Introduction
Glioblastoma (GBM) is the most common and lethal
form of primary brain tumor affecting humans (1). It originates from astrocytic cells
and belongs to grade IV of diffuse glioma according to the WHO
classification (1). The incidence
rate for GBM is 3.2 per 100, 000 individuals, which ranks first
among all brain tumors (2).
Notably, GBM comprises up to 45.2% of malignant primary brain
tumors and 54% of all gliomas (2). It mainly occurs in older patients,
and the average age at diagnosis is 64 years. GBM is associated
with a very poor prognosis, with a median survival rate as low as
15 months (3). The current
standard therapy for patients with GBM includes surgical resection,
radiation and temozolomide (TMZ) chemotherapy (4). Despite these aggressive treatments,
the majority of patients with GBM develop recurrence within 1 year
following treatment, and the 5-year survival rate is <5%
(5). TMZ, an oral DNA alkylating
agent, can inhibit cell cycle progression at the G2/M phase,
ultimately leading to apoptosis. Acquired resistance to TMZ often
occurs in patients with GBM and this is a predictor of a poor
prognosis. Thus, it is crucial to clarify the mechanisms through
which GBM cells acquire resistance to TMZ.
Exosomes are extracellular vesicles derived from the
endosome. The diameter of exosomes is between 40 to 160 nm, with an
average size of almost 100 nm (6). Exosomes can transfer cytoplasmic or
membrane components, such as DNA, RNA, proteins or other bioactive
molecules to nearby cells, thus playing a vital role in
cell-to-cell communications (7).
Tumor-derived exosomes are involved in cancer microenvironment
remodeling, angiogenesis, metastasis and drug resistance (8). For example, Skog et al
(9) demonstrated that exosomes
derived from GBM cells facilitated the tubule formation of
endothelial cells and the proliferation of glioma cells. The
immunoglobulin superfamily protein, L1 cell adhesion molecule
(L1CAM) has been shown to be associated with the increased growth
and metastasis of GBM cells. Pace et al (10) proved that exosomes carrying L1CAM
significantly promoted the growth and metastasis of glioma cells.
Furthermore, Zhang et al (11) found that long non-coding RNA
SBF2-AS1 delivered by exosomes from TMZ-resistant cells promoted
the tolerance of sensitive GBM cells to TMZ. Although exosomes are
crucial for cancer progression and drug resistance in GBM, the
underlying molecular mechanism are not yet fully understood.
MicroRNAs (miRNAs/miRs) are a small subset of
non-coding RNAs with a length of 19-25 nucleotides. Notably, miRNAs
can be delivered from cell to cell via exosomes, thus possessing
the function of the genome-wide regulation of gene expression to a
larger extent (12). Exosomes are
enriched with miRNAs. miRNAs transferred by exosomes are vital for
tumor immunity, metastasis and drug resistance (12). For example, as previously
demonstrated, miR-1910-3p transferred by exosomes increased the
growth, metastasis and autophagy of breast cancer cells by
activating NF-κB and the Wnt/β-catenin pathway (13). Exosmal miRNAs also play a vital
role in the progression and drug resistance of GBM (14). For example, it was previously
demonstrated that the delivery of miR-1238 by exosomes from
TMZ-resistant GBM cells facilitated TMZ tolerance to other GBM
cells by regulating the CAV1/EGFR axis (15).
miR-25-3p is well-characterized in cancers. Of note,
miR-25-3p is upregulated and plays an oncogenic role in various
types of cancer, such as pancreatic (16), breast (17), esophageal (18), gastric cancer (19) and osteosarcoma (20). The present study investigated
exosomal miRNAs from TMZ-resistant GBM cells. Exosomal miRNAs were
evaluated by high-throughput RNA sequencing. miR-25-3p was
identified as an exosomal miRNA associated with TMZ resistance. The
possible role of exosomal miR-25-3p was thus evaluated in in
vitro and in vivo experiments. F-box and WD repeat
domain-containing-7 (FBXW7) was proven to be a direct target of
miR-25-3p. The exosomal transfer of miR-25-3p regulated the
proliferation and TMZ resistance of GBM cells by inhibiting FBXW7,
thus promoting the expression of oncoproteins, such as c-Myc and
cyclin E. The findings of the present study provide insight into
the role of exosomal miRNAs in the acquired resistance of GBM cells
to TMZ.
Materials and methods
Patient samples
All patients and healthy donors signed written
informed consent forms. The collection and use of serum samples
were approved by the Ethics Committee of Henan Provincial People's
Hospital [SYXK (Yu) 2018-0004]. Serum samples from 42 patients with
GBM treated with TMZ, 34 untreated patients with GBM and 15 healthy
donors were obtained at Henan Provincial People's Hospital from
June, 2018 to July, 2019. The demographic and clinicopathological
data of the patients with GBM were extracted from the hospital
database. The patients and healthy donors did not exhibit any
significant differences as regards age, sex or nationality
(Table I). All patients in the
present study were Chinese. The serum samples were maintained at
−80°C until use.
 | Table IAssociation between serum miR-25-3p
levels and clinicopathological characteristics of patients with
glioblastoma. |
Table I
Association between serum miR-25-3p
levels and clinicopathological characteristics of patients with
glioblastoma.
|
Characteristics | No. of
patients | Serum miR-25-3p
level
| P-value |
|---|
| Low | High |
|---|
| Age (years) | | | | 0.845 |
| ≤60 | 35 | 16 | 19 | |
| >60 | 41 | 23 | 18 | |
| Sex | | | | 0.895 |
| Male | 39 | 18 | 21 | |
| Female | 37 | 21 | 16 | |
| Tumor size
(mm) | | | | 0.029 |
| ≥30 | 49 | 19 | 30 | |
| <30 | 27 | 20 | 7 | |
| Tumor location | | | | 0.748 |
|
Supratentorial | 53 | 28 | 25 | |
|
Infratentorial | 23 | 11 | 12 | |
| Temozolomide
response | | | | |
| Sensitive | 29 | 23 | 6 | 0.001 |
| Resistant | 47 | 16 | 31 | |
| KPS score | | | | 0.338 |
| <80 | 51 | 24 | 27 | |
| ≥80 | 25 | 15 | 10 | |
Cells, cell culture and reagents
293T cells, and the GBM cells, U87 (ATCC #HTB-14)
and A172 (ATCC #CRL-1620), were obtained from the American Type
Culture Collection (ATCC). The U87 cell line used in the present
study is considered is probably a GBM cell line; however, its true
origin is unknown. All cell lines were authenticated by STR
profiling. All cells were maintained in Dulbecco's modified Eagle's
medium (DMEM, HyClone; Cytiva). The culture medium was supplemented
with 10% fetal bovine serum (HyClone; Cytiva), 100 U/ml penicillin
and 100 µg/ml streptomycin. The cells were cultured at 37°C
with 5% CO2 under a humidified condition. The A172R cell
line was constructed by treating the A172 cells with stepwise
increasing concentrations of TMZ (20 to 200 µM) for 4 months
until a stable resistant phenotype was achieved (data not shown).
These TMZ-resistant A172 cell populations were dispersed as single
cells in 96-well plates and exposed to 200 µM TMZ to obtain
TMZ-resistant subclones. Finally, 5 subclones were examined and
they exhibited a similar morphology and growth rates. Thus, one
subclone of these was designated as A172R cells and subjected to
miRNA sequencing analysis. TMZ (#S1237) was purchased from Selleck
Chemicals. Dimethyl sulfoxide (DMSO) was used to dissolve TMZ.
Plasmid constructs, lentiviral packaging
and infection
Anti-miR-25-3p was constructed by inserting 8
repeats of antisense miR-25-3p (5′-TCA GAC CGA GAC AAG TGC AAT
G-3′) into the pGLV3/H1/GFP vector (Shanghai GenePharma Co., Ltd.)
as previously described (21).
Anti-miR-ctrl was constructed by cloning 8 repeats of artificial
miRNA (5′-AAG TTT TCA GAA AGC TAA CA-3′) into the pGLV3/H1/GFP
vector. Ectopic miR-25-3p expression was achieved by inserting the
miR-25-3p mature sequence (5′-CAT TGC ACT TGT CTC GGT CTG A-3′)
into the pCMV-MIR vector (#PCMVMIR; OriGene Technologies, Inc.).
Empty pCMV-MIR vector was regarded as miR-ctrl. Single-guide RNA
(sgRNA) plasmids targeting human FBXW7 (sgFBXW7-1 and sgFBXW7-2)
were constructed by cloning these sgRNAs into lentiCRISPRv2 plasmid
(plasmid#52961; Addgene, Inc.). A non-targeting sequence was
inserted into the lentiCRISPRv2 vector as a sgNC control. The DNA
sequences for sgRNAs were as follows: sgFBXW7-1, 5′-ACC TAC TCT AAA
CCA TGG CT-3′; sgFBXW7-2, 5′-AGC ACA GAA TTG ATA CTA AC-3′; sgNC,
5′-ACG GAG GCT AAG CGT CGC AA-3′. miRNA antagomirs (100 nM) for
miR-670-5p, miR-3194-5p, miR-211-5p, miR-4423-5p, miR-25-3p,
miR-599, miR-486-5p, miR-221-3p, miR-10a-5p and miR-155-5p, and an
antagomir control were synthesized by Guangzhou RiboBio Co., Ltd.
and introduced into GBM cells (2×106) using
Lipofectamine 3000® (Invitrogen; Thermo Fisher
Scientific, Inc.). A lentiviral package (3rd generation system) was
conducted by introducing lentiviral vectors and helper virus
packaging plasmids (pCMVΔR8.9 and pHCMV-VSV-G; 10:10:1) into 293T
cells using Lipofectamine 3000® (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C overnight. Culture medium
containing virus was collected at 24, 48 and 72 h following
transfection, and then maintained at -80°C until further use.
Lentivirus infection was achieved by incubating (1×106)
cells with 1 ml virus-containing medium (~5×107 viral
particles) and 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA)
overnight. The cells were then used in subsequent experiments at 24
h post-infection. Transient transfection was conducted according to
the standard protocol provided with Lipofectamine 3000®
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were introduced
with the indicated plasmids at 37°C overnight and used in
subsequent experiments at 24 h post-transfection.
Exosomal isolation
Exosomes were extracted from the culture medium of
GBM cells as previously described (15). Briefly, serum samples or culture
media were centrifuged at 10,000 × g for 30 min at 4°C to remove
large microvesicles. The supernatants were then filtered using a
0.22-µm filter, and centrifuged at 100,000 × g for 3 h at
4°C. The pellets were dissolved and centrifuged again at 100,000 ×
g for 3 h at 4°C. Exosomes were dissolved in PBS, and maintained at
−80°C until further use. A BCA kit (Thermo Fisher Scientific, Inc.)
was used to examine the concentration of exosomes.
Cell viability assay
Cell viability was determined using a Cell Counting
Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Inc.) as per the
manufacturer's protocol. At each time point, the cells were
incubated with 10 µl CCK-8 solution per well in 96-well
plates at 37°C for 3 h. The absorbance at 450 nm was detected using
a microplate reader (Multiskan SkyHigh; Thermo Fisher Scientific,
Inc.).
Exosome internalization assay
The PKH67 Green Fluorescent Cell Linker kit
(Sigma-Aldrich, Merck KGaA) was used for the labeling of exosomes
according to the manufacturer's instructions. Cells
(1×106) were stained with PKH67-labeled exosomes (2
µg) for 2 h at 37°C in exosome-free medium. The cells were
fixed by 4% PFA for 20 min at 4°C, then washed with cold PBS on ice
for three times. The nuclei were then stained with DAPI
(Sigma-Aldrich, Merck KGaA) for 15 min at 4°C. The cells were kept
on ice and viewed under a laser scanning microscope (Zeiss AG).
Colony formation and soft agar assay
For colony formation assay, cells were seeded
(3,000/well) in 6-well plates for 3 weeks until colonies reached an
appropriate diameter (≥50 µm). The colonies were then fixed
with 4% paraformaldehyde for 15 min at room temperature and then
incubated with 0.5% crystal violet (Beyotime Institute of
Biotechnology) for 1 h at room temperature. For soft agar assay,
cells were seeded (8,000 cells/well) in 0.4% top agar in 6-well
plates. The colonies were allowed to grow for 3 weeks. The colonies
were then incubated with 1 mg/ml thiazolyl blue tetrazolium bromide
(MTT; Sigma-Aldrich; Merck KGaA) at 37°C for 3 h. Images for
colonies were obtained using a scanner (TMA 1600III; Shanghai
Microtek Technology Co., Ltd.).
Western blot analysis
Cells were digested using RIPA buffer (Beyotime
Institute of Biotechnology, Inc.) supplemented with protease
inhibitors (Sigma-Aldrich; Merck KGaA). The protein concentration
was examined using a BCA kit (Thermo Fisher Scientific, Inc.).
Proteins (25 µg) were separated by 10 or 15% SDS-PAGE, then
transferred onto nitrogen membranes. Blocking was performed by
incubating the membranes with 5% non-fat milk for 1 h at room
temperature. Target proteins were detected by staining with
specific primary antibodies at 4°C overnight and corresponding
secondary antibody at room temperature for 1 h. The protein signals
were determined using an ECL plus kit (#32132X3; Thermo Fisher
Scientific, Inc.) on a ChemiDoc Touch Imaging System (Bio-Rad
Laboratories, Inc.). Relative protein quantification was performed
using Image Quant TL 8.1 (GE Healthcare; Cytiva) software. The
specific antibodies used in the present study were as follows:
Cleaved PARP (Asp214) rabbit monoclonal antibody (mAb; 1:1,000;
cat. no. 5625), full-length PARP rabbit mAb (1:1,000; cat. no.
9532), GAPDH (D16H11) rabbit mAb (1:1,000; cat. no. 5174) (all from
Cell Signaling Technology, Inc.), anti-FBXW7 (1:500; cat. no.
ab109617, Abcam), anti-Ago (1:500; cat. no. ab279392, Abcam), c-Myc
(E5Q6W) rabbit mAb (1:1,000; cat. no. 18583), cyclin E1 rabbit mAb
(1:1,000; cat. no. 20808), cleaved caspase-3 (Asp175) rabbit mAb
(1:1,000; cat. no. 9664), caspase-3 rabbit antibody (1:1,000; cat.
no. 9662) and anti-rabbit IgG HRP-linked antibody (1:4,000; cat.
no. 7074) (all from Cell Signaling Technology, Inc.). The anti-CD63
rabbit mAb (1:1,000, cat. no. ab134045) and anti-CD81 rabbit mAb
(1:1,000, ab219209) were purchased from Abcam.
Flow cytometry
Cells were dispersed as single cells using 0.05%
trypsin. Subsequently, 1×106 cells were collected and
suspended in 300 µl PBS. Cell apoptosis was evaluated by
staining the cells using the Annexin V-FITC detection kit (Beyotime
Instittue of Biotechnology, Inc.) as per the manufacturer's
protocol. Propidium iodide (PI) was used to stain the nuclei. A
Guava EasyCyte 6HT-2L flow cytometer (EMD Millipore) was used to
determine apoptotic cells. The results of flow cytometry were
analyzed using FlowJo 7.6 software (BD Biosciences).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The level of
miR-25-3p was measured using TaqMan fast advanced master mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Complementary
DNA was synthesized using the RevertAid First Strand cDNA Synthesis
kit (Thermo Fisher Scientific, Inc.). The amplification reactions
for RT-PCR were as follows: 25°C for 10 min, 42°C for 15 min and
85°C for 50 sec. The level of FBXW7 was detected using Express
SYBR®-GreenER™ qPCR SuperMix (Thermo Fisher Scientific,
Inc.). qPCR was performed on the ABI 7900 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
amplification reactions for qPCR were as follows: 94°C for 10 min,
29 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec.
GAPDH and U6 were used as internal controls. Relative RNA
expression was measured using the 2−ΔΔCq method
(22). The primer sequences used
were as follows: FBXW7 forward, 5′-TGG TCA GCA GTC ACA GGC AAA T-3′
and reverse, 5′-GCA TAC AAC GCA CAG TGG AAG T-3′; GAPDH forward,
5′-TGC ACC ACC AAC TGC TTA GC-3′ and reverse, 5′-GGC ATG GAC TGT
GGT CAT GAG-3′; and U6 forward, 5′-CGC TTC GGC AGC ACA TAT ACT A-3′
and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC A-3′.
BrdU incorporation assay
Cells were grown on coverslips. The cells were then
stained with 10 µM BrdU (Beyotime Institute of
Biotechnology) for 4 h at 37°C and were then incubated with BrdU
mouse mAb (1:300; cat. no. 5292; Cell Signaling Technology, Inc.)
for 1 h at room temperature. The cells were then stained with
anti-mouse IgG Alexa Fluor 488 (1:300; cat. no. 4408, Cell
Signaling Technology, Inc.) for 1 h at room temperature avoiding
light. The nuclei were stained with DAPI for 5 min at room
temperature. The cells were viewed using a confocal microscope
(FV10i; Olympus Corporation).
Tumor xenograft model
The animal experiments were reviewed and approved by
the Animal Care and Experimental Committee of Henan Provincial
People's Hospital. Six-week old female BALB\c nude mice (n=20;
weighing ~20 g) were used in the present study. The BABL/c nude
mice were housed in individually ventilated cages under specific
pathogen-free conditions under 24-h light/dark cycle, 20-26°C and
50-80% humidity. Mice were allowed access to sterilized water and
feed ad libitum. The U87 cells (3×106) infected
with miR-25-3p or miR-ctrl were subcutaneously injected into the
nude mice for 2 weeks until visible tumors were observed. The mice
were then randomly assigned into the TMZ (TMZ) or vehicle (Veh)
group (n=10 for each group). The mice in the TMZ group received
daily treatment with 40 mg/kg TMZ for 3 weeks, while those in the
Veh group received an equal volume of PEG-400: PBS (1:1)
simultaneously. TMZ was dissolved in PEG-400: PBS (1:1) and
administered by oral gavage. Tumor size was evaluated every 3 days
using calipers and determined using the following formula: Length x
width2/2. Finally, the mice were anesthetized by
inhalation with 3% isoflurane and sacrificed by cervical
dislocation. The tumors were then dissected and weighed. In the
present study, the largest tumor diameter was observed <2 cm and
the largest tumor volume was <2,000 mm3.
Luciferase reporter assay
The target of miR-25-3p was predicted using
TargetScan 7.2 (http://www.targetscan.org/vert_72/). The 3′UTR of
FBXW7 with miR-25-3p binding sites was inserted into the
pMIR-REPORT vector (#AM5795; Thermo Fisher Scientific, Inc.). The
QuickChange Site-Directed Mutagenesis kit (#210518, Agilent
Technologies, Inc.) was used to generate mutations in the predicted
binding sites for FBXW7 as per the manufacturer's protocol. The
293T cells were co-transfected with miR-25-3p, anti-miR-25-3p or
control plasmids, pMIR-REPORT vector containing the 3′UTR of FBXW7
or mutant, and a Renilla luciferase plasmid at a ratio of
2:2:1 using Lipofectamine 3000® (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C overnight. Dual-luciferase
reporter assay system (Promega Corporation) was performed to
determine the luciferase activity at 48 h post-transfection.
RNA immunoprecipitation (RNA-IP)
RNA immunoprecipitation was performed according to
the protocol provided with the Magna RIP RNA-Binding Protein
Immunoprecipitation kit (EMD Millipore #17-700). Briefly, cells
were washed with ice-cold PBS, then lysed with RIP lysis buffer for
5 min at 4°C. Subsequently, 50 µl Magnetic Beads Protein A/G
(#CS203178, EMD Millipore) beads were washed with 0.5 ml RIP wash
buffer. The beads (100 µl) were then incubated with
anti-Ago1 (5 µg, cat. no. ab279392, Abcam) or anti-IgG (5
µg; cat. no. ab238004, Abcam) for 30 min at room
temperature. The beads were then washed the with 0.5 ml RIP Wash
Buffer. The cell lysates (100 µl) were then incubated with
antibody-labeled beads in 900 µl RIP immunoprecipitation
buffer at 4°C overnight. The beads were then washed with RIP wash
buffer for 5 times, and treated with Proteinase K for 30 min with
shaking at 55°C. Immunoprecipitated RNA in the precipitates was
purified using TRIzol® RNA reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and analyzed by RT-qPCR.
miRNA sequencing analysis
miRNA sequencing was carried out as previously
described (23). In brief, total
RNA was extracted using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Small RNA libraries were prepared
according to the NEBNext Small RNA Library Prep Set for Illumina
(New England BioLabs) as indicated in the manufacturer's protocol.
The libraries were then sequenced on a NextSeq 500 Sequencer
(Illumina, Inc.). Small RNA-seq reads were trimmed using Cutadpt
(https://cutadapt.readthedocs.io/en/stable/). miRNA
expression was quantified using ncPRO-seq (version v1.5.1)
(https://sourceforge.net/projects/ncproseq/files/)
based on miRbase v19. Differentially expressed miRNAs were defined
as those with a |log2 (FC)| >2 and FDR <0.01
change in expression.
Statistical analysis
SPSS 19.0 (SPSS, Inc.) and GraphPad Prism 8.0
(GraphPad Software, Inc.) were used for statistical analysis. A
two-tailed unpaired or paired Student's t-test was used to compare
differences between 2 groups. One-way ANOVA (with Tukey's post-hoc
test) was used to compare differences between multiple groups. The
half maximal inhibitory concentration (IC50) of TMZ was evaluated
using GraphPad Prism 8.0. Pearson's Chi-squared test was applied to
analyze clinical variables. Data are presented as the mean ±
standard deviation (mean ± SD). P≤0.05 was considered to indicate a
statistically significant difference.
Results
Exosomes from TMZ-resistant GBM cells
promote the proliferation and TMZ resistance of sensitive GBM
cells
To examine the exosomes of TMZ-resistant GBM cells,
a TMZ-resistant GBM cell line (A172R) was constructed by exposing
parental A172 cells to various concentrations of TMZ for 4 months
(data not shown). A172 is a frequently used TMZ-sensitive GBM cell
line (24). It has been proven
that long-term exposure to TMZ can confer TMZ resistance to A172
cells (24). In the present
study, it was found that the A172R cells exhibited increased
resistance to TMZ compared with the parental A172 cells, with the
TMZ IC50 changing from 114.3 to 1345.8 nM (A172 vs. A172R, Fig. 1A). The resistance of A172R cells
to TMZ was also demonstrated by colony formation assay. It was
found that TMZ significantly suppressed the colony formation of
parental A172 cells, although this effect was diminished in
resistant A172R cells (Fig. 1B and
C). The exosomes from resistant A172R cells and parental A172
cells were then extracted from the culture medium of these cells.
CD81 and CD63 are markers for exosomes (25). It was verified that CD81 and CD63
were expressed only in exosomes extracted from the culture medium
of A172R (A172R-exo) and A172 cells (A172-exo), but not in cell
lysates (Fig. 1D). To validate
the successful internalization of A172R-exo or A172-exo by GBM
cells, A172R-exo and A172-exo were labeled with the green
fluorescent dye, PKH67, then added into the culture medium of A172
cells. It was found that the A172 cells treated with A172R-exo or
A172-exo exhibited green fluorescent signals; however, the A172
cells treated with PBS did not exhibit any fluorescent signals,
suggesting that A172R-exo and A172-exo were successfully
internalized by the GBM cells (Fig.
1E and F). Subsequently, the effects of exosomes of A172R and
A172 cells on the GBM cells were examined by cell viability assay
and soft agar assay. U87 is a frequently used TMZ-sensitive GBM
cell line (24). It was found
that exosomes of A172R cells (A172R-exo) evidently facilitated the
growth of U87 and A172 cells compared with exosomes derived from
A172 cells (A172-exo) (Fig. 1G).
In addition, the U87 and A172 cells treated with A172R-exo formed
larger colonies in soft agar compared with the cells treated with
A172-exo (Fig. 1H and I). The
effects of A172R-exo and A172-exo on the resistance of GBM cells to
TMZ were then evaluated. It was found that the U87 and A172 cells
exposed to A172R-exo exhibited an increased TMZ IC50 compared with
those treated with PBS or A172-exo, indicating that the exosomes of
TMZ-resistant A172R cells spread TMZ resistance to sensitive GBM
cells (Fig. 1J). In colony
formation assay, the U87 and A172 cells co-treated with TMZ and
A172R-exo formed more colonies than the cells co-treated with TMZ
and PBS or A172-exo (Fig. 1K and
L). Moreover, the U87 and A172 cells co-treated with TMZ and
A172R-exo (A172R-exo + TMZ) exhibited a decreased apoptosis
compared with the cells co-treated with TMZ and A172-exo (A172-exo
+ TMZ) (Fig. 1M and N). Western
blot analysis revealed that the U87 and A172 cells co-treated with
TMZ and A172R-exo (A172R-exo + TMZ) exhibited low levels of cleaved
PARP and cleaved caspase-3 compared with the cells co-treated with
TMZ and A172-exo (A172-exo + TMZ) (Fig. 1O and P). Taken together, these
data demonstrated that the exosomes of TMZ-resistant GBM cells
promoted the proliferation and TMZ resistance of sensitive GBM
cells.
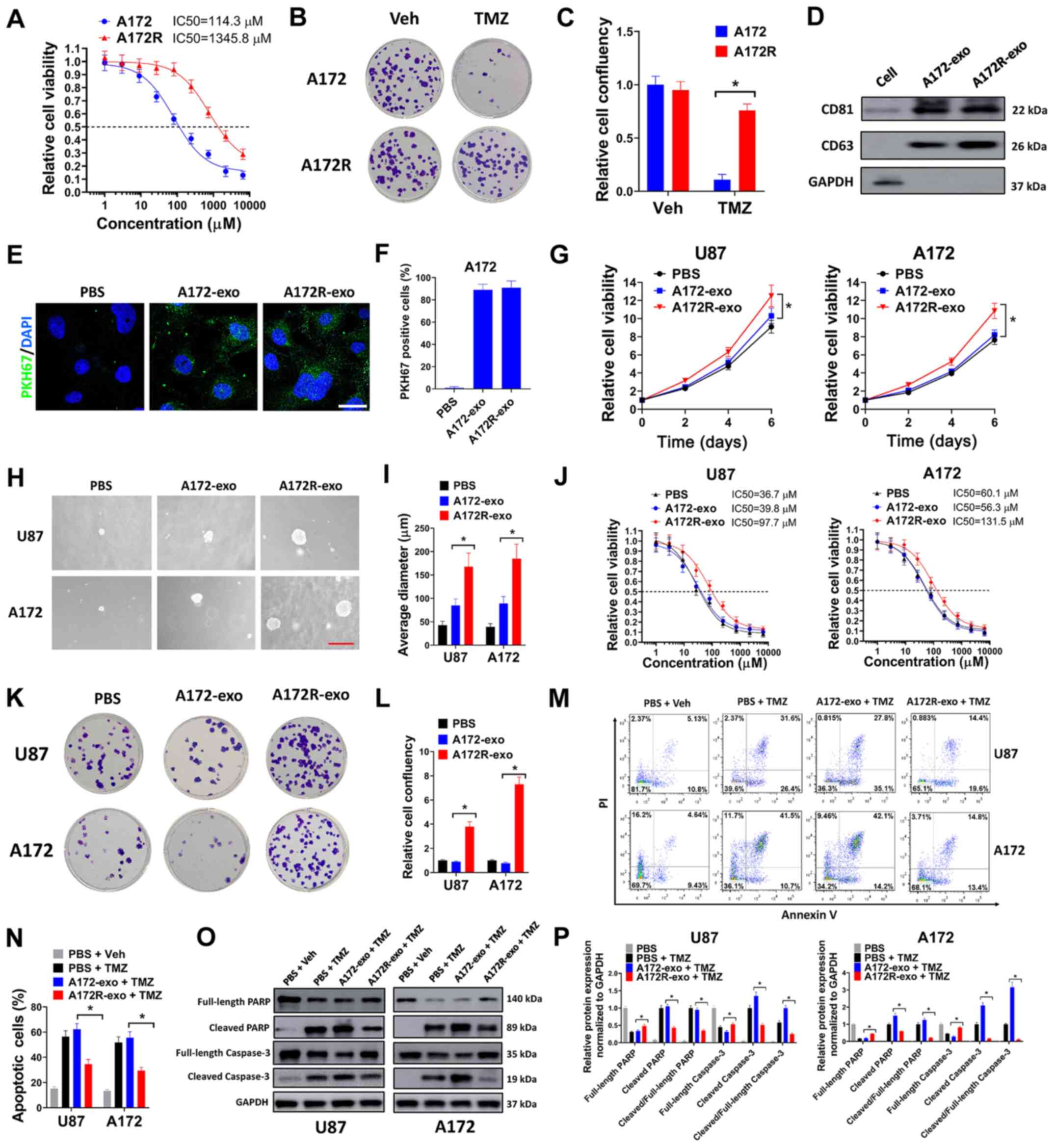 | Figure 1Exosomes from TMZ-resistant GBM cells
promote the proliferation and TMZ resistance of sensitive GBM
cells. (A) A172R or A172 cells were seeded in 96-well plates (5,000
cells/well), then exposed to 0, 1, 3, 9, 27, 81, 243, 729, 2,181 or
6,543 µM TMZ for 6 days, then evaluated by cell viability
assay. (B and C) A172R or A172 cells were seeded in 6-well plates
(3,000 cells/well) and exposed to 50 µM TMZ (TMZ group) or
an equal volume of DMSO (Veh group) for colony formation assay. (B)
Representative plates and (C) relative cell confluency are shown.
(D) Expression of the exosomal markers, CD81 and CD63, was
evaluated by western blot analysis. (E) A172 cells were incubated
with PKH67-labeled A172-exo or A172R-exo, or PBS for exosome
internalization assay. DAPI was used to stain the nuclei. Scale
bar, 10 µm. (F) The percentages of PKH67-positive cells are
shown. (G) U87 or A172 cells were treated with A172R-exo (50
µg/ml), A172-exo (50 µg/ml) or an equal volume of
PBS, and cell viability was evaluated on days 2, 4 and 6. (H and I)
U87 or A172 cells were seeded at 8,000 cells/well for soft agar
assay, and simultaneously treated with A172R-exo (50 µg/ml),
A172-exo (50 µg/ml) or an equal volume of PBS. Represent
images of (H) colonies (scale bar, 200 µm) and (I) the
average diameter of colonies are shown. (J) U87 or A172 cells were
seeded in 96-well plates (5,000 cells/well), then exposed to 0, 1,
3, 9, 27, 81, 243, 729, 2,181 or 6,543 µM TMZ in combination
with A172R-exo (50 µg/ml), A172-exo (50 µg/ml) or an
equal volume of PBS for 6 days, then evaluated by cell viability
assay. (K and L) U87 or A172 cells were seeded in 6-well plates
(3,000 cells/well) and treated with 50 µM TMZ in combination
with A172R-exo (50 µg/ml), A172-exo (50 µg/ml) or an
equal volume of PBS for colony formation assay. Represent (K)
plates and (L) relative cell confluency are shown. (M-P) U87 or
A172 cells were treated with 50 µM TMZ or an equal volume of
DMSO (Veh) in combination with A172R-exo (50 µg/ml),
A172-exo (50 µg/ml) or an equal volume of PBS for 72 h, then
cells were used for (M) flow cytometric analysis or (O) western
blot analysis. (N) Percentages of apoptotic cells and (P) relative
protein expression normalized to GAPDH are shown. All experiments
were performed in triplicate. *P≤0.05. TMZ,
temozolomide; GBM, glioblastoma; A172R, TMZ-resistant A172 cells;
A172-exo or A172R-exo, exosomes from A172 and A172R cells,
respectively. |
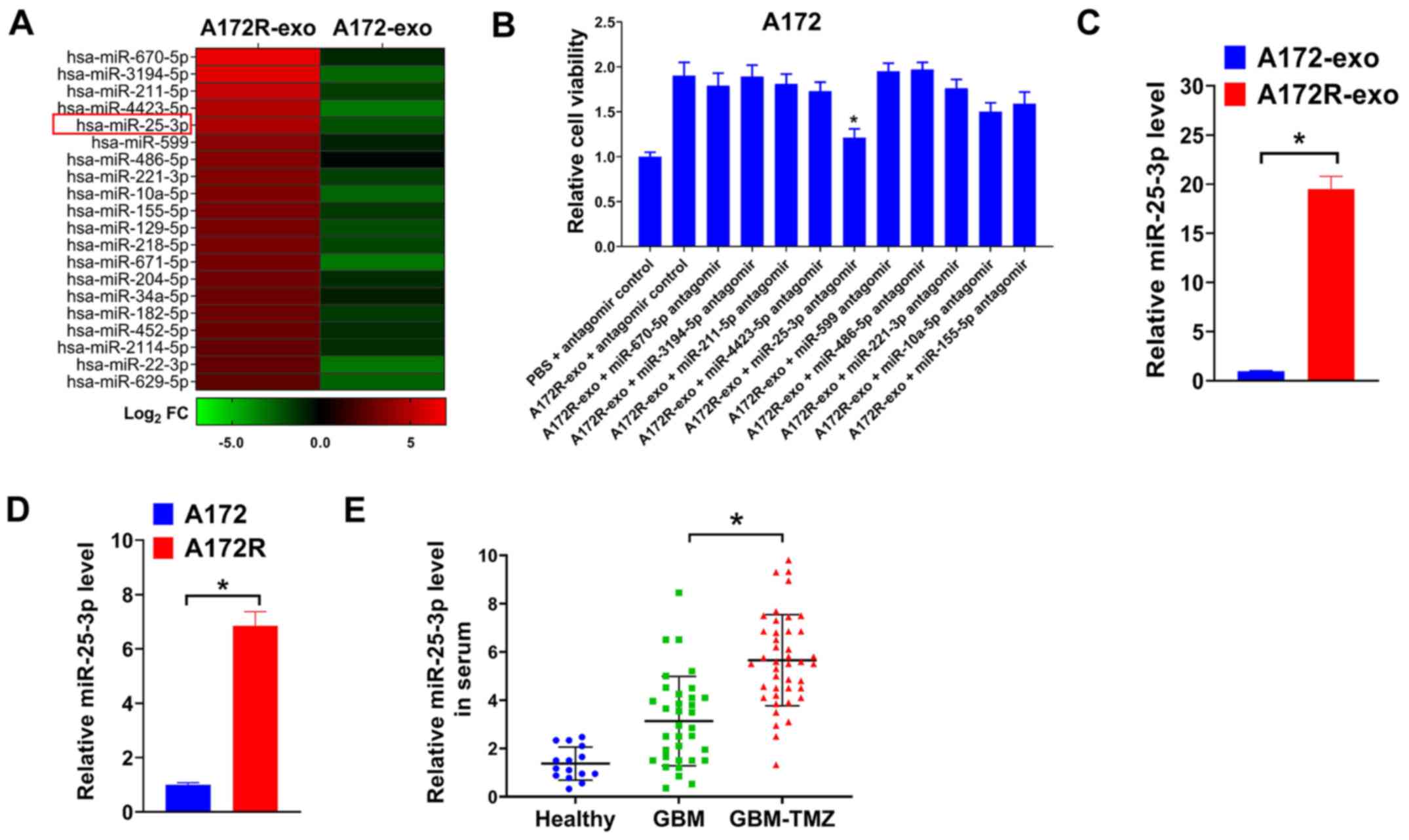
miR-25-3p is upregulated in exosomes of
TMZ-resistant GBM cells and in serum of patients with GBM treated
with TMZ
Exosomes are enriched with miRNAs (8). Previous studies have indicated that
exosomal miRNAs are crucial for acquired TMZ resistance in GBM
(11,14,15). In the present study, to explore
the possible exosomal miRNAs involved in the TMZ resistance of
A172R cells, miRNA sequencing data for A172R-exo and A172-exo were
obtained using Illumina NextSeq 500 sequencing. The dysregulated
miRNAs between A172R-exo and A172-exo were defined as |log2 fold
change| ≥2. The top 20 upregulated miRNAs in A172R-exo compared
with A172-exo are depicted in Fig.
2A. To search for exosomal miRNAs that involved in the effects
of A172R-exo, A172 cells were treated with A172R-exo in combination
with antagomirs for indicated miRNAs, and cell viability was then
evaluated. As shown in Fig. 2B,
it was found that the depletion of exosomal miR-25-3p significantly
impaired the effects of A172R-exo on the viability of A172 cells.
Moreover, previous studies have indicated that miR-25-3p plays a
key role in the tumorigenesis of GBM (26,27). In the present study, miR-25-3p
expression in A172R cells and exosomes was determined by RT-qPCR.
It was found that miR-25-3p expression was upregulated in A172R
cells, as well as exosomes derived from these cells (A72R-exo)
(Fig. 2C and D). As miR-25-3p was
overexpressed in A172R-exo, it was hypothesized that miR-25-3p
expression may be increased in serum from patients with GBM treated
with TMZ. Thus, circulating miR-25-3p in healthy donors and
patients with GBM treated with or without TMZ was assayed by
RT-qPCR. As was expected, miR-25-3p expression was upregulated in
patients with GBM treated with TMZ (GBM-TMZ) compared with
untreated patients with GBM or healthy donors (Fig. 2E). Furthermore, the association of
circulating miR-25-3p with the clinicopathological characteristics
of patients with GBM was evaluated. Patients with GBM were divided
into the serum miR-25-3p high expression group and the serum
miR-25-3p low expression group using the median of miR-25-3p
expression as the cut-off value. It was found that a high
circulating miR-25-3p level was positively associated with a larger
tumor size and TMZ resistance in patients with GBM (Table I). On the whole, these data
suggested that miR-25-3p expression was increased in exosomes from
TMZ-resistant GBM cells and in serum of patients with GBM treated
with TMZ.
Knockdown of exosomal miR-25-3p partially
abrogates the effects induced by exsomes transferred from
TMZ-resistant GBM cells
To explore the possible role of exosomal miR-25-3p
in TMZ-resistant GBM cells, miR-25-3p expression was depleted by
transduction with anti-miR-25-3p. It was found that the
transduction of anti-miR-25-3p significantly decreased the
expression levels of miR-25-3p in the A172R, A172 and U87 cells
(Fig. 3A). Moreover, exosomal
miR-25-3p expression was significantly downregulated in the A172R
cells transfected with anti-miR-25-3p (A172R-exo + anti-miR-25-3p
group) compared with the cells transfected with anti-miR-ctrl
(A172R-exo + anti-miR-ctrl group) (Fig. 3B). In the cell viability assay
(Fig. 3C) and soft agar assay
(Fig. 3D and E), it was observed
that the depletion of miR-25-3p attenuated the effects of A172R-exo
on the growth and colony formation of U87 and A172R cells.
Moreover, the U87 and A172 cells treated with A172R-exo exhibited
an increased TMZ IC50 compared with the cells treated with PBS;
however, these effects were attenuated by miR-25-3p knockdown
(Fig. 3F). In the colony
formation assay, the U87 and A172 cells treated with A172R-exo
(A172R-exo + anti-miR-ctrl) exhibited increased colony formation
compared with the cells treated with PBS; however, these effects
were partially attenuated by miR-25-3p knockdown (A172R-exo +
anti-miR-25-3p) (Fig. 3G and H).
In flow cytometric analysis, the U87 and A172 cells co-treated with
TMZ and A172R-exo (A172R-exo + anti-miR-ctrl + TMZ) exhibited a
decreased apoptosis compared with the cells co-treated with PBS and
TMZ (PBS + TMZ); however, these effects were partially attenuated
by miR-25-3p knockdown (A172R-exo + anti-miR-25-3p + TMZ) (Fig. 3I and J). In western blot analysis,
the U87 and A172 cells co-treated with TMZ and A172R-exo (A172R-exo
+ anti-miR-ctrl + TMZ) exhibited low levels of cleaved PARP and
cleaved caspase-3 compared with the cells co-treated with PBS and
TMZ (PBS + TMZ); however, these effects were diminished by
miR-25-3p knockdown (A172R-exo + anti-miR-25-3p + TMZ) (Fig. 3K and L). Collectively, these data
indicated that the knockdown of exosomal miR-25-3p partially
abrogated the effects induced by exsomes transferred from
TMZ-resistant GBM cells.
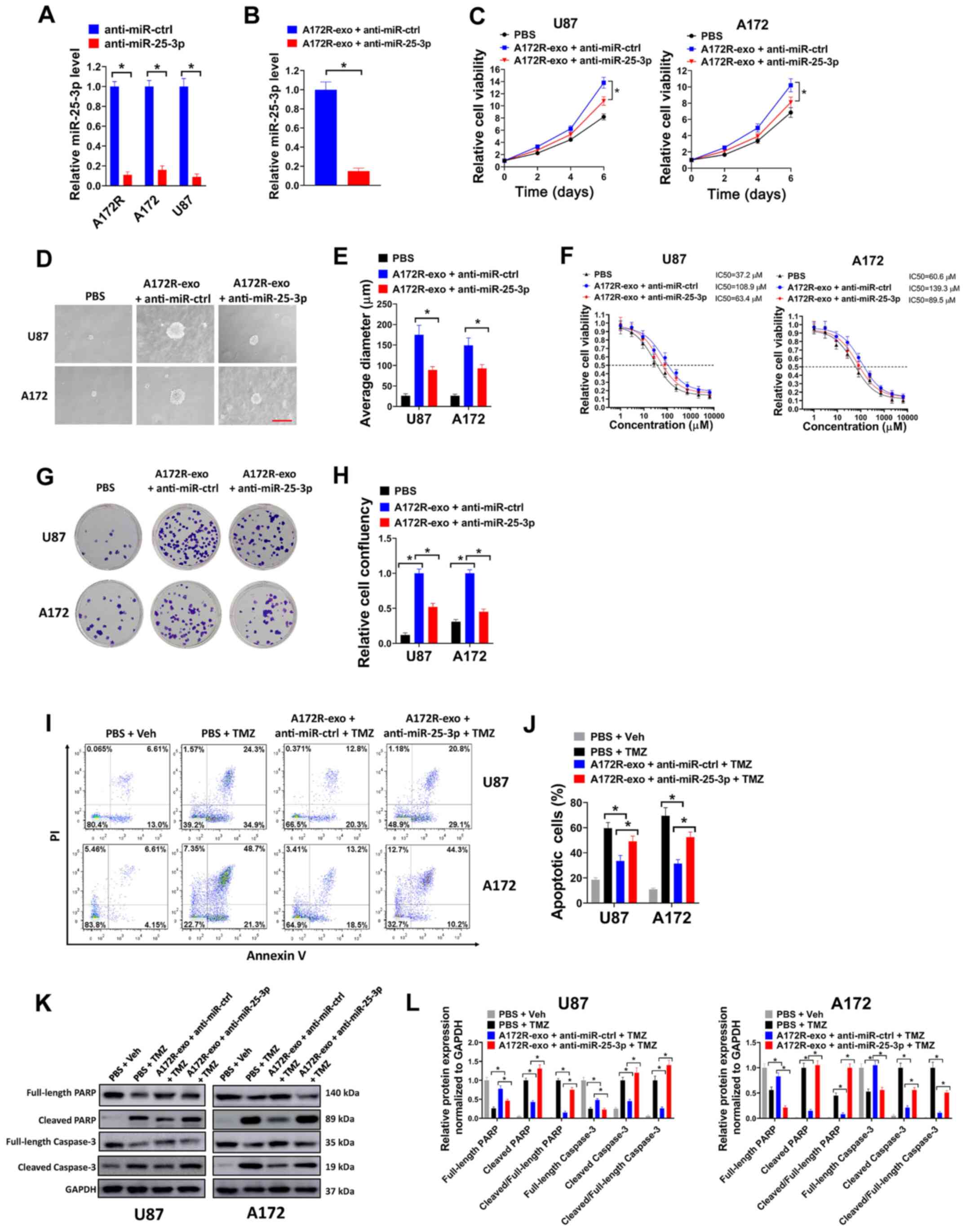 | Figure 3Knockdown of exosomal miR-25-3p
partially abrogates the effects caused by exsomes transferred from
TMZ-resistant GBM cells. (A) Relative miR-25-3p expression in
A172R, A172 and U87 cells transduced with anti-miR-25-3p or
anti-miR-ctrl was evaluated by RT-qPCR. (B) Relative miR-25-3p
expression in exosomes derived from A172R cells transduced with
anti-miR-25-3p (A172R-exo + anti-miR-25-3p group) or exosomes
derived from A172R cells transduced with anti-miR-ctrl (A172R-exo +
anti-miR-ctrl group) was evaluated by RT-qPCR. (C) U87 or A172
cells were treated with exosomes (50 µg/ml) derived from
A172R cells transduced with anti-miR-25-3p or miR-ctrl, or PBS, and
cell viability was then evaluated on days 2, 4 and 6. (D and E) U87
or A172 cells were seeded at 8,000 cells/well for soft agar assay,
and simultaneously treated with exosomes (50 µg/ml) derived
from A172R cells transduced with anti-miR-25-3p or miR-ctrl, or
PBS. (D) Representative images of colonies (scale bar, 200
µm) and (E) average diameter of colonies are shown. (F) U87
or A172 cells were seeded in 96-well plates (5,000 cells/well),
then exposed to 0, 1, 3, 9, 27, 81, 243, 729, 2,181 or 6,543
µM TMZ in combination with exosomes (50 µg/ml)
derived from A172R cells transduced with anti-miR-25-3p or
miR-ctrl, or PBS for 6 days, and cell viability assay was then
performed. (G and H) U87 or A172 cells were seeded in 6-well plates
(3,000 cells/well) and treated with 50 µM TMZ in combination
with exosomes (50 µg/ml) derived from A172R cells transduced
with anti-miR-25-3p or miR-ctrl, or PBS for colony formation assay.
(G) Represent plates and (H) relative cell confluency are shown.
(I-L) U87 or A172 cells were treated with 50 µM TMZ or an
equal volume of DMSO (Veh) in combination with exosomes (50
µg/ml) derived from A172R cells transduced with
anti-miR-25-3p or miR-ctrl, or PBS for 72 h, then cells were used
for (I) flow cytometry or (K) western blot analysis. (J)
Percentages of apoptotic cells and (L) relative protein expression
normalized to GAPDH are shown. All experiments were performed in
triplicate. *P≤0.05. TMZ, temozolomide; GBM,
glioblastoma; A172R, TMZ-resistant A172 cells; A172-exo or
A172R-exo, exosomes from A172 and A172R cells, respectively. |
miR-25-3p overexpression promotes the
proliferation and TMZ resistance of GBM cells
The effects of miR-25-3p overexpression on GBM cells
were evaluated in the present study. miR-25-3p overexpression was
achieved by transfecting the cells with lentivirus expressing
miR-25-3p (Fig. 4A). It was found
that the enforced expression of miR-25-3p promoted the growth of
U87 and A172 cells compared with the cells transfected with
miR-ctrl (Fig. 4B). In BrdU
incorporation assay (Fig. 4C and
D) and soft agar assay (Fig. 4E
and F), miR-25-3p overexpression increased the number of
BrdU-positive cells and the diameter of the colonies of U87 and
A172 cells. These data suggested that miR-25-3p overexpression
increased the proliferation of GBM cells. The effects of miR-25-3p
on the TMZ resistance of GBM cells were then evaluated. In cell
viability assay, miR-25-3p overexpression markedly increased the
TMZ IC50 of U87 and A172 cells compared with the miR-ctrl (Fig. 4G). Flow cytometric analysis
indicated that miR-25-3p overexpression decreased the number of
apoptotic cells treated with TMZ (Fig. 4H and I). In the tumor xenograft
model, U87 cells infected with miR-25-3p or miR-ctrl were
subcutaneously injected into nude mice and the mice were then
treated with TMZ or the vehicle control for 3 weeks. It was found
that TMZ treatment significantly reduced the tumor growth of U87
cells transduced with miR-ctrl (TMZ + miR-ctrl group) compared with
the Veh + miR-ctrl group, with reduced tumor volume and weight
(Fig. 4J-M). However, the U87
cells transduced with miR-25-3p (TMZ + miR-25-3p group) led to
increased tumor growth, volume and weight compared with the cells
transduced with miR-ctrl when the mice were treated with TMZ (TMZ +
miR-ctrl group) (Fig. 4J-M).
Taken together, these findings demonstrated that miR-25-3p
overexpression promoted the proliferation and TMZ resistance of GBM
cells.
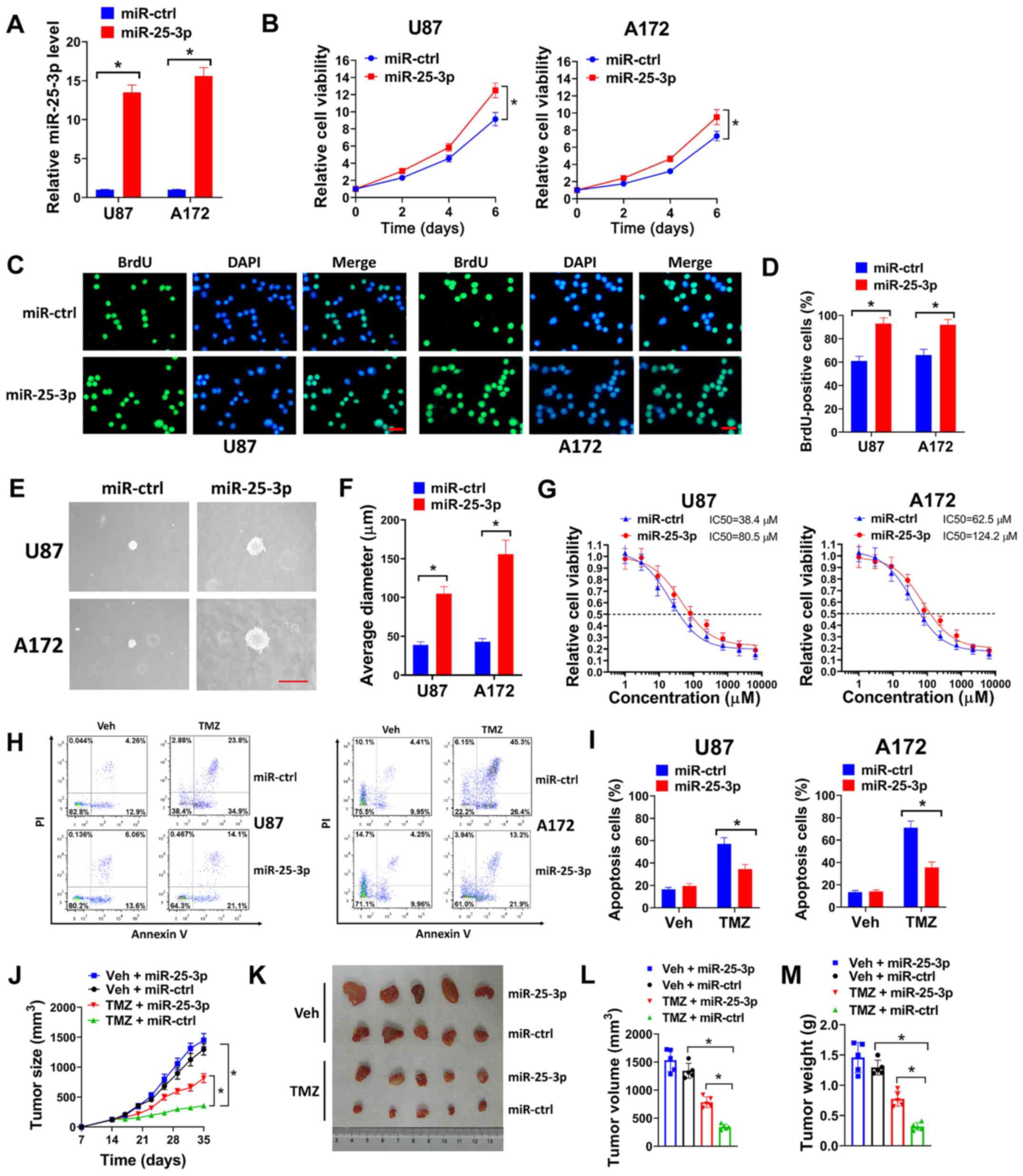 | Figure 4miR-25-3p overexpression promotes the
proliferation and TMZ resistance of GBM cells. (A) U87 or A172
cells were transduced with miR-25-3p expression lentivirus or
miR-ctrl, and the relative miR-25-3p expression was then evaluated
by RT-qPCR. (B) U87 or A172 cells transduced with miR-25-3p or
miR-ctrl were seeded in 96-well plates (5,000 cells/well), and cell
viability was then evaluated on days 2, 4 and 6. (C and D) U87 or
A172 cells were transduced with miR-25-3p expression lentivirus or
miR-ctrl, then seeded on coverslips for BrdU incorporation assay.
(C) Represent images (scale bar, 50 µm) and (D) percentages
of BrdU-positive cells are shown. (E and F) U87 or A172 cells
transduced with miR-25-3p expression lentivirus or miR-ctrl were
seeded in 6-well plates (8,000 cells/well) for soft agar assay. (E)
Represent images of colonies (scale bar, 200 µm) and (F) the
average diameter of colonies are shown. (G) U87 or A172 cells
transduced with miR-25-3p expression lentivirus or miR-ctrl were
seeded in 96-well plates (5,000 cells/well), then exposed to 0, 1,
3, 9, 27, 81, 243, 729, 2,181 or 6,543 µM TMZ for 6 days and
cell viability assay was then performed. (H and I) U87 or A172
cells transduced with miR-25-3p expression lentivirus or miR-ctrl
were treated with 50 µM TMZ for 72 h, and (H) the cells were
then used for flow cytometry. (I) Percentages of apoptotic cells
are shown. (J-M) U87 cells (3×106) transduced with
miR-25-3p or miR-ctrl were subcutaneously injected into nude mice
for 2 weeks, and the mice were then treated with 40 mg/kg TMZ (TMZ
group) or an equal volume of PEG-400: PBS (1:1) (Veh group) daily
for 3 weeks. (J) Tumor growth, (K) representative images of tumors,
(L) tumor volume and (M) tumor weight are shown for each group. All
experiments were performed in triplicate. *P≤0.05. TMZ,
temozolomide; GBM, glioblastoma. |
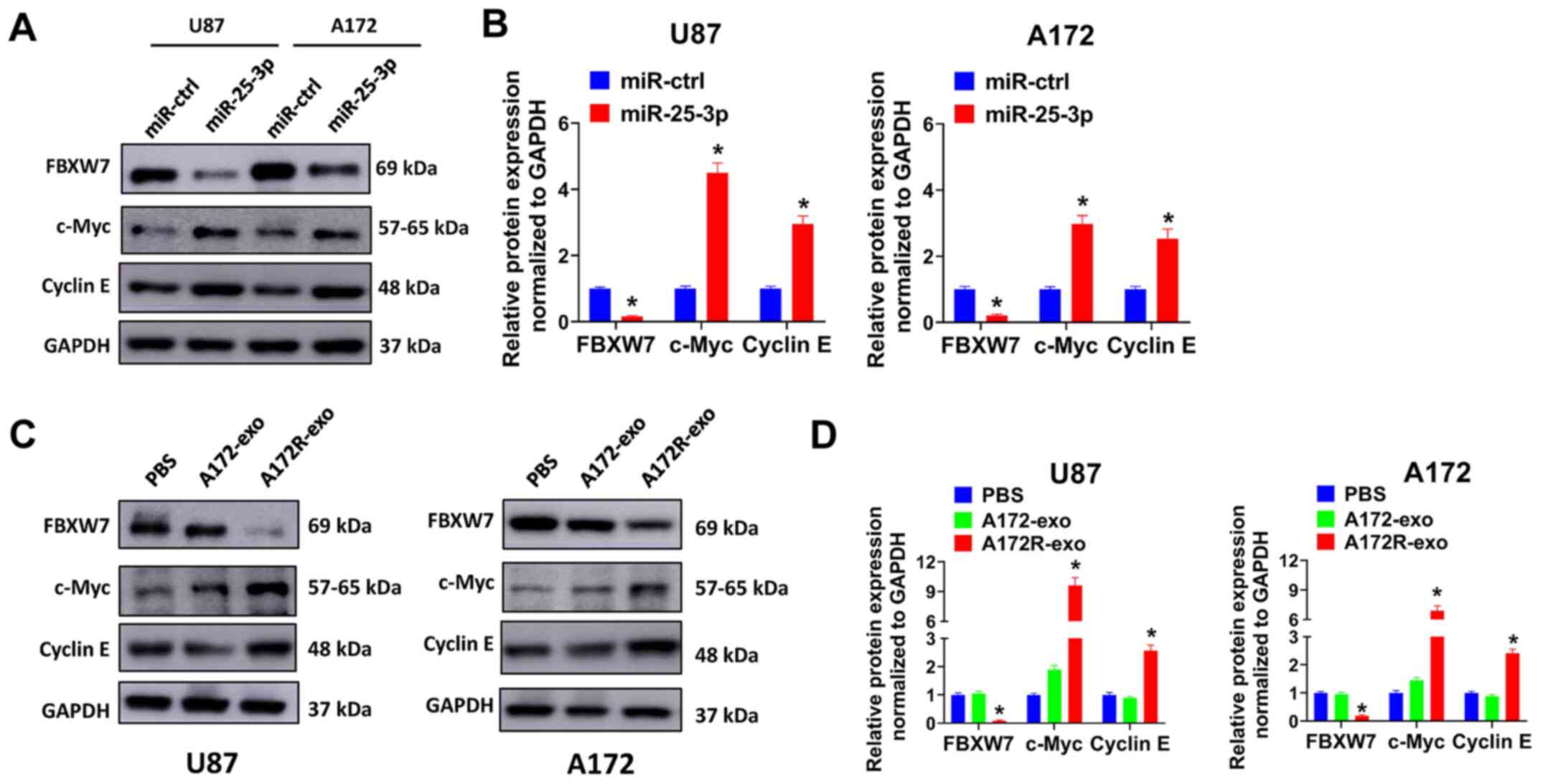
FBXW7 is a direct target of miR-25-3p in
GBM cells
To clarify the underlying mechanisms of miR-25-3p in
GBM, the target of miR-25-3p was predicted using TargetScan 7.2.
FBXW7 was predicted to interact with miR-25-3p in the present study
(Fig. 5A). FBXW7 is a
well-characterized tumor suppressor with a role in tumorigenesis
and the TMZ tolerance of GBM (28,29). In the present study, the
interaction between miR-25-3p and FBXW7 was validated by luciferase
reporter assay. A 3-bp mutation at the binding sites of miR-25-3p
and FBXW7 was generated at the 3′UTR of FBXW7 (mt FBXW7) (Fig. 5A). It was found that miR-25-3p
overexpression markedly inhibited the luciferase activity of the
3′UTR of wt FBXW7, while miR-25-3p knockdown evidently increased
the luciferase activity of the 3′UTR of wt FBXW7 in U87 and A172
cells (Fig. 5B). Furthermore, a
3-bp mutation at the binding sites of miR-25-3p and FBXW7 abolished
the effects induced by miR-25-3p overexpression or knockdown
(Fig. 5B). Furthermore, it was
found that miR-25-3p overexpression decreased FBXW7 expression in
the U87 and A172 cells, while miR-25-3p knockdown exerted opposite
effects (Fig. 5C and D).
Subsequently, RNA-IP assay was performed to identify mRNA levels in
the Ago/RNA-induced silencing complex (RISC) following miR-25-3p
overexpression (Fig. 5E-G).
Firstly, it was found that miR-25-3p overexpression had no effect
on the protein expression of Pan-Ago in the U87 and A172 cells
(Fig. 5E and F). It was then
proven that miR-25-3p overexpression evidently increased the levels
of miR-25-3p incorporated into the RISC (Fig. 5G, left panel). Moreover, the
levels of FBXW7 incorporated into the RISC were markedly reduced by
the enforced expression of miR-25-3p (Fig. 5G, right panel). As miR-25-3p
expression was upregulated in exosomes derived from A72R cells
(A172R-exo), the FBXW7 level we also evaluated in U87 and A172
cells treated with A172R-exo or A172-exo. It was proven that the
mRNA and protein levels of FBXW7 were markedly decreased by
treatment with A172R-exo (Fig.
5H-J). These data demonstrated that FBXW7 was targeted by
miR-25-3p in GBM cells.
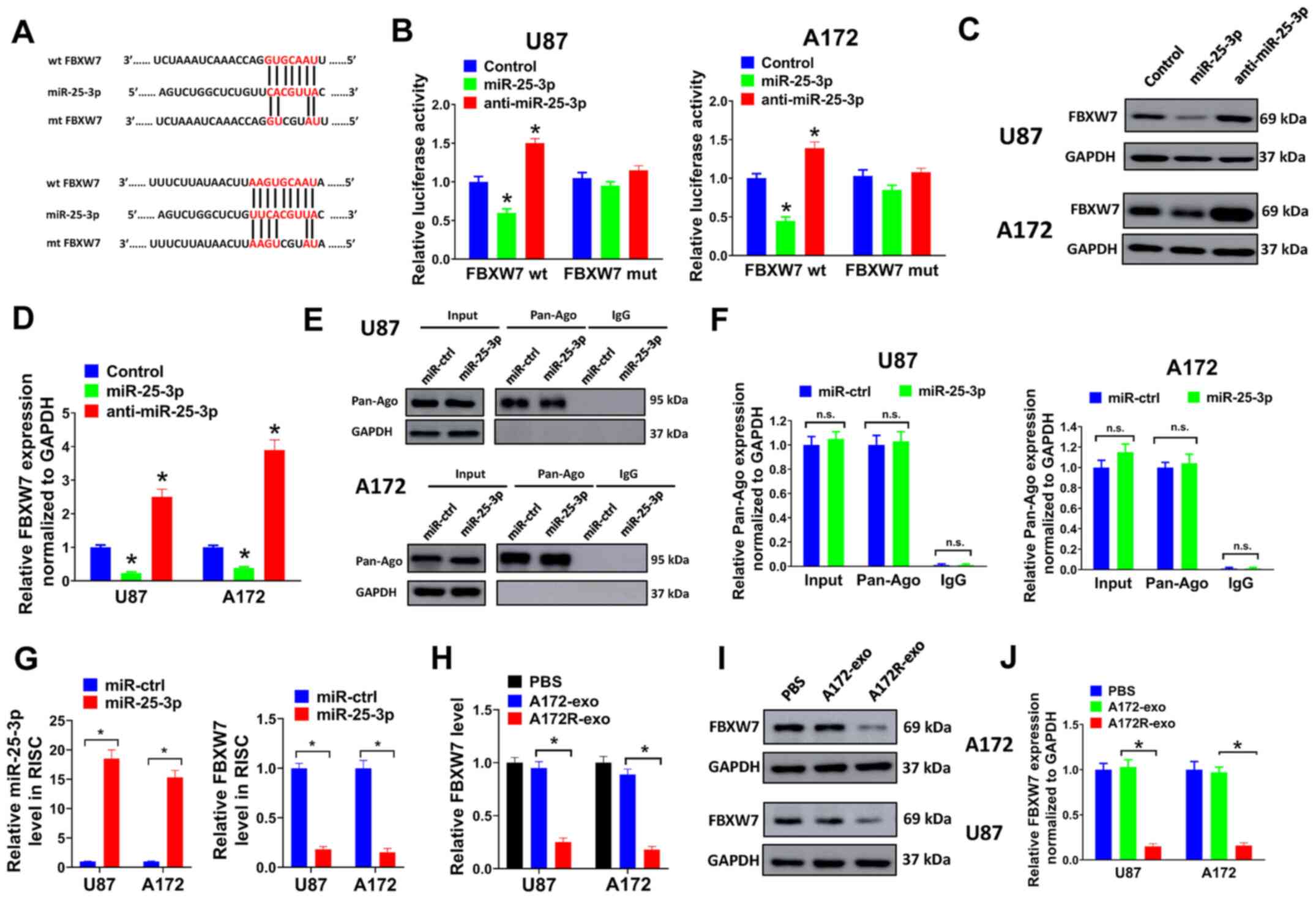 | Figure 5FBXW7 is a direct target of miR-25-3p
in GBM cells. (A) Binding sites for miR-25-3p and FBXW7 were
predicted using TargetScan 7.2. A 3-bp mutation at the binding
sites of miR-25-3p and FBXW7 was generated at the 3′UTR of FBXW7
using a QuickChange site-directed mutagenesis kit. (B) 293T cells
co-transfected with miR-25-3p, anti-miR-25-3p or control plasmids,
pMIR-REPORT vector containing the 3′UTR of FBXW7 or mutant, or a
Renilla luciferase plasmid for luciferase reporter assay. (C
and D) U87 or A172 cells were transduced with miR-25-3p,
anti-miR-25-3p or control plasmids, and the protein expression of
FBXW7 was then evaluated by western blot analysis. (D) Relative
protein expression levels normalized to GAPDH are shown. (E-G) U87
or A172 cells transduced with miR-25-3p expression lentivirus or
miR-ctrl were used for RNA-IP analysis. (E and F) Protein
expression of Pan-Ago was evaluated by western blot analysis. The
antibodies of anti-Ago1 and anti-IgG cannot pull down GAPDH; thus,
no bands are shown here. (F) Relative protein levels normalized to
GAPDH are shown. (G) Relative expression of miR-25-3p or FBXW7 was
evaluated by RT-qPCR. (H-J) U87 or A172 cells were treated with
A172R-exo (50 µg/ml), A172-exo (50 µg/ml) or an equal
volume of PBS, and the relative (H) mRNA or (I-J) protein
expression of FBXW7 was evaluated by RT-qPCR or western blot
analysis, respectively. (J) Relative protein expression levels
normalized to GAPDH are shown. All experiments were performed in
triplicate. *P≤0.05. FBXW7, F-box and WD repeat domain
containing 7; GBM, glioblastoma. |
FBXW7 knockdown promotes the
proliferation and TMZ resistance of GBM cells
To clarify the function of FBXW7 in GBM, FBXW7 was
knocked down in GBM cells and cell viability, BrdU incorporation
and colony formation assays were then performed. Two sgRNAs of
FBXW7 (sgFBXW7-1 and sgFBXW7-2) were constructed and introduced
into the U87 and A172 cells. It was found that FBXW7 was
successfully knock down by these two sgRNAs (Fig. 6A and B). In cell viability assay,
FBXW7 knockdown promoted the growth of U87 and A172 cells (Fig. 6C). In BrdU incorporation assay,
the knockdown of FBXW7 evidently increased the percentage of
BrdU-positive cells (Fig. 6D and
E). Moreover, it was found that FBXW7 knockdown increased the
TMZ IC50 in U87 and A172 cells (Fig.
6F). In colony formation assay, FBXW7 knockdown facilitated the
colony formation of U87 and A172 cells co-treated with TMZ
(Fig. 6G and H). Collectively,
our results indicated that FBXW7 knockout promoted proliferation
and TMZ tolerance of GBM cells.
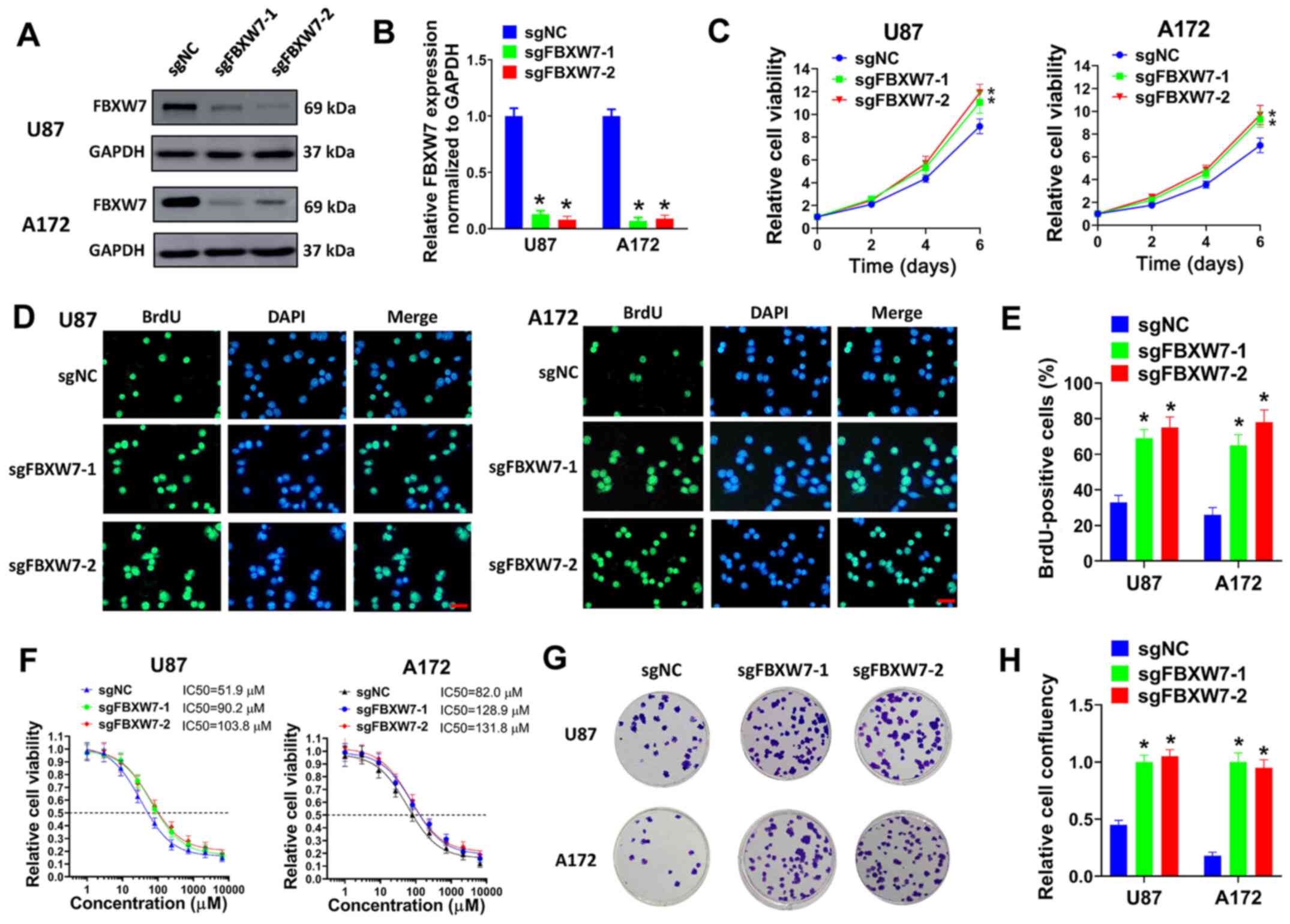 | Figure 6FBXW7 knockdown promotes the
proliferation and TMZ resistance of GBM cells. (A and B) U87 or
A172 cells were transduced with sgFBXW7-1, sgFBXW7-2 or sgNC, and
(A) the protein expression of FBXW7 was evaluated by western blot
analysis. (B) Relative protein levels normalized to GAPDH are
shown. (C) U87 or A172 cells transduced with sgFBXW7-1, sgFBXW7-2
or sgNC were seeded in 96-well plates (5,000 cells/well), and cell
viability was then evaluated on days 2, 4 and 6. (D and E) U87 or
A172 cells were transduced with sgFBXW7-1, sgFBXW7-2 or sgNC, and
then seeded on coverslips for BrdU incorporation assay. (D)
Represent images (scale bar, 50 µm) and (E) percentages of
BrdU-positive cells are shown. (F) U87 or A172 cells transduced
with sgFBXW7-1, sgFBXW7-2 or sgNC were seeded in 96-well plates
(5,000 cells/well), then exposed to 0, 1, 3, 9, 27, 81, 243, 729,
2,181 or 6,543 µM TMZ for 6 days and evaluated by cell
viability assay. (G and H) U87 or A172 cells transduced with
sgFBXW7-1, sgFBXW7-2 or sgNC were seeded in 6-well plates (3,000
cells/well) and treated with 50 µM TMZ for colony formation
assay. (G) Represent plates and (H) relative cell confluency are
shown. All experiments were performed in triplicate.
*P≤0.05. FBXW7, F-box and WD repeat domain containing 7;
GBM, glioblastoma. |
Exosomal transfer of miR-25-3p increases
c-Myc and cyclin E expression by regulating FBXW7
FBXW7 is a key regulator of the cell cycle and
cancer stemness (28).
Accumulating evidence has indicated that c-Myc and cyclin E are
downstream targets of FBXW7 (30-32). This was also evaluated in the
present study. It was proven that miR-25-3p overexpression
decreased FBXW7 expression, and simultaneously increased the
expression of c-Myc and cyclin E in the U87 and A172 cells
(Fig. 7A and B). These results
indicated that the enforced expression of miR-25-3p promoted c-Myc
and cyclin E expression by decreasing FBXW7 expression. Thus, it
was hypothesized exosomes derived from TMZ-resistant A172R cells
may also promote c-Myc and cyclin E expression. As was expected,
the U87 and A172 cells treated with A172R-exo exhibited a decreased
expression of FBXW7 and an increased expression of c-Myc and cyclin
E compared with the cells treated with A172-exo or PBS (Fig. 7C and D). These results proved that
the exosomal transfer of miR-25-3p increased c-Myc and cyclin E
expression by regulating FBXW7 in GBM cells, and this may be one of
the mechanisms involved in the promotion of the proliferation and
TMZ resistance of GBM cells by exosomal miR-25-3p.
Discussion
Exosomes play a key role in the tolerance of GBM to
TMZ. The exosomal transfer of proteins and RNAs may confer TMZ
resistance to recipient cells. For example, the exosomal transfer
of PTPRZ1-MET fusion protein has been shown to facilitate the
migration, invasion, neurosphere growth and TMZ resistance of GBM
cells (33). CircNFIX has also
been shown to be increased in serum samples of patients with
TMZ-resistant GBM. The exosomal transfer of CircNFIX enhances the
migration, invasion and inhibits apoptosis of GBM cells under TMZ
exposure (34). As previously
demonstrated, lncRNA HOTAIR is overexpressed in TMZ-resistant
glioma cells and promotes proliferation, metastasis and
epithelial-mesenchymal transition. The exosomal transfer of HOTAIR
induces TMZ resistance in target cells via the miR-519a/RRM1 axis
(35). miR-221 expression has
been shown to be increased in tissue samples and exosomes of glioma
patients. Exosomal miR-221 promotes tumorigenesis and TMZ
resistance of glioma cells by targeting DNM3 (36). In the present study, a
TMZ-resistant GBM cell line (A172R) was established. It was found
that exosomes of A172R cells (A172R-exo) promoted the growth and
TMZ resistance of sensitive GBM cells. Moreover, miRNA sequencing
data for A172R-exo identified miR-25-3p as a miRNA associated with
TMZ resistance. The depletion of miR-25-3p in A172R-exo partially
abrogated the effects caused by A172R-exo transfer. These data
proved that the exosomal transfer of miR-25-3p may confer TMZ
resistance to recipient cells.
The present study has some potential limitations
which should be mentioned. Only one TMZ-resistant cell line was
established and used to verify the effects of exosomes and the
exosomal miR-25-3p of this TMZ-resistant cell line. The data would
be more valid if the results could be duplicate in other
TMZ-resistant cell lines. Moreover, the results were mainly
conducted on GBM cell lines in vitro and nude mice in
vivo; thus, there are a lack of data on patient samples, such
as patient-derived tumor xenografts or tumor cells. Finally, the
number of patients in the present study were small, and the
association of exosomal miR-25-3p with TMZ resistance and the
prognosis of patients with GBM was not fully elucidated.
miR-25-3p is overexpressed and functions as an
oncogene in various cancers, including GBM. For example, as
previously demonstrated, a high level of miR-25-3p facilitates the
malignant progression of pancreatic cancer cells by decreasing
PHLPP2 and subsequently activating AKT/p70S6K signaling (16). Moreover, miR-25-3p has been shown
to facilitate the proliferation and tumor xenograft growth of
breast cancer cells by suppressing BTG2 and indirectly activating
AKT and ERK/MAPK signaling (17).
miR-25 has been shown to be evidently overexpressed in astrocytoma
and GBM cells lines. The ectopic expression of miR-25 has also been
shown to increase the proliferation and invasion of GBM cells by
targeting NEFL (27). In another
study, miR-25-3p was reported to regulate the growth and migration
of glioma cells by targeting FBXW7 and DKK3 (26). In the present study, it was found
that the enforced expression of miR-25-3p promoted the
proliferation and TMZ resistance of GBM cells, indicating that
miR-25-3p functioned as an oncogene in GBM, in accordance with the
findings of previous studies (26,27). It was also revealed that miR-25-3p
was overexpressed in the serum of TMZ-treated patients with GBM and
exosomes of TMZ-resistant GBM cells. Indeed, circulating miR-25-3p
may be a predictor of prognosis in cancers, such as osteosarcoma
(37) and gastric cancer
(38). Ebrahimkhani et al
(39) reported that miR-25-3p
expression was increased in the serum of patients with GBM,
suggesting that miR-25-3p was a potential diagnostic marker for
GBM. The present study found that the exosomal delivery of
miR-25-3p spread TMZ tolerance to other sensitive GBM cells.
Notably, the exosomal delivery of miR-25-3p has exhibited
cancer-promoting activity in other types of cancers as well. For
example, in a previous study, the exosomal delivery of miR-25-3p
induced a pre-metastatic nice by inhibiting KLF2 and KLF4, thus
facilitating vascular permeability and the angiogenesis of
colorectal cancer (40). In
liposarcoma, miR-25-3p has been shown to promote the IL-6
production of tumor-associated macrophages, thus stimulating the
proliferation and metastasis of liposarcoma cells (41). These studies and the findings of
the present study indicate an important function of exosomal
miR-25-3p in the tumorigenesis and drug resistance of cancer.
FBXW7 is a part of the Skp1-Cullin1-F-box (SCF)
complex. The SCF complex plays a role in the degradation of
oncoproteins, such as c-Myc, Notch, cyclin E and c-Jun (30). FBXW7 functions as a tumor
suppressor in GBM. For example, FBXW7 has been proven to be
negatively associated with glioma histology and positively with
patient survival, while FBXW7 knockdown regulates proliferation,
metastasis and the TMZ resistance of GBM cells (29). FBXW7 has been shown to be
downregulated in GBM tissues, and the overexpression of FBXW7
suppresses the proliferation of GBM cells in vitro (42). In a previous study, in a
genetic-engineered mouse model, p53 mutation was proven to promote
gliomagenesis by inhibiting FBXW7 expression, thus increasing c-Myc
expression and protecting against c-Myc induced apoptosis (43). The present study revealed that
FBXW7 was targeted by miR-25-3p. FBXW7 knockdown facilitated the
proliferation and TMZ tolerance of GBM cells. Moreover, c-Myc and
cyclin E expression levels were increased by FBXW7 knockdown. The
results proved that the effects of the exosomal transfer of
miR-25-3p may be due to the inhibition of the tumor suppressor,
FBXW7, thus increasing the levels of oncoproteins, such as c-Myc
and cyclin E. Indeed, there are several studies demonstrating that
FBXW7 is a direct target of miR-25-3p. For example, Wang et
al (44) reported that miR-25
regulates cardiomyocyte growth by inhibiting FBXW7. In prostatic
small cell neuroendocrine carcinoma, p53 mutation has been proven
to inhibit FBXW7 expression by upregulating miR-25 (45). mR-25 has also been reported to
exert cancer-promoting effects in esophageal squamous cell
carcinoma and glioma by suppressing FBXW7 (26,46). Although it is well-established
that FBXW7 is targeted by miR-25-3p, the novelty of the present
study is that it mainly focuses on the function of exosomes of
TMZ-resistant GBM cells. The present study proved that the exosomal
transfer of miR-25-3p may spread TMZ resistance to sensitive GBM
cells, thus revealing an important role of tumor-derived exosomes
in drug resistance. Moreover, the present study also demonstrated,
for the first time, to the best of our knowledge, that exosomal
miR-25-3p was involved in acquired TMZ resistance in GBM.
In conclusion, the present study found that exosomes
of TMZ-resistant GBM cells promoted the proliferation and spread
TMZ resistance to other GBM cells. miRNA sequencing data identified
miR-25-3p as a significantly upregulated exosomal miRNA. The
depletion of exosomal miR-25-3p partially abolished the effects
induced by exosomes derived from TMZ-resistant GBM cells. Moreover,
miR-25-3p overexpression promoted the proliferation and TMZ
resistance of GBM cells in vitro and in vivo. In
addition, FBXW7 was proven to be a direct target of miR-25-3p. The
knockdown of FBXW7 increased the proliferation and TMZ resistance
of GBM cells. Furthermore, the exosomal transfer of miR-25-3p
decreased c-Myc and Cyclin E expression by inhibiting FBXW7. The
results of the present study provide new insight into exosomal
miRNAs in the acquired TMZ resistance of GBM cells. Exosomal
miR-25-3p may thus be a potential prognostic marker for patients
with GBM.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors guarantee the integrity of the entire
study. The experiments were conducted by JW, TL and BW. Clinical
analyses were conducted by BW. Data were analyzed by TL. The
manuscript was prepared and reviewed by JW. All authors have read
and approved the manuscript. JW and TL confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
All patients and healthy donors signed the written
informed consents. The collection and use of serum samples were
approved by the Ethics Committee of the Henan Provincial People's
Hospital [SYXK (Yu) 2018-0004]. The animal experiments were
reviewed and approved by the Animal Care and Experimental Committee
of Henan Provincial People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Gittleman H, Fulop J, Liu M,
Blanda R, Kromer C, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2008-2012. Neuro Oncol.
17(Suppl 4(Suppl 4)): iv1–iv62. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: Effects of radiotherapy with concomitant and
adjuvant temozolomide versus radiotherapy alone on survival in
glioblastoma in a randomised phase III study: 5-year analysis of
the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davis ME: Glioblastoma: Overview of
disease and treatment. Clin J Oncol Nurs. 20(Suppl 5): S2–S8. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar :
|
|
7
|
Valadi H, Ekstrom K, Bossios A, Sjostrand
M, Lee JJ and Lotvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mashouri L, Yousefi H, Aref AR, Ahadi AM,
Molaei F and Alahari SK: Exosomes: Composition, biogenesis, and
mechanisms in cancer metastasis and drug resistance. Mol Cancer.
18:752019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Skog J, Wurdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pace KR, Dutt R and Galileo DS: Exosomal
L1CAM stimulates glioblastoma cell motility, proliferation, and
invasiveness. Int J Mol Sci. 20:39822019. View Article : Google Scholar :
|
|
11
|
Zhang Z, Yin J, Lu C, Wei Y, Zeng A and
You Y: Exosomal transfer of long non-coding RNA SBF2-AS1 enhances
chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer
Res. 38:1662019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer. 17:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang B, Mao JH, Wang BY, Wang LX, Wen HY,
Xu LJ, Fu JX and Yang H: Exosomal miR-1910-3p promotes
proliferation, metastasis, and autophagy of breast cancer cells by
targeting MTMR3 and activating the NF-kB signaling pathway. Cancer
Lett. 489:87–99. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ghaemmaghami AB, Mahjoubin-Tehran M,
Movahedpour A, Morshedi K, Sheida A, Taghavi SP, Mirzaei H and
Hamblin MR: Role of exosomes in malignant glioma: MicroRNAs and
proteins in pathogenesis and diagnosis. Cell Commun Signal.
18:1202020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yin J, Zeng A, Zhang Z, Shi Z, Yan W and
You Y: Exosomal transfer of miR-1238 contributes to
temozolomide-resistance in glioblastoma. EBioMedicine. 42:238–251.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C,
Li S, Tan L, Mai D, Li G, et al: Excessive miR-25-3p maturation via
N6− methyladenosine stimulated by cigarette smoke
promotes pancreatic cancer progression. Nat Commun. 10:18582019.
View Article : Google Scholar
|
|
17
|
Chen H, Pan H, Qian Y, Zhou W and Liu X:
MiR-25-3p promotes the proliferation of triple negative breast
cancer by targeting BTG2. Mol Cancer. 17:42018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang L, Tong Z, Sun Z, Zhu G, Shen E and
Huang Y: MiR-25-3p targets PTEN to regulate the migration,
invasion, and apoptosis of esophageal cancer cells via the PI3K/AKT
pathway. Biosci Rep. 40:BSR202019012020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ning L, Zhang M, Zhu Q, Hao F, Shen W and
Chen D: MiR-25-3p inhibition impairs tumorigenesis and invasion in
gastric cancer cells in vitro and in vivo. Bioengineered. 11:81–90.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rao HC, Wu ZK, Wei SD, Jiang Y, Guo QX,
Wang JW, Chen CX and Yang HY: MiR-25-3p serves as an oncogenic
MicroRNA by downregulating the expression of merlin in
osteosarcoma. Cancer Manag Res. 12:8989–9001. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ebert MS and Sharp PA: MicroRNA sponges:
Progress and possibilities. RNA. 16:2043–2050. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R,
Cao Z, Singh B, Franklin JL, Wang J, Hu H, et al: lncRNA
MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance
via Wnt/β-catenin signaling. Nat Med. 23:1331–1341. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee SY: Temozolomide resistance in
glioblastoma multiforme. Genes Dis. 3:198–210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim DK, Nishida H, An SY, Shetty AK,
Bartosh TJ and Prockop DJ: Chromatographically isolated CD63+CD81+
extracellular vesicles from mesenchymal stromal cells rescue
cognitive impairments after TBI. Proc Natl Acad Sci USA.
113:170–175. 2016. View Article : Google Scholar :
|
|
26
|
Peng G, Yang C, Liu Y and Shen C:
MiR-25-3p promotes glioma cell proliferation and migration by
targeting FBXW7 and DKK3. Exp Ther Med. 18:769–778. 2019.PubMed/NCBI
|
|
27
|
Peng G, Yuan X, Yuan J, Liu Q, Dai M, Shen
C, Ma J, Liao Y and Jiang W: MiR-25 promotes glioblastoma cell
proliferation and invasion by directly targeting NEFL. Mol Cell
Biochem. 409:103–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Davis RJ, Welcker M and Clurman BE: Tumor
suppression by the Fbw7 ubiquitin ligase: Mechanisms and
opportunities. Cancer Cell. 26:455–464. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin J, Ji A, Qiu G, Feng H, Li J, Li S,
Zou Y, Cui Y, Song C, He H and Lu Y: FBW7 is associated with
prognosis, inhibits malignancies and enhances temozolomide
sensitivity in glioblastoma cells. Cancer Sci. 109:1001–1011. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sailo BL, Banik K, Girisa S, Bordoloi D,
Fan L, Halim CE, Wang H, Kumar AP, Zheng D, Mao X, et al: FBXW7 in
cancer: What has been unraveled thus far? Cancers (Basel).
11:2462019. View Article : Google Scholar
|
|
31
|
Ibusuki M, Yamamoto Y, Shinriki S, Ando Y
and Iwase H: Reduced expression of ubiquitin ligase FBXW7 mRNA is
associated with poor prognosis in breast cancer patients. Cancer
Sci. 102:439–445. 2011. View Article : Google Scholar
|
|
32
|
Iwatsuki M, Mimori K, Ishii H, Yokobori T,
Takatsuno Y, Sato T, Toh H, Onoyama I, Nakayama KI, Baba H and Mori
M: Loss of FBXW7, a cell cycle regulating gene, in colorectal
cancer: Clinical significance. Int J Cancer. 126:1828–1837. 2010.
View Article : Google Scholar
|
|
33
|
Zeng AL, Yan W, Liu YW, Wang Z, Hu Q, Nie
E, Zhou X, Li R, Wang XF, Jiang T and You YP: Tumour exosomes from
cells harbouring PTPRZ1-MET fusion contribute to a malignant
phenotype and temozolomide chemoresistance in glioblastoma.
Oncogene. 36:5369–5381. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z,
Zhang G, Gu J and Kang D: Exosome-mediated transfer of circRNA
CircNFIX enhances temozolomide resistance in glioma. Cancer Lett.
479:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan Z, Yang Z, Li W, Wu A, Su Z and Jiang
B: Exosome-mediated transfer of long noncoding RNA HOTAIR regulates
temozolomide resistance by miR-519a-3p/RRM1 axis in glioblastoma.
Cancer Biother Radiopharm. 24–Jul;2020.Epub ahead of print.
View Article : Google Scholar
|
|
36
|
Yang JK, Yang JP, Tong J, Jing SY, Fan B,
Wang F, Sun GZ and Jiao BH: Exosomal miR-221 targets DNM3 to induce
tumor progression and temozolomide resistance in glioma. J
Neurooncol. 131:255–265. 2017. View Article : Google Scholar
|
|
37
|
Fujiwara T, Uotani K, Yoshida A, Morita T,
Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, et al:
Clinical significance of circulating miR-25-3p as a novel
diagnostic and prognostic biomarker in osteosarcoma. Oncotarget.
8:33375–33392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
ZiaSarabi P, Sorayayi S, Hesari A and
Ghasemi F: Circulating microRNA-133, microRNA-17 and microRNA-25 in
serum and its potential diagnostic value in gastric cancer. J Cell
Biochem. 120:12376–12381. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ebrahimkhani S, Vafaee F, Hallal S, Wei H,
Lee MYT, Young PE, Satgunaseelan L, Beadnall H, Barnett MH,
Shivalingam B, et al: Deep sequencing of circulating exosomal
microRNA allows non-invasive glioblastoma diagnosis. NPJ Precis
Oncol. 2:282018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J,
Zhou K, Liu X, Ren X, Wang F, et al: Cancer-derived exosomal
miR-25-3p promotes pre-metastatic niche formation by inducing
vascular permeability and angiogenesis. Nat Commun. 9:53952018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Casadei L, Calore F, Creighton CJ,
Guescini M, Batte K, Iwenofu OH, Zewdu A, Braggio DA, Bill KL,
Fadda P, et al: Exosome-derived miR-25-3p and miR-92a-3p stimulate
liposarcoma progression. Cancer Res. 77:3846–3856. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hagedorn M, Delugin M, Abraldes I, Allain
N, Belaud-Rotureau MA, Turmo M, Prigent C, Loiseau H, Bikfalvi A
and Javerzat S: FBXW7/hCDC4 controls glioma cell proliferation in
vitro and is a prognostic marker for survival in glioblastoma
patients. Cell Div. 2:92007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim HS, Woolard K, Lai C, Bauer PO, Maric
D, Song H, Li A, Kotliarova S, Zhang W and Fine HA: Gliomagenesis
arising from Pten- and Ink4a/Arf-deficient neural progenitor cells
is mediated by the p53-Fbxw7/Cdc4 pathway, which controls c-Myc.
Cancer Res. 72:6065–6075. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang B, Xu M, Li M, Wu F, Hu S, Chen X,
Zhao L, Huang Z, Lan F, Liu D and Wang Y: MiR-25 promotes
cardiomyocyte proliferation by targeting FBXW7. Mol Ther Nucleic
Acids. 19:1299–1308. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Z, Sun Y, Chen X, Squires J,
Nowroozizadeh B, Liang C and Huang J: p53 mutation directs AURKA
overexpression via miR-25 and FBXW7 in prostatic small cell
neuroendocrine carcinoma. Mol Cancer Res. 13:584–591. 2015.
View Article : Google Scholar :
|
|
46
|
Hua Y, Zhao K, Tao G, Dai C and Su Y:
MiR-25 promotes metastasis via targeting FBXW7 in esophageal
squamous cell carcinoma. Oncol Rep. 38:3030–3038. 2017. View Article : Google Scholar : PubMed/NCBI
|