Introduction
Nasopharyngeal carcinoma (NPC) is a cancer with
distinct ethnic and geographic distribution. The cancer is common
in Southern China, North Africa and South East Asia, including
Malaysia (1). Approximately 95% of
NPC cases (2) are associated with
latent infection of the Epstein-Barr virus (EBV), a γherpesvirus of
the Lymphocryptovirus genus. EBV-infected NPC cells exhibit
type II latency and express EBV-encoded RNAs (EBERs; small,
non-coding, nonpolyadenylated), EBV-associated nuclear antigen-1
(EBNA1), latent membrane proteins 1 and 2 (LMP1, 2A and 2B),
BamH1 A RNA transcripts (BART) and BARF1 protein. LMP1,
BARF1 and EBNA1 are oncogenic (1).
EBER-1 and EBER-2 are highly transcribed nuclear
RNAs, composed of 167 and 172 nucleotides, respectively (3). Their abundance in infected cells has
led to the use of in situ hybridisation (ISH) for the
detection of EBV infection in tissue specimens (3). Numerous studies have analysed the
role of EBERs in Burkitts’ lymphoma (4–7). In
addition, a small number of reports have described the role of
EBERs in epithelial malignancies. These include reports that EBERs
confer resistance to pIC-induced apoptosis (8) and induce insulin-like growth factor 1
(IGF-1) (9,10).
To understand the role of EBERs in NPC, we
established stable expression of EBERs in HK1, an EBV-negative NPC
cell line, by transfecting HK1 with an expression vector containing
10 tandem repeats of EBER-1 and-2 (4). Our findings indicated that EBERs
enhanced cell proliferation. Gene expression profiling by
microarray revealed that members of the cellular lipid metabolic
process were aberrantly expressed. We found that EBER-positive
cells demonstrated the upregulation of low-density lipoprotein
receptor (LDLR) and fatty acid synthase (FASN). NPC cells were
identified to undergo low-density lipoprotein (LDL)-dependent cell
proliferation and were sensitive to a polyphenolic compound,
quercetin.
Materials and methods
Cell lines and culture
HK1, EBV-negative (11) and C666-1, EBV-positive NPC cell
lines (12) and all stable cell
lines established from HK1 thereafter, were maintained in RPMI-1640
medium supplemented with 10% heat-inactivated foetal calf serum
(FCS), 10 U/ml of penicillin and 10 μg/ml streptomycin (all
obtained from Gibco-BRL, Carlsbad, CA, USA) and cultured at 37˚C in
a 5% CO2 humidified atmosphere. The identity of HK1 and
C666-1 were validated by DNA fingerprinting using the AmpFISTR
Identifiler® polymerase chain reaction (PCR)
amplification kit (Applied Biosystems, Foster City, CA, USA). The
short tandem repeat profiles were consistent with published data
(13). Tests for Mycoplasma
using e-myco™ Mycoplasma PCR Detection kit (Intron
Biotechnology Co. Ltd., Gyeonggi-do, Korea) were conducted
routinely and contamination-free cells were used throughout the
study.
Plasmids and transfection
The EBER plasmid (designated as pcDNA 3 Eks10)
contained 10 tandem repeats of the EBER-1 and -2 subfragments
inserted into a pcDNA 3 vector (4). Empty vector pcDNA 3.1 was used as a
control. Plasmids were transfected into HK1 cells using
Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA), according to the manufacturer’s instructions. Stable
transfectants were selected by culturing the cells in medium
supplemented with 10% FCS containing G418 (Sigma-Aldrich, St.
Louis, MO, USA) for 6 days. Resistant colonies were pooled and used
for subsequent experiments.
Confirmation of EBER expression by
reverse transcription (RT)-PCR
Total RNA was extracted from each cell line using
TRIzol® reagent (Invitrogen). DNase I (Promega
Corporation, Madison, WI, USA) treatment was performed according to
the manufacturer’s instructions. cDNA was synthesized from 2 μg
total RNA using the High Capacity cDNA Reverse Transcription kit
(Applied Biosystems) followed by PCR amplification, using DyNAzyme™
II PCR Master Mix (Finnzymes, Espoo, Finland) in a thermal cycler.
Primer sequences were as follows: EBER-1 forward,
5′-AGGACCTACGCTGCCCTAGA-3′; reverse, 5′-AAAACATGCGGACCACCAGC-3′;
EBER-2 forward, 5′-CAACGCTCAGTGCGGTGCTA-3′; reverse,
5′-CAGCGGACAAGCCGAATACC-3′. β-actin (ACTB), a housekeeping gene,
was amplified as the internal control using the primers: ACTB
forward, 5′-TCATCACCATTGGCAATGAG-3′ and reverse,
5′-CACTGTGTTGGCGTACAGGT-3′. PCR products were separated on 2%
agarose gels and visualized by ethidium bromide staining.
MTS assay and light microscopy
Cells were seeded at 5,000 cells/well into 96-well
microtiter plates in 100 μl culture medium supplemented with 1 or
10% FCS for 5 days at 37˚C in a 5% CO2 atmosphere. The
number of viable cells at each time point was measured using the
CellTiter 96® AQueous One Solution Cell
Proliferation (MTS) assay (Promega Corporation), according to the
manufacturer’s instructions. Absorbance at 490 nm was read using
the EnVision Multilabel Plate Reader (Perkin-Elmer, Waltham, MA,
USA) and subtracted with non-specific absorbance measured at 630
nm. Wells containing the appropriate medium but without cells
served as the blank. Population growth was calculated as % of
viable vs. cells on Day 0 (arbitrarily assigned viability = 100%).
Experiments were performed in triplicate. Results were expressed as
the mean ± SEM.
To observe the effects of EBERs on the cells by
light microscopy, 1.8×105 cells/well were seeded into
6-well plates in 2.5 ml culture medium supplemented with 1 or 10%
FCS for 2 days at 37˚C in a 5% CO2 atmosphere. Cell
morphology was observed under a Leica DM IRB (Leica Microsystems,
Wetzlar, Germany) inverted microscope equipped with HC PLAN S
10x/22 ocular lens and PH1 N PLAN 10x/0.25 objective lens.
To determine the effects of human LDL
supplementation, 96-well microtiter plates were seeded with 5,000
cells/well in 100 μl serum-free culture medium only or added with
50–100 μg/ml LDL (Sigma-Aldrich) for 6 days at 37˚C in a 5%
CO2 atmosphere. The MTS assay was performed as described
above. For blockade of receptor-ligand interaction, 5 μg/ml goat
anti-human LDLR antibody (R&D Systems, Minneapolis, MN, USA)
was added to the cells and incubated for 24 h. Following this,
culture medium was aspirated and replaced with serum-free culture
medium supplemented with LDL or serum-free culture medium only and
cultured for 6 days, following which the MTS assay was performed.
Population growth was calculated as % of viable vs. control cells
(arbitrarily assigned viability = 100%). Experiments were performed
in triplicate. Results were expressed as the mean ± SEM.
Apoptosis analysis assay
HK1 cells (1.2×106) were seeded in 10-cm
culture dishes and were allowed to adhere overnight. Following
this, cells were treated with 50–100 μg/ml cisplatin or left
untreated (as control). Culture dishes were re-incubated for an
additional 24 h. Apoptosis was determined using a FACSCalibur flow
cytometer (BD Biosciences, San Jose, CA, USA) and the BD Pharmingen
FITC Annexin V apoptosis detection kit (San Diego, CA, USA),
according to the manufacturer’s instructions.
Scratch-wound assay
A 6-well plate was seeded with 1×106
cells/well and incubated overnight until attachment and confluence
were achieved, followed by treatment with 3 μg/ml Mitomycin C
(Sigma-Aldrich) for 2 h. Three wells were seeded per stable cell
line. A scratch mimicking a wound was made using a 200 μl pipette
tip. Wells were washed thoroughly with PBS to remove cells detached
by scratching. Zero hour images were captured using a Leica DM IRB
inverted microscope. The migration pattern was captured 24 h later.
Using TScratch, a software tool to analyse wound healing assays
developed by the Koumoutsakos group (CSE Lab), at ETH Zürich
(14), open (area unoccupied by
cells) and the closed image areas (area where cells have migrated),
were calculated.
Invasion assay
Cells were grown to near confluence overnight
following which cell suspensions in serum-free medium were
prepared. A total of 1.8×105 cells/well were seeded in
the apical chamber of BD Falcon FluoroBlok 24-Multiwell Insert
System uncoated or coated with BD Matrigel Basement Membrane
Matrix. Differential in vitro invasive properties of the
cells were assessed using the BD BioCoat Tumor Invasion System (all
obtained from BD Biosciences), according to the manufacturer’s
instructions. Culture medium supplemented with 10% FCS was used as
the chemoattractant in the basal chamber. Cells were labelled for
quantification post-invasion with calcein AM fluorescent dye.
Fluorescence readings were obtained using a bottom-reading
fluorescent BioTek H4 (Winooski, VT, USA) plate reader.
Microarray analysis
Quality of total RNA isolated from stable HK1
transfectants was determined using the RNA 6000 Nano kit in a 2100
Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Total RNA
was labelled using GeneChip® Whole Transcript Sense
Target Labelling assay (Affymetrix, Santa Clara, CA, USA) and then
hybridized onto the GeneChip® Human Gene 1.0 ST array
(Affymetrix), in triplicate. Microarray scanning and data
acquisition were performed using an Affymetrix 3000 7G scanner.
Minimum Information About a Microarray Experiment compliant
microarray data were deposited in the NCBI’s Gene Expression
Omnibus (15) (http://www.ncbi.nlm.nih.gov/geo/; accession no.
GSE 29123). Differential expression of candidate genes was
identified using the unpaired t-test and GeneSpring GX 10 (Agilent
Technologies). P<0.05 was considered to indicate a statistically
significant difference and the threshold was set at ≥2-fold change.
Results were categorised using the Gene Ontology (GO) database to
identify involvement in biological processes, cell components and
molecular functions.
Quantitative real time PCR (qRT-PCR)
cDNA generated from total RNA (as described) was
amplified using Power SYBR-Green Master Mix (Applied Biosystems) in
the Applied Biosystems 7500 Fast Real-Time PCR system and analysed
with 7500 software v.2.0.4. A series of diluted cDNA samples were
used to generate standard curves to determine primer efficiency and
melting curves to verify the presence of a single amplicon. Primer
sets were as follows: sterol regulatory element binding protein
(SREBF) 1 forward, 5′-AGTGACTCGGAGCCTGACA-3′; reverse,
5′-TATGGTAGACGCTGGTGGTATC-3′; SREBF2 forward,
5′-GCAGTGGTGGTAGTGGTAGCA-3′; reverse, 5′-GTGGGAACTGAGGTGGGAGAAA-3′;
FASN forward, 5′-CAAAGAAGCCCATCTCCCG-3′; reverse,
5′-GCACCTCCTTGGCAAACAC-3′; LDLR forward,
5′-AGAAGAAGCCCAGTAGCGTGA-3′; reverse, 5′-TTGTGGCAAATGTGGACCTC-3′;
peptidylprolyl isomerise A (PPIA) forward,
5′-GGCCAGGCTCGTGCCGTTTT-3′ and reverse,
5′-TGCTGTCTTTGGGACCTTGTCTGC-3′. ACTB and PPIA were consistently
expressed at similar levels in HK1-vector and HK1-EBER. Therefore,
these genes were used to normalize the expression levels of all
other candidate genes. Experiments were performed in quadruplicates
and control reactions were performed in parallel in the absence of
reverse transcriptase or by substituting template cDNA with
ultrapure water.
For RT-PCR of xenografts, snap frozen tumour was
removed from liquid nitrogen and pulverized in 1 ml
TRIzol® reagent with a power homogenizer. The extracted
total RNA was dissolved in DEPC-treated water and subjected to the
procedure described for cell culture followed by PCR amplification
to detect EBER-1 and -2. In addition, qRT-PCR was performed for
SREBF1 and 2, FASN and LDLR.
Western blot analysis
Cells (1.2×106) were seeded in 10-cm
plates and cultured in medium containing 10% FCS overnight in the
absence or presence of 100 μM quercetin. Cells were lysed in 1X
RIPA lysis buffer and boiled for 10 min. Quantity of protein in the
cell lysate was determined by protein assay (Bio-Rad, Hercules, CA,
USA). A total amount of 10 μg of protein was resolved in
NuPAGE® Novex® Bis-Tris Mini Gels
(Invitrogen) and electrotransferred to polyvinylidene fluoride
membranes (Millipore, Bedford, MA, USA). Membranes were blocked
using 5% skimmed milk and incubated overnight in primary antibodies
diluted in 5% skimmed milk. Primary antibodies against FASN
(Sigma-Aldrich), LDLR (Abcam, Cambridge, UK), β-actin (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) and monoclonal mouse
anti-human Ki67 antigen (Dako Denmark A/S, Glostrup, Denmark) were
utilised. The secondary antibody reaction was performed using
anti-mouse or anti-rabbit horseradish peroxidase- conjugated IgG
(Promega Corporation). Western Lighting® Plus ECL
substrate (Perkin Elmer) and autoradiography were used to visualize
the protein expression. Densitometric analysis of X-ray films was
performed on Alpha Imager System (ProteinSimple, Santa Clara, CA,
USA) using Alpha View software.
xCELLigence cell proliferation assay
Cells were seeded at a density of 5,000 cells/well
into three E-Plate 16 (ACEA Biosciences, Inc., San Diego, CA, USA)
containing 100 μl culture medium supplemented with 10% FCS/well.
Following 48 h, 1 mM quercetin
(C15H10O7.xH2O, 302.24
kDa, anhydrous basis, purity ≥95%; obtained from Sigma-Aldrich)
dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) was added to
the culture medium to yield a final concentration of 100 μM. Cells
were monitored for ~170 h at 37˚C in a 5% CO2
atmosphere, with one change of freshly-prepared 100 μM quercetin
and medium at 72-h post-seeding. Dynamic real-time monitoring of
the growth inhibition pattern was determined by the impedance-based
xCELLigence system (Roche Diagnostics GmbH, Mannheim, Germany). The
cell index was derived from measured cell-electrode impedance which
correlates with number of cells, viability and/or cytotoxicity.
Control cultures received DMSO alone. The final concentration of
DMSO in the cultures was ≤0.5%.
Generation of xenografts
Stable HK1 transfectants were resuspended in 1:1
serum-free medium and BD Matrigel Basement Membrane Matrix at a
concentration of 1.0×106 cells. Nude mice (4–6
weeks-old) were subcutaneously injected on the upper right flank
under mild anaesthesia. Living conditions of the animals were
monitored routinely. Following 28 days of inoculation, the animals
were euthanized and tumours were excised. A section of the excised
tumour was snap frozen in liquid nitrogen and the remainder fixed
in 10% formalin and paraffin-embedded (FFPE). All procedures were
conducted in accordance to the appropriate ethics guidelines of the
Ministry of Health (MOH; Malaysia) and approved by the Animal Care
and Use Committee (MOH).
EBV ISH
FFPE (4 μm) tissue sections were placed on silanized
glass slides. ISH was performed using the Epstein-Barr Virus
Fluorescein-conjugated Probe for ISH kit (Novocastra,
Newcastle-upon-Tyne, UK), according to the manufacturer’s
instructions. Sections from the known EBV-positive xenograft, C15
(16), maintained in nude mice,
were used as positive controls. The negative control was performed
by substituting the EBV probe with a control probe consisting of a
fluorescein-labelled random oligonucleotide cocktail.
Statistical analysis
Calculations were performed using SPSS 13.0
statistical software for Windows (SPSS Inc., Chicago, IL, USA).
Differences between mean values were evaluated with the Student’s
t-test or one-way analysis of variance and Tukey’s post hoc
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Establishment of vector- and
EBER-transfected HK1 stable cell lines
To investigate the role of EBERs in NPC,
EBER-plasmid or vector alone (control) was introduced via
liposome-mediated transfection in NPC HK1 cells. Stable
transfectants were generated and examined for the presence of EBERs
by RT-PCR. The cell line generated by this approach (designated as
HK1-EBER) successfully expressed EBER-1 and -2, while no EBERs were
detected in HK1 cells transfected with vector plasmid (HK1-vector),
which served as the control (Fig.
1).
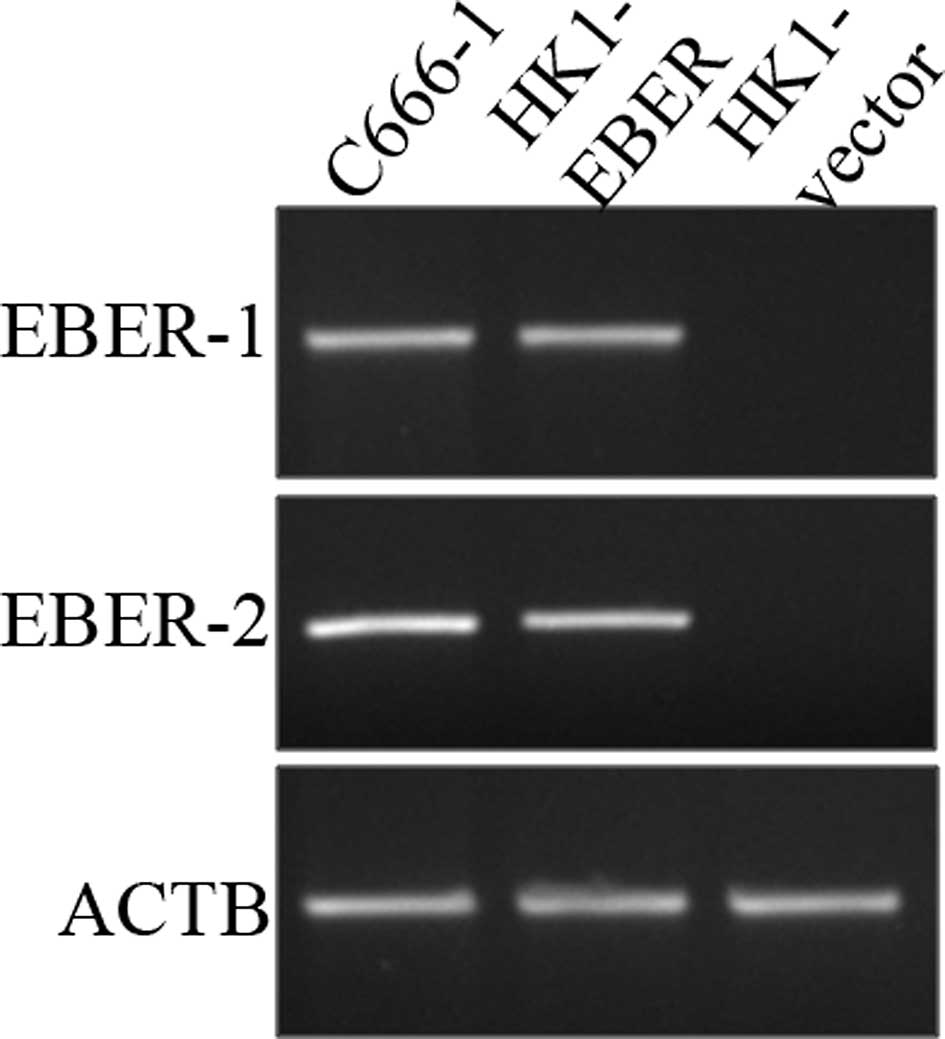 | Figure 1Establishment of HK1 stable cell line
harbouring EBER plasmid or control vector. HK1, an EBV-negative NPC
cell line, was transfected with EBER plasmid or empty vector and
expression of EBER-1 and -2 was determined by RT-PCR. C666-1, an
EBV-positive NPC cell line, was used as a positive control, whereas
ACTB served as the housekeeping gene. EBV, Epstein-Barr virus;
EBERs, EBV-encoded RNAs; RT-PCR, reverse transcription polymerase
chain reaction; NPC, nasopharyngeal carcinoma; ACTB, β-actin. |
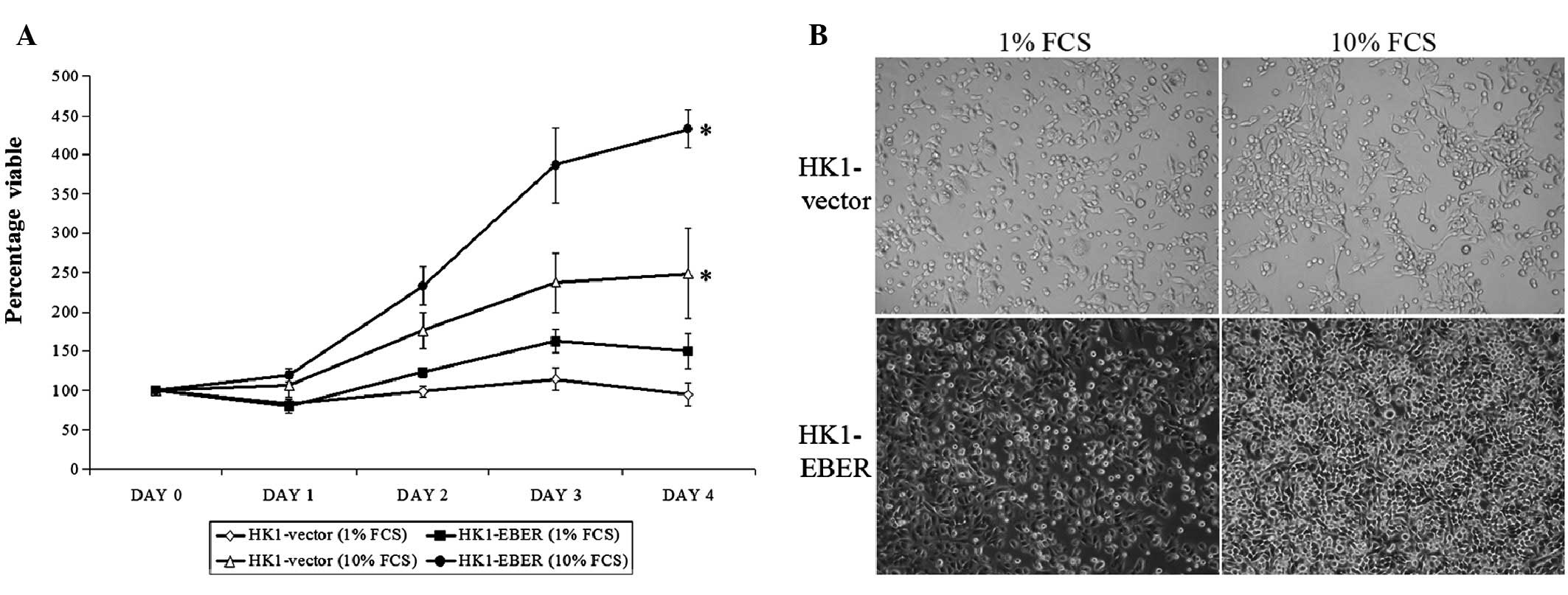
Effect of EBERs on cell proliferation and
morphology
Stable cell lines were assessed for growth. On Days
1–4, HK1-EBER exhibited a higher proliferation rate compared with
the control. On Day 4, the proliferation capacity of HK1-EBER in
full medium (10% FCS) was observed as significantly greater than
the control (Fig. 2A). In low
serum, lack of growth factors affected the growth of HK1-EBER and
the higher proliferation rate observed in full medium was not found
to be statistically significant compared with HK1-vector under
these conditions. Growth patterns and morphological changes in
culture medium supplemented with 1 and 10% FCS were observed using
light microscopy (Fig. 2B).
HK1-vector and -EBER cells were observed under light microscopy as
adherent with similar morphologies. In addition, microscopic
examination clearly showed that HK1-EBER cells reached confluence
more rapidly than the control under normal growth conditions. These
observations were consistent with growth patterns obtained by MTS
assay (Fig. 2A).
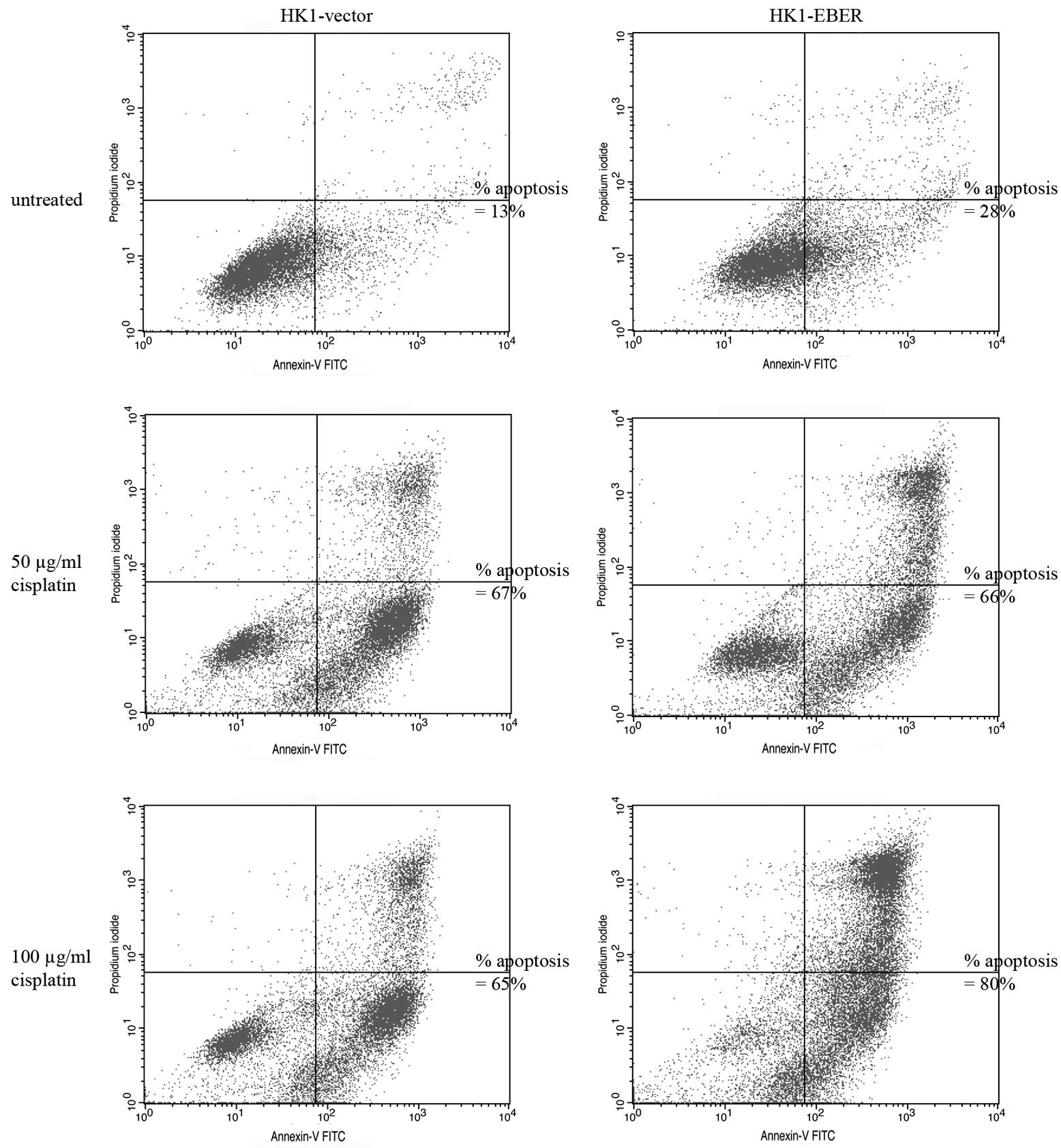
EBERs do not alter sensitivity of NPC
cells to cisplatin
To determine whether EBERs confer resistance to
apoptosis-induction, stable cell lines were cultured in the
presence of 50–100 μg/ml cisplatin, a drug commonly used for
treatment of NPC. Apoptotic cells were identified by flow
cytometry. Cisplatin clearly induced apoptosis in HK1-vector and
-EBER stable transfectants (Fig.
3). Treatment with 100 μg/ml cisplatin for 24 h caused ~50%
cell death following correction of background apoptosis (Fig. 3). Under these conditions, no
difference in the percentage of dead cells between the two stable
transfectants was found.
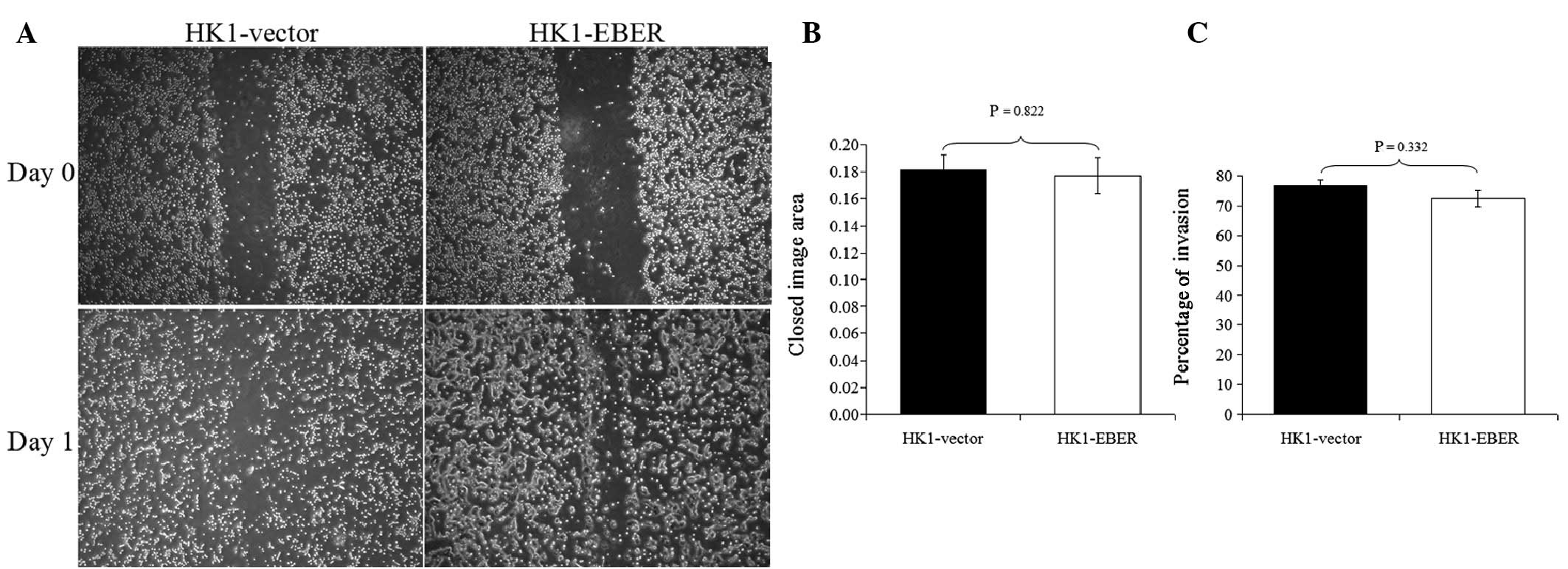
EBERs do not alter migration and invasion
of NPC cells
To determine whether EBERs affect cell migration and
invasion, HK1-EBER and -vector were subjected to scratch-wound
(17) and FluoroBlok invasion
assays. HK1-EBER and -vector cells exhibited similar migration and
invasion patterns (Fig. 4). EBER
expression did not affect migration and invasion in HK1 NPC
cells.
Deregulation of the cellular lipid
metabolic pathway
To determine transcriptome-wide gene expression
profiles associated with EBERs, we performed microarray analysis in
RNA extracted from HK1-EBER and -vector cells. Following stable
transfection, RNA was prepared from the cell lines and hybridized
to oligonucleotide microarray chips. Significant differential gene
expression was defined using the unpaired t-test (P<0.05;
threshold, ≥2-fold change). Under these criteria, a total of 54
gene transcripts were observed as significantly upregulated and 155
were significantly downregulated by the EBERs. GO analysis
indicated that 19 genes upregulated by EBERs were significantly
overexpressed in the cellular lipid metabolic process (Fig. 5A and Table I). Hierarchical clustering was
performed and a heat map was generated using Genesis IGB-TUG
Software (18). FASN,
LDLR and SREBF1 and 2 were selected from our
gene chip data for further validation. Consistent with microarray
findings, qRT-PCR confirmed significant overexpression of
FASN, LDLR and SREBF1 and 2 in HK1-EBER
cells compared with control (Fig.
5B). FASN and LDLR protein levels were identified by western
blot analysis as markedly elevated in HK1-EBER (Fig. 5C). Together with microarray and
qRT-PCR results, these observations indicate upregulation of the
cellular lipid metabolic process in HK1-EBER cells.
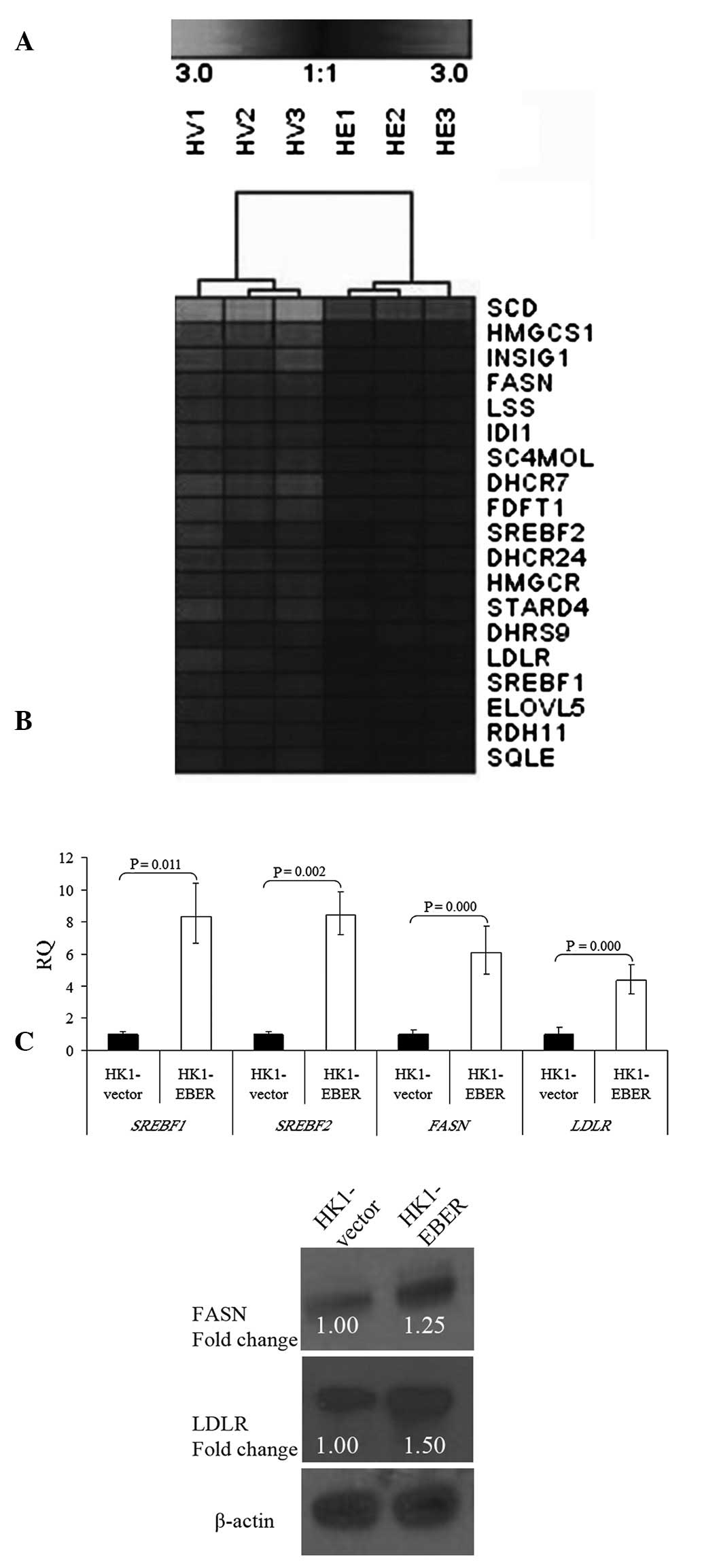 | Figure 5Hierarchical clustering of
transcripts and validation of gene expression. (A) Selected gene
expression clusters of functionally annotated gene sets
representing the cellular lipid metabolic process. Varying shades
represent downregulation and upregulation (P<0.05). HV,
HK1-vector; HE, HK1-EBER. (B) qRT-PCR of SREBF1 and
2, FASN and LDLR in HK1-EBER and HK1-vector.
mRNA levels of SREBF1 and 2, FASN and
LDLR were found to be significantly increased in HK1-EBER
(P<0.05). ACTB and PPIA were used as multiple controls for
normalization. Relative expression levels were calculated by
2-ΔΔCt and reported as RQ. (C) Overexpression of FASN
and LDLR in HK1-EBER demonstrated by western blot analysis. ACTB
was used as the loading control. Similar results were obtained in
two subsequent independent experiments. EBERs, Epstein-Barr
virus-encoded RNAs; qRT-PCR, quantitative reverse transcription
polymerase chain reaction; SREBF, sterol regulatory element binding
protein; FASN, fatty acid synthase; LDLR, low-density lipoprotein
receptor; ACTB, β-actin; PPIA, peptidylprolyl isomerise A; RQ,
relative quantification. |
 | Table IGO classification of upregulated
genes in HK1-EBER. |
Table I
GO classification of upregulated
genes in HK1-EBER.
| GO (biological
process) | No. of genes
upregulated | P-value |
|---|
| Cellular lipid
metabolic | 19 | 1.09E-15 |
| Steroid
metabolic | 15 | 1.48E-15 |
| Lipid
metabolic | 19 | 2.34E-14 |
| Sterol
metabolic | 12 | 2.34E-14 |
| Lipid
biosynthetic | 13 | 2.42E-11 |
| Steroid
biosynthetic | 10 | 2.64E-11 |
| Cholesterol
metabolic | 10 | 3.97E-11 |
| Sterol
biosynthetic | 8 | 4.54E-11 |
| Cellular
biosynthetic | 18 | 1.19E-09 |
| Alcohol
metabolic | 12 | 3.08E-08 |
| Cholesterol
biosynthetic | 6 | 1.16E-07 |
| Isoprenoid
biosynthetic | 4 | 8.11E-04 |
| Biosynthetic | 18 | 0.004 |
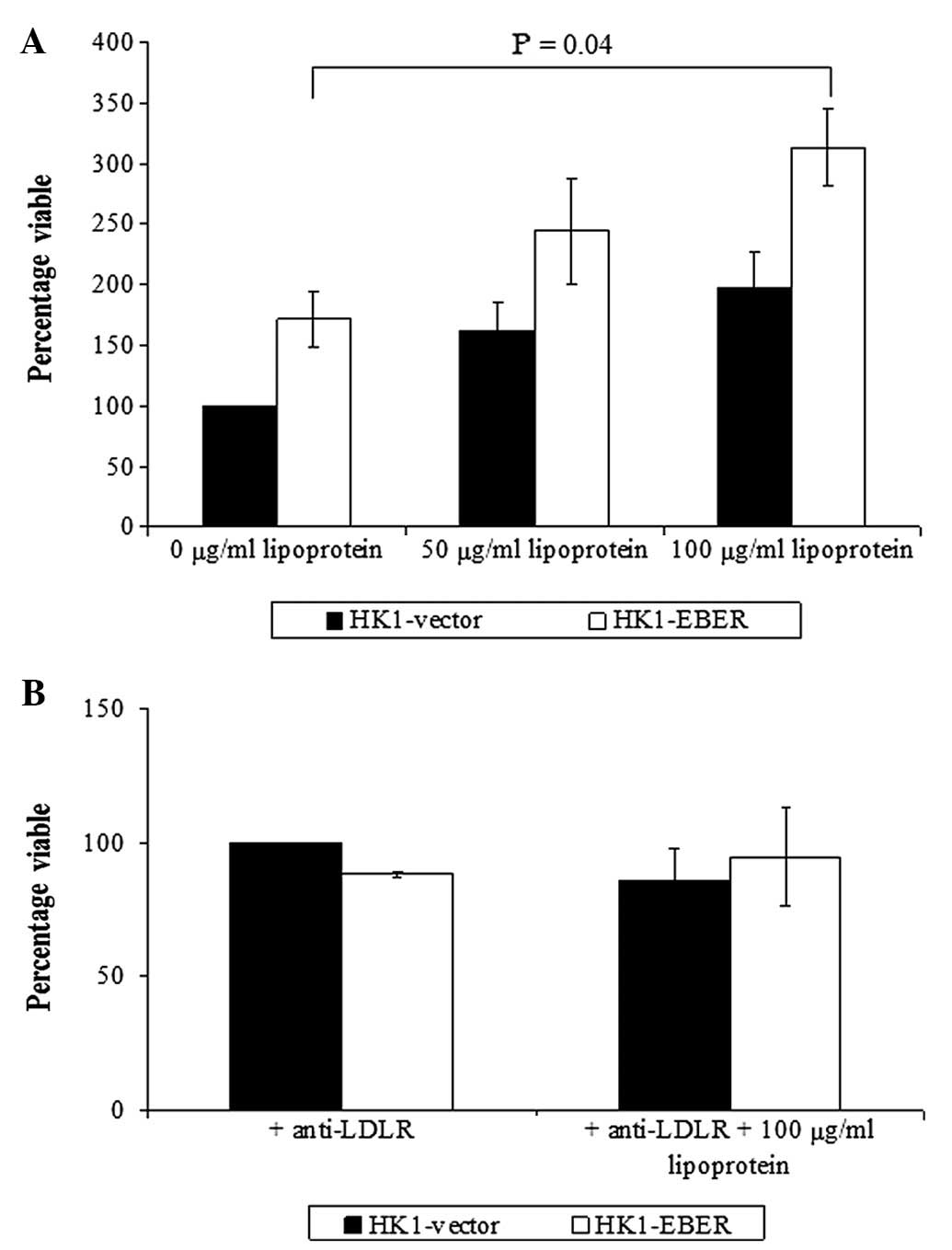
The role of LDL receptor in NPC
As LDLR was elevated in HK1-EBER, we investigated
whether supplementation of LDL stimulates NPC cell growth. Although
stimulation of cell growth was observed in HK1-EBER and -vector,
significantly increased growth was identified in HK1-EBER cells
treated with 100 μg/ml LDL only (Fig.
6A). Inhibition of the LDL receptor with anti-human LDLR
antibody blocked LDL-induced cell proliferation (Fig. 6B).
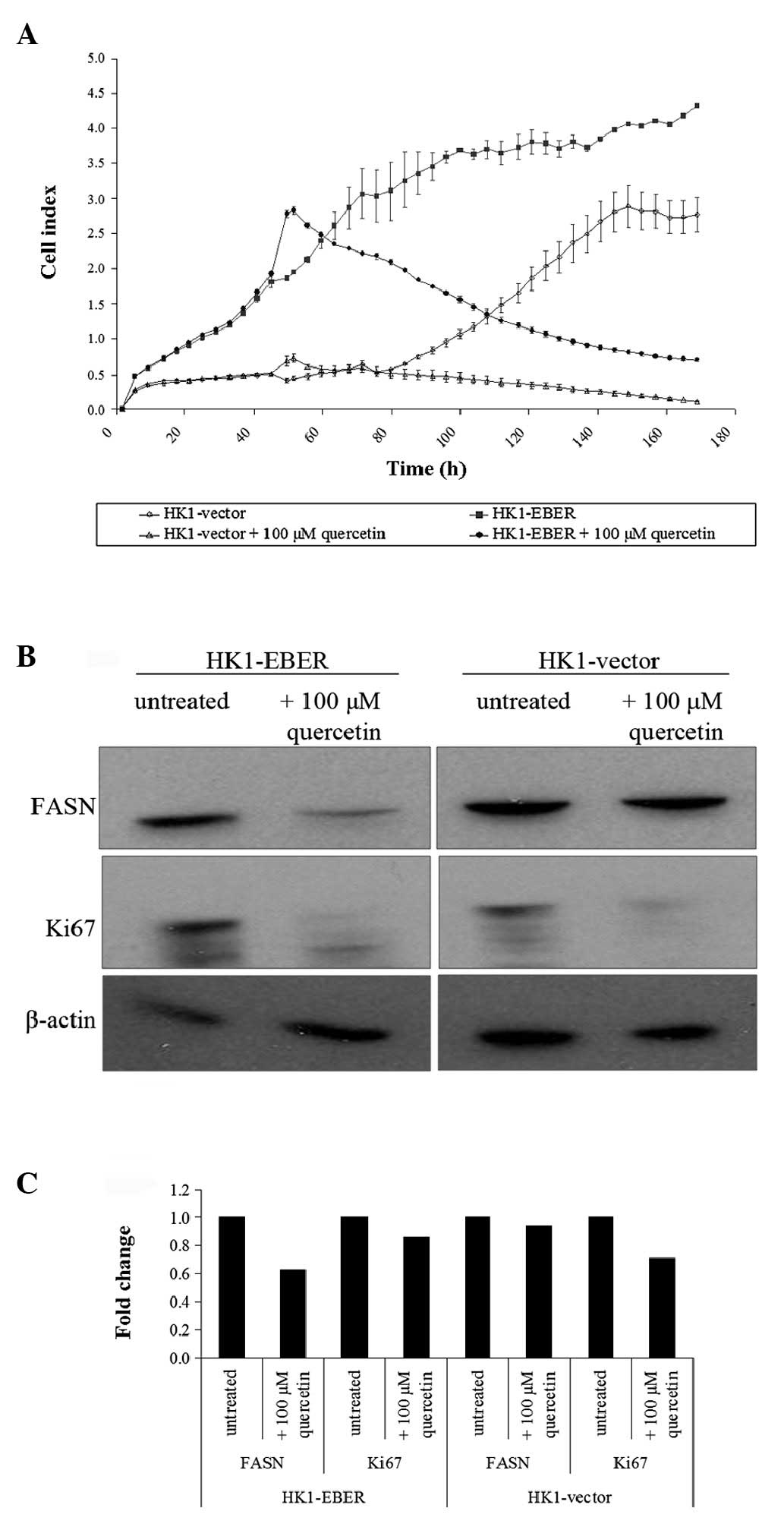
Effects of quercetin on HK1-vector and
-EBER cells
Quercetin was previously reported to inhibit FASN
(19). To determine the effects of
quercetin on EBER-expressing cells, HK1 stable transfectants
treated (or untreated) with quercetin were tested by RT xCELLigence
cell proliferation assay. The cell index demonstrated represents
growth over time. When untreated, HK1-EBER cells proliferated
faster than control (Fig. 7A),
consistent with the MTS assay performed (Fig. 2A). Quercetin treatment inhibited
HK1-EBER and -vector cell proliferation (Fig. 7A), consistent with a previous
report that quercetin inhibited NPC cells (20).
Using western blot analysis, we demonstrated that
quercetin-treatment resulted in the downregulation of FASN and Ki67
proliferation antigen expression (Fig.
7B and C). This observation was consistent with Fig. 7A which revealed that cell growth
halted in the presence of quercetin. The connection between FASN
expression and the Ki67 marker of proliferation indicates that FASN
may be associated with cell proliferation.
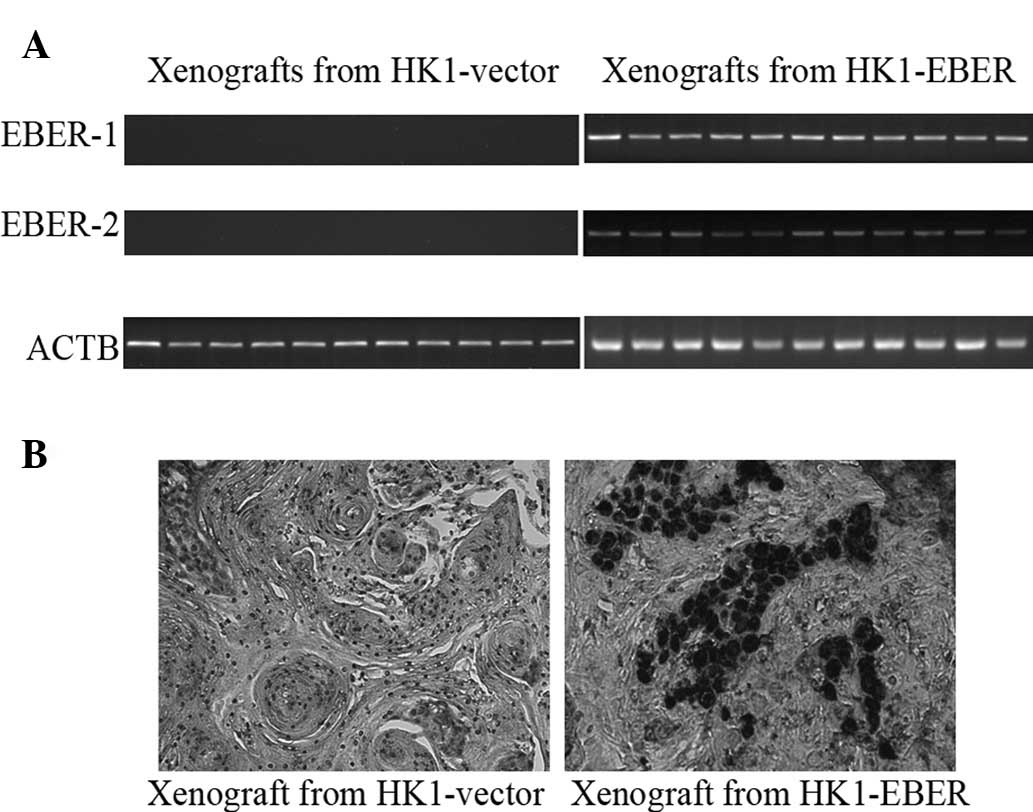
EBER-1 and -2, FASN, LDLR and SREBF1 and
2 expression in xenografts
HK1-EBER-transfected xenografts and controls (11
each) were tested for expression of EBERs by RT-PCR and EBV ISH.
All HK1-EBER-transfected xenografts were positive for EBERs and all
controls were negative (Fig. 8A and
B). All 11 pairs of xenografts were then analysed by real-time
qPCR for FASN, LDLR, SREBF1 and 2
expression. EBER-positive xenografts had variable, although a
generally higher expression of genes associated with the cellular
lipid metabolic process (Fig. 9),
in agreement with in vitro data (Fig. 5B).
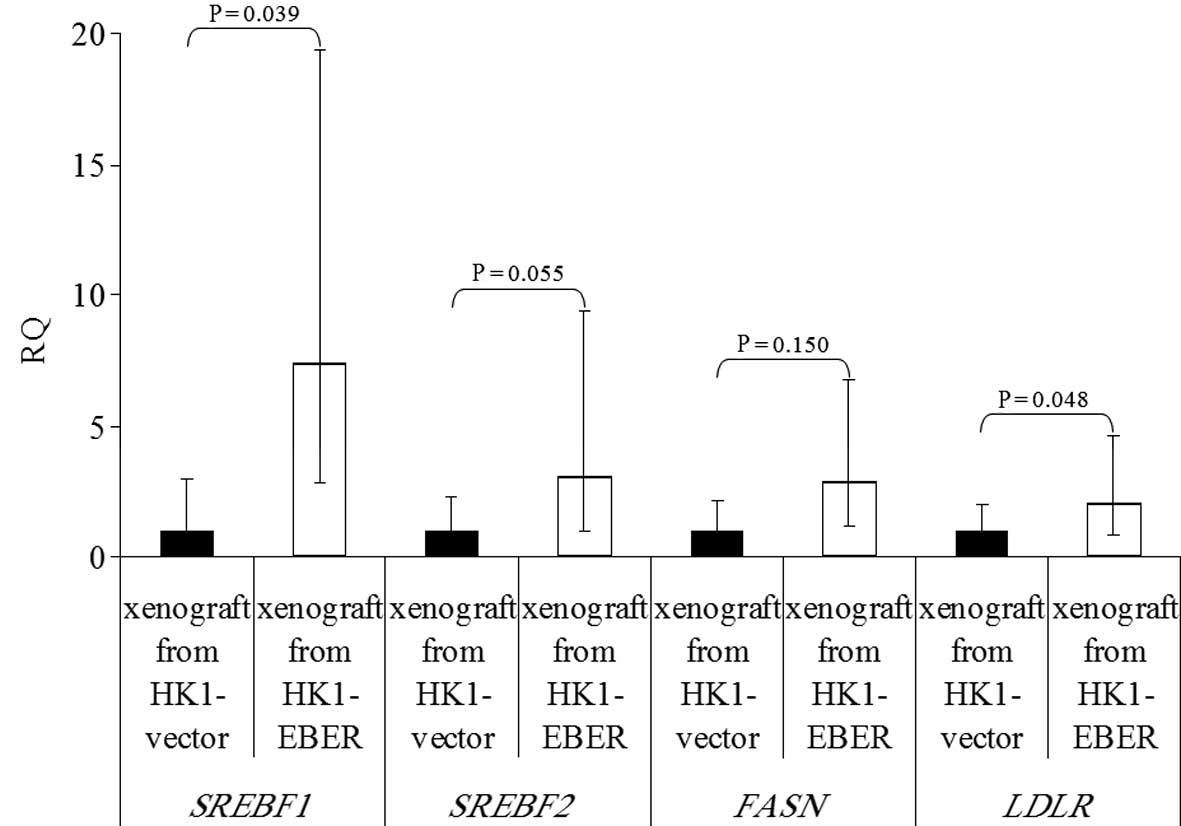 | Figure 9Expression of SREBF1 and
2, FASN and LDLR in xenografts formed from
HK1-EBER and the control, quantified by qRT-PCR. The results are
expressed as RQ to the control xenografts following normalization
to the endogenous control ACTB. The trend in differential gene
expression in vivo was consistent with that observed in
vitro. P>0.05, HK1-EBER overexpression of SREBF2 and
FASN vs. control. P≤0.05, HK1-EBER overexpression of
SREBF1 and LDLR vs. control. Limited significance may
be explained by a large variation in expression levels. SREBF,
sterol regulatory element binding protein; FASN, fatty acid
synthase; LDLR, low-density lipoprotein receptor; EBERs,
Epstein-Barr virus-encoded RNAs; qRT-PCR, quantitative reverse
transcription polymerase chain reaction; RQ, relative
quantification; ACTB, β-actin. |
Discussion
A number of studies have reported an association
between malignancies and metabolic syndromes, including
atherosclerosis, diabetes and obesity. Hirsch et al reported
common gene networks linking cancer with lipid metabolism. The
authors of that study noted that multiple genes involved in lipid
metabolism, cholesterol biosynthesis and atherosclerosis, including
OLR1 (oxidized LDL receptor 1), SCD1, SREBP1, SNAP23 and VAMP4 were
deregulated in cell transformation. OLR1, GLRX and SNAP23 were
overexpressed in human prostate and breast cancer tissues and high
expression levels of these molecules are associated with aggressive
phenotype and metastatic stage (21).
The present study demonstrates that EBERs deregulate
the cellular lipid metabolic process, contributing to elevation of
the FASN enzyme and LDLR. EBERs increased proliferation of NPC
cells. In addition, NPC cells demonstrated LDL-dependent cell
proliferation. The ability of a polyphenolic flavonoid, quercetin,
to inhibit FASN and cell proliferation was also revealed in NPC
cells.
LDLR is a cell surface glycoprotein that mediates
the uptake of LDL via endocytosis to be delivered to extrahepatic
tissues for membrane synthesis (22). In specific cancer cells, including
acute myelogenous leukemia (23),
prostate (24,25) and colorectal cancer (26), feedback regulation of LDLR is
lacking, leading to excess energy sources and subsequent
stimulation of uncontrolled growth. Cell growth, differentiation
and neoplastic transformation alter LDLR levels (27). Increased LDLR activity of tumour
cells may occur to facilitate increased LDL uptake to satisfy the
high cholesterol demand for cell growth or mechanisms associated
with cell transformation (28).
Consistent with the hypothesis that LDLR promotes proliferation, we
demonstrated that HK1-EBER, which exhibited accelerated cell
proliferation (Fig. 2), also
expressed higher levels of LDLR (Fig.
5B and C).
Lum et al studied the effect of LDL on growth
and gene regulation in DiFi colorectal cancer cells (26). When cultured in
lipoprotein-deficient serum, the growth of DiFi cells was reduced
compared with culture with LDL. In the presence of excess LDL,
uptake of LDL continued indicating a lack of normal feedback
regulation. However, when LDLR blocking antibody was added to
growing cells, growth was observed to be significantly reduced. As
demonstrated in Fig. 6A, when
exogenous LDL was added to serum-free culture medium, the growth of
HK1-EBER was significantly increased. Serum-free culture medium was
necessary to study the effect of exogenous LDL supplementation as
FCS contains lipoproteins, insulin and additional growth factors
that affect LDLR activity (29).
NPC cells were able to uptake LDL via endocytosis and utilised LDL
to boost growth in the absence of FCS. However, cell proliferation
was not induced by the addition of lipoprotein following inhibition
of LDLR-ligand interaction. We hypothesise that this was due to
blocked receptors being unable to uptake lipoprotein for
proliferation. These results confirm LDL-dependent cell
proliferation and indicate that LDLR may be crucial for
proliferation of NPC cells.
SREBP1 and 2 are an important family of
transcription factors for cholesterol and fatty acid synthesis.
SREBP1 is encoded by the SREBF1 gene and is involved in
fatty acid metabolism. SREBP2 is encoded by the SREBF2 gene
and is involved in cholesterol synthesis. SREBPs activate >30
genes, including FASN, LDLR, 3-hydroxy-3-methyl-glutaryl (HMG)-CoA
reductase and synthase and stearoyl-CoA desaturase (30,31),
all of which were upregulated by EBERs in the current study
(Fig. 5A). Previously, EBERs were
reported to induce IGF-1 in the EBV-positive C666-1 NPC cell line
and in EBER-transfected NPC-derived EBV-negative CNE1 and HONE1
cell lines (10), as well as in
EBER-transfected gastric carcinoma NU-GC-3 cells (9). SREBP1 is associated with the insulin
and IGF-1 signal transduction pathway, leading to activation of the
LDLR gene promoter (32).
An isoform of SREBP1, SREBP1c, was previously identified as a major
mediator of insulin action on the hepatic expression of
lipogenesis-related genes (33).
Streicher et al studied the mechanisms that stimulate the
promoter activity of LDLR in HepG2 cells (32). Those authors observed that
hormones, including insulin and IGF-1 increase LDLR mRNA
concentration in the presence of repressive concentrations of LDL.
In a study by LaVoie et al, 100 ng/ml IGF-1 significantly
stimulated LDLR expression in serum-free cultures of swine
granulosa cells. IGF-1 and follicle stimulating hormone increased
LDLR binding and internalization and utilisation of LDL (34). Therefore, upregulation of lipid
metabolism genes is consistent with the upregulation of IGF-1 by
EBERs.
In normal cells, FASN produces a 16-carbon saturated
fatty acid at the expense of nicotinamide adenine dinucleotide
phosphate (35). Human cancer
cells have elevated levels of FASN and undergo exacerbated
endogenous fatty acid synthesis independent of regulatory signals
that downregulate fatty acid synthesis in normal cells (36). Increased de novo fatty acid
synthesis has been recognised as a hallmark of cancer (36). In cancer cells, endogenously
synthesized fatty acids are stored as phospholipids (compared with
triglycerides in normal cells) which are incorporated into
biological membranes of proliferating tumour cells (37). As fatty acid synthesis is necessary
to maintain a constant supply of lipids and lipid precursors for
membrane production in a highly-proliferating population (36), it is assumed that increased fatty
acid synthesis aids tumour growth. Elevated FASN was identified in
human breast, bladder, colon, head and neck, endometrium, lung,
prostate, oesophagus, ovary, stomach, tongue and thyroid cancers
(35,37).
Quercetin (3,3′,4′,5,7-pentahydroxyflavone) is a
polyphenolic flavonoid widely distributed in fruits and vegetables.
Previously, quercetin was identified to inhibit enzymatic activity
of FASN and arrest cell growth (19). We recently demonstrated that
quercetin inhibited cell proliferation and decreased the expression
of FASN in NPC cells (20). In
addition, quercetin was demonstrated to effectively inhibit growth
in EBV-negative NPC HEN1 (38),
human gastric cancer HGC-27 (39)
and human leukaemia T-cells (40).
Naturally occurring polyphenols, including the flavonoids luteolin,
quercetin and kaempferol, were the most effective FASN inhibitors
in breast and prostate cancer cells and were associated with cell
growth arrest and apoptosis induction, indicating that flavonoids
may exert anti-carcinogenic effects via FASN inhibition (41). In the present study, FASN was
inhibited by 100 μM quercetin treatment while tumour cell
proliferation ceased and the bands obtained in western blot
analysis were less intense for the Ki67 proliferation antigen in
response to quercetin (Fig. 7),
demonstrating that lipid metabolism may be associated with cell
proliferation. FASN expression has been reported to correlate with
Ki67 labelling index in human endometrial carcinomas (42). This close and direct link between
FASN expression and Ki67 marker of proliferation indicates that
FASN is associated with cell proliferation. According to Kuhajda
(35,37), a rapid decline occurs in fatty acid
synthesis following FASN inhibition. In addition, cell cycle arrest
was induced, leading to a decrease in tumour cell proliferation and
ultimately apoptosis, indicating that cancer cells are dependent on
fatty acid synthesis for survival. Inhibition of FASN may be
responsible for cancer cell death as FASN inhibition introduces
changes in the synthesis of membrane phospholipids (36). Findings of the present study
demontstrated that quercetin was able to inhibit cell proliferation
and may present a means to block uncontrolled growth of cancer
cells expressing increased levels of fatty acid synthase. These
preliminary results provide additional evidence for the potential
of quercetin in NPC.
The data generated in this study suggest that EBERs
may contribute to the deregulation of the cellular lipid metabolic
process pathway in NPC cells. Aggressive growth was associated with
elevated FASN and LDLR expression induced by EBERs in the cell
lines tested. Results demonstrate that the cells exhibited
LDL-dependent cell proliferation. In addition, cell growth may be
inhibited by quercetin treatment.
The present study was primarily performed in
EBV-negative HK1 NPC cells which were stably transfected with
EBERs. The aim of this study was to determine whether EBERs alter
the biology of a pre-existing cancer cells. While lipid metabolism
was reported to share specific common gene signatures with cell
transformation, our data were performed on pre-existing cancer
cells and analysed lipid metabolism genes which had been previously
associated with prognosis (36,37),
indicating that lipid metabolism may share specific common features
with cancer progression as well.
In the present study, we generated stable
transfectants expressing EBERs, enabling multiple assays to be
performed in similar cells. However, the use of stable
transfectants requires selection of clones by antibiotics (G418).
It is plausible that the cells which were selected had incidental
pre-existing difference in biological properties not associated
with EBERs. Pooling of resistant clones was performed to minimize
the effects of selected spurious clones. In this preliminary study,
we did not distinguish whether the altered biology of cells was due
to direct or indirect effects of EBERs. However, as discussed, the
effects of EBERs on the deregulation of lipid metabolism genes are
likely to be indirect.
Lipid metabolism genes may be affected by several
pathways. There is wide variability in the expression levels of the
genes found in xenografts generated from these cells, although the
trend is consistent with the gene expression studies obtained from
in vitro experiments. Gene expression levels in xenografts
may be altered by additional factors, including tumour
microenvironmental factors.
We hypothesise that the effect of EBERs was mediated
through LDLR and FASN as those genes were confirmed to be
upregulated at the transcript and protein levels. It is plausible
that the increased effect of quercetin observed in EBER-positive
cells was simply due to a higher proliferative rate than
EBER-negative cells. Nevertheless, it is noteworthy that LDL and
quercetin had an effect on NPC cells and that this effect was more
marked in EBER-positive cells, which were of higher proliferative
rate.
Further validation of the association between EBERs
and lipid metabolism pathway in additional cell types is important.
In addition, the effect of knocking down EBERs in EBV-positive
cells may provide useful information. Understanding the effects of
deregulation of lipid metabolism on cancer progression is likely to
be of significant interest as manipulation of these pathways may
lead to new approaches of therapeutic targeting in a number of
different types of cancer, including NPC.
Acknowledgements
The authors would like to thank the Director General
of Health (Malaysia) for permission to publish this article and the
Director of the Institute for Medical Research for her support. The
authors also thank G.S.W. Tsao (University of Hong Kong), K.W. Lo
(Chinese University of Hong Kong) and Pierre Busson (Institut de
Cancérologie Gustave Roussy) for kindly providing HK1, C666-1 and
C15 cells, respectively. Many thanks to Dr Tan Lu Ping for her
helpful intellectual guidance. The present study was supported by
the Ministry of Health (Malaysia).
References
|
1
|
Tao Q and Chan AT: Nasopharyngeal
carcinoma: molecular pathogenesis and therapeutic developments. Exp
Rev Mol Med. 9:1–24. 2007.PubMed/NCBI
|
|
2
|
Chou J, Lin YC, Kim J, You L, Xu Z, He B
and Jablons DM: Nasopharyngeal carcinoma - review of the molecular
mechanisms of tumorigenesis. Head Neck. 30:946–963. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takada K and Nanbo A: The role of EBERs in
oncogenesis. Semin Cancer Biol. 11:461–467. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Komano J, Maruo S, Kurozumi K, Oda T and
Takada K: Oncogenic role of Epstein-Barr virus-encoded RNAs in
Burkitt’s lymphoma cell line Akata. J Virol. 73:9827–9831.
1999.
|
|
5
|
Yamamoto N, Takizawa T, Iwanaga Y, Shimizu
N and Yamamoto N: Malignant transformation of B lymphoma cell line
BJAB by Epstein-Barr virus-encoded small RNAs. FEBS Lett.
484:153–158. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kitagawa N, Goto M, Kurozumi K, Maruo S,
Fukayama M, Naoe T, Yasukawa M, Hino K, Suzuki T, Todo S and Takada
K: Epstein-Barr virus-encoded poly(A)(-) RNA supports Burkitt’s
lymphoma growth through interleukin-10 induction. EMBO J.
19:6742–6750. 2000.PubMed/NCBI
|
|
7
|
Yang L, Aozasa K, Oshimi K and Takada K:
Epstein-Barr virus (EBV)-encoded RNA promotes growth of
EBV-infected T cells through interleukin-9 induction. Cancer Res.
64:5332–5337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wong HL, Wang X, Chang RC, et al: Stable
expression of EBERs in immortalized nasopharyngeal epithelial cells
confers resistance to apoptotic stress. Mol Carcinog. 44:92–101.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iwakiri D, Eizuru Y, Tokunaga M and Takada
K: Autocrine growth of Epstein-Barr virus-positive gastric
carcinoma cells mediated by an Epstein-Barr virus-encoded small
RNA. Cancer Res. 63:7062–7067. 2003.PubMed/NCBI
|
|
10
|
Iwakiri D, Sheen TS, Chen JY, Huang DP and
Takada K: Epstein-Barr virus-encoded small RNA induces insulin-like
growth factor 1 and supports growth of nasopharyngeal
carcinoma-derived cell lines. Oncogene. 24:1767–1773. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang DP, Ho JH, Poon YF, Chew EC, Saw D,
Lui M, Li CL, Mak LS, Lai SH and Lau WH: Establishment of a cell
line (NPC/HK1) from a differentiated squamous carcinoma of the
nasopharynx. Int J Cancer. 26:127–132. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW,
Tsang YS, Wong N, Whitney BM and Lee JC: Nasopharyngeal carcinoma
cell line (C666-1) consistently harbouring Epstein-Barr virus. Int
J Cancer. 83:121–126. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chan SY, Choy KW, Tsao SW, Tao Q, Tang T,
Chung GT and Lo KW: Authentication of nasopharyngeal carcinoma
tumor lines. Int J Cancer. 122:2169–2171. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gebäck T, Schulz MM, Koumoutsakos P and
Detmar M: TScratch: a novel and simple software tool for automated
analysis of monolayer wound healing assays. Biotechniques.
46:265–274. 2008.PubMed/NCBI
|
|
15
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Busson P, Ganem G, Flores P, Mugneret F,
Clausse B, Caillou B, Braham K, Wakasugi H, Lipinski M and Tursz T:
Establishment and characterization of three transplantable
EBV-containing nasopharyngeal carcinomas. Int J Cancer. 42:599–606.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cory G: Scratch-wound assay. Cell
Migration: Developmental Methods and Protocols, Methods in
Molecular Biology. Wells CM and Parsons M: 769. Springer
Science+Business Media, LLC; New York, NY: pp. 25–30. 2011,
View Article : Google Scholar
|
|
18
|
Sturn A, Quackenbush J and Trajanoski Z:
Genesis: cluster analysis of microarray data. Bioinformatics.
18:207–208. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brusselmans K, Vrolix R, Verhoeven G and
Swinnen JV: Induction of cancer cell apoptosis by flavonoids is
associated with their ability to inhibit fatty acid synthase
activity. J Biol Chem. 280:5636–5645. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Daker M, Ahmad M and Khoo ASB:
Quercetin-induced inhibition and synergistic activity with
cisplatin - a chemotherapeutic strategy for nasopharyngeal
carcinoma cells. Cancer Cell Int. 12:342012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hirsch HA, Iliopoulos D, Joshi A, Zhang Y,
Jaeger SA, Bulyk M, Tsichlis PN, Shirley Liu X and Struhl K: A
transcriptional signature and common gene networks link cancer with
lipid metabolism and diverse human diseases. Cancer Cell.
17:348–361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chung NS and Wasan KM: Potential role of
the low-density lipoprotein receptor family as mediators of
cellular drug uptake. Adv Drug Deliv Rev. 56:1315–1334. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tatidis L, Gruber A and Vitols S:
Decreased feedback regulation of low density lipoprotein receptor
activity by sterols in leukemic cells from patients with acute
myelogenous leukemia. J Lipid Res. 38:2436–2445. 1997.
|
|
24
|
Chen Y and Hughes-Fulford M: Human
prostate cancer cells lack feedback regulation of low-density
lipoprotein receptor and its regulator, SREBP2. Int J Cancer.
91:41–45. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sekine Y, Koike H, Nakano T, Nakajima K,
Takahashi S and Suzuki K: Remnant lipoproteins induced
proliferation of human prostate cancer cell, PC-3 but not LNCaP,
via low density liporprotein receptor. Cancer Epidemiol. 33:16–23.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lum DF, McQuaid KR, Gilbertson VL and
Hughes-Fulford M: Coordinate up-regulation of low-density
lipoprotein receptor and cyclo-oxygenase-2 gene expression in human
colorectal cells and in colorectal adenocarcinoma biopsies. Int J
Cancer. 83:162–166. 1999. View Article : Google Scholar
|
|
27
|
Rao KN: The significance of the
cholesterol biosynthetic pathway in cell growth and carcinogenesis
(review). Anticancer Res. 15:309–314. 1995.PubMed/NCBI
|
|
28
|
Gueddari N, Favre G, Hachem H, Marek E, Le
Gaillard F and Soula G: Evidence for up-regulated low density
lipoprotein receptor in human lung adenocarcinoma cell line A549.
Biochimie. 75:811–819. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shiroeda O, Yamaguchi N and Kawai K:
Stimulation of low density lipoprotein receptor activity by
conditioned medium from a human cancer cell line. Cancer Res.
47:4630–4633. 1987.PubMed/NCBI
|
|
30
|
Shimano H: Sterol regulatory
element-binding proteins (SREBPs): transcriptional regulators of
lipid synthetic genes. Prog Lipid Res. 40:439–452. 2001. View Article : Google Scholar
|
|
31
|
Weber LW, Boll M and Stampfl A:
Maintaining cholesterol homeostasis: sterol regulatory
element-binding proteins. World J Gastroenterol. 10:3081–3087.
2004.PubMed/NCBI
|
|
32
|
Streicher R, Kotzka J, Müller-Wieland D,
Siemeister G, Munck M, Avci H and Krone W: SREBP-1 mediates
activation of the low density lipoprotein receptor promoter by
insulin and insulin-like growth factor-1. J Biol Chem.
271:7128–7133. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Foretz M, Guichard C, Ferré P and Foufelle
F: Sterol regulatory element binding protein-1c is a major mediator
of insulin action on the hepatic expression of glucokinase and
lipogenesis-related genes. Proc Natl Acad Sci USA. 12737–12742.
1999. View Article : Google Scholar
|
|
34
|
LaVoie HA, Garmey JC, Day RN and Veldhuis
JD: Concerted regulation of low density lipoprotein receptor gene
expression by follicle-stimulating hormone and insulin-like growth
factor 1 in porcine granulosa cells: promoter activation, messenger
ribonucleic acid stability and sterol feedback. Endocrinology.
140:178–186. 1999.
|
|
35
|
Kuhajda FP: Fatty-acid synthase and human
cancer: new perspectives on its role in tumor biology. Nutrition.
16:202–208. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Menendez JA and Lupu R: Fatty acid
synthase and the lipogenic phenotype in cancer pathogenesis. Nat
Rev Cancer. 7:763–777. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kuhajda FP: Fatty acid synthase and
cancer: new application of an old pathway. Cancer Res.
66:5977–5980. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang F, Cui Y and Cao P: Effect of
quercetin on proliferation and apoptosis of human nasopharyngeal
carcinoma HEN1 cells. J Huazhong Univ Sci Technolog Med Sci.
28:369–372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshida M, Sakai T, Hosokawa N, Marui N,
Matsumoto K, Fujioka A, Nishino H and Aoike A: The effect of
quercetin on cell cycle progression and growth of human gastric
cancer cells. FEBS Lett. 260:10–13. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoshida M, Yamamoto M and Nikaido T:
Quercetin arrests human leukemic T-cells in late G1 phase of the
cell cycle. Cancer Res. 52:6676–6681. 1992.PubMed/NCBI
|
|
41
|
Lupu R and Menendez JA: Pharmacological
inhibitors of Fatty Acid Synthase (FASN)-catalyzed endogenous fatty
acid biogenesis: a new family of anti-cancer agents? Curr Pharm
Biotechnol. 7:483–493. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pizer ES, Chrest FJ, DiGiuseppe JA and Han
WF: Pharmacological inhibitors of mammalian fatty acid synthase
suppress DNA replication and induce apoptosis in tumor cell lines.
Cancer Res. 58:4611–4615. 1998.PubMed/NCBI
|