Osteosarcoma (OS) is considered to be the most
commonly occurring type of primary bone tumor, with an estimated
worldwide incidence of 3-4 new cases per million, which accounts
for approximately 60% of the total number of cases of bone
malignancy (1,2). OS is derived from the transformation
of primitive mesenchymal cells, and typically occurs in the
metaphyseal region of long bones, with a peak incidence among young
individuals (3,4). OS exhibits a range of aggressive
behaviors, including early metastasis potential, rapid progression,
poor clinical prognosis and insensitivity to chemoradiotherapy,
which collectively lead to a poor overall survival rate (5,6).
Prior to the 1970s, surgical resection was the preferred treatment
for OS, although, as the 5-year survival rate was <20%, it was
insufficient as a means of therapy for numerous patients (7,8).
The current therapeutic strategy for OS includes
neoadjuvant chemotherapy after surgical removal of the tumor and
adjuvant chemotherapy with or without lesion metastasis, which has
led to a marked increase in the survival rate to approximately 65%
over the course of the last 30 years (9). However, despite notable improvements
achieved in terms of surgical techniques and neoadjuvant
chemotherapy, the overall survival time of patients with distant
metastasis or multi-drug resistance cannot be effectively prolonged
(10-12). Therefore, it is crucial to
elucidate the underlying molecular mechanisms that are involved in
the tumorigenesis and progression of OS, and to identify novel
biomarkers for developing alternative therapies or improving the
efficiency of existing treatments.
After having mapped out the transcriptional
'landscape' of the mammalian genome, findings showed that
protein-coding mRNAs only account for 1.4% of the genome (13,14). Furthermore, by comparing the
number of protein-coding genes with the genome size among different
species, it was shown that the more complex eukaryotes carry a
larger proportion of non-coding RNA (ncRNA) (15). These phenomena led to the
suggestion that the ncRNAs in the human genome may contribute
towards complex physiological and pathological processes. Based on
a cutoff at 200 bases of length, ncRNAs are standardly categorized
as short ncRNAs (sncRNAs) and long ncRNAs (lncRNAs) (16). MicroRNAs (miRNAs/miRs), as a
typical class of sncRNAs, have been extensively studied. It has
been confirmed that miRNAs exercise a regulatory role on the
expression of protein-coding mRNAs, which leads to the initiation
or progression of numerous diseases, including cancer (17,18). In addition, there are other types
of ncRNAs, including small interfering RNAs (siRNAs), small nuclear
RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). The lncRNAs, as
observed from a wide spectrum of profiling in mammals, are a class
of transcripts >200 nucleotides (nt) in length (13,19,20). Increasing evidence has
demonstrated that lncRNAs fulfill functional roles in the
occurrence and development of tumors (21,22). Along with the development of
high-throughput sequencing technology and novel computational
approaches, a set of circular RNAs (circRNAs) have recently been
identified. It was revealed that circRNAs are involved in diverse
pathological processes, including oncogenesis and tumor progression
(23).
With the aim of encouraging further studies on these
RNA species, the present review article provides a concise summary
of ncRNAs and their biogenesis, their underlying molecular
mechanisms and their potential clinical applications in OS. It is
the authors' hope that associated research in the future may lead
to a more comprehensive understanding of OS and present a
reasonable perspective on the potential diagnostic and therapeutic
application of ncRNAs in patients with OS.
The biogenesis of miRNAs is a complex process,
including nuclear synthesis and cytoplasmic synthesis with the
involvement of a specific set of enzymes. First, in the nucleus, a
primary miRNA (pri-miRNA) with special hairpin structures (AAAAA
and 7MGpppG) is synthesized according to the transcriptional gene
that encodes the miRNA by RNA polymerase II. Subsequently, the
hairpin domain of the pri-miRNA is cleaved by an RNA-specific
nuclease termed Drosha (ribonuclease III) to produce precursor
miRNAs (pre-miRNAs) that possess a stem-ring structure and are
70-80 nt in length (28). Then,
with the help of cytoplasmic transporter exportin-5, the pre-miRNAs
are translocated from the nucleus to the cytoplasm. Within the
cytoplasm, these pre-miRNAs are further cleaved into a miRNA duplex
of ~19-23 nt by ribonuclease III (Dicerase) (29). The miRNA duplex consists of two
strands: A mature miRNA strand and a passenger miRNA strand. After
strand unwinding, the mature miRNA strand is transformed into a
mature miRNA via its interaction with Argonaute protein, whereas
the passenger miRNA is usually degraded (30).
Aberrant expression of miRNAs has been reported as a
common phenomenon occurring in a diversity of cancer types,
including breast, lung, hepatocellular, colon and cervical cancer
(31,32). miRNAs exert their regulatory role
through interacting with their mRNA target genes. Normally, there
are two mechanisms by which mature miRNAs are able to form
RNA-induced silencing complex (RISC). In the first scenario, in
cases where the miRNA is fully complementary to the target gene,
the miRNA degrades the target gene. In the second scenario, where
the miRNA is not fully complementary to target gene, miRNA combines
with the 3′-untranslated region to inhibit translation of the
target gene (33). In addition,
it has been confirmed that one single miRNA may affect multiple
mRNAs, or conversely, multiple miRNAs may affect one single mRNA
(27). Through the mechanism
described above, miRNAs are heavily involved in multiple instances
of cancer occurrence and development, including proliferation,
apoptosis and metastasis (34,35).
Similarly, certain miRNAs have been demonstrated to
regulate malignancy by serving either as an oncogene or as an
onco-suppressor in OS. miR-210-5p was recently shown to be
upregulated in human OS tissues and cell lines, closely correlating
with the advanced tumor-node-metastasis (TNM) stage, tumor size and
pulmonary metastasis. Overexpressed miR-210-5p led to an increase
in the rates of tumor invasion, migration and autophagy by
suppressing the downstream target, phosphoinositide-3-kinase
regulatory subunit 5 (PIK3R5). Of note, miR-210-5p-mediated
autophagy facilitates miR-210-5p-induced tumor invasion and
migration promotion via inhibiting the AKT/mTOR pathway (36). Another upregulated miRNA,
miR-624-5p, has been identified in clinical OS specimens and cell
lines. Further functional analysis suggested that miR-624-5p may
promote cell proliferation, migration and invasion both in
vitro and in vivo. Identified as the target gene of
miR-624-5p, protein tyrosine phosphatase receptor type B (PTPRB)
was found to be negatively correlated with miR-624-5p. Furthermore,
PTPRB restored the effects of miR-624-5p on OS migration and
invasion (37). miR-627-3p was
shown to be downregulated in OS tissues using biochip analysis, and
was also shown to be expressed at a lower level in OS cell lines
compared with human osteoblastic cells. Moreover, miR-627-3p
significantly suppressed the expression and activity of
pleiotrophin (PTN), and PTN affected the proliferation and
migration of OS cells via regulation of a range of different
proteins, including cyclin D1 and matrix metalloproteinase-2
(38). Also through microarray
analysis, miR-15b was found to be markedly downregulated in the
doxorubicin-resistance cell lines, KHOS and U-2OS (39). Furthermore, patients with OS who
had high expression levels of miR-15b received a significantly
improved clinical prognosis compared with those with low expression
levels. Wee1, the direct target of miR-15b, was shown to mediate
both the cytotoxic effect of doxorubicin and the
multidrug-resistance capabilities in OS (39).
Although the participation of miRNAs in OS is a
complex and multifactorial process, emerging evidence has strongly
supported the involvement of numerous miRNAs in the processes of OS
initiation and progression, including cell proliferation,
apoptosis, immigration, invasion and drug resistance. These miRNAs
and their roles are shown in Table
I (36-51).
MiRNAs have consistently attracted a great deal of
attention from researchers as putative diagnostic or prognostic
biomarkers for OS due to their stability in the plasma and serum
(52). miR-21 overexpression has
been shown to be strongly associated with advanced Enneking stage,
chemotherapeutic resistance and an unfavorable prognosis, and
therefore miR-21 may be used as an individual marker for OS staging
and prognosis (53). Low levels
of miR-101 have been observed in serum samples from patients with
OS; however, the miR-101 expression levels reverted to
significantly higher levels following treatment (54). miR-195-5p and miR-199a-3p have
been shown to have remarkable potential in terms of distinguishing
between metastatic and non-metastatic statuses in patients with OS,
whereas miR-320a and miR-199a-3p were associated with the
histological subtype (55).
At present, the standard clinical treatments mainly
comprise surgical resection and neoadjuvant therapy. The
involvement of miRNAs in the initiation and progression of OS
renders miRNAs suitable as possible therapeutic targets. The
corresponding approach would involve the use of miRNA mimics to
substitute for the loss of expression of a tumor-suppressor miRNA
or to block the expression of an oncomiR using oligonucleotides or
anti-viral constructs (56). A
mimic of miR-34 as a tumor suppressor for cancer treatment was
entered into Phase I clinical trials (57). Furthermore, it was identified that
a miR-34 mimic significantly suppressed lung metastasis in OS mouse
models, strongly suggesting that miR-34 may serve as a potential
therapeutic target (58).
lncRNAs form a large subgroup of ncRNAs with
transcripts >200 bases in length that lack the ability to encode
proteins. For the most part, they are transcribed by RNA polymerase
II, the same as mRNAs, and they are often 5′-capped, polyadenylated
and spliced without a translated open reading frame (59,60). Based on their location with the
neighboring protein-coding genes, they can be divided into the
following five classes: Sense, antisense, bidirectional, intronic
and intergenic lncRNAs (61-63). lncRNAs are also characterized by
their low abundance and through the tissue- and developmental
stage-specific manner of their expression (64,65).
Similar to miRNAs, numerous lncRNAs have been
demonstrated to have key roles as contributors to tumor initiation
or progression. The manner in which a given lncRNA exerts its
regulatory effect depends on its subcellular localization. lncRNAs
in the cytoplasm that share miRNA response elements with mRNAs
contain similar sequences to these target-coding RNAs and inhibit
the interactions between miRNAs and mRNAs. These lncRNAs, which are
termed competing endogenous RNAs (ceRNAs), act as 'sponges' for
miRNAs and regulate the process of translation mediated by miRNAs
on their target mRNAs (66).
lncRNAs in the nucleus mainly act at the epigenetic and genetic
levels by binding to the transcription preinitiation complex at the
promoter (67).
The lncRNA DANCR has been shown to be elevated in OS
tissue specimens and cell lines and is closely correlated with poor
prognosis among clinical patients. DANCR serves as an oncogene,
regulating ROCK1-mediated proliferation and metastasis through
sequestering both miR-335-5p and miR-1972 as a ceRNA (68). The lncRNA HIF1A-AS2 has also been
shown to be upregulated in OS, and is associated with poor
survival. HIF1A-AS2 regulates the tumorigenesis of OS, as
demonstrated by its effects on cell proliferation, cell cycle
progression and invasion, through 'sponging' miR-129-5p (69). In OS, the lncRNA TTN-AS1 has been
shown to facilitate cell growth, apoptosis and drug resistance via
the miR-134-5p/MBTD1 axis (70).
By contrast, the lncRNA TTN-AS1 also acts as a ceRNA on miRNA-376a,
enhancing the malignancy of OS via upregulating dickkopf-1
(71). Other confirmed lncRNAs
are presented in Table II
(68-79).
At present, the main surveillance methods of OS are
limited to physical examination, blood biochemistry and
radiographic examination (80).
Due to the lack of an effective and noninvasive measure to monitor
patient status or predict overall survival, lncRNAs are considered
as potential candidates for prognosis prediction and treatment
guidance. The level of metastasis-associated lung adenocarcinoma
transcript 1 (MALAT1) has been shown to be upregulated in 162 OS
tissues, closely correlating with advanced clinical stage, distant
metastasis and shorter survival times. Therefore, MALAT1 is able to
serve as an independent prognostic factor for OS (81). The clinical potential of
taurine-upregulated gene 1 (TUG1) has also been demonstrated
through its marked elevation in patients with OS progression and
relapse. Furthermore, the serum levels of TUG1 were shown to be
decreased after surgical resection of OS tissues in postoperative
patients (82). The most common
obstacles in the treatment of OS are the poor therapeutic response
to traditional chemo- and radio-therapies and the emergence of
resistance during treatment (83). lncRNAs that mediate acquired
resistance are potential candidates as targets for OS
therapies.
CircRNAs, a novel class of non-protein-coding RNAs,
were first discovered in the 1970s, and were then considered as
'junk' molecules with little functional potential (84). They are generated from pre-mRNAs
through back-splicing, and are expressed in a tissue- and
developmental stage-specific manner (85,86). In addition, they are
evolutionarily conserved and highly abundant in the brain. Unlike
traditional linear RNAs, circRNAs are characterized by a continuous
covalently closed loop structure lacking either a 5′-cap or a
3′-polyadenylated tail, which gives them stronger resistance to
ribonucleases compared with their corresponding linear counterparts
(87,88).
Emerging evidence has shown that circRNAs fulfill
essential roles in both physiological and pathological processes,
including oncogenesis and tumor progression (23). circRNAs have multiple functions,
such as regulating gene expression at the transcriptional or
post-transcriptional level by interacting with miRNAs as 'sponges',
binding to RNA-binding protein and initiating protein translation
in a splicing-dependent, cap-independent manner (89,90). The circRNA circTADA2A has been
reported to be highly upregulated in OS, and acts as a sponge for
the miRNA miR-203a-3p, which regulates CREB3 expression to promote
the proliferation, migration and invasion of OS cells in
vitro (91). The circRNA
hsa_circ_0001564, detected through circRNA microarray analysis, has
been shown to be upregulated in OS tissues. A further study
revealed that hsa_circ_001564 aggravates OS proliferation and
apoptosis via sponging miR-29c-3p (92). circPVT1 facilitates the
doxorubicin and cisplatin resistance of OS via increasing the
expression of the classical drug resistance-associated gene,
ABCB1 (93). In patients
with OS, circNASP expression was found to be positively correlated
with tumor size and lung metastasis. Upregulated circNASP acts as a
sponge of miR-1253, targeting the transcription factor FOXF1 to
markedly promote the proliferation, cell cycle progression and
invasion of OS cells (94). Other
novel circRNAs are listed in Table
III (91-103).
CircRNAs can be secreted in body fluids where they
are circulated, and the structures of circRNAs are geared towards a
high level of resistance to cleavage by RNA exonucleases or
ribonuclease R. These features, along with high specificity and
sensitivity, demonstrate that circRNAs may serve as good candidates
for OS (104,105). For example, the high expression
level of serum circPVT1 enabled patients with OS to be
distinguished from healthy individuals, suggesting that circPVT1
may be more reliable as a diagnostic biomarker compared to the
traditional biomarker alkaline phosphatase (93). In another study, evaluated
expression levels of circUBAP2 were found via Kaplan-Meier survival
analysis to be correlated with reduced survival and poor prognosis,
and they were also significantly correlated with the tumor stages
(106).
CircRNAs, as ceRNAs, are natural miRNA inhibitors
that bind to their corresponding miRNA to regulate the malignant
behavior of OS. This property ensures that circRNAs have great
potential in terms of therapeutic strategies. Recently, a newly
designed artificial miRNA sponge has been developed. This
artificial circRNA can sponge multiple miR-21 molecules, and has
been reported to upregulate the expression of the tumor suppressor
gene DAXX to inhibit the proliferation of gastric cancer
cells (107).
The identification of ncRNAs and their role in
cancer initiation and progression has provided revolutionary
insights into how the research efforts for OS may be directed.
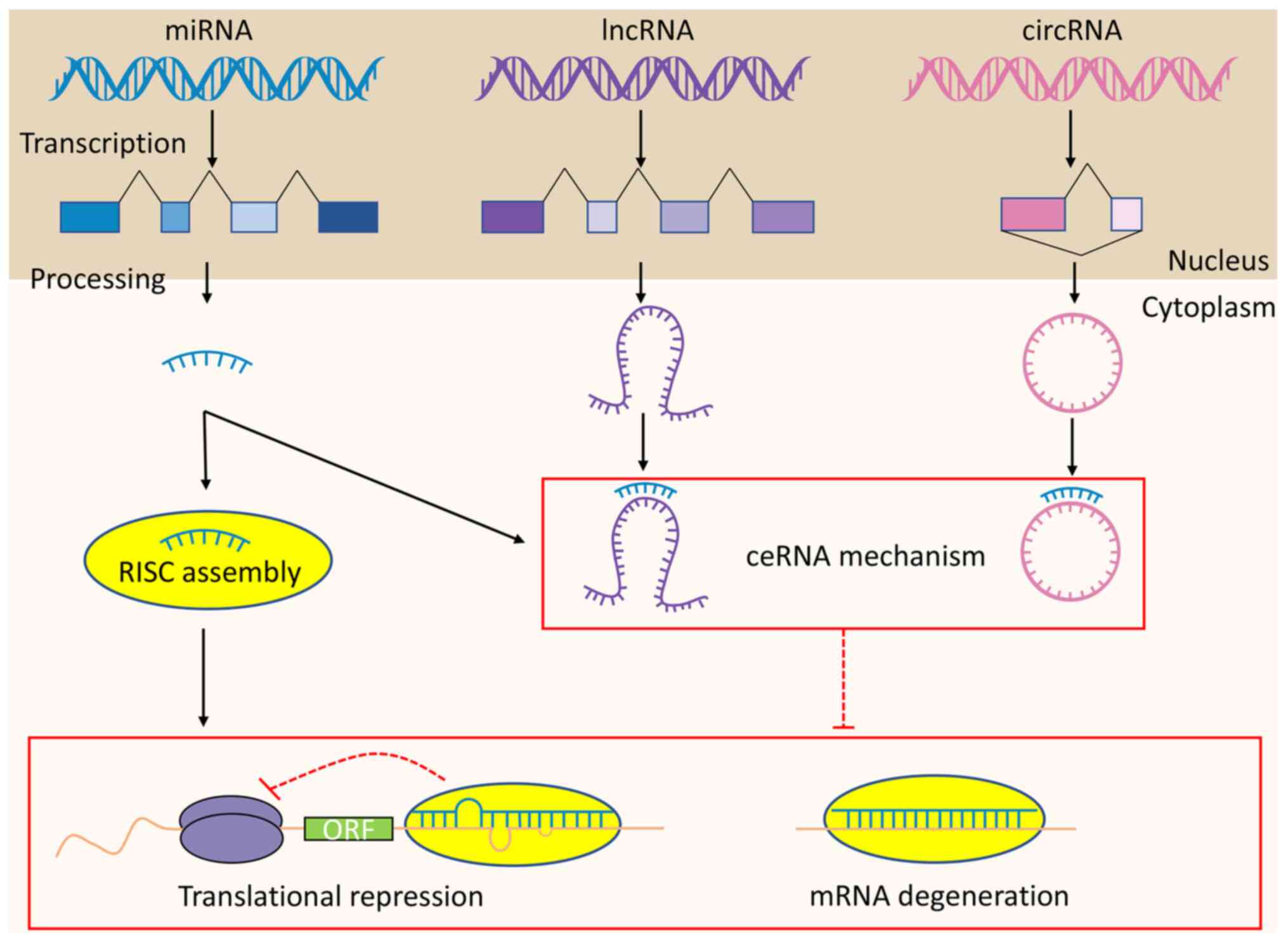
MiRNAs exert their regulatory functions via RISC, whereas lncRNAs
and circRNAs function according to mechanisms involving ceRNAs
(Fig. 1). However, since the
majority of these studies have focused on miRNAs, lncRNAs and
circRNAs, rather than other types of ncRNA (such as snoRNAs), the
data remain incomplete. Of the numerous human ncRNAs, only a few
have been thoroughly studied, and only a limited number of these
have an important biological impact. The RNA world is much more
complicated than was once considered to be the case, and the
clinical environment for OS progression should be greatly improved
if more research is devoted to the study of ncRNAs and their
involvement in this type of cancer.
Although the significant clinical potential of
ncRNAs as biomarkers in OS has been acknowledged, there exist
several limitations. In most of the studies that have been
performed to date, the cohort of patients with OS was relatively
small; thus, long-term, controlled and large-sample experiments are
required. In addition, the detected ncRNA biomarkers do not perform
entirely consistently even for a particular cancer type, which is
an important obstacle in terms of their usefulness as biomarkers.
Therefore, it is necessary to identify combinations of several
ncRNAs with a high degree of specificity and sensitivity, and to
develop a standardized approach in methodology to normalize the
expression of ncRNAs.
NcRNAs possess a unique advantage in terms of their
clinical application in OS treatment. A single ncRNA simultaneously
targets multiple downstream factors and is involved in multiple
signaling pathways, which brings important benefits for refractory
cancers with genomic heterogeneity. However, there are certain
disadvantages or challenges associated with ncRNA-targeted
treatment strategy. First, they may break the balance of gene
expression profiles in cells due to unrelated genes being targeted
by the same ncRNAs. It has been observed that miRNAs may exert
different effects, which means that miRNA antagonists or mimics
must be carefully selected according to different conditions.
Furthermore, the existence of off-target effects for ncRNA
antagonists cannot be ignored. Therefore, extensive toxicity
studies and preclinical safety requirements should be assessed
before an ncRNA-based therapeutic approach may be considered as
being appropriate for patients with OS (108). At present, researchers have also
made efforts to develop effective drug delivery systems for
chemotherapy. The reduction-responsive polypeptide micelles were
developed as multifunctional nanoparticle-based drug delivery
systems based on methoxy poly-block-poly copolymers. These micelles
can selectively accumulate in OS tumors, which induces antitumor
effects with less systematic toxicity (109). As for nucleic acid therapeutics,
approaches of delivery systems for targeting ncRNAs have been
developing at a rapid pace. There are some general problems of
nucleic acid delivery strategies including short half-life,
off-target effects and low transfection efficiency in RNA delivery,
which makes nucleic acid drugs remain at a low bioavailability
in vivo (110,111). To overcome the aforementioned
obstacles, a variety of ncRNA carriers or systems have been
investigated, including nanoparticles, ncRNA modification, and
oncolytic adenovirus strategy (112). Several of the delivery
strategies have been applied in the research of hepatic carcinoma
(113,114). Despite the lack of nucleic acid
drug delivery strategies and associated research in OS, nucleic
acid therapy may prove beneficial for OS treatment through further
study.
In the present review, we have made detailed
predictions on the future research directions:
One single ncRNA is able to affect multiple
downstream target molecules associated with cancer development, and
one single downstream target can be regulated by multiple upstream
molecules. For instance, the lncRNA DANCR promotes cell
proliferation and metastasis via sponging miR-335-5p and miR-1972
in OS (68). Therefore,
understanding the complicated connections between the ncRNA
regulatory networks, as well as determining some important core
ncRNAs in OS, requires further investigation.
Typically, researchers obtain tissue samples for
early diagnosis and prognosis via surgery resections, which is a
difficult and inconvenient procedure. Findings have shown that the
serum expression of the lncRNA UCA1 was significantly higher in
patients with OS compared with healthy controls. In addition, the
upregulation of UCA1 was correlated with clinical stage and
metastasis (115). Although the
data reported are only preliminary, it is possible to predict that
liquid biopsy, such as human peripheral blood, is a promising
non-invasive technique for OS diagnosis and prognosis in clinical
practice. Furthermore, clinical specimens can vary from puncture
fluid to sputum if patients have lung metastasis.
At present, a limited number of proteins or peptides
encoded by ncRNAs have been verified, and these have important
biological and/or pathological functions in the occurrence and
development of different tumors. A conserved 53-amino-acid peptide
encoded by the lncRNA HOXB-AS3 was shown to suppress the
proliferation, migration, invasion of colon cancer cells and tumor
growth both in vitro and in vivo (116). FBXW7-185aa is encoded by the
circRNA FBXW7, and inhibits glioma growth (117). Therefore, it is important to
distinguish whether ncRNAs exert functions by acting directly as
RNA molecules, or through encoding peptides or proteins.
Avoiding immune surveillance is an important
hallmark of tumor initiation and progression (118). Recent findings have shown that
ncRNAs are able to facilitate tumor immune escape to enhance
malignant behaviors. The PD-1/PD-L1 pathway provides a key
immunosuppressive mechanism for cancer cells, and miR-140 is
associated with anti-tumor immunity via its effects on the
PD-L1/PD-1 immune checkpoint signaling pathway in OS (119). At present, however, there are
only limited numbers of studies on ncRNAs and their association
with the tumor immune response in OS. Thus, further investigation
to develop promising immunotherapies for patients with OS is
necessary.
From a technical perspective, the research
technology of circular RNA (circRNA) is similar to the classic RNA
research technology, such as RT-PCR, qPCR, FISH (RNA positioning)
and NB (northern blot) (RNA expression). However, with the
emergence of new technologies, the research methods of circular RNA
(circRNA) have become more abundant, such as using CRISPR/Cas13
system to efficiently knock down the expression of circular RNA
without affecting the expression of its parental linear mRNA
(120).
In conclusion, the investigation of emerging
functional ncRNAs has led to a deeper understanding of the
pathologies that control initiation and progression in OS.
Moreover, the potential applications of ncRNAs in the diagnosis,
prognosis and therapy of OS have been revealed in recent years.
Further efforts, however, are required to elucidate the
ncRNA-associated regulatory mechanisms and to establish
ncRNA-targeted therapeutic options.
Not applicable.
GY, YW, HS and HL made substantial contributions to
the concept and design of the review, collected information and
wrote the manuscript. RW and WH collected references, and reviewed
and edited the manuscript. HS was the major contributor in drafting
and revising the manuscript. The authenticity of all the raw data
have been assessed by GY and YW to ensure its legitimacy. All
authors read and approved the final version of this manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar
|
|
2
|
Ma O, Cai WW, Zender L, Dayaram T, Shen J,
Herron AJ, Lowe SW, Man TK, Lau CC and Donehower LA: MMP13, Birc2
(cIAP1), and Birc3 (cIAP2), amplified on chromosome 9, collaborate
with p53 deficiency in mouse osteosarcoma progression. Cancer Res.
69:2559–2567. 2009. View Article : Google Scholar
|
|
3
|
Sampson VB, Kamara DF and Kolb EA:
Xenograft and genetically engineered mouse model systems of
osteosarcoma and Ewing's sarcoma: Tumor models for cancer drug
discovery. Expert Opin Drug Discov. 8:1181–1189. 2013. View Article : Google Scholar
|
|
4
|
Moore DD and Luu HH: Osteosarcoma. Cancer
Treat Res. 162:65–92. 2014. View Article : Google Scholar
|
|
5
|
Shi ZW, Wang JL, Zhao N, Guan Y and He W:
Single nucleotide polymorphism of hsa-miR-124a affects risk and
prognosis of osteosarcoma. Cancer Biomark. 17:249–257. 2016.
View Article : Google Scholar
|
|
6
|
He F, Zhang W, Shen Y, Yu P, Bao Q, Wen J,
Hu C and Qiu S: Effects of resection margins on local recurrence of
osteosarcoma in extremity and pelvis: Systematic review and
meta-analysis. Int J Surg. 36:283–292. 2016. View Article : Google Scholar
|
|
7
|
Marcove RC, Miké V, Hajek JV, Levin AG and
Hutter RV: Osteogenic sarcoma under the age of twenty-one. A review
of one hundred and forty-five operative cases. J Bone Joint Surg
Am. 52:411–423. 1970. View Article : Google Scholar
|
|
8
|
Dahlin DC and Coventry MB: Osteogenic
sarcoma. A study of six hundred cases. J Bone Joint Surg Am.
49:101–110. 1967. View Article : Google Scholar
|
|
9
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar
|
|
10
|
Wang B, Xu M, Zheng K and Yu X: Effect of
unplanned therapy on the prognosis of patients with extremity
osteosarcoma. Sci Rep. 6:387832016. View Article : Google Scholar
|
|
11
|
Liu K, Huang J, Ni J, Song D, Ding M, Wang
J, Huang X and Li W: MALAT1 promotes osteosarcoma development by
regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle.
16:578–587. 2017. View Article : Google Scholar
|
|
12
|
Chen L, Wang Q, Wang GD, Wang HS, Huang Y,
Liu XM and Cai XH: miR-16 inhibits cell proliferation by targeting
IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS
Lett. 587:1366–1372. 2013. View Article : Google Scholar
|
|
13
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
The transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005. View Article : Google Scholar
|
|
14
|
Baltimore D: Our genome unveiled. Nature.
409:814–816. 2001. View
Article : Google Scholar
|
|
15
|
Mattick JS: The genetic signatures of
noncoding RNAs. PLoS Genet. 5:e10004592009. View Article : Google Scholar
|
|
16
|
Kour S and Rath PC: Long noncoding RNAs in
aging and age-related diseases. Ageing Res Rev. 26:1–21. 2016.
View Article : Google Scholar
|
|
17
|
Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman
SB, Bi W, Xu M, Jiao S, Maloney WJ and Wang Y: miR-223-3p inhibits
human osteosarcoma metastasis and progression by directly targeting
CDH6. Mol Ther. 26:1299–1312. 2018. View Article : Google Scholar
|
|
18
|
Andersen GB, Knudsen A, Hager H, Hansen LL
and Tost J: miRNA profiling identifies deregulated miRNAs
associated with osteosarcoma development and time to metastasis in
two large cohorts. Mol Oncol. 12:114–131. 2018. View Article : Google Scholar
|
|
19
|
ENCODE Project Consortium; Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar
|
|
20
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar
|
|
21
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View
Article : Google Scholar
|
|
22
|
Lin C and Yang L: Long noncoding RNA in
cancer: Wiring signaling circuitry. Trends Cell Biol. 28:287–301.
2018. View Article : Google Scholar
|
|
23
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016. View Article : Google Scholar
|
|
24
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811.
1998. View Article : Google Scholar
|
|
25
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar
|
|
26
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar
|
|
27
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
28
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar
|
|
29
|
Foulkes WD, Priest JR and Duchaine TF:
DICER1: Mutations, microRNAs and mechanisms. Nat Rev Cancer.
14:662–672. 2014. View
Article : Google Scholar
|
|
30
|
Stappert L, Roese-Koerner B and Brüstle O:
The role of microRNAs in human neural stem cells, neuronal
differentiation and subtype specification. Cell Tissue Res.
359:47–64. 2015. View Article : Google Scholar
|
|
31
|
Bottai G, Pasculli B, Calin GA and
Santarpia L: Targeting the microRNA-regulating DNA damage/repair
pathways in cancer. Expert Opin Biol Ther. 14:1667–1683. 2014.
View Article : Google Scholar
|
|
32
|
Adams BD, Kasinski AL and Slack FJ:
Aberrant regulation and function of microRNAs in cancer. Curr Biol.
24:R762–R776. 2014. View Article : Google Scholar
|
|
33
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar
|
|
34
|
Ell B and Kang Y: MicroRNAs as regulators
of bone homeostasis and bone metastasis. Bonekey Rep. 3:5492014.
View Article : Google Scholar
|
|
35
|
Nugent M: MicroRNA function and
dysregulation in bone tumors: The evidence to date. Cancer Manag
Res. 6:15–25. 2014. View Article : Google Scholar
|
|
36
|
Liu W, Jiang D, Gong F, Huang Y, Luo Y,
Rong Y, Wang J, Ge X, Ji C, Fan J and Cai W: miR-210-5p promotes
epithelial-mesenchymal transition by inhibiting PIK3R5 thereby
activating oncogenic autophagy in osteosarcoma cells. Cell Death
Dis. 11:932020. View Article : Google Scholar
|
|
37
|
Luo Y, Liu W, Tang P, Jiang D, Gu C, Huang
Y, Gong F, Rong Y, Qian D, Chen J, et al: miR-624-5p promoted
tumorigenesis and metastasis by suppressing hippo signaling through
targeting PTPRB in osteosarcoma cells. J Exp Clin Cancer Res.
38:4882019. View Article : Google Scholar
|
|
38
|
He M, Shen P, Qiu C and Wang J: miR-627-3p
inhibits osteosarcoma cell proliferation and metastasis by
targeting PTN. Aging (Albany NY). 11:5744–5756. 2019. View Article : Google Scholar
|
|
39
|
Duan Z, Gao Y, Shen J, Choy E, Cote G,
Harmon D, Bernstein K, Lozano-Calderon S, Mankin H and Hornicek FJ:
miR-15b modulates multidrug resistance in human osteosarcoma in
vitro and in vivo. Mol Oncol. 11:151–166. 2017. View Article : Google Scholar
|
|
40
|
Xu M, Jin H, Xu CX, Sun B, Mao Z, Bi WZ
and Wang Y: miR-382 inhibits tumor growth and enhance
chemosensitivity in osteosarcoma. Oncotarget. 5:9472–9483. 2014.
View Article : Google Scholar
|
|
41
|
Wu P, Liang J, Yu F, Zhou Z, Tang J and Li
K: miR-145 promotes osteosarcoma growth by reducing expression of
the transcription factor friend leukemia virus integration 1.
Oncotarget. 7:42241–42251. 2016. View Article : Google Scholar
|
|
42
|
Hirahata M, Osaki M, Kanda Y, Sugimoto Y,
Yoshioka Y, Kosaka N, Takeshita F, Fujiwara T, Kawai A, Ito H, et
al: PAI-1, a target gene of miR-143, regulates invasion and
metastasis by upregulating MMP-13 expression of human osteosarcoma.
Cancer Med. 5:892–902. 2016. View Article : Google Scholar
|
|
43
|
Lu J, Song G, Tang Q, Yin J, Zou C, Zhao
Z, Xie X, Xu H, Huang G, Wang J, et al: MiR-26a inhibits stem
cell-like phenotype and tumor growth of osteosarcoma by targeting
Jagged1. Oncogene. 36:231–241. 2017. View Article : Google Scholar
|
|
44
|
Zhu K, Liu L, Zhang J, Wang Y, Liang H,
Fan G, Jiang Z, Zhang CY, Chen X and Zhou G: MiR-29b suppresses the
proliferation and migration of osteosarcoma cells by targeting
CDK6. Protein Cell. 7:434–444. 2016. View Article : Google Scholar
|
|
45
|
Jin H and Wang W: MicroRNA-539 suppresses
osteosarcoma cell invasion and migration in vitro and targeting
matrix metallopeptidase-8. Int J Clin Exp Pathol. 8:8075–8082.
2015.
|
|
46
|
Xu B, Xia H, Cao J, Wang Z, Yang Y and Lin
Y: MicroRNA-21 inhibits the apoptosis of osteosarcoma cell line
SAOS-2 via targeting caspase 8. Oncol Res. 25:1161–1168. 2017.
View Article : Google Scholar
|
|
47
|
Zhang H, Guo X, Feng X, Wang T, Hu Z, Que
X, Tian Q, Zhu T, Guo G, Huang W and Li X: MiRNA-543 promotes
osteosarcoma cell proliferation and glycolysis by partially
suppressing PRMT9 and stabilizing HIF-1α protein. Oncotarget.
8:2342–2355. 2017. View Article : Google Scholar
|
|
48
|
Salah Z, Arafeh R, Maximov V, Galasso M,
Khawaled S, Abou-Sharieha S, Volinia S, Jones KB, Croce CM and
Aqeilan RI: miR-27a and miR-27a* contribute to metastatic
properties of osteosarcoma cells. Oncotarget. 6:4920–4935. 2015.
View Article : Google Scholar
|
|
49
|
Huang YZ, Zhang J, Shao HY, Chen JP and
Zhao HY: MicroRNA-191 promotes osteosarcoma cells proliferation by
targeting checkpoint kinase 2. Tumour Biol. 36:6095–6101. 2015.
View Article : Google Scholar
|
|
50
|
Wang C, Ba X, Guo Y, Sun D, Jiang H, Li W,
Huang Z, Zhou G, Wu S, Zhang J and Chen J: MicroRNA-199a-5p
promotes tumour growth by dual-targeting PIAS3 and p27 in human
osteosarcoma. Sci Rep. 7:414562017. View Article : Google Scholar
|
|
51
|
Zhu SW, Li JP, Ma XL, Ma JX, Yang Y, Chen
Y and Liu W: miR-9 modulates osteosarcoma cell growth by targeting
the GCIP tumor suppressor. Asian Pac J Cancer Prev. 16:4509–4513.
2015. View Article : Google Scholar
|
|
52
|
Zhou S, Wang B, Hu J, Zhou Y, Jiang M, Wu
M, Qin L and Yang X: miR-421 is a diagnostic and prognostic marker
in patients with osteosarcoma. Tumour Biol. 37:9001–9007. 2016.
View Article : Google Scholar
|
|
53
|
Yuan J, Chen L, Chen X, Sun W and Zhou X:
Identification of serum microRNA-21 as a biomarker for
chemosensitivity and prognosis in human osteosarcoma. J Int Med
Res. 40:2090–2097. 2012. View Article : Google Scholar
|
|
54
|
Yao ZS, Li C, Liang D, Jiang XB, Tang JJ,
Ye LQ, Yuan K, Ren H, Yang ZD, Jin DX, et al: Diagnostic and
prognostic implications of serum miR-101 in osteosarcoma. Cancer
Biomark. 22:127–133. 2018. View Article : Google Scholar
|
|
55
|
Lian F, Cui Y, Zhou C, Gao K and Wu L:
Identification of a plasma four-microRNA panel as potential
noninvasive biomarker for osteosarcoma. PLoS One. 10:e01214992015.
View Article : Google Scholar
|
|
56
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar
|
|
57
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar
|
|
58
|
Jian C, Tu MJ, Ho PY, Duan Z, Zhang Q, Qiu
JX, DeVere White RW, Wun T, Lara PN, Lam KS, et al: Co-targeting of
DNA, RNA, and protein molecules provides optimal outcomes for
treating osteosarcoma and pulmonary metastasis in spontaneous and
experimental metastasis mouse models. Oncotarget. 8:30742–30755.
2017. View Article : Google Scholar
|
|
59
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar
|
|
60
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar
|
|
61
|
Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX
and Hong W: The ways of action of long non-coding RNAs in cytoplasm
and nucleus. Gene. 547:1–9. 2014. View Article : Google Scholar
|
|
62
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772. 2016.
View Article : Google Scholar
|
|
63
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar
|
|
64
|
Sun J, Lin Y and Wu J: Long non-coding RNA
expression profiling of mouse testis during postnatal development.
PLoS One. 8:e757502013. View Article : Google Scholar
|
|
65
|
Ríos-Barrera LD, Gutiérrez-Pérez I,
Domínguez M and Riesgo-Escovar JR: acal is a long non-coding RNA in
JNK signaling in epithelial shape changes during drosophila dorsal
closure. PLoS Genet. 11:e10049272015. View Article : Google Scholar
|
|
66
|
Tao F, Tian X, Ruan S, Shen M and Zhang Z:
miR-211 sponges lncRNA MALAT1 to suppress tumor growth and
progression through inhibiting PHF19 in ovarian carcinoma. FASEB J.
fj201800495RR2018.Online ahead of print.
|
|
67
|
Schaukowitch K and Kim TK: Emerging
epigenetic mechanisms of long non-coding RNAs. Neuroscience.
264:25–38. 2014. View Article : Google Scholar
|
|
68
|
Wang Y, Zeng X, Wang N, Zhao W, Zhang X,
Teng S, Zhang Y and Lu Z: Long noncoding RNA DANCR, working as a
competitive endogenous RNA, promotes ROCK1-mediated proliferation
and metastasis via decoying of miR-335-5p and miR-1972 in
osteosarcoma. Mol Cancer. 17:892018. View Article : Google Scholar
|
|
69
|
Wang X, Peng L, Gong X, Zhang X and Sun R:
LncRNA HIF1A-AS2 promotes osteosarcoma progression by acting as a
sponge of miR-129-5p. Aging (Albany NY). 11:11803–11813. 2019.
View Article : Google Scholar
|
|
70
|
Fu D, Lu C, Qu X, Li P, Chen K, Shan L and
Zhu X: LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and
drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY).
11:8374–8385. 2019. View Article : Google Scholar
|
|
71
|
Li S, Liu F, Pei Y, Wang W, Zheng K and
Zhang X: Long noncoding RNA TTN-AS1 enhances the malignant
characteristics of osteosarcoma by acting as a competing endogenous
RNA on microRNA-376a thereby upregulating dickkopf-1. Aging (Albany
NY). 11:7678–7693. 2019. View Article : Google Scholar
|
|
72
|
Ba Z, Gu L, Hao S, Wang X, Cheng Z and Nie
G: Downregulation of lncRNA CASC2 facilitates osteosarcoma growth
and invasion through miR-181a. Cell Prolif. 51:e124092018.
View Article : Google Scholar
|
|
73
|
Wang Z, Liu Z and Wu S: Long non-coding
RNA CTA sensitizes osteosarcoma cells to doxorubicin through
inhibition of autophagy. Oncotarget. 8:31465–31477. 2017.
View Article : Google Scholar
|
|
74
|
Ye K, Wang S, Zhang H, Han H, Ma B and Nan
W: Long noncoding RNA GAS5 suppresses cell growth and
epithelial-mesenchymal transition in osteosarcoma by regulating the
miR-221/ARHI pathway. J Cell Biochem. 118:4772–4781. 2017.
View Article : Google Scholar
|
|
75
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma
cells through down-regulating ABCB1 and ABCC1. Oncotarget.
8:71881–71893. 2017. View Article : Google Scholar
|
|
76
|
Zhao J and Ma ST: Downregulation of lncRNA
H19 inhibits migration and invasion of human osteosarcoma through
the NF-κB pathway. Mol Med Rep. 17:7388–7394. 2018.
|
|
77
|
Zhou S, Yu L, Xiong M and Dai G: LncRNA
SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by
upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res
Commun. 495:1822–1832. 2018. View Article : Google Scholar
|
|
78
|
Ji S, Wang S, Zhao X and Lv L: Long
noncoding RNA NEAT1 regulates the development of osteosarcoma
through sponging miR-34a-5p to mediate HOXA13 expression as a
competitive endogenous RNA. Mol Genet Genomic Med. 7:e6732019.
View Article : Google Scholar
|
|
79
|
Guan H, Shang G, Cui Y, Liu J, Sun X, Cao
W, Wang Y and Li Y: Long noncoding RNA APTR contributes to
osteosarcoma progression through repression of miR-132-3p and
upregulation of yes-associated protein 1. J Cell Physiol.
234:8998–9007. 2019. View Article : Google Scholar
|
|
80
|
Bielack S, Carrle D and Casali PG; ESMO
Guidelines Working Group: Osteosarcoma: ESMO clinical
recommendations for diagnosis, treatment and follow-up. Ann Oncol.
20(Suppl): S137–S139. 2009. View Article : Google Scholar
|
|
81
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar
|
|
82
|
Ma B, Li M, Zhang L, Huang M, Lei JB, Fu
GH, Liu CX, Lai QW, Chen QQ and Wang YL: Upregulation of long
non-coding RNA TUG1 correlates with poor prognosis and disease
status in osteosarcoma. Tumour Biol. 37:4445–4455. 2016. View Article : Google Scholar
|
|
83
|
Holohan C, Van Schaeybroeck S, Longley DB
and Johnston PG: Cancer drug resistance: An evolving paradigm. Nat
Rev Cancer. 13:714–726. 2013. View Article : Google Scholar
|
|
84
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar
|
|
85
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar
|
|
86
|
Lei K, Bai H, Wei Z, Xie C, Wang J, Li J
and Chen Q: The mechanism and function of circular RNAs in human
diseases. Exp Cell Res. 368:147–158. 2018. View Article : Google Scholar
|
|
87
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar
|
|
88
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar
|
|
89
|
de Almeida RA, Fraczek MG, Parker S,
Delneri D and O'Keefe RT: Non-coding RNAs and disease: The
classical ncRNAs make a comeback. Biochem Soc Trans. 44:1073–1078.
2016. View Article : Google Scholar
|
|
90
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar
|
|
91
|
Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y,
Huang K, Wang G, Wang J, Ma J, et al: Circular RNA circTADA2A
promotes osteosarcoma progression and metastasis by sponging
miR-203a-3p and regulating CREB3 expression. Mol Cancer. 18:732019.
View Article : Google Scholar
|
|
92
|
Song YZ and Li JF: Circular RNA
hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis
by acting miRNA sponge. Biochem Biophys Res Commun. 495:2369–2375.
2018. View Article : Google Scholar
|
|
93
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330. 2018.
View Article : Google Scholar
|
|
94
|
Huang L, Chen M, Pan J and Yu W: Circular
RNA circNASP modulates the malignant behaviors in osteosarcoma via
miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 500:511–517.
2018. View Article : Google Scholar
|
|
95
|
Wu Z, Shi W and Jiang C: Overexpressing
circular RNA hsa_ circ_0002052 impairs osteosarcoma progression via
inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis.
Biochem Biophys Res Commun. 502:465–471. 2018. View Article : Google Scholar
|
|
96
|
Ren C, Liu J, Zheng B, Yan P, Sun Y and
Yue B: The circular RNA circ-ITCH acts as a tumour suppressor in
osteosarcoma via regulating miR-22. Artif Cells Nanomed Biotechnol.
47:3359–3367. 2019. View Article : Google Scholar
|
|
97
|
Li H, Lan M, Liao X, Tang Z and Yang C:
Circular RNA cir-ITCH promotes osteosarcoma migration and invasion
through cir-ITCH/miR-7/EGFR pathway. Technol Cancer Res Treat.
19:15330338198987282020.
|
|
98
|
Xiao-Long M, Kun-Peng Z and Chun-Lin Z:
Circular RNA circ_HIPK3 is down-regulated and suppresses cell
proliferation, migration and invasion in osteosarcoma. J Cancer.
9:1856–1862. 2018. View Article : Google Scholar
|
|
99
|
Jin Y, Li L, Zhu T and Liu G: Circular RNA
circ_0102049 promotes cell progression as ceRNA to target MDM2 via
sponging miR-1304-5p in osteosarcoma. Pathol Res Pract.
215:1526882019. View Article : Google Scholar
|
|
100
|
Li L, Guo L, Yin G, Yu G, Zhao Y and Pan
Y: Upregulation of circular RNA circ_0001721 predicts unfavorable
prognosis in osteosarcoma and facilitates cell progression via
sponging miR-569 and miR-599. Biomed Pharmacother. 109:226–232.
2019. View Article : Google Scholar
|
|
101
|
Li JF and Song YZ: Circular RNA GLI2
promotes osteosarcoma cell proliferation, migration, and invasion
by targeting miR-125b-5p. Tumour Biol. 39:10104283177099912017.
View Article : Google Scholar
|
|
102
|
Li S, Pei Y, Wang W, Liu F, Zheng K and
Zhang X: Circular RNA 0001785 regulates the pathogenesis of
osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2.
Cell Cycle. 18:1281–1291. 2019. View Article : Google Scholar
|
|
103
|
Cao J and Liu XS: Circular RNA 0060428
sponges miR-375 to promote osteosarcoma cell proliferation by
upregulating the expression of RPBJ. Gene. 740:1445202020.
View Article : Google Scholar
|
|
104
|
Li S, Sun X, Miao S, Lu T, Wang Y, Liu J
and Jiao W: hsa_circ_0000729, a potential prognostic biomarker in
lung adenocarcinoma. Thorac Cancer. 9:924–930. 2018. View Article : Google Scholar
|
|
105
|
Li XM, Ge HM, Yao J, Zhou YF, Yao MD, Liu
C, Hu HT, Zhu YX, Shan K, Yan B and Jiang Q: Genome-wide
identification of circular RNAs as a novel class of putative
biomarkers for an ocular surface disease. Cell Physiol Biochem.
47:1630–1642. 2018. View Article : Google Scholar
|
|
106
|
Zhang H, Wang G, Ding C, Liu P, Wang R,
Ding W, Tong D, Wu D, Li C, Wei Q, et al: Increased circular RNA
UBAP2 acts as a sponge of miR-143 to promote osteosarcoma
progression. Oncotarget. 8:61687–61697. 2017. View Article : Google Scholar
|
|
107
|
Liu X, Abraham JM, Cheng Y, Wang Z, Wang
Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al:
Synthetic circular RNA functions as a miR-21 sponge to suppress
gastric carcinoma cell proliferation. Mol Ther Nucleic Acids.
13:312–321. 2018. View Article : Google Scholar
|
|
108
|
Xu S, Gong Y, Yin Y, Xing H and Zhang N:
The multiple function of long noncoding RNAs in osteosarcoma
progression, drug resistance and prognosis. Biomed Pharmacother.
127:1101412020. View Article : Google Scholar
|
|
109
|
Yin F, Wang Z, Jiang Y, Zhang T, Wang Z,
Hua Y, Song Z, Liu J, Xu W, Xu J, et al: Reduction-responsive
polypeptide nanomedicines significantly inhibit progression of
orthotopic osteosarcoma. Nanomedicine. 23:1020852020. View Article : Google Scholar
|
|
110
|
Matsui M and Corey DR: Non-coding RNAs as
drug targets. Nat Rev Drug Discov. 16:167–179. 2017. View Article : Google Scholar
|
|
111
|
Li Z and Rana TM: Therapeutic targeting of
microRNAs: Current status and future challenges. Nat Rev Drug
Discov. 13:622–638. 2014. View Article : Google Scholar
|
|
112
|
Wang WT, Han C, Sun YM, Chen TQ and Chen
YQ: Noncoding RNAs in cancer therapy resistance and targeted drug
development. J Hematol Oncol. 12:552019. View Article : Google Scholar
|
|
113
|
Huang KW, Lai YT, Chern GJ, Huang SF, Tsai
CL, Sung YC, Chiang CC, Hwang PB, Ho TL, Huang RL, et al: Galactose
derivative-modified nanoparticles for efficient siRNA delivery to
hepatocellular carcinoma. Biomacromolecules. 19:2330–2339. 2018.
View Article : Google Scholar
|
|
114
|
Nair JK, Willoughby JL, Chan A, Charisse
K, Alam MR, Wang Q, Hoekstra M, Kandasamy P, Kel'in AV, Milstein S,
et al: Multivalent N-acetylgalactosamine-conjugated siRNA localizes
in hepatocytes and elicits robust RNAi-mediated gene silencing. J
Am Chem Soc. 136:16958–16961. 2014. View Article : Google Scholar
|
|
115
|
Wen JJ, Ma YD, Yang GS and Wang GM:
Analysis of circulating long non-coding RNA UCA1 as potential
biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med
Pharmacol Sci. 21:498–503. 2017.
|
|
116
|
Huang JZ, Chen M, Chen D, Gao XC, Zhu S,
Huang H, Hu M, Zhu H and Yan GR: A peptide encoded by a putative
lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell.
68:171–184.e6. 2017. View Article : Google Scholar
|
|
117
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:3042018. View Article : Google Scholar
|
|
118
|
Ribatti D: The concept of immune
surveillance against tumors. The first theories Oncotarget.
8:7175–7180. 2017. View Article : Google Scholar
|
|
119
|
Ji X, Wang E and Tian F: MicroRNA-140
suppresses osteosarcoma tumor growth by enhancing anti-tumor immune
response and blocking mTOR signaling. Biochem Biophys Res Commun.
495:1342–1348. 2018. View Article : Google Scholar
|
|
120
|
Li S, Li X, Xue W, Zhang L, Yang LZ, Cao
SM, Lei YN, Liu CX, Guo SK, Shan L, et al: Screening for functional
circular RNAs using the CRISPR-Cas13 system. Nat Methods. 18:51–59.
2021. View Article : Google Scholar
|