The term leukemia collectively describes a group of
malignant clonal diseases of hematopoietic stem-progenitor cells,
which present with various diverse and biological subtypes
(1). Due to chromosomal
abnormalities and genetic alterations, these cells expand in an
oligoclonal manner and invade the bloodstream and extramedullary
tissues (2). Epidemiologic
cross-sectional research performed in 2012 revealed that the
worldwide age-standardized incidence of leukemia was 5.6 per
100,000 in men and 3.9 in women, ranking it as the 11th most
prevalent with the 10th highest mortality among all cancers, with
even higher numbers for specific subtypes among young and elderly
patients (3). According to the
World Health Organization standard classification (4), four subtypes of leukemia are
recognized, based on their progression state and the affected cell
lineage: Acute myeloid leukemia (AML), chronic myeloid leukemia
(CML), acute lymphoblastic leukemia (ALL) and chronic lymphocytic
leukemia (CLL).
For decades, genetic aberrations have been
considered to serve an essential role in the pathogenesis of
leukemia (5-7). These mutations can be categorized
into three main functional groups regulating cellular activities:
Mutation genes encoding transcription factors, epigenetic modifiers
regulating gene expression and genes associated with signaling
pathway activation. In AML, pro-proliferative signaling pathways,
such as the RAS/RAF, Janus kinase/STAT, PI3K/AKT signaling
pathways, are aberrantly activated as a result of gene mutations,
including mutations of fms related receptor tyrosine kinase 3, KIT
proto-oncogene, receptor tyrosine kinase, RAS family members and
serine/threonine kinases (7). In
lymphoid leukemia, the most commonly mutated gene is NOTCH1, and
this contributes to NOTCH1 signaling pathway activation (8,9).
BCR activator of RhoGEF and GTPase-ABL proto-oncogene 1,
non-receptor tyrosine kinase, the key fusion gene of CML, leads to
tyrosine kinases deregulation (6). Apart from gene mutation, epigenetic
regulators also serve essential roles in leukaemogenesis. For
example, DNA methyltransferase 3α, tet methylcytosine dioxygenase
2, isocitrate dehydrogenase [NADP(+)]1, methyltransferase 3,
N6-adenosine-methyltransferase complex catalytic subunit and FTO
α-ketoglutarate dependent dioxygenase, have been reported to be
involved in pathological DNA methylation and mRNA modification in
AML (10,11). These efforts have been well
described in other reviews. Although progress has been made in the
treatment of leukemia, especially in terms of the use of tyrosine
kinase inhibitors (12) and
immunotherapy (13,14), the disease remains incurable,
either due to frequent relapse or refractory cases, and the
best-practice treatment regiments are still being identified.
Extrinsic signals from the bone marrow (BM)
microenvironment promoting leukaemogenesis provide novel mechanisms
in treating leukemia (15,16).
Inflammation mediator-related genes, and specifically expressed
proteins, serve a vital role in the pathogenesis of various tumor
diseases, including breast cancer, gastrointestinal tumors and
genitourinary cancers (17,18). It is widely accepted that the
activity of inflammatory factors, especially when causing chronic
inflammation, can result in a pro-tumor microenvironment, promoting
tumor survival, proliferation and metastasis (17-19). Among these, macrophage migration
inhibitory factor (MIF) is one of the pro-inflammatory cytokines,
which is upregulated in a number of autoimmune diseases (20), as well as in cancer (21), including leukemia (22). Its multiple functions are
necessary for cell proliferation, survival and invasion (23), suggesting this protein could be a
promising candidate therapeutic target. This review focuses on the
function of MIF in general and its role in cancer, and on how these
functions influence the development of leukemia.
MIF is a soluble symmetrical homotrimer (37.5 kDa),
consisting of three small (115 amino acids long) 12.5 kDa monomers
(24). The protein is
evolutionary highly conserved, resulting in homologies >80%
among protein sequences of different species, including bacteria,
plants, protozoa and other non-mammals (25). Notably, MIF executes tautomerase
activity and catalyzes the conversion of D-isomer of
2-carboxy-2,3-dihydroindole-5,6-quinone (D-dopachrome) to
5,6-dihydroxyindole-2-carboxylic acid (26). Its main, although not sole,
receptor is CD74 (27). Binding
depends on the protein-protein interaction between the N-terminal
proline residue of the active site of MIF and the type II
transmembrane CD74 receptor (27).
MIF has been characterized as a pleiotropic,
multifunctional, pro-inflammatory factor (28). First identified in the 1930s
(29), MIF was recognized as a
soluble immune cell-derived factor in 1966 and was first cloned in
1989 (26,30). Notably, MIF acts as an endogenous
regulator of glucocorticoids (31). Under normal conditions, MIF can be
detected in the serum at a range of 2-6 ng/ml, following the
circadian rhythm of glucocorticoids (31). The main sources of MIF are
anterior pituitary cells, where a pre-secreted form is stored in
the cytoplasm (31). Serum levels
of MIF peak 2-3 h before relative serum levels of steroids reach
their peak (32). Apart from
pituitary cells, different types of cells, including
monocytes/macrophages, granulocytes, dendritic cells, endothelial
cells and mesenchymal cells, can secret MIF in response to
inflammatory stimuli (33-36).
The MIF protein lacks an N-terminal secretion signal (37,38). Instead, its release is partly
dependent on Golgi-associated protein p115 (38) or exosomes (39).
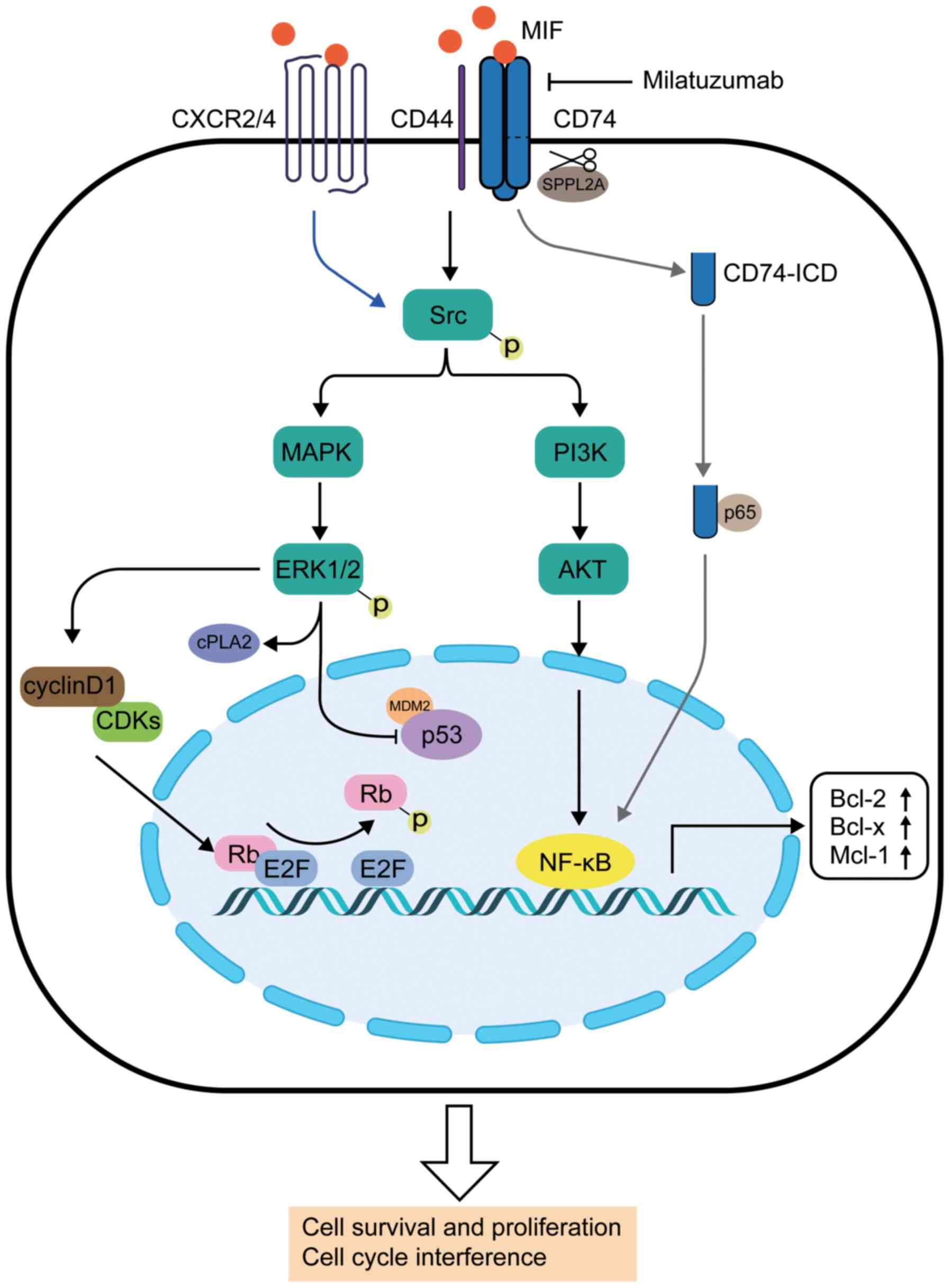
Several signaling pathways in which MIF is involved
have been identified in the past decades (Fig. 1). The interaction between MIF and
the CD74/CD44 complex was a landmark discovery (40,41). CD74, which is also known as
constant chain protein, is a molecular marker expressed on the cell
surface (40). It belongs to the
major histocompatibility complex (MHC) II invariant chain and
facilitates the interaction of MHC II-antigen peptides for antigen
presentation (42). Multiple
studies have demonstrated that CD74 is upregulated in different
types of cancer cells (43-45). CD44 is an adhesion molecule that
mediates the activation of SRC proto-oncogene, non-receptor
tyrosine kinase (Src) family proteins (46). Notably, half of the exons of the
gene encoding CD44 can be spliced into different subtypes, to
generate different protein ectodomains (46). As a result, MIF-activated CD44 is
expressed in cells with dynamic proliferation, such as epithelial
and tumor cells (46). CD44 can
be recruited by CD74 to form a CD74/CD44 complex, which is involved
in the activation of downstream signaling pathways (47).
First, the interaction of MIF-CD74/CD44 results in
phosphorylation of Src family proteins (41). Subsequently, the phosphorylated
Src proteins activate the ERK1/2 MAPK signaling pathway by
phosphorylation (41),
accompanied by the activation of cytosolic phospholipase A2 (cPLA2)
and the inhibition of p53, which is associated with anti-apoptosis
and proliferation effects (48,49). MIF acts as a negative regulator of
p53, probably via binding to p53 and MDM2 proto-oncogene (an E3
ubiquitin ligase), to form a ternary compound (50,51). As a result, cell cycle arrest is
repressed, increasing the risk of malignant transformation
(52). MIF also affects the
retinoblastoma protein-adenoviral early region 2 binding factor
complex by antagonizing Rb-mediated suppression of DNA replication
by upregulating expression of cyclin D1 (53,54), which progresses the cell cycle
from the G1 phase into the S phase, thus promoting cell
proliferation (54). In addition,
the PI3K/AKT and NF-κB signaling pathways are involved in the
downstream signaling, promoting cell survival and proliferation
(55). Secondly, MIF can also
initiate downstream signals in a non-covalent manner following
binding to C-X-C chemokine receptor type 2 (CXCR2)/CXCR4 (56), which is associated with cell
migration and inflammation (57)
(blue arrow; Fig. 1). Thirdly,
MIF can promote the cleavage of the intermembrane part of CD74 via
sPPLA2 protease, resulting in a 42 amino acid peptide (CD74-ICD)
(58). Subsequently, CD74-ICD
migrates into the cytosol and binds to p65 (an NF-κB family
member), regulating the transcription of NF-κB in the nucleus (grey
arrows; Fig. 1) (59). It has been identified that the
cleavage of CD74-ICD and NF-κB activation occurs in B cell
maturation via upregulation of TAp63 (59). In addition, the tyrosine kinase
receptor c-Met is involved, as it contributes to B cell
proliferation and survival (60).
Lastly, research suggests that a soluble form of CD74 is involved
in the regulation of MIF activation (61); however, its mechanism needs to be
further elucidated.
Hematopoietic homeostasis is maintained by the
hematopoietic stem cells (HSCs) and the hematopoietic
microenvironment (62). HSCs stay
in the BM niche, a special structure within the BM that can be
considered as a complex ecological system (62). The niche is composed of different
types of cells that interact with HSCs, providing signals by
secretion of supporting factors to regulate blood cell production
(63). For example, stem cell
factor, TGF-β1, platelet factor 4 [also referred to as chemokine
(C-X-C motif) ligand 4] and angiopoietin 1 are all factors that
maintain HSC quiescent status (63), whereas stromal-derived factor 1
(also referred to as C-X-C motif chemokine ligand 12) and its
receptor CXCR4 (64,65), or adhesion molecules, such as
vascular cell adhesion protein 1 (66), are necessary for cell migration
and homing. In addition, IL-7 (67) and erythropoietin (68) facilitate HSC proliferation and
differentiation.
CD74 is an important regulator involved in the
maturation and differentiation of B cells, and MIF participates in
regulation of B cell differentiation and survival. Gore et
al (55) reported that the
CD74/CD44 complex was found in the membrane of murine B cells,
activating downstream signaling in the classical MIF-CD74
interaction described in the previous section. Furthermore,
dendritic cells in the BM facilitate B cell survival in a
MIF-dependent manner (69).
However, to the best of our knowledge, whether MIF is involved in
the differentiation and proliferation of HSCs has not yet been
established and this requires further study.
Hypoxia-induced factors (HIFs) include a
heterodimeric transcription factor whose classical activation is
oxygen concentration-dependent (70). BM is distinguished by high
cellularity and low oxygen concentrations, albeit being supplied by
a complex vascular network (71).
Extrinsic factors, such as stem cell factors, further promote
increased levels of HIF proteins (72). In the leukemic BM, increased
cellularity and high metabolic activity of proliferating cells
further reduce oxygen concentrations and are associated with
increased expression levels of HIF factors, mainly HIF-1α (73-75), which are involved in a number of
pro-tumor processes, such as cell proliferation and
differentiation, metabolism, and angiogenesis (76-79). Hypoxia is an important factor in
the upregulation of MIF (80),
and HIF-1α can induce MIF expression (81) in a p53-dependent manner (82), while the secretion of MIF can in
turn promote the activation of HIF-related signaling pathways,
forming a positive feedback loop (83).
In addition, the leukemic BM niche allows clonal
proliferation of pre-leukemia HSCs and leukaemic stem cells, while
reducing the capacity of supporting normal hematopoiesis (15). This partly results from BM
structure changes, such as endostral stroma remodeling and fibrosis
(84). Additionally, the
increased inflammatory signaling also contributes to
leukaemogenesis (85), again
resulting in MIF signaling. The functions of MIF in the different
subtypes of leukemia are reviewed in the next section.
CLL comprises a group of chronic lymphoproliferative
disorders. Its prevalence is higher in Caucasians compared with
Asian, Caribbean or African populations (9). It is characterized by malignant
mature B cell proliferation and accumulation (9).
AML is a heterogeneous group of diseases
characterized by myeloid progenitor cells with abnormal
proliferation and differentiation (96). Similar to CLL, the serum levels of
MIF are increased in AML compared with healthy bodies (97). This indicates that the presence of
MIF in the microenvironment may serve an important role in the
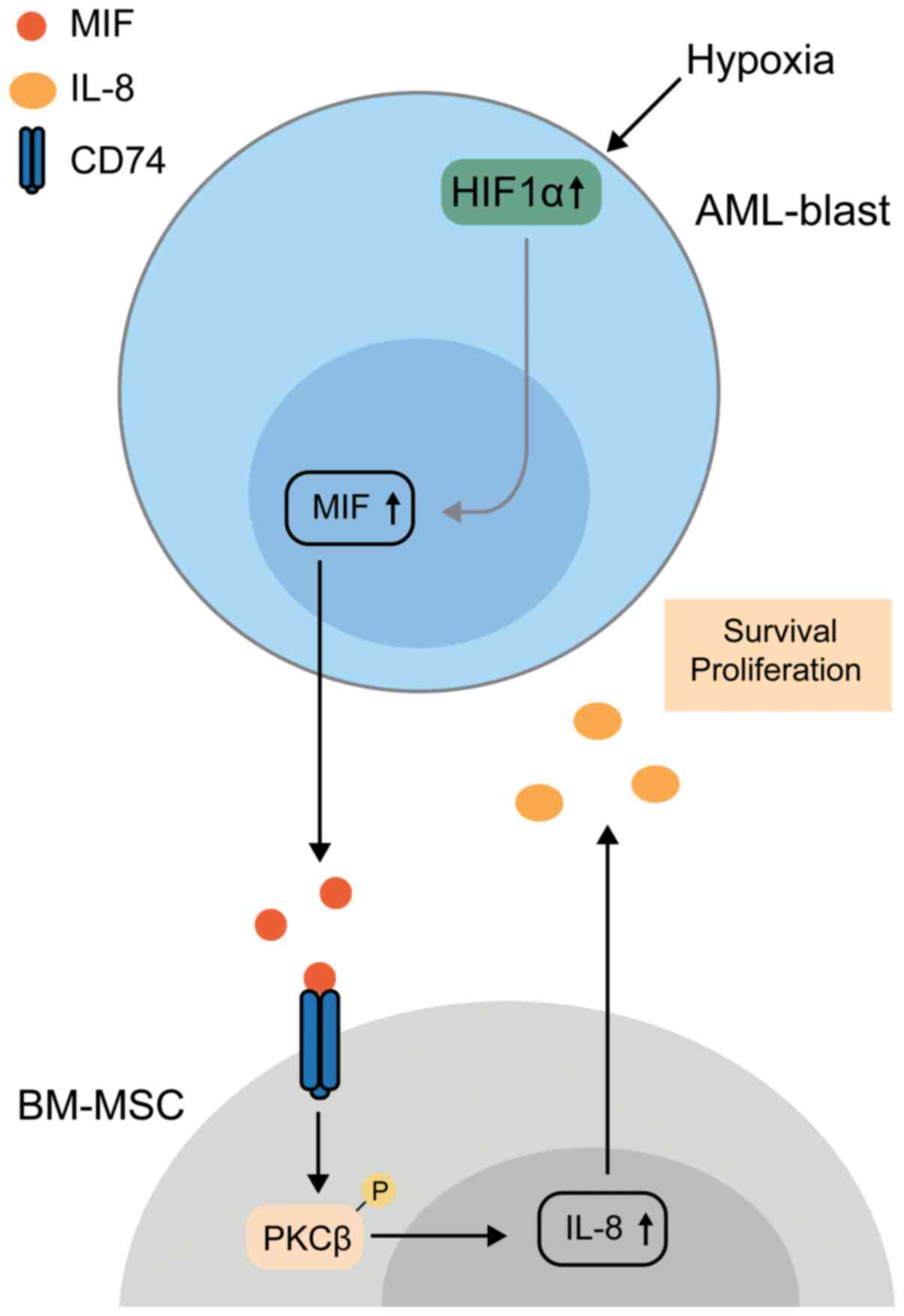
pathogenesis of AML. In 2014, by studying BM samples from 85
patients with AML or myelodysplastic syndromes, Falantes et
al (80) demonstrated that
MIF was highly expressed in BM, which was consistent with the
levels in peripheral blood. Higher MIF expression was associated
with a poorer prognosis and less sensitivity to azacitidine
(80), a first-line therapeutic
drug of AML. A mechanistic explanation was provided by Abdul-Aziz
et al (45), whose in
vitro work deepened the understanding of the role of MIF in
AML. They described that MIF is secreted by AML blasts, after which
it interacts with CD74 via protein kinase C β, but not CXCR2, and
thus, this induces IL-8 expression in BM mesenchymal stromal cells,
which may then promote AML survival (45). Subsequently, they demonstrated
that HIF modulates MIF expression in response to a hypoxic BM
microenvironment. Indeed, knockdown of HIF1α or MIF prolongs the
life of xenograft mice, suggesting that HIF1α promotes MIF
expression and enhances AML blast survival (98). This process is shown in Fig. 2.
Somatic mutations have been identified in different
AML phenotypes, and are associated with response to therapy and
subsequent relapse (99). MIF
promotes the survival of AML-blasts carrying the lysine
methyltransferase 2A-MLLT3 super elongation complex subunit
mutation (99). Future studies
are required to identify the association between other mutations or
subtypes of AML and MIF expression, as their identification would
have potential in precision medical care.
ALL is characterized by proliferation of malignant
lymphoid precursor cells, mainly caused by genetic alterations
(8). Two types are recognized,
T-cell acute lymphoblastic leukaemia (T-ALL) and B-cell acute
lymphoblastic leukemia, depending on the lymphoid precursor cells
involved (8). During treatment of
ALL, the administration of glucocorticoids is important in all
phases (8). However,
glucocorticoid resistance weakens the effects of treatment
(100). MIF counteracts the
function of steroids by suppressing NF-κB inhibitor IκB and
reversing cPLA2 activity (48,101). In vitro data suggest that
MIF expression in a CEM cell line was not affected by treatment
with glucocorticoids (102). A
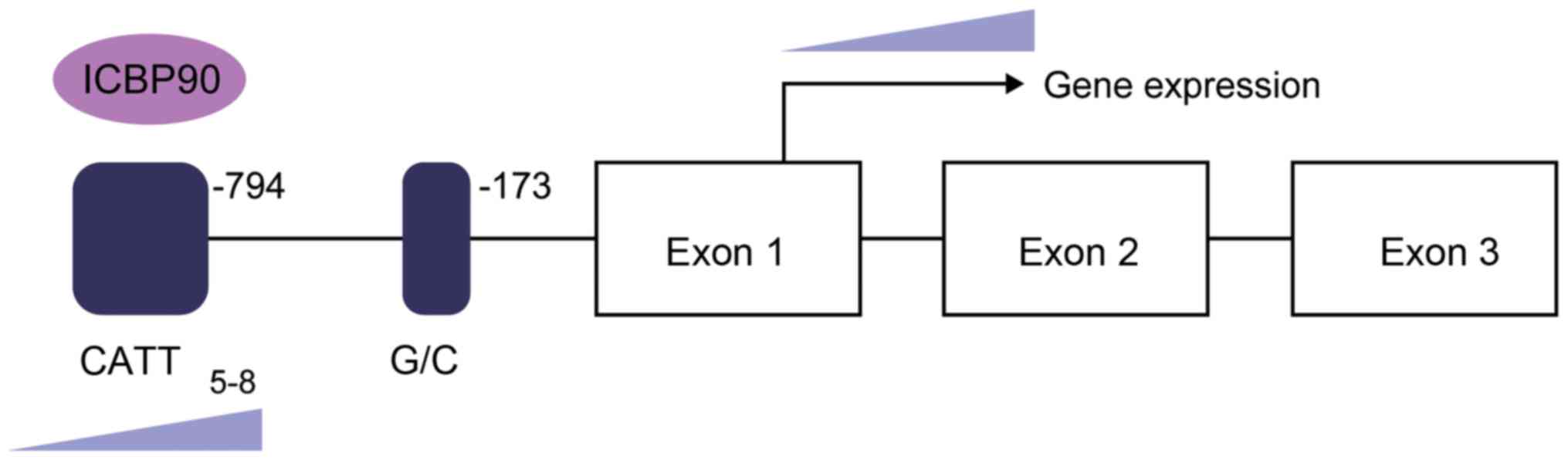
polymorphism near the MIF promoter (details provided in the next
section) is associated with ALL prognosis, and its mechanism
remains to be elucidated.
In the past few decades, the treatment of leukemia
has greatly improved and developments are still ongoing (122). Taking CLL as an example, a
clinical trial (CALGB 9712) demonstrated that the combination of
rituximab and fludarabine improved the rate of complete response,
due to cytotoxic synergism (123). Although various types of drugs,
such as anti-CD20 monoclonal antibody (mAb) (124,125), B cell receptor signaling kinase
inhibitors (126) and BCL-2
antagonists (127), have been
applied in clinical practice, other possible targets remain to be
identified in order to improve treatment response and efficacy.
Three main types of drugs target the MIF/CD74
signaling pathway: MIF inhibitors, mAb targeting MIF and CD74
(128). In hematopoietic tumors,
anti-CD74 mAbs exhibited promising therapeutic potential.
Milatuzumab, an anti-CD74 humanized murine mAb, is generated by
grafting of antigen-recognizing variable regions of LL1 onto human
IgG1 (129). Hertlein et
al (130) demonstrated that
milatuzumab mediates cytotoxicity on CLL directly via CD74
expression. Furthermore, clinical data described promising results
for treatment of refractory patients with CLL (131). The data from a phase I trial
conducted by Martin et al (132) revealed an improvement of WBC
count (usually elevated in leukemia) from an average of
91×109 cells/l to a nadir of 32×109 cells/l,
despite short clinical benefits. A phase I-II study from Israel
revealed that milatuzumab improved the treatment response in 62.5%
(5/8) of patients, with a decreased spleen size and a decreased
requirement of packed red cell transfusion (133). Researchers have also identified
that the amounts of lymphocytes and platelets are increased, while
circulating levels of BCL-2 are decreased, as a result of treatment
with milatuzumab (133). For
safety, neutropenia, thrombocytopenia and rash are the most common
treatment-related adverse events in a dose-dependent manner
(132). The Israel study
indicated that infection was the most common adverse event but was
not associated with milatuzumab (133). It may have resulted from the
generation situation of enrolled individuals (133). The efficacy of the drug has also
been demonstrated in multiple myeloma (134). More evidence is required based
on larger, randomized clinical trials, as well as trials in other
subtypes of leukemia.
Although treatment options have greatly improved
over time, AML treatment remains a great challenge, due to the
complicated genetic alterations and immunophenotypes responsible
for this disease (135). Recent
studies have provided novel insights on combination treatments with
immune checkpoint inhibitors and hypomethylating agents (136), targeting tumor-associated
metabolic and energetic signaling pathways (137), although more clinical data are
required to support such a treatment strategy.
Notably, in hematopoietic tumors, UHRF1 expression
is associated with tumor aggression (138). Alhosin et al (139) reported that thymoquinone could
induce apoptosis in ALL cells, at least in vitro, in a
p73-dependent manner. Other research suggests that UHRF1
facilitates the degradation of promyelocytic leukemia (PML) protein
(140). Knockdown of UHRF1 could
restore PML protein expression and inhibit cell migration and
capillary formation in vitro (140). Furthermore, UHRF1 stabilizes
receptor tyrosine kinase-like orphan receptor 1 in pre-B cells of
ALL, which decreases the sensitivity to chemotherapy (141). Our previous study also suggested
that UHRF1 acts as pro-tumor factor by promoting T-ALL cell
survival (121). Further
investigations could focus on whether UHRF1 can be used as a
potential therapeutic target.
The various functions of MIF go far beyond its
initial description as a pro-inflammatory chemical kinase-like
protein in the early 1930s. This improved understanding of its
complex and multiple functions is enabled and supported by in
vitro experiments and investigations using transgenic animal
models, often in combination with MIF inhibitors. An improved
understanding of the relevant MIF signaling mechanisms in leukemia
can be obtained by studying the complex MIF interactions with
various receptors and their downstream signaling pathways, which
may eventually provide a novel platform for therapeutic strategies
in the future.
Not applicable.
JY contributed to the concept of the review. YL and
XW searched the associated studies. XW drafted the document and YL
wrote the study. JS and JY supervised the study. Data
authentication is not applicable. All authors read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
The authors would like to thank Dr Yu Zhao
(Department of Hematology, The Third Affiliated Hospital of
Southern Medical University, Guangzhou, China) and Dr Zhangfang Li
(Department of Endocrinology and Metabolism, The Third Affiliated
Hospital of Southern Medical University, Guangzhou, China) for
their constructive comments.
This study was supported by Scientific Research Start Plan of
Shunde Hospital, Southern Medical University (grant no.
SRSP2019013) and The National Natural Scientific Foundation of
China (grant no. 81770148).
|
1
|
Saracci R and Wild CP: Fifty years of the
international agency for research on cancer (1965 to 2015). Int J
Cancer. 138:1309–1311. 2016. View Article : Google Scholar
|
|
2
|
Chen J, Odenike O and Rowley JD:
Leukaemogenesis: More than mutant genes. Nat Rev Cancer. 10:23–36.
2010. View Article : Google Scholar :
|
|
3
|
Miranda-Filho A, Pineros M, Ferlay J,
Soerjomataram I, Monnereau A and Bray F: Epidemiological patterns
of leukaemia in 184 countries: A population-based study. Lancet
Haematol. 5:e14–e24. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cazzola M: Introduction to a review
series: The 2016 revision of the WHO classification of tumors of
hematopoietic and lymphoid tissues. Blood. 127:2361–2364. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Landau DA, Tausch E, Taylor-Weiner AN,
Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M,
Böttcher S, et al: Mutations driving CLL and their evolution in
progression and relapse. Nature. 526:525–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Apperley JF: Chronic myeloid leukaemia.
Lancet. 385:1447–1459. 2015. View Article : Google Scholar
|
|
7
|
Bullinger L, Döhner K and Döhner H:
Genomics of acute myeloid leukemia diagnosis and pathways. J Clin
Oncol. 35:934–946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malard F and Mohty M: Acute lymphoblastic
leukaemia. Lancet. 395:1146–1162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bosch F and Dalla-Favera R: Chronic
lymphocytic leukaemia: From genetics to treatment. Nat Rev Clin
Oncol. 16:684–701. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Figueroa ME, Lugthart S, Li Y,
Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J,
van Putten W, Skrabanek L, et al: DNA methylation signatures
identify biologically distinct subtypes in acute myeloid leukemia.
Cancer Cell. 17:13–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vu LP, Cheng Y and Kharas MG: The biology
of m6A RNA methylation in normal and malignant
hematopoiesis. Cancer Discov. 9:25–33. 2019. View Article : Google Scholar
|
|
12
|
Sasaki K, Strom SS, O'Brien S, Jabbour E,
Ravandi F, Konopleva M, Borthakur G, Pemmaraju N, Daver N, Jain P,
et al: Relative survival in patients with chronic-phase chronic
myeloid leukaemia in the tyrosine-kinase inhibitor era: Analysis of
patient data from six prospective clinical trials. Lancet Haematol.
2:e186–e193. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Porter DL, Hwang WT, Frey NV, Lacey SF,
Shaw PA, Loren AW, Bagg A, Marcucci KT, Shen A, Gonzalez V, et al:
Chimeric antigen receptor T cells persist and induce sustained
remissions in relapsed refractory chronic lymphocytic leukemia. Sci
Transl Med. 7:303ra1392015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Turtle CJ, Hanafi LA, Berger C, Gooley TA,
Cherian S, Hudecek M, Sommermeyer D, Melville K, Pender B, Budiarto
TM, et al: CD19 CAR-T cells of defined
CD4+:CD8+ composition in adult B cell ALL
patients. J Clin Invest. 126:2123–2138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schepers K, Campbell TB and Passegue E:
Normal and leukemic stem cell niches: Insights and therapeutic
opportunities. Cell Stem Cell. 16:254–267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamashita M, Dellorusso PV, Olson OC and
Passegué E: Dysregulated haematopoietic stem cell behaviour in
myeloid leukaemogenesis. Nat Rev Cancer. 20:365–382. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Elinav E, Nowarski R, Thaiss CA, Hu B, Jin
C and Flavell RA: Inflammation-induced cancer: Crosstalk between
tumours, immune cells and microorganisms. Nat Rev Cancer.
13:759–771. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Greten FR and Grivennikov SI: Inflammation
and cancer: Triggers, mechanisms, and consequences. Immunity.
51:27–41. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Coussens LM, Zitvogel L and Palucka AK:
Neutralizing tumor-promoting chronic inflammation: A magic bullet?
Science. 339:286–291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kang I and Bucala R: The immunobiology of
MIF: Function, genetics and prospects for precision medicine. Nat
Rev Rheumatol. 15:427–437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bucala R and Donnelly SC: Macrophage
migration inhibitory factor: A probable link between inflammation
and cancer. Immunity. 26:281–285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benjamin D, Aderka D, Livni E, Joshua H,
Shaklai M and Pinkhas J: Migration inhibition factor activity in
sera of patients with chronic lymphatic leukemia. J Natl Cancer
Inst. 63:1175–1177. 1979.PubMed/NCBI
|
|
23
|
Nobre CC, de Araújo JM, Fernandes TA,
Cobucci RN, Lanza DC, Andrade VS and Fernandes JV: Macrophage
migration inhibitory factor (MIF): Biological activities and
relation with cancer. Pathol Oncol Res. 23:235–244. 2017.
View Article : Google Scholar
|
|
24
|
Sun HW, Bernhagen J, Bucala R and Lolis E:
Crystal structure at 2.6-A resolution of human macrophage migration
inhibitory factor. Proc Natl Acad Sci USA. 93:5191–5196. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sparkes A, De Baetselier P, Roelants K, De
Trez C, Magez S, Van Ginderachter JA, Raes G, Bucala R and
Stijlemans B: The non-mammalian MIF superfamily. Immunobiology.
222:473–482. 2017. View Article : Google Scholar :
|
|
26
|
Bloom BR and Bennett B: Mechanism of a
reaction in vitro associated with delayed-type hypersensitivity.
Science. 153:80–82. 1966. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pantouris G, Syed MA, Fan C, Rajasekaran
D, Cho TY, Rosenberg EM Jr, Bucala R, Bhandari V and Lolis EJ: An
Analysis of MIF structural features that control functional
activation of CD74. Chem Biol. 22:1197–1205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Harris J, VanPatten S, Deen NS, Al-Abed Y
and Morand EF: Rediscovering MIF: New tricks for an old cytokine.
Trends Immunol. 40:447–462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rich AR and Lewis MR: The nature of
allergy in tuberculosis as revealed by tissue culture studies. Bull
Johns Hopkins Hosp. 50:115–131. 1932.
|
|
30
|
David JR: Delayed hypersensitivity in
vitro: Its mediation by cell-free substances formed by lymphoid
cell-antigen interaction. Proc Natl Acad Sci USA. 56:72–77. 1966.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bernhagen J, Calandra T, Mitchell RA,
Martin SB, Tracey KJ, Voelter W, Manogue KR, Cerami A and Bucala R:
MIF is a pituitary-derived cytokine that potentiates lethal
endotoxaemia. Nature. 365:756–759. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Petrovsky N, Socha L, Silva D, Grossman
AB, Metz C and Bucala R: Macrophage migration inhibitory factor
exhibits a pronounced circadian rhythm relevant to its role as a
glucocorticoid counter-regulator. Immunol Cell Biol. 81:137–143.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Calandra T, Bernhagen J, Mitchell RA and
Bucala R: The macrophage is an important and previously
unrecognized source of macrophage migration inhibitory factor. J
Exp Med. 179:1895–1902. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bacher M, Metz CN, Calandra T, Mayer K,
Chesney J, Lohoff M, Gemsa D, Donnelly T and Bucala R: An essential
regulatory role for macrophage migration inhibitory factor in
T-cell activation. Proc Natl Acad Sci USA. 93:7849–7854. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Daryadel A, Grifone RF, Simon HU and
Yousefi S: Apoptotic neutrophils release macrophage migration
inhibitory factor upon stimulation with tumor necrosis
factor-alpha. J Biol Chem. 281:27653–27661. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Calandra T and Roger T: Macrophage
migration inhibitory factor: A regulator of innate immunity. Nat
Rev Immunol. 3:791–800. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mitchell R, Bacher M, Bernhagen J,
Pushkarskaya T, Seldin MF and Bucala R: Cloning and
characterization of the gene for mouse macrophage migration
inhibitory factor (MIF). J Immunol. 154:3863–3870. 1995.PubMed/NCBI
|
|
38
|
Merk M, Baugh J, Zierow S, Leng L, Pal U,
Lee SJ, Ebert AD, Mizue Y, Trent JO, Mitchell R, et al: The
Golgi-associated protein p115 mediates the secretion of macrophage
migration inhibitory factor. J Immunol. 182:6896–6906. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Costa-Silva B, Aiello NM, Ocean AJ, Singh
S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et
al: Pancreatic cancer exosomes initiate pre-metastatic niche
formation in the liver. Nat Cell Biol. 17:816–826. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Leng L, Metz CN, Fang Y, Xu J, Donnelly S,
Baugh J, Delohery T, Chen Y, Mitchell RA and Bucala R: MIF signal
transduction initiated by binding to CD74. J Exp Med.
197:1467–1476. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shi X, Leng L, Wang T, Wang W, Du X, Li J,
McDonald C, Chen Z, Murphy JW, Lolis E, et al: CD44 is the
signaling component of the macrophage migration inhibitory
factor-CD74 receptor complex. Immunity. 25:595–606. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Henne C, Schwenk F, Koch N and Möller P:
Surface expression of the invariant chain (CD74) is independent of
concomitant expression of major histocompatibility complex class II
antigens. Immunology. 84:177–182. 1995.PubMed/NCBI
|
|
43
|
Starlets D, Gore Y, Binsky I, Haran M,
Harpaz N, Shvidel L, Becker-Herman S, Berrebi A and Shachar I:
Cell-surface CD74 initiates a signaling cascade leading to cell
proliferation and survival. Blood. 107:4807–4816. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Choi JW, Kim Y, Lee JH and Kim YS: CD74
expression is increased in high-grade, invasive urothelial
carcinoma of the bladder. Int J Urol. 20:251–255. 2013. View Article : Google Scholar
|
|
45
|
Abdul-Aziz AM, Shafat MS, Mehta TK, Di
Palma F, Lawes MJ, Rushworth SA and Bowles KM: MIF-induced stromal
PKCβ/IL8 is essential in human acute myeloid leukemia. Cancer Res.
77:303–311. 2017. View Article : Google Scholar
|
|
46
|
Ponta H, Sherman L and Herrlich PA: CD44:
From adhesion molecules to signalling regulators. Nat Rev Mol Cell
Biol. 4:33–45. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yoo SA, Leng L, Kim BJ, Du X, Tilstam PV,
Kim KH, Kong JS, Yoon HJ, Liu A, Wang T, et al: MIF
allele-dependent regulation of the MIF coreceptor CD44 and role in
rheumatoid arthritis. Proc Natl Acad Sci USA. 113:E7917–E7926.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mitchell RA, Metz CN, Peng T and Bucala R:
Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic
phospholipase A2 activation by macrophage migration inhibitory
factor (MIF). Regulatory role in cell proliferation and
glucocorticoid action. J Biol Chem. 274:18100–18106. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mitchell RA, Liao H, Chesney J,
Fingerle-Rowson G, Baugh J, David J and Bucala R: Macrophage
migration inhibitory factor (MIF) sustains macrophage
proinflammatory function by inhibiting p53: Regulatory role in the
innate immune response. Proc Natl Acad Sci USA. 99:345–350. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jung H, Seong HA and Ha H: Critical role
of cysteine residue 81 of macrophage migration inhibitory factor
(MIF) in MIF-induced inhibition of p53 activity. J Biol Chem.
283:20383–20396. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jankauskas SS, Wong DWL, Bucala R, Djudjaj
S and Boor P: Evolving complexity of MIF signaling. Cell Signal.
57:76–88. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hafner A, Bulyk ML, Jambhekar A and Lahav
G: The multiple mechanisms that regulate p53 activity and cell
fate. Nat Rev Mol Cell Biol. 20:199–210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liao H, Bucala R and Mitchell RA:
Adhesion-dependent signaling by macrophage migration inhibitory
factor (MIF). J Biol Chem. 278:76–81. 2003. View Article : Google Scholar
|
|
54
|
Petrenko O and Moll UM: Macrophage
migration inhibitory factor MIF interferes with the Rb-E2F pathway.
Mol Cell. 17:225–236. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gore Y, Starlets D, Maharshak N,
Becker-Herman S, Kaneyuki U, Leng L, Bucala R and Shachar I:
Macrophage migration inhibitory factor induces B cell survival by
activation of a CD74-CD44 receptor complex. J Biol Chem.
283:2784–2792. 2008. View Article : Google Scholar
|
|
56
|
Bernhagen J, Krohn R, Lue H, Gregory JL,
Zernecke A, Koenen RR, Dewor M, Georgiev I, Schober A, Leng L, et
al: MIF is a noncognate ligand of CXC chemokine receptors in
inflammatory and atherogenic cell recruitment. Nat Med. 13:587–596.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Schneppenheim J, Dressel R, Hüttl S,
Lüllmann-Rauch R, Engelke M, Dittmann K, Wienands J, Eskelinen EL,
Hermans-Borgmeyer I, Fluhrer R, et al: The intramembrane protease
SPPL2a promotes B cell development and controls endosomal traffic
by cleavage of the invariant chain. J Exp Med. 210:41–58. 2013.
View Article : Google Scholar :
|
|
59
|
Lantner F, Starlets D, Gore Y, Flaishon L,
Yamit-Hezi A, Dikstein R, Leng L, Bucala R, Machluf Y, Oren M and
Shachar I: CD74 induces TAp63 expression leading to B-cell
survival. Blood. 110:4303–4311. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gordin M, Tesio M, Cohen S, Gore Y,
Lantner F, Leng L, Bucala R and Shachar I: c-Met and its ligand
hepatocyte growth factor/scatter factor regulate mature B cell
survival in a pathway induced by CD74. J Immunol. 185:2020–2031.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Assis DN, Leng L, Du X, Zhang CK, Grieb G,
Merk M, Garcia AB, McCrann C, Chapiro J, Meinhardt A, et al: The
role of macrophage migration inhibitory factor in autoimmune liver
disease. Hepatology. 59:580–591. 2014. View Article : Google Scholar
|
|
62
|
Morrison SJ and Scadden DT: The bone
marrow niche for haematopoietic stem cells. Nature. 505:327–334.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wilson A, Laurenti E and Trumpp A:
Balancing dormant and self-renewing hematopoietic stem cells. Curr
Opin Genet Dev. 19:461–468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sugiyama T, Kohara H, Noda M and Nagasawa
T: Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4
chemokine signaling in bone marrow stromal cell niches. Immunity.
25:977–988. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ding L and Morrison SJ: Haematopoietic
stem cells and early lymphoid progenitors occupy distinct bone
marrow niches. Nature. 495:231–235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Papayannopoulou T, Craddock C, Nakamoto B,
Priestley GV and Wolf NS: The VLA4/VCAM-1 adhesion pathway defines
contrasting mechanisms of lodgement of transplanted murine
hemopoietic progenitors between bone marrow and spleen. Proc Natl
Acad Sci USA. 92:9647–9651. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cordeiro Gomes A, Hara T, Lim VY,
Herndler-Brandstetter D, Nevius E, Sugiyama T, Tani-Ichi S,
Schlenner S, Richie E, Rodewald HR, et al: Hematopoietic stem cell
niches produce lineage-instructive signals to control multipotent
progenitor differentiation. Immunity. 45:1219–1231. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Koury MJ and Bondurant MC: Erythropoietin
retards DNA breakdown and prevents programmed death in erythroid
progenitor cells. Science. 248:378–381. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Sapoznikov A, Pewzner-Jung Y, Kalchenko V,
Krauthgamer R, Shachar I and Jung S: Perivascular clusters of
dendritic cells provide critical survival signals to B cells in
bone marrow niches. Nat Immunol. 9:388–395. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang GL and Semenza GL: Purification and
characterization of hypoxia-inducible factor 1. J Biol Chem.
270:1230–1237. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Parmar K, Mauch P, Vergilio JA, Sackstein
R and Down JD: Distribution of hematopoietic stem cells in the bone
marrow according to regional hypoxia. Proc Natl Acad Sci USA.
104:5431–5436. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Semenza GL: Oxygen sensing,
hypoxia-inducible factors, and disease pathophysiology. Annu Rev
Pathol. 9:47–71. 2014. View Article : Google Scholar
|
|
73
|
Wellmann S, Guschmann M, Griethe W, Eckert
C, von Stackelberg A, Lottaz C, Moderegger E, Einsiedel HG, Eckardt
KU, Henze G and Seeger K: Activation of the HIF pathway in
childhood ALL, prognostic implications of VEGF. Leukemia.
18:926–933. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Frolova O, Samudio I, Benito JM, Jacamo R,
Kornblau SM, Markovic A, Schober W, Lu H, Qiu YH, Buglio D, et al:
Regulation of HIF-1α signaling and chemoresistance in acute
lymphocytic leukemia under hypoxic conditions of the bone marrow
microenvironment. Cancer Biol Ther. 13:858–870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen H, Shen Y, Gong F, Jiang Y and Zhang
R: HIF-α promotes chronic myelogenous leukemia cell proliferation
by upregulating p21 expression. Cell Biochem Biophys. 72:179–183.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kim JW, Tchernyshyov I, Semenza GL and
Dang CV: HIF-1-mediated expression of pyruvate dehydrogenase
kinase: A metabolic switch required for cellular adaptation to
hypoxia. Cell Metab. 3:177–185. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Han ZB, Ren H, Zhao H, Chi Y, Chen K, Zhou
B, Liu YJ, Zhang L, Xu B, Liu B, et al: Hypoxia-inducible factor
(HIF)-1 alpha directly enhances the transcriptional activity of
stem cell factor (SCF) in response to hypoxia and epidermal growth
factor (EGF). Carcinogenesis. 29:1853–1861. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Rey S and Semenza GL: Hypoxia-inducible
factor-1-dependent mechanisms of vascularization and vascular
remodelling. Cardiovasc Res. 86:236–242. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Magliulo D and Bernardi R: HIF-α factors
as potential therapeutic targets in leukemia. Expert Opin Ther
Targets. 22:917–928. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Falantes JF, Trujillo P, Piruat JI,
Calderón C, Márquez-Malaver FJ, Martín-Antonio B, Millán A, Gómez
M, González J, Martino ML, et al: Overexpression of GYS1, MIF, and
MYC is associated with adverse outcome and poor response to
azacitidine in myelodysplastic syndromes and acute myeloid
leukemia. Clin Lymphoma Myeloma Leuk. 15:236–244. 2015. View Article : Google Scholar
|
|
81
|
Baugh JA, Gantier M, Li L, Byrne A,
Buckley A and Donnelly SC: Dual regulation of macrophage migration
inhibitory factor (MIF) expression in hypoxia by CREB and HIF-1.
Biochem Biophys Res Commun. 347:895–903. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Oda S, Oda T, Nishi K, Takabuchi S,
Wakamatsu T, Tanaka T, Adachi T, Fukuda K, Semenza GL and Hirota K:
Macrophage migration inhibitory factor activates hypoxia-inducible
factor in a p53-dependent manner. PLoS One. 3:e22152008. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Gaber T, Schellmann S, Erekul KB, Fangradt
M, Tykwinska K, Hahne M, Maschmeyer P, Wagegg M, Stahn C, Kolar P,
et al: Macrophage migration inhibitory factor counterregulates
dexamethasone-mediated suppression of hypoxia-inducible factor-1
alpha function and differentially influences human CD4+
T cell proliferation under hypoxia. J Immunol. 186:764–774. 2011.
View Article : Google Scholar
|
|
84
|
Schepers K, Pietras EM, Reynaud D, Flach
J, Binnewies M, Garg T, Wagers AJ, Hsiao EC and Passegué E:
Myeloproliferative neoplasia remodels the endosteal bone marrow
niche into a self-reinforcing leukemic niche. Cell Stem Cell.
13:285–299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Meisel M, Hinterleitner R, Pacis A, Chen
L, Earley ZM, Mayassi T, Pierre JF, Ernest JD, Galipeau HJ, Thuille
N, et al: Microbial signals drive pre-leukaemic myeloproliferation
in a Tet2-deficient host. Nature. 557:580–584. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Richard V, Kindt N and Saussez S:
Macrophage migration inhibitory factor involvement in breast cancer
(review). Int J Oncol. 47:1627–1633. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Soumoy L, Kindt N, Ghanem G, Saussez S and
Journe F: Role of macrophage migration inhibitory factor (MIF) in
melanoma. Cancers (Basel). 11:5292019. View Article : Google Scholar
|
|
88
|
Penticuff JC, Woolbright BL, Sielecki TM,
Weir SJ and Taylor JA III: MIF family proteins in genitourinary
cancer: Tumorigenic roles and therapeutic potential. Nat Rev Urol.
16:318–328. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhang H, Duan J and Wu O: The expression
of macrophage migration inhibitory factor in the non-small cell
lung cancer. Saudi J Biol Sci. 27:1527–1532. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Binsky I, Haran M, Starlets D, Gore Y,
Lantner F, Harpaz N, Leng L, Goldenberg DM, Shvidel L, Berrebi A,
et al: IL-8 secreted in a macrophage migration-inhibitory factor-
and CD74-dependent manner regulates B cell chronic lymphocytic
leukemia survival. Proc Natl Acad Sci USA. 104:13408–13413. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Cohen S, Shoshana OY, Zelman-Toister E,
Maharshak N, Binsky-Ehrenreich I, Gordin M, Hazan-Halevy I,
Herishanu Y, Shvidel L, Haran M, et al: The cytokine midkine and
its receptor RPTPζ regulate B cell survival in a pathway induced by
CD74. J Immunol. 188:259–269. 2012. View Article : Google Scholar
|
|
92
|
Binsky-Ehrenreich I, Marom A, Sobotta MC,
Shvidel L, Berrebi A, Hazan-Halevy I, Kay S, Aloshin A, Sagi I,
Goldenberg DM, et al: CD84 is a survival receptor for CLL cells.
Oncogene. 33:1006–1016. 2014. View Article : Google Scholar
|
|
93
|
Reinart N, Nguyen PH, Boucas J, Rosen N,
Kvasnicka HM, Heukamp L, Rudolph C, Ristovska V, Velmans T, Mueller
C, et al: Delayed development of chronic lymphocytic leukemia in
the absence of macrophage migration inhibitory factor. Blood.
121:812–821. 2013. View Article : Google Scholar
|
|
94
|
Barthel R, Fedorchenko O, Velmans T, Rosen
N, Nguyen PH, Reinart N, Florin A, Herling M, Hallek M and
Fingerle-Rowson G: CD74 is dispensable for development of chronic
lymphocytic leukemia in Eµ-TCL1 transgenic mice. Leuk Lymphoma.
61:2799–2810. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Binsky I, Lantner F, Grabovsky V, Harpaz
N, Shvidel L, Berrebi A, Goldenberg DM, Leng L, Bucala R, Alon R,
et al: TAp63 regulates VLA-4 expression and chronic lymphocytic
leukemia cell migration to the bone marrow in a CD74-dependent
manner. J Immunol. 184:4761–4769. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Short NJ, Rytting ME and Cortes JE: Acute
myeloid leukaemia. Lancet. 392:593–606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Islam M, Mohamed EH, Esa E, Kamaluddin NR,
Zain SM, Yusoff YM, Assenov Y, Mohamed Z and Zakaria Z: Circulating
cytokines and small molecules follow distinct expression patterns
in acute myeloid leukaemia. Br J Cancer. 117:1551–1556. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Abdul-Aziz AM, Shafat MS, Sun Y, Marlein
CR, Piddock RE, Robinson SD, Edwards DR, Zhou Z, Collins A, Bowles
KM and Rushworth SA: HIF1α drives chemokine factor pro-tumoral
signaling pathways in acute myeloid leukemia. Oncogene.
37:2676–2686. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hyrenius-Wittsten A, Pilheden M, Sturesson
H, Hansson J, Walsh MP, Song G, Kazi JU, Liu J, Ramakrishan R,
Garcia-Ruiz C, et al: De novo activating mutations drive clonal
evolution and enhance clonal fitness in KMT2A-rearranged leukemia.
Nat Commun. 9:17702018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Polak R, de Rooij B, Pieters R and den
Boer ML: B-cell precursor acute lymphoblastic leukemia cells use
tunneling nanotubes to orchestrate their microenvironment. Blood.
126:2404–2414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Daun JM and Cannon JG: Macrophage
migration inhibitory factor antagonizes hydrocortisone-induced
increases in cytosolic IkappaBalpha. Am J Physiol Regul Integr Comp
Physiol. 279:R1043–R1049. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Leng L, Wang W, Roger T, Merk M, Wuttke M,
Calandra T and Bucala R: Glucocorticoid-induced MIF expression by
human CEM T cells. Cytokine. 48:177–185. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhong XB, Leng L, Beitin A, Chen R,
McDonald C, Hsiao B, Jenison RD, Kang I, Park SH, Lee A, et al:
Simultaneous detection of microsatellite repeats and SNPs in the
macrophage migration inhibitory factor (MIF) gene by thin-film
biosensor chips and application to rural field studies. Nucleic
Acids Res. 33:e1212005. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Baugh JA, Chitnis S, Donnelly SC, Monteiro
J, Lin X, Plant BJ, Wolfe F, Gregersen PK and Bucala R: A
functional promoter polymorphism in the macrophage migration
inhibitory factor (MIF) gene associated with disease severity in
rheumatoid arthritis. Genes Immun. 3:170–176. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Donn RP, Shelley E, Ollier WE and Thomson
W; British Paediatric Rheumatology Study Group: A novel 5′-flanking
region polymorphism of macrophage migration inhibitory factor is
associated with systemic-onset juvenile idiopathic arthritis.
Arthritis Rheum. 44:1782–1785. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Shi J, Fu H, Jia Z, He K, Fu L and Wang W:
High expression of CPT1A predicts adverse outcomes: A potential
therapeutic target for acute myeloid leukemia. EBioMedicine.
14:55–64. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Sharaf-Eldein M, Elghannam D, Elderiny W
and Abdel-Malak C: Prognostic implication of MIF gene expression in
childhood acute lymphoblastic leukemia. Clin Lab. 64:1429–1437.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Sharaf-Eldein M, Elghannam D and
Abdel-Malak C: MIF-173G/C (rs755622) polymorphism as a risk factor
for acute lymphoblastic leukemia development in children. J Gene
Med. 20:e30442018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Xue Y, Xu H, Rong L, Lu Q, Li J, Tong N,
Wang M, Zhang Z and Fang Y: The MIF -173G/C polymorphism and risk
of childhood acute lymphoblastic leukemia in a Chinese population.
Leuk Res. 34:1282–1286. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ramireddy L, Lin CY, Liu SC, Lo WY, Hu RM,
Peng YC and Peng CT: Association study between macrophage migration
inhibitory factor-173 polymorphism and acute myeloid leukemia in
Taiwan. Cell Biochem Biophys. 70:1159–1165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Donnelly SC, Haslett C, Reid PT, Grant IS,
Wallace WA, Metz CN, Bruce LJ and Bucala R: Regulatory role for
macrophage migration inhibitory factor in acute respiratory
distress syndrome. Nat Med. 3:320–323. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Donn RP, Plant D, Jury F, Richards HL,
Worthington J, Ray DW and Griffiths CE: Macrophage migration
inhibitory factor gene polymorphism is associated with psoriasis. J
Invest Dermatol. 123:484–487. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
De la Cruz-Mosso U, Bucala R,
Palafox-Sánchez CA, Parra-Rojas I, Padilla-Gutiérrez JR,
Pereira-Suárez AL, Rangel-Villalobos H, Vázquez-Villamar M,
Angel-Chávez LI and Muñoz-Valle JF: Macrophage migration inhibitory
factor: association of -794 CATT5-8 and -173 G>C polymorphisms
with TNF-α in systemic lupus erythematosus. Hum Immunol.
75:433–439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Radstake TR, Sweep FC, Welsing P, Franke
B, Vermeulen SH, Geurts-Moespot A, Calandra T, Donn R and van Riel
PL: Correlation of rheumatoid arthritis severity with the genetic
functional variants and circulating levels of macrophage migration
inhibitory factor. Arthritis Rheum. 52:3020–3029. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Vivarelli M, D'Urbano LE, Insalaco A, Lunt
M, Jury F, Tozzi AE, Ravelli A, Martini A, Donn R and De Benedetti
F: Macrophage migration inhibitory factor (MIF) and oligoarticular
juvenile idiopathic arthritis (o-JIA): Association of MIF promoter
polymorphisms with response to intra-articular glucocorticoids.
Clin Exp Rheumatol. 25:775–781. 2007.PubMed/NCBI
|
|
116
|
Mousli M, Hopfner R, Abbady AQ, Monté D,
Jeanblanc M, Oudet P, Louis B and Bronner C: ICBP90 belongs to a
new family of proteins with an expression that is deregulated in
cancer cells. Br J Cancer. 89:120–127. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Liu YZ, Jiang YY, Wang BS, Hao JJ, Shang
L, Zhang TT, Cao J, Xu X, Zhan QM and Wang MR: A panel of protein
markers for the early detection of lung cancer with bronchial
brushing specimens. Cancer Cytopathol. 122:833–841. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Fu H, Xing F, Lv Y, Zeng B, You P and Liu
J: ICBP90 mediates Notch signaling to facilitate human
hepatocellular carcinoma growth. Tissue Cell. 54:65–71. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Hopfner R, Mousli M, Jeltsch JM, Voulgaris
A, Lutz Y, Marin C, Bellocq JP, Oudet P and Bronner C: ICBP90, a
novel human CCAAT binding protein, involved in the regulation of
topoisomerase IIalpha expression. Cancer Res. 60:121–128.
2000.PubMed/NCBI
|
|
120
|
Yao J, Leng L, Sauler M, Fu W, Zheng J,
Zhang Y, Du X, Yu X, Lee P and Bucala R: Transcription factor
ICBP90 regulates the MIF promoter and immune susceptibility locus.
J Clin Invest. 126:732–744. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Yao J, Luo Y, Zeng C, He H and Zhang X:
UHRF1 regulates the transcriptional repressor HBP1 through MIF in T
acute lymphoblastic leukemia. Oncol Rep. 46:1312021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wolach O and Stone RM: Optimal therapeutic
strategies for mixed phenotype acute leukemia. Curr Opin Hematol.
27:95–102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Woyach JA, Ruppert AS, Heerema NA,
Peterson BL, Gribben JG, Morrison VA, Rai KR, Larson RA and Byrd
JC: Chemoimmunotherapy with fludarabine and rituximab produces
extended overall survival and progression-free survival in chronic
lymphocytic leukemia: Long-term follow-up of CALGB study 9712. J
Clin Oncol. 29:1349–1355. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Cartron G, de Guibert S, Dilhuydy MS,
Morschhauser F, Leblond V, Dupuis J, Mahe B, Bouabdallah R, Lei G,
Wenger M, et al: Obinutuzumab (GA101) in relapsed/refractory
chronic lymphocytic leukemia: Final data from the phase 1/2 GAUGUIN
study. Blood. 124:2196–2202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Byrd JC, Flynn JM, Kipps TJ, Boxer M,
Kolibaba KS, Carlile DJ, Fingerle-Rowson G, Tyson N, Hirata J and
Sharman JP: Randomized phase 2 study of obinutuzumab monotherapy in
symptomatic, previously untreated chronic lymphocytic leukemia.
Blood. 127:79–86. 2016. View Article : Google Scholar :
|
|
126
|
Chang BY, Francesco M, De Rooij MF,
Magadala P, Steggerda SM, Huang MM, Kuil A, Herman SE, Chang S,
Pals ST, et al: Egress of CD19(+)CD5(+) cells into peripheral blood
following treatment with the Bruton tyrosine kinase inhibitor
ibrutinib in mantle cell lymphoma patients. Blood. 122:2412–2424.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Souers AJ, Leverson JD, Boghaert ER,
Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH,
Fairbrother WJ, et al: ABT-199, a potent and selective BCL-2
inhibitor, achieves anti-tumor activity while sparing platelets.
Nat Med. 19:202–208. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Bilsborrow JB, Doherty E, Tilstam PV and
Bucala R: Macrophage migration inhibitory factor (MIF) as a
therapeutic target for rheumatoid arthritis and systemic lupus
erythematosus. Expert Opin Ther Targets. 23:733–744. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Stein R, Qu Z, Cardillo TM, Chen S,
Rosario A, Horak ID, Hansen HJ and Goldenberg DM: Antiproliferative
activity of a humanized anti-CD74 monoclonal antibody, hLL1, on
B-cell malignancies. Blood. 104:3705–3711. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hertlein E, Triantafillou G, Sass EJ,
Hessler JD, Zhang X, Jarjoura D, Lucas DM, Muthusamy N, Goldenberg
DM, Lee RJ and Byrd JC: Milatuzumab immunoliposomes induce cell
death in CLL by promoting accumulation of CD74 on the surface of B
cells. Blood. 116:2554–2558. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Berkova Z, Tao RH and Samaniego F:
Milatuzumab-a promising new immunotherapeutic agent. Expert Opin
Investig Drugs. 19:141–149. 2010. View Article : Google Scholar
|
|
132
|
Martin P, Furman RR, Rutherford S, Ruan J,
Ely S, Greenberg J, Coleman M, Goldsmith SJ and Leonard JP: Phase I
study of the anti-CD74 monoclonal antibody milatuzumab (hLL1) in
patients with previously treated B-cell lymphomas. Leuk Lymphoma.
56:3065–3070. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Haran M, Mirkin V, Braester A, Harpaz N,
Shevetz O, Shtreiter M, Greenberg S, Mordich O, Amram O,
Binsky-Ehrenreich I, et al: A phase I-II clinical trial of the
anti-CD74 monoclonal antibody milatuzumab in frail patients with
refractory chronic lymphocytic leukaemia: A patient based approach.
Br J Haematol. 182:125–128. 2018. View Article : Google Scholar
|
|
134
|
Kaufman J, Niesvizky R, Stadtmauer EA,
Chanan-Khan A, Siegel D, Horne H, Teoh N, Leoni MJ, Wegener W and
Goldenberg DM: First trial of humanized anti-CD74 monoclonal
antibody (MAb), milatuzumab, in multiple myeloma. Blood.
112:36972008. View Article : Google Scholar
|
|
135
|
Döhner H, Wei AH and Löwenberg B: Towards
precision medicine for AML. Nat Rev Clin Oncol. May 18–2021.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Stahl M and Goldberg AD: Immune checkpoint
inhibitors in acute myeloid leukemia: Novel combinations and
therapeutic targets. Curr Oncol Rep. 21:372019. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Rashkovan M and Ferrando A: Metabolic
dependencies and vulnerabilities in leukemia. Genes Dev.
33:1460–1474. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Simonetti G, Padella A, do Valle IF,
Fontana MC, Fonzi E, Bruno S, Baldazzi C, Guadagnuolo V, Manfrini
M, Ferrari A, et al: Aneuploid acute myeloid leukemia exhibits a
signature of genomic alterations in the cell cycle and protein
degradation machinery. Cancer. 125:712–725. 2019. View Article : Google Scholar
|
|
139
|
Alhosin M, Razvi SSI, Sheikh RA, Khan JA,
Zamzami MA and Choudhry H: Thymoquinone and difluoromethylornithine
(DFMO) synergistically induce apoptosis of human acute T
lymphoblastic leukemia jurkat cells through the modulation of
epigenetic pathways. Technol Cancer Res Treat.
19:15330338209474892020. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Guan D, Factor D, Liu Y, Wang Z and Kao
HY: The epigenetic regulator UHRF1 promotes ubiquitination-mediated
degradation of the tumor-suppressor protein promyelocytic leukemia
protein. Oncogene. 32:3819–3828. 2013. View Article : Google Scholar :
|
|
141
|
Chow M, Gao L, MacManiman JD, Bicocca VT,
Chang BH, Alumkal JJ and Tyner JW: Maintenance and pharmacologic
targeting of ROR1 protein levels via UHRF1 in t(1;19) pre-B-ALL.
Oncogene. 37:5221–5232. 2018. View Article : Google Scholar : PubMed/NCBI
|