Recent research has revealed that RNA-binding
proteins (RBPs) have a special function in regulating the contents
of exosomes. RBPs are a diverse class of proteins capable of
binding to RNA molecules, including mRNAs, microRNAs (miRNAs/miRs)
and others (4). This family of
proteins consists of >2,000 members and plays a critical role in
all aspects of RNA-driven processes, ranging from RNA transcription
and maturation to translation processes (17,18).
RBPs affect these processes by forming single protein-RNA element
interactions or recruiting multiple RBPs to form protein-RNA
complexes. Critical RBP functions include the regulation of RNA
metabolism, including RNA splicing (19), mRNA stability (20), translation process to proteins
(21), intracellular localization
(22) and co-operation with
non-coding RNAs (23,24). The ectopic expression of RBPs
contributes to the development of various diseases, particularly in
tumorigenesis by stabilizing mRNAs or inducing alternative splicing
(25,26). Recently, RBPs were also reported to
be localized in EVs and to be associated with disease progression.
For example, insulin like growth factor 2 mRNA binding protein
(IGF2BP)1 has been reported to be overexpressed in EVs of
colorectal cancer and may serve as a biomarker for tumor diagnosis
(27). Mancarella et al
(28) found that IGF2BP3 affected
the miRNA cargo profile of EVs in Ewing sarcoma and then
contributed to cell migration by regulating the PI3K/Akt pathway in
neighboring cells. Furthermore, RBPs have been reported to regulate
the sorting process of RNA molecules into EVs. Zhang et al
(29) reported that hnRNP,
heterogeneous nuclear ribonucleoprotein (hnRNP)A1 regulated the
transfer of miR-522 to the EVs of cancer-associated fibroblast
(CAFs) and promoted the progression of gastric cancer. hnRNPA2B1
mediates the loading of lymph node metastasis-associated transcript
2 (LNMAT2) into bladder cancer cell-secreted EVs and promotes tumor
lymphatic metastasis (30).
Furthermore, it has been shown that hnRNPH1 is upregulated in
castration-resistant prostate cancer cells, and that the inhibition
of hnRNPH1 contributes to the reduced EV biogenesis and secretion
(31). These results indicate a
novel function of EV-associated RBPs in regulating tumorigenesis,
and these RBPs may serve as potential targets for cancer diagnosis
and treatment.
The present review aimed to provide an overview of
the current understanding of the role of EV-associated RBPs in
tumorigenesis. Specifically, the EV-transported RBPs and their
functions in recipient cells, the function of these EV-associated
RBPs in the sorting processes of RNAs into EVs, as well as the
specific function of these RBPs in tumorigenesis and their clinical
value in cancer diagnosis, are summarized herein. Finally, the
potential value of engineered EVs in cancer treatment is discussed.
The EVs discussed herein are referred to as exosomes, as they did
not involve apoptosis bodies. Information regarding apoptosis
bodies has been previously discussed (32–34).
Studies have reported that some RBPs can localize in
EVs and be transported to recipient cells, and subsequently affect
the biological process of recipient cells and contribute to
tumorigenesis (35–38). These are summarized in Table I.
First, the EVs deliver some RBPs to recipient cells,
which then induce oncogene expression. For example, it has been
found that Y-box binding protein 1 (YBX1) can be transferred by
gastric cancer exosomes and promote angiogenesis by enhancing the
expression of angiogenic factors in receipt vascular endothelial
cells (35). Wang et al
(36) reported that embryonic stem
cell-derived EVs containing hnRNPU were transferred into human
coronary artery endothelial cells, and hnRNPU promoted VEGF
expression in human coronary artery endothelial cells. In addition,
Qin et al (37)
demonstrated that gallbladder cancer cell-derived EVs promoted
macrophage M2 polarization, induced the malignant behavior of
gallbladder cancer cells by carrying IGF2BP3 and increased the
expression level of p-STAT3.
Secondly, EVs transport RBPs to recipient cells and
regulate the stability of targeted mRNA. Fang et al
(38) reported that IGF2BP2 was
secreted by lung squamous cell carcinoma cell exosomes and absorbed
by endothelial cells, thereby improving the stability of
Fms-related receptor tyrosine kinase 4 mRNA, activating the
PI3K-Akt signaling pathway, and eventually promoting angiogenesis
and metastasis. Furthermore, colon cancer cell-derived EVs
containing human antigen R (HuR) have been found to promote the
proliferation of lung cells by stabilizing c-Myc mRNA and HuR
associated with lung metastasis in patients with colon cancer
(39). In addition, serum-derived
EVs have been shown to deliver hnRNPC to non-small lung cancer
cells, wherein hnRNPC recognizes the m6A modification of DLG
associated protein 5 mRNA, ultimately promoting cancer cell growth
and metastasis (40). In addition,
hnRNPA1 can be SUMOylated, and then packaged and transported to
lymphatic endothelial cells, thus stabilizing prospero homeobox 1
mRNA and promoting lymphangiogenesis and lymph node metastasis in
pancreatic cancer (41).
Finally, EVs can deliver some RBPs that function as
splicing factors to recipient cells and regulate the alterative
splicing of target genes. For example, Pavlyukov et al
(42) found that RBM11 RNA binding
motif protein 11 could be transferred by EVs and promoted the
malignancy of glioblastoma by switching the splicing of MDM2
homolog MDMX and cyclin D1. Moreover, Zhang et al (43) reported that multiple
myeloma-derived exosomes delivered splicing factor SWAP homolog
(SFRS8) into osteoclasts, and SFRS8 promoted multiple myeloma
malignancy and bone lesion by alterative splicing of
calcyclin-binding protein.
RBPs in EVs directly affect the functions of
recipient cells to promote tumor progression; however, recent
studies have also highlighted that the critical role of RBPs is to
determine the enrichment of selected RNA transcripts into EVs
(44–48). These RBPs include members of the
hnRNP family, YBX1, IGF2BPs and HuR. The functions of these
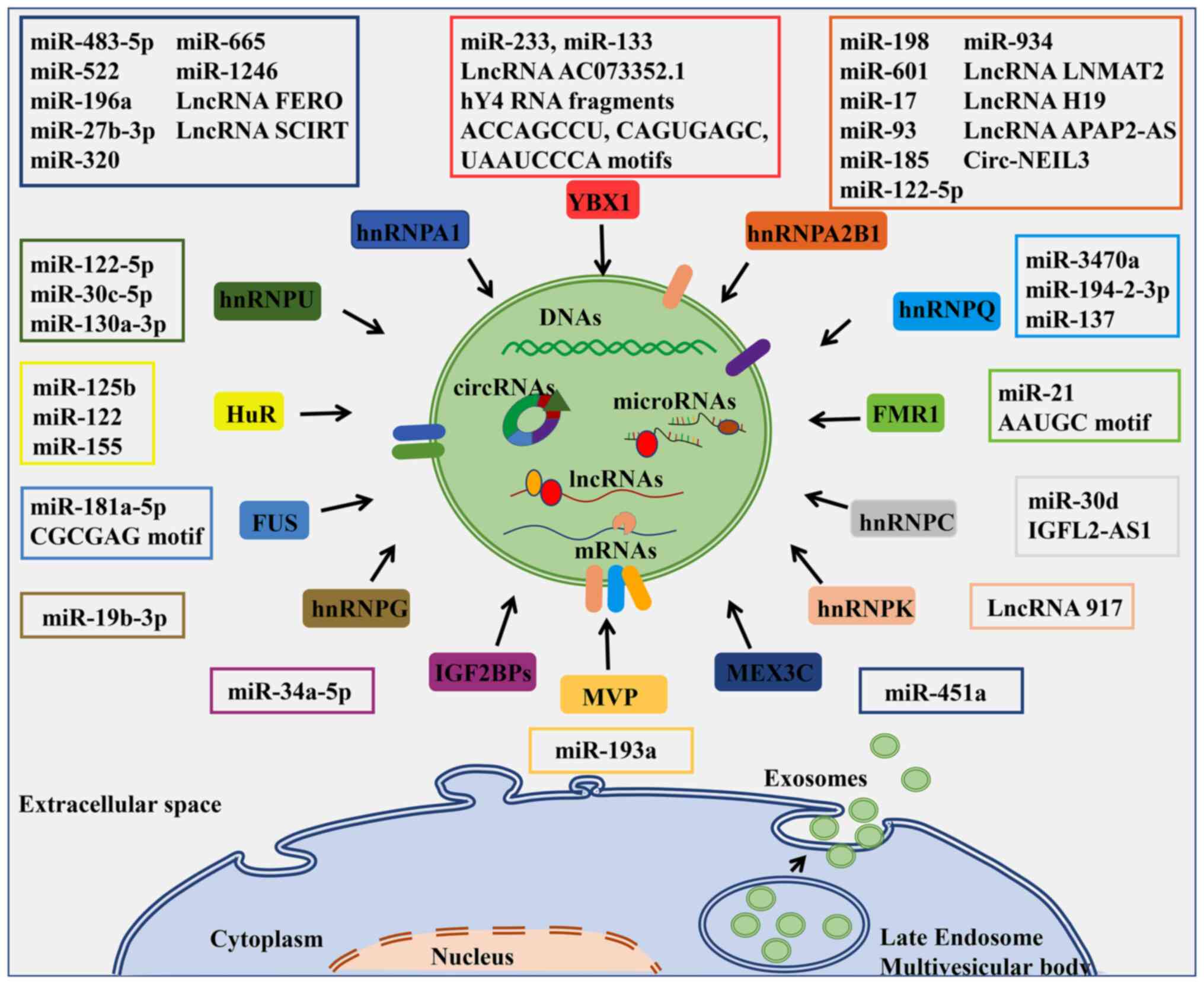
EV-localized RBPs in transcript sorting are summarized in Fig. 1 and Table II.
Human hnRNPs consist of 20 proteins with
differential RNA-binding capacities. The RNA recognition motif
(RRM) of hnRNPs allows them to bind with RNAs and modulate RNA
metabolisms. Several RBPs participate in the sorting process of
specific RNA molecules into EVs (44–48).
Firstly, hnRNPA2B1 has been reported to be
associated with the recruitment of RNA into EVs and to play a
crucial role in herpes simplex virus 1 release from infected cells
(44). In addition,
Villarroya-Beltri et al (45) revealed that hnRNPA2B1 controlled
the sorting of miR-198 and miR-601 into EVs by binding to specific
GGAG/UGCA motifs. The promotion of the sorting of miR-17 and miR-93
into EVs by hnRNPA2B1 has been shown to be dependent on its binding
with AGG/UAG motifs (46,47).
Secondly, hnRNPA1 is another key hnRNP involved in
packaging RNAs into EVs. It regulates the transfer of miR-27b-3p to
human umbilical vein endothelial cells, increasing blood vessel
permeability and generating circulating tumor cells (48). Other hnRNPs have also been reported
as a secreted factor in EVs. hnRNPQ has been reported to regulate
the exosomal sorting of miRNAs, such as miR-3470a and miR-194-2-3p
(49). Hobor et al
(50) demonstrated that the
N-terminal of hnRNPQ may mediate the recognition and exosomal
partitioning of miRNA targets. Kim et al (51) reported that hnRNPQ also regulated
the sorting of miR-137 into EVs in the dorsal striatum. Moreover,
Robinson et al (52)
reported that hnRPNK could recruit AsUGnA motif-containing miRNAs
and cause their release within EVs. Gao et al (53) revealed a physical interaction
between hnRNPK and lncRNA 91H, indicating that hnRNPK may mediate
the EV-induced sorting of lncRNA 91H. Leidal et al (54) reported the involvement of hnRNPK in
the specific LC3-conjugated EV loading and secretion machinery.
Furthermore, Statello et al (55) reported that hnRNPH1 facilitated the
transport of RNAs into EVs and the maintenance of RNAs inside EVs.
Hosen et al (56) reported
that hnRNPU encapsulated miR-122-5p into EVs and then regulated the
viability and apoptosis of cardiomyocytes. Balaguer et al
(57) reported that hnRNPC1 may
control miR-30d levels in endometrial exosomes.
YBX1, also known as YB1, is a protein that functions
as both a DNA and RBP. Recent studies have revealed that it plays a
crucial role in the regulation of mRNA packaging into EVs (58–61).
Kossinova et al (58)
demonstrated that YBX-1 binds specifically to potential RNA sorting
motifs, including ACCAGCCU, CAGUGAGC and UAAUCCCA in EVs derived
from 293 cells. Furthermore, Shurtleff et al (62) found that YBX-1 was required for the
sorting of miRNAs into EVs, such as miR-233. They also reported
that YBX-1 mediates miRNA sorting in a phase separation-dependent
manner, promoting the local enrichment of YBX-1 and its cognate
RNAs, enabling their targeting and packaging by vesicles (63).
IGF2BPs consist of three members: IGF2BP1, IGF2BP2
and IGF2BP3. The RRM domains and hnRNPK homology domains allow them
to bind with RNAs (64). Recently,
Chen et al (65) reported
that EVs carry distinct proteo-transcriptomic signatures, including
IGF2BP2, that differ from their cancer cell of origin. Mizutani
et al (66) also revealed
the interaction between IGF2BP3 and exosome using
co-immunoprecipitation analysis, indicating the potential role of
IGF2BPs in sorting of RNAs into EVs. IGF2BP3 is involved in the
methyltransferase 3, N6-adenosine-methyltransferase complex
catalytic subunit-mediated m6A modification of pre-miR-34A and the
secretion of exosomal miR-34a-5p in mesenchymal stem cells
(67).
The human FET family of RBPs includes FUS, TATA-box
binding protein associated factor 15 and EWS RNA-binding protein 1
(68), with FUS containing a
Gly-rich domain, an RRM domain, two arginine/glycine-rich domains
and a zinc finger motif. It has been reported that FUS is present
in amyotrophic lateral sclerosis muscle vesicles and can induce
cellular toxicity in recipient cells (69). Additionally, FUS has been found in
EVs and can bind to RNAs containing enriched GUGGU or GUU motifs
(70,71), suggesting its role in RNA exosomal
sorting. Recently, Garcia-Martin et al (72) demonstrated that FUS and Aly/REF
export factor (ALYREF) were involved in the exporting miRNAs
carrying CGGGAG motifs. FUS and ALYREF have been reported to
regulate the distribution of miRNAs into EVs, and this regulation
is associated with senescence and aging (73).
HuR belongs to the embryonic lethal abnormal vision
family of RBPs and has three RRM domains. It increases the
stability of target mRNAs by binding to the AU-rich elements. It
was recently reported that HuR may have a function in EVs, with
diabetic milieu stimulating HuR nuclear-to-cytoplasmic
translocation and EV transfer in cardiac- and cultured bone
marrow-derived macrophages (74).
HuR has also been shown to enhance the exosomal export of miR-125b
in response to ultraviolet irradiation (75) and accelerate the EV-mediated export
of miR-122 in starved human hepatic cells (76). Furthermore, Li et al
(77) developed a novel strategy
for enhanced RNA cargo encapsulation into engineered EVs using the
fused CD-9-HuR to successfully enrich miR-155 into EVs. In
addition, lysosome-associated membrane protein 2 (LAMP2)-HuR fusion
protein-engineered exosomes have been shown to recruit specific RNA
to lysosomes for targeted degradation (78).
Fragile X messenger ribonucleoprotein 1 (FMR1) is an
RBP involved in mRNA transport from the nucleus to the cytoplasm.
Recently, Wozniak et al (79) found that FMR1 controlled the EV
loading of miRNAs with the AAUGC motif during inflammation. Muscle
excess 3 RNA binding family member C has been found to promote the
exosomal sorting of miR-451a (80). Major vault protein (MVP) has been
shown to facilitate the transport of RNAs into EVs (55). Luo et al (81) found that MVP expression was higher
in astrocytes than in neurons and regulated the sorting of miRNAs
with a GUAC motif into astrocytic EVs. These data suggest that RBPs
play key roles in mediating RNA exosomal sorting and indirectly
affect the function of recipient cells.
RBPs in tumor cells regulate the cargo sorting of
RNA molecules into tumor derived EVs. For example, Li et al
(82) found that hnRNPA2B1
mediated the sorting of miR-122-5p into lung cancer cell-derived
EVs, promoting tumor progression. hnRNPA2B1 has also been shown to
mediate the packaging of miR-934 into EVs of in cancer cells,
inducing macrophage M2 polarization and liver metastasis (83). Furthermore, hnRNPA2B1 regulates the
loading of LNMAT2 into bladder cancer cell-secreted EVs, promoting
lymphatic metastasis (30). Zheng
et al (84) revealed that
hnRNPA2B1 mediated the secretion of lncRNA AGAP2 antisense RNA 1
outside of cells by EVs, inhibiting trastuzumab-induced cell
cytotoxicity in breast cancer cells. hnRNPA2B1 has also been found
to mediate the packaging process of lncRNA H19 into tumor-derived
EVs, inducing gefitinib resistance in non-small cell lung cancer
(NSCLC) (85). Moreover, hnRNPA2B1
has been shown to promote the packaging of circ-nei like DNA
glycosylase 3 into EVs, which are then transmitted to infiltrated
tumor-associated macrophages, inducing immunosuppressive properties
and glioma progression (86). In
addition, hnRNPA2B1 mediates the EV secretion of circ-cell cycle
and apoptosis regulator 1 of hepatocellular carcinoma cells and
promotes CD8+ T-cell dysfunction and anti-PD1 resistance
(87). Wang et al (88) reported that hnRNPA2B1 regulated the
enrichment of miR-378a-3p in tumor-derived EVs and then promoted
bone metastasis of prostate cancer by activating the
Dyrk1a/Nfatc1/Angptl2 axis in bone marrow macrophages.
It has been demonstrated that hnRNPA1 expression is
upregulated by chemotoxicity and is involved in
ferroptosis-associated lncRNA packaging into EVs, leading to
enhanced stemness and acquired chemoresistance in gastric cancer
cells (89). Zheng et al
(90) found that hnRNPA1 regulated
the sorting of lnc-brain cytoplasmic RNA 1 into tumor-derived
exosomes and contributed to lymphatic metastasis of bladder cancer
by activating the WNT5A/VEGF-C/VEGFR3 axis. Moreover, Jiang et
al (91) reported that long
intergenic non-protein coding RNA, regulator of reprogramming was
packaged into EVs in an hnRNPA1-dependent manner and then
disseminated the docetaxel resistance phenotype to receipt cells in
prostate cancer. Furthermore, Wang et al (92) demonstrated that hnRNPA1 assisted
the exosomal loading of lncRNA stem cell inhibitory RNA transcript
and miR-665 in lung cancer cells, promoting cancer cell metastasis.
Moreover, hnRNPA1 has been shown to promote the selective packaging
of miR-1246 into glioma-derived EVs, driving the differentiation
and activation of myeloid-derived suppressor cells (93).
hnRNPG promotes the packaging of miR-19b-3p into
lung adenocarcinoma cell-derived exosomes, facilitating M2
macrophage polarization and tumor metastasis (94). Pan et al (95) reported that hnRNPC regulated the
packaging of IGFL2 antisense RNA 1 (IGFL2-AS1) into EVs and
contributed to the sunitinib resistance in renal cell carcinoma.
Moreover, Guo et al (96)
found that hnRNPC contributed to the exosome transfer of ANL-210 to
macrophages, and subsequently promoted macrophage polarization and
stimulated the growth of head and neck squamous cell carcinoma.
Furthermore, YBX1 has been shown to promote the metastasis and
angiogenesis of breast cancer by binding and packaging the lncRNA
AC073352.1 into EVs (97). Li
et al (98) demonstrated
that YBX1 promoted the progression of lung cancer by selectively
sorting hY4 RNA fragments into EVs. Ghoshal et al (99) suggested that IGF2BP1 was intimately
involved in regulating the cargo of EVs, affecting the
pro-metastatic function of melanoma-derived EVs. Of note, Latifkar
et al (100) recently
reported that IGF2BP2 promoted tumor cell survival and invasiveness
by increasing the release of EVs enriched in ubiquitinated protein
cargo and soluble hydrolases. Furthermore, FUS has been shown to
mediate the packaging of miR-181a-5p into colorectal cancer
cell-derived EVs, which in turn persistently activate hepatic
stellate cells, remodeling the tumor microenvironment and promoting
liver metastasis (101). In
addition, Teng et al (102) revealed that MVP regulated the
exosomal sorting of miR-193a and promoted colon cancer
progression.
In the tumor microenvironment, EVs mediate
communications among cancer cells, tumor-associated fibroblasts,
tumor-associated macrophages, tumor-associated endothelial cells
and tumor-associated adipocytes (103). Therefore, in addition to RBPs in
tumor cells, RBPs in tumor-associated cells can also affect the
functions of tumor cells by regulating RNA sorting into EVs
originating from the tumor microenvironment. A previous study
demonstrated that the exosomal transfer of miR-185 from vascular
smooth muscle cells to endothelial cells was controlled by
hnRNPA2B1 (104). Furthermore,
hnRNPA1 has been found to mediate the exosomal sorting of
miR-483-5p out of renal tubular epithelial cells (105), and to promote the transfer of
miR-522 into the EVs of CAFs, suppressing ferroptosis and promoting
chemotherapy resistance in gastric cancer (29). Furthermore, hnRNPA1 facilitates the
packaging of miR-196a into CAF-derived EVs, conferring cisplatin
resistance in head and neck cancer by regulating cyclin dependent
kinase inhibitor 1B and inhibitor of growth family member 5
(106). hnRNPA1 has also been
found to be involved in the regulation of the exosomal transfer of
miR-320 from leukemia cells to bone marrow stromal cells and is a
critical mediator of leukemia progression (107). Moreover, hnRNPU has been reported
to retain miR-30c-5p, miR-130a-3p and other miRNAs, preventing
their export into large EVs in endothelial cells (108,109). YBX-1 mediates the sorting of
miR-133 into EVs derived from hypoxia/reoxygenation-induced human
endothelial progenitor cells, which increases fibroblast
angiogenesis and mesenchymal-endothelial transition (110). Furthermore, Shaban et al
(111) reported that the
expression of miR-21 in EVs from senescent endothelial cells was
associated with elevated FMR1 expression. In addition, Brossa et
al (112) reported that RBP
Annexin A2H existed on the surface of EVs isolated from human liver
stem cells. Annexin A2H bound to miR-145, protecting it from
ribonuclease digestion, and then more effectively inhibited the
invasive properties of cancer stem cells (112).
As aforementioned, RBPs can be delivered by EVs to
recipient cells and can play critical roles in non-coding RNA
sorting in EVs, contributing to tumorigenesis. The contained RBPs
are derived from tumor cells or other tumor
microenvironment-associated cells, and these characteristics
suggest the possibility of the development of EV-RBP-based
strategies for diagnosis and therapy. Tumor-derived EVs containing
specific RBPs could function as novel cancer biomarkers, and
targeting the RBP-mediated RNA molecule sorting process may be
beneficial for cancer treatment.
For a number of years, studies focused on non-coding
RNAs of EVs as novel biomarkers for cancer diagnosis. However, the
critical function of exosomal proteins has been neglected (113). Melo et al (114) identified glypican-1, a cell
surface proteoglycan, specifically enriched on cancer cell-derived
EVs, which may serve as a potential non-invasive diagnostic marker
for the detection of early stages of pancreatic cancer. Recently,
using liquid chromatography-mass spectrometry, Yeung et al
(115) found that
circadian-synchronized tendon fibroblasts release small EVs
enriched in RBPs (115).
Furthermore, Uzbekova et al (116) found numerous RBPs within
follicular fluid EVs that originated from follicular and other
cells; the different expression patterns of these RBPs may affect
oocyte competence. Kuhn et al (27) reported that IGF2BP1 could directly
enter EVs in a transformation-dependent manner, regulate the
progression of colorectal cancer and serve as a
diagnostic/prognostic circulating tumor biomarker. Xing et
al (117) analyzed the
proteins of circulating tumor-derived EVs from 50 µl serum and
revealed the potential application of nucleolin+ EVs for
nasopharyngeal carcinoma cancer diagnosis. Zhang et al
(118) found that poly(A) binding
protein cytoplasmic 1 (PABPC1) bound to miR-21-5p via an ACUGAUG
sequence to direct miR-21-5p packaging into EVs, and that an
elevated PABPC1 expression was associated with tumor cell
differentiation and a poor prognosis of patients. Furthermore,
tumor-secreted proteins are often degraded or diluted in the
circulating blood. However, these proteins can be well-enriched and
protected within tumor-derived EVs, enabling the easy detection and
in-depth analysis of relevant tumors (119,120). Proteins in EVs can be more
straightforward and representative, and may provide comprehensive
information about the distal primary parent tumor cells compared
with miRNAs (113,121). These features suggest that
proteins in EVs have potential value for use in disease diagnosis.
These proteins and their potential clinical applications in cancer
diagnosis and target treatments are summarized in Table III.
It has been well-illustrated that EVs can function
as novel delivery platforms of RNAs, particularly miRNAs and/or
siRNAs for cancer therapy. However, their loading efficiency is
limited. Recent studies have reported that the loading efficiency
of EVs can be improved by constructing engineered EVs with a fusion
protein of exosomal membrane proteins and RBPs that select and sort
specific RNAs into EVs. For example, Li et al (77) fused the exosomal membrane protein
CD9 with the RBP HuR, and successfully enriched miR-155, functional
miRNA inhibitor, or CRISPR/dCas9, which transport the AU-rich
elements into EVs. They also reported that HuR could be fused to
the C-terminus of LAMP2B (77).
Another study demonstrated that EVs engineered with LAMP2B-HuR
successfully decreased the abundance of RNA targets and
significantly reduced liver fibrosis in a mouse model of
CCl4-induced liver injury (78).
Furthermore, EVs engineered with MS2 bacteriophage coat protein, a
fusion protein, have been shown to successfully enrich the low
density lipoprotein receptor (LDLR) mRNA into EVs and efficiently
deliver LDLR to the liver cells, restore LDLR expression and
ameliorate the phenotype of high LDL cholesterol, atherosclerosis
and steatosis in the LDLR−/− mouse model (122). Wang et al (123) demonstrated that aptamer
AS1411-modified EVs could deliver lethal-7 to breast cancer cells
and inhibit cell proliferation by targeting binding nucleolin,
which is highly expressed on the surface membrane of breast cancer
cells. Furthermore, Es-Haghi et al (124) developed a fusion protein of the
EV membrane protein CD9 and the RBP argonaute RISC catalytic
component 2 (AGO2), and revealed that the engineered EVs exhibited
significantly higher levels of miRNA or short hairpin RNA (shRNA;
miR-466c or shRNA-451, respectively). These results suggest that
RBP-fusion protein-engineered EVs can potentially resolve the issue
of the inefficient endogenous loading of cargo and have significant
value for the future development of targeted cancer treatment.
As described above, RBPs play a crucial role in the
sorting of non-coding RNAs into EVs. It has also been proven that
RBPs play a critical role in the formation of circRNAs and
maturation of miRNAs, and then these non-coding RNAs are sorted
into EVs. Recent studies have demonstrated that some small
molecules can inhibit the functions of RBPs, which may serve as a
potential strategy for cancer treatment. For example, MO-460 can
bind to the C-terminal glycine-rich domain of hnRNPA2B1 and inhibit
its binding with targeted transcripts (125). Pérez-Boza et al (126) reported that epirubicin disrupted
the interaction between hnRNPA2B1 and miR-503, thereby affecting
the exosomal sorting of miR-503. In addition, the expression of
hnRNPA1 has been shown to be suppressed by the natural compound,
quercetin (127), and the
ubiquitin-proteasome-dependent degradation of hnRNPK has been found
to be accelerated by the natural compound, nujiangexathone A
(128). Furthermore, it has been
demonstrated that the HuR inhibitor, MS-444, inhibits HuR
dimerization and blocks the nucleocytoplasmic transport of targeted
mRNA (129,130). Wallis et al (131) reported that a small molecule
inhibited the binding of IGF2BP1 with the target mRNAs by
interacting with the hydrophobic surface at the boundary of IGF2BP1
KH3 and KH4 domains.
On the other hand, certain RNA molecules can
competitively bind with RBPs and subsequently suppress their
carcinogenic function. Yu et al (132) reported that circ-TNPO3
competitively interacted with IGF2BP3; thus, the role of IGF2BP3 in
stabilizing MYC mRNA was weakened, which inhibited the expression
of MYC and its target snail family transcriptional repressor 1,
thereby suppressing the proliferation and metastasis of GC. It has
been reported that circ_0000079 decoys FMR1 autosomal homolog 1
(FXR1) to interrupt the formation of the FXR1/protein kinase C,
iota complex, and suppresses the cell invasion and drug resistance
of NSCLC (133). Wang et
al (134) found that the
administration of RNA decoys specifically targeting YB-1 in a mouse
xenograft model of glioblastoma resulted in slower tumor growth and
an improved survival. Furthermore, Barbagallo et al
(135) identified a GAUGAA motif
which could function as a decoy for serine and arginine rich
splicing factor (SRSF)1, decrease the binding between SRSF2 and
tumor suppressor circ-SWI/SNF related, matrix associated, actin
dependent regulator of chromatin, subfamily A, member 5, and could
subsequently regulate the migration and angiogenesis of
glioblastoma multiforme. These results indicate that both RBP
inhibitors and RBP decoy nucleic acids can suppress the function of
RBPs and may be used for targeted cancer treatment.
The number of studies on EVs have markedly
increased over the last decade since they were identified in the
1980s (1). EVs have been proven to
play a crucial role in cellular communication and regulate the
phenotype of recipient cells by delivering specific contents. A
number of studies have focused on non-coding RNAs in EVs and the
regulatory mechanisms whereby cells can selectively control their
non-coding RNA cargo. According to current research, there are four
potential modes of miRNA sorting into EVs: The neural
sphingomyelinase 2-dependent pathway, the miRNA-induced silencing
complex-related pathway, the 3′ miRNA sequence-dependent pathway,
and the miRNA motif-dependent pathway (136,137). However, the specifics of these
mechanisms remain largely unclear. Studies have demonstrated that
RBPs play a critical role in selectively sorting non-coding RNAs
and facilitating their transfer into EVs (45,72).
Of note, the selective shuttling of non-coding RNAs into EVs
directly influences the pathological process of various diseases.
Moreover, the dysregulation of RBPs has been proven to be
associated with the development of diseases, particularly cancer
(138). These studies have
indicated that RBPs may influence the pathological process of
disease by regulating the selective sorting of non-coding RNAs into
EVs (136–138).
RBPs can be used for cancer diagnosis and targeted
cancer treatment due to their specific functions in EVs. First,
RBPs in EVs can provide more straightforward, representative and
comprehensive information about the distal primary parent tumor
cells, as compared with non-coding RNAs. These proteins are
enriched and protected in EVs, enabling the easy detection and
in-depth analysis of relevant tumors, and thus rendering them ideal
biomarkers for cancer diagnosis (65). Hu et al (113) reported that exosomal proteins
have potential value as novel cancer biomarkers for liquid biopsy.
Fuji et al (139) revealed
that the detection of serum-derived AGO2 exosomes could monitor the
tumor dynamics of colorectal cancer patients during chemotherapy.
Secondly, the sorting of tumor suppressor non-coding RNAs could be
modulated by the overexpression of specific RBPs, and the sorting
of oncogenic non-coding RNAs could be suppressed by RBPs
inhibitors. Furthermore, a number of RNA motifs have been reported
to be recognized by RBPs and sorted into EVs, which can be used for
RNA interference-dependent gene therapy. In addition, the loading
efficiency could be improved by constructing engineered EVs in
which a special RBP is overexpressed.
In conclusion, the present review underlines the
current knowledge of RBPs in EVs, and discusses their potential
value in cancer diagnosis and target treatment. RBPs have a
specific function in regulating the EV sorting of RNA molecules and
affecting tumorigenesis, and have notable clinical value for cancer
diagnosis and treatment. Specifically, RBPs in EVs have potential
applications for liquid biopsy. To the best of our knowledge, this
topic is novel, and herein, EVs, RBPs and tumorigenesis were
discussed in combination for the first time. However, their
functions and regulatory mechanisms remain complex and are not yet
completely understood. Further studies will hopefully enable their
clinical use. It is considered that following further research,
more accurate conclusions will be drawn.
Not applicable.
The present study was supported by funds from the National
Natural Sciences Foundation of China (grant no. 82003126), the
Sichuan Science and Technology Program (grant no. 2022NSFSC1368),
the Shenzhen High-level Hospital Construction Fund (grant no.
1801024), the Shenzhen Science and Technology Projects (grant nos.
JCYJ20190806165001761, JCYJ20210324103604013 and
JCYJ20190807102601647), the Luzhou Science and Technology Program
(grant no. 2021-JYJ-71) and the Scientific Research Foundation of
Southwest Medical University (grant no. 2021ZKMS009).
Not applicable.
WeichaoS was responsible for writing the original
draft, writing, reviewing and editing the final manuscript, and
funding acquisition. HC was responsible for writing the original
draft and funding acquisition. TX revised the manuscript, tables
and figure. JY, JL and WY were responsible for editing the
manuscript. WeiS was responsible for the conceptualization,
writing, reviewing and editing of the manuscript. QY was
responsible for the conceptualization, writing, reviewing and
editing of the manuscript, and funding acquisition. WeichaoS and HC
contributed equally to the study. All authors have read and
approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Meng W, He C, Hao Y, Wang L, Li L and Zhu
G: Prospects and challenges of extracellular vesicle-based drug
delivery system: Considering cell source. Drug Deliv. 27:585–598.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Görgens A, Bremer M, Ferrer-Tur R, Murke
F, Tertel T, Horn PA, Thalmann S, Welsh JA, Probst C, Guerin C, et
al: Optimisation of imaging flow cytometry for the analysis of
single extracellular vesicles by using fluorescence-tagged vesicles
as biological reference material. J Extracell Vesicles.
8:15875672019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang T, Song C, Zheng L, Xia L, Li Y and
Zhou Y: The roles of extracellular vesicles in gastric cancer
development, microenvironment, anti-cancer drug resistance, and
therapy. Mol Cancer. 18:622019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yi Q, Deng Z, Yue J, He J, Xiong J and Sun
W and Sun W: RNA binding proteins in osteoarthritis. Front Cell Dev
Biol. 10:9543762022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeppesen DK, Fenix AM, Franklin JL,
Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q,
Evans R, et al: Reassessment of exosome composition. Cell.
177:428–445.e18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sil S, Dagur RS, Liao K, Peeples ES, Hu G,
Periyasamy P and Buch S: Strategies for the use of extracellular
vesicles for the delivery of therapeutics. J Neuroimmune Pharmacol.
15:422–442. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schiera G, Di Liegro CM and Di Liegro I:
Extracellular membrane vesicles as vehicles for brain cell-to-cell
interactions in physiological as well as pathological conditions.
Biomed Res Int. 2015:1529262015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang C, Wang XY, Zhang P, He TC, Han JH,
Zhang R, Lin J, Fan J, Lu L, Zhu WW, et al: Cancer-derived exosomal
HSPC111 promotes colorectal cancer liver metastasis by
reprogramming lipid metabolism in cancer-associated fibroblasts.
Cell Death Dis. 13:572022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qiu Y, Yang Y, Yang R, Liu C, Hsu JM,
Jiang Z, Sun L, Wei Y, Li CW, Yu D, et al: Activated T cell-derived
exosomal PD-1 attenuates PD-L1-induced immune dysfunction in
triple-negative breast cancer. Oncogene. 40:4992–5001. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang Y, Duan L, Lu J and Xia J:
Engineering exosomes for targeted drug delivery. Theranostics.
11:3183–3195. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andreu Z and Yáñez-Mó M: Tetraspanins in
extracellular vesicle formation and function. Front Immunol.
5:4422014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang W, Ou X and Wu X: Proteomics
profiling of plasma exosomes in epithelial ovarian cancer: A
potential role in the coagulation cascade, diagnosis and prognosis.
Int J Oncol. 54:1719–1733. 2019.PubMed/NCBI
|
|
13
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: The new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seimiya T, Otsuka M, Iwata T, Shibata C,
Tanaka E, Suzuki T and Koike K: Emerging roles of exosomal circular
RNAs in cancer. Front Cell Dev Biol. 8:5683662020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin LY, Yang L, Zeng Q, Wang L, Chen ML,
Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, et al: Tumor-originated
exosomal lncUEGC1 as a circulating biomarker for early-stage
gastric cancer. Mol Cancer. 17:842018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou B, Mo Z, Lai G, Chen X, Li R, Wu R,
Zhu J and Zheng F: Targeting tumor exosomal circular RNA cSERPINE2
suppresses breast cancer progression by modulating MALT1-NF-κB-IL-6
axis of tumor-associated macrophages. J Exp Clin Cancer Res.
42:482023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bohnsack KE and Bohnsack MT: RNA-binding
proteins chaperone ribonucleoprotein complex assembly to solve the
RNA-folding problem. Cell. 179:1248–1250. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seufert L, Benzing T, Ignarski M and
Müller RU: RNA-binding proteins and their role in kidney disease.
Nat Rev Nephrol. 18:153–170. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rachez C, Legendre R, Costallat M, Varet
H, Yi J, Kornobis E and Muchardt C: HP1γ binding pre-mRNA intronic
repeats modulates RNA splicing decisions. EMBO Rep. 22:e523202021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Wang X, Zhang X, Wang J, Ma Y,
Zhang L and Cao X: RNA-binding protein YTHDF3 suppresses
interferon-dependent antiviral responses by promoting FOXO3
translation. Proc Natl Acad Sci USA. 116:976–981. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo L, Kim HJ, Wang H, Monaghan J,
Freyermuth F, Sung JC, O'Donovan K, Fare CM, Diaz Z, Singh N, et
al: Nuclear-import receptors reverse aberrant phase transitions of
RNA-binding proteins with prion-like domains. Cell.
173:677–692.e20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yao ZT, Yang YM, Sun MM, He Y, Liao L,
Chen KS and Li B: New insights into the interplay between long
non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun
(Lond). 42:117–140. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Corley M, Burns MC and Yeo GW: How
RNA-binding proteins interact with RNA: Molecules and mechanisms.
Mol Cell. 78:9–29. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liao Y, Feng J, Sun W, Wu C, Li J, Jing T,
Liang Y, Qian Y, Liu W and Wang H: CIRP promotes the progression of
non-small cell lung cancer through activation of Wnt/β-catenin
signaling via CTNNB1. J Exp Clin Cancer Res. 40:2752021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ullah I, Liao Y, Wan R, Tang L and Feng J:
Alternative splicing of SMAD4 and its function in HaCaT cells in
response to UVB irradiation. J Cancer. 9:3177–3186. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kuhn M, Zhang Y, Favate J, Morita M,
Blucher A, Das S, Liang S, Preet R, Parham LR, Williams KN, et al:
IMP1/IGF2BP1 in human colorectal cancer extracellular vesicles. Am
J Physiol Gastrointest Liver Physiol. 323:G571–G585. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mancarella C, Giusti V, Caldoni G,
Laginestra MA, Parra A, Toracchio L, Giordano G, Roncuzzi L, Piazzi
M, Blalock W, et al: Extracellular vesicle-associated IGF2BP3 tunes
Ewing sarcoma cell migration and affects PI3K/Akt pathway in
neighboring cells. Cancer Gene Ther. Jun 23–2023.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang H, Deng T, Liu R, Ning T, Yang H,
Liu D, Zhang Q, Lin D, Ge S, Bai M, et al: CAF secreted miR-522
suppresses ferroptosis and promotes acquired chemo-resistance in
gastric cancer. Mol Cancer. 19:432020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu
H, Zhong G, Li Y, Li J, Huang J, et al: Exosomal long noncoding RNA
LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin
Invest. 130:404–421. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Datta A, Kim H, Lal M, McGee L, Johnson A,
Moustafa AA, Jones JC, Mondal D, Ferrer M and Abdel-Mageed AB:
Manumycin A suppresses exosome biogenesis and secretion via
targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in
castration-resistant prostate cancer cells. Cancer Lett. 408:73–81.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X and Liu Y, Liu X, Du J, Bhawal UK, Xu
J, Guo L and Liu Y: Advances in the therapeutic effects of
apoptotic bodies on systemic diseases. Int J Mol Sci. 23:82022022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu X, Lai Y and Hua ZC: Apoptosis and
apoptotic body: Disease message and therapeutic target potentials.
Biosci Rep. 39:BSR201809922019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akers JC, Gonda D, Kim R, Carter BS and
Chen CC: Biogenesis of extracellular vesicles (EV): Exosomes,
microvesicles, retrovirus-like vesicles, and apoptotic bodies. J
Neurooncol. 113:1–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xue X, Huang J, Yu K, Chen X, He Y, Qi D
and Wu Y: YB-1 transferred by gastric cancer exosomes promotes
angiogenesis via enhancing the expression of angiogenic factors in
vascular endothelial cells. BMC Cancer. 20:9962020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang H, Liu H, Zhao X and Chen X:
Heterogeneous nuclear ribonucleoprotein U-actin complex derived
from extracellular vesicles facilitates proliferation and migration
of human coronary artery endothelial cells by promoting RNA
polymerase II transcription. Bioengineered. 13:11469–11486. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qin Y, Zhang M, Lei H, Wu H, Huang C, Zhou
X, Fu Y, Weng M and Ma M: Knockdown of IGF2BP3 inhibits the
tumorigenesis of gallbladder cancer and modifies tumor
microenvironment. Drug Dev Res. 83:1831–1844. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fang H, Sun Q, Zhou J, Zhang H, Song Q,
Zhang H, Yu G, Guo Y, Huang C, Mou Y, et al: m6A
methylation reader IGF2BP2 activates endothelial cells to promote
angiogenesis and metastasis of lung adenocarcinoma. Mol Cancer.
22:992023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xiao H, Ye X, Vishwakarma V, Preet R and
Dixon DA: CRC-derived exosomes containing the RNA binding protein
HuR promote lung cell proliferation by stabilizing c-Myc mRNA.
Cancer Biol Ther. 23:139–149. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi S, Wu T, Ma Z, Zhang X, Xu K, Tian Q,
Gao L, Yin X, Xu S and Yang S: Serum-derived extracellular vesicles
promote the growth and metastasis of non-small cell lung cancer by
delivering the m6A methylation regulator HNRNPC through the
regulation of DLGAP5. J Cancer Res Clin Oncol. 149:4639–4651. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Luo Y, Li Z, Kong Y, He W, Zheng H, An M,
Lin Y, Zhang D, Yang J, Zhao Y, et al: KRAS mutant-driven
SUMOylation controls extracellular vesicle transmission to trigger
lymphangiogenesis in pancreatic cancer. J Clin Invest.
132:e1576442022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pavlyukov MS, Yu H, Bastola S, Minata M,
Shender VO, Lee Y, Zhang S, Wang J, Komarova S, Wang J, et al:
Apoptotic cell-derived extracellular vesicles promote malignancy of
glioblastoma via intercellular transfer of splicing factors. Cancer
Cell. 34:119–135.e10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Yu X, Sun R, Min J, Tang X, Lin
Z, Xie S, Li X, Lu S, Tian Z, et al: Splicing factor
arginine/serine-rich 8 promotes multiple myeloma malignancy and
bone lesion through alternative splicing of CACYBP and
exosome-based cellular communication. Clin Transl Med.
12:e6842022.PubMed/NCBI
|
|
44
|
Zhou X, Wang L, Zou W, Chen X, Roizman B
and Zhou GG: hnRNPA2B1 associated with recruitment of RNA into
exosomes plays a key role in herpes simplex virus 1 release from
infected cells. J Virol. 94:e00367–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Villarroya-Beltri C, Gutiérrez-Vázquez C,
Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N,
Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M and
Sánchez-Madrid F: Sumoylated hnRNPA2B1 controls the sorting of
miRNAs into exosomes through binding to specific motifs. Nat
Commun. 4:29802013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu B, Su S, Patil DP, Liu H, Gan J,
Jaffrey SR and Ma J: Molecular basis for the specific and
multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun. 9:4202018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lee H, Li C, Zhang Y, Zhang D, Otterbein
LE and Jin Y: Caveolin-1 selectively regulates microRNA sorting
into microvesicles after noxious stimuli. J Exp Med. 216:2202–2220.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dou R, Liu K, Yang C, Zheng J, Shi D, Lin
X, Wei C, Zhang C, Fang Y, Huang S, et al: EMT-cancer cells-derived
exosomal miR-27b-3p promotes circulating tumour cells-mediated
metastasis by modulating vascular permeability in colorectal
cancer. Clin Transl Med. 11:e5952021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Santangelo L, Giurato G, Cicchini C,
Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A
and Tripodi M: The RNA-binding protein SYNCRIP is a component of
the hepatocyte exosomal machinery controlling MicroRNA sorting.
Cell Rep. 17:799–808. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hobor F, Dallmann A, Ball NJ, Cicchini C,
Battistelli C, Ogrodowicz RW, Christodoulou E, Martin SR, Castello
A, Tripodi M, et al: A cryptic RNA-binding domain mediates Syncrip
recognition and exosomal partitioning of miRNA targets. Nat Commun.
9:8312018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kim B, Tag SH, Nam E, Ham S, Ahn S, Kim J,
Cho DW, Lee S, Yang YS, Lee SE, et al: SYNCRIP controls miR-137 and
striatal learning in animal models of methamphetamine abstinence.
Acta Pharm Sin B. 12:3281–3297. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Robinson H, Ruelcke JE, Lewis A, Bond CS,
Fox AH, Bharti V, Wani S, Cloonan N, Lai A, Margolin D, et al:
Caveolin-1-driven membrane remodelling regulates hnRNPK-mediated
exosomal microRNA sorting in cancer. Clin Transl Med. 11:e3812021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gao T, Liu X, He B, Nie Z, Zhu C, Zhang P
and Wang S: Exosomal lncRNA 91H is associated with poor development
in colorectal cancer by modifying HNRNPK expression. Cancer Cell
Int. 18:112018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Leidal AM, Huang HH, Marsh T, Solvik T,
Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, et al: The
LC3-conjugation machinery specifies the loading of RNA-binding
proteins into extracellular vesicles. Nat Cell Biol. 22:187–199.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Statello L, Maugeri M, Garre E, Nawaz M,
Wahlgren J, Papadimitriou A, Lundqvist C, Lindfors L, Collén A,
Sunnerhagen P, et al: Identification of RNA-binding proteins in
exosomes capable of interacting with different types of RNA:
RBP-facilitated transport of RNAs into exosomes. PLoS One.
13:e01959692018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hosen MR, Goody PR, Zietzer A, Xiang X,
Niepmann ST, Sedaghat A, Tiyerili V, Chennupati R, Moore JB IV,
Boon RA, et al: Circulating MicroRNA-122-5p is associated with a
lack of improvement in left ventricular function after
transcatheter aortic valve replacement and regulates viability of
cardiomyocytes through extracellular vesicles. Circulation.
146:1836–1854. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Balaguer N, Moreno I, Herrero M, González
M, Simón C and Vilella F: Heterogeneous nuclear ribonucleoprotein
C1 may control miR-30d levels in endometrial exosomes affecting
early embryo implantation. Mol Hum Reprod. 24:411–425. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kossinova OA, Gopanenko AV, Tamkovich SN,
Krasheninina OA, Tupikin AE, Kiseleva E, Yanshina DD, Malygin AA,
Ven'yaminova AG, Kabilov MR and Karpova GG: Cytosolic YB-1 and
NSUN2 are the only proteins recognizing specific motifs present in
mRNAs enriched in exosomes. Biochim Biophys Acta Proteins Proteom.
1865:664–673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yanshina DD, Kossinova OA, Gopanenko AV,
Krasheninina OA, Malygin AA, Venyaminova AG and Karpova GG:
Structural features of the interaction of the 3′-untranslated
region of mRNA containing exosomal RNA-specific motifs with YB-1, a
potential mediator of mRNA sorting. Biochimie. 144:134–143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gopanenko AV, Malygin AA, Kossinova OA,
Tupikin AE, Kabilov MR and Karpova GG: Degenerate consensus
sequences in the 3′-untranslated regions of cellular mRNAs as
specific motifs potentially involved in the YB-1-mediated packaging
of these mRNAs. Biochimie. 170:152–162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shurtleff MJ, Yao J, Qin Y, Nottingham RM,
Temoche-Diaz MM, Schekman R and Lambowitz AM: Broad role for YBX1
in defining the small noncoding RNA composition of exosomes. Proc
Natl Acad Sci USA. 114:E8987–E8995. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shurtleff MJ, Temoche-Diaz MM, Karfilis
KV, Ri S and Schekman R: Y-box protein 1 is required to sort
microRNAs into exosomes in cells and in a cell-free reaction.
Elife. 5:e192762016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu XM, Ma L and Schekman R: Selective
sorting of microRNAs into exosomes by phase-separated YBX1
condensates. Elife. 10:e719822021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Publisher
correction: Recognition of RNA N6-methyladenosine by
IGF2BP proteins enhances mRNA stability and translation. Nat Cell
Biol. 22:12882020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen TY, Gonzalez-Kozlova E, Soleymani T,
La Salvia S, Kyprianou N, Sahoo S, Tewari AK, Cordon-Cardo C,
Stolovitzky G and Dogra N: Extracellular vesicles carry distinct
proteo-transcriptomic signatures that are different from their
cancer cell of origin. iScience. 25:1044142022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mizutani R, Imamachi N, Suzuki Y, Yoshida
H, Tochigi N, Oonishi T, Suzuki Y and Akimitsu N: Oncofetal protein
IGF2BP3 facilitates the activity of proto-oncogene protein eIF4E
through the destabilization of EIF4E-BP2 mRNA. Oncogene.
35:3495–3502. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li YJ, Xu QW, Xu CH and Li WM: MSC
promotes the secretion of exosomal miR-34a-5p and improve
intestinal barrier function through METTL3-mediated Pre-miR-34A
m6A modification. Mol Neurobiol. 59:5222–5235. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lindén M, Thomsen C, Grundevik P, Jonasson
E, Andersson D, Runnberg R, Dolatabadi S, Vannas C, Luna Santamarίa
M, Fagman H, et al: FET family fusion oncoproteins target the
SWI/SNF chromatin remodeling complex. EMBO Rep. 20:e457662019.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Le Gall L, Duddy WJ, Martinat C, Mariot V,
Connolly O, Milla V, Anakor E, Ouandaogo ZG, Millecamps S, Lainé J,
et al: Muscle cells of sporadic amyotrophic lateral sclerosis
patients secrete neurotoxic vesicles. J Cachexia Sarcopenia Muscle.
13:1385–1402. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kamelgarn M, Chen J, Kuang L, Arenas A,
Zhai J, Zhu H and Gal J: Proteomic analysis of FUS interacting
proteins provides insights into FUS function and its role in ALS.
Biochim Biophys Acta. 1862:2004–2014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lagier-Tourenne C, Polymenidou M, Hutt KR,
Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur
C, et al: Divergent roles of ALS-linked proteins FUS/TLS and TDP-43
intersect in processing long pre-mRNAs. Nat Neurosci. 15:1488–1497.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Garcia-Martin R, Wang G, Brandão BB,
Zanotto TM, Shah S, Kumar Patel S, Schilling B and Kahn CR:
MicroRNA sequence codes for small extracellular vesicle release and
cellular retention. Nature. 601:446–451. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Brázda V and Mergny JL: Quadruplexes and
aging: G4-binding proteins regulate the presence of miRNA in small
extracellular vesicles (sEVs). Biochimie. S0300-9084(23)00014-7.
2023.(Epub ahead of print). View Article : Google Scholar
|
|
74
|
Govindappa PK, Patil M, Garikipati VNS,
Verma SK, Saheera S, Narasimhan G, Zhu W, Kishore R, Zhang J and
Krishnamurthy P: Targeting exosome-associated human antigen R
attenuates fibrosis and inflammation in diabetic heart. FASEB J.
34:2238–2251. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Goswami B, Ahuja D, Pastré D and Ray PS:
p53 and HuR combinatorially control the biphasic dynamics of
microRNA-125b in response to genotoxic stress. Commun Biol.
6:1102023. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mukherjee K, Ghoshal B, Ghosh S,
Chakrabarty Y, Shwetha S, Das S and Bhattacharyya SN: Reversible
HuR-microRNA binding controls extracellular export of miR-122 and
augments stress response. EMBO Rep. 17:1184–1203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Z, Zhou X, Wei M, Gao X, Zhao L, Shi R,
Sun W, Duan Y, Yang G and Yuan L: In vitro and in vivo RNA
inhibition by CD9-HuR functionalized exosomes encapsulated with
miRNA or CRISPR/dCas9. Nano Lett. 19:19–28. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Li Z, Zhou X, Gao X, Bai D, Dong Y, Sun W,
Zhao L, Wei M, Yang X, Yang G and Yuan L: Fusion protein engineered
exosomes for targeted degradation of specific RNAs in lysosomes: A
proof-of-concept study. J Extracell Vesicles. 9:18167102020.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wozniak AL, Adams A, King KE, Dunn W,
Christenson LK, Hung WT and Weinman SA: The RNA binding protein
FMR1 controls selective exosomal miRNA cargo loading during
inflammation. J Cell Biol. 219:e2019120742020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lu P, Li H, Li N, Singh RN, Bishop CE,
Chen X and Lu B: MEX3C interacts with adaptor-related protein
complex 2 and involves in miR-451a exosomal sorting. PLoS One.
12:e01859922017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Luo X, Jean-Toussaint R, Sacan A and Ajit
SK: Differential RNA packaging into small extracellular vesicles by
neurons and astrocytes. Cell Commun Signal. 19:752021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li C, Qin F, Wang W, Ni Y, Gao M, Guo M
and Sun G: hnRNPA2B1-mediated extracellular vesicles sorting of
miR-122-5p potentially promotes lung cancer progression. Int J Mol
Sci. 22:128662021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu
Y, Zhang Z, Cai S, Xu Y, Li X, et al: Tumor-derived exosomal
miR-934 induces macrophage M2 polarization to promote liver
metastasis of colorectal cancer. J Hematol Oncol. 13:1562020.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zheng Z, Chen M, Xing P, Yan X and Xie B:
Increased expression of exosomal AGAP2-AS1 (AGAP2 antisense RNA 1)
in breast cancer cells inhibits trastuzumab-induced cell
cytotoxicity. Med Sci Monit. 25:2211–2220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lei Y, Guo W, Chen B, Chen L, Gong J and
Li W: Tumor-released lncRNA H19 promotes gefitinib resistance via
packaging into exosomes in non-small cell lung cancer. Oncol Rep.
40:3438–3446. 2018.PubMed/NCBI
|
|
86
|
Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q,
Zhang S, Zhao S, Xu H, Li M, et al: EWSR1-induced circNEIL3
promotes glioma progression and exosome-mediated macrophage
immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer.
21:162022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y,
Jiang J, Wang L, Mang Y, Gao Y, et al: Exosome-derived circCCAR1
promotes CD8 + T-cell dysfunction and anti-PD1 resistance in
hepatocellular carcinoma. Mol Cancer. 22:552023. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang J, Du X, Wang X, Xiao H, Jing N, Xue
W, Dong B, Gao WQ and Fang YX: Tumor-derived miR-378a-3p-containing
extracellular vesicles promote osteolysis by activating the
Dyrk1a/Nfatc1/Angptl2 axis for bone metastasis. Cancer Lett.
526:76–90. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhang H, Wang M, He Y, Deng T, Liu R, Wang
W, Zhu K, Bai M, Ning T, Yang H, et al: Chemotoxicity-induced
exosomal lncFERO regulates ferroptosis and stemness in gastric
cancer stem cells. Cell Death Dis. 12:11162021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zheng H, Chen C, Luo Y, Yu M, He W, An M,
Gao B, Kong Y, Ya Y, Lin Y, et al: Tumor-derived exosomal BCYRN1
activates WNT5A/VEGF-C/VEGFR3 feedforward loop to drive lymphatic
metastasis of bladder cancer. Clin Transl Med. 11:e4972021.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jiang X, Xu Y, Liu R and Guo S: Exosomal
lincROR promotes docetaxel resistance in prostate cancer through a
β-catenin/HIF1α positive feedback loop. Mol Cancer Res. 21:472–482.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang Z, Lin M, He L, Qi H, Shen J and Ying
K: Exosomal lncRNA SCIRT/miR-665 transferring promotes lung cancer
cell metastasis through the inhibition of HEYL. J Oncol.
2021:98137732021.PubMed/NCBI
|
|
93
|
Qiu W, Guo X, Li B, Wang J, Qi Y, Chen Z,
Zhao R, Deng L, Qian M, Wang S, et al: Exosomal miR-1246 from
glioma patient body fluids drives the differentiation and
activation of myeloid-derived suppressor cells. Mol Ther.
29:3449–3464. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen J, Zhang K, Zhi Y, Wu Y, Chen B, Bai
J and Wang X: Tumor-derived exosomal miR-19b-3p facilitates M2
macrophage polarization and exosomal LINC00273 secretion to promote
lung adenocarcinoma metastasis via Hippo pathway. Clin Transl Med.
11:e4782021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Pan Y, Lu X, Shu G, Cen J, Lu J, Zhou M,
Huang K, Dong J, Li J, Lin H, et al: Extracellular vesicle-mediated
transfer of LncRNA IGFL2-AS1 confers sunitinib resistance in renal
cell carcinoma. Cancer Res. 83:103–116. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Guo E, Mao X, Wang X, Guo L, An C, Zhang
C, Song K, Wang G, Duan C, Zhang X, et al: Alternatively spliced
ANLN isoforms synergistically contribute to the progression of head
and neck squamous cell carcinoma. Cell Death Dis. 12:7642021.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kong X, Li J, Li Y, Duan W, Qi Q, Wang T,
Yang Q, Du L, Mao H and Wang C: A novel long non-coding RNA
AC073352.1 promotes metastasis and angiogenesis via interacting
with YBX1 in breast cancer. Cell Death Dis. 12:6702021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Li C, Wang W, Sun Y, Ni Y, Qin F, Li X,
Wang T, Guo M and Sun G: Selective sorting and secretion of hY4 RNA
fragments into extracellular vesicles mediated by methylated YBX1
to promote lung cancer progression. J Exp Clin Cancer Res.
41:1362022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ghoshal A, Rodrigues LC, Gowda CP, Elcheva
IA, Liu Z, Abraham T and Spiegelman VS: Extracellular
vesicle-dependent effect of RNA-binding protein IGF2BP1 on melanoma
metastasis. Oncogene. 38:4182–4196. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Latifkar A, Wang F, Mullmann JJ, Panizza
E, Fernandez IR, Ling L, Miller AD, Fischbach C, Weiss RS, Lin H,
et al: IGF2BP2 promotes cancer progression by degrading the RNA
transcript encoding a v-ATPase subunit. Proc Natl Acad Sci USA.
119:e22004771192022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhao S, Mi Y, Zheng B, Wei P, Gu Y, Zhang
Z, Xu Y, Cai S, Li X and Li D: Highly-metastatic colorectal cancer
cell released miR-181a-5p-rich extracellular vesicles promote liver
metastasis by activating hepatic stellate cells and remodelling the
tumour microenvironment. J Extracell Vesicles. 11:e121862022.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Teng Y, Ren Y, Hu X, Mu J, Samykutty A,
Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, et al:
MVP-mediated exosomal sorting of miR-193a promotes colon cancer
progression. Nat Commun. 8:144482017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Clancy JW and D'Souza-Schorey C:
Tumor-derived extracellular vesicles: Multifunctional entities in
the tumor microenvironment. Annu Rev Pathol. 18:205–229. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Si Y, Liu F, Wang D, Fang C, Tang X, Guo
B, Shi Z, Dong Z, Guo D, Yue J and Fu W: Exosomal transfer of
miR-185 is controlled by hnRNPA2B1 and impairs
re-endothelialization after vascular injury. Front Cell Dev Biol.
9:6194442021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Liu D, Liu F, Li Z, Pan S, Xie J, Zhao Z
and Liu Z, Zhang J and Liu Z: HNRNPA1-mediated exosomal sorting of
miR-483-5p out of renal tubular epithelial cells promotes the
progression of diabetic nephropathy-induced renal interstitial
fibrosis. Cell Death Dis. 12:2552021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Qin X, Guo H, Wang X, Zhu X, Yan M, Wang
X, Xu Q, Shi J, Lu E, Chen W and Zhang J: Exosomal miR-196a derived
from cancer-associated fibroblasts confers cisplatin resistance in
head and neck cancer through targeting CDKN1B and ING5. Genome
Biol. 20:122019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Gao X, Wan Z, Wei M, Dong Y, Zhao Y, Chen
X, Li Z, Qin W, Yang G and Liu L: Chronic myelogenous leukemia
cells remodel the bone marrow niche via exosome-mediated transfer
of miR-320. Theranostics. 9:5642–5656. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zietzer A, Hosen MR, Wang H, Goody PR,
Sylvester M, Latz E, Nickenig G, Werner N and Jansen F: The
RNA-binding protein hnRNPU regulates the sorting of microRNA-30c-5p
into large extracellular vesicles. J Extracell Vesicles.
9:17869672020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zietzer A, Steffen E, Niepmann S, Düsing
P, Hosen MR, Liu W, Jamme P, Al-Kassou B, Goody PR, Zimmer S, et
al: MicroRNA-mediated vascular intercellular communication is
altered in chronic kidney disease. Cardiovasc Res. 118:316–333.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lin F, Zeng Z, Song Y, Li L, Wu Z, Zhang
X, Li Z, Ke X and Hu X: YBX-1 mediated sorting of miR-133 into
hypoxia/reoxygenation-induced EPC-derived exosomes to increase
fibroblast angiogenesis and MEndoT. Stem Cell Res Ther. 10:2632019.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Shaban SA, Rezaie J and Nejati V: Exosomes
derived from senescent endothelial cells contain distinct
pro-angiogenic miRNAs and proteins. Cardiovasc Toxicol. 22:592–601.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Brossa A, Tapparo M, Fonsato V,
Papadimitriou E, Delena M, Camussi G and Bussolati B: Coincubation
as miR-Loading strategy to improve the anti-tumor effect of stem
cell-derived EVs. Pharmaceutics. 13:762021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hu C, Jiang W, Lv M, Fan S, Lu Y, Wu Q and
Pi J: Potentiality of exosomal proteins as novel cancer biomarkers
for liquid biopsy. Front Immunol. 13:7920462022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Yeung CC, Dondelinger F, Schoof EM, Georg
B, Lu Y, Zheng Z, Zhang J, Hannibal J, Fahrenkrug J and Kjaer M:
Circadian regulation of protein cargo in extracellular vesicles.
Sci Adv. 8:eabc90612022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Uzbekova S, Almiñana C, Labas V,
Teixeira-Gomes AP, Combes-Soia L, Tsikis G, Carvalho AV, Uzbekov R
and Singina G: Protein cargo of extracellular vesicles from bovine
follicular fluid and analysis of their origin from different
ovarian cells. Front Vet Sci. 7:5849482020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Xing S, Lu Z, Huang Q, Li H, Wang Y, Lai
Y, He Y, Deng M and Liu W: An ultrasensitive hybridization chain
reaction-amplified CRISPR-Cas12a aptasensor for extracellular
vesicle surface protein quantification. Theranostics.
10:10262–10273. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhang Y, Chen C, Liu Z, Guo H, Lu W, Hu W
and Lin Z: PABPC1-induced stabilization of IFI27 mRNA promotes
angiogenesis and malignant progression in esophageal squamous cell
carcinoma through exosomal miRNA-21-5p. J Exp Clin Cancer Res.
41:1112022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhou B, Xu K, Zheng X, Chen T, Wang J,
Song Y, Shao Y and Zheng S: Application of exosomes as liquid
biopsy in clinical diagnosis. Signal Transduct Target Ther.
5:1442020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Li W, Li C, Zhou T, Liu X, Liu X, Li X and
Chen D: Role of exosomal proteins in cancer diagnosis. Mol Cancer.
16:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yang Z, Ji P, Li Z, Zhang R, Wei M, Yang
Y, Yuan L, Han Y and Yang G: Improved extracellular vesicle-based
mRNA delivery for familial hypercholesterolemia treatment.
Theranostics. 13:3467–3479. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Wang Y, Chen X, Tian B, Liu J, Yang L,
Zeng L, Chen T, Hong A and Wang X: Nucleolin-targeted extracellular
vesicles as a versatile platform for biologics delivery to breast
cancer. Theranostics. 7:1360–1372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Es-Haghi M, Neustroeva O, Chowdhury I,
Laitinen P, Väänänen MA, Korvenlaita N, Malm T, Turunen MP and
Turunen TA: Construction of fusion protein for enhanced small RNA
loading to extracellular vesicles. Genes (Basel). 14:2612023.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Soung NK, Kim HM, Asami Y, Kim DH, Cho Y,
Naik R, Jang Y, Jang K, Han HJ, Ganipisetti SR, et al: Mechanism of
the natural product moracin-O derived MO-460 and its targeting
protein hnRNPA2B1 on HIF-1α inhibition. Exp Mol Med. 51:1–14. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Pérez-Boza J, Boeckx A, Lion M, Dequiedt F
and Struman I: hnRNPA2B1 inhibits the exosomal export of miR-503 in
endothelial cells. Cell Mol Life Sci. 77:4413–4428. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Pham TND, Stempel S, Shields MA, Spaulding
C, Kumar K, Bentrem DJ, Matsangou M and Munshi HG: Quercetin
enhances the anti-tumor effects of BET inhibitors by suppressing
hnRNPA1. Int J Mol Sci. 20:42932019. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhang L, Feng J, Kong S, Wu M, Xi Z, Zhang
B, Fu W, Lao Y, Tan H and Xu H: Nujiangexathone A, a novel compound
from Garcinia nujiangensis, suppresses cervical cancer growth by
targeting hnRNPK. Cancer Lett. 380:447–456. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Blanco FF, Preet R, Aguado A, Vishwakarma
V, Stevens LE, Vyas A, Padhye S, Xu L, Weir SJ, Anant S, et al:
Impact of HuR inhibition by the small molecule MS-444 on colorectal
cancer cell tumorigenesis. Oncotarget. 7:74043–74058. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Nie Y, Xu W, Tian GG, Li X, Guo Y, Liu X,
He L, Shao Z, Li X and Wu J: Mechanistic insights into HuR
inhibitor MS-444 arresting embryonic development revealed by
low-input RNA-seq and STORM. Cell Biol Toxicol. 38:1175–1197. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wallis N, Oberman F, Shurrush K, Germain
N, Greenwald G, Gershon T, Pearl T, Abis G, Singh V, Singh A, et
al: Small molecule inhibitor of Igf2bp1 represses Kras and a
pro-oncogenic phenotype in cancer cells. RNA Biol. 19:26–43. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Yu T, Ran L, Zhao H, Yin P, Li W, Lin J,
Mao H, Cai D, Ma Q, Pan X, et al: Circular RNA circ-TNPO3
suppresses metastasis of GC by acting as a protein decoy for
IGF2BP3 to regulate the expression of MYC and SNAIL. Mol Ther
Nucleic Acids. 26:649–664. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen C, Zhang M and Zhang Y: Circ_0000079
decoys the RNA-binding protein FXR1 to interrupt formation of the
FXR1/PRCKI complex and decline their mediated cell invasion and
drug resistance in NSCLC. Cell Transplant. 29:9636897209610702020.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Wang JZ, Zhu H, You P, Liu H, Wang WK, Fan
X, Yang Y, Xu K, Zhu Y, Li Q, et al: Upregulated YB-1 protein
promotes glioblastoma growth through a YB-1/CCT4/mLST8/mTOR
pathway. J Clin Invest. 132:e1465362022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Barbagallo D, Caponnetto A, Barbagallo C,
Battaglia R, Mirabella F, Brex D, Stella M, Broggi G, Altieri R,
Certo F, et al: The GAUGAA motif is responsible for the binding
between circSMARCA5 and SRSF1 and related downstream effects on
glioblastoma multiforme cell migration and angiogenic potential.
Int J Mol Sci. 22:16782021. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Groot M and Lee H: Sorting mechanisms for
MicroRNAs into extracellular vesicles and their associated
diseases. Cells. 9:10442020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhang L, Zhang Y, Shen D, Chen Y, Feng J,
Wang X, Ma L, Liao Y and Tang L: RNA binding motif protein 3
promotes cell metastasis and epithelial-mesenchymal transition
through STAT3 signaling pathway in hepatocellular carcinoma. J
Hepatocell Carcinoma. 9:405–422. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Fuji T, Umeda Y, Nyuya A, Taniguchi F,
Kawai T, Yasui K, Toshima T, Yoshida K, Fujiwara T, Goel A and
Nagasaka T: Detection of circulating microRNAs with Ago2 complexes
to monitor the tumor dynamics of colorectal cancer patients during
chemotherapy. Int J Cancer. 144:2169–2180. 2019. View Article : Google Scholar : PubMed/NCBI
|