Introduction
Hepatocellular carcinoma (HCC) is the sixth most
frequently diagnosed cancer and the third leading cause of
cancer-related mortality worldwide (1,2).
Exosomes are cell-derived extracellular vesicles (EVs) with a
diameter of 40-100 nm (3,4) that contain a large number of
biologically active molecules, such as microRNAs (miRNAs), proteins
and messenger RNAs (mRNAs) (5). A
study has shown that tumor cell-derived exosomes are involved in
several cancer processes (6). In
addition, miRNAs within exosomes have become the focus of modern
research because of their potential roles in tumor development and
progression (7).
miRNAs are unstable biomolecules and most miRNAs
need to bind to specific RNA-binding proteins (RBPs) to form
miRNA-protein complexes in order to exist stably in cells (8); the dynamic association of miRNAs
with RBPs occurs throughout the entire life cycle of miRNA
transcription, synthesis, processing, modification, intracellular
translocation, functional exertion and degradation (9,10).
Accumulating evidence has indicated that miRNA packaging in
exosomes is selective (11).
However, the molecular mechanisms regulating the profiles of
exosomal miRNAs remain to be elucidated. On one hand, HCC cells
selectively excrete some endogenous miRNAs to the outside of cells
through exosomes. On the other hand, HCC cell-derived exosomes, as
essential carriers, can shuttle miRNAs into recipient cells (HCC
cells) and promote the proliferation, migration and invasion of
recipient cells (12). Therefore,
it was hypothesized that some RBPs, as specific proteins bound to
miRNAs, may be involved in regulating the profile of exosomal
miRNAs in HCC cells.
Ribosomal proteins (RPs) are the components of
ribosomes and are RBPs (13). In
addition to stabilizing specific ribosomal (r)RNA structures in
ribosomal subunits and promoting the correct folding of rRNA, RPs
have functions other than protein biosynthesis, known as
extra-ribosomal functions (14,15). RPs have also been associated with
the occurrence and development of malignant tumors (16).
Ribosomal protein L9 (RPL9) is a member of the 60S
subunit of the L6P family of ribosomal proteins (17). However, until recently, only a few
studies have reported the relationship between RPL9 and exosomes.
The present study focused on the extra-ribosomal functions of RPL9.
RPL9 is a cancer-promoting RBP that binds to specific miRNAs and
translocates them into exosomes, thereby affecting the profiles of
miRNAs within the exosome. Additionally, these miRNAs can enter
recipient cells via exosomes and regulate the biological
progression of cells. These results initially elucidated the
regulatory mechanisms of RBPs in the targeted transport of miRNAs
into exosomes and lay the theoretical foundation for exploring
exosomal miRNAs interfering with tumor progression.
Materials and methods
Mass spectrometry analysis
Serum exosomes samples were collected from three HCC
patients diagnosed with TNM stage I/II, two HCC patients diagnosed
with TNM stage III/IV, and three patients with benign liver
disease. The proteins in those exosomes were electrophoresed by
SDS-PAGE. The gel was stained with Coomassie Blue. The gel with the
sample was cut off, digested and analyzed by LC-MS/MS by
Bioclouds.
The peptide samples were separated by nano-liter
liquid chromatograph (Shimadzu Corporation; cat. no. LC-20AD) at a
flow rate gradient of 300 nl/min. The peptides were then
transferred to the ESI-tandem mass spectrometer (TripleTOF 5600
LC-MS/MS system; SCIEX). The ion source (Nanospray III source;
SCIEX) used a negative ionization mode. The data were collected at
an ion source spray voltage of 2,300 V, a nitrogen pressure of 30
psi, and a spray interface temperature of 150°C. The details of the
multiple reaction monitoring transitions evaluated were: i) m/z
range of 350-1250 Da; ii) the number of charges is 2-5 charges; and
iii) the dynamic exclusion of precursor ion was set as follows: The
same precursor ion should not undergo fragmentation more than twice
within half of the peak-out time, which is approximately 12
sec.
MicroRNA microarrays
Total RNA was isolated from the exosomes of the
control group and the RPL9 gene knockdown group. Subsequently,
reverse transcriptase was employed to convert RNA into cDNA. The
resulting labeled cDNA was then subjected to hybridization with a
pre-designed microarray chip. Following scanning of the chip using
a scanner, fluorescence signal intensity on each probe was
obtained. Specialized software was utilized for data analysis,
quality assessment, and differential expression analysis of the
scanning results.
Cell lines and culture
SNU387, SNU182 and Hep3B cells were provided by Stem
Cell Bank, Chinese Academy of Sciences. MHCC97H cells were
purchased from the BeNa Culture Collection (cat. no. BNCC359345)
and Huh-7 cells were obtained from Procell Life Science &
Technology Co., Ltd. (cat. no. CL-0120). These two cells passed the
short tandem repeat analysis and mycoplasma contamination
detection. All HCC cell lines were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% fetal bovine serum (FBS; Corning, Inc.) at 37°C with 5%
CO2. The FBS used for exosome isolation was depleted of
EVs by ultracentrifugation for 12 h at 120,000 × g at 4°C (Optima
L-100XP; Beckman Coulter, Inc.).
In vivo tumorigenesis assay
During the experiments, all animals were cared for
humanely according to the standards outlined in the 'Guide for the
Care and Use of Laboratory Animals' (18). Animal experimental procedures were
approved by the Ethics Committee of Sun Yat-Sen University
(Guangzhou, China; approval number 2023001498). A total of 21 male
athymic nude mice (BALB/c-nu/nu; 5 weeks old; weight ~30 g) were
obtained from the Guangdong Medical Laboratory Animal Center. The
mice were maintained at a temperature of 26-28°C and relative
humidity was maintained at 40-60% with a 10-h/14-h light/dark
cycle. Then 1×107 MHCC97H-Control, MHCC97H-RPL9-short
hairpin (sh), Huh7-Control and Huh7-RPL9-sh cells were resuspended
in 100 µl of Matrigel Matrix (Xiamen Wintop) and injected
subcutaneously into the right axilla of nude mice to establish the
xenograft tumors.
The specific criteria (i.e., humane endpoints) used
to determine whether animals should be sacrificed were: i) Animals
on the verge of death, unable to move, or exhibiting no response to
gentle stimulation; ii) difficulty in breathing (typical symptoms
being salivation and/or cyanosis); iii) diarrhea or urinary
incontinence; iv) weight loss of 20% of the pre-experiment weight;
v) inability to eat or drink; vi) animals with obvious anxiety,
irritability or tumor weight >10% of the animal's own body
weight, or tumor rupture; vii) paralysis, persistent epilepsy or
stereotyped behavior; viii) skin injury area of the animal
accounting for >30% of the whole body, or infection and
suppuration; ix) other situations requiring humane end points
determined by veterinarians. The experiment lasted for 28 days.
Animal health and behavior was monitored once a day.
Of the 21 mice used in the experiment, 20 were
sacrificed and one found dead accidentally, the cause unknown.
Animal welfare refers to the process of raising, management and use
of experimental animals, to take effective measures to make the
experimental animals from unnecessary injury, hunger, thirst,
discomfort, panic, torture, disease and pain, to ensure that the
animals can achieve natural behavior, by excellent management and
care, to provide a clean and comfortable living environment, to
provide adequate, ensure the health of food and water, to avoid or
reduce pain and suffering. The method of sacrifice was cervical
dislocation under deep anesthesia with pentobarbital sodium (30-90
mg/kg; IP). Mortality was verified when the laboratory animal lost
consciousness, accompanied by cessation of heartbeat and
breathing.
Western blotting
The HCC cells or exosomes were lysed in Western and
IP buffers (Key GEN Bio TECH). BCA protein assay kit (Thermo Fisher
Scientific, Inc.) was used to measure the protein concentration.
Then, the same protein mass (cellular proteins 20 µg per
lane; exosome protein 40 µg per lane) was separated by
SDS-PAGE on an 11% gel and transferred to polyvinylidene difluoride
membranes (Merck KGaA). The primary and secondary antibodies were
diluted with universal antibody diluent (cat. no. GF1600-02;
Genefist, Inc.). After blocking with 5% skimmed milk for 60 min at
room temperature, the membranes were incubated with primary
antibody overnight at 4°C. Finally, the membranes were incubated
with the secondary antibody for 60 min at room temperature to
visualize the membrane using an enhanced chemiluminescence system
(ImageQuant; Cytiva). Western blots were quantified using ImageJ
2.1.0/1.53c (National Institutes of Health) The antibodies were:
Apoptosis related gene 2-interacting protein X (Alix; Cell
Signaling Technology, Inc.; cat. no. 92880S), tumor susceptibility
gene 101 (TSG101; ABclonal Biotech Co., Ltd.; cat. no. A1692),
cluster of differentiation (CD)63 (MilliporeSigma; cat. no.
SAB4301607), heat shock protein 90 homolog (HSP90; Santa Cruz
Biotechnology; cat. no. SC-13119), β-actin (Cell Signaling
Technology, Inc.; cat. no. 4970S), GAPDH (Cell Signaling
Technology, Inc.; cat. no. 2118S), β-Tubulin (Cell Signaling
Technology, Inc.; cat. no. 2146S), RPL9 (Abcam.; cat. no.
ab182556), cystathionine β-synthase (CBS; Proteintech Group, Inc.;
cat. no. 14787-1-AP), horse anti-mouse IgG horseradish peroxidase
(HRP) conjugated secondary antibody (Cell Signaling Technology,
Inc.; cat. no. 7076S) and goat anti-rabbit IgG HRP-conjugated
secondary antibody (Cell Signaling Technology, Inc.; cat. no.
7074S).
Cytoplasmic and nuclear protein
extraction
The nuclear/cytoplasm protein extraction kit
(Fudebio-tech; cat. no. FD0199) was used to extract proteins from
the nucleus and cytoplasm, respectively, in accordance with the
manufacturer's protocols.
RNA isolation and reverse
transcription-quantitative (RT-q) PCR
TRIzol® reagent (Thermo Fisher
Scientific, Inc) was used for RNA extraction when cell confluence
was 70-80%. Prime Script RT Master Mix (Toyobo Life Science) was
used for mRNA reverse transcription. Reverse transcription of miRNA
was performed using miRNA first-strand cDNA synthesis (tailing
method) kit (Sangon Biotech, China). RT-qPCR was performed using
SYBR Premix EX Taq TMII (Toyobo Life Science). RNA extraction, cDNA
synthesis and qPCR were performed according to the manufacturer's
protocols. PCR cycle conditions: Denaturation (95°C, 30 sec);
annealing (95°C, 5 sec) and extension (60°C, 30 sec) for 40 cycles.
Quantification method was ΔCt=Ct (target gene)-Ct (internal
reference gene), ΔΔCt=ΔCt (experiment group)-ΔCt (control group)
and final results were shown as 2ΔΔCq (12). These experiments were repeated at
least 3 times. The internal references for mRNA and miRNA extracted
from cells were GAPDH and U6, respectively. However, miRNAs
extracted from exosomes used miR16-5p as an internal reference. The
primer sequences for RT-qPCR are given in Table SI.
Immunohistochemistry (IHC)
The present study analyzed samples from 38 patients
who underwent hepatectomy at Sun Yat-sen Memorial Hospital
affiliated with Sun Yat-sen University (Guangzhou, China). The
patients, ranging in age from 31-72, consisted of 19 individuals
with HCC and a male:female ratio of 14:5. In addition, there were
19 patients with benign liver disease and a male:female ratio of
9:10. From the in vivo tumorigenicity assay, 20 xenograft
tumor tissue samples were obtained in nude mice (MHCC97H-Control
tumors, n=4; MHCC97H-RPL9-sh tumors, n=5; Huh7-Control tumors, n=5;
Huh7-RPL9-sh tumors, n=6). The antibody used for
immunohistochemical detection was anti-RPL9 (Sino Biological; cat.
no. 202771-T44). diluted to 1:200 and incubated overnight at 4°C.
Selected tissue images were randomly selected from five fields
using the cell imaging system (EVOS FL Auto; Thermo Fisher
Scientific, Inc.) and the integrated optical density (IOD) score
was assessed using ImageJ 2.1.0/1.53c (National Institutes of
Health, USA).
Enzyme linked immunosorbent assay
(ELISA)
A total of 56 serum samples were collected from HCC
patients (age 24-78; male:female ratio 48:8) and 23 from patients
with benign liver disease (BLD) (age 27-72; male:female ratio 9:14)
at Sun Yat-sen Memorial Hospital affiliated with Sun Yat-sen
University (Guangzhou, China), between 2016 and 2018. First,
isolated exosome pellets from 100 µl serum were lysed in 100
µl nondenatured protein solubilization reagent (Invent
Biotechnologies Inc.) containing a phosphatase-inhibitor and
protease-inhibitor (MilliporeSigma). Then the levels were
determined using the Human RPL9 ELISA Kit (cat. no. OM516307;
Omnimabs, Inc.) according to the manufacturer's instructions.
Patients and specimens
All the human samples were obtained from patients
with the written informed consent. The study protocols were
approved by the Ethics Committee of Sun Yat-sen Memorial Hospital,
Sun Yat-sen University (Guangzhou, China; approval number
2023001498). All research was conducted in accordance with both the
Declarations of Helsinki and Istanbul. Patients with a prior
diagnosis of primary HCC had not undergone any cancer treatment
prior to surgery.
Establishment of stable and transient
cell lines
Based on the GV493 vector (Shanghai GeneChem Co.,
Ltd.), lentiviruses were constructed with knockdown of RPL9
(NM_000661; RPL9-sh-gcGFP; target sequence: gaTG GTA TCT ATG TCT
CTG AA) and control lentiviruses expressing only gcGFP
(RPL9-Control-gcGFP; target sequence: TTC TCC GAA CGT GTC ACG T).
When the cell confluence reached 30%, lentivirus was used to infect
MHCC97H and Huh7 cell lines. After transfection for 48 h at 37°C,
puromycin was used to screen with 2 µg/ml. The screening was
completed when all cells showed green fluorescence.
Human cDNAs encoding N-terminal Flag-tagged RPL9
(genomic fragments encompassing the CDS sequence 579 bp) were
cloned into a pLVX-AcGFP-N1 lentiviral vector to construct the RPL9
expression vector. The primer sequences for cloning were 5'-CCC TCG
AGA TGG ATT ACA AGG ATG ACG ACG ATA AGA AGA CTA TTC TCA GCA ATC
AG-3' (forward) and 5'-CGG GAT CCC GTT CAT CAG CCT GCT GAA CAG TTC
C-3' (reverse). The resultant plasmid was designated as
pLVX-Flag-RPL9-AcGFP. The human cDNA encoding miR-24-3p (5'-TGG CTC
AGT TCA GCA GGA AC AG-3') and miR-185-5p (5'-TGG AGA GAA AGG CAG
TTC CTG A-3') were cloned into the pCMV-mChery-SV40-neo vector to
construct the vectors pCMV-miR-24-3p-mCherry and
pCMV-miR-185-5p-mCherry (fluorescence observation only).
Finally, hsa-miR-24-3p-mimics-Cy5 (5'-UGG CUC AGU
UCA GCA GGA ACA G-3'), hsa-miR-185-5p-mimics-Cy5.5 (5'-UGG AGA GAA
AGG CAG UUC CUG A-3') and the negative control group for the two
miRNA-mimics (5'-UUU GUA CUA CAC AAA AGU ACU G-3') were designed
and constructed by Nanjing KeyGen Biotech Co., Ltd.
The short interfering (si)RNA sequence sense (5'-3')
of human Alix was: GCU CCU GAG AUA UUA UGA UCA TT. The siRNA
sequence sense (5'-3') of human VPS4A was: CGA GAA GCU GAA GGA UUA
UUU TT. The siRNA sequence sense (5'-3') of human TSG101 was: CAG
UCU UCU CUC GUC CUA UUU TT. The control group used for the three
siRNAs sequence sense (5'-3') was: UUC UCC GAA CGU GUC ACG UdT
dT.
The transient transfection was conducted in MHCC97H
and Huh7 cell lines. Plasmids (1 µg/ml) were introduced when
the cell confluence reached 60-80%; miRNA mimics or siRNA (10-50
nmol/l) were added when the cell confluence reached 50%. The
transfection process was carried out at 37°C for 24 h using
jetPRIME transfection reagent (Polyplus-transfection SA). Laser
confocal microscopy or RNA extraction was typically performed after
48 h of transfection, while protein extraction was conducted after
72 h of transfection.
Exosome isolation
The HCC cells were seeded in a 10 cm dish and
cultured at 37°C for 2-3 days in a vesicle-depleted medium. The
collected media was centrifuged at 300 × g for 10 min, 2,000 × g
for 20 min and 10,000 × g for 30 min at 4°C. The supernatant was
filtered through a 0.22 µm filter (Millex-GP;
MilliporeSigma) to remove large vesicles. The resulting supernatant
was further ultracentrifuged at 100,000 × g for 2 h. Alternatively,
an appropriate amount of ExoQuick-TC Tissue Culture Media Exosome
Precipitation Solution (System Biosciences) was added to isolate
exosomes according to the manufacturer's protocols. The exosomes
extracted by ultracentrifugation were used for transmission
electron microscopy (TEM; JEM-1,400; JEOL, Ltd.) and the
reagent-extracted exosomes used for a nanoparticle tracking
analyzer (Particle Metrix GmbH), western blotting, PCR and cell
co-production cultivation.
Nanoparticle tracking analyzer
The exosome particles, separated from a 10 ml
culture medium, were resuspended in 100 µl of PBS.
Subsequently, a 10 µl aliquot was taken and diluted to 30
µl with PBS. Operate according to the instructions of the
particle size analyzer (N30E; NanoFCM Co., Ltd.). The performance
of the instrument was tested with standard substances before the
exosome samples were loaded. Note that gradient dilution was
required to avoid the blockage of the needle. Following detection,
the information regarding the particle size and concentration of
exosomes detected by the instrument could be obtained.
TEM
Isolated exosome pellets from 10 ml culture medium
were resuspended in 10 µl PBS and fixed with 10 µl
paraformaldehyde (4%), absorbed onto the formvar-carbon-coated
copper grids for 10 min and dried by filter paper; pellets were
then stained with 10 µl uranyl acetate (2%) for 90 sec and
dried before examination using TEM operated at 120 kV.
Co-localization experiments with
miRNA-mimics transfection
First, RPL9-GFP plasmids, miR-24-3p-mimics-cy5 and
miR-185-5p-mimics-cy5.5 were cotransfected with MHCC97H and Huh7
cells, respectively at 37°C for 24 h using jetPRIME transfection
reagent (Polyplus-transfection SA). The cells were labeled with
PKH26 (PKH26 Red Fluorescent Cell Linker Kit; Beijing Fluorescence
Biotechnology Co., Ltd.), which has a fluorescence region of
551-567 nm. According to the manufacturer's protocols, the cells
were placed in PKH26 solution and rinsed with serum and complete
culture medium after incubation at 25°C for 2-5 min. These cells
were cultured in confocal special dishes for 24 h and finally
directly observed with a confocal fluorescence microscope (Olympus
LV3000; Olympus Corporation).
Confocal laser scanning microscopy
Following transfection of RPL9-GFP and miRNA-mCherry
plasmids at 37°C for 24-48 h, the cells were transferred to a glass
culture dish and grown for 12 h. The cells were fixed with 4%
paraformaldehyde for 15 min at room temperature, treated with 0.3%
Triton X-100 for 15 min, stained with DAPI for 10 min and directly
observed with a laser confocal microscope (Olympus LV3000, Olympus
Corporation).
Exosome and cell co-culture
experiments
First, 2x107 cells were treated with
PKH26, seeded in several 10 cm diameter dishes and cultured at 37°C
for 12 h with a vesicle-free medium. When the cell confluence
reached 60%, flag-RPL9-AcGFP (1 µg/ml), miR-24-3p-mimics-Cy5
(50 nmol/l) and miR-185-5p-mimics-Cy5.5 (50 nmol/l) were
transfected into cells using jetPRIME transfection reagent
(Polyplus-transfection SA) and cultured at 37°C for 48 h. Finally,
10 µg of the isolated exosomes was added to a glass culture
dish to co-culture with the cells for 12 h and observed under a
laser confocal microscope (Olympus LV3000; Olympus
Corporation).
RNA immunoprecipitation (RIP)
RIP assays were performed using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (MilliporeSigma; cat.
no. 17-700) according to the manufacturer's protocols. i) Lysate
preparation: serum exosomes isolated from 500 µl of patient
serum were utilized as the quantity of exosome lysate for each IP
reaction. For MHCC97H and Huh7 cell lines, a total of
2x107 cells were employed as the amount of lysate per IP
reaction. ii) Preparation of magnetic beads for
immunoprecipitation: 50 µl of magnetic beads protein A/G
suspension (the magnetic beads were coated with Protein A or G;
MilliporeSigma; cat. no. CS203178) was mixed with RPL9 antibodies
(5 µg per IP; Sino Biological; cat. no. 202771-T44) or
negative control IgG (5 µg per IP; MilliporeSigma) and
incubated for 30 min at room temperature. iii) Immunoprecipitation
of RNA-binding Protein-RNA complexes: the lysate obtained from step
1 was mixed with the magnetic beads-antibody complex prepared in
step 2 and subjected to overnight incubation at a temperature of
4°C. Next, the immunoprecipitated RNA was extracted, separated and
purified. The total RNA under co-immunoprecipitation of RPL9
protein and IgG in the experiment were transformed into cDNA by
using the miRNA first-strand cDNA synthesis (tailing method) kit
(Sangon Biotech Co., Ltd.). Finally, the enrichment of binding
miRNAs was identified by RT-qPCR.
Co-immunoprecipitation assay (Co-IP)
Cells were lysed with IP lysis buffer (Beyotime
Institute of Biotechnology) containing protease inhibitors and
phosphatase inhibitors (MilliporeSigma). The protein sample (30
µg) was used as input group and the remaining samples were
0.5 mg of cell lysate diluted to 300 µl 1X PBS. Then, 4
µg of anti-RPL9 (Sino Biological; cat. no. 202771-T44) and
IgG (cat. no. PP64B; MilliporeSigma) were added respectively,
incubated at 4°C overnight. Then, 50 µl protein A/G magnetic
beads (MedChem Express; cat. no. HY-K0202) were added and incubated
at room temperature for 4 h. The magnetic beads were washed with
1XPBS with 0.5% Triton, separated by magnetic separation magnetic
frame. Subsequently, they were treated with 2X loading buffer (20
µl) and subjected to heat at 95°C for 5 min in a metal bath.
The magnetic beads were isolated by magnetic separation. The
interaction between RPL9 protein and other proteins was detected by
western blotting.
In order to avoid the influence of IgG heavy chain,
rabbit anti-RPL9 and IgG, mouse anti-TSG101 (MilliporeSigma; cat.
no. SAB2702167) and VPS4A (Santa Cruz Biotechnology, Inc.; cat. no.
SC-393428) and anti-heavy chain secondary antibody (Abbkine
Scientific Co., Ltd.; Anti-Mouse; cat. no. A25012; Anti-Rabbit;
cat. no. A25022;) were used. Other antibodies were: Alix (Cell
Signaling Technology, Inc.; cat. no. 92880S), CD63 (MilliporeSigma;
cat. no. SAB4301607), HSP90 (Santa Cruz Biotechnology; cat. no.
SC-13119),
Immunofluorescence (IF)
Cells were grown on glass dishes for 24 h then fixed
for 15 min in 4% paraformaldehyde and permeabilized with 0.3%
Triton X-100 for 15 min at room temperature. Next, cells were
blocked with 10% goat serum for 1 h at room temperature, followed
by overnight incubation with primary antibodies at 4°C. After being
washed with PBS, the cells were incubated with Alexa-conjugated
secondary antibody (Cell Signaling Technology, Inc.) at 37°C for 1
h and then incubated with DAPI at 37°C for 5 min. The cells were
visualized and images captured on a confocal microscope (Olympus
LV3000; Olympus Corporation). Primary antibodies were: RPL9
(dilution: 1:50; Abcam; cat. no. ab182556), TSG101 (dilution:
3:100; MilliporeSigma; cat. no. SAB2702167), VPS4A (dilution: 1:50;
Santa Cruz Biotechnology; cat. no. SC-393428) and Alix (dilution:
1:50; Santa Cruz Biotechnology; cat. no. sc-53540).
In vitro cell proliferation, colony
formation, migration and invasion assay
The exosomes were diluted to a protein concentration
of 10 µg/ml for co-culture with cells.
Cell Counting Kit-8 (CCK-8)
Cells (1×103) were seeded in 96-well
plates. On days 1, 3, 5 and 7 (cells cocultured with exosomes at
12, 24, 48 and 72 h), the cells were treated using Cell Counting
Kit-8 (MilliporeSigma) according to the manufacturer's
instructions. The OD values of each well were then measured at a
wavelength of 450 nm using a multifunctional enzyme marker (Thermo
Fisher Scientific, Inc.).
Wound healing assay
Cells were cultured on 6-well plates and scratched
with a 10-µl pipette tip when the confluence reached
>90%. They were washed three times with PBS and serum-free
medium was added and incubate at 37°C for 3 days. The images were
captured under the microscope at 0, 24 and 48 h. Selected images
were randomly selected from five fields using the cell imaging
system (EVOS FL Auto; Thermo Fisher Scientific, Inc.) at
magnification x10.
Transwell migration and invasion
assays
The lower chamber was supplemented with 400
µl of DMEM complete medium (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS (Corning, Inc.), followed by loading the 8
µm pore size Transwell chambers (Corning, Inc.) and seeding
1×105 cells in the upper chamber. For migration assays,
the upper chamber was filled with 200 µl of FBS-free DMEM
medium. In invasion experiments, a mixture of Matrigel Matrix
(Xiamen Wintop) and FBS-free medium (200 µl) was added to
the upper chamber. Subsequently, the cells were incubated at 37°C
for 24 h in a cell incubator. Then, cells remaining on the upper
surface of the membrane were removed and cells in the lower surface
that had passed through the 8 µm hole were fixed with 4%
paraformaldehyde and stained with 0.1% crystal violet at 37°C for
15 min. Finally, those cells were washed and dried with PBS and
observed with a transmission light microscope (EVOS FL Auto; Thermo
Fisher Scientific, Inc.).
Statistical analysis
The RPL9 expression levels in the HCC and the benign
liver disease (BLD) were compared using a paired sample t-test. The
relationships between RPL9 expression with the relevant patient and
tumor characteristics were analyzed using the χ2 test.
The continuous overall survival (OS) time started from the date of
tumor resection until the patient succumbed or was lost to
follow-up. Univariate Cox regression analysis was used to
understand the relationship between OS and influence factors.
Significant prognostic factors were further evaluated by
multivariate Cox regression analysis. The Kaplan-Meier curve was
used to analyze the effect of a single factor on OS. The
statistical analyses were completed by SPSS v23 (IBM Corp.), ImageJ
v2.1.0/1.53c (National Institutes of Health) and GraphPad Prism
7.04 (Dotmatics). P<0.05 was considered to indicate a
statistically significant difference.
Results
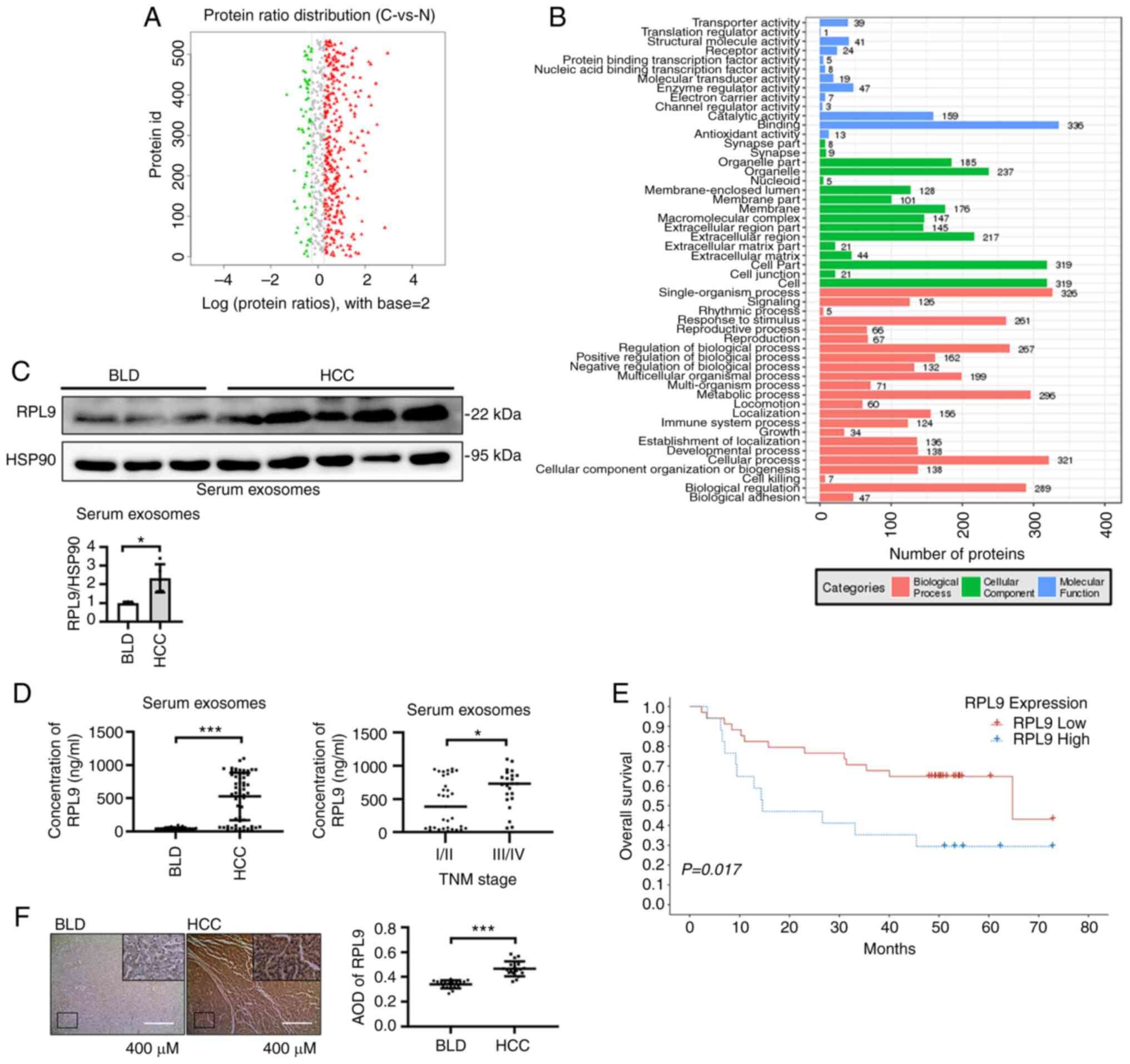
RPL9 is upregulated in serum exosomes and
clinical samples of patients with HCC
Serum samples were collected from five patients with
HCC (C1-C5) and three patients with BLD (N1-N3). Exosomes were
extracted and subjected to protein profiling. First, the samples
were divided into group C (C1-C5) and group N (N1-N3) and the
differentially contained proteins in exosomes of HCC patients and
BLD patients were obtained after statistical analysis (Fig. 1A; Table SII). Then, proteins with
'RNA-binding' function were screened out from the 'binding'
classification of cluster analysis (Fig. 1B; Table SIII). Finally, seven RBPs (RPL9,
RPL28, RPS13, RPS14, RPS4X, PSMA6 and SNU13) were found to be
highly contained in serum exosomes of HCC patients. Since C1-C2 was
derived from HCC patients with TNM stage I/II and C3-C5 was derived
from HCC patients with TNM stage III/IV, the samples were again
divided into CA (C1-C2), CB (C3-C5) and N (N1-N3) groups. By
analyzing the content of RNA binding protein in the three groups
(Fig. S1A; Tables SIV, SV and
SVI), the present study screened RPL9 genes, which was not only
highly contained in the serum exosomes of HCC patients, but also
had a higher content in the serum exosomes of TNM stage III/IV HCC
patients (Fig. S1B).
Western blotting experiments initially verified the
content of RPL9 in serum exosomes of five HCC patients and three
BLD patients (Fig. 1C). To
further verify the difference in RPL9 content, the sample
collection was expanded. ELISA showed that the content of RPL9 in
the serum exosomes of patients with HCC (56 cases) was
significantly higher than that in the serum exosomes of patients
with BLD (23 cases). In patients with HCC, the serum exosome RPL9
was generally higher in TNM stage III/IV (23 cases) compared with
TNM stage I/II (33 cases; Fig.
1D). Data on 56 patients with HCC was then gathered and
followed up, finally obtaining prognostic information on 51
patients with HCC after excluding patients who had missed
follow-up. Based on the optimal cut-off value, patients with HCC
were divided into high-risk (RPL9 high) and low-risk (RPL9 low)
groups (their clinicopathological characteristics are presented in
Table SVII). The analysis
revealed that higher serum exosome RPL9 levels were a risk factor
for postoperative survival (Fig.
1E; Tables SVIII and SIX).
In addition, 19 HCC and 19 BLD tissue samples were selected and
their IHC analyses showed that RPL9 was more highly expressed in
HCC tissues than in BLD tissues (Fig.
1F). By examining clinical samples, it was determined that the
RBP RPL9 was upregulated in serum exosomes and cancer tissues,
suggesting that this gene may be associated with cancer-promoting
effects in humans.
Downregulation of RPL9 suppresses the
ability of HCC cells to proliferate, migrate and invade
To evaluate the role of RPL9 in HCC more fully, the
expression of RPL9 in different hepatoma cell lines (SNU387,
SNU182, Hep3B, Huh7 and MHCC97H) and their secreted exosomes was
examined (Fig. 2A). As the RPL9
protein was strongly expressed, lentiviruses were designed with the
knocked-down RPL9 gene and control viruses. MHCC97H and Huh7 were
then selected to construct stably transfected cell lines and the
cells divided into control (MHCC97H-control and HuH7-control) and
RPL9-sh groups (MHCC97H-RPL9-sh and HuH7-RPL9-sh) for comparison
(Fig. 2B).
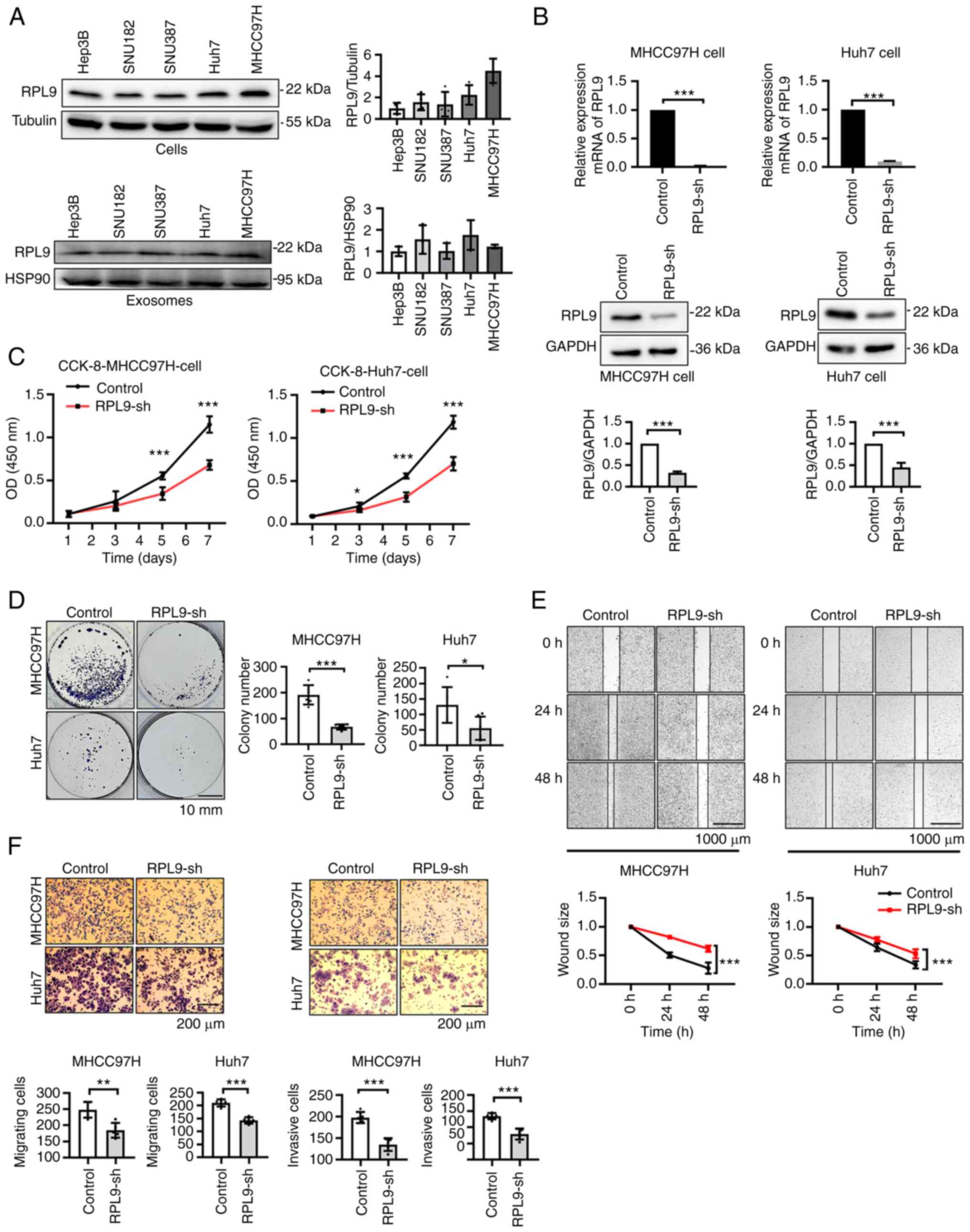 | Figure 2Downregulation of RPL9 suppresses the
ability of HCC cells to proliferate, migrate and invade. (A)
Western blotting to detect the expression of RPL9 in five human HCC
cell lines (Hep3B, SNU182, SNU387, Huh7, MHCC97H) and their
secreted exosomes. (B) Stably expressed RPL9 knockdown (RPL9-sh)
and control cell lines were constructed in MHCC97H and Huh7 cells.
Western blotting and reverse transcription-quantitative PCR
experiments were used to verify the expression level of RPL9
(***P<0.001). (C) CCK-8 (n=5;
***P<0.001) and (D) cell colony formation assays
(n=6; *P<0.05, ***P<0.001; scale bar=10
mm) indicated that RPL9 downregulation hindered cell proliferation.
(E) Wound healing (n=6; ***P<0.001; scale bar=1,000
µm) revealed that RPL9 silence reduced cell migration
capacity. (F) Transwell migration and Transwell invasion experiment
(n=5; **P<0.01, ***P<0.001; scale
bar=200 µm) suggested that the migration and invasion
abilities were suppressed after RPL9 knockdown. RPL9, ribosomal
protein L9; HCC, hepatocellular carcinoma; sh, short hairpin. |
Next, CCK-8 (Fig.
2C), clone formation (Fig.
2D), wound healing (Fig. 2E)
and Transwell assays (Fig. 2F)
were performed and it was found that the proliferation, migration
and invasion abilities of MHCC97H and Huh7 cells were significantly
inhibited when RPL9 was knocked down. These results indicated that
downregulation of RPL9 suppressed the progression of HCC,
suggesting that RPL9 may function as an oncogene.
Downregulation of RPL9 affects the
profile of miRNAs in the exosomes of HCC cells
EVs were extracted from the culture medium
supernatant of the HCC cells. Nanoparticle tracking analysis showed
that most of the EVs were in the range of 40-100 nm (Fig. 3A). TEM revealed a smooth,
cup-shaped envelope (Fig. 3B).
Meanwhile, western blotting showed that EVs contained typical
exosome markers (Alix, HSP90, TSG101 and CD63; Fig. 3C). These results suggest that the
isolated EVs have typical characteristics of exosomes. It was also
found that downregulation of RPL9 significantly reduced RPL9
protein content at the exosome level (Fig. 3D).
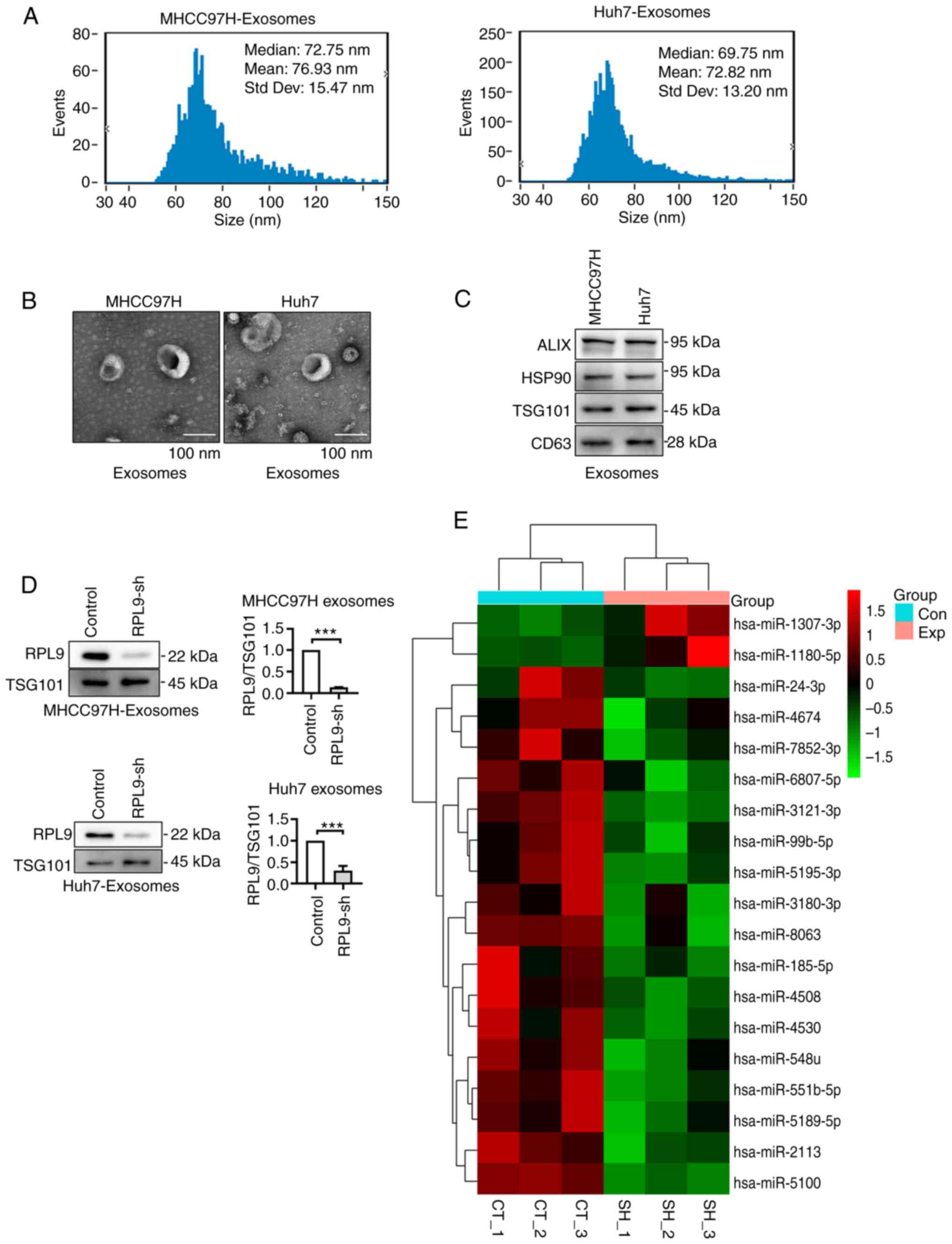 | Figure 3Downregulation of RPL9 affects the
profile of miRNAs in the exosomes of HCC cells. (A) NTA results
indicated that the size of the isolated exosomes was mainly
distributed at 40-100 nm. (B) Transmission electron microscopy
showed the classic cup-like structure of exosomes secreted by
MHCC97H and Huh7 cells (scale bar=100 nm). (C) The isolated
exosomes contained representative exosome markers (Alix, HSP90,
TSG101 and CD63) in western blotting. (D) Western blotting showed
that downregulation of RPL9 in the cells resulted in decreased
secretion of RPL9 in exosomes (***P<0.001). (E) Heat
map revealed the differentially content miRNAs into exosomes
released from control cells (CT_1-3) and RPL9 knockdown (SH_1-3)
cells (P<0.05). RPL9, ribosomal protein L9; miRNA/miR, microRNA;
HCC, hepatocellular carcinoma; sh, short hairpin; Con, control;
Exp, expression decrease; CT, MHCC97H control cell-derived
exosomes; SH, MHCC97H-RPL9-sh cell-derived exosomes. |
As an RBP, RPL9 is widely involved in the
biosynthesis, transport and binding of miRNAs and is abundant in
exosomes. The present study examined the effect of RPL9 on the
miRNA profile in HCC exosomes and screened miRNAs that were
significantly altered with the reduction of RPL9 protein in
exosomes using miRNA microarray technology (Fig. 3E; Tables SX and SXI).
Downregulation of RPL9 results in
differential expression of miR-24-3p and miR-185-5p and affects the
biological activity of exosomes derived from HCC cells
To further verify the reliability of the selected
miRNAs, the profile changes of some miRNAs after RPL9 knockdown
were examined (Fig. S2).
However, it was found that the content of these miRNAs was not
consistent between cells and exosomes. Most of the miRNAs were
increased or unchanged in the cells, but were clearly reduced in
the exosomes. In particular, miR-24-3p and miR-185-5p showed
significantly decrease in cells and exosomes (Fig. 4A). It was hypothesized that the
downregulation of RPL9 would inhibit the transfer of several miRNAs
into exosomes and that there could be interactions between RPL9 and
miR-24-3p or miR-185-5p.
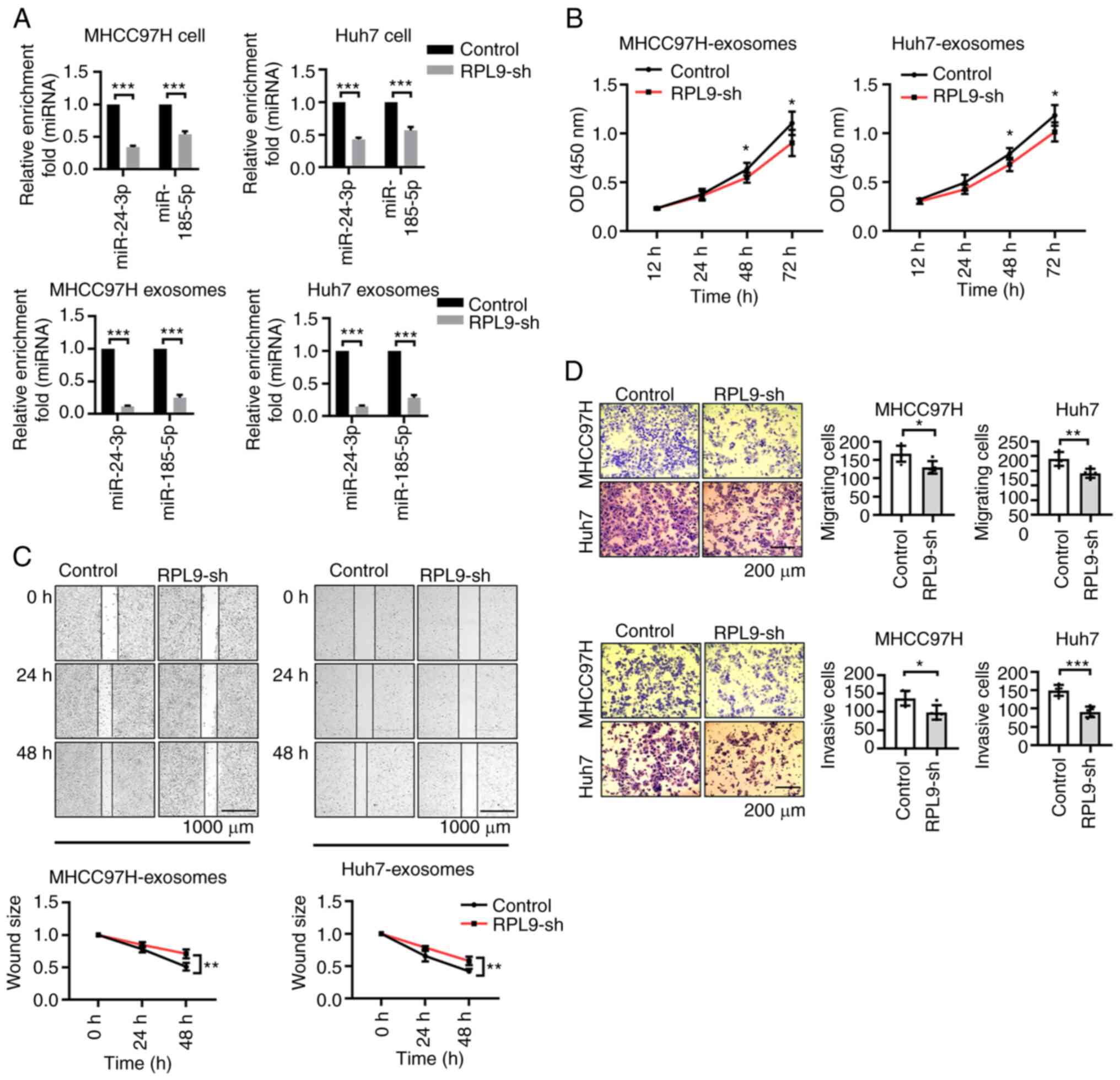 | Figure 4Downregulation of RPL9 results in
differential expression of miR-24-3p and miR-185-5p and affects the
biological activity of exosomes derived from HCC cells. (A) Reverse
transcription-quantitative PCR showed that the content of miR-24-3p
and miR-185-5p in cells and exosomes were distinctly decreased with
the knockdown of RPL9 (***P<0.001). The present study
co-cultured exosomes (10 µg/ml, exosomes were diluted to a
protein concentration of 10 µg/ml) from the RPL9-control and
RPL9-sh groups with recipient HCC cells, respectively. From (B)
CCK-8 (n=6; *P<0.05), (C) wound healing (n=6;
**P<0.01; scale bar=1,000 µm) and (D)
Transwell assays (n=5; *P<0.05,
**P<0.01, ***P<0.001; scale bar=200
µm), recipient cell proliferation, migration, invasion
abilities were relatively reduced after receiving exosomes from the
RPL9-sh group. RPL9, ribosomal protein L9; miRNA/miR, microRNA;
HCC, hepatocellular carcinoma; sh, short hairpin. |
Our previous experiments revealed that HCC
cell-derived exosomes promote the biological behavior of parent HCC
cells (12). As RPL9 can affect
the profile of exosomal miRNAs, it was hypothesized whether RPL9
affects the biological activity of exosomes. After the exosomes
secreted by the cells in the RPL9-control group and the RPL9-sh
group were extracted, the exosomes were diluted to a protein
concentration of 10 µg/ml and co-incubated with the
recipient cells, changing the fluid every 24 h. Using CCK-8, wound
healing and Transwell assays (Fig.
4B-D), it was found that knockdown of RPL9 inhibited the
proliferation, migration and invasion abilities of the recipient
cells. This suggested that RPL9 downregulation markedly suppressed
the biological activity of HCC cell-derived exosomes.
Overexpression of RPL9 facilitates
miR-24-3p and miR-185-5p entry into the recipient cells via the
exosome pathway
It was hypothesized that RPL9 may be closely related
to miR-24-3p and miR-185-5p. To further explore this relationship,
the present study first transfected the pLVX-Flag-RPL9-AcGFP
plasmid into MHCC97H and Huh7 cells to construct an RPL9
overexpression cell line. The RPL9 control cell line was
constructed by transfecting cells with the empty plasmid
pLVX-Flag-AcGFP. After verifying the expression of RPL9 (Fig. 5A), it was found that the content
of miR-24-3p and miR-185-5p were elevated in both cells and
exosomes when RPL9 protein expression was enhanced (Fig. 5B). Based on the consistency
between RPL9 and miRNA changes, it was hypothesized that RPL9 may
promote the biological properties of tumors in an miRNA-dependent
manner.
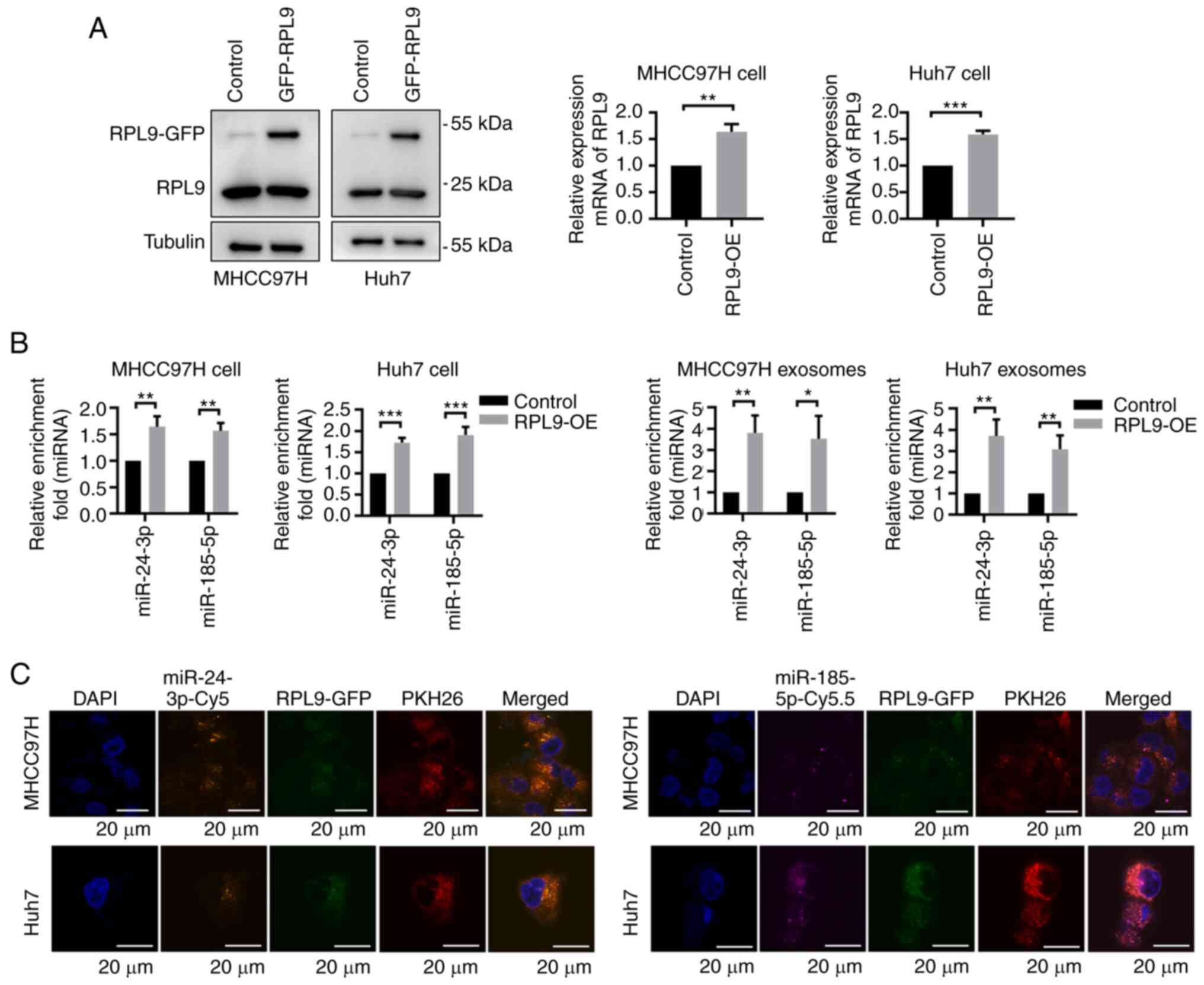 | Figure 5Overexpression of RPL9 facilitates
miR-24-3p and miR-185-5p entry into the recipient cells via the
exosome pathway. (A) In the RPL9 overexpression cell lines, reverse
transcription-quantitative PCR and western blot experiments were
performed to verify their expression effect
(**P<0.01, ***P<0.001). (B) When RPL9
protein expression was enhanced, miR-24-3p and miR-185-5p were
subsequently elevated in cells and exosomes (*P<0.05,
**P<0.01, ***P<0.001). (C) The cells
were treated with PKH26 (red) and transfected with fusion plasmids
Flag-RPL9-AcGFP (green) and miR-24-3p-mimics-Cy5 (orange),
miR-185-5p-mimics-Cy5.5 (purple), their exosomes were extracted and
co-cultured with fresh original cells. The fluorescence could be
observed in the recipient cells (scale bar=20 µm). RPL9,
ribosomal protein L9; miRNA/miR, microRNA; RPL9-OE, RPL9-over
expressed; GFP, green fluorescent protein; DAPI,
4',6-diamidino-2-phenylindole; Cy, cyanine. |
While it was hypothesized that changes in miRNA
profiles in exosomes affected the biological progression of
recipient cells, it was unclear whether RPL9 and miRNAs can enter
cells via exosomes. To test this, cells were stained with the
membrane structural marker PKH26 (red) and then successively
transfected them with the fusion plasmids pLVX-Flag-RPL9-AcGFP
(green), miR-24-3p-mimics-cy5 (orange) and miR-185-5p-mimics-cy5.5
(purple); the collected exosomes were co-cultured with the original
cells. The fluorescence was visualized inside the recipient cells
using laser confocal microscopy (Fig.
5C), suggesting that RPL9, miR-24-3p and miR-185-5p were
present in the exosomes and were then transferred to fresh cells
with exosomes. These data indicated that RPL9 may bind to and carry
miRNAs that play a role through exosomes.
RPL9 directly binds to miR-24-3p and
miR-185-5p
To verify whether RPL9 protein and miRNA bind to
each other in cells and exosomes, an RIP assay was performed using
serum exosomes from patients with HCC, MHCC97H cells and Huh7 cells
as lysates. Subsequently, miRNAs screened by the miRNA microarray
were selected for validation (Fig.
S3A-C) and it was found that miR-24-3p and miR-185-5p could
directly bind RPL9 in both HCC cells and exosomes (Fig. 6A-C).
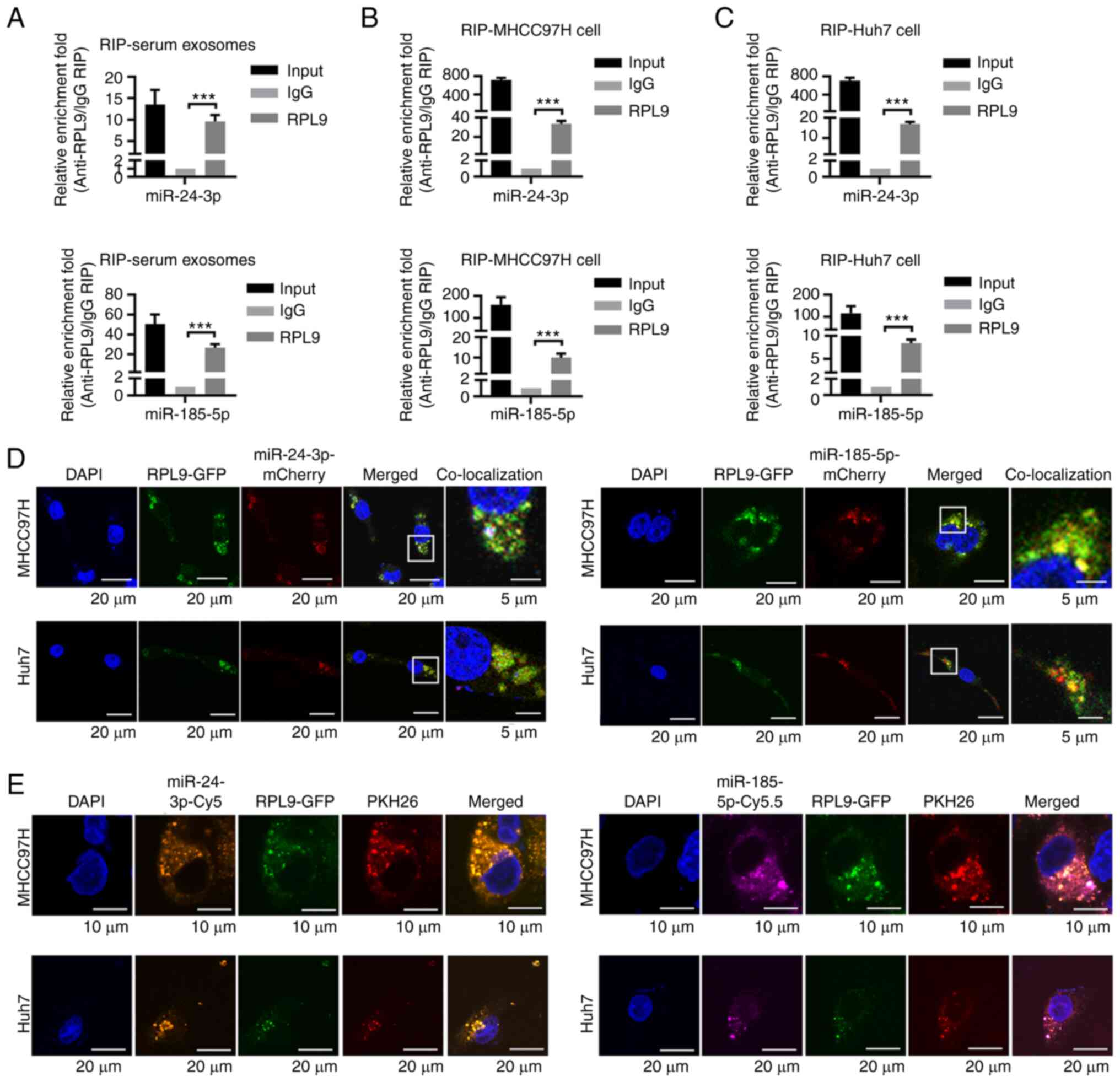 | Figure 6RPL9 directly binds to miR-24-3p and
miR-185-5p. RIP assay was performed using (A) serum exosomes from
HCC patients, (B) MHCC97H cells and (C) Huh7 cells as lysates.
Reverse transcription-quantitative PCR results showed that among
these three substrates, miR-24-3p and miR-185-5p could bind
directly to RPL9 (***P<0.001). (D) Confocal laser
scanning microscopy revealed the colocalization of RPL9-GFP with
miR-24-3p-mCherry and miR-185-5p-mCherry (scale bar=20 µm;
scale bar=5 µm). (E) Subsequently, cells were transfected
with pLVX-Flag-RPL9-AcGFP (green), miR-24-3p-mimics-cy5 (orange)
and miR-185-5p-mimics-cy5.5 (purple), followed by staining the cell
membrane with PKH26 (red). RPL9, miR-24-3p and miR-185-5p were
enriched in the intracellular spherical membrane structure (scale
bar=10 µm; scale bar=20 µm). RPL9, ribosomal protein
L9; miRNA/miR, microRNA; RIP, RNA immunoprecipitation; HCC,
hepatocellular carcinoma. |
To further determine the interactions among RPL9,
miR-24-3p and miR-185-5p, fusion plasmids were designed
(pLVX-Flag-RPL9-AcGFP, pCMV-miR24-3p-mCherry and
pCMV-miR-185-5p-mCherry) and transfected into MHCC97H and Huh7
cells. Co-localization experiments showed that the green light
emitted by RPL9 could fuse with the red light emitted by miR-24-3p
or miR-185-5p (Fig. 6D). This
suggested the possible mutual binding of RPL9 with miR-24-3p or
miR-185-5p. It was also observed that plasmid-generated
fluorescence tended to aggregate within the cell; therefore, it was
hypothesized that this may be a manifestation of RPL9 and miRNA
enrichment in multivesicular bodies. Next, the cells were
transfected with pLVX-Flag-RPL9-AcGFP (green), miR-24-3p-mimics-cy5
(orange) and miR-185-5p-mimics-cy5.5 (purple), followed by PKH26
(red) staining of the cell membrane. Using laser confocal
microscopy, the present study not only visualized the
co-localization phenomenon, but also more precisely observed the
enrichment of RPL9, miR-24-3p and miR-185-5p in the intracellular
spherical membrane structure (Fig.
6E). These results indicated that RPL9 may bind directly to
miR-24-3p and miR-185-5p in both cells and exosomes.
Downregulation of RPL9 suppresses the
growth of HCC tumors and reduces miR-24-3p and miR-185-5p content
in vivo
To investigate the role of RPL9 in vivo, the
present study performed subcutaneous graft tumor formation
experiments in naked mice. During cancer growth, it was noted that
the neoplasm volume (Fig. 7A) and
mass (Fig. 7B) of mice injected
with RPL9-sh cells were distinctly smaller than those of mice
injected with control cells. An IHC assay was used to detect the
expression level of RPL9 in tumor tissues (Fig. 7C). RT-qPCR found that miR-24-3p
and miR-185-5p in tissues decreased when RPL9 was downregulated
(Fig. 7D). Finally, to understand
the changes in human-derived miRNAs in mice, exosomes were
extracted from the circulating blood of mice for RT-qPCR. The
results showed that RPL9 inhibition led to a reduction in the
amount of miR-24-3p and miR-185-5p in the exosomes released by
cancer cells (Fig. 7E). The in
vivo study revealed that RPL9 is oncogenic and modulates the
profiles of miR-24-3p and miR-185-5p in exosomes.
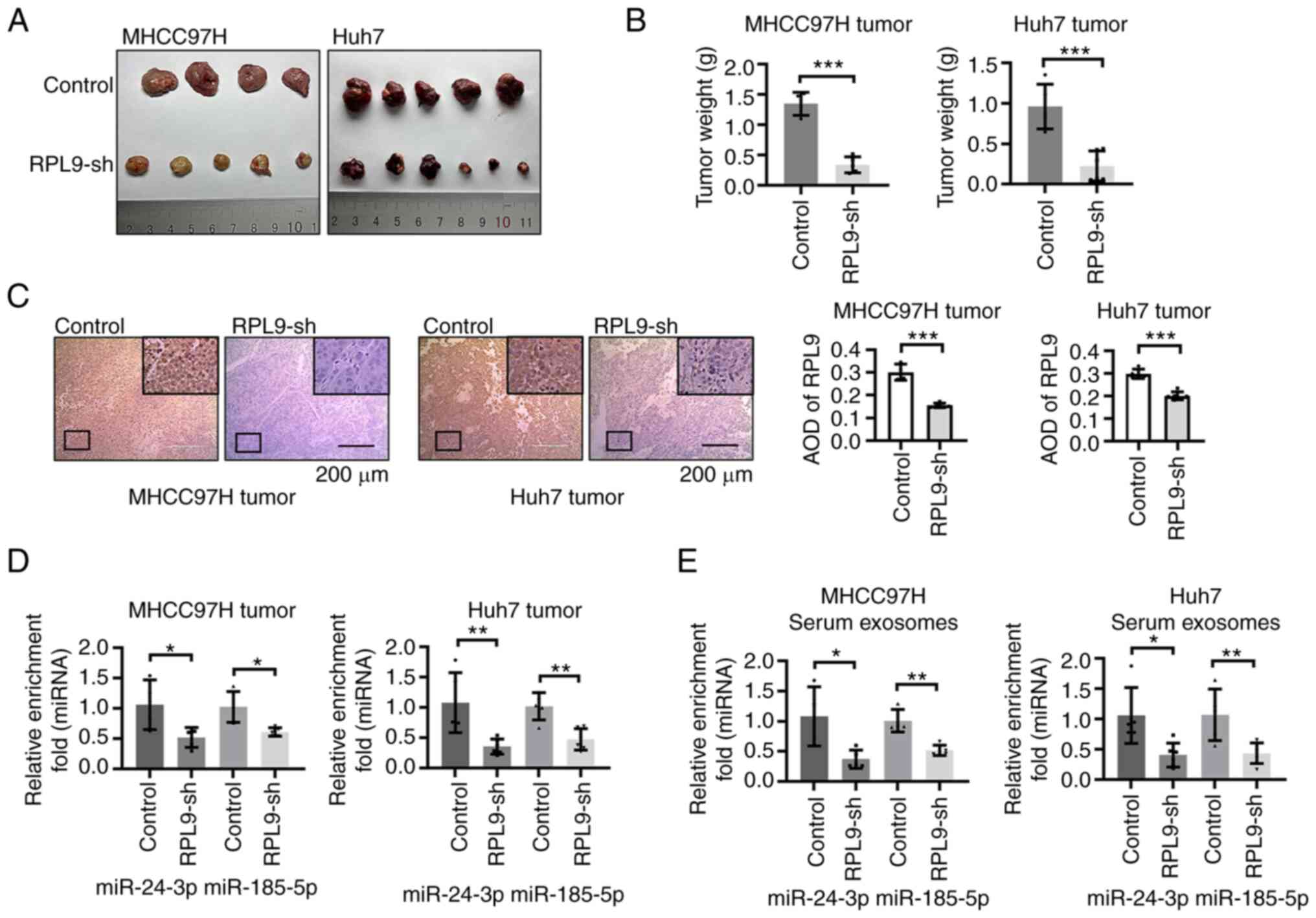 | Figure 7Downregulation of RPL9 suppresses the
growth of HCC tumors and reduces miR-24-3p and miR-185-5p content
in vivo. (A) Images of xenografts. HCC cells were
subcutaneously injected into the right axilla of 5-week-old male
BALB/c-nu/nu mice. One mouse died accidentally leaving 20
tumor-bearing mice (MHCC97H-Control tumors, n=4; MHCC97H-RPL9-sh
tumors, n=5; Huh7-Control tumors, n=5; Huh7-RPL9-sh tumors, n=6).
(B) At 28 days after HCC cell implantation, the tumor weight of
RPL9-sh group was significantly less than that of the RPL9-control
group (***P<0.001). (C) Immunohistochemistry
demonstrated that the expression level of RPL9 in the RPL9-sh group
were clearly lower than control (five fields were selected for each
tissue; ***P<0.001; scale bar=200 µm). Reverse
transcription-quantitative PCR showed that the number of miR-24-3p
and miR-185-5p in (D) tumor cells and (E) mouse serum exosomes
decreased with RPL9 knockdown (*P<0.05,
**P<0.01). RPL9, ribosomal protein L9; HCC,
hepatocellular carcinoma; miRNA/miR, microRNA; sh, short
hairpin. |
Overexpression of miR-24-3p promotes the
biological activity of exosomes and causes an upregulation of RPL9
in exosomes
It was hypothesized that RPL9 probably plays a role
in regulating miRNA transport via the exosomal pathway in HCC
cells. When miR-24-3p was overexpressed in RPL9 control HCC cells,
the content of miR-24-3p in exosomes secreted by HCC cells was
clearly increased. However, overexpression of the same dose of
miR-24-3p in RPL9 knockdown cells did not significantly alter the
amount of miR-24-3p in exosomes (Fig.
8A). After overexpression of miR-24-3p in RPL9 control cells,
RPL9 protein was increased in both cytoplasm and exosomes to
varying degrees. Nevertheless, after overexpression of miR-24-3p in
RPL9 knockdown cells, the changes of RPL9 protein and miR-24-3p in
exosomes tended to be normal (Fig.
8B). It was hypothesized that excessive miR-24-3p could
stimulate the transport of RPL9 to exosomes, resulting in the
secretion of more RPL9-miR-24-3p complexes into the exosomes;
however, when the intracellular level of RPL9 was low, this process
might be inhibited. The exosomes secreted by the RPL9 control cells
miR-24-3p-control group, the RPL9 control cells miR-24-3p
overexpression group and the RPL9 knockdown cells miR-24-3p
overexpression group were extracted. After co-culturing exosomes
(10 µg/ml) with cells, CCK-8 (Fig. 8C), wound healing (Fig. 8D) and Transwell assays (Fig. 8E) were used and it was found that
the increase of miR-24-3p in exosomes promoted the proliferation,
migration and invasion of receptor cells compared with that of
negative control cells. However, the exosomes secreted by HCC cells
after simultaneous downregulation of RPL9 and upregulation of
miR-24-3p did not cause phenotypic changes in recipient cells.
Therefore, it was hypothesized that the biological progression of
recipient HCC cells after co-culturing with exosomes may be related
to difference miRNA content in exosomes.
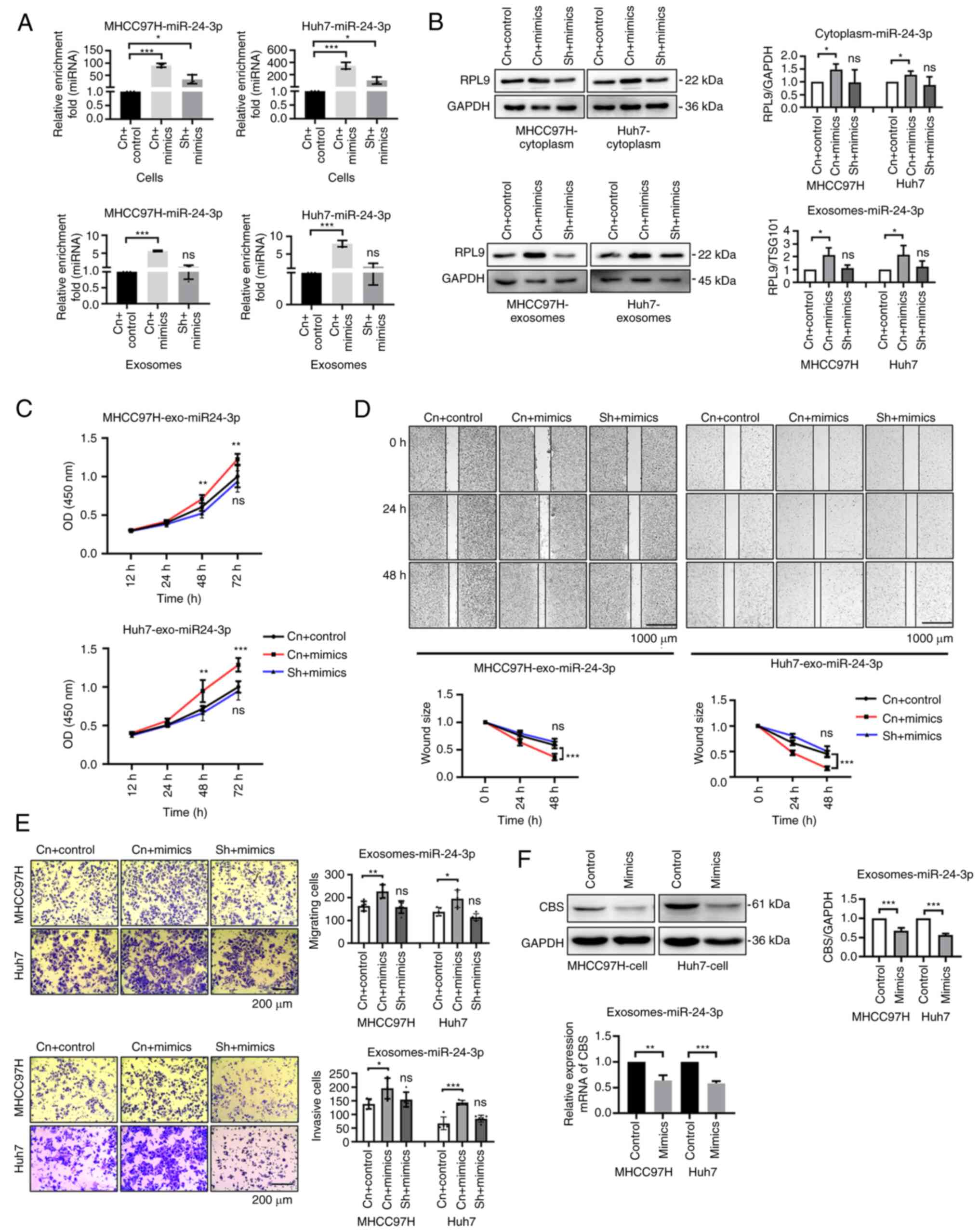 | Figure 8Overexpression of miR-24-3p promotes
the biological activity of exosomes and causes an upregulation of
RPL9 in exosomes. Following transfection with miR-24-3p-mimics into
RPL9 control and RPL9 silenced HCC cells, (A) reverse
transcription-quantitative PCR showed that miR-24-3p was
significantly increased in cells and exosomes of RPL9 control
cells. Meanwhile, the miR-24-3p exhibited a slight increase in
cells with RPL9 silenced, while exosomes showed little alteration
(nsP>0.05; *P<0.05; ***P<0.001). (B)
Western blotting results showed a relative increase in RPL9 protein
content in both the cytoplasm and exosomes of cells in the RPL9
control group, while no significant difference was observed between
the cytoplasm and exosomes of cells in the RPL9 silencing group
(nsP>0.05; *P<0.05). RPL9 control cells were
transfected with the miR-24-3p-control and the miR-24-3p mimic,
followed by transfection of the miR-24-3p mimic into RPL9 knockdown
cells. Finally, recipient HCC cells were co-cultured with extracted
exosomes (10 µg/ml). From (C) CCK-8 (n=6; nsP>0.05;
**P<0.01, ***P<0.001), (D)
wound-healing (n=6; nsP>0.05; ***P<0.001; scale
bar=1,000 µm) and (E) Transwell assays (n=5; nsP>0.05;
*P<0.05, **P<0.01,
***P<0.001; scale bar=200 µm), it was observed
the exosomes, secreted by RPL9 control cells receiving
miR-24-3p-mimics, caused a relative enhancement in the
proliferation, migration, and invasion of recipient cells. However,
the exosomes secreted by RPL9 knockdown cells receiving
miR-24-3p-mimics did not caused significant differences in those
abilities of recipient cells. Overexpression of miR-24-3p resulted
in CBS downregulation in the recipient cell via the exosome
pathway. (F) Exosomes secreted by the miR-24-3p-control group and
the overexpression group were extracted and co-cultured (10
µg/ml) with the recipient cells. Western blotting and
reverse transcription-quantitative qPCR experiments showed that CBS
gene expression in target cells was substantially downregulated
after receiving more miR-24-3p exosomes (**P<0.01,
***P<0.001). miRNA/miR, microRNA; RPL9, ribosomal
protein L9; HCC, hepatocellular carcinoma; CBS, cystathionine
β-synthase; Cn + control, the miR-24-3p-control group into RPL9
control cells; Cn + mimics, the miR-24-3p-mimics group into RPL9
control cells; sh + mimics, the miR-24-3p-mimics group into RPL9
knockdown cells; ns, no significance; Control, the
miR-24-3p-control group into HCC cells; Mimics, the
miR-24-3p-mimics group into HCC cells. |
Overexpression of miR-24-3p results in
CBS downregulation in the recipient cell via the exosome
pathway
It was hypothesized that exosomal miR-24-3p
transported into recipient HCC cells via RPL9 may facilitate their
biological progression by directly affecting downstream targets.
One study (19) demonstrated that
miR-24-3p is a direct upstream regulator of CBS. As a tumor
suppressor, CBS exerts anti-liver cancer effects by inhibiting the
PRRX2/IL-6/STAT3 pathway. Therefore, the exosomes (10 µg/ml)
secreted from cells in the miR-24-3p overexpression and control
groups were co-cultured with the recipient cells. The results
revealed that CBS gene expression in the target cells was
downregulated after receiving more miR-24-3p-containing exosomes
(Fig. 8F). This suggested that
miRNAs in exosomes can be delivered to another cell and function at
the new location.
Discussion
Abnormal miRNA profiles are important in the
development and occurrence of liver cancer. Exosomes are essential
for HCC cells to regulate the profile of their miRNAs, which can
enter and function in recipient cells via exosomes (20,21). By contrast, the mechanism of
selective entry of miRNAs into exosomes was previously unknown;
however, because of the specificity of the RNA structure, it was
hypothesized that there must be a transport mechanism in cells that
can load the miRNA cargo into the exosomes and that RNA transport
to exosomes probably occurs through the action of certain RBPs. Our
previous study (12) showed that
the exosome regulatory factor Vps4A, an accessory molecule
(22,23) of endosomal sorting complexes
required for transport (ESCRT), affects the secretion of miRNAs in
HCC cells and the uptake of miRNAs by recipient cells through
exosomes. Altering the integrity of ESCRT usually interferes with
the RBP GW182, consequently impairing miRNA function (24). GW182 protects mature miRNAs from
degradation by interacting with argonaute and knockdown of GW182
reduces miRNA secretion via exosomes (25). In addition, some RBPs, such as
YBX1 and hnRNPA2B1, are capable of directly sorting miRNAs into
exosomes (26,27). Therefore, it was hypothesized that
RBPs may be involved in regulating the process of miRNA packaging
into exosomes in HCC cells and that this process may be one of the
mechanisms leading to the anomalous profile of miRNAs in HCC
cells.
Thus, mass spectrometry analysis was performed on
serum exosome samples from patients with BLD and HCC and several
differentially expressed RBPs were screened. Among these, RPL9,
RPL28, RPS13, RPS14 and RPS4X are ribosome-binding proteins.
Ribosome is a stable and ancient protein-RNA complex (28) and ribosome-binding proteins have
both RNA-binding properties and protein-protein interaction
domains, providing natural advantages in RNA transport and
regulation (29). RPL9, which was
highly contained in the serum exosomes of patients with HCC, was
selected to continue the present study. RPL9 is an abundant RBP
with various extra-ribosomal functions. For example, defects in
ribosome synthesis lead to cell cycle arrest or apoptosis (30); genetic variants of RPL9 promote
the development of certain diseases or types of cancer (31); and RPL9 interacts with Gag
proteins and affects viral assembly (32). In addition, after long-term
follow-up of patients with HCC, the increase of RPL9 protein in
serum exosomes was an influencing factor for poor prognosis. Thus,
the level of RPL9 in circulating exosomes could be used as a
biomarker to predict the prognosis of patients with HCC. By
suppressing the expression of RPL9 in HCC cells, their
proliferation, migration and invasion abilities were inhibited. Its
role in regulating the biological behavior of cancer cells has
previously been demonstrated in colon cancer (33). Therefore, it was hypothesized that
RPL9 promotes HCC progression and that RPL9 may carry miRNAs into
exosomes in the form of RPL9-miRNAs interaction similar to some
RBPs. Based on this hypothesis, miRNA microarray technology was
used to screen for miRNAs that were significantly altered in the
exosomes of MHCC97H cells following RPL9 knockdown. The results
showed that RPL9 affects the profile of exosomal miRNAs and most of
these altered miRNAs have oncogenic effects on HCC cells. Next, the
present study confirmed by RIP and co-localization experiments that
RPL9, miR-24-3p and miR-185-5p can bind to each other in HCC
exosomes and cells. It was also observed that RPL9, miR-24-3p and
miR-185-5p accumulated in intracellular membrane structures. By
co-culturing exosomes containing RPL9, miR-24-3p and miR-185-5p
with receptor cells, it was demonstrated that the exosomes could
ensure the integrity of the carried miRNAs (34) and shuttle miRNAs to recipient
cells (35,36). It was suggested that RPL9 had a
carrying and protective effect on miRNAs. In MHCC97H cells, an
interesting phenomenon was also discovered: RPL9 could bind to
miR-5100 and after downregulation of RPL9, the miR-5100 increased
in cells but decreased significantly in exosomes. It was
hypothesized that RPL9 mainly plays a transport role in this
process; RPL9 binds to miR-5100 and translocates it into exosomes.
When RPL9 expression was reduced, miR-5100 aggregated in the cells
and decreased in exosomes.
RPL9, miR-24-3p and miR-185-5p are jointly involved
in intercellular communication and the present study showed that
RPL9 and miR-24-3p can affect the biological activity of exosomes
and affect the recipient cells through exosomes to regulate tumor
progression. miR-24-3p and miR-185-5p positively correlated with
changes in RPL9 expression in cells. Studies have reported that
miR-24-3p and miR-185-5p are enriched in patients with HCC
(37,38) and can promote cancer progression
(19,39). Therefore, it was hypothesized that
RPL9 may be similar to other RBPs (40-42) in maintaining the stability of
certain miRNAs by binding to each other and acting as a
pro-oncogenic factor in an miRNA-dependent manner within cells.
RPL9 was found to bind directly to miR-24-3p in cells and exosomes.
Downregulation of RPL9 caused a reduction in miR-24-3p levels in
exosomes and inhibited the biological activity of exosomes.
Similarly, increasing miR-24-3p not only increased the content of
RPL9 in exosomes but also enhanced the biological activity of
exosomes and promoted the proliferation, migration and invasion of
recipient cells. The present study showed that RPL9 functions as an
oncogenic factor in an miRNA-dependent manner. Future studies will
focus on exploring the regulatory mechanisms of RPL9 packaging of
miRNAs into exosomes (Figs. S4 and
S5).
In summary, the present study showed that RPL9 has a
pro-cancer effect, can carry miR-24-3p and miR-185-5p to exosomes
in a mutually binding manner and modulates the biological
properties of receptor HCC cells by changing the profile of
exosomal miRNAs. These findings may lead to the identification of
new biomarkers and development of therapeutic strategies.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
AL and JYX conceived the study, performed most of
the experiments and was a major contributor in writing the
manuscript. LHL designed the study, analyzed data and revised the
manuscript. WBY and WFZ collected the clinical samples and acquired
data. ZHZ and SJY performed part of the experiments. DKC and JXD
acquired data. PQL, JM and JXW and designed the study, analyzed
data, obtained funding and supervised the study. AL and JXW confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Tissue and serum samples were collected from
patients after obtaining informed consent in accordance with a
protocol approved by the Ethics Committee of Sun Yat-sen Memorial
Hospital (Guangzhou, China; approval number 2023001498). All
experimental procedures involving animals were performed according
to the Guide for the Care and Use of Laboratory Animals (National
Institutes of Health publication No. 85-23, revised 2011) and in
accordance with the institutional ethical guidelines for animal
experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
miRNAs
|
microRNAs
|
|
RBPs
|
RNA-binding proteins
|
|
RPL9
|
ribosomal protein L9
|
|
EVs
|
extracellular vesicles
|
|
mRNAs
|
messenger RNAs
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
IHC
|
immunohistochemistry
|
|
BLD
|
benign liver disease
|
|
RIP
|
RNA immunoprecipitation
|
|
CCK-8
|
Cell Counting Kit-8
|
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81672420, 81702406,
U21A20419 and 81372563), the Natural Science Foundation of
Guangdong Province of China (grant no. 2016A030310207) and from the
Guangdong Provincial Key Laboratory of Construction Foundation
(grant no. 2017B030314030).
References
|
1
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang L and Yu D: Exosomes in cancer
development, metastasis, and immunity. Biochim Biophys Acta Rev
Cancer. 1871:455–468. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tan S, Xia L, Yi P, Han Y, Tang L, Pan Q,
Tian Y, Rao S, Oyang L, Liang J, et al: Exosomal miRNAs in tumor
microenvironment. J Exp Clin Cancer Res. 39:672020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Corley M, Burns MC and Yeo GW: How
RNA-binding proteins interact with RNA: Molecules and mechanisms.
Mol Cell. 78:9–29. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nussbacher JK and Yeo GW: Systematic
discovery of RNA binding proteins that regulate MicroRNA levels.
Mol Cell. 69:1005–1016. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fabbiano F, Corsi J, Gurrieri E, Trevisan
C, Notarangelo M and D'Agostino VG: RNA packaging into
extracellular vesicles: An orchestra of RNA-binding proteins? J
Extracell Vesicles. 10:e120432020. View Article : Google Scholar
|
|
12
|
Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin
HM, Zhou R, Shang CZ, Cao J, He H, et al: Vps4A functions as a
tumor suppressor by regulating the secretion and uptake of exosomal
microRNAs in human hepatoma cells. Hepatology. 61:1284–1294. 2015.
View Article : Google Scholar
|
|
13
|
de la Cruz J, Karbstein K and Woolford JL
Jr: Functions of ribosomal proteins in assembly of eukaryotic
ribosomes in vivo. Annu Rev Biochem. 84:93–129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou X, Liao WJ, Liao JM, Liao P and Lu H:
Ribosomal proteins: Functions beyond the ribosome. J Mol Cell Biol.
7:92–104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luan Y, Tang N, Yang J, Liu S, Cheng C,
Wang Y, Chen C, Guo YN, Wang H, Zhao W, et al: Deficiency of
ribosomal proteins reshapes the transcriptional and translational
landscape in human cells. Nucleic Acids Res. 50:6601–6617. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ebright RY, Lee S, Wittner BS,
Niederhoffer KL, Nicholson BT, Bardia A, Truesdell S, Wiley DF,
Wesley B, Li S, et al: Deregulation of ribosomal protein expression
and translation promotes breast cancer metastasis. Science.
367:1468–1473. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bork iewicz L, Mołoń M, Molesta k E, Grela
P, Horbowicz-Drożdżal P, Wawiórka L and Tchórzewski M: Functional
analysis of the ribosomal uL6 protein of saccharomyces cerevisiae.
Cells. 8:7182019. View Article : Google Scholar
|
|
18
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals. 8th
edition. Washington (DC): National Academies Press (US); 2011
|
|
19
|
Zhou YF, Song SS, Tian MX, Tang Z, Wang H,
Fang Y, Qu WF, Jiang XF, Tao CY, Huang R, et al: Cystathionine
β-synthase mediated PRRX2/IL-6/STAT3 inactivation suppresses Tregs
infiltration and induces apoptosis to inhibit HCC carcinogenesis. J
Immunother Cancer. 9:e0030312021. View Article : Google Scholar
|
|
20
|
Liu J, Fan L, Yu H, Zhang J, He Y, Feng D,
Wang F, Li X, Liu Q, Li Y, et al: Endoplasmic reticulum stress
causes liver cancer cells to release exosomal miR-23a-3p and
up-regulate programmed death ligand 1 expression in macrophages.
Hepatology. 70:241–258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang B, Feng X, Liu H, Tong R, Wu J, Li C,
Yu H, Chen Y, Cheng Q, Chen J, et al: High-metastatic cancer cells
derived exosomal miR92a-3p promotes epithelial-mesenchymal
transition and metastasis of low-metastatic cancer cells by
regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene.
39:6529–6543. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McCullough J, Frost A and Sundquist WI:
Structures, functions, and dynamics of ESCRT-III/Vps4 membrane
remodeling and fission complexes. Annu Rev Cell Dev Biol.
34:85–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pfitzner AK, Mercier V, Jiang X, Moser
VFJ, Baum B, Šarić A and Roux A: An ESCRT-III polymerization
sequence drives membrane deformation and fission. Cell.
182:1140–1155. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gibbings DJ, Ciaudo C, Erhardt M and
Voinnet O: Multivesicular bodies associate with components of miRNA
effector complexes and modulate miRNA activity. Nat Cell Biol.
11:1143–1149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao B, La LB, Chen YC, Chang LJ and Chan
EK: Defining a new role of GW182 in maintaining miRNA stability.
EMBO Rep. 13:1102–1108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Villarroya-Beltri C, Gutiérrez-Vázquez C,
Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N,
Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M and
Sánchez-Madrid F: Sumoylated hnRNPA2B1 controls the sorting of
miRNAs into exosomes through binding to specific motifs. Nat
Commun. 4:29802013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shurtleff MJ, Yao J, Qin Y, Nottingham RM,
Temoche-Diaz MM, Schekman R and Lambowitz AM: Broad role for YBX1
in defining the small noncoding RNA composition of exosomes. Proc
Natl Acad Sci USA. 114:E8987–E8995. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hoffman DW, Davies C, Gerchman SE, Kycia
JH, Porter SJ, White SW and Ramakrishnan V: Crystal structure of
prokaryotic ribosomal protein L9: A bi-lobed RNA-binding protein.
EMBO J. 13:205–212. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Warner JR and McIntosh KB: How common are
extraribosomal functions of ribosomal proteins? Mol Cell. 34:3–11.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang T, Jiang C, Yang M, Xiao H, Huang X,
Wu L and Yao M: Salmonella enterica serovar Typhimurium inhibits
the innate immune response and promotes apoptosis in a
ribosomal/TRP53-dependent manner in swine neutrophils. Vet Res.
51:1052020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lezzerini M, Penzo M, O'Donohue MF,
Marques DSVC, Saby M, Elfrink HL, Diets IJ, Hesse AM, Couté Y,
Gastou M, et al: Ribosomal protein gene RPL9 variants can
differentially impair ribosome function and cellular metabolism.
Nucleic Acids Res. 48:770–787. 2020. View Article : Google Scholar :
|
|
32
|
Beyer AR, Bann DV, Rice B, Pultz IS, Kane
M, Goff SP, Golovkina TV and Parent LJ: Nucleolar trafficking of
the mouse mammary tumor virus gag protein induced by interaction
with ribosomal protein L9. J Virol. 87:1069–1082. 2013. View Article : Google Scholar :
|
|
33
|
Baik IH, Jo GH, Seo D, Ko MJ, Cho CH, Lee
MG and Lee YH: Knockdown of RPL9 expression inhibits colorectal
carcinoma growth via the inactivation of Id-1/NF-κB signaling axis.
Int J Oncol. 49:1953–1962. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kanada M, Bachmann MH, Hardy JW,
Frimannson DO, Bronsart L, Wang A, Sylvester MD, Schmidt TL, Kaspar
RL, Butte MJ, et al: Differential fates of biomolecules delivered
to target cells via extracellular vesicles. Proc Natl Acad Sci USA.
112:E1433–E1442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kogure T, Lin WL, Yan IK, Braconi C and
Patel T: Intercellular nanovesicle-mediated microRNA transfer: A
mechanism of environmental modulation of hepatocellular cancer cell
growth. Hepatology. 54:1237–1248. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Montecalvo A, Larregina AT, Shufesky WJ,
Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G,
Wang Z, et al: Mechanism of transfer of functional microRNAs
between mouse dendritic cells via exosomes. Blood. 119:756–766.
2012. View Article : Google Scholar :
|
|
37
|
Fan JC, Zeng F, Le YG and Xin L: LncRNA
CASC2 inhibited the viability and induced the apoptosis of
hepatocellular carcinoma cells through regulating miR-24-3p. J Cell
Biochem. 119:6391–6397. 2018. View Article : Google Scholar
|
|
38
|
Wen Y, Han J, Chen J, Dong J, Xia Y, Liu
J, Jiang Y, Dai J, Lu J, Jin G, et al: Plasma miRNAs as early
biomarkers for detecting hepatocellular carcinoma. Int J Cancer.
137:1679–1690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zou L, Chai J, Gao Y, Guan J, Liu Q and Du
JJ: Down-regulated PLAC8 promotes hepatocellular carcinoma cell
proliferation by enhancing PI3K/Akt/GSK3β/Wnt/β-catenin signaling.
Biomed Pharmacother. 84:139–146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bitaraf A, Razmara E, Bakhshinejad B,
Yousefi H, Vatanmakanian M, Garshasbi M, Cho WC and Babashah S: The
oncogenic and tumor suppressive roles of RNA-binding proteins in
human cancers. J Cell Physiol. 236:6200–6224. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mukohyama J, Shimono Y, Minami H, Kakeji Y
and Suzuki A: Roles of microRNAs and RNA-binding proteins in the
regulation of colorectal cancer stem cells. Cancers (Basel).
9:1432017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vos PD, Leedman PJ, Filipovska A and
Rackham O: Modulation of miRNA function by natural and synthetic
RNA-binding proteins in cancer. Cell Mol Life Sci. 76:3745–3752.
2019. View Article : Google Scholar : PubMed/NCBI
|