Cancer accounts for more than 10 million mortalities
and is the second most leading cause of death worldwide, and it is
estimated that nearly 30 million patients will succumb due to
cancer each year by 2040 (1).
Cancer is a disease that can start occurring in any tissue of the
body where cell growth is uncontrollable and starts spreading to
other parts of the body, leading to metastasis, a major cause of
death in patients with cancer. Inability to contain physiological
apoptosis leads to cancer development and is a reason for
resistance to radiotherapy and chemotherapy (2). Certain of the common types of cancer
in women are cervical, breast, colorectal, thyroid and lung, while
men commonly have cancers such as prostate, colorectal, lungs,
liver and stomach (3). At present,
numerous treatments, including radiation, chemotherapeutic drugs
and surgical operations, have been introduced to treat cancer,
which often leads to the damage of healthy cells and enhances the
toxicity in the patients. Thus, currently, researchers emphasize
eliminating only cancerous cells without hampering the normal
functioning of the body. Application of chemotherapy and
radiotherapy are the primary major interruptions of most cancers,
but due to the presence of acquired and intrinsic resistance, their
therapeutic efficiencies have been retarded. Several limitations
are making current therapies ineffective against cancer which
includes, firstly, non-specific mechanisms of action, eliminating
not only the cancer cells but also the normal cells; secondly, the
metastatic ability of cancer cells to other tissues other than the
primary cancer site, and lastly, non-availability of efficient
diagnosis system due to lack of effective biomarkers (4). Hence, researchers nowadays are looking
for more biochemically relevant alternatives to cancer cells that
can be targeted for their reliable specificity and are participants
in the regulatory mechanism of cancerous cells.
In previous studies, it has been found that circular
RNAs (circRNAs) play a major role in tissue homeostasis and
cellular differentiation, which even leads to the development of
diseases (5,6). Previously, circRNAs were considered
peculiarities having no specific biological functions and being a
result of an error in the process of splicing (5). It has been found that there is often
no correlation between linear expressions of host gene circRNAs.
This leads to an understanding that circRNAs are not just a product
of normal mRNA splicing but a product of alternative splicing,
which is finely regulated (6). The
circRNA sequence was analyzed, and it was found that they are
conserved and have certain biological importance. Biological
functions of circRNAs have recently become fascinating in the
scientific world and the scientific community is very much curious
to investigate it. Therefore, more therapeutic approaches are being
developed by knowing the function of circRNAs, and their specific
role in diseases has been revealed with the help of the advancement
of science and technology. CircRNAs are non-coding RNAs present in
the genome having different functions of regulating various
molecules such as mRNAs, DNAs, non-coding RNAs and proteins to
regulate cell functioning and physiology of an organism (7). Structures of circRNAs have a
covalently closed circle. Regions of introns involved in circle
formation are more likely to consist of inverted complementary Alu
repeats, and these regions of Alu repeats act as transposons within
the genome. With the help of base pairing between Alu repeats,
circularization becomes markedly easier as it facilitates splice
site recognition (8,9). CircRNAs are highly stable in the body
due to their circular structure, which safeguards them from the
effect of enzymes like exonucleases.
In the development of tumors, circRNAs play a vital
role and enhance the effectiveness and sensitivity in chemotherapy
and radiation (10). Several
studies have shown that certain circRNAs are expressed abnormally
in tumor cells, but it does not always promote the cause of
malignancy; nevertheless, it takes part in the regulation of
tumorigenesis (11,12). Multiple varieties of tumors,
particularly those originating from the gastrointestinal tract and
ovary, impart a stronger preference to the peritoneal cavity as the
site of metastasis. A balance mediates the spread of malignancy in
the intraperitoneal region between normal residential peritoneal
cells and cancer cells that are actively invading (10). Numerous genes produce stable and
conserved closed circRNA with a high potential for gene regulation
(12). This knowledge may lead to
targeting specific circRNA to cure different types of cancer. In
the pathogenesis of human cancers such as lung, liver, ovarian,
breast, prostate, including tumors and malignancies in the central
nervous system, circRNAs are being found to play a crucial role in
the development of cancers. CircRNAs are very much tissue-specific,
having distinct expressions for different diseases and they are
specific hallmarks of cancer (13,14).
CircRNAs are detectable in fluids such as saliva and blood
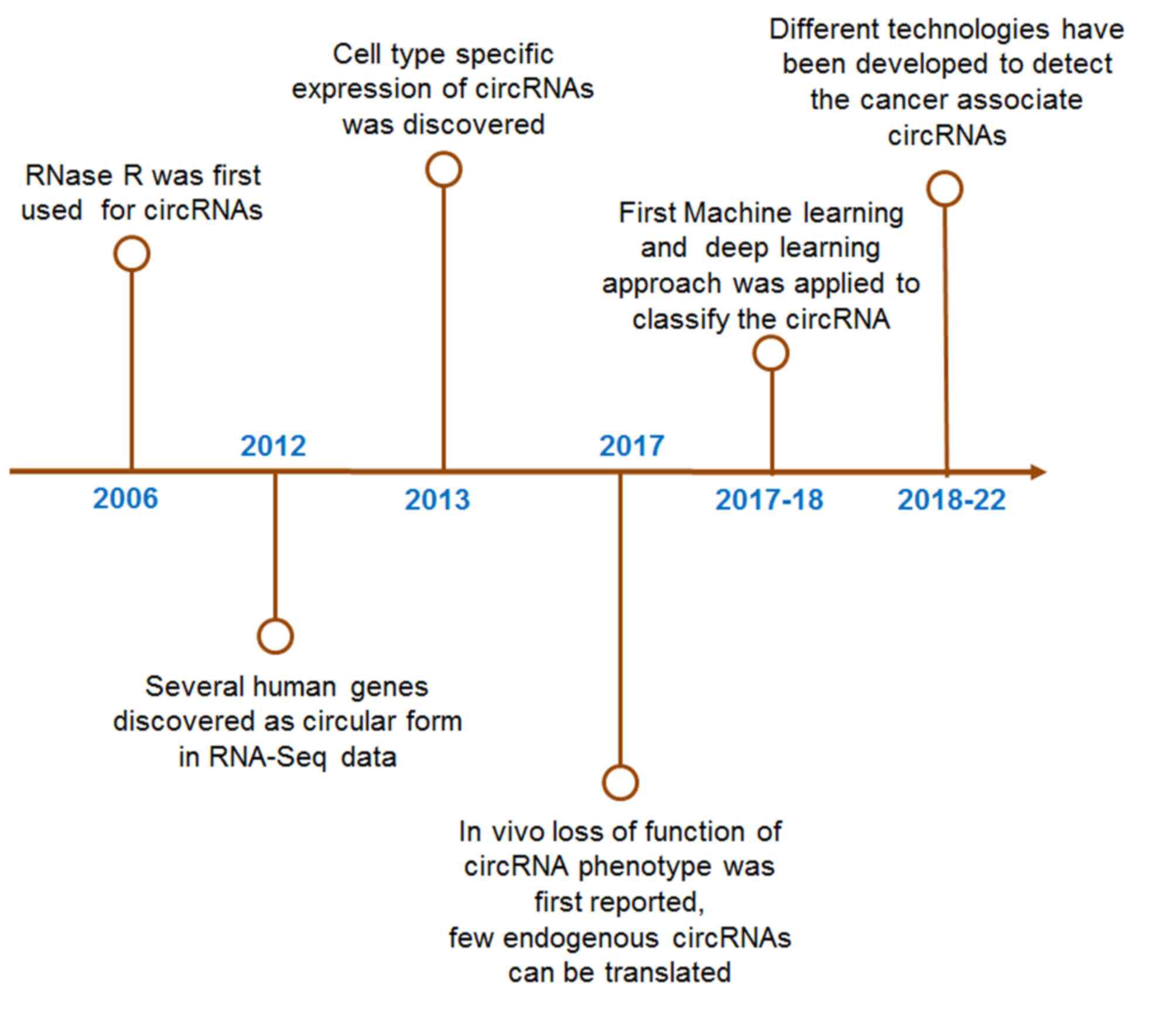
(15–18). Recent research is progressing
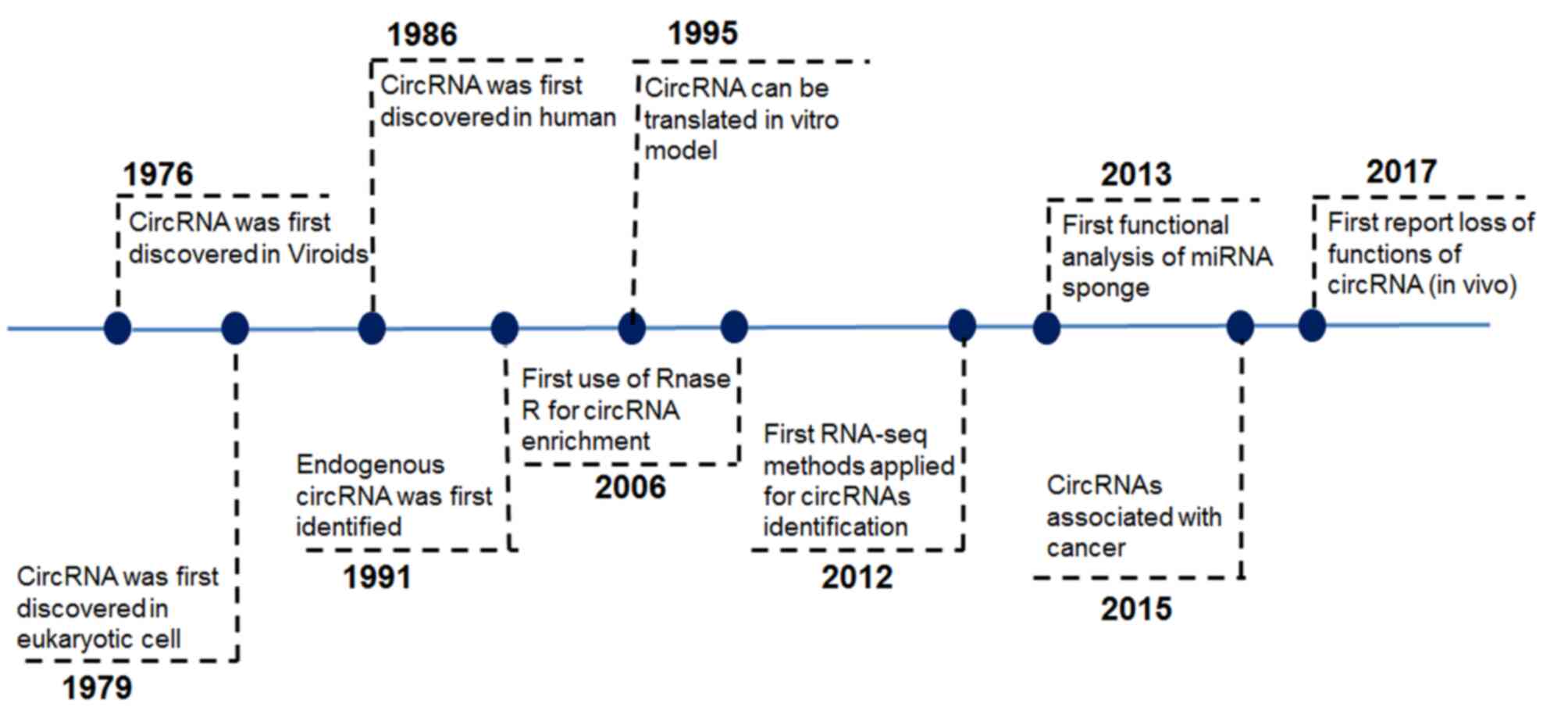
rapidly in recent times on circRNAs after discovering this
non-coding RNA in 1976 (Fig. 1).
Furthermore, scientists are trying to discover the biological
mechanisms behind cancer development regulated by circRNAs and
assessing them as biomarkers, making the process of diagnosis
easier and promoting personalized medicine.
Thus, in the present article the role of recently
discovered circRNAs in cancer development has been illustrated and
how they may offer therapeutic potential for the cure of cancer has
been reviewed. Moreover, technological advances in the field of
discovery and annotation of circRNAs, clinical relevance and
manifestations, and commercial biopharmaceutical aspects (patents)
have also been discussed elaborately.
In most eukaryotic genes, intronic sequences between
the exons are removed by the spliceosome from the nascent precursor
mRNA. As per the literature available, it has been considered for
long that after transcription, most introns are removed
sequentially and rapidly to allow exons to link to form a
functional linear mRNA covalently. The well-regulated splicing
process generates a diversity of functional spliced transcripts
with distinctive exon arrangements (19). CircRNAs also undergo this splicing
mechanism to form a covalently linked structure. Though information
regarding the regulation of circRNA biogenesis is not very clear,
it has the characteristic of covalently joining of 3 end of one
exon to upstream of 5′ end by the process of back splicing to form
circRNA with the help of flanking Alu repeats in circularized exon
(20–24). Back splicing can be regulated by
binding splicing factors to regulatory elements of the cis-acting
splicing site (25,26). An alternative process for circRNA
biogenesis includes lariat precursors in exons which can be spliced
internally, and introns can be removed to produce mature circRNA
(27). Studies have shown that
outcomes of back-splicing are related to the elongation rate of RNA
Polymerase II and are under the tight control of cis-elements
(28,29). Moreover, circRNAs are mainly
processed posttranscriptionally and are stable. Strikingly,
endogenously the productivity of circRNA from pre-mRNA is
exceptionally low (28).
Nevertheless, certain circRNAs are in higher concentration than
their related linear mRNAs, particularly in the nervous system, and
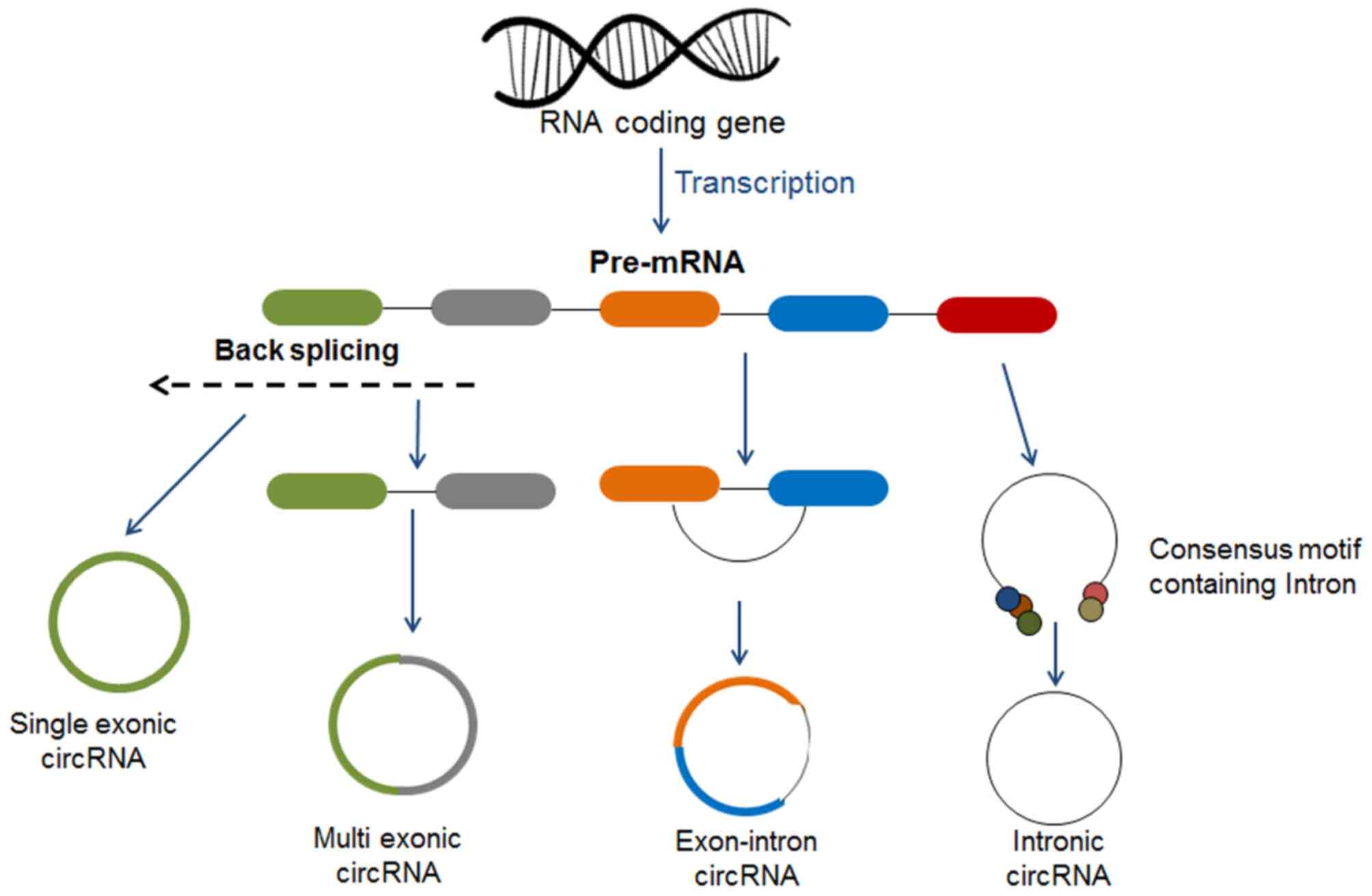
can play a substantial role in organismal phenotypes (29). A majority (>80%) of circRNAs are
derived either from single or several exons in humans and are
termed exonic circRNAs (ecircRNAs). Other than ecircRNAs,
high-throughput sequencing has identified three other types of
circRNAs, which are exon-intron circular RNAs (EIciRNAs), having
both introns and exons; circular intronic RNAs (ciRNAs), having
introns only; and tRNA intronic circular RNAs (tricRNAs), makes
stable circRNA via pretRNA splicing (Fig. 2) (19,30).
Previous studies showed that the length of circRNAs
generally consists of 1–5 exons, and it has also been suggested
that length of exons may be crucial in deciding whether the exon
will circularize or not (6,31). The size of exons in circRNAs varies,
and it can be three times longer as compared with the average exons
that are expressed. Introns that are present adjacent to the exons
which get circularised are also three times longer as compared with
introns that are not present in the flanking regions of pre-circle
exons (31). During the process of
circRNA formation, mostly exon 2 is considered as the acceptor
exon, and it is present upstream (32).
The detection of circRNA is an arduous task as
reverse-transcription quantitative PCR (RT-qPCR) assays are unable
to differentiate between normal linear RNA and circRNA in the
presence of a template of the linear genome for designing primers.
The presence of poly-A tail makes rRNA purification easier, but
circRNAs lack poly-A tail in them, thus it becomes even more
challenging to detect them (33).
In addition, detection issues such as high false discovery rate,
underestimation of back-spliced junction reads, RNase R treatment
efficiency, and uneven rRNA depletion further complicates the
identification of circRNAs (34).
Thus, various computational approaches having stronger algorithms
(having efficient alignment-based strategies) are being employed
for the characterizing of circRNAs.
Computational biology has immensely contributed to
identifying the different novel circRNAs in cancer. Several
scientists are developing various new bioinformatics tools and
continuing their research in this direction (35,36).
Multiple databases have been made available to researchers for
circRNA analysis utilizing these developing techniques with
continuing efforts. Certain bioinformatics tools are
pcircRNA_finder (37),
CircRNAFisher (38), find_circ, and
circTools (39). These databases
can be utilized to analyze various aspects of circRNAs, including
anticipating probable interactions of circRNAs with desirable
molecules, their ability to translate and make proteins, and
evaluating any correlation with diseases. A comprehensive list of
databases to identify the circRNA in various diseases, including
cancer, has been listed in Table I.
Though numerous circRNAs have been identified recently, limited
information is available for their translation in vivo and
probable biological functions.
Recent advances in high-throughput sequencing
techniques and the development of various bioinformatics algorithms
have facilitated the annotation and quantification process for
circRNAs (40). Few studies have
highlighted certain crucial functions of circRNAs, including
protein sponges or decoys, microRNA (miRNA or miR) sponges, and DNA
replication regulators.
There is a presence of multiple or single RNA
binding protein sites within the circRNAs, which act as protein
sponges. From the locus of mannose-binding lectin (MBL), the
protein sponge of circRNA has been derived, which itself has the
MBL protein binding site (21). MBL
proteins can attach themselves with introns, flanking from
circularized exons, and enhance their biogenesis process with
autoregulation. It also leads to the prevention of MBL protein from
binding to other potential targets when they are already bound with
circRNAs. In the presence of excess MBL protein, their production
can be regulated by forming circRNAs by decreasing their mRNA
production. Human antigen R (HuR) binds to both mRNA of PABPN1 and
circRNA from the gene PABPN1 itself, and the translation
process gets enhanced (17).
Translation of HuR mRNA is controlled by binding of HuR to
circPABPN1. mRNAs of several cancer-causing genes and tumor
suppressor genes such as BCL2, VHL, MYC, TP53 and
HIF1A are the efficient targets for HuR. Information
regarding the role of HuR directly in cancer remains unclear, and
it needs to be investigated further (17,41).
By modulating its expression of binding proteins through
protein-protein interactions, circFOXO3 has been hypothesized to be
involved in cancer (42). It was
revealed that circFOXO3 could bind to p53 and MDM2 to sensitize the
breast cancer cells to cisplatin and doxorubicin. Ubiquitination of
p53 is also mediated by circFOXO3 and gradually gets degraded by
the proteasome. More circRNAs that can function as protein
scaffolds need to be detected to understand their molecular
mechanisms in an improved way (43). At present, it can only be
hypothesized that circRNA may form a complex with the proteins
having tumor suppressor activity for preventing normal activities
of the cells. However, to prove this hypothesis, further studies
and analysis are required in the near future.
miRNA sponges or decoys depend upon the interaction
between miRNAs and protein-coding RNAs or non-coding RNAs. Due to
the predominant location of circRNAs in the cytoplasm, circRNAs may
either compete for miRNA-binding sites to modulate the activity of
miRNAs or act as competitive endogenous RNAs (44). circRNAs having the ability to
regulate the activities of miRNAs have an indispensable role in the
pathogenesis of cancers in humans (45,46).
It was found that circRNAs have specific binding sites for miRNAs,
and there is the absence of any single nucleotide polymorphism,
which indicates the presence of conserved sequences within the
organism (47). CircRNAs may
contain multiple or single miRNA binding sites, and due to
conserved sequences, miRNA regulation does not require multiple
miRNA binding sites. However, circCCDC66 from the CCDC66
gene provides more than one binding site for miRNAs, targeting
oncogenes other than genes for tumor suppressors (48). mir-7 was the first circRNA sponge
whose gene regulatory function was revealed, and it has 70
conserved sites for miR-7 binding and is present on the opposite
strand of the CDR1 gene, thus it is considered antisense
CDR1 (6,44). Oncogenes are the target of miR-7,
and its expression and stability were found to be higher in
specific tissues of humans, as a result of which miR-7 targeting
gene's expression will be increased by reducing the activity of
miR-7 (49). CircRNAs possess the
property of miRNA sponges along with their other molecular
functions in the biological system (50). CircRNA, circRNAHIPK3, functions as
the miRNA sponge for cancer, and binding sites for miRNA are being
predicted and verified from the data derived from Argonaute
HITS-CLIP of circRNA (51).
Similarly, circPVT1 was found responsible for stimulating the
growth of cells by sponging miR-125 family members (52,53).
Another example is circRNA itchy E3 ubiquitin-protein ligase
(circ-ITCH), which acts as a miRNA sponge to suppress tumor growth
and increases the level of ITCH (54). It has been observed that the
formation of miRNA and circRNA complex will not result in miRNA
suppression, and circRNAs with binding sites for miRNA will
function as the potential reservoir for miRNAs and facilitate the
transportation of miRNA to exhibit other molecular functions.
CircRNAs present in the nucleus can interfere in the
transcription process or help initiate alternative splicing. Still,
primarily circRNAs are present in the cytoplasm, where they may act
as protein or miRNA decoys, scaffolds, or transporters. There is an
association between RNA pol II and intron-exon circular RNA and U1
snRNP to enhance parental gene transcription (55). The formation of circRNA and
functional mRNA are generally incompatible due to competition among
linear and circular splicing of most of the genes from the host.
Thus, a deviation in the balance between linear and circular
splicing will lead to divergence in the normal transcription
process of genes related to oncogenes or tumor-suppressor.
ci-Ankyrin Repeat Domain 52 (ci-ANKRD52), an abundant RNA,
stimulates transcription of its parental gene, ANKRD52, by
interacting with the elongation pol II complex and getting
accumulated at the transcription sites (56). It has been reported that a special
class of circRNAs, EIciRNA-U1 (small nuclear ribonucleoprotein
(snRNP) complexes amid U1 snRNP and EIciRNAs), may interact with
factors like U1 snRNP through RNA-RNA interaction. To stimulate
gene expression of its parental genes, EIciRNAs-U1 snRNP complexes
may further interact with the RNA pol II transcription complex at
the promoter site (55).
By contrast, circRNA-7 positively affects cell cycle
progression by inhibiting miR-7 activity and enhancing the
mitogen-activated protein kinase pathway (65). On the other hand, the expression of
cyclin-dependent kinase 6 (CDK6) is increased by circSLC30A7, which
leads to the enhancement of cell cycle progression (66). The effect of circRNAs like
circCCDC66, circZKSCAN1, circHIAT1 and circKCNH1 in metastasis has
been identified in vivo and in vitro studies
(62,63,66,67).
Numerous circRNA expressions usually deviate from their normal
functioning in the initial cancer stage but are even detectable at
the later stages of diagnosis (63,68).
It is evident from previous studies that specific
circRNAs are being translated in certain tissues under particular
conditions only (69,70). It has been observed that different
types of human cancer have been regulated by several circRNAs
(Table II). Currently, the circRNA
translation procedure in vivo is not well known. Still,
there is evidence of the presence of elements like ribosomal entry
sites and AUG sites within the circRNAs (31). The functioning of circRNAs is not
dependent upon their host gene, and instead, they are regulated
independently. For detecting and mapping miRNA binding sites and
RNA binding proteins on human circRNA, a new web tool has been
established named Circlnteractome to make understanding more
accessible and quicken analysis (157). Another database named MiOncoCirc
has been made available for the progress of circRNAs as diagnostic
or therapeutic candidates in various types of cancer (158).
Detection of cancer at the initial stages remains a
challenge in the fight against cancer. A scarcity of reliable
biomarkers has always been a hindrance in this cause. Early
diagnosis of cancer and treatment at the initial stages can save
the life of the patient. Thus, the advances in next-generation
biomarkers are an urgent prerequisite to address this issue
(159). Next-generation biomarkers
could help to detect diseases very quickly and efficiently. To
diagnose cancer at an early stage, scientists are trying to develop
the next-generation biomarkers. Specifically, circRNAs are emerging
as reliable biomarkers for detecting different types of cancer
(160,161). Several circRNAs-based biomarker
development incentives are listed in Table III. However, more studies are
required to develop the next-generation circRNAs-based
biomarker.
With the help of the RNA-seq analysis approach, it
has been found that the expression of circRNAs varies from
different tissues and at various stages of development (173). Platelets are translationally
competent circulating blood cells with a significant circRNA
reservoir. Though several circRNAs have been reported from cultured
platelets, the number of circRNAs in cultured cells is quite
smaller than in matured platelets (71,174,175). Hence, further research is required
in platelets derived from patient samples to understand the role of
circRNAs. Another aspect is the circRNAs present in the vesicles
(exosomes and microvesicles) which are generated from platelets. An
abundant amount of circRNAs are present in exosomes compared with
platelets. By varying the miRNAs associated with the sorting and
loading of circRNAs into exosomes, circRNAs may be made to transfer
in recipient cells as required. Nearly ~1,000 circRNAs have been
reported from serum exosomes of patients with colon cancer.
Identified circRNAs may be utilized to distinguish between patients
with colon cancer and healthy normal individuals (14). In KRAS mutant colon cancer cells,
ample circRNAs were observed in secreted extracellular vesicles and
then exosomes compared with cells (176). From 170 patients and 45 healthy
controls, a RT-qPCR analysis of circulatory exosomes revealed that
hsa_circ_0004771 could be used to differentiate patients with CRC
stage I/II and benign intestinal diseases (177). In liver-metastatic pancreatic
ductal adenocarcinoma (PDAC) tissues, a high expression of
circPDE8A was reported and was shown to be closely related to
lymphatic infiltration, TNM stage, and tumor status. Further
studies on the plasma of PDAC patients further confirmed that the
tumor cells release exosomes with circPDE8A (178).
In patients with acute myeloid leukemia (AML),
hsa_circ_0009910 is overexpressed in the bone marrow. It was found
to sponge miR-20a-5p and associated with poor overall survival of
patients. Eliminating hsa_circ_0009910 resulted in the induction of
apoptosis in AML cells (179). On
the other hand, expression of hsa_circ_0004277 was found to be
downregulated in patients with AML. Expression of hsa_circ_0004277
appeared to be restored after chemotherapy and thus can be
considered a potential therapeutic target for AML (180). In patients with AML, circRNA,
circMYBL2, is overexpressed, having FLT3-ITD mutations. Knockdown
of circMYBL2 inhibits growth and promotes differentiation of
FLT3-ITD AML (181). In the case
of chronic myeloid leukemia (CML), overexpression of
hsa_circ_0009910 sponges miR-34a-5p to induce imatinib resistance.
Elimination of hsa_circ_0009910 decreases imatinib resistance and
inhibits cell growth by inducing autophagy and apoptosis (182). In chronic lymphocytic leukemia
(CLL), upregulated circRPL15 sponges miR-146b-3p to activate the
RAS/RAF1/MEK/ERK signaling pathway axis and induce the development
of CLL (183).
Human peripheral whole blood contains a substantial
amount of circRNAs, which may project them as diagnostic tools for
cancer. In human peripheral blood mononuclear cells (PBMCs), a
comprehensive and abundant expression of circRNAs has been
observed. It was observed that the expression of circRNAs in PBMCs
of patients with active tuberculosis was different than in healthy
controls (184). Another analysis
revealed that five circRNAs were increased in the blood of patients
with coronary artery disease (CAD) compared with control (healthy
individuals) (185). Among these
five circular miRNAs, the highly expressed was hsa_circ_0124644. A
study on plasma circRNAs to diagnose patients with hepatocellular
carcinoma (HCC) with hepatitis B virus (HBV) observed that
hsa_circ_0007750, hsa_circ_0000976 and hsa_circ_0139897 were
significantly higher in patients with HCC than in patients with
HBV-related liver cirrhosis and in healthy controls (186). The circ-STIL, circ-ABCC1, and
circ-CCDC66 in the plasma of patients with colorectal cancer (CRC)
were substantially decreased than in the control group. For CRC
diagnosis, together, these three circRNAs demonstrated a
specificity of 85.2% and sensitivity of 64% (187). It was noted from the aformentioned
study that f-circRNAs (from fusion genes) have unique
characteristics as tumor biomarkers due to their highly
cancer-specific expression.
BC remains one of the primary causes of
cancer-related deaths in women worldwide (1.7 million breast cancer
cases and 521,900 breast cancer-related deaths were reported in
2012) (188). Several treatments
such as hormone therapy, targeted therapy and chemotherapy are
being implemented to cure one of the most expensive malignancies
such as breast cancer. ~1/5 of breast cancers worldwide are
diagnosed as ductal carcinoma in situ (DCIS) (189). Though it is treatable, it becomes
a life-threatening type as invasive ductal cancer (IDC) in a few
cases. The hsa_circ_0122662 and hsa_circ_0001358A were found in the
screening of the circRNAs in five patients with DCIS/IDC and MCF-7
(breast cancer cell line) (189).
From the Starbase human pan-cancer tool it was established that
hsa_circ_0001358 was related to five miRNAs (miR-376a-3p,
miR-200c-3p, miR-429, miR-376b-3p and miR-200b-3p). Since a
differential surge in the dynamic expression of circRNAs was
observed in DCIS or IDC, more future studies are prerequisites to
explore the function of these circRNAs in breast cancer
prognosis.
A high-throughput circRNA microarray from samples of
patients with triple-negative breast cancer (TNBC) revealed that
circEPSTI1 (EPSTI1 gene) was a highly expressed circRNA
among the overexpressed circRNAs in the majority of TNBC cells and
tissues, and its deletion induced apoptosis and suppressed cell
proliferation. Additionally, increased expression of circEPSTI1 was
found to be associated with lymph node infiltration, tumor size and
TNM stage (190). A downregulation
of hsa_circ_0025202 has been observed in breast cancer cells and
tissues. Moreover, it was found to be in an inverse correlation
with lymphatic metastasis and histological grade, which decreased
cell proliferation, migration, colony formation, increased cell
apoptosis and rendered cells sensitive to tamoxifen (TAM)
treatment. Hsa_circ_0025202 regulates the expression and activity
of FOXO3a by absorbing miR-182-5p which then affects inhibition of
tumor and TAM sensitization effects (191). Thus, hsa_circ_0025202 can be
considered as a therapeutic target in HR-positive breast cancers
patients, particularly undergoing TAM therapy.
It has been observed that circFOXO3 and tumor
suppressor genes are downregulated in breast tumor samples compared
with non-tumor cells and tissues (18). An increased expression of circFOXO3
has been found to be associated with reduced cell viability and
cell proliferation in the MDA-MB-231, a breast cancer cell line
(42). It was observed that
circFOXO3 could act as a miRNA sponge and play a critical role in
cancer progression.
The circSMARCA5 expression was reduced in breast
cancer tissue compared with its host gene SMARCA5 (83). An induced expression of circSMARCA5
was observed to illicit drug sensitivity in breast cancer cell
lines. The circSMARCA5 can prevent the expression of its host gene
and can enhance the drug sensitivity of breast cancer cells to
cytotoxic drugs, implicating therapeutic potential of it in
drug-resistant breast cancer cells.
One of the prominent leading causes of health
hazards and cancer-related mortality is lung cancer (192). With the increase in the number of
patients with cancer, a greater number of patients are being
diagnosed with multiple primary lung cancer. Diagnostic cases with
similar histologies for intrapulmonary metastasis and multiple
primary lung cancers become more complicated to be adequately
detected with complete assurance of specificity. Due to this,
designing a strategy for the treatment takes a longer time, and by
that time, the condition of the patient has worsened. With the
advancement of technology, targeted and specific drugs are being
applied to treat lung cancers in the advanced stage. This approach
shows positive and effective results in patients with lung cancer
in advanced stages. However, the major challenge is detection at an
early stage to make the process of treatment easier and more
effective for the health of the patient. It is evident from
previous studies that specific types of circRNAs are present
abundantly in specific tissues. The presence of conserved sequences
in circRNAs may contribute significantly to the occurrence of
cancer, and it has noticeable clinical values for the development
of cancerous cells in the lung (33,193).
Of late, a significant increase in the circARHGAP10
expression was reported in non-small cell lung cancer cells and
tissues associated with poor prognosis. Knockdown of this circRNA
resulted in decreased proliferation and metastasis, implicating its
potential role in the treatment of lung cancer. CircRNA from the
gene circ-ITCH was downregulated in lung cancer tissues compared
with non-cancerous tissues (194).
The expression of the parental cancer-suppressive gene,
ITCH, was markedly elevated by the ectopic expression of
circ-ITCH, inhibiting the proliferation of lung cancer cells by
suppressing the Wnt signaling pathway. It was observed that
circ-ITCH may have suppressed miR-214 and miR-7 by acting as a
sponge in lung cancer cells (194). Furthermore, circRNA_100876
expression was also found to be upregulated in non-small cell lung
cancer tissues compared with normal lung tissues (91). A decreased survival rate of patients
was observed with elevated expression of circRNA_100876 compared
with those with reduced expression. Thus, circRNA_100876 may be
considered as a biomarker for lung cancer. In lung squamous cell
carcinoma (LUSC), increased expression of circTP63 was found to
sponge miR-873-3p, eliminating its suppressive effect and elevating
the expression of FOXM1. This event, in turn, upregulated CENPB and
CENPA and caused cell cycle progression. Elevated expression of
circTP63 was associated with higher TNM stage and increased tumor
size in LUSC patients implicating its role in cancer prognosis. In
lung adenocarcinoma, the expression levels of circRNA from gene
ACP6 were found elevated and associated with tumor growth
and metastasis, implicating a novel target for lung cancer
treatment (68). Application of
circRNAs in the treatment of cancer, including lung cancer, may be
possible after developing suitable vector systems in the coming
future. However, concern related to the negative effect of circRNAs
due to mistargeting is true and disturbing. This issue can be
resolved by smartly designing the small interfering (si)RNAs to
target specific circRNAs (in a localized tumor environment).
HCC prognosis is very poor due to the aggressiveness
and reoccurrence rate of cancer. With the progress of HCC, several
circRNAs have been reported to participate in the development and
invasion of hepatoma. Bioinformatic data analysis has revealed that
several circRNAs can regulate the expression of miR-181a-3p. The
miR-181a-3p can regulate the enzyme O(6)-methylguanine-DNA methyltransferase that
is associated with DNA disruption. This possible link between HCC
progression and circRNAs through the regulation of miRNA clearly
implicates a relationship between the two (195). The expression levels of
circβ-catenin were found to increase in liver cancer tissues than
the nearby normal tissues (196).
The circβ-catenin encodes a novel β-catenin isoform that promotes
proliferation and migration of liver cancer cells in vitro
and activates the Wnt signaling pathway that results in the
reduction of tumorigenesis and metastasis in vivo (195). In addition, the hsa_circ_0005075
is more highly expressed in large liver cancer tumors than in
smaller tumors, indicating its role in regulating tumor growth
(95). Furthermore, the expression
of hsa_circ_0005075 was found to be associated with the
pathogenesis of patients with HCC. Analysis of pathways and gene
ontology studies revealed it to be associated with cell adhesion, a
crucial factor for cell proliferation and metastasis. The
hsa_circ_0005075 decreases the miR-23b-5p expression in cancer by
acting as a miRNA sponge (196).
In HCC tissues and portal vein tumor thrombus metastasis, the
expression of circ0003998 was upregulated (95), which acts as a sponge to miR-143-3p
to reduce the suppressive effect of FOSL2 [epithelial-mesenchymal
transition (EMT) related stimulator]. Moreover, to enhance the
expression levels of EMT-related gene CD44v6, circ0003998
can bind to PCBP1 (94).
In comparison with adjacent liver tissues, HCC
tissues show suppressed expression of hsa_circ_0001649 (104). siRNA-mediated inhibition of
hsa_circ_0001649 increased the expression levels of pro-metastatic
matrix metalloproteinases (MMP)9, MMP10 and MMP13, implicating its
negative correlation with metastasis of HCC (164). Likewise, a suppressed expression
of circHIAT1 was observed in HCC cell lines and tissues (197). The overexpression of circHIAT1
decreased HCC progression by regulating the miR-3171/PTEN axis. In
HCC tissues, a significant increase in the expression of ciRS-7 has
been reported (198). A release of
miR-7 is known to be associated with HCC cell proliferation and
metastasis (198,199). Therefore, it is likely that ciRS-7
may sponge miR-7 and regulate its downstream targeted genes. A
recent study showed that the hsa_circ_0079929 expression was less
in HCC tissues (97). The
PI3K/AKT/mTOR signaling pathway was found to be responsible for the
inhibitory effect of hsa_circ_0079299. Another circRNA,
circ-0051443, was significantly downregulated in plasma exosomes
and tissues (96). It was reported
that circ-0051443 from exosomes was able to act as a sponge for
miR-331-3p to promote BAK1 expression resulting in the inhibition
of the progression of HCC (96).
CircRNA derived from the genes SHPRH and ZKSCAN1 were
found to be downregulated in HCC cells as compared with
non-cancerous cells (78,200).
An abnormal expression of circRNA has been reported
in patients suffering from osteosarcoma, and the overall survival
rate is relatively low. An increased expression of circLRP6 was
observed in osteosarcoma, causing a shorter disease-free and
overall survival (105). CircLRP6
binds to LSD1 and EZH2 and acts as an oncogene to suppress KLF2 and
APC expression levels. The downregulation of circ-PVT1 can reduce
the encoding of a classical multidrug resistance-associated gene
(ATP binding cassette subfamily B member 1), implying circ-PVT1 to
be a more efficient diagnostic marker than alkaline phosphatase for
osteosarcoma (124).
High-throughput analysis revealed abnormal expression of
circ-5′-nucleotidase cytosolic II in osteosarcoma, and it was found
to be involved in regulating miR-488 and stimulating tumor cell
growth and metabolism (201).
Samples from patients with osteosarcoma show
comparatively more upregulated expressions of circUBAP2 than
non-cancerous cells. Higher expression of circUBAP2 results in
inhibition of apoptosis (202). It
was revealed to enhance the expression of SEMA6D to improve the
cisplatin resistance by sponging miR-506-3p and activating the Wnt
signaling pathway (103).
During the differential expression screening of
circRNAs in CRC tissues and normal tissue samples, circCCDC66 was
observed to regulate various pathological processes, such as
invasion, migration and anchorage-independent growth (48). High expression of hsa_circ_001569
has been observed in CRC compared with non-cancerous tissues
(116,203). It was suggested that
hsa_circ_001569 via miR-145 could promote cell proliferation and
invasion in CRC via blocking the downregulation of E2F5/BAG4/FMNL2.
Likewise, in lung cancer, a reduced expression of circITCH was
observed in colorectal tissues compared with adjacent normal
tissues (127). Previously, it has
been shown that circ-ITCH possesses sponge activity for miR-7,
miR-214 and miR-20a in CRC (127).
These three miRNAs have been reported to be involved in regulating
the cell cycle, including cyclin D1 (a proliferative target
gene). The circ-ITCH expression may block the gene expression of
the Wnt signaling pathway like cyclin D1 and c-Myc, implicating its
role in regulating the Wnt signaling pathway and thus in cell
proliferation and migration (127).
An increased expression of hsa_circ_0000069 was
reported in CRC. It was identified that siRNA-mediated inhibition
of hsa_circ_0000069 suppressed the cell proliferation, migration
and invasion, and exerted cell cycle arrest in HT-29 cells.
Similarly, a high expression level of the circ-BTG3 associated
nuclear protein (BANP) was observed in CRC (123). A siRNA-mediated knockdown of
circ-BANP reduced the cell proliferation of CRC. In addition, the
knockout resulted in the downregulation of phosphorylated
(p)-protein kinase B (Akt), involved in the regulation of cell
cycle and survival. This points toward the involvement of circ-BANP
in the phosphoinositide-3 kinase (PI3K)/Akt signaling pathway in
CRC (123).
In CRC cell lines, a downregulation of
hsa_circ_001988 was observed compared with normal colon mucosa
(120). The expression level was
found to be associated with the perineural invasion and cancer cell
differentiation. The characteristic of perineural invasion is a
marker for CRC prognosis in patients, suggesting hsa_circ_001988 as
a potential biomarker for CRC prognosis.
In case of morbidity and mortality related to
cancer, glioma is one of the most common primary form of cancer. By
performing deep sequencing in 10 pathologically diagnosed
glioblasts, an upregulation of circ-FBXW7 was observed (107). The circ-FBXW7 was revealed to
regulate the expression of a novel 21-kDa protein, designated as
FBXW7-185aa. Overexpression of FBXW7-185aa suppressed the
proliferation and acceleration of cell cycle in glioblast cell
lines while promoting malignant phenotypes on the deletion of
FBXW7-185aa, implicating a positive correlation with the overall
survival of patients with glioblastoma (107). In glioma tissues and cell lines, a
significant increase in circSCAF11 expression was observed
(108). Moreover, a poor clinical
outcome in glioma patients was found to be related to the ectopic
overexpression of circSCAF11. The study revealed that
miR-421/SP1/VEGFA axis is utilized by circSCAF11 to exert its
tumorigenic effect in glioma (108). Gliomagenesis is enhanced in
glioblastoma and oligodendroglioma due to the increased expression
of a circRNA from the gene VCAN (204). However, cirZNF292 was found to be
downregulated in patients with glioma (58). In a few cases, circRNAs can absorb
proteins to affect tumorigenesis, apart from acting as miRNA
sponges. For example, highly expressed circCPA4 in glioma tissues
is proposed to function as a sponge for let-7 and regulate the
translation of CPA4 and glioma progression. In human glioma cells,
an upregulated expression of circTTBK2 was reported (115). Increased expression of circTTBK2
was found to be responsible for enhanced cell proliferation,
migration and invasion. circTTBK2 acted as a sponge for miR-217,
and the overexpression of miR-217 into the glioma cells was able to
reverse the circ-TTBK2-induced progression of glioma. CircRNAs have
also been reported to encode certain proteins that are involved in
tumor progression. circ-SHPRH was shown to regulate the expression
of SHPRH-146aa (a 17 kDa truncated protein), and the expression of
both is low in glioma (205).
Increased expression of SHPRH-146aa prevents the degradation of
full-length SHPRH protein from proteasomal degradation. Stabilized
SHPRH protein ubiquitinates proliferating cell nuclear antigen as
an E3 ligase, resulting in inhibition of cell proliferation and
tumorigenicity (205).
From the data of RNA-seq analysis, it was found that
expression of circRNAs varies at different stages of ovarian
cancer, and it has been observed that the expression of circRNA is
higher in ovarian cancer cell lines than the surface epithelial
cells of the ovary that are made immortalized normally (63). A faster cell proliferation rate was
found in the surface epithelium of ovarian cells that are made
immortalized than the cell lines of ovarian cancer, which
facilitates the accumulation of circRNAs within a short time
(60). In ovarian cancer tissues
resistant to cisplatin, the expression levels of circRNA, CDR1as
were found substantially downregulated (206). It was observed that CDR1as
utilizes the miR-1270/SCAI signaling pathway to exert a sensitizing
effect in cisplatin-resistant ovarian cancer. Another circRNA,
circPLEKHM3, was also found suppressed in ovarian cancer tissues
compared with non-cancerous tissues (113). CircPLEKHM3 was reported to act as
a sponge for miR-9 and thus inactivating AKT1 signaling by
regulating the expression of BRCA1, DNAJB6 and KLF4. It was
suggested as a prognostic marker as well as a therapeutic target
for ovarian cancer. In ovarian cancer cell lines, upregulated
expression of hsa_circ_0061140 was observed. A deletion of
hsa_circ_0061140 inhibited the cell proliferation and process of
migration in ovarian cancer. It was shown that hsa_circ_0061140
could sponge miR-370 to inhibit the expression of FOXM1 (114). Another circRNA, hsa_circ_0051240,
was also reported to be significantly upregulated (207). hsa_circ_0051240 was found to
induce cell proliferation and migration in ovarian cancer by
inhibiting the miR-637/KLK4 axis. An increased expression of
circRNA, circWHSC1, was found to be associated with proliferation,
migration and invasion of ovarian cancer cells. This possibly
occurs due to the fact that circWHSC1 can sponge miR-1182 and
miR-145 to enhance the MUC1 and hTERT expressions (115).
A number of studies have been performed to analyze
circRNA expression in bladder cancer. Through RNA-sequencing
analysis of bladder cancer tissues, the expression levels of
circRNA, circSLC8A1, were found downregulated (132). It was reported that circSLC8A1 may
act as a tumor suppressor by sponging miR-130b and miR-494 and
regulating the downstream target gene, PTEN. During bladder
cancer progression, androgen receptor-mediated regulation of
circRNA, circFNTA was reported (134). Androgen receptor-mediated
increased expression of circFNTA was related to the sponging of
miR-370-3p and inducing the expression of its host gene
FNTA. This causes activation of KRAS signaling in bladder
cancer cells, rendering them sensitive to cisplatin. It was
suggested that to suppress metastasis and induce chemo-sensitivity
in bladder cancer cells, circFNTA may be considered as a
therapeutic target. Microarray analysis of bladder carcinoma
reported dysregulation of 469 circular transcripts compared with
normal tissues (208). Among them,
the aforementioned study reported that circRNA, circTCF25, can
enhance cell proliferation, migration, and invasion by sponging
miR-103a-3p and miR-107. miR-103a-3p and miR-107 regulate the
expression of CDK6, involved in cell proliferation and migration,
thus implanting the regulatory role of circTCF25 in bladder
carcinoma. Another microarray analysis of bladder carcinoma
revealed that the expression level of circRNA-MYLK was upregulated
(61). CircRNA-MYLK was reported to
sponge miR-29a and enhance the expression of its target gene,
VEGFA, promoting EMT and progression of bladder cancer
(61). In urothelial carcinoma of
the bladder, advanced clinical stage and survival rate of patients
were positively associated with the upregulation of circRNA,
circPRMT5 (209). The
circPRMT5/miR-30c/SNAIL1/E-cadherin axis was found to be critical
for the progression of urothelial carcinoma of the bladder.
Significant downregulation of circRNA,
hsa_circ_0018069, was observed in bladder cancer tissue (131). Bioinformatics analysis revealed
that it may have an anticancer role by modulating focal adhesion
and calcium signaling pathways. It was shown to have an interaction
with miR-181b-5p, miR-23c, miR-34a-5p, miR-3666 and miR-454-3p.
Similarly, a downregulated expression of circLPAR1
(hsa_circ_0087960) was observed in muscle-invasive bladder cancer
(135). The circLPAR1 was revealed
to act as a sponge for miR-762, and its knockdown increased
invasion and metastasis of muscle-invasive bladder cancer. A
downregulation of circRNA, circMTO1, was found to be positively
associated with the metastasis of bladders cancer and poor survival
of patients (210). circMTO1 was
shown to sponge miR-221, and its overexpression suppressed the
E-cadherin/N-cadherin pathway to inhibit EMT in bladder cancer
cells. Through qPCR analysis, a substantial decrease in the
expression of circRNA, hsa_circ_0000285, was reported in bladder
cancer tissues compared with normal tissues (130). Moreover, patients with
cisplatin-resistant bladder cancer had low expression of
hsa_circ_0000285 compared with cisplatin-sensitive bladder cancer
patients. Its expression was found to be associated with lymph node
metastasis, tumor size, distant metastasis and TNM stage. The
expression of circRNA, circBCRC3, is suppressed in bladder cancer
tissues (128). Ectopic expression
of circBCRC3 causes the proliferation of bladder cancer cells.
CircBCRC3 was shown to sponge miR-182-5p and regulate the
expression of cyclin-dependent kinase inhibitor 1B (p27).
Gastric cancer is considered the fifth most deadly
form of cancer due to associated high morbidity and mortality rates
(738,000 individuals succumb every year as reported in 2014)
(211). Several studies have
highlighted the dysregulation of circRNAs in gastric cancer
progression. Bioinformatics analysis revealed that the expression
level of circRNA, circ-DONSON, was upregulated in gastric cancer
tissues compared with normal tissues (122). The elevated expression of
circ-DONSON was reported to be associated with lymphoid metastasis
and advanced TNM stage. Due to the nuclear localization of
circ-DONSON, circ-DONSON was hypothesized to act as a sponge for
miRNA. It was reported to involve the NURF complex to the promoter
site of SOX4 and regulate its transcription, involved in gastric
cancer malignancy (122). Analysis
of two circRNA databases and qPCR analysis revealed that circRNA,
hsa_circ_002059, is considerably downregulated in gastric cancer
tissues compared with normal tissues (141). The plasma of postoperative
patients with gastric cancer showed different levels of
hsa_circ_002059 compared with plasma from preoperative patients
with gastric cancer, suggesting it to be a potential biomarker for
gastric cancer. An overexpression of circRNA, ciRS-7, was observed
in 102 gastric cancer tissues compared with normal tissues, and its
upregulation was found to be associated with poor patient survival
(142). Furthermore, miR-7-induced
tumor suppression was inhibited by the overexpression of ciRS-7 in
gastric cancer cell lines. ciRS-7 involves the PTEN/PI3K/AKT
signaling pathway to mediate aggressive oncogenic phenotype in
gastric cancer cells. Similarly, the expression level of another
circRNA, circNRIP1, was found to be elevated in gastric cancer
tissues (142). A deletion of
circNRIP1 in gastric cancer cells resulted in suppressed
proliferation, migration, invasion, and decreased the levels of
AKT1. The circNRIP1 was found to positively affect gastric cancer
development by utilizing the circNRIP1-miR-149-5p-AKT1/mTOR
axis.
Another circRNA that has been found to be
upregulated in gastric cancer cells is circPVT1 (53). The circPVT1 has been revealed to
sponge the member of the miR-125 family and promote cell
proliferation in gastric cancer cells, making it a potential
biomarker for gastric carcinoma. From the analysis of the GEO
database, a novel circRNA, circFAT1(e2) (hsa_circ_0001461), was
identified (144). A downregulated
expression of circFAT1(e2) was observed in gastric cancer tissues,
and it was found to be related to the overall survival rate of
patients with gastric cancer. CircFAT1(e2) was found to regulate
the expression of tumor suppressor gene RUNX1 by acting as a
sponge for miR-548g in gastric cancer cells. By controlling the
expression of miR-548g in the cytoplasm and interacting with Y-box
binding protein-1 in the nucleus, circFAT1(e2) was able to inhibit
gastric cancer progression. Another circRNA, circPSMC3, was found
to be downregulated in gastric cells and plasma from patients with
gastric cancer (212). A
suppressed expression of circPSMC3 was found to be associated with
a shorter survival rate of patients and higher TNM stage. CircPSMC3
could modulate the expression of Phosphatase and Tensin Homolog by
sponging miR-296-5p and further suppressing the gastric cancer
progression. Screening for potential circRNAs from 17 gastric
cancer tissues by RT-qPCR and 80 paired gastric cancer tissues by
fluorescence in situ hybridization (FISH) revealed
significantly lower expression of circRNA, circYAP1, in gastric
cancer tissues compared with adjacent normal tissues (213). CircYAP1 inhibited the gastric
cancer progression by sponging miR-367-5p to inhibit the expression
of p27Kip1, suppressing the progression of gastric
cancer. In gastric cancer tissues and cell lines compared with
normal gastric tissues, the expression levels of hsa_circ_0000096
(circHIAT1), were found to be downregulated (67). On deletion of circHIAT1 in gastric
cancer cells, the cell proliferation and migration were inhibited,
and the expression levels of cyclin D1, CDK6, MMP-2, and MMP-9 were
substantially reduced. It was revealed that the expression of Ki67
and VEGF were decreased in gastric cancer xenograft nude mouse
model in a dose-dependent manner after the knockdown of circHIAT1.
An analysis of gastric cancer tissues and cell lines by RT-qPCR
showed that circRNA_0023642 was significantly upregulated (147).
A suppressed expression of circRNA_0023642 was
found to be associated with tumor inhibitory effects such as
suppressed cell proliferation, migration, invasion and induction of
apoptosis. CircRNA_0023642 was found to regulate the EMT signaling
pathway and act as a promoter for metastasis in gastric cancer. RNA
sequencing and bioinformatics analysis of gastric cancer tissues
identified circRNA_LARP4 (circLARP4) as a sponge for miR-424
(148). Ectopic expression of
miR-424 was shown to promote proliferation and invasion in gastric
cancer cells by modulating the large tumor suppressor kinase 1
gene. Utilizing FISH in tissues of patients with gastric cancer, an
elevated expression of circRNAs, circDLST, was observed (149). A deletion of circDLST inhibited
cell proliferation, cell invasion and metastasis, while
overexpression of circDLST rendered opposite effects. circDLST was
shown to favour tumorigenesis and metastasis in gastric cancer
cells by activating the NRAS/MEK1/ERK1/2 signaling via sponging of
miR-502-5p. Activating HuR-derived circRNA, circ-HuR
(hsa_circ_0049027) was found to be downregulated in gastric cancer
cells (150). Circ-HuR
downregulates the expression Hur gene and suppression of
gastric cancer progression by interacting with CCHC-type zinc
finger nucleic acid-binding protein.
Next-generation sequencing profiling showed
abundant circRNA, circOSBPL10 (derived from the OSBPL10
gene), in gastric cancer cells (151). A knockdown of circOSBPL10
inhibited proliferation, migration and invasion of gastric cancer
cells. circOSBPL10 was found to promote tumor growth and metastasis
by utilizing the circOSBPL10-miR-136-5p-WNT2 axis.
A downregulation of circRNA, hsa_circ_0000745, was
observed in gastric cancer tissues and plasma samples (152). In gastric cancer tissues, the
expression level of hsa_circ_0000745 was associated with tumor
differentiation, while in plasma, it was found to be associated
with the tumor-node-metastasis stage. The expression level of
hsa_circ_0000745 in plasma along with carcinoembryonic antigen was
suggested as a potential diagnostic marker for gastric cancer
malignancy. Another circRNA that has been projected as a biomarker
for gastric cancer is hsa_circ_0000520 (153). A significantly reduced expression
of hsa_circ_0000520 was observed in gastric cancer tissues, plasma
and gastric cancer cell lines. Moreover, hsa_circ_0000520 was
reported to have an interaction with a total of 9 miRNAs.
The current outlook is to understand the role of
the immune system in tumor biology and how it participates in
identifying and destroying cancer cells. A total of two types of
tumor antigens are expressed by tumor cells which are
tumor-associated antigens (TAAs) and tumor-specific antigens
(TSAs). Generally, both tumor and normal cells express TAAs, but
TSAs are only expressed in tumor cells (214). The immune system responds to the
tumor cells by targeting TSAs or TAAs. Malignant tumor cells are
recognized by the immune system as a foreign body during
immunosurveillance and are eliminated (215). Previous studies have revealed that
tumor cells adopt certain immune evasive mechanisms to avoid immune
surveillance and establish a microenvironment analogous to normal
tissue (215,216). The tumor microenvironment (TME) is
a complex, dynamically regulated ecosystem harboring cancerous
cells, surrounding stromal cells and immune cells (217). Immune cells play a critical role
during cancer progression and are the most significant cell
population in TME. The population of immune cells in TME usually
comprises CD4+ T cells, CD8+ T cells,
neutrophils, T regulatory cells, B cells, dendritic cells, natural
killer cells (NK), tumor-associated neutrophils, macrophages and
myeloid-derived suppressor cells. A complex composition at the
cellular and molecular level, including signaling molecules from
the immune cells, forms a tumor immune microenvironment (TIME),
closely associated with an adaptive or innate immune response to
cancer cells (218,219). Previous studies have highlighted
that cancer cells can modulate the immune cells in their favor and
assist in immune escape and cancer progression. Thus, targeting the
cross-talk between cancer and immune cells in TME may be a
potential approach to treat cancer (217,218).
Recently, the roles of circRNAs in the TME have
been recognized, appreciated and reviewed (220,221). CircRNAs have been shown to
modulate the function of immune cells like macrophages, neutrophils
and NK cells in various types of cancer (222), also discussed under circRNAs in
various types of cancer in the present review. Moreover, promoting
immunosuppression and inducing resistance against cancer therapies
has also been attributed to the circRNAs (223,224). In pancreatic cancer tissues,
overexpression of circPTPN22 was found to be associated with cancer
size. circPTPN22 was shown to stimulate STAT3 acetylation,
resulting in immunosuppression. A knockdown of circPTPN22 was shown
to induce high immune cell infiltration (NK cells, CD8+
T cells, CD4+ T cells and γδT cells) of tumor and
suppress tumor growth. CircRNAs (circ0000831, circ005019,
circ0031584 circ0006935 and circ0001730) have been revealed to
induce changes in the TME of recurrent nasopharyngeal carcinoma.
Altered distribution of immune cells and a decline in the ratio of
CD4+/CD8+ T cells were observed in the
presence of these circRNAs (225).
Targeting T cell activation by inhibiting the checkpoints such as
the PD-1/PD-L1 pathway is termed checkpoint immunotherapy (226). In lung cancer tissues, the
expression of circ-CPA4 and PD-L1 is reportedly high, while the
expression of miR-let-7 is low. High expression of PD-L1 has been
observed to induce tumor immune escape, resistance to therapy and
cancer progression (227).
circ-CPA4 was revealed to regulate the activation of
CD8+ T cells in TIME by let-7 miRNA/PD-L1 axis. Elevated
expression of circ-CPA4 was shown to inhibit miR-let-7 and promote
exosomal PD-L1 secretion, resulting in the inactivation of
CD8+ T cells and increased resistance against cisplatin
(228). In lung adenocarcinoma,
circRNA, circRNA002178 (contained in tumor exosomes) can induce the
expression of PD-1 receptors in CD8+ T cells,
inactivating CD8+ T cells and promoting tumorigenesis
(229). CircRNA, circ-LAMP1, is
overexpressed in T cell lymphoblastic lymphoma and inhibits cell
apoptosis. It promotes cell growth by sponging miR-615-5p [targets
domain receptor tyrosine kinase 2 (DDR2)] and activating DDR2, a
family member of receptor tyrosine kinase and closely associated
with tumor progression (230).
Tertiary lymphoid structures (TLS), closely
associated with B cells, are present in a wide number of cancer
tissues (231). A number of
studies have acknowledged the importance of TLS in tumor
immunotherapy. CircRNAs have been shown to mediate the antibody
response directly or indirectly. CircRNA
NC_006099.4:15993284|16006290 and circRNA NC_006099.4:1
6132825|16236906 related to parental gene, NFATC2 was shown
to mediate B cell proliferation via Foxp1 pathway (232). A study in plasma of patients with
HCC revealed that hsa_circ_0064428 was associated with high
tumor-infiltrating lymphocytes (TILs), and it may function as a key
regulator of TIL formation (233).
The role of circRNAs in regulating NK cells has also been reported.
The expression of the inflammatory gene, MMP2, is regulated
by hsa_circ_0008433 by sponging miR-181c-5p and miR-181b-5p,
leading to attack of NK cells on arterial fibers causing
progression of the aneurysm (234). In pancreatic cancer, the
overexpression of circ_0000977 results in sponging of miR-153,
resulting in the accumulation of HI1FA. This causes inhibiton of NK
cell-mediated lysis, promoting immune escape properties in
pancreatic cancer cells (235).
Recent discoveries revealed that circRNAs regulate the activation
of macrophages. A circRNA microarray analysis for the expression of
circRNAs in two different polarizing macrophages (M1 and M2)
activation revealed differential expression of circRNAs. CircRNAs,
circRNA003780 circRNA010056, and circRNA010231 were highly
expressed in M1 macrophages, while the expression of
circRNA-003424, circRNA-013630, circRNA-001489 and circRNA-018127
was higher in M2 macrophages. Differential expression of circRNAs
in two subtypes of macrophages having a distinct function in tumor
progression demonstrates the role of circRNAs in the
differentiation and polarization of macrophages and thus a
potential in tumor immunotherapy (236). Higher expression of another
circRNA, circ-CDR1as, was found to be associated with a higher
ratio of M2 macrophages implicating a role of circ-CDR1as in TME
via M2 macrophages (237).
TME has to be reprogrammed for a potent therapy
against cancer, and thus identification of novel therapeutic which
can influence TME is a prerequisite. CircRNAs have been found to
regulate the TIME by influencing the regulatory mechanism of immune
cells such as T cells, B cells, macrophages and NK cells. Though
tumor immunotherapy plays a substantial role in the treatment of
cancer, the application of circRNAs in tumor immunotherapy remains
in its fancy state. To date, no preclinical trials have been
reported for circRNAs alone as therapeutic targets or along with
vectors in cancer. However, it is likely that with the ongoing
efforts in the field of circRNAs and TME, novel immunotherapy
involving circRNAs can be expected.
Different novel bioinformatics algorithms and high
throughput technologies have been developed to detect the novel
circRNAs in cancer (158).
High-throughput RNA-seq technologies can be used for the discovery
of circRNAs. Like, RNA-seq technologies are being employed and
being further developed properly for the discovery of circRNAs,
which can profile non-polyadenylated transcripts (238). At present, it has been observed
that Ribo-Zero and RNase R are two standard gold techniques for
discovery of circRNAs (158). This
first gold standard technique, RNase R, has been developed for the
enrichment of circRNAs from the sample, which is a significant
milestone for degrading linear RNAs and enriching circRNAs within
samples (31). At the same time,
Ribo-Zero has also helped to enrich circRNAs. It can deplete the
rRNAs and preserve linear transcripts (239). At the same time, bioinformatics
technology has developed several tools and servers to classify
circRNA from other noncoding RNA. In this direction, machine
learning and deep learning approaches have been used to classify
the circRNAs (240,241).
Scientists are developing new circRNA-based
technology and applying the patents for these developed
technologies (Fig. 5) (158). Recently, Wesselhoeft et al
(5) developed a technology for
circRNA-related transfer vehicles. The technologies have been
developed by a team of researchers from MIT, and patented in the
USA (USA patient no: US20210371494A1). Researchers from China have
filed Chinese patents claiming prostate cancer therapy can be
treated using one circRNA (circCRKL). Jinshan Hospital of Fudan
University has also applied for the patent (Chinese patent no.
CN109908369A). Again, technology was developed to detect
triple-negative breast cancer using specific circRNA (Chinese
patent no. CN107254519B). Another patent was filed from China to
develop a new technology that describes circRNA (hsa _ circ
_0004872) for use in gastric cancer treatment. Shandong University
has filed the patent (Chinese patent no. CN110117658B).
For pharmacological applications, the therapeutic
RNA products should possess certain desirable characteristics such
as stability, quality, safety and the ability to retain the
biological activity until it reaches the site of action (specific
organelles, cytoplasm, or nucleus) (242–244). Rapidly expanding functional
abilities of RNAs have been projecting them as next-generation
therapeutics for numerous diseases. With the progress in the field
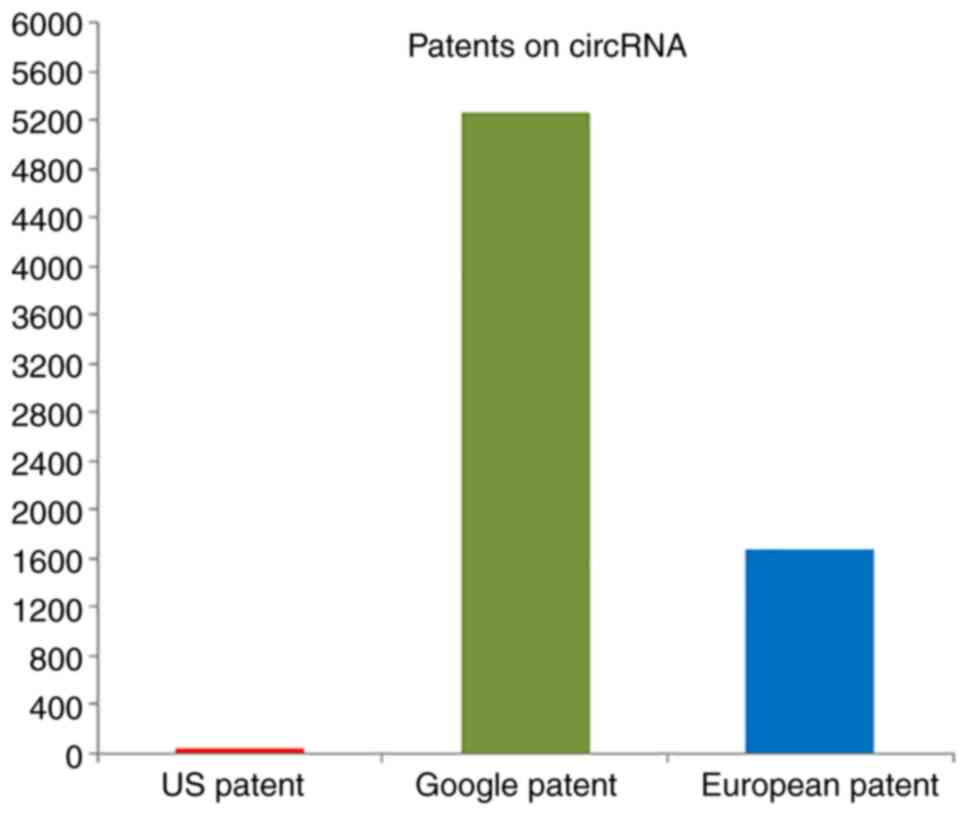
of circRNAs in cancer biology, a number of patents have been issued
in several countries (Fig. 6). A
list of patents for circRNAs in cancer has been summarized in
Table IV. Moreover, with the
advent of new technologies on RNA delivery systems like lipid-based
nanoparticles, polymer-based nanoparticles, and metal
nanoparticles, the delivery of circRNAs to the site of action can
be made more feasible (244). It
is expected that in the near future this next generation of
therapeutics will be available in the commercial market for the
treatment of several types of cancer.
At the initial stage of research, circRNAs were
considered to be the errors in the process of RNA splicing.
Gradually with the progression of research in the field of
bioinformatics, several endogenous circRNAs were found and
identified with the involvement of RNA-seq approaches to reveal the
mystery of circRNAs, but it needs more research to find out the
specific mechanism behind the biosynthesis of circRNA and its
significant role in the pathogenesis of cancer. By the development
of certain tools like CIRI (51),
find-circ (61), MapSplice
(208), circRNA finder (53), Acfs (245) and CIRCexplorer (6) has made the work of finding circRNA
markedly easier and faster, and their role in specific tissues was
also discovered. It has been found that circRNAs are highly
conserved in their sequences within the cells of mammals. As
circRNAs are found to be tissue-specific, thus ideally, they can be
targeted in such a way that there will be no interference with the
liner mRNA (246). CircRNAs are
recognized as the biomarkers for cancer, thus with the involvement
of a larger clinical sample size, their sensitivity and efficiency
can be figured out as the potential targets in different
individuals at different stages of cancer for getting a validated
result (19,52). With more in-depth analysis and the
progress of advanced scientific technologies, the biological
functions of circRNAs would be elucidated in the near future.
Moreover, circRNAs have the potential role in influencing the
choice of therapy as they may be favored for cancer resistance to
chemotherapy. As discussed earlier, in various types of cancer
resistant to certain drugs, the expression of certain circRNAs has
a regulatory role in cancer resistance to drugs. However, advanced
research is required for their site-specific delivery and
understanding the complete mechanism of regulation of tumor
biology. It would be interesting to decipher the role of circRNAs
as biomarkers and or therapeutic targets for the treatment of
cancer.
It can be summarized that with the advancement of
research and technology, the role of circRNAs in cancer treatment
has come into recognition which may help design therapeutic and
diagnostic strategies to combat the problem of cancer. But in the
current scenario, very little is known regarding functions of
circRNAs, and a lot of research must be carried out to project them
as therapeutic targets for cancer treatment. However, it is a very
new and promising field of cancer research. The aspect of
post-transcriptional modification has not been revealed properly
yet in circRNA biology. Activity and stability of RNA vary due to
chemical modifications such as inosine to adenine,
N6-methyladenosine, 5-methylcytosine, N1-methyladenosine and
5-hydroxymethyladenosine. Thus, the activity of circRNA is likely
to get hampered due to aberrant modification of RNA and thus needs
further validation for safe efficacy in clinical trials. According
to the data that has been revealed to date, circRNAs appear to be a
promising structural element as an efficient biomarker for cancer
and potential and valuable therapeutic targets for cancer
treatment.
Not applicable.
The present study was supported by the Hallym University
Research Fund and by Basic Science Research Program through the
National Research Foundation of Korea (NRF) funded by the Ministry
of Education (NRF-2020R1C1C1008694 and NRF-2020R1I1A3074575).
All data generated or analyzed during this study
are included in this published article.
ARS researched data for the article and wrote the
manuscript. SB substantially contributed to the discussion of the
topic and contributed to writing of the article. MB and CC
contributed to the editing, generating the figures and tables of
the manuscript. AS reviewed the manuscript. SSL and CC reviewed and
edited the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mok SW, Fu SC, Cheuk YC, Chu IM, Chan KM,
Qin L, Yung SH and Kevin Ho KW: Intra-articular delivery of
quercetin using thermosensitive hydrogel attenuate cartilage
degradation in an osteoarthritis rat model. Cartilage. 11:490–499.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carbone A: Cancer classification at the
crossroads. Cancers (Basel). 12:9802020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chakraborty S and Rahman T: The
difficulties in cancer treatment. Ecancermedicalscience.
6:ed162012.PubMed/NCBI
|
|
5
|
Wesselhoeft RA, Kowalski PS and Anderson
DG: Engineering circular RNA for potent and stable translation in
eukaryotic cells. Nat Commun. 9:26292018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharma AR, Bhattacharya M, Bhakta S, Saha
A, Lee SS and Chakraborty C: Recent research progress on circular
RNAs: Biogenesis, properties, functions, and therapeutic potential.
Mol Ther Nucleic Acids. 25:355–371. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jeck R, Jeck WR, Sorrentino JA, Wang K,
Slevin MK, Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular
RNAs are abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilusz JE and Sharp PA: A circuitous route
to noncoding RNA. Science. 340:440–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui C, Yang J, Li X, Liu D, Fu L and Wang
X: Functions and mechanisms of circular RNAs in cancer radiotherapy
and chemotherapy resistance. Mol Cancer. 19:582020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen T and Yang Y: Role of Circular RNA in
diagnosis, development and durg resistance of lung cancer. Chin J
Lung Cancer. 22:532–536. 2019.(In Chinese). PubMed/NCBI
|
|
12
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zamarin D, Holmgaard RB, Subudhi SK, Park
JS, Mansour M, Palese P, Merghoub T, Wolchok JD and Allison JP:
Localized oncolytic virotherapy overcomes systemic tumor resistance
to immune checkpoint blockade immunotherapy. Sci Transl Med.
6:226ra2322014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hanan M, Soreq H and Kadener S: CircRNAs
in the brain. RNA Biol. 14:1028–1034. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim KM, Abdelmohsen K, Mustapic M,
Kapogiannis D and Gorospe M: RNA in extracellular vesicles. Wiley
Interdiscip Rev RNA. 82017.doi: 10.1002/wrna.1413. PubMed/NCBI
|
|
18
|
Yang W, Du W, Li X, Yee A and Yang B:
Foxo3 activity promoted by non-coding effects of circular RNA and
Foxo3 pseudogene in the inhibition of tumor growth and
angiogenesis. Oncogene. 35:3919–3931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang G, Li S, Yang N, Zou Y, Zheng D and
Xiao T: Recent progress in circular RNAs in human cancers. Cancer
Lett. 404:8–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Venø MT, Hansen TB, Venø ST, Clausen BH,
Grebing M, Finsen B, Holm IE and Kjems J: Spatio-temporal
regulation of circular RNA expression during porcine embryonic
brain development. Genome Biol. 16:2452015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y and Wang Z: Efficient backsplicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barrett SP, Wang PL and Salzman J:
Circular RNA biogenesis can proceed through an exon-containing
lariat precursor. Elife. 4:e075402015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Xue W, Li X, Zhang J, Chen S,
Zhang JL, Yang L and Chen LL: The biogenesis of nascent circular
RNAs. Cell Rep. 15:611–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wilusz JE: Circular RNAs: Unexpected
outputs of many protein-coding genes. RNA Biol. 14:1007–1017. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bach DH, Lee SK and Sood AK: Circular RNAs
in cancer. Mol Ther Nucleic Acids. 16:118–129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mu Y and Xie F: Research progress of
circular RNA in lung cancer. Chin J Lung Cancer.
21:5432018.PubMed/NCBI
|
|
34
|
Zhang J, Chen S, Yang J and Zhao F:
Accurate quantification of circular RNAs identifies extensive
circular isoform switching events. Nat Commun. 11:902020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu YP, Lin XD, Chen SH, Ke ZB, Lin F, Chen
DN, Xue XY, Wei Y, Zheng QS, Wen YA and Xu N: Identification of
prostate cancer-related circular RNA through bioinformatics
analysis. Front Genet. 11:8922020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang J, Wu A, Yang B, Zhu X, Teng Y and Ai
Z: Profiling and bioinformatics analyses reveal differential
circular RNA expression in ovarian cancer. Gene. 724:1441502020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen L, Yu Y, Zhang X, Liu C, Ye C and Fan
L: PcircRNA_finder: A software for circRNA prediction in plants.
Bioinformatics. 32:3528–3529. 2016.PubMed/NCBI
|
|
38
|
Jia GY, Wang DL, Xue MZ, Liu YW, Pei YC,
Yang YQ, Xu JM, Liang YC and Wang P: CircRNAFisher: A systematic
computational approach for de novo circular RNA identification.
Acta Pharmacol Sinica. 40:55–63. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jakobi T, Uvarovskii A and Dieterich C:
circtools-a one-stop software solution for circular RNA research.
Bioinformatics. 35:2326–2328. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jakobi T and Dieterich C: Computational
approaches for circular RNA analysis. Wiley Interdiscip Rev RNA.
10:e15282019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lebedeva S, Jens M, Theil K, Schwanhäusser
B, Selbach M, Landthaler M and Rajewsky N: Transcriptome-wide
analysis of regulatory interactions of the RNA-binding protein HuR.
Mol Cell. 43:340–352. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:35032020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discovery. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kohlhapp FJ, Mitra AK, Lengyel E and Peter
ME: MicroRNAs as mediators and communicators between cancer cells
and the tumor microenvironment. Oncogene. 34:5857–5868. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Thomas LF and Sætrom P: Circular RNAs are
depleted of polymorphisms at microRNA binding sites.
Bioinformatics. 30:2243–2246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kristensen L, Hansen T, Venø M and Kjems
J: Circular RNAs in cancer: Opportunities and challenges in the
field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Boeckel JN, Jaé N, Heumüller AW, Chen W,
Boon RA, Stellos K, Zeiher AM, John D, Uchida S and Dimmeler S:
Identification and characterization of hypoxia-regulated
endothelial circular RNA. Circ Res. 117:884–890. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang H, Wang G, Ding C, Liu P, Wang R,
Ding W, Tong D, Wu D, Li C, Wei Q, et al: Increased circular RNA
UBAP2 acts as a sponge of miR-143 to promote osteosarcoma
progression. Oncotarget. 8:61687–61697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhong Z, Huang M, Lv M, He Y, Duan C,
Zhang L and Chen J: Circular RNA MYLK as a competing endogenous RNA
promotes bladder cancer progression through modulating VEGFA/VEGFR2
signaling pathway. Cancer Lett. 403:305–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yao Z, Luo J, Hu K, Lin J, Huang H, Wang
Q, Zhang P, Xiong Z, He C, Huang Z, et al: ZKSCAN1 gene and its
related circular RNA (circZKSCAN1) both inhibit hepatocellular
carcinoma cell growth, migration, and invasion but through
different signaling pathways. Mol Oncol. 11:422–437. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jin H, Jin X, Zhang H and Wang W: Circular
RNA hsa-circ-0016347 promotes proliferation, invasion and
metastasis of osteosarcoma cells. Oncotarget. 8:25571–25581. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Weng W, Wei Q, Toden S, Yoshida K,
Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y and Goel A: Circular RNA
ciRS-7-a promising prognostic biomarker and a potential therapeutic
target in colorectal cancer. Clin Cancer Res. 23:3918–3928. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: circRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li P, Chen H, Chen S, Mo X, Li T, Xiao B,
Yu R and Guo J: Circular RNA 0000096 affects cell growth and
migration in gastric cancer. Br J cancer. 116:626–633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan
X, Han S and Wu G: hsa_circ_0013958: A circular RNA and potential
novel biomarker for lung adenocarcinoma. FEBS J. 284:2170–2182.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Alhasan AA, Izuogu OG, Al-Balool HH, Steyn
JS, Evans A, Colzani M, Ghevaert C, Mountford JC, Marenah L,
Elliott DJ, et al: Circular RNA enrichment in platelets is a
signature of transcriptome degradation. Blood. 127:e1–e11. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J,
Wang Z, Wang Q, Li A, Marks JR, et al: CircIRAK3 sponges miR-3607
to facilitate breast cancer metastasis. Cancer Lett. 430:179–192.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R,
Yang SY, Yang DC and Wang XL: Circular RNA hsa_circ_0001982
promotes breast cancer cell carcinogenesis through decreasing
miR-143. DNA Cell Biol. 36:901–908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang
L, Xu Y and Zhang F: Circular RNA profile indicates circular RNA
VRK1 is negatively related with breast cancer stem cells.
Oncotarget. 8:95704–95718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang H, Xiao Y, Wu L and Ma D:
Comprehensive circular RNA profiling reveals the regulatory role of
the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J
Oncol. 52:743–754. 2018.PubMed/NCBI
|
|
76
|
Liu Y, Lu C, Zhou Y, Zhang Z and Sun L:
Circular RNA hsa_circ_0008039 promotes breast cancer cell
proliferation and migration by regulating miR-432-5p/E2F3 axis.
Biochem Biophys Res Commun. 502:358–363. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gao D, Zhang X, Liu B, Meng D, Fang K, Guo
Z and Li L: Screening circular RNA related to chemotherapeutic
resistance in breast cancer. Epigenomics. 9:1175–1188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liang HF, Zhang XZ, Liu BG, Jia GT and Li
WL: Circular RNA circ-ABCB10 promotes breast cancer proliferation
and progression through sponging miR-1271. Am J Cancer Res.
7:1566–1576. 2017.PubMed/NCBI
|
|
79
|
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M,
Liu X, Sun H, Pan Y and Wang S: The pro-metastasis effect of
circANKS1B in breast cancer. Mol Cancer. 17:1602018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ,
Ouyang YX, Feng J, Zhang F, Huang WH, Ying Q, et al: circTADA2As
suppress breast cancer progression and metastasis via targeting
miR-203a-3p/SOCS3 axis. Cell Death Dis. 10:1752019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang Y, Lu T, Wang Q, Liu J and Jiao W:
Circular RNAs: Crucial regulators in the human body. Oncol Rep.
40:3119–3135. 2018.PubMed/NCBI
|
|
82
|
Li H, Li Q and He S: Hsa_circ_0025202
suppresses cell tumorigenesis and tamoxifen resistance via
miR-197-3p/HIPK3 axis in breast cancer. World J Surg Oncol.
19:392021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen
W, Yuan X, Yin W, Xu J, Chen K, et al: CircRNA inhibits DNA damage
repair by interacting with host gene. Mol Cancer. 19:1282020.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li
D and Zhang L: Increased expression of circRNA_102231 in lung
cancer and its clinical significance. Biomed Pharmacother.
102:639–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li X, Zhang Z, Jiang H, Li Q, Wang R, Pan
H, Niu Y, Liu F, Gu H, Fan X and Gao J: Circular RNA circPVT1
promotes proliferation and invasion through sponging miR-125b and
activating E2F2 signaling in non-small cell lung cancer. Cell
Physiol Biochem. 51:2324–2340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z,
Xu W, Zhang E, Wang J, Fang T, et al: The circular RNA circPRKCI
promotes tumor growth in lung adenocarcinoma. Cancer Res.
78:2839–2851. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu T, Song Z and Gai Y: Circular RNA
circ_0001649 acts as a prognostic biomarker and inhibits NSCLC
progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys
Res Commun. 503:1503–1509. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang B, Chen M, Jiang N, Shi K and Qian
R: A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the
proliferation of lung adenocarcinoma. Cancer Biol Ther.
20:1127–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang Z, Xie L, Han L, Qu X, Yang Y, Zhang
Y, He Z, Wang Y and Li J: Circular RNAs: Regulators of
cancer-related signaling pathways and potential diagnostic
biomarkers for human cancers. Theranostics. 7:3106–3117. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Jin M, Shi C, Yang C, Liu J and Huang G:
Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in
NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Mol Ther
Nucleic Acids. 18:219–231. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX,
Liao ZJ and Nan KJ: Over-expression of CircRNA_100876 in non-small
cell lung cancer and its prognostic value. Pathol Res Pract.
213:453–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang L, Long H, Zheng Q, Bo X, Xiao X and
Li B: Circular RNA circRHOT1 promotes hepatocellular carcinoma
progression by initiation of NR2F6 expression. Mol Cancer.
18:1192019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Liang G, Liu Z, Tan L, Su A, Jiang WG and
Gong C: HIF1α-associated circDENND4C promotes proliferation of
breast cancer cells in hypoxic environment. Anticancer Res.
37:4337–4343. 2017.PubMed/NCBI
|
|
94
|
Song LN, Qiao GL, Yu J, Yang CM, Chen Y,
Deng ZF, Song LH, Ma LJ and Yan HL: Hsa_circ_0003998 promotes
epithelial to mesenchymal transition of hepatocellular carcinoma by
sponging miR-143-3p and PCBP1. J Exp Clin Cancer Res. 39:1142020.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular crcinoma development. Medicine (Baltimore).
95:e38112016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen W, Quan Y, Fan S, Wang H, Liang J,
Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular
RNA hsa_circ_0051443 suppresses hepatocellular carcinoma
progression. Cancer Lett. 475:119–128. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zheng H, Chen T, Li C, Xu C, Ding C, Chen
J, Ju S, Zhang Z, Liang Z, Cui Z and Zhao J: A circular RNA
hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma.
Cancer Manag Res. 11:443–454. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang
S, Song W, Li X, Li L, Du Z, et al: A novel FLI1 exonic circular
RNA promotes metastasis in breast cancer by coordinately regulating
TET1 and DNMT1. Genome Biol. 19:2182018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lv S, Li Y, Ning H, Zhang M, Jia Q and
Wang X: CircRNA GFRA1 promotes hepatocellular carcinoma progression
by modulating the miR-498/NAP1L3 axis. Sci Rep. 11:3862021.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX,
Yan PW and Wen J: Emerging roles of circular RNA hsa_circ_0000064
in the proliferation and metastasis of lung cancer. Biomed
Pharmacother. 96:892–898. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang Y, Zhao H and Zhang L:
Identification of the tumor-suppressive function of circular RNA
FOXO3 in non-small cell lung cancer through sponging miR-155. Mol
Med Rep. 17:7692–7700. 2018.PubMed/NCBI
|
|
102
|
Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T and
Zhang L: Analyzing the interactions of mRNAs and ncRNAs to predict
competing endogenous RNA networks in osteosarcoma chemo-resistance.
Mol Ther. 27:518–530. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Gong X, Li W, Dong L and Qu F: CircUBAP2
promotes SEMA6D expression to enhance the cisplatin resistance in
osteosarcoma through sponging miR-506-3p by activating
Wnt/β-catenin signaling pathway. J Mol Histol. 51:329–340. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Song YZ and Li JF: Circular RNA
hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis
by acting miRNA sponge. Biochem Biophys Res Commun. 495:2369–2375.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zheng S, Qian Z, Jiang F, Ge D, Tang J,
Chen H, Yang J, Yao Y, Yan J, Zhao L, et al: CircRNA LRP6 promotes
the development of osteosarcoma via negatively regulating KLF2 and
APC levels. Am J Transl Res. 11:4126–4138. 2019.PubMed/NCBI
|
|
106
|
Deng N, Li L, Gao J, Zhou J, Wang Y, Wang
C and Liu Y: Hsa_circ_0009910 promotes carcinogenesis by promoting
the expression of miR-449a target IL6R in osteosarcoma. Biochem
Biophys Res Commun. 495:189–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Meng Q, Li S, Liu Y, Zhang S, Jin J, Zhang
Y, Guo C, Liu B and Sun Y: Circular RNA circSCAF11 accelerates the
glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol Ther
Nucleic Acids. 17:669–677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yang M, Li G, Fan L, Zhang G, Xu J and
Zhang J: Circular RNA circ_0034642 elevates BATF3 expression and
promotes cell proliferation and invasion through miR-1205 in
glioma. Biochem Biophys Res Commun. 508:980–985. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Li X and Diao H: Circular RNA circ_0001946
acts as a competing endogenous RNA to inhibit glioblastoma
progression by modulating miR-671-5p and CDR1. J Cell Physiol.
234:13807–13819. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen Z and Duan X:
Hsa_circ_0000177-miR-638-FZD7-Wnt signaling cascade contributes to
the malignant behaviors in glioma. DNA Cell Biol. 37:791–797. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Duan X, Liu D, Wang Y and Chen Z: Circular
RNA hsa_circ_0074362 promotes glioma cell proliferation, migration,
and invasion by attenuating the inhibition of miR-1236-3p on HOXB7
expression. DNA Cell Biol. 37:917–924. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J,
Li X, Lu B, Cheng X, Liu P, et al: CircPLEKHM3 acts as a tumor
suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1
axis in ovarian cancer. Mol Cancer. 18:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chen Q, Zhang J, He Y and Wang Y:
Hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and
metastasis in ovarian cancer through miR-370 sponge activity. Mol
Ther Nucleic Acids. 13:55–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zong ZH, Du YP, Guan X, Chen S and Zhao Y:
CircWHSC1 promotes ovarian cancer progression by regulating MUC1
and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer
Res. 38:4372019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhong L, Wang Y, Cheng Y, Wang W, Lu B,
Zhu L and Ma Y: Circular RNA circC3P1 suppresses hepatocellular
carcinoma growth and metastasis through miR-4641/PCK1 pathway.
Biochem Biophys Res Commun. 499:1044–1049. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Chen Z, Ren R, Wan D, Wang Y, Xue X, Jiang
M, Shen J, Han Y, Liu F, Shi J, et al: Hsa_circ_101555 functions as
a competing endogenous RNA of miR-597-5p to promote colorectal
cancer progression. Oncogene. 38:6017–6034. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Tu FL, Guo XQ, Wu HX, He ZY, Wang F, Sun
AJ and Dai XD: Circ-0001313/miRNA-510-5p/AKT2 axis promotes the
development and progression of colon cancer. Am J Transl Res.
2:281–291. 2020.PubMed/NCBI
|
|
119
|
Guo JN, Li J, Zhu CL, Feng WT, Shao JX,
Wan L, Huang MD and He JD: Comprehensive profile of differentially
expressed circular RNAs reveals that hsa_circ_0000069 is
upregulated and promotes cell proliferation, migration, and
invasion in colorectal cancer. Onco Targets Ther. 9:7451–7458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S and Zheng W: Decreased expression
of hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025.
2015.PubMed/NCBI
|
|
121
|
Chen HY, Li XN, Ye CX, Chen ZL and Wang
ZJ: Circular RNA circHUWE1 is upregulated and promotes cell
proliferation, migration and invasion in colorectal cancer by
sponging miR-486. Onco Targets Ther. 13:423–434. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhou C, Liu HS, Wang FW, Hu T, Liang ZX,
Lan N, He XW, Zheng XB, Wu XJ, Xie D, et al: circCAMSAP1 promotes
tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol
Ther. 28:914–928. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhu M, Xu Y, Chen Y and Yan F: Circular
BANP, an upregulated circular RNA that modulates cell proliferation
in colorectal cancer. Biomed Pharmacother. 88:138–144. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z,
Que Z and Liu Y: TTBK2 circular RNA promotes glioma malignancy by
regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol.
10:522017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/β-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Xie F, Li Y, Wang M, Huang C, Tao D, Zheng
F, Zhang H, Zeng F, Xiao X and Jiang G: Circular RNA BCRC-3
suppresses bladder cancer proliferation through miR-182-5p/p27
axis. Mol Cancer. 17:1442018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF,
Bi S, Gui SL, Zhou SB, Qin WB, Wu DM and Wang SQ: Downregulation of
hsa_circ_0000285 serves as a prognostic biomarker for bladder
cancer and is involved in cisplatin resistance. Neoplasma.
66:197–202. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Li M, Wang Y, Liu Y, Zhang X, Liu J and
Wang P: Low expression of hsa_circ_0018069 in human bladder cancer
and its clinical significance. Biomed Res Int.
2019:96818632019.PubMed/NCBI
|
|
132
|
Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen
W, Jiang B, Qin H, Guo X, Liu M, et al: Circular RNA circSLC8A1
acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer
progression via regulating PTEN. Mol Cancer. 18:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the Bladder through
Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Chen J, Sun Y, Ou Z, Yeh S, Huang CP, You
B, Tsai YC, Sheu TJ, Zu X and Chang C: Androgen receptor-regulated
circFNTA activates KRAS signaling to promote bladder cancer
invasion. EMBO Rep. 21:e484672020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Lin G, Sheng H, Xie H, Zheng Q, Shen Y,
Shi G and Ye D: circLPAR1 is a novel biomarker of prognosis for
muscle-invasive bladder cancer with invasion and metastasis by
miR-762. Oncol Lett. 17:3537–3547. 2019.PubMed/NCBI
|
|
136
|
Zhang J, Zhao X, Zhang J, Zheng X and Li
F: Circular RNA hsa_circ_0023404 exerts an oncogenic role in
cervical cancer through regulating miR-136/TFCP2/YAP pathway.
Biochem Biophys Res Commun. 501:428–433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Wang L, Shen J and Jiang Y:
Circ_0027599/PHDLA1 suppresses gastric cancer progression by
sponging miR-101-3p.1. Cell Biosci. 8:582018. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhong S, Wang J, Hou J, Zhang Q, Xu H, Hu
J, Zhao J and Feng J: Circular RNA hsa_circ_0000993 inhibits
metastasis of gastric cancer cells. Epigenomics. 10:1301–1313.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J,
Jin H, Zheng W, Tang W, Cao H and Cao X: Circ-SFMBT2 promotes the
proliferation of gastric cancer cells through sponging miR-182-5p
to enhance CREB1 expression. Cancer Manag Res. 10:5725–5734. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu
X, Li Z, Wei J, Liu M and Li G: Circular RNA circ-DONSON
facilitates gastric cancer growth and invasion via NURF complex
dependent activation of transcription factor SOX4. Mol Cancer.
18:452019. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang
Z, Yu H and Kong D: Overexpression of circular RNA ciRS-7 abrogates
the tumor suppressive effect of miR-7 on gastric cancer via
PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 119:440–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin
M, Liang H and Gao L: A novel circular RNA, circFAT1(e2), inhibits
gastric cancer progression by targeting miR-548g in the cytoplasm
and interacting with YBX1 in the nucleus. Cancer Lett. 442:222–232.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang
W and Cao H: CircPSMC3 suppresses the proliferation and metastasis
of gastric cancer by acting as a competitive endogenous RNA through
sponging miR-296-5p. Mol Cancer. 18:252019. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Liu H, Liu Y, Bian Z, Zhang J, Zhang R,
Chen X, Huang Y, Wang Y and Zhu J: Circular RNA YAP1 inhibits the
proliferation and invasion of gastric cancer cells by regulating
the miR-367-5p/p27 Kip1 axis. Mol Cancer. 17:1512018. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
148
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhang J, Hou L, Liang R, Chen X, Zhang R,
Chen W and Zhu J: CircDLST promotes the tumorigenesis and
metastasis of gastric cancer by sponging miR-502-5p and activating
the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 18:802019. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y,
Li H, Chen Y, Wang X, Huang K, et al: Circ-HuR suppresses HuR
expression and gastric cancer progression by inhibiting CNBP
transactivation. Mol Cancer. 18:1582019. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Wang S, Zhang X, Li Z, Wang W, Li B, Huang
X, Sun G, Xu J, Li Q, Xu Z, et al: Circular RNA profile identifies
circOSBPL10 as an oncogenic factor and prognostic marker in gastric
cancer. Oncogene. 38:6985–7001. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang
W, Liu Z, Cao H and Cao X: Hsa_circ_0000520, a potential new
circular RNA biomarker, is involved in gastric carcinoma. Cancer
Biomark. 21:299–306. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Lai Z, Yang Y, Yan Y, Li T, Li Y, Wang Z,
Shen Z, Ye Y, Jiang K and Wang S: Analysis of co-expression
networks for circular RNAs and mRNAs reveals that circular RNAs
hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are
candidate oncogenes in gastric cancer. Cell Cycle. 16:2301–2311.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Ma HB, Yao YN, Yu JJ, Chen XX and Li HF:
Extensive profiling of circular RNAs and the potential regulatory
role of circRNA-000284 in cell proliferation and invasion of
cervical cancer via sponging miR-506. Am J Transl Res. 10:592–604.
2018.PubMed/NCBI
|
|
156
|
Xu Y, Leng K, Yao Y, Kang P, Liao G, Han
Y, Shi G, Ji D, Huang P, Zheng W, et al: A circular RNA,
Cholangiocarcinoma-Associated Circular RNA 1, contributes to
cholangiocarcinoma progression, induces angiogenesis, and disrupts
vascular endothelial barriers. Hepatology. 73:1419–1435. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Rodrigues PM, Vogel A, Arrese M,
Balderramo DC, Valle JW and Banales JM: Next-generation biomarkers
for cholangiocarcinoma. Cancers. 13:32222021. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Huang L, Rong Y, Tang X, Yi K, Wu J and
Wang F: Circular RNAs are promising biomarkers in liquid biopsy for
the diagnosis of non-small cell lung cancer. Front Mol Biosci.
8:6257222021. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Arias I: Mechanisms and consequences of
ion transport in the liver. Prog Liver Dis. 8:145–159.
1986.PubMed/NCBI
|
|
162
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Sun X, Kong S, Jiang C, Jing R, Ju S and
Cong H: Diagnostic value of circular RNA hsa_circ_0002874
expression in peripheral blood of patients with gastric cancer. Lab
Med. 53:65–70. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang
L, Deng Q and Zheng F: Using circular RNA SMARCA5 as a potential
novel biomarker for hepatocellular carcinoma. Clin Chim Acta.
492:37–44. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Ji W, Qiu C, Wang M, Mao N, Wu S and Dai
Y: Hsa_circ_0001649: A circular RNA and potential novel biomarker
for colorectal cancer. Biochem Biophys Res Commun. 497:122–126.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Li S, Sun X, Miao S, Lu T, Wang Y, Liu J
and Jiao W: Hsa_circ_0000729, a potential prognostic biomarker in
lung adenocarcinoma. Thoracic Cancer. 9:924–930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Ning L, Long B, Zhang W, Yu M, Wang S, Cao
D, Yang J, Shen K, Huang Y and Lang J: Circular RNA profiling
reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers
in epithelial ovarian cancer. Int J Oncol. 53:2637–2646.
2018.PubMed/NCBI
|
|
169
|
Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H,
Shi P, Li J, Gao L and Tian X: Circular RNA profiling reveals
exosomal circ_0006156 as a novel biomarker in papillary thyroid
cancer. Mol Ther Nucleic Acids. 19:1134–1144. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S
and Yuan H: Microarray profile of circular RNAs identifies
hsa_circ_0014130 as a new circular RNA biomarker in non-small cell
lung cancer. Sci Rep. 8:28782018. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W
and Zhang RP: Circulating circular RNA hsa_circ_0001785 acts as a
diagnostic biomarker for breast cancer detection. Clin Chim Acta.
487:363–368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Zhang X, Yang S, Chen W, Dong X, Zhang R,
Ye H, Mei X, Liu H, Fang Y and He C: Circular RNA circYPEL2: A
novel biomarker in cervical cancer. Genes (Basel). 13:382022.
View Article : Google Scholar
|
|
173
|
Bonizzato A, Gaffo E, Te Kronnie G and
Bortoluzzi S: CircRNAs in hematopoiesis and hematological
malignancies. Blood Cancer J. 6:e483. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Ahmed I, Karedath T, Andrews SS, Al-Azwani
IK, Mohamoud YA, Querleu D, Rafii A and Malek JA: Altered
expression pattern of circular RNAs in primary and metastatic sites
of epithelial ovarian carcinoma. Oncotarget. 7:36366–36381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Haemmerle M, Stone RL, Menter DG,
Afshar-Kharghan V and Sood AK: The platelet lifeline to cancer:
Challenges and opportunities. Cancer Cell. 33:965–983. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Pan B, Qin J, Liu X, He B, Wang X, Pan Y,
Sun H, Xu T, Xu M, Chen X, et al: Identification of serum exosomal
hsa-circ-0004771 as a novel diagnostic biomarker of colorectal
cancer. Front Genet. 10:10962019. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L
and Ming Z: Silencing of circ_0009910 inhibits acute myeloid
leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol
Dis. 75:41–47. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C
and Ma D: Characterization of hsa_circ_0004277 as a new biomarker
for acute myeloid leukemia via circular RNA profile and
bioinformatics analysis. Int J Mol Sci. 18:5972017. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C,
Pan Q, Huang W, Fang K, Sun LY, Zhou YF, et al: circMYBL2, a
circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1
to promote FLT3-ITD AML progression. Blood. 134:1533–1546. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Cao HX, Miao CF, Sang LN, Huang YM, Zhang
R, Sun L and Jiang ZX: Circ_0009910 promotes imatinib resistance
through ULK1-induced autophagy by sponging miR-34a-5p in chronic
myeloid leukemia. Life Sci. 243:1172552020. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Wu Z, Sun H, Liu W, Zhu H, Fu J, Yang C,
Fan L, Wang L, Liu Y, Xu W, et al: Circ-RPL15: A plasma circular
RNA as novel oncogenic driver to promote progression of chronic
lymphocytic leukemia. Leukemia. 34:919–923. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Qian Z, Liu H, Li M, Shi J, Li N, Zhang Y,
Zhang X, Lv J, Xie X, Bai Y, et al: Potential diagnostic power of
blood circular RNA expression in active pulmonary tuberculosis.
EBioMedicine. 27:18–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L
and Li M: Peripheral blood circular RNA hsa_circ_0124644 can be
used as a diagnostic biomarker of coronary artery disease. Sci Rep.
7:399182017. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu
QG, Yang Y, Sun SH, Liu JF, Qin LX, et al: Plasma circular RNA
panel to diagnose hepatitis B virus-related hepatocellular
carcinoma: A large-scale, multicenter study. Int J Cancer.
146:1754–1763. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Lin J, Cai D, Li W, Yu T, Mao H, Jiang S
and Xiao B: Plasma circular RNA panel acts as a novel diagnostic
biomarker for colorectal cancer. Clin Biochem. 74:60–68. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
DeSantis CE, Bray F, Ferlay J,
Lortet-Tieulent J, Anderson BO and Jemal A: International variation
in female breast cancer incidence and mortality rates. Cancer
Epidemiol Biomark Prev. 24:1495–1506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Galasso M, Costantino G, Pasquali L,
Minotti L, Baldassari F, Corrà F, Agnoletto C and Volinia S:
Profiling of the predicted circular RNAs in ductal in situ and
invasive breast cancer: A pilot study. Int J Genomics.
2016:45038402016. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Chen B, Wei W, Huang X and Xie X, Kong Y,
Dai D, Yang L, Wang J, Tang H and Xie X: circEPSTI1 as a prognostic
marker and mediator of triple-negative breast cancer progression.
Theranostics. 8:4003–4015. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Sang Y, Chen B, Song X, Li Y, Liang Y, Han
D, Zhang N, Zhang H, Liu Y, Chen T, et al: circRNA_0025202
regulates tamoxifen sensitivity and tumor progression via
regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther.
27:1638–1652. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Douillard JY, Tribodet H, Aubert D,
Shepherd FA, Rosell R, Ding K, Veillard AS, Seymour L, Le Chevalier
T, Spiro S, et al: Adjuvant cisplatin and vinorelbine for
completely resected non-small cell lung cancer: Subgroup analysis
of the Lung Adjuvant Cisplatin Evaluation. J Thorac Oncol.
5:220–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Romaszko AM: Multiple primary lung cancer.
A literature review. 27:725–730. 2018.PubMed/NCBI
|
|
194
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH Suppresses lung cancer proliferation
via inhibiting the Wnt/β-catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Caiment F, Gaj S, Claessen S and Kleinjans
J: High-throughput data integration of RNA-miRNA-circRNA reveals
novel insights into mechanisms of benzo[a]pyrene-induced
carcinogenicity. Nucleic Acids Res. 43:2525–2534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circbeta-catenin promotes liver cancer cell
growth through activation of the Wnt pathway. Genome Biol.
20:842019. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Wang Z, Zhao Y, Wang Y and Jin C: Circular
RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by
regulating miR-3171/PTEN axis. Biomed Pharmacother. 116:1089322019.
View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The Circular RNA Cdr1as Act as an oncogene in hepatocellular
carcinoma through targeting miR-7 Expression. PLoS One.
11:e01583472016. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Xu L, Zhang M, Zheng X, Yi P, Lan C and Xu
M: The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of
hepatic microvascular invasion in hepatocellular carcinoma. J
Cancer Res Clin Oncol. 143:17–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Lu WY: Roles of the circular RNA
circ-Foxo3 in breast cancer progression. Cell Cycle. 16:589–590.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Liu X, Zhong Y, Li J and Shan A: Circular
RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation
and metastasis through targeting miR-448. Oncotarget.
8:114829–114838. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Gong L, Li W, Dong L and Qu F: Correction
to: CircUBAP2 promotes SEMA6D expression to enhance the cisplatin
resistance in osteosarcoma through sponging miR-506-3p by
activating Wnt/β-catenin signaling pathway. J Mol Histol.
51:4712020. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Song X, Zhang N, Han P, Moon BS, Lai RK,
Wang K and Lu W: Circular RNA profile in gliomas revealed by
identification tool UROBORUS. Nucleic Acids Res. 44:e872016.
View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Zhao Z, Ji M, Wang Q, He N and Li Y:
Circular RNA Cdr1as Upregulates SCAI to suppress cisplatin
resistance in ovarian cancer via miR-1270 suppression. Mol Ther
Nucleic Acids. 18:24–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Zhang M, Xia B, Xu Y, Zhang Y, Xu J and
Lou G: Circular RNA (hsa_circ_0051240) promotes cell proliferation,
migration and invasion in ovarian cancer through miR-637/KLK4 axis.
Artif Cells Nanomed Biotechnol. 47:1224–1233. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: Correction: PRMT5
Circular RNA promotes metastasis of urothelial carcinoma of the
bladder through sponging miR-30c to induce epithelial-mesenchymal
transition. Clin Cancer Res. 27:26642021. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Li Y, Wan B, Liu L, Zhou L and Zeng Q:
Circular RNA circMTO1 suppresses bladder cancer metastasis by
sponging miR-221 and inhibiting epithelial-to-mesenchymal
transition. Biochem Biophys Res Commun. 508:991–996. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang
W and Cao H: Correction to: CircPSMC3 suppresses the proliferation
and metastasis of gastric cancer by acting as a competitive
endogenous RNA through sponging miR-296-5p. Mol Cancer. 19:1402020.
View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Liu H, Liu Y, Bian Z, Zhang J, Zhang R,
Chen X, Huang Y, Wang Y and Zhu J: Correction to: Circular RNA YAP1
inhibits the proliferation and invasion of gastric cancer cells by
regulating the miR-367-5p/p27 Kip1 axis. Mol Cancer. 18:1172019.
View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Calabrese L and Velcheti V: Checkpoint
immunotherapy: Good for cancer therapy, bad for rheumatic diseases.
Ann Rheum Dis. 76:1–3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Finn OJ: Cancer immunology. N Engl J Med.
358:2704–2715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Cervantes-Villagrana RD, Albores-García D,
Cervantes-Villagrana AR and García-Acevez SJ: Tumor-induced
neurogenesis and immune evasion as targets of innovative
anti-cancer therapies. Signal Transduct Target Ther. 5:992020.
View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Arneth B: Tumor microenvironment. Medicina
(Kaunas). 56:152020. View Article : Google Scholar
|
|
218
|
Tavakoli F, Sartakhti JS, Manshaei MH and
Basanta D: Cancer immunoediting: A game theoretical approach. In
Silico Biology. 14:1–2. 2020. View Article : Google Scholar
|
|
219
|
Wilkinson K, Ng W, Roberts TL, Becker TM,
Lim SH, Chua W and Lee CS: Tumour immune microenvironment
biomarkers predicting cytotoxic chemotherapy efficacy in colorectal
cancer. J Clin Pathol. 74:625–634. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Song H, Liu Q and Liao Q: Circular RNA and
tumor microenvironment. Cancer Cell Int. 20:2112020. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Ma Z, Shuai Y, Gao X, Wen X and Ji J:
Circular RNAs in the tumour microenvironment. Mol Cancer. 19:82020.
View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Carlos-Reyes Á, Romero-Garcia S,
Contreras-Sanzón E, Ruiz V and Prado-Garcia H: Role of circular
RNAs in the regulation of immune cells in response to cancer
therapies. Front Genet. 13:8232382022. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Katopodi T, Petanidis S, Domvri K,
Zarogoulidis P, Anestakis D, Charalampidis C, Tsavlis D, Bai C,
Huang H, Freitag L, et al: Kras-driven intratumoral heterogeneity
triggers infiltration of M2 polarized macrophages via the
circHIPK3/PTK2 immunosuppressive circuit. Sci Rep. 11:154552021.
View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng
B, Chen C, Chang W, Ping Y, Ji P, et al: Exosomal circPACRGL
promotes progression of colorectal cancer via the
miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 19:1172020.
View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Wang Y, Peng Z, Wang Y, Yang Y, Fan R, Gao
K, Zhang H, Xie Z and Jiang W: Immune microenvironment change and
involvement of circular RNAs in TIL cells of recurrent
nasopharyngeal carcinoma. Front Cell Dev Biol. 9:7222242021.
View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Haanen JB: Immune checkpoint inhibitors
for the treatment of cancer. Ann Oncol. 26:vii72015. View Article : Google Scholar
|
|
227
|
Incorvaia L, Fanale D, Badalamenti G,
Barraco N, Bono M, Corsini LR, Galvano A, Gristina V, Listì A,
Vieni S, et al: Programmed death ligand 1 (PD-L1) as a predictive
biomarker for pembrolizumab therapy in patients with advanced
non-small-cell lung cancer (NSCLC). Adv Ther. 36:2600–2617. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Hong W, Xue M, Jiang J, Zhang Y and Gao X:
Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell
growth, stemness, drug resistance and immune evasion in non-small
cell lung cancer (NSCLC). J Exp Clin Cancer Res. 39:1492020.
View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Deng L, Liu G, Zheng C, Zhang L, Kang Y
and Yang F: Circ-LAMP1 promotes T-cell lymphoblastic lymphoma
progression via acting as a ceRNA for miR-615-5p to regulate DDR2
expression. Gene. 701:146–151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Pitzalis C, Jones GW, Bombardieri M and
Jones SA: Ectopic lymphoid-like structures in infection, cancer and
autoimmunity. Nat Rev Immunol. 14:447–462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Zheng L, Liu L, Lin L, Tang H, Fan X, Lin
H and Li X: Cecal CircRNAs are associated with the response to
salmonella enterica serovar enteritidis inoculation in the chicken.
Front Immunol. 10:11862019. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Weng Q, Chen M, Li M, Zheng YF, Shao G,
Fan W, Xu XM and Ji J: Global microarray profiling identified
hsa_circ_0064428 as a potential immune-associated prognosis
biomarker for hepatocellular carcinoma. J Med Genet. 56:32–38.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Huang Q, Huang QY, Sun Y and Wu SY:
High-throughput data reveals novel circular RNAs via competitive
endogenous RNA networks associated with human intracranial
aneurysms. Med Sci Monit. 25:4819–4830. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Ou ZL, Luo Z, Wei W, Liang S, Gao TL and
Lu YB: Hypoxia-induced shedding of MICA and HIF1A-mediated immune
escape of pancreatic cancer cells from NK cells: Role of
circ_0000977/miR-153 axis. RNA Biol. 16:1592–1603. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Zhang Y, Zhang Y, Li X, Zhang M and Lv K:
Microarray analysis of circular RNA expression patterns in
polarized macrophages. Int J Mol Med. 39:373–379. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Zou Y, Zheng S, Deng X, Yang A and Xie X,
Tang H and Xie X: The role of circular RNA CDR1as/ciRS-7 in
regulating tumor microenvironment: A pan-cancer analysis.
Biomolecules. 9:4292019. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Shi H, Zhou Y, Jia E, Liu Z, Pan M, Bai Y,
Zhao X and Ge Q: Comparative analysis of circular RNA enrichment
methods. RNA Biol. 19:55–67. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Giannoukos G, Ciulla DM, Huang K, Haas BJ,
Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV, et al:
Efficient and robust RNA-seq process for cultured bacteria and
complex community transcriptomes. Genome Biol. 13:R232012.
View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Chaabane M, Williams RM, Stephens AT and
Park JW: circDeep: Deep learning approach for circular RNA
classification from other long non-coding RNA. Bioinformatics.
36:73–80. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Amin N, McGrath A and Chen YP: Evaluation
of deep learning in non-coding RNA classification. Nat Machine
Intelligence. 1:246–256. 2019. View Article : Google Scholar
|
|
242
|
Chakraborty C, Sharma AR, Sharma G, Doss
CGP and Lee SS: Therapeutic miRNA and siRNA: Moving from bench to
clinic as next generation medicine. Mol Ther Nucleic Acids.
8:132–143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Baptista B, Riscado M, Queiroz J, Pichon C
and Sousa F: Non-coding RNAs: Emerging from the discovery to
therapeutic applications. Biochem Pharmacol. 189:1144692021.
View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Damase TR, Sukhovershin R, Boada C,
Taraballi F, Pettigrew RI and Cooke JP: The limitless future of RNA
therapeutics. Front Bioeng Biotechnol. 9:6281372021. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
You X and Conrad TO: Acfs: Accurate
circRNA identification and quantification from RNA-Seq data. Sci
Rep. 6:388202016. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Wang K, Singh D, Zeng Z, Coleman SJ, Huang
Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al:
MapSplice: Accurate mapping of RNA-seq reads for splice junction
discovery. Nucleic Acids Res. 38:e1782010. View Article : Google Scholar : PubMed/NCBI
|