1. Introduction
Hepatocellular carcinoma (HCC) is the most common
type of primary liver cancer (1,2)
and the second leading cause of cancer-associated mortality
worldwide (3). HCC occurs most
frequently in patients with chronic liver diseases, including
cirrhosis caused by hepatitis B or C infection, which accounts for
~50% of cases (4,5). The risk of HCC, which is the most
common type of liver cancer, is higher in patients with long-term
liver diseases as well as in patients infected with hepatitis B or
C (6,7). HCC is also more common in
individuals who consume large quantities of alcohol and in those
with accumulated fat in the liver (8). Other factors, such as tobacco smoke
inhalation and intake of aflatoxin B1, are also well-described
contributors to HCC (8).
As HCC is one of the most common malignant tumors in
clinical practice, its diagnosis, treatment and prognosis attract
significantly considerable attention. Surgical interventions,
including resection and transplantation, are the most common
methods for early HCC therapy (9). However, since HCC is difficult to
diagnose at an early stage, and the majority of patients tend to be
diagnosed at advanced stages and are not ideal for resection,
conservative treatment strategies are generally useful for advanced
HCC (2). However, radiotherapy,
including stereotactic body radiation therapy and radiofrequency
ablation, is affected by increased radioresistance, indicating that
HCC therapy should be improved due to the high morbidity, mortality
and recurrence of this disease (10,11). Radioresistance is a complex
biological process associated with multiple factors, such as
abnormal DNA damage response, gene mutations, cell autophagy,
apoptosis, cell cycle checkpoint and other dysregulated signaling
pathways (12). These mechanisms
lead to poor prognosis in patients and cause major clinical
obstacles for radiotherapy, ultimately leading to tumor metastasis
and relapse (13).
Radioresistance is also highly associated with the tumor
microenvironment (14,15), which is an important factor
associated with tumor progression and therapeutic response
(16,17) and is actively achieved by
exosome-regulated cell-cell communication.
Exosomes are single-membrane secreted organelles
40-150 nm in diameter. Exosomes have similar topologies with cells,
and are enriched in nucleic acids, selected proteins, lipids and
glycoconjugates. The biogenesis of exosomes relies on a mechanism
of protein quality control, and exosomes display activities in
remodeling the extracellular matrix and transmitting signals and
molecules from the contributor cells to other cells. This pathway
plays important functions in numerous aspects of human homeostasis
and disease, such as development, tissue homeostasis, immunity,
neurodegenerative diseases and cancer (18,19). Increasing evidence suggests that
cancer-derived exosomes play multiple and critical functions in
cancer. Exosomes, as well as their components, may serve as cancer
prognostic markers, therapeutic targets or anticancer drug carriers
(20,21). Regarding the advantages of
exosomes in cancer diagnosis and their newly suggested application,
previous studies have indicated that radiation-derived
extracellular vesicles, particularly exosomes, decrease survival,
increase tumor burden, cause radiation-induced bystander effects
and promote radioresistance. The present study reviews recent
reports on radiation-induced changes in extracellular vesicles
(particularly exosomes) in HCC, and discusses the molecular
mechanisms of exosome-regulated HCC progression and radioresistance
for the development of novel therapeutic strategies.
2. Biological components of exosomes in
HCC
The majority of commonly studied cells are
characterized by the production of a set of lipid membrane-coated
vesicles that can be secreted to the extracellular space. They are
named extracellular vesicles, and the most well-known ones are
exosomes (22). Exosomes, which
were first identified in the late 1990s, are a set of membranous
vesicles released into the extracellular space by contributor cells
after multiple intracellular vesicles fuse into the cell membrane
(23). The majority of known cell
types, including normal and pathogenic cells, are capable of
secreting exosomes, and numerous studies suggest that, compared
with that of normal epithelial cells, the release of exosomes by
tumor cells is more active (24).
Exosomes are present in almost all types of body fluids, including
urine, saliva, blood, breast milk, bile, synovial fluid, seminal
fluid and amniotic liquid, indicating their critical functions in
intercellular communication by transducing both genomic and
proteomic materials among cells and subsequently modulating
physiological responses (25).
However, the mechanisms of the biological formation of exosomes
remain to be clarified. The functions and characteristics of
exosomes are mostly based on their sizes, expression of surface
markers and composition, including DNA, RNA, lipids, metabolites
and surface proteins (18,26).
This variety of characteristic protein components on the outer
membrane of exosomes, which serve as surface markers, includes CD9,
CD81, CD63, CD53, CD82, CD37, TSG101, Hsp70 and Alix (26,27). These proteins are commonly used as
markers to confirm the presence of exosomes, since they are
detectable by western blotting. In addition, exosomes contain
numerous lipid molecules, which participate in multiple biological
processes and play critical roles in the morphological stability of
exosomes in the extracellular fluid (28).
In the tumor microenvironment, cancer cells secret
numerous exosomes, which are transferred from cancer cells to other
cell types to participate in signaling transduction, and in the
processes of tumor formation and progression (29). In addition, exosomes derived from
both tumor and stromal cells have been reported to be implicated in
all stages of cancer progression and to exhibit crucial functions
in tumor therapy resistance. Due to their characteristics as
mediators of cell-cell communication, exosomes are integral to
tumor microenvironment-dependent therapy resistance (30). Furthermore, exosomes are also able
to deliver oncogenes to normal epithelial cells under pathological
conditions, which has been reported to be one of the mechanisms
involved in tumor invasion and metastasis (31). Numerous previous studies have
demonstrated that exosomes can deliver various proteins and
ribonucleic acids to recipient cells (18). These studies are validated by the
activation and expansion of exosome-producing immune cells in
murine models of acute or chronic inflammation (32).
Exosomes are of different phenotypes in different
body fluids and contain a variety of biologically active molecules,
which leads to diverse and heterogenous exosome types (26). These molecules are divided into
lipids (including sphingomyelin, phosphatidylserine and
cholesterol) (33), proteins
(including tumor-suppressor proteins, oncoproteins and
transcription regulators) (34),
DNA (including genomic DNA, single-strand DNA, and retrotransposon
elements) (35,36) and RNAs [including long non-coding
RNAs (lncRNAs), microRNAs (miRNAs), circular RNAs (circRNAs),
messenger RNAs (mRNAs) and other non-coding RNAs (ncRNAs)]
(37). Multiple studies have
confirmed that these exosomal active components exhibit critical
functions in the resistance of a variety of tumors, particularly in
HCC radioresistance (18).
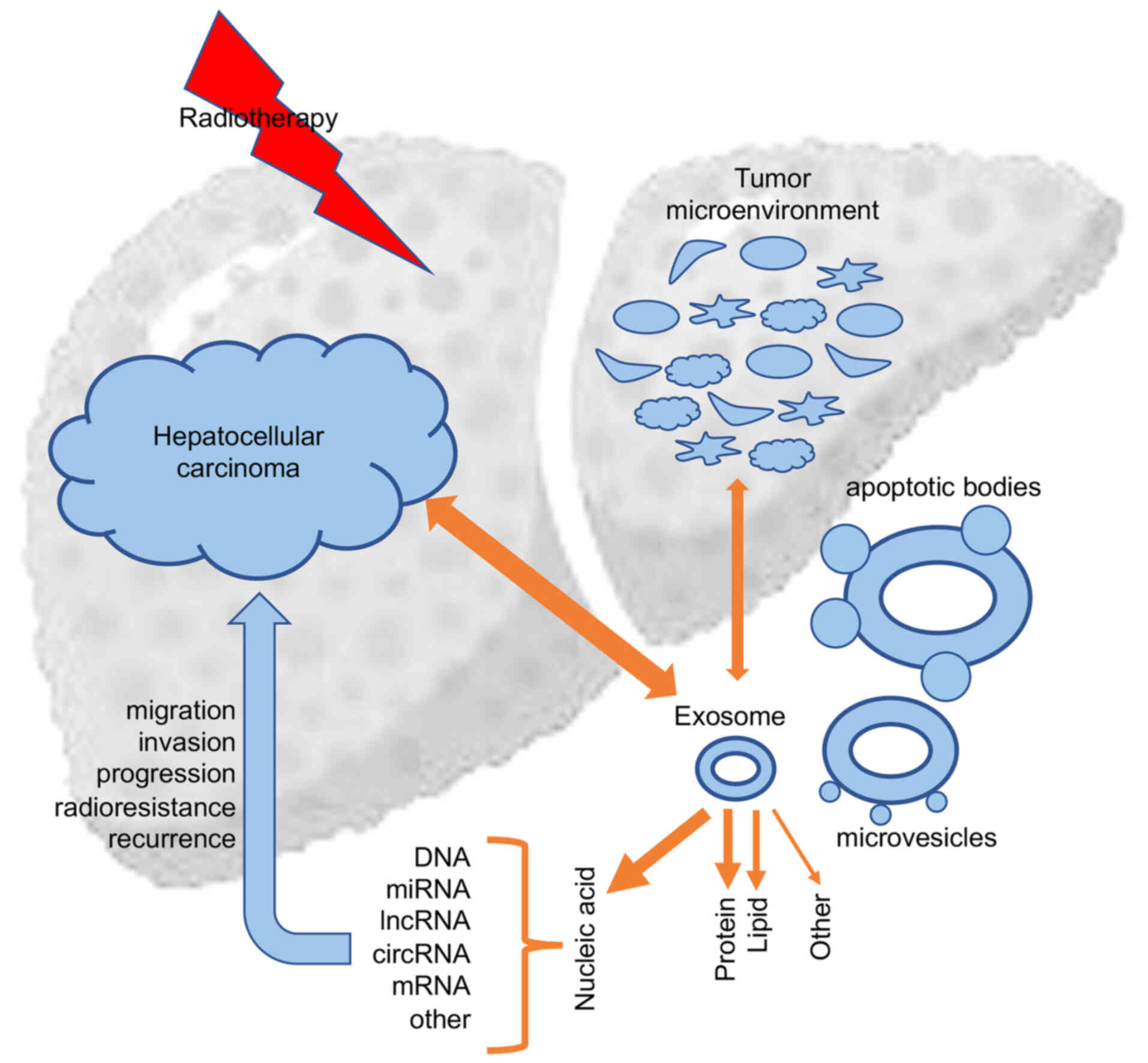
Therapeutic interventions such as chemotherapy and radiotherapy,
could affect exosome uptake and subsequent tumor biological
responses, which in return could influence the secretion and
components of exosomes (13)
(Fig. 1).
3. Protein transduction by HCC-derived
exosomes
Exosomes contain abundant proteins, which include
not only self-proteins, but also proteins derived from contributor
cells.
Exosomal proteins as HCC biomarkers
Exosomal proteins derived from cancer cells are
becoming novel biomarkers for cancer monitoring and efficacy
evaluation, and are also becoming a popular research topic in
studies on cancer radioresistance (38), particularly in HCC. Mass
spectrometry (MS) analysis demonstrated that 213 proteins could be
detected in exosomes secreted by an HCC cell line, and, among them,
158 proteins were specifically expressed in exosomes secreted by
highly malignant HCC cells compared with the exosomal proteome of
each cell line (39). Exosomes
secreted by metastatic HCC cell lines were found to carry several
protumorigenic RNAs and proteins, including MET proto-oncogene,
caveolins and S100 family members. Notably, exosomes from motile
HCC cell lines significantly enhanced the migratory and invasive
abilities of non-motile MIHA cells. Uptake of exosomal molecules
may trigger signaling pathways, including the PI3K/AKT and MAPK
signaling pathways, in MIHA cells with increased secretion of
active matrix metalloproteinases MMP-2 and MMP-9. These findings
demonstrated that HCC-derived exosomes had the potential of
mobilizing normal hepatocytes, which may have implications in
promoting the protrusive activities of HCC cells through the liver
parenchyma during the process of metastasis (39). Another study using MS investigated
exosomes isolated from the HCC cell line HepG2 and identified a
total of 1,428 proteins in HepG2 cell exosomes. These proteins were
found to facilitate the activation of several kinases and the NF-κB
signaling pathway in HCC cells (39). Eukaryotic translation initiation
factor 3 subunit C (EIF3C), a protein associated with poor patient
survival that is upregulated during HCC tumor progression, was
found to significantly increase extracellular exosome secretion,
exosome size and exosome marker expression diversity.
EIF3C-overexpressing exosomes were oncogenic, and enhanced tumor
angiogenesis and blood vessel growth. Subcutaneous inoculation of
EIF3C-enriched exosomes together with Huh7 HCC cells increased the
growth of vessels and upregulated the expression of EIF3C in
tumors. Furthermore, EIF3C activated the expression of S100A11,
which is involved in EIF3C-enriched exosome-mediated increased tube
formation in angiogenesis (40).
A previous study performed protein profiling of exosomes derived
from different HCC cell types, and identified 129 proteins, among
which, adenylyl cyclase-associated protein 1 (CAP1), a protein
implicated in HCC metastasis, was significantly enriched in
exosomes secreted by HCC cells with high motile abilities (41). Similarly, super-stable isotope
labeling by/with amino acids in cell culture-based MS analysis on
exosomes secreted by three human HCC cell lines, non-motile Hep3B
cells, and motile 97H and LM3 cells quantified >1,400 exosomal
proteins in each MS analysis with highly biological
reproducibility. In total, 469 and 443 exosomal proteins,
respectively, represented differentially expressed proteins in
LM3/Hep3B vs. 97H/Hep3B cells. These proteins were involved in
sugar metabolism-centric canonical pathways according to signaling
pathway analysis. Specifically, these pathways included the
glycolysis I, gluconeogenesis I and pentose phosphate pathways, and
the proteins enriched in these signaling pathways could form a
tightly connected network. The above findings suggest that motile
HCC cells have the potential to preferentially export more sugar
metabolism-associated proteins by exosomes that facilitate the
differentiation of motile HCC from non-motile HCC cells (42).
Exosomal proteins in HCC metastasis and
therapy resistance
Another study investigated the proteome of
macrovesicles present in urine samples acquired from liver injury
experimental models as a method to identify potential biomarkers of
hepatic diseases. The biochemical and proteomic characterization of
highly purified exosomes identified 28 previously unreported
proteins, a number of which were closely associated with liver
diseases. This analysis suggested that urinary exosomes could
potentially be a sample source for identifying biomarkers of liver
injury, and certain proteins such as CD26, CD81, solute carrier
family 3 member 1 (SLC3A1) and CD10 were differentially expressed
in urinary exosomes derived from experimental models, thus serving
as potential biomarkers for liver injury (43). HCC cells were reported to promote
cancer cell proliferation and lung metastasis formation in a
paracrine/endocrine manner, which was achieved by primary
HCC-derived exosomes. In the presence of these malignant exosomes,
enhanced cell adhesion was observed. Notably, attached HCC cells
released exosomes containing both mothers against decapentaplegic
homolog 3 (SMAD3) protein and mRNA, and delivered them to detached
HCC cells to subsequently facilitate their adhesion. These exosomes
contained activated SMAD3 signaling in the recipient HCC cells and
improved their adhesive capacities. Clinically, SMAD3-containing
exosomes were detected in the peripheral blood of patients with
HCC, and their levels were closely associated with disease stage
and SMAD3 expression in primary tumors. That study suggested a
potential mechanism by which exosome SMAD3 mediated communication
between primary and circulating HCC cells (44). Tumor-derived exosomes were found
to activate B cells, which expressed high levels of T-cell
immunoglobulin and mucin domain 1 (TIM-1) protein and suppressed
the activities of CD8+ T cells in a similar manner to
TIM-1+ regulatory B (Breg) cells isolated from HCC
tissues. Enrichment of TIM-1+ Breg cells in HCC tissues
was involved in advanced disease stage, and predicted early
recurrence in HCC as well as reduced patient survival in HCC. High
mobility group box protein 1 (HMGB1) derived from exosomes
activated B cells and promoted the expansion of TIM-1+
Breg cells through the Toll-like receptor 2/4 (TLR2/4) and MAPK
signaling pathways (45). T cells
could uptake exosomes containing 14-3-3ζ secreted by HCC cells,
which suggested that 14-3-3ζ is delivered from HCC cells to
tumor-infiltrating T lymphocytes by exosomes. In tumorinfiltrating
T cells of the HCC microenvironment, 14-3-3ζ was upregulated and
inhibited their antitumor functions. Specifically, 14-3-3ζ is
potentially transferred from HCC cells to T cells partially through
exosomes (46). Exosomes with
increased C-X-C motif chemokine receptor 4 (CXCR4) levels secreted
by high metastatic mouse hepatocarcinoma Hca-F cells were confirmed
to increase the migration and invasion of paired syngeneic Hca-P
cells with low metastatic capacities. The increase in cell
migratory and invasive capacities of Hca-P cells was facilitated by
the internalization of exosomes isolated from Hca-F cells and
subsequent transferring of CXCR4 via exosomes. Hca-F cell migration
and invasion were increased by lymphatic endothelial cells via
upregulation of stromal cell-derived factor-1α (SDF-1α) binding to
C-X-C motif chemokine receptor 4 (CXCR4) in Hca-F cells and
subsequently upregulating the secretion of vascular endothelial
growth factor (VEGF)-C, MMP-9 and MMP-2. In addition, CXCR4 derived
from Hca-F exosomes significantly increased the abilities of
lymphatic endothelial cell proliferation and lymphatic tube
formation (47). C-type lectin
domain family 3 member B (CLEC3B) downregulation in HCC was found
to suggest a poor prognosis. Exosomes secreted by HCC with
downregulated CLEC3B significantly increased the
epithelial-mesenchymal transition (EMT), migration and invasion of
tumor cells. CLEC3B downregulation in HCC exosomes decreased VEGF
secretion in HCC cells and inhibited angiogenesis. Mechanistically,
VEGF expression mediated by CLEC3B in tumor cells relied on the
activation of the AMP-activated protein kinase signaling pathway
(48). Both in vivo and
in vitro, overexpression of chitinase 3 like 1 (CHI3L1), a
cancer poor prognosis-associated secreted glycoprotein,
significantly facilitated the growth, migration and invasion of
liver cancer cells. Overexpression of CHI3L1 influenced cell-cell
adhesion, extracellular exosomes and adherent junction-related
genes via TGF-β signaling pathway activation (49). The expression of golgi membrane
protein 1 (GOLM1) was inhibited by miR-145 via targeting a coding
sequence of the GOLM1 gene. The expression of GOLM1 and
miR-145 showed an inverse correlation in human HCC tissues.
Mechanistically, exosomes with GOLM1 enrichment activated the
GSK-3β/MMP signaling axis of recipient cells, and accelerated HCC
migration and proliferation (50).
4. Lipid transduction by HCC-derived
exosomes
Exosome structures and contents consist of proteins,
nucleic acids and lipids, which are mostly derived from contributor
cells. Similar to structures such as cell membranes, exosomes
contain a lipid bilayer membrane that protects the encapsulated
material (nucleic acids, proteins, metabolites and lipids) from the
extracellular environment (33,51). Lipids not only act as essential
components of exosomal membranes, which protect exosome contents
from various stimuli in the circulating body fluids, but also are
necessary players in exosome formation and release to the
extracellular environment (33).
Increasing evidence suggests that specific lipid components are
enriched in exosome particles compared with their parental
cells.
In contrast to those of proteins and nucleic acids,
the functions of exosomal lipids in HCC development, progression,
radioresistance and chemoresistance are less established.
Increasing data suggest that lipid molecules in exosomes may serve
as potential cancer diagnostic biomarkers, as confirmed by data
obtained in urine (52) and
cancer cell lines (53). Exosomes
derived from the oligodendroglial precursor cell line Oli-neu were
enriched in cholesterol, and contained higher levels of
sphingolipids (sphingomyelin and hexosylceramide) and lower levels
of phosphatidylcholine than those found in the cell membrane
(54). In previous studies on
hepatitis and HCC, exosomes were found to play key roles in
hepatitis E virus (HEV) egress; however, HEV infection did not
modify the characteristics of exosomes produced by infected cells.
Enrichment in phosphatidylserines and cholesterol in these exosomes
could be critical for HEV entry into cells. Meanwhile, HEV
particles in exosomes are protected from the attack of the immune
system, which leads to wide spreading of HEV in the circulatory
system of the host (55). A
previous high-resolution lipidomic and proteomic study reported the
analysis of exosomes and extracellular vesicles obtained by
differential ultracentrifugation from the glioblastoma cell line
U87, the HCC cell line Huh7 and human bone marrow-derived
mesenchymal stem cells (MSCs). Exosomes enriched in glycolipids and
characterized by the presence of free fatty acids, as well as
exosomes from Huh7 cells and MSCs were specifically enriched in
cardiolipins (56). Unlike
circulating tumor cells, exosomes are highly abundant in biofluids
and cells, and are diverse in vesicle type and lipid composition.
Different from other single-molecule circulating biomarkers,
exosomes protect their molecular cargo from degradation and may
carry molecular signatures involved in specific phenotypes
(57). Increased understanding of
the role of exosomal lipids in HCC progression and radioresistance
suggests that exosome-derived lipids have a considerable potential
as HCC biomarkers and a tool to monitor radiotherapy in the future
(26).
5. Nucleic acids derived from HCC
exosomes
Exosome-derived nucleic acids are diverse, but the
majority of the currently identified exosome-derived nucleic acids
are RNAs, and few studies have focused on exosomal DNA in HCC.
DNA
Analyses of circulating cell-free tumor DNA and
circulating tumor cells have paved the way for novel diagnostic
approaches, and are the basis of diagnostic techniques based on
liquid biopsies. The application of circulating cell-free tumor DNA
on early cancer detection is of high public interest (58). Exosomal DNA has attracted broad
interest in cancer research; however, few studies on HCC have been
conducted to date (36,59). The majority of DNA associated with
tumor exosomes has been confirmed to be double-stranded in three
different cancer models: human chronic myeloid leukemia cell model
K-562; human colorectal carcinoma cell model HCT116; and murine
melanoma cell model B16-F10. This double-stranded DNA in exosomes
represents the whole genomic DNA. Exosomal DNA has been confirmed
to be used to identify mutations in parental tumor cells, which
suggests its translational potential as a circulating biomarker of
cancer in the clinic (36). A
recent study on liquid biopsy samples of 194 patients, who were
undergoing treatment for localized or metastatic pancreatic
adenocarcinoma, showed a marked increase in exosomal DNA levels
after neoadjuvant therapy, which suggested a close association with
disease progression. Significantly shorter times of
progression-free and overall survival were found in patients with
metastases and detectable circulating tumor DNA (ctDNA) at baseline
status compared with those of patients without detectable ctDNA
(60). Plasma cell-free DNA
(cfDNA) levels in patients with HCC and hepatitis B virus-related
liver fibrosis were closely associated with the degree of liver
inflammation, body mass index and α-fetoprotein level, but showed
no association with liver fibrosis stage. Significantly elevated
plasma cfDNA levels were detected in patients with HCC compared
with those found in patients without HCC. The HCC index, which is
generated by a combination model including age, and cfDNA and
α-fetoprotein levels, had an area of 0.98 under the receiver
operating characteristics curve for the diagnosis of HCC,
suggesting that the diagnostic power of the HCC index was more
promising than that of cfDNA or α-fetoprotein levels alone
(61).
miRNA
Exosomal RNAs include miRNAs, lncRNAs, mRNAs,
circRNAs and other non-coding ncRNAs (37). ncRNA families, including miRNAs,
lncRNAs, circRNAs and other ncRNAs, are not functional to encode
proteins, but function as specific RNA inhibitors by degradation or
transformation to regulate post-transcriptional gene expression,
which is associated with the control of nuclear architecture,
transcription in the nucleus, and modulation of mRNA stability,
translation and post-translational modifications in the cell
cytoplasm (62,63).
miRNAs are small ncRNAs of 17-24 nt, which mediate
post-transcriptional gene silencing by binding to the
3′untranslated or open reading frame region of target mRNAs
(64). miRNAs have been detected
in exosomes, which have been found to be taken up by neighbor or
distant cells, and then modulate the activities of recipient cells.
Accumulating evidence has shown that miRNAs can be stable in body
fluids, including saliva, urine, breast milk and blood. Besides
being packed into exosomes or other microvesicles, extracellular
miRNAs can also be loaded into high-density lipoprotein (65) or be bound by argonaute-2 protein
outside these microvesicles (66). All these three modes of action
protect miRNAs from degradation and ensure their stability
(67). Changes in the expression
levels of miRNAs contribute to the pathogenesis of numerous human
malignancies. These changes are potentially caused by multiple
different mechanisms, such as gene mutations, amplifications or
deletions involving miRNA loci, epigenetic silencing or
dysregulation of transcription factors targeting specific miRNAs.
Malignant cells depend on the dysregulated expression of multiple
miRNAs, and in turn control or are controlled by dysregulation of
multiple protein-coding oncogenes or tumor-suppressor genes, which
suggests that these small RNAs may provide opportunities for the
development of novel miRNA-based therapies (68). Circulating exosomal miRNA-21 has
been associated with TNM stages and other prognostic factors,
including T stage and portal vein thrombosis, in HCC. High miRNA-21
level was reported to be a promising predictor of increased
mortality and disease progression, as well as larger tumor size and
higher C-reactive protein levels. Patients with higher circulating
levels of exosomal miRNA-21 exhibited significantly lower overall
and progression-free survival (69). Another study found that the
migration, invasion and proliferation of recipient HCC cells were
significantly increased after treatment with exosomes secreted by
HCC cells cultured in acidic medium. miR-21 and miR-10b were the
most critically functional miRNAs in acidic HCC cell-derived
exosomes. Activation of hypoxia-inducible factor (HIF)-1α and
HIF-2α was facilitated by the acidic microenvironment, which also
stimulated the expression and secretion of exosomal miR-21 and
miR-10b, substantially promoting HCC cell proliferation, invasion,
migration and proliferation both in vivo and in
vitro. Advanced tumor stage, and HIF-1α and HIF-2α expression
in patients with early HCC were closely associated with serum
exosomal miR-21 and miR-10b levels, suggesting that exosomal miR-21
and miR-10b were independent prognostic factors for disease-free
survival of patients with early HCC (70). A previous study demonstrated that
the levels of 19 miRNAs were markedly upregulated in the sera of
patients with HCC (71). A
following study demonstrated that HCC cells secreted miR-210 via
exosomes. The in vitro tubulogenesis of endothelial cells
and the in vivo angiogenesis of HCC were significantly
promoted by exosomal miR-210 by inhibiting the expression levels of
SMAD4 and STAT6 in the recipient endothelial cells (72). Another study focused on the
functions of exosomal miR-224 in the development of HCC, and
assessed its diagnostic and prognostic value in HCC. Patients with
HCC who had high expression levels of serum exosomal miR-224
exhibited a lower overall survival, suggesting that exosomal
miR-224 is potentially a tumor-promoting gene, and may serve as a
biomarker for diagnosis and prognosis in patients with HCC
(73). In addition to these
studies, a number of other exosome-derived miRNAs have been found
to be closely associated with the pathogenesis and progression of
HCC. Their potential roles include being biomarkers for HCC
diagnosis and serve as therapeutic targets for HCC treatment.
lncRNAs
lncRNAs, another type of ncRNAs with a size >200
nt that lack protein-coding functions, have been reported to be
critical modulators of intercellular communications (74). Cancer cell-derived exosomes have
been confirmed to contain various lncRNAs, usually defined exosomal
lncRNAs, which regulate the cancer microenvironment by modulating
multiple cellular functions, including regulation of the
transcription of certain critical genes that determine cancer
growth and progression, thus defining the biological functions of
cancer exosomes (75,76). Furthermore, deregulation of lncRNA
expression has been identified in multiple human tumors, and
lncRNAs may serve as attractive diagnostic biomarkers to predict
different cancer types (77).
Similar to miRNAs, exosomal lncRNAs are also
important contributors to the occurrence, development and
transferring of HCC. A stress-responsive lncRNA, long intergenic
non-protein coding RNA, regulator of reprogramming (linc-ROR), was
significantly upregulated in HCC cells and was also found to be
enriched in tumor cell-derived extracellular vesicles. Incubation
with HCC-derived extracellular vesicles significantly increased the
expression of linc-ROR and blunt chemotherapy-induced cell death in
recipient cells (78). Incubation
with linc-very low density lipoprotein receptor (VLDLR)-enriched
exosomes reduced chemotherapy-induced cell death in HCC cells, and
increased the expression of linc-VLDLR in recipient cells (79). The expression levels of
lncRNA-focally amplified lncRNA on chromosome 1 (lnc-FAL1) were
significantly upregulated in HCC tissues, and lnc-FAL1 was found to
function as an oncogene in HCC (80). In serum exosomes of patients with
HCC, the levels of lnc-FAL1 were upregulated, and transferring
lnc-FAL1 to HCC cells increased their cell migration and
proliferation abilities (81).
Numerous studies have reported HCC-derived exosomes
as cargo of novel lncRNA biomarkers. Exosomes derived from non-HCC
serum contained a large number of lncRNAs with a high level of
alternative splicing compared with those found in patients with
other hepatic diseases. Exosomal lncRNAs, including lnc-G
protein-coupled receptor 89B-15 (lnc-GPR89B-15), lnc-family with
sequence similarity 72 member D-3 (lnc-FAM72D-3), lnc-enhancer of
polycomb homolog1-4 (lnc-EPC1-4) and lnc-zinc finger E-box binding
homeobox 2-19 (lncZEB2-19), showed differential expression in the
serum of patients with HCC (82).
Another study revealed that five lncRNAs, namely CTD-2116N20.1,
AC012074.2, RP11-538D16.2, long intergenic non-protein coding RNA
(LINC) 00501 and RP11-136I14.5, showed significantly different
expression. Co-expression and competing endogenous (ce)RNA network
analyses revealed possible mechanisms of CTD-2116N20.1 and
RP11-538D16.2 as exosome-related lncRNAs, and CTD-2116N20.1 and
RP11-538D16.2 were correlated with poor prognosis in patients with
HCC (83). The expression of
lncRNA-HEIH (lnc-HEIH) was increased in the serum and exosomes of
patients with hepatitis C virus-related HCC compared with the
findings in patients with chronic hepatitis C (84). High levels of ENSG00000258332.1 in
HCC were associated with overall survival, portal vein tumor
emboli, TNM stage and lymph node metastasis, and high levels of
LINC00635 were closely associated with overall survival, TNM stage
and lymph node metastasis (85).
Another study evaluated the relative expression levels of 8
selected serum lncRNAs in the training and validation sets of
patients with HCC and matched healthy controls. The expression of
LINC00161 was markedly upregulated, and exhibited high stability
and specificity in serum samples of patients with HCC, indicating
the biomarker potential of LINC00161 in HCC diagnosis and prognosis
(86). The expression level of
X-inactive-specific transcript (Xist), another lncRNA, was
significantly increased in peripheral blood mononuclear cells and
granulocytes of patients with HCC. Another lncRNA, Jpx, also showed
increased upregulation in mononuclear cells, granulocytes and
exosomes of female patients with HCC. Delivery of Jpx from HCC
cells to blood cells via exosomes activated Xist expression in
blood cells, possibly by inhibiting the trans-regulatory
effects of CCCTC-binding factor (87). A previous study also identified
Xist as a mediator of miRNA-92b, which promoted HCC progression by
targeting SMAD7 (88). Another
commonly reported HCC-associated lncRNA is TUC339. The most highly
significantly expressed lncRNA in extracellular vesicles derived
from HCC cells was identified as TUC339 in a previous study.
Functionally, TUC339 was found to be involved in regulating tumor
cell adhesion and growth (89).
Compared with those from healthy controls, exosomes derived from
HCC cells contained upregulated levels of TUC339, and HCC-secreted
exosomes could be taken up by THP-1 cells. These THP-1 cells also
showed a significant increase in pro-inflammatory cytokine
production, increased co-stimulatory molecule expression and
enhanced phagocytosis upon inhibition of TUC339 (90). LncRNA acetylserotonin
O-methyltransferase like antisense RNA 1 (ASMTL-AS1) was found to
be upregulated in HCC tissues, and exhibited increased expression
in tumors after radiofrequency ablation (RFA) insufficiency. The
expression levels of ASMTL-AS1 were found to be closely associated
with disease stage, metastasis and prognosis of HCC, and were found
to contribute to the malignancy of HCC cells. ASMTL-AS1 could be
packaged by exosomes and then contribute to the malignancy of HCC
cells via the nemo-like kinase (NLK)/yes-associated protein (YAP)
regulatory axis between cells even in residual HCC after RFA
insufficiency (91). High
expression of exosomal H19, an exosomal lncRNA, could enhance the
motility and proliferation of HCC cells while reducing their
apoptosis through binding to and sponging miR-520a-3p (92).
lncRNAs may play a role in HCC recurrence. A set of
deregulated lncRNAs showed potential to differentiate HCC from
cirrhotic tissue. Among these lncRNAs, cancer susceptibility 9
(CASC9) and lung cancer associated transcript 1 (LUCAT1) showed
upregulation in a subset of HCC-derived cell lines and in certain
HCC tissues, whose donors exhibited decreased recurrence after
surgery. LUCAT1 was found to directly sponge the onco-miR-181d-5p,
a novel regulatory element of liver cancer stem/progenitor cells.
Both lncRNAs, LUCAT1 and CASC9, were detected in secreted exosomes,
and upregulated circulating CASC9 levels were closely associated
with tumor size and HCC recurrence after surgery in human patients
(93).
circRNAs
circRNAs, first identified in RNA viruses in 1976 by
electron microscopy (94), are
another type of ncRNAs that form a covalently circled continuous
loop via the process of back-splicing, during which the downstream
splice donor site is joined with the upstream splice acceptor site
(95). Different to common linear
RNAs, circRNAs are characterized by featuring a covalently circled
continuous loops without 5′ or 3′polarities (96). Previous studies have demonstrated
that circRNAs are capable of absorbing miRNAs by stably
competitively binding and play a role as sponges of miRNAs to
regulate gene expression (97).
Numerous studies have demonstrated that circRNAs are abundant,
stable and one of the major components of exosomes (98). Increasing evidence has indicated
that dysregulation of exosomal circRNAs contributes to
tumorigenesis and progression in human liver cancer (99).
circRNAs secreted by adipose tissue were found to
regulate deubiquitination in HCC cells to facilitate cell growth.
Exosome circ-de-ubiquitination (circ-DB) levels were significantly
upregulated in patients with HCC who had higher body fat ratios.
circ-DB has been demonstrated to promote HCC cell growth and
inhibit DNA damage by suppressing miR-34a expression and
upregulating the de-ubiquitination of ubiquitin specific protease 7
(USP7). The effects of adipose-secreted exosomes on HCC cells could
be reversed by downregulation of circ-DB. These findings indicated
that exosome circRNAs secreted from adipocytes promoted tumor
growth and reduced DNA damage via miR-34a suppression and
activation of the USP7/cyclin A2 signaling pathway axis in HCC
cells (100). CircRNA
circ-0051443 was significantly downregulated in the tissues and
plasma exosomes of patients with HCC compared with the levels found
in those of healthy controls, and was mainly packaged into exosomes
in HCC cells. HCC cells could be distinguished from healthy control
cells by the presence of circ-0051443 in exosomes. Transmitting
circ-0051443 from healthy normal cells to HCC cells via exosomes
significantly suppressed the malignancy of HCC cells via cell
apoptosis increase and cell cycle arrest (101).
Numerous studies have reported critical roles of
exosomal circRNAs in HCC progression. The level of another circRNA,
circ-tripartite motif containing 33-12 (TRIM33-12), was also
decreased significantly in HCC tissues and cell lines.
circ-TRIM33-12 downregulation in HCC was closely associated with
HCC malignant behavior, and served as a promising risk factor of
recurrence-free and overall survival in patients with HCC after
surgery (102). A recent study
characterized the facilitation of MET expression by exosome
circ-prostaglandin reductase 1 (PTGR1) competing with the seed
sequence of miR-449a, a miRNA with roles in the inhibition of tumor
growth and metastasis in HCC (103). Further mechanistic analyses
confirmed that three different isoforms of exosomal circPTGR1
promoted HCC metastasis through the miR-449a/MET pathway (104). The level of circTMEM45A was
markedly reduced in HCC. circTMEM45A levels were closely associated
with clinicopathological features as well as decreased prognosis of
patients with HCC. Clinically, circTMEM45A expression levels were
markedly increased in exosomes from patients with HCC (105). Several studies have demonstrated
that exosomal circRNAs, particularly those derived from pathogenic
exosomes, may have an important influence on the pathophysiological
stage and processes of HCC; however, only a limited number of
circRNAs have been reported to establish critical functions or
clinical applications (26).
Exosomal circRNAs are likely to attract more interest in HCC
research.
mRNAs
mRNAs are mediators of genetic information
transferred from the cell nucleus to ribosomes in the cytoplasm, in
which mRNAs serve as templates for protein coding and synthesis
(106). Although relatively few
studies have focused on exosomal mRNAs, recent reports have been
published on the promising and increasing functions of exosomal
mRNAs in cancer development and prognosis (18,107), particularly in liver disease and
cancer (108). An increasing
number of exosomal mRNAs have been reported to be potential
biomarkers in HCC diagnosis. By using bioinformatics, a recent
study identified an HCC-exosomal RNA-based biomarker selection
panel, which was mostly associated with RAB11A gene expression and
its competing endogenous network, including lncRNA-RP11-513I15.6
and miR-1262. The expression levels of this network were validated
in the sera of 60 patients with HCC, and were compared with those
found in 42 patients with chronic hepatitis C virus infection and
18 healthy donors. Panels of three novel exosomal RNA-based genes,
including RAB11A, miR-1262 and lncRNA-RP11-513I15.6, displayed good
sensitivity and specificity in predicting and differentiating
patients with HCC from patients with chronic hepatitis C virus
infection and healthy donors. Of these three RNAs, the RAB11A mRNA
levels in serum were the most independent and promising prognosis
factor (109). Another cohort
study on patients with HCC, liver cirrhosis, chronic hepatitis B
and healthy controls quantified serum exosomal heterogeneous
nuclear ribonucleoprotein H1 (hnRNPH1) mRNA, and found that the
hnRNPH1 mRNA levels in serum exosomes from patients with HCC were
markedly upregulated compared with those in the other groups. The
mRNA levels of hnRNPH1 discriminated patients with HCC from
patients with chronic hepatitis B. Furthermore, the hnRNPH1 mRNA
levels in serum exosomes from patients with HCC were closely
associated with portal vein tumor emboli, lymph node metastasis,
Child-Pugh classification, overall survival and TNM stage. These
data indicated that hnRNPH1 mRNA levels in serum exosomes were
potentially a promising biomarker for HCC diagnosis in areas with a
high prevalence of patients with hepatitis B virus infection
(110). A recent study enrolled
a cohort consisting of healthy donors (n=159), as well as patients
with benign hepatic tumors (n=24), hepatitis (n=11), hepatic
cirrhosis (n=8), HCC (n=104), and breast (n=10), gastric (n=9),
kidney (n=15) and colorectal cancer (n=12). The authors detected
>10,000 extracellular vesicle-derived long RNAs, including
mRNAs, circRNAs and lncRNAs, in plasma. Among these RNAs, a
majority of the total mapped reads were mRNAs. Blood extracellular
vesicles contained a substantial fraction of intact mRNAs, and 8
extracellular vesicle-derived long RNAs were suggested to serve as
potential biomarkers for HCC diagnosis in areas with high
diagnostic efficiency (108).
6. Exosomes in the tumor microenvironment of
HCC
Tumor progression depends on the complexity and
heterogeneity of the tumor microenvironment, which consists of a
network of cellular and acellular constituents. Increasing evidence
reveals that this process is also impacted by exosome-mediated
cross-talks or communications within the tumor microenvironment
(111). Communications between
HCC cells and their microenvironmental cell components are
necessary mechanisms that modulate tumor development and
progression. As one of the major components of the ncRNA exosomal
cargo, miRNAs play multiple critical roles in modulating cellular
pathways in targeted cells and microenvironments, and in regulating
multiple processes associated with tumor progression, invasion and
metastasis, including EMT, invasion, angiogenesis, multi-drug
resistance and immune escape. Increasing evidence suggests that
exosomal miRNAs function as important players in the dynamic
crosstalk among cancerous, stromal and immune cells to establish
the microenvironment of tumorigenesis (112). In the HCC microenvironment,
exosomes participate in multiple steps during tumor progression and
radioresistance, and numerous cell types (particularly stromal and
inflammatory cells) within the microenvironment are involved in
these processes (113). Exosomes
have been reported to be critical in mediating the regulation of
the inflammatory microenvironment to promote cancer progression and
metastasis (114). HCC cells
exposed to arsenite secreted miR-155-rich exosomes that enhanced
inflammation and were positively correlated with IL-6 or IL-8
levels (115). The bidirectional
communication between HCC and its exosome-mediated inflammatory
microenvironment was also confirmed by a previous study, which
reported that the inflammatory microenvironment promoted
tumorigenesis while the tumor also created an inflammatory
environment that promoted its own development. The
β1-integrin/NF-κB signaling pathway in cancer-associated
fibroblasts (CAFs) was activated by exosomes with miR-1247-3p
secreted by high-metastatic HCC cells via directly targeting
β-1,4-galactosyltransferase 3. These activated CAFs further
promoted pleiotropic actions, including the secretion of
proinflammatory cytokines, such as IL-6 and IL-8, that governed
tumor progression (116).
Exosomes with miR-320a overexpression derived from CAFs showed
potential to inhibit tumorigenesis, thus suggesting a possible
cause of HCC progression mediated by CAFs by deficiency in
antitumor miR-320a in CAF-derived exosomes (117). CAFs could transfer miR-320a to
HCC cells via exosomes and then inhibit EMT, while loss of
antitumor miR-320a in CAF-derived exosomes could induce EMT and
promote tumor progression. In solid HCC tumors, a number of the
main components of the extracellular matrix, such as collagens,
laminins, fibronectin, glycosaminoglycans and proteoglycans, play
crucial roles in changing the phenotypic and functional
characteristics of HCC and stroma cells via various mechanisms,
including exosome-involved interaction (118). Beside these studies, additional
evidence in recent years has implied the roles of exosomes in
establishing and modifying the HCC microenvironment in a manner
that enhances tumor progression, metastasis and tumor
resistance.
7. Other extracellular vesicle subtypes in
HCC progression and radioresistance
Besides exosomes, which are well studied, the other
two major extracellular vesicle subtypes, microvesicles and
apoptotic bodies (which are differentiated based upon their
biogenesis, release pathways, size, content and function) (119), are less studied in the context
of the development, progression and radioresistance of HCC.
Microvesicles derived from human liver stem cells induced the in
vitro proliferation and apoptosis resistance of hepatocytes by
transporting mRNA into hepatocytes (120). Microvesicles derived from human
adult liver stem cells may reprogram in vitro HepG2 liver
cancer and primary HCC cells by inhibiting their growth and
survival. In vivo intratumor administration of microvesicles
induced the regression of ectopic tumors developed in SCID mice by
a mechanism involving the delivery of miRNAs from human adult liver
stem cell-derived microvesicles to tumor cells according to various
studies. The antitumor effect of adult liver stem cell-derived
microvesicles was also observed in tumors other than liver, such as
lymphoblastoma and glioblastoma (121). These studies suggest that the
delivery of selected miRNAs by microvesicles derived from stem
cells may inhibit tumor growth and stimulate apoptosis. Apoptotic
bodies, another major class of extracellular vesicles released as a
product of apoptotic cell disassembly as well as insufficient
apoptosis, have been associated with the development and
progression of tumors of the liver and biliary tree (122). Accumulating evidence suggests a
defective apoptotic process in human HCC, and numerous potential
therapeutic strategies that induce the apoptotic process in various
ways have the potential to aid the management of HCC (123).
8. Conclusions and perspectives
Exosomes are emerging as novel modes of
intercellular communications in human homeostasis and diseases.
Exosomes act as bioactive components that transfer proteins,
lipids, DNA, mRNAs, ncRNAs and recently identified molecules from
contributor to recipient cells, which leads to the exchange of
genetic information and the transcriptional re-programming of
recipient cells (22,124). Circulating exosomes, as well as
their main components, may potentially be used as liquid biopsies,
and are promising biomarkers for early detection, diagnosis and
multi-therapy in patients with cancer (125), such as HCC (59,126). Clinically, HCC is considered a
radioresistant tumor, and increasing evidence suggests a critical
role of HCC-derived exosomes in radioresistance (13,127), due to their influence on tumor
initiation, growth, progression, metastasis, drug radioresistance
and recurrence. Despite these adverse conditions in HCC therapy, as
increasing novel technologies are developed and to monitor and
modify exosomes in cancer progression and radiotherapy,
exosome-associated HCC diagnosis, therapies and post-surgery
prediction are becoming more promising, and exosome-containing
molecules are valuable in HCC research as novel biomarkers.
Availability of data and materials
Not applicable.
Authors' contributions
QF, YY and CW designed and wrote the manuscript. YZ
and SZ collected and summarized the related papers and references.
CW revised and conceived the final approval of the version to be
submitted and obtaining of the funding.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The current study was funded by the Natural Science Foundation
of Liaoning Province (2021-MS-181 to CW) and the Young and
Middle-Aged Scientific and Technological Talents Support Program of
Shenyang City (RC200554 to CW).
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
mRNA
|
messenger RNA
|
|
miRNA
|
micro-RNA
|
|
lncRNA
|
long non-coding RNA
|
|
circRNA
|
circular RNA
|
|
ncRNA
|
non-coding RNA
|
|
HEV
|
hepatitis E virus
|
|
ctDNA
|
circulating tumor DNA
|
|
EMT
|
epithelial-mesenchymal transition
|
|
TNM
|
tumor, node, metastasis
|
|
ceRNA
|
competitive endogenous RNA
|
References
|
1
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nair H, Simoes EA, Rudan I, Gessner BD,
Azziz-Baumgartner E, Zhang JSF, Feikin DR, Mackenzie GA, Moiïsi JC,
Roca A, et al: Global and regional burden of hospital admissions
for severe acute lower respiratory infections in young children in
2010: A systematic analysis. Lancet. 381:1380–1390. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Global Burden of Disease Liver Cancer
Collaboration; Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu
MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, et al: The
burden of primary liver cancer and underlying etiologies from 1990
to 2015 at the global, regional, and national level: Results from
the global burden of disease study 2015. JAMA Oncol. 3:1683–1691.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J and Fan D: Hepatitis B in China.
Lancet. 369:1582–1583. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laursen L: A preventable cancer. Nature.
516:S2–S3. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
European Association for the study of the
Liver and European Organisation for research and treatment of
Cancer: EASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol. 56:908–943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ishizawa T, Hasegawa K, Aoki T, Takahashi
M, Inoue Y, Sano K, Imamura H, Sugawara Y, Kokudo N and Makuuchi M:
Neither multiple tumors nor portal hypertension are surgical
contraindications for hepatocellular carcinoma. Gastroenterology.
134:1908–1916. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Y and Feng M: Radiotherapy for
hepatocellular carcinoma. Semin Radiat Oncol. 28:277–287. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bang A and Dawson LA: Radiotherapy for
HCC: Ready for prime time? JHEP Rep. 1:131–137. 2019. View Article : Google Scholar
|
|
12
|
Ohri N, Dawson LA, Krishnan S, Seong J,
Cheng JC, Sarin SK, Kinkhabwala M, Ahmed MM, Vikram B, Coleman CN
and Guha C: Radiotherapy for hepatocellular carcinoma: New
indications and directions for future study. J Natl Cancer Inst.
108:djw1332016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ni J, Bucci J, Malouf D, Knox M, Graham P
and Li Y: Exosomes in cancer radioresistance. Front Oncol.
9:8692019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barker HE, Paget JT, Khan AA and
Harrington KJ: The tumour microenvironment after radiotherapy:
Mechanisms of resistance and recurrence. Nat Rev Cancer.
15:409–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chan R, Sethi P, Jyoti A, McGarry R and
Upreti M: Investigating the radioresistant properties of lung
cancer stem cells in the context of the tumor microenvironment.
Radiat Res. 185:169–181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Swartz MA, Iida N, Roberts EW, Sangaletti
S, Wong MH, Yull FE, Coussens LM and DeClerck YA: Tumor
microenvironment complexity: Emerging roles in cancer therapy.
Cancer Res. 72:2473–2480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Son B, Lee S, Youn H, Kim E, Kim W and
Youn B: The role of tumor microenvironment in therapeutic
resistance. Oncotarget. 8:3933–3945. 2017. View Article : Google Scholar :
|
|
18
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar :
|
|
19
|
Jeppesen DK, Fenix AM, Franklin JL,
Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q,
Evans R, et al: Reassessment of exosome composition. Cell.
177:428–445.e18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tai YL, Chen KC, Hsieh JT and Shen TL:
Exosomes in cancer development and clinical applications. Cancer
Sci. 109:2364–2374. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dai J, Su Y, Zhong S, Cong L, Liu B, Yang
J, Tao Y, He Z, Chen C and Jiang Y: Exosomes: Key players in cancer
and potential therapeutic strategy. Signal Transduct Target Ther.
5:1452020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sato S, Zhu XL and Sly WS: Carbonic
anhydrase Isozymes IV and II in urinary membranes from carbonic
anhydrase II-deficient patients. Proc Natl Acad Sci USA.
87:6073–6076. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang L and Yu D: Exosomes in cancer
development, metastasis, and immunity. Biochim Biophys Acta Rev
Cancer. 1871:455–468. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen W, Mao Y, Liu C, Wu H and Chen S:
Exosome in hepatocellular carcinoma: An update. J Cancer.
12:2526–2536. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang HB, Lu ZM and Zhao XX: Tumorigenesis,
diagnosis, and therapeutic potential of exosomes in liver cancer. J
Hematol Oncol. 12:1332019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Keerthikumar S, Chisanga D, Ariyaratne D,
Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M,
Chilamkurti N, et al: ExoCarta: A Web-Based compendium of exosomal
cargo. J Mol Biol. 428:688–692. 2016. View Article : Google Scholar :
|
|
29
|
Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon
T, Kim BC, Benci JL, DeMichele AM, Tchou J, Marcotrigiano J and
Minn AJ: Exosome RNA unshielding couples stromal activation to
pattern recognition receptor signaling in cancer. Cell.
170:352–366.e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li I and Nabet BY: Exosomes in the tumor
microenvironment as mediators of cancer therapy resistance. Mol
Cancer. 18:322019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Conigliaro A, Costa V, Lo Dico A, Saieva
L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M,
et al: CD90+liver cancer cells modulate endothelial cell phenotype
through the release of exosomes containing H19 lncRNA. Mol Cancer.
14:1552015. View Article : Google Scholar
|
|
32
|
Ridder K, Keller S, Dams M, Rupp AK,
Schlaudraff J, Del Turco D, Starmann J, Macas J, Karpova D, Devraj
K, et al: Extracellular vesicle-mediated transfer of genetic
information between the hematopoietic system and the brain in
response to inflammation. PLoS Biol. 12:–e1001874. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Skotland T, Sandvig K and Llorente A:
Lipids in exosomes: Current knowledge and the way forward. Prog
Lipid Res. 66:30–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li WH, Li CY, Zhou T, Liu X, Liu X, Li X
and Chen D: Role of exosomal proteins in cancer diagnosis. Mol
Cancer. 16:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cai J, Han Y, Ren HM, Chen C, He D, Zhou
L, Eisner GM, Asico LD, Jose PA and Zeng C: Extracellular
vesicle-mediated transfer of donor genomic DNA to recipient cells
is a novel mechanism for genetic influence between cells. J Mol
Cell Biol. 5:227–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thakur BK, Zhang H, Becker A, Matei I,
Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et
al: Double-stranded DNA in exosomes: A novel biomarker in cancer
detection. Cell Res. 24:766–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wong CM, Tsang FHC and Ng IOL: Non-coding
RNAs in hepatocellular carcinoma: Molecular functions and
pathological implications. Nat Rev Gastro Hepat. 15:137–151. 2018.
View Article : Google Scholar
|
|
38
|
He M, Qin H, Poon TC, Sze SC, Ding X, Co
NN, Ngai SM, Chan TF and Wong N: Hepatocellular carcinoma-derived
exosomes promote motility of immortalized hepatocyte through
transfer of oncogenic proteins and RNAs. Carcinogenesis.
36:1008–1018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang S, Xu M, Li X, Su X, Xiao X, Keating
A and Zhao RC: Exosomes released by hepatocarcinoma cells endow
adipocytes with tumor-promoting properties. J Hematol Oncol.
11:822018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee HY, Chen CK, Ho CM, Lee SS, Chang CY,
Chen KJ and Jou YS: EIF3C-enhanced exosome secretion promotes
angiogenesis and tumorigenesis of human hepatocellular carcinoma.
Oncotarget. 9:13193–13205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang S, Chen G, Lin X, Xing X, Cai Z, Liu
X and Liu J: Role of exosomes in hepatocellular carcinoma cell
mobility alteration. Oncol Lett. 14:8122–8131. 2017.PubMed/NCBI
|
|
42
|
Zhang J, Lu S, Zhou Y, Meng K, Chen Z, Cui
Y, Shi Y, Wang T and He QY: Motile hepatocellular carcinoma cells
preferentially secret sugar metabolism regulatory proteins via
exosomes. Proteomics. 17:17001032017. View Article : Google Scholar
|
|
43
|
Conde-Vancells J, Rodriguez-Suarez E,
Gonzalez E, Berisa A, Gil D, Embade N, Valle M, Luka Z, Elortza F,
Wagner C, et al: Candidate biomarkers in exosome-like vesicles
purified from rat and mouse urine samples. Proteom Clin Appl.
4:416–425. 2010. View Article : Google Scholar
|
|
44
|
Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen
Q, Chen Y, Zhang J, Wang J, Wei T, et al: Primary tumor-derived
exosomes facilitate metastasis by regulating adhesion of
circulating tumor cells via SMAD3 in liver cancer. Oncogene.
37:6105–6118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ye L, Zhang Q, Cheng Y, Chen X, Wang G,
Shi M, Zhang T, Cao Y, Pan H, Zhang L, et al: Tumor-derived
exosomal HMGB1 fosters hepatocellular carcinoma immune evasion by
promoting TIM-1 + regulatory B cell expansion. J Immunother Cancer.
6:1452018. View Article : Google Scholar
|
|
46
|
Wang X, Shen H, Zhangyuan G, Huang R,
Zhang W, He Q, Jin K, Zhuo H, Zhang Z, Wang J, et al: 14-3-3ζ
delivered by hepatocellular carcinoma-derived exosomes impaired
anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death
Dis. 9:1592018. View Article : Google Scholar
|
|
47
|
Li M, Lu Y, Xu Y, Wang J, Zhang C, Du Y,
Wang L, Li L, Wang B, Shen J, et al: Horizontal transfer of
exosomal CXCR4 promotes murine hepatocarcinoma cell migration,
invasion and lymphangiogenesis. Gene. 676:101–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dai WJ, Wang YL, Yang TX, Wang J, Wu WC
and Gu JX: Downregulation of exosomal CLEC3B in hepatocellular
carcinoma promotes metastasis and angiogenesis via AMPK and VEGF
signals. Cell Commun Signal. 17:1132019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qiu QC, Wang L, Jin SS, Liu GF, Liu J, Ma
L, Mao RF, Ma YY, Zhao N, Chen M and Lin BY: CHI3L1 promotes tumor
progression by activating TGF-β signaling pathway in hepatocellular
carcinoma. Sci Rep. 8:150292018. View Article : Google Scholar
|
|
50
|
Gai X, Tang B, Liu F, Wu Y, Wang F, Jing
Y, Huang F, Jin D, Wang L and Zhang H: mTOR/miR-145-regulated
exosomal GOLM1 promotes hepatocellular carcinoma through augmented
GSK-3 beta/MMPs. J Genet Genomics. 46:235–245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Skotland T, Sagini K, Sandvig K and
Llorente A: An emerging focus on lipids in extracellular vesicles.
Adv Drug Deliv Rev. 159:308–321. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Skotland T, Ekroos K, Kauhanen D, Simolin
H, Seierstad T, Berge V, Sandvig K and Llorente A: Molecular lipid
species in urinary exosomes as potential prostate cancer
biomarkers. Eur J Cancer. 70:122–132. 2017. View Article : Google Scholar
|
|
53
|
Lydic TA, Townsend S, Adda CG, Collins C,
Mathivanan S and Reid GE: Rapid and comprehensive 'Shotgun'
lipidome profiling of colorectal cancer cell derived exosomes.
Methods. 87:83–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Trajkovic K, Hsu C, Chiantia S, Rajendran
L, Wenzel D, Wieland F, Schwille P, Brügger B and Simons M:
Ceramide triggers budding of exosome vesicles into multivesicular
Endosomes. Science. 319:1244–1247. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chapuy-Regaud S, Dubois M,
Plisson-Chastang C, Bonnefois T, Lhomme S, Bertrand-Michel J, You
B, Simoneau S, Gleizes PE, Flan B, et al: Characterization of the
lipid envelope of exosome encapsulated HEV particles protected from
the immune response. Biochimie. 141:70–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Haraszti RA, Didiot MC, Sapp E, Leszyk J,
Shaffer SA, Rockwell HE, Gao F, Narain NR, DiFiglia M, Kiebish MA,
et al: High-resolution proteomic and lipidomic analysis of exosomes
and microvesicles from different cell sources. J Extracell
Vesicles. 5:325702016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sadovska L, Eglitis J and Line A:
Extracellular vesicles as biomarkers and therapeutic targets in
breast cancer. Anticancer Res. 35:6379–6390. 2015.PubMed/NCBI
|
|
58
|
Alix-Panabieres C and Pantel K: Clinical
applications of circulating tumor cells and circulating tumor DNA
as liquid biopsy. Cancer Discov. 6:479–491. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li X, Li C, Zhang L, Wu M, Cao K, Jiang F,
Chen D, Li N and Li W: The significance of exosomes in the
development and treatment of hepatocellular carcinoma. Mol Cancer.
19:12020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bernard V, Kim DU, San Lucas FA, Castillo
J, Allenson K, Mulu FC, Stephens BM, Huang J, Semaan A, Guerrero
PA, et al: Circulating nucleic acids are associated with outcomes
of patients with pancreatic cancer. Gastroenterology.
156:108–118.e4. 2019. View Article : Google Scholar
|
|
61
|
Yan LL, Chen YH, Zhou JY, Zhao H, Zhang HH
and Wang GQ: Diagnostic value of circulating cell-free DNA levels
for hepatocellular carcinoma. Int J Infect Dis. 67:92–97. 2018.
View Article : Google Scholar
|
|
62
|
Slack FJ and Chinnaiyan AM: The role of
Non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Vickers KC, Palmisano BT, Shoucri BM,
Shamburek RD and Remaley AT: MicroRNAs are transported in plasma
and delivered to recipient cells by high-density lipoproteins. Nat
Cell Biol. 13:423–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK,
Pritchard CC, Gibson DF, Mitchell PS, Bennett CF,
Pogosova-Agadjanyan EL, Stirewalt DL, et al: Argonaute2 complexes
carry a population of circulating microRNAs independent of vesicles
in human plasma. Proc Natl Acad Sci USA. 108:5003–5008. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal MicroRNA: Trafficking, sorting, and
function. Genom Proteom Bioinf. 13:17–24. 2015. View Article : Google Scholar
|
|
68
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lee YR, Kim G, Tak WY, Jang SY, Kweon YO,
Park JG, Lee HW, Han YS, Chun JM, Park SY and Hur K: Circulating
exosomal noncoding RNAs as prognostic biomarkers in human
hepatocellular carcinoma. Int J Cancer. 144:1444–1452. 2019.
View Article : Google Scholar
|
|
70
|
Tian XP, Wang CY, Jin XH, Li M, Wang FW,
Huang WJ, Yun JP, Xu RH, Cai QQ and Xie D: Acidic microenvironment
Up-Regulates exosomal miR-21 and miR-10b in early-stage
hepatocellular carcinoma to promote cancer cell proliferation and
metastasis. Theranostics. 9:1965–1979. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lin XJ, Chong YT, Guo ZW, Xie C, Yang XJ,
Zhang Q, Li SP, Xiong Y, Yuan Y, Min J, et al: A serum microRNA
classifier for early detection of hepatocellular carcinoma: A
multicentre, retrospective, longitudinal biomarker identification
study with a nested case-control study. Lancet Oncol. 16:804–815.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y,
Zheng L and Zhuang SM: Hepatocellular carcinoma cell-secreted
exosomal MicroRNA-210 promotes angiogenesis in vitro and in vivo.
Mol Ther Nucleic Acids. 11:243–252. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y
and Cang SD: Mechanism of exosomal microRNA-224 in development of
hepatocellular carcinoma and its diagnostic and prognostic value.
World J Gastroentero. 25:1890–1898. 2019. View Article : Google Scholar
|
|
74
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar
|
|
75
|
Pathania AS and Challagundla KB: Exosomal
long Non-coding RNAs: Emerging players in the tumor
microenvironment. Mol Ther Nucleic Acids. 23:1371–1383. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Prensner JR and Chinnaiyan AM: The
Emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bolha L, Ravnik-Glavac M and Glavac D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Takahashi K, Yan IK, Kogure T, Haga H and
Patel T: Extracellular vesicle-mediated transfer of long non-coding
RNA ROR modulates chemosensitivity in human hepatocellular cancer.
FEBS Open Bio. 4:458–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Takahashi K, Yan IK, Wood J, Haga H and
Patel T: Involvement of extracellular vesicle long noncoding RNA
(linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer
Res. 12:1377–1387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chen SY, Teng SC, Cheng TH and Wu KJ:
miR-1236 regulates hypoxia-induced epithelial-mesenchymal
transition and cell migration/invasion through repressing SENP1 and
HDAC3. Cancer Lett. 378:59–67. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li BG, Mao R, Liu CF, Zhang WH, Tang Y and
Guo Z: LncRNA FAL1 promotes cell proliferation and migration by
acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells.
Life Sci. 197:122–129. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yao ZC, Jia CC, Tai Y, Liang H, Zhong Z,
Xiong Z, Deng M and Zhang Q: Serum exosomal long noncoding RNAs
lnc-FAM72D3 and lnc-EPC14 as diagnostic biomarkers for
hepatocellular carcinoma. Aging (Albany NY). 12:11843–11863. 2020.
View Article : Google Scholar
|
|
83
|
Hou YC, Yu Z, Tam NL, Huang S, Sun C, Wang
R, Zhang X, Wang Z, Ma Y, He X and Wu L: Exosome-related lncRNAs as
predictors of HCC patient survival: A prognostic model. Am J Transl
Res. 10:1648–1662. 2018.PubMed/NCBI
|
|
84
|
Zhang C, Yang X, Qi Q, Gao YH, Wei Q and
Han SW: lncRNA-HEIH in serum and exosomes as a potential biomarker
in the HCV-related hepatocellular carcinoma. Cancer Biomark.
21:651–659. 2018. View Article : Google Scholar
|
|
85
|
Xu H, Chen YM, Dong XY and Wang XJ: Serum
exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for
the diagnosis and prognosis of hepatocellular carcinoma. Cancer
Epidem Biomar. 27:710–716. 2018. View Article : Google Scholar
|
|
86
|
Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y,
Guo Z, Bai T, Dong L, Wei C, et al: Serum and exosome long non
coding RNAs as potential biomarkers for hepatocellular carcinoma. J
Cancer. 9:2631–2639. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ma X, Yuan T, Yang C, Wang Z, Zang Y, Wu L
and Zhuang L: X-inactive-specific transcript of peripheral blood
cells is regulated by exosomal Jpx and acts as a biomarker for
female patients with hepatocellular carcinoma. Ther Adv Med Oncol.
9:665–677. 2017. View Article : Google Scholar
|
|
88
|
Zhuang LK, Yang YT, Ma X, Han B, Wang ZS,
Zhao QY, Wu LQ and Qu ZQ: MicroRNA-92b promotes hepatocellular
carcinoma progression by targeting Smad7 and is mediated by long
non-coding RNA XIST. Cell Death Dis. 7:e22032016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kogure T, Yan IK, Lin WL and Patel T:
Extracellular vesicle-mediated transfer of a novel long noncoding
RNA TUC339: A mechanism of intercellular signaling in human
hepatocellular cancer. Genes Cancer. 4:261–272. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li X, Lei Y, Wu M and Li N: Regulation of
macrophage activation and polarization by HCC-Derived Exosomal
lncRNA TUC339. Int J Mol Sci. 19:29582018. View Article : Google Scholar :
|
|
91
|
Ma D, Gao X, Liu Z, Lu X, Ju H and Zhang
N: Exosome-transferred long non-coding RNA ASMTL-AS1 contributes to
malignant phenotypes in residual hepatocellular carcinoma after
insufficient radiofrequency ablation. Cell Prolif. 53:e127952020.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang D, Xing N, Yang T, Liu J, Zhao H, He
J, Ai Y and Yang J: Exosomal lncRNA H19 promotes the progression of
hepatocellular carcinoma treated with Propofol via
miR-520a-3p/LIMK1 axis. Cancer Med. 9:7218–7230. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ji JF, Yamashita T, Budhu A, Forgues M,
Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al:
Identification of MicroRNA-181 by genome-wide screening as a
critical player in EpCAM-Positive hepatic cancer stem cells.
Hepatology. 50:472–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Seimiya T, Otsuka M, Iwata T, Shibata C,
Tanaka E, Suzuki T and Koike K: Emerging roles of exosomal circular
RNAs in cancer. Front Cell Dev Biol. 8:5683662020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Li SS, Yao JP, Xie MJ, Liu YN and Zheng M:
Exosomal miRNAs in hepatocellular carcinoma development and
clinical responses. J Hematol Oncol. 11:542018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Geng X, Lin X, Zhang Y, Li Q, Guo Y, Fang
C and Wang H: Exosomal circular RNA sorting mechanisms and their
function in promoting or inhibiting cancer. Oncol Lett.
19:3369–3380. 2020.PubMed/NCBI
|
|
100
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu
K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted
from adipocytes promotes the growth of hepatocellular carcinoma by
targeting deubiquitination-related USP7. Oncogene. 38:2844–2859.
2019. View Article : Google Scholar :
|
|
101
|
Chen W, Quan Y, Fan S, Wang H, Liang J,
Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular
RNA hsa_ circ_0051443 suppresses hepatocellular carcinoma
progression. Cancer Lett. 475:119–128. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang PF, Wei CY, Huang XY, Peng R, Yang
X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, et al: Circular RNA
circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress
hepatocellular carcinoma progression. Mol Cancer. 18:1052019.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Chen SP, Liu BX, Xu J, Pei XF, Liao YJ,
Yuan F and Zheng F: MiR-449a suppresses the epithelial-mesenchymal
transition and metastasis of hepatocellular carcinoma by multiple
targets. BMC Cancer. 15:7062015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang GY, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang TT, Jing B, Bai YX, Zhang Y and Yu
HY: Circular RNA circTMEM45A Acts as the Sponge of MicroRNA-665 to
promote hepatocellular carcinoma progression. Mol Ther Nucleic
Acids. 22:285–297. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Shyu AB, Wilkinson MF and van Hoof A:
Messenger RNA regulation: To translate or to degrade. Embo J.
27:471–481. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yu SL, Li YC, Liao Z, Wang Z, Wang Z, Li
Y, Qian L, Zhao J, Zong H, Kang B, et al: Plasma extracellular
vesicle long RNA profiling identifies a diagnostic signature for
the detection of pancreatic ductal adenocarcinoma. Gut. 69:540–550.
2020. View Article : Google Scholar
|
|
108
|
Li Y, Zhao J, Yu S, Wang Z, He X, Su Y,
Guo T, Sheng H, Chen J, Zheng Q, et al: Extracellular vesicles long
RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human
blood as potential biomarkers for cancer diagnosis. Clin Chem.
65:798–808. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Abd El Gwad A, Matboli M, El-Tawdi A,
Habib EK, Shehata H, Ibrahim D and Tash F: Role of exosomal
competing endogenous RNA in patients with hepatocellular carcinoma.
J Cell Biochem. 119:8600–8610. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Xu H, Dong XY, Chen YM and Wang XJ: Serum
exosomal hnRNPH1 mRNA as a novel marker for hepatocellular
carci-noma. Clin Chem Lab Med. 56:479–484. 2018. View Article : Google Scholar
|
|
111
|
He R, Wang Z, Shi W, Yu L, Xia H, Huang Z,
Liu S, Zhao X, Xu Y, Yam JWP and Cui Y: Exosomes in hepatocellular
carcinoma microenvironment and their potential clinical application
value. Biomed Pharmacother. 138:1115292021. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Pascut D, Pratama MY, Vo NVT, Masadah R
and Tiribelli C: The Crosstalk between tumor cells and the
microenvironment in hepatocellular carcinoma: The role of exosomal
microRNAs and their clinical implications. Cancers (Basel).
12:8232020. View Article : Google Scholar
|
|
113
|
Wu Q, Zhou L, Lv D, Zhu X and Tang H:
Exosome-mediated communication in the tumor microenvironment
contributes to hepatocellular carcinoma development and
progression. J Hematol Oncol. 12:532019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chen X, Chi H, Zhao X, Pan R, Wei Y and
Han Y: Role of exosomes in immune microenvironment of
hepatocellular carcinoma. J Oncol. 2022:25210252022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chen C, Luo F, Liu X, Lu L, Xu H, Yang Q,
Xue J, Shi L, Li J, Zhang A and Liu Q: NF-kB-regulated exosomal
miR-155 promotes the inflammation associated with arsenite
carcinogenesis. Cancer Lett. 388:21–33. 2017. View Article : Google Scholar
|
|
116
|
Fang T, Lv H, Lv G, Li T, Wang C, Han Q,
Yu L, Su B, Guo L, Huang S, et al: Tumor-derived exosomal
miR-1247-3p induces cancer-associated fibroblast activation to
foster lung metastasis of liver cancer. Nat Commun. 9:1912018.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhang Z, Li X, Sun W, Yue S, Yang J, Li J,
Ma B, Wang J, Yang X, Pu M, et al: Loss of exosomal miR-320a from
cancer-associated fibroblasts contributes to HCC proliferation and
metastasis. Cancer Lett. 397:33–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Goessler UR, Hormann K and Riedel F:
Tissue engineering with chondrocytes and function of the
extracellular matrix (Review). Int J Mol Med. 13:505–513.
2004.PubMed/NCBI
|
|
119
|
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles. 4:270662015.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Herrera MB, Fonsato V, Gatti S, Deregibus
MC, Sordi A, Cantarella D, Calogero R, Bussolati B, Tetta C and
Camussi G: Human liver stem cell-derived microvesicles accelerate
hepatic regeneration in hepatectomized rats. J Cell Mol Med.
14:1605–1618. 2010. View Article : Google Scholar
|
|
121
|
Fonsato V, Collino F, Herrera MB,
Cavallari C, Deregibus MC, Cisterna B, Bruno S, Romagnoli R,
Salizzoni M, Tetta C and Camussi G: Human liver stem cell-derived
microvesicles inhibit hepatoma growth in SCID mice by delivering
antitumor microRNAs. Stem Cells. 30:1985–1998. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Fabregat I, Roncero C and Fernandez M:
Survival and apoptosis: A dysregulated balance in liver cancer.
Liver Int. 27:155–162. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kountouras J, Zavos C and Chatzopoulos D:
Apoptosis in hepatocellular carcinoma. Hepatogastroenterology.
50:242–249. 2003.PubMed/NCBI
|
|
124
|
Thery C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extra-cellular vesicles 2018 (MISEV2018): A position statement of
the International Society for Extracellular Vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar
|
|
125
|
McAndrews KM and Kalluri R: Mechanisms
associated with biogenesis of exosomes in cancer. Mol Cancer.
18:522019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sasaki R, Kanda T, Yokosuka O, Kato N,
Matsuoka S and Moriyama M: Exosomes and hepatocellular carcinoma:
From bench to bedside. Int J Mol Sci. 20:14062019. View Article : Google Scholar :
|
|
127
|
Hennequin C, Quero L and Rivera S:
Radiosensitivity of hepatocellular carcinoma. Cancer Radiother.
15:39–42. 2011.In French. View Article : Google Scholar : PubMed/NCBI
|