo C, Liu S and Sun M-Z: Potential role
of Anxa1 in cancer. Future Oncol. 9:1773–1793. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Swa HLF, Shaik AA, Lim LHK and Gunaratne
J: Mass spectrometry based quantitative proteomics and integrative
network analysis accentuates modulating roles of annexin-1 in
mammary tumorigenesis. Proteomics. 15:408–418. 2015. View Article : Google Scholar
|
|
55
|
Okano M, Kumamoto K, Saito M, Onozawa H,
Saito K, Abe N, Ohtake T and Takenoshita S: Upregulated Annexin A1
promotes cellular invasion in triple-negative breast cancer. Oncol
Rep. 33:1064–1070. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tu Y, Johnstone CN and Stewart AG: Annexin
A1 influences in breast cancer: Controversies on contributions to
tumour, host and immunoediting processes. Pharmacol Res.
119:278–288. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liao S-H, Zhao X-Y, Han Y-H, Zhang J, Wang
L-S, Xia L, Zhao K-W, Zheng Y, Guo M and Chen G-Q: Proteomics-based
identification of two novel direct targets of hypoxia-inducible
factor-1 and their potential roles in migration/invasion of cancer
cells. Proteomics. 9:3901–3912. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu Y, Wang J, Xu Y, Xiao H, Li J and Wang
Z: Screening critical genes associated with malignant glioma using
bioinformatics analysis. Mol Med Rep. 16:6580–6589. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kang H, Ko J and Jang S-W: The role of
annexin A1 in expression of matrix metalloproteinase-9 and invasion
of breast cancer cells. Biochem Biophys Res Commun. 423:188–194.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Grindheim AK, Saraste J and Vedeler A:
Protein phosphorylation and its role in the regulation of Annexin
A2 function. Biochim Biophys Acta, Gen Subj. 1861A:A2515–A2529.
2017. View Article : Google Scholar
|
|
61
|
Christensen MV, Høgdall CK, Jochumsen KM
and Høgdall EVS: Annexin A2 and cancer: A systematic review. Int J
Oncol. 52:5–18. 2018.
|
|
62
|
Maule F, Bresolin S, Rampazzo E, Boso D,
Della Puppa A, Esposito G, Porcù E, Mitola S, Lombardi G, Accordi
B, et al: Annexin 2A sustains glioblastoma cell dissemination and
proliferation. Oncotarget. 7:54632–54649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu Y, Li H, Ban Z, Nai M, Yang L, Chen Y
and Xu Y: Annexin A2 inhibition suppresses ovarian cancer
progression via regulating β-catenin/EMT. Oncol Rep. 37:3643–3650.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rocha MR, Barcellos-de-Souza P,
Sousa-Squiavinato ACM, Fernandes PV, de Oliveira IM, Boroni M and
Morgado-Diaz JA: Annexin A2 overexpression associates with
colorectal cancer invasiveness and TGF-β induced epithelial
mesenchymal transition via Src/ANXA2/STAT3. Sci Rep. 8:112852018.
View Article : Google Scholar
|
|
65
|
Yoneura N, Takano S, Yoshitomi H, Nakata
Y, Shimazaki R, Kagawa S, Furukawa K, Takayashiki T, Kuboki S,
Miyazaki M, et al: Expression of annexin II and stromal tenascin C
promotes epithelial to mesenchymal transition and correlates with
distant metastasis in pancreatic cancer. Int J Mol Med. 42:821–830.
2018.PubMed/NCBI
|
|
66
|
Zhang Q, Zhao Z, Ma Y, Wang H, Ma J, He X
and Zhang D: Combined expression of S100A4 and Annexin A2 predicts
disease progression and overall survival in patients with
urothelial carcinoma. Urol Oncol. 32:798–805. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang Q, Ye Z, Yang Q, He X, Wang H and
Zhao Z: Upregulated expression of annexin II is a prognostic marker
for patients with gastric cancer. World J Surg Oncol. 10:1032012.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Korwar AM, Bhonsle HS, Chougale AD, Kote
SS, Gawai KR, Ghole VS, Koppikar CB and Kulkarni MJ: Analysis of
AGE modified proteins and RAGE expression in HER2/neu negative
invasive ductal carcinoma. Biochem Biophys Res Commun. 419:490–494.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Sheng SH and Zhu HL: Proteomic analysis of
pleural effusion from lung adenocarcinoma patients by shotgun
strategy. Clin Transl Oncol. 16:153–157. 2014. View Article : Google Scholar
|
|
70
|
Ricciardelli C, Lokman NA, Ween MP and
Oehler MK: WOMEN IN CANCER THEMATIC REVIEW: Ovarian
cancer-peritoneal cell interactions promote extracellular matrix
processing. Endocr Relat Cancer. 23:T155–T168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
van den Brûle FA, Buicu C, Berchuck A,
Bast RC, Deprez M, Liu F-T, Cooper DNW, Pieters C, Sobel ME and
Castronovo V: Expression of the 67-kD laminin receptor, galectin-1,
and galectin-3 in advanced human uterine adenocarcinoma. Hum
Pathol. 27:1185–1191. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Camby I, Belot N, Rorive S, Lefranc F,
Maurage C-A, Lahm H, Kaltner H, Hadari Y, Ruchoux MM, Brotchi J, et
al: Galectins are differentially expressed in supratentorial
pilocytic astrocytomas, astrocytomas, anaplastic astrocytomas and
glioblastomas, and significantly modulate tumor astrocyte
migration. Brain Pathol. 11:12–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Astorgues-Xerri L, Riveiro ME,
Tijeras-Raballand A, Serova M, Neuzillet C, Albert S, Raymond E and
Faivre S: Unraveling galectin-1 as a novel therapeutic target for
cancer. Cancer Treat Rev. 40:307–319. 2014. View Article : Google Scholar
|
|
74
|
Cousin JM and Cloninger MJ: The role of
galectin-1 in cancer progression, and synthetic multivalent systems
for the study of galectin-1. Int J Mol Sci. 17:15662016. View Article : Google Scholar :
|
|
75
|
Bhat R, Belardi B, Mori H, Kuo P, Tam A,
Hines WC, Le Q-T, Bertozzi CR and Bissell MJ: Nuclear
repartitioning of galectin-1 by an extracellular glycan switch
regulates mammary morphogenesis. Proc Natl Acad Sci USA.
113:E4820–E4827. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Shen K-H, Li C-F, Chien L-H, Huang C-H, Su
C-C, Liao AC and Wu T-F: Role of galectin-1 in urinary bladder
urothelial carcinoma cell invasion through the JNK pathway. Cancer
Sci. 107:1390–1398. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chong Y, Tang D, Xiong Q, Jiang X, Xu C,
Huang Y, Wang J, Zhou H, Shi Y, Wu X, et al: Galectin-1 from
cancer-associated fibroblasts induces epithelial-mesenchymal
transition through β1 integrin-mediated upregulation of Gli1 in
gastric cancer. J Exp Clin Cancer Res. 35:1752016. View Article : Google Scholar
|
|
78
|
Chong Y, Tang D, Gao J, Jiang X, Xu C,
Xiong Q, Huang Y, Wang J, Zhou H, Shi Y, et al: Galectin-1 induces
invasion and the epithelial-mesenchymal transition in human gastric
cancer cells via non-canonical activation of the hedgehog signaling
pathway. Oncotarget. 7:83611–83626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang P-F, Li K-S, Shen YH, Gao P-T, Dong
Z-R, Cai J-B, Zhang C, Huang X-Y, Tian M-X, Hu Z-Q, et al:
Galectin-1 induces hepatocellular carcinoma EMT and sorafenib
resistance by activating FAK/PI3K/AKT signaling. Cell Death Dis.
7:e22012016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Qian D, Lu Z, Xu Q, Wu P, Tian L, Zhao L,
Cai B, Yin J, Wu Y, Staveley-O'Carroll KF, et al: Galectin-1-driven
upregulation of SDF-1 in pancreatic stellate cells promotes
pancreatic cancer metastasis. Cancer Lett. 397:43–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Noda Y, Kishino M, Sato S, Hirose K, Sakai
M, Fukuda Y, Murakami S and Toyosawa S: Galectin-1 expression is
associated with tumour immunity and prognosis in gingival squamous
cell carcinoma. J Clin Pathol. 70:126–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Song L, Tang JW, Owusu L, Sun M-Z, Wu J
and Zhang J: Galectin-3 in cancer. Clin Chim Acta. 431:185–191.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ajani JA, Estrella JS, Chen Q, Correa AM,
Ma L, Scott AW, Jin J, Liu B, Xie M, Sudo K, et al: Galectin-3
expression is prognostic in diffuse type gastric adenocarcinoma,
confers aggressive phenotype, and can be targeted by YAP1/BET
inhibitors. Br J Cancer. 118:52–61. 2018. View Article : Google Scholar :
|
|
84
|
Ruvolo PP: Galectin 3 as a guardian of the
tumor microenvi-ronment. Biochim Biophys Acta. 1863:427–437. 2016.
View Article : Google Scholar
|
|
85
|
Cardoso AC, Andrade LN, Bustos SO and
Chammas R: Galectin-3 determines tumor cell adaptive strategies in
stressed tumor microenvironments. Front Oncol. 6:1272016.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mackay A, Jones C, Dexter T, Silva RL,
Bulmer K, Jones A, Simpson P, Harris RA, Jat PS, Neville AM, et al:
cDNA microarray analysis of genes associated with ERBB2 (HER2/neu)
overexpression in human mammary luminal epithelial cells. Oncogene.
22:2680–2688. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Nomura T and Katunuma N: Involvement of
cathepsins in the invasion, metastasis and proliferation of cancer
cells. J Med Invest. 52:1–9. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Derocq D, Prébois C, Beaujouin M,
Laurent-Matha V, Pattingre S, Smith GK and Liaudet-Coopman E:
Cathepsin D is partly endocytosed by the LRP1 receptor and inhibits
LRP1-regulated intramembrane proteolysis. Oncogene. 31:3202–3212.
2012. View Article : Google Scholar
|
|
89
|
Dubey V and Luqman S: Cathepsin D as a
promising target for the discovery of novel anticancer agents. Curr
Cancer Drug Targets. 17:404–422. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Brown KD: Transglutaminase 2 and NF-κB: An
odd couple that shapes breast cancer phenotype. Breast Cancer Res
Treat. 137:329–336. 2013. View Article : Google Scholar
|
|
91
|
Yang P, Yu D, Zhou J, Zhuang S and Jiang
T: TGM2 interference regulates the angiogenesis and apoptosis of
colorectal cancer via Wnt/β-catenin pathway. Cell Cycle.
18:1122–1134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Eble JA and Niland S: The extracellular
matrix in tumor progression and metastasis. Clin Exp Metastasis.
36:171–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Garamszegi N, Garamszegi SP, Shehadeh LA
and Scully SP: Extracellular matrix-induced gene expression in
human breast cancer cells. Mol Cancer Res. 7:319–329. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Giehl K and Menke A: Microenvironmental
regulation of E-cadherin-mediated adherens junctions. Front Biosci.
13:3975–3985. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yang S, Shin J, Park KH, Jeung H-C, Rha
SY, Noh SH, Yang WI and Chung HC: Molecular basis of the
differences between normal and tumor tissues of gastric cancer.
Biochim Biophys Acta. 1772:1033–1040. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Liang Y, Diehn M, Bollen AW, Israel MA and
Gupta N: Type I collagen is overexpressed in medulloblastoma as a
component of tumor microenvironment. J Neurooncol. 86:133–141.
2008. View Article : Google Scholar
|
|
97
|
Montgomery H, Rustogi N, Hadjisavvas A,
Tanaka K, Kyriacou K and Sutton CW: Proteomic profiling of breast
tissue collagens and site-specific characterization of
hydroxyproline residues of collagen alpha-1-(I). J Proteome Res.
11:5890–5902. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Mitsuhashi A, Goto H, Saijo A, Trung VT,
Aono Y, Ogino H, Kuramoto T, Tabata S, Uehara H, Izumi K, et al:
Fibrocyte-like cells mediate acquired resistance to anti-angiogenic
therapy with bevacizumab. Nat Commun. 6:87922015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Rong L, Huang W, Tian S, Chi X, Zhao P and
Liu F: COL1A2 is a novel biomarker to improve clinical prediction
in human gastric cancer: Integrating bioinformatics and
meta-analysis. Pathol Oncol Res. 24:129–134. 2018. View Article : Google Scholar
|
|
100
|
Yang X, Staren ED, Howard JM, Iwamura T,
Bartsch JE and Appert HE: Invasiveness and MMP expression in
pancreatic carcinoma. J Surg Res. 98:33–39. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Parmo-Cabañas M, Molina-Ortiz I,
Matías-Román S, García-Bernal D, Carvajal-Vergara X, Valle I,
Pandiella A, Arroyo AG and Teixidó J: Role of metalloproteinases
MMP-9 and MT1-MMP in CXCL12-promoted myeloma cell invasion across
basement membranes. J Pathol. 208:108–118. 2006. View Article : Google Scholar
|
|
102
|
Ren F, Tang R, Zhang X, Madushi WM, Luo D,
Dang Y, Li Z, Wei K and Chen G: Overexpression of MMP family
members functions as prognostic biomarker for breast cancer
patients: A systematic review and meta-analysis. PLoS One. 10:pp.
e01355442015, View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Liu H-Y, Gu W-J, Wang C-Z, Ji X-J and Mu
Y-M: Matrix metalloproteinase-9 and -2 and tissue inhibitor of
matrix metalloproteinase-2 in invasive pituitary adenomas: A
systematic review and meta-analysis of case-control trials.
Medicine (Baltimore). 95:pp. e39042016, View Article : Google Scholar
|
|
104
|
Delassus GS, Cho H, Park J and Eliceiri
GL: New pathway links from cancer-progression determinants to gene
expression of matrix metalloproteinases in breast cancer cells. J
Cell Physiol. 217:739–744. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Franco-Barraza J, Valdivia-Silva JE,
Zamudio-Meza H, Castillo A, García-Zepeda EA, Benítez-Bribiesca L
and Meza I: Actin cytoskeleton participation in the onset of
IL-1beta induction of an invasive mesenchymal-like phenotype in
epithelial MCF-7 cells. Arch Med Res. 41:170–181. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Matsuura I, Lai C-Y and Chiang K-N:
Functional interaction between Smad3 and S100A4 (metastatin-1) for
TGF-beta-mediated cancer cell invasiveness. Biochem J. 426:327–335.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Oskarsson T: Extracellular matrix
components in breast cancer progression and metastasis. Breast.
22(Suppl 2): S66–S72. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kaplan RN, Riba RD, Zacharoulis S, Bramley
AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et
al: VEGFR1-positive haematopoietic bone marrow progenitors initiate
the pre-metastatic niche. Nature. 438:820–827. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Topalovski M and Brekken RA: Matrix
control of pancreatic cancer: New insights into fibronectin
signaling. Cancer Lett. 381:252–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Liao Y-X, Zhang Z-P, Zhao J and Liu J-P:
Effects of fibronectin 1 on cell proliferation, senescence and
apoptosis of human glioma cells through the PI3K/AKT signaling
pathway. Cell Physiol Biochem. 48:1382–1396. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Sercu S, Zhang L and Merregaert J: The
extracellular matrix protein 1: Its molecular interaction and
implication in tumor progression. Cancer Invest. 26:375–384. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Bergamaschi A, Tagliabue E, Sørlie T,
Naume B, Triulzi T, Orlandi R, Russnes HG, Nesland JM, Tammi R,
Auvinen P, et al: Extracellular matrix signature identifies breast
cancer subgroups with different clinical outcome. J Pathol.
214:357–367. 2008. View Article : Google Scholar
|
|
113
|
Lal G, Hashimi S, Smith BJ, Lynch CF,
Zhang L, Robinson RA and Weigel RJ: Extracellular matrix 1 (ECM1)
expression is a novel prognostic marker for poor long-term survival
in breast cancer: A Hospital-based Cohort Study in Iowa. Ann Surg
Oncol. 16:2280–2287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Lee KM, Nam K, Oh S, Lim J, Kim RK, Shim
D, Choi JH, Lee S-J, Yu J-H, Lee JW, et al: ECM1 regulates tumor
metastasis and CSC-like property through stabilization of
β-catenin. Oncogene. 34:6055–6065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chen H, Jia W and Li J: ECM1 promotes
migration and invasion of hepatocellular carcinoma by inducing
epithelial-mesen-chymal transition. World J Surg Oncol. 14:1952016.
View Article : Google Scholar
|
|
116
|
Gómez-Contreras P, Ramiro-Díaz JM, Sierra
A, Stipp C, Domann FE, Weigel RJ and Lal G: Extracellular matrix 1
(ECM1) regulates the actin cytoskeletal architecture of aggressive
breast cancer cells in part via S100A4 and Rho-family GTPases. Clin
Exp Metastasis. 34:37–49. 2017. View Article : Google Scholar :
|
|
117
|
Gan L, Meng J, Xu M, Liu M, Qi Y, Tan C,
Wang Y, Zhang P, Weng W, Sheng W, et al: Extracellular matrix
protein 1 promotes cell metastasis and glucose metabolism by
inducing integrin β4/FAK/SOX2/HIF-1α signaling pathway in gastric
cancer. Oncogene. 37:744–755. 2018. View Article : Google Scholar
|
|
118
|
Troup S, Njue C, Kliewer EV, Parisien M,
Roskelley C, Chakravarti S, Roughley PJ, Murphy LC and Watson PH:
Reduced expression of the small leucine-rich proteoglycans,
lumican, and decorin is associated with poor outcome in
node-negative invasive breast cancer. Clin Cancer Res. 9:207–214.
2003.PubMed/NCBI
|
|
119
|
Vuillermoz B, Khoruzhenko A, D'Onofrio
M-F, Ramont L, Venteo L, Perreau C, Antonicelli F, Maquart F-X and
Wegrowski Y: The small leucine-rich proteoglycan lumican inhibits
melanoma progression. Exp Cell Res. 296:294–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Radwanska A, Litwin M, Nowak D, Baczynska
D, Wegrowski Y, Maquart F-X and Malicka-Blaszkiewicz M:
Overexpression of lumican affects the migration of human colon
cancer cells through up-regulation of gelsolin and filamentous
actin reorganization. Exp Cell Res. 318:2312–2323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Coulson-Thomas VJ, Coulson-Thomas YM,
Gesteira TF, Andrade de Paula CA, Carneiro CR, Ortiz V, Toma L, Kao
WW and Nader HB: Lumican expression, localization and antitumor
activity in prostate cancer. Exp Cell Res. 319:967–981. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
de Wit M, Carvalho B, Delis-van Diemen PM,
van Alphen C, Beliën JAM, Meijer GA and Fijneman RJA: Lumican and
versican protein expression are associated with colorectal
adenoma-to-carcinoma progression. PLoS One. 12:pp. e01747682017,
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Farace C, Oliver JA, Melguizo C, Alvarez
P, Bandiera P, Rama AR, Malaguarnera G, Ortiz R, Madeddu R and
Prados J: Microenvironmental modulation of decorin and lumican in
Temozolomide-resistant glioblastoma and neuroblastoma cancer
stem-like cells. PLoS One. 10:pp. e01341112015, View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Jeanne A, Untereiner V, Perreau C, Proult
I, Gobinet C, Boulagnon-Rombi C, Terryn C, Martiny L, Brézillon S
and Dedieu S: Lumican delays melanoma growth in mice and drives
tumor molecular assembly as well as response to matrix-targeted
TAX2 therapeutic peptide. Sci Rep. 7:77002017. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Gritsenko PG, Ilina O and Friedl P:
Interstitial guidance of cancer invasion. J Pathol. 226:185–199.
2012. View Article : Google Scholar
|
|
126
|
Ruan K, Bao S and Ouyang G: The
multifaceted role of periostin in tumorigenesis. Cell Mol Life Sci.
66:2219–2230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Kim G-E, Lee JS, Park MH and Yoon JH:
Epithelial periostin expression is correlated with poor survival in
patients with invasive breast carcinoma. PLoS One. 12:pp.
e01876352017, View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Mino M, Kanno K, Okimoto K, Sugiyama A,
Kishikawa N, Kobayashi T, Ono J, Izuhara K, Kobayashi T, Ohigashi
T, et al: Periostin promotes malignant potential by induction of
epithelial-mesenchymal transition in intrahepatic
cholangiocar-cinoma. Hepatol Commun. 1:1099–1109. 2017. View Article : Google Scholar
|
|
129
|
Sid B, Sartelet H, Bellon G, El Btaouri H,
Rath G, Delorme N, Haye B and Martiny L: Thrombospondin 1: A
multifunctional protein implicated in the regulation of tumor
growth. Crit Rev Oncol Hematol. 49:245–258. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Okada K, Hirabayashi K, Imaizumi T,
Matsuyama M, Yazawa N, Dowaki S, Tobita K, Ohtani Y, Tanaka M,
Inokuchi S, et al: Stromal thrombospondin-1 expression is a
prognostic indicator and a new marker of invasiveness in
intraductal papillary-mucinous neoplasm of the pancreas. Biomed
Res. 31:13–19. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Firlej V, Mathieu JRR, Gilbert C,
Lemonnier L, Nakhlé J, Gallou-Kabani C, Guarmit B, Morin A,
Prevarskaya N, Delongchamps NB, et al: Thrombospondin-1 triggers
cell migration and development of advanced prostate tumors. Cancer
Res. 71:7649–7658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Radziwon-Balicka A, Santos-Martinez MJ,
Corbalan JJ, O'Sullivan S, Treumann A, Gilmer JF, Radomski MW and
Medina C: Mechanisms of platelet-stimulated colon cancer invasion:
Role of clusterin and thrombospondin 1 in regulation of the
P38MAPK-MMP-9 pathway. Carcinogenesis. 35:324–332. 2014. View Article : Google Scholar
|
|
133
|
Joshi R, Goihberg E, Ren W, Pilichowska M
and Mathew P: Proteolytic fragments of fibronectin function as
matrikines driving the chemotactic affinity of prostate cancer
cells to human bone marrow mesenchymal stromal cells via the α5β1
integrin. Cell Adhes Migr. 11:305–315. 2017. View Article : Google Scholar
|
|
134
|
Lebdai S, Verhoest G, Parikh H, Jacquet
SF, Bensalah K, Chautard D, Rioux Leclercq N, Azzouzi AR and Bigot
P: Identification and validation of TGFBI as a promising prognosis
marker of clear cell renal cell carcinoma. Urol Oncol. 33:pp.
69.e11–69.e18. 2015, View Article : Google Scholar
|
|
135
|
Nummela P, Lammi J, Soikkeli J, Saksela O,
Laakkonen P and Hölttä E: Transforming growth factor beta-induced
(TGFBI) is an anti-adhesive protein regulating the invasive growth
of melanoma cells. Am J Pathol. 180:1663–1674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Klamer SE, Kuijk CG, Hordijk PL, van der
Schoot CE, von Lindern M, van Hennik PB and Voermans C: BIGH3
modulates adhesion and migration of hematopoietic stem and
progenitor cells. Cell Adhes Migr. 7:434–449. 2013. View Article : Google Scholar
|
|
137
|
Kontostathi G, Zoidakis J, Makridakis M,
Lygirou V, Mermelekas G, Papadopoulos T, Vougas K, Vlamis-Gardikas
A, Drakakis P, Loutradis D, et al: Cervical cancer cell line
secretome highlights the roles of transforming growth
factor-beta-induced protein ig-h3, peroxiredoxin-2, and NRF2 on
cervical carcinogenesis. BioMed Res Int. 2017:41807032017.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Mathias RA, Wang B, Ji H, Kapp EA, Moritz
RL, Zhu HJ and Simpson RJ: Secretome-based proteomic profiling of
Ras-transformed MDCK cells reveals extracellular modulators of
epithelial-mesenchymal transition. J Proteome Res. 8:2827–2837.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Lunter PC, van Kilsdonk JWJ, van Beek H,
Cornelissen IMHA, Bergers M, Willems PHGM, van Muijen GNP and Swart
GWM: Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD),
a novel actor in invasive growth, controls matrix metalloproteinase
activity. Cancer Res. 65:8801–8808. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ofori-Acquah SF and King JA: Activated
leukocyte cell adhesion molecule: A new paradox in cancer. Transl
Res. 151:122–128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Weidle UH, Eggle D, Klostermann S and
Swart GWM: ALCAM/CD166: Cancer-related issues. Cancer Genomics
Proteomics. 7:231–243. 2010.PubMed/NCBI
|
|
142
|
von Lersner A, Droesen L and Zijlstra A:
Modulation of cell adhesion and migration through regulation of the
immunoglobulin superfamily member ALCAM/CD166. Clin Exp Metastasis.
36:87–95. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Glentis A, Gurchenkov V and Matic
Vignjevic D: Assembly, heterogeneity, and breaching of the basement
membranes. Cell Adhes Migr. 8:236–245. 2014. View Article : Google Scholar
|
|
144
|
Pozzi A, Yurchenco PD and Iozzo RV: The
nature and biology of basement membranes. Matrix Biol. 57–58:1–11.
2017. View Article : Google Scholar
|
|
145
|
Randles MJ, Humphries MJ and Lennon R:
Proteomic definitions of basement membrane composition in health
and disease. Matrix Biol. 57–58:12–28. 2017. View Article : Google Scholar
|
|
146
|
Zhou Y, Zhu Y, Fan X, Zhang C, Wang Y,
Zhang L, Zhang H, Wen T, Zhang K, Huo X, et al: NID1, a new
regulator of EMT required for metastasis and chemoresistance of
ovarian cancer cells. Oncotarget. 8:33110–33121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Pedrola N, Devis L, Llauradó M, Campoy I,
Martinez-Garcia E, Garcia M, Muinelo-Romay L, Alonso-Alconada L,
Abal M, Alameda F, et al: Nidogen 1 and Nuclear Protein 1: Novel
targets of ETV5 transcription factor involved in endometrial cancer
invasion. Clin Exp Metastasis. 32:467–478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
McMahon B and Kwaan HC: The plasminogen
activator system and cancer. Pathophysiol Haemost Thromb.
36:184–194. 2008. View Article : Google Scholar
|
|
149
|
Durand MKV, Bødker JS, Christensen A,
Dupont DM, Hansen M, Jensen JK, Kjelgaard S, Mathiasen L, Pedersen
KE, Skeldal S, et al: Plasminogen activator inhibitor-I and tumour
growth, invasion, and metastasis. Thromb Haemost. 91:438–449. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Sitaram RT, Mallikarjuna P, Landström M
and Ljungberg B: Transforming growth factor-β promotes
aggressiveness and invasion of clear cell renal cell carcinoma.
Oncotarget. 7:35917–35931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Rabi ZA, Todorović-Raković N, Vujasinović
T, Milovanović J and Nikolić-Vukosavljević D: Markers of
progression and invasion in short term follow up of untreated
breast cancer patients. Cancer Biomark. 15:745–754. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Rhone P, Ruszkowska-Ciastek B, Bielawski
K, Brkic A, Zarychta E, Góralczyk B, Roszkowski K and Rość D:
Comprehensive analysis of haemostatic profile depending on
clinicopathological determinants in breast cancer patients. Biosci
Rep. 38:BSR201716572018. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Li S, Wei X, He J, Tian X, Yuan S and Sun
L: Plasminogen activator inhibitor-1 in cancer research. Biomed
Pharmacother. 105:83–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Ochieng J, Nangami G, Sakwe A, Moye C,
Alvarez J, Whalen D, Thomas P and Lammers P: Impact of Fetuin-A
(AHSG) on tumor progression and type 2 diabetes. Int J Mol Sci.
19:22112018. View Article : Google Scholar :
|
|
155
|
Nangami GN, Watson K, Parker-Johnson K,
Okereke KO, Sakwe A, Thompson P, Frimpong N and Ochieng J: Fetuin-A
(α2HS-glycoprotein) is a serum chemo-attractant that also promotes
invasion of tumor cells through Matrigel. Biochem Biophys Res
Commun. 438:660–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Watson K, Koumangoye R, Thompson P, Sakwe
AM, Patel T, Pratap S and Ochieng J: Fetuin-A triggers the
secretion of a novel set of exosomes in detached tumor cells that
mediate their adhesion and spreading. FEBS Lett. 586:3458–3463.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Niu L, Song X, Wang N, Xue L, Song X and
Xie L: Tumor-derived exosomal proteins as diagnostic biomarkers in
non-small cell lung cancer. Cancer Sci. 110:433–442. 2019.
View Article : Google Scholar
|
|
158
|
Adams GN, Rosenfeldt L, Frederick M,
Miller W, Waltz D, Kombrinck K, McElhinney KE, Flick MJ, Monia BP,
Revenko AS, et al: Colon cancer growth and dissemination relies
upon thrombin, stromal PAR-1, and fibrinogen. Cancer Res.
75:4235–4243. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Honda K-I, Asada R, Kageyama K, Fukuda T,
Terada H, Yasui T, Sumi T, Koyama M, Ishiko O and Sugawa T: Protein
complex of fibrinogen gamma chain and complement factor H in
ovarian cancer patient plasma. Anticancer Res. 37:2861–2866.
2017.PubMed/NCBI
|
|
160
|
Duan S, Gong B, Wang P, Huang H, Luo L and
Liu F: Novel prognostic biomarkers of gastric cancer based on gene
expression microarray: COL12A1, GSTA3, FGA and FGG. Mol Med Rep.
18:3727–3736. 2018.PubMed/NCBI
|
|
161
|
Zhang X, Wang F, Huang Y, Ke K, Zhao B,
Chen L, Liao N, Wang L, Li Q, Liu X, et al: FGG promotes migration
and invasion in hepatocellular carcinoma cells through activating
epithelial to mesenchymal transition. Cancer Manag Res.
11:1653–1665. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Brakebusch C and Fässler R: beta 1
integrin function in vivo: Adhesion, migration and more. Cancer
Metastasis Rev. 24:403–411. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Pan B, Guo J, Liao Q and Zhao Y: β1 and β3
integrins in breast, prostate and pancreatic cancer: A novel
implication (Review). Oncol Lett. 15:5412–5416. 2018.PubMed/NCBI
|
|
164
|
Sun Q, Zhou C, Ma R, Guo Q, Huang H, Hao
J, Liu H, Shi R and Liu B: Prognostic value of increased
integrin-beta 1 expression in solid cancers: A meta-analysis.
OncoTargets Ther. 11:1787–1799. 2018. View Article : Google Scholar
|
|
165
|
Albrektsen T, Richter HE, Clausen JT and
Fleckner J: Identification of a novel integral plasma membrane
protein induced during adipocyte differentiation. Biochem J.
359:393–402. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Pessentheiner AR, Huber K, Pelzmann HJ,
Prokesch A, Radner FPW, Wolinski H, Lindroos-Christensen J, Hoefler
G, Rülicke T, Birner-Gruenberger R, et al: APMAP interacts with
lysyl oxidase-like proteins, and disruption of Apmap leads to
beneficial visceral adipose tissue expansion. FASEB J.
31:4088–4103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Gabriele C, Cantiello F, Nicastri A,
Crocerossa F, Russo GI, Cicione A, Vartolomei MD, Ferro M, Morgia
G, Lucarelli G, et al: High-throughput detection of low abundance
sialylated glycoproteins in human serum by TiO2
enrichment and targeted LC-MS/MS analysis: Application to a
prostate cancer sample set. Anal Bioanal Chem. 411:755–763. 2019.
View Article : Google Scholar
|
|
168
|
Jiang S, Wang X, Song D, Liu X, Gu Y, Xu
Z, Wang X, Zhang X, Ye Q, Tong Z, et al: Cholesterol induces
epithelial-to-mesenchymal transition of prostate cancer cells by
suppressing degradation of EGFR through APMAP. Cancer Res.
79:3063–3075. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Moriyama-Kita M, Endo Y, Yonemura Y,
Heizmann CW, Miyamori H, Sato H, Yamamoto E and Sasaki T: S100A4
regulates E-cadherin expression in oral squamous cell carcinoma.
Cancer Lett. 230:211–218. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Shin J, Song I-S, Pak JH and Jang S-W:
Upregulation of annexin A1 expression by butyrate in human melanoma
cells induces invasion by inhibiting E-cadherin expression. Tumour
Biol. 37:14577–14584. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Wong SHM, Fang CM, Chuah L-H, Leong CO and
Ngai SC: E-cadherin: Its dysregulation in carcinogenesis and
clinical implications. Crit Rev Oncol Hematol. 121:11–22. 2018.
View Article : Google Scholar
|
|
172
|
Yu W, Wu J, Ning ZL, Liu QY and Quan RL:
High expression of peroxiredoxin 1 is associated with
epithelial-mesenchymal transition marker and poor prognosis in
gastric cancer. Med Sci Monit. 24:2259–2270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Kim S-Y: Cancer energy metabolism:
Shutting power off cancer factory. Biomol Ther (Seoul). 26:39–44.
2018. View Article : Google Scholar
|
|
174
|
Warburg O, Posener K and Negelein E: Über
den stoffwechsel der carcinomzelle. Biochem Zeitschr. 152:309–344.
1924.
|
|
175
|
Chen T, Huang Z, Tian Y, Wang H, Ouyang P,
Chen H, Wu L, Lin B and He R: Role of triosephosphate isomerase and
downstream functional genes on gastric cancer. Oncol Rep.
38:1822–1832. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Lincet H and Icard P: How do glycolytic
enzymes favour cancer cell proliferation by nonmetabolic functions?
Oncogene. 34:3751–3759. 2015. View Article : Google Scholar
|
|
177
|
Lone SN, Maqbool R, Parray FQ and Ul
Hussain M: Triose-phosphate isomerase is a novel target of miR-22
and miR-28, with implications in tumorigenesis. J Cell Physiol.
233:8919–8929. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Menendez JA and Lupu R: Fatty acid
synthase and the lipogenic phenotype in cancer pathogenesis. Nat
Rev Cancer. 7:763–777. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Madigan AA, Rycyna KJ, Parwani AV, Datiri
YJ, Basudan AM, Sobek KM, Cummings JL, Basse PH, Bacich DJ and
O'Keefe DS: Novel nuclear localization of fatty acid synthase
correlates with prostate cancer aggressiveness. Am J Pathol.
184:2156–2162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Wang H, Xi Q and Wu G: Fatty acid synthase
regulates invasion and metastasis of colorectal cancer via Wnt
signaling pathway. Cancer Med. 5:1599–1606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Wen S, Niu Y, Lee SO, Yeh S, Shang Z, Gao
H, Li Y, Chou F and Chang C: Targeting fatty acid synthase with
ASC-J9 suppresses proliferation and invasion of prostate cancer
cells. Mol Carcinog. 55:2278–2290. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Menendez JA and Lupu R: Fatty acid
synthase (FASN) as a therapeutic target in breast cancer. Expert
Opin Ther Targets. 21:1001–1016. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Zaidi N, Swinnen JV and Smans K:
ATP-citrate lyase: A key player in cancer metabolism. Cancer Res.
72:3709–3714. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Cheng Y, Jia B, Wang Y and Wan S: miR-133b
acts as a tumor suppressor and negatively regulates ATP citrate
lyase via PPARγ in gastric cancer. Oncol Rep. 38:3220–3226. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Granchi C: ATP citrate lyase (ACLY)
inhibitors: An anti-cancer strategy at the crossroads of glucose
and lipid metabolism. Eur J Med Chem. 157:1276–1291. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Icard P and Lincet H: The reduced
concentration of citrate in cancer cells: An indicator of cancer
aggressiveness and a possible therapeutic target. Drug Resist
Updat. 29:47–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Nicolussi A, D'Inzeo S, Capalbo C,
Giannini G and Coppa A: The role of peroxiredoxins in cancer.
(Review) Mol Clin Oncol. 6:pp. 139–153. 2017, View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Veal E, Jackson T and Latimer H: Role/s of
'Antioxidant' enzymes in ageing. Subcell Biochem. 90:425–450. 2018.
View Article : Google Scholar
|
|
189
|
Hampton MB, Vick KA, Skoko JJ and Neumann
CA: Peroxiredoxin involvement in the initiation and progression of
human cancer. Antioxid Redox Signal. 28:591–608. 2018. View Article : Google Scholar
|
|
190
|
Kang SW, Lee S and Lee JHS:
Cancer-associated function of 2-Cys peroxiredoxin subtypes as a
survival gatekeeper. Antioxidants. 7:1612018. View Article : Google Scholar :
|
|
191
|
Forshaw TE, Holmila R, Nelson KJ, Lewis
JE, Kemp ML, Tsang AW, Poole LB, Lowther WT and Furdui CM:
Peroxiredoxins in cancer and response to radiation therapies.
Antioxidants. 8:112019. View Article : Google Scholar :
|
|
192
|
Kim E-K, Lee SY, Kim Y, Ahn S-M and Jang
HH: Peroxiredoxin 1 post-transcriptionally regulates snoRNA
expression. Free Radic Biol Med. 141:1–9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Dayton TL, Jacks T and Vander Heiden MG:
PKM2, cancer metabolism, and the road ahead. EMBO Rep.
17:1721–1730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Méndez-Lucas A, Li X, Hu J, Che L, Song X,
Jia J, Wang J, Xie C, Driscoll PC, Tschaharganeh DF, et al: Glucose
catabolism in liver tumors induced by c-MYC can be sustained by
various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer
Res. 77:4355–4364. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Zhao Y, Butler EB and Tan M: Targeting
cellular metabolism to improve cancer therapeutics. Cell Death Dis.
4:pp. e5322013, View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Zhou W, Liotta LA and Petricoin EF: Cancer
metabolism and mass spectrometry-based proteomics. Cancer Lett.
356A. pp. A176–A183. 2015, View Article : Google Scholar
|
|
197
|
Cheng T-Y, Yang Y-C, Wang H-P, Tien Y-W,
Shun C-T, Huang H-Y, Hsiao M and Hua K-T: Pyruvate kinase M2
promotes pancreatic ductal adenocarcinoma invasion and metastasis
through phosphorylation and stabilization of PAK = 2 protein.
Oncogene. 37:1730–1742. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Marchitti SA, Brocker C, Stagos D and
Vasiliou V: Non-P450 aldehyde oxidizing enzymes: The aldehyde
dehydrogenase superfamily. Expert Opin Drug Metab Toxicol.
4:697–720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Xu X, Chai S, Wang P, Zhang C, Yang Y,
Yang Y and Wang K: Aldehyde dehydrogenases and cancer stem cells.
Cancer Lett. 369:50–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Zhou C and Sun B: The prognostic role of
the cancer stem cell marker aldehyde dehydrogenase 1 in head and
neck squamous cell carcinomas: A meta-analysis. Oral Oncol.
50:1144–1148. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Wei D, Peng J-J, Gao H, Zhang T, Tan Y and
Hu Y-H: ALDH1 expression and the prognosis of lung cancer: A
systematic review and meta-analysis. Heart Lung Circ. 24:780–788.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Li J, Zhang B, Yang YF, Jin J and Liu YH:
Aldehyde dehydrogenase 1 as a predictor of the neoadjuvant
chemotherapy response in breast cancer: A meta-analysis. Medicine
(Baltimore). 97:pp. e120562018, View Article : Google Scholar
|
|
203
|
Dvorakova M, Nenutil R and Bouchal P:
Transgelins, cytoskeletal proteins implicated in different aspects
of cancer development. Expert Rev Proteomics. 11:149–165. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Rao D, Kimler BF, Nothnick WB, Davis MK,
Fan F and Tawfik O: Transgelin: A potentially useful diagnostic
marker differentially expressed in triple-negative and
non-triple-negative breast cancers. Hum Pathol. 46:876–883. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Tan VY, Lewis SJ, Adams JC and Martin RM:
Association of fascin-1 with mortality, disease progression and
metastasis in carcinomas: A systematic review and meta-analysis.
BMC Med. 11:522013. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Iancu-Rubin C and Atweh GF: p27(Kip1) and
stathmin share the stage for the first time. Trends Cell Biol.
15:346–348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Belletti B and Baldassarre G: Stathmin: A
protein with many tasks. New biomarker and potential target in
cancer. Expert Opin Ther Targets. 15:1249–1266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Biaoxue R, Hua L, Wenlong G and Shuanying
Y: Overexpression of stathmin promotes metastasis and growth of
malignant solid tumors: A systemic review and meta-analysis.
Oncotarget. 7:78994–79007. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Ding Z, Bae YH and Roy P: Molecular
insights on context-specific role of profilin-1 in cell migration.
Cell Adhes Migr. 6:442–449. 2012. View Article : Google Scholar
|
|
210
|
Alkam D, Feldman EZ, Singh A and Kiaei M:
Profilin1 biology and its mutation, actin(g) in disease. Cell Mol
Life Sci. 74:967–981. 2017. View Article : Google Scholar :
|
|
211
|
Jiang C, Ding Z, Joy M, Chakraborty S, Kim
SH, Bottcher R, Condeelis J, Singh S and Roy P: A balanced level of
profilin-1 promotes stemness and tumor-initiating potential of
breast cancer cells. Cell Cycle. 16:2366–2373. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Wang Z, Shi Z, Zhang L, Zhang H and Zhang
Y: Profilin 1, negatively regulated by microRNA-19a-3p, serves as a
tumor suppressor in human hepatocellular carcinoma. Pathol Res
Pract. 215:499–505. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Nakamura F, Stossel TP and Hartwig JH: The
filamins: Organizers of cell structure and function. Cell Adhes
Migr. 5:160–169. 2011. View Article : Google Scholar
|
|
214
|
Savoy RM and Ghosh PM: The dual role of
filamin A in cancer: Can't live with (too much of) it, can't live
without it. Endocr Relat Cancer. 20:R341–R356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Shao Q-Q, Zhang T-P, Zhao W-J, Liu Z-W,
You L, Zhou L, Guo J-C and Zhao Y-P: Filamin A: Insights into its
exact role in cancers. Pathol Oncol Res. 22:245–252. 2016.
View Article : Google Scholar
|
|
216
|
Wang Y, Liu S, Zhang Y and Yang J: Myosin
heavy chain 9: Oncogene or tumor suppressor gene? Med Sci Monit.
25:888–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Desouza-Armstrong M, Gunning PW and Stehn
JR: Tumor suppressor tropomyosin Tpm2.1 regulates sensitivity to
apoptosis beyond anoikis characterized by changes in the levels of
intrinsic apoptosis proteins. Cytoskeleton (Hoboken). 74:233–248.
2017. View Article : Google Scholar
|
|
218
|
Ma Y, Xiao T, Xu Q, Shao X and Wang H:
iTRAQ-based quantitative analysis of cancer-derived secretory
proteome reveals TPM2 as a potential diagnostic biomarker of
colorectal cancer. Front Med. 10:278–285. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Zhang J, Zhang J, Xu S, Zhang X, Wang P,
Wu H, Xia B, Zhang G, Lei B, Wan L, et al: Hypoxia-induced TPM2
methylation is associated with chemoresistance and poor prognosis
in breast cancer. Cell Physiol Biochem. 45:692–705. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Shin H, Kim D and Helfman DM: Tropomyosin
isoform Tpm2.1 regulates collective and amoeboid cell migration and
cell aggregation in breast epithelial cells. Oncotarget.
8:95192–95205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Mitchell CB, Black B, Sun F, Chrzanowski
W, Cooper-White J, Maisonneuve B, Stringer B, Day B, Biro M and
O'Neill GM: Tropomyosin Tpm 2.1 loss induces glioblastoma spreading
in soft brain-like environments. J Neurooncol. 141:303–313. 2019.
View Article : Google Scholar
|
|
222
|
Shishkin S, Eremina L, Pashintseva N,
Kovalev L and Kovaleva M: Cofilin-1 and other ADF/Cofilin
superfamily members in human malignant cells. Int J Mol Sci.
18:E102016. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Gasparski AN, Ozarkar S and Beningo KA:
Transient mechanical strain promotes the maturation of invadopodia
and enhances cancer cell invasion in vitro. J Cell Sci.
130:1965–1978. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Tsai C-H, Lin L-T, Wang C-Y, Chiu Y-W,
Chou Y-T, Chiu S-J, Wang H-E, Liu R-S, Wu C-Y, Chan P-C, et al:
Over-expression of cofilin-1 suppressed growth and invasion of
cancer cells is associated with up-regulation of let-7 microRNA.
Biochim Biophys Acta. 1852:851–861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Curto M and McClatchey AI: Ezrin...a
metastatic detERMinant? Cancer Cell. 5:113–114. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Haase G, Gavert N, Brabletz T and
Ben-Zé'ev A: The Wnt target gene L1 in colon cancer invasion and
metastasis. Cancers (Basel). 8:482016. View Article : Google Scholar
|
|
227
|
Cihan YB: Does ezrin play a predictive
role in cancer patients undergoing radiotherapy and/or
chemotherapy? Hum Pathol. 80:247–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Mierke CT: The role of vinculin in the
regulation of the mechanical properties of cells. Cell Biochem
Biophys. 53:115–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Goldmann WH, Auernheimer V, Thievessen I
and Fabry B: Vinculin, cell mechanics and tumour cell invasion.
Cell Biol Int. 37:397–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Gao Y, Wang Z, Hao Q, Li W, Xu Y, Zhang J,
Zhang W, Wang S, Liu S, Li M, et al: Loss of ERα induces
amoeboid-like migration of breast cancer cells by downregulating
vinculin. Nat Commun. 8:144832017. View Article : Google Scholar
|
|
231
|
Colombo E, Alcalay M and Pelicci PG:
Nucleophosmin and its complex network: A possible therapeutic
target in hematological diseases. Oncogene. 30:2595–2609. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Andersen JS, Wilkinson CJ, Mayor T,
Mortensen P, Nigg EA and Mann M: Proteomic characterization of the
human centrosome by protein correlation profiling. Nature.
426:570–574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Box JK, Paquet N, Adams MN, Boucher D,
Bolderson E, O'Byrne KJ and Richard DJ: Nucleophosmin: From
structure and function to disease development. BMC Mol Biol.
17:192016. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Werner MT, Zhao C, Zhang Q and Wasik MA:
Nucleophosmin- anaplastic lymphoma kinase: The ultimate oncogene
and therapeutic target. Blood. 129:823–831. 2017. View Article : Google Scholar
|
|
235
|
Arrigo A-P: Mammalian HspB1 (Hsp27) is a
molecular sensor linked to the physiology and environment of the
cell. Cell Stress Chaperones. 22:517–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Xie Y-H, Li L-Y, He J-Z, Xu X-E, Liao L-D,
Zhang Q, Xie J-J, Xu L-Y and Li E-M: Heat shock protein family B
member 1 facilitates ezrin activation to control cell migration in
esophageal squamous cell carcinoma. Int J Biochem Cell Biol.
112:79–87. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Honda K, Yamada T, Endo R, Ino Y, Gotoh M,
Tsuda H, Yamada Y, Chiba H and Hirohashi S: Actinin-4, a novel
actin-bundling protein associated with cell motility and cancer
invasion. J Cell Biol. 140:1383–1393. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Hayashida Y, Honda K, Idogawa M, Ino Y,
Ono M, Tsuchida A, Aoki T, Hirohashi S and Yamada T: E-cadherin
regulates the association between beta-catenin and actinin-4.
Cancer Res. 65:8836–8845. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Thomas DG and Robinson DN: The fifth
sense: Mechanosensory regulation of alpha-actinin-4 and its
relevance for cancer metastasis. Semin Cell Dev Biol. 71:68–74.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Yamaguchi H, Ito Y, Miura N, Nagamura Y,
Nakabo A, Fukami K, Honda K and Sakai R: Actinin-1 and actinin-4
play essential but distinct roles in invadopodia formation by
carcinoma cells. Eur J Cell Biol. 96:685–694. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Keeling MC, Flores LR, Dodhy AH, Murray ER
and Gavara N: Actomyosin and vimentin cytoskeletal networks
regulate nuclear shape, mechanics and chromatin organization. Sci
Rep. 7:52192017. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Battaglia RA, Delic S, Herrmann H and
Snider NT: Vimentin on the move: new developments in cell
migration. F1000 Res 7 (F1000 Faculty Rev). 17962018.
|
|
243
|
Rao J and Li N: Microfilament actin
remodeling as a potential target for cancer drug development. Curr
Cancer Drug Targets. 4:345–354. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Piktel E, Levental I, Durnaś B, Janmey PA
and Bucki R: Plasma gelsolin: Indicator of inflammation and its
potential as a diagnostic tool and therapeutic target. Int J Mol
Sci. 19:25162018. View Article : Google Scholar :
|
|
245
|
Krishnakumar S, Sundaram A, Abhyankar D,
Krishnamurthy V, Shanmugam MP, Gopal L, Sharma T and Biswas J:
Major histocompatibility antigens and antigen-processing molecules
in retinoblastoma. Cancer. 100:1059–1069. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Okayama A, Miyagi Y, Oshita F, Nishi M,
Nakamura Y, Nagashima Y, Akimoto K, Ryo A and Hirano H: Proteomic
analysis of proteins related to prognosis of lung adenocarcinoma. J
Proteome Res. 13:4686–4694. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Ryan D, Carberry S, Murphy AC, Lindner AU,
Fay J, Hector S, McCawley N, Bacon O, Concannon CG, Kay EW, et al:
Calnexin, an ER stress-induced protein, is a prognostic marker and
potential therapeutic target in colorectal cancer. J Transl Med.
14:1962016. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Vogiatzi F, Brandt DT, Schneikert J, Fuchs
J, Grikscheit K, Wanzel M, Pavlakis E, Charles JP, Timofeev O, Nist
A, et al: Mutant p53 promotes tumor progression and metastasis by
the endoplasmic reticulum UDPase ENTPD5. Proc Natl Acad Sci USA.
113:E8433–E8442. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Chen Y, Ma D, Wang X, Fang J, Liu X, Song
J, Li X, Ren X, Li Q, Li Q, et al: Calnexin impairs the antitumor
immunity of CD4+ and CD8+ T cells. Cancer
Immunol Res. 7:123–135. 2018. View Article : Google Scholar
|
|
250
|
Dudek J, Benedix J, Cappel S, Greiner M,
Jalal C, Müller L and Zimmermann R: Functions and pathologies of
BiP and its interaction partners. Cell Mol Life Sci. 66:1556–1569.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Wang J, Lee J, Liem D and Ping P: HSPA5
Gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum.
Gene. 618:14–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Herroon MK, Rajagurubandara E, Diedrich
JD, Heath EI and Podgorski I: Adipocyte-activated oxidative and ER
stress pathways promote tumor survival in bone via upregulation of
Heme Oxygenase 1 and Survivin. Sci Rep. 8:402018. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Chen W, Do KC, Saxton B, Leng S, Filipczak
P, Tessema M, Belinsky SA and Lin Y: Inhibition of the hexosamine
biosynthesis pathway potentiates cisplatin cytotoxicity by
decreasing BiP expression in non-small-cell lung cancer cells. Mol
Carcinog. 58:1046–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
254
|
Sauk JJ, Nikitakis N and Siavash H: Hsp47
a novel collagen binding serpin chaperone, autoantigen and
therapeutic target. Front Biosci. 10:107–118. 2005. View Article : Google Scholar
|
|
255
|
Duarte BDP and Bonatto D: The heat shock
protein 47 as a potential biomarker and a therapeutic agent in
cancer research. J Cancer Res Clin Oncol. 144:2319–2328. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Vatolin S, Phillips JG, Jha BK, Govindgari
S, Hu J, Grabowski D, Parker Y, Lindner DJ, Zhong F, Distelhorst
CW, et al: Novel protein disulfide isomerase inhibitor with
anticancer activity in multiple myeloma. Cancer Res. 76:3340–3350.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Kuo T-F, Chen T-Y, Jiang S-T, Chen K-W,
Chiang Y-M, Hsu Y-J, Liu Y-J, Chen H-M, Yokoyama KK, Tsai K-C, et
al: Protein disulfide isomerase a4 acts as a novel regulator of
cancer growth through the procaspase pathway. Oncogene.
36:5484–5496. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Fucikova J, Kasikova L, Truxova I, Laco J,
Skapa P, Ryska A and Spisek R: Relevance of the chaperone-like
protein calreticulin for the biological behavior and clinical
outcome of cancer. Immunol Lett. 193:25–34. 2018. View Article : Google Scholar
|
|
259
|
Sheng W, Chen C, Dong M, Wang G, Zhou J,
Song H, Li Y, Zhang J and Ding S: Calreticulin promotes EGF-induced
EMT in pancreatic cancer cells via Integrin/EGFR-ERK/MAPK signaling
pathway. Cell Death Dis. 8:pp. e31472017, View Article : Google Scholar : PubMed/NCBI
|
|
260
|
Schcolnik-Cabrera A, Oldak B, Juárez M,
Cruz-Rivera M, Flisser A and Mendlovic F: Calreticulin in
phagocytosis and cancer: Opposite roles in immune response
outcomes. Apoptosis. 24:245–255. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
261
|
Wang C, Jin G, Jin H, Wang N, Luo Q, Zhang
Y, Gao D, Jiang K, Gu D, Shen Q, et al: Clusterin facilitates
metastasis by EIF3I/Akt/MMP13 signaling in hepatocellular
carcinoma. Oncotarget. 6:2903–2916. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Shapiro B, Tocci P, Haase G, Gavert N and
Ben-Ze'ev A: Clusterin, a gene enriched in intestinal stem cells,
is required for L1-mediated colon cancer metastasis. Oncotarget.
6:34389–34401. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Liu Y, Men C, Xu Y, Zhao K, Luo L, Dong D
and Yu Q: Clusterin promotes growth and invasion of clear cell
renal carcinoma cell by upregulation of S100A4 expression. Cancer
Biomark. 21:915–923. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Tew KD, Manevich Y, Grek C, Xiong Y, Uys J
and Townsend DM: The role of glutathione S-transferase P in
signaling pathways and S-glutathionylation in cancer. Free Radic
Biol Med. 51:299–313. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Wang Z, He W, Yang G, Wang J, Wang Z,
Nesland JM, Holm R and Suo Z: Decreased expression of GST pi is
correlated with a poor prognosis in human esophageal squamous
carcinoma. BMC Cancer. 10:3522010. View Article : Google Scholar : PubMed/NCBI
|
|
266
|
Zannis-Hadjopoulos M, Yahyaoui W and
Callejo M: 14-3-3 cruciform-binding proteins as regulators of
eukaryotic DNA replication. Trends Biochem Sci. 33:44–50. 2008.
View Article : Google Scholar
|
|
267
|
Bortner JD Jr, Das A, Umstead TM, Freeman
WM, Somiari R, Aliaga C, Phelps DS and El-Bayoumy K:
Down-regulation of 14-3-3 isoforms and annexin A5 proteins in lung
adenocarcinoma induced by the tobacco-specific nitrosamine NNK in
the A/J mouse revealed by proteomic analysis. J Proteome Res.
8:4050–4061. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Leal MF, Calcagno DQ, Demachki S,
Assumpção PP, Chammas R, Burbano RR and Smith MA: Clinical
implication of 14-3-3 epsilon expression in gastric cancer. World J
Gastroenterol. 18:1531–1537. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Qiu Y, Zhou Z, Li Z, Lu L, Li L, Li X,
Wang X and Zhang M: Pretreatment 14-3-3 epsilon level is predictive
for advanced extranodal NK/T cell lymphoma therapeutic response to
asparaginase-based chemotherapy. Proteomics Clin Appl. 11:3–4.
2017. View Article : Google Scholar
|
|
270
|
Bavelloni A, Piazzi M, Raffini M, Faenza I
and Blalock WL: Prohibitin 2: At a communications crossroads. IUBMB
Life. 67:239–254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
271
|
Mishra S and Nyomba BG: Prohibitin - At
the crossroads of obesity-linked diabetes and cancer. Exp Biol Med
(Maywood). 242:1170–1177. 2017. View Article : Google Scholar
|
|
272
|
Taniguchi K, Matsumura K, Kageyama S, Ii
H, Ashihara E, Chano T, Kawauchi A, Yoshiki T and Nakata S:
Prohibitin-2 is a novel regulator of p21WAF1/CIP1 induced by
depletion of γ-glutamylcyclotransferase. Biochem Biophys Res
Commun. 496:218–224. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
273
|
Zhou Z, Ai H, Li K, Yao X, Zhu W, Liu L,
Yu C, Song Z, Bao Y, Huang Y, et al: Prohibitin 2 localizes in
nucleolus to regulate ribosomal RNA transcription and facilitate
cell proliferation in RD cells. Sci Rep. 8:14792018. View Article : Google Scholar : PubMed/NCBI
|
|
274
|
Shen Y, Gao Y, Yuan H, Cao J, Jia B, Li M,
Peng Y, Du X, Zhang J and Shi J: Prohibitin-2 negatively regulates
AKT2 expression to promote prostate cancer cell migration. Int J
Mol Med. 41:1147–1155. 2018.
|
|
275
|
Yan C, Gong L, Chen L, Xu M, Abou-Hamdan
H, Tang M, Désaubry L and Song Z: PHB2 (prohibitin 2) promotes
PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis.
Autophagy. 16:419–434. 2020. View Article : Google Scholar :
|
|
276
|
Jubran R, Kocsis J, Garam N, Maláti É,
Gombos T, Barabás L, Gráf L, Prohászka Z and Fishelson Z:
Circulating mitochondrial stress 70 protein/mortalin and cytosolic
Hsp70 in blood: Risk indicators in colorectal cancer. Int J Cancer.
141:2329–2335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Cruz IN, Coley HM, Kramer HB, Madhuri TK,
Safuwan NAM, Angelino AR and Yang M: Proteomics analysis of ovarian
cancer cell lines and tissues reveals drug resistance-associated
proteins. Cancer Genomics Proteomics. 14:35–51. 2017. View Article : Google Scholar :
|
|
278
|
Niu X, Su L, Qi S, Gao Z, Zhang Q and
Zhang S: GRP75 modulates oncogenic Dbl-driven endocytosis derailed
via the CHIP-mediated ubiquitin degradation pathway. Cell Death
Dis. 9:9712018. View Article : Google Scholar : PubMed/NCBI
|
|
279
|
Chang HJ, Lee MR, Hong S-H, Yoo BC, Shin
Y-K, Jeong JY, Lim S-B, Choi HS, Jeong S-Y and Park J-G:
Identification of mitochondrial FoF1-ATP synthase involved in liver
metastasis of colorectal cancer. Cancer Sci. 98:1184–1191. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Kühlbrandt W: Structure and mechanisms of
F-type ATP synthases. Annu Rev Biochem. 88:515–549. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Liu J, Zhan X, Li M, Li G, Zhang P, Xiao
Z, Shao M, Peng F, Hu R and Chen Z: Mitochondrial proteomics of
nasopha-ryngeal carcinoma metastasis. BMC Med Genomics. 5:622012.
View Article : Google Scholar
|
|
282
|
Chen W-L, Kuo K-T, Chou T-Y, Chen C-L,
Wang C-H, Wei Y-H and Wang L-S: The role of cytochrome c oxidase
subunit Va in non-small cell lung carcinoma cells: Association with
migration, invasion and prediction of distant metastasis. BMC
Cancer. 12:2732012. View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Wang Y, Hu L, Zhang X, Zhao H, Xu H, Wei
Y, Jiang H, Xie C, Zhou Y and Zhou F: Downregulation of
mitochondrial single stranded DNA binding protein (SSBP1) induces
mitochondrial dysfunction and increases the radiosensitivity in
non-small cell lung cancer cells. J Cancer. 8:1400–1409. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
284
|
Rajapakse A, Suraweera A, Boucher D, Naqi
A, O'Byrne K, Richard DJ and Croft LV: Redox Regulation in the Base
Excision Repair Pathway: Old and New Players as Cancer Therapeutic
Targets. Curr Med Chem. 27:1901–1921. 2020. View Article : Google Scholar
|
|
285
|
Croft LV, Bolderson E, Adams MN, El-Kamand
S, Kariawasam R, Cubeddu L, Gamsjaeger R and Richard DJ: Human
single-stranded DNA binding protein 1 (hSSB1, OBFC2B), a critical
component of the DNA damage response. Semin Cell Dev Biol.
86:121–128. 2019. View Article : Google Scholar
|
|
286
|
Bozlu M, Orhan D, Baltaci S, Yaman O,
Elhan AH, Tulunay O and Müftüoğlu YZ: The prognostic value of
proliferating cell nuclear antigen, Ki-67 and nucleolar organizer
region in transitional cell carcinoma of the bladder. Int Urol
Nephrol. 33:59–66. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
287
|
Maeda K, Chung Y-S, Onoda N, Ogawa M, Kato
Y, Nitta A, Arimoto Y, Kondo Y, Arakawa T and Sowa M: Association
of tumor cell proliferation with lymph node metastasis in early
gastric cancer. Oncology. 53:1–5. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
288
|
Ma S, Yang J, Li J and Song J: The
clinical utility of the proliferating cell nuclear antigen
expression in patients with hepatocellular carcinoma. Tumour Biol.
37:7405–7412. 2016. View Article : Google Scholar
|
|
289
|
Wang L, Kong W, Liu B and Zhang X:
Proliferating cell nuclear antigen promotes cell proliferation and
tumorigenesis by up-regulating STAT3 in non-small cell lung cancer.
Biomed Pharmacother. 104:595–602. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
290
|
Kato K, Kawashiri S, Yoshizawa K, Kitahara
H, Okamune A, Sugiura S, Noguchi N and Yamamoto E: Expression form
of p53 and PCNA at the invasive front in oral squamous cell
carcinoma: Correlation with clinicopathological features and
prognosis. J Oral Pathol Med. 40:693–698. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
291
|
Dahl JA and Collas P: Q2ChIP, a quick and
quantitative chromatin immunoprecipitation assay, unravels
epigenetic dynamics of developmentally regulated genes in human
carcinoma cells. Stem Cells. 25:1037–1046. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
292
|
Mahen R, Hattori H, Lee M, Sharma P,
Jeyasekharan AD and Venkitaraman AR: A-type lamins maintain the
positional stability of DNA damage repair foci in mammalian nuclei.
PLoS One. 8:pp. e618932013, View Article : Google Scholar : PubMed/NCBI
|
|
293
|
Sakthivel KM and Sehgal P: A novel role of
lamins from genetic disease to cancer biomarkers. Oncol Rev.
10:3092016.PubMed/NCBI
|
|
294
|
Kim J-K, Louhghalam A, Lee G, Schafer BW,
Wirtz D and Kim D-H: Nuclear lamin A/C harnesses the perinuclear
apical actin cables to protect nuclear morphology. Nat Commun.
8:21232017. View Article : Google Scholar : PubMed/NCBI
|
|
295
|
Taheri F, Isbilir B, Müller G, Krieger JW,
Chirico G, Langowski J and Tóth K: Random motion of chromatin is
influenced by lamin A interconnections. Biophys J. 114:2465–2472.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
296
|
Zuo L, Zhao H, Yang R, Wang L, Ma H, Xu X,
Zhou P and Kong L: Lamin A/C might be involved in the EMT
signalling pathway. Gene. 663:51–64. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
297
|
Dmello C, Srivastava SS, Tiwari R,
Chaudhari PR, Sawant S and Vaidya MM: Multifaceted role of keratins
in epithelial cell differentiation and transformation. J Biosci.
44:332019. View Article : Google Scholar : PubMed/NCBI
|
|
298
|
Sharma P, Alsharif S, Fallatah A and Chung
BM: Intermediate filaments as effectors of cancer development and
metastasis: A focus on keratins, vimentin, and nestin. Cells.
8:4972019. View Article : Google Scholar :
|
|
299
|
Awe JA, Saranchuk J, Drachenberg D and Mai
S: Filtration-based enrichment of circulating tumor cells from all
prostate cancer risk groups. Urol Oncol. 35:300–309. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
300
|
Erlandsson A, Forssell-Aronsson E, Seidal
T and Bernhardt P: Binding of TS1, an anti-keratin 8 antibody, in
small-cell lung cancer after 177Lu-DOTA-Tyr3-octreotate treatment:
A histological study in xenografted mice. EJNMMI Res. 1:192011.
View Article : Google Scholar
|
|
301
|
Sawant S, Vaidya M, Chaukar D, Gangadaran
P, Singh AK, Rajadhyax S, Kannan S, Kane S, Pagare S and Kannan R:
Clinicopathological features and prognostic implications of loss of
K5 and gain of K1, K8 and K18 in oral potentially malignant lesions
and squamous cell carcinomas: An immunohistochemical analysis.
Edorium J Tumor Biol. 1:1–22. 2014.
|
|
302
|
Alam H, Kundu ST, Dalal SN and Vaidya MM:
Loss of keratins 8 and 18 leads to alterations in
α6β4-integrin-mediated signalling and decreased neoplastic
progression in an oral-tumour-derived cell line. J Cell Sci.
124:2096–2106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
303
|
Sun L and Fang J: Epigenetic regulation of
epithelial-mesenchymal transition. Cell Mol Life Sci. 73:4493–4515.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
304
|
Zhou B-R and Bai Y: Chromatin structures
condensed by linker histones. Essays Biochem. 63:75–87. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
305
|
Sridharan R, Gonzales-Cope M, Chronis C,
Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini
M, et al: Proteomic and genomic approaches reveal critical
functions of H3K9 methylation and heterochromatin protein-1γ in
reprogramming to pluripotency. Nat Cell Biol. 15:872–882. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
306
|
Tzivion G, Gupta VS, Kaplun L and Balan V:
14-3-3 proteins as potential oncogenes. Semin Cancer Biol.
16:203–213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
307
|
Kurpińska A, Suraj J, Bonar E, Zakrzewska
A, Stojak M, Sternak M, Jasztal A and Walczak M: Proteomic
characterization of early lung response to breast cancer metastasis
in mice. Exp Mol Pathol. 107:129–140. 2019. View Article : Google Scholar
|
|
308
|
Kawahara T, Hotta N, Ozawa Y, Kato S, Kano
K, Yokoyama Y, Nagino M, Takahashi T and Yanagisawa K: Quantitative
proteomic profiling identifies DPYSL3 as pancreatic ductal
adenocarcinoma-associated molecule that regulates cell adhesion and
migration by stabilization of focal adhesion complex. PLoS One.
8:e796542013. View Article : Google Scholar : PubMed/NCBI
|
|
309
|
Matsunuma R, Chan DW, Kim B-J, Singh P,
Han A, Saltzman AB, Cheng C, Lei JT, Wang J, Roberto da Silva L, et
al: DPYSL3 modulates mitosis, migration, and
epithelial-to-mesenchymal transition in claudin-low breast cancer.
Proc Natl Acad Sci USA. 115:E11978–E11987. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
310
|
Yang Y, Jiang Y, Xie D, Liu M, Song N, Zhu
J, Fan J and Zhu C: Inhibition of cell-adhesion protein DPYSL3
promotes metastasis of lung cancer. Respir Res. 19:412018.
View Article : Google Scholar : PubMed/NCBI
|
|
311
|
Zarogoulidis P, Tsakiridis K, Karapantzou
C, Lampaki S, Kioumis I, Pitsiou G, Papaiwannou A,
Hohenforst-Schmidt W, Huang H, Kesisis G, et al: Use of proteins as
biomarkers and their role in carcinogenesis. J Cancer. 6:9–18.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
312
|
Frauchiger AL, Dummer R and Mangana J:
Serum S100B levels in melanoma. Methods Mol Biol. 1929:691–700.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
313
|
Yang T, Cheng J, You J, Yan B, Liu H and
Li F: S100B promotes chemoresistance in ovarian cancer stem cells
by regulating p53. Oncol Rep. 40:1574–1582. 2018.PubMed/NCBI
|
|
314
|
Darlix A, Lamy P-J, Lopez-Crapez E,
Braccini AL, Firmin N, Romieu G, Thezenas S and Jacot W: Serum HER2
extra-cellular domain, S100β and CA 15-3 levels are independent
prognostic factors in metastatic breast cancer patients. BMC
Cancer. 16:4282016. View Article : Google Scholar
|
|
315
|
Gao H, Zhang IY, Zhang L, Song Y, Liu S,
Ren H, Liu H, Zhou H, Su Y, Yang Y, et al: S100B suppression alters
polarization of infiltrating myeloid-derived cells in gliomas and
inhibits tumor growth. Cancer Lett. 439:91–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
316
|
Sadeghi M, Ordway B, Rafiei I, Borad P,
Fang B, Koomen JL, Zhang C, Yoder S, Johnson J and Damaghi M:
Integrative analysis of breast cancer cells reveals an
epithelial-mesenchymal transition role in adaptation to acidic
microenvironment. Front Oncol. 10:3042020. View Article : Google Scholar : PubMed/NCBI
|
|
317
|
Pampalakis G, Zingkou E, Sidiropoulos KG,
Diamandis EP, Zoumpourlis V, Yousef GM and Sotiropoulou G:
Biochemical pathways mediated by KLK6 protease in breast cancer.
Mol Oncol. 13:2329–2343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
318
|
Seguella L, Capuano R, Pesce M, Annunziata
G, Pesce M, de Conno B, Sarnelli G, Aurino L and Esposito G: S100B
protein stimulates proliferation and angiogenic mediators release
through RAGE/pAkt/mTOR pathway in human colon adenocarcinoma Caco-2
cells. Int J Mol Sci. 20:32402019. View Article : Google Scholar :
|
|
319
|
Méndez O, Peg V, Salvans C, Pujals M,
Fernández Y, Abasolo I, Pérez J, Matres A, Valeri M, Gregori J, et
al: Extracellular HMGA1 promotes tumor invasion and metastasis in
triple-negative breast cancer. Clin Cancer Res. 24:6367–6382. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
320
|
Wu X-J, Chen Y-Y, Gong C-C and Pei D-S:
The role of high-mobility group protein box 1 in lung cancer. J
Cell Biochem. 119:6354–6365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
321
|
Sharma AK, Sharma VR, Gupta GK, Ashraf GM
and Kamal MA: Advanced glycation end products (AGEs), glutathione
and breast cancer: Factors, mechanism and therapeutic
interventions. Curr Drug Metab. 20:65–71. 2019. View Article : Google Scholar
|
|
322
|
Schröter D and Höhn A: Role of advanced
glycation end products in carcinogenesis and their therapeutic
implications. Curr Pharm Des. 24:5245–5251. 2018. View Article : Google Scholar
|
|
323
|
Palanissami G and Paul SFD: RAGE and its
ligands: Molecular interplay between glycation, inflammation, and
hallmarks of cancer - a review. Horm Cancer. 9:295–325. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
324
|
Ahmad S, Khan H, Siddiqui Z, Khan MY,
Rehman S, Shahab U, Godovikova T, Silnikov V and Moinuddin: AGEs,
RAGEs and s-RAGE; friend or foe for cancer. Semin Cancer Biol.
49:44–55. 2018. View Article : Google Scholar
|
|
325
|
Thimmulappa RK, Mai KH, Srisuma S, Kensler
TW, Yamamoto M and Biswal S: Identification of Nrf2-regulated genes
induced by the chemopreventive agent sulforaphane by
oligonucleotide microarray. Cancer Res. 62:5196–5203.
2002.PubMed/NCBI
|
|
326
|
Sova M and Saso L: Design and development
of Nrf2 modulators for cancer chemoprevention and therapy: A
review. Drug Des Devel Ther. 12:3181–3197. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
327
|
Taguchi K and Yamamoto M: The KEAP1-NRF2
system in cancer. Front Oncol. 7:852017. View Article : Google Scholar : PubMed/NCBI
|
|
328
|
Rojo de la Vega M, Chapman E and Zhang DD:
NRF2 and the hallmarks of cancer. Cancer Cell. 34:21–43. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
329
|
Lignitto L, LeBoeuf SE, Homer H, Jiang S,
Askenazi M, Karakousi TR, Pass HI, Bhutkar AJ, Tsirigos A,
Ueberheide B, et al: Nrf2 activation promotes lung cancer
metastasis by inhibiting the degradation of Bach1. Cell.
178:316–329e18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
330
|
Wiel C, Le Gal K, Ibrahim MX, Jahangir CA,
Kashif M, Yao H, Ziegler DV, Xu X, Ghosh T, Mondal T, et al: BACH1
stabilization by antioxidants stimulates lung cancer metastasis.
Cell. 178:330–345e22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
331
|
Ryoo IG, Choi BH, Ku S-K and Kwak M-K:
High CD44 expression mediates p62-associated NFE2L2/NRF2 activation
in breast cancer stem cell-like cells: Implications for cancer stem
cell resistance. Redox Biol. 17:246–258. 2018. View Article : Google Scholar : PubMed/NCBI
|