Introduction
Hypertension is a crucial risk factor for
cardiovascular disease. It causes cardiac remodeling, accompanied
by fibrosis, inflammation, hypertrophy, pump failure and myocyte
degeneration (1). Despite recent
advances in the understanding of the molecular and cellular
processes of cardiac remodeling, further development of effective
therapies for the prevention of heart failure is required.
MicroRNAs (miRNAs) are small, endogenous, non-coding
RNAs that have emerged as a new set of modulators of gene
expression (2). It is estimated
that >1,000 unique miRNAs are encoded within the human genome
(3). These miRNAs bind with
imperfect complementarity to their target mRNAs and regulate a
variety of diseases, such as cancer, Alzheimer’s disease and
cardiovascular diseases (4–6).
Recent studies have revealed that miRNAs modulate the pathogenesis
of cardiac remodeling (7). The
expression profile of miRNAs in failing myocardium has been
explored by performing microarrays on various murine models of
cardiac hypertrophy, including calcineurin overexpression (6), Akt overexpression (8), coronary artery ligation,
ischemia-reperfusion (9) and
thoracic aortic constriction (10). Based on these murine models, a
variety of miRNAs involved in cardiac myocyte hypertrophy have been
identified (11–14). However, little information is
available with regard to miRNAs in the renovascular hypertensive
murine model. The roles and expression profiles of miRNAs in the
left ventricular myocardium of renovascular hypertensive rats are
poorly understood.
This study aims to investigate the miRNA expression
profile associated with left ventricular remodeling in two-kidney
one-clip (2K1C) hypertensive rats. Target genes and biological
networks of the differentially expressed miRNAs were also
identified. Furthermore, the possible mechanisms of miRNA-mediated
signaling were analyzed using ingenuity pathway analysis (IPA) for
the first time.
Materials and methods
Animals
Male Wistar rats (weight, 120–150 g), purchased from
Vital River Lab Animal Technology Co., Ltd. (Beijing, China), were
housed individually in temperature- (20±2°C) and humidity
(55±5%)-controlled rooms, with a 12-hour light/dark cycle. The rats
were given free access to food and tap water. Animal studies were
performed under conditions approved by the Local Animal Care and
Use Committee.
Induction of 2K1C hypertension
Following one week of acclimatization, the rats were
randomly divided into two groups. As described in detail previously
(15), the male Wistar rats (n=14)
were anesthetized with sodium pentobarbital (50 mg/kg body weight,
i.p.) and the left kidney was exposed by a flank incision. After
separating the renal arteries, a silver clip was placed around the
left renal artery. Sham surgery rats (n=12) underwent an identical
surgical procedure with the exception of the placing of the
arterial clip. The systolic blood pressure (SBP) of the rats in
each group was measured weekly using a non-invasive computerized
tail-cuff system (HX-II, Hunan Medical Univ. Medical Instruments
Inc., Changsha, China). Eight weeks after the initial surgery, the
rats were anesthetized with sodium pentobarbital following
overnight fasting. Organs of interest (the heart and kidneys) were
immediately removed, blotted dry, weighed and stored at −80°C until
RNA extraction. Organ indices were calculated using the following
formula: organ index = organ weight (g)/body weight (g) × 100.
Histologic analysis
Heart samples from each rat were fixed in 10%
buffered formalin and embedded in paraffin. Heart sections of 5–6
μm were cut and stained with hematoxylin-eosin (HE) and van
Gieson’s (VG). Histopathological changes were examined under the
microscope.
RNA isolation and miRNA microarray
Total RNA was extracted from 30 mg of tissues.
Samples were mechanically disrupted and simultaneously homogenized
in the presence of QIAzol Lysis reagent (Qiagen, Valencia, CA, USA)
using a Mikro-Dismembrator (Braun Biotech International, Melsungen,
Germany). RNA was extracted using the miRNeasy Mini kit (Qiagen)
according to the manufacturer’s instructions. RNA concentrations
were measured using the NanoDrop 2000c Spectrophotometer (NanoDrop
Technologies, Wilmington, DE, USA), while RNA quality was assessed
using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa
Clara, CA, USA) using the RNA 6000 Nano kit (Agilent Technologies).
Total RNA (100 ng) was dephosphorylated at 37°C for 30 min with
calf intestinal phosphatase and denatured using 100% DMSO at 100°C
for 5 min. Samples were labeled with Cyanine 3-pCp using T4 DNA
ligase (Invitrogen, Grand Island, NY, USA) by incubation at 16°C
for 1 h and by hybridization on a 8×15 K format Agilent rat miRNA
array G2534A (Agilent Technologies). Arrays were washed according
to the manufacturer’s instructions and scanned at a resolution of 5
μm using an Agilent G2565CA microarray scanner (Agilent
Technologies). Data were acquired and the quantile was normalized
using Agilent Feature Extraction software version 9.5.3 (Agilent
Technologies).
Bioinformatics analysis
Datasets representing miRNAs with altered expression
profiles derived from microarray analyses were imported into the
IPA program (http://www.ingenuity.com). The basis
of the IPA tool consists of the ingenuity pathway knowledge base
(IPKB) derived from the known functions and interactions of miRNAs
published in the literature. In this study, the target genes,
biological networks and functional pathways of the differentially
expressed miRNAs were identified.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
(SPSS Inc., Somers, New York, USA). The differences between groups
were compared using the Student’s t-test. Correlations among the
expression profiles of miRNAs were calculated using Pearson’s
correlation. P<0.05 was considered to indicate a statistically
significant difference. All values are expressed as the mean ±
SEM.
Results
Establishment of 2K1C hypertensive
rats
We developed a rat model of 2K1C hypertension as
previously described (15). Eight
weeks after renal artery clipping, the 2K1C rats exhibited atrophy
of the clipped (left) kidney and hypertrophy of the non-clipped
(right) kidney (P<0.05; Fig.
1). 2K1C rats exhibited an increase in SBP (P<0.05), while
the SBP of the sham surgery rats remained unaltered (Fig. 1). Furthermore, 2K1C caused a
significant increase in the weight of the heart, the cardiac index
and the right renal index when compared with the sham surgery group
(P<0.05; Table I).
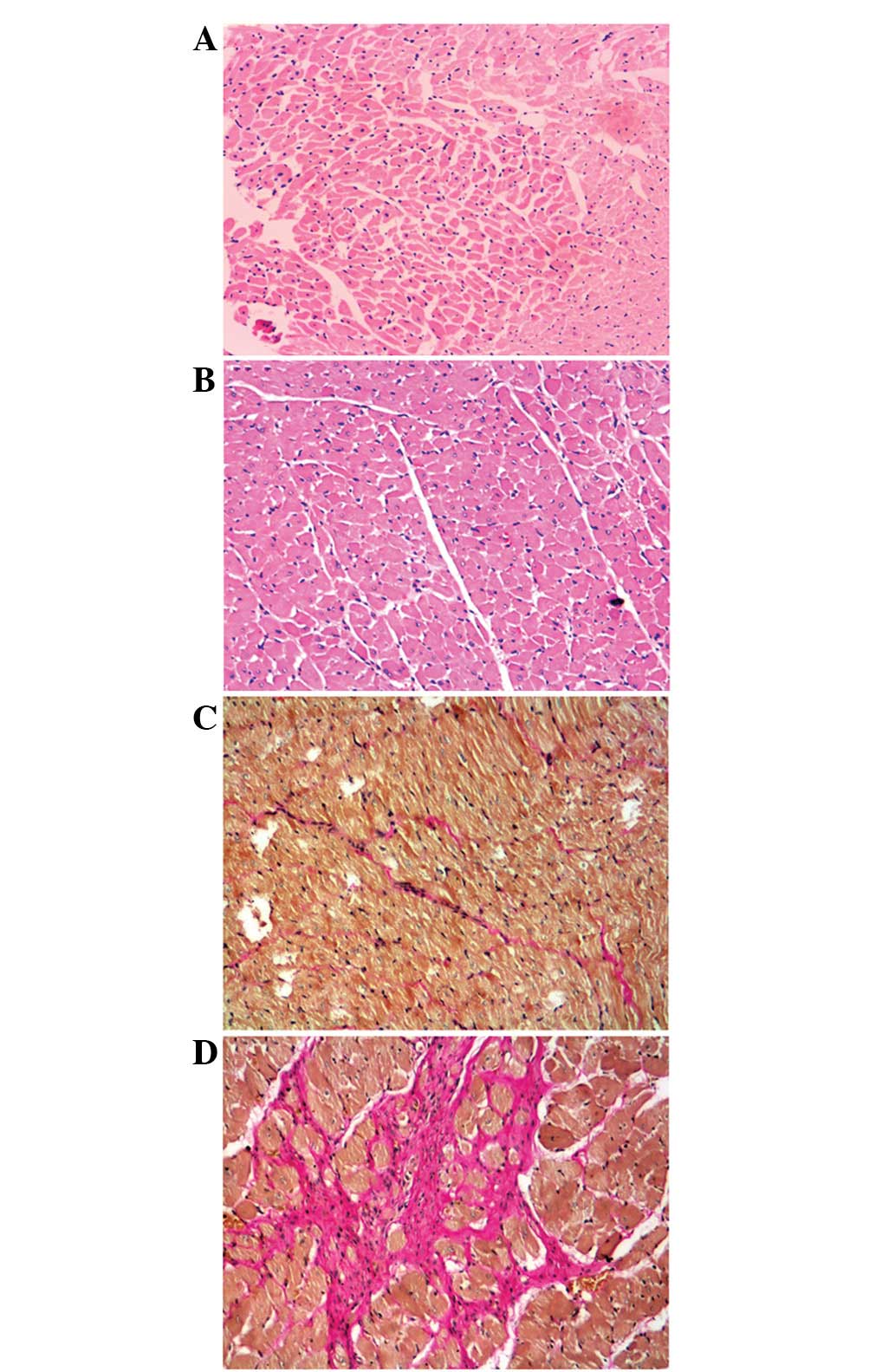
Additionally, our results demonstrated that 2K1C promoted
cardiomyocyte hypertrophy and increased interstitial collagen
deposition and infiltration of the myocardium (Fig. 2).
 | Table IComparison of organ indices in sham
surgery (n=12) and 2K1C rats (n=14). |
Table I
Comparison of organ indices in sham
surgery (n=12) and 2K1C rats (n=14).
| Group | Heart weight (g) | Cardiac index | Right renal
index |
|---|
| Sham surgery | 1.08±0.06 |
(2.8±0.3)×10−3 |
(3.3±0.5)×10−3 |
| 2K1C | 1.20±0.14a |
(3.2±0.3)×10−3a |
(4.9±1.1)×10−3a |
Identification of the miRNA expression
profile
In order to identify differentially expressed miRNAs
in 2K1C rats, we performed miRNA microarray analyses. Of the miRNAs
investigated, our results identified 48 that were differentially
expressed between the sham surgery group and the 2K1C group, of
which 25 were upregulated and 23 were downregulated in the left
ventricular myocardium of the 2K1C hypertensive rats (Table II). A 2.0-fold change cut-off was
applied to all array data sets.
 | Table IIDifferentially expressed miRNAs in
2K1C rats. |
Table II
Differentially expressed miRNAs in
2K1C rats.
| Downregulated
expression |
|---|
|
|---|
| Systematic name | Fold change |
|---|
| rno-miR-139-5p | −38.4905200 |
| rno-miR-224 | −26.9225270 |
| rno-miR-129-2* | −17.7938580 |
| rno-miR-129-1* | −17.4628870 |
| rno-miR-92b | −15.4211950 |
| rno-miR-505* | −10.1010240 |
| rno-miR-127 | −6.6837770 |
| rno-miR-141 | −4.8718386 |
| rno-miR-224* | −4.8661530 |
| rno-miR-455 | −4.3821400 |
| rno-miR-384-5p | −4.2651870 |
| rno-miR-362 | −3.9236674 |
| rno-miR-122 | −3.6222723 |
| rno-miR-363 | −3.3113322 |
| rno-miR-434 | −3.2692702 |
| rno-miR-425* | −3.1358542 |
| rno-miR-872* | −3.1162152 |
| rno-miR-582 | −3.0495758 |
| rno-miR-667 | −2.9269574 |
| rno-miR-547* | −2.8845766 |
| rno-miR-483 | −2.8532393 |
| rno-miR-1188-3p | −2.1270647 |
| rno-miR-10b | −2.0181155 |
|
| Upregulated
expression |
|
| Systematic name | Fold change |
|
| rno-miR-3544 | +2.0963676 |
| rno-miR-298 | +2.1669054 |
| rno-miR-196c | +2.2632172 |
| rno-miR-181a-1* | +2.3843806 |
| rno-miR-598-3p | +2.3982275 |
| rno-miR-188 | +2.4536152 |
| rno-miR-760-5p | +2.4810490 |
| rno-miR-335 | +2.4852781 |
| rno-miR-31 | +2.7537080 |
| rno-miR-190* | +2.7569172 |
| rno-miR-484 | +2.7729888 |
| rno-miR-672 | +2.8125286 |
| rno-miR-874 | +2.8486352 |
| rno-miR-455* | +2.8730786 |
| rno-miR-3584-5p | +3.1480370 |
| rno-miR-132 | +3.5094187 |
| rno-miR-345-3p | +3.6336637 |
| rno-miR-136 | +3.6917052 |
| rno-miR-3588 | +3.7029164 |
| rno-miR-31* | +3.7426915 |
| rno-miR-20a* | +5.3322973 |
| rno-miR-32* | +6.2950974 |
| rno-miR-3562 | +9.9319600 |
| rno-miR-338* | +18.3044530 |
| rno-miR-139-3p | +49.2387500 |
The target genes and biological networks
of differentially expressed miRNAs
The target genes of differentially expressed miRNAs
were analyzed using the IPA tool, which combined the sources of
TarBase, TargetScan, miRecords and Ingenuity Expert. A total of
10,156 target genes were identified; of these, 1,140 presented in
the rats. In order to eliminate the false positive rates of the
target prediction databases, only the target genes validated by
biological experiments were selected for further analysis.
Following the analysis of published data, 56 target genes were
identified (Table III). Highly
rated networks of 56 target genes were analyzed and the results
revealed five significant pathways (Table IV). Of these networks,
‘cardiovascular disease, cardiovascular system development and
function, organ morphology’ was the highest rated network with a
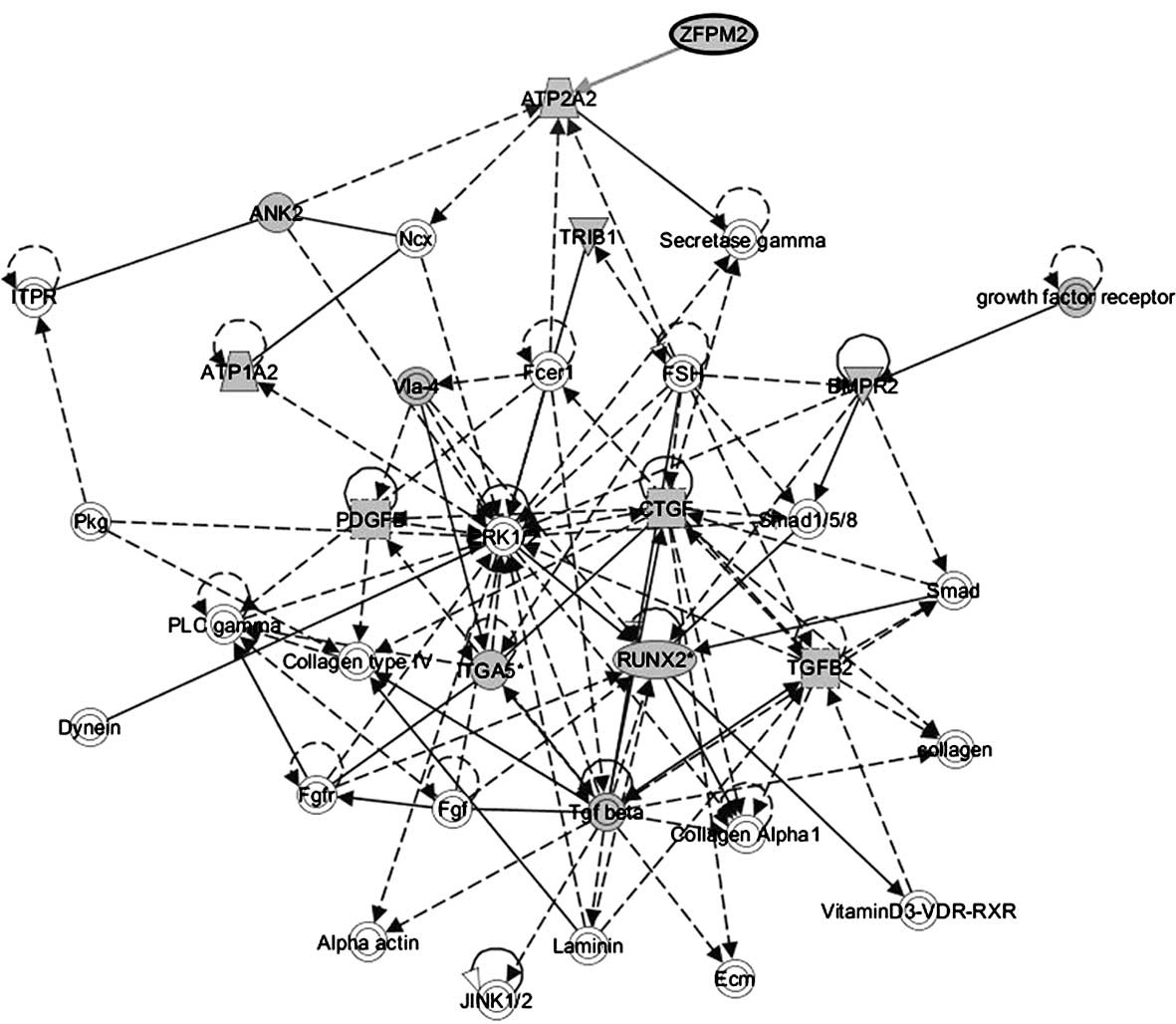
significance score of 22. Target genes ANK2, ATP1A2,
ATP1A2, BMPR2, CTGF, ITGA5*,
PDGFB, RUNX2*, TGFB2, TRIB1 and
ZFPM2 were involved in this network (Fig. 3). Moreover, the genes associated
with the top toxicology list are provided in Table V. ‘Cardiac hypertrophy’ and
‘cardiac necrosis/cell death’ were identified as the top two
toxicology pathways.
 | Table IIITarget genes for differentially
expressed miRNAs in 2K1C rats. |
Table III
Target genes for differentially
expressed miRNAs in 2K1C rats.
| Target genes |
|---|
| ABL1 | ADAM17 | AKT3 | ALOX5AP | ANK2 | ANXA1 | AP3M2 | ATP1A2 |
| ATP2A2 | BCL2L11 | BCL6* | BECN1 | BMPR2 | CDCP1 | CDKN1A | CDKN2A |
| CERS6 | CTGF | CTNNB1 | EGLN3 | F2 | FBXW7 | FOXP1 | FOXP3 |
| GEMIN2 | GNAI2 | HIF1A | IDH1 | IKBKB | IL11 | ITGA5* | JAK2 |
| JUN | MAP2K4* | P38MAPK | MMP9 | NCEH1 | NFATC1 | NPR3 | NT5E |
| NTRK3 | PDGFB | PPP3CA | PTEN | RB1 | RHOA | RUNX2* | S100A9 |
| SOCS3 | STX7 | TGFB2 | TP53 | TRIB1 | VSNL1 | XBP1 | ZFPM2 |
 | Table IVTop five networks with their
respective scores obtained from IPA. |
Table IV
Top five networks with their
respective scores obtained from IPA.
| ID | Associated network
functions | Score |
|---|
| 1 | Cardiovascular
disease, cardiovascular system development and function, organ
morphology | 22 |
| 2 | Cancer,
hematological disease, neurological disease | 15 |
| 3 | Cellular
development, cellular growth and proliferation, hematological
system development and function | 13 |
| 4 | Cell death and
survival, cancer, hematological disease | 12 |
| 5 | Hematological
system development and function, tissue morphology, organismal
injury and abnormalities | 10 |
 | Table VTop five toxicology pathways obtained
from IPA. |
Table V
Top five toxicology pathways obtained
from IPA.
| Pathway | P-value | Ratio |
|---|
| Cardiac
hypertrophy |
6.39×10−19 | 19/333 (0.057) |
| Cardiac
necrosis/cell death |
1.71×10−14 | 14/227 (0.062) |
| Liver
proliferation |
5.38×10−11 | 11/201 (0.032) |
| Renal necrosis/cell
death |
1.21×10−10 | 14/437 (0.032) |
| P53 signaling |
9.18×10−10 | 8/95 (0.084) |
Potential mechanisms of the
differentially expressed miRNAs in the development of left
ventricular remodeling
Signal transduction pathways associated with target
genes of the differentially expressed miRNAs were investigated. IPA
analysis revealed that the 56 target genes participated in pathways
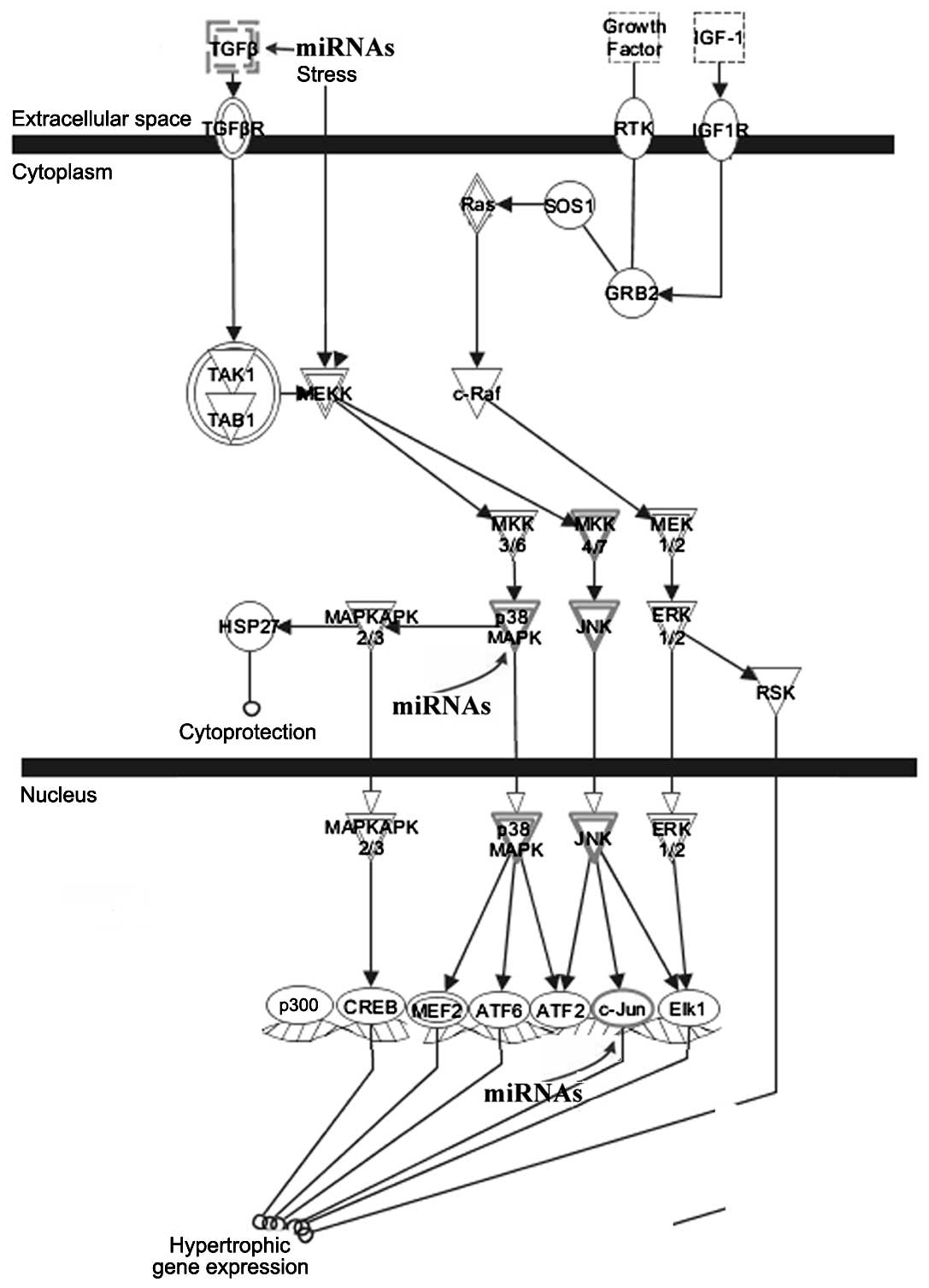
linked to cardiac hypertrophy (Fig.
4). Certain target genes, including TGFβ, p38MAPK
and c-Jun, were involved in the cardiac hypertrophy
signaling pathway. We also examined the roles of the target genes
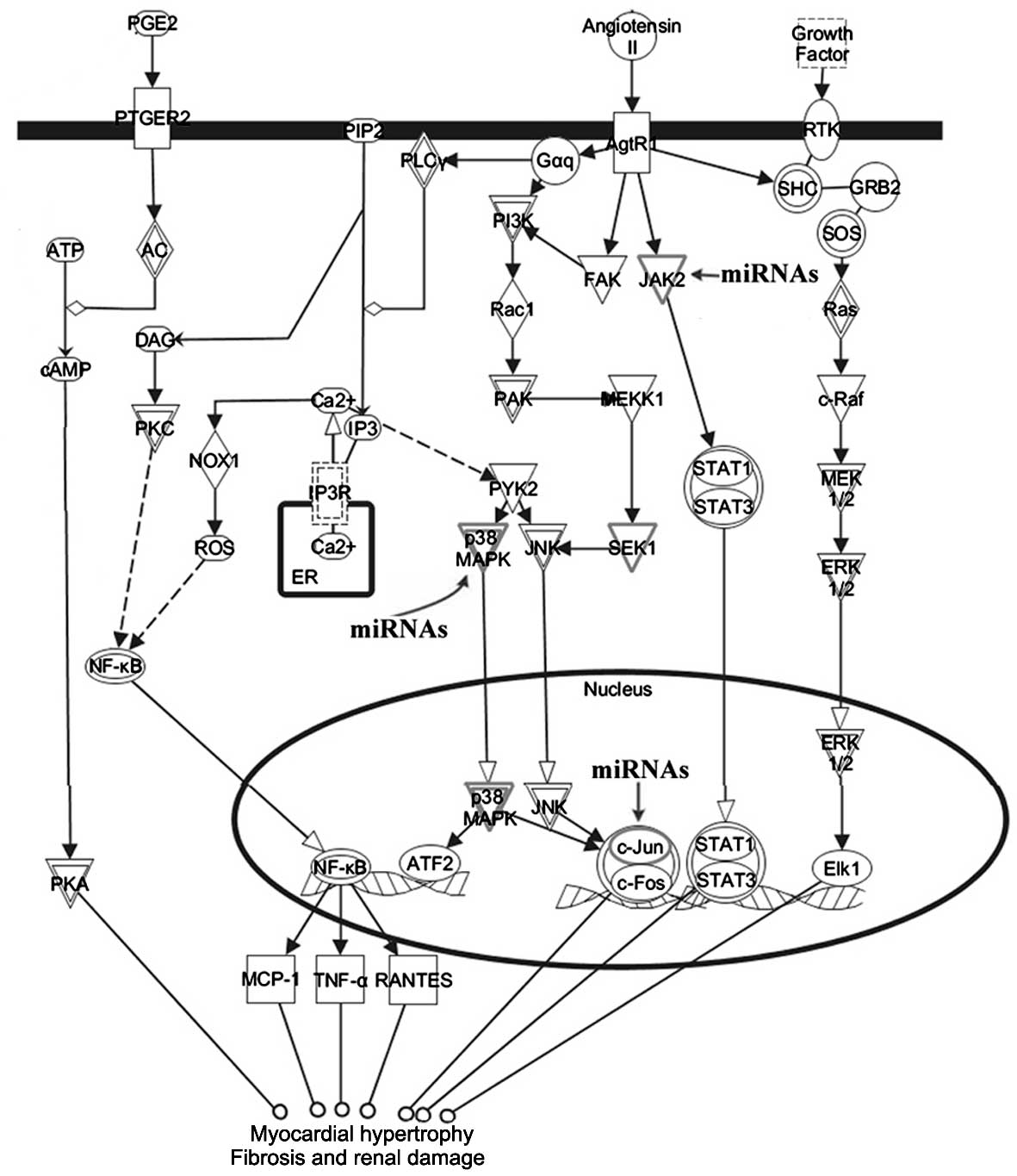
as mediators of renin-angiotensin signaling. We discovered that
p38MAPK, c-Jun and JAK2 were involved in renin-angiotensin
signaling in response to left ventricular remodeling (Fig. 5).
Discussion
The 2K1C rat model is useful for studying the
molecular mechanisms involved in hypertension-related cardiac
remodeling. In an attempt to identify the expression profiles of
the miRNAs in this experimental model, we undertook systematic
profiling using a commercially available microarray platform. We
discovered that 48 miRNAs were differentially expressed between the
2K1C and sham surgery groups. Notably, cardiac remodeling
represents collective changes in the biology of the cardiac
myocyte, the volume of myocyte and non-myocyte components of the
myocardium, in addition to the geometry of the left ventricular
chamber. Therefore, the altered miRNA levels should be attributed
to cardiomyocytes in addition to other myocardial cell types, such
as fibroblasts and endothelial cells.
To understand the involvement of miRNAs in left
ventricular remodeling, we selected differentially expressed miRNAs
and performed target gene analysis. Due to the complex nature of
cardiac remodeling and since existing computational algorithms
remain challenging, the target prediction analysis was unable to
reveal specific sets of genes involved in defined biological
aspects. Thus, only 56 target genes that had been validated by
biological experiments were selected for further analysis in the
current study. Moreover, IPA analysis revealed ‘cardiovascular
disease, cardiovascular system development and function, organ
morphology’ to be the most favored associated network function in
2K1C rats. The two most significant pathways ‘cardiac hypertrophy’
and ‘cardiac necrosis/cell death’ were observed. These results
suggested that alterations to the expression of miRNAs observed in
this study represented the physiological responses to an altered
cardiac workload.
To gain an understanding of the molecular mechanisms
of left ventricular remodeling, signal transduction pathways
associated with the differentially expressed miRNAs and their
target genes were investigated. This study revealed that the target
genes participated in pathways linked to cardiac hypertrophy.
Signaling molecules, such as TGFβ and p38MAPK, were
validated target genes. Our results suggested that differentially
expressed miRNAs were mediators of the TGFβ-TAK1-p38MAPK signaling
pathway, which is crucial for myocyte hypertrophy, as described
previously (16). Renovascular
hypertensive rats were used in this experiment. The
renin-angiotensin system contributes to left ventricular
hypertrophy and fibrosis (17).
Therefore, the correlations between the renin-angiotensin signaling
pathway and the target genes of differentially expressed miRNAs
were evaluated. In addition to a p38MAPK-dependent pathway
contributing to left ventricular hypertrophy, we observed that the
target gene JAK2 is a signaling molecule, suggesting that the
differentially expressed miRNAs were also sensitive to the
JAK2-STAT signaling pathway, which is crucial for cardiac fibrosis
(18).
In conclusion, the miRNA expression profiles in the
left ventricular myocardium of 2K1C hypertensive rats were
identified. Further analysis of the roles of the differentially
expressed miRNAs, their target genes and signaling pathways would
provide greater insight into the molecular mechanisms of left
ventricular remodeling.
Acknowledgements
This work was supported by the National Natural
Science Foundation of China (no. 81160033). We thank Mr. J. C. Lin
of CloudScientific Technology Co., Ltd. for performing the
ingenuity pathway analyses.
References
|
1
|
Soares ER, Lima WG, Machado RP, et al:
Cardiac and renal effects induced by different exercise workloads
in renovascular hypertensive rats. Braz J Med Biol Res. 44:573–582.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lecellier CH, Dunoyer P, Arar K, et al: A
cellular microRNA mediates antiviral defense in human cells.
Science. 308:557–560. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Berezikov E, Guryev V, van de Belt J, et
al: Phylogenetic shadowing and computational identification of
human microRNA genes. Cell. 120:21–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jost D, Nowojewski A and Levine E: Small
RNA biology is systems biology. BMB Rep. 44:11–21. 2011. View Article : Google Scholar
|
|
5
|
Lukiw WJ: Micro-RNA speciation in fetal,
adult and Alzheimer’s disease hippocampus. Neuroreport. 18:297–300.
2007.PubMed/NCBI
|
|
6
|
van Rooij E, Sutherland LB, Liu N, et al:
A signature pattern of stress-responsive microRNAs that can evoke
cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA.
103:18255–18260. 2006.PubMed/NCBI
|
|
7
|
Yang B, Lin H, Xiao J, et al: The
muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic
potential by targeting GJA1 and KCNJ2. Nat Med. 13:486–491. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carè A, Catalucci D, Felicetti F, et al:
MicroRNA-133 controls cardiac hypertrophy. Nat Med. 13:613–618.
2007.
|
|
9
|
van Rooij E, Sutherland LB, Thatcher JE,
et al: Dysregulation of microRNAs after myocardial infarction
reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci
USA. 105:13027–13032. 2008.PubMed/NCBI
|
|
10
|
Sayed D, Hong C, Chen IY, Lypowy J and
Abdellatif M: MicroRNAs play an essential role in the development
of cardiac hypertrophy. Circ Res. 100:416–424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Elia L, Contu R, Quintavalle M, et al:
Reciprocal regulation of microRNA-1 and insulin-like growth
factor-1 signal transduction cascade in cardiac and skeletal muscle
in physiological and pathological conditions. Circulation.
120:2377–2385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Horie T, Ono K, Nishi H, et al:
MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15
and is involved in metabolic control in cardiac myocytes. Biochem
Biophys Res Commun. 389:315–320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thum T, Gross C, Fiedler J, et al:
MicroRNA-21 contributes to myocardial disease by stimulating MAP
kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin Z, Murtaza I, Wang K, Jiao J, Gao J
and Li PF: miR-23a functions downstream of NFATc3 to regulate
cardiac hypertrophy. Proc Natl Acad Sci USA. 106:12103–12108. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wiesel P, Mazzolai L, Nussberger J and
Pedrazzini T: Two-kidney, one clip and one-kidney, one clip
hypertension in mice. Hypertension. 29:1025–1030. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsumoto-Ida M, Takimoto Y, Aoyama T,
Akao M, Takeda T and Kita T: Activation of TGF-beta1-TAK1-p38 MAPK
pathway in spared cardiomyocytes is involved in left ventricular
remodeling after myocardial infarction in rats. Am J Physiol Heart
Circ Physiol. 290:H709–H715. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mascareno E, Galatioto J, Rozenberg I, et
al: Cardiac lineage protein-1 (CLP-1) regulates cardiac remodeling
via transcriptional modulation of diverse hypertrophic and fibrotic
responses and angiotensin II-transforming growth factor β (TGF-β1)
signaling axis. J Biol Chem. 287:13084–13093. 2012.PubMed/NCBI
|
|
18
|
Liao W, Yu C, Wen J, et al: Adiponectin
induces interleukin-6 production and activates STAT3 in adult mouse
cardiac fibroblasts. Biol Cell. 101:263–272. 2009. View Article : Google Scholar : PubMed/NCBI
|