Introduction
Colorectal cancer (CRC) is a leading cause of
cancer-related deaths (1). A total
of ~35% of patients with CRC are initially diagnosed with
metastasis and 20-50% of patients with non-metastatic CRC develop
metastasis during the course of disease (2,3).
Although efforts have been made to elucidate molecular pathways
associated with CRC progression (4-6),
metastasis of CRC is a major factor responsible for poor prognosis
and its treatment remains challenging. Therefore, better
understanding of the molecular mechanisms underlying CRC metastasis
is required.
Ring1 and YY-1 binding protein (RYBP) was originally
identified as a member of the polycomb repressive complex (7). RYBP epigenetically regulates gene
expression and is involved in embryonic development, stem cell
self-renewal, cell differentiation, and X chromosome inactivation
(7-9). RYBP interacts with several
transcription factors, including YY-1, and serves as a bridge
factor that mediates the formation of transcription factor
complexes, thereby regulating gene expression independent of
polycomb group functions (10,11).
RYBP mediates the interaction of YY-1 with the E2F family of genes,
which encode transcription factors that serve an important role in
G1/S phase transition (11). Our
previous study demonstrated that YY-1 acts as a tumour suppressor
and high YY-1 expression contributes to improved CRC prognosis
(4). RYBP may also act as a tumour
suppressor. Previous studies have demonstrated that RYBP plays
tumour-suppressive roles in several types of cancer, such as
hepatocellular carcinoma (12) and
breast (13), lung (14) and oesophageal cancer (15). To the best of our knowledge,
however, there have been no functional analyses or
clinicopathological studies of RYBP with regards to CRC. Therefore,
the present study aimed to evaluate the role and expression of RYBP
in CRC, its association with clinical outcomes and underlying
molecular mechanisms.
Materials and methods
Patients and specimens
Primary CRC tissues were obtained from 140
consecutive patients aged 27-91 years (49 females and 91 males who
underwent surgical resection between January 2012 and December 2013
at Chiba University Hospital (Chiba, Japan). Corresponding liver
metastasis tissue was obtained from 11 matched patients who
underwent surgical resection at Chiba University Hospital. These
patients ranged in age from 48 to 76 years, and consisted of 5
females and 6 males. Cases in which preoperative chemotherapy
resulted in the total disappearance of tumour cells were excluded.
Patients who underwent two-stage hepatectomy were excluded. The
clinicopathological characteristics of patients are shown in
Table SI. The Ethics Committees
of the Department of General Surgery, Chiba University Hospital
(Chiba, Japan) approved the study protocol (approval no. M10101)
and written informed consent to participate was obtained from each
patient before surgery. Union for International Cancer Control TNM
classification of 8th edition (16) was used to assess the clinical
outcome of the patients with CRC.
Immunohistochemistry (IHC)
Paraffin-embedded tissue samples after 10% formalin
fixed for 24 h at room temperature were cut to obtain
4-μm-thick slices and de-paraffinized with xylene and
rehydrated with descending ethanol series. Antigen retrieval was
performed by microwaving (500 W) slides in citric acid buffer (0.01
M; pH, 6.0) for 25 min at 100°C. Subsequently, endogenous
peroxidase activity was blocked with hydrogen peroxide (3% in
methanol) for 15 min at room temperature. Non-specific proteins
were blocked with 5% bovine serum albumin (BSA; cat. no. 01860-36;
Nacalai Tesque, Inc.) for 10 min at room temperature. Slides were
incubated overnight at 4°C with the following primary antibodies:
Anti-RYBP polyclonal (1:200; cat. no. HPA053357; Sigma-Aldrich;
Merck KGaA) and anti-p53 Ab (1:100; cat. no. M7001; Dako; Agilent
Technologies, Inc.). Secondary antibodies (undiluted; EnVision™
kits; cat. nos. K4001 and K4003; Dako; Agilent Technologies, Inc.)
were applied for 30 min at room temperature, followed by staining
with peroxidase DAB kit (cat. no. 25985-50; Nacalai Tesque, Inc.).
Counterstaining was performed with haematoxylin before dehydration
for 1 min at room temperature, penetration and mounting.
Using an inverted light microscope (cat. no. BX40;
Olympus Corporation), the expression levels of RYBP and p53 were
independently evaluated by two investigators and a pathologist, all
of whom were blinded to clinical information. A 1%
BSA/phosphate-buffered saline (PBS) solution without primary
antibody served as a negative control. As a positive control,
staining of normal liver tissue that originated from the tissue at
least 1cm away from the tumour in a resected specimen from a
patient with colorectal liver metastasis was confirmed because RYBP
is expressed in the cytoplasm of normal hepatocytes (12). The evaluations were performed after
establishing an inter-observer consensus using samples from
preliminary experiments. In primary CRC, RYBP expression was
evaluated by the percentage of positively stained nuclei in tumour
cells relative to the total number of malignant cells in three
positive high-power fields which were randomly selected
(magnification, ×400); low and high expression were defined as
<20 and ≥20%, respectively. The expression of p53 was evaluated
by considering the distribution pattern of positive cells and the
intensity of nuclear staining and categorised as wild-type (wt;
absent and weakly and sporadically positive staining) and mutant
p53 (strongly mosaic and diffuse staining).
Human colon cancer cell lines and
culture
The human colon cancer cell lines HCT116
(TP53wt/wt) and HCT116
(TP53−/−) were kindly provided by Dr Mamoru
Takada (Chiba University, Chiba, Japan). SW48, DLD-1, HT29, COLO201
and SW620 were purchased from American Type Culture Collection.
HCT116 cells were cultured in McCoy's 5A medium (cat. no. 16600082)
with 10% foetal bovine serum (FBS; both Gibco; Thermo Fisher
Scientific, Inc.) and incubated at 37°C with 5% CO2.
SW48 and SW620 cells were cultured in Leibovitz's L-15 medium (cat.
no. 11415064; Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS
and incubated at 37°C without CO2. DLD-1 and COLO201
cells were cultured in RPMI 1640 medium (cat. no. 22400089; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and
incubated at 37°C with 5% CO2.
Small interfering (si)RNA and plasmid
transfection
HCT116 (TP53wt/wt), HCT116
(TP53−/−), SW48 and DLD-1 cells
(2-5×105/well) were transfected with siRNA-1 and siRNA-2
[ON-TARGETplus Human RYBP (23429) siRNA, cat.no. J-015936-06
(target; 5′-GAA AGA UCC UCC UAG UGA A-3′) and J-015936-07 (target;
5′-CGA CAU GUC AGC AGU CAA U-3′); both GE Healthcare Dharmacon,
Inc.] at a final concentration of 10 nmol/l or an equimolar
concentration of control siRNA (AllStars Negative Control siRNA;
sequence not available; cat. no. 1027280; Qiagen GmbH.) using
Lipofectamine™ RNAiMAX transfection reagent (cat. no. 13778075;
Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 24 h
according to the manufacturer's recommendations. The expression
(pcDNA3.1-RYBP) and empty vector (pcDNA3.1) were purchased from
GenScript. Plasmid transfection was performed using 1 μg
pcDNA and Lipofectamine® 3000 transfection reagent (cat.
no. L3000015; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
for 24 h incubation, according to the manufacturer's instructions.
Changes in RYBP levels were monitored using western blotting 48 h
after transfection. The cells were used for subsequent assays 24 h
after transfection.
Western blot analysis
Proteins were extracted from cultured cells using
RIPA buffer (cat. no. 16488-34; Nacalai Tesque, Inc.). Each protein
sample was lysed in Laemmli Sample Buffer (cat. no. 1610737;
Bio-Rad Laboratories, Inc.) containing 5% 2-mercaptoethanol and
incubated at 97°C for 10 min. After measuring the protein
concentration of each sample using the Pierce™ BCA Protein Assay
kit (cat. no. 23225; Thermo Scientific; Thermo Fisher Scientific,
Inc.), 20 μg/lane protein was separated by electrophoresis
on 10% XV PANTERA Gels (cat. no. NXV-224P; DRC) and transferred
onto a polyvinylidene difluoride membrane. The membranes were
blocked in 5% skimmed milk diluted in 0.1% Tris-buffered saline
with Tween-20 or Blocking One-P (cat. no. 05999-84; Nacalai Tesque,
Inc.) in the case of phosphorylated protein at room temperature
(15-25°C) for 60 min. The membranes were incubated at 4°C overnight
with primary antibodies against RYBP (1:1,000; cat. no. HPA053357;
Sigma-Aldrich; Merck KGaA), p53 (1:1,000; cat. no. M7001; Dako;
Agilent Technologies, Inc.), p21 (1:1,000; cat. no. 2947; Cell
Signaling Technology, Inc.), cyclin D1 (1:1,000; cat. MA5-14512;
Thermo Fisher Scientific, Inc.), Bax (1:1,000; cat. no. 5023; Cell
Signaling Technology, Inc.), p-cyclin D1 (1:1,000; cat. no. 3300;
Cell Signaling Technology, Inc.), YY-1 (1:1,000; cat. no. ab109228;
Abcam) and β-actin (1:2,000; cat. no. 5125; Cell Signaling
Technology, Inc.). Subsequently, the membranes were incubated with
secondary antibodies at room temperature (15-25°C) for 60 min as
follows: Anti-mouse (1:2,000; cat. no. SC516102; Santa Cruz
Biotechnology, Inc.) and anti-rabbit HRP conjugated secondary
antibodies (1:2,000; cat. no. 7074; Cell Signaling Technology,
Inc.). The membranes were incubated with an enhanced
chemiluminescence detection reagent (Chemi-Lumi One Ultra; Nacalai
Tesque, Inc.) and developed using a LAS-4000UV mini luminescent
image analyser (FUJIFILM Wako Pure Chemical Corporation). Band
intensities were quantified by densitometric analysis using the
ImageJ software version 1.53 (National Institutes of Health) to
calculate the relative protein levels normalised to β-actin.
Cell proliferation assay
HCT116 (TP53wt/wt), HCT116
(TP53−/−), SW48 and DLD-1 cells were seeded in
96-well microplates at a density of 3,000 cells/well. At 0, 24, 48,
72, or 96 h, 10 μl Cell Count Reagent SF (cat. no. 07553044;
Nacalai Tesque, Inc.) was added to 100 μl culture medium.
After 2 h incubation at 37°C, the absorbance at 450 nm was measured
using a microplate reader.
Cell cycle assay
HCT116 (TP53wt/wt), HCT116
(TP53−/−) cells were collected 48 h after
transfection, with or without 24 h pre-treatment with 4 or 10
μM oxaliplatin (Nippon Kayaku Co., Ltd.) at 37°C, washed
with ice-cold PBS and fixed in 70% ethanol at -30°C overnight.
Prior to staining, cells were washed with PBS. Pellets of the cells
were mixed with 500 μl FxCycle™ PI/RNAse Solution (cat. no.
10797; Invitrogen; Thermo Fisher Scientific, Inc.) and incubated
for 30 min at room temperature. Samples were analysed by
fluorescence-activated cell sorting using FACS Canto II (BD
Biosciences). All data were analysed using FlowJo v10.7.1 software
(BD Biosciences).
Cell apoptosis assessment
Cell apoptosis was assessed using the FITC Annexin
V/PI Apoptosis Detection kit (cat. no. 15342-54; Nacalai Tesque,
Inc.), according to the manufacturer's instructions. Briefly,
HCT116 (TP53wt/wt), HCT116
(TP53−/−) cells were collected, washed with
Binding Buffer at 4°C and resuspended in 100 μl Binding
Buffer. A total of 1×105 cells were incubated with 5
μl Annexin V-FITC and PI at room temperature in the dark for
15 min. In total, 10,000-20,000 cells were analysed on a FACS Canto
II (BD Biosciences) using FlowJo v10.7.1 (BD Biosciences).
Oxaliplatin-treated cells were used as a positive control. The
percentage of apoptotic cells was calculated based on the number of
early apoptotic cells which were Annexin V-positive and PI
negative.
Cell viability assessment
HCT116 (TP53wt/wt), HCT116
(TP53−/−) cells were seeded in 96-well
microplates at a density of 5,000 cells/well and 100 μl of
cell-free McCoy's 5A medium was prepared as a blank. After 24 h
pre-incubation at 37°C, oxaliplatin was added at concentrations of
0.001, 0.010, 0.1, 0.5, 1, 2, 4 and 10 μM and cells were
incubated for 2 days at 37°C. A total of 10 μl Cell Count
Reagent SF (cat. no. 07553044; Nacalai Tesque, Inc.) was added to
each well. After 2 h incubation, absorbance at 450 nm was measured
using a microplate reader. The cell viability was calculated as
follows: Cell viability (%)=(absorbance of treated
sample-absorbance of cell-free sample)/(absorbance of control
sample-absorbance of cell-free sample) ×100.
The half-maximal inhibitory concentration
(IC50) was calculated with a non-linear regression fit
to a sigmoidal dose-response curve and compared using the
extra-sum-of-squares F test in Prism 9 (GraphPad Software, Inc.;
Dotmatics). Experiments were performed six times independently.
Statistical analysis
Data are expressed as mean ± standard error of the
mean or median ± standard deviation. The association between RYBP
or p53 staining and patient characteristics were evaluated using
χ2 for categorical variables, unpaired Student's t for
parametric continuous variables or Mann-Whitney U test for
nonparametric continuous variables. Survival rates were calculated
using Kaplan-Meier analysis and assessed using the log-rank test.
Survival data were evaluated using univariate and multivariate Cox
proportional hazards regression analysis. When analysing the
association between RYBP expression in primary tumour and long-term
outcomes, cancer-specific survival (CSS) and disease-free survival
(DFS) were calculated from the date of primary tumour resection.
When analysing the association between RYBP expression in liver
metastases and long-term outcome, CSS and DFS were calculated from
the date of initial hepatectomy. The in vitro experiments
were performed at least three times independently and data were
analysed using unpaired Student's t test or one-way or multivariate
analysis of variance followed by Tukey's post hoc test. P≤0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using the JMP® Pro15
software (SAS Institute Inc.).
Results
RYBP expression in primary tumour is
associated with better prognosis
To investigate RYBP expression and subcellular
localisation in CRC, immunohistochemical staining of primary CRC
and liver metastases was performed. In primary CRC, RYBP was
predominantly expressed in the nuclei of both tumour and adjacent
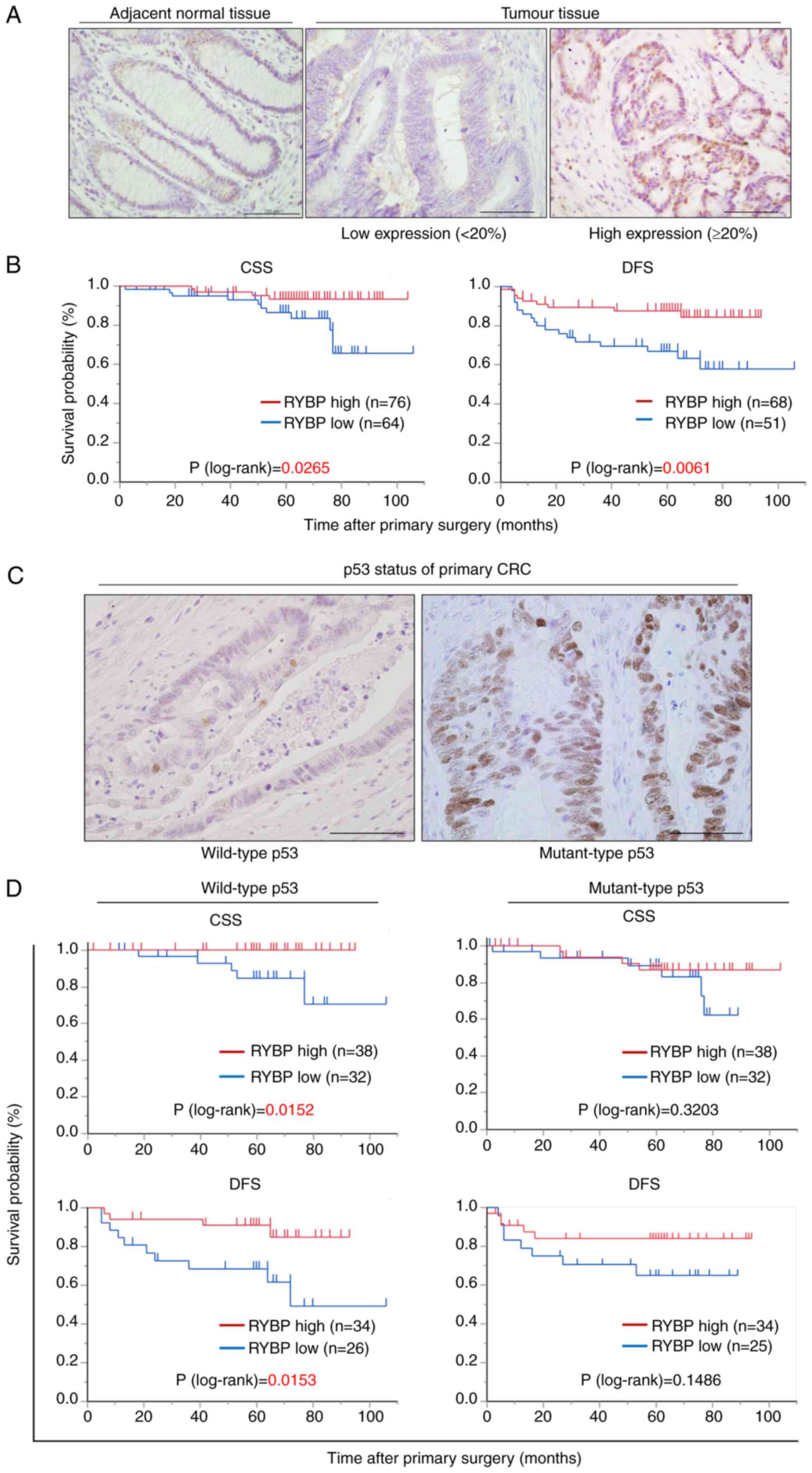
normal tissue (Fig. 1A). High RYBP
expression was observed in 76 (54.3%) patients. The association
between RYBP expression profiles and clinicopathological features
is shown in Table I. The rate of
distant metastases and recurrence after primary surgery were
significantly higher in patients with low RYBP expression than in
those with high RYBP expression (Table
I). Kaplan-Meier analysis showed that patients with high RYBP
expression had a significantly longer CSS and DFS after primary
surgery than those with low RYBP expression (Fig. 1B). Univariate and multivariate
analyses revealed that poor pathological differentiation, severe
lymphatic invasion and low RYBP expression were independent
prognostic factors associated with CSS (Table II).
 | Table IAssociation between RYBP expression
and clinicopathological features of patients with colorectal
cancer. |
Table I
Association between RYBP expression
and clinicopathological features of patients with colorectal
cancer.
| Characteristic | RYBP expression
| P-value |
|---|
| High (n=76) | Low (n=64) |
|---|
| Median age, years
(range) | 70 (29-91) | 69 (27-91) | 0.5608 |
| Sex,
female/male | 25/51 | 24/40 | 0.5693 |
| Primary lesion
site, colon/rectum | 42/34 | 40/24 | 0.3865 |
| Primary lesion
site, right colon/left colon/rectum | 9/33/34 | 3/37/24 | 0.1385 |
| CEA, <5/≥5
ng/ml | 50/26 | 33/31 | 0.0878 |
| CA19-9, <37/≥37
U/ml | 60/16 | 54/10 | 0.4107 |
| Postoperative
chemotherapy, no/yes | 53/23 | 42/22 | 0.6038 |
| T stagea, 1-3/4 | 60/16 | 45/19 | 0.1616 |
| Lymph node
metastasis, no/yes | 51/25 | 33/31 | 0.0615 |
| Ly, 0/1-3 | 34/42 | 27/37 | 0.7619 |
| V, 0/1-3 | 33/43 | 26/38 | 0.7386 |
| Degree of
differentiation, pap or tub/por or muc | 72/3 | 63/1 | 0.3916 |
| KRAS,
wild-type/mutant | 6/13 | 10/16 | 0.6338 |
| BRAF,
wild-type/mutant | 0/19 | 2/24 | 0.2162 |
| Distant metastasis,
no/yes | 63/13 | 38/26 | 0.0019b |
| Recurrence after
primary surgery, no/yes | 63/5 | 38/13 | 0.0062b |
 | Table IIUnivariate and multivariate analysis
of cancer-specific survival in patients with colorectal cancer. |
Table II
Univariate and multivariate analysis
of cancer-specific survival in patients with colorectal cancer.
| Variable | n | 5-year survival,
% | Univariate
P-value | Multivariate
|
|---|
| HR (95%CI) | P-value |
|---|
| Age, 65</≥65
years | 43/97 | 86.7/92.2 | 0.307 | | |
| Sex,
male/female | 91/49 | 85.1/100.0 | 0.155 | | |
| CEA, <5/≥5
ng/ml | 83/57 | 93.1/86.0 | 0.181 | | |
| CA19-9, 37</≥37
U/ml | 114/26 | 93.6/74.5 | 0.050 | | |
| Site of primary
tumour, left/right colon | 128/12 | 90.5/88.9 | 0.867 | | |
| T stagea, 1-3/4 | 105/35 | 91.0/87.5 | 0.337 | | |
| Degree of
differentiation, tub or pap/por or muc | 135/4 | 91.8/50.0 | 0.004c | 7.46
(1.32-42.26) | 0.023b |
| Ly, 0-1/2-3 | 121/19 | 95.6/60.8 | <0.001d | 5.17
(1.63-16.42) | 0.005c |
| V, 0-1/2-3 | 96/44 | 94.7/81.1 | 0.027b | 2.46
(0.82-7.41) | 0.109 |
| Lymph node
metastasis, no/yes | 84/56 | 96.6/81.7 | <0.001d | 2.98
(0.56-15.78) | 0.199 |
| RAS,
wild-type/mutant | 29/16 | 75.2/67.1 | 0.448 | | |
| Expression of RYBP,
high/low | 76/64 | 93.3/86.8 | 0.027b | 3.77
(1.02-13.90) | 0.046b |
Association between RYBP expression and
better prognosis depends on p53 expression status
Wt p53 expression was observed in 70 patients (50%),
whereas mutant p53 expression was observed in 70 patients (50.0%;
Fig. 1C). The status of p53
expression was not associated with clinicopathological features or
prognosis (Table III). In
patients with wt p53, CSS and DFS were significantly longer in
those with higher RYBP expression (Fig. 1D). By contrast, in patients with
mutant p53 expression, there were no significant differences in CSS
and DFS (Fig. 1D). These data
indicated that RYBP contributed to better prognosis in a
TP53 status-dependent manner.
 | Table IIIAssociation between p53 expression
and clinicopathological features of patients with colorectal
cancer. |
Table III
Association between p53 expression
and clinicopathological features of patients with colorectal
cancer.
| Characteristic | p53
| P-value |
|---|
| Wild-type
(n=70) | Mutant (n=70) |
|---|
| Median age, years
(range) | 71 (30-91) | 68 (27-91) | 0.1768 |
| Sex,
female/male | 26/44 | 23/47 | 0.5949 |
| Primary lesion
site, colon/rectum | 43/27 | 39/31 | 0.4924 |
| Primary lesion
site, right colon/left colon/rectum | 8/35/27 | 4/35/31 | 0.4415 |
| CEA, <5/≥5
ng/ml | 44/26 | 39/31 | 0.3895 |
| CA19-9, <37/≥37
U/ml | 57/13 | 57/13 | 1.0000 |
| Postoperative
chemotherapy, no/yes | 50/20 | 45/25 | 0.3652 |
| T stagea, 1-3/4 | 54/16 | 51/19 | 0.5580 |
| Lymph node
metastasis, no/yes | 45/25 | 39/31 | 0.3885 |
| Ly, 0/1-3 | 29/41 | 32/38 | 0.6091 |
| V, 0/1-3 | 31/39 | 28/42 | 0.6076 |
| Degree of
differentiation, pap or tub/por or muc | 68/1 | 67/3 | 0.3063 |
| KRAS,
wild-type/mutant | 7/11 | 9/18 | 0.7034 |
| BRAF,
wild-type/mutant | 1/17 | 1/26 | 0.7699 |
| Distant metastasis,
no/yes | 51/19 | 50/20 | 0.8505 |
| Recurrence after
primary surgery, no/yes | 51/9 | 50/9 | 0.9691 |
| Expression of RYBP,
high/low | 38/32 | 38/32 | 1.0000 |
RYBP expression is downregulated in
matched liver metastases
Nuclear expression of RYBP was not observed in 11
patients with patient-matched liver metastases. In all adjacent
normal liver tissue samples, RYBP was moderately expressed in the
cytoplasm (Fig. S1B). In nine of
the 11 liver metastases, RYBP was also expressed in the cytoplasm,
with a lower staining intensity than in adjacent normal tissue.
RYBP regulates tumour cell proliferation
in TP53 wt cell lines
As the aforementioned data indicated that RYBP may
have a tumour-suppressive role in CRC, in vitro experiments
were performed to elucidate the molecular mechanisms by which RYBP
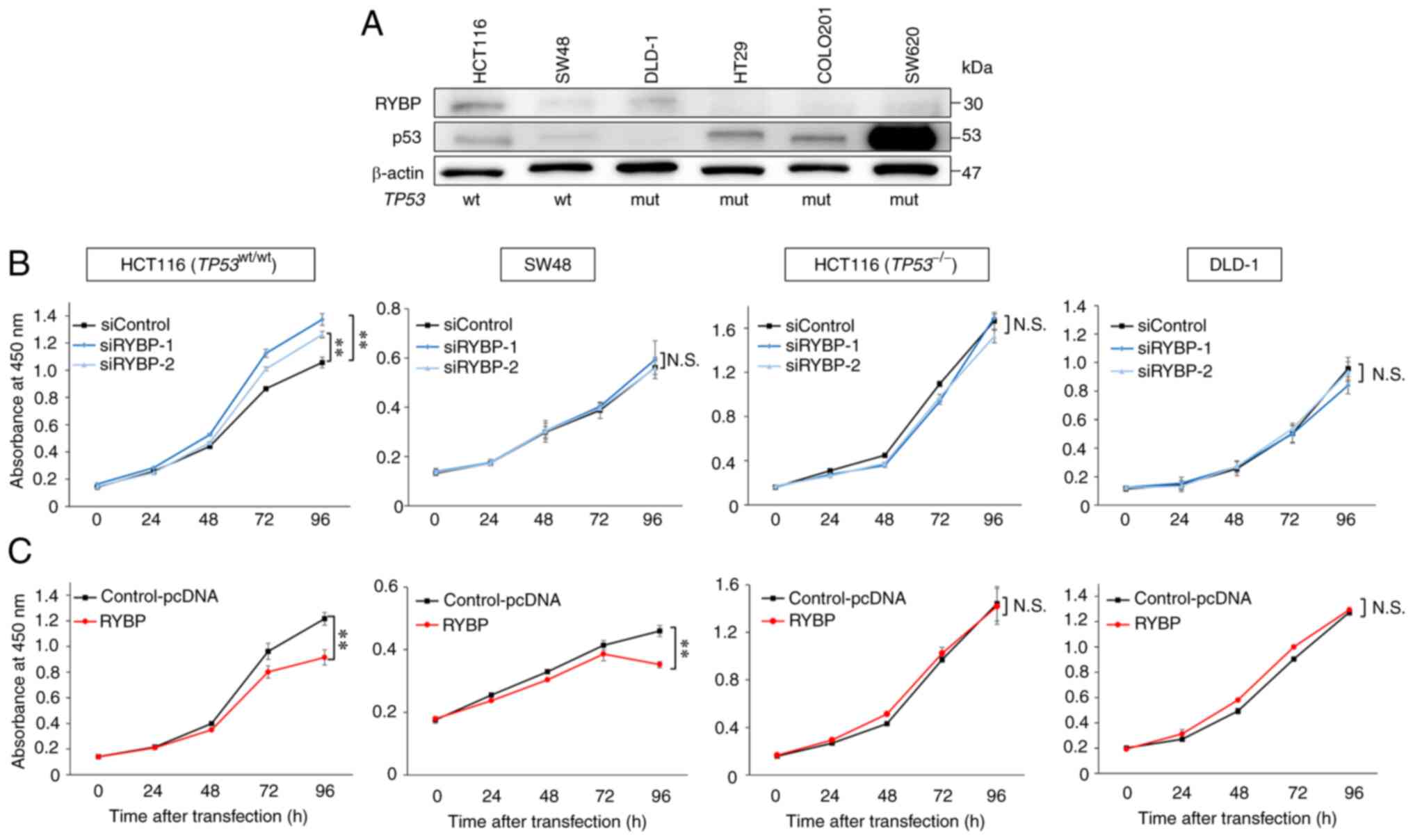
regulates behaviour of CRC cells. Western blot analyses showed that
the expression of both RYBP and p53 was high in HCT116 cells
(TP53wt/wt), whereas expression was low in SW48
cells. However, no association between RYBP and p53 expression was
observed in cell lines with TP53 mutation, such as DLD-1,
HT29, COLO201 and SW620 (Fig. 2A).
To evaluate the effects of RYBP on TP53 wt colon cancer
cells, HCT116 (TP53wt/wt) and SW48 cells and
TP53-mutant DLD-1 and TP53-null HCT116
(TP53−/−) cells were used as the control groups
for subsequent experiments.
To verify the function of RYBP, cell proliferation
assay was performed following knockdown of RYBP using siRNAs and
overexpression of RYBP using pcDNA3.1-RYBP plasmid. RYBP knockdown
significantly increased proliferation of HCT116
(TP53wt/wt) cells. However, knockdown did not
alter the proliferation of SW48 cells, in which RYBP expression was
low, and in TP53-mutant DLD-1 and TP53-null HCT116
(TP53−/−) cells (Fig. 2B). Overexpression of RYBP
significantly decreased proliferation of HCT116
(TP53wt/wt) and SW48, but not TP53-mutant
DLD-1 and TP53-null HCT116 (TP53−/−) cells
(Fig. 2C). These results suggested
that RYBP may regulate tumour cell proliferation via a
p53-associated pathway.
RYBP induces p53-associated cell cycle
arrest and apoptosis
To elucidate the tumour-suppressive mechanisms
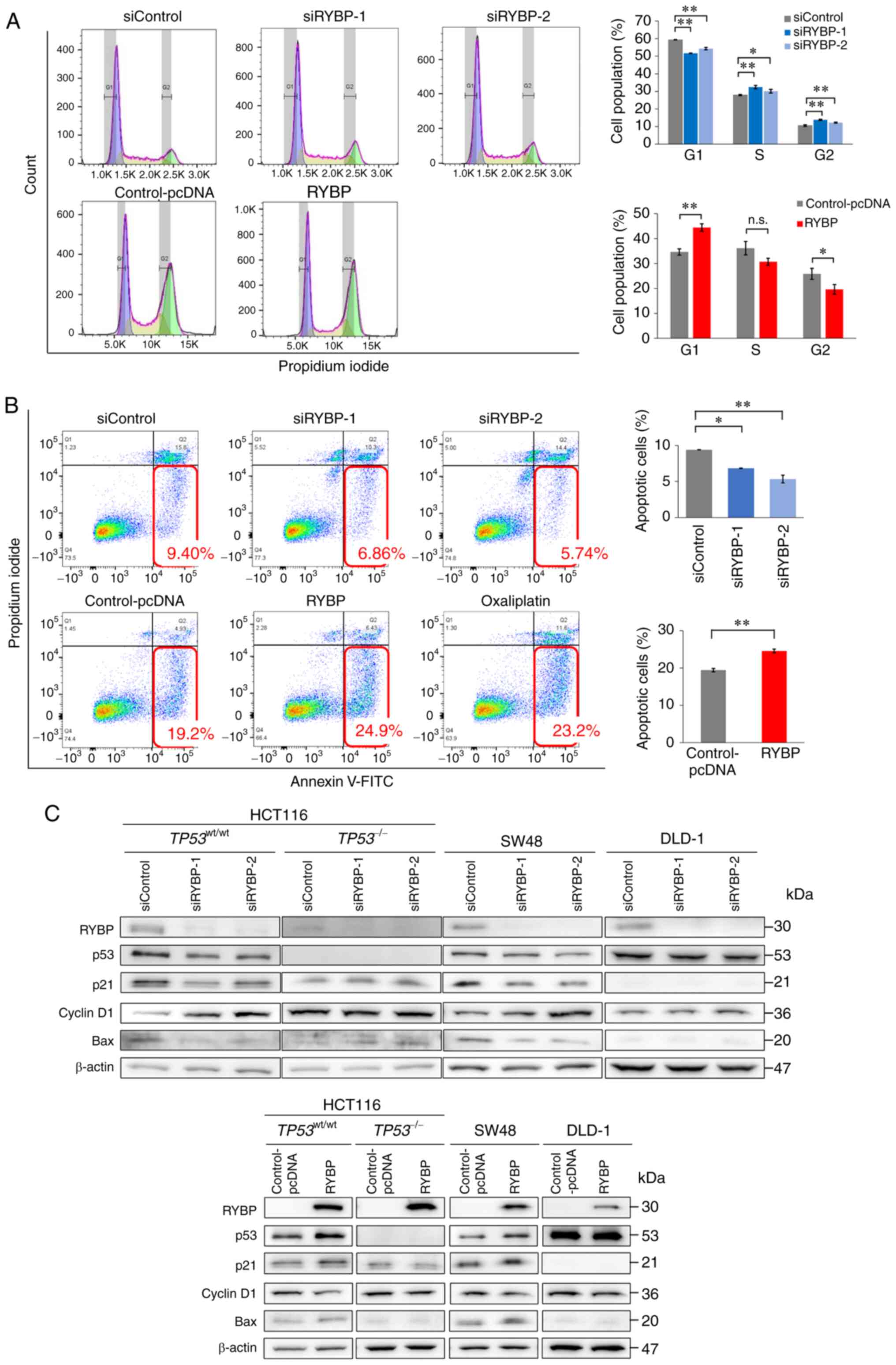
associated with p53, cell cycle and apoptosis assays were
performed. In HCT116 cells (TP53wt/wt), RYBP
knockdown significantly decreased the number of cells in G1/0 phase
and increased the number of cells in S phase, whereas RYBP
overexpression significantly increased the number of cells in G1/0
and decreased the number of cells in S phase (Fig. 3A). However, the number of HCT116
(TP53−/−) cells in each cell cycle phase was not
affected by modulation of RYBP expression (Fig. S2A). This also indicated that RYBP
did not directly damage DNA. Western blot analysis was performed to
evaluate expression of p21, a protein downstream of p53, and cyclin
D1, a cell cycle G1-S checkpoint regulator (17). In HCT116
(TP53wt/wt) cells, RYBP knockdown significantly
decreased expression of p53 and p21 but induced cyclin D1
expression. Conversely, RYBP overexpression significantly induced
expression of p53 and p21 but decreased cyclin D1 expression.
However, neither knockdown nor overexpression had any effect on
p53, p21 or cyclin D1 expression in TP53-mutant DLD-1 and
TP53-null HCT116 (TP53−/−) cells (Fig. 3C). We confirmed that RYBP bands
were visible in cells transfected with the empty vector in
preliminary experiments (Fig.
S1C). Protein expression normalised to β-actin is shown in
Fig. S3A-C. Corresponding β-actin
from same membrane is shown in Fig.
S4A and B.
In the apoptosis assay, knockdown of RYBP
significantly decreased the number of HCT116
(TP53wt/wt) cells in early apoptosis, whereas
RYBP overexpression significantly increased the number of apoptotic
cells (Fig. 3B). However, the
number of apoptotic HCT116 (TP53−/−) cells was
not affected by modulation of RYBP expression (Fig. S2B). Western blot analysis revealed
that knockdown of RYBP significantly reduced expression of Bax, a
p53-related pro-apoptotic protein, and conversely, RYBP
overexpression induced Bax expression (Figs. 3C and S3D).
RYBP enhances oxaliplatin sensitivity by
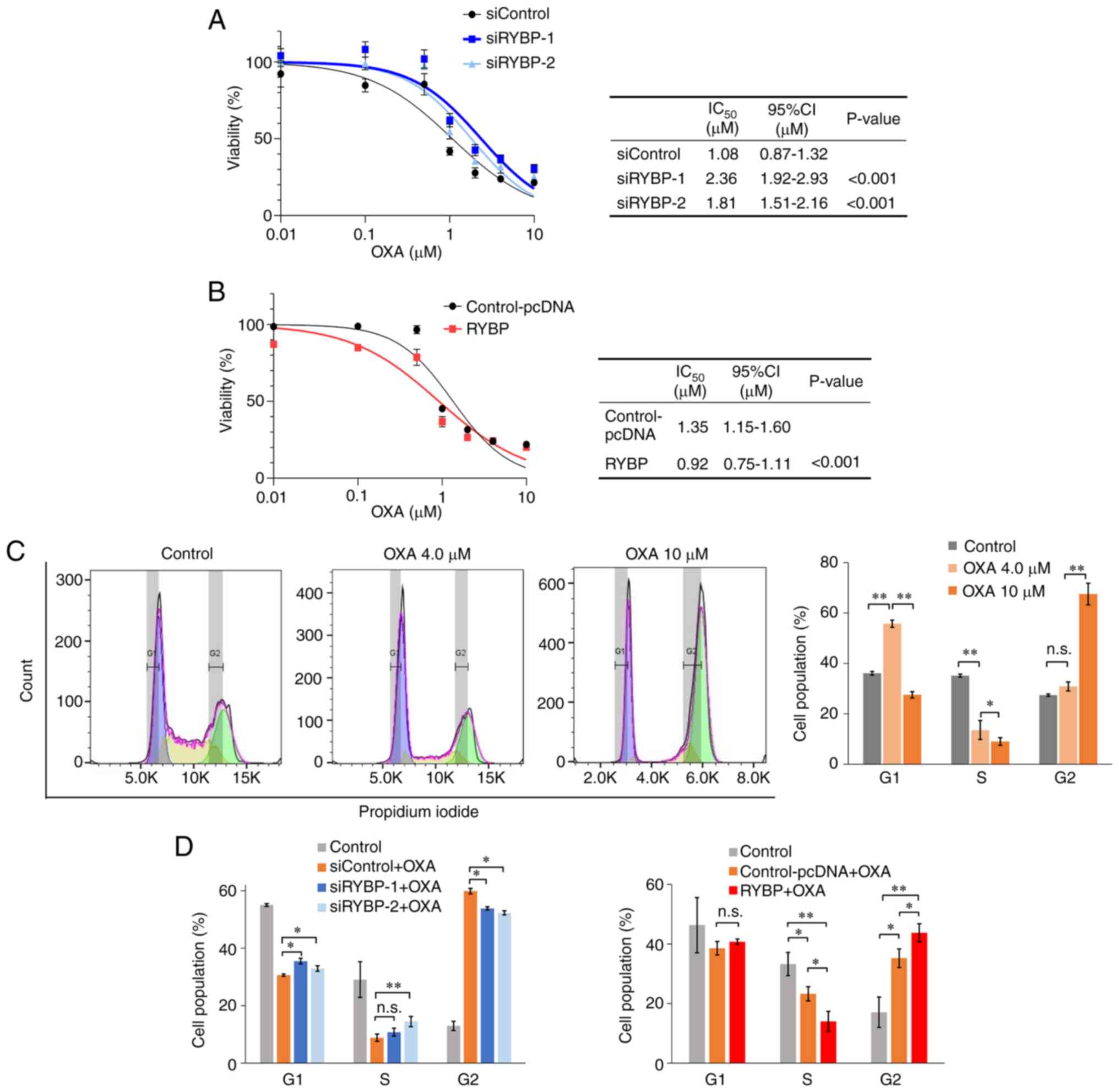
regulating p53-mediated cell cycle arrest
To evaluate the effect of RYBP on oxaliplatin
sensitivity, cell viability assay was used to calculate
IC50 value. In HCT116 (TP53wt/wt)
cells, RYBP knockdown significantly increased viability and the
IC50 value of oxaliplatin was significantly higher in
cells treated with siRNA-1 and siRNA-2 than in the control
(Fig. 4A). By contrast, RYBP
overexpression decreased cell viability and the IC50 was
significantly lower in cells treated with pcDNA-RYBP than in the
control (Fig. 4B). Low-dose (4.0
μM)oxaliplatin increased the number of cells in G1 phase and
decreased the number of cells in the S phase. Higher doses (10
μM) of oxaliplatin further decreased the number of cells in
S phase, indicating that disruption of DNA replication leads to
anti-tumour effects (Fig. 4C). The
results from the cell cycle assay following treatment of cells with
10 μM oxaliplatin showed that RYBP knockdown significantly
increased the number of cells in the S phase, whereas RYBP
overexpression further decreased the number of cells in S phase
(Fig. 4D). These data suggested
that RYBP enhanced the sensitivity of CRC cells to oxaliplatin.
Discussion
To the best of our knowledge, the present study is
the first to demonstrate that RYBP is an inhibitory protein that
protects against distant metastases and recurrence following
primary tumour resection in CRC. RYBP stabilises p53 by decreasing
mouse double minute 2-mediated ubiquitination and increasing p53
activity (18). The present
clinical data showed that low RYBP expression was significantly
associated with a higher rate of distant metastases and recurrence,
which lead to a poor prognosis in patients with CRC. The
tumour-suppressive effect of RYBP was observed in patients with
TP53 wt CRC, but not in those with TP53 mutant CRC.
These data indicated that RYBP plays a role in modulating
TP53 activity. The present in vitro data support
these clinical data. Using CRC cell lines with or without p53
mutations, the present study demonstrated that RYBP has an effect
on cell apoptosis and cell cycle progression by regulating the
p53/p21 signalling pathway. Moreover, RYBP increased the
sensitivity of CRC cells to oxaliplatin, a key therapeutic drug for
CRC (19).
RYBP was originally identified as a member of the
non-canonical polycomb repressive complex 1 (7). Studies have demonstrated a key role
of RYBP in cancer biology: RYBP expression is downregulated in
cancer cells and low RYBP expression is associated with poor
prognosis (12,13,20-23).
The present clinical data showed the tumour-suppressive role of
RYBP in CRC, consistent with the aforementioned studies. To explore
the molecular mechanisms underlying these clinical data, the
present study performed in vitro experiments using CRC cell
lines with and without TP53 mutation. A novel finding of the
present study is that RYBP induced apoptosis and G1/S cell cycle
arrest by regulating the p53/p21 signalling pathway in TP53
wt CRC. As a downstream factor of p53, p21 serves a critical role
in cell cycle regulation (16,24).
DNA damage activates the p53/p21 pathway and induced p21 binds to
cyclin D1/cyclin-dependent kinase 4 (CDK4) and cyclin E/CDK2
complexes at the G1 phase checkpoint (17). The binding of p21 to cyclin D1
prevents nuclear transport of the complex, thereby inducing the
nuclear accumulation of cyclin D1 and inhibiting its
phosphorylation (24). Here,
expression of p-cyclin D1 was not changed by knockdown or
overexpression of RYBP with or without TP53 mutation. Since
p-cyclin D1 undergoes rapid proteasomal degradation, it is
difficult to determine its protein expression by western blotting
(25). Independent of the p53/p21
pathway, it is known that p53 directly activates the pro-apoptotic
Bcl-2 protein Bax (26). The
present data indicated that RYBP-induced p21 suppressed endogenous
cyclin D1 expression and lead to cell cycle arrest at G1 phase,
resulting in suppression of tumour growth. Additionally, the
present data indicated that RYBP-induced Bax upregulation may
induce the endogenous apoptotic pathway in CRC.
Although RYBP did not regulate YY-1 expression
directly in the present study, previous studies have reported the
significant interaction between RYBP and YY-1 (10,11).
YY-1 may be associated with the p53 signalling pathway. Our recent
study demonstrated downregulated expression of p53 pathway-related
genes, such as CDK2, Damage Specific DNA Binding Protein 2,
p53 apoptosis effector related to PMP22) and methyl malonyl
aciduria cobalamin deficiency B type (MMAB), following YY-1
knockdown in colon cancer cell lines using microarray and gene
enrichment analyses (4). These
data suggest that YY-1 modulates the p53 signalling pathway
upstream of YY-1.
In the present study, nine of 11 metastatic tumours
did not show nuclear expression of RYBP, although nuclear
expression was predominantly observed in primary CRC. The clinical
significance of this differential expression pattern is not clear
owing to the small sample size. Regarding the subcellular
localisation of RYBP, previous studies demonstrated that RYBP
predominantly interacts with DED-containing DNA-binding protein
(27) or apoptin (28). However, RYBP is also reported to
promote the formation of the death-inducing signalling complex in
the cytoplasm and induce apoptosis of tumour cells by activating
the p53 signalling pathway (29).
Our previous study demonstrated that the expression pattern of the
critical transcription factor differs between primary and
metastatic tumours during tumour progression (5). Similarly, previous studies have
demonstrated that the expression pattern of PD-L1 in non-small cell
lung cancer (30) and Ki-67 and
cyclin D1 in colon cancer (31)
differ between primary tumours and matched liver metastases. This
may be explained by 'intratumour heterogeneity in space and time'
and 'actionable mutation', as proposed by Swanton (32). Tumours actively develop, resulting
in metastatic lesions that may be genetically distinct. This tumour
heterogeneity facilitates resistance to systemic chemotherapy
(1,33). Taken together, the present results
suggested that the expression pattern of RYBP may be altered during
tumour progression, and this alteration may impact tumour biology.
Moreover, RYBP may have different functions depending on the host
organ and/or subcellular localisation, even in the same cancer
type. These hypotheses need to be tested in future.
Although p53 IHC is used as a surrogate for
TP53 mutations, its accuracy has not been established.
Mutant p53 can generally be identified as being overexpressed by
immunostaining because of its delayed degradation time and
accumulation in the nucleus. By contrast, the absence of p53 in
tumour cells indicate a loss-of-function (LOF) mutation in
TP53. However, no detectable p53 could also indicate
TP53 wt, because wt p53 has a short half-life and is rarely
detected by immunostaining, meaning cells with no immunostaining of
p53 are considered a mixture of both genotypes (34,35).
In a previous study using next-generation sequencing (NGS), 53% of
cells with no expression of p53 had LOF mutations and the rest had
wt TP53 (36). Therefore,
it is difficult to divide cells into those with LOF mutation and wt
TP53 using IHC only. More accurate assessment of p53
expression using NGS should be conducted in future studies.
Notably, the present data demonstrated that RYBP was
associated with chemosensitivity in CRC cells. For advanced-stage
CRC, particularly with distant metastases, multidisciplinary
treatment in combination with surgical resection and systemic
chemotherapy is needed to achieve a cure or long-term survival. An
optimal response to systemic chemotherapy is key to maximising the
benefits of multidisciplinary treatment strategies. During
treatment with successive regimens of chemotherapy, the sensitivity
for anticancer drugs may change and tumour cells could become drug
resistant by acquiring an embryonic-like and quiescent state
epigenetically (37,38). In addition to oxaliplatin,
overexpression of RYBP has been shown to enhance sensitivity to
cisplatin in hepatocellular carcinoma and anaplastic thyroid cancer
(12,21). In addition, TP53 status has
been reported to affect the sensitivity of not only CRC but also
esophageal, breast and numerous other types of cancer to
chemotherapeutic drugs (39-41).
To the best of our knowledge, there are no reports showing the
effect of RYBP on 5-fluorouracil or irinotecan, key drugs in the
treatment of CRC. However, both drugs induce anticancer effects by
inhibiting DNA replication, and it is possible that RYBP enhances
their efficacy via the cell cycle and apoptosis (42,43).
Therefore, modulation of RYBP expression and RYBP/p53 pathway
signalling may be a new target to increase chemosensitivity of
oxaliplatin (and other drugs), which could lead to improved
clinical outcomes in CRC.
The present data demonstrate a significant
association between RYBP and long-term outcome. RYBP may be a
useful prognostic predictor in patients with CRC. Moreover, RYBP
may be a potential candidate as an effective modulator of systemic
chemotherapy for CRC (and other types of cancers).
Accumulating evidence suggests that p53 regulates
innate and adaptive immune responses (44,45).
Inflammation is involved in the initiation and development of CRC
(46). Elevated levels of
cytokines such as tumour necrosis factor-α and chemokines such as
CXC chemokine ligand 1 and 2 in the serum of patients with CRC are
associated with cancer development and progression (47). Therefore, the RYBP/p53 pathway is
involved in tumour immunity and chronic inflammation.
The present study has limitations. First, the
background of patients was heterogeneous because all data were
collected retrospectively. Several prognostic factors such as T
stage, degree of differentiation, lymph node metastasis and tumour
marker were not randomised for analyses of long-term outcomes.
Second, in vitro experiments only evaluated RYBP in primary
CRC. However, clinical data suggested that its function at
metastatic sites may be altered by different pathways and studies
in other experimental systems, including in vivo
experiments, are warranted. Third, the present study evaluated cell
proliferation in vitro but not ongoing DNA synthesis, which
is typically assessed using a BrdU incorporation assay. To evaluate
the effect of RYBP on cell proliferation through its regulation of
the cell cycle, BrdU assay should be performed. Lastly, in
experiments using oxaliplatin, although no significant effect of
RYBP was observed on the cell cycle following treatment with 4
μM oxaliplatin (Fig. S2C),
effects may vary depending on exposure time and timing of
administration, and further investigation is warranted. In
conclusion, high RYBP expression in CRC cells was an independent
predictor of improved prognosis in patients with CRC. The
tumour-suppressive role of RYBP was associated with cell cycle
arrest, apoptosis and increased oxaliplatin sensitivity. Further
studies are warranted to explore the detailed molecular mechanisms
by which RYBP is regulated and to identify factors that enhance
RYBP expression in CRC, which could lead to a novel, effective and
safe therapeutic approach for CRC.
Supplementary Data
Availability of data and materials
The datasets generated and/or analysed during the
current study are available from the corresponding author upon
reasonable request.
Authors' contributions
TM and NS designed and performed the experiments.
TT, GO, SK, ST, MO and HM made substantial contributions to
conception and design of the present study and analysis and
interpretation of data. MO and HM confirm the authenticity of all
the raw data. TM wrote the manuscript. NS, MO and HM edited the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All study participants provided written informed
consent to participate. The study was approved by the Ethics
Committees of the Department of General Surgery, Chiba University
Hospital (Chiba, Japan; approval number: M10101). The present study
was conducted in accordance with the Declaration of Helsinki.
Patient consent for publication
Written informed consent for publication of clinical
details and/or clinical images was obtained from all patients.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
CSS
|
cancer-specific survival
|
|
DFS
|
disease-free survival
|
|
IHC
|
immunohistochemistry
|
|
RYBP
|
ring1 and YY-1 binding protein
|
Acknowledgments
The authors would like to thank Dr Mamoru Takada
(Chiba University, Chiba, Japan) for providing the HCT116 cell
lines used in this study.
Funding
The present study was supported by the Japan Society for the
Promotion of Science KAKENHI (grant no. JP22K08889).
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zacharakis M, Xynos ID, Lazaris A, Smaro
T, Kosmas C, Dokou A, Felekouras E, Antoniou E, Polyzos A,
Sarantonis J, et al: Predictors of survival in stage IV metastatic
colorectal cancer. Anticancer Res. 30:653–660. 2010.PubMed/NCBI
|
|
3
|
Manfredi S, Lepage C, Hatem C, Coatmeur O,
Faivre J and Bouvier AM: Epidemiology and management of liver
metastases from colorectal cancer. Ann Surg. 244:254–259. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sato N, Sakai N, Furukawa K, Takayashiki
T, Kuboki S, Takano S, Ohira G, Matsubara H and Ohtsuka M: Yin Yang
1 regulates ITGAV and ITGB1, contributing to improved prognosis of
colorectal cancer. Oncol Rep. 47:872022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takagi Y, Sakai N, Yoshitomi H, Furukawa
K, Takayashiki T, Kuboki S, Takano S, Suzuki D, Kagawa S, Mishima
T, et al: High expression of Krüppel-like factor 5 is associated
with poor prognosis in patients with colorectal cancer. Cancer Sci.
111:2078–2092. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sato N, Sakai N, Furukawa K, Takayashiki
T, Kuboki S, Takano S, Ohira G, Miyauchi H, Matsubara H and Ohtsuka
M: Tumor-suppressive role of Smad ubiquitination regulatory factor
2 in patients with colorectal cancer. Sci Rep. 12:54952022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
García E, Marcos-Gutiérrez C, del Mar
Lorente M, Moreno JC and Vidal M: RYBP, a new repressor protein
that interacts with components of the mammalian polycomb complex,
and with the transcription factor YY1. EMBO J. 18:3404–3418. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tavares L, Dimitrova E, Oxley D, Webster
J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et
al: RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb
target sites independently of PRC2 and H3K27me3. Cell. 148:664–678.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Simoes da Silva CJ, Simón R and Busturia
A: Epigenetic and non-epigenetic functions of the RYBP protein in
development and disease. Mech Ageing Dev. 174:111–120. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sawa C, Yoshikawa T, Matsuda-Suzuki F,
Deléhouzée S, Goto M, Watanabe H, Sawada J, Kataoka K and Handa H:
YEAF1/RYBP and YAF-2 are functionally distinct members of a
cofactor family for the YY1 and E4TF1/hGABP transcription factors.
J Biol Chem. 277:22484–22490. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schlisio S, Halperin T, Vidal M and Nevins
JR: Interaction of YY1 with E2Fs, mediated by RYBP, provides a
mechanism for specificity of E2F function. EMBO J. 21:5775–5786.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang W, Cheng J, Qin JJ, Voruganti S, Nag
S, Fan J, Gao Q and Zhang R: RYBP expression is associated with
better survival of patients with hepatocellular carcinoma (HCC) and
responsiveness to chemotherapy of HCC cells in vitro and in vivo.
Oncotarget. 5:11604–11619. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou H, Li J, Zhang Z, Ye R, Shao N,
Cheang T and Wang S: RING1 and YY1 binding protein suppresses
breast cancer growth and metastasis. Int J Oncol. 49:2442–2452.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dinglin X, Ding L, Li Q, Liu Y, Zhang J
and Yao H: RYBP inhibits progression and metastasis of lung cancer
by suppressing EGFR signaling and epithelial-mesenchymal
transition. Transl Oncol. 10:280–287. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ke Y, Guo W, Huang S, Li Y, Guo Y, Liu X,
Jin Y and Ma H: RYBP inhibits esophageal squamous cell carcinoma
proliferation through downregulating CDC6 and CDC45 in G1-S phase
transition process. Life Sci. 250:1175782020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brierley JD, Gospodarowicz MK and
Wittekind C (eds): TNM classification of malignant tumours. 8th
edition. Wiley-Blackwel; 2016
|
|
17
|
He G, Siddik ZH, Huang Z, Wang R, Koomen
J, Kobayashi R, Khokhar AR and Kuang J: Induction of p21 by p53
following DNA damage inhibits both Cdk4 and Cdk2 activities.
Oncogene. 24:2929–2943. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen D, Zhang J, Li M, Rayburn ER, Wang H
and Zhang R: RYBP stabilizes p53 by modulating MDM2. EMBO Rep.
10:166–172. 2009. View Article : Google Scholar :
|
|
19
|
Meyerhardt JA and Mayer RJ: Systemic
therapy for colorectal cancer. N Engl J Med. 352:476–487. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang C, Wang H, Deng M, He L, Ping F, He
Y, Fan Z, Cheng B and Xia J: Upregulated miR-411-5p levels promote
lymph node metastasis by targeting RYBP in head and neck squamous
cell carcinoma. Int J Mol Med. 47:362021. View Article : Google Scholar :
|
|
21
|
Voruganti S, Xu F, Qin JJ, Guo Y, Sarkar
S, Gao M, Zheng Z, Wang MH, Zhou J, Qian B, et al: RYBP predicts
survival of patients with non-small cell lung cancer and regulates
tumor cell growth and the response to chemotherapy. Cancer Lett.
369:386–395. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tong AH, Tan J, Zhang JH, Xu FJ, Li FY and
Cao CY: Overexpression of RYBP inhibits proliferation, invasion,
and chemoresistance to cisplatin in anaplastic thyroid cancer cells
via the EGFR pathway. J Biochem Mol Toxicol. 33:e222412019.
View Article : Google Scholar
|
|
23
|
Zhu X, Yan M, Luo W, Liu W, Ren Y, Bei C,
Tang G, Chen R and Tan S: Expression and clinical significance of
PcG-associated protein RYBP in hepatocellular carcinoma. Oncol
Lett. 13:141–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alt JR, Gladden AB and Diehl JA: p21(Cip1)
promotes cyclin D1 nuclear accumulation via direct inhibition of
nuclear export. J Biol Chem. 277:8517–8523. 2002. View Article : Google Scholar
|
|
25
|
Diehl JA, Zindy F and Sherr CJ: Inhibition
of cyclin D1 phosphorylation on threonine-286 prevents its rapid
degradation via the ubiquitin-proteasome pathway. Genes Dev.
11:957–972. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chipuk JE, Kuwana T, Bouchier-Hayes L,
Droin NM, Newmeyer DD, Schuler M and Green DR: Direct activation of
Bax by p53 mediates mitochondrial membrane permeabilization and
apoptosis. Science. 303:1010–1014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan K, Zhang X, Cong X, Huang B, Chen H
and Chen D: Tumor suppressor RYBP harbors three nuclear
localization signals and its cytoplasm-located mutant exerts more
potent anti-cancer activities than corresponding wild type. Cell
Signal. 29:127–137. 2017. View Article : Google Scholar
|
|
28
|
Danen-van Oorschot AA, Voskamp P, Seelen
MC, van Miltenburg MH, Bolk MW, Tait SW, Boesen-de Cock JG, Rohn
JL, Borst J and Noteborn MH: Human death effector domain-associated
factor interacts with the viral apoptosis agonist apoptin and
exerts tumor-preferential cell killing. Cell Death Differ.
11:564–573. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng L, Schickling O, Peter ME and
Lenardo MJ: The death effector domain-associated factor plays
distinct regulatory roles in the nucleus and cytoplasm. J Biol
Chem. 276:31945–31952. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ilie M, Long-Mira E, Bence C, Butori C,
Lassalle S, Bouhlel L, Fazzalari L, Zahaf K, Lalvée S, Washetine K,
et al: Comparative study of the PD-L1 status between surgically
resected specimens and matched biopsies of NSCLC patients reveal
major discordances: A potential issue for anti-PD-L1 therapeutic
strategies. Ann Oncol. 27:147–153. 2016. View Article : Google Scholar
|
|
31
|
Ganepola GAP, Mazziotta RM, Weeresinghe D,
Corner GA, Parish CJ, Chang DH, Tebbutt NC, Murone C, Ahmed N,
Augenlicht LH and Mariadason JM: Gene expression profiling of
primary and metastatic colon cancers identifies a reduced
proliferative rate in metastatic tumors. Clin Exp Metastasis.
27:1–9. 2010. View Article : Google Scholar
|
|
32
|
Swanton C: Intratumor heterogeneity:
Evolution through space and time. Cancer Res. 72:4875–4882. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Burrell RA, McGranahan N, Bartek J and
Swanton C: The causes and consequences of genetic heterogeneity in
cancer evolution. Nature. 501:338–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rogel A, Popliker M, Webb CG and Oren M:
p53 cellular tumor antigen: Analysis of mRNA levels in normal adult
tissues, embryos, and tumors. Mol Cell Biol. 5:2851–2855.
1985.PubMed/NCBI
|
|
35
|
Finlay CA, Hinds PW, Tan TH, Eliyahu D,
Oren M and Levine AJ: Activating mutations for transformation by
p53 produce a gene product that forms an hsc70-p53 complex with an
altered half-life. Mol Cell Biol. 8:531–539. 1988.PubMed/NCBI
|
|
36
|
Oh HJ, Bae JM, Wen X, Jung S, Kim Y, Kim
KJ, Cho NY, Kim JH, Han SW, Kim TY and Kang GH: p53 expression
status is associated with cancer-specific survival in stage III and
high-risk stage II colorectal cancer patients treated with
oxaliplatin-based adjuvant chemotherapy. Br J Cancer. 120:797–805.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rehman SK, Haynes J, Collignon E, Brown
KR, Wang Y, Nixon AML, Bruce JP, Wintersinger JA, Singh Mer A, Lo
EBL, et al: Colorectal cancer cells enter a diapause-like DTP state
to survive chemotherapy. Cell. 184:226–242.e21. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ketkar M and Dutt S: Epigenetic regulation
towards acquired drug resistance in cancer. Subcell Biochem.
100:473–502. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kobayashi T, Makino T, Yamashita K, Saito
T, Tanaka K, Takahashi T, Kurokawa Y, Yamasaki M, Nakajima K, Morii
E, et al: APR-246 induces apoptosis and enhances chemo-sensitivity
via activation of ROS and TAp73-Noxa signal in oesophageal squamous
cell cancer with TP53 missense mutation. Br J Cancer.
125:1523–1532. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Geisler S, Børresen-Dale AL, Johnsen H,
Aas T, Geisler J, Akslen LA, Anker G and Lønning PE: TP53 gene
mutations predict the response to neoadjuvant treatment with
5-fluorouracil and mitomycin in locally advanced breast cancer.
Clin Cancer Res. 9:5582–5588. 2003.PubMed/NCBI
|
|
41
|
Chen X, Zhang T, Su W, Dou Z, Zhao D, Jin
X, Lei H, Wang J, Xie X, Cheng B, et al: Mutant p53 in cancer: from
molecular mechanism to therapeutic modulation. Cell Death Dis.
13:9742022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu Y and Villalona-Calero MA: Irinotecan:
Mechanisms of tumor resistance and novel strategies for modulating
its activity. Ann Oncol. 13:1841–1851. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Blagih J, Buck MD and Vousden KH: p53,
cancer and the immune response. J Cell Sci. 133:jcs2374532020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Levine AJ: P53 and the immune response: 40
Years of exploration-a plan for the future. Int J Mol Sci.
21:5412020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Oshima H, Nakayama M, Han TS, Naoi K, Ju
X, Maeda Y, Robine S, Tsuchiya K, Sato T, Sato H, et al:
Suppressing TGFβ signaling in regenerating epithelia in an
inflammatory microenvironment is sufficient to cause invasive
intestinal cancer. Cancer Res. 75:766–776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Goodla L and Xue X: The role of
inflammatory mediators in colorectal cancer hepatic metastasis.
Cells. 11:23132022. View Article : Google Scholar : PubMed/NCBI
|