Introduction
Hepatocellular carcinoma (HCC) is a challenging
malignancy of global importance associated with high prevalence and
mortality (1). Previously, HCC was
predominant in undeveloped or less-developed countries; however,
incidence of HCC has increased recently in developed regions,
including Western Europe, the United States and Japan (2–3). HCC
has several noteworthy epidemiological features, including oncogene
mutation, ethnic groups and the presence of several well-documented
environmental potentially preventable risk factors, such as
hepatitis C virus infection (4,5).
Although there has been a growing understanding of the molecular
mechanisms underlying hepatocarcinogenesis in recent years
(1), the causes and biology of HCC
are not yet fully understood. Moreover, the efficacies of current
treatment for HCC are unsatisfactory (6) resulting in poor prognosis with a
median survival time of four months (2).
Sirtuins, mammalian homologs of the yeast protein
silent information regulator 2 (Sir2), are a unique subclass of
deacetylases and mono-ADP-ribosyltransferases that use
NAD+ as a cosubstrate (7). There are seven members of the sirtuin
family in mammals (Sirt1-Sirt7) (7) These proteins influence a wide array
of pathophysiological processes, including DNA repair, cell
survival, stress responses, metabolic homeostasis and aging
(7). Sirt1, the most studied
member of the family, has been implicated in carcinogenesis and
cancer progression, and is considered to be a therapeutic target
(8). Sirt1 induces histone
deacetylation and methylation, promoter CpG island methylation,
transcriptional repression and deacetylation of tumor suppressor
proteins (8). Sirt1 was reported
to have a bidirectional effect on tumor initiation, progression and
drug resistance; it can operate as a tumor suppressor or as an
oncogenic factor, depending on the context and the study conditions
(8). Sirt3 is another member of
the sirtuin family, which preferentially localizes to mitochondria
and is involved in mitochondrial energy production and substrate
oxidation (9–10). Unlike Sirt1, accumulating evidence
suggests that Sirt3 is a tumor suppressor in breast cancer
(11), oral cancer (12) and HCC (13).
Sirt6 is a less-studied nuclear sirtuin member. Thus
far, Sirt6 has been found to regulate glucose homeostasis in the
liver (14) and maintain genome
stability (15–16). In 2012, Min et al (17) showed that Sirt6-dependent
inhibition of survivin contributed to activator protein-1 binding
site-induced tumor suppression. Another group confirmed that
deletion of Sirt6 in vivo increased the number, size and
aggressiveness of tumors (18).
However, evidence from other studies indicates that Sirt6 may be
tumorigenic in pancreatic cancer (19) and breast cancer (20). Consequently, the exact role of
Sirt6 in cancer is still being analyzed. In the present study, the
expression of Sirt6 in human HCC tissues and the potential role of
Sirt6 in HCC were investigated.
Materials and Methods
Reagents
Antibodies against Sirt6 and β-actin were purchased
from Sigma (St. Louis, MO, USA). Antibodies against cleaved
caspase-3, phosphorylated extracellular signal-regulated kinase
(ERK1/2), total-ERK1/2 and U0126 were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA). An immunofluorescence terminal
deoxynucleotidyl transferase-mediated dUTP nick end labeling
(TUNEL) kit was purchased from Promega Corporation (Madison, WI,
USA). DAPI and dichlorofluorescein (DCF) were purchased from
Invitrogen Life Technologies (Carslbad, CA, USA). Enhanced
chemiluminescence and protease/phosphatase inhibitors were
purchased from Pierce (Rockford, IL, USA).
Human HCC tissue
Four pairs of HCC and matched normal adjacent tissue
extracts were obtained from Chinese patients who underwent surgical
resection for diagnosis and therapy in the Provincial Hospital
Affiliated to Shandong University (Jinan, China). Samples were
obtained after receiving informed consent according to an
established protocol approved by the Ethics Committee of Shandong
University (Jinan, China).
Cell culture
HepG2 human HCC cells and HEK293 cells were obtained
from the American Type Culture Collection (Manassas, VA, USA).
Cells were cultured in Dulbecco’s modified Eagle’s medium
supplemented with 10% (v/v) fetal bovine serum (FBS) in 95%
O2 and 5% CO2 (21).
Plasmid construction
The adenovirus expressing Sirt6 (Ad-Sirt6) and the
control adenovirus expressing green fluorescent protein (Ad-GFP)
were generated using the Adenoviral Expression system (Cell
Biolabs, Inc., San Diego, CA, USA) according to the manufacturer’s
instructions as described previously (22). Mouse Sirt6 mRNA was extracted from
mouse liver tissue using TRIzol (Invitrogen Life Technologies) and
was reverse transcribed with AMV to obtain cDNA. The full length of
the mouse Sirt6 cDNA was amplified using polymerase chain reaction
(PCR) with specific primers. Full length PCR products of Sir6 were
then subcloned into the pacAd5 CMV-IRES vector (Cell Biolabs,
Inc.). Next, pacAd5 CMV-IRES-Sirt6 and pacAd5 backbone vectors were
linearized by PacI (New England Biolabs, Ipswich, MA,
USA).
The short hairpin-RNA (shRNA)-induced RNA
interference (RNAi) was achieved using the RAPAd® shRNA
Adenoviral Expression system (Cell Biolabs, Inc.) according to the
manufacturer’s instructions. The nucleotide sequence for shRNA was
designed using BLOCK-iT™ RNAi Online Designer tool (Invitrogen Life
Technologies). The following nucleotide sequence against human
Sirt6 was used in this study: 5′-GGTCTGGCAGTCTTCCAGTGT-3′.
Generation of adenovirus
HEK293 cells were used to produce adenovirus. The
purified linearized DNAs or plasmids were transfected into HEK293
cells using Lipofectamine 2000 reagent (Invitrogen Life
Technologies). At 6 h after transfection, the medium containing
Lipofectamine 2000 was removed and novel medium was added.
Adenovirus-containing HEK293 cells and media were harvested on the
6th day post-transfection. Viruses were released by three
freeze/thaw cycles and stored at −80°C. For virus transfection, 30
μl viral stock solution was added into culture medium (2 ml) of
HepG2 cells for 6 h.
Cell proliferation assay
Cell counting kit-8 (CCK-8; Dojindo Laboratories,
Kumamoto, Japan) was used to detect cell proliferation as described
previously (13,23). Ad-GFP and Ad-Sirt6-transfected
cells (3×104) were seeded into 48-well plates and
cultured overnight to allow attachment. On the second day, cells
were serum-starved for 8 h, and FBS was added into medium. At 12,
24, 36 and 48 h, cells were incubated with 10 μl CCK-8 solution for
2 h, and the optical density at 450 nm was analyzed (Tecan Ultra
384 reader; Tecan, Männedorf, Switzerland). Experiments were
performed in duplicate.
TUNEL assay
TUNEL staining was performed as described previously
(13,24). Cells were transfected with Ad-GFP
or Ad-Sirt6 for 6 h. At 24 h after treatment, cells were incubated
in TUNEL reaction buffer in a humidified chamber for 1 h at 37°C in
the dark, then rinsed four times with phosphate-buffered saline
(PBS) and incubated with DAPI (1 mg/ml) for 15 min. The stained
cells were visualized using a fluorescence microscope (IX-71,
Olympus, Tokyo, Japan). TUNEL-positive cells (green) were counted
as apoptotic cells. Images were acquired digitally from a randomly
selected pool of 10–15 fields under each condition (25).
Quantitative PCR analysis
qPCR analysis was performed on an OpticonDNA engine
(MJ Research, Inc., St. Bruno, QC, Canada) using
PrimerScript® RT Reagent kit (Takara Bio, Inc., Shiga,
Japan) and normalized as described previously (23,26).
The total RNA was extracted form human tissue using TRIzol
(Invitrogen Life Technologies) and 10 μg RNA was used for first
strand cDNA synthesis. cDNA (100 ng) was amplified using primers as
follows: Sense: 5′-CGCAGTACGTCCGAGACACAGT-3′ and antisense:
5′-TTGGTAGCCAGCGGCAGGTT-3′ for Sirt6; and sense:
5′-GCACTCTTCCAGCCTTCCTTCC-3′ and antisense:
5′-CCGCCAGACAGCACTGTGTT-3′ for β-actin. The mRNA level of
housekeeping gene β-actin served as a control.
Immunoblotting
Immunoblotting analyses of cell-extracts were
performed as described previously (27–28).
Human tissues or cells were lysed in 50 mM Tris-HCl (pH 7.5) and 1%
SDS with protease/phosphatase inhibitor cocktail (Pierce), and then
heated at 95°C for 10 min. Samples were subjected to 10% SDS-PAGE,
and transferred onto polyvinylidene fluoride membranes (Millipore,
Milford, MA, USA) at 100V for 90 min. Following blocking in 5%
skimmed milk and PBS containing 0.1% Tween-20, membranes were
incubated with primary antibodies (cleaved caspase-3 and Sirt6)
followed by horseradish peroxidase-labeled secondary antibodies
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The
membranes were then detected using an enhanced chemiluminescence
kit (Pierce).
ROS measurement
A DCF assay was used for the quantification of
intracellular ROS as previously described (29). Ad-GFP and Ad-Sirt6 transfected
cells were plated onto 96-well plates (5×104 cells/well)
and loaded with 100 μM DCF (Invitrogen Life Technologies) for 1 h
at 37°C. Cells were subsequently washed using PBS buffer.
Fluorescence was measured using a fluorescence microplate reader
(Tecan) with an excitation filter of 485 nm and an emission filter
at 530 nm.
Superoxide anion measurement
Ad-GFP- and Ad-Sirt6-transfected cells were seeded
in 96-well plates and grown overnight. Following removal of the
cell culture medium, the cells were washed with PBS, and 200 μl of
25 μM dihydroethidium (DHE) dissolved in PBS was added to each well
for 1 h. The fluorescence (DHE ex/em: 530/620 nm) was measured
using a multiwell plate reader (Tecan) for 20 min at 37°C.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean. Differences were evaluated by two-tailed Student’s t-test
or analysis of variance followed by Tukey’s post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
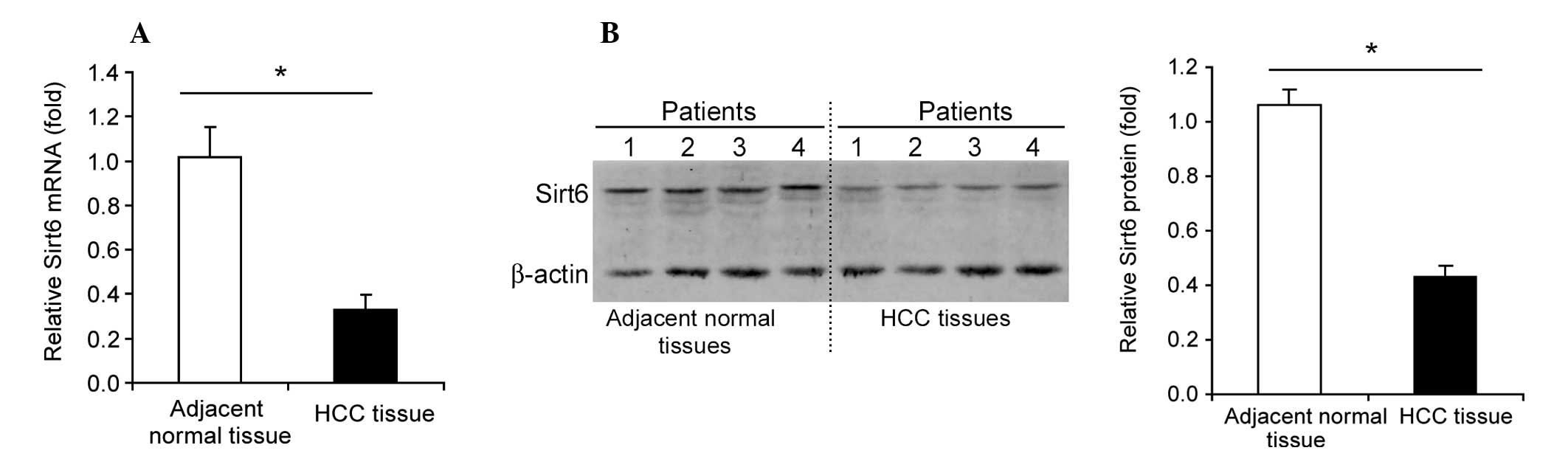
Downregulation of Sirt6 in human HCC
tissue
Sirt6 mRNA levels were significantly downregulated
(35–40%, P<0.05) in human HCC tissue compared with normal
adjacent tissue (Fig. 1A). A
marked downregulation of Sirt6 protein levels was also observed in
human HCC tissue compared with normal adjacent tissue (Fig. 1B).
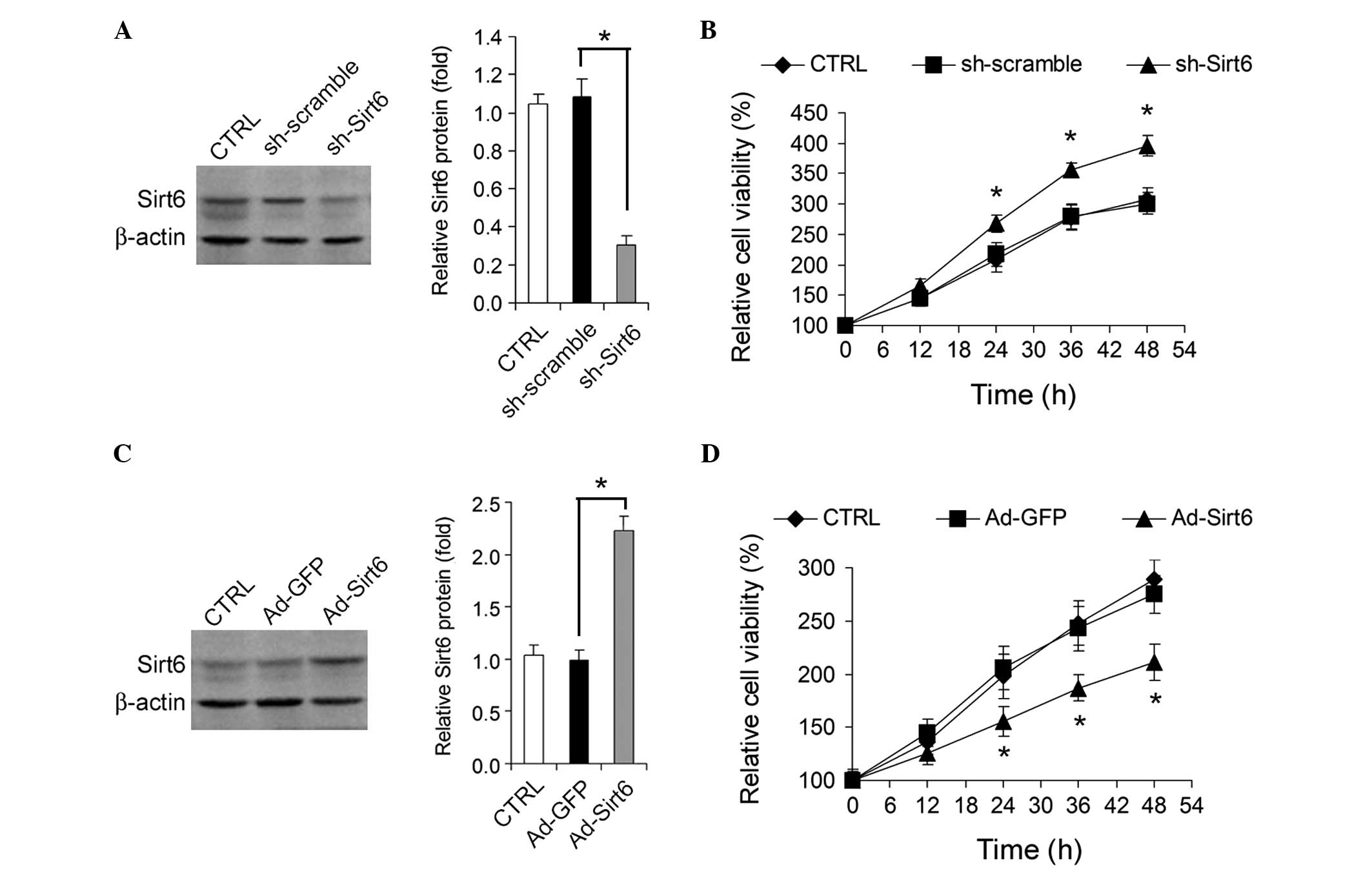
Sirt6 regulates HepG2 cell growth
The effect of knockdown or overexpression of Sirt6
on HepG2 HCC cell growth in vitro was analyzed.
Adenovirus-mediated knockdown of Sirt6 (Fig. 2A) by shRNA promoted HepG2 cell
growth (Fig. 2B), whereas
adenovirus-mediated overexpression of Sirt6 (Fig. 2C) significantly inhibited HepG2
cell growth (Fig. 2D).
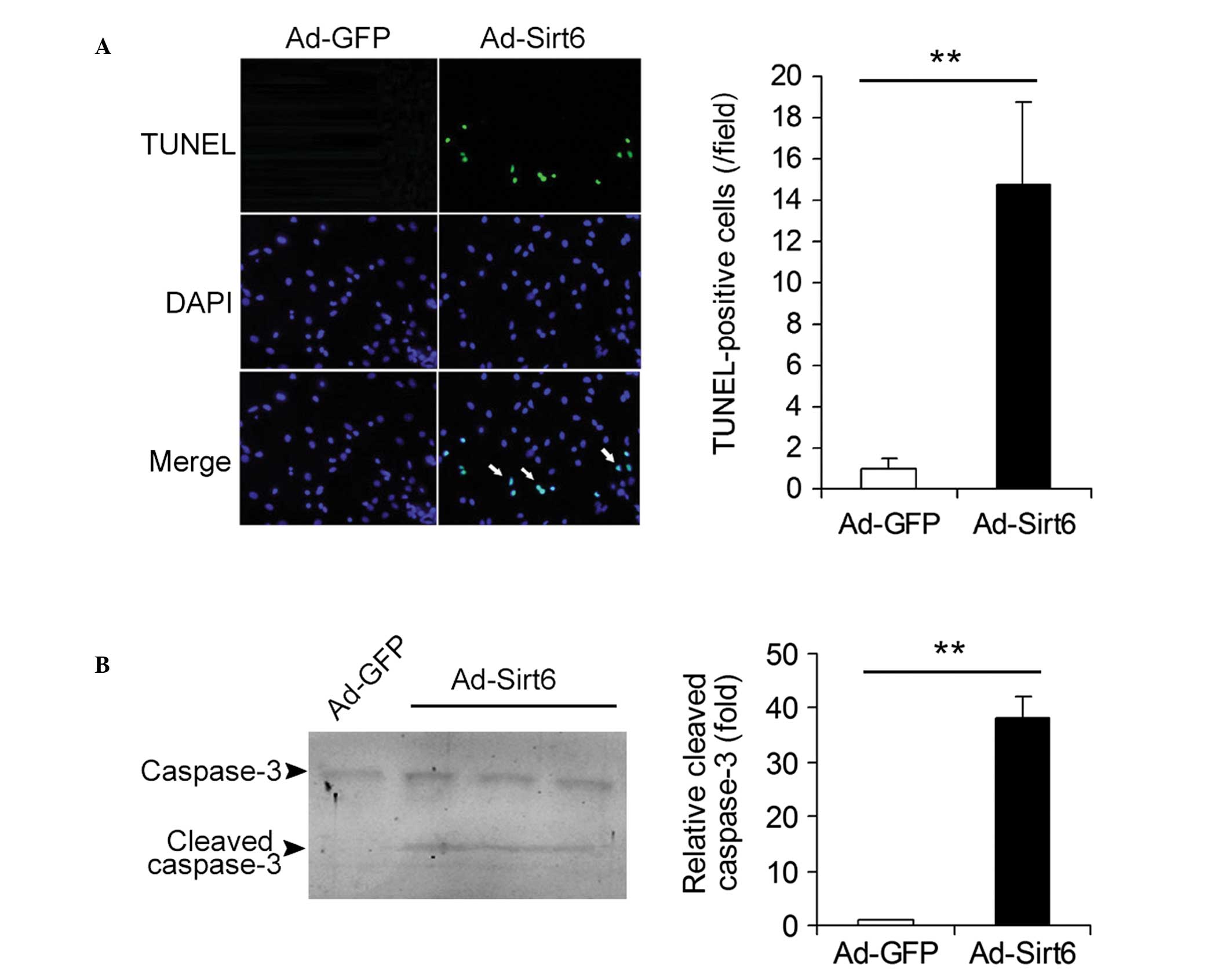
Overexpression of Sirt6 induces apoptosis
in HepG2 HCC cells
The effects of overexpression of Sirt6 on the
apoptosis of HCC cells were studied. Ad-GFP- and
Ad-Sirt6-transfected cells were analyzed using the TUNEL assay. The
apoptotic cells (TUNEL-positive cells, green) were detected in
Ad-Sirt6 cells but not in control Ad-GFP cells (Fig. 3A). The protein levels of cleaved
caspase-3, a key mediator and marker protein of apoptosis were also
analyzed. Cleaved caspase-3 expression was detected in cells
overexpressing Sirt6 but not in control cells (Fig. 3B). These results indicate that
overexpression of Sirt6 induces apoptosis in HepG2 HCC cells.
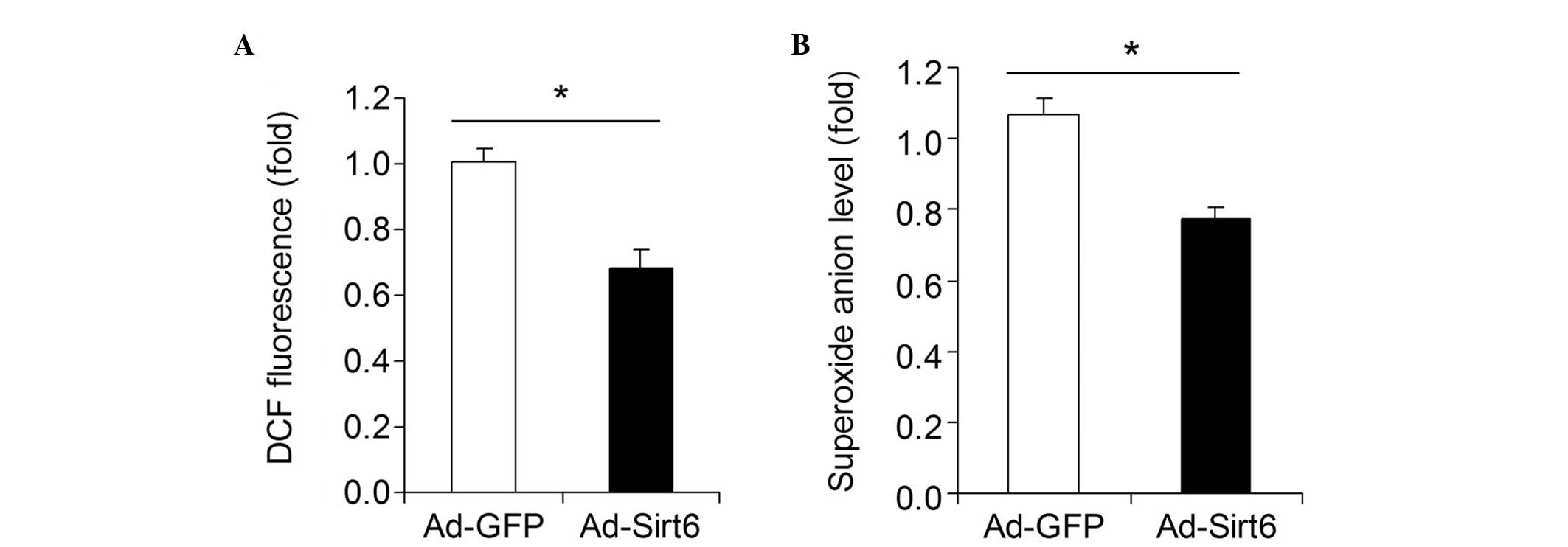
Overexpression of Sirt6 decreases
oxidative stress in HCC cells
The influence of Sirt6 overexpression on oxidative
stress in HepG2 HCC cells was investigated. A DCF assay showed that
overexpression of Sirt6 significantly decreased the total ROS level
in HepG2 cells (Fig. 4A).
Moreover, overexpression of Sirt6 downregulated the superoxide
anion level in HepG2 cells (Fig.
4B).
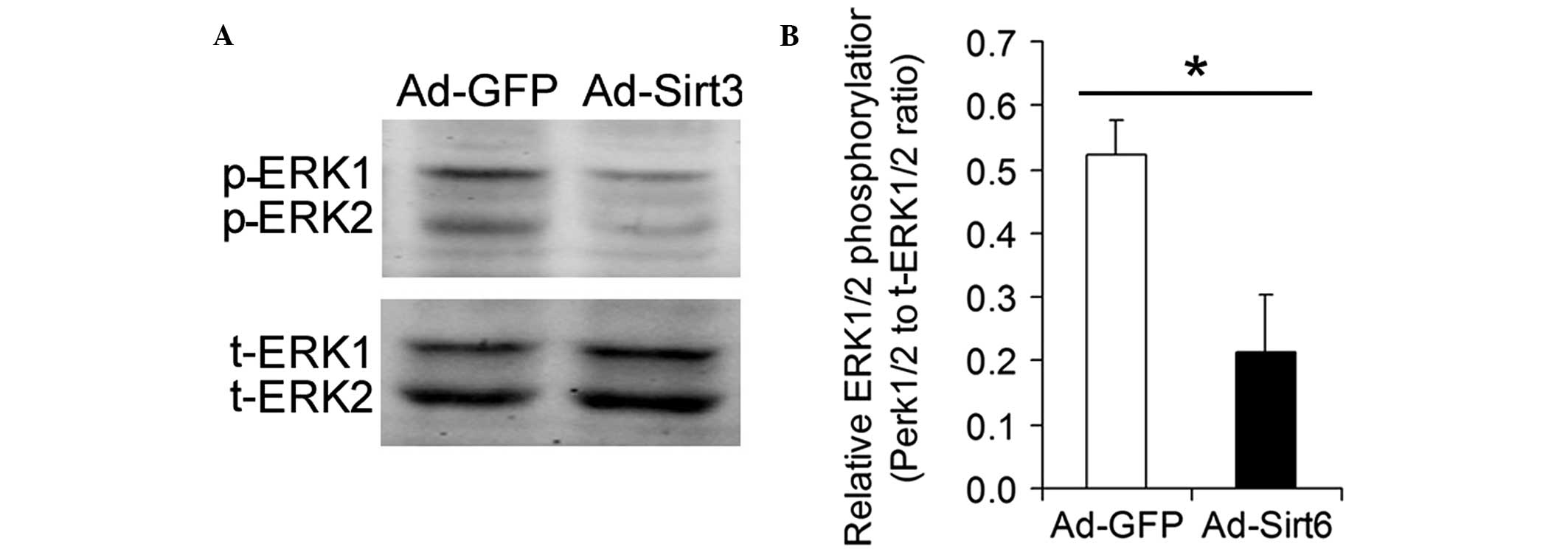
Overexpression of Sirt6 inhibits ERK1/2
phosphorylation in HCC cells
ERK1/2 is a major transducer of extracellular
mitogenic signals that promote cell proliferation. The influence of
the overexpression of Sirt6 on the phosphorylation of ERK1/2 was
analyzed. Overexpression of Sirt6 significantly inhibited ERK1/2
phosphorylation in HCC cells (Fig.
5).
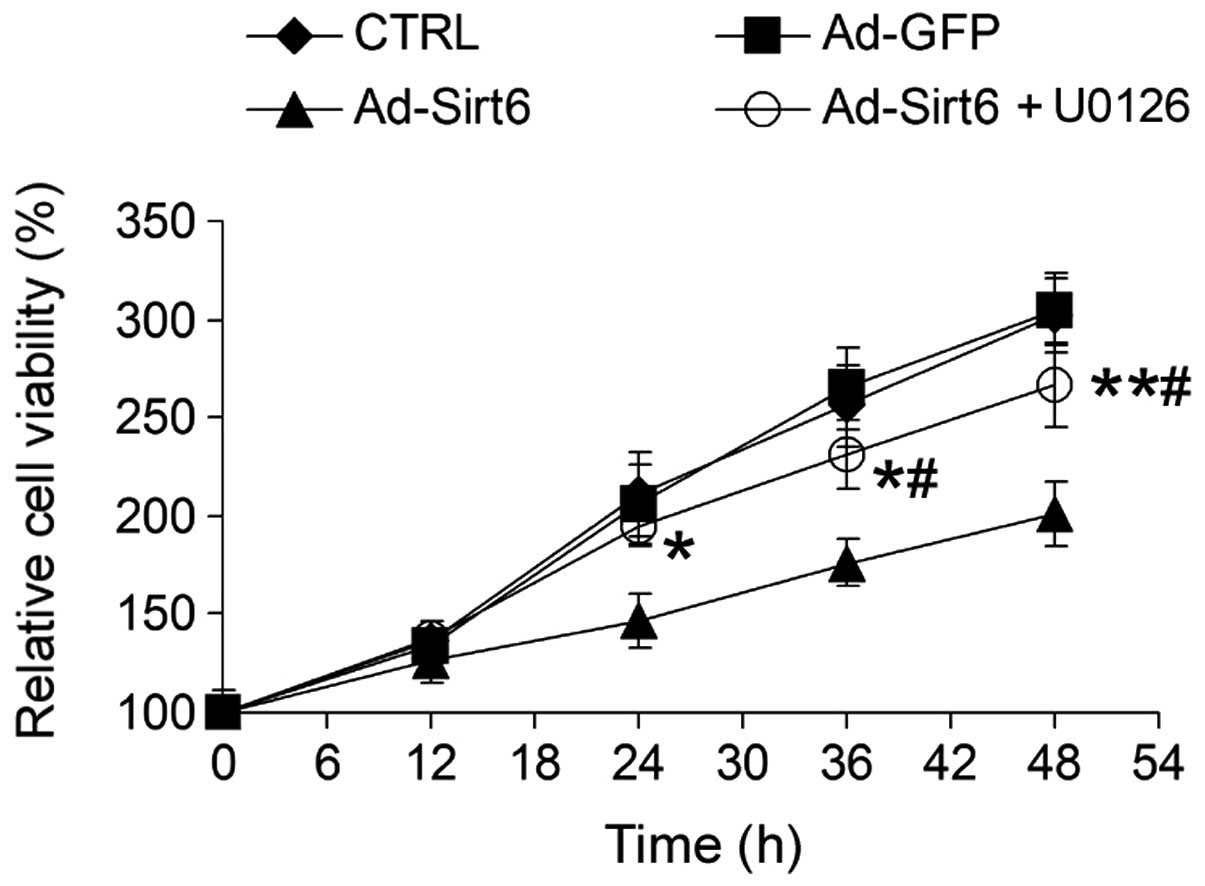
Sirt6 suppresses HCC cell growth via
regulation of the ERK1/2 signaling pathway
To examine whether the altered ERK1/2 signaling
pathway induced by Sirt6 overexpression was important for
Sirt6-mediated tumor suppression, HepG2 cells were treated with
U0126, a chemical inhibitor of the ERK1/2 pathway. U0126 markedly
attenuated the inhibitory effect of Sirt6 on HCC cell growth
(Fig. 6), suggesting that Sirt6
suppresses HCC cell growth via inhibition of the ERK1/2 signaling
pathway.
Discussion
In the present study, Sirt6 was markedly
downregulated in HCC tissue compared with normal adjacent tissue.
Using adenovirus-mediated knockdown and overexpression, modulation
of Sirt6 affected the growth of HepG2 HCC cells. Further analyses,
including a TUNEL assay and cleaved caspase-3 immunoblotting,
revealed that Sirt6 overexpression promoted apoptosis in HepG2 HCC
cells. In addition, Sirt6 overexpression decreased ROS/superoxide
anion levels in HepG2 HCC cells. Finally, Sirt6 overexpression was
found to inhibit phosphorylation of ERK1/2 in HepG2 HCC cells.
Blocking the ERK1/2 pathway with U0126 attenuated the inhibitory
effect of Sirt6 overexpression on HepG2 cell growth. These results
suggest that Sirt6 is a tumor suppressor in HCC cells.
Several previous studies have demonstrated a change
in Sirt6 expression in tumors. In a gene expression screening of
endometrial carcinoma samples, Colas et al (30) revealed that Sirt6 was marginally
upregulated. By contrast, in a recent study, Sebastián et al
(18) reported that the Sirt6
locus was deleted in 35% of ~1,000 cancer cell lines and in 62.5%
and 29% of pancreatic and colorectal cancer cell lines
respectively. Moreover, this group demonstrated that Sirt6
expression was downregulated in 36 pancreatic ductal
adenocarcinomas and in 55 colorectal carcinomas. In head and neck
squamous cell carcinomas, Lai et al (31) confirmed that Sirt6 was
downregulated in cancerous tissues when compared with noncancerous
tissues. To the best of our knowledge, the present study provides
the first evidence of Sirt6 downregulation in human HCC tissue. A
report showed impaired Sirt6 expression in c-Jun-deficient livers
during tumor initiation in mice and Sirt6 expression in human HCC
tissue (17); however, whether
Sirt6 expression was different in human HCC tissues compared to
normal tissues was not investigated in this study.
In the present study, shRNA-mediated knockdown of
Sirt6 was identified to promote HepG2 cell growth, which was
consistent with a previous report (18). The tumor suppressive effect of
Sirt6 has also been demonstrated in cervical carcinoma,
fibrosarcoma, primary breast tumor and metastatic breast tumor cell
lines (32). In concordance with
these results, the results of the present study provide evidence
for the tumor suppressive effect of Sirt6 in HCC cells. Notably,
Sirt6 overexpression appeared to induce apoptosis in a variety of
cancer cell lines but not in normal, non-transformed cells
(32). Additionally,
mono-ADP-ribosyltransferase, but not deacetylase activity, was
required for the tumor suppressive effect of Sirt6 (32).
As a member of mitogen-activated protein kinases,
ERK1/2 mediates intracellular signaling pathways involved in
proliferative functions, including meiosis, mitosis and postmitotic
differentiation in cells (33). A
number of different stimuli, including cytokines, growth factors,
transforming agents and carcinogens, activate the ERK1/2 pathway to
promote cell proliferation (33).
Activated ERK1/2 is an important feature in HCC and multiple
anticancer agents inhibit HCC cell growth via inhibition of ERK1/2
signaling (34). Previous studies
revealed that Sirt1 activated the ERK1/2 pathway (35–36),
whereas Sirt3 repressed the ERK1/2 signaling pathway (13). To the best of our knowledge, there
has been no report on the association between ERK1/2 and Sirt6.
Using immunoblotting, Sirt6 overexpression was found to inhibit the
phosphorylation of ERK1/2 in HCC cells. In addition, blocking the
ERK1/2 signaling pathway with the specific chemical inhibitor
U0126, markedly attenuated the tumor suppressive effect of Sirt6.
As ERK1/2 localizes to the cytoplasm and is translocated to the
nucleus following phosphorylation (33), it is not known how Sirt6, a nuclear
protein, regulates ERK1/2 phosphorylation. This is an important
question that needs to be addressed in future studies.
In conclusion, the present study demonstrated that
the expression of Sirt6 was decreased in human HCC tissue.
Overexpression of Sirt6 in the HepG2 HCC cell line exhibited
antitumor effects through the induction of apoptosis and the
inhibition of the ERK1/2 signaling pathway. These findings on the
regulation of HCC cell growth by Sirt6 may provide an improved
understanding of HCC and aid in the possible development of
therapeutic interventions.
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
3
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: worldwide incidence and trends.
Gastroenterology. 127(5 Suppl 1): S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moradpour D, Penin F and Rice CM:
Replication of hepatitis C virus. Nat Rev Microbiol. 5:453–463.
2007. View Article : Google Scholar
|
|
5
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kozyreva ON, Chi D, Clark JW, et al: A
multicenter retrospective study on clinical characteristics,
treatment patterns, and outcome in elderly patients with
hepatocellular carcinoma. Oncologist. 16:310–318. 2011. View Article : Google Scholar
|
|
7
|
Haigis MC and Guarente LP: Mammalian
sirtuins - emerging roles in physiology, aging, and calorie
restriction. Genes Dev. 20:2913–2921. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu T, Liu PY and Marshall GM: The
critical role of the class III histone deacetylase SIRT1 in cancer.
Cancer Res. 69:1702–1705. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alhazzazi TY, Kamarajan P, Verdin E and
Kapila YL: SIRT3 and cancer: tumor promoter or suppressor? Biochim
Biophys Acta. 1816:80–88. 2011.PubMed/NCBI
|
|
10
|
Hirschey MD, Shimazu T, Goetzman E, et al:
SIRT3 regulates mitochondrial fatty-acid oxidation by reversible
enzyme deacetylation. Nature. 464:121–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Finley LW, Carracedo A, Lee J, et al:
SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α
destabilization. Cancer Cell. 19:416–428. 2011.PubMed/NCBI
|
|
12
|
Alhazzazi TY, Kamarajan P, Joo N, et al:
Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral
cancer. Cancer. 117:1670–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang YY and Zhou LM: Sirt3 inhibits
hepatocellular carcinoma cell growth through reducing Mdm2-mediated
p53 degradation. Biochem Biophys Res Commun. 423:26–31. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhong L, D’Urso A, Toiber D, et al: The
histone deacetylase Sirt6 regulates glucose homeostasis via
Hif1alpha. Cell. 140:280–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mao Z, Hine C, Tian X, et al: SIRT6
promotes DNA repair under stress by activating PARP1. Science.
332:1443–1446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaidi A, Weinert BT, Choudhary C and
Jackson SP: Human SIRT6 promotes DNA end resection through CtIP
deacetylation. Science. 329:1348–1353. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Min L, Ji Y, Bakiri L, et al: Liver cancer
initiation is controlled by AP-1 through SIRT6-dependent inhibition
of survivin. Nat Cell Biol. 14:1203–1211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sebastián C, Zwaans BM, Silberman DM, et
al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012.PubMed/NCBI
|
|
19
|
Bauer I, Grozio A, Lasigliè D, et al: The
NAD+-dependent histone deacetylase SIRT6 promotes
cytokine production and migration in pancreatic cancer cells by
regulating Ca2+ responses. J Biol Chem. 287:40924–40937.
2012.PubMed/NCBI
|
|
20
|
Khongkow M, Olmos Y, Gong C, et al: SIRT6
modulates paclitaxel and epirubicin resistance and survival in
breast cancer. Carcinogenesis. 34:1476–1486. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang P, Guan YF, Du H, Zhai QW, Su DF and
Miao CY: Induction of autophagy contributes to the neuroprotection
of nicotinamide phosphoribosyltransferase in cerebral ischemia.
Autophagy. 8:77–87. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Filippi BM, Yang CS, Tang C and Lam TK:
Insulin activates Erk1/2 signaling in the dorsal vagal complex to
inhibit glucose production. Cell Metab. 16:500–510. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang P, Xu TY, Guan YF, Su DF, Fan GR and
Miao CY: Perivascular adipose tissue-derived visfatin is a vascular
smooth muscle cell growth factor: role of nicotinamide
mononucleotide. Cardiovasc Res. 81:370–380. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nezis IP, Shravage BV, Sagona AP, Johansen
T, Baehrecke EH and Stenmark H: Autophagy as a trigger for cell
death: autophagic degradation of inhibitor of apoptosis dBruce
controls DNA fragmentation during late oogenesis in Drosophila.
Autophagy. 6:1214–1215. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang P, Tian WW, Song J, Guan YF and Miao
CY: Deficiency of NG2+ cells contributes to the
susceptibility of stroke-prone spontaneously hypertensive rats. CNS
Neurosci Ther. 17:327–332. 2011.
|
|
26
|
Wang P, Xu TY, Guan YF, et al:
Nicotinamide phosphoribosyltransferase protects against ischemic
stroke through SIRT1-dependent adenosine monophosphate-activated
kinase pathway. Ann Neurol. 69:360–374. 2011. View Article : Google Scholar
|
|
27
|
Wang P, Yang FJ, Du H, et al: Involvement
of leptin receptor long isoform (LepRb)-STAT3 signaling pathway in
brain fat mass- and obesity-associated (FTO) downregulation during
energy restriction. Mol Med. 17:523–532. 2011.PubMed/NCBI
|
|
28
|
Wang P, Zhang RY, Song J, et al: Loss of
AMP-activated protein kinase-α2 impairs the insulin-sensitizing
effect of calorie restriction in skeletal muscle. Diabetes.
61:1051–1061. 2012.
|
|
29
|
Devarajan A, Grijalva VR, Bourquard N, et
al: Macrophage paraoxonase 2 regulates calcium homeostasis and cell
survival under endoplasmic reticulum stress conditions and is
sufficient to prevent the development of aggravated atherosclerosis
in paraoxonase 2 deficiency/apoE−/− mice on a Western diet. Mol
Genet Metab. 107:416–427. 2012.PubMed/NCBI
|
|
30
|
Colas E, Perez C, Cabrera S, et al:
Molecular markers of endometrial carcinoma detected in uterine
aspirates. Int J Cancer. 129:2435–2444. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lai CC, Lin PM, Lin SF, et al: Altered
expression of SIRT gene family in head and neck squamous cell
carcinoma. Tumour Biol. 34:1847–1854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Van Meter M, Mao Z, Gorbunova V and
Seluanov A: SIRT6 overexpression induces massive apoptosis in
cancer cells but not in normal cells. Cell Cycle. 10:3153–3158.
2011.PubMed/NCBI
|
|
33
|
Nishimoto S and Nishida E: MAPK
signalling: ERK5 versus ERK1/2. EMBO Rep. 7:782–786. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wiesenauer CA, Yip-Schneider MT, Wang Y
and Schmidt CM: Multiple anticancer effects of blocking MEK-ERK
signaling in hepatocellular carcinoma. J Am Coll Surg. 198:410–421.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Xu W, McBurney MW and Longo VD:
SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and
protects neurons. Cell Metab. 8:38–48. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Y, Luo P, Guo Q, et al: Interactions
between SIRT1 and MAPK/ERK regulate neuronal apoptosis induced by
traumatic brain injury in vitro and in vivo. Exp Neurol.
237:489–498. 2012. View Article : Google Scholar : PubMed/NCBI
|