Prostate cancer (PCa) is one of the most common
cancers in men worldwide and there is a great need for a method of
accurately identifying drugs for individualized therapy, especially
for those with existing drug-resistant prostate cancer. The
American Cancer Society estimates >190,000 new cases and
>33,000 mortalities from PCa in 2020 (1). Of newly diagnosed PCa, ~81% will be
localized (2). The primary
recommended treatment for newly diagnosed localized cancer is
radiation, radical prostatectomy, brachytherapy, or active
monitoring for those with low risk (3). Patients with localized but high-risk
disease are recommended to have surgery with lymph node dissection
or androgen deprivation therapy (ADT) with radiation. Those with
metastatic disease are recommended ADT and possibly radiation
(3). Although treatment with ADT
has high initial response rate, resistance will most likely occur
within a year (4). Subsequently,
these metastatic castrate resistant prostate cancers (mCRPC) are
usually treated with androgen receptor targeted therapy,
chemotherapy, immunotherapy or bone targeted therapy such as
bisphosphonates, RANKL antibody, alpha-emitting calcium mimetic for
osseous metastases (3–5).
One approach of improving treatment can be a
personalized medicine treatment plan based on specific genetic
lesions. In the past few decades, genotypic screening for mutated
genes that lead to inherited predisposition to cancer have been
developed for several conditions (9–11). For
PCa, germline genetic testing is currently recommended if there is
personal and family history, Ashkenazi Jewish ancestry and BRCA 1/2
and Lynch syndrome mutation (3).
However, genetic testing in the absence of family history will
provide a low yield for PCa (3).
Furthermore, patients may not benefit from screening due to
variants with uncertain significance, missed mutations, or lack of
mutation-specific intervention (7).
Testing for somatic mutation in tumors is used in the hope of
identifying possible targets for therapy. However, this approach is
limited by the uncertainty in predicting response based on
mutations and the availability of interventions (8).
Another approach for personalized or individualized
therapy is explored in the current review: Phenotypic or empiric
drug screening. This is similar to the concept of antimicrobial
susceptibility testing, in which specific antimicrobial agents that
will be most effective for individual patients are identified
(12). In cancer therapy, a living
organoid biobank that matched the wide varieties of breast cancer
phenotypes, including histopathology, hormone receptor status and
oncogene activation, such as HER2, was described by Sachs et
al (13). Such an organoid
biobank may allow in vitro phenotypic screening for cancer
treatment. In addition, an organoid biobank can be a promising
model for drug discovery, biological insights and translational and
clinical research (14,15). Thus, a PCa model that can accurately
represent an individual's tumor and its microenvironment will be
useful for screening for effective therapies. Such a model is
especially applicable for PCa clinical therapeutic strategy since
it is a relatively slowly growing cancer.
In the current review, three potential screening
models for prostate cancer will be reviewed and the ideal model
that can apply to the treatment of mCRPC will be discussed:
Conditional reprogramming cells, patient derived xenografts and
organoids. Their experimental conditions, advantages and
disadvantages and what could be improved to achieve the most
effective screening for individualized PCa therapy are discussed
below.
Individualizing anticancer chemotherapy is best
facilitated via a model that can accurately represent the tumor of
the patient and its progression, such as from being localized to
exhibiting metastases. Such cancer screening models need to take
into consideration the complexity of the tumor and its
microenvironment to best replicate the in vivo environment
in vitro. It is impossible with the current in vitro
technology to completely mimic the in vivo conditions.
However, the following features are considered essential and should
be included in a representative in vitro model.
PCa tumors are heterogeneous with variations in
genetic abnormalities, gene expression, epigenetic regulation and
responses to therapeutics (7).
Prostate cells are also normally dependent on testosterone and have
androgen receptors (AR) (16–19).
Normal development as well as cancer growth and survival is
effected by AR (20). While
including all these variations in a dynamic manner is impossible
with current in vitro technology, a good in vitro
model should incorporate as a number of these characteristics as
possible.
The prostate gland is made of epithelial and stromal
cell types. Epithelial cells include luminal and basal cells
(21,22). Secretory products are released into
the gland lumen by luminal cells. Luminal cells and ductal
structure are derived from basal cells (23). Studies have suggested that luminal
and basal cells are sporadically bipotent and have stem cell-like
properties (24–26). Basal cells are more efficient at
forming organoids and providing self-renewal, whereas luminal cells
are more ready to differentiate into ducts and acini. PCa can be
derived from both cell types (7,27,28)
and also, rarely, from neuroendocrine cells (29–34).
While each model may not need all of these cells, it is important
that the type of cells that correspond to each individual's unique
cancer is exhibited in a model.
There is a fibromuscular stroma consisting of smooth
muscle, fibroblasts and elastic fibers. In addition, there are
blood vessels, peripheral nerves, macrophages and white blood cells
(29,34). A co-evolution is undergone by tumor
cells and the associated stroma. Growth factors and proteases are
released by cancer cells, causing angiogenesis and inflammation,
activating the surrounding stroma, which in turn secretes
additional growth factors, proteases and pro-migratory
extracellular matrix (ECM) components (Table II) (35,36).
The stromal cells are influenced by cancer cells to become more
supportive of tumor progression, which can in turn, increase the
malignancy of cancer cells or progression to becoming drug- and
castration-resistant (35–37). Cancer cells are highly heterogeneous
due to genomic instability and selection pressure from the
microenvironment (37). Another
phenomenon based in interaction with the immune system and the
microenvironment is immune escape. It is the failure of the immune
system to eliminate cancer, allowing it to continue developing into
metastatic cancer (38). Tumor
cells may lose their antigenicity through immune selection, lose
their immunogenicity through expressing immunoinhibitory molecules,
or by creating a suppressive microenvironment for immune cells
(38,39). While it may not be currently
possible to replicate all the stromal interactions with cancer
cells, the components representing key signals and relevant factors
released by the stromal cells should be added to the growth
medium.
Primary cells in culture usually undergo senescence,
a cessation of proliferation and changes in the metabolism and
phenotype of the cells after a few passages (40). Thus, unlike microorganisms, direct
culture and sensitivity approach utilized for identifying specific
antimicrobial agents may not work for cancer therapeutic screening.
Although numerous immortal cancer cell lines, including PCa cell
lines such as PC3, LnCap, DU145 have been developed and have been
routinely used for initial drug development purposes, they may not
be representative of the primary tumor and lack 3D structure and
broad representation of different tumor types and subtypes
(7,16–18,41–49).
In the past decade, long term proliferation without
changing most of the fundamental genetic makeup and expression of
cells has been made possible by a newly developed approach called
conditional reprogramming (CR) (44,48,50).
Establishment of cultures of almost all types of epithelial cells
is enabled by CR. Considerable interest has been generated in its
possible applications such as establishment of disease models,
therapeutic drug assessment and new platforms for basic and
translational research (48).
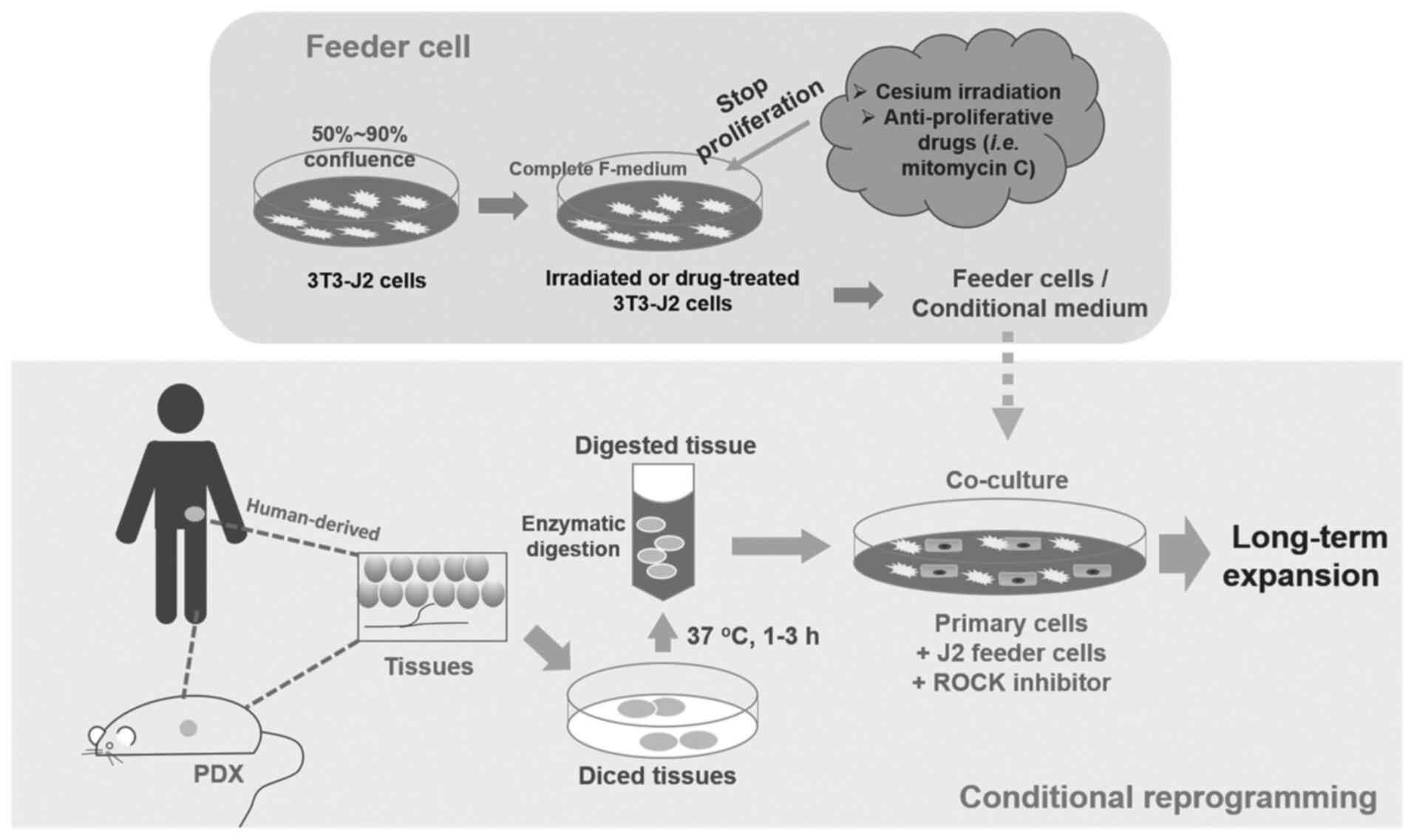
CR cells, which can be disaggregated human or
patient derived xenograft (PDX) primary cells (48), are co-cultured with inactivated
mouse 3T3-J2 fibroblasts as feeder cells and RHO-related protein
kinase (ROCK) inhibitor Y-27632 (Fig.
1). [The digested cells will also need pathological evaluation
to distinguish cancer cells from normal cells (48)]. J2 fibroblasts are given a dose of
irradiation or treated with mitomycin C (2–4 µg/ml) to stop their
proliferation. These cells are used as a feeder cell layer for
physically contact with primary cells (51). Alternatively, a conditional medium
with J2 feeder cell secreted factors can be used (44). Apoptosis and differentiation is
inhibited by ROCK inhibitor Y-27632 (48). Stem-like characteristics and the
ability to fully differentiate can be observed from this co-culture
(40). The establishment of tissue
cultures with this method appears to be highly efficient (40,44).
The advantages of CR is that it is simple, rapid and
has a high rate of success (88–100% success rate for some cells)
(40,44,52–54).
For example, the primary tumor or normal epithelial cells obtained
from prostate surgery biopsy and undergoing CR culture can generate
2×106 cells or reach confluence in ~5 days (40). Cells can also proliferate for long
periods while still maintaining much of the genetic makeup, gene
expression and heterogeneity of the primary cells from the biopsy
(40,55–57).
Thus CR cells can allow for drug sensitivity screening and testing
after 5 days (57–59). Culturing cells of carcinoma of the
lung in a patient using conditional reprogramming and then
determination of drug sensitivity, leading to the selection of an
effective therapeutic agent, was reported by Yuan et al
(60). Such an approach can be an
important advance for individualized therapy (48).
A drawback is that CR cells, when in a stem-like
state, do not have the same characteristics as the primary cancers.
Normally, prostate tissue has basal cell marker P63 and AR, but
these are not expressed in CR primary human prostate cells
(41,64). Some of the irradiated 3T3-J2 feeder
cells are not arrested in cell proliferation, as they should be,
and can transform to become malignant and gain cancer-like
characteristics in vivo (65–67).
ROCK inhibitor Y-27632 can also alter the actin cytoskeleton, which
is involved in migration and invasion of tumor cells (55). Instead of irradiation, some studies
inactivate 3T3-J2 feeder cells using mitomycin C, so there may be
biological differences in CR (51,68,69).
Thus, at present, CR cells are not suitable for modeling PCa due to
the culture components having unwanted influences. More research
could be done to optimize take rates as well.
In the future, several improvements will need to be
made. To improve the take rate in vitro, combining CR with
3D culture to provide the best conditions for improved
differentiation and recognition of normal cells was suggested by
Liu et al (70). In
addition, prevention of normal epithelial cell over proliferation,
which could outcompete cancer cell proliferation, can be improved
by using human fibroblasts instead of mouse fibroblasts in CR.
Using human fibroblasts does not support long term in vitro
proliferation of non-cancer cells, such as normal airway and lung
epithelial cells (48), so further
research on this technique for PCa may improve PCa cell take
rates.
A PDX model is established by transplanting patient
tissue into immunocompromised mice, which can be athymic nude mice,
severe compromised immunodeficient (SCID) mice, non-obese diabetic
(NOD)-SCID mice and recombination-activating gene 2 (Rag2)-knockout
mice (16,71). Recently, NSG (NOD.Cg-Prkdcscid
Il2rgtm1Wjl/SzJ) mice are preferred because there is also a
deficiency in innate immunity (72).
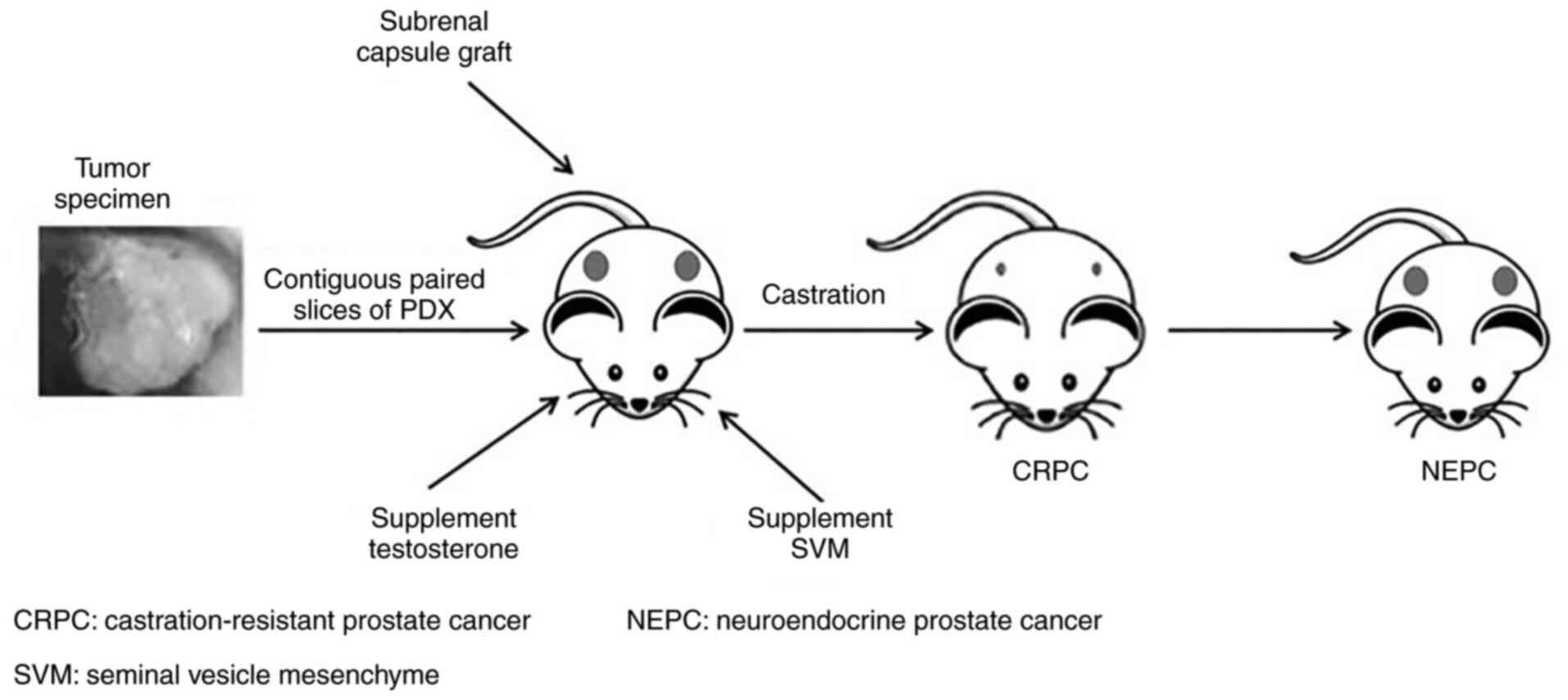
Culture or growth condition. Prostate tissue samples
are usually transplanted sub-renal capsule and supplemented with
testosterone and mouse seminal vesicle mesenchyme (16) (Fig.
2). Mouse seminal vesicle mesenchyme implanted along with PCa
tissue is able to mimic the stromal microenvironment and androgen
secretion to support prostate cell differentiation and
proliferation (73,74). The ideal transplantation site is one
similar to the origin of the tissue, but mice have limited capacity
in the pelvic region (16).
Instead, the tissue is transplanted under the renal capsule due to
its rich vasculature (37,75,76).
Similar tissue histology, heterogeneity, major
markers and genetic profile and expression to individual human PCa
are shown in PDX models. Due to these similar features, PDX can
also predict metastatic potential and drug response of the human
tumor, deeming it suitable for drug screening and validation
(16,44,77–79).
Addition of testosterone can increase the establishment and help
growth of PCa (80); although
establishment rates (10%) are still low (80,81).
With testosterone supplementation and transplantation with seminal
vesicle mesenchyme, increases of graft establishment to >90%
have been reported (77,82). In addition, aggressiveness and
growth of grafted tumors has been found to correlate with worse
clinical outcomes (77).
Genome changes and the phenotypic markers similar to
the primary cancers obtained from clinical PCa patients can be
shown in PDX models. Loss of PTEN and RB1, amplification of AR and
TMPRSS2-ERG fusion gene are often seen in PDX models (16). In addition, similar to the original
tumor, AR, prostate specific antigen, prostate-specific membrane
antigen and alpha-methylacyl-CoA-racemase are expressed in these
models (16). Stromal and vascular
components (7) and some
interactions within the tumor microenvironment (16) between stromal components and
epithelial tumor cells are also displayed in PDX models (44). Hormone dependence or independence
can be partly simulated and the transition from hormone dependence
to independence can be simulated (16,77,83,84).
Most importantly, similar treatment responses in patients have been
shown in PDX models (16,78,85,86).
For example, the LuCaP PDX model series, which has 21 successfully
established PDXs, is been shown to display similar responses to the
corresponding clinical patients (81). While there are a number of articles
finding that PDX models correlate with clinical responses (86–88),
there are few that describe PDX screening followed by a clinical
trial, which is a co-clinical trial. Reviews by Gao and Chen
(85) describe a good correlation
in treatment responses between initial PDX screening and
subsequently individual patients for a wide variety of cancers, but
not prostate cancer. Some studies also include integration of
genomic data with PDX model to identify precision treatment
(89,90). For example, xenografts from patients
with BRAF wild-type metastatic melanoma recapitulate the treatment
response to digoxin plus trametinib in such patients (125).
Due to its similarities to clinical PCa, PDX can be
a useful model for studying the biology and progression of PCa,
validation of screening for effective therapies for individual
cancers and drug development. PDX can be grown and passaged for
long periods, allowing tumor progression and stages to be observed
and used for drug and other therapy testing (16,81).
Despite the advantages described above, one
important disadvantage of the PDX model is the expense due to the
high maintenance cost of mice and long (2–5 month) ‘incubation’
period (16,44).
The second disadvantage is that PDX models are not
easily genetically modified or used for high-throughput drug
screening assays (85). Development
of cell lines from PDX models would allow easier high-throughput
drugs screenings and genetic modification, but this is challenging
due to overgrowth of stroma and limited differentiation potential,
especially if it is under CR conditions (44,57).
There is also a replacement of human stromal components with those
of the mice (91) with time. Since
PDX lacks human immune cells, the model is not suitable for
immunotherapy studies.
To address the problem of immunotherapy study in the
PDX models, human hematopoietic stem cells have been transplanted
into the immunodeficient mice to create a human-like immune system
(16,92,93).
There are various approaches to improve the generation of the human
hematopoietic/immune system and the reduction of graft vs. host
disease (94). The technology is
still evolving and the cost of producing humanized mice is
substantial.
The third disadvantage is that fresh surgical or
biopsy material is needed to establish a PDX (16,95).
In the future, the circulating tumor cells (CTCs) from the tumors
that are released into the vasculature may be utilized. Collecting
CTCs is like a ‘liquid biopsy’ that is safe and does not require an
invasive procedure (96). However,
there are a number of concerns which will need to be resolved. For
example, CTCs often have undergone epithelial-to-mesenchymal
transition, which has a downregulation of epithelial markers. In
addition, epithelial cell adhesion molecule, which is used to
isolate CTCs, may not be expressed in subtypes of cancer (16,97,98).
Last, it is often difficult to isolate sufficient number of CTC for
PDX and the lack of associated stromal cells may reduce engraftment
potential.
The final disadvantage is that although PDXs can
predict metastatic potential, the occurrence and development of PCa
metastases cannot be simulated. Most models do not spontaneously
metastasize (16). It is also well
recognized that some primary tumors only have a minority of cells
with metastatic ability (77). Shi
et al and Nguyen et al further suggested that PDX
models should match different stages of a patient's disease and
that they be treated in parallel (16,81).
Careful evaluation of histological cell types, signalers and
receptors to match PDX and patient disease stage can be explored in
future research.
In summary, despite the a number of suitable
features with the PDX model, its long growth time, costly upkeep,
biopsy sample requirement and the absence of a human stroma and a
human immune system (unless humanized) are disadvantages. The model
can be used for individualized evaluation of drug treatment but not
for high throughput screening for therapeutic discovering and
development.
Organoids are grown in Matrigel, a 3D gel made of
basement membrane proteins, to support cells growth and
differentiation. Generic serum-free media for all organoids with
prostate-specific growth factors are also used (7) (Tables
III, IV and V) (103–114). The complex matrix environment is
set to be similar to that in vivo, to allow easy propagation
of cells. Such conditions should provide more reliable drug testing
responses (16,115).
The utility of organoids in PCa research and therapy
assessment not only allow high throughput drug testing and
screening (95,116), but also can be useful to study
basic biology in early stage cancers, identify drug targets and
study drug resistance (44).
Additionally, further research options can be explored in
cryopreservation of organoids (19).
The key advantage is that the diverse histological
and genetic features in primary noncancer or PCa tissues can be
recapitulated in organoids. Specifically, the 3D culture system
allows the self-organization of organoids and luminal and basal
cell architecture and AR signaling can be retained (27). This model also can be genetically
modified using several genome editing techniques (27,41,117).
It can be used for in vitro studies and can be transplanted
in vivo for xenografts studies (27,100,118). Notably, organoids can produce
similar responses to therapies as those in patients (99).
Organoids derived from PDX tissues can be
conveniently transplanted into mice for long term growth study
(119,120). One specific advantage of
PDX-derived organoids is improved cost efficiency, especially for
high throughput drug screens, while PDX models are not practical
(7,99). However, PDX tissue can be an
additional source of tissue for organoid establishment when the
primary tissue, is scarce. Lastly, the PDX-derived organoids are
easier to genetically modify when compared to PDX (85).
Another practical advantage is that organoids can be
detected within 2–3 days of plating while small cystic organoids
are observed from luminal cells after 5–7 days (118). Organoids can have a high culture
take rate for certain types of cells, e.g., more advanced or
aggressive PCa, which allows side by side comparison and evaluation
between organoids and human primary tumor (119).
Despite the studies showing that organoids and
patient cancers have similar responses to therapies (121–123), the translation organoid studies to
clinical cases needs to be further verified (16). The first disadvantage of organoids
is its lack of blood vessels, immune cells and its need for ECM
substitutes (16). Since cancer
cell growth is effected by the microenvironment, lack of such
microenvironment could affect cell polarity, organization,
migration and invasion (120).
To improve the microenvironment, there have been
developments to include immune cells in the microenvironment with
organoids. In addition, Richards et al co-cultured PCa with
prostate fibromuscular stroma, increases organoid formation and
directs organoid growth into branched acinar structure, similar to
that seen in vivo (112).
In addition, light-mediated patterning technologies are used to
create gradients of these biochemical cues to imitate the
spatio-temporal patterns seen in vivo (124). It is hoped that these will be
applicable to prostate organoids in the future (103–105).
A possible second disadvantage is having foreign
factors such as the ECM substitute (16) and not having elements similar to the
human microenvironment which may affect drug screening results.
Although media components are able to effect organoid growth and
allow recapitulation of important features of the original tumor,
there may be an underrepresentation of biochemical signals, such as
growth factors. Due to this, organoids may not completely reflect
real-life growth. There is variability of components between fetal
bovine batches (106). The role
and differences of components between batches of Matrigel (107,108) is still unknown. Matrigel matrix
and FBS also may have components that effect experimental outcomes.
Steps are being made to more closely imitate the microenvironment
and to improve the matrix. There is development of Matrigel matrix
alternatives (109,110) and medium alternatives (111).
The third disadvantage is that although organoid
cultures from mouse and human prostate tissue can be established
with >95% efficiency (19). Due
to having the small amount of starting tissue and normal epithelial
overgrowth, the establishment rates are only 15–20% for advanced
PCa (19,118). Puca et al (95) also established 3/30 organoids. Among
the prostate basal cell and luminal cell cancers, the establishment
rates are ~70% from basal cell and 1–2% from luminal cell PCa
(27). Long term propagation may be
difficult for certain organoids, e.g., those basal cell-derived
(41,99,100,118). To improve and maintain long term
growth, fresh medium <2 weeks old with well tested and stored
growth factors and chemicals should be used. Histological
examinations may also help detect the overgrowth of normal
epithelial cells.
The fourth disadvantage at present is the problem
with high heterogeneity. Growth rate and morphology of organoids
from advanced PCa can vary between tumors of different patients as
well as tumors from the same patient (118). There is a need to match patient,
tumor and model, such as by using genomic analysis. A larger
organoid biobank (7) can be useful
in the future to provide genomic analysis improve the
stratification of the organoids and correlate them with patient
samples and data.
Although the ideal drug-testing model for
individualized cancer therapy screening is difficult to construct
or establish, of the current 3 models, the PDX model and organoid
model are more suited for use in culture and drug sensitivity
testing for individualized PCa therapy (Table VI). There is continuing research to
mimic the microenvironment more closely, which includes immune
cells and fibromuscular stroma. Using light-mediated patterning
technologies to mimic patterns of biochemical cues can further
improve these models (112,124).
Cell cultures could be the first step to explore if the immune
cells are able to react to the target cells (by killing and/or
elaboration of cytokines) that are supposed to express the
appropriate antigen.
Despite the existing shortcomings associated with
the PDX and organoid models, further development of a biobank with
cryopreserved organoids can be an important step to overcome the
shortcomings for practical application. Together with careful
histological examination, DNA sequencing and tumor
immunophenotyping can be used to characterize the tumor sample of a
given patient and match it with a specific organoid in the biobank.
Once the organoid is matched and selected, screening of several
potentially effective compounds can be performed to identify the
best therapeutic candidate. Afterwards, further verification of
in vivo efficacy can be conducted with the PDX models. This
approach can be applied to existing approved drugs or to
investigational agents. This combined organoid-PDX approach should
be especially applicable for PCa, which is usually a more slowly
progressing cancer that can benefit from such culture, drug
sensitivity and verification testing before selecting the drug or
combination for patient treatment. Such culture/drug sensitivity
testing may be especially useful for drug resistant cancer. While
there is no published success story of the proposed approach from
organoid to PDX and subsequent confirmation with a clinical trial,
there are successes from PDX to patient efficacy as well as
potential use of genetic mouse for drug screening (13,86–88,125–129). In view of the lack of practicality
of using PDX or other mouse models for rapid drug screening,
incorporation of the initial step using organoids can be a distinct
advantage. Future exploration with pre-clinical trials using
combined organoid and PDX models may pave the way toward
discovering novel agents, repurposing FDA approved drugs or
specific combinations or new sequencing of agents for precision
treatment of resistant prostate cancer.
The authors thank Ms. Yuna Kwon, College of
Pharmacy, Western University of Health Sciences for her assistance
on manuscript submission.
This work was supported by the Western University of Health
Sciences Summer Research Fellowship (grant no. 20/SSR/011),
2020.
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
JC gathered articles regarding tumor models and
evaluated their possible use as a cancer model for improving
individualized therapy. WCC read, suggested edits and approved the
final manuscript. MSSC provided the concept of the topic for review
and edit each draft. Data authentication is not applicable. All
authors reviewed and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stanford JL, Feng Z, Hamilton AS,
Gilliland FD, Stephenson RA, Eley JW, Albertsen PC, Harlan LC and
Potosky AL: Urinary and sexual function after radical prostatectomy
for clinically localized prostate cancer: The prostate cancer
outcomes study. JAMA. 283:354–360. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mohler JL, Antonarakis ES, Armstrong AJ,
D'Amico AV, Davis BJ, Dorff T, Eastham JA, Enke CA, Farrington TA,
Higano CS, et al: Prostate cancer, version 2.2019, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
17:479–505. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nuhn P, De Bono JS, Fizazi K, Freedland
SJ, Grilli M, Kantoff PW, Sonpavde G, Sternberg CN,
Yegnasubramanian S and Antonarakis ES: Update on systemic prostate
cancer therapies: Management of metastatic castration-resistant
prostate cancer in the era of precision oncology. Eur Urol.
75:88–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dorff TB and Agarwal N: Bone-targeted
therapies to reduce skeletal morbidity in prostate cancer. Asian J
Androl. 20:215–220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kelly SP, Anderson WF, Rosenberg PS and
Cook MB: Past, current, and future incidence rates and burden of
metastatic prostate cancer in the United States. Eur Urol Focus.
4:121–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gleave AM, Ci X, Lin D and Wang Y: A
synopsis of prostate organoid methodologies, applications, and
limitations. Prostate. 80:518–526. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng HH, Sokolova AO, Schaeffer EM, Small
EJ and Higano CS: Germline and somatic mutations in prostate cancer
for the clinician. J Natl Compr Canc Netw. 17:515–521. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lucas AL, Frado LE, Hwang C, Kumar S,
Khanna LG, Levinson EJ, Chabot JA, Chung WK and Frucht H: BRCA1 and
BRCA2 germline mutations are frequently demonstrated in both
high-risk pancreatic cancer screening and pancreatic cancer
cohorts. Cancer. 120:1960–1967. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Couch FJ, Nathanson KL and Offit K: Two
decades after BRCA: Setting paradigms in personalized cancer care
and prevention. Science. 343:1466–1470. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nishikawa T, Maemura K, Hirata I, Matsuse
R, Morikawa H, Toshina K, Murano M, Hashimoto K, Nakagawa Y, Saitoh
O, et al: A simple method of detecting K-ras point mutations in
stool samples for colorectal cancer screening using one-step
polymerase chain reaction/restriction fragment length polymorphism
analysis. Clin Chim Acta. 318:107–112. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bayot ML and Bragg BN: Antimicrobial
Susceptibility Testing. StatPearls [Internet] Treasure Island, FL:
StatPearls Publishing; 2020, [cited Jun 26, 2020]. Available from.
http://www.ncbi.nlm.nih.gov/books/NBK539714/
|
|
13
|
Sachs N, de Ligt J, Kopper O, Gogola E,
Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H,
et al: A living biobank of breast cancer organoids captures disease
heterogeneity. Cell. 172:373–386.e10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weeber F, Ooft SN, Dijkstra KK and Voest
EE: Tumor organoids as a pre-clinical cancer model for drug
discovery. Cell Chem Biol. 24:1092–1100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Muthuswamy SK: Organoid models of cancer
explode with possibilities. Cell Stem Cell. 22:290–291. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi C, Chen X and Tan D: Development of
patient-derived xenograft models of prostate cancer for maintaining
tumor heterogeneity. Transl Androl Urol. 8:519–528. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Namekawa T, Ikeda K, Horie-Inoue K and
Inoue S: Application of prostate cancer models for preclinical
study: Advantages and limitations of cell lines, patient-derived
xenografts, and three-dimensional culture of patient-derived cells.
Cells. 8:742019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cunha GR, Donjacour AA, Cooke PS, Mee S,
Bigsby RM, Higgins SJ and Sugimura Y: The endocrinology and
developmental biology of the prostate. Endocr Rev. 8:338–362. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Drost J, Karthaus WR, Gao D, Driehuis E,
Sawyers CL, Chen Y and Clevers H: Organoid culture systems for
prostate epithelial tissue and prostate cancer tissue. Nat Protoc.
11:347–358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heinlein CA and Chang C: Androgen receptor
in prostate cancer. Endocr Rev. 25:276–308. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robinson EJ, Neal DE and Collins AT: Basal
cells are progenitors of luminal cells in primary cultures of
differentiating human prostatic epithelium. Prostate. 37:149–160.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barclay WW, Woodruff RD, Hall MC and
Cramer SD: A system for studying epithelial-stromal interactions
reveals distinct inductive abilities of stromal cells from benign
prostatic hyperplasia and prostate cancer. Endocrinology.
146:13–18. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kurita T, Medina RT, Mills AA and Cunha
GR: Role of p63 and basal cells in the prostate. Development.
131:4955–4964. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kwon OJ, Zhang L and Xin L: Stem Cell
Antigen-1 identifies a distinct androgen-independent murine
prostatic luminal cell lineage with bipotent potential. Stem Cells.
34:191–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shibata M, Epsi NJ, Xuan S, Mitrofanova A
and Shen MM: Bipotent progenitors do not require androgen receptor
for luminal specification during prostate organogenesis. Stem Cell
Reports. 15:1026–1036. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ousset M, Van Keymeulen A, Bouvencourt G,
Sharma N, Achouri Y, Simons BD and Blanpain C: Multipotent and
unipotent progenitors contribute to prostate postnatal development.
Nat Cell Biol. 14:1131–1138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karthaus WR, Iaquinta PJ, Drost J,
Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel
H, Sachs N, et al: Identification of multipotent luminal progenitor
cells in human prostate organoid cultures. Cell. 159:163–175. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi N, Zhang B, Zhang L, Ittmann M and
Xin L: Adult murine prostate basal and luminal cells are
self-sustained lineages that can both serve as targets for prostate
cancer initiation. Cancer Cell. 21:253–265. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu AY and True LD: Characterization of
prostate cell types by CD cell surface molecules. Am J Pathol.
160:37–43. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hudson DL: Epithelial stem cells in human
prostate growth and disease. Prostate Cancer Prostatic Dis.
7:188–194. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zenzmaier C, Untergasser G and Berger P:
Aging of the prostate epithelial stem/progenitor cell. Exp
Gerontol. 43:981–985. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Sant'Agnese PA: Neuroendocrine cells of
the prostate and neuroendocrine differentiation in prostatic
carcinoma: A review of morphologic aspects. Urology. 51 (5A
Suppl):S121–S124. 1998. View Article : Google Scholar
|
|
33
|
Abrahamsson PA: Neuroendocrine
differentiation in prostatic carcinoma. Prostate. 39:135–148. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prostate gland [Internet]. Kenhub. [cited
Sep 7, 2020]. Available from. https://www.kenhub.com/en/library/anatomy/the-prostate-gland
|
|
35
|
Chung LW, Baseman A, Assikis V and Zhau
HE: Molecular insights into prostate cancer progression: The
missing link of tumor microenvironment. J Urol. 173:10–20. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mueller MM and Fusenig NE: Friends or
foes-bipolar effects of the tumour stroma in cancer. Nat Rev
Cancer. 4:839–849. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang R, Xu J, Juliette L, Castilleja A,
Love J, Sung SY, Zhau HE, Goodwin TJ and Chung LW:
Three-dimensional co-culture models to study prostate cancer
growth, progression, and metastasis to bone. Semin Cancer Biol.
15:353–364. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Igney FH and Krammer PH: Immune escape of
tumors: Apoptosis resistance and tumor counterattack. J Leukoc
Biol. 71:907–920. 2002.PubMed/NCBI
|
|
39
|
Beatty GL and Gladney WL: Immune escape
mechanisms as a guide for cancer immunotherapy. Clin Cancer Res.
21:687–692. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu X, Ory V, Chapman S, Yuan H, Albanese
C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al:
ROCK inhibitor and feeder cells induce the conditional
reprogramming of epithelial cells. Am J Pathol. 180:599–607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Drost J, van Jaarsveld RH, Ponsioen B,
Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus
GJ, Begthel H, et al: Sequential cancer mutations in cultured human
intestinal stem cells. Nature. 521:43–47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Toivanen R, Taylor RA, Pook DW, Ellem SJ
and Risbridger GP: Breaking through a roadblock in prostate cancer
research: An update on human model systems. J Steroid Biochem Mol
Biol. 131:122–131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nupponen NN, Hyytinen ER, Kallioniemi AH
and Visakorpi T: Genetic alterations in prostate cancer cell lines
detected by comparative genomic hybridization. Cancer Genet
Cytogenet. 101:53–57. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Palechor-Ceron N, Krawczyk E, Dakic A,
Simic V, Yuan H, Blancato J, Wang W, Hubbard F, Zheng YL, Dan H, et
al: Conditional reprogramming for patient-derived cancer models and
next-generation living biobanks. Cells. 8:13272019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dasgupta P, Baade PD, Aitken JF, Ralph N,
Chambers SK and Dunn J: Geographical variations in prostate cancer
outcomes: A systematic review of International evidence. Front
Oncol. 9:2382019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Parrinello S, Samper E, Krtolica A,
Goldstein J, Melov S and Campisi J: Oxygen sensitivity severely
limits the replicative lifespan of murine fibroblasts. Nat Cell
Biol. 5:741–747. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Panchision DM: The role of oxygen in
regulating neural stem cells in development and disease. J Cell
Physiol. 220:562–568. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wu X, Wang S, Li M, Li J, Shen J, Zhao Y,
Pang J, Wen Q, Chen M, Wei B, et al: Conditional reprogramming:
Next generation cell culture. Acta Pharm Sin B. 10:1360–1381. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sharpless NE and DePinho RA: The mighty
mouse: Genetically engineered mouse models in cancer drug
development. Nat Rev Drug Discov. 5:741–754. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chapman S, Liu X, Meyers C, Schlegel R and
McBride AA: Human keratinocytes are efficiently immortalized by a
Rho kinase inhibitor. J Clin Invest. 120:2619–2626. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hynds RE, Ben Aissa A, Gowers KHC, Watkins
TBK, Bosshard-Carter L, Rowan AJ, Veeriah S, Wilson GA, Quezada SA,
Swanton C, et al: Expansion of airway basal epithelial cells from
primary human non-small cell lung cancer tumors. Int J Cancer.
143:160–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Suprynowicz FA, Upadhyay G, Krawczyk E,
Kramer SC, Hebert JD, Liu X, Yuan H, Cheluvaraju C, Clapp PW,
Boucher RC Jr, et al: Conditionally reprogrammed cells represent a
stem-like state of adult epithelial cells. Proc Natl Acad Sci USA.
109:20035–20040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Suprynowicz FA, Kamonjoh CM, Krawczyk E,
Agarwal S, Wellstein A, Agboke FA, Choudhury S, Liu X and Schlegel
R: Conditional cell reprogramming involves non-canonical β-catenin
activation and mTOR-mediated inactivation of Akt. PLoS One.
12:e01808972017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sugaya M, Takenoyama M, Osaki T, Yasuda M,
Nagashima A, Sugio K and Yasumoto K: Establishment of 15 cancer
cell lines from patients with lung cancer and the potential tools
for immunotherapy. Chest. 122:282–288. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu X, Krawczyk E, Suprynowicz FA,
Palechor-Ceron N, Yuan H, Dakic A, Simic V, Zheng YL, Sripadhan P,
Chen C, et al: Conditional reprogramming and long-term expansion of
normal and tumor cells from human biospecimens. Nat Protoc.
12:439–451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Timofeeva OA, Palechor-Ceron N, Li G, Yuan
H, Krawczyk E, Zhong X, Liu G, Upadhyay G, Dakic A, Yu S, et al:
Conditionally reprogrammed normal and primary tumor prostate
epithelial cells: A novel patient-derived cell model for studies of
human prostate cancer. Oncotarget. 8:22741–22758. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Borodovsky A, McQuiston TJ, Stetson D,
Ahmed A, Whitston D, Zhang J, Grondine M, Lawson D, Challberg SS,
Zinda M, et al: Generation of stable PDX derived cell lines using
conditional reprogramming. Mol Cancer. 16:1772017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Saeed K, Rahkama V, Eldfors S, Bychkov D,
Mpindi JP, Yadav B, Paavolainen L, Aittokallio T, Heckman C,
Wennerberg K, et al: Comprehensive drug testing of patient-derived
conditionally reprogrammed cells from castration-resistant prostate
cancer. Eur Urol. 71:319–327. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vondálová Blanářová O, Šafaříková B,
Herůdková J, Krkoška M, Tománková S, Kahounová Z, Anděra L, Bouchal
J, Kharaishvili G, Král M, et al: Cisplatin or LA-12 enhance
killing effects of TRAIL in prostate cancer cells through
Bid-dependent stimulation of mitochondrial apoptotic pathway but
not caspase-10. PLoS One. 12:e01885842017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yuan H, Myers S, Wang J, Zhou D, Woo JA,
Kallakury B, Ju A, Bazylewicz M, Carter YM, Albanese C, et al: Use
of reprogrammed cells to identify therapy for respiratory
papillomatosis. N Engl J Med. 367:1220–1227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Brown DD, Dabbs DJ, Lee AV, McGuire KP,
Ahrendt GM, Bhargava R, Davidson NE, Brufsky AM, Johnson RR,
Oesterreich S and McAuliffe PF: Developing in vitro models of human
ductal carcinoma in situ from primary tissue explants. Breast
Cancer Res Treat. 153:311–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ellis L, Ku S, Li Q, Azabdaftari G,
Seliski J, Olson B, Netherby CS, Tang DG, Abrams SI, Goodrich DW
and Pili R: Generation of a C57BL/6 MYC-Driven Mouse Model and Cell
Line of Prostate Cancer. Prostate. 76:1192–1202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jensen TJ, Foster C, Sayej W and Finck CM:
Conditional reprogramming of pediatric human esophageal epithelial
cells for use in tissue engineering and disease investigation. J
Vis Exp. 121:e552432017.
|
|
64
|
Tricoli L, Naeem A, Parasido E, Mikhaiel
JP, Choudhry MU, Berry DL, Abdelgawad IA, Lee RJ, Feldman AS,
Ihemelandu C, et al: Characterization of the effects of defined,
multidimensional culture conditions on conditionally reprogrammed
primary human prostate cells. Oncotarget. 9:2193–2207. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Serrano-Heras G, Domínguez-Berzosa C,
Collantes E, Guadalajara H, García-Olmo D and García-Olmo DC:
NIH-3T3 fibroblasts cultured with plasma from colorectal cancer
patients generate poorly differentiated carcinomas in mice. Cancer
Lett. 316:85–90. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yu F, Lu Y, Tao L, Jiang YY, Lin DC, Wang
L, Petersson F, Yoshiyama H, Koeffler PH, Goh BC and Loh KS:
Non-malignant epithelial cells preferentially proliferate from
nasopharyngeal carcinoma biopsy cultured under conditionally
reprogrammed conditions. Sci Rep. 7:173592017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yu F, Hsieh W, Petersson F, Yang H, Li Y,
Li C, Low SW, Liu J, Yan Y, Wang DY and Loh KS: Malignant cells
derived from 3T3 fibroblast feeder layer in cell culture for
nasopharyngeal carcinoma. Exp Cell Res. 322:193–201. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhao W, Liu K, Sun Z, Wang L, Liu B, Liu
L, Qu X, Cao Z, Sun J and Chai J: Application research of
individualized conditional reprogramming system to guide treatment
of gastric cancer. Front Oncol. 11:7095112021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Dong Y, Wang J, Ji W, Zheng M, Wang P, Liu
L and Li S: Establishment and preclinical application of
conditional reprogramming culture system for laryngeal and
hypopharyngeal carcinoma. Front Cell Dev Biol. 9:7449692021.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu X, Krawczyk E, Timofeeva O,
Palechor-Ceron N, Dakic A, Simic V, Kallakury B, Dritschilo A and
Schlegel R: Functional analysis for cancer precision medicine using
patient-derived 2D and 3D cell models. Cancer Res. 76 (Suppl
14):S42562016.
|
|
71
|
Morton CL and Houghton PJ: Establishment
of human tumor xenografts in immunodeficient mice. Nat Protoc.
2:247–250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cao X, Shores EW, Hu-Li J, Anver MR,
Kelsail BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET,
et al: Defective lymphoid development in mice lacking expression of
the common cytokine receptor γ chain. Immunity. 2:223–238. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Govindaraj V, Arya SV and Rao AJ:
Differential action of glycoprotein hormones: Significance in
cancer progression. Horm Cancer. 5:1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Lawrence MG, Taylor RA, Toivanen R,
Pedersen J, Norden S, Pook DW, Frydenberg M; Australian Prostate
Cancer BioResource, ; Papargiris MM, Niranjan B, et al: A
preclinical xenograft model of prostate cancer using human tumors.
Nat Protoc. 8:836–848. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
McLean DT, Strand DW and Ricke WA:
Prostate cancer xenografts and hormone induced prostate
carcinogenesis. Differentiation. 97:23–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lam HM, Nguyen HM and Corey E: Generation
of prostate cancer patient-derived xenografts to investigate
mechanisms of novel treatments and treatment resistance. Methods
Mol Biol. 1786:1–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lin D, Wyatt AW, Xue H, Wang Y, Dong X,
Haegert A, Wu R, Brahmbhatt S, Mo F, Jong L, et al: High fidelity
patient-derived xenografts for accelerating prostate cancer
discovery and drug development. Cancer Res. 74:1272–1283. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Williams SA, Anderson WC, Santaguida MT
and Dylla SJ: Patient-derived xenografts, the cancer stem cell
paradigm, and cancer pathobiology in the 21st century. Lab Invest.
93:970–982. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wu CH, Yang CY, Wang L, Gao HX,
Rakhshandehroo T, Afghani S, Pincus L, Balassanian R, Rubenstein J,
Gill R, et al: Cutaneous T-cell lymphoma PDX drug screening
platform identifies cooperation between inhibitions of PI3Kα/δ and
HDAC. J Invest Dermatol. 141:364–373. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Russell PJ, Russell P, Rudduck C, Tse BW,
Williams ED and Raghavan D: Establishing prostate cancer patient
derived xenografts: Lessons learned from older studies. Prostate.
75:628–636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nguyen HM, Vessella RL, Morrissey C, Brown
LG, Coleman IM, Higano CS, Mostaghel EA, Zhang X, True LD, Lam HM,
et al: LuCaP prostate cancer patient-derived xenografts reflect the
molecular heterogeneity of advanced disease and serve as models for
evaluating cancer therapeutics. Prostate. 77:654–671. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang Y, Revelo MP, Sudilovsky D, Cao M,
Chen WG, Goetz L, Xue H, Sadar M, Shappell SB, Cunha GR and Hayward
SW: Development and characterization of efficient xenograft models
for benign and malignant human prostate tissue. Prostate.
64:149–159. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yoshikawa T, Kobori G, Goto T, Akamatsu S,
Terada N, Kobayashi T, Tanaka Y, Jung G, Kamba T, Ogawa O and Inoue
T: An original patient-derived xenograft of prostate cancer with
cyst formation. Prostate. 76:994–1003. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Gao D and Chen Y: Organoid development in
cancer genome discovery. Curr Opin Genet Dev. 30:42–48. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Owonikoko TK, Zhang G, Kim HS, Stinson RM,
Bechara R, Zhang C, Chen Z, Saba NF, Pakkala S, Pillai R, et al:
Patient-derived xenografts faithfully replicated clinical outcome
in a phase II co-clinical trial of arsenic trioxide in relapsed
small cell lung cancer. J Transl Med. 14:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Williams JA: Using PDX for preclinical
cancer drug discovery: The evolving field. J Clin Med. 7:412018.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gao H, Korn JM, Ferretti S, Monahan JE,
Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, et al:
High-throughput screening using patient-derived tumor xenografts to
predict clinical trial drug response. Nat Med. 21:1318–1325. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ni J, Ramkissoon SH, Xie S, Goel S, Stover
DG, Guo H, Luu V, Marco E, Ramkissoon LA, Kang YJ, et al:
Combination inhibition of PI3K and mTORC1 yields durable remissions
in orthotopic patient-derived xenografts of HER2-positive breast
cancer brain metastases. Nat Med. 22:723–726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Corcoran RB, Atreya CE, Falchook GS, Kwak
EL, Ryan DP, Bendell JC, Hamid O, Messersmith WA, Daud A, Kurzrock
R, et al: Combined BRAF and MEK inhibition with dabrafenib and
trametinib in BRAF V600-Mutant colorectal cancer. J Clin Oncol.
33:4023–4031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lai Y, Wei X, Lin S, Qin L, Cheng L and Li
P: Current status and perspectives of patient-derived xenograft
models in cancer research. J Hematol Oncol. 10:1062017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Bartucci M, Ferrari AC, Kim IY, Ploss A,
Yarmush M and Sabaawy HE: Personalized medicine approaches in
prostate cancer employing patient derived 3D organoids and
humanized mice. Front Cell Dev Biol. 4:642016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ito R, Takahashi T and Ito M: Humanized
mouse models: Application to human diseases. J Cell Physiol.
233:3723–3728. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Puca L, Bareja R, Prandi D, Shaw R,
Benelli M, Karthaus WR, Hess J, Sigouros M, Donoghue A, Kossai M,
et al: Patient derived organoids to model rare prostate cancer
phenotypes. Nat Commun. 9:24042018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Praharaj PP, Bhutia SK, Nagrath S, Bitting
RL and Deep G: Circulating tumor cell-derived organoids: Current
challenges and promises in medical research and precision medicine.
Biochim Biophys Acta Rev Cancer. 1869:117–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Gorges TM, Tinhofer I, Drosch M, Röse L,
Zollner TM, Krahn T and von Ahsen O: Circulating tumour cells
escape from EpCAM-based detection due to epithelial-to-mesenchymal
transition. BMC Cancer. 12:1782012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Armstrong AJ, Marengo MS, Oltean S, Kemeny
G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ and
Garcia-Blanco MA: Circulating tumor cells from patients with
advanced prostate and breast cancer display both epithelial and
mesenchymal markers. Mol Cancer Res. 9:997–1007. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Beshiri ML, Tice CM, Tran C, Nguyen HM,
Sowalsky AG, Agarwal S, Jansson KH, Yang Q, McGowen KM, Yin J, et
al: A PDX/organoid biobank of advanced prostate cancers captures
genomic and phenotypic heterogeneity for disease modeling and
therapeutic screening. Clin Cancer Res. 24:4332–4345. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Chua CW, Shibata M, Lei M, Toivanen R,
Barlow LJ, Bergren SK, Badani KK, McKiernan JM, Benson MC,
Hibshoosh H and Shen MM: Single luminal epithelial progenitors can
generate prostate organoids in culture. Nat Cell Biol. 16:951–961.
1–4. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Clevers H: Modeling development and
disease with organoids. Cell. 165:1586–1597. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Allard WJ: Tumor cells circulate in the
peripheral blood of all major carcinomas but not in healthy
subjects or patients with nonmalignant diseases. Clin Cancer Res.
10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tsai S, McOlash L, Palen K, Johnson B,
Duris C, Yang Q, Dwinell MB, Hunt B, Evans DB, Gershan J and James
MA: Development of primary human pancreatic cancer organoids,
matched stromal and immune cells and 3D tumor microenvironment
models. BMC Cancer. 18:3352018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Neal JT, Li X, Zhu J, Giangarra V,
Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, et
al: Organoid modeling of the tumor immune microenvironment. Cell.
175:1972–1988.e16. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Dijkstra KK, Cattaneo CM, Weeber F,
Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL,
Kaing S, Kelderman S, et al: Generation of Tumor-Reactive T Cells
by Co-culture of peripheral blood lymphocytes and tumor organoids.
Cell. 174:1586–1598.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Gstraunthaler G, Lindl T and van der Valk
J: A plea to reduce or replace fetal bovine serum in cell culture
media. Cytotechnology. 65:791–793. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Hughes CS, Postovit LM and Lajoie GA:
Matrigel: A complex protein mixture required for optimal growth of
cell culture. Proteomics. 10:1886–1890. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Patel R and Alahmad AJ: Growth-factor
reduced Matrigel source influences stem cell derived brain
microvascular endothelial cell barrier properties. Fluids Barriers
CNS. 13:62016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Nguyen EH, Daly WT, Le NNT, Farnoodian M,
Belair DG, Schwartz MP, Lebakken CS, Ananiev GE, Saghiri MA,
Knudsen TB, et al: Versatile synthetic alternatives to Matrigel for
vascular toxicity screening and stem cell expansion. Nat Biomed
Eng. 1:00962017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhang S, Gelain F and Zhao X: Designer
self-assembling peptide nanofiber scaffolds for 3D tissue cell
cultures. Semin Cancer Biol. 15:413–420. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Stingl J, Rowbotham D, Thomas TE, Eaves AC
and Louis SA: Expansion of mouse prostate epithelial stem cells in
serum-free ProstaCult Organoid Growth Medium. Cancer Res. 78 (13
Suppl):S31112018.
|
|
112
|
Richards Z, McCray T, Marsili J, Zenner
ML, Manlucu JT, Garcia J, Kajdacsy-Balla A, Murray M, Voisine C,
Murphy AB, et al: Prostate stroma increases the viability and
maintains the branching phenotype of human prostate organoids.
iScience. 12:304–317. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
von Amsberg G and Merseburger AS:
Treatment of metastatic, castration-resistant prostate cancer.
Urologe A. 59:673–679. 2020.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Kantoff PW, Higano CS, Shore ND, Berger
ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims
RB, et al: Sipuleucel-T immunotherapy for castration-resistant
prostate cancer. N Engl J Med. 363:411–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Kleinman HK and Martin GR: Matrigel:
Basement membrane matrix with biological activity. Semin Cancer
Biol. 15:378–386. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Pauli C, Hopkins BD, Prandi D, Shaw R,
Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, et al:
Personalized in vitro and in vivo cancer models to guide precision
medicine. Cancer Discov. 7:462–477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Koo BK, Stange DE, Sato T, Karthaus W,
Farin HF, Huch M, van Es JH and Clevers H: Controlled gene
expression in primary Lgr5 organoid cultures. Nat Methods. 9:81–83.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Gao D, Vela I, Sboner A, Iaquinta PJ,
Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora
VK, et al: Organoid cultures derived from patients with advanced
prostate cancer. Cell. 159:176–187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Risbridger GP, Toivanen R and Taylor RA:
Preclinical models of prostate cancer: Patient-derived xenografts,
organoids, and other explant models. Cold Spring Harb Perspect Med.
8:a0305362018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Lawrence MG, Obinata D, Sandhu S, Selth
LA, Wong SQ, Porter LH, Lister N, Pook D, Pezaro CJ, Goode DL, et
al: Patient-derived models of abiraterone- and
enzalutamide-resistant prostate cancer reveal sensitivity to
ribosome-directed therapy. Eur Urol. 74:562–572. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ooft SN, Weeber F, Dijkstra KK, McLean CM,
Kaing S, van Werkhoven E, Schipper L, Hoes L, Vis DJ, van de Haar
J, et al: Patient-derived organoids can predict response to
chemotherapy in metastatic colorectal cancer patients. Sci Transl
Med. 11:eaay25742019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Tiriac H, Belleau P, Engle DD, Plenker D,
Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche
RE, Jang GH, et al: Organoid profiling identifies common responders
to chemotherapy in pancreatic cancer. Cancer Discov. 8:1112–1129.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ganesh K, Wu C, O'Rourke KP, Szeglin BC,
Zheng Y, Sauvé CG, Adileh M, Wasserman I, Marco MR, Kim AS, et al:
A rectal cancer organoid platform to study individual responses to
chemoradiation. Nat Med. 25:1607–1614. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Sawicki LA and Kloxin AM: Light-mediated
formation and patterning of hydrogels for cell culture
applications. J Vis Exp. 115:e544622016.PubMed/NCBI
|
|
125
|
Koga Y and Ochiai A: Systematic review of
Patient-Derived xenograft models for preclinical studies of
anti-cancer drugs in solid tumors. Cells. 8:4182019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Nardella C, Lunardi A, Patnaik A, Cantley
LC and Pandolfi PP: The APL paradigm and the ‘co-clinical trial’
project. Cancer Discov. 1:108–116. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Clohessy JG and Pandolfi PP: Mouse
hospital and co-clinical trial project-from bench to bedside. Nat
Rev Clin Oncol. 12:491–498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chen M and Pandolfi PP: Preclinical and
coclinical studies in prostate cancer. Cold Spring Harb Perspect
Med. 8:a0305442018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lunardi A, Ala U, Epping MT, Salmena L,
Clohessy JG, Webster KA, Wang G, Mazzucchelli R, Bianconi M, Stack
EC, et al: A co-clinical approach identifies mechanisms and
potential therapies for androgen deprivation resistance in prostate
cancer. Nat Genet. 45:747–755. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
van Moorselaar RJA and Voest EE:
Angiogenesis in prostate cancer: its role in disease progression
and possible therapeutic approaches. Mol Cell Endocrinol.
197:239–250. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zittermann SI and Issekutz AC: Basic
fibroblast growth factor (bFGF, FGF-2) potentiates leukocyte
recruitment to inflammation by enhancing endothelial adhesion
molecule expression. Am J Pathol. 168:835–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Lail-Trecker M, Gulati R and Peluso JJ: A
role for hepatocyte growth factor/scatter factor in regulating
normal and neoplastic cells of reproductive tissues. J Soc Gynecol
Investig. 5:114–121. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Blanchère M, Saunier E, Mestayer C,
Broshuis M and Mowszowicz I: Alterations of expression and
regulation of transforming growth factor beta in human cancer
prostate cell lines. J Steroid Biochem Mol Biol. 82:297–304. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Royuela M, Ricote M, Parsons MS,
García-Tuñón I, Paniagua R and de Miguel MP: Immunohistochemical
analysis of the IL-6 family of cytokines and their receptors in
benign, hyperplasic, and malignant human prostate. J Pathol.
202:41–49. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Planz B, Wang Q, Kirley SD, Marberger M
and McDougal WS: Regulation of keratinocyte growth factor receptor
and androgen receptor in epithelial cells of the human prostate. J
Urol. 166:678–683. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Francis JC, Thomsen MK, Taketo MM and
Swain A: β-catenin is required for prostate development and
cooperates with pten loss to drive invasive carcinoma. PLoS Genet.
9:e10031802013. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Cook C, Vezina CM, Allgeier SH, Shaw A, Yu
M, Peterson RE and Bushman W: Noggin is required for normal lobe
patterning and ductal budding in the mouse prostate. Dev Biol.
312:217–230. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Jarrard DF, Blitz BF, Smith RC, Patai BL
and Rukstalis DB: Effect of epidermal growth factor on prostate
cancer cell line PC3 growth and invasion. Prostate. 24:46–53. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Sastry KS, Karpova Y and Kulik G:
Epidermal growth factor protects prostate cancer cells from
apoptosis by inducing BAD phosphorylation via redundant signaling
pathways. J Biol Chem. 281:27367–27377. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Watanabe K, Ueno M, Kamiya D, Nishiyama A,
Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S,
Muguruma K and Sasai Y: A ROCK inhibitor permits survival of
dissociated human embryonic stem cells. Nat Biotechnol. 25:681–686.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Tojo M, Hamashima Y, Hanyu A, Kajimoto T,
Saitoh M, Miyazono K, Node M and Imamura T: The ALK-5 inhibitor
A-83-01 inhibits Smad signaling and epithelial-to-mesenchymal
transition by transforming growth factor-beta. Cancer Sci.
96:791–800. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Zhang F, Lau SS and Monks TJ: The
Cytoprotective Effect of N-acetyl-L-cysteine against ROS-induced
cytotoxicity is independent of its ability to enhance glutathione
synthesis. Toxicol Sci. 120:87–97. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Gu Y, Fu J, Lo PK, Wang S, Wang Q and Chen
H: The Effect of B27 Supplement on Promoting In Vitro Propagation
of Her2/neu-Transformed mammary tumorspheres. J Biotech Res.
3:7–18. 2011.
|
|
144
|
Sato T, Stange DE, Ferrante M, Vries RG,
Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J,
Siersema PD and Clevers H: Long-term expansion of epithelial
organoids from human colon, adenoma, adenocarcinoma, and Barrett's
Epithelium. Gastroenterology. 141:1762–1772. 2011. View Article : Google Scholar : PubMed/NCBI
|