Protein arginine methylation is a common PTM
catalysed by protein arginine methyltransferases (PRMTs) (2). In 1967, Paik and Kim (3) first discovered methylated arginine in
a nuclear protein of calf thymocytes. The first member of the PRMT
family, PRMT1, was identified in 1996, followed by other members
(4). Protein arginine methylation
has a role in the maintenance of key cellular processes, such as
tissue homeostasis and disease phenotype (2).
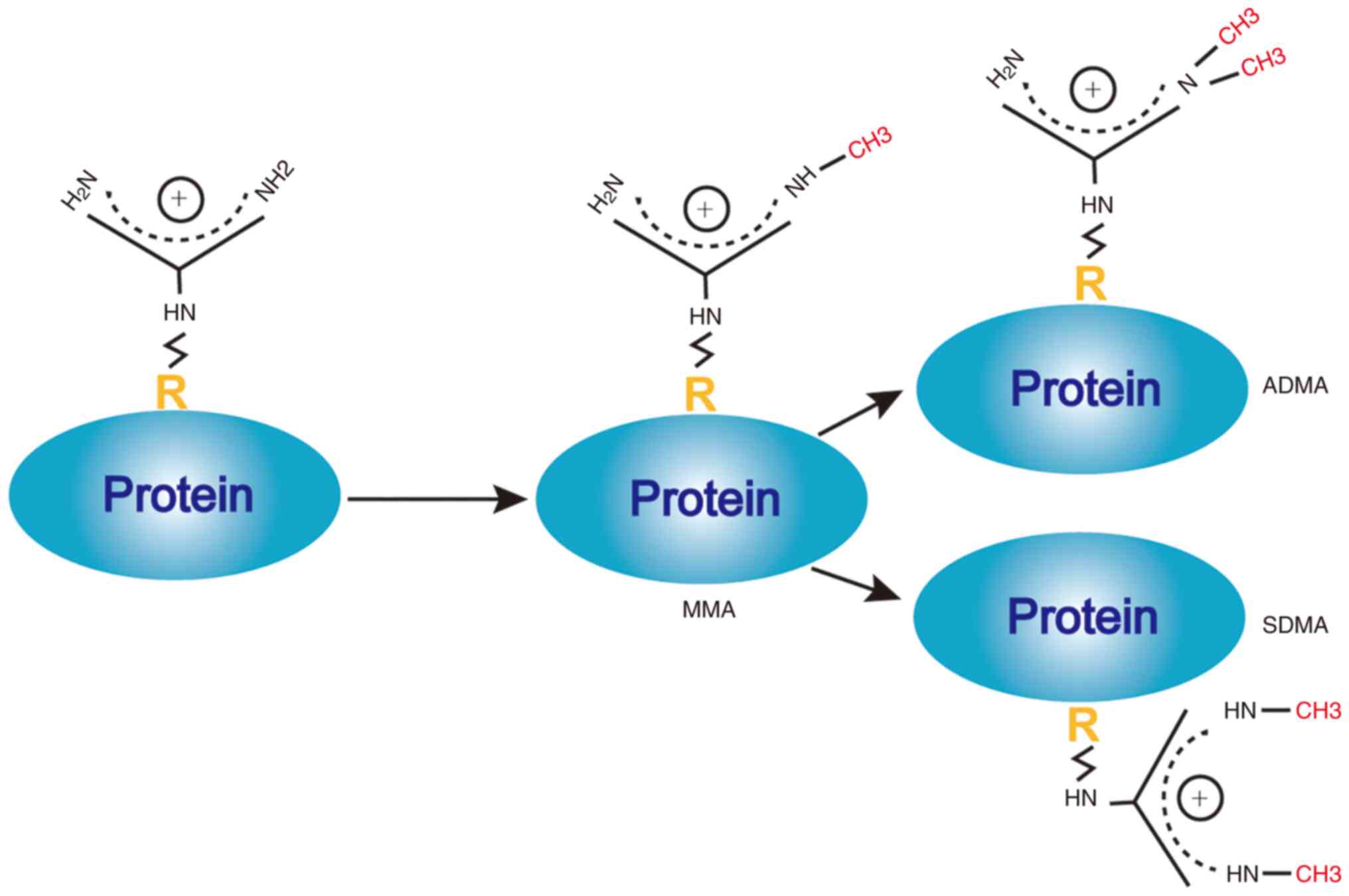
Based on the number and position of methyl groups on
the ω-guanidino nitrogen atom of the protein arginine, arginine
methylation modification may be divided into
ω-NG-monomethylarginine (MMA),
ω-NG,NG-asymmetric dimethylarginine (ADMA)
and ω-NG,N'G-symmetric dimethylarginine
(SDMA). The process by which PRMTs methylate arginine to produce
MMA, ADMA and SDMA is provided in Fig.
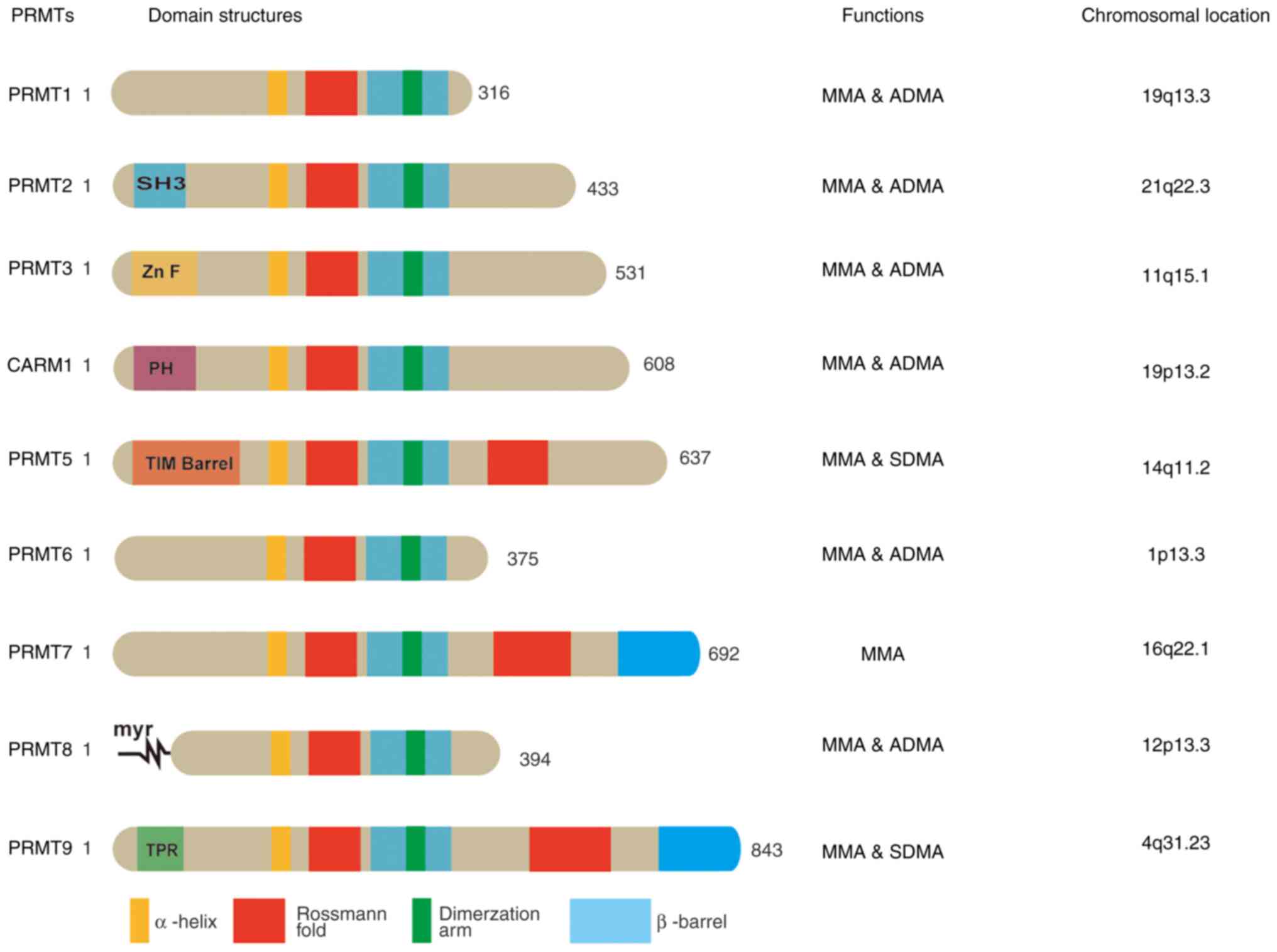
1. Among them, type I PRMTs include PRMT1-4, -6 and -8, which
methylate MMA and ADMA; type II PRMTs include PRMT5 and -9, which
methylate MMA and SDMA; and type III PRMTs include PRMT7, which
methylates MMA (5). The protein
structure, modulation function and chromosomal location of each
member of the PRMTs are provided in Fig. 2.
Arginine is a basic amino acid with a positively
charged guanidine group containing five potential hydrogen bond
donors, which may interact with negatively charged molecules
(6). PRMTs transfer the methyl
group from S-adenosine methionine (SAM) to the guanidino group of
arginine in protein substrates, resulting in S-adenosyl
homocysteine and methylated proteins. After methylation of arginine
residues, the distance between side chains increases and the
molecular configuration changes. At the same time, the addition of
methyl groups reduces the number of potential hydrogen bond donors,
resulting in enhanced hydrophobicity of methylated arginine, which
in turn affects intramolecular and intermolecular interactions,
such as protein-to-protein, protein-to-nucleic acid, as well as
protein structure and stability, ultimately affecting the
biological function of the modified protein (7).
PRMT1 is the most widely studied PRMT enzyme due to
its powerful methyl transfer function, accounting for >85% of
all modifications of PRMTs (8).
The residues M48, E100, E144, E153, M155 and H293 on the active
site of PRMT1 are critical for substrate and cofactor interactions
(9). Mutations in these active
sites weaken or result in the loss of the catalytic activity of
PRMT1, leading to ADMA synthesis disorders (10). The interaction between PRMT2 and RB
downregulates the activity of E2F through multiple mechanisms,
including histone methylation, transcription factor (TF)
methylation and RNA splicing (11). PRMT3 is localised in the cytoplasm
and its protein structure includes a catalytic core and a zinc
finger domain. Of note, zinc finger domains not only help to
recognise RNA substrates but also regulate catalytic activity by
recruiting interacting proteins (12). PRMT4, also known as
coactivator-associated arginine methyltransferase 1 (CARM1), is
mainly located in the nucleus and normally promotes transcription.
CARM1 consists of a unique N-terminal EVH1 domain (residues 28-140)
that binds to a proline-rich sequence that is essential for
substrate recognition and catalytic activity (13). PRMT5 is a major type II PRMT
consisting of four domains: An N-terminal TIM-barrel domain, an
intermediate Rossmann fold, a C-terminal β-barrel subunit and a
dimerization arm (14). The
conserved F379 residue of PRMT5 methylates the production of SDMA.
The F379M mutation not only increased the methylation activity of
PRMT5 but also altered product specificity by generating SDMA and
ADMA of H4R3. By contrast, both F379G and F379A mutations
significantly reduced PRMT5 activity (14). PRMT6, a signature tag for
epigenetic transcriptional repression, specifically methylates ADMA
of histone H3 (15). PRMT7 is the
only type III PRMT, and compared with other types of PRMTs, the
protein structure of PRMT7 includes a narrower substrate-binding
site, which may lead to catalytic production of MMA (16). PRMT8 is significantly and
specifically highly expressed in brain and neuronal tissues. The
N-terminal region of PRMT8 is able to bind to the plasma membrane,
and there is a substrate binding site in the middle, which is
crucial for PRMT8 methylation activity (17). PRMT9 is a recently identified type
II PRMT that includes MTase domains and forms a pseudodimer for
substrate binding (18).
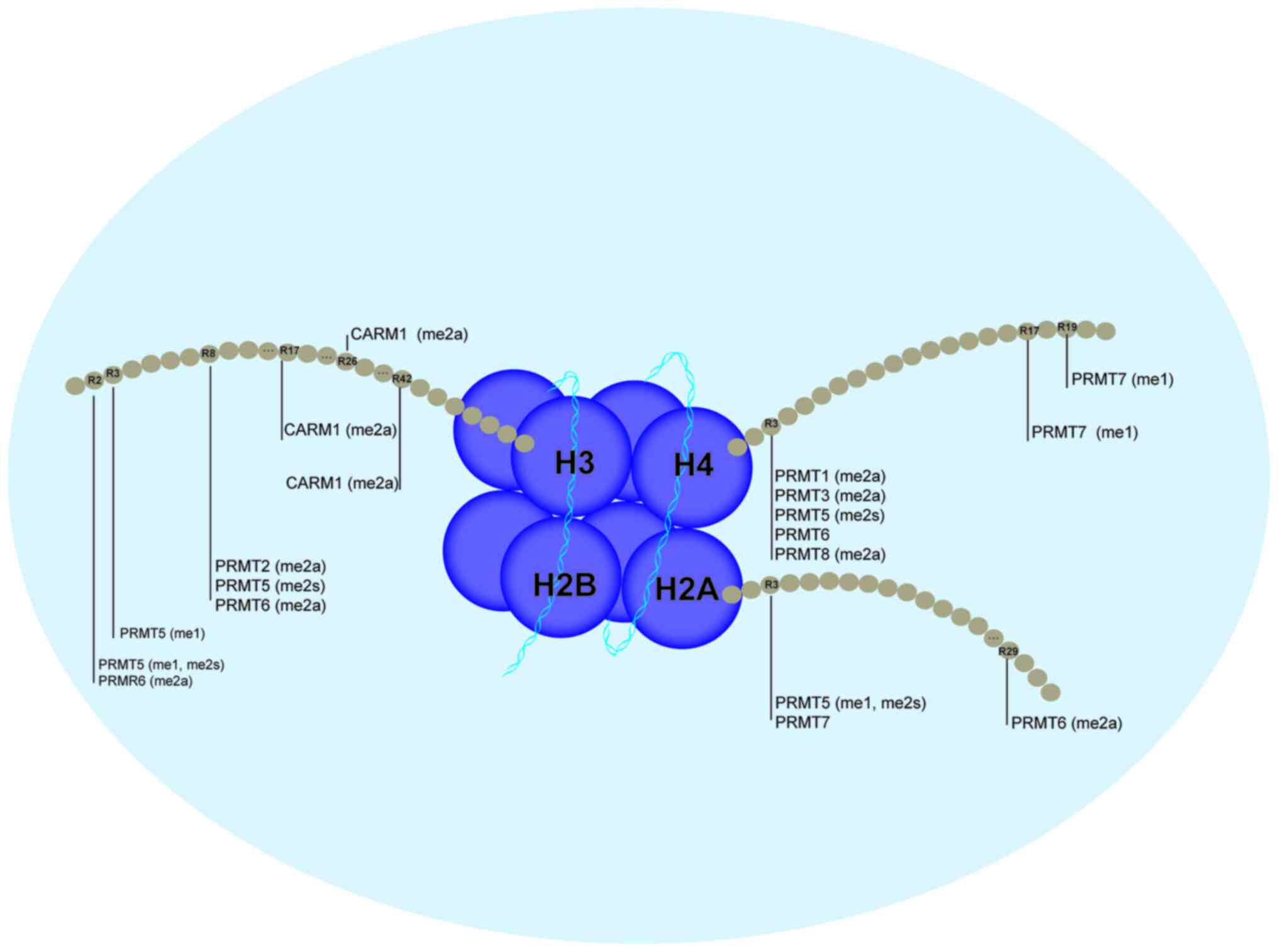
Arginine methylation of histone tails is an
epigenetic modification catalysed by PRMTs and regulates gene
expression. PRMTs may methylate H2AR3, H2AR29, H3R2, H3R8,
H3R17/26/42, H4R3 and H4R17/19, as presented in Fig. 3. Structural analysis indicated that
electrostatic interactions have a mechanistic role in the process
of substrate methylation catalysed by PRMTs. It has been indicated
that PRMT1, -3, -5 and -8 preferentially methylate histone H4,
while PRMT4/CARM1 preferentially methylates histone H3 (19). The detailed functions of the
PRMT-methylated histones H2A, H3 and H4 are presented in Table I.
PRMT5-7 are able to methylate H2A. Since the first
five residues of H2A and H4 are identical, it is likely that most
of the methylation of H4R3 also applies to H2AR3 (20). A systematic analysis of the H2A
methylation status revealed that H2AR29me2 was specifically
enriched in genes inhibited by PRMT6, suggesting that H2AR29me2 is
involved in transcriptional repression (21). The haemagglutinin-PRMT5 complex was
also able to monomethylate and symmetrically dimethylate bovine
histone H2A (22). Chromatin
immunoprecipitation revealed that PRMT7 dimethylates H2AR3 and H4R3
and is enriched at target DNA repair genes in parental cells
(23). Studies have indicated that
PRMTs are able to methylate H2A and mainly take part in
transcriptional repression, but the underlying regulatory mechanism
remains to be elucidated.
H3 methylation exerts transcriptional activation or
transcriptional inhibition by PRMT2, CARM1 and PRMT5-7. For
instance, PRMT2 is responsible for generating H3R8me2a. H3R8me2a
enrichment at the BCL2 promoter may increase its accessibility to
STAT3, promoting Bcl2 gene expression (24). PRMT2 acts as a transcriptional
coactivator for oncogenic gene expression programs in glioblastoma
multiforme (GBM) pathogenesis. PRMT2-mediated H3R8me2a enrichment
at promoters and enhancers is closely associated with known active
histone marks and is required for the maintenance of target gene
expression (25). CARM1 may
methylate H3R17, H3R26 and H3R42 and is recognised by tudor domain
containing 3 to function as a coactivator (26). It is necessary that PRMT5
accumulation activates H3R2me1/me2s and recruitment of the WD
repeat domain 5 (WDR5)/mixed linage leukemia (MLL) complex to
promote H3K4me3, which in turn activates transcription (27-30).
Recruitment of PRMT5 to the forkhead box (FOX)P1 promoter may
increase H3R2me2s and H3K4me3 (29). The potential interaction of
PRMT5-mediated H3R2me1 with MLL complexes (absent, small, or
homologous 2 and WDR5) may activate the expression of
metastasis-related genes, such as vimentin, snail family
transcriptional repressor 1, snail family transcriptional repressor
2 and cadherin 2 (28). Genotoxic
stress induces interactions among β-catenin, ATM phosphorylated Jun
isomerization protein 2 and PRMT5, promoting redox-related gene
transcription. During this process, PRMT5-mediated recruitment of
H3R2me1/H3R2me2s to the WDR5/MLL complex leads to transcriptional
activation of H3K4me3 and redox-related genes (30). H3R2me2a acts as a repressive mark
that antagonises H3K4me3, but H3R2 is also symmetrically
dimethylated (H3R2me2s) by PRMT5 and PRMT7 and is present in
euchromatic regions (31).
Profiling of H3-tail interactors indicated that H3R2me2s excludes
binding of RBBP7, a central component of the co-repressor complexes
SIN3 transcription regulator family member A, nucleosome remodeling
and deacetylase and polycomb repressive complex 2. Conversely,
H3R2me2s may enhance binding of WDR5, which is a common component
of the coactivator complexes MLL, nuclear localization signals 1,
Ada two-A containing, SET-domain-containing 1A and
SET-domain-containing 1B (31).
PRMT6-mediated H3R2me2a inhibits transcription by preventing
H3K4me3 readers from binding (32). Of note, H3R8me2s and H4R3me2s are
mainly considered repressive markers, but they have also been
implicated in the transcriptional activation of certain genes, such
as fibroblast growth factor receptor (FGFR)3 and eukaryotic
initiation factor 4E expression in colorectal cancer (33) and androgen receptor expression in
prostate cancer (34). Overall,
for the regulation of H3 methylation, the symmetric arginine
dimethylation of H3 generally has a role of transcriptional
activation, while the asymmetric arginine dimethylation frequently
has a role in transcriptional inhibition.
The methylation of H4 also has a key role in
regulating the activation and repression of transcription. For
instance, SWI/SNF related, matrix associated, actin dependent
regulator of chromatin, subfamily a, member 4, the ATPase subunit
of the switch/sucrose nonfermentable (SWI/SNF) chromatin
remodelling complex, acts as a binder for PRMT1 to methylate
H4R3me2a and upregulate epidermal growth factor receptor (EGFR) in
colorectal cancer (35).
PRMT5-mediated H4R3me2s have been found on promoters of tumour
suppressor and cyclin-dependent kinase (CDK) suppressor genes,
which silence cancer-cell proliferation (36,37).
Menin (MEN1) has an essential role in both repressing and
activating gene expression. In MEN1-excised cells, the levels of
both PRMT5 binding and H4R3m2s were decreased at the GLI1 promoter
(38). MEN1 is a crucial factor
for binding of the Sonic Hedgehog ligand to its receptor Patched 1
and subsequent activation of the Hedgehog signalling pathway. Of
note, MEN1 mutants have reduced binding to PRMT5 and fail to impart
the repressive H4R3m2s mark at the growth arrest specific 1
promoter, resulting in its elevated expression (38,39).
Pharmacologic inhibition of Hedgehog signalling significantly
reduces the proliferation of insulinoma cells and promotes the
expression of Hedgehog signalling targets (39). Zinc finger E-box binding homeobox 1
(ZEB1), a zinc finger TF, is a key factor for epithelial to
mesenchymal transition (EMT) (40). PRMT1 impacted the EMT process by
mediating the asymmetric dimethylation of H4R3me2as at the ZEB1
promoter to activate its transcription, indicating the essential
roles of this epigenetic control in EMT (41). Otherwise, exogenous TGFβ promotes
EMT through PRMT5-MEP50 catalysing arginine monomethylation and
dimethylation (42). PHD finger
protein 1 (PHF1) recognizes H4R3me2s and recruits CUL4Bring E3
ligase through PHF to form a complex, silencing the expression of
E-cad and FBXW7 to promote cell growth and migration (43). In addition, in 293 cells,
overexpression of PRMT3 may increase the level of H4R3me2a, but the
specific mechanism remains to be clarified (44). In conclusion, these findings reveal
a novel link between PRMTs and H4, whereby PRMTs epigenetically
regulate tumour signalling pathways, revealing them as targets for
treating tumours.
Dynamic crosstalk between different histone
modification types may affect gene expression. PRMT7 methylates
H4R17me1 and allosterically enhances H4R3me2 of PRMT5, which in
turn inhibits subsequent H3K4me3, H3Ac and H4Ac (45). Deacetylation of H3K9 by H3R8me2s is
a transcriptional repression marker (46), whereas H4R3me2s is associated with
H4K5 acetylation and may serve as a transcriptional activation
marker (47). Of note, an
increasing number of studies have demonstrated the existence of
nonlinear crosstalk between different histone modification types,
leading to the diversity of protein functions.
PRMTs may also affect protein tail modification by
targeting microRNA (miRNA/miR) and regulating tumour gene
expression. PRMT5 repressed the transcription of the miR-99 family
by symmetrical dimethylation of H4R3, which increased FGFR3
expression and in turn activated Erk1/2 and Akt, leading to cell
growth and metastasis in lung cancer (48). PRMT5 knockdown results in miR33b,
miR96 and miR503 derepression through loss of repressive complex
recruitment targeting miRNA promoters. PRMT5 is overexpressed in
B-cell lymphoma and promotes the binding of miR33b, miR96 and
miR503 to the 3'-untranslated region of cyclin D1 and c-MYC mRNAs,
indirectly leading to enhanced cyclin D1 and c-MYC expression,
which reinforces the relevance of PRMT5 in promoting lymphoma cell
growth and survival (49).
According to the function of non-histones after
methylation, PRMTs may be divided into five types, including TFs,
RNA-binding proteins (RBPs), DNA damage repair proteins, RNA
splicing proteins and functional proteins in cell signalling
pathways. The detailed functions of PRMT-methylated nonhistones are
provided in Table II.
TFs may combine with RNA polymerase to form a
transcription initiation complex, which jointly participates in the
process of transcription initiation. E2F-1 may regulate
transcription in a methylation-dependent manner (50). PRMT5 and -1 methylate E2F-1 to
generate functionally opposite effects. The DNA damage response
(DDR) induces E2F-1 methylation by PRMT1, increases E2F-1
expression levels and activates apoptotic gene transcription.
Conversely, PRMT5 methylation of E2F-1 is recognised by Tudor
domain protein, p100/tudor-SN (TSN), which reduces the E2F-1
half-life and increases cell viability. During cell cycle
progression, the binding of E2F-1 to cyclin A masks the PRMT1
methylation of E2F-1, thereby inhibiting apoptosis (50). Sterol regulatory element binding
protein 1 (SREBP1) is a TF regulated for de novo fatty acid
synthesis. PRMT5 methylation of SREBP1 prevents its phosphorylation
by glycogen synthase kinase (GSK)3β, but it is subsequently
ubiquitinated by F-box and WD repeat domain containing 7 (FBXW7),
increasing adipogenesis and promoting tumour growth in
hepatocellular carcinoma (HCC) (51). BCL6 is a transcriptional repressor
and master regulator of normal germinal centre (GC) formation and
GC-derived B-cell lymphomas. PRMT5 methylation of BCL6 at R305
downregulates the activity of BCL6 target genes, inhibiting the
growth of diffuse large B-cell lymphoma (52).
ICP27 is an RBP that has a crucial role in the gene
expression and replication of herpes simplex virus type 1 (HSV-1)
(53). Methylation of ICP27 at
residues R138, R148 and R150 by PRMT1 is responsible for the
formation of ICP27 nuclear foci, RNA-binding affinity and SRPK
interaction. Hypomethylation of ICP27 may significantly inhibit
HSV-1 replication, suggesting that PRMT inhibitors have an
important role in HSV-2 therapy (53). RNA and transporters compete for
binding to nuclear poly(A)-binding protein 1 (PABPN1), but
methylation of PABPN1 at R289 reduces the binding capacity between
PABPN1 and transporters by ~10-fold, resulting in promotion of the
PABPN1-RNA interaction (54).
Although eIF4A1 shares >80% sequence similarity with eIF4A2 and
-3, only residue R368 of eIF4A1 protein is selectively methylated
by PRMT1, whereas other eIF4A isotypes are not (55).
Phosphorylation and ubiquitination are key
components of the DDR and arginine methylation is no exception.
Deficiency of PRMT1 and PRMT5 leads to spontaneous DNA damage,
checkpoint defects and genomic instability in mouse embryonic
fibroblasts (56,57). PRMT1 is involved in the methylation
of DNA repair proteins, including meiotic recombination 11 (MRE11),
p53 binding protein 1, heterogeneous nuclear ribonucleoprotein U
like 1, breast cancer 1 (BRCA1) and Separase, which ensure the
maintenance of genome stability through homologous recombination
(HR) repair and nonhomologous end-joining (NHEJ) (2,58).
MRE11 is an essential component of the MRE11-RAD50-NBS1 (MRN)
complex that activates the DNA repair pathway. Although methylation
of MRE11 does not regulate MRN complex formation, it anchors MRE11
to double-strand breaks (DSBs), preventing nucleases from
activating DNA repair. In addition, the TF growth factor
independent 1 interacts with PRMT1 and promotes MRE11 methylation
(59). Furthermore, CARM1 promotes
mitotic arrest deficient 2 like 2 (MAD2L2) silencing by driving the
switch from the SWI/SNF complex to EZH2 by methylating the BAF155
subunit of the SWI/SNF complex on the MAD2L2 promoter. EZH2
inhibition upregulates MAD2L2 to decrease DNA end resection, which
increases NHEJ and chromosomal abnormalities, ultimately causing
mitotic catastrophe in PARP inhibitor-treated HR-proficient cells
(60). PRMT5-deficient HeLa cells
are sensitive to radiotherapy and accumulate DNA damage (61). Stress-responsive activator of p300
may be recruited to the p53 complex in the DDR, which recruits
PRMT5 and promotes methylation of the p53 oligomerization domain to
reduce oligomerization and increase nuclear retention (62), while increasing target expression
of p21 and p53-up-regulated modulator of apoptosis (63).
Upregulation of key DNA repair genes is the main
mechanism of chemotherapy and radiation therapy resistance in
tumour cells (64). PRMT5 may
promote the survival of tumour cells in the context of genetic
damage; therefore, the combination of PRMT5 inhibitors and
chemotherapy may be a new strategy to treat cancer resistance. In
patients with BRCA1-mutated breast cancer, the HR repair pathway is
missing, so the cells rely mainly on PARP-mediated DSB repair.
Therefore, Olaparib treatment is effective (65). However, the majority of patients
with triple-negative breast cancer (TNBC) do not have BRCA1
mutations and targeting PRMT5 inhibition is similar to HR
deficiency in BRCA1 mutations; therefore, targeting PRMT5
inhibition is beneficial for improving patient outcomes (66). Regulator of chromosome condensation
1 (RCC1), as a guanylate exchange factor of RAN, localises in the
nucleus and binds to chromatin to regulate DNA damage repair. The
methylation of RCC1 at R214 by PRMT6 is necessary for RCC1 to bind
to chromatin and activate RAN (67). Inhibition of PRMT6 reduces the
tumorigenicity of the cells in GBM and improves the effects of
radiation therapy on GBM growth in mice (67).
Interactions between different posttranslational
modifications are critical for the DNA damage response. Arginine
methylation has an essential role in maintaining genome stability,
and arginine methylation and ubiquitination crosstalk control DNA
end resection and HR repair (68).
Mass spectrometry analysis of PRMT1-interacting proteins revealed
that ubiquitin specific peptidase 11 (USP11) has a key role in the
early stage of DSB repair by regulating the activity of the
PRMT1-MRE11 pathway. USP11 is a substrate of PRMT1 and methylation
of USP11 promotes DNA end resection and DSB repair of DNA through
HR. PRMT1 is also a ubiquitinated protein that acts as a target of
de-ubiquitination to regulate the binding and methylation of PRMT1
to MRE11 (68).
Methylation of arginine-specific proteins may modify
the structure or activity of the protein, alter the interaction
between specific molecules and then affect tumour cell signalling
pathways. Recent cancer-related studies on the role of PRMTs are
summarized in Table III. In
TNBC, PRMT1 regulates the EGFR and the Wnt signalling pathways
(80). Type I PRMT inhibitors
decrease breast cancer cell proliferation and have anti-tumour
activity. These inhibitors display synergistic interactions with
certain chemotherapies used to treat TNBC, as well as erlotinib, an
EGFR inhibitor. Therefore, targeting PRMT1 in combination with
these chemotherapies may improve existing treatments for TNBC
(80). Inhibition of PRMT5, the
predominant type II PRMT, produces synergistic cancer-cell growth
inhibition when combined with GSK3368715, which is a potent and
reversible type I PRMT inhibitor (81). Of note, deletion of the
methylthioadenosine phosphorylase gene (MTAP) results in
accumulation of the metabolite 2-methylthioadenosine, an endogenous
inhibitor of PRMT5, and is associated with sensitivity to
GSK3368715. Overall, the MTAP status may serve as a biomarker for
patient selection (81). NF-κB has
an important role in tumorigenesis and PRMT5 activates NF-κB
through methylation of the p65 subunit (82). Although TNFα-induced intracellular
signalling pathways have been well studied, the TRAIL signalling
pathway remains to be fully elucidated. PRMT5, a novel TRAIL
receptor-binding protein, contributes to TRAIL-induced activation
of inhibitor of κB kinase (IKK) and NF-κB, leading to induction of
several NF-κB target genes (83).
PRMT5 methylation of TRIM21 induces selective autophagy, which
inhibits TRIM21-dependent monoubiquitination and degradation of
IKKβ and activates the NF-κB signalling pathway. Thus, PRMT5
inhibition blocks the NF-κB signalling pathway (84). SKI is a transcriptional repressor
that interacts with SMAD and may be methylated by the
PRMT5-methylosome protein 50 (MEP50)-SHANK-associated RH domain
interactor (SHARPIN) complex, altering transcriptional regulation
of the TGFβ signalling pathway (52). In HCC, PRMT9 activates the
PI3K/Akt/GSK3β/Snail signalling pathway to regulate Snail,
increasing cell migration and invasion through EMT (85). SHARPIN, an adaptor for the linear
ubiquitin chain assembly complex, has an important role in the
NF-κB signalling pathway. Activated PRMT5 controls the expression
of SRY-box transcription factor 10 and melanocyte inducing
transcription factor and inhibition of the transcriptional
corepressor SKI by SHARPIN-dependent arginine demethylation,
contributing to the occurrence of melanomagenesis (86). The expression of PRMT3 is
upregulated in colorectal cancer and may stabilise the protein
structure of c-MYC, and PRMT3 promotes the expression of c-MYC by
interacting with c-MYC through the SAM-dependent MTase-PRMT domain
(87). PRMT3 methylates
hypoxia-inducible factor (HIF)1α at R282 and stabilizes the
structure of HIF1α, while activating the HIF1/VEGFA signalling
pathway to promote tumorigenesis (88). PRMT1, -5 and -7 regulate
glioma-associated oncogene 1 (GLI1) and GLI2 activity (89). Methylation of GLI1 by PRMT1
upregulates its activity and promotes target gene expression. PRMT5
methylates GLI1 in the cytoplasm and increases GLI1 protein
stability. Conversely, nuclear PRMT5 interacts with MENIN to
inhibit the expression of growth arrest-specific protein 1, which
facilitates Hedgehog ligand binding to Patched and indirectly
downregulates GLI1 activity. PRMT7 methylates GLI2 to upregulate
its activity through GLI2 dissociation and fusion inhibitors
(89). PRMT1 expression is
upregulated and promotes tumour cell growth in pancreatic ductal
adenocarcinoma (PDAC). PRMT1 promotes β-catenin expression by
binding -699 to -874 bp and -1,191 to -1,413 bp of the β-catenin
promoter (90). Tumour suppressor
protein von Hippel-Lindau interacts with PRMT3 and then forms a
protein complex with Auxin response factor and regulates the
methylation of p53 (91). PRMT3
protein expression is upregulated in patients with gemcitabine
(GEM)-resistant pancreatic cancer. ATP binding cassette subfamily G
member 2 (ABCG2) is a newly discovered target of PRMT3, and PRMT3
overexpression increases the methylation of heterogeneous nuclear
ribonucleoprotein A1 (hnRNPA1) at R31, resulting in enhanced
RNA-binding activity of hnRNPA1 and increased expression of ABCG2
mRNA. Therefore, PRMT3 methylates the RNA recognition motif of
hnRNPA1 to promote the binding of hnRNPA1 and ABCG2 to enhance the
resistance of pancreatic cancer to GEM (92). PRMT5 methylation of hnRNPA1
promotes the interaction of hnRNPA1 with internal ribosome entry
site (IRES) RNA to promote IRES-dependent translation of cyclin D1
and c-MYC (93). The PRMT type 1
inhibitor MS023 is a potent inducer of colon cancer-cell
differentiation with a wide therapeutic window. This finding may
lead to the development of clinically effective anti-cancer drugs
based on the mechanism of cancer cell differentiation (94).
PRMTs can modulate Toll-like receptor and interferon
(IFN) activation at multiple levels to modulate immune responses
(95). PRMT5 expression was
observed to be negatively associated with antitumor immunity. After
PRMT5 inhibition, the number of infiltrating immune cells increased
and antitumour immunity was enhanced in immunocompetent mice
(96). PRMT5 promotes antitumour
immunity through two different intertumoral pathways. First,
PRMT5-mediated interferon gamma inducible protein 16 (IFI16)/IFI204
methylation attenuates dsDNA-induced TANK binding kinase 1
(TBK1)-interferon regulatory factor 3 (IRF3) activation and
chemokine production. dsDNA induced activation of TBK1-IRF3, as
reflected by the levels of STING phosphorylation, dimerization and
polymerization. PRMT5 methylation of IFI16/IFI204 impacts cyclic
GMP-AMP synthase-stimulator of interferon genes (STING) signalling.
Ectopically expressed IFI204 in B16 cells activated TBK1-IRF3
signalling and increased the expression of IFNB1 and C-C motif
chemokine ligand 5 following dsDNA treatment. IFI204Mt1 (R12A)
expression increased STING dimerization and polymerization
following dsDNA stimuli, suggesting a critical role of IFI204
methylation on Arg12 in the dsDNA-stimulated STING pathway
activation. PRMT5-mediated IFI16/IFI204 methylation attenuates
dsDNA-induced TBK1-IRF3 activation and type I interferon and
chemokine production. In addition, NLR family CARD domain
containing 5, a master regulator of inflammasomes and antigen
presentation pathways, was inversely correlated with PRMT5
expression. PRMT5 inhibits immune cell recruitment and activation
as well as tumour recognition, thereby influencing tumour immune
evasion. Likewise, inhibition of PRMT5 is expected to enhance the
response of cold tumours to immune checkpoint therapy (96). MS023, a type I PRMT inhibitor,
causes splicing of modulatory drugs, treatment alterations with
intron retention and exon skipping, and these alterations result in
substantial enrichment of major histocompatibility complex
I-binding peptides. Of note, a fairly large proportion (up to 43%)
of these putative neoantigens are immunogenic, resulting in
neoantigen-specific CD8+ T-cell activation (97).
Controlling PRMT5 activity is a promising strategy
for cancer therapy when host immunity against tumours occurs in a
FOXP3-dependent manner (98).
Arginine methylation occurs frequently at R27, R51 and R146 of
FOXP3, but pharmacological inhibition of PRMT5 by DS-437 may reduce
T-regulatory cell (Treg) functions and inhibits the methylation of
FOXP3. Furthermore, DS-437 significantly enhanced the anti-tumour
effects of anti-erbB2/neu monoclonal antibody targeted therapy in
BALB/c mice, which bore CT26Her2 tumours, by inhibiting Treg
function and induction of tumour immunity (98). Of note, FOXP3 also undergoes
methylation on R48 and R51 by interacting with PRMT1. The
inhibition of arginine methylation confers gene expression profiles
representing type I helper T cells to FOXP3+ T cells, which
resulted in attenuated suppressive activity (99). Otherwise, knockout of PRMT1 may
enhance anti-programmed cell death receptor-1 immunotherapy in
MC38-derived tumours in isogenic C57BL/6 mice (100). Of note, the PRMT1 polymorphism
rs975484 modulates programmed cell death ligand-1 (PD-L1) and PD-L2
levels and serves as a predictor of immune checkpoint blockade
efficiency in HCC (101). CARM1
was identified as a negative regulator of tumour-specific T cells
in B16F10 melanoma-resistant C57BL/6 mice (102).
Furthermore, studies have indicated that type I PRMT
inhibitors may enhance the effect of immunotherapy. PT1001B
enhances antitumor immunity, and combining it with anti-PD-L1
checkpoint inhibitors provides a potential strategy to overcome
anti-PD-L1 resistance in PDAC (103). MS023 treatment significantly
improved anti-PD1 therapy in C57BL/6 tumour-bearing mice (104). Therefore, further studies are
needed to determine the effect of PRMT inhibition, not only on
tumour cells, but also on other cell types in the tumour
microenvironment, including immune cells and stromal cells.
Metabolic reprogramming is an important process by
which cancer cells adapt to high energy demands and supplement
biosynthetic needs, and numerous cancers switch their tumour cell
metabolism to the glycolytic pathway under oxygen-rich conditions.
Therefore, inhibition of metabolic reprogramming by modulating
different metabolic pathways in tumours provides a new strategy for
cancer therapy (105,106).
In pancreatic cancer, PRMT3 may methylate GAPDH at
R248 to enhance cancer glycolysis and mitochondrial respiration.
PRMT3-overexpressing cancer cells were addicted to GAPDH-mediated
metabolism and sensitive to the inhibition of GAPDH and
mitochondrial respiration. Both intermediates in the glycolytic
pathway and the tricarboxylic acid cycle are enriched in
PRMT3-expressed cells. In addition, these cells exhibit an
increased extracellular acidification rate and oxygen consumption
rate. Double blockade of GAPDH and mitochondrial respiration will
be a novel strategy for the treatment of PRMT3-overexpressing
pancreatic cancer (107).
Esophageal squamous cell carcinoma (ESCC) is associated with
elevated asymmetric and systemic arginine dimethylarginine. PRMT1,
PRMT5, ornithine decarboxylase 1 and nitric oxide (NO) synthase 2
are overexpressed, and arginase 1, arginase 2 and dimethylarginine
dimethylaminohydrolase 1 are downregulated in tumours compared to
adjacent tissues. Arginine bioavailability increased and citrulline
decreased along with ESCC advancement. In short, metabolic
reprogramming in ESCC manifests as alterations in the L-arginine/NO
pathway (108). In HCC, PRMT3
mediates ADMA modification of lactate dehydrogenase A (LDHA) at
R112. LDHA-R112K-mutant-expressing cells exhibited a decrease in
LDH activity, HCC cell glycolysis and proliferation (109). In chronic myeloid leukemia (CML),
loss of PRMT7 resulted in reduced expression of glycine
decarboxylase, leading to the reprograming of glycine metabolism to
generate methylglyoxal, which is detrimental to leukemia stem cells
(LSCs). These findings link histone arginine methylation with
glycine metabolism, while suggesting PRMT7 as a potential
therapeutic target for the eradication of LSCs in CML (109).
Despite the relevance of PRMTs for signal
transduction, metabolism, transcription and other cellular
phenotypes, the methylation profiles of protein arginine remain
understudied. The function of PRMTs under physiological and
pathological conditions often depends on methyltransferase
activity. Therefore, comprehensively revealing the substrates of
PRMTs is the key to exploring their functions and underlying
molecular mechanisms.
Combining two newly developed methylation sequencing
methods, immunoaffinity purification and high pH strong cation
exchange, may improve the coverage of protein methylation and
reveal new PRMT1 targets (110).
After knockout of PRMT1, 127 arginine methylation sites on 78
proteins were significantly changed. In contrast, only one lysine
methylation site was significantly changed after PRMT1 knockdown,
indicating that amino acid methylation was not affected by PRMT1
knockdown. In PRMT1-knockout cells, 114 MMA sites were found to be
significantly altered on proteins enriched in the mRNA metabolic
process. A high-confidence list of 18 PRMT1 substrates and 12
methylation sites scavenged by other PRMTs in the absence of PRMT1
activity was found through integrative analysis of MMA and DMA.
Most importantly, the methylation site hnRNPA1 R206 switched from
ADMA to SDMA after PRMT1 knockout (110).
High-resolution mass spectrometry combined with
SILAC technology was used to analyse the arginine methylation
regulated by PRMT7 in 293 cells (111). A total of 1,031 MMA sites of 513
proteins were detected and a two-fold decrease in monomethylation
levels at 297 arginine sites in 174 proteins was found, termed the
PRMT7 methylome. During this process, the methylation of 176 MMA
sites in 108 proteins disappeared completely. After treatment with
the PRMT7-specific inhibitor SGC3027, 503 MMA sites of 274 proteins
were more than two-fold reduced, and ~60% of PRMT7-regulated
substrates were also inhibited. The same method was also used to
identify the methylated substrates of PRMT4 and -5, representing
type I and II PRMTs, respectively. The PRMT4 methyl group (two-fold
decrease) includes 301 proteins with 660 methylation sites, while
the PRMT5 methyl group (two-fold decrease) includes 244 proteins
with 429 methylation sites. PRMT4 substrates, such as mediator
complex subunit 12, SWI/SNF related, matrix associated, actin
dependent regulator of chromatin subfamily c member 1 and E1A
binding protein p300, and PRMT5 substrates, such as FUS RNA binding
protein, hnRNPA1 and survival of motor neuron 1, have also been
identified. Most importantly, PRMT4, -5 and -7 coregulate
alternative splicing in an enzyme-dependent manner and share a
number of RNA splicing factors, such as hnRNPA1; furthermore,
hnRNPA1 arginine methylation is required for the growth of various
cancer cells (116). Taken
together, the methylation profiles of PRMTs indicated that hnRNAPA1
has a role of co-RNA splicing factor in various arginine
methylation modification processes and may also promote the
transformation of ADMA to SDMA. Further study of its mechanism will
reveal the types of arginine methylation.
While much important progress has been made in
research on PRMT function, the complexity and significance of gene
post-transcriptional processing determine the diversity of the PRMT
regulatory mechanisms. The regulatory effect of PRMTs on the
function of RBP clearly exemplifies the important regulatory role
of arginine methylation in post-transcriptional processing.
Although numerous important advances have been made in the study of
PRMTs regulating the function of RBP, various questions remain to
be answered in the field of cancer, such as how a single arginine
methylation site determines the function of specific proteins, how
different PRMT family members synergistically regulate the
occurrence of specific substrate methylation and how arginine
methylation modifications are recognized and removed. In addition,
the sequence characteristic of the target RNA directly affected by
RBP and how arginine methylation regulates the interaction between
RBP and its target RNA at the genome-wide level are also key
questions to be solved. In addition, PRMTs directly methylate
numerous proteins to control their subcellular localization,
protein-protein interactions, stability or activity. Many of these
contribute to oncogenic transformation, and thus, evaluation of
potential PRMT inhibitors is warranted. What remains to be
established is how the inhibition of arginine methylation may be
integrated with immunotherapeutic approaches to achieve a maximal,
long-lasting therapeutic effect.
Not applicable.
KW, CN and HL were involved in the conception of the
review. KW wrote the manuscript and performed the literature
search. KW and LF reviewed and edited the final manuscript. All
authors have read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This study was supported by grants from the National Natural
Science Foundation of China (grant nos. 81672637, 81872164 and
82173344).
|
1
|
Vu LD, Gevaert K and De Smet I: Protein
language: Post-translational modifications talking to each other.
Trends Plant Sci. 23:1068–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jarrold J and Davies CC: PRMTs and
arginine methylation: Cancer's best-kept secret? Trends Mol Med.
25:993–1009. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paik WK and Kim S: Enzymatic methylation
of protein fractions from calf thymus nuclei. Biochem Biophys Res
Commun. 29:14–20. 1967. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin WJ, Gary JD, Yang MC, Clarke S and
Herschman HR: The mammalian immediate-early TIS21 protein and the
leukemia-associated BTG1 protein interact with a protein-arginine
N-methyltransferase. J Biol Chem. 271:15034–15044. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blanc RS and Richard S: Arginine
methylation: The coming of age. Mol Cell. 65:8–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bedford MT and Clarke SG: Protein arginine
methylation in mammals: Who, what, and why. Mol Cell. 33:1–13.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hughes RM and Waters ML: Arginine
methylation in a beta-hairpin peptide: Implications for Arg-pi
interactions, DeltaCp(o), and the cold denatured state. J Am Chem
Soc. 128:12735–12742. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Obianyo O, Causey CP, Jones JE and
Thompson PR: Activity-based protein profiling of protein arginine
methyltransferase 1. ACS Chem Biol. 6:1127–1135. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gui S, Wooderchak WL, Daly MP, Porter PJ,
Johnson SJ and Hevel JM: Investigation of the molecular origins of
protein-arginine methyltransferase I (PRMT1) product specificity
reveals a role for two conserved methionine residues. J Biol Chem.
286:29118–29126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gui S, Gathiaka S, Li J, Qu J, Acevedo O
and Hevel JM: A remodeled protein arginine methyltransferase 1
(PRMT1) generates symmetric dimethylarginine. J Biol Chem.
289:9320–9327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshimoto T, Boehm M, Olive M, Crook MF,
San H, Langenickel T and Nabel EG: The arginine methyltransferase
PRMT2 binds RB and regulates E2F function. Exp Cell Res.
312:2040–2053. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Frankel A and Clarke S: PRMT3 is a
distinct member of the protein arginine N-methyltransferase family.
Conferral of substrate specificity by a zinc-finger domain. J Biol
Chem. 275:32974–32982. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shishkova E, Zeng H, Liu F, Kwiecien NW,
Hebert AS, Coon JJ and Xu W: Global mapping of CARM1 substrates
defines enzyme specificity and substrate recognition. Nat Commun.
8:155712017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S,
Gong W and Xu RM: Structural insights into protein arginine
symmetric dimethylation by PRMT5. Proc Natl Acad Sci USA.
108:20538–20543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okuno K, Akiyama Y, Shimada S, Nakagawa M,
Tanioka T, Inokuchi M, Yamaoka S, Kojima K and Tanaka S: Asymmetric
dimethylation at histone H3 arginine 2 by PRMT6 in gastric cancer
progression. Carcinogenesis. 40:15–26. 2019. View Article : Google Scholar
|
|
16
|
Hasegawa M, Toma-Fukai S, Kim JD, Fukamizu
A and Shimizu T: Protein arginine methyltransferase 7 has a novel
homodimer-like structure formed by tandem repeats. FEBS Lett.
588:1942–1948. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee J, Sayegh J, Daniel J, Clarke S and
Bedford MT: PRMT8, a new membrane-bound tissue-specific member of
the protein arginine methyltransferase family. J Biol Chem.
280:32890–32896. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hadjikyriacou A, Yang Y, Espejo A, Bedford
MT and Clarke SG: Unique features of human protein arginine
methyltransferase 9 (PRMT9) and its substrate RNA splicing factor
SF3B2. J Biol Chem. 290:16723–16743. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fulton MD, Cao M, Ho MC, Zhao X and Zheng
YG: The macromolecular complexes of histones affect protein
arginine methyltransferase activities. J Biol Chem. 297:1011232021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Osborne TC, Obianyo O, Zhang X, Cheng X
and Thompson PR: Protein arginine methyltransferase 1: Positively
charged residues in substrate peptides distal to the site of
methylation are important for substrate binding and catalysis.
Biochemistry. 46:13370–13381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Waldmann T, Izzo A, Kamieniarz K, Richter
F, Vogler C, Sarg B, Lindner H, Young NL, Mittler G, Garcia BA and
Schneider R: Methylation of H2AR29 is a novel repressive PRMT6
target. Epigenetics Chromatin. 4:112011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Branscombe TL, Frankel A, Lee JH, Cook JR,
Yang Z, Pestka S and Clarke S: PRMT5 (Janus kinase-binding protein
1) catalyzes the formation of symmetric dimethylarginine residues
in proteins. J Biol Chem. 276:32971–32976. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Karkhanis V, Wang L, Tae S, Hu YJ,
Imbalzano AN and Sif S: Protein arginine methyltransferase 7
regulates cellular response to DNA damage by methylating promoter
histones H2A and H4 of the polymerase δ catalytic subunit gene,
POLD1. J Biol Chem. 287:29801–29814. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu G, Yan C, Xie P, Cao Y, Shao J and Ge
J: PRMT2 accelerates tumorigenesis of hepatocellular carcinoma by
activating Bcl2 via histone H3R8 methylation. Exp Cell Res.
394:1121522020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dong F, Li Q, Yang C, Huo D, Wang X, Ai C,
Kong Y, Sun X, Wang W, Zhou Y, et al: PRMT2 links histone H3R8
asymmetric dimethylation to oncogenic activation and tumorigenesis
of glioblastoma. Nat Commun. 9:45522018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang
S and Bedford MT: TDRD3 is an effector molecule for
arginine-methylated histone marks. Mol Cell. 40:1016–1023. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu J and Richard S: Cellular pathways
influenced by protein arginine methylation: Implications for
cancer. Mol Cell. 81:4357–4368. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu T, Lv X, Kong Q and Yuan C: A novel
SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell
invasion. Oncotarget. 8:54809–54820. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chiang K, Zielinska AE, Shaaban AM,
Sanchez-Bailon MP, Jarrold J, Clarke TL, Zhang J, Francis A, Jones
LJ, Smith S, et al: PRMT5 is a critical regulator of breast cancer
stem cell function via histone methylation and FOXP1 expression.
Cell Rep. 21:3498–3513. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao L, Wu G, Zhu J, Tan Z, Shi D, Wu X,
Tang M, Li Z, Hu Y, Zhang S, et al: Genotoxic stress-triggered
β-catenin/JDP2/PRMT5 complex facilitates reestablishing glutathione
homeostasis. Nat Commun. 10:37612019. View Article : Google Scholar
|
|
31
|
Migliori V, Müller J, Phalke S, Low D,
Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, et al:
Symmetric dimethylation of H3R2 is a newly identified histone mark
that supports euchromatin maintenance. Nat Struct Mol Biol.
19:136–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mitchell LH, Drew AE, Ribich SA, Rioux N,
Swinger KK, Jacques SL, Lingaraj T, Boriack-Sjodin PA, Waters NJ,
Wigle TJ, et al: Aryl pyrazoles as potent inhibitors of arginine
methyltransferases: Identification of the first PRMT6 tool
compound. ACS Med Chem Lett. 6:655–659. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang B, Dong S, Zhu R, Hu C, Hou J, Li Y,
Zhao Q, Shao X, Bu Q, Li H, et al: Targeting protein arginine
methyltransferase 5 inhibits colorectal cancer growth by decreasing
arginine methylation of eIF4E and FGFR3. Oncotarget. 6:22799–22811.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deng X, Shao G, Zhang HT, Li C, Zhang D,
Cheng L, Elzey BD, Pili R, Ratliff TL, Huang J and Hu CD: Protein
arginine methyltransferase 5 functions as an epigenetic activator
of the androgen receptor to promote prostate cancer cell growth.
Oncogene. 36:1223–1231. 2017. View Article : Google Scholar :
|
|
35
|
Yao B, Gui T, Zeng X, Deng Y, Wang Z, Wang
Y, Yang D, Li Q, Xu P, Hu R, et al: PRMT1-mediated H4R3me2a
recruits SMARCA4 to promote colorectal cancer progression by
enhancing EGFR signaling. Genome Med. 13:582021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kaushik S, Liu F, Veazey KJ, Gao G, Das P,
Neves LF, Lin K, Zhong Y, Lu Y, Giuliani V, et al: Genetic deletion
or small-molecule inhibition of the arginine methyltransferase
PRMT5 exhibit anti-tumoral activity in mouse models of
MLL-rearranged AML. Leukemia. 32:499–509. 2018. View Article : Google Scholar
|
|
37
|
Yang L, Ma DW, Cao YP, Li DZ, Zhou X, Feng
JF and Bao J: PRMT5 functionally associates with EZH2 to promote
colorectal cancer progression through epigenetically repressing
CDKN2B expression. Theranostics. 11:3742–3759. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gurung B, Feng Z and Hua X: Menin directly
represses Gli1 expression independent of canonical Hedgehog
signaling. Mol Cancer Res. 11:1215–1222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gurung B, Feng Z, Iwamoto DV, Thiel A, Jin
G, Fan CM, Ng JM, Curran T and Hua X: Menin epigenetically
represses Hedgehog signaling in MEN1 tumor syndrome. Cancer Res.
73:2650–2658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Krebs AM, Mitschke J, Lasierra Losada M,
Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D,
Reichardt W, Bronsert P, et al: The EMT-activator Zeb1 is a key
factor for cell plasticity and promotes metastasis in pancreatic
cancer. Nat Cell Biol. 19:518–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao Y, Zhao Y, Zhang J, Lu Y, Liu X, Geng
P, Huang B, Zhang Y and Lu J: The dual function of PRMT1 in
modulating epithelial-mesenchymal transition and cellular
senescence in breast cancer cells through regulation of ZEB1. Sci
Rep. 6:198742016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen H, Lorton B, Gupta V and Shechter D:
A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through
histone H3 and H4 arginine methylation coupled transcriptional
activation and repression. Oncogene. 36:373–386. 2017. View Article : Google Scholar
|
|
43
|
Liu R, Gao J, Yang Y, Qiu R, Zheng Y,
Huang W, Zeng Y, Hou Y, Wang S, Leng S, et al: PHD finger protein 1
(PHF1) is a novel reader for histone H4R3 symmetric dimethylation
and coordinates with PRMT5-WDR77/CRL4B complex to promote
tumorigenesis. Nucleic Acids Res. 46:6608–6626. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Siarheyeva A, Senisterra G, Allali-Hassani
A, Dong A, Dobrovetsky E, Wasney GA, Chau I, Marcellus R, Hajian T,
Liu F, et al: An allosteric inhibitor of protein arginine
methyltransferase 3. Structure. 20:1425–1435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jain K and Clarke SG: PRMT7 as a unique
member of the protein arginine methyltransferase family: A review.
Arch Biochem Biophys. 665:36–45. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pal S, Vishwanath SN, Erdjument-Bromage H,
Tempst P and Sif S: Human SWI/SNF-associated PRMT5 methylates
histone H3 arginine 8 and negatively regulates expression of ST7
and NM23 tumor suppressor genes. Mol Cell Biol. 24:9630–9645. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Scaglione A, Patzig J, Liang J, Frawley R,
Bok J, Mela A, Yattah C, Zhang J, Teo SX, Zhou T, et al:
PRMT5-mediated regulation of developmental myelination. Nat Commun.
9:28402018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jing P, Zhao N, Ye M, Zhang Y, Zhang Z,
Sun J, Wang Z, Zhang J and Gu Z: Protein arginine methyltransferase
5 promotes lung cancer metastasis via the epigenetic regulation of
miR-99 family/FGFR3 signaling. Cancer Lett. 427:38–48. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Karkhanis V, Alinari L, Ozer HG, Chung J,
Zhang X, Sif S and Baiocchi RA: Protein arginine methyltransferase
5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1
and c-MYC expression in aggressive B-cell lymphoma. J Biol Chem.
295:1165–1180. 2020. View Article : Google Scholar :
|
|
50
|
Cho EC, Zheng S, Munro S, Liu G, Carr SM,
Moehlenbrink J, Lu YC, Stimson L, Khan O, Konietzny R, et al:
Arginine methylation controls growth regulation by E2F-1. EMBO J.
31:1785–1797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu L, Zhao X, Zhao L, Li J, Yang H, Zhu
Z, Liu J and Huang G: Arginine methylation of SREBP1a via PRMT5
promotes de novo lipogenesis and tumor growth. Cancer Res.
76:1260–1272. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lu X, Fernando TM, Lossos C, Yusufova N,
Liu F, Fontá L, Durant M, Geng H, Melnick J, Luo Y, et al: PRMT5
interacts with the BCL6 oncoprotein and is required for germinal
center formation and lymphoma cell survival. Blood. 132:2026–2039.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu J, Shin B, Park ES, Yang S, Choi S,
Kang M and Rho J: Protein arginine methyltransferase 1 regulates
herpes simplex virus replication through ICP27 RGG-box methylation.
Biochem Biophys Res Commun. 391:322–328. 2010. View Article : Google Scholar
|
|
54
|
Fronz K, Güttinger S, Burkert K, Kühn U,
Stöhr N, Schierhorn A and Wahle E: Arginine methylation of the
nuclear poly(a) binding protein weakens the interaction with its
nuclear import receptor, transportin. J Biol Chem. 286:32986–32994.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Katsuno Y, Qin J, Oses-Prieto J, Wang H,
Jackson-Weaver O, Zhang T, Lamouille S, Wu J, Burlingame A, Xu J
and Derynck R: Arginine methylation of SMAD7 by PRMT1 in
TGF-β-induced epithelial-mesenchymal transition and epithelial
stem-cell generation. J Biol Chem. 293:13059–13072. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Clarke TL, Sanchez-Bailon MP, Chiang K,
Reynolds JJ, Herrero-Ruiz J, Bandeiras TM, Matias PM, Maslen SL,
Skehel JM, Stewart GS and Davies CC: PRMT5-dependent methylation of
the TIP60 coactivator RUVBL1 is a key regulator of homologous
recombination. Mol Cell. 65:900–916.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hamard PJ, Santiago GE, Liu F, Karl DL,
Martinez C, Man N, Mookhtiar AK, Duffort S, Greenblatt S, Verdun RE
and Nimer SD: PRMT5 regulates DNA repair by controlling the
alternative splicing of histone-modifying enzymes. Cell Rep.
24:2643–2657. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hellmuth S, Gutiérrez-Caballero C, Llano
E, Pendás AM and Stemmann O: Local activation of mammalian separase
in interphase promotes double-strand break repair and prevents
oncogenic transformation. EMBO J. 37:e991842018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vadnais C, Chen R, Fraszczak J, Yu Z,
Boulais J, Pinder J, Frank D, Khandanpour C, Hébert J, Dellaire G,
et al: GFI1 facilitates efficient DNA repair by regulating PRMT1
dependent methylation of MRE11 and 53BP1. Nat Commun. 9:14182018.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Karakashev S, Fukumoto T, Zhao B, Lin J,
Wu S, Fatkhutdinov N, Park PH, Semenova G, Jean S, Cadungog MG, et
al: EZH2 inhibition sensitizes CARM1-high, homologous recombination
proficient ovarian cancers to PARP inhibition. Cancer Cell.
37:157–167.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wei X, Yang J, Adair SJ, Ozturk H, Kuscu
C, Lee KY, Kane WJ, O'Hara PE, Liu D, Demirlenk YM, et al: Targeted
CRISPR screening identifies PRMT5 as synthetic lethality
combinatorial target with gemcitabine in pancreatic cancer cells.
Proc Natl Acad Sci USA. 117:28068–28079. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li Y, Chitnis N, Nakagawa H, Kita Y,
Natsugoe S, Yang Y, Li Z, Wasik M, Klein-Szanto AJ, Rustgi AK and
Diehl JA: PRMT5 is required for lymphomagenesis triggered by
multiple oncogenic drivers. Cancer Discov. 5:288–303. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jansson M, Durant ST, Cho EC, Sheahan S,
Edelmann M, Kessler B and La Thangue NB: Arginine methylation
regulates the p53 response. Nat Cell Biol. 10:1431–1439. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rocha CRR, Silva MM, Quinet A, Cabral-Neto
JB and Menck CFM: DNA repair pathways and cisplatin resistance: An
intimate relationship. Clinics (Sao Paulo). 73(Suppl 1): e478s2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
McCabe N, Turner NC, Lord CJ, Kluzek K,
Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka
MZ, et al: Deficiency in the repair of DNA damage by homologous
recombination and sensitivity to poly(ADP-ribose) polymerase
inhibition. Cancer Res. 66:8109–8115. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Peshkin BN, Alabek ML and Isaacs C:
BRCA1/2 mutations and triple negative breast cancers. Breast Dis.
32:25–33. 2010. View Article : Google Scholar
|
|
67
|
Huang T, Yang Y, Song X, Wan X, Wu B,
Sastry N, Horbinski CM, Zeng C, Tiek D, Goenka A, et al: PRMT6
methylation of RCC1 regulates mitosis, tumorigenicity, and
radiation response of glioblastoma stem cells. Mol Cell.
81:1276–1291.e9. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sanchez-Bailon MP, Choi SY, Dufficy ER,
Sharma K, McNee GS, Gunnell E, Chiang K, Sahay D, Maslen S, Stewart
GS, et al: Arginine methylation and ubiquitylation crosstalk
controls DNA end-resection and homologous recombination repair. Nat
Commun. 12:63132021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Dvinge H, Kim E, Abdel-Wahab O and Bradley
RK: RNA splicing factors as oncoproteins and tumour suppressors.
Nat Rev Cancer. 16:413–430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Bonnal SC, López-Oreja I and Valcárcel J:
Roles and mechanisms of alternative splicing in cancer-implications
for care. Nat Rev Clin Oncol. 17:457–474. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fong JY, Pignata L, Goy PA, Kawabata KC,
Lee SC, Koh CM, Musiani D, Massignani E, Kotini AG, Penson A, et
al: Therapeutic targeting of RNA splicing catalysis through
inhibition of protein arginine methylation. Cancer Cell.
36:194–209.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Guccione E and Richard S: The regulation,
functions and clinical relevance of arginine methylation. Nat Rev
Mol Cell Biol. 20:642–657. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Radzisheuskaya A, Shliaha PV, Grinev V,
Lorenzini E, Kovalchuk S, Shlyueva D, Gorshkov V, Hendrickson RC,
Jensen ON and Helin K: PRMT5 methylome profiling uncovers a direct
link to splicing regulation in acute myeloid leukemia. Nat Struct
Mol Biol. 26:999–1012. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Musiani D, Bok J, Massignani E, Wu L,
Tabaglio T, Ippolito MR, Cuomo A, Ozbek U, Zorgati H, Ghoshdastider
U, et al: Proteomics profiling of arginine methylation defines
PRMT5 substrate specificity. Sci Signal. 12:eaat83882019.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cai T, Cinkornpumin JK, Yu Z, Villarreal
OD, Pastor WA and Richard S: Deletion of RBMX RGG/RG motif in
Shashi-XLID syndrome leads to aberrant p53 activation and neuronal
differentiation defects. Cell Rep. 36:1093372021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Gerhart SV, Kellner WA, Thompson C,
Pappalardi MB, Zhang XP, Montes de Oca R, Penebre E, Duncan K,
Boriack-Sjodin A, Le B, et al: Activation of the p53-MDM4
regulatory axis defines the anti-tumour response to PRMT5
inhibition through its role in regulating cellular splicing. Sci
Rep. 8:97112018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
AbuHammad S, Cullinane C, Martin C,
Bacolas Z, Ward T, Chen H, Slater A, Ardley K, Kirby L, Chan KT, et
al: Regulation of PRMT5-MDM4 axis is critical in the response to
CDK4/6 inhibitors in melanoma. Proc Natl Acad Sci USA.
116:17990–18000. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Roworth AP, Carr SM, Liu G, Barczak W,
Miller RL, Munro S, Kanapin A, Samsonova A and La Thangue NB:
Arginine methylation expands the regulatory mechanisms and extends
the genomic landscape under E2F control. Sci Adv. 5:eaaw46402019.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yang Y, Hadjikyriacou A, Xia Z, Gayatri S,
Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG and Bedford
MT: PRMT9 is a type II methyltransferase that methylates the
splicing factor SAP145. Nat Commun. 6:64282015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Suresh S, Huard S, Brisson A, Némati F,
Dakroub R, Poulard C, Ye M, Martel E, Reyes C, Silvestre DC, et al:
PRMT1 regulates EGFR and Wnt signaling pathways and is a promising
target for combinatorial treatment of breast cancer. Cancers
(Basel). 14:3062022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Fedoriw A, Rajapurkar SR, O'Brien S,
Gerhart SV, Mitchell LH, Adams ND, Rioux N, Lingaraj T, Ribich SA,
Pappalardi MB, et al: Anti-tumor activity of the type I PRMT
inhibitor, GSK3368715, synergizes with PRMT5 inhibition through
MTAP loss. Cancer Cell. 36:100–114.e25. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wei H, Wang B, Miyagi M, She Y, Gopalan B,
Huang DB, Ghosh G, Stark GR and Lu T: PRMT5 dimethylates R30 of the
p65 subunit to activate NF-κB. Proc Natl Acad Sci USA.
110:13516–13521. 2013. View Article : Google Scholar
|
|
83
|
Tanaka H, Hoshikawa Y, Oh-hara T, Koike S,
Naito M, Noda T, Arai H, Tsuruo T and Fujita N: PRMT5, a novel
TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis
via nuclear factor-kappaB activation. Mol Cancer Res. 7:557–569.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Gullà A, Hideshima T, Bianchi G, Fulciniti
M, Kemal Samur M, Qi J, Tai YT, Harada T, Morelli E, Amodio N, et
al: Protein arginine methyltransferase 5 has prognostic relevance
and is a druggable target in multiple myeloma. Leukemia.
32:996–1002. 2018. View Article : Google Scholar
|
|
85
|
Jiang H, Zhou Z, Jin S, Xu K, Zhang H and
Xu J, Sun Q, Wang J and Xu J: PRMT9 promotes hepatocellular
carcinoma invasion and metastasis via activating
PI3K/Akt/GSK-3β/Snail signaling. Cancer Sci. 109:1414–1427. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Tamiya H, Kim H, Klymenko O, Kim H, Feng
Y, Zhang T, Han JY, Murao A, Snipas SJ, Jilaveanu L, et al:
SHARPIN-mediated regulation of protein arginine methyltransferase 5
controls melanoma growth. J Clin Invest. 128:517–530. 2018.
View Article : Google Scholar :
|
|
87
|
Hu Y, Su Y, He Y, Liu W and Xiao B:
Arginine methyltransferase PRMT3 promote tumorigenesis through
regulating c-MYC stabilization in colorectal cancer. Gene.
791:1457182021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang X, Wang K, Feng X, Wang J, Chu Y,
Jia C, He Q and Chen C: PRMT3 promotes tumorigenesis by methylating
and stabilizing HIF1α in colorectal cancer. Cell Death Dis.
12:10662021. View Article : Google Scholar
|
|
89
|
Abe Y and Tanaka N: Fine-tuning of GLI
activity through arginine methylation: its mechanisms and function.
Cells. 9:19732020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Song C, Chen T, He L, Ma N, Li JA, Rong
YF, Fang Y, Liu M, Xie D and Lou W: PRMT1 promotes pancreatic
cancer growth and predicts poor prognosis. Cell Oncol (Dordr).
43:51–62. 2020.
|
|
91
|
Lai Y, Song M, Hakala K, Weintraub ST and
Shiio Y: Proteomic dissection of the von Hippel-Lindau (VHL)
interactome. J Proteome Res. 10:5175–5182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Hsu MC, Pan MR, Chu PY, Tsai YL, Tsai CH,
Shan YS, Chen LT and Hung WC: Protein arginine methyltransferase 3
enhances chemoresistance in pancreatic cancer by methylating
hnRNPA1 to increase ABCG2 expression. Cancers (Basel). 11:82018.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gao G, Dhar S and Bedford MT: PRMT5
regulates IRES-dependent translation via methylation of hnRNP A1.
Nucleic Acids Res. 45:4359–4369. 2017.PubMed/NCBI
|
|
94
|
Plotnikov A, Kozer N, Cohen G, Carvalho S,
Duberstein S, Almog O, Solmesky LJ, Shurrush KA, Babaev I, Benjamin
S, et al: PRMT1 inhibition induces differentiation of colon cancer
cells. Sci Rep. 10:200302020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sengupta S, Kennemer A, Patrick K,
Tsichlis P and Guerau-de-Arellano M: Protein arginine
methyltransferase 5 in T lymphocyte biology. Trends Immunol.
41:918–931. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kim H, Kim H, Feng Y, Li Y, Tamiya H,
Tocci S and Ronai ZA: PRMT5 control of cGAS/STING and NLRC5
pathways defines melanoma response to antitumor immunity. Sci
Transl Med. 12:eaaz56832020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Elliott K, Nilsson J and Van den Eynden J:
Pharmacologic RNA splicing modulation: A novel mechanism to enhance
neoantigen-directed anti-tumor immunity and immunotherapy response.
Signal Transduct Target Ther. 6:3732021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Nagai Y, Ji MQ, Zhu F, Xiao Y, Tanaka Y,
Kambayashi T, Fujimoto S, Goldberg MM, Zhang H, Li B, et al: PRMT5
associates with the FOXP3 homomer and when disabled enhances
targeted p185erbB2/neu tumor immunotherapy. Front
Immunol. 10:1742019. View Article : Google Scholar
|
|
99
|
Kagoya Y, Saijo H, Matsunaga Y, Guo T,
Saso K, Anczurowski M, Wang CH, Sugata K, Murata K, Butler MO, et
al: Arginine methylation of FOXP3 is crucial for the suppressive
function of regulatory T cells. J Autoimmun. 97:10–21. 2019.
View Article : Google Scholar
|
|
100
|
Hou J, Wang Y, Shi L, Chen Y, Xu C, Saeedi
A, Pan K, Bohat R, Egan NA, McKenzie JA, et al: Integrating
genome-wide CRISPR immune screen with multi-omic clinical data
reveals distinct classes of tumor intrinsic immune regulators. J
Immunother Cancer. 9:e0018192021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Schonfeld M, Zhao J, Komatz A, Weinman SA
and Tikhanovich I: The polymorphism rs975484 in the protein
arginine methyltransferase 1 gene modulates expression of immune
checkpoint genes in hepatocellular carcinoma. J Biol Chem.
295:7126–7137. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kumar S, Zeng Z, Bagati A, Tay RE, Sanz
LA, Hartono SR, Ito Y, Abderazzaq F, Hatchi E, Jiang P, et al:
CARM1 inhibition enables immunotherapy of resistant tumors by dual
action on tumor cells and T cells. Cancer Discov. 11:2050–2071.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zheng NN, Zhou M, Sun F, Huai MX, Zhang Y,
Qu CY, Shen F and Xu LM: Combining protein arginine
methyltransferase inhibitor and anti-programmed death-ligand-1
inhibits pancreatic cancer progression. World J Gastroenterol.
26:3737–3749. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Lu SX, De Neef E, Thomas JD, Sabio E,
Rousseau B, Gigoux M, Knorr DA, Greenbaum B, Elhanati Y, Hogg SJ,
et al: Pharmacologic modulation of RNA splicing enhances anti-tumor
immunity. Cell. 184:4032–4047.e31. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Tennant DA, Durán RV and Gottlieb E:
Targeting metabolic transformation for cancer therapy. Nat Rev
Cancer. 10:267–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ganapathy-Kanniappan S and Geschwind JF:
Tumor glycolysis as a target for cancer therapy: Progress and
prospects. Mol Cancer. 12:1522013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Hsu MC, Tsai YL, Lin CH, Pan MR, Shan YS,
Cheng TY, Cheng SH, Chen LT and Hung WC: Protein arginine
methyltransferase 3-induced metabolic reprogramming is a vulnerable
target of pancreatic cancer. J Hematol Oncol. 12:792019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Bednarz-Misa I, Fortuna P, Fleszar MG,
Lewandowski Ł, Diakowska D, Rosińczuk J and Krzystek-Korpacka M:
Esophageal squamous cell carcinoma is accompanied by local and
systemic changes in L-arginine/NO pathway. Int J Mol Sci.
21:62822020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lei Y, Han P, Chen Y, Wang H, Wang S, Wang
M, Liu J, Yan W, Tian D and Liu M: Protein arginine
methyltransferase 3 promotes glycolysis and hepatocellular
carcinoma growth by enhancing arginine methylation of lactate
dehydrogenase A. Clin Transl Med. 12:e6862022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hartel NG, Chew B, Qin J, Xu J and Graham
NA: Deep protein methylation profiling by combined chemical and
immunoaffinity approaches reveals novel PRMT1 targets. Mol Cell
Proteomics. 18:2149–2164. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Li WJ, He YH, Yang JJ, Hu GS, Lin YA, Ran
T, Peng BL, Xie BL, Huang MF, Gao X, et al: Profiling PRMT
methylome reveals roles of hnRNPA1 arginine methylation in RNA
splicing and cell growth. Nat Commun. 12:19462021. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Iberg AN, Espejo A, Cheng D, Kim D,
Michaud-Levesque J, Richard S and Bedford MT: Arginine methylation
of the histone H3 tail impedes effector binding. J Biol Chem.
283:3006–3010. 2008. View Article : Google Scholar
|
|
113
|
Neault M, Mallette FA, Vogel G,
Michaud-Levesque J and Richard S: Ablation of PRMT6 reveals a role
as a negative transcriptional regulator of the p53 tumor
suppressor. Nucleic Acids Res. 40:9513–9521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Lorton BM, Harijan RK, Burgos ES, Bonanno
JB, Almo SC and Shechter D: A binary arginine methylation switch on
histone H3 arginine 2 regulates its interaction with WDR5.
Biochemistry. 59:3696–3708. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Guccione E, Bassi C, Casadio F, Martinato
F, Cesaroni M, Schuchlautz H, Lüscher B and Amati B: Methylation of
histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually
exclusive. Nature. 449:933–937. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Dacwag CS, Ohkawa Y, Pal S, Sif S and
Imbalzano AN: The protein arginine methyltransferase Prmt5 is
required for myogenesis because it facilitates ATP-dependent
chromatin remodeling. Mol Cell Biol. 27:384–394. 2007. View Article : Google Scholar :
|
|
117
|
Tarighat SS, Santhanam R, Frankhouser D,
Radomska HS, Lai H, Anghelina M, Wang H, Huang X, Alinari L, Walker
A, et al: The dual epigenetic role of PRMT5 in acute myeloid
leukemia: Gene activation and repression via histone arginine
methylation. Leukemia. 30:789–799. 2016. View Article : Google Scholar
|
|
118
|
Zhang Z, Nikolai BC, Gates LA, Jung SY,
Siwak EB, He B, Rice AP, O'Malley BW and Feng Q: Crosstalk between
histone modifications indicates that inhibition of arginine
methyltransferase CARM1 activity reverses HIV latency. Nucleic
Acids Res. 45:9348–9360. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Cheung N, Fung TK, Zeisig BB, Holmes K,
Rane JK, Mowen KA, Finn MG, Lenhard B, Chan LC and So CW: Targeting
aberrant epigenetic networks mediated by PRMT1 and KDM4C in acute
myeloid leukemia. Cancer Cell. 29:32–48. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Min Z, Xiaomeng L, Zheng L, Yangge D,
Xuejiao L, Longwei L, Xiao Z, Yunsong L, Ping Z and Yongsheng Z:
Asymmetrical methyltransferase PRMT3 regulates human mesenchymal
stem cell osteogenesis via miR-3648. Cell Death Dis. 10:5812019.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhang Y, van Haren MJ and Martin NI:
Peptidic transition state analogues as PRMT inhibitors. Methods.
175:24–29. 2020. View Article : Google Scholar
|
|
122
|
Hamey JJ, Rakow S, Bouchard C, Senst JM,
Kolb P, Bauer UM, Wilkins MR and Hart-Smith G: Systematic
investigation of PRMT6 substrate recognition reveals broad
specificity with a preference for an RG motif or basic and bulky
residues. FEBS J. 288:5668–5691. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhao Q, Rank G, Tan YT, Li H, Moritz RL,
Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al:
PRMT5-mediated methylation of histone H4R3 recruits DNMT3A,
coupling histone and DNA methylation in gene silencing. Nat Struct
Mol Biol. 16:304–311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Jain K, Jin CY and Clarke SG: Epigenetic
control via allosteric regulation of mammalian protein arginine
methyltransferases. Proc Natl Acad Sci USA. 114:10101–10106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Di Lorenzo A and Bedford MT: Histone
arginine methylation. FEBS Lett. 585:2024–2031. 2011. View Article : Google Scholar
|
|
126
|
Avasarala S, Van Scoyk M, Karuppusamy
Rathinam MK, Zerayesus S, Zhao X, Zhang W, Pergande MR, Borgia JA,
DeGregori J, Port JD, et al: PRMT1 is a novel regulator of
epithelial-mesenchymal-transition in non-small cell lung cancer. J
Biol Chem. 290:13479–13489. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhao Z, Rahman MA, Chen ZG and Shin DM:
Multiple biological functions of Twist1 in various cancers.
Oncotarget. 8:20380–20393. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Jobert L, Argentini M and Tora L: PRMT1
mediated methylation of TAF15 is required for its positive gene
regulatory function. Exp Cell Res. 315:1273–1286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Mizutani S, Yoshida T, Zhao X, Nimer SD,
Taniwaki M and Okuda T: Loss of RUNX1/AML1 arginine-methylation
impairs peripheral T cell homeostasis. Br J Haematol. 170:859–873.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Yamagata K, Daitoku H, Takahashi Y, Namiki
K, Hisatake K, Kako K, Mukai H, Kasuya Y and Fukamizu A: Arginine
methylation of FOXO transcription factors inhibits their
phosphorylation by Akt. Mol Cell. 32:221–231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Liu LM, Sun WZ, Fan XZ, Xu YL, Cheng MB
and Zhang Y: Methylation of C/EBPα by PRMT1 inhibits its
tumor-suppressive function in breast cancer. Cancer Res.
79:2865–2877. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Fronz K, Otto S, Kölbel K, Kühn U,
Friedrich H, Schierhorn A, Beck-Sickinger AG, Ostareck-Lederer A
and Wahle E: Promiscuous modification of the nuclear
poly(A)-binding protein by multiple protein-arginine
methyltransferases does not affect the aggregation behavior. J Biol
Chem. 283:20408–20420. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Boisvert FM, Rhie A, Richard S and Doherty
AJ: The GAR motif of 53BP1 is arginine methylated by PRMT1 and is
necessary for 53BP1 DNA binding activity. Cell Cycle. 4:1834–1841.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Boisvert FM, Déry U, Masson JY and Richard
S: Arginine methylation of MRE11 by PRMT1 is required for DNA
damage checkpoint control. Genes Dev. 19:671–676. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Guendel I, Carpio L, Pedati C, Schwartz A,
Teal C, Kashanchi F and Kehn-Hall K: Methylation of the tumor
suppressor protein, BRCA1, influences its transcriptional cofactor
function. PLoS One. 5:e113792010. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Huang L, Wang Z, Narayanan N and Yang Y:
Arginine methylation of the C-terminus RGG motif promotes TOP3B
topoisomerase activity and stress granule localization. Nucleic
Acids Res. 46:3061–3074. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Friesen WJ, Paushkin S, Wyce A, Massenet
S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M and Dreyfuss G:
The methylosome, a 20S complex containing JBP1 and pICln, produces
dimethylarginine-modified Sm proteins. Mol Cell Biol. 21:8289–8300.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhang L, Tran NT, Su H, Wang R, Lu Y, Tang
H, Aoyagi S, Guo A, Khodadadi-Jamayran A, Zhou D, et al: Cross-talk
between PRMT1-mediated methylation and ubiquitylation on RBM15
controls RNA splicing. Elife. 4:e079382015. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Li Z, Wang D, Lu J, Huang B, Wang Y, Dong
M, Fan D, Li H, Gao Y, Hou P, et al: Methylation of EZH2 by PRMT1
regulates its stability and promotes breast cancer metastasis. Cell
Death Differ. 27:3226–3242. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Le Romancer M, Treilleux I, Leconte N,
Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S,
Gobert-Gosse S and Corbo L: Regulation of estrogen rapid signaling
through arginine methylation by PRMT1. Mol Cell. 31:212–221. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Nakai K, Xia W, Liao HW, Saito M, Hung MC
and Yamaguchi H: The role of PRMT1 in EGFR methylation and
signaling in MDA-MB-468 triple-negative breast cancer cells. Breast
Cancer. 25:74–80. 2018. View Article : Google Scholar
|
|
142
|
Choucair A, Pham TH, Omarjee S,
Jacquemetton J, Kassem L, Trédan O, Rambaud J, Marangoni E, Corbo
L, Treilleux I and Le Romancer M: The arginine methyltransferase
PRMT1 regulates IGF-1 signaling in breast cancer. Oncogene.
38:4015–4027. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wang L, Jia Z, Xie D, Zhao T, Tan Z, Zhang
S, Kong F, Wei D and Xie K: Methylation of HSP70 orchestrates its
binding to and stabilization of BCL2 mRNA and renders pancreatic
cancer cells resistant to therapeutics. Cancer Res. 80:4500–4513.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Chuang CY, Chang CP, Lee YJ, Lin WL, Chang
WW, Wu JS, Cheng YW, Lee H and Li C: PRMT1 expression is elevated
in head and neck cancer and inhibition of protein arginine
methylation by adenosine dialdehyde or PRMT1 knockdown
downregulates proliferation and migration of oral cancer cells.
Oncol Rep. 38:1115–1123. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Zhou W, Yue H, Li C, Chen H and Yuan Y:
Protein arginine methyltransferase 1 promoted the growth and
migration of cancer cells in esophageal squamous cell carcinoma.
Tumour Biol. 37:2613–2619. 2016. View Article : Google Scholar
|
|
146
|
Zhong J, Cao RX, Zu XY, Hong T, Yang J,
Liu L, Xiao XH, Ding WJ, Zhao Q, Liu JH and Wen GB: Identification
and characterization of novel spliced variants of PRMT2 in breast
carcinoma. FEBS J. 279:316–335. 2012. View Article : Google Scholar
|
|
147
|
Ou CY, LaBonte MJ, Manegold PC, So AY,
Ianculescu I, Gerke DS, Yamamoto KR, Ladner RD, Kahn M, Kim JH and
Stallcup MR: A coactivator role of CARM1 in the dysregulation of
β-catenin activity in colorectal cancer cell growth and gene
expression. Mol Cancer Res. 9:660–670. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
El Messaoudi S, Fabbrizio E, Rodriguez C,
Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT
and Sardet C: Coactivator-associated arginine methyltransferase 1
(CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl
Acad Sci USA. 103:13351–13356. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wang L, Zhao Z, Meyer MB, Saha S, Yu M,
Guo A, Wisinski KB, Huang W, Cai W, Pike JW, et al: CARM1
methylates chromatin remodeling factor BAF155 to enhance tumor
progression and metastasis. Cancer Cell. 30:179–180. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Liu J, Feng J, Li L, Lin L, Ji J, Lin C,
Liu L, Zhang N, Duan D, Li Z, et al: Arginine methylation-dependent
LSD1 stability promotes invasion and metastasis of breast cancer.
EMBO Rep. 21:e485972020. View Article : Google Scholar
|
|
151
|
Al-Dhaheri M, Wu J, Skliris GP, Li J,
Higashimato K, Wang Y, White KP, Lambert P, Zhu Y, Murphy L and Xu
W: CARM1 is an important determinant of ERα-dependent breast cancer
cell differentiation and proliferation in breast cancer cells.
Cancer Res. 71:2118–2128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Wang YP, Zhou W, Wang J, Huang X, Zuo Y,
Wang TS, Gao X, Xu YY, Zou SW, Liu YB, et al: Arginine methylation
of MDH1 by CARM1 inhibits glutamine metabolism and suppresses
pancreatic cancer. Mol Cell. 64:673–687. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zakrzewicz D, Didiasova M, Krüger M,
Giaimo BD, Borggrefe T, Mieth M, Hocke AC, Zakrzewicz A, Schaefer
L, Preissner KT and Wygrecka M: Protein arginine methyltransferase
5 mediates enolase-1 cell surface trafficking in human lung
adenocarcinoma cells. Biochim Biophys Acta Mol Basis Dis.
1864:1816–1827. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Zhang S, Ma Y, Hu X, Zheng Y and Chen X:
Targeting PRMT5/Akt signalling axis prevents human lung cancer cell
growth. J Cell Mol Med. 23:1333–1342. 2019. View Article : Google Scholar :
|
|
155
|
Rengasamy M, Zhang F, Vashisht A, Song WM,
Aguilo F, Sun Y, Li S, Zhang W, Zhang B, Wohlschlegel JA and Walsh
MJ: The PRMT5/WDR77 complex regulates alternative splicing through
ZNF326 in breast cancer. Nucleic Acids Res. 45:11106–11120. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Wang Z, Kong J, Wu Y, Zhang J, Wang T, Li
N, Fan J, Wang H, Zhang J and Ling R: PRMT5 determines the
sensitivity to chemotherapeutics by governing stemness in breast
cancer. Breast Cancer Res Treat. 168:531–542. 2018. View Article : Google Scholar
|
|
157
|
Beketova E, Fang S, Owens JL, Liu S, Chen
X, Zhang Q, Asberry AM, Deng X, Malola J, Huang J, et al: Protein
arginine methyltransferase 5 promotes pICln-dependent androgen
receptor transcription in castration-resistant prostate cancer.
Cancer Res. 80:4904–4917. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Liu M, Yao B, Gui T, Guo C, Wu X, Li J, Ma
L, Deng Y, Xu P, Wang Y, et al: PRMT5-dependent transcriptional
repression of c-Myc target genes promotes gastric cancer
progression. Theranostics. 10:4437–4452. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Liu X, Zhang J, Liu L, Jiang Y, Ji J, Yan
R, Zhu Z and Yu Y: Protein arginine methyltransferase 5-mediated
epigenetic silencing of IRX1 contributes to tumorigenicity and
metastasis of gastric cancer. Biochim Biophys Acta Mol Basis Dis.
1864:2835–2844. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Jeon JY, Lee JS, Park ER, Shen YN, Kim MY,
Shin HJ, Joo HY, Cho EH, Moon SM, Shin US, et al: Protein arginine
methyltransferase 5 is implicated in the aggressiveness of human
hepatocellular carcinoma and controls the invasive activity of
cancer cells. Oncol Rep. 40:536–544. 2018.PubMed/NCBI
|
|
161
|
Jiang H, Zhu Y, Zhou Z and Xu J, Jin S, Xu
K, Zhang H, Sun Q, Wang J and Xu J: PRMT5 promotes cell
proliferation by inhibiting BTG2 expression via the ERK signaling
pathway in hepatocellular carcinoma. Cancer Med. 7:869–882. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Chung J, Karkhanis V, Baiocchi RA and Sif
S: Protein arginine methyltransferase 5 (PRMT5) promotes survival
of lymphoma cells via activation of WNT/β-catenin and AKT/GSK3β
proliferative signaling. J Biol Chem. 294:7692–7710. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Zhu F, Guo H, Bates PD, Zhang S, Zhang H,
Nomie KJ, Li Y, Lu L, Seibold KR, Wang F, et al: PRMT5 is
upregulated by B-cell receptor signaling and forms a
positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia.
33:2898–2911. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Hu G, Wang X, Han Y and Wang P: Protein
arginine methyltransferase 5 promotes bladder cancer growth through
inhibiting NF-kB dependent apoptosis. EXCLI J. 17:1157–1166.
2018.
|
|
165
|
Avasarala S, Wu PY, Khan SQ, Yanlin S, Van
Scoyk M, Bao J, Di Lorenzo A, David O, Bedford MT, Gupta V, et al:
PRMT6 promotes lung tumor progression via the alternate activation
of tumor-associated macrophages. Mol Cancer Res. 18:166–178. 2020.
View Article : Google Scholar :
|
|
166
|
Jiang N, Li QL, Pan W, Li J, Zhang MF, Cao
T, Su SG and Shen H: PRMT6 promotes endometrial cancer via AKT/mTOR
signaling and indicates poor prognosis. Int J Biochem Cell Biol.
120:1056812020. View Article : Google Scholar
|
|
167
|
Baldwin RM, Haghandish N, Daneshmand M,
Amin S, Paris G, Falls TJ, Bell JC, Islam S and Côté J: Protein
arginine methyltransferase 7 promotes breast cancer cell invasion
through the induction of MMP9 expression. Oncotarget. 6:3013–3032.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Cheng D, He Z, Zheng L, Xie D, Dong S and
Zhang P: PRMT7 contributes to the metastasis phenotype in human
non-small-cell lung cancer cells possibly through the interaction
with HSPA5 and EEF2. Onco Targets Ther. 11:4869–4876. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
View Article : Google Scholar
|