Cancer develops from genetically altered cells with
a high proliferation rate and the ability to disseminate from a
primary location to invade distant sites (1). In the past, scientists assumed that
cancer progression and invasiveness were solely determined by
factors within tumor cells (1,2).
However, focus is now put on cancer-supporting components, which
have been demonstrated to aid tumor cells in manifesting the
disease (3,4). It is now widely established that the
tumor microenvironment (TME) components contribute to different
cancer hallmarks and are thus recognized as possible cancer therapy
targets (2,5–7).
These components include cells of the stroma [cancer-associated
fibroblasts (CAFs), endothelial cells (ECs), pericytes and immune
cells] in addition to non-cellular components, such as the
extracellular matrix (ECM), extracellular vesicles (EVs) or
exosomes, and the microbiome, collectively forming the TME
(5,8,9).
Oxygen levels, metabolites, nutrients and pH have
also been acknowledged as factors that may be controlled by the TME
(10,11). The immunosuppressive and
metabolically stressed nature of the TME serves an instrumental
role in exacerbating the aggressiveness of cancer cells (11). For instance, interactions between
tumor and stromal components may result in additional modifications
of the TME cells, ECM remodeling and angiogenesis, thus leading to
metastasis (12).
Cross-talk between cancer cells and TME components
may also decrease the efficacy of antitumor treatments,
contributing to drug resistance (13). Accordingly, an improved
understanding of the biological and chemical nature of the TME
paves the way for the development of therapeutic strategies for
more efficient targeted cancer therapy. The present review aims to
discuss TME components and their molecular features, and how they
modulate cancer hallmarks. It also reviews key factors of the TME
for targeted cancer treatment, with a focus on current TME pathways
and mediators targeted in interventional clinical trials.
TME refers to all non-cancer cellular components
surrounding tumor cells, and non-cellular components exerting
tumor-supporting roles (2,6).
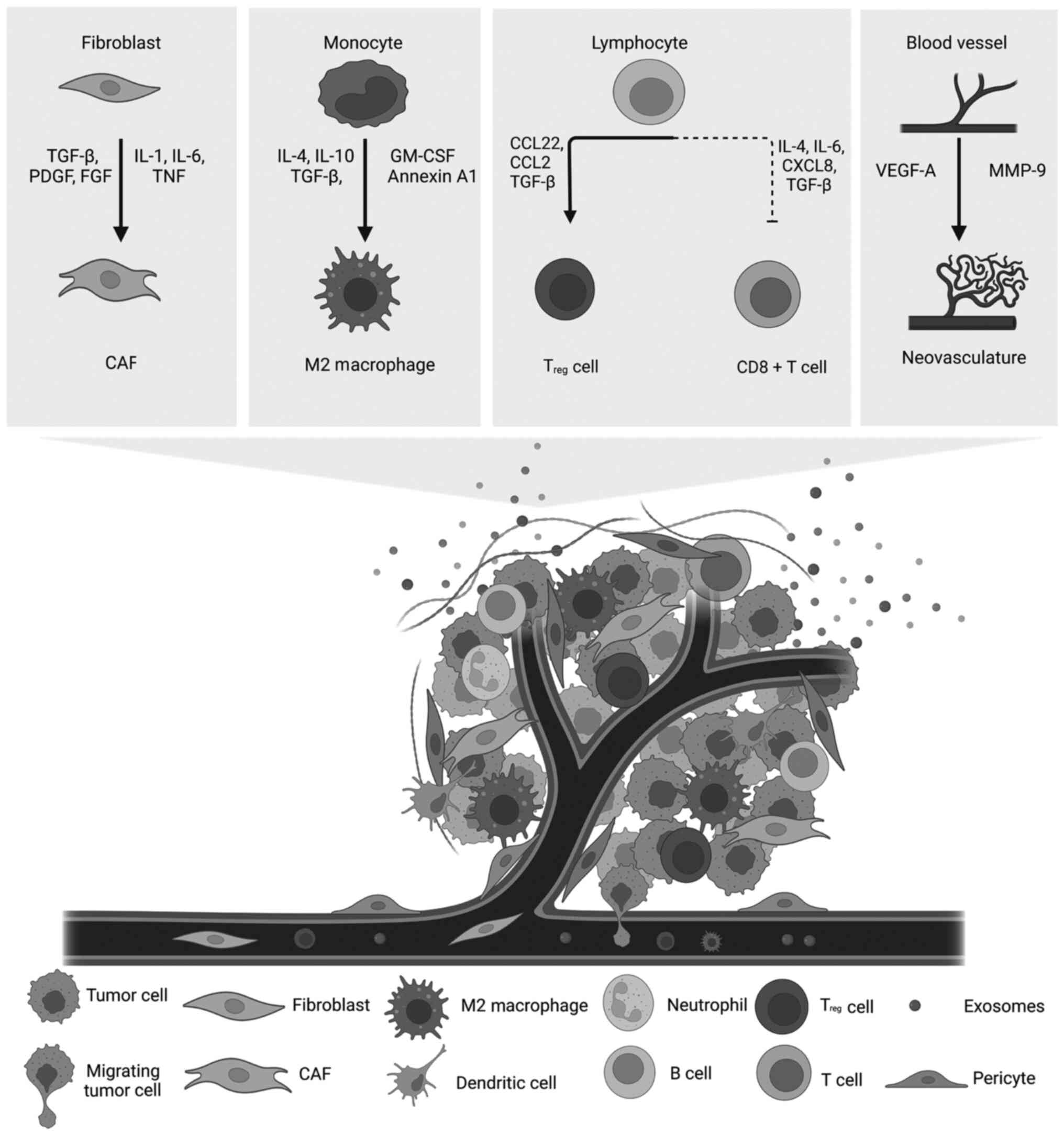
Stromal cells of the TME include CAFs, ECs lining the blood vessels
and immune cells (Fig. 1) that
are recruited by cancer cells from neighboring tissue stroma
(5). Interactions of TME stromal
cells with cancer cells create a protective environment that
promotes tumor growth in both cases (2,5).
Tumor-associated stromal cells not only physically support cancer
cells but also secrete growth factors, cytokines, chemokines and
ECM proteins with tumor-promoting properties (14,15). In addition to stromal cells,
scientists have identified the ECM, the microbiome and cell
messengers referred to as EVs as non-cancerous constituents of the
TME (10,16–18).
Fibroblasts are the prevalent cell type in
connective tissue stroma and the primary source for ECM and
basement membrane proteins (15,19). Most fibroblasts within tumors
differentiate into CAFs (15,20). CAF activation is driven by
different stimuli, such as inflammation (Fig. 1), ECM stiffness and other
physiological stresses (19,21). CAFs are a highly heterogeneous
cell population and, to the best of our knowledge, their origin
remains unclear (15). While it
was earlier hypothesized that most CAFs originate from local
fibroblasts that are activated and reprogrammed to support tumor
growth (22), some groups have
demonstrated that CAFs originate from mesenchymal stem cells,
specifically bone marrow-derived stem cells located in the bones
(23–25). Others attribute their origin to
the human adipose tissue-derived stem cells found in the adipose
tissues (26–28). This emphasizes the remarkable
plasticity of cancer, enabling it to employ different sources to
promote growth and progression (27).
Given their role in angiogenesis, ECs form the inner
lining of the blood vessels and remain the most extensively studied
cells of the TME (36).
Typically, blood vessels enable the exchange of oxygen, nutrients,
wastes and immune cells between the circulatory system and body
tissues (36). Due to the
increased metabolic and nutritional requirements of tumor cells,
ECs branch from pre-existing vasculature to form new blood vessels
(37). Newly formed vessels are
structurally and functionally abnormal because of their leaky
nature and dissimilar chaotic branching that increase the
interstitial fluid pressure rendering a hypoxic and acidic
environment (38). Tumor
vascularization, caused by hypoxia, involves vascular ECs and other
TME cell types, including pericytes and bone marrow-derived
precursor cells (39,40). The pro- and anti-angiogenic
molecules secreted by cells determine the transformation of normal
angiogenic processes to tumor angiogenesis (37,41). While thrombospondin-1 and
Endostatin are major anti-angiogenic factors (42), VEGF-A secreted by cancer cells can
stimulate the formation of new vasculature that, in turn, supplies
tumor cells (Fig. 1) (37). In addition, platelet-derived
growth factor β (PDGFβ) recruits pericytes to the tumor vasculature
aiding blood vessel formation and maturation (43).
Tumor cells and CAFs secrete chemoattractant factors
that recruit various immune cells to their niche (6). TME IICs include immunosuppressive
and antitumor immune cells of myeloid lineages, such as
macrophages, myeloid-derived suppressor cells (MDSCs), neutrophils,
mast cells and dendritic cells (DCs), and lymphoid lineages, such
as B and T lymphocytes, and natural killer cells (NKs) (14,44).
Activated macrophages can be polarized into two main
subtypes: Pro-inflammatory M1 and anti-inflammatory M2 (45). Tumor-recruited macrophages
infiltrating the TME constitute the tumor-associated macrophages
(TAMs), the most abundant immune cells of the TME (46,47). Data indicate that TAMs are mainly
of the M2 subtype, and thus, are tumor-promoting (45,48). TAMs secrete cytokines and soluble
factors that contribute to tumor progression by influencing
angiogenesis, cell migration, invasion and metastasis (2). The presence of TAMs is associated
with poor prognosis in most cancer types (49).
MDSCs represent a unique category of
immunosuppressive myeloid cells that are abundant in the TME
(50,51). Chronic inflammation in cancer
disturbs normal myelopoiesis, and thus, differentiation and
maturation of immature myeloid cells (IMCs) are impaired (52). This disturbance drives the
generation of MDSCs from IMCs (52). MDSC are subdivided in two main
subsets according to their origin and phenotypical and
morphological characteristics. These subsets are the granulocytic
polymorphonuclear neutrophils-MDSCs and monocytic MDSCs (53).
Neutrophils, also referred to as tumor-associated
neutrophils, possess immunosuppressive activity (54). Neutrophils are also polarized into
two subsets: Antitumor N1 and tumor-promoting N2 (54). IICs include mast cells capable of
releasing soluble factors that enhance EC proliferation and promote
tumor angiogenesis (50). DCs are
antigen-presenting cells (APCs) capable of producing
pro-inflammatory cytokines and chemokines and promoting T cell
stimulation (50,55). However, DCs in the tumor exhibit
abnormal antigen-presenting capabilities, and thus, are
dysfunctional (56).
Among the lymphoid lineage of IICs, different types
of T cell populations infiltrate the TME (44). Cytotoxic T cells can eliminate
malignant cells, and thus, are associated with a good cancer
prognosis (44,57). This is the case of the antigen
recognizing cytotoxic memory T cells that are positive for CD8 and
produce IL-2 and IFNγ (2).
CD4+ T helper (Th) cells are divided into different
subtypes: The pro-inflammatory Th1 lineage, anti-inflammatory Th2
cells and the immunosuppressive regulatory T (Treg)
cells (2). The ratio of Th1 to
Th2 cells in cancer is associated with tumor stage and grade
(50,58,59). B cells are also present at the
invasive borders of tumors and in the lymph nodes and lymphoid
structures neighboring the TME (2). In breast and ovarian cancer, the
presence of B cells is associated with a good prognosis (60). On the other hand, the presence of
immunosuppressive regulatory B cells is associated with skin cancer
and may promote lung metastasis, suggesting a type-specific effect
of B cells on cancer (61).
Finally, NKs are cytotoxic lymphocytes capable of killing tumor
cells without antigen presentation (50). NKs control tumor growth by
providing innate immunity to the sites of transformed tumor cells
and inducing cytotoxicity (62,63).
Overall, TME stromal cells, namely CAFs, ECs and
IICs, and their secretome contribute to the growth and development
of tumors (14,15). Notably, the complexity of
interactions between cancer and TME cells demonstrates remarkable
tumor mass heterogeneity (64).
In their review, Koppensteiner et al (65) discussed how negative anticancer
immune responses may result from the interactions between CAFs and
T cells. On the other hand, Mun et al (66) reviewed the positive and negative
relationships between immune and stromal cells of the TME. Despite
the advancement in technologies capable of studying the TME at the
single-cell level (67), a
detailed understanding of all tumor-TME connections remains largely
lacking. Therefore, anticancer strategies that only target one cell
population are inadequate, and need to be fully updated in line
with such rapid discoveries in TME biology.
The ECM is a dynamic network of interconnected
macromolecules in which the cells reside (10,68). It comprises minerals, an array of
extracellular proteins, glycosaminoglycans, and other proteoglycans
(PGs) and polysaccharides (10,68). The main components include
collagens, elastins, fibronectins, laminins, hyaluronic acid (HA),
heparan sulfate, chondroitin sulfate and keratan sulfate (10,68). This intricately organized
structure forms a supportive substrate that serves as a biological
scaffold for surrounding cells and as an anchor for cell attachment
to the ECM (at focal adhesions and hemidesmosomes) (69–71). The ECM also regulates cell-cell
and cell-matrix bidirectional signal transduction, including
transport and mechano-transduction (16,72). This is partly due to the ECM being
a reservoir for EVs and soluble bioactive effectors, such as
cytokines, growth factors, chemokines and enzymes (73–75). ECM components and ECM-associated
factors collectively make up the ECM ‘matrisome’, which is
responsible for regulating transport, proliferation, motility,
survival, homeostasis and other fundamental cellular mechanisms
(76–79).
The ECM is present in all tissues and organs in the
body, including tumors and the TME (68). Its organization is both
cell-specific and tissue-specific (80). The ECM composition within tumors
is heterogeneous and accounts for up to 60% of the tumor mass
(10). Both cancer cells and
stromal cells contribute to the production of the tumor ECM
(77,81). However, CAFs remain the primary
source of ECM in the TME (10,20,74,82). Cancer ECM differs from normal
tissue ECM in composition, organization, density, and physical and
biochemical properties (68).
These differences are also noted across tumors of different
metastatic potentials (83,84). For instance, primary and
pre-metastatic cancers increase ECM production (74,85–88). This TME fibrotic response,
clinically termed desmoplasia, results in a substantial
accumulation of collagens, fibronectins and PGs in benign and
malignant tumors (89,90). Collagen and collagen-processing
enzymes, laminins, integrins, MMPs and HA are among the most
enriched ECM proteins in tumors (10,62,78,81,91,92).
On the other hand, the shift between low and high
molecular weight PGs in different solid tumors illustrates the
association between the composition of the ECM and cancer grade
(93). Similarly, the tumor
environment favors the increase in collagen type I, III or V at the
expense of collagen type IV in breast, ovarian, lung and ductal
carcinoma (94–96).
Finally, the crosslinking of collagen, and other
fibrillary proteins, such as elastins, renders the ECM denser and
stiffer (10,74,91,97–99). Changes in the ECM may be induced
by proteases (MMPs and cathepsins) or nonproteolytic enzymes
(heparanases and hyaluronidases) secreted by tumor and stromal
cells, by oxygen free radicals produced by IICs, or as a response
to hypoxia and acidosis (16,77,100).
The upsurge in the production of ECM components with
altered properties, in turn, reduces the diffusion of nutrients and
metabolites, and modulates cytokine secretion (79,83,101). This creates a hypoxic
tumor-promoting environment capable of stimulating proliferation,
tumor growth, epithelial-to-mesenchymal transition (EMT),
aggressiveness, resistance to cell death, evasion from the immune
system, and invasion and tumor dissemination, among others
(10,97,102). Overall, this highlights the need
for potent ECM-targeting therapies.
EVs are cell messengers, which mediate the signaling
cross-talk between a cell and its environment (103–105). Cancerous and non-cancerous
cellular constituents of the TME, including the microbiome,
communicate with one another by secreting soluble factors and/or
releasing EVs (75,103,105–110). Therefore, EVs are an integral
and functional non-cellular component of the TME (75,103,111,112). Briefly, EVs are
membrane-enclosed particles subdivided into exosomes, microvesicles
and oncosomes depending on their size, biogenesis, function, etc.
(105,113). Exosomes are intraluminal
vesicles destined for exocytosis (105). They exhibit a classic dish or
saucer-like morphology with diameters ranging between 30 and 100 nm
(17,114,115). As the name suggests,
intraluminal vesicles are formed by the inward budding of endosomal
membranes inside the lumen of endosomes (17,114–116). Unlike exosomes, microvesicles
are the products of the outward budding and fission of the plasma
membrane (105,117,118). Their diameters range between 100
and 1,000 nm (105). By
contrast, oncosomes are cancer-specific large EVs with diameters
ranging between 1 and 10 µm (105,118,119). They shed off the ‘non-apoptotic
membrane blebs’ of amoeboid cancer cells (105,120). All aforementioned biogenesis
processes simultaneously result in the packaging of various
cytosolic materials inside the EVs (105). Therefore, the cargo of EVs can
comprise lipids, proteins and nucleic acids, such as DNA, mRNA,
microRNA (miRNA), long non-coding RNA (lncRNA) and circular RNA
(circRNA) (17,114–116,121,122).
EV cargo commonly includes type-specific and or
stage-specific cancer biomarkers (117). For instance, EVs isolated from
patients with ovarian cancer express distinct protein and miRNA
sets compared with those found in cancer-free individuals (114). Similarly, specific RNA classes
are particularly abundant in EVs of patients with triple-negative
breast cancer compared with those with hormone receptor-positive
breast cancer (124,135). Several other non-coding RNAs
have been demonstrated to serve a role in tumor development
(136). The long non-coding
lymph enhancer-binding factor 1-antisense RNA1 has been found to
act as a tumor promoter in a number of malignant tumors; however,
it acts as a tumor suppressor in myeloid cancer (137). Furthermore, circ0021205 is a
non-coding circRNA that promotes cancer progression in
cholangiocarcinoma and non-small cell lung cancer (NSCLC) (138). In patients with colorectal
cancer (CRC), upregulated miRNA-7062-5p inhibits G protein-coupled
receptor 65, thus promoting osteoclast genesis during bone
metastasis (139). Long
intergenic non-coding RNA (LINC)02257 is a survival-associated
enhancer RNA serving important immunotherapy roles in a number of
cancer types (140). In lung
adenocarcinoma, the LINC00987/A2M axis acts as an effective tumor
suppressor, as well as a biomarker for the evaluation of the tumor
immune microenvironment or the prognostic and therapeutic potential
(141).
Further observations have revealed that the
expression of the polymerase I and the transcript release factor
glioma biomarker in EVs is positively associated with tumor grade
(142). The clinical expression
of programmed cell death ligand-1 (PD-L1) in EVs is associated with
diverse cancer types, including melanoma and colon cancer (143–145). Finally, researchers have
reported that both surgery and radiation treatments change the
composition of EVs, thus demonstrating the close association
between tumors and their TME (132,142). These data collectively highlight
the growing interest in EVs as promising targets cancer for
diagnosis, prognosis and treatment.
The microbiome is a component of the TME, which has
become a subject of interest in cancer research recently (18,146). By definition, the microbiome
represents ‘the characteristic microbial community occupying a
reasonably well-defined habitat, which has distinct
physico-chemical properties’ (146). In principle, only the microbial
community present in or around the tumor tissues can strictly be
labeled as the microbiome of the TME (18). However, microbes at distant sites
from tumors (such as the gut) have also emerged as critical
modulators of cancer onset and progression (147–153). Therefore, this section reviews
the composition and role of all microbes with direct or indirect
effects on cancer to overcome the shortcomings of the TME-centric
view and highlight the importance of the different layers of cancer
environment beyond the immediate spatial boundaries of tumors
(i.e., at the level of what is now known as the tumor organismal
environment) (154). The
microbiome is: i) A fingerprint for tumors; ii) a major factor in
the pathology of the disease; and iii) a diagnostic tool to predict
the response to treatment of patients (150,151,155–164). Researchers have reported
substantial differences in the microbiome composition of normal
tissues compared with tumor tissues (162,163,165–173). These observations have revealed
tumor-type specific, tumor subtype-specific and grade-specific
bacteria spanning several major phyla (162,163,165–173). Furthermore, the data suggest
that the majority of bacteria of the tumor microbiome of different
solid tumors are intracellular (165).
The hypoxic nature of tumors contributes to the
abundance of anaerobic bacteria, which are unable to survive in an
oxygen-rich environment (174).
The anaerobic Fusobacterium genus is particularly abundant
in various carcinomas, including oral, colorectal and bladder
cancer (168,171,173–175). For instance, several species of
Fusobacterium, namely, F. nucleatum, F. mortiferum
and F. necrophorum, have been identified in metastatic colon
cancer tissues, while F. nucleatum and F.
periodonticum are a signature of oral cancers (174,175). Further highlighting the
tumor-specificity, the Bacteroides depleted in colon cancers
were, by contrast, profuse in rectal tumors (175,176). In addition, rectal tissue sample
analysis revealed a distinct microbiome in cancer tissues compared
with normal tissues (176,177). Phascolarctobacterium,
Parabacteroides, Desulfovibrio and Odoribacter species
are abundant in tumors, whereas Pseudomonas, Escherichia,
Acinetobacter, Lactobacillus and Bacillus species are
primarily found in healthy tissues (176). Similarly, the microbiome of the
breast cancer TME indicated the enrichment in specific microbes
(Actinobacteria, Listeria spp, Haemophilus influenzae,
Anaerococcus, Caulobacter and Streptococcus) and the
depletion of others (Propionibacterium and
Staphylococcus) (164,177). Interestingly, in lung cancer,
the genera Streptococcus and Staphylococcus exhibit
the same expression trend as that observed in breast cancer
(163). Specifically,
Streptococcus and Neisseria genera thrive in cancer,
unlike Staphylococcus and Dialister, which favor
normal tissues (163). Different
microbes, including Fusobacterium, Streptococcus and
Bacteroides, are also differentially expressed in bladder
cancer, prostate cancer and other cancer types (171,172,178).
The mutational capabilities of cancer cells have
been recognized as the core factor for sustaining cancer
proliferative signaling (187).
Stromal cells serve an important role in augmenting oncogenic
mutations, and thus, the hyperproliferation of cancer cells by
driving mitogenic signals (Fig.
2) (6,7,187).
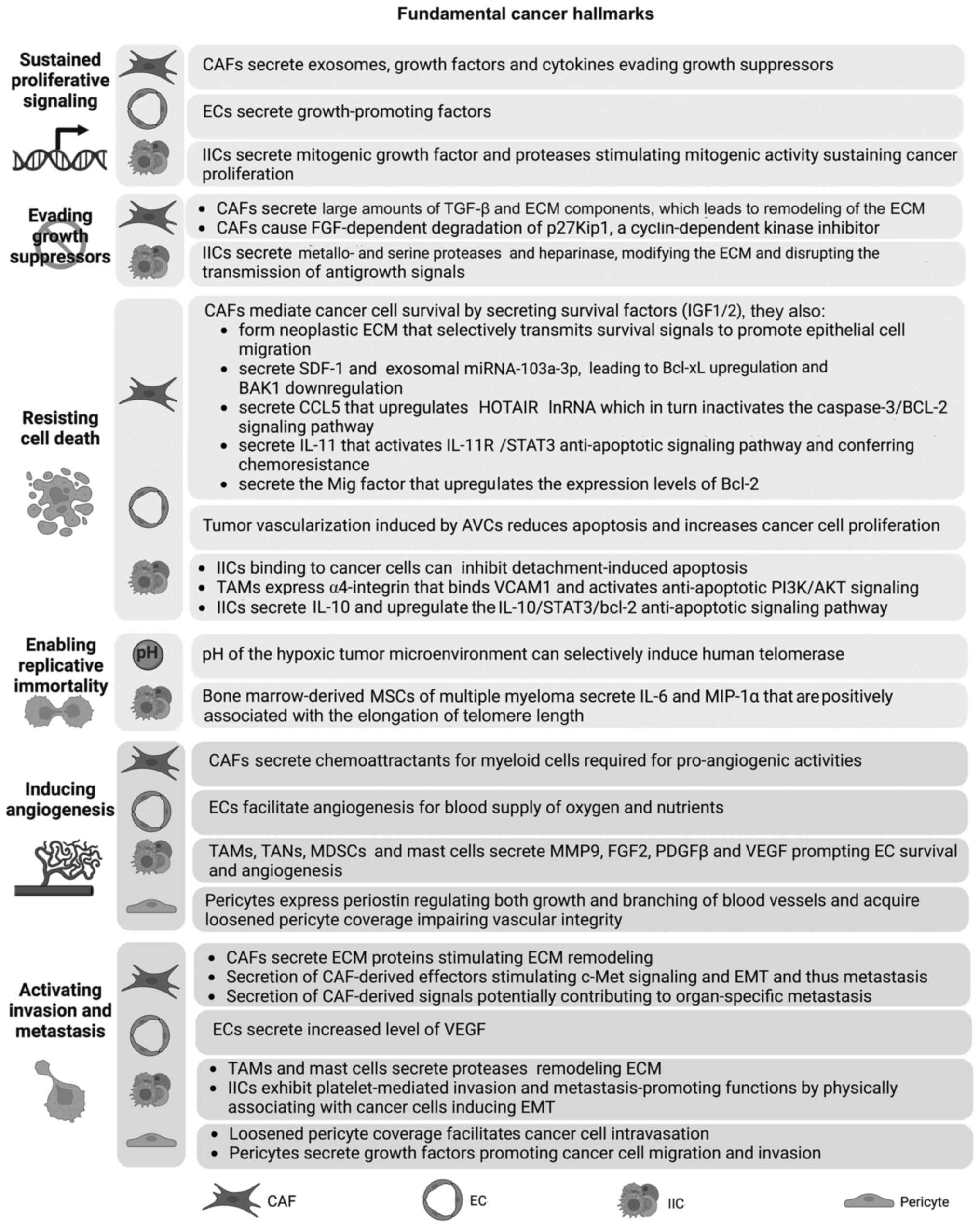
Different CAF subtypes exhibit diverse functions
and affect multiple cancer hallmark capabilities (30). CAFs secrete mitogenic epithelial
growth factors (EGF), fibroblast growth factors (FGF), hepatocyte
growth factors (HGF) and other signaling proteins that drive cancer
cell proliferation (189–191).
HGF secretion results in the activation of mesenchymal epithelial
transition factor (a HGF receptor), and thus, the activation of the
MAPK and PI3K/AKT survival signaling pathways (192). These pathways are also activated
by CAF-secreted vascular cell adhesion molecule 1 (VCAM1) and
promote the proliferation of lung cancer cells (193). Furthermore, leptin, a cytokine
secreted by CAFs, binds to its receptor, activates MAPK/ERK1/2 and
PI3K/AKT signaling pathways, and promotes proliferation of cancer
cells in NSCLC (194).
Angiogenic vascular cells (AVCs) also directly
support cancer hyperproliferation. Experimentally stimulating
angiogenesis results in increased proliferation of cancer cells
(195,196). These ECs secrete
growth-promoting factors that influence multiple hallmarks,
including proliferation, invasion and metastasis (further described
subsequently) (197). Similarly,
IICs stimulate neoplastic cell proliferation by secreting mitogenic
growth factors, such as TNF-α, ILs, chemokines, heparins and
histamine, in addition to EGF, FGF and TGF-β [reviewed in (198)]. Additionally, IICs secrete
metallo- and serine proteases that cleave and modify the ECM
leading to chronic paracrine and juxtacrine mitogenic activity
sustaining cell proliferation (199).
Adhesion molecules at cell-cell and cell-ECM
connections transmit extrinsic growth-suppressing signals to cancer
cell cycle machinery (5,187). As aforementioned, disruption of
adhesion molecules is induced by IIC-secreted metallo- and serine
proteases and heparinase, which cleave and modify the ECM (10). ECM modifications disrupt the
transmission of antigrowth signals and the formation of
growth-suppressing adhesion complexes (200–202).
Notably, fibroblasts naturally exhibit extrinsic
growth-suppressing capabilities to maintain epithelial homeostasis
(15). In the TME, CAFs secrete
high amounts TGF-β and ECM components [reviewed in (203)], thus stimulating mechanical
remodeling of the ECM. Therefore, it was hypothesized that CAFs may
acquire a ‘loss-of-function’ phenotype as they are reprogrammed to
sustain cancer hallmarks (5). TME
components can also affect tumor growth-suppressing signals by
regulating cell cycle check points (204). The TME of renal cell carcinoma
(RCC) exhibits FGF-dependent degradation of p27Kip1, a
cyclin-dependent kinase inhibitor (204). This leads to enhanced tumor cell
proliferation (Fig. 2) (204).
Cancer cells foster an intrinsic ability to resist
cell death programs, mainly apoptosis (187). Stromal cells of the TME confer
an additional protective mechanism for cancer cells to resist cell
death and targeted cytotoxic therapy (Fig. 2) (6). CAFs mediate cancer cell survival by
secreting survival factors [insulin-like growth factor 1 (IGF-1)
and insulin-like growth factor 2 (IGF-2)] (15). CAFs also form neoplastic ECM that
selectively transmits survival signals and promotes epithelial cell
migration (199).
The prominent role of CAFs in resisting cell death
is demonstrated by the contribution of CAFs to chemoresistance
(15). CAFs co-cultured with
NSCLC cell lines have been demonstrated to resist apoptosis and
enhance chemoresistance (205,206). This is achieved by the secretion
of stromal cell-derived factor-1 (SDF-1) and the expression of
exosomal miRNA-103a-3p, which lead to Bcl-xL upregulation and BCL2
antagonist/killer 1 downregulation, respectively (205,206). In a separate study, Sun and Chen
(207) demonstrated that
CAF-secreted C-C motif chemokine ligand (CCL)5 upregulated HOX
transcript antisense RNA (HOTAIR) lncRNA expression. HOTAIR, in
turn, inactivated the caspase-3/BCL-2 signaling pathway in these
cells conferring chemotherapy resistance in NSCLC cells (207). Additionally, the
chemotherapeutic drug, cisplatin, induces CAF-secreted IL-11 in
lung adenocarcinoma (208).
IL-11 activates the IL-11 receptor/STAT3 anti-apoptotic signaling
pathway (208). The monokine
induced by IFN-γ factor is a CAFs-secreted chemokine that has also
been demonstrated to upregulate the expression levels of Bcl-2 and
protect Tca8113tongue squamous cell carcinoma cells from
heat-induced apoptosis (209).
Tumor vascularization induced by AVCs reduces
apoptosis, thus increasing cancer cell proliferation (5). This phenotype is altered by the
administration of vascular disrupting agents that increase cell
death in treated cancer (210).
Binding of IICs to cancer cells can also inhibit detachment-induced
apoptosis (5). TAMs, on the other
hand, express α4-integrin that binds VCAM1 expressed on breast
cancer cells (211). This
interaction initiates a signaling pathway that activates
anti-apoptotic PI3K/AKT signaling and resists apoptosis (211). In addition, IICs secrete
cytokines, leading to cell death resistance (212). For instance, TAM-secreted IL-10
is associated with elevated Bcl-2 expression via upregulation of
the IL-10/STAT3/bcl-2 anti-apoptotic signaling pathway (212). This leads to increased
proliferation and is associated with drug resistance of breast
cancer (212).
Shortening of telomeric DNA obstructs cellular
replication and triggers senescence or apoptosis (5). Cancer cells, however, need to insure
limitless replication as a defense mechanisms to overcome normal
senescence caused by telomere shortening (4,5).
Therefore, cancer cells activate telomerases that stabilize
telomere length and confer replicative immortality (4,213). This critical trait occurs in
>90% of cancers (213).
Telomerase activation is enhanced by the upregulation of the human
telomerase reverse transcription (hTERT) gene (214). The pH of the hypoxic TME can
selectively induce human telomerase (215). At present, there is little
evidence for the contribution of TME stromal cells to stabilizing
telomeres in cancer cells (5).
For instance, bone marrow-derived mesenchymal stem cells (MSCs) of
multiple myeloma secrete IL-6 and macrophage inflammatory protein
(MIP)-1α (216). Li et al
(216) provided evidence of a
positive association between IL-6 and MIP-1α secretion and the
elongation of telomere length. MSCs may thus facilitate multiple
myeloma development (216).
Further research is required to investigate whether other TME
cellular components can regulate telomerase activity and enable
replicative immortality.
Angiogenesis in chronic inflammation is illustrated
by constitutive activation of pro-angiogenic factors (37). In tumors, it is regulated by
different components of the TME, such as CAFs, ECs, different IICs
and pericytes, which secrete angiogenesis-inducing factors
(Fig. 2) (2,5,6,37).
Myeloid cells secrete soluble mediators that impact EC survival and
new vessel remodeling (50). For
instance, TAMs control tumor angiogenesis by producing VEGF-A
(50,217), the bioavailability of which
depends on TAM-secreted MMP-9 (218). Mast cells, on the other hand,
secrete VEGF, histamine and heparin, thus regulating tumor
angiogenesis (50). Mast cells
also secrete proteases, such as MMP-9 and tryptase, which in turn
activate pro-angiogenic signaling pathways (218–220). In addition to the secretion of
pro-angiogenic factors, CAFs synthesize ECM proteins that sequester
angiogenic growth factors and ECM-degrading enzymes (15). For example, in hepatocellular
carcinoma, CAFs secrete VEGF, regulating the enhancer of zeste
homolog-2/vasohibin 1 pathway, thus promoting angiogenesis
(221). CAFs also regulate tumor
angiogenesis by secreting chemoattractants for myeloid cells
(5,222). Additionally, pericytes promote
angiogenesis in glioma by expressing periostin, which regulates
both growth and branching of blood vessels (223).
A key feature of cancer cells is the ability to
spread throughout the body by invasion and metastasis (187). Cancer cells and tumor stromal
cells can mediate local invasive growth or seeding metastases at
distant sites (2,6,7).
Tumor vasculature upregulation of VEGF loosens tight junctions
between ECs and reduces pericyte coverage (62,224). This impairs vascular integrity
and facilitates cancer cell intravasation into circulation
(62). Hypoxia, induced by
hypoxia-inducible factors, then triggers tumor dissemination and
metastasis (224,225). Furthermore, pericytes in the TME
activate TGF-β receptors (226).
The subsequent TGF-β response initiates an autocrine activation
loop (226). Analysis of the
secretome of these activated pericytes has revealed upregulation of
IGF-binding protein-3, a key paracrine factor that has been
demonstrated to promote cancer cell migration and invasion
(226). Additionally, proteases
secreted by TAMs and mast cells remodel ECM components, promoting
tissue invasion and dissemination (227,228). Soluble factors secreted by IICs
also contribute to this hallmark. For instance, TNF-α, secreted by
IICs, activates downstream JNK and NF-κB signaling cascades,
ultimately enhancing MMP-2 and MMP-9 activity (229). Equally importantly, IICs mediate
cancer metastasis by inhibiting the expression of metastasis
suppressor genes. For example, IICs inhibit maspin, a serine
protease inhibitor, which normally acts as a tumor suppressor by
increasing cell adhesion to extracellular matrix. Thus, maspin
inhibition negatively regulates tumor migration and invasion
(230,231).
Platelets also exhibit invasion and
metastasis-promoting functions by physically associating with
cancer cells, inducing EMT, enabling extravasation and forming
secondary tumors at metastatic sites (232). Different components within the
ECM might also initiate or enhance EMT-like processes (10). Collagen reorganization, PG
expression and protease-mediated ECM macromolecule degradation
affect cell invasion and metastasis (16). Finally, CAFs are also implicated
in activating invasion and metastasis (15). CAF-derived effectors, such as HGF
and TGF-β, trigger/activate c-Met signaling and EMT, respectively,
mediating tumor invasion and metastasis (233). In breast cancer cells, IGF-1 and
CXCL12 secreted by CAFs stimulate cancer metastasis to the bone
(234). ECM proteins and
remodeling enzymes produced by CAFs are also considered to support
cancer invasion by modifying the structure and function of the ECM
(7). One study revealed that
cancer cells circulate in the blood alongside CAFs derived from the
primary tumor (235). It has
also been suggested that CAF-derived signals may control
organ-specific metastasis of breast tumors (236). Therefore, CAFs are promising
targets in pre-clinical therapeutic strategies in patients with
breast cancer (236).
Effective destruction of cancer cells necessitates
an influx of immune cells, including Treg cells, NKs and
NK T cells (5,187). However, tumor vasculature is
considered to attenuate the influx of immune cells, rendering them
incapable of killing cancer cells (5). This is partly mediated by high
endothelial venules (HEVs), which typically support the homeostatic
trafficking of immune cells during routine immune surveillance
(43). Absence of HEVs in tumor
vasculature allows cancer cells to evade immune destruction
(237–239).
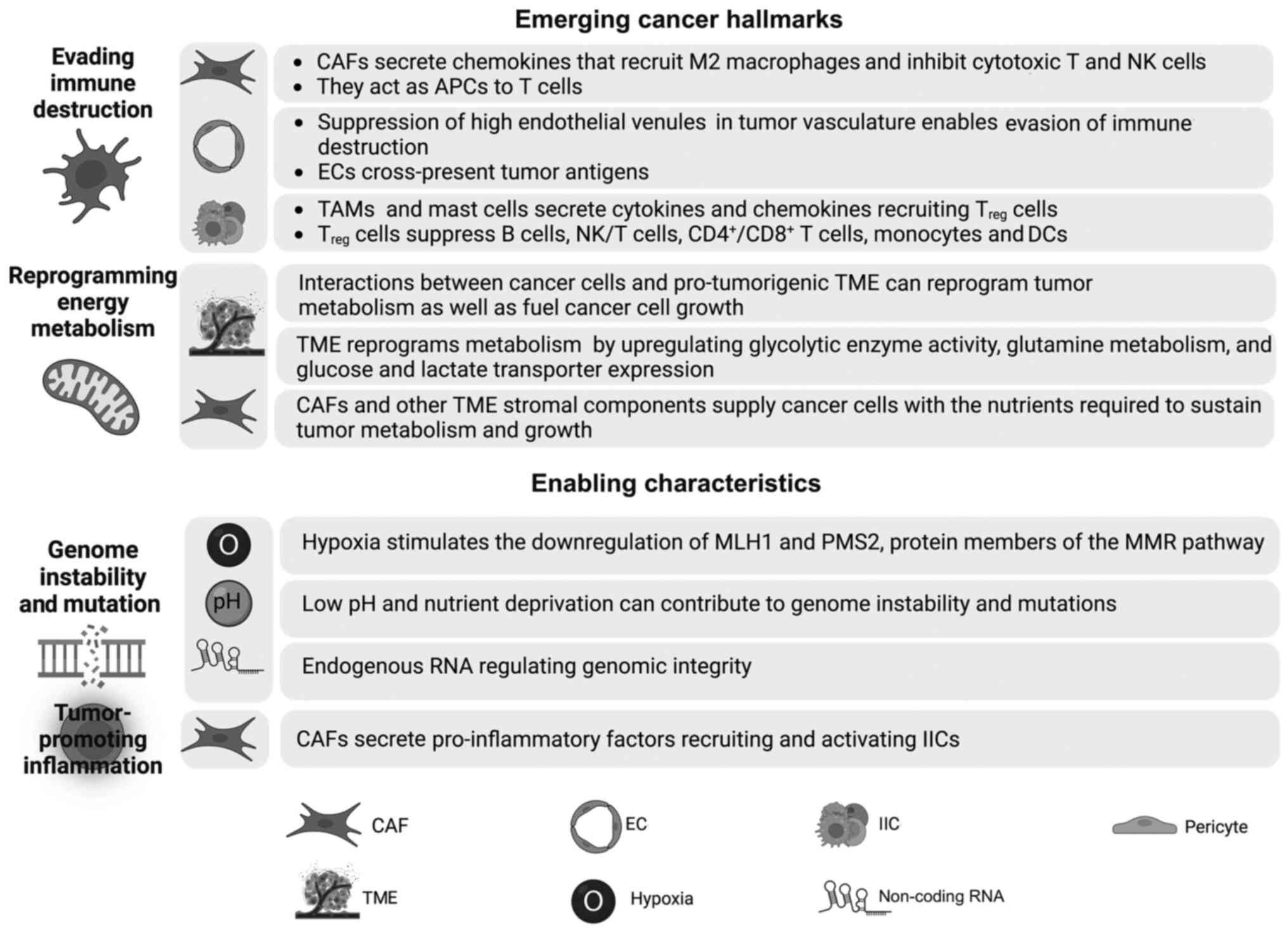
In the TME stroma, ECs cross-present tumor antigens
and stimulate the development of a tolerizing, hence
immunosuppressive, environment (240). CAFs are also essential factors
that allow the tumor to evade immune destruction (Fig. 3). CAF-derived interleukins (IL-4
and IL-6) and chemokines (TGF-β and CXCL8) recruit M2 macrophages
and inhibit cytotoxic T lymphocytes (CTLs) and NK cells (240,241). Among the intricate
tumor-promoting roles of CAFs is the ability to act as APCs to T
cells (20). In addition to ECs
and CAFs, IICs serve an important role in enabling cancer to avoid
immune destruction (44). IICs
prompt immunosuppressive activity that blocks the antitumor effect
of CTLs and NK cells (14). Among
these IICs are TAMs, which lack cytotoxic activity, and release
immunosuppressive factors and suppress CD8+ T cell
proliferation (242–244). CCL22 secreted by TAMs recruits
Treg cells, which enable cancer cells to evade immune
destruction (245).
Treg cells can also be recruited by other cytokines,
such as CCL2 and TGF-β, secreted by mast cells and other
immunosuppressive IICs (50,246). Treg cells can
suppress B cells, NK/T cells, CD4+/CD8+ T
cells, monocytes and DCs (247).
Treg cells suppression can be direct through cell
contact or immunosuppressive soluble mediators or indirect by
suppressing APCs (14). In
particular, Treg-induced inhibition of CD4+ T
cells is mediated by inhibition of receptor-induced calcium,
nuclear factor of activated T-cells and NFκB signaling (50,247). Tumors expressing high levels of
immunosuppressive cytokines are often associated with decreased
CD8+ T cell populations and poor survival (242). Reciprocal communication occurs
between IICs and other TME stromal cells where M2 macrophages
secrete EGF, FGF and TGF-β to support CAF survival and activation
(240). Finally, TAM, MDSC and
mast cell secretion of MMP9, FGF2, PDGFβ and VEGF prompts EC
survival and angiogenesis (240).
Advancements in physiologic magnetic resonance
imaging of tumors have demonstrated that metabolic phenotypes
remain flexible and can switch depending on the surrounding TME
conditions and the nature of the exchanged signaling molecules
(254). Therefore, the
plasticity is contingent on the availability of nutrients and
anaplerotic molecules (255).
Consequently, interactions between cancer cells and pro-tumorigenic
TMEs can reprogram tumor metabolism and fuel cancer cell
proliferation (Fig. 3) (255).
Substantial evidence indicates that tumors can
reshape TME metabolism and even the patients' organismal metabolism
and homeostasis (256–258). For instance, cancer cells can
induce aerobic glycolysis in TME CAFs and stromal cells through the
reverse Warburg effect (259).
Tumor-derived metabolites can also exert immunosuppressive effects
that block the activation and differentiation of various immune
cells of the TME (260,261). In return, CAFs and other TME
stromal components supply cancer cells with the nutrients required
to sustain tumor metabolism and growth (260).
In summary, most findings highlight the ability of
the metabolism to reshape the TME, whereby cancer cells turn the
normal TME into a permissive tumor-promoting environment and a
nutrient factory to be used for efficient energy production
(259,262). Therefore, it is crucial to
review the metabolic features of TME components and to identify
critical factors behind metabolic reprogramming.
Cell metabolism refers to the set of chemical
reactions that sustain the normal functions of cells, including: i)
Production of macromolecule building blocks; ii) energy harvesting;
and iii) the elimination of metabolic wastes (248,263). Cells rely on nutrient uptake and
degradation to generate ATP, the energy currency of cells (248,263). Cancer cells are fast replicating
cells with high anabolic and catabolic energy requirements
(4). These cells can rewire their
metabolism to maintain sufficient ATP production and ensure
proliferation and survival (264–266). Metabolism reprogramming is
achieved by a complex interplay of various signaling pathways,
which can be intrinsically triggered by oncogenes or extrinsically
influenced by the inhospitable TME (264,267–269). One such mechanism is the
significant increase in glucose uptake, typically observed in
positron emission tomography scans of patients (270). Another major metabolic
difference between normal and cancer cells is the fate of glucose.
Normal cells harvest glucose energy through aerobic respiration, a
four-stage process that combines glycolysis, pyruvate oxidation,
the tricarboxylic acid (TCA) cycle and oxidative phosphorylation
(OXPHOS) (248,263). In this route, glucose is
oxidized into carbon dioxide and water.
On the other hand, cancer cells switch to aerobic
glycolysis, which converts glucose into pyruvate and then lactate
via the lactate dehydrogenase enzyme (248,265). This phenomenon, also known as
the Warburg effect or the Warburg phenotype, is commonly stimulated
by energy demands of rapid proliferation and/or hypoxia (251,270,271). To the best of our knowledge, it
is still unclear whether the Warburg effect is a cause or
consequence of carcinogenesis (272). However, this phenotype allows
faster ATP production than OXPHOS and may provide a selective
advantage for cells, specifically in the hypoxic TME (273,274).
Another important benefit of aerobic glycolysis is
that the generated glycolytic metabolites facilitate the
biosynthesis of sugars, amino acids, nucleotides and fatty acids
critical for rapid cell proliferation (248). These advantages explain why high
amounts of aerobic glycolysis have been detected in proliferating
cells and progressive cancer types (270,275). The TCA cycle is also active in
these cells, with the resulting substrates rerouted for use in
de novo synthesis pathways, particularly lipogenesis
(276).
Cancer cells heavily rely on the uptake and
metabolism of glutamine as an alternative carbon source to glucose
(276). Glutamine, the amide
derivative of glutamate, is one of the most abundant nutrients in
the plasma (255,276). Glutaminolysis converts glutamine
to lactate and NADPH (255,276). In normal cells, glutamine is
used as a nitrogen source to synthesize nucleotides and other
non-essential amino acids (264). However, in cancer cells,
glutamine metabolism exceeds the needs of cells for de novo
proteins and nucleotides production (276). Instead, the data suggest that
glutamine metabolism allows the cells to use glucose-derived carbon
and TCA cycle intermediates as precursors for fatty acid synthesis
(276). This is achieved by
continuous replenishment of the TCA cycle intermediates (mainly
oxaloacetate) through a set of five chemical reactions, which
combined, form the anaplerosis process (276).
Metabolic reprogramming in the TME is driven by
oncogenic alterations in cancer cells, as well as by changes in the
signaling of normal cells (269). Typically, these modifications
impact the dynamics underlying nutrient uptake and bioenergetic
gene expression (269). For
instance, constitutive activation of the PI3K-AKT-mTOR signaling
pathway has been directly linked to glycolysis stimulation in
cancer cells and in CAFs (269,277,278).
Similarly, there is a clear association between Myc
transcription factor and the expression of various metabolic genes,
glycolytic enzymes, and glucose and glutamine transporters
(279). Specifically, Myc
activates glucose and glutamine metabolism, as well as purine,
pyrimidine, fatty acid and cholesterol synthesis (269,280–282). Oncogenic KRAS also triggers TME
metabolic reprogramming by upregulating glycolytic enzyme activity,
glutamine metabolism, and glucose and lactate transporter
expression (283–285). In addition, KRAS stimulates
nucleotide biosynthesis by channeling glycolytic metabolite
intermediates to the pentose phosphate pathway (284–286). Furthermore, KRAS sustains
autocrine and paracrine signaling by inducing the expression of
cell surface receptors responsible for upregulating type I cytokine
receptors (269,287,288). KRAS promotes micropinocytosis
and autophagy processes for nutrient scavenging by cancer cells
(289,290). On the other hand, loss of p53
function in cancer cells increases glycolysis, glucose transporters
and lipid metabolism, among others (291–294). Overall, cells of the TME have
several distinct metabolic signatures that are directly associated
with tumor growth and represent promising targets for cancer
therapy.
Genome maintenance in normal cells results in a low
rate of spontaneous mutations (4). Compromised check points and
sensitivity to mutagenic agents increase the rate of mutation
(4). The TME is characterized by
hypoxia, low pH and nutrient deprivation (295). These conditions contribute to
genome instability and mutations (Fig. 3) (295). For instance, hypoxia stimulates
the downregulation of mutL homolog 1 and PMS1 homolog 2, mismatch
repair system component, which are protein members of the mismatch
repair pathway and required to rectify DNA mismatch errors
(296,297). Therefore, hypoxia is a major
factor inducing substantial DNA damage leading to genetic
instability of solid tumors (295).
In addition to hypoxia, it has been recently shown
that a novel competing network of competing endogenous RNA can
regulate genomic integrity (298). Therefore, these genome
instability-related lncRNAs may act as biomarkers for genetic
instability, immunetherapy prognosis and therapeutic sensitivity in
colon adenocarcinoma and colon cancer (298,299).
Chronic inflammation is a major contributor to
cancer, and the inflammatory response can be triggered by various
factors, including pathogens, carcinogen exposure and imbalanced
immune regulation (Fig. 3)
(300). Immune cells can either
exert an antitumor or protumor activity depending on the
polarization state. For example, Th1 cells act as antitumor agents,
whereas Th17 subsets of CD4+ T cells act as
tumor-promoters (300,301). On the other hand,
anti-inflammatory M2 macrophages and N2 neutrophils are both
tumor-promoting cells that secrete cytokines, proteases and growth
factors, contributing to tissue remodeling and angiogenesis,
eventually leading to the conversion of cells into malignant cells
and cancer formation (300,301). TAMs, an M2 subtype, produce
VEGF-C and VEGF-D, which leads to peritumoral inflammation and
lymphangiogenesis in human cervical cancer (302).
CAFs exert pleiotropic functions in
immunomodulation mainly by secreting a range of pro-inflammatory
factors, which recruit and activate IICs (191,303,304). In 2010, Erez et al
(191) revealed that the CAF
secretome causes tumor-promoting inflammation in a NF-κB-dependent
manner. iCAFs secrete various chemokines and cytokines, such as
CXC-chemokine ligand (CXCL)1, cyclooxygenase-2, IL-1, IL-6 and
SDF-1, and receptors, such as IL 6 receptor α and IL-6 cytokine
family signal transducer, which add to the tumor-promoting
inflammatory milieu of the TME (305–308). In addition to TGF-β production,
CAFs secrete thymic stromal lymphopoietin, favoring Th2 cell
polarization, which is associated with poor prognosis (309). Overall, TME-mediated
inflammation influences tumor development, invasion, angiogenesis,
metastasis and immunosuppression (191,305,306). Therefore, targeting inflammation
may be a promising tool for cancer treatment.
Therefore, cancer development is mediated by TME
components that contribute to major cancer hallmarks, including
tumor proliferation, survival, angiogenesis, invasion and
metastasis (2,66,240). Overall, these findings have
prompted researchers to target TME cells for cancer treatment alone
or in combination with conventional therapeutic modalities
(9,67,310).
Clinical anticancer therapeutic efficiency is
limited due to several factors, including tumor heterogeneity and
the ability of cancer cells to develop multidrug resistance
(11). Another reason for this
observation is that cancer cells exhibit different responses when
moving from bench to bedside translational medicine (311). The cross-talk among tumor cells,
stromal cells and other TME components adds to the complexity of
efficient treatment (8).
Increasing awareness of the role of the TME in tumor development
brought about novel cancer therapy strategies targeting TME
components (310). Additionally,
combined therapies targeting more than one cell type or signaling
pathway are also being investigated (11). The present review highlights the
TME cells, signaling pathways and soluble factors that are targeted
for cancer treatment. It also reviews anticancer drugs that are
currently in clinical trials or show promising results for drug
development.
Membrane-bound serine protease FAP expression
distinguishes tumor tissues from healthy tissues (30). Inhibitors selectively targeting
FAP (FAPi) are currently in phase I and II clinical trials
(http://clinicaltrials.gov). For
instance, 68Ga-DOTA-FAPI is being studied for FAP-based
imaging and therapy using gallium-68 (312,313). FAP is also targeted by CD40
agonist (RO7300490) or 4-1BB agonist (RO7122290; phase I/II;
NCT04826003; Table I). The latter
resulted in activation of T and NK cells in the first-in-human
phase I study, suggesting potential antitumor activity (314). An early phase I clinical trial
is investigating the effect of combining FAPi with anti-neoplastic
monoclonal antibodies (NCT01722149). More studies are warranted to
determine the efficiency of targeting FAP-positive CAFs and tumor
cells.
One main mechanism for inhibiting angiogenesis is
targeting VEGF or VEGFR, alone or in combination with
chemotherapeutic drugs (315).
More than 400 interventional clinical trials are investigating the
anticancer potential of VEGF targeting (based on http://clinicaltrials.gov; accessed, January 6,
2022). Promising results have been reported with the administration
of the U.S. Food and Drug Administration (FDA)-approved bevacizumab
(Avastin), an anti-angiogenic antibody that targets VEGF, and
Bevacizumab-IRDye800CW, its fluorescent form (316,317). Combined administration with
chemotherapeutic agents resulted in increased overall survival or
progression-free survival (PFS) in CRC, NSCLC and breast cancer
(315). Additionally,
therapeutic strategies are currently considering the administration
of two anti-angiogenic agents. For instance, a phase III trial
carried out on patients with NSCLC is comparing the efficacy of two
anti-VEGF antibodies, LY1008 and bevacizumab (Avastin), combined
with the chemotherapeutic drugs paclitaxel and carboplatin
(NCT03533127). In addition to VEGF or VEGFR targeting,
anti-angiogenic strategies include using multi-receptor tyrosine
kinase inhibitors that stimulate the inhibition of VEGF, VGEFR,
PDGFR or c-Kit (NCT03533127). The FDA-approved pazopanib (Votrient)
is one example that inhibits VEGFR1/2/3 and c-Kit for the treatment
of patients with soft tissue sarcoma and RCC (318,319). There are currently three phase
IV clinical trials that target the VEGF pathway. Two of these
trials studied the effect of everolimus (RAD001), an mTOR
inhibitor, for the treatment of patients with advanced or
metastatic RCC and (NCT01206764 and NCT01266837). Another study
assessed endostatin, an inhibitor of tyrosine phosphorylation of
VEGFR2 (320) in combination
with the anti-mitotic vinorelbine and cisplatin for the treatment
of patients with NSCLC (NCT02497118). Therapeutic agents targeting
vascular ECs in interventional clinical trials (in phases III and
IV) are listed in Table I.
Given the pivotal role of the immune system in
cancer, several anti-inflammatory drugs have been designed to
inhibit tumor-promoting inflammation (50). TME immune cells within the tumor
are targets of several clinical phase trials (8,51,315). One approach is the inhibition of
macrophage recruitment and activation in tumors (9). This involves targeting and
inhibiting colony-stimulating factor-1 (CSF-1), a
macrophage-recruiting mediator, and its receptor (CSF-1R) (51). This approach is associated with
reduced infiltration of TAMs and MDSCs, and inhibiting tumor
progression and metastasis (51).
At present, there are >50 clinical trials targeting CSF-1 and 16
clinical trials targeting CSF-1R, according to https://clinicaltrials.gov (date accessed, January 6,
2022). One promising drug, vimseltinib, a CSF-1R inhibitor also
referred to as DCC-3014, has reached a phase III clinical trial and
is being assessed for its efficacy in treating patients with
tenosynovial giant cell tumor (NCT05059262). TAMs are characterized
by the high expression of CD204 (macrophage scavenger receptor
class A) and folate receptor β (FRβ) on their surface (321). TAMs were successfully eliminated
using an anti-CD204 antibody and a targeted FRβ-immunotoxin in mice
and rat models, respectively (322,323).
Secondly, a promising approach is to target
pro-tumorigenic factors secreted by IICs (310). The use of TGF-β inhibitors and
immune checkpoint inhibitors (ICIs), such as PD-L1 antibodies, and
cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) antibodies has
been reported in a number of clinical phase trials (310,324). These strategies increase the
infiltration of T cells into the tumor vicinity and the inhibition
of Treg cells (8,51,325,326). Furthermore, signal transducers
and transcription factors that mediate tumor growth and survival,
such as STAT3 and NF-κB, are targeted (50,51). Prolonged inhibition of NFκB may
lead to immune deficiency and enhanced acute inflammation (315). Consequently, the progress of
NF-κB inhibitor development is obstructed in clinical trials
(315,327). Pro-inflammatory chemokines and
cytokines are also targeted. In in vivo studies, receptor
antagonists are used to inhibit C-C chemokine receptor 2 and CXC
chemokine receptor 4 (229).
Clinical trials are also evaluating inhibitors targeting other
cytokines, such as IL-1, IL-6 and TNFα (51,315). One important example is the
FDA-approved anakinra, an IL-1 receptor antagonist used to treat
patients with pancreatic cancer and metastatic breast, colon and
prostate cancer (NCT02550327/NCT03233776).
Enhancing the antitumor activity by increasing the
infiltration of pro-inflammatory cells is also a promising approach
(51). For instance, embelin, a
small-molecule inhibitor of X-linked inhibitor of apoptosis
protein, induces apoptosis and suppresses gastric carcinoma and
pancreatic cancer in vivo (328,329). Mechanistically, this is achieved
by increasing the infiltration of pro-inflammatory immune cells,
such as Th1 cells, NKs and NK T cells, while decreasing the
infiltration of immunosuppressive MDSCs and IL-8- and IL-6-positive
immune cells (8,328,329). Therefore, it would be
interesting to move this research forward in clinical trials.
Another approach is the use of pro-inflammatory
cytokines for tumor treatment. One example is the cytokine
granulocyte-macrophage colony-stimulating factor (GM-CSF), which
stimulates antigen presentation on macrophages and DCs, thus
enhancing antibody-dependent cellular cytotoxicity (330). GM-CSF has been evaluated in a
number of clinical trials, both as a monotherapy or adjuvant
(NCT02451488 and NCT03686683 for example). A phase IV clinical
study is testing the neoadjuvant effect of GM-CSF in cutaneous
stage L-III melanoma (NCT02451488; Table III). In addition, one active
non-recruiting phase III clinical trial is investigating the
therapeutic potential of sipuleucel-T in patients with prostate
adenocarcinoma (NCT03686683). Sipuleucel-T is an autologous cell
product comprising APCs loaded with a recombinant fusion protein,
PA2024, composed of prostatic acid phosphatase linked to GM-CSF
(NCT03686683). Drugs targeting CSFs in phase III and IV clinical
trials are presented in Table
II.
In addition to cytokine therapies and ICIs,
immunity of the TME can be also triggered by adoptive cell therapy
(ACT) and cancer vaccines (50).
During ACT, autologous T lymphocytes with antitumor activity are
isolated from a patient, expanded ex vivo, and then
amplified tumor-resident or engineered T cells are transferred back
to patients (331,332). One promising ACT approach is the
chimeric antigen receptor (CAR) gene therapy where CAR modified T
cells recognize various types of antigens regardless of their
presentation on MHC molecules (333). T cells then mediate tumor
killing via: i) The perforin and granzyme axis; ii) cytokine
secretion; or iii) Fas-Fas ligand axis (334). Currently, there are 48 completed
clinical trials that used CAR-T cell therapy on different
malignancies (based on http://clinicaltrials.gov; accessed, January 6,
2022). One study evaluated CAR-engineered autologous primary human
CD8+ T lymphocytes against IL13 receptor α2 in 3
patients with recurrent glioblastoma (NCT00730613), and reported
promising anti-glioma activity (335). CAR-T cell immunotherapy has
shown promise in terms of efficacy, while causing minimal toxicity
(334,336). However, limitations such as
tumor heterogeneity and antigen heterogeneous expression, as well
as the function of T lymphocytes at tumor sites, make tumor
eradication difficult (336).
In addition to ACT, cancer vaccines are currently
intensively studied as a promising therapeutic approach that
activates the humoral and cellular immunity of patients with cancer
(337–339). An efficient cancer vaccine
design depends on a good antigen selection, where an ideal antigen
should be specifically expressed and presented on all cancer cells
but not normal cells, highly immunogenic and essential for the
survival of cancer cells (340).
After antigen delivery, DCs will uptake these antigens and present
relevant antigens on MHC I and MHC II to CD8+ and
CD4+ T cells, respectively (337). Effector T cells then migrate to
the TME, recognize and kill cancer cells by releasing cytotoxic
particles, including perforin, granzymes, IFN-γ or TNF-α, or by
directly inducing apoptosis (341). In addition to T cells, B
lymphocytes, NK cells and macrophages promote tumor eradication
(341). Personalized vaccines
are also gaining interest. There are currently three clinical
trials that evaluated personalized cancer vaccination in patients
with glioblastoma (active, not recruiting, NCT00045968; and
completed, NCT01280552 and NCT00643097) (324). These studies reported increased
numbers of infiltrating T cells with improved PFS (338,339,342). These studies show that
immunization with vaccines has a promising effect in patients with
cancer.
In addition to targeting of the cellular components
of the TME and their soluble factors, TME non-cellular features are
also targeted and evaluated in clinical trials. ECM remodeling and
increased stiffness (desmoplasia), for instance, are targeted to
reduce mortality in different cancer types (310). FDA-approved angiotensin II
receptor antagonists, such as losartan and candesartan, increased
the survival of patients with gastro-esophageal cancer by
inhibiting the TGF-β signaling pathway and consequently reducing
collagen I secretion and desmoplasia (343). Losartan has also shown clinical
benefits in pancreatic cancer phase II trials (NCT01821729) when
combined with FOLFIRINOX and chemoradiation with fluorouracil or
capecitabine (344). The ECM may
be alternatively modified by targeting integrins or focal adhesion
kinase (FAK) proteins using the FAK inhibitor defactinib
(NCT01870609) (345). MMP
inhibitors target MMPs. However, trials failed clinically mainly
because these inhibitors exhibit a broad-spectrum activity that may
result in secondary side effects (346), and ECM degradation may boost
cancer progression instead of inhibiting it (310,347,348).
Exosomes are: i) Targeted for reducing vesicle
trafficking in cancer cells; ii) used as biomarkers for cancer
diagnosis; or iii) used as vehicles of small interfering RNA for
targeted therapy (51).
Furthermore, the association between non-coding RNA and the TME is
gaining interest, especially with respect to the TME immune
environment (136,349). In this regard, Huang et
al (350) developed a novel
TME-related lncRNA risk model that could be used as a predictor of
ICIs and a prognostic biomarker in patients with hepatocellular
carcinoma.
Published research has linked the gut and
intratumoral microbiota to response and toxicity in a variety of
treatments, including chemotherapy (351). For instance, commensal microbes
interact with chemotherapeutics primarily by modulating drug
metabolism and host immunity (351,352). Drug activity can either be
directly driven by microbes or indirectly driven by microbe-derived
metabolites (352). Therefore,
targeting the microbiome may hold promise for improving
chemotherapeutic efficacy and lowering toxicity (18). Retrospective clinical studies on
patients with PDAC demonstrated that administration of antibiotics
to target bacteria that produce a long isoform of cytidine
deaminase resulted in improved gemcitabine response, and thus,
overcame the intratumoral bacterial-induced chemoresistance
(353–355).
The microbiome has also been recognized for its
intricate interaction with host immunity, and thus, is considered a
potential therapeutic target to optimize immunotherapy responses
(310). Gut microbiota, in
particular, serve a role in modulating immune checkpoint blockade
responses in multiple cancer types (356–359). A recent recruiting observational
study aims to evaluate the effect of the microbiome in terms of
efficacy and toxicity of ICIs in patients with advanced cancer
(NCT04107168). The search for biomarkers in the gut microbiome has
resulted in the identification of microbiome signatures that aid in
determining when ICIs are effective (360,361). According to a study that looked
at phase II neoadjuvant trials of anti-programmed cell death 1
(PD-1)/anti-CTLA-4 antibodies for melanoma, NSCLC and sarcoma,
patients with high abundance of Ruminococcus were reported
as responders with a marked increase in B cell signatures (362). Given that the favorable
microbiota signatures result in enhanced intratumoral immune
infiltrates (357–359), creating an ideal combination of
bacteria is a potential therapeutic approach to be administered in
combination with checkpoint blockade.
In addition to targeting the gut microbiome,
efforts are now being made to target the tumor microbiome in order
to slow cancer progression and improve the response to cancer
therapy. For instance, targeting the tumoral microbiome with
antibiotics results in enhanced response to both chemotherapy and
ICIs in CRC and pancreatic cancer (181,363,364). The intratumoral microbiome can
also be targeted by bioengineered bacteria that can either kill
tumor cells directly, or create an immune microenvironment that
encourages antitumor immune responses (18). In mice for example, attenuated
Salmonella strains expressing Vibrio-derived toll-like
receptor 5 ligand flagellin elicited an immune response that
recruited an antitumor immune responses against orthotopic human
CRC87 lines (365).
A growing body of evidence suggests that the
microbiome serves a role in determining cancer therapeutic efficacy
and toxicity (18,351). Laboratory research and clinical
trials have also shown that microbiota modulation can help with
cancer treatment (18,366–368). Therefore, understanding the
microbiome and its interactions with cancer is critical in
personalized medicine. Manipulation of the gut microbiota may yield
novel cancer treatment insights for enhanced cancer therapeutic
responses. Since the microbiome exhibits complex interactions with
both the host immunity and cancer cells, it would be challenging to
identify an optimal bacterial consortia and metabolites to affect
the TME, as well as to introduce it for cancer treatment (18).
In addition to the aforementioned targeted
therapeutic strategies, combination therapies have gained
popularity as they result in enhanced efficacy, reduced drug
resistance and lowered toxicity compared with monotherapy (369). Studies are investigating
combination regimens that simultaneously affect several targets,
thus achieving cooperative and synergistic effects (369,370). In this context, therapeutic
agents targeting multiple stromal cells of the TME have been
evaluated in interventional clinical trials (Table IV) (51,371). For example, the PD-1/PD-L1
signaling pathway is targeted with anti-angiogenic interventions
targeting VEGFR1/2/3, PDGFR or c-Kit (235), or with tyrosine kinase
inhibitors (372–374). Additionally, anti-stromal
interventions are also combined with chemotherapeutics and
radiation agents (51,315,369,370). Furthermore, to better decide on
a combination therapy, researchers should fully understand the
stage of the tumor, as well as the specific state of the TME and
its immune markers.
Cancer is a complex disease caused by malignant
cells and a supporting TME. The cross-talk between these two main
entities embodied by bidirectional mediators governs tumor
progression. Low levels of both oxygen and pH create local stress
within the TME, triggering a response from, thereby activated,
stromal cells and the infiltration of more immune cells (136). Such communications are not only
governed by cytokines, chemokines and metabolic products secreted
by TME stromal cells but other factors such as epigenetic factors
(such as miRNA), methylation DNA and histone modification are also
critical (375). Furthermore, an
increasing number of studies have highlighted the important effects
of metabolism on the activities of immune cells, and thus, their
effect on cancer progression (257,260,376). The orchestration of autocrine
and paracrine communications within the tumor environment may
expedite ECM stiffness, inflammation and angiogenesis, and possibly
cancer cell dissemination and metastasis (4). These results warrant examining the
effect of TME components on the outcome of the disease. Such
findings urged research to investigate the relevance of TME
targeting for more efficient therapeutic methods. While most
studies focus mainly on the stromal composition of the TME
(64,66), the present review provides a
comprehensive examination of not only the stromal components but
also the non-cellular TME components, including the ECM, exosomes
and microbiome. The present discusses the contribution of cellular
and non-cellular TME components to fundamental cancer hallmarks as
well as emerging hallmarks and enabling characteristics. The
present review also provides a detailed report on TME cells,
signaling pathways and soluble factors that can be targeted for
cancer therapy, highlighting TME components that are currently
targeted in interventional clinical trials.
TME targeting provides promising strategies to
overcome the chemotherapeutic resistance of tumor cells. Research
efforts have resulted in the development of FDA-approved or newly
developed TME-targeted drugs, including anti-angiogenic and
anti-inflammatory agents. Clinical implementation of these drugs
also shows promising successful clinical results (9,51,315). In addition, and with the help of
large-scale data mining and bioinformatics analysis, several
immune-related gene signatures serving as predictors for
therapeutic outcomes or biomarkers for prognosis in several cancer
types have been constructed (136,377–380).
TME-targeted strategies may soon become mainstream
for cancer therapy and can be used in combination with conventional
antitumor methods. However, further research is required to address
the time of TME-targeted drug administration and the treatment
strategies, since certain studies have indicated that TME
components may augment tumor resistance to cancer therapy (97,181,200,381). For instance, CAFs promote
resistance to chemotherapy primarily by mediating EMT, maintaining
the stemness of cancer stem cells and promoting metabolic
reprogramming (382). The
augmented ECM deposition and increased cytokine secretions mediated
by CAFs may aid tumor cells in resisting cancer-therapies and, in
particular, chemotherapy (383).
Furthermore, a growing body of evidence suggests that
hypoxia-driven residual VEGF and other pro-angiogenic factors cause
resistance to VEGF receptor inhibition (381,384). Therefore, combinations of
medicines targeting these factors may enhance treatment outcomes
compared with single VEGF pathway blocking alone (385).
In addition to the aforementioned concerns, the
pharmacokinetics and biodistribution of TME-targeted drugs is not
yet well investigated due to the difficulty of detecting the exact
state of the TME (386). One way
to overcome this limitation is using 3D cell culture systems to
recapitulate the complexity of tumor architecture and simulate the
TME (387). Another method is
using animal models that facilitate the recreation of a developed
tumor in an improved pathophysiologic environment (324,388–390). Tumor tissues obtained from a
patient are processed into patient-derived organoids or
patient-derived xenografts, which are then functionally and
quantitively analyzed after treatment [reviewed in (391)]. These methods may help identify
clinically relevant immune checkpoints and predict treatment
efficacy (391). Such efforts
allow for an enhanced pre-clinical validation of novel cancer
methodologies towards full integration of immunotherapeutic
prediction tools (391,392). In addition to combination
therapy, nanotechnology also promises good therapeutic prospects
(324,393). Regarding all discussed
interventions and their limitations, and arriving at the era of the
comprehensive cancer model treatment, it is important to treat
tumors as a multifactorial disease in a stage, tissue/organ and
patient-specific manner.
Not applicable.
Funding: No funding was received.
Not applicable.
RN and IF equally designed the review article and
wrote the majority of the article. AEF researched references and
contributed to the writing. RAH and MES wrote the final draft and
edited the manuscript. Data authentication is not applicable. All
authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Cooper GM: The development and causes of
cancer, The cell: A molecular approach. 2nd edition. Sunderland
(MA): Sinauer Associates; 2000
|
|
2
|
Balkwill FR, Capasso M and Hagemann T: The
tumor microenvironment at a glance. J Cell Sci. 125:5591–5596.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu P, Weaver VM and Werb Z: The
extracellular matrix: A dynamic niche in cancer progression. J Cell
Biol. 196:395–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li H, Fan X and Houghton J: Tumor
microenvironment: The role of the tumor stroma in cancer. J Cell
Biochem. 101:805–815. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pietras K and Ostman A: Hallmarks of
cancer: Interactions with the tumor stroma. Exp Cell Res.
316:1324–1331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li H, Zhou L, Zhou J, Li Q and Ji Q:
Underlying mechanisms and drug intervention strategies for the
tumour microenvironment. J Exp Clin Cancer Res. 40:972021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Joyce JA: Therapeutic targeting of the
tumor microenvironment. Cancer Cell. 7:513–520. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Henke E, Nandigama R and Ergün S:
Extracellular matrix in the tumor microenvironment and its impact
on cancer therapy. Front Mol Biosci. 6:1602020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsai MJ, Chang WA, Huang MS and Kuo PL:
Tumor microenvironment: A new treatment target for cancer. ISRN
Biochem. 2014:e3519592014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Willumsen N, Thomsen LB, Bager CL, Jensen
C and Karsdal MA: Quantification of altered tissue turnover in a
liquid biopsy: A proposed precision medicine tool to assess chronic
inflammation and desmoplasia associated with a pro-cancerous niche
and response to immuno-therapeutic anti-tumor modalities. Cancer
Immunol Immunother. 67:1–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi
Y, Hu G and Sun Y: New horizons in tumor microenvironment biology:
Challenges and opportunities. BMC Med. 13:452015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X and Song E: Turning foes to
friends: Targeting cancer-associated fibroblasts. Nat Rev Drug
Discov. 18:99–115. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brassart-Pasco S, Brézillon S, Brassart B,
Ramont L, Oudart JB and Monboisse JC: Tumor microenvironment:
Extracellular matrix alterations influence tumor progression. Front
Oncol. 10:3972020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Chen JQ, Liu JL and Tian L:
Exosomes in tumor microenvironment: Novel transporters and
biomarkers. J Transl Med. 14:2972016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Helmink BA, Khan MAW, Hermann A,
Gopalakrishnan V and Wargo JA: The microbiome, cancer, and cancer
therapy. Nat Med. 25:377–388. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sahai E, Astsaturov I, Cukierman E,
DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR,
Hunter T, et al: A framework for advancing our understanding of
cancer-associated fibroblasts. Nat Rev Cancer. 20:174–186. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ganguly D, Chandra R, Karalis J, Teke M,
Aguilera T, Maddipati R, Wachsmann MB, Ghersi D, Siravegna G, Zeh
HJ III, et al: Cancer-associated fibroblasts: Versatile players in
the tumor microenvironment. Cancers (Basel). 12:26522020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao Z, Tan ZW, Zhu P and Tan NS:
Cancer-associated fibroblasts in tumor microenvironment-accomplices
in tumor malignancy. Cell Immunol. 343:1037292019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arina A, Idel C, Hyjek EM, Alegre ML, Wang
Y, Bindokas VP, Weichselbaum RR and Schreiber H: Tumor-associated
fibroblasts predominantly come from local and not circulating
precursors. Proc Natl Acad Sci USA. 113:7551–7556. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jung Y, Kim JK, Shiozawa Y, Wang J, Mishra
A, Joseph J, Berry JE, McGee S, Lee E, Sun H, et al: Recruitment of
mesenchymal stem cells into prostate tumours promotes metastasis.
Nat Commun. 4:17952013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mishra PJ, Mishra PJ, Humeniuk R, Medina
DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW and Banerjee D:
Carcinoma-associated fibroblast-like differentiation of human
mesenchymal stem cells. Cancer Res. 68:4331–4339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu Q, Zhang X, Zhang L, Li W, Wu H, Yuan
X, Mao F, Wang M, Zhu W, Qian H and Xu W: The IL-6-STAT3 axis
mediates a reciprocal crosstalk between cancer-derived mesenchymal
stem cells and neutrophils to synergistically prompt gastric cancer
progression. Cell Death Dis. 5:e12952014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jotzu C, Alt E, Welte G, Li J, Hennessy
BT, Devarajan E, Krishnappa S, Pinilla S, Droll L and Song YH:
Adipose tissue-derived stem cells differentiate into
carcinoma-associated fibroblast-like cells under the influence of
tumor-derived factors. Anal Cell Pathol (Amst). 33:61–79. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kidd S, Spaeth E, Watson K, Burks J, Lu H,
Klopp A, Andreeff M and Marini FC: Origins of the tumor
microenvironment: Quantitative assessment of adipose-derived and
bone marrow-derived stroma. PLoS One. 7:e305632012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miyazaki Y, Oda T, Mori N and Kida YS:
Adipose-derived mesenchymal stem cells differentiate into
pancreatic cancer-associated fibroblasts in vitro. FEBS Open Bio.
10:2268–2281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nurmik M, Ullmann P, Rodriguez F, Haan S
and Letellier E: In search of definitions: Cancer-associated
fibroblasts and their markers. Int J Cancer. 146:895–905. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Simon T and Salhia B: Cancer-Associated
fibroblast subpopulations with diverse and dynamic roles in the
tumor microenvironment. Mol Cancer Res. 20:183–192. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sebastian A, Hum NR, Martin KA, Gilmore
SF, Peran I, Byers SW, Wheeler EK, Coleman MA and Loots GG:
Single-cell transcriptomic analysis of tumor-derived fibroblasts
and normal tissue-resident fibroblasts reveals fibroblast
heterogeneity in breast cancer. Cancers (Basel). 12:13072020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Öhlund D, Handly-Santana A, Biffi G,
Elyada E, Almeida AS, Ponz-Sarvise M, Corbo V, Oni TE, Hearn SA,
Lee EJ, et al: Distinct populations of inflammatory fibroblasts and
myofibroblasts in pancreatic cancer. J Exp Med. 214:579–596. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Elyada E, Bolisetty M, Laise P, Flynn WF,
Courtois ET, Burkhart RA, Teinor JA, Belleau P, Biffi G, Lucito MS,
et al: Cross-species single-cell analysis of pancreatic ductal
adenocarcinoma reveals antigen-presenting cancer-associated
fibroblasts. Cancer Discov. 9:1102–1123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cohen N, Shani O, Raz Y, Sharon Y, Hoffman
D, Abramovitz L and Erez N: Fibroblasts drive an immunosuppressive
and growth-promoting microenvironment in breast cancer via
secretion of Chitinase 3-like 1. Oncogene. 36:4457–4468. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Amatangelo MD, Bassi DE, Klein-Szanto AJP
and Cukierman E: Stroma-derived three-dimensional matrices are
necessary and sufficient to promote desmoplastic differentiation of
normal fibroblasts. Am J Pathol. 167:475–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Carmeliet P and Jain RK: Principles and
mechanisms of vessel normalization for cancer and other angiogenic
diseases. Nat Rev Drug Discov. 10:417–427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nishida N, Yano H, Nishida T, Kamura T and
Kojiro M: Angiogenesis in cancer. Vasc Health Risk Manag.
2:213–219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jain RK: Normalization of tumor
vasculature: An emerging concept in antiangiogenic therapy.
Science. 307:58–62. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Semenza GL: Cancer-stromal cell
interactions mediated by hypoxia-inducible factors promote
angiogenesis, lymphangiogenesis, and metastasis. Oncogene.
32:4057–4063. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Weis SM and Cheresh DA: Tumor
angiogenesis: Molecular pathways and therapeutic targets. Nat Med.
17:1359–1370. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hillen F and Griffioen AW: Tumour
vascularization: Sprouting angiogenesis and beyond. Cancer
Metastasis Rev. 26:489–502. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Italiano JE Jr, Richardson JL, Patel-Hett
S, Battinelli E, Zaslavsky A, Short S, Ryeom S, Folkman J and
Klement GL: Angiogenesis is regulated by a novel mechanism: Pro-
and antiangiogenic proteins are organized into separate platelet
alpha granules and differentially released. Blood. 111:1227–1233.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Armulik A, Genové G and Betsholtz C:
Pericytes: Developmental, physiological, and pathological
perspectives, problems, and promises. Dev Cell. 21:193–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Balta E, Wabnitz GH and Samstag Y:
Hijacked immune cells in the tumor microenvironment: Molecular
mechanisms of immunosuppression and cues to improve T cell-based
immunotherapy of solid tumors. Int J Mol Sci. 22:57362021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Biswas SK and Mantovani A: Macrophage
plasticity and interaction with lymphocyte subsets: Cancer as a
paradigm. Nat Immunol. 11:889–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Franklin RA, Liao W, Sarkar A, Kim MV,
Bivona MR, Liu K, Pamer EG and Li MO: The cellular and molecular
origin of tumor-associated macrophages. Science. 344:921–925. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu K, Lin K, Li X, Yuan X, Xu P, Ni P and
Xu D: Redefining tumor-associated macrophage subpopulations and
functions in the tumor microenvironment. Front Immunol.
11:17312020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dehne N, Mora J, Namgaladze D, Weigert A
and Brüne B: Cancer cell and macrophage cross-talk in the tumor
microenvironment. Curr Opin Pharmacol. 35:12–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cassetta L, Fragkogianni S, Sims AH,
Swierczak A, Forrester LM, Zhang H, Soong DYH, Cotechini T, Anur P,
Lin EY, et al: Human tumor-associated macrophage and monocyte
transcriptional landscapes reveal cancer-specific reprogramming,
biomarkers, and therapeutic targets. Cancer Cell. 35:588–602.e10.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Costa AC, Santos JMO, Gil da Costa RM and
Medeiros R: Impact of immune cells on the hallmarks of cancer: A
literature review. Crit Rev Oncol Hematol. 168:1035412021.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20:8402019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ostrand-Rosenberg S and Sinha P:
Myeloid-derived suppressor cells: Linking inflammation and cancer.
J Immunol. 182:4499–4506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Veglia F, Sanseviero E and Gabrilovich DI:
Myeloid-derived suppressor cells in the era of increasing myeloid
cell diversity. Nat Rev Immunol. 21:485–498. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Masucci MT, Minopoli M and Carriero MV:
Tumor associated neutrophils. Their role in tumorigenesis,
metastasis, prognosis and therapy. Front Oncol. 9:11462019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ye Y, Gaugler B, Mohty M and Malard F:
Plasmacytoid dendritic cell biology and its role in immune-mediated
diseases. Clin Transl Immunology. 9:e11392020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Karthaus N, Torensma R and Tel J:
Deciphering the message broadcast by tumor-infiltrating dendritic
cells. Am J Pathol. 181:733–742. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Benavente S, Sánchez-García A, Naches S,
LLeonart ME and Lorente J: Therapy-induced modulation of the tumor
microenvironment: New opportunities for cancer therapies. Front
Oncol. 10:5828842020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Campbell DJ and Koch MA: Treg cells:
Patrolling a dangerous neighborhood. Nat Med. 17:929–930. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hsieh CS, Lee HM and Lio CWJ: Selection of
regulatory T cells in the thymus. Nat Rev Immunol. 12:157–167.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Coronella JA, Telleman P, Kingsbury GA,
Truong TD, Hays S and Junghans RP: Evidence for an antigen-driven
humoral immune response in medullary ductal breast cancer. Cancer
Res. 61:7889–7899. 2001.PubMed/NCBI
|
|
61
|
Schioppa T, Moore R, Thompson RG, Rosser
EC, Kulbe H, Nedospasov S, Mauri C, Coussens LM and Balkwill FR: B
regulatory cells and the tumor-promoting actions of TNF-α during
squamous carcinogenesis. Proc Natl Acad Sci USA. 108:10662–10667.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wu J and Lanier LL: Natural killer cells
and cancer. Adv Cancer Res. 90:127–156. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hong J, Jin JO, Chen WY, Poggi A and
Cheong JH: Editorial: Emerging roles and mechanisms of stromal
cells in carcinomas at the molecular level. Front Immunol.
13:10258382022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Koppensteiner L, Mathieson L, O'Connor RA
and Akram AR: Cancer associated fibroblasts-an impediment to
effective anti-cancer T cell immunity. Front Immunol.
13:8873802022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mun JY, Leem SH, Lee JH and Kim HS: Dual
relationship between stromal cells and immune cells in the tumor
microenvironment. Front Immunol. 13:8647392022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Belli C, Antonarelli G, Repetto M, Boscolo
Bielo L, Crimini E and Curigliano G: Targeting cellular components
of the tumor microenvironment in solid malignancies. Cancers
(Basel). 14:42782022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Theocharis AD, Skandalis SS, Gialeli C and
Karamanos NK: Extracellular matrix structure. Adv Drug Deliv Rev.
97:4–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Badylak SF: The extracellular matrix as a
biologic scaffold material. Biomaterials. 28:3587–3593. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fuhrmann A and Engler AJ: The Cytoskeleton
regulates cell attachment strength. Biophys J. 109:57–65. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kechagia JZ, Ivaska J and Roca-Cusachs P:
Integrins as biomechanical sensors of the microenvironment. Nat Rev
Mol Cell Biol. 20:457–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Romani P, Valcarcel-Jimenez L, Frezza C
and Dupont S: Crosstalk between mechanotransduction and metabolism.
Nat Rev Mol Cell Biol. 22:22–38. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cichon MA and Radisky DC: Extracellular
matrix as a contextual determinant of transforming growth factor-β
signaling in epithelial-mesenchymal transition and in cancer. Cell
Adhes Migr. 8:588–594. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Karamanos NK, Piperigkou Z, Passi A, Götte
M, Rousselle P and Vlodavsky I: Extracellular matrix-based cancer
targeting. Trends Mol Med. 27:1000–1013. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Rilla K, Mustonen AM, Arasu UT, Härkönen
K, Matilainen J and Nieminen P: Extracellular vesicles are integral
and functional components of the extracellular matrix. Matrix Biol.
75–76. 201–219. 2019.PubMed/NCBI
|
|
76
|
Apte MV, Yang L, Phillips PA, Xu Z, Kaplan
W, Cowley M, Pirola RC and Wilson JS: Extracellular matrix
composition significantly influences pancreatic stellate cell gene
expression pattern: Role of transgelin in PSC function. Am J
Physiol Gastrointest Liver Physiol. 305:G408–G417. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Naba A, Clauser KR, Hoersch S, Liu H, Carr
SA and Hynes RO: The matrisome: In silico definition and in vivo
characterization by proteomics of normal and tumor extracellular
matrices. Mol Cell Proteomics. 11:M111.014647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Musiime M, Chang J, Hansen U, Kadler KE,
Zeltz C and Gullberg D: Collagen assembly at the cell surface:
Dogmas revisited. Cells. 10:6622021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Eble JA and Niland S: The extracellular
matrix in tumor progression and metastasis. Clin Exp Metastasis.
36:171–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yue B: Biology of the extracellular
matrix: An overview. J Glaucoma. 23 (8 Suppl 1):S20–S23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Naba A, Pearce OMT, Del Rosario A, Ma D,
Ding H, Rajeeve V, Cutillas PR, Balkwill FR and Hynes RO:
Characterization of the extracellular matrix of normal and diseased
tissues using proteomics. J Proteome Res. 16:3083–3091. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Erdogan B and Webb DJ: Cancer-associated
fibroblasts modulate growth factor signaling and extracellular
matrix remodeling to regulate tumor metastasis. Biochem Soc Trans.
45:229–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Najafi M, Farhood B and Mortezaee K:
Extracellular matrix (ECM) stiffness and degradation as cancer
drivers. J Cell Biochem. 120:2782–2790. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Muncie JM and Weaver VM: The physical and
biochemical properties of the extracellular matrix regulate cell
fate. Curr Top Dev Biol. 130:1–37. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Provenzano PP, Inman DR, Eliceiri KW,
Knittel JG, Yan L, Rueden CT, White JG and Keely PJ: Collagen
density promotes mammary tumor initiation and progression. BMC Med.
6:112008. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mammoto T, Jiang A, Jiang E, Panigrahy D,
Kieran MW and Mammoto A: Role of collagen matrix in tumor
angiogenesis and glioblastoma multiforme progression. Am J Pathol.
183:1293–1305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Peinado H, Zhang H, Matei IR, Costa-Silva
B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang
Y, et al: Pre-metastatic niches: Organ-specific homes for
metastases. Nat Rev Cancer. 17:302–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Moreira AM, Pereira J, Melo S, Fernandes
MS, Carneiro P, Seruca R and Figueiredo J: The extracellular
matrix: An accomplice in gastric cancer development and
progression. Cells. 9:3942020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Høgdall D, Lewinska M and Andersen JB:
Desmoplastic tumor microenvironment and immunotherapy in
cholangiocarcinoma. Trends Cancer. 4:239–255. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ho WJ, Jaffee EM and Zheng L: The tumour
microenvironment in pancreatic cancer-clinical challenges and
opportunities. Nat Rev Clin Oncol. 17:527–540. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Levental KR, Yu H, Kass L, Lakins JN,
Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, et
al: Matrix crosslinking forces tumor progression by enhancing
integrin signaling. Cell. 139:891–906. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Damodarasamy M, Vernon RB, Chan CK,
Plymate SR, Wight TN and Reed MJ: Hyaluronan in aged collagen
matrix increases prostate epithelial cell proliferation. In Vitro
Cell Dev Biol Anim. 51:50–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Suhovskih AV, Mostovich LA, Kunin IS,
Boboev MM, Nepomnyashchikh GI, Aidagulova SV and Grigorieva EV:
Proteoglycan expression in normal human prostate tissue and
prostate cancer. ISRN Oncol. 2013:6801362013.PubMed/NCBI
|
|
94
|
Ajeti V, Nadiarnykh O, Ponik SM, Keely PJ,
Eliceiri KW and Campagnola PJ: Structural changes in mixed Col
I/Col V collagen gels probed by SHG microscopy: Implications for
probing stromal alterations in human breast cancer. Biomed Opt
Express. 2:2307–2316. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Fang S, Dai Y, Mei Y, Yang M, Hu L, Yang
H, Guan X and Li J: Clinical significance and biological role of
cancer-derived type I collagen in lung and esophageal cancers.
Thorac Cancer. 10:277–288. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Miskolczi Z, Smith MP, Rowling EJ,
Ferguson J, Barriuso J and Wellbrock C: Collagen abundance controls
melanoma phenotypes through lineage-specific microenvironment
sensing. Oncogene. 37:3166–3182. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Rossow L, Veitl S, Vorlová S, Wax JK, Kuhn
AE, Maltzahn V, Upcin B, Karl F, Hoffmann H, Gätzner S, et al:
LOX-catalyzed collagen stabilization is a proximal cause for
intrinsic resistance to chemotherapy. Oncogene. 37:4921–4940. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Guzman A, Ziperstein MJ and Kaufman LJ:
The effect of fibrillar matrix architecture on tumor cell invasion
of physically challenging environments. Biomaterials. 35:6954–6963.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Xu S, Xu H, Wang W, Li S, Li H, Li T,
Zhang W, Yu X and Liu L: The role of collagen in cancer: From bench
to bedside. J Transl Med. 17:3092019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Gilkes DM, Semenza GL and Wirtz D: Hypoxia
and the extracellular matrix: Drivers of tumour metastasis. Nat Rev
Cancer. 14:430–439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Allen SC, Widman JA, Datta A and Suggs LJ:
Dynamic extracellular matrix stiffening induces a phenotypic
transformation and a migratory shift in epithelial cells. Integr
Biol (Camb). 12:161–174. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Fischer KR, Durrans A, Lee S, Sheng J, Li
F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al:
Epithelial-to-mesenchymal transition is not required for lung
metastasis but contributes to chemoresistance. Nature. 527:472–476.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Han L, Lam EWF and Sun Y: Extracellular
vesicles in the tumor microenvironment: Old stories, but new tales.
Mol Cancer. 18:592019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Kanada M, Bachmann MH and Contag CH:
Signaling by extracellular vesicles advances cancer hallmarks.
Trends Cancer. 2:84–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Minciacchi VR, Freeman MR and Di Vizio D:
Extracellular vesicles in cancer: Exosomes, microvesicles and the
emerging role of large oncosomes. Semin Cell Dev Biol. 40:41–51.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chronopoulos A and Kalluri R: Emerging
role of bacterial extracellular vesicles in cancer. Oncogene.
39:6951–6960. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Morad G and Moses MA: Brainwashed by
extracellular vesicles: The role of extracellular vesicles in
primary and metastatic brain tumour microenvironment. J Extracell
Vesicles. 8:16271642019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Parayath NN, Padmakumar S and Amiji MM:
Extracellular vesicle-mediated nucleic acid transfer and
reprogramming in the tumor microenvironment. Cancer Lett.
482:33–43. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Patras L and Banciu M: Intercellular
crosstalk via extracellular vesicles in tumor milieu as emerging
therapies for cancer progression. Curr Pharm Des. 25:1980–2006.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Xie C, Ji N, Tang Z, Li J and Chen Q: The
role of extracellular vesicles from different origin in the
microenvironment of head and neck cancers. Mol Cancer. 18:832019.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Caicedo-Carvajal CE, Liu Q and Goy A:
Three-dimensional cell culture models for biomarker discoveries and
cancer research. Transl Med. 1:1–8. 2012.
|
|
112
|
Wang HX and Gires O: Tumor-derived
extracellular vesicles in breast cancer: From bench to bedside.
Cancer Lett. 460:54–64. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Gould SJ and Raposo G: As we wait: Coping
with an imperfect nomenclature for extracellular vesicles. J
Extracell Vesicles. 2:2013. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Beach A, Zhang HG, Ratajczak MZ and Kakar
SS: Exosomes: An overview of biogenesis, composition and role in
ovarian cancer. J Ovarian Res. 7:142014. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Mashouri L, Yousefi H, Aref AR, Ahadi A
mohammad, Molaei F and Alahari SK: Exosomes: Composition,
biogenesis, and mechanisms in cancer metastasis and drug
resistance. Mol Cancer. 18:752019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
117
|
Chang WH, Cerione RA and Antonyak MA:
Extracellular Vesicles and their roles in cancer progression.
Methods Mol Biol. 2174:143–170. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Menck K, Sivaloganathan S, Bleckmann A and
Binder C: Microvesicles in cancer: Small size, large potential. Int
J Mol Sci. 21:53732020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Al-Nedawi K, Meehan B, Micallef J, Lhotak
V, May L, Guha A and Rak J: Intercellular transfer of the oncogenic
receptor EGFRvIII by microvesicles derived from tumour cells. Nat
Cell Biol. 10:619–624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Di Vizio D, Kim J, Hager MH, Morello M,
Yang W, Lafargue CJ, True LD, Rubin MA, Adam RM, Beroukhim R, et
al: Oncosome formation in prostate cancer: Association with a
region of frequent chromosomal deletion in metastatic disease.
Cancer Res. 69:5601–5609. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Gurunathan S, Kang MH, Jeyaraj M, Qasim M
and Kim JH: Review of the isolation, characterization, biological
function, and multifarious therapeutic approaches of exosomes.
Cells. 8:3072019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Hessvik NP and Llorente A: Current
knowledge on exosome biogenesis and release. Cell Mol Life Sci.
75:193–208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Baig AM, Khaleeq A, Ali U and Syeda H:
Evidence of the COVID-19 virus targeting the CNS: Tissue
distribution, host-virus interaction, and proposed neurotropic
mechanisms. ACS Chem Neurosci. 11:995–998. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Li I and Nabet BY: Exosomes in the tumor
microenvironment as mediators of cancer therapy resistance. Mol
Cancer. 18:322019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Peinado H, Alečković M, Lavotshkin S,
Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M,
Williams C, García-Santos G, Ghajar C, et al: Melanoma exosomes
educate bone marrow progenitor cells toward a pro-metastatic
phenotype through MET. Nat Med. 18:883–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Walbrecq G, Margue C, Behrmann I and Kreis
S: Distinct cargos of small extracellular vesicles derived from
hypoxic cells and their effect on cancer cells. Int J Mol Sci.
21:50712020. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Azulay EE, Cooks T and Elkabets M:
Potential oncogenic roles of mutant-p53-derived exosomes in the
tumor-host interaction of head and neck cancers. Cancer Immunol
Immunother. 69:285–292. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Pavlakis E, Neumann M and Stiewe T:
Extracellular vesicles: Messengers of p53 in tumor-stroma
communication and cancer metastasis. Int J Mol Sci. 21:96482020.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
de Jong OG, Verhaar MC, Chen Y, Vader P,
Gremmels H, Posthuma G, Schiffelers RM, Gucek M and van Balkom BW:
Cellular stress conditions are reflected in the protein and RNA
content of endothelial cell-derived exosomes. J Extracell Vesicles.
1:2012. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Drake RR and Kislinger T: The proteomics
of prostate cancer exosomes. Expert Rev Proteomics. 11:167–177.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Hamzah RN, Alghazali KM, Biris AS and
Griffin RJ: Exosome traceability and cell source dependence on
composition and cell-cell cross talk. Int J Mol Sci. 22:53462021.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Jelonek K, Widlak P and Pietrowska M: The
influence of ionizing radiation on exosome composition, secretion
and intercellular communication. Protein Pept Lett. 23:656–663.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Jiao YJ, Jin DD, Jiang F, Liu JX, Qu LS,
Ni WK, Liu ZX, Lu CH, Ni RZ, Zhu J and Xiao MB: Characterization
and proteomic profiling of pancreatic cancer-derived serum
exosomes. J Cell Biochem. 120:988–999. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Hornung T, O'Neill HA, Logie SC, Fowler
KM, Duncan JE, Rosenow M, Bondre AS, Tinder T, Maher V, Zarkovic J,
et al: ADAPT identifies an ESCRT complex composition that
discriminates VCaP from LNCaP prostate cancer cell exosomes.
Nucleic Acids Res. 48:4013–4027. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon
T, Kim BC, Benci JL, DeMichele AM, Tchou J, Marcotrigiano J and
Minn AJ: Exosome RNA unshielding couples stromal activation to
pattern recognition receptor signaling in cancer. Cell.
170:352–366.e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Luo N: Editorial: Tumor microenvironment
in cancer hallmarks and therapeutics. Front Mol Biosci.
9:10198302022. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zheng Q, Yu X, Zhang M, Zhang S, Guo W and
He Y: Current research progress of the role of LncRNA LEF1-AS1 in a
variety of tumors. Front Cell Dev Biol. 9:7500842021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Tu J, Chen W, Zheng L, Fang S, Zhang D,
Kong C, Yang Y, Qiu R, Zhao Z, Lu C, et al: Circular RNA
Circ0021205 promotes cholangiocarcinoma progression through
MiR-204-5p/RAB22A axis. Front Cell Dev Biol. 9:6532072021.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Chen L, Wang Y, Lu X, Zhang L and Wang Z:
miRNA-7062-5p promoting bone resorption after bone metastasis of
colorectal cancer through inhibiting GPR65. Front Cell Dev Biol.
9:6819682021. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Xiao J, Liu Y, Yi J and Liu X: LINC02257,
an enhancer RNA of prognostic value in colon adenocarcinoma,
correlates with multi-omics immunotherapy-related analysis in 33
cancers. Front Mol Biosci. 8:6467862021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Ma J, Lin X, Wang X, Min Q, Wang T and
Tang C: Reconstruction and analysis of the immune-related
LINC00987/A2M axis in lung adenocarcinoma. Front Mol Biosci.
8:6445572021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Huang K, Fang C, Yi K, Liu X, Qi H, Tan Y,
Zhou J, Li Y, Liu M, Zhang Y, et al: The role of PTRF/Cavin1 as a
biomarker in both glioma and serum exosomes. Theranostics.
8:1540–1557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Ayala-Mar S, Donoso-Quezada J and
González-Valdez J: Clinical implications of exosomal PD-L1 in
cancer immunotherapy. J Immunol Res. 2021:88399782021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Chen G, Huang AC, Zhang W, Zhang G, Wu M,
Xu W, Yu Z, Yang J, Wang B, Sun H, et al: Exosomal PD-L1
contributes to immunosuppression and is associated with anti-PD-1
response. Nature. 560:382–386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Rai A, Greening DW, Chen M, Xu R, Ji H and
Simpson RJ: Exosomes derived from human primary and metastatic
colorectal cancer cells contribute to functional heterogeneity of
activated fibroblasts by reprogramming their proteome. Proteomics.
19:e18001482019. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Berg G, Rybakova D, Fischer D, Cernava T,
Vergès MC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, et
al: Microbiome definition re-visited: Old concepts and new
challenges. Microbiome. 8:1032020. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
AlHilli MM and Bae-Jump V: Diet and gut
microbiome interactions in gynecologic cancer. Gynecol Oncol.
159:299–308. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Anfossi S and Calin GA: Gut microbiota: A
new player in regulating immune- and chemo-therapy efficacy. Cancer
Drug Resist. 3:356–370. 2020.PubMed/NCBI
|
|
149
|
De Almeida CV, de Camargo MR, Russo E and
Amedei A: Role of diet and gut microbiota on colorectal cancer
immunomodulation. World J Gastroenterol. 25:151–162. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Di Modica M, Gargari G, Regondi V, Bonizzi
A, Arioli S, Belmonte B, De Cecco L, Fasano E, Bianchi F,
Bertolotti A, et al: Gut microbiota condition the therapeutic
efficacy of trastuzumab in HER2-positive breast cancer. Cancer Res.
81:2195–2206. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Liu L, Tabung FK, Zhang X, Nowak JA, Qian
ZR, Hamada T, Nevo D, Bullman S, Mima K, Kosumi K, et al: Diets
that promote colon inflammation associate with risk of colorectal
carcinomas that contain Fusobacterium nucleatum. Clin
Gastroenterol Hepatol. 16:1622–1631.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Ponziani FR, Nicoletti A, Gasbarrini A and
Pompili M: Diagnostic and therapeutic potential of the gut
microbiota in patients with early hepatocellular carcinoma. Ther
Adv Med Oncol. 11:17588359198481842019. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Rodríguez-García C, Sánchez-Quesada C,
Algarra I and Gaforio JJ: The high-fat diet based on extra-virgin
olive oil causes dysbiosis linked to colorectal cancer prevention.
Nutrients. 12:17052020. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Laplane L, Duluc D, Bikfalvi A, Larmonier
N and Pradeu T: Beyond the tumour microenvironment. Int J Cancer.
145:2611–2618. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Chakladar J, Kuo SZ, Castaneda G, Li WT,
Gnanasekar A, Yu MA, Chang EY, Wang XQ and Ongkeko WM: The
pancreatic microbiome is associated with carcinogenesis and worse
prognosis in males and smokers. Cancers (Basel). 12:26722020.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Chandel D, Sharma M, Chawla V, Sachdeva N
and Shukla G: Isolation, characterization and identification of
antigenotoxic and anticancerous indigenous probiotics and their
prophylactic potential in experimental colon carcinogenesis. Sci
Rep. 9:147692019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Clanton R, Saucier D, Ford J and Akabani
G: Microbial influences on hormesis, oncogenesis, and therapy: A
review of the literature. Environ Res. 142:239–256. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Guo W, Zhang Y, Guo S, Mei Z, Liao H, Dong
H, Wu K, Ye H, Zhang Y, Zhu Y, et al: Tumor microbiome contributes
to an aggressive phenotype in the basal-like subtype of pancreatic
cancer. Commun Biol. 4:10192021. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Huang H, Ren Z, Gao X, Hu X, Zhou Y, Jiang
J, Lu H, Yin S, Ji J, Zhou L and Zheng S: Integrated analysis of
microbiome and host transcriptome reveals correlations between gut
microbiota and clinical outcomes in HBV-related hepatocellular
carcinoma. Genome Med. 12:1022020. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Ingman WV: The gut microbiome: A new
player in breast cancer metastasis. Cancer Res. 79:3539–3541. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Jenkins SV, Robeson MS II, Griffin RJ,
Quick CM, Siegel ER, Cannon MJ, Vang KB and Dings RPM:
Gastrointestinal tract dysbiosis enhances distal tumor progression
through suppression of leukocyte trafficking. Cancer Res.
79:5999–6009. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Li L, Deng X, Zou Y, Lv X and Guo Y:
Characterization of the nasopharynx microbiota in patients with
nasopharyngeal carcinoma vs healthy controls. Braz J Microbiol.
52:1873–1880. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Liu HX, Tao LL, Zhang J, Zhu YG, Zheng Y,
Liu D, Zhou M, Ke H, Shi MM and Qu JM: Difference of lower airway
microbiome in bilateral protected specimen brush between lung
cancer patients with unilateral lobar masses and control subjects.
Int J Cancer. 142:769–778. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Tzeng A, Sangwan N, Jia M, Liu CC, Keslar
KS, Downs-Kelly E, Fairchild RL, Al-Hilli Z, Grobmyer SR and Eng C:
Human breast microbiome correlates with prognostic features and
immunological signatures in breast cancer. Genome Med. 13:602021.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Livyatan I, Nejman D, Shental N and
Straussman R: Characterization of the human tumor microbiome
reveals tumor-type specific intra-cellular bacteria.
Oncoimmunology. 9:18009572020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Burns MB, Lynch J, Starr TK, Knights D and
Blekhman R: Virulence genes are a signature of the microbiome in
the colorectal tumor microenvironment. Genome Med. 7:552015.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Burns MB, Montassier E, Abrahante J, Priya
S, Niccum DE, Khoruts A, Starr TK, Knights D and Blekhman R:
Colorectal cancer mutational profiles correlate with defined
microbial communities in the tumor microenvironment. PLoS Genet.
14:e10073762018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Lee JA, Yoo SY, Oh HJ, Jeong S, Cho NY,
Kang GH and Kim JH: Differential immune microenvironmental features
of microsatellite-unstable colorectal cancers according to
Fusobacterium nucleatum status. Cancer Immunol Immunother.
70:47–59. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Liu W, Zhang R, Shu R, Yu J, Li H, Long H,
Jin S, Li S, Hu Q, Yao F, et al: Study of the relationship between
microbiome and colorectal cancer susceptibility using 16SrRNA
sequencing. Biomed Res Int. 2020:78283922020.PubMed/NCBI
|
|
170
|
Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng
Y, Liu F, Yan C, Li L and Ling Z: Alterations of gastric mucosal
microbiota across different stomach microhabitats in a cohort of
276 patients with gastric cancer. EBioMedicine. 40:336–348. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Oresta B, Braga D, Lazzeri M, Frego N,
Saita A, Faccani C, Fasulo V, Colombo P, Guazzoni G, Hurle R and
Rescigno M: The microbiome of catheter collected urine in males
with bladder cancer according to disease stage. J Urol. 205:86–93.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Wu P, Zhang G, Zhao J, Chen J, Chen Y,
Huang W, Zhong J and Zeng J: Profiling the urinary microbiota in
male patients with bladder cancer in China. Front Cell Infect
Microbiol. 8:1672018. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Yin J, Dong L, Zhao J, Wang H, Li J, Yu A,
Chen W and Wei W: Composition and consistence of the bacterial
microbiome in upper, middle and lower esophagus before and after
Lugol's iodine staining in the esophagus cancer screening. Scand J
Gastroenterol. 55:1467–1474. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Minarovits J: Anaerobic bacterial
communities associated with oral carcinoma: Intratumoral,
surface-biofilm and salivary microbiota. Anaerobe. 68:1023002021.
View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Kostic AD, Gevers D, Pedamallu CS, Michaud
M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et
al: Genomic analysis identifies association of Fusobacterium
with colorectal carcinoma. Genome Res. 22:292–298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Thomas AM, Jesus EC, Lopes A, Aguiar S Jr,
Begnami MD, Rocha RM, Carpinetti PA, Camargo AA, Hoffmann C,
Freitas HC, et al: Tissue-associated bacterial alterations in
rectal carcinoma patients revealed by 16S rRNA community profiling.
Front Cell Infect Microbiol. 6:1792016. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Thompson KJ, Ingle JN, Tang X, Chia N,
Jeraldo PR, Walther-Antonio MR, Kandimalla KK, Johnson S, Yao JZ,
Harrington SC, et al: A comprehensive analysis of breast cancer
microbiota and host gene expression. PLoS One. 12:e01888732017.
View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Cavarretta I, Ferrarese R, Cazzaniga W,
Saita D, Lucianò R, Ceresola ER, Locatelli I, Visconti L, Lavorgna
G, Briganti A, et al: The microbiome of the prostate tumor
microenvironment. Eur Urol. 72:625–631. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Duan H, Chen L, Qu L, Yang H, Song SW, Han
Y, Ye M, Chen W, He X and Shou C: Mycoplasma hyorhinis
infection promotes NF-κB-dependent migration of gastric cancer
cells. Cancer Res. 74:5782–5794. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Duan H, Qu L and Shou C: Mycoplasma
hyorhinis induces epithelial-mesenchymal transition in gastric
cancer cell MGC803 via TLR4-NF-κB signaling. Cancer Lett.
354:447–454. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Geller LT, Barzily-Rokni M, Danino T,
Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee
K, et al: Potential role of intratumor bacteria in mediating tumor
resistance to the chemotherapeutic drug gemcitabine. Science.
357:1156–1160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Huang S, Li JY, Wu J, Meng L and Shou CC:
Mycoplasma infections and different human carcinomas. World J
Gastroenterol. 7:266–269. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Liu D, Hu Y, Guo Y, Zhu Z, Lu B, Wang X
and Huang Y: Mycoplasma-associated multidrug resistance of
hepatocarcinoma cells requires the interaction of P37 and Annexin
A2. PLoS One. 12:e01845782017. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Liu X, Rong Z and Shou C: Mycoplasma
hyorhinis infection promotes gastric cancer cell motility via
β-catenin signaling. Cancer Med. 8:5301–5312. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Xu Y, Li H, Chen W, Yao X, Xing Y, Wang X,
Zhong J and Meng G: Mycoplasma hyorhinis activates the NLRP3
inflammasome and promotes migration and invasion of gastric cancer
cells. PLoS One. 8:e779552013. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Gedye C, Cardwell T, Dimopoulos N, Tan BS,
Jackson H, Svobodová S, Anaka M, Behren A, Maher C, Hofmann O, et
al: Mycoplasma infection alters cancer stem cell properties in
vitro. Stem Cell Rev Rep. 12:156–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Franco OE, Shaw AK, Strand DW and Hayward
SW: Cancer associated fibroblasts in cancer pathogenesis. Semin
Cell Dev Biol. 21:33–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Spaeth EL, Dembinski JL, Sasser AK, Watson
K, Klopp A, Hall B, Andreeff M and Marini F: Mesenchymal stem cell
transition to tumor-associated fibroblasts contributes to
fibrovascular network expansion and tumor progression. PLoS One.
4:e49922009. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Erez N, Truitt M, Olson P, Arron ST and
Hanahan D: Cancer-associated fibroblasts are activated in incipient
neoplasia to orchestrate tumor-promoting inflammation in an
NF-kappaB-dependent manner. Cancer Cell. 17:135–147. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Straussman R, Morikawa T, Shee K,
Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J,
Frederick DT, et al: Tumour micro-environment elicits innate
resistance to RAF inhibitors through HGF secretion. Nature.
487:500–504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Zhou Z, Zhou Q, Wu X, Xu S, Hu X, Tao X,
Li B, Peng J, Li D, Shen L, et al: VCAM-1 secreted from
cancer-associated fibroblasts enhances the growth and invasion of
lung cancer cells through AKT and MAPK signaling. Cancer Lett.
473:62–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Li F, Zhao S, Guo T, Li J and Gu C: The
nutritional cytokine leptin promotes NSCLC by activating the
PI3K/AKT and MAPK/ERK pathways in NSCLC cells in a paracrine
manner. Biomed Res Int. 2019:25857432019.PubMed/NCBI
|
|
195
|
Folkman J, Watson K, Ingber D and Hanahan
D: Induction of angiogenesis during the transition from hyperplasia
to neoplasia. Nature. 339:58–61. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Hanahan D and Folkman J: Patterns and
emerging mechanisms of the angiogenic switch during tumorigenesis.
Cell. 86:353–364. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Butler JM, Kobayashi H and Rafii S:
Instructive role of the vascular niche in promoting tumour growth
and tissue repair by angiocrine factors. Nat Rev Cancer.
10:138–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Balkwill F, Charles KA and Mantovani A:
Smoldering and polarized inflammation in the initiation and
promotion of malignant disease. Cancer Cell. 7:211–217. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Lu P, Takai K, Weaver VM and Werb Z:
Extracellular matrix degradation and remodeling in development and
disease. Cold Spring Harb Perspect Biol. 3:a0050582011. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Pontiggia O, Sampayo R, Raffo D, Motter A,
Xu R, Bissell MJ, Joffé EB and Simian M: The tumor microenvironment
modulates tamoxifen resistance in breast cancer: A role for soluble
stromal factors and fibronectin through β1 integrin. Breast Cancer
Res Treat. 133:459–471. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Mohamed MM and Sloane BF: Cysteine
cathepsins: Multifunctional enzymes in cancer. Nat Rev Cancer.
6:764–775. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Xu R, Boudreau A and Bissell MJ: Tissue
architecture and function: Dynamic reciprocity via extra- and
intra-cellular matrices. Cancer Metastasis Rev. 28:167–176. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Goulet CR and Pouliot F: TGFβ signaling in
the tumor microenvironment. Adv Exp Med Biol. 1270:89–105. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Hou W, Kaczorowski A, Lantwin P,
Kippenberger M, Schütz V, Franke D, Dieffenbacher SC, Hohenfellner
M and Duensing S: Microenvironment-derived FGF-2 stimulates renal
cell carcinoma cell proliferation through modulation of p27Kip1:
Implications for spatial niche formation and functional
intratumoral heterogeneity. Pathobiology. 87:114–124. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Zou F, Zhang ZH, Zhang YT, Zhao JQ, Zhang
XL, Wen CL, Song XY and Zhou WM: Cancer-associated-fibroblasts
regulate the chemoresistance of lung cancer cell line A549 via
SDF-1 secretion. Zhonghua Zhong Liu Za Zhi. 39:339–343. 2017.(In
Chinese). PubMed/NCBI
|
|
206
|
Wang H, Huang H, Wang L, Liu Y, Wang M,
Zhao S, Lu G and Kang X: Cancer-associated fibroblasts secreted
miR-103a-3p suppresses apoptosis and promotes cisplatin resistance
in non-small cell lung cancer. Aging (Albany NY). 13:14456–14468.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Sun X and Chen Z: Cancer-associated
fibroblast-derived CCL5 contributes to cisplatin resistance in A549
NSCLC cells partially through upregulation of lncRNA HOTAIR
expression. Oncol Lett. 22:6962021. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Tao L, Huang G, Wang R, Pan Y, He Z, Chu
X, Song H and Chen L: Cancer-associated fibroblasts treated with
cisplatin facilitates chemoresistance of lung adenocarcinoma
through IL-11/IL-11R/STAT3 signaling pathway. Sci Rep. 6:384082016.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Bian L, Sun X, Jin K and He Y: Oral
cancer-associated fibroblasts inhibit heat-induced apoptosis in
Tca8113 cells through upregulated expression of Bcl-2 through the
Mig/CXCR3 axis. Oncol Rep. 28:2063–2068. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Daenen LG, Shaked Y, Man S, Xu P, Voest
EE, Hoffman RM, Chaplin DJ and Kerbel RS: Low-dose metronomic
cyclophosphamide combined with vascular disrupting therapy induces
potent antitumor activity in preclinical human tumor xenograft
models. Mol Cancer Ther. 8:2872–2881. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Chen Q, Zhang XHF and Massagué J:
Macrophage binding to receptor VCAM-1 transmits survival signals in
breast cancer cells that invade the lungs. Cancer Cell. 20:538–549.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Yang C, He L, He P, Liu Y, Wang W, He Y,
Du Y and Gao F: Increased drug resistance in breast cancer by
tumor-associated macrophages through IL-10/STAT3/bcl-2 signaling
pathway. Med Oncol. 32:3522015. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Harley CB, Kim NW, Prowse KR, Weinrich SL,
Hirsch KS, West MD, Bacchetti S, Hirte HW, Counter CM, Greider CW,
et al: Telomerase, cell immortality, and cancer. Cold Spring Harb
Symp Quant Biol. 59:307–315. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Jäger K and Walter M: Therapeutic
targeting of telomerase. Genes (Basel). 7:392016. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Ge Y, Wu S, Xue Y, Tao J, Li F, Chen Y,
Liu H, Ma W, Huang J and Zhao Y: Preferential extension of short
telomeres induced by low extracellular pH. Nucleic Acids Res.
44:8086–8096. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Li S, Jiang Y, Li A, Liu X, Xing X, Guo Y,
Xu Y, Hao Y and Zheng C: Telomere length is positively associated
with the expression of IL-6 and MIP-1α in bone marrow mesenchymal
stem cells of multiple myeloma. Mol Med Rep. 16:2497–2504. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Lin EY, Li JF, Bricard G, Wang W, Deng Y,
Sellers R, Porcelli SA and Pollard JW: Vascular endothelial growth
factor restores delayed tumor progression in tumors depleted of
macrophages. Mol Oncol. 1:288–302. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Bergers G, Brekken R, McMahon G, Vu TH,
Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z and
Hanahan D: Matrix metalloproteinase-9 triggers the angiogenic
switch during carcinogenesis. Nat Cell Biol. 2:737–744. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Coussens LM, Raymond WW, Bergers G,
Laig-Webster M, Behrendtsen O, Werb Z, Caughey GH and Hanahan D:
Inflammatory mast cells up-regulate angiogenesis during squamous
epithelial carcinogenesis. Genes Dev. 13:1382–1397. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Khazaie K, Blatner NR, Khan MW, Gounari F,
Gounaris E, Dennis K, Bonertz A, Tsai FN, Strouch MJ, Cheon E, et
al: The significant role of mast cells in cancer. Cancer Metastasis
Rev. 30:45–60. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Huang B, Huang M and Li Q:
Cancer-associated fibroblasts promote angiogenesis of
hepatocellular carcinoma by VEGF-mediated EZH2/VASH1 pathway.
Technol Cancer Res Treat. 18:15330338198799052019. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Räsänen K and Vaheri A: Activation of
fibroblasts in cancer stroma. Exp Cell Res. 316:2713–2722. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Huizer K, Zhu C, Chirifi I, Krist B,
Zorgman D, van der Weiden M, van den Bosch TPP, Dumas J, Cheng C,
Kros JM and Mustafa DA: Periostin is expressed by pericytes and is
crucial for angiogenesis in glioma. J Neuropathol Exp Neurol.
79:863–872. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Xian X, Håkansson J, Ståhlberg A, Lindblom
P, Betsholtz C, Gerhardt H and Semb H: Pericytes limit tumor cell
metastasis. J Clin Invest. 116:642–651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Branco-Price C, Zhang N, Schnelle M, Evans
C, Katschinski DM, Liao D, Ellies L and Johnson RS: Endothelial
cell HIF-1α and HIF-2α differentially regulate metastatic success.
Cancer Cell. 21:52–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Navarro R, Tapia-Galisteo A, Martín-García
L, Tarín C, Corbacho C, Gómez-López G, Sánchez-Tirado E, Campuzano
S, González-Cortés A, Yáñez-Sedeño P, et al: TGF-β-induced IGFBP-3
is a key paracrine factor from activated pericytes that promotes
colorectal cancer cell migration and invasion. Mol Oncol.
14:2609–2628. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Pirilä E, Ramamurthy NS, Sorsa T, Salo T,
Hietanen J and Maisi P: Gelatinase A (MMP-2), collagenase-2
(MMP-8), and laminin-5 gamma2-chain expression in murine
inflammatory bowel disease (ulcerative colitis). Dig Dis Sci.
48:93–98. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Vasiljeva O, Papazoglou A, Krüger A,
Brodoefel H, Korovin M, Deussing J, Augustin N, Nielsen BS, Almholt
K, Bogyo M, et al: Tumor cell-derived and macrophage-derived
cathepsin B promotes progression and lung metastasis of mammary
cancer. Cancer Res. 66:5242–5250. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Balkwill F: Tumour necrosis factor and
cancer. Nat Rev Cancer. 9:361–371. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Abraham S, Zhang W, Greenberg N and Zhang
M: Maspin functions as tumor suppressor by increasing cell adhesion
to extracellular matrix in prostate tumor cells. J Urol.
169:1157–1161. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Gorden DL, Fingleton B, Crawford HC,
Jansen DE, Lepage M and Matrisian LM: Resident stromal cell-derived
MMP-9 promotes the growth of colorectal metastases in the liver
microenvironment. Int J Cancer. 121:495–500. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Labelle M, Begum S and Hynes RO: Direct
signaling between platelets and cancer cells induces an
epithelial-mesenchymal-like transition and promotes metastasis.
Cancer Cell. 20:576–590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Zhang XHF, Jin X, Malladi S, Zou Y, Wen
YH, Brogi E, Smid M, Foekens JA and Massagué J: Selection of bone
metastasis seeds by mesenchymal signals in the primary tumor
stroma. Cell. 154:1060–1073. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Duda DG, Duyverman AMMJ, Kohno M, Snuderl
M, Steller EJA, Fukumura D and Jain RK: Malignant cells facilitate
lung metastasis by bringing their own soil. Proc Natl Acad Sci USA.
107:21677–21682. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Lappano R, Rigiracciolo DC, Belfiore A,
Maggiolini M and De Francesco EM: Cancer associated fibroblasts:
Role in breast cancer and potential as therapeutic targets. Expert
Opin Ther Targets. 24:559–572. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Onrust SV, Hartl PM, Rosen SD and Hanahan
D: Modulation of L-selectin ligand expression during an immune
response accompanying tumorigenesis in transgenic mice. J Clin
Invest. 97:54–64. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Fisher DT, Chen Q, Skitzki JJ, Muhitch JB,
Zhou L, Appenheimer MM, Vardam TD, Weis EL, Passanese J, Wang WC,
et al: IL-6 trans-signaling licenses mouse and human tumor
microvascular gateways for trafficking of cytotoxic T cells. J Clin
Invest. 121:3846–3859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Manzur M, Hamzah J and Ganss R: Modulation
of the ‘blood-tumor’ barrier improves immunotherapy. Cell Cycle.
7:2452–2455. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Turley SJ, Cremasco V and Astarita JL:
Immunological hallmarks of stromal cells in the tumour
microenvironment. Nat Rev Immunol. 15:669–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Stover DG, Bierie B and Moses HL: A
delicate balance: TGF-beta and the tumor microenvironment. J Cell
Biochem. 101:851–861. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
DeNardo DG, Brennan DJ, Rexhepaj E,
Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD,
Junaid SA, et al: Leukocyte complexity predicts breast cancer
survival and functionally regulates response to chemotherapy.
Cancer Discov. 1:54–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Qian BZ and Pollard JW: Macrophage
diversity enhances tumor progression and metastasis. Cell.
141:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Topalian SL, Drake CG and Pardoll DM:
Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor
immunity. Curr Opin Immunol. 24:207–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Curiel TJ, Coukos G, Zou L, Alvarez X,
Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L,
Burow M, et al: Specific recruitment of regulatory T cells in
ovarian carcinoma fosters immune privilege and predicts reduced
survival. Nat Med. 10:942–949. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
246
|
van der Vliet HJJ, Koon HB, Atkins MB,
Balk SP and Exley MA: Exploiting regulatory T-cell populations for
the immunotherapy of cancer. J Immunother. 30:591–595. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Schmidt A, Oberle N and Krammer P:
Molecular mechanisms of treg-mediated T cell suppression. Front
Immunol. 3:512012. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Kalyanaraman B: Teaching the basics of
cancer metabolism: Developing antitumor strategies by exploiting
the differences between normal and cancer cell metabolism. Redox
Biol. 12:833–842. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Hensley CT, Faubert B, Yuan Q, Lev-Cohain
N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al:
Metabolic heterogeneity in human lung tumors. Cell. 164:681–694.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Yu TJ, Ma D, Liu YY, Xiao Y, Gong Y, Jiang
YZ, Shao ZM, Hu X and Di GH: Bulk and single-cell transcriptome
profiling reveal the metabolic heterogeneity in human breast
cancers. Mol Ther. 29:2350–2365. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Hao X, Ren Y, Feng M, Wang Q and Wang Y:
Metabolic reprogramming due to hypoxia in pancreatic cancer:
Implications for tumor formation, immunity, and more. Biomed
Pharmacother. 141:1117982021. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Kim J and DeBerardinis RJ: Mechanisms and
implications of metabolic heterogeneity in cancer. Cell Metab.
30:434–446. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Xiao Z, Dai Z and Locasale JW: Metabolic
landscape of the tumor microenvironment at single cell resolution.
Nat Commun. 10:37632019. View Article : Google Scholar : PubMed/NCBI
|
|
254
|
Stadlbauer A, Oberndorfer S, Zimmermann M,
Renner B, Buchfelder M, Heinz G, Doerfler A, Kleindienst A and
Roessler K: Physiologic MR imaging of the tumor microenvironment
revealed switching of metabolic phenotype upon recurrence of
glioblastoma in humans. J Cereb Blood Flow Metab. 40:528–538. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Gupta S, Roy A and Dwarakanath BS:
Metabolic cooperation and competition in the tumor
microenvironment: Implications for therapy. Front Oncol. 7:682017.
View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Martinez-Outschoorn U, Sotgia F and
Lisanti MP: Tumor microenvironment and metabolic synergy in breast
cancers: Critical importance of mitochondrial fuels and function.
Semin Oncol. 41:1952014. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Ocaña MC, Martínez-Poveda B, Quesada AR
and Medina MÁ: Metabolism within the tumor microenvironment and its
implication on cancer progression: An ongoing therapeutic target.
Med Res Rev. 39:70–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Wang YA, Li XL, Mo YZ, Fan CM, Tang L,
Xiong F, Guo C, Xiang B, Zhou M, Ma J, et al: Effects of tumor
metabolic microenvironment on regulatory T cells. Mol Cancer.
17:1682018. View Article : Google Scholar : PubMed/NCBI
|
|
259
|
Pavlides S, Whitaker-Menezes D,
Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro
MC, Wang C, Fortina P, Addya S, et al: The reverse Warburg effect:
Aerobic glycolysis in cancer associated fibroblasts and the tumor
stroma. Cell Cycle. 8:3984–4001. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
260
|
Reina-Campos M, Moscat J and Diaz-Meco M:
Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol.
48:47–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
261
|
Dietl K, Renner K, Dettmer K, Timischl B,
Eberhart K, Dorn C, Hellerbrand C, Kastenberger M, Kunz-Schughart
LA, Oefner PJ, et al: Lactic acid and acidification inhibit TNF
secretion and glycolysis of human monocytes. J Immunol.
184:1200–1209. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Whitaker-Menezes D, Martinez-Outschoorn
UE, Lin Z, Ertel A, Flomenberg N, Witkiewicz AK, Birbe RC, Howell
A, Pavlides S, Gandara R, et al: Evidence for a stromal-epithelial
‘lactate shuttle’ in human tumors: MCT4 is a marker of oxidative
stress in cancer-associated fibroblasts. Cell Cycle. 10:1772–1783.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Alberts B, Johnson A, Lewis J, Raff M,
Roberts K and Walter P: Molecular biology of the cell. 4th edition.
New York: Garland Science; 2002
|
|
264
|
Deberardinis RJ, Sayed N, Ditsworth D and
Thompson CB: Brick by brick: Metabolism and tumor cell growth. Curr
Opin Genet Dev. 18:54–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Yoshida GJ: Metabolic reprogramming: the
emerging concept and associated therapeutic strategies. J Exp Clin
Cancer Res. 34:1112015. View Article : Google Scholar : PubMed/NCBI
|
|
266
|
Koundouros N and Poulogiannis G:
Reprogramming of fatty acid metabolism in cancer. Br J Cancer.
122:4–22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
267
|
Michalopoulou E, Bulusu V and Kamphorst
JJ: Metabolic scavenging by cancer cells: When the going gets
tough, the tough keep eating. Br J Cancer. 115:635–640. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Dey P, Kimmelman AC and DePinho RA:
Metabolic codependencies in the tumor microenvironment. Cancer
Discov. 11:1067–1081. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
270
|
Bailey KM, Wojtkowiak JW, Hashim AI and
Gillies RJ: Targeting the metabolic microenvironment of tumors. Adv
Pharmacol. 65:63–107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
271
|
Sormendi S and Wielockx B: Hypoxia pathway
proteins as central mediators of metabolism in the tumor cells and
their microenvironment. Front Immunol. 9:402018. View Article : Google Scholar : PubMed/NCBI
|
|
272
|
Devic S: Warburg effect-a consequence or
the cause of carcinogenesis? J Cancer. 7:817–822. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
273
|
Cairns RA: Drivers of the Warburg
phenotype. Cancer J. 21:56–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
274
|
El Hassouni B, Granchi C, Vallés-Martí A,
Supadmanaba IGP, Bononi G, Tuccinardi T, Funel N, Jimenez CR,
Peters GJ, Giovannetti E and Minutolo F: The dichotomous role of
the glycolytic metabolism pathway in cancer metastasis: Interplay
with the complex tumor microenvironment and novel therapeutic
strategies. Semin Cancer Biol. 60:238–248. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
275
|
Chang CH, Qiu J, O'Sullivan D, Buck MD,
Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ,
et al: Metabolic competition in the tumor microenvironment is a
driver of cancer progression. Cell. 162:1229–1241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
276
|
DeBerardinis RJ, Mancuso A, Daikhin E,
Nissim I, Yudkoff M, Wehrli S and Thompson CB: Beyond aerobic
glycolysis: Transformed cells can engage in glutamine metabolism
that exceeds the requirement for protein and nucleotide synthesis.
Proc Natl Acad Sci USA. 104:19345–19350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Carito V, Bonuccelli G,
Martinez-Outschoorn UE, Whitaker-Menezes D, Caroleo MC, Cione E,
Howell A, Pestell RG, Lisanti MP and Sotgia F: Metabolic remodeling
of the tumor microenvironment: Migration factor (MSF) reprograms
myofibroblasts toward lactate production, fueling anabolic tumor
growthstimulating. Cell Cycle. 11:3403–3414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
278
|
Manning BD and Toker A: AKT/PKB signaling:
Navigating the network. Cell. 169:381–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
279
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, metabolism, and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Osthus RC, Shim H, Kim S, Li Q, Reddy R,
Mukherjee M, Xu Y, Wonsey D, Lee LA and Dang CV: Deregulation of
glucose transporter 1 and glycolytic gene expression by c-Myc. J
Biol Chem. 275:21797–21800. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
282
|
Zhao X, Petrashen AP, Sanders JA, Peterson
AL and Sedivy JM: SLC1A5 glutamine transporter is a target of MYC
and mediates reduced mTORC1 signaling and increased fatty acid
oxidation in long-lived Myc hypomorphic mice. Aging Cell.
18:e129472019. View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Sasaki H, Shitara M, Yokota K, Hikosaka Y,
Moriyama S, Yano M and Fujii Y: Overexpression of GLUT1 correlates
with Kras mutations in lung carcinomas. Mol Med Rep. 5:599–602.
2012.PubMed/NCBI
|
|
284
|
Ying H, Kimmelman AC, Lyssiotis CA, Hua S,
Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff
JL, et al: Oncogenic Kras maintains pancreatic tumors through
regulation of anabolic glucose metabolism. Cell. 149:656–670. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
285
|
Son J, Lyssiotis CA, Ying H, Wang X, Hua
S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et
al: Glutamine supports pancreatic cancer growth through a
KRAS-regulated metabolic pathway. Nature. 496:101–105. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
286
|
Baek G, Tse YF, Hu Z, Cox D, Buboltz N,
McCue P, Yeo CJ, White MA, DeBerardinis RJ, Knudsen ES and
Witkiewicz AK: MCT4 defines a glycolytic subtype of pancreatic
cancer with poor prognosis and unique metabolic dependencies. Cell
Rep. 9:2233–2249. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
287
|
Dey P, Li J, Zhang J, Chaurasiya S, Strom
A, Wang H, Liao WT, Cavallaro F, Denz P, Bernard V, et al:
Oncogenic KRAS-driven metabolic reprogramming in pancreatic cancer
cells utilizes cytokines from the tumor microenvironment. Cancer
Discov. 10:608–625. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
288
|
Amendola CR, Mahaffey JP, Parker SJ,
Ahearn IM, Chen WC, Zhou M, Court H, Shi J, Mendoza SL, Morten MJ,
et al: KRAS4A directly regulates hexokinase 1. Nature. 576:482–486.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
289
|
Zhang Y and Commisso C: Macropinocytosis
in cancer: A complex signaling network. Trends Cancer. 5:332–334.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
290
|
Commisso C, Davidson SM, Soydaner-Azeloglu
RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin
JA, Thompson CB, et al: Macropinocytosis of protein is an amino
acid supply route in Ras-transformed cells. Nature. 497:633–637.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
291
|
Schwartzenberg-Bar-Yoseph F, Armoni M and
Karnieli E: The tumor suppressor p53 down-regulates glucose
transporters GLUT1 and GLUT4 gene expression. Cancer Res.
64:2627–2633. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
292
|
Wang L, Xiong H, Wu F, Zhang Y, Wang J,
Zhao L, Guo X, Chang LJ, Zhang Y, You MJ, et al: Hexokinase
2-mediated Warburg effect is required for PTEN- and
p53-deficiency-driven prostate cancer growth. Cell Rep.
8:1461–1474. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
293
|
Yahagi N, Shimano H, Matsuzaka T, Najima
Y, Sekiya M, Nakagawa Y, Ide T, Tomita S, Okazaki H, Tamura Y, et
al: p53 activation in adipocytes of obese mice. J Biol Chem.
278:25395–25400. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
294
|
Bensaad K, Tsuruta A, Selak MA, Vidal MN,
Nakano K, Bartrons R, Gottlieb E and Vousden KH: TIGAR, a
p53-inducible regulator of glycolysis and apoptosis. Cell.
126:107–120. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
295
|
Sonugür FG and Akbulut H: The role of
tumor microenvironment in genomic instability of malignant tumors.
Front Genet. 10:10632019. View Article : Google Scholar : PubMed/NCBI
|
|
296
|
Nakamura H, Tanimoto K, Hiyama K, Yunokawa
M, Kawamoto T, Kato Y, Yoshiga K, Poellinger L, Hiyama E and
Nishiyama M: Human mismatch repair gene, MLH1, is transcriptionally
repressed by the hypoxia-inducible transcription factors, DEC1 and
DEC2. Oncogene. 27:4200–4209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
297
|
Rodríguez-Jiménez FJ, Moreno-Manzano V,
Lucas-Dominguez R and Sánchez-Puelles JM: Hypoxia causes
downregulation of mismatch repair system and genomic instability in
stem cells. Stem Cells. 26:2052–2062. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
298
|
Ren Z, Wang Z, Gu D, Ma H, Zhu Y, Cai M
and Zhang J: Genome instability and long noncoding RNA reveal
biomarkers for immunotherapy and prognosis and novel competing
endogenous RNA mechanism in colon adenocarcinoma. Front Cell Dev
Biol. 9:7404552021. View Article : Google Scholar : PubMed/NCBI
|
|
299
|
Guo JN, Xia TY, Deng SH, Xue WN, Cui BB
and Liu YL: Prognostic immunity and therapeutic sensitivity
analyses based on differential genomic instability-associated
LncRNAs in left- and right-sided colon adenocarcinoma. Front Mol
Biosci. 8:6688882021. View Article : Google Scholar : PubMed/NCBI
|
|
300
|
Fridlender ZG, Sun J, Kim S, Kapoor V,
Cheng G, Ling L, Worthen GS and Albelda SM: Polarization of
tumor-associated neutrophil phenotype by TGF-beta: ‘N1’ versus ‘N2’
TAN. Cancer Cell. 16:183–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
301
|
DeNardo DG, Barreto JB, Andreu P, Vasquez
L, Tawfik D, Kolhatkar N and Coussens LM: CD4(+) T cells regulate
pulmonary metastasis of mammary carcinomas by enhancing protumor
properties of macrophages. Cancer Cell. 16:91–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
302
|
Schoppmann SF, Birner P, Stöckl J, Kalt R,
Ullrich R, Caucig C, Kriehuber E, Nagy K, Alitalo K and Kerjaschki
D: Tumor-associated macrophages express lymphatic endothelial
growth factors and are related to peritumoral lymphangiogenesis. Am
J Pathol. 161:947–956. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
303
|
Celis JE, Gromov P, Cabezón T, Moreira JM,
Ambartsumian N, Sandelin K, Rank F and Gromova I: Proteomic
characterization of the interstitial fluid perfusing the breast
tumor microenvironment: A novel resource for biomarker and
therapeutic target discovery. Mol Cell Proteomics. 3:327–344. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
304
|
Dirat B, Bochet L, Dabek M, Daviaud D,
Dauvillier S, Majed B, Wang YY, Meulle A, Salles B, Le Gonidec S,
et al: Cancer-associated adipocytes exhibit an activated phenotype
and contribute to breast cancer invasion. Cancer Res. 71:2455–2465.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
305
|
Ershaid N, Sharon Y, Doron H, Raz Y, Shani
O, Cohen N, Monteran L, Leider-Trejo L, Ben-Shmuel A, Yassin M, et
al: NLRP3 inflammasome in fibroblasts links tissue damage with
inflammation in breast cancer progression and metastasis. Nat
Commun. 10:43752019. View Article : Google Scholar : PubMed/NCBI
|
|
306
|
Wu X, Tao P, Zhou Q, Li J, Yu Z, Wang X,
Li J, Li C, Yan M, Zhu Z, et al: IL-6 secreted by cancer-associated
fibroblasts promotes epithelial-mesenchymal transition and
metastasis of gastric cancer via JAK2/STAT3 signaling pathway.
Oncotarget. 8:20741–20750. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
307
|
Davidson S, Efremova M, Riedel A, Mahata
B, Pramanik J, Huuhtanen J, Kar G, Vento-Tormo R, Hagai T, Chen X,
et al: Single-cell RNA sequencing reveals a dynamic stromal niche
that supports tumor growth. Cell Rep. 31:1076282020. View Article : Google Scholar : PubMed/NCBI
|
|
308
|
Erez N, Glanz S, Raz Y, Avivi C and
Barshack I: Cancer associated fibroblasts express pro-inflammatory
factors in human breast and ovarian tumors. Biochem Biophys Res
Commun. 437:397–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
309
|
De Monte L, Reni M, Tassi E, Clavenna D,
Papa I, Recalde H, Braga M, Di Carlo V, Doglioni C and Protti MP:
Intratumor T helper type 2 cell infiltrate correlates with
cancer-associated fibroblast thymic stromal lymphopoietin
production and reduced survival in pancreatic cancer. J Exp Med.
208:469–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
310
|
Bejarano L, Jordāo MJC and Joyce JA:
Therapeutic targeting of the tumor microenvironment. Cancer Discov.
11:933–959. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
311
|
Lieu CH, Tan AC, Leong S, Diamond JR and
Eckhardt SG: From bench to bedside: Lessons learned in translating
preclinical studies in cancer drug development. J Natl Cancer Inst.
105:1441–1456. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
312
|
Syed M, Flechsig P, Liermann J, Windisch
P, Staudinger F, Akbaba S, Koerber SA, Freudlsperger C, Plinkert
PK, Debus J, et al: Fibroblast activation protein inhibitor (FAPI)
PET for diagnostics and advanced targeted radiotherapy in head and
neck cancers. Eur J Nucl Med Mol Imaging. 47:2836–2845. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
313
|
Chen H, Pang Y, Wu J, Zhao L, Hao B, Wu J,
Wei J, Wu S, Zhao L, Luo Z, et al: Comparison of
[68Ga]Ga-DOTA-FAPI-04 and [18F] FDG PET/CT
for the diagnosis of primary and metastatic lesions in patients
with various types of cancer. Eur J Nucl Med Mol Imaging.
47:1820–1832. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
314
|
Melero I, Sanmamed MF, Calvo E, Moreno I,
Moreno V, Hernandez Guerrero TC, Martinez-Garcia M, Rodriguez-Vida
A, Tabernero J, Azaro Pedrazzoli AB, et al: 1025MO first-in-human
(FIH) phase I study of RO7122290 (RO), a novel FAP-targeted 4-1BB
agonist, administered as single agent and in combination with
atezolizumab (ATZ) to patients with advanced solid tumours. Ann
Oncol. 31 (Suppl 4):S7072020. View Article : Google Scholar
|
|
315
|
Sounni NE and Noel A: Targeting the tumor
microenvironment for cancer therapy. Clin Chem. 59:85–93. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
316
|
Lamberts LE, Koch M, de Jong JS, Adams
ALL, Glatz J, Kranendonk MEG, Terwisscha van Scheltinga AGT, Jansen
L, de Vries J, Lub-de Hooge MN, et al: Tumor-specific uptake of
fluorescent bevacizumab-IRDye800CW microdosing in patients with
primary breast cancer: A phase I feasibility study. Clin Cancer
Res. 23:2730–2741. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
317
|
Harlaar NJ, Koller M, de Jongh SJ, van
Leeuwen BL, Hemmer PH, Kruijff S, van Ginkel RJ, Been LB, de Jong
JS, Kats-Ugurlu G, et al: Molecular fluorescence-guided surgery of
peritoneal carcinomatosis of colorectal origin: A single-centre
feasibility study. Lancet Gastroenterol Hepatol. 1:283–290. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
318
|
Nakano K, Funauchi Y, Hayakawa K, Tanizawa
T, Ae K, Matsumoto S and Takahashi S: Relative dose intensity of
induction-phase pazopanib treatment of soft tissue sarcoma: Its
relationship with prognoses of pazopanib responders. J Clin Med.
8:602019. View Article : Google Scholar : PubMed/NCBI
|
|
319
|
Noda S, Yoshida T, Hira D, Murai R, Tomita
K, Tsuru T, Kageyama S, Kawauchi A, Ikeda Y, Morita SY and Terada
T: Exploratory investigation of target pazopanib concentration
range for patients with renal cell carcinoma. Clin Genitourin
Cancer. 17:e306–e313. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
320
|
Wang S, Lu J, You Q, Huang H, Chen Y and
Liu K: The mTOR/AP-1/VEGF signaling pathway regulates vascular
endothelial cell growth. Oncotarget. 7:53269–53276. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
321
|
Komohara Y, Fujiwara Y, Ohnishi K and
Takeya M: Tumor-associated macrophages: Potential therapeutic
targets for anti-cancer therapy. Adv Drug Deliv Rev. 99:180–185.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
322
|
Bak SP, Walters JJ, Takeya M,
Conejo-Garcia JR and Berwin BL: Scavenger receptor-A-targeted
leukocyte depletion inhibits peritoneal ovarian tumor progression.
Cancer Res. 67:4783–4789. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
323
|
Nagai T, Tanaka M, Tsuneyoshi Y, Xu B,
Michie SA, Hasui K, Hirano H, Arita K and Matsuyama T: Targeting
tumor-associated macrophages in an experimental glioma model with a
recombinant immunotoxin to folate receptor beta. Cancer Immunol
Immunother. 58:1577–1586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
324
|
Naser R, Dilabazian H, Bahr H, Barakat A
and El-Sibai M: A guide through conventional and modern cancer
treatment modalities: A specific focus on glioblastoma cancer
therapy (review). Oncol Rep. 48:1902022. View Article : Google Scholar : PubMed/NCBI
|
|
325
|
Liu Y and Zheng P: Preserving the CTLA-4
checkpoint for safer and more effective cancer immunotherapy.
Trends Pharmacol Sci. 41:4–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
326
|
Tauriello DVF, Palomo-Ponce S, Stork D,
Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M,
Ibiza S, Cañellas A, Hernando-Momblona X, et al: TGFβ drives immune
evasion in genetically reconstituted colon cancer metastasis.
Nature. 554:538–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
327
|
Greten FR, Arkan MC, Bollrath J, Hsu LC,
Goode J, Miething C, Göktuna SI, Neuenhahn M, Fierer J, Paxian S,
et al: NF-kappaB is a negative regulator of IL-1beta secretion as
revealed by genetic and pharmacological inhibition of IKKbeta.
Cell. 130:918–931. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
328
|
Marsh JL, Jackman CP, Tang SN, Shankar S
and Srivastava RK: Embelin suppresses pancreatic cancer growth by
modulating tumor immune microenvironment. Front Biosci (Landmark
Ed). 19:113–125. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
329
|
Xu CL, Zheng B, Pei JH, Shen SJ and Wang
JZ: Embelin induces apoptosis of human gastric carcinoma through
inhibition of p38 MAPK and NF-κB signaling pathways. Mol Med Rep.
14:307–312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
330
|
Waldmann TA: Cytokines in cancer
immunotherapy. Cold Spring Harb Perspect Biol. 10:a0284722018.
View Article : Google Scholar : PubMed/NCBI
|
|
331
|
Rosenberg SA, Restifo NP, Yang JC, Morgan
RA and Dudley ME: Adoptive cell transfer: A clinical path to
effective cancer immunotherapy. Nat Rev Cancer. 8:299–308. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
332
|
Redeker A and Arens R: Improving adoptive
T cell therapy: The particular role of T cell costimulation,
cytokines, and post-transfer vaccination. Front Immunol. 7:3452016.
View Article : Google Scholar : PubMed/NCBI
|
|
333
|
Chruściel E, Urban-Wójciuk Z, Arcimowicz
Ł, Kurkowiak M, Kowalski J, Gliwiński M, Marjański T, Rzyman W,
Biernat W, Dziadziuszko R, et al: Adoptive cell therapy-harnessing
antigen-specific T cells to target solid tumours. Cancers (Basel).
12:6832020. View Article : Google Scholar : PubMed/NCBI
|
|
334
|
Benmebarek MR, Karches CH, Cadilha BL,
Lesch S, Endres S and Kobold S: Killing mechanisms of chimeric
antigen receptor (CAR) T cells. Int J Mol Sci. 20:12832019.
View Article : Google Scholar : PubMed/NCBI
|
|
335
|
Brown CE, Badie B, Barish ME, Weng L,
Ostberg JR, Chang WC, Naranjo A, Starr R, Wagner J, Wright C, et
al: Bioactivity and safety of IL13Rα2-redirected chimeric antigen
receptor CD8+ T cells in patients with recurrent glioblastoma. Clin
Cancer Res. 21:4062–4072. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
336
|
Xu S, Tang L, Li X, Fan F and Liu Z:
Immunotherapy for glioma: Current management and future
application. Cancer Lett. 476:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
337
|
Liu J, Fu M, Wang M, Wan D, Wei Y and Wei
X: Cancer vaccines as promising immuno-therapeutics: Platforms and
current progress. J Hematol Oncol. 15:282022. View Article : Google Scholar : PubMed/NCBI
|
|
338
|
Keskin DB, Anandappa AJ, Sun J, Tirosh I,
Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E,
et al: Neoantigen vaccine generates intratumoral T cell responses
in phase Ib glioblastoma trial. Nature. 565:234–239. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
339
|
Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur
V, Stevanović S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V,
van der Burg SH, et al: Actively personalized vaccination trial for
newly diagnosed glioblastoma. Nature. 565:240–245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
340
|
Hollingsworth RE and Jansen K: Turning the
corner on therapeutic cancer vaccines. NPJ Vaccines. 4:72019.
View Article : Google Scholar : PubMed/NCBI
|
|
341
|
Rüttinger D, Winter H, van den Engel NK,
Hatz R, Jauch KW, Fox BA and Weber JS: Immunotherapy of cancer: Key
findings and commentary on the third tegernsee conference.
Oncologist. 15:112–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
342
|
AIVITA, Biomedical Inc., . AIVITA
biomedical's phase 2 glioblastoma trial shows improved progression
free survival. 2021.
|
|
343
|
Busby J, McMenamin Ú, Spence A, Johnston
BT, Hughes C and Cardwell CR: Angiotensin receptor blocker use and
gastro-oesophageal cancer survival: A population-based cohort
study. Aliment Pharmacol Ther. 47:279–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
344
|
Murphy JE, Wo JY, Ryan DP, Clark JW, Jiang
W, Yeap BY, Drapek LC, Ly L, Baglini CV, Blaszkowsky LS, et al:
Total neoadjuvant therapy with FOLFIRINOX in combination with
losartan followed by chemoradiotherapy for locally advanced
pancreatic cancer: A phase 2 clinical trial. JAMA Oncol.
5:1020–1027. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
345
|
Fennell DA, Baas P, Taylor P, Nowak AK,
Gilligan D, Nakano T, Pachter JA, Weaver DT, Scherpereel A,
Pavlakis N, et al: Maintenance defactinib versus placebo after
first-line chemotherapy in patients with merlin-stratified pleural
mesothelioma: COMMAND-A double-blind, randomized, phase II study. J
Clin Oncol. 37:790–798. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
346
|
Wang-Gillam A: Targeting stroma: A tale of
caution. J Clin Oncol. 37:1041–1043. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
347
|
Ulisse S, Baldini E, Sorrenti S and
D'Armiento M: The urokinase plasminogen activator system: A target
for anti-cancer therapy. Curr Cancer Drug Targets. 9:32–71. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
348
|
Feng S, Agoulnik IU, Bogatcheva NV, Kamat
AA, Kwabi-Addo B, Li R, Ayala G, Ittmann MM and Agoulnik AI:
Relaxin promotes prostate cancer progression. Clin Cancer Res.
13:1695–1702. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
349
|
Raue R, Frank AC, Syed SN and Brüne B:
Therapeutic targeting of MicroRNAs in the tumor microenvironment.
Int J Mol Sci. 22:22102021. View Article : Google Scholar : PubMed/NCBI
|
|
350
|
Huang S, Zhang J, Lai X, Zhuang L and Wu
J: Identification of novel tumor microenvironment-related long
noncoding RNAs to determine the prognosis and response to
immunotherapy of hepatocellular carcinoma patients. Front Mol
Biosci. 8:7813072021. View Article : Google Scholar : PubMed/NCBI
|
|
351
|
Ting NLN, Lau HCH and Yu J: Cancer
pharmacomicrobiomics: Targeting microbiota to optimise cancer
therapy outcomes. Gut. 71:1412–1425. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
352
|
Spanogiannopoulos P, Bess EN, Carmody RN
and Turnbaugh PJ: The microbial pharmacists within us: A
metagenomic view of xenobiotic metabolism. Nat Rev Microbiol.
14:273–287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
353
|
Imai H, Saijo K, Komine K, Otsuki Y,
Ohuchi K, Sato Y, Okita A, Takahashi M, Takahashi S, Shirota H, et
al: Antibiotic therapy augments the efficacy of
gemcitabine-containing regimens for advanced cancer: A
retrospective study. Cancer Manag Res. 11:7953–7965. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
354
|
Nakano S, Komatsu Y, Kawamoto Y, Saito R,
Ito K, Nakatsumi H, Yuki S and Sakamoto N: Association between the
use of antibiotics and efficacy of gemcitabine plus nab-paclitaxel
in advanced pancreatic cancer. Medicine (Baltimore). 99:e222502020.
View Article : Google Scholar : PubMed/NCBI
|
|
355
|
Sunakawa Y, Arai H, Izawa N, Mizukami T,
Horie Y, Doi A, Hirakawa M, Ogura T, Tsuda T and Nakajima TE:
Antibiotics may enhance the efficacy of gemcitabine treatment for
advanced pancreatic cancer. Ann Oncol. 29 (Suppl
8):viii251–viii252. 2018. View Article : Google Scholar
|
|
356
|
Frankel AE, Coughlin LA, Kim J, Froehlich
TW, Xie Y, Frenkel EP and Koh AY: Metagenomic shotgun sequencing
and unbiased metabolomic profiling identify specific human gut
microbiota and metabolites associated with immune checkpoint
therapy efficacy in melanoma patients. Neoplasia. 19:848–855. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
357
|
Routy B, Le Chatelier E, Derosa L, Duong
CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C,
Roberti MP, et al: Gut microbiome influences efficacy of PD-1-based
immunotherapy against epithelial tumors. Science. 359:91–97. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
358
|
Matson V, Fessler J, Bao R, Chongsuwat T,
Zha Y, Alegre ML, Luke JJ and Gajewski TF: The commensal microbiome
is associated with anti-PD-1 efficacy in metastatic melanoma
patients. Science. 359:104–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
359
|
Gopalakrishnan V, Spencer CN, Nezi L,
Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman
K, Wei SC, et al: Gut microbiome modulates response to anti-PD-1
immunotherapy in melanoma patients. Science. 359:97–103. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
360
|
Vétizou M, Pitt JM, Daillère R, Lepage P,
Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong
CP, et al: Anticancer immunotherapy by CTLA-4 blockade relies on
the gut microbiota. Science. 350:1079–1084. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
361
|
Sivan A, Corrales L, Hubert N, Williams
JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B,
Alegre ML, et al: Commensal bifidobacterium promotes antitumor
immunity and facilitates anti-PD-L1 efficacy. Science.
350:1084–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
362
|
Khalesi S, Bellissimo N, Vandelanotte C,
Williams S, Stanley D and Irwin C: A review of probiotic
supplementation in healthy adults: Helpful or hype? Eur J Clin
Nutr. 73:24–37. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
363
|
Pushalkar S, Hundeyin M, Daley D,
Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres
LE, et al: The pancreatic cancer microbiome promotes oncogenesis by
induction of innate and adaptive immune suppression. Cancer Discov.
8:403–416. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
364
|
Yu T, Guo F, Yu Y, Sun T, Ma D, Han J,
Qian Y, Kryczek I, Sun D, Nagarsheth N, et al: Fusobacterium
nucleatum promotes chemoresistance to colorectal cancer by
modulating autophagy. Cell. 170:548–563.e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
365
|
Zheng JH, Nguyen VH, Jiang SN, Park SH,
Tan W, Hong SH, Shin MG, Chung IJ, Hong Y, Bom HS, et al: Two-step
enhanced cancer immunotherapy with engineered Salmonella
typhimurium secreting heterologous flagellin. Sci Transl Med.
9:eaak95372017. View Article : Google Scholar : PubMed/NCBI
|
|
366
|
Juul FE, Garborg K, Bretthauer M, Skudal
H, Øines MN, Wiig H, Rose Ø, Seip B, Lamont JT, Midtvedt T, et al:
Fecal microbiota transplantation for primary clostridium difficile
infection. N Engl J Med. 378:2535–2536. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
367
|
van Nood E, Vrieze A, Nieuwdorp M, Fuentes
S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF,
Tijssen JG, et al: Duodenal infusion of donor feces for recurrent
clostridium difficile. N Engl J Med. 368:407–415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
368
|
Arbel LT, Hsu E and McNally K:
Cost-effectiveness of fecal microbiota transplantation in the
treatment of recurrent clostridium difficile infection: A
literature review. Cureus. 9:e15992017.PubMed/NCBI
|
|
369
|
Bayat Mokhtari R, Homayouni TS, Baluch N,
Morgatskaya E, Kumar S, Das B and Yeger H: Combination therapy in
combating cancer. Oncotarget. 8:38022–38043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
370
|
Lane D: Designer combination therapy for
cancer. Nat Biotechnol. 24:163–164. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
371
|
Allen E, Jabouille A, Rivera LB,
Lodewijckx I, Missiaen R, Steri V, Feyen K, Tawney J, Hanahan D,
Michael IP and Bergers G: Combined antiangiogenic and anti-PD-L1
therapy stimulates tumor immunity through HEV formation. Sci Transl
Med. 9:eaak96792017. View Article : Google Scholar : PubMed/NCBI
|
|
372
|
Aparicio LMA, Fernandez IP and Cassinello
J: Tyrosine kinase inhibitors reprogramming immunity in renal cell
carcinoma: Rethinking cancer immunotherapy. Clin Transl Oncol.
19:1175–1182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
373
|
Duchnowska R, Loibl S and Jassem J:
Tyrosine kinase inhibitors for brain metastases in HER2-positive
breast cancer. Cancer Treat Rev. 67:71–77. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
374
|
Ntanasis-Stathopoulos I, Fotopoulos G,
Tzanninis IG and Kotteas EA: The emerging role of tyrosine kinase
inhibitors in ovarian cancer treatment: A systematic review. Cancer
Invest. 34:313–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
375
|
Bożyk A, Wojas-Krawczyk K, Krawczyk P and
Milanowski J: Tumor microenvironment-a short review of cellular and
interaction diversity. Biology (Basel). 11:9292022.PubMed/NCBI
|
|
376
|
Russo M and Nastasi C: Targeting the tumor
microenvironment: A close up of tumor-associated macrophages and
neutrophils. Front Oncol. 12:8715132022. View Article : Google Scholar : PubMed/NCBI
|
|
377
|
Mao D, Xu R, Chen H, Chen X, Li D, Song S,
He Y, Wei Z and Zhang C: Cross-talk of focal adhesion-related gene
defines prognosis and the immune microenvironment in gastric
cancer. Front Cell Dev Biol. 9:7164612021. View Article : Google Scholar : PubMed/NCBI
|
|
378
|
Qian H, Li H, Xie J, Lu X, Li F, Wang W,
Tang X, Shi M, Jiang L, Li H, et al: Immunity-related gene
signature identifies subtypes benefitting from adjuvant
chemotherapy or potentially responding to PD1/PD-L1 blockage in
pancreatic cancer. Front Cell Dev Biol. 9:6822612021. View Article : Google Scholar : PubMed/NCBI
|
|
379
|
Zhou S, Sun Y, Chen T, Wang J, He J, Lyu
J, Shen Y, Chen X and Yang R: The landscape of the tumor
microenvironment in skin cutaneous melanoma reveals a prognostic
and immunotherapeutically relevant gene signature. Front Cell Dev
Biol. 9:7395942021. View Article : Google Scholar : PubMed/NCBI
|
|
380
|
Wu J, Zhou J, Xu Q, Foley R..Guo J, Zhang
X, Tian C, Mu M, Xing Y, Liu Y, et al: Identification of key genes
driving tumor associated macrophage migration and polarization
based on immune fingerprints of lung adenocarcinoma. Front Cell Dev
Biol. 9:7518002021. View Article : Google Scholar : PubMed/NCBI
|
|
381
|
Haibe Y, Kreidieh M, El Hajj H, Khalifeh
I, Mukherji D, Temraz S and Shamseddine A: Resistance mechanisms to
anti-angiogenic therapies in cancer. Front Oncol. 10:2212020.
View Article : Google Scholar : PubMed/NCBI
|
|
382
|
Navab R, Strumpf D, To C, Pasko E, Kim KS,
Park CJ, Hai J, Liu J, Jonkman J, Barczyk M, et al: Integrin α11β1
regulates cancer stromal stiffness and promotes tumorigenicity and
metastasis in non-small cell lung cancer. Oncogene. 35:1899–1908.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
383
|
Junttila MR and de Sauvage FJ: Influence
of tumour micro-environment heterogeneity on therapeutic response.
Nature. 501:346–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
384
|
Giuliano S and Pagès G: Mechanisms of
resistance to anti-angiogenesis therapies. Biochimie. 95:1110–1119.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
385
|
Flaherty KT, Manola JB, Pins M, McDermott
DF, Atkins MB, Dutcher JJ, George DJ, Margolin KA and DiPaola RS:
BEST: A randomized phase II study of vascular endothelial growth
factor, RAF kinase, and mammalian target of rapamycin combination
targeted therapy with bevacizumab, sorafenib, and temsirolimus in
advanced renal cell carcinoma-a trial of the ECOG-ACRIN cancer
research group (E2804). J Clin Oncol. 33:2384–2391. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
386
|
Wang J, Li Y, Nie G and Zhao Y: Precise
design of nanomedicines: Perspectives for cancer treatment. Natl
Sci Rev. 6:1107–1110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
387
|
Atat OE, Farzaneh Z, Pourhamzeh M, Taki F,
Abi-Habib R, Vosough M and El-Sibai M: 3D modeling in cancer
studies. Hum Cell. 35:23–36. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
388
|
Selek L, Seigneuret E, Nugue G, Wion D,
Nissou MF, Salon C, Seurin MJ, Carozzo C, Ponce F, Roger T and
Berger F: Imaging and histological characterization of a human
brain xenograft in pig: the first induced glioma model in a large
animal. J Neurosci Methods. 221:159–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
389
|
Khoshnevis M, Carozzo C, Bonnefont-Rebeix
C, Belluco S, Leveneur O, Chuzel T, Pillet-Michelland E, Dreyfus M,
Roger T, Berger F and Ponce F: Development of induced glioblastoma
by implantation of a human xenograft in Yucatan minipig as a large
animal model. J Neurosci Methods. 282:61–68. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
390
|
Khoshnevis M, Carozzo C, Brown R, Bardiès
M, Bonnefont-Rebeix C, Belluco S, Nennig C, Marcon L, Tillement O,
Gehan H, et al: Feasibility of intratumoral 165Holmium siloxane
delivery to induced U87 glioblastoma in a large animal model, the
Yucatan minipig. PLoS One. 15:e02347722020. View Article : Google Scholar : PubMed/NCBI
|
|
391
|
Mackenzie NJ, Nicholls C, Templeton AR,
Perera MP, Jeffery PL, Zimmermann K, Kulasinghe A, Kenna TJ, Vela
I, Williams ED and Thomas PB: Modelling the tumor immune
microenvironment for precision immunotherapy. Clin Transl
Immunology. 11:e14002022. View Article : Google Scholar : PubMed/NCBI
|
|
392
|
Mendes N, Dias Carvalho P, Martins F,
Mendonça S, Malheiro AR, Ribeiro A, Carvalho J and Velho S: Animal
models to study cancer and its microenvironment. Adv Exp Med Biol.
1219:389–401. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
393
|
Hsu JF, Chu SM, Liao CC, Wang CJ, Wang YS,
Lai MY, Wang HC, Huang HR and Tsai MH: Nanotechnology and
nanocarrier-based drug delivery as the potential therapeutic
strategy for glioblastoma multiforme: An update. Cancers (Basel).
13:1952021. View Article : Google Scholar : PubMed/NCBI
|