The incidence of prostate cancer (PCa) remained
stable from 2014 to 2018 but contributed to 27% of all cancer cases
in the USA (1). However, the
incidence of advanced PCa in the USA has been increasing by 4-6%
annually since 2011. PCa has remained the second leading cause of
death in males with cancer over the past decade, and this has been
attributed to the development of castration-resistant prostate
cancer (CRPC) (1). At present,
the treatment and management of CRPC is challenging and most
patients have a poor prognosis (2,3).
Like normal prostate cells, PCa cells require androgens for
continued growth (4). Therefore,
the primary treatment for advanced or metastatic PCa is androgen
deprivation therapy (ADT) by surgical or pharmacological castration
(5).
The androgen receptor (AR), a ligand-dependent
nuclear transcription factor, binds to testosterone or
dihydrotestosterone (DHT), leading to the transcription of
AR-responsive genes, which drive the proliferation and survival of
prostate cells (6). Compared with
benign prostatic hyperplasia (BPH), the AR is upregulated in
primary PCa and is even upregulated throughout the progression to
CRPC during ADT (7). The
mechanisms by which androgen-dependent prostate cancer (ADPC)
progresses to CRPC remain largely unknown. However, the AR is the
most researched molecular factor in the context of PCa research and
is reported to promote CRPC. The mechanisms underlying the
development of CRPC are divided into AR-dependent and
AR-independent pathways (also termed bypass ways) (8).
The AR-dependent pathways include: i) High affinity
for ligands by translated AR gene mutants; ii) AR splice variants
that are constitutively active without ligand; iii) AR gene locus
amplification; iv) ectopic biosynthesis of androgens from adrenal
steroids and cholesterol or paracrine biosynthesis from mesenchymal
cells; and v) non-canonical induction of AR signaling, such as the
IL-6/STAT3 pathways, in the absence of ligand (9-12).
Including AR V7 and Arv567es, >20 AR variants have been
identified. A dedicated review that discusses these AR mutations is
already available (13), and
therefore, AR variants will not be discussed in the present review.
In addition, alternative signaling pathways supporting the growth
and viability of CRPC cells have been demonstrated to bypass the AR
absolutely (14). For instance,
the glucocorticoid receptor (GR) binds to androgen response
elements to sustain PCa cell proliferation. In previous studies, it
was demonstrated that inhibiting the GR or glucocorticoid-regulated
kinase 1, a target gene of both the AR and GR, delayed
castrate-resistant tumor formation (15-17).
Regardless of the aforementioned mechanisms,
proteins are the executors of the biological functions in CRPC
progression (18). Therefore,
identifying new proteins or studying their functions in CRPC will
lead to the identification of new treatment targets, biomarkers for
early stages of PCa and markers for predicting recurrent or
treatment response. This will lead to an improvement in prognosis
for patients with CRPC. Mass spectrometry is a powerful method that
enables increasingly comprehensive insights into changes in the
proteome, allowing for high-throughput analysis of clinical patient
samples (19). In addition,
studies involving large-scale, mass spectrometry-based proteomics
of human cancer have recently been published (20,21). Researchers have found and
validated many differentially expressed proteins from primary PCa
and CRPC clinical tissue samples (22). Emerging proteins in CRPC have been
identified from various sources, including human tissues, mouse
xenografts, cell lines and human serum or urine samples (23). Therefore, the present review
focuses on the expression and function of proteins in patients with
CRPC, particularly whether they can regulate AR expression or
translation activity. In addition, their clinical applications in
the management of CRPC are outlined, which were identified or
validated using immunohistochemistry (IHC). These proteins are
expected to be potential diagnostic markers, therapeutic monitoring
indicators and therapeutic targets for CRPC.
Although CRPC is androgen-independent, AR gene
amplification or upregulation is observed in up to 80% of samples
from patients with CRPC(24).
Various proteins, such as epigenetic modification factors, promote
ectopic AR expression, contributing to CRPC (25,26). Protein arginine methyltransferase
5 (PRMT5), an epigenetic activator, is upregulated in CRPC and
activates AR transcription by recruiting pICln to the AR promoter
(25). In addition, PRMT5 or
pICln induces CRPC tumor growth in mice (25). Moreover, 4-1BB ligand (4-1BBL,
also termed CD137L), a transmembrane glycoprotein belonging to the
tumor necrosis factor family, has been found to be upregulated
during the progression of PCa to CRPC, thereby promoting the
expression of AR (26). However,
the process by which 4-1BBL promotes AR expression is yet to be
identified. In addition, 4-1BBL augments the proliferation and
invasion abilities of PCa cells in an androgen-deprived environment
(26).
Similarly, several transcription factors that induce
AR expression at the transcriptional level are upregulated in
patients with CPRC (27). Y-box
binding protein-1 (YB-1) is one of the transcription factors that
regulate AR transcription by binding to the Y-box in the AR
promoter. In addition, YB-1 modulates the expression of AR variants
at the transcription and splicing levels (28-30). Moreover, another transcription
factor, twist basic helix-loop-helix transcription factor 1
(Twist1) was reported to upregulate AR gene expression by binding
to E-boxes in the AR promoter region (31). Notably, Twist1 enhances CRPC cell
proliferation, leading to cisplatin and taxane resistance by
increasing YB-1 expression. YB-1 also regulates the expression of
Twist1, suggesting a strong functional crosstalk between the two
proteins (32).
Kinases responsible for the phosphorylation of
transcription factors also play critical roles in CRPC progression.
During ADT, YB-1 phosphorylation is induced by ribosomal protein S6
kinase A1 (RSK1) phosphorylation, which regulates full-length AR
and AR V7 splicing (28,29). In addition, Twist1 is stabilized
by LIM-domain kinase-2 (LIMK2) via phosphorylation to prevent its
degradation (33). The feedback
loop between Twist1 and LIMK2 increases PCa cell migration and
promotes epithelial-to-mesenchymal transition (EMT) (33). Moreover, LIMK2 degradation is
decreased by its phosphorylation via aurora A kinase (AURKA). It
has also been shown that AURKA is upregulated by LIMK2 and is also
positively regulated by the androgen-induced AR, which binds in its
intronic region (34). In
conclusion, YB-1, Twist1, RSK1, LIMK2, AURKA and AR are
significantly upregulated in CRPC samples, and are involved in a
positive feedback loop that synergistically promotes CRPC
progression.
In addition to alterations in AR expression or
structure, many factors contribute to AR activation despite
castrate levels of serum androgens. These alterations include
changes in AR stability, steroid metabolism, coactivator
expression/activity and cell signaling (35). Furthermore, the modulation of AR
activity through various posttranslational modifications, such as
ubiquitination and phosphorylation, has been extensively studied
(36-38).
As a transcription factor, the AR is regulated by E3
ubiquitin ligases of the ubiquitin-proteasome pathway, such as
mouse double minute-2 (Mdm2), seven in absentia homolog 2 (Siah2)
and ring finger protein 6 (RNF6), which have been implicated in the
control of AR stability and activity (36,39,40). The expression level of Siah2 is
significantly upregulated in CRPC tissues and is required for the
growth of CRPC tumors in mice (39). Notably, Siah2 binds to the
corepressor, NCOR1, to remove the transcriptionally inactive AR
from chromatin, enhancing AR transcriptional activity (39). In addition, the expression of
another ubiquitin E3 ligase, RNF6, was found to be upregulated
during PCa progression (36).
Furthermore, RNF6 assumes a critical role in promoting PCa cell
growth under androgen-depleted conditions by ubiquitinating AR at
K845. This ubiquitination event serves as a scaffold for the
recruitment of coactivators such as ARA54 (36).
The expression of non-receptor tyrosine kinase is
upregulated in CRPC samples and correlates with AR Y534
phosphorylation. Studies have shown that phosphorylation of the AR,
induced by Src interacting with the AR through its Src homology 2
domain, profoundly affects the stability and turnover of the AR. As
a result, this interaction may prevent the association of the AR
with Mdm2 (37). FK506 binding
protein 4 (FKBP4, also termed FKBP52), is another protein that
promotes phosphorylation of the AR to enhance its transcriptional
activity. A previous study analyzing >500 PCa samples revealed
that FKBP4 expression is upregulated in CRPC, when compared with
hormone-sensitive prostate cancer (HNPC) (38). Growth factor receptor bound
protein 10 (GRB10) also phosphorylates the AR at S81, which is
critical for AR transcriptional activity (41). GRB10 is the most significantly and
consistently upregulated gene during CRPC progression and markedly
induces PCa cell growth (42).
Several studies have demonstrated that steroidogenic
enzymes involved in androgen biosynthesis are upregulated in PCa
tissues, promoting CRPC development (43-45). The AR becomes more sensitive due
to the tumor's own de novo androgen production, thereby
promoting the progression of PCa. Notably, increased expression of
lipocalin 2 (LCN2) was detected in CRPC tissues, when compared with
patients with PCa or BPH (43).
LCN2 upregulation leads to upregulation of the AR downstream gene,
SLC45A3, without affecting AR levels, suggesting that LCN2 enhances
AR transcriptional activity and thereby contributes to CRPC
progression (43). Similarly,
ido-keto reductase family 1 member C3 (AKR1C3) is a critical enzyme
for catalyzing the biochemical reduction of 5α-Adione to DHT in PCa
cells (44). In addition, AKR1C3
promotes EMT during PCa metastasis by activating extracellular
regulated protein kinase (ERK) signaling (44). Moreover, the expression of AKR1C3
is significantly higher in CRPC tissues than in HNPC tissues of the
same patients (46).
Estrogen-related receptor α(ERRα) was also reported to be
upregulated in metastatic CRPC (mCRPC) (45). In addition, ERRα enhances
intra-tumoral androgen biosynthesis by regulating the transcription
of AKR1C3 (45). ETS-related gene
1 (ERG) and AKR1C3 are co-expressed in human prostate tumor tissue
specimens, and they predict a lower probability of survival
(47).
Evidence from previous studies has indicated that
the dysregulation of AR cofactors contributes to the development
and progression of CRPC (48-52). DEAH-box RNA helicase family member
15 (DHX15) is an AR coactivator that forms a complex with the AR
and Siah2 to increase their stability and enhance the E3 ubiquitin
ligase activity of Siah2 (48).
The expression of DHX15 is also upregulated in human CRPC tissues
compared with HNPC tissues (53).
Thioredoxin domain-containing protein 5 (TXNDC5) is another AR
cofactor upregulated in CRPC. TXNDC5 directly interacts with the AR
protein to increase its stability, and thus enhances its
transcriptional activity through hypoxia inducible factor-1α in a
miR-200b-dependent manner (49).
Moreover, octamer transcription factor 1 (OCT1) is an
AR-interacting protein that regulates target gene expression in PCa
cells and has been shown to enhance AR transcriptional activity to
induce the growth and migration of 22Rv1 cells (50). Notably, the expression of OCT1 and
disks large-associated protein 5 was found to be upregulated in
CRPC specimens (50). Nuclear
receptor coactivator 2 (NCoA2, also termed SRC-2), is a
well-studied coactivator of the AR. NCoA2 is often found to be
upregulated in patients with metastatic PCa and plays a key role in
driving the development of CRPC (51). When androgen levels are reduced
due to deprivation therapy, NCoA2 levels increase. This heightened
NCoA2 expression, in turn, triggers activation of the
phosphatidylinositol-3 kinase (PI3K) signaling pathway, thereby
promoting the metastasis of prostate cancer (51). Based on quantitative protein
results, forkhead box protein A1 (FOXA1) was identified to be
elevated in PCa tumor-node-metastasis stage 3 (including both
Gleason grade 3 and Gleason grade uncertain) and CRPC despite ADT
treatment (54,55). FOXA1 is a pioneer factor
facilitating AR transcription and PCa growth (52,56) and possesses an AR-independent role
in regulating EMT (52). Notably,
the expression of Yes-associated protein 1 (YAP1) is upregulated
and activated in CRPC and enzalutamide-resistant cells but is
downregulated in neuroendocrine prostate cancer (NEPC) (57,58). YAP1 binding to the AR in the
nucleus is regulated by macrophage stimulating 1 (MST1) signaling,
which may play a prominent role in the emergence of advanced PCa
(59). Moreover, YAP1 silencing
attenuates the growth and invasion of PCa cells in vitro
(59). Functional analyses have
uncovered that YAP1 positively regulates numerous genes involved in
cancer stemness and lipid metabolism (60). In addition, YAP1 interacts with
chicken ovalbumin upstream promoter transcription factor 2 to form
a transcriptional complex.
Studies have shown that several growth factors and
cytokines, such as epidermal growth factor, transforming growth
factor (TGF)α, IL-6 and their downstream tyrosine kinases,
including erbB2, Src and focal adhesion kinase, can activate the AR
and minimize or possibly even negate the requirement for ligand
(61-63). During ADT, the AR is inactive,
however, in compensation, IL-6 and STAT3 induce activation of the
AR in a ligand-independent manner (64). Moreover, the upregulation of
nuclear AR expression by IL-6 has been demonstrated (65). In addition, both IL-6 and
phosphorylated STAT3 (pSTAT3) are upregulated in bone metastases
tissues from patients who died from CRPC (66). The downstream target genes and
relevant signaling pathways regulated by IL-6 interweave into a
vast signaling network that could promote the progression of CRPC.
For instance, inositol-requiring enzyme 1α (IRE1α), a key regulator
of the unfolded protein response, is associated with CRPC
development and promotes the castration-resistant growth of PCa
cells in an IL-6/AR-mediated manner (67).
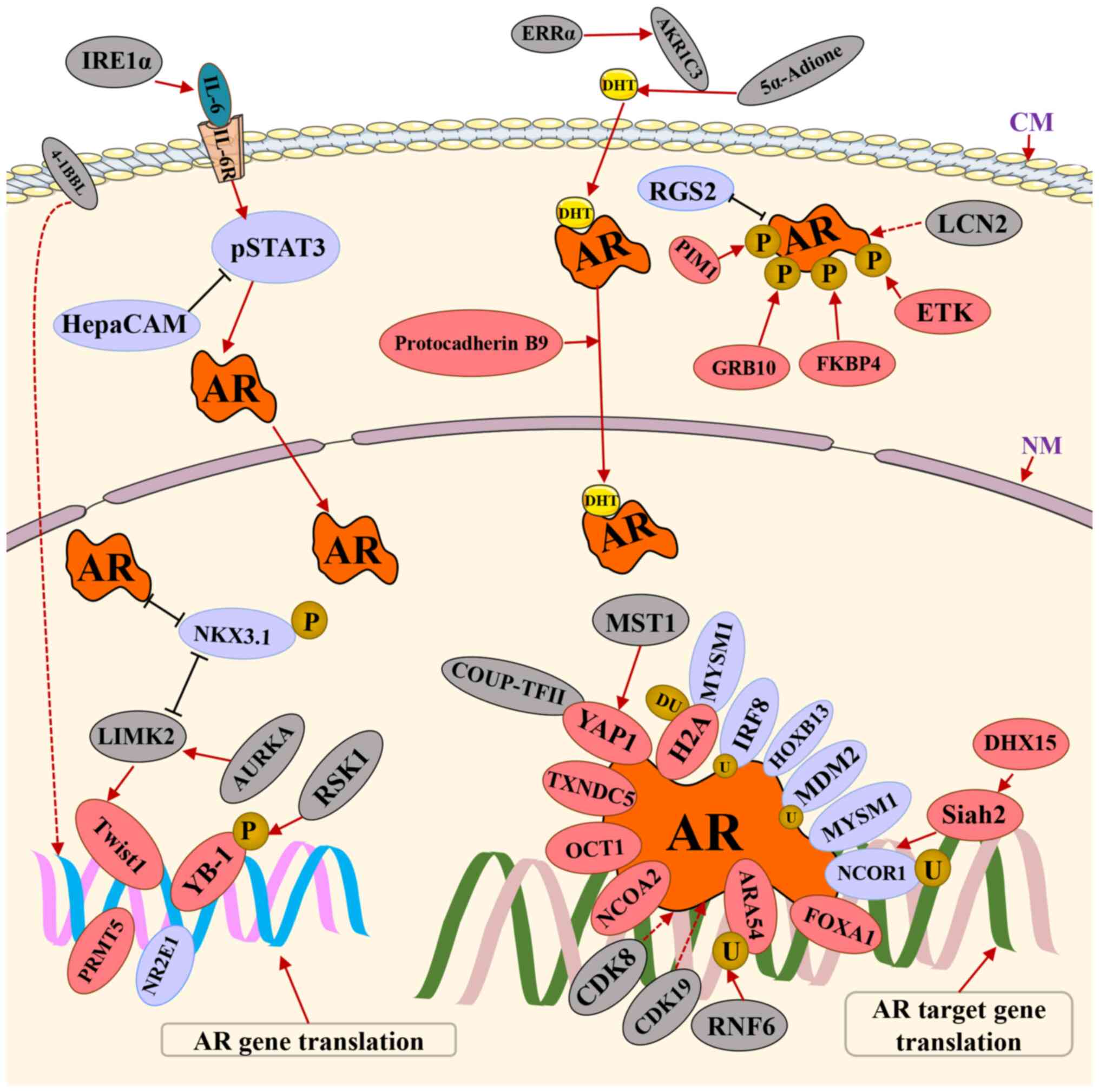
Protocadherin B9, which is involved in cell adhesion
and migration, promotes nuclear AR translocation in LNCaP cells
(68). The expression of
protocadherin B9 is associated with the preoperative prostate
specific antigen (PSA) concentration, the Gleason score, lymphatic
invasion and seminal vesicle invasion in PCa cases (68). In addition, the expression of
nuclear CDK19 and CDK8 is upregulated during PCa progression to
CRPC (69). A study demonstrated
that CDK8/CDK19 inhibition reduced cell migration and increased
collagen I-dependent adhesion (70). It also demonstrated that combining
CDK8/CDK19 inhibitors with anti-androgens lead to synergistic
antiproliferative effects and sensitized androgen-independent cells
to bicalutamide. It was therefore suggested that CDK8/CDK19
partially mediates its pro-oncogenic effects via the AR axis.
Proteins that upregulate AR expression or activity
have been extensively examined, but there are limited reports on
proteins that suppress AR expression or activity. The expression of
orphan nuclear receptor, TLX (also termed nuclear receptor
subfamily 2, group E, member 1), is upregulated in mCRPC and it
directly binds to the AR promoter and represses AR transcription by
recruiting histone modifiers, including histone deacetylase
(HDAC)1, HDAC3 and lysine-specific demethylase 1, inducing
resistance to androgen deprivation in PCa cells (71). NK3 homeobox 1 (NKX3.1), a
prostate-specific homeodomain-containing transcription factor, is
negatively associated with the initiation and progression of PCa
and the progression of CRPC (72). NKX3.1 downregulates Ak strain
transforming (AKT) activation and decreases AR and ARv7 levels in
CRPC cells, and NKX3.1 can be degraded following phosphorylation
via LIMK2 in CRPC (72).
A study has suggested that Mdm2, an E3 ubiquitin
ligase, can induce polyubiquitination of the AR, which results in
AR nuclear degradation (40). In
addition, Mdm2 is downregulated in CRPC cell lines compared with
the hormone sensitive prostate cancer cell lines (40,73). Unlike Mdm2, interferon regulatory
factor 8 (IRF8) directly combines with the AR and promotes its
degradation by activating the ubiquitin/proteasome systems
(74). It is also of note that
IRF8 expression was upregulated in primary PCa tissues but
downregulated in CRPC tissues, compared with normal prostate
tissues (74). By contrast,
G-protein signaling protein 2 (RGS2) regulators are downregulated
at the early stages of PCa (75).
However, late or advanced stages of PCa are associated with RGS2
upregulation, which correlates with a poor survival rate and high
metastasis (75-77). Both the AR and RGS2 inhibit each
other and RGS2 may suppress androgen-independent AR activity by
inhibiting ERK activity in PCa cells (77). In addition, Myb-like SWIRM and MPN
domains 1 (MYSM1) acts as a histone H2A deubiquitinase and a study
has shown that MYSM1 expression is downregulated in CRPC compared
with localized PCa (78). In
addition, MYSM1 interacts with the AR and reduces AR activity by
inhibiting AKT/c-Raf/GSK-3β signaling (78). Homeobox B13 (HOXB13) is one of the
39 HOX homeodomain proteins. A reporter transcription assay
demonstrated that HOXB13 significantly suppressed hormone-mediated
AR activity in a dose-responsive manner, and suppression was
specific to the AR, with which HOXB13 physically interacts
(79). Another study has shown
that HOXB13 regulates AR action on endogenous target genes
(80). HOXB13 is a bifunctional
regulator of AR transcriptional activity, demonstrating the
hallmarks of both an activator and a repressor (80). Notably, the upregulation of HOXB13
led to the suppressed proliferation of LNCaP cells (79). In addition, interference with
HOXB13 expression with small interfering RNA also resulted in the
inhibition of LNCaP cell proliferation (80). Collectively, these observations
suggest a pivotal role of HOXB13 in LNCaP cell proliferation
(80). The relationship between
AR and the aforementioned proteins is outlined in Fig. 1 and Table I.
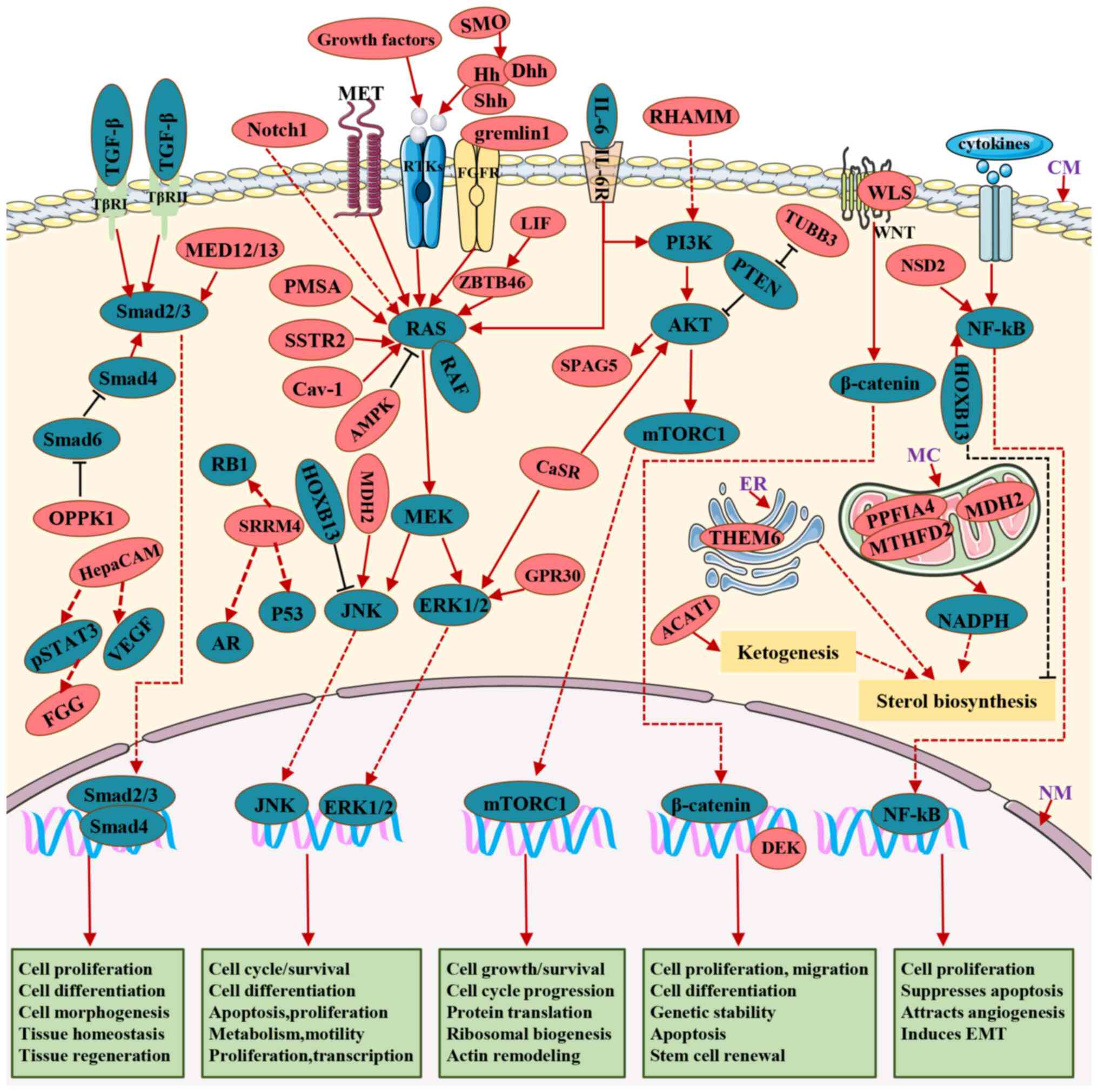
Once PCa cells undergo androgen targeted therapies
(ATTs), either AR signaling is reactivated as previously described
or AR pathways are bypassed, probably by transdifferentiating
neuroendocrine (NE) or by switching to an AR-null NE-null phenotype
termed double-negative prostate cancer (DNPC) (81,82). The process by which AR bypassing
promotes PCa progression to CRPC includes dysregulated receptor
tyrosine kinases (RTKs; receptors of growth factors) and downstream
effectors comprising mitogen-activated protein kinase (MAPK),
mitogen-activated extracellular signal-regulated kinase (MEK) and
ERK, TGF-β/mothers against decapentaplegic (SMAD) signaling, the
PI3K/AKT/mTOR pathway, Wnt/β-catenin signaling, activation of the
NF-κB pathway by cytokines and GR upregulation (83-86). This process was reviewed by Makino
et al (83) and Saraon
et al (84). Some pathways
form intricate connections with AR signaling by directly regulating
AR, while others completely circumvent AR to facilitate the
survival, proliferation, migration and invasion of CRPC cells
(84). Consequently, the abnormal
expression of proteins involved in these pathways plays a
significant role in driving the progression of CRPC.
The RTKs/MAPK/MEK/ERK signaling network is a
canonical pathway that is essential to the carcinogenesis of
various human tumors since it is closely related to cell growth,
survival, differentiation, invasion, metastasis, extracellular
matrix degradation and angiogenesis (85). Consequently, any RTK ligand, such
as growth factors or proteins, can regulate any of the signal
factors that lead to CRPC (87-89). A study has shown that gremlin1, a
fibroblast growth factor receptor 1 (FGFR1) ligand, promotes CRPC
by activating the FGFR1/MAPK signaling pathway, and that the
transcription of GREM1 is suppressed by AR and released following
ADT (90). MET, the hepatocyte
growth factor receptor, is almost exclusively expressed in CRPC
(91). Moreover, MET
overexpression in DU145 cells enhances cell migration, cell
invasion and the acquisition of a stem-like phenotype (91). The hedgehog (Hh) ligands, sonic
and desert, have been reported to be elevated in castration-induced
CRPC (92). The expression of Hh
promotes CRPC progression by eliciting paracrine effects on
epithelial growth and differentiation (93). Target genes of Hh signaling
include several growth factors, such as insulin-like growth factor
binding protein (Igfbp)-6 and Igfbp-3 (93), indicating that Hh signaling may
activate RTK pathways in CRPC.
Caveolin-1 (Cav-1) expression has been shown to be
upregulated in immunohistochemical assays of biopsies from patients
with CRPC, compared with primary PCa samples (94). Moreover, Cav-1 promotes the
invasion and migration of CRPC cells by activating the
H-Ras/phospholipase Cε signaling pathway in the cell membrane
caveolae (94). IL-6 exhibits the
capability to enhance not only the activity and expression of the
AR but also to activate the ERK1/2-MAPK signaling pathway (64,65,95). Upon binding to IL-6, Janus kinase
(JAK) phosphorylates Src homology 2 domain-containing tyrosine
phosphatase 2 (95). This event
triggers the activation of Ras, setting off a cascade of reactions
that lead to the sequential activation of Raf, followed by MEK and
culminating in the activation of ERK. Additionally, the levels of
zinc finger and BTB domain-containing protein 46 (ZBTB46) and
leukemia inhibitory factor (LIF) are associated with PCa
progression (24,96). LIF-induced androgen-independent
proliferation, invasion and NE transdifferentiation via activation
of ZBTB46 expression further activates the JAK/STAT and Ras/MAPK
pathways in CRPC cells (24,96,97). Although diminished or lost
somatostatin receptor 2 (SSTR2) expression is consistent with
advanced tumor grade (98), SSTR2
expression is elevated following hormone depletion in PCa and
contributes to NEPC via modulating MAPK through G protein-dependent
mechanisms (99). In addition,
patients with mCRPC have higher G protein coupled receptor 30
(GPR30) expression compared with primary PCa (100). Moreover, GPR30 is also
upregulated in stromal cells, promoting PCa cell invasion (100,101). Notably, GPR30 induces the growth
of breast and ovarian cancer cells, whereas the activation of GPR30
by a selective agonist, G-1, inhibited the growth of
androgen-dependent and androgen-independent PCa cells in
vitro and in vivo via continuous activation of ERK1/2
and c-jun/c-fos (102). GPR30
can also promote the proliferation and migration of PCa cells in a
paracrine manner (101).
Considering the contradictory effect of GPR30 in stromal and
epithelial cells on PCa development, there is a need for further
studies to investigate the underlying mechanism.
A study has shown that the tumor suppressor gene,
PTEN, is downregulated in PCa, inhibiting the activation of the
PI3K/AKT/mTOR pathway (112).
Consequently, the expression levels of class III β-tubulin (TUBB3)
and PTEN are inversely regulated, suggesting that TUBB3 is related
to PTEN deficiency, and that TUBB3 may activate the PI3K/AKT
pathway (112). TUBB3, a
primarily neural isoform of β-tubulin, is significantly upregulated
when ADPC progresses to CRPC (113). In addition, TUBB3 is an adverse
prognostic factor in patients with mCRPC treated with docetaxel
(114). Moreover, the TUBB3
protein is stabilized by Src-mediated tyrosine phosphorylation,
promoting the stabilization of mitotic spindles in dividing cells
and resulting in resistance to taxane therapy (115).
The hyaluronan-mediated motility receptor (RHAMM)
signal transduction pathway not only activates cell cycle genes but
also plays a fundamental role in cell growth, differentiation and
motility (116). ADT upregulates
the expression of RHAMM in patients with PCa (116). In addition, the expression of
RHAMM is upregulated when tumor cells progress to a castration
resistant stage. Hyaluronan binds to RHAMM and activates its
downstream proteins, including ROK1, GRB2-associated binding
protein-1, PI3K*p110α and eukaryotic translation initiation factor
4E family member 3, to facilitate cell motility and accelerate cell
invasion and metastasis of CRPC cells, compared with ADPC cells
(117). IL-6 possesses the
ability to not only activate ERK but also initiate signal
transduction via the PI3K signaling pathway (95). Activation of PI3K leads to the
recruitment of the protein kinase, AKT, to the plasma membrane and
subsequent binding, and the complex crosstalk between the
PI3K/AKT/mTOR pathway and multiple interacting cell signaling
cascades can further promote CRPC progression (86,95).
The calcium-sensing receptor (CaSR) is a receptor
for several ligands, including Ca2+, amino acids,
vitamin D and IL-6 (118). The
expression of CaSR is upregulated in mCRPC and NEPC and is
associated with shorter overall survival (119,120). CaSR is a potential NE marker
that promotes NE differentiation in PCa (119). CaSR enhances the proliferation
and migration of PCa cells by both ERK and AKT signaling pathways
(121,122). Similarly, sperm-associated
antigen 5 (SPAG5) expression is markedly upregulated in primary PCa
(compared with normal tissues), metastatic PCa (compared with
primary PCa), CRPC (compared with HNPC) and NEPC (compared with
prostate adenocarcinoma) (123).
SPAG5 promotes colony formation, migration and invasion of PCa
cells and increases both the tumor volume and weight in mice
xenograft models (123). In
addition, targeting the PI3K/AKT/mTOR signaling pathway leads to
the downregulation of SPAG5 in LNCaP cells, indicating that SPAG5
is involved in the AKT/mTOR pathway (124).
Both Mediator complex subunit 12 (MED12) and MED15
are components of the Mediator complex, which modulates TGF-β
receptor signaling (125). A
study has shown that nuclear MED12 is upregulated in 40% of distant
mCRPC and 21% of local recurrent CRPC, inconsistent with the low
frequencies (11%) in HNPC and the lack of expression in BPH tissues
(126). Similar to MED12, MED15
is upregulated in both distant mCRPC (76%) and local recurrent CRPC
(70%), compared with HNPC or BPH tissues (127). In addition, expression of
nuclear MED12 was significantly correlated with the nuclear
localization of phosphorylated SMAD3, whereas MED12 knockdown
reduced levels of the TGF-β target gene, vimentin, and promoted the
expression of p27 (126). It was
also found that MED15 expression is upregulated in CRPC tissues
after ADT (72%) and the CRPC cell line, PC3 (128). Moreover, inhibition of MED15
expression reduced viability and induced apoptosis in LNCaP cells
after ADT. The expression of MED15 is positive correlated with the
phosphorylation level of both AKT and SMAD3 (128).
A study has shown that the transmembrane
co-receptor, neuropilin-1 (NRP1), promotes cancer progression via
TGF-β/SMAD signaling (129).
NRP1, which is repressed by androgen, is upregulated in mCRPC, and
the inhibition of NRP1 expression significantly restores the
invasive and metastatic ability of PC3 cells (130). Similarly, the κ-type opioid
receptor (OPRK1) is a G protein-coupled receptor repressed by the
AR in androgen-containing medium (131). OPRK1 expression is significantly
induced by ADT and is upregulated during CRPC progression. OPRK1
supports the androgen-independent growth of VCaP cells, and the
SMAD6 pathway is downregulated by OPRK1 blockade under castrated
conditions (131).
A study has shown that the Wnt secretion mediator,
Wntless (WLS), is a significant driver of NEPC and promotes the
growth of NEPC cells by activating the receptor tyrosine
kinase-like orphan receptor 2/protein kinase Cδ/ERK signaling
pathway (132). However, the
expression of WLS is repressed by the AR in HNPC cells. It was also
revealed in the same study that the expression of WLS is enhanced
in both CRPC and NEPC tumors.
It is well-known that histone modifications are
essential in gene transcription and participate in tumor
progression. Nuclear receptor binding SET domain 2 (NSD2, also
termed MMSET), a histone methyltransferase, catalyzes the mono- and
di-methylation of H3K36 (133).
Analysis of clinical samples demonstrated that NSD2 expression was
increased in CRPC samples compared with HNPC (134). In addition, NF-κB is
constitutively activated by the cytokine autocrine loop mediated
via NSD2 binding to the chromatin of NF-κB, which improves survival
and promotes the proliferative capacity of tumor cells (135).
HOXB13 was significantly upregulated in
hormone-refractory tumors compared with tumors without PSA after
initial treatment (136).
Additionally, heightened expression of HOXB13 was correlated with
an increased growth advantage in PCa cells under conditions of low
or absent androgen levels. This effect was linked to the activation
of the retinoblastoma tumor suppressor (RB)/E2F signaling pathway
and the suppression of c-Jun N-terminal kinase (JNK)/c-Jun
expression, which was achieved through the inhibition of the
p21waf tumor suppressor (136,137) Furthermore, another study found
that HOXB13 promotes PCa invasion and metastasis by reducing
intracellular zinc levels, consequently stimulating NF-κB signaling
(138). This suggests that
HOXB13 plays a pivotal role in enhancing the malignant
characteristics of PCa. By contrast, in ~30% of mCRPC cases,
hypermethylation and subsequent downregulation of the HOXB13 gene
were observed (139). The loss
of HOXB13 was associated with lipid accumulation in PCa cells,
leading to increased cell motility and enhanced xenograft tumor
metastasis. Therefore, the impact of HOXB13 on the proliferation
and migration of PCa cells in various cellular contexts and under
different androgen level environments has yielded conflicting
research findings. These conflicting results underscore the
complexity of the involvement of HOXB13 in PCa and emphasize the
importance of further research to gain a comprehensive
understanding of its effects.
Numerous studies have demonstrated that some PCa
cells can survive ATTs via lineage transition, such as transforming
to NEPC cells for survival (140-142). NEPC is an aggressive subtype of
CRPC with poor overall survival (142). Several proteins have been
reported to participate in NE lineage transition, such as the
previously mentioned SSTR2 (99),
CaSR (118) and SPAG5 (123). However, the mechanisms of
certain other proteins, such as splicing factor serine/arginine
repetitive matrix 4 (SRRM4) (143) and the DNA topology modulator,
DEK (24), detected in NEPC are
unknown or do not involve the aforementioned RTKs/MAPK/MEK/ERK,
PI3K/AKT/mTOR, TGF-β/SMAD and Wnt/β-catenin, NF-κB pathways. The
expression of SRRM4 and DEK, are both upregulated in NEPC (143,144). SRRM4 is a vital driver gene that
not only promotes PCa cell survival, proliferation and
tumorigenesis but also alters cellular morphology and transforms
ADPC cells into NEPC xenografts in vivo (24). SRRM4 crosstalks with other
signaling pathways, such as the AR, p53 and RB1 pathways, to
modulate the phenotypical reprogramming of PCa cells (145). DEK induces tumorigenesis and
neoplastic progression by promoting cell division, inhibiting cell
differentiation, senescence and apoptosis, and cooperating with
transforming oncogenes (146).
Inhibition of DEK significantly reduces cell proliferation,
migration and invasion in PC3 cells (143).
The development of treatment resistance in cancer
cells is accompanied by metabolic adaptations that enhance their
survival under stress-inducing conditions (147). Both the mitochondria and the
endoplasmic reticulum (ER) play a pivotal role in modulating
stress-signaling pathways. In a previous study, a fraction of
tyrosine phosphatase receptor type F polypeptide interacting
protein α4 (PPFIA4) interacted with methylenetetrahydrofolate
dehydrogenase 2 (MTHFD2), a critical enzyme for one-carbon
metabolism (148). PPFIA4 was
located in the mitochondria, and its expression was significantly
upregulated in CRPC samples compared with localized PCa. ADT
induced PPFIA4 translocation into the mitochondria, where it
subsequently bound to MTHFD2, promoting the phosphorylation of
MTHFD2 (148). Subsequently, the
production of NADPH was upregulated, promoting the survival of
tumor cells in androgen deprivation-induced mitochondrial
dysfunction. The expression of malate dehydrogenase 2 (MDH2),
another mitochondrial tricarboxylic acid cycle enzyme, is
significantly upregulated in CRPC compared with PCa and BPH
(149). Moreover, inhibition of
MDH2 further increases the docetaxel-induced phosphorylation of
JNK, activating transcription factor (ATF) 2 and c-Jun (150). Consequently, the anti-apoptotic
protein, B-cell lymphoma 2, is inactivated by the phosphorylated
JNK, which promotes the initiation of mitochondria-based apoptosis.
In addition, the ER membrane-associated protein, thioesterase
superfamily member 6 (THEM6), is an ADT-induced protein that is
significantly increased in CRPC cells and that alters ER function,
promoting de novo sterol biosynthesis and mediating
lipid-mediated activation of ATF4 to maintain the growth and
survival of CRPC cells (151).
Metabolic pathways, such as lipogenesis, cholesterol biosynthesis
and ketogenesis, play essential roles in PCa progression (152-154). The expression of acetyl-coenzyme
A acetyltransferase 1 (ACAT1) is upregulated in patients receiving
ADT and during CRPC progression, with metastatic bone lesions
containing the most prominent expression patterns (154,155).
The three oncogenic PIM family kinases, PIM1-3,
have been implicated in the development of PCa. In CRPC biopsies,
both PIM1 and PIM2 expression levels were significantly upregulated
compared with primary PCa samples (156). The expression of the PIM family
members was also positively correlated with ERG and MYC
oncoproteins. Notably, ERG directly binds to the promoter of all
PIM genes, upregulating both gene and protein expression levels of
the PIMs (156). Serum
fibrinogen γ (FGG) is a downstream target gene regulated by the
IL-6/STAT3 pathway. The expression of FGG was significantly higher
in patients with CRPC than in patients with localized PCa (157). In addition, FGG knockdown
resulted in the inhibition of proliferation, migration and invasion
capabilities while inducing the apoptosis of PCa cells (157). Nonetheless, there is a
requirement for additional research to explore the downstream
pathways associated with PIM and FGG that contribute to the
progression of CRPC. Expression of the nuclear Notch homolog 1,
translocation-associated (Notch1) receptor intracellular domain, is
significantly upregulated in high Gleason score (8-10)
cases of HNPC and in almost all mCRPC samples, but not in benign
samples or low Gleason score (<8) localized PCa (158). Furthermore, there is a
synergistic effect among Notch1 with the AKT, Myc and Ras/Raf/MAPK
pathways, promoting the prostate castration-resistant phenotype
(158). However, Notch1
contradictorily plays both a suppressive and oncogenic role in PCa
development, which requires further investigation (159).
Repressors, unlike the extensively examined
proteins that drive CRPC through AR-independent pathways, have not
received adequate research attention. Two examples of repressors
are adenosine monophosphate-activated protein kinase (AMPK) and
aconitase 2 (ACO2). AMPK restrains CRPC progression by inhibiting
both fatty acid and cholesterol synthesis (160). In a previous study, AMPK was
found to be downregulated in CRPC specimens compared with HNPC
specimens due to phosphorylation, which promoted CRPC progression
(161). Notably, ACO2 expression
was higher in PCa than in BPH but lower in CRPC than in PCa
(149). ACO2 promoted in
vivo prostate cancer progression through promoting
mitochondrial citrate synthesis to facilitate de novo
lipogenesis. Sirtuin 3, which acetylates ACO2, is upregulated
following ATT (162). However,
the role and exact mechanism by which ACO2 expression is
downregulated in CRPC remains unknown.
The G protein-coupled receptor smoothened (SMO)
plays an important role in the Hh pathway and the loss of SMO was
observed in all NEPC specimens but only in 9% (2 of 22) of
high-grade ADPC samples (163).
Moreover, the loss of SMO attenuated AR signaling, indicating that
the Hh pathway is inhibited during the pathogenesis of NEPC.
Previously, it was reported that activation of the IL-6/STAT3
pathway and the downstream target genes of this pathway could
promote the progression of CRPC (64). However, hepatocyte cell adhesion
molecule (HepaCAM) is a tumor suppressor that is downregulated in
CRPC tissues compared with matched primary PCa tissues (164), and it suppresses the
proliferation, migration and invasion of PCa cells by decreasing
the expression of pSTAT3, G1/S-specific cyclin-D1, MYC
proto-oncogene bHLH transcription factor, matrix metallopeptidase
(MMP) 2, MMP9 and vascular endothelial growth factor (165). More notably, HepaCAM inhibits
the metastasis of CRPC cells from the prostate to the lungs
(165). These AR-independent
signaling proteins and their associated pathways are described in
Fig. 2 and Table II.
In an earlier discussion, the proteins exhibiting
changes in expression within CRPC clinical samples were explored,
highlighting their potential clinical significance, including roles
as diagnostic markers, indicators for therapeutic monitoring and
targets for CRPC treatment, as detailed in Tables I and II. Conventional methods like elevated
PSA, bone scans, biopsies and positron emission tomography (PET)
imaging are commonly employed for detecting CRPC recurrence or new
metastases (166). However, the
diagnostic accuracy of these methods is low and there are
relatively few diagnostic markers for CRPC. Since all of the
aforementioned proteins are significantly increased or decreased in
CRPC tissues, they can serve as diagnostic markers for CRPC.
However, unfortunately most of these proteins have low specificity
for CRPC. PSMA-PET, a new sensitive imaging tool for PCa, has been
developed to help clinicians determine the appropriate treatment
strategy for advanced PCa (111). In addition, CRPC samples from a
tissue microarray exhibited elevated FKBP4 protein expression
levels with an average FKBP4 histoscore of 87.1, compared with a
score of only 14.4 for HNCP (Wilcoxon rank sum test,
P=2.301×10−7) (38),
suggesting that FKBP4 may serve as a CRPC diagnostic marker.
Additional proteins that could be used as markers for the diagnosis
of NEPC include WLS (132),
SRRM4 (144), DEK (143), SSTR2 (98), CaSR (120), SPAG5 (167) and SMO (163). Among them, CaSR has been found
to be highly expressed in all cases of NEPC (119).
Protocadherin B9 and NRP1 serve not only as
biomarkers for predicting the overall survival of patients with PCa
but also as predictive indicators for non-recurrence in those
undergoing ATT (68,130). Moreover, patients exhibiting low
TUBB3 expression experience a significant decline in PSA levels of
at least 10% in 89% of cases, whereas this reduction is seen in
only 65% of patients with high TUBB3 expression (P=0.0267). It is
suggested that patients with PCa with low TUBB3 expression will
have a good response to ATT (114).
Collectively, the present review summarizes the
proteins dysregulated in CRPC tissues and highlights the expression
levels and distribution patterns of these proteins in HRPC,
compared with CRPC, together with the mechanisms they regulate in
CRPC development. The expression levels of these proteins were
verified through IHC tests on tissues from clinical patients.
Proteins that were only detected in cell lines, xenograft mice,
serum or succus prostaticus were not included in the present
review. In addition, fusion proteins generated by gene
rearrangements, such as PTEN and ER and protein variants, including
AR-splice V7 were not described in the present study.
Certain proteins may exert their influence during
the progression of CRPC through both AR-dependent and
AR-independent molecular mechanisms. For instance, IL-6 and its
downstream tyrosine kinases not only directly activate the AR but
also promote CRPC through the RTKs/MAPK/MEK/ERK and PI3K/AKT/mTOR
AR-independent pathways. Another example is HOXB13, which is
strategically positioned at the reprogrammed AR binding sites
within PCa tissues (80). HOXB13
serves as a multifaceted regulator of AR biology, either activating
or inhibiting the transcription of distinct AR target genes via the
AR-dependent pathway, thereby impacting disease progression.
Simultaneously, HOXB13 has been reported to promote the progression
of CRPC through the AR-independent NF-κB and JNK/c-Jun pathways.
However, it is evident that there exist conflicting research
findings regarding the precise impact of HOXB13 on CRPC
progression. These discrepancies underscore the need for a more
comprehensive and in-depth research approach to provide a clearer
understanding of the role of HOXB13 in CRPC. Furthermore, certain
proteins influence not just a single signaling pathway, but they
concurrently engage multiple pathways, resulting in a complex
tumorigenesis regulation network. For instance, IL-6 activates the
Ras and PI3K signaling pathways in the development of CRPC, while
CaSR activates the ERK and AKT signaling pathways in CRPC
progression.
Targeting a single protein might not yield an
effective treatment for CRPC, as inhibiting one pathway could
potentially trigger compensation through another pathway. Notably,
studies have illustrated that ADT plays a role in regulating cancer
cell adaptation through the modulation of protein expression and
epigenetic modifications. Cancer cells activate novel pathways in
an ongoing process of adaptation and evolution, which consequently
results in the development of drug resistance, an almost inevitable
outcome (169). Certain studies
have unveiled an array of distinct mechanisms underpinning cancer
drug resistance. Resulting mutations can arise within the same
protein or across different proteins (170,171), as well as within the same
pathway or parallel pathways (172), effectively circumventing
intercepted signaling cascades (173). Additionally, ADT triggers the
activation of various proteins, including Gremlin1, MET, ZBTB46,
SSTR2, RHAMM, NRP1, OPRK1, ACAT1 and ACO2. These proteins
contribute to tumor cells acquiring heightened capabilities in
proliferation, invasion and migration, along with increased
resistance to apoptosis, ultimately culminating in the progression
towards CRPC.
Not applicable.
KF and CL wrote the manuscript and abstract; PK
wrote the conclusion section and revised the paragraph structure of
the manuscript; WW completed the figures and tables; ZT
participated in revising and editing the manuscript; WL provided
constructive feedback and guidance, completed critical revisions
and proofread the manuscript. Data authentication is not
applicable. All authors have read and approved the final version of
the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by grants from The National
Natural Science Foundation of China Youth Science Foundation
Project (grant no. 81802571), Zhejiang Medical and Health Science
and Technology Project (grant no. 2019RC039) and Keqiao District
Scientific Research Project (grant no. 2021KZ42).
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Attard G, Parker C, Eeles RA, Schröder F,
Tomlins SA, Tannock I, Drake CG and de Bono JS: Prostate cancer.
Lancet. 387:70–82. 2016. View Article : Google Scholar
|
|
3
|
Mitsuzuka K and Arai Y: Metabolic changes
in patients with prostate cancer during androgen deprivation
therapy. Int J Urol. 25:45–53. 2018. View Article : Google Scholar
|
|
4
|
Huggins C and Hodges CV: Studies on
prostatic cancer. I. The effect of castration, of estrogen and
androgen injection on serum phosphatases in metastatic carcinoma of
the prostate. CA Cancer J Clin. 22:232–240. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sartor O: Androgen deprivation therapy in
prostate cancer: New findings and questions for the future. Lancet
Oncol. 20:176–177. 2019. View Article : Google Scholar
|
|
6
|
Bennett NC, Gardiner RA, Hooper JD,
Johnson DW and Gobe GC: Molecular cell biology of androgen receptor
signalling. Int J Biochem Cell Biol. 42:813–827. 2010. View Article : Google Scholar
|
|
7
|
Ruizeveld de Winter JA, Janssen PJ,
Sleddens HM, Verleun-Mooijman MC, Trapman J, Brinkmann AO, Santerse
AB, Schröder FH and van der Kwast TH: Androgen receptor status in
localized and locally progressive hormone refractory human prostate
cancer. Am J Pathol. 144:735–746. 1994.PubMed/NCBI
|
|
8
|
Crona DJ and Whang YE: Androgen
receptor-dependent and -independent mechanisms involved in prostate
cancer therapy resistance. Cancers (Basel). 9:672017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waltering KK, Helenius MA, Sahu B, Manni
V, Linja MJ, Jänne OA and Visakorpi T: Increased expression of
androgen receptor sensitizes prostate cancer cells to low levels of
androgens. Cancer Res. 69:8141–8149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grasso CS, Wu YM, Robinson DR, Cao X,
Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC,
et al: The mutational landscape of lethal castration-resistant
prostate cancer. Nature. 487:239–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai C and Balk SP: Intratumoral androgen
biosynthesis in prostate cancer pathogenesis and response to
therapy. Endocr Relat Cancer. 18:R175–R182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cao XL, Song XM, Yu WC, Chen YQ, Wei YY,
Liu YL and Lu KQ: Expression of pituitary tumor-transforming gene 1
during the development of androgen-independent prostate cancer.
Zhonghua Nan Ke Xue. 22:686–691. 2016.In Chinese. PubMed/NCBI
|
|
13
|
Cao S, Zhan Y and Dong Y: Emerging data on
androgen receptor splice variants in prostate cancer. Endocr Relat
Cancer. 23:T199–T210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoang DT, Iczkowski KA, Kilari D, See W
and Nevalainen MT: Androgen receptor-dependent and -independent
mechanisms driving prostate cancer progression: Opportunities for
therapeutic targeting from multiple angles. Oncotarget.
8:3724–3745. 2017. View Article : Google Scholar :
|
|
15
|
Kumar R: Emerging role of glucocorticoid
receptor in castration resistant prostate cancer: A potential
therapeutic target. J Cancer. 11:696–701. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Isikbay M, Otto K, Kregel S, Kach J, Cai
Y, Vander Griend DJ, Conzen SD and Szmulewitz RZ: Glucocorticoid
receptor activity contributes to resistance to androgen-targeted
therapy in prostate cancer. Horm Cancer. 5:72–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu W, Wang X, Liu Z, Wang Y, Yin B, Yu P,
Duan X, Liao Z, Chen Y, Liu C, et al: SGK1 inhibition induces
autophagy-dependent apoptosis via the mTOR-Foxo3a pathway. Br J
Cancer. 117:1139–1153. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Crick F: Central dogma of molecular
biology. Nature. 227:561–563. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Macklin A, Khan S and Kislinger T: Recent
advances in mass spectrometry based clinical proteomics:
Applications to cancer research. Clin Proteomics. 17:172020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Intasqui P, Bertolla RP and Sadi MV:
Prostate cancer proteomics: Clinically useful protein biomarkers
and future perspectives. Expert Rev Proteomics. 15:65–79. 2018.
View Article : Google Scholar
|
|
21
|
Chen YT, Tsai CH, Chen CL, Yu JS and Chang
YH: Development of biomarkers of genitourinary cancer using mass
spectrometry-based clinical proteomics. J Food Drug Anal.
27:387–403. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mantsiou A, Vlahou A and Zoidakis J:
Tissue proteomics studies in the investigation of prostate cancer.
Expert Rev Proteomics. 15:593–611. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kong P, Zhang L, Zhang Z, Feng K, Sang Y,
Duan X, Liu C, Sun T, Tao Z and Liu W: Emerging proteins in CRPC:
Functional roles and clinical implications. Front Oncol.
12:8738762022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen WY, Tsai YC, Siu MK, Yeh HL, Chen CL,
Yin JJ, Huang J and Liu YN: Inhibition of the androgen receptor
induces a novel tumor promoter, ZBTB46, for prostate cancer
metastasis. Oncogene. 36:6213–6224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beketova E, Fang S, Owens JL, Liu S, Chen
X, Zhang Q, Asberry AM, Deng X, Malola J, Huang J, et al: Protein
arginine methyltransferase 5 promotes pICln-dependent androgen
receptor transcription in castration-resistant prostate cancer.
Cancer Res. 80:4904–4917. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu H, Wang M, Du Y, Liu X, Weng X and Li
C: 4-1BBL has a possible role in mediating castration-resistant
conversion of prostate cancer via up-regulation of androgen
receptor. J Cancer. 10:2464–2471. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Özturan D, Morova T and Lack NA: Androgen
receptor-mediated transcription in prostate cancer. Cells.
11:8982022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shiota M, Fujimoto N, Imada K, Yokomizo A,
Itsumi M, Takeuchi A, Kuruma H, Inokuchi J, Tatsugami K, Uchiumi T,
et al: Potential role for YB-1 in castration-resistant prostate
cancer and resistance to enzalutamide through the androgen receptor
V7. J Natl Cancer Inst. 108:djw0052016. View Article : Google Scholar
|
|
29
|
Shiota M, Sekino Y, Tsukahara S, Abe T,
Kinoshita F, Imada K, Ueda S, Ushijima M, Nagakawa S, Matsumoto T,
et al: Gene amplification of YB-1 in castration-resistant prostate
cancer in association with aberrant androgen receptor expression.
Cancer Sci. 112:323–330. 2021. View Article : Google Scholar
|
|
30
|
Shiota M, Takeuchi A, Song Y, Yokomizo A,
Kashiwagi E, Uchiumi T, Kuroiwa K, Tatsugami K, Fujimoto N, Oda Y
and Naito S: Y-box binding protein-1 promotes castration-resistant
prostate cancer growth via androgen receptor expression. Endocr
Relat Cancer. 18:505–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shiota M, Yokomizo A, Tada Y, Inokuchi J,
Kashiwagi E, Masubuchi D, Eto M, Uchiumi T and Naito S: Castration
resistance of prostate cancer cells caused by castration-induced
oxidative stress through Twist1 and androgen receptor
overexpression. Oncogene. 29:237–250. 2010. View Article : Google Scholar
|
|
32
|
Shiota M, Izumi H, Onitsuka T, Miyamoto N,
Kashiwagi E, Kidani A, Yokomizo A, Naito S and Kohno K: Twist
promotes tumor cell growth through YB-1 expression. Cancer Res.
68:98–105. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nikhil K, Chang L, Viccaro K, Jacobsen M,
McGuire C, Satapathy SR, Tandiary M, Broman MM, Cresswell G, He YJ,
et al: Identification of LIMK2 as a therapeutic target in
castration resistant prostate cancer. Cancer Lett. 448:182–196.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kivinummi K, Urbanucci A, Leinonen K,
Tammela TLJ, Annala M, Isaacs WB, Bova GS, Nykter M and Visakorpi
T: The expression of AURKA is androgen regulated in
castrationresistant prostate cancer. Sci Rep. 7:179782017.
View Article : Google Scholar
|
|
35
|
Shafi AA, Yen AE and Weigel NL: Androgen
receptors in hormone-dependent and castration-resistant prostate
cancer. Pharmacol Ther. 140:223–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu K, Shimelis H, Linn DE, Jiang R, Yang
X, Sun F, Guo Z, Chen H, Li W, Chen H, et al: Regulation of
androgen receptor transcriptional activity and specificity by
RNF6-induced ubiquitination. Cancer Cell. 15:270–282. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dai B, Chen H, Guo S, Yang X, Linn DE, Sun
F, Li W, Guo Z, Xu K, Kim O, et al: Compensatory upregulation of
tyrosine kinase Etk/BMX in response to androgen deprivation
promotes castration-resistant growth of prostate cancer cells.
Cancer Res. 70:5587–5596. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Federer-Gsponer JR, Quintavalle C, Müller
DC, Dietsche T, Perrina V, Lorber T, Juskevicius D, Lenkiewicz E,
Zellweger T, Gasser T, et al: Delineation of human prostate cancer
evolution identifies chromothripsis as a polyclonal event and FKBP4
as a potential driver of castration resistance. J Pathol.
245:74–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qi J, Tripathi M, Mishra R, Sahgal N,
Fazli L, Ettinger S, Placzek WJ, Claps G, Chung LW, Bowtell D, et
al: The E3 ubiquitin ligase Siah2 contributes to
castration-resistant prostate cancer by regulation of androgen
receptor transcriptional activity. Cancer Cell. 23:332–346. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin HK, Wang L, Hu YC, Altuwaijri S and
Chang C: Phosphorylation-dependent ubiquitylation and degradation
of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J.
21:4037–4048. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hao J, Ci X, Wang Y, Choi SYC, Sullivan
SE, Xue H, Wu R, Dong X, Haegert AM, Collins CC, et al: GRB10
sustains AR activity by interacting with PP2A in prostate cancer
cells. Int J Cancer. 148:469–480. 2021. View Article : Google Scholar
|
|
42
|
Hao J, Ci X, Xue H, Wu R, Dong X, Choi
SYC, He H, Wang Y, Zhang F, Qu S, et al: Patient-derived
hormone-naive prostate cancer xenograft models reveal growth factor
receptor bound protein 10 as an androgen receptor-repressed gene
driving the development of castration-resistant prostate cancer.
Eur Urol. 73:949–960. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ding G, Wang J, Feng C, Jiang H, Xu J and
Ding Q: Lipocalin 2 over-expression facilitates progress of
castration-resistant prostate cancer via improving androgen
receptor transcriptional activity. Oncotarget. 7:64309–64317. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang B, Gu Y, Hui K, Huang J, Xu S, Wu S,
Li L, Fan J, Wang X, Hsieh JT, et al: AKR1C3, a crucial androgenic
enzyme in prostate cancer, promotes epithelial-mesenchymal
transition and metastasis through activating ERK signaling. Urol
Oncol. 36:472.e11–472.e20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu Z, Ma T, Zhou J, Gao W, Li Y, Yu S,
Wang Y and Chan FL: Nuclear receptor ERRα contributes to
castration-resistant growth of prostate cancer via its regulation
of intratumoral androgen biosynthesis. Theranostics. 10:4201–4216.
2020. View Article : Google Scholar :
|
|
46
|
Miyazaki Y, Teramoto Y, Shibuya S, Goto T,
Okasho K, Mizuno K, Uegaki M, Yoshikawa T, Akamatsu S, Kobayashi T,
et al: Consecutive prostate cancer specimens revealed increased
aldoketo reductase family 1 member C3 expression with progression
to castration-resistant prostate cancer. J Clin Med. 8:6012019.
View Article : Google Scholar
|
|
47
|
Powell K, Semaan L, Conley-LaComb MK,
Asangani I, Wu YM, Ginsburg KB, Williams J, Squire JA, Maddipati
KR, Cher ML and Chinni SR: ERG/AKR1C3/AR constitutes a feed-forward
loop for AR signaling in prostate cancer cells. Clin Cancer Res.
21:2569–2579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jing Y, Nguyen MM, Wang D, Pascal LE, Guo
W, Xu Y, Ai J, Deng FM, Masoodi KZ, Yu X, et al: DHX15 promotes
prostate cancer progression by stimulating Siah2-mediated
ubiquitination of androgen receptor. Oncogene. 37:638–650. 2018.
View Article : Google Scholar :
|
|
49
|
Wang L, Song G, Chang X, Tan W, Pan J, Zhu
X, Liu Z, Qi M, Yu J and Han B: The role of TXNDC5 in
castration-resistant prostate cancer-involvement of androgen
receptor signaling pathway. Oncogene. 34:4735–4745. 2015.
View Article : Google Scholar
|
|
50
|
Yamamoto S, Takayama KI, Obinata D,
Fujiwara K, Ashikari D, Takahashi S and Inoue S: Identification of
new octamer transcription factor 1-target genes upregulated in
castration-resistant prostate cancer. Cancer Sci. 110:3476–3485.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qin J, Lee HJ, Wu SP, Lin SC, Lanz RB,
Creighton CJ, DeMayo FJ, Tsai SY and Tsai MJ: Androgen
deprivation-induced NCoA2 promotes metastatic and
castration-resistant prostate cancer. J Clin Invest. 124:5013–5026.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Teng M, Zhou S, Cai C, Lupien M and He HH:
Pioneer of prostate cancer: Past, present and the future of FOXA1.
Protein Cell. 12:29–38. 2021. View Article : Google Scholar :
|
|
53
|
Xu Y, Song Q, Pascal LE, Zhong M, Zhou Y,
Zhou J, Deng FM, Huang J and Wang Z: DHX15 is up-regulated in
castration-resistant prostate cancer and required for androgen
receptor sensitivity to low DHT concentrations. Prostate.
79:657–666. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Na AY, Choi S, Yang E, Liu KH, Kim S, Jung
HJ, Choe Y, Ha YS, Kwon TG, Lee JN and Lee S: Characterization of
novel progression factors in castration-resistant prostate cancer
based on global comparative proteome analysis. Cancers (Basel).
13:34322021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jain RK, Mehta RJ, Nakshatri H, Idrees MT
and Badve SS: High-level expression of forkhead-box protein A1 in
metastatic prostate cancer. Histopathology. 58:766–772. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhao Y, Tindall DJ and Huang H: Modulation
of androgen receptor by FOXA1 and FOXO1 factors in prostate cancer.
Int J Biol Sci. 10:614–619. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cheng S, Prieto-Dominguez N, Yang S,
Connelly ZM, StPierre S, Rushing B, Watkins A, Shi L, Lakey M,
Baiamonte LB, et al: The expression of YAP1 is increased in
high-grade prostatic adenocarcinoma but is reduced in
neuroendocrine prostate cancer. Prostate Cancer Prostatic Dis.
23:661–669. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang L, Yang S, Chen X, Stauffer S, Yu F,
Lele SM, Fu K, Datta K, Palermo N, Chen Y and Dong J: The hippo
pathway effector YAP regulates motility, invasion, and
castration-resistant growth of prostate cancer cells. Mol Cell
Biol. 35:1350–1362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kuser-Abali G, Alptekin A, Lewis M,
Garraway IP and Cinar B: YAP1 and AR interactions contribute to the
switch from androgen-dependent to castration-resistant growth in
prostate cancer. Nat Commun. 6:81262015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lee HC, Ou CH, Huang YC, Hou PC, Creighton
CJ, Lin YS, Hu CY and Lin SC: YAP1 overexpression contributes to
the development of enzalutamide resistance by induction of cancer
stemness and lipid metabolism in prostate cancer. Oncogene.
40:2407–2421. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lyons LS, Rao S, Balkan W, Faysal J,
Maiorino CA and Burnstein KL: Ligand-independent activation of
androgen receptors by Rho GTPase signaling in prostate cancer. Mol
Endocrinol. 22:597–608. 2008. View Article : Google Scholar
|
|
62
|
Craft N, Shostak Y, Carey M and Sawyers
CL: A mechanism for hormone-independent prostate cancer through
modulation of androgen receptor signaling by the HER-2/neu tyrosine
kinase. Nat Med. 5:280–285. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
63
|
Figel S and Gelman IH: Focal adhesion
kinase controls prostate cancer progression via intrinsic kinase
and scaffolding functions. Anticancer Agents Med Chem. 11:607–616.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lee MH, Kundu JK, Keum YS, Cho YY, Surh YJ
and Choi BY: Resveratrol inhibits IL-6-induced transcriptional
activity of AR and STAT3 in human prostate cancer LNCaP-FGC cells.
Biomol Ther (Seoul). 22:426–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lee SO, Lou W, Hou M, de Miguel F, Gerber
L and Gao AC: Interleukin-6 promotes androgen-independent growth in
LNCaP human prostate cancer cells. Clin Cancer Res. 9:370–376.
2003.PubMed/NCBI
|
|
66
|
Don-Doncow N, Marginean F, Coleman I,
Nelson PS, Ehrnström R, Krzyzanowska A, Morrissey C, Hellsten R and
Bjartell A: Expression of STAT3 in prostate cancer metastases. Eur
Urol. 71:313–316. 2017. View Article : Google Scholar :
|
|
67
|
Yang F, Yuan C, Wu D, Zhang J and Zhou X:
IRE1α expedites the progression of castration-resistant prostate
cancers via the positive feedback loop of IRE1α/IL-6/AR. Front
Oncol. 11:6711412021. View Article : Google Scholar
|
|
68
|
Sekino Y, Oue N, Mukai S, Shigematsu Y,
Goto K, Sakamoto N, Sentani K, Hayashi T, Teishima J, Matsubara A
and Yasui W: Protocadherin B9 promotes resistance to bicalutamide
and is associated with the survival of prostate cancer patients.
Prostate. 79:234–242. 2019. View Article : Google Scholar
|
|
69
|
Becker F, Joerg V, Hupe MC, Roth D, Krupar
R, Lubczyk V, Kuefer R, Sailer V, Duensing S, Kirfel J, et al:
Increased mediator complex subunit CDK19 expression associates with
aggressive prostate cancer. Int J Cancer. 146:577–588. 2020.
View Article : Google Scholar
|
|
70
|
Offermann A, Joerg V, Becker F, Roesch MC,
Kang D, Lemster AL, Tharun L, Behrends J, Merseburger AS, Culig Z,
et al: Inhibition of cyclin-dependent kinase 8/cyclin-dependent
kinase 19 suppresses its pro-oncogenic effects in prostate cancer.
Am J Pathol. 192:813–823. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Jia L, Wu D, Wang Y, You W, Wang Z, Xiao
L, Cai G, Xu Z, Zou C, Wang F, et al: Orphan nuclear receptor TLX
contributes to androgen insensitivity in castration-resistant
prostate cancer via its repression of androgen receptor
transcription. Oncogene. 37:3340–3355. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sooreshjani MA, Nikhil K, Kamra M, Nguyen
DN, Kumar D and Shah K: LIMK2-NKX3.1 engagement promotes
castration-resistant prostate cancer. Cancers (Basel). 13:23242021.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lv S, Song Q, Chen G, Cheng E, Chen W,
Cole R, Wu Z, Pascal LE, Wang K, Wipf P, et al: Regulation and
targeting of androgen receptor nuclear localization in
castration-resistant prostate cancer. J Clin Invest.
131:e1413352021. View Article : Google Scholar :
|
|
74
|
Wu H, You L, Li Y, Zhao Z, Shi G, Chen Z,
Wang Z, Li X, Du S, Ye W, et al: Loss of a negative feedback loop
between IRF8 and AR promotes prostate cancer growth and
enzalutamide resistance. Cancer Res. 80:2927–2939. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wolff DW, Xie Y, Deng C, Gatalica Z, Yang
M, Wang B, Wang J, Lin MF, Abel PW and Tu Y: Epigenetic repression
of regulator of G-protein signaling 2 promotes androgen-independent
prostate cancer cell growth. Int J Cancer. 130:1521–1531. 2012.
View Article : Google Scholar
|
|
76
|
Linder A, Larsson K, Welén K and Damber
JE: RGS2 is prognostic for development of castration resistance and
cancer-specific survival in castration-resistant prostate cancer.
Prostate. 80:799–810. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Cao X, Qin J, Xie Y, Khan O, Dowd F,
Scofield M, Lin MF and Tu Y: Regulator of G-protein signaling 2
(RGS2) inhibits androgen-independent activation of androgen
receptor in prostate cancer cells. Oncogene. 25:3719–3734. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Sun J, Hu X, Gao Y, Tang Q, Zhao Z, Xi W,
Yang F, Zhang W, Song Y, Song B, et al: MYSM1-AR complex-mediated
repression of Akt/c-Raf/GSK-3β signaling impedes
castration-resistant prostate cancer growth. Aging (Albany NY).
11:10644–10663. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Jung C, Kim RS, Zhang HJ, Lee SJ and Jeng
MH: HOXB13 induces growth suppression of prostate cancer cells as a
repressor of hormone-activated androgen receptor signaling. Cancer
Res. 64:9185–9192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Norris JD, Chang CY, Wittmann BM, Kunder
RS, Cui H, Fan D, Joseph JD and McDonnell DP: The homeodomain
protein HOXB13 regulates the cellular response to androgens. Mol
Cell. 36:405–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Puca L, Vlachostergios PJ and Beltran H:
Neuroendocrine differentiation in prostate cancer: Emerging
biology, models, and therapies. Cold Spring Harb Perspect Med.
9:a0305932019. View Article : Google Scholar
|
|
82
|
Bluemn EG, Coleman IM, Lucas JM, Coleman
RT, Hernandez-Lopez S, Tharakan R, Bianchi-Frias D, Dumpit RF,
Kaipainen A, Corella AN, et al: Androgen receptor
pathway-independent prostate cancer is sustained through FGF
signaling. Cancer Cell. 32:474–489.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Makino T, Izumi K and Mizokami A:
Undesirable status of prostate cancer cells after intensive
inhibition of AR signaling: Post-AR Era of CRPC treatment.
Biomedicines. 9:4142021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Saraon P, Jarvi K and Diamandis EP:
Molecular alterations during progression of prostate cancer to
androgen independence. Clin Chem. 57:1366–1375. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y and
Hu LL: ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med.
19:1997–2007. 2020.PubMed/NCBI
|
|
86
|
Shorning BY, Dass MS, Smalley MJ and
Pearson HB: The PI3K-AKT-mTOR pathway and prostate cancer: At the
crossroads of AR, MAPK, and WNT signaling. Int J Mol Sci.
21:45072020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Mukherjee R, McGuinness DH, McCall P,
Underwood MA, Seywright M, Orange C and Edwards J: Upregulation of
MAPK pathway is associated with survival in castrate-resistant
prostate cancer. Br J Cancer. 104:1920–1928. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhong S, Peng S, Chen Z, Chen Z and Luo
JL: Choosing kinase inhibitors for androgen deprivation
therapy-resistant prostate cancer. Pharmaceutics. 14:4982022.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Cheung S, Jain P, So J, Shahidi S, Chung S
and Koritzinsky M: p38 MAPK inhibition mitigates hypoxia-induced AR
signaling in castration-resistant prostate cancer. Cancers (Basel).
13:8312021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Cheng C, Wang J, Xu P, Zhang K, Xin Z,
Zhao H, Ji Z, Zhang M, Wang D, He Y, et al: Gremlin1 is a
therapeutically targetable FGFR1 ligand that regulates lineage
plasticity and castration resistance in prostate cancer. Nat
Cancer. 3:565–580. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Verhoef EI, Kolijn K, De Herdt MJ, van der
Steen B, Hoogland AM, Sleddens HF, Looijenga LH and van Leenders
GJ: MET expression during prostate cancer progression. Oncotarget.
7:31029–31036. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ibuki N, Ghaffari M, Pandey M, Iu I, Fazli
L, Kashiwagi M, Tojo H, Nakanishi O, Gleave ME and Cox ME: TAK-441,
a novel investigational smoothened antagonist, delays
castration-resistant progression in prostate cancer by disrupting
paracrine hedgehog signaling. Int J Cancer. 133:1955–1966. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yu M, Gipp J, Yoon JW, Iannaccone P,
Walterhouse D and Bushman W: Sonic hedgehog-responsive genes in the
fetal prostate. J Biol Chem. 284:5620–5629. 2009. View Article : Google Scholar :
|
|
94
|
Gao Y, Li L, Li T, Ma L, Yuan M, Sun W,
Cheng HL, Niu L, Du Z, Quan Z, et al: Simvastatin delays
castration-resistant prostate cancer metastasis and androgen
receptor antagonist resistance by regulating the expression of
caveolin-1. Int J Oncol. 54:2054–2068. 2019.PubMed/NCBI
|
|
95
|
Nguyen DP, Li J and Tewari AK:
Inflammation and prostate cancer: The role of interleukin 6 (IL-6).
BJU Int. 113:986–992. 2014. View Article : Google Scholar
|
|
96
|
Niwa H, Burdon T, Chambers I and Smith A:
Self-renewal of pluripotent embryonic stem cells is mediated via
activation of STAT3. Genes Dev. 12:2048–2060. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Liu YN, Niu S, Chen WY, Zhang Q, Tao Y,
Chen WH, Jiang KC, Chen X, Shi H, Liu A, et al: Leukemia inhibitory
factor promotes castration-resistant prostate cancer and
neuroendocrine differentiation by activated ZBTB46. Clin Cancer
Res. 25:4128–4140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Cariaga-Martinez AE, Lorenzati MA, Riera
MA, Cubilla MA, De La Rossa A, Giorgio EM, Tiscornia MM, Gimenez
EM, Rojas ME, Chaneton BJ, et al: Tumoral prostate shows different
expression pattern of somatostatin receptor 2 (SSTR2) and
phosphotyrosine phosphatase SHP-1 (PTPN6) according to tumor
progression. Adv Urol. 2009:7238312009. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Bakht MK, Derecichei I, Li Y, Ferraiuolo
RM, Dunning M, Oh SW, Hussein A, Youn H, Stringer KF, Jeong CW, et
al: Neuroendocrine differentiation of prostate cancer leads to PSMA
suppression. Endocr Relat Cancer. 26:131–146. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lam HM, Ouyang B, Chen J, Ying J, Wang J,
Wu CL, Jia L, Medvedovic M, Vessella RL and Ho SM: Targeting GPR30
with G-1: A new therapeutic target for castration-resistant
prostate cancer. Endocr Relat Cancer. 21:903–914. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang R, Zong J, Peng Y, Shi J, Du X, Liu
H, Shen Y, Cao J, Jia B, Liu F and Zhang J: GPR30 knockdown weakens
the capacity of CAF in promoting prostate cancer cell invasion via
reducing macrophage infiltration and M2 polarization. J Cell
Biochem. May 3–2021.Epub ahead of print. View Article : Google Scholar
|
|
102
|
Chan QKY, Lam HM, Ng CF, Lee AYY, Chan ES,
Ng HK, Ho SM and Lau KM: Activation of GPR30 inhibits the growth of
prostate cancer cells through sustained activation of Erk1/2,
c-jun/c-fos-dependent upregulation of p21, and induction of G(2)
cell-cycle arrest. Cell Death Differ. 17:1511–1523. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Bitting RL and Armstrong AJ: Targeting the
PI3K/Akt/mTOR pathway in castration-resistant prostate cancer.
Endocr Relat Cancer. 20:R83–R99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Edlind MP and Hsieh AC: PI3K-AKT-mTOR
signaling in prostate cancer progression and androgen deprivation
therapy resistance. Asian J Androl. 16:378–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Staniszewska M, Fragoso Costa P, Eiber M,
Klose JM, Wosniack J, Reis H, Szarvas T, Hadaschik B, Lückerath K,
Herrmann K, et al: Enzalutamide enhances PSMA expression of
PSMA-low prostate cancer. Int J Mol Sci. 22:74312021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Caromile LA and Shapiro LH: PSMA redirects
MAPK to PI3K-AKT signaling to promote prostate cancer progression.
Mol Cell Oncol. 4:e13211682017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Meller B, Bremmer F, Sahlmann CO, Hijazi
S, Bouter C, Trojan L, Meller J and Thelen P: Alterations in
androgen deprivation enhanced prostate-specific membrane antigen
(PSMA) expression in prostate cancer cells as a target for
diagnostics and therapy. EJNMMI Res. 5:662015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Rodríguez-Fraile M, Tamayo Alonso P,
Rosales JJ, de Arcocha-Torres M, Caresia-Aróztegui AP,
Cózar-Santiago MP, Orcajo-Rincon J, Simó Perdigó M, Delgado Bolton
RC and Artigas Guix C; Oncology Working Group of the Spanish
Society of Nuclear Medicine and Molecular Imaging: The role of PSMA
radioligands in the diagnosis and treatment of prostate carcinoma.
Rev Esp Med Nucl Imagen Mol (Engl Ed). 41:126–135. 2022.PubMed/NCBI
|
|
109
|
Chen R, Wang Y, Zhu Y, Shi Y, Xu L, Huang
G and Liu J: The added value of 18F-FDG PET/CT compared
with 68Ga-PSMA PET/CT in patients with
castration-resistant prostate cancer. J Nucl Med. 63:69–75. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Weber M, Hadaschik B, Ferdinandus J,
Rahbar K, Bögemann M, Herrmann K, Fendler WP and Kesch C:
Prostate-specific membrane antigen-based imaging of
castration-resistant prostate cancer. Eur Urol Focus. 7:279–287.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Paschalis A, Sheehan B, Riisnaes R,
Rodrigues DN, Gurel B, Bertan C, Ferreira A, Lambros MBK, Seed G,
Yuan W, et al: Prostate-specific membrane antigen heterogeneity and
DNA repair defects in prostate cancer. Eur Urol. 76:469–478. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Sekino Y, Han X, Kawaguchi T, Babasaki T,
Goto K, Inoue S, Hayashi T, Teishima J, Shiota M, Yasui W and
Matsubara A: TUBB3 reverses resistance to docetaxel and cabazitaxel
in prostate cancer. Int J Mol Sci. 20:39362019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Terry S, Ploussard G, Allory Y, Nicolaiew
N, Boissière-Michot F, Maillé P, Kheuang L, Coppolani E, Ali A,
Bibeau F, et al: Increased expression of class III beta-tubulin in
castration-resistant human prostate cancer. Br J Cancer.
101:951–956. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Maahs L, Sanchez BE, Gupta N, Van Harn M,
Barrack ER, Reddy PV and Hwang C: Class III β-tubulin expression as
a predictor of docetaxel-resistance in metastatic
castration-resistant prostate cancer. PLoS One. 14:e02225102019.
View Article : Google Scholar
|
|
115
|
Alfano A, Xu J, Yang X, Deshmukh D and Qiu
Y: SRC kinase-mediated tyrosine phosphorylation of TUBB3 regulates
its stability and mitotic spindle dynamics in prostate cancer
cells. Pharmaceutics. 14:9322022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Korkes F, de Castro MG, de Cassio Zequi S,
Nardi L, Del Giglio A and de Lima Pompeo AC: Hyaluronan-mediated
motility receptor (RHAMM) immunohistochemical expression and
androgen deprivation in normal peritumoral, hyperplasic and
neoplastic prostate tissue. BJU Int. 113:822–829. 2014. View Article : Google Scholar
|
|
117
|
Lin SL, Chang D and Ying SY: Hyaluronan
stimulates transformation of androgen-independent prostate cancer.
Carcinogenesis. 28:310–320. 2007. View Article : Google Scholar
|
|
118
|
Hannan FM, Kallay E, Chang W, Brandi ML
and Thakker RV: The calcium-sensing receptor in physiology and in
calcitropic and noncalcitropic diseases. Nat Rev Endocrinol.
15:33–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Bery F, Cancel M, Chantôme A, Guibon R,
Bruyère F, Rozet F, Mahéo K and Fromont G: The calcium-sensing
receptor is a marker and potential driver of neuroendocrine
differentiation in prostate cancer. Cancers (Basel). 12:8602020.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Ahearn TU, Tchrakian N, Wilson KM, Lis R,
Nuttall E, Sesso HD, Loda M, Giovannucci E, Mucci LA, Finn S and
Shui IM: Calcium-sensing receptor tumor expression and lethal
prostate cancer progression. J Clin Endocrinol Metab.
101:2520–2527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Yamamura A, Nayeem MJ and Sato M:
Calcilytics inhibit the proliferation and migration of human
prostate cancer PC-3 cells. J Pharmacol Sci. 139:254–257. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Liao J, Schneider A, Datta NS and McCauley
LK: Extracellular calcium as a candidate mediator of prostate
cancer skeletal metastasis. Cancer Res. 66:9065–9073. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang H, Li S, Yang X, Qiao B, Zhang Z and
Xu Y: miR-539 inhibits prostate cancer progression by directly
targeting SPAG5. J Exp Clin Cancer Res. 35:602016. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Ebhardt HA, Root A, Liu Y, Gauthier NP,
Sander C and Aebersold R: Systems pharmacology using mass
spectrometry identifies critical response nodes in prostate cancer.
NPJ Syst Biol Appl. 4:262018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Huang S, Hölzel M, Knijnenburg T,
Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C,
Prahallad A, et al: MED12 controls the response to multiple cancer
drugs through regulation of TGF-β receptor signaling. Cell.
151:937–950. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Adler D, Offermann A, Braun M, Menon R,
Syring I, Nowak M, Halbach R, Vogel W, Ruiz C, Zellweger T, et al:
MED12 overexpression is a frequent event in castration-resistant
prostate cancer. Endocr Relat Cancer. 21:663–675. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Adler D, Menon R, Braun M, Offermann A,
Queisser A, Boehm D, Vogel W, Rüenauver K, Ruiz C, Zellweger T, et
al: MED15, encoding a subunit of the mediator complex, is
overexpressed at high frequency in castration-resistant prostate
cancer. Int J Cancer. 135:19–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Offermann A, Vlasic I, Syring I, Vogel W,
Ruiz C, Zellweger T, Rentsch CA, Hagedorn S, Behrends J, Nowak M,
et al: MED15 overexpression in prostate cancer arises during
androgen deprivation therapy via PI3K/mTOR signaling. Oncotarget.
8:7964–7976. 2017. View Article : Google Scholar :
|
|
129
|
Chen Z, Gao H, Dong Z, Shen Y, Wang Z, Wei
W, Yi J, Wang R, Wu N and Jin S: NRP1 regulates radiation-induced
EMT via TGF-β/Smad signaling in lung adenocarcinoma cells. Int J
Radiat Biol. 96:1281–1295. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Tse BWC, Volpert M, Ratther E, Stylianou
N, Nouri M, McGowan K, Lehman ML, McPherson SJ, Roshan-Moniri M,
Butler MS, et al: Neuropilin-1 is upregulated in the adaptive
response of prostate tumors to androgen-targeted therapies and is
prognostic of metastatic progression and patient mortality.
Oncogene. 36:3417–3427. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Makino Y, Kamiyama Y, Brown JB, Tanaka T,
Murakami R, Teramoto Y, Goto T, Akamatsu S, Terada N, Inoue T, et
al: Comprehensive genomics in androgen receptor-dependent
castration-resistant prostate cancer identifies an adaptation
pathway mediated by opioid receptor kappa 1. Commun Biol.
5:2992022. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Bland T, Wang J, Yin L, Pu T, Li J, Gao J,
Lin TP, Gao AC and Wu BJ: WLS-Wnt signaling promotes neuroendocrine
prostate cancer. iScience. 24:1019702021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Sengupta D, Zeng L, Li Y, Hausmann S,
Ghosh D, Yuan G, Nguyen TN, Lyu R, Caporicci M, Morales Benitez A,
et al: NSD2 dimethylation at H3K36 promotes lung adenocarcinoma
pathogenesis. Mol Cell. 81:4481–4492.e9. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Filon M, Gawdzik J, Truong A, Allen G,
Huang W, Khemees T, Machhi R, Lewis P, Yang B, Denu J and Jarrard
D: Tandem histone methyltransferase upregulation defines a unique
aggressive prostate cancer phenotype. Br J Cancer. 125:247–254.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Yang P, Guo L, Duan ZJ, Tepper CG, Xue L,
Chen X, Kung HJ, Gao AC, Zou JX and Chen HW: Histone
methyltransferase NSD2/MMSET mediates constitutive NF-κB signaling
for cancer cell proliferation, survival, and tumor growth via a
feed-forward loop. Mol Cell Biol. 32:3121–3131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Kim YR, Oh KJ, Park RY, Xuan NT, Kang TW,
Kwon DD, Choi C, Kim MS, Nam KI, Ahn KY and Jung C: HOXB13 promotes
androgen independent growth of LNCaP prostate cancer cells by the
activation of E2F signaling. Mol Cancer. 9:1242010. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Kim YR, Kang TW, To PK, Xuan Nguyen NT,
Cho YS, Jung C and Kim MS: HOXB13-mediated suppression of
p21WAF1/CIP1 regulates JNK/c-Jun signaling in prostate cancer
cells. Oncol Rep. 35:2011–2016. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Kim YR, Kim IJ, Kang TW, Choi C, Kim KK,
Kim MS, Nam KI and Jung C: HOXB13 downregulates intracellular zinc
and increases NF-κB signaling to promote prostate cancer
metastasis. Oncogene. 33:4558–4567. 2014. View Article : Google Scholar
|
|
139
|
Lu X, Fong KW, Gritsina G, Wang F, Baca
SC, Brea LT, Berchuck JE, Spisak S, Ross J, Morrissey C, et al:
HOXB13 suppresses de novo lipogenesis through HDAC3-mediated
epigenetic reprogramming in prostate cancer. Nat Genet. 54:670–683.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Fine SW: Neuroendocrine tumors of the
prostate. Mod Pathol. 31(S1): S122–S132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Carceles-Cordon M, Kelly WK, Gomella L,
Knudsen KE, Rodriguez-Bravo V and Domingo-Domenech J: Cellular
rewiring in lethal prostate cancer: The architect of drug
resistance. Nat Rev Urol. 17:292–307. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wang Y and Wang Y, Ci X, Choi SYC, Crea F,
Lin D and Wang Y: Molecular events in neuroendocrine prostate
cancer development. Nat Rev Urol. 18:581–596. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Lin D, Dong X, Wang K, Wyatt AW, Crea F,
Xue H, Wang Y, Wu R, Bell RH, Haegert A, et al: Identification of
DEK as a potential therapeutic target for neuroendocrine prostate
cancer. Oncotarget. 6:1806–1820. 2015. View Article : Google Scholar :
|
|
144
|
Li Y, Zhang Q, Lovnicki J, Chen R, Fazli
L, Wang Y, Gleave M, Huang J and Dong X: SRRM4 gene expression
correlates with neuroendocrine prostate cancer. Prostate.
79:96–104. 2019. View Article : Google Scholar
|
|
145
|
Lee AR, Che N, Lovnicki JM and Dong X:
Development of neuroendocrine prostate cancers by the Ser/Arg
repetitive matrix 4-mediated RNA splicing network. Front Oncol.
8:932018. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Kavanaugh GM, Wise-Draper TM, Morreale RJ,
Morrison MA, Gole B, Schwemberger S, Tichy ED, Lu L, Babcock GF,
Wells JM, et al: The human DEK oncogene regulates DNA damage
response signaling and repair. Nucleic Acids Res. 39:7465–7476.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Marine JC, Dawson SJ and Dawson MA:
Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev
Cancer. 20:743–756. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Zhao R, Feng T, Gao L, Sun F, Zhou Q, Wang
X, Liu J, Zhang W, Wang M, Xiong X, et al: PPFIA4 promotes
castration-resistant prostate cancer by enhancing mitochondrial
metabolism through MTHFD2. J Exp Clin Cancer Res. 41:1252022.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Latonen L, Afyounian E, Jylhä A, Nättinen
J, Aapola U, Annala M, Kivinummi KK, Tammela TTL, Beuerman RW,
Uusitalo H, et al: Integrative proteomics in prostate cancer
uncovers robustness against genomic and transcriptomic aberrations
during disease progression. Nat Commun. 9:11762018. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Liu Q, Harvey CT, Geng H, Xue C, Chen V,
Beer TM and Qian DZ: Malate dehydrogenase 2 confers docetaxel
resistance via regulations of JNK signaling and oxidative
metabolism. Prostate. 73:1028–1037. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Blomme A, Peter C, Mui E, Rodriguez Blanco
G, An N, Mason LM, Jamieson LE, McGregor GH, Lilla S, Ntala C, et
al: THEM6-mediated reprogramming of lipid metabolism supports
treatment resistance in prostate cancer. EMBO Mol Med.
14:e147642022. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Zadra G, Ribeiro CF, Chetta P, Ho Y,
Cacciatore S, Gao X, Syamala S, Bango C, Photopoulos C, Huang Y, et
al: Inhibition of de novo lipogenesis targets androgen receptor
signaling in castration-resistant prostate cancer. Proc Natl Acad
Sci USA. 116:631–640. 2019. View Article : Google Scholar :
|
|
153
|
El-Kenawi A, Dominguez-Viqueira W, Liu M,
Awasthi S, Abraham-Miranda J, Keske A, Steiner KK, Noel L, Serna
AN, Dhillon J, et al: Macrophage-derived cholesterol contributes to
therapeutic resistance in prostate cancer. Cancer Res.
81:5477–5490. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Labanca E, Bizzotto J, Sanchis P,
Anselmino N, Yang J, Shepherd PDA, Paez A, Antico-Arciuch V,
Lage-Vickers S, Hoang AG, et al: Prostate cancer castrate resistant
progression usage of non-canonical androgen receptor signaling and
ketone body fuel. Oncogene. 40:6284–6298. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Saraon P, Cretu D, Musrap N, Karagiannis
GS, Batruch I, Drabovich AP, van der Kwast T, Mizokami A, Morrissey
C, Jarvi K and Diamandis EP: Quantitative proteomics reveals that
enzymes of the ketogenic pathway are associated with prostate
cancer progression. Mol Cell Proteomics. 12:1589–1601. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Eerola SK, Kohvakka A, Tammela TLJ,
Koskinen PJ, Latonen L and Visakorpi T: Expression and ERG
regulation of PIM kinases in prostate cancer. Cancer Med.
10:3427–3436. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Peng HH, Wang JN, Xiao LF, Yan M, Chen SP,
Wang L and Yang K: Elevated serum FGG levels prognosticate and
promote the disease progression in prostate cancer. Front Genet.
12:6516472021. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Stoyanova T, Riedinger M, Lin S,
Faltermeier CM, Smith BA, Zhang KX, Going CC, Goldstein AS, Lee JK,
Drake JM, et al: Activation of Notch1 synergizes with multiple
pathways in promoting castration-resistant prostate cancer. Proc
Natl Acad Sci USA. 113:E6457–E6466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Su Q and Xin L: Notch signaling in
prostate cancer: Refining a therapeutic opportunity. Histol
Histopathol. 31:149–157. 2016.
|
|
160
|
Babcook MA, Shukla S, Fu P, Vazquez EJ,
Puchowicz MA, Molter JP, Oak CZ, MacLennan GT, Flask CA, Lindner
DJ, et al: Synergistic simvastatin and metformin combination
chemotherapy for osseous metastatic castration-resistant prostate
cancer. Mol Cancer Ther. 13:2288–2302. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Babcook MA, Akgul M, Margevicius S,
MacLennan GT, Fu P, Abouassaly R and Gupta S: Ser-486/491
phosphorylation and inhibition of AMPKα activity is positively
associated with Gleason score, metastasis, and
castration-resistance in prostate cancer: A retrospective clinical
study. Prostate. 78:714–723. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Sawant Dessai A, Dominguez MP, Chen UI,
Hasper J, Prechtl C, Yu C, Katsuta E, Dai T, Zhu B, Jung SY, et al:
Transcriptional repression of SIRT3 potentiates mitochondrial
aconitase activation to drive aggressive prostate cancer to the
bone. Cancer Res. 81:50–63. 2021.
|
|
163
|
Wang L, Li H, Li Z, Li M, Tang Q, Wu C and
Lu Z: Smoothened loss is a characteristic of neuroendocrine
prostate cancer. Prostate. 81:508–520. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Du Z, Li L, Sun W, Wang X, Zhang Y, Chen
Z, Yuan M, Quan Z, Liu N, Hao Y, et al: HepaCAM inhibits the
malignant behavior of castration-resistant prostate cancer cells by
downregulating Notch signaling and PF-3084014 (a γ-secretase
inhibitor) partly reverses the resistance of refractory prostate
cancer to docetaxel and enzalutamide in vitro. Int J Oncol.
53:99–112. 2018.PubMed/NCBI
|
|
165
|
Tan B, Chen X, Fan Y, Yang Y, Yang J and
Tan L: STAT3 phosphorylation is required for the HepaCAM-mediated
inhibition of castration-resistant prostate cancer cell viability
and metastasis. Prostate. 81:603–611. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Vellky JE and Ricke WA: Development and
prevalence of castration-resistant prostate cancer subtypes.
Neoplasia. 22:566–575. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Liu JY, Zeng QH, Cao PG, Xie D, Yang F, He
LY, Dai YB, Li JJ, Liu XM, Zeng HL, et al: SPAG5 promotes
proliferation and suppresses apoptosis in bladder urothelial
carcinoma by upregulating Wnt3 via activating the AKT/mTOR pathway
and predicts poorer survival. Oncogene. 37:3937–3952. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Farolfi A, Calderoni L, Mattana F, Mei R,
Telo S, Fanti S and Castellucci P: Current and emerging clinical
applications of PSMA PET diagnostic imaging for prostate cancer. J
Nucl Med. 62:596–604. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Nussinov R, Tsai CJ and Jang H: Anticancer
drug resistance: An update and perspective. Drug Resist Updat.
59:1007962021. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Genovese I, Ilari A, Assaraf YG, Fazi F
and Colotti G: Not only P-glycoprotein: Amplification of the
ABCB1-containing chromosome region 7q21 confers multidrug
resistance upon cancer cells by coordinated overexpression of an
assortment of resistance-related proteins. Drug Resist Updat.
32:23–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Shahar N and Larisch S: Inhibiting the
inhibitors: Targeting anti-apoptotic proteins in cancer and therapy
resistance. Drug Resist Updat. 52:1007122020. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Assaraf YG, Brozovic A, Gonçalves AC,
Jurkovicova D, Linē A, Machuqueiro M, Saponara S, Sarmento-Ribeiro
AB, Xavier CPR and Vasconcelos MH: The multi-factorial nature of
clinical multidrug resistance in cancer. Drug Resist Updat.
46:1006452019. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Harbinski F, Craig VJ, Sanghavi S, Jeffery
D, Liu L, Sheppard KA, Wagner S, Stamm C, Buness A,
Chatenay-Rivauday C, et al: Rescue screens with secreted proteins
reveal compensatory potential of receptor tyrosine kinases in
driving cancer growth. Cancer Discov. 2:948–959. 2012. View Article : Google Scholar : PubMed/NCBI
|