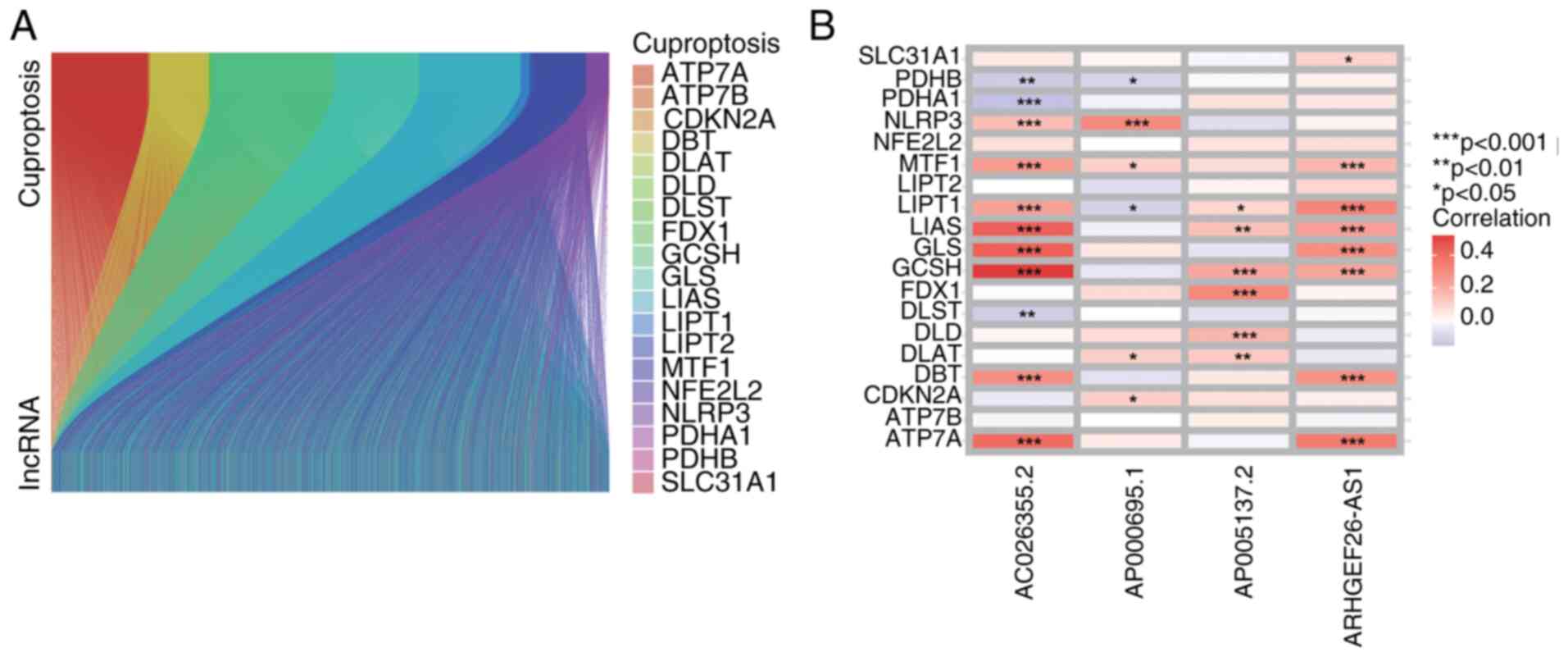
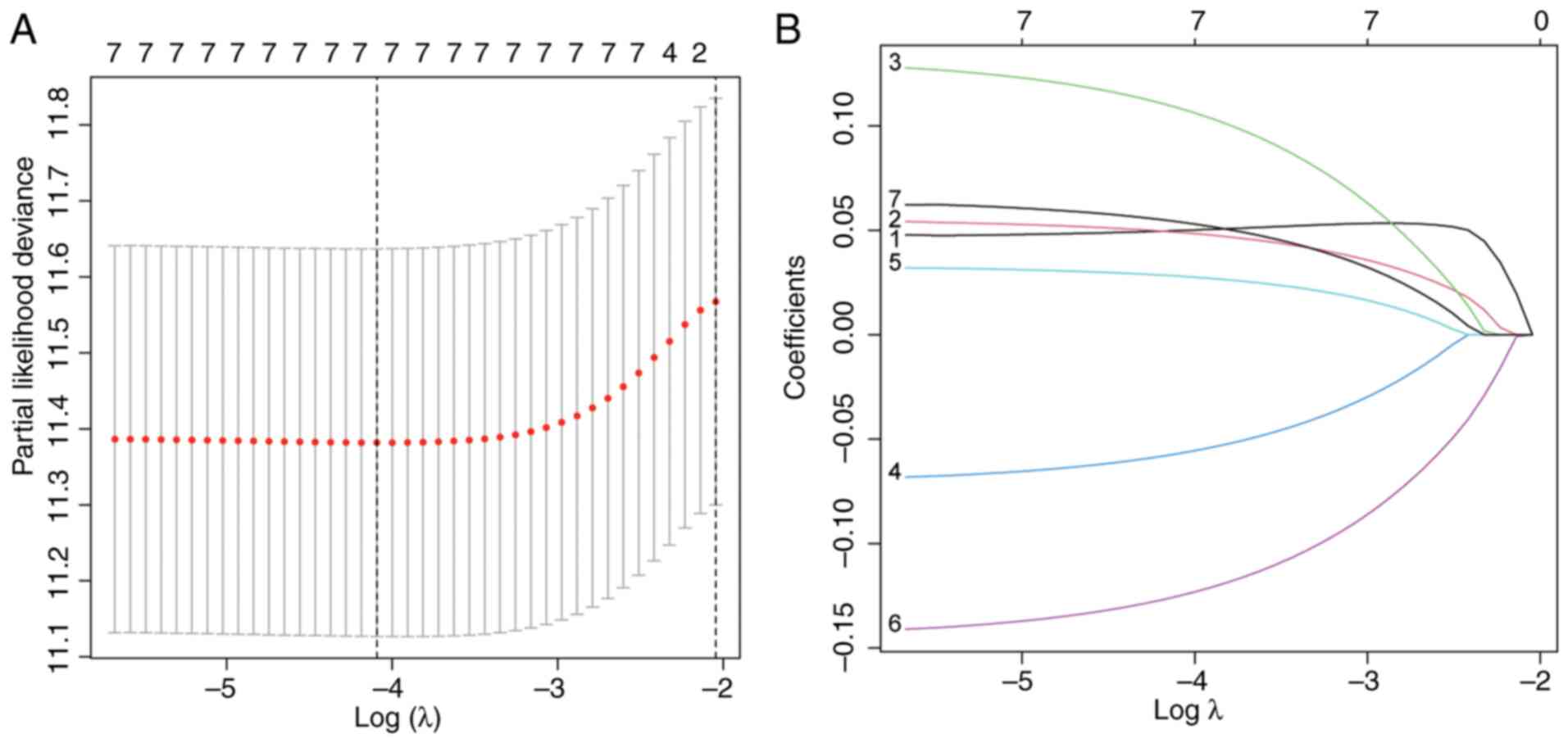
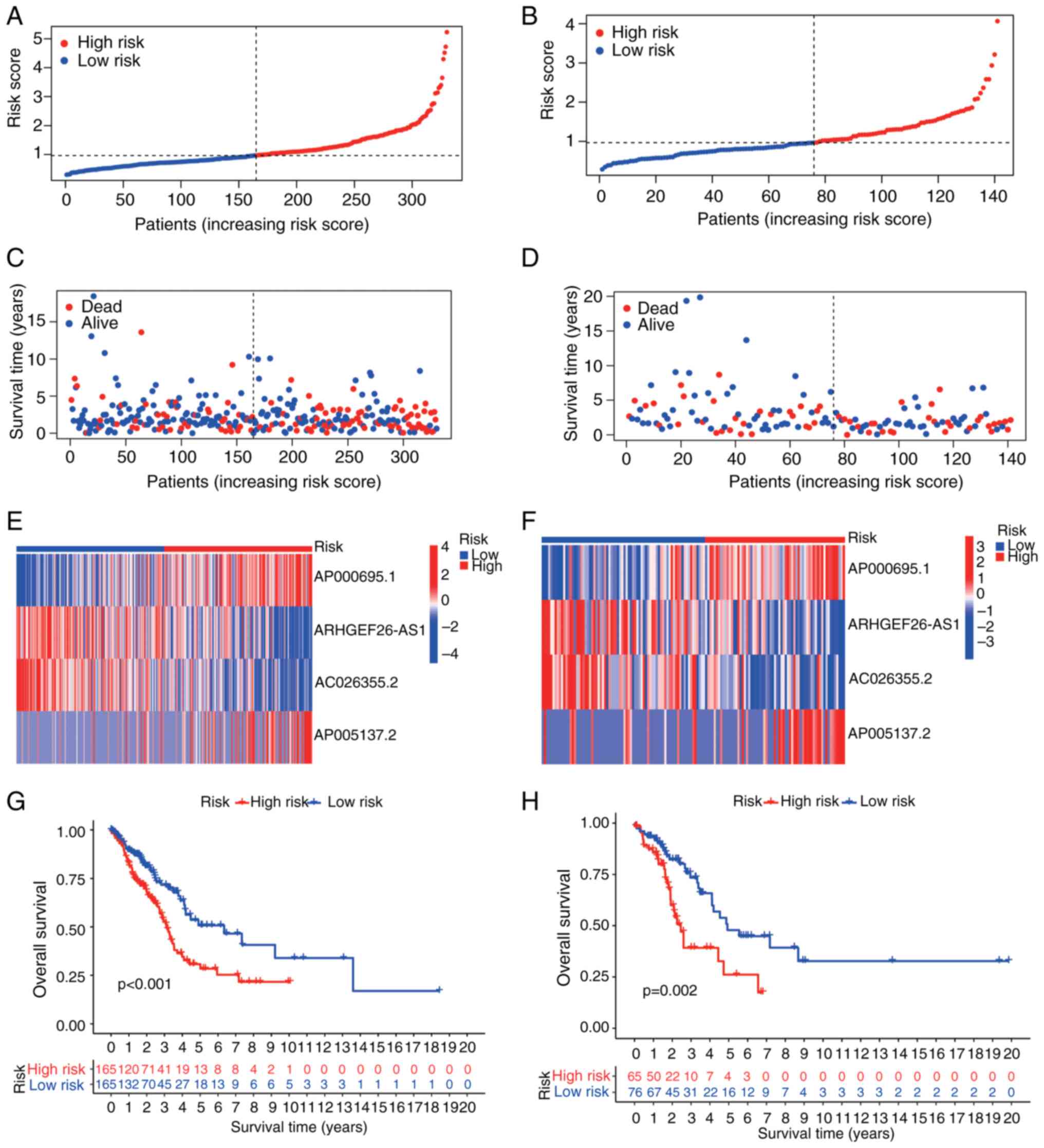
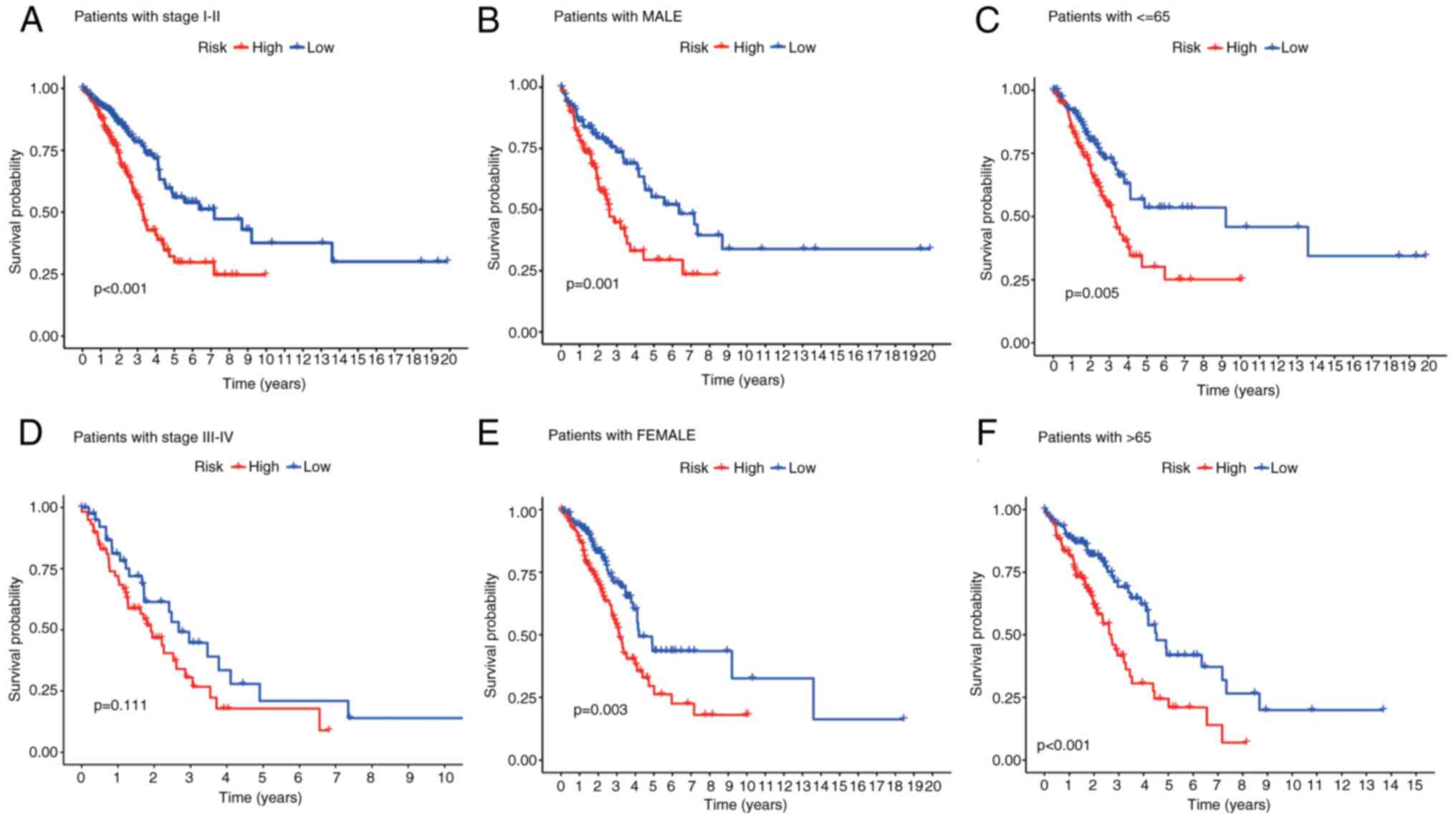
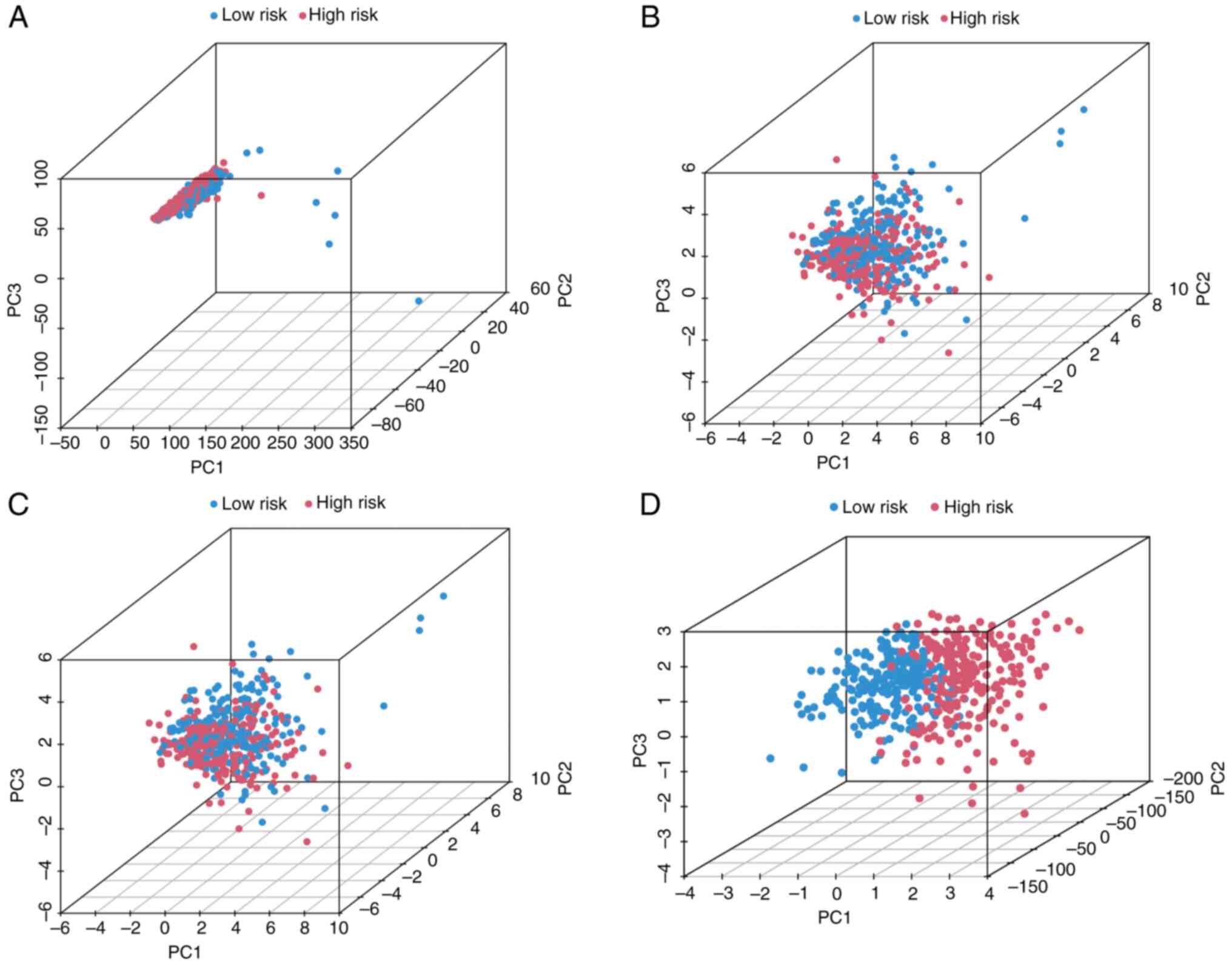
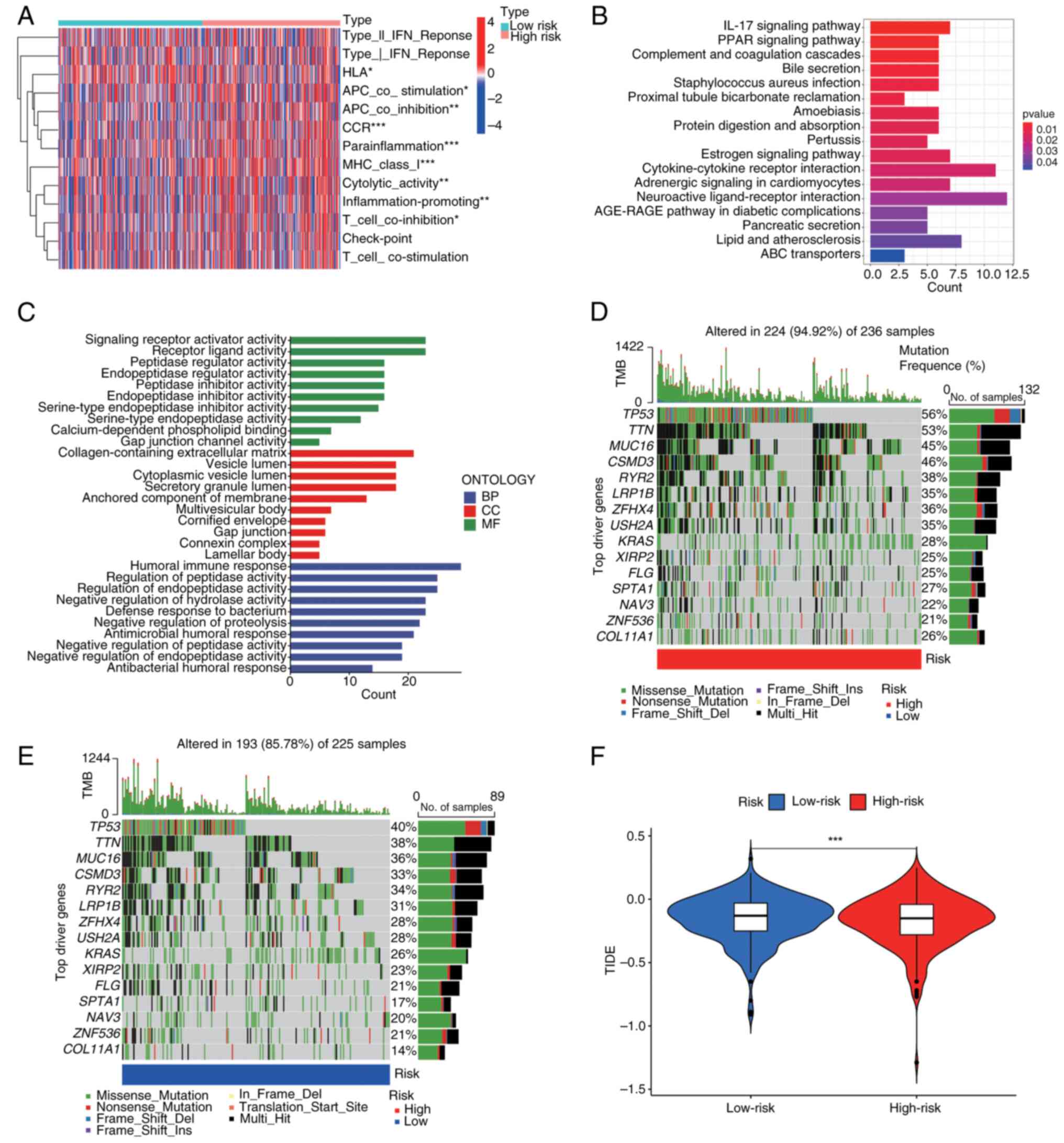
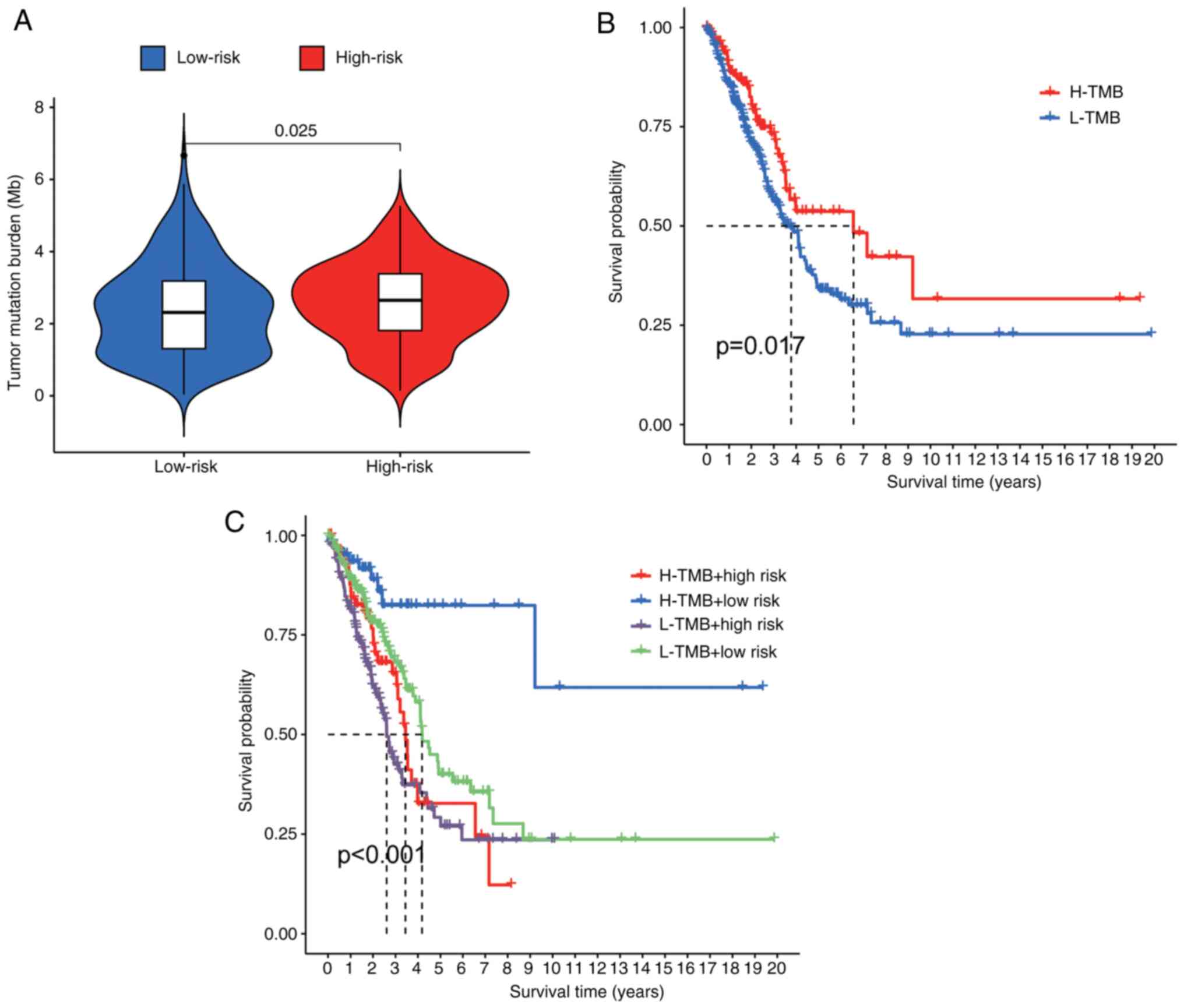
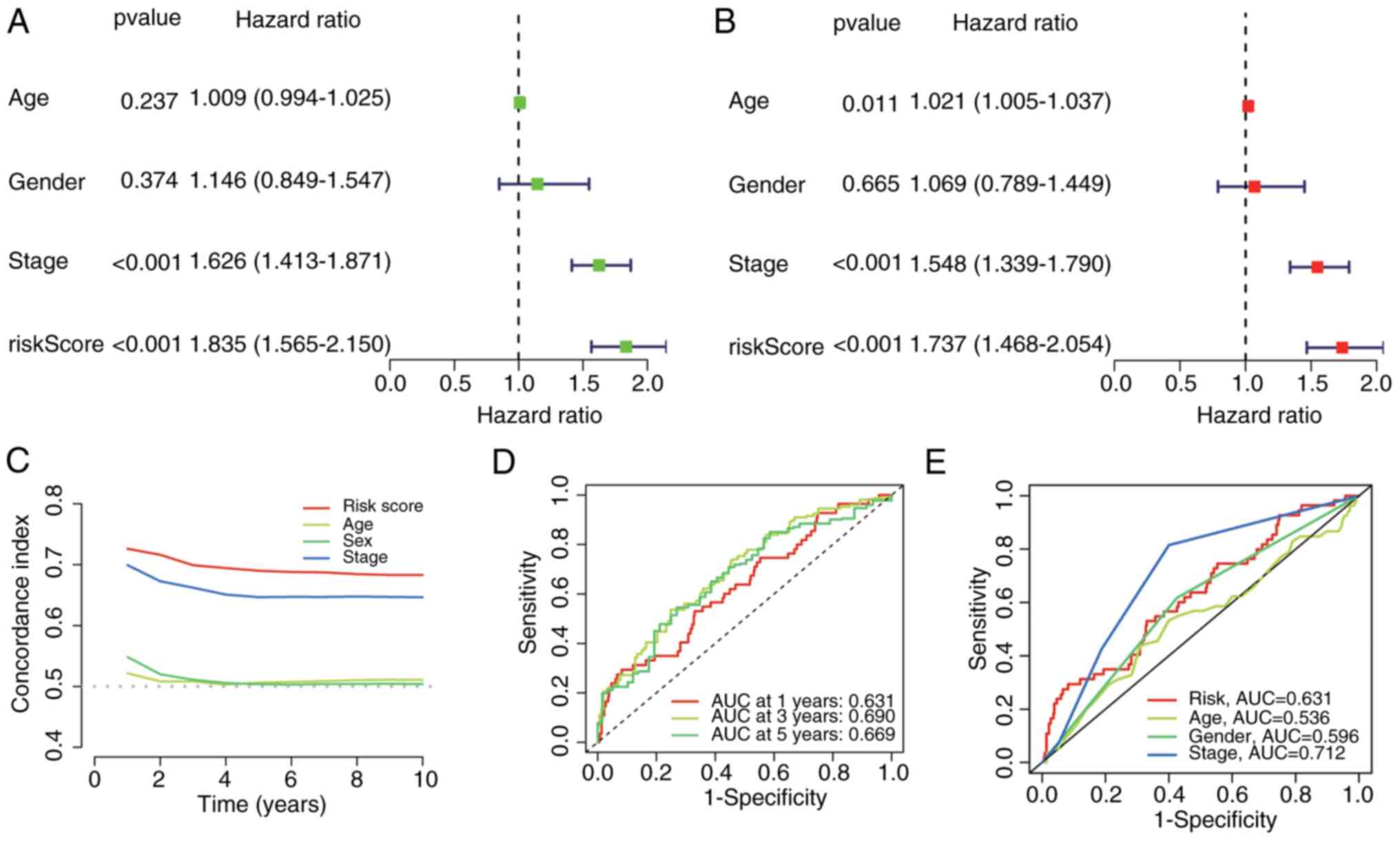
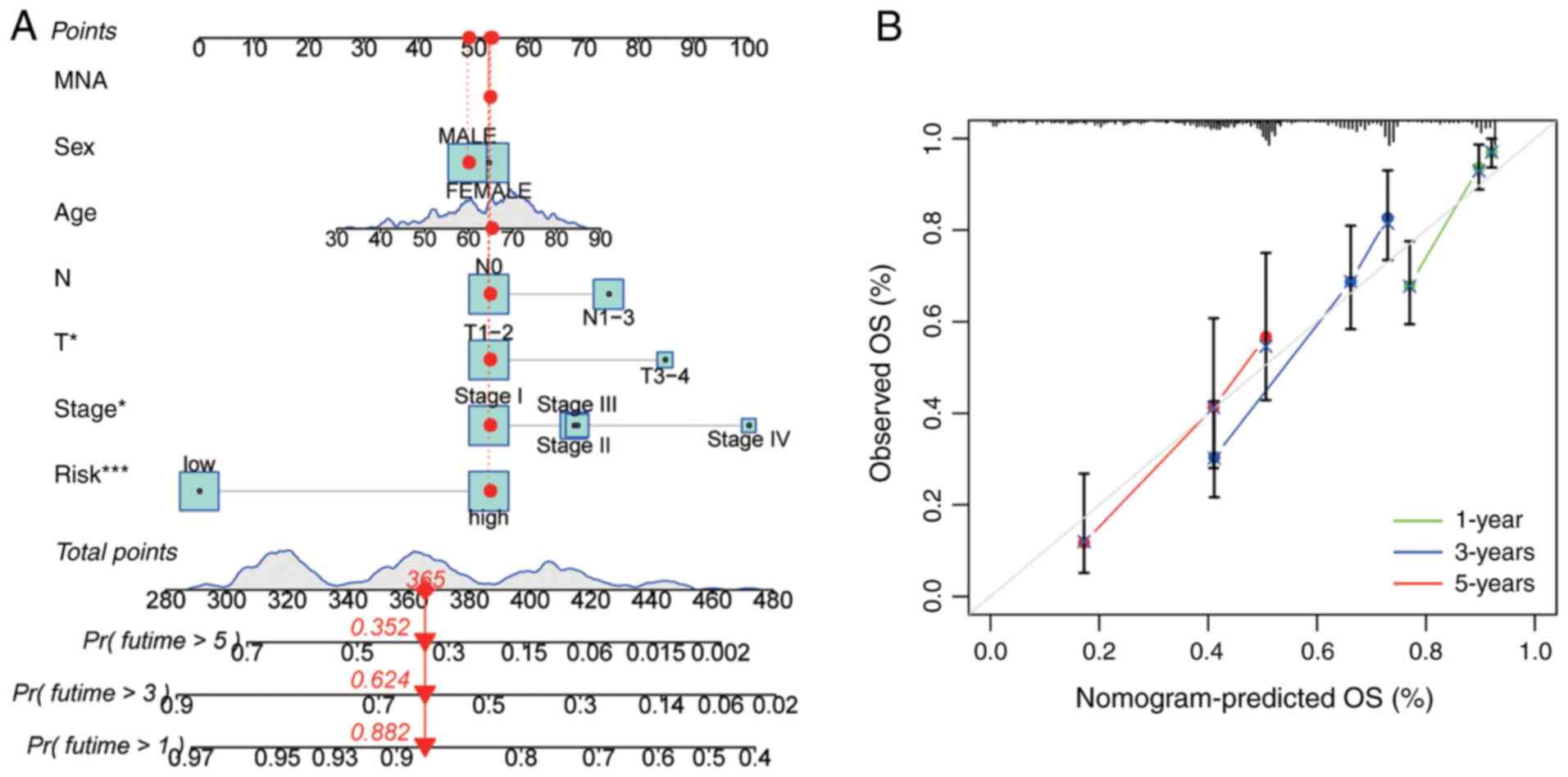
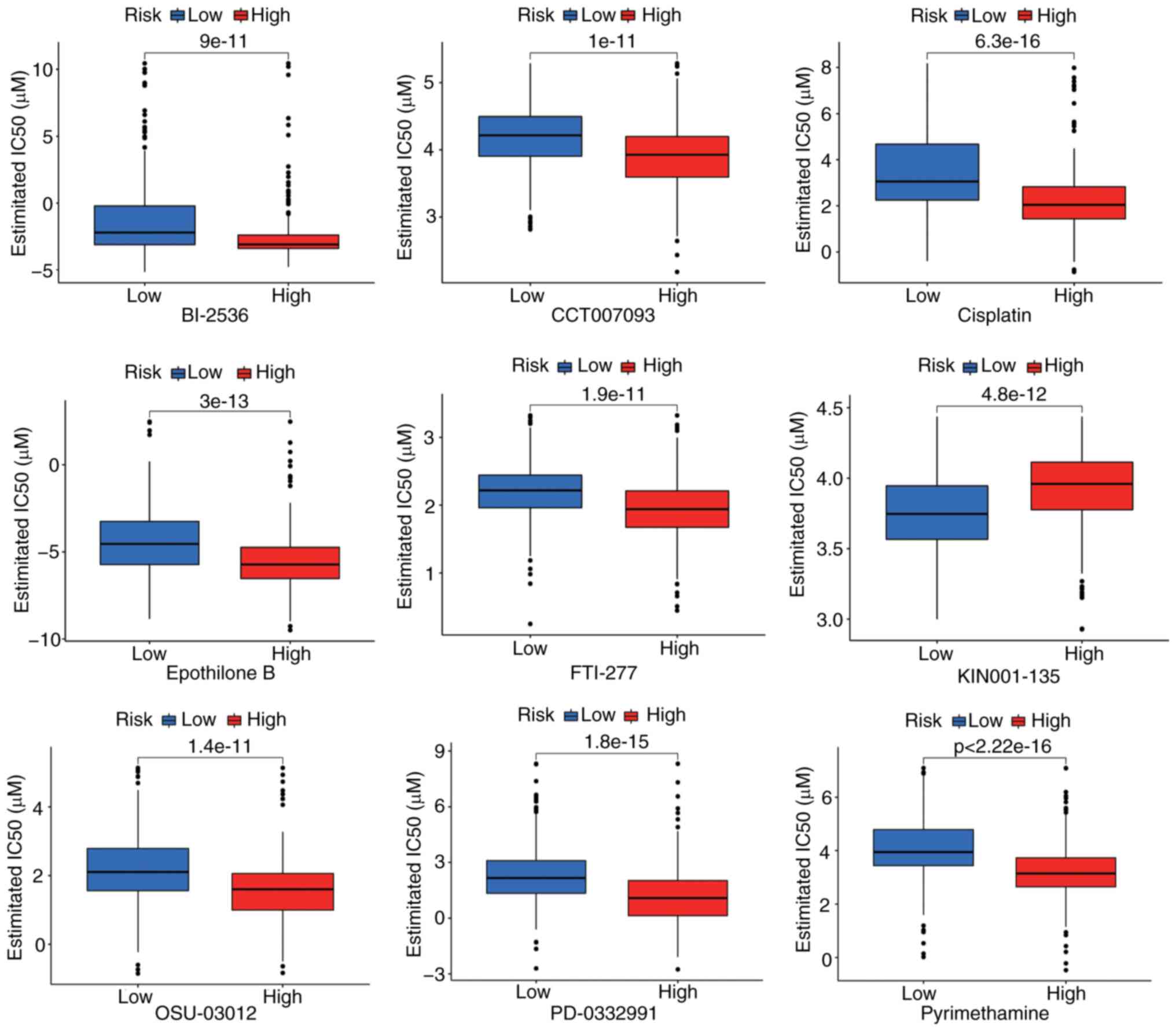
|
1
|
Cao M, Li H, Sun D and Chen W: Cancer
burden of major cancers in China: A need for sustainable actions.
Cancer Commun (Lond). 40:205–210. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jurisic V, Vukovic V, Obradovic J,
Gulyaeva LF, Kushlinskii NE and Djordjević N: EGFR
polymorphism and survival of NSCLC patients treated with TKIs: A
systematic review and meta-analysis. J Oncol.
2020(1973241)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhang C, Zhang J, Xu FP, Wang YG, Xie Z,
Su J, Dong S, Nie Q, Shao Y, Zhou Q, et al: Genomic landscape and
immune microenvironment features of preinvasive and early invasive
lung adenocarcinoma. J Thorac Oncol. 14:1912–1923. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhao L, Li M, Shen C and Luo Y, Hou X, Qi
Y, Huang Z, Li W, Gao L, Wu M and Luo Y: Nano-assisted radiotherapy
strategies: New opportunities for treatment of non-small cell lung
cancer. Research (Wash D C). 7(0429)2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liu C, Chen Y, Xu X, Yin M, Zhang H and Su
W: Utilizing macrophages missile for sulfate-based nanomedicine
delivery in lung cancer therapy. Research (Wash D C).
7(0448)2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang Y, Shen C, Wu C, Zhan Z, Qu R, Xie Y
and Chen P: Self-assembled DNA machine and selective complexation
recognition enable rapid homogeneous portable quantification of
lung cancer CTCs. Research (Wash D C). 7(0352)2024.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tsvetkov P, Coy S, Petrova B, Dreishpoon
M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R,
Spangler RD, et al: Copper induces cell death by targeting
lipoylated TCA cycle proteins. Science. 375:1254–1261.
2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tsvetkov P, Detappe A, Cai K, Keys HR,
Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, et al:
Mitochondrial metabolism promotes adaptation to proteotoxic stress.
Nat Chem Biol. 15:681–689. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tang D, Kroemer G and Kang R: Targeting
cuproplasia and cuproptosis in cancer. Nat Rev Clin Oncol.
21:370–388. 2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kahlson MA and Dixon SJ: Copper-induced
cell death. Science. 375:1231–1232. 2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Aubert L, Nandagopal N, Steinhart Z,
Lavoie G, Nourreddine S, Berman J, Saba-El-Leil MK, Papadopoli D,
Lin S, Hart T, et al: Copper bioavailability is a KRAS-specific
vulnerability in colorectal cancer. Nat Commun.
11(3701)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Dong J, Wang X, Xu C, Gao M, Wang S, Zhang
J, Tong H, Wang L and Han Y, Cheng N and Han Y: Inhibiting NLRP3
inflammasome activation prevents copper-induced neuropathology in a
murine model of Wilson's disease. Cell Death Dis.
12(87)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ren X, Li Y, Zhou Y, Hu W, Yang C, Jing Q,
Zhou C, Wang X, Hu J, Wang L, et al: Overcoming the compensatory
elevation of NRF2 renders hepatocellular carcinoma cells more
vulnerable to disulfiram/copper-induced ferroptosis. Redox Biol.
46(102122)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Polishchuk EV, Merolla A, Lichtmannegger
J, Romano A, Indrieri A, Ilyechova EY, Concilli M, De Cegli R,
Crispino R, Mariniello M, et al: Activation of autophagy, observed
in liver tissues from patients with Wilson disease and from
ATP7B-deficient animals, protects hepatocytes from copper-induced
apoptosis. Gastroenterology. 156:1173–1189.e5. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xu F, Lin H, He P, He L, Chen J, Lin L and
Chen Y: A TP53-associated gene signature for prediction of
prognosis and therapeutic responses in lung squamous cell
carcinoma. Oncoimmunology. 9(1731943)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hong W, Liang L, Gu Y, Qi Z, Qiu H, Yang
X, Zeng W, Ma L and Xie J: Immune-related lncRNA to construct novel
signature and predict the immune landscape of human hepatocellular
carcinoma. Mol Ther Nucleic Acids. 22:937–947. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao X, Liu X and Cui L: Development of a
five-protein signature for predicting the prognosis of head and
neck squamous cell carcinoma. Aging (Albany NY). 12:19740–19755.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dong HX, Wang R, Jin XY, Zeng J and Pan J:
LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via
inhibiting hsa-mir-22-3p. J Cell Physiol. 233:4126–4136.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tian Y, Yu M, Sun L, Liu L, Wang J, Hui K,
Nan Q, Nie X, Ren Y and Ren X: Distinct patterns of mRNA and lncRNA
expression differences between lung squamous cell carcinoma and
adenocarcinoma. J Comput Biol. 27:1067–1078. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang L, Wu Y, Xu H, Zhang J, Zheng X,
Zhang L, Wang Y, Chen W and Wang K: Identification and validation
of a novel six-lncRNA-based prognostic model for lung
adenocarcinoma. Front Oncol. 11(775583)2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang H, Guo L and Chen J: Rationale for
lung adenocarcinoma prevention and drug development based on
molecular biology during carcinogenesis. Onco Targets Ther.
13:3085–3091. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sun Q, Qin X, Zhao J, Gao T, Xu Y, Chen G,
Bai G, Guo Z and Liu J: Cuproptosis-related LncRNA signatures as a
prognostic model for head and neck squamous cell carcinoma.
Apoptosis. 28:247–262. 2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cao Q, Dong Z, Liu S, An G, Yan B and Lei
L: Construction of a metastasis-associated ceRNA network reveals a
prognostic signature in lung cancer. Cancer Cell Int.
20(208)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang P, Pei S, Liu J, Zhang X, Feng Y,
Gong Z, Zeng T, Li J and Wang W: Cuproptosis-related lncRNA
signatures: Predicting prognosis and evaluating the tumor immune
microenvironment in lung adenocarcinoma. Front Oncol.
12(1088931)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bian M, Wang W, Song C, Pan L, Wu Y and
Chen L: Autophagy-related genes predict the progression of
periodontitis through the ceRNA network. J Inflamm Res.
15:1811–1824. 2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang G, Ye Q, Ning S, Yang Z, Chen Y,
Zhang L, Huang Y, Xie F, Cheng X, Chi J, et al: LncRNA MEG3
promotes endoplasmic reticulum stress and suppresses proliferation
and invasion of colorectal carcinoma cells through the
MEG3/miR-103a-3p/PDHB ceRNA pathway. Neoplasma. 68:362–374.
2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xuan J, Zhu D, Cheng Z, Qiu Y, Shao M,
Yang Y, Zhai Q, Wang F and Qin F: Crocin inhibits the activation of
mouse hepatic stellate cells via the lnc-LFAR1/MTF-1/GDNF pathway.
Cell Cycle. 19:3480–3490. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mafra ACP and Dias SMG: Several faces of
glutaminase regulation in cells. Cancer Res. 79:1302–1304.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang T, Zhang XD and Hua KQ: A ceRNA
network of BBOX1-AS1-hsa-miR-125b-5p/hsa-miR-125a-5p-CDKN2A shows
prognostic value in cervical cancer. Taiwan J Obstet Gynecol.
60:253–261. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu J, Liu Q, Shen H, Liu Y, Wang Y, Wang
G and Du J: Identification and validation of a three
pyroptosis-related lncRNA signature for prognosis prediction in
lung adenocarcinoma. Front Genet. 13(838624)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun X, Song J, Lu C, Sun X, Yue H, Bao H,
Wang S and Zhong X: Characterization of cuproptosis-related lncRNA
landscape for predicting the prognosis and aiding immunotherapy in
lung adenocarcinoma patients. Am J Cancer Res. 13:778–801.
2023.PubMed/NCBI
|
|
35
|
Zhang S, Li X, Tang C and Kuang W:
Inflammation-related long non-coding RNA signature predicts the
prognosis of gastric carcinoma. Front Genet.
12(736766)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou D, Wang J and Liu X: Development of
six immune-related lncRNA signature prognostic model for
smoking-positive lung adenocarcinoma. J Clin Lab Anal.
36(e24467)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Qiu Y, Yao J, Li L, Xiao M, Meng J, Huang
X, Cai Y, Wen Z, Huang J, Zhu M, et al: Machine learning identifies
ferroptosis-related genes as potential diagnostic biomarkers for
osteoarthritis. Front Endocrinol (Lausanne).
14(1198763)2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang D, Tian Z, Zhang P, Zhen L, Meng Q,
Sun B, Xu X, Jia T and Li S: The molecular mechanisms of
cuproptosis and its relevance to cardiovascular disease. Biomed
Pharmacother. 163(114830)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wang F, Lin H, Su Q and Li C:
Cuproptosis-related lncRNA predict prognosis and immune response of
lung adenocarcinoma. World J Surg Oncol. 20(275)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yalimaimaiti S, Liang X, Zhao H, Dou H,
Liu W, Yang Y and Ning L: Establishment of a prognostic signature
for lung adenocarcinoma using cuproptosis-related lncRNAs. BMC
Bioinformatics. 24(81)2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zheng J, Zhao Z, Wan J, Guo M, Wang Y,
Yang Z, Li Z, Ming L and Qin Z: N-6 methylation-related lncRNA is
potential signature in lung adenocarcinoma and influences tumor
microenvironment. J Clin Lab Anal. 35(e23951)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu L, Wang T, Huang D and Song D:
Comprehensive analysis of differentially expressed genes in
clinically diagnosed irreversible pulpitis by multiplatform data
integration using a robust rank aggregation approach. J Endod.
47:1365–1375. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yang L, He YT, Dong S, Wei XW, Chen ZH,
Zhang B, Chen WD, Yang XR, Wang F, Shang XM, et al: Single-cell
transcriptome analysis revealed a suppressive tumor immune
microenvironment in EGFR mutant lung adenocarcinoma. J Immunother
Cancer. 10(e003534)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Franzese O, Palermo B, Frisullo G, Panetta
M, Campo G, D'Andrea D, Sperduti I, Taje R, Visca P and Nisticò P:
ADA/CD26 axis increases intra-tumor
PD-1+CD28+CD8+ T-cell fitness and
affects NSCLC prognosis and response to ICB. Oncoimmunology.
13(2371051)2024.PubMed/NCBI View Article : Google Scholar
|