Introduction
Immune system activation is involved in the
pathogenesis of numerous common complex diseases, including
cardiovascular disease, diabetes and cancer (1). Ongoing inflammatory insults may
contribute to the development of such diseases. Inflammatory
activation, a result of cellular exposure to stress conditions, is
reflected by increases in levels of circulating proinflammatory
cytokines (2). Sepsis is an
increasingly common cause of morbidity and mortality, particularly
in elderly, immunocompromised and critically ill patients (3). The most common cause of sepsis is an
exposure to the structural component of a gram-negative bacterial
membrane lipopolysaccharide (LPS), with key symptoms including
hypotension and vasoplegia, which may lead to multiple organ
dysfunction and, ultimately, mortality (4–6).
Bacterial LPS in the bloodstream induces the overexpression of
various inflammatory mediators, including interleukin-1 (IL-1),
tumor necrosis factor-α (TNF-α), and a large quantity of
inflammatory mediators produced in the body are hypothesized to
contribute to the LPS-induced symptoms of septic shock and
mortality (7).
Selenoprotein Sl (SEPS1) has previously been
identified as an endoplasmic reticulum stress response protein that
is likely to correlate with an inflammatory response (8–10).
Genetic variation in the SEPS1 gene was observed to correlate with
circulating levels of pro-inflammatory cytokines in human
populations and SEPS1 may regulate cytokine production in cultured
macrophage cells (11,12). The protective effect of SEPS1 on
mice with sepsis is unknown. In order to investigate the role of
SEPS1 in mice with sepsis, a LPS-induced sepsis model was used in
the present study. SEPS1 small interfering RNA (siRNA) was used to
silence the SEPS1 gene. The associated biochemical indicators as
well as gene and protein expression were examined.
Materials and methods
Reagents
SEPS1, p38 mitogen-activated protein kinase (p38
MAPK), phosphorylated p38 mitogen-activated protein kinase (p-p38
MAPK) and β-actin antibodies were obtained from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). Oligonucleotide primers
and dNTP mix were purchased from Bio Basic Inc. (Bio Basic,
Toronto, Canada). Protein extraction and BCA protein assay kits
were purchased from Promega Co. (Madison, WI, USA). Pyrogen-free
water was purchased from Shanghai Yihua Clinical Medicine
Technologies Company (Shanghai, China). LPS (from E. coli
strain O55:B5) and all other chemicals were purchased from
Sigma-Aldrich (St. Louis, MO, USA).
Animals
Inbred BALB/c mice (male, 17–20 g), were purchased
from the Institute of Medical Animal Experimental Center, Peking
Union Medical College (Beijing, China). The mice were kept at
standard laboratory conditions of temperature and humidity with a
12-h light/dark cycle. All experiments were performed according to
the Guide for the Care and Use of Laboratory Animals (Institute of
Laboratory Animal Resources, 1996) and were treated ethically. The
study was approved by the Ethics Committee of PLA General Hospital
(Beijing, China).
Animal transfection
siRNAs for SEPS1 and the scrambled control were
synthesized in vitro using a kit from Ambion (Austin, TX,
USA; Silencer® Negative Control No. 1 siRNA). Primer
sequences are provided in Table I
(Beijing AuGCT DNA-SYN Biotechnology Co., Ltd., Beijing, China).
The mice were transfected with siRNA oligonucleotides (0.5 μg)
formulated by Lipofector 2000 (Beyotime Institute of Biotechnology,
Jiangsu, China) via tail vein injection. In total, 30 mice were
randomly assigned to three groups: i) H group (LPS-induced sepsis
group; n=10): Mice with intraperitoneal injection of LPS (10
mg/kg); ii) K group (scrambled siRNA group; n=10): Mice transfected
with scrambled control siRNA 12 h prior to injection with LPS; iii)
L group (SEPS1 siRNA group; n=10): Mice transfected with SEPS1
siRNA 12 h prior to injection with LPS. The mice in all groups
fasted with free access to water following LPS injection and were
continuously monitored over a 24-h period. For each time-point (12
and 24 h), five animals were used.
 | Table IPrimer sequences of SEPS1 and
scrambles. |
Table I
Primer sequences of SEPS1 and
scrambles.
| Primer name | Primer sequence
5′-3′ |
|---|
| SEPS1 antisense |
AAGATCTAAATGCCCAAGTTGCCTGTCTC |
| SEPS1 sense |
AACAACTTGGGCATTTAGATCCCTGTCTC |
| Scrambled
antisense |
AAGTATCTAGGTACACACTCACCTGTCTC |
| Scrambled sense |
AATGAGTGTACCTAGATACCCTGTCTC |
Assessment of blood biochemical
parameters
Following the LPS injections at 12 and 24 h, the
blood was collected in heparinized tubes by extirpating the left
eyeball of five mice in each group. Serum was isolated by
centrifugation at 4,000 × g, 4°C for 15 min and maintained at −70°C
until it was required for further analysis. The levels of serum
alanine transaminase (ALT), aspartate aminotransferase (AST), serum
creatinine (Cr), blood urea nitrogen (BUN), myocardial kinase
(CK-MB), creatine kinase (CK) and lactic dehydrogenase (LDH) were
assessed using an automatic biochemical analyzer (HITACHI 7170,
Hitachi, Ltd., Tokyo, Japan) with commercial kits (LDH-Cytotoxicity
Assay Kit II; Biosino Biotechnology Co., Ltd., Beijing, China).
Assessment of TNF-α and IL-6 in the liver
homogenate
At each time-point of assessment, the mice were
sacrificed once the blood had been obtained. Under strict aseptic
conditions, 0.1–0.2 g of liver tissue was removed and placed into a
pyrogen-free homogenizer. Three volume equivalents of pyrogen-free
saline was added and the liver tissue was homogenized in an ice
bath. The liver homogenate supernatant was prepared by
centrifugation at 10,000 × g at 4°C for 15 min and maintained at
−30°C until it was required for further analysis. The samples were
analyzed for TNF-α and IL-6 using an ELISA kit (Endogen Inc.,
Woburn, MA, USA) according to the manufacturer’s instructions. The
absorbance was measured at 450 nm. The concentrations of TNF-α and
IL-6 in the experimental samples were extrapolated from a standard
curve.
Western blot analysis
Once the mice had been sacrificed, the liver tissue
samples were maintained in liquid nitrogen for western blot
analysis. The liver tissue samples were cut up and ground with
pre-cooled lysis buffer. Equal quantities of protein were separated
by SDS-PAGE (10% gel) and transferred to nitrocellulose (CN)
membranes (Hybond Inc., Escondido, CA, USA). The membranes were
blocked for 1 h at room temperature with 10% non-fat dry milk and
subsequently incubated at 4°C overnight with anti-mouse antibody
(1:300 dilution in blocking buffer). Once the membranes had been
washed, they were incubated with a 1:1,000 dilution of horseradish
peroxidase-conjugated anti-rabbit immunoglobulin G secondary
antibody (ZSGB-Bio Co., Beijing, China) for 1 h at room
temperature. The blots were visualized using an Enhanced
Chemiluminescence kit (Pierce Biotechnology Inc., Rockford, IL,
USA), and data were quantified using the Gel Doc EQ system
(Bio-Rad, Hercules, CA, USA)
RNA extractions and quantitative
polymerase chain reaction (qPCR)
Total cellular RNA was extracted with
TRIzol® reagent (Takara, Seoul, Korea) according to the
manufacturer’s instructions. Reverse transcription was performed at
37°C for 1 h in a reaction mixture containing 2 mg total RNA, 0.5
mg oligo-dT primer, 10 mM dNTPs, 25 U RNase inhibitor and 200 U
M-MLV reverse transcriptase (Promega). qPCR was performed using a
SYBR FAST qPCR kit (KAPA, Boston, Massachusetts, USA) with 10 ng
cDNA according to the manufacturer’s instructions. Cycling
conditions were as follows: 95°C for 1.5 min and 40 subsequent
cycles of 95°C for 3 sec, 56.4°C for 40 sec and 72°C for 10 sec.
Upon the completion of 40 PCR amplification cycles, melting curve
analysis was performed. SEPS1 mRNA levels were normalized to actin,
a stable housekeeping gene.
Immunohistochemical detection
Liver and lung tissue samples were deparaffinized
three times with xylene for 5 min each, followed by rehydration
with a series of absolute, 70% and 50% ethanol for three min each
and washed under running tap water for 5 min. Tissue samples were
subsequently blocked with 3% hydrogen peroxide for 5 min, placed in
distilled water for 5 min, followed by 10 min incubation with a
1:20 dilution of goat serum. Subsequently, tissue samples were
incubated with anti-SEPS1 polyclonal serum at 4°C overnight and
washed five times with phosphate-buffered saline and Tween 20
(PBST), for two min each. Tissue samples were then incubated with a
1:1,000 dilution of horseradish peroxidase-conjugated antibody
(Sigma-Aldrich) for 30 min and washed again with PBST. Following
washing, the samples were developed with 3,3′-diaminobenzidine
substrate solution for 3 min and washed again with PBST. Finally,
the tissue samples were counterstained with Harris’ hematoxylin
(Sigma-Aldrich) for 1 min, followed by washing, differentiation
with ethanol containing 1% acid alcohol solution, bluing with
ammonia in water, another washing step, dehydration with increasing
series of alcohols, clearance with xylene and subsequent mounting
with dibutyl phthalate xylene (DPX).
Pathological examination
Processed liver and lung tissue samples were
deparaffinized three times with xylene for two min each, rehydrated
with a series of absolute, 95% and 80% ethanol for two min each,
followed by a wash with running tap water for 5 min. Subsequently,
the tissue samples were stained with Harris’ haematoxylin
(Sigma-Aldrich) for 5 min and washed with running tap water.
Differentiation with 1% acid alcohol solution was performed for 10
sec, followed by washing and bluing by placing the tissue samples
into a solution of ammonia in water for 10 sec. Following a washing
step, the samples were counterstained with eosin Y (Sigma-Aldrich)
for two min, dehydrated with an increasing series of ethanol for
two min each, cleared by three washes with xylene for two min each
and finally mounted with DPX.
Statistical analysis
Values are expressed as the mean ± standard
deviation from a minimum of three independent experiments. The
statistical significance of the results between each treated group
was analyzed by Student’s t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
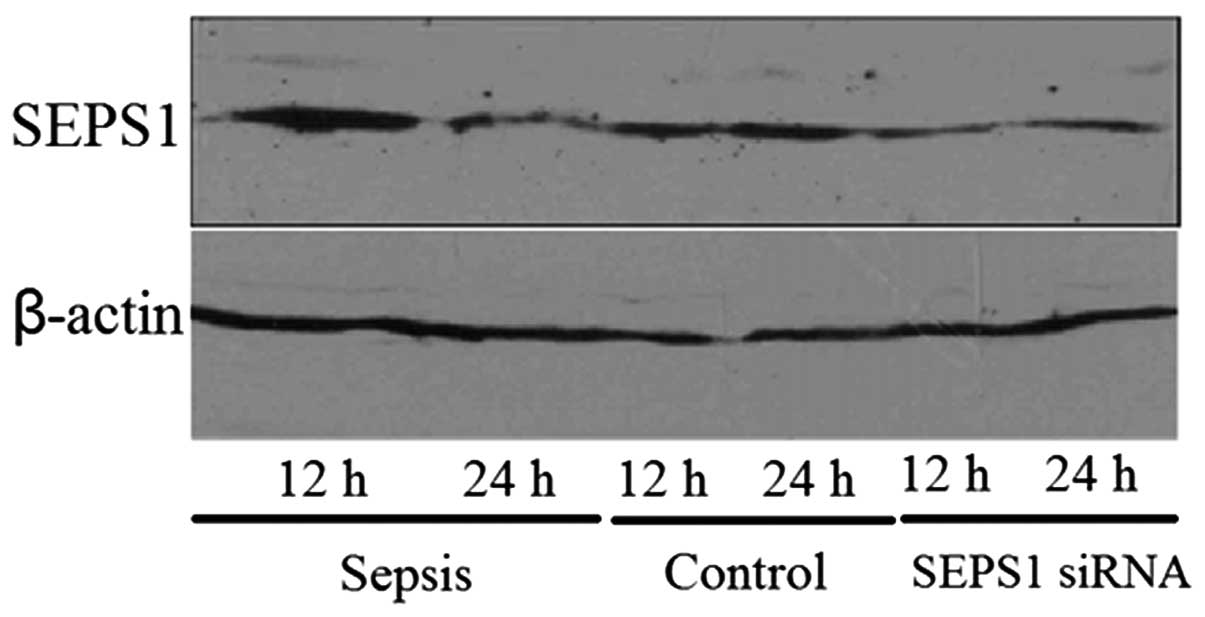
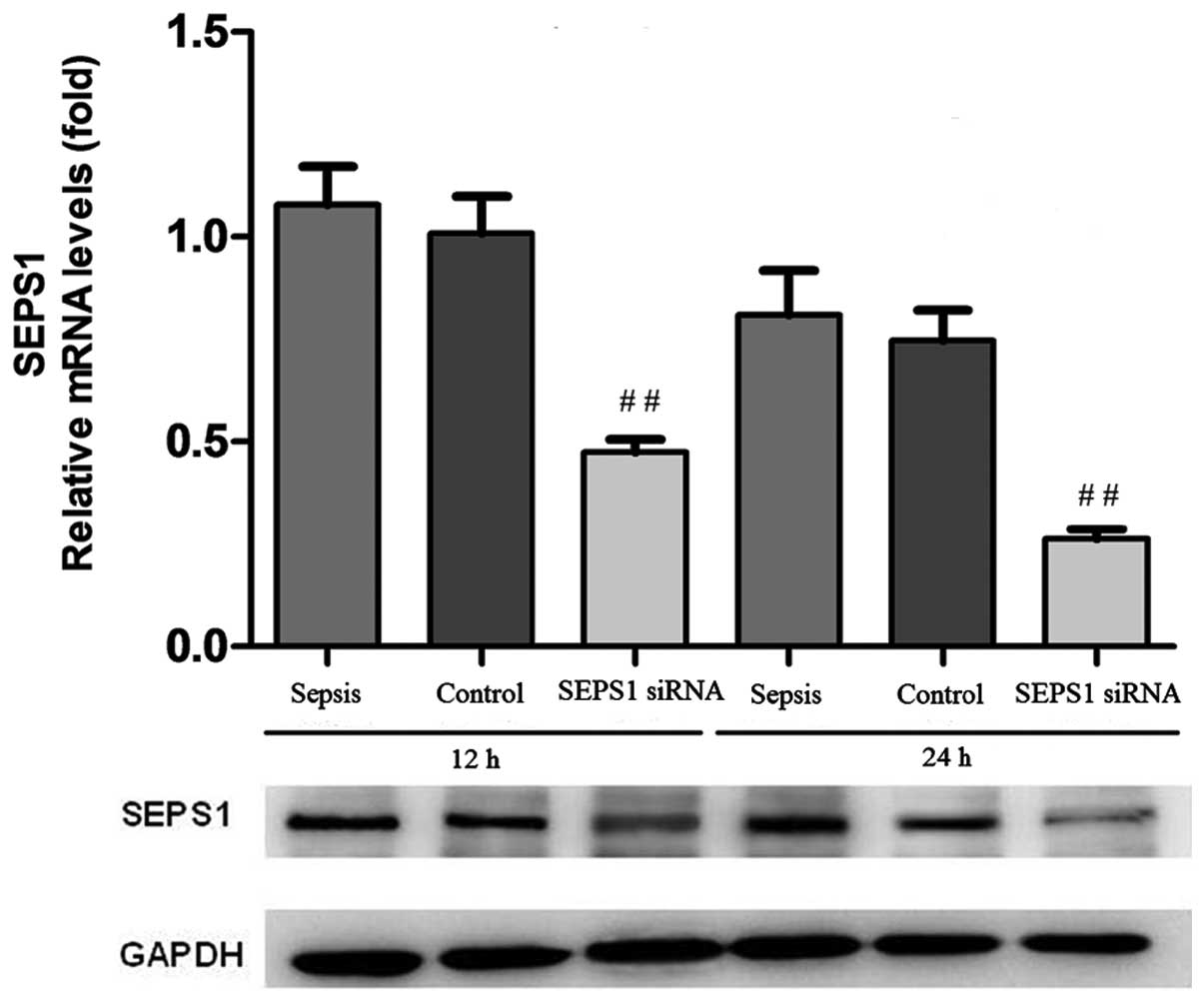
Confirmation of SEPS1 silencing
In order to examine whether the SEPS1 gene was
silenced, the expression of the SEPS1 gene was detected by qPCR and
western blot analysis. SEPS1 protein expression in liver tissue
decreased 12 and 24 h following transfection with SEPS1 siRNA
(Fig. 1). The results of the
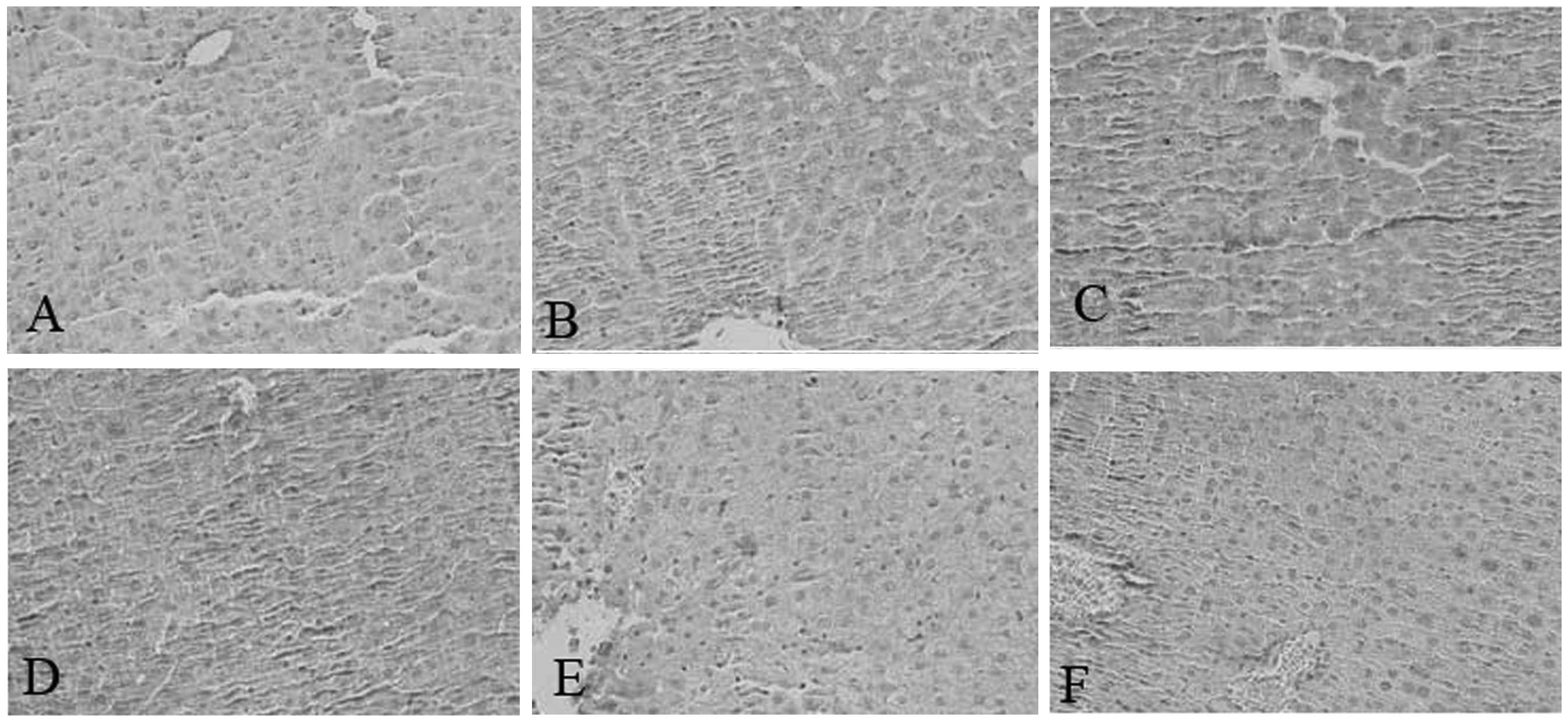
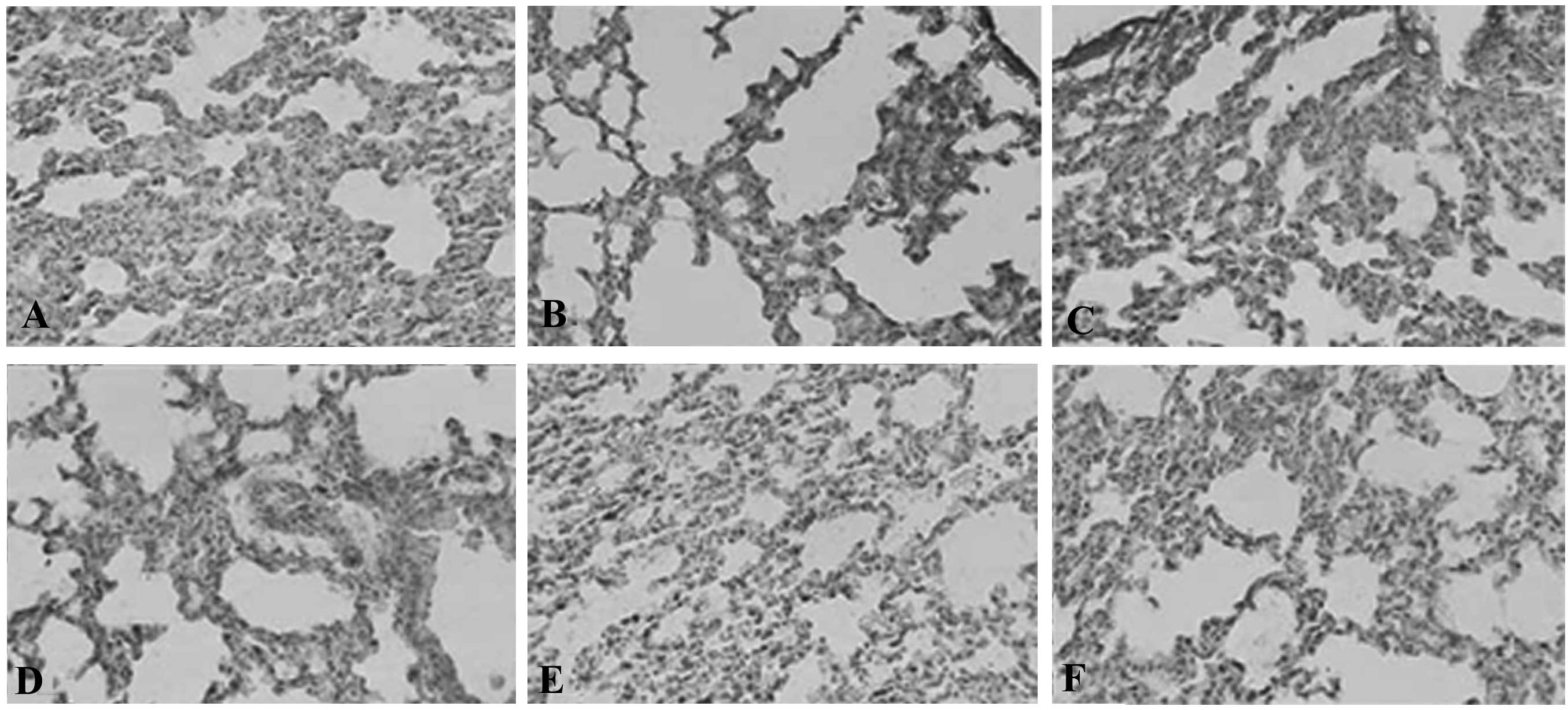
immunohistochemical analysis also revealed that SEPS1 protein
expression in liver and lung tissue decreased in the SEPS1 siRNA
group (Figs. 2 and 3). qPCR results demonstrated that SEPS1
gene expression in liver tissue decreased significantly compared
with the control group (Fig.
4).
SEPS1 silencing in mice with sepsis
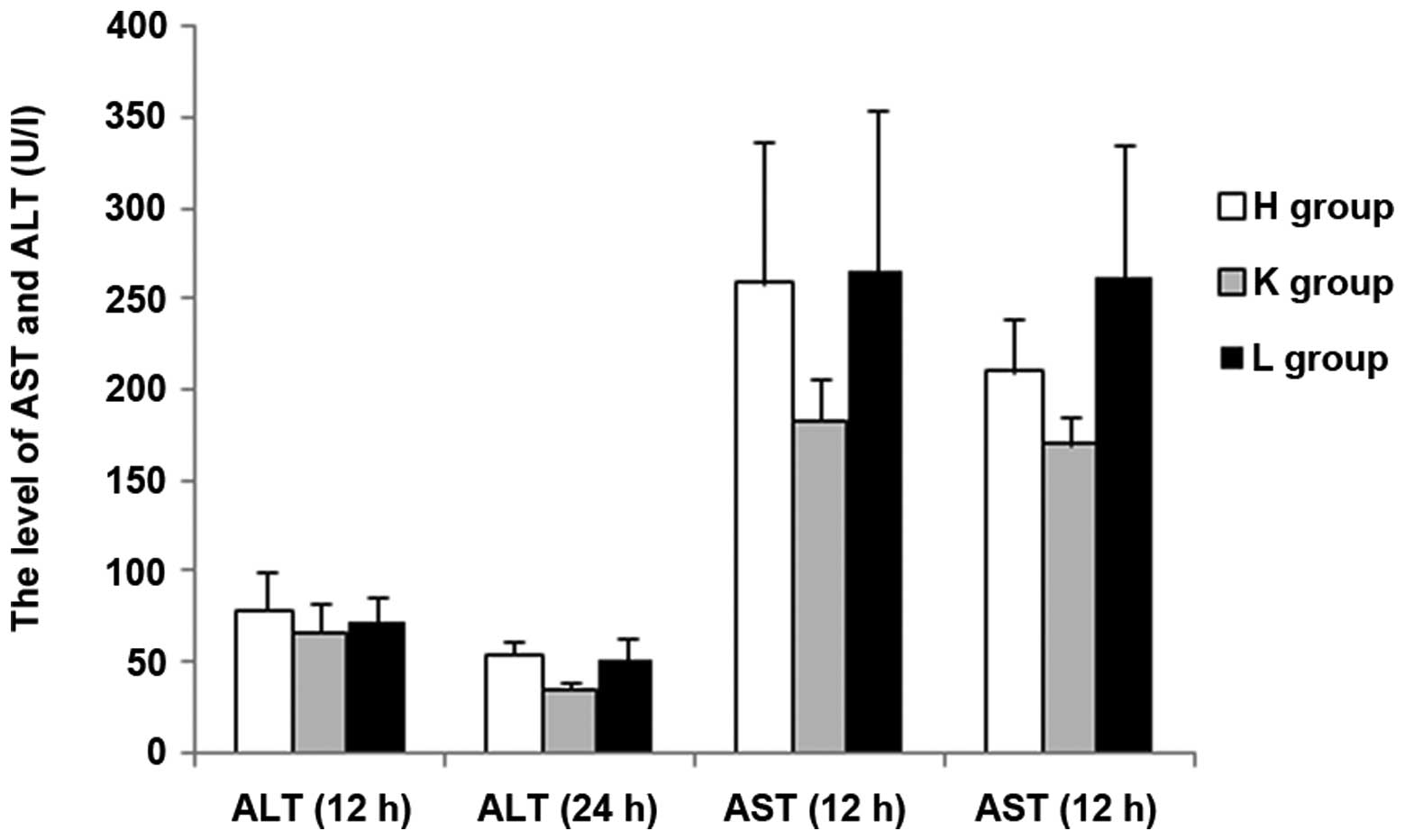
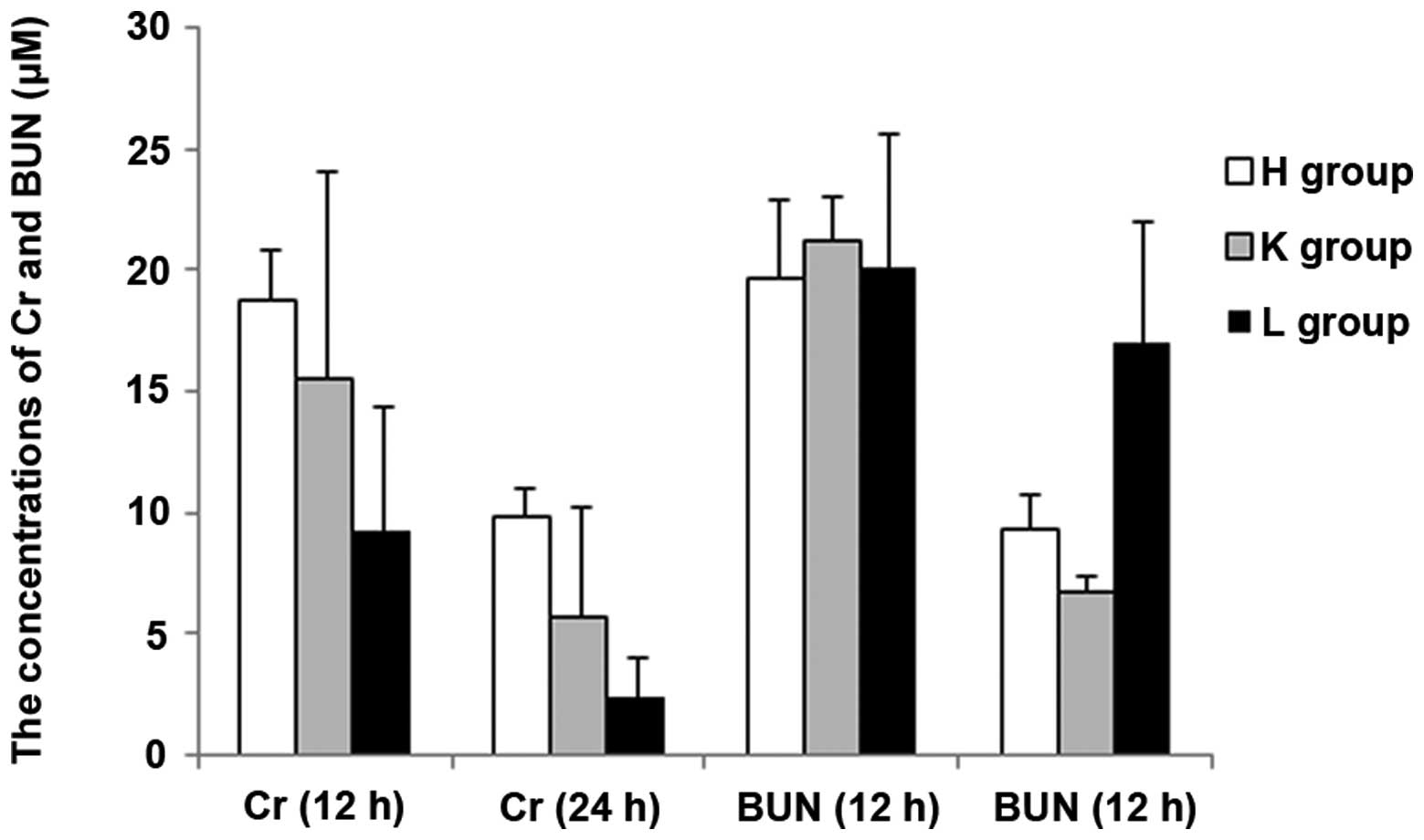
increases ALT, AST, BUN, LDH, CK and CK-MB levels
In the present study, important biochemical
indicators were detected in order to reflect the effects on the
liver, kidney and heart following siRNA silencing of the SEPS1
gene. At 12 h following injection with LPS, the concentrations of
ALT and BUN exhibited no statistical difference among the three
groups (P>0.05); AST and LDH levels exhibited significant
differences between the groups except for a non-statistical
difference between the H and L groups (P>0.05); the Cr levels
showed no statistical difference among the groups except for a
significant difference between the H and L groups (P<0.05); CK
and CK-MB concentrations exhibited no statistical difference among
the groups except for a significant difference between the K and L
groups (P<0.05). At 24 h following injection with LPS, the
concentration of ALT was significantly higher in the H group than
in the other two groups (P<0.05), and demonstrated no
statistical difference between the K and L groups; there were
significant differences among the three groups in the
concentrations of AST and BUN (P<0.05); there was no significant
difference among the three groups in LDH levels (P>0.05); the
statistical results of difference in Cr, CK and CK-MB
concentrations among the three groups at 24 h were the same as that
at 12 h. There was a significant increase in the ALT levels and a
significant decrease in the Cr levels at 24 h compared with 12 h
among the groups (P<0.05). Furthermore, there was a significant
increase in the AST levels and a significant decrease in the BUN
levels at 24 h compared with 12 h in the H and K groups
(P<0.05); however, the difference in group L was not
significant. There was no significant difference in CK, LDH and
CK-MB concentrations at 24 h compared with 12 h among the groups
(P<0.05) (Figs. 5, 6 and 7).
The results demonstrated that the blood levels of ALT, AST, BUN,
CK-MB, CK and LDH all increased in the SEPS1 siRNA group following
silencing of the SEPS1 gene with SEPS1 siRNA (Tables II and III).
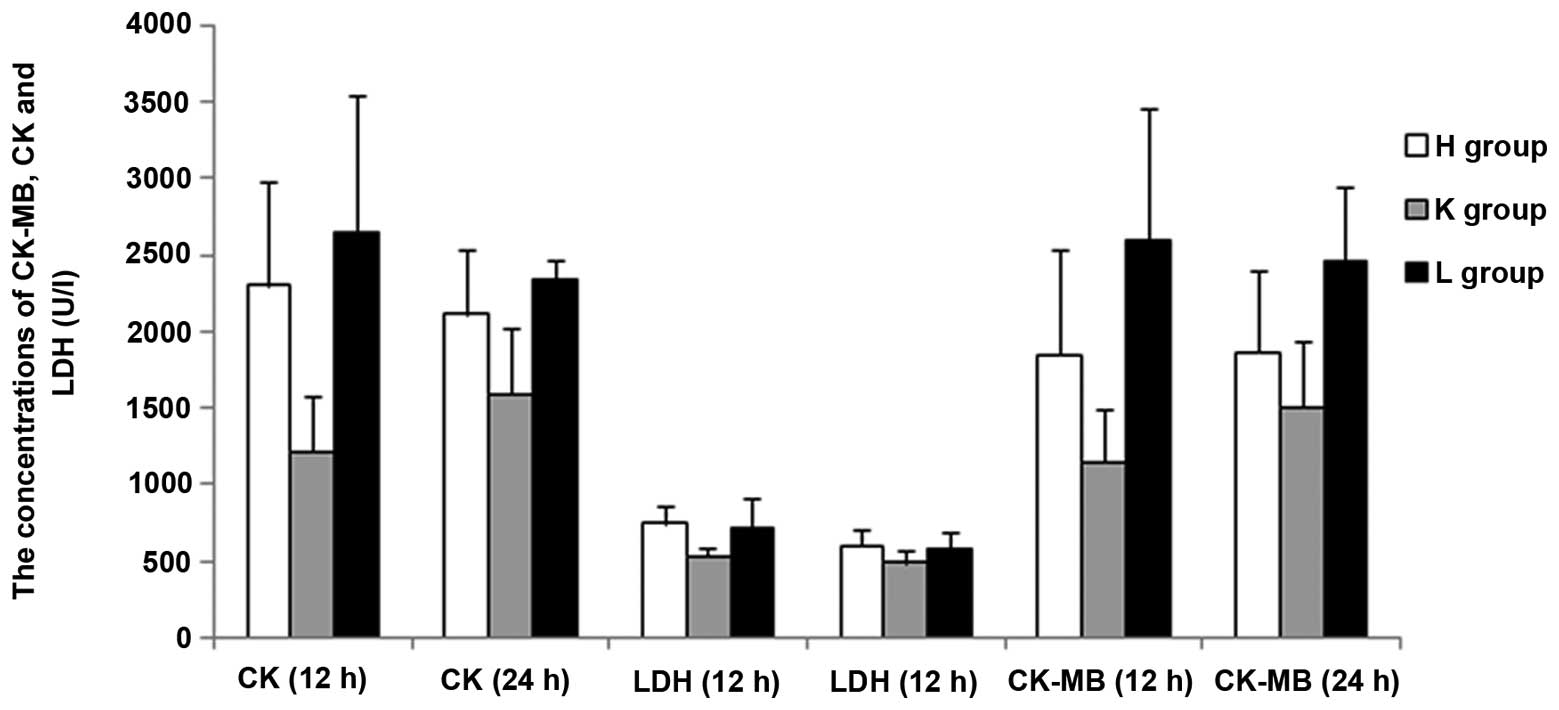 | Figure 7The level of CK-MB, CK and LDH at 24 h
compared with 12 h in the H, K and L groups. H group, (LPS-induced
sepsis group; n=10): Mice with intraperitoneal injection of LPS (10
mg/kg); K group, (scrambled siRNA group; n=10): Mice transfected
with scrambled control siRNA 12 h prior to injection with LPS; L
group, (SEPS1 siRNA group; n=10): Mice transfected with SEPS1 siRNA
12 h prior to injection with LPS; CK-MB, myocardial kinase; Cr,
creatinine; LDH, lactic dehydrogenase; SEPS1, selenoprotein S;
siRNA, small interfering RNA. |
 | Table IIEffect of SEPS1 siRNA on the levels of
ALT, AST and BUN (n=6). |
Table II
Effect of SEPS1 siRNA on the levels of
ALT, AST and BUN (n=6).
| ALT (U/l) | AST (U/l) | BUN (mmol/l) |
|---|
|
|
|
|
|---|
| Group | 12 h | 24 h | 12 h | 24 h | 12 h | 24 h |
|---|
| Sepsis | 66.7±15.2 | 34.4±4.1 | 19.7±3.2 | 9.3±1.5 | 182.2±23.7 | 169.7±15.9 |
| Control | 71.3±14.6 | 51.1±12.3 | 21.2±1.9 | 6.7±0.7 | 258.5±78.2 | 209.8±28.1 |
| SEPS1 siRNA | 77.8±22.1a | 53.5±7.3b | 20.0±5.7 | 16.9±5.1a,c | 264.8±89.3a | 261.0±73.4b |
 | Table IIIEffect of SEPS1 siRNA on the levels of
CK, LDH and CK-MB (n=5, U/l). |
Table III
Effect of SEPS1 siRNA on the levels of
CK, LDH and CK-MB (n=5, U/l).
| CK (U/l) | LDH (U/l) | CK-MB (U/l) |
|---|
|
|
|
|
|---|
| Group | 12 h | 24 h | 12 h | 24 h | 12 h | 24 h |
|---|
| Sepsis | 2302.9±1378.4 | 2118.4±923.5 | 743.6±123.7 | 604.8±114.7 | 1848.5±1383.0 | 1871.1±829.9 |
| Control | 1219.0±358.6 | 1593.7±439.7 | 535.2±52.4 | 488.7±90.1 | 1148.0±348.4 | 1509.2±425.6 |
| SEPS1 siRNA |
2657.6±882.9b |
2349.2±127.4a | 725.7±186.3a | 584.7±114.7 |
2602.2±856.6b |
2465.0±477.7b |
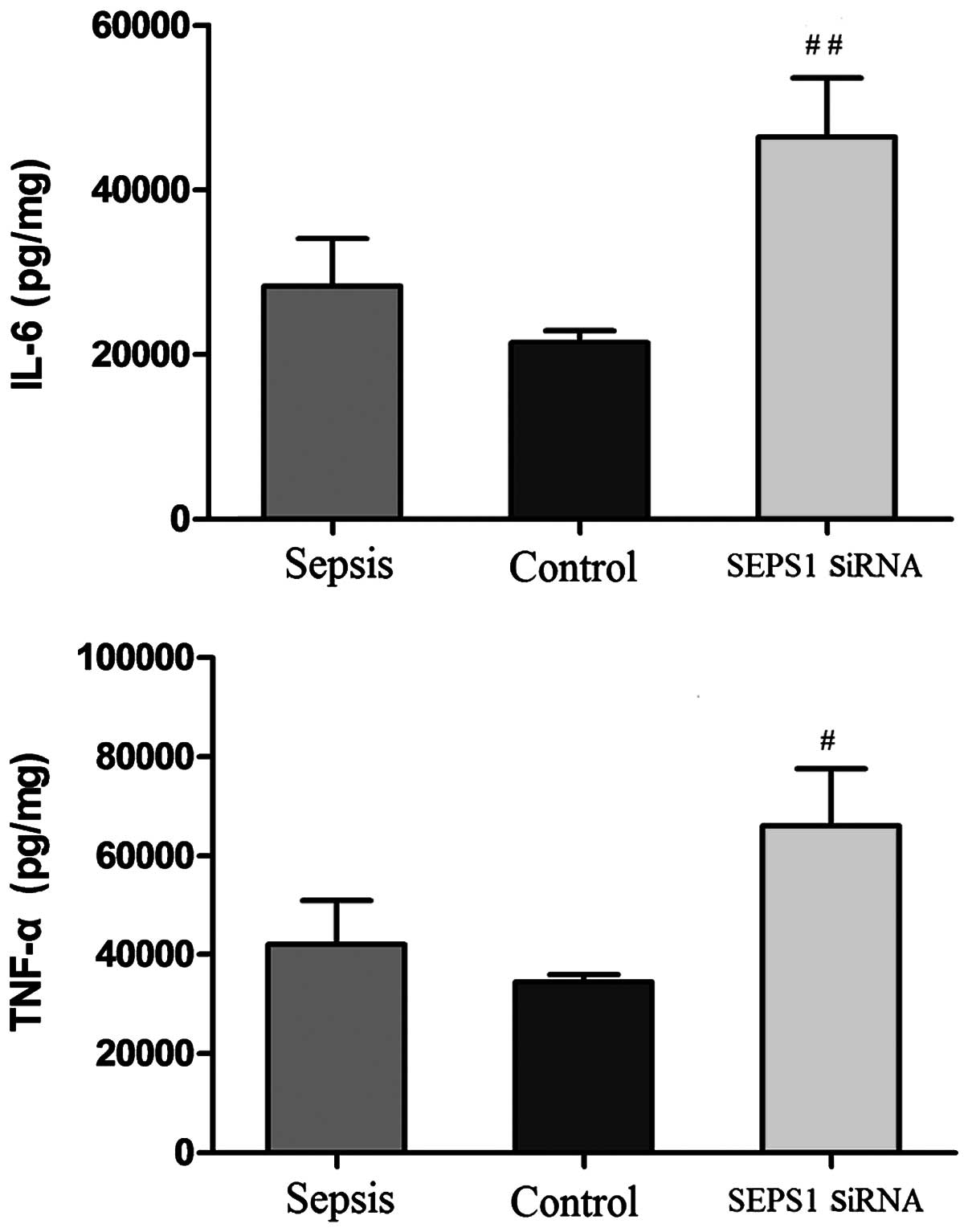
SEPS1 silencing in mice with sepsis
increases TNF-α and IL-6 levels
To investigate the proinflammatory cytokine levels
in mice with sepsis, the effect of SEPS1 siRNA on the circulating
levels of primary (TNF-α) and secondary (IL-6) cytokines was
examined. At 12 h following LPS injection, IL-6 and TNF-α levels
were highest in group K and lowest in group H. However, neither
IL-6 nor TNF-α levels exhibited a statistical difference among the
three groups (P>0.05). At 24 h, following LPS injection, IL-6
and TNF-α levels were highest in group L and lowest in group K.
There was a significant difference in IL-6 levels between all
groups except for the difference between groups H and K, whereas no
significant difference in TNF-α levels was present between the
groups except for a statistically significant difference between
groups K and L. There was a significant decrease in the IL-6 and
TNF-α levels at 24 h compared with 12 h in the K group (P<0.05).
However, there was no significant difference in IL-6 and TNF-α
levels at 24 h compared with 12 h in the other groups
(P>0.05)(Fig. 8).
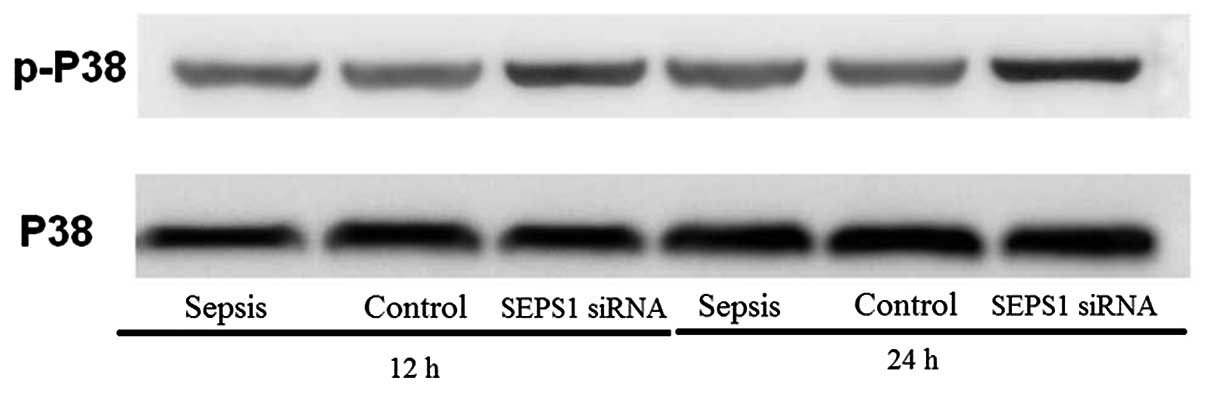
SEPS1 silencing in mice with sepsis
increases p38 MAPK phosphorylation
MAPKs are involved in signal transduction pathways
leading to the regulation of inflammatory mediators. In the present
study, the SEPS1 siRNA group enhanced p-p38 protein levels at 12
and 24 h following LPS stimulation (Fig. 9), suggesting that SEPS1 siRNA
treatment activated the phosphorylation of p38 MAPK.
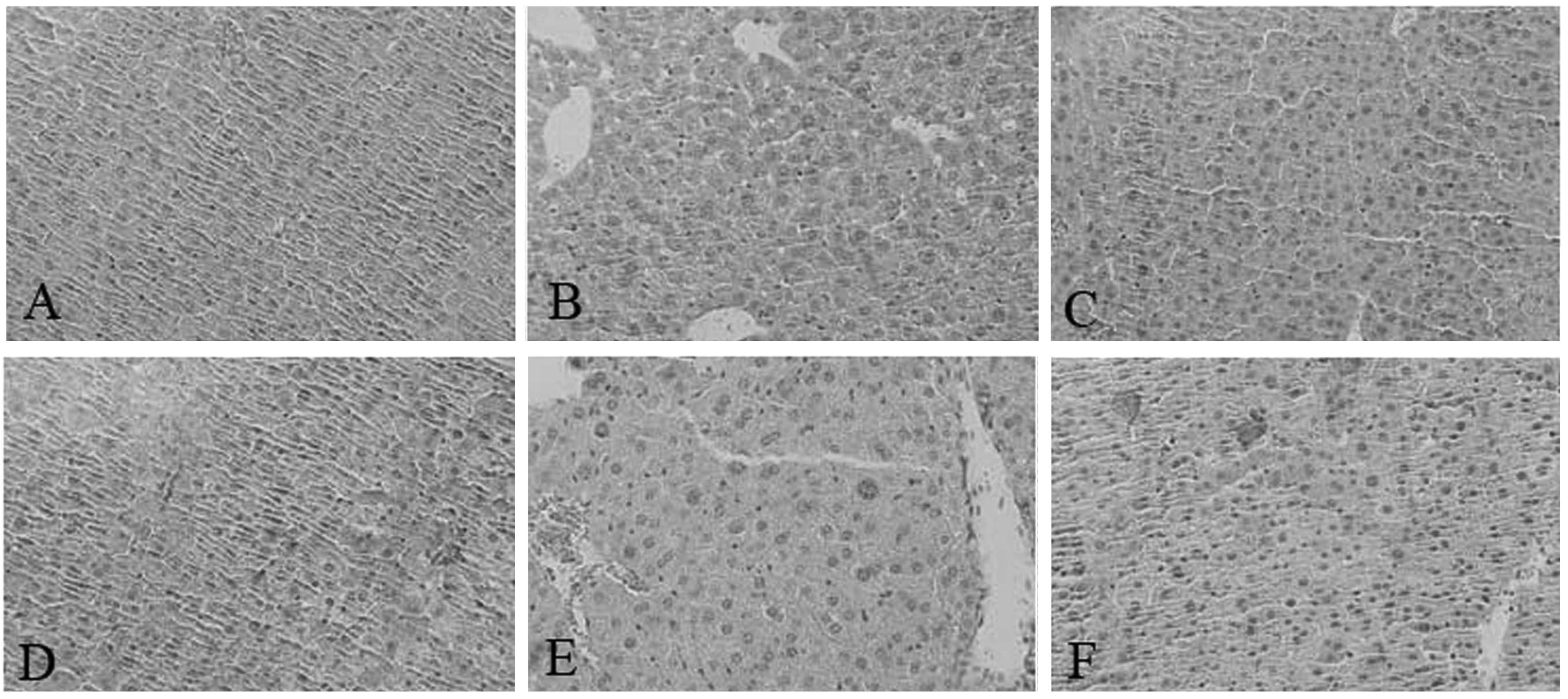
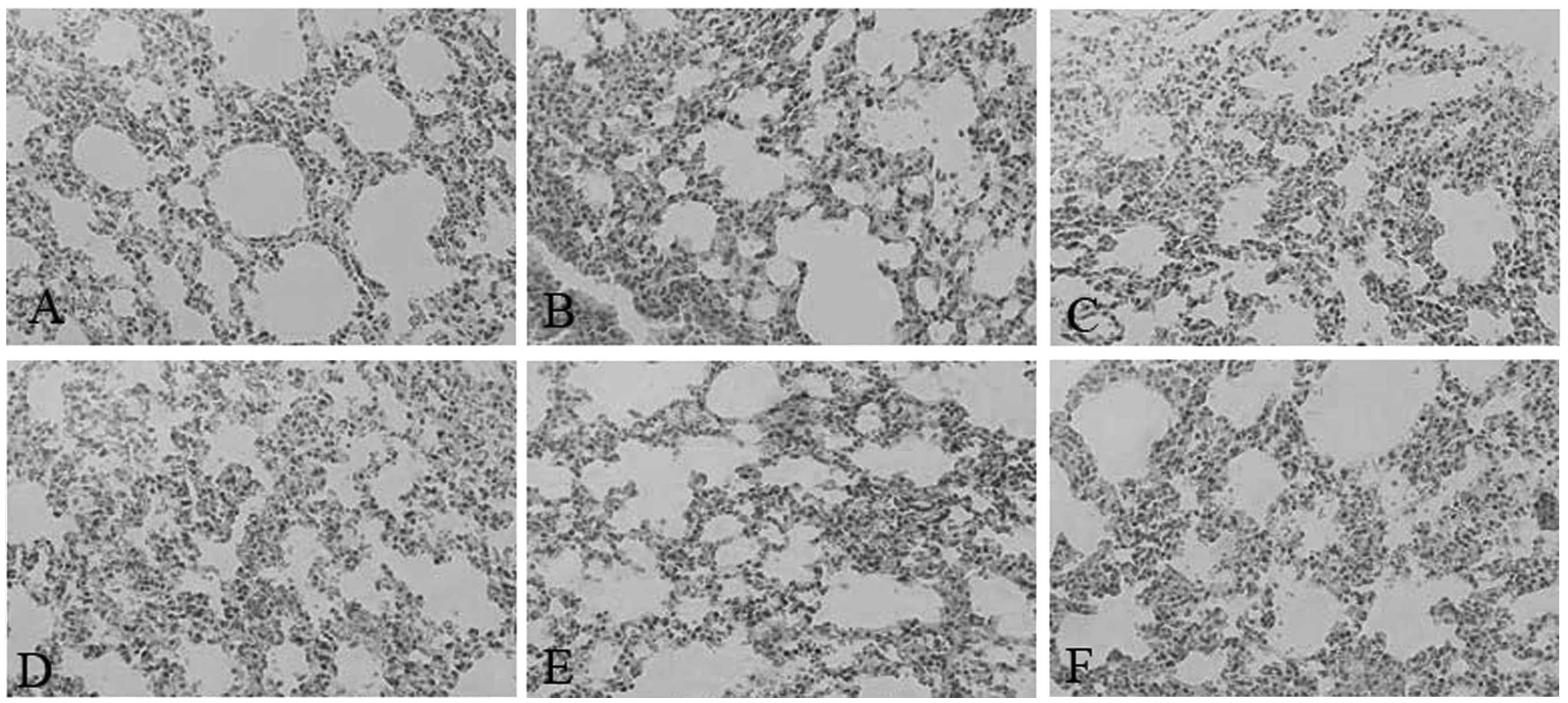
Sepsis in SEPS1-silenced mice causes
tissue damage
Pathological results demonstrated that the liver and
lung cells swelled significantly and the inflammatory cells
infiltrated the portal area, indicating that cell lesion markedly
occurred in the SEPS1 siRNA group compared with the control group,
which may correlate with the decrease in SEPS1 gene expression
(Figs. 10 and 11).
Discussion
SEPS1 is a newly identified member of the
selenoprotein family that contains enzymes, including thioredoxin
reductase and glutathione peroxidase (13). The human gene SEPS1 is located on
chromosome 15q26.3, consists of six exons and encodes a 189-amino
acid protein. This region of chromosome 15 was previously suggested
to contain quantitative trait loci that influence inflammatory
disorders, including insulin-dependent diabetes mellitus,
Alzheimer’s disease and celiac disease (14–17).
It has been observed that genetic variation in the SEPS1 gene
correlated strongly with circulating levels of pro-inflammatory
cytokines in human populations and that SEPS1 may regulate cytokine
production in cultured macrophage cells (11,18).
It has also been reported that cytokines have a direct impact on
SEPS1 levels (19). Thus, there
appears to be a regulatory loop whereby cytokines stimulate the
expression of SEPS1, which in turn suppresses cytokine production.
In order to clarify the importance of SEPS1 in multiple organ
dysfunction in mice with sepsis, siRNA was used to inhibit SEPS1
gene expression and observe the impact on organ function and
prognosis.
RNA interference (RNAi) is an RNA-dependent gene
silencing process controlled by the RNA-induced silencing complex
(RISC) and is initiated by short double-stranded RNA molecules in
the cell cytoplasm, where they interact with the catalytic RISC
component, argonaute. RNAi is widely used in the study of gene
function and biological genetic improvement. Synthetic
double-stranded (ds) siRNA in mammalian cells exerted a target
gene-silencing effect. siRNA is favored as a gene therapy drug due
to its specific efficiency in addition to its enormous potential.
Although it is difficult to introduce long dsRNA strands into
mammalian cells due to the interferon response, the use of siRNA
mimics has been more successful (20). In the present study, siRNAs for
SEPS1 and a scrambled control were synthesized. In order to examine
whether the SEPS1 gene was silenced, SEPS1 gene and protein
expression were assessed. The results demonstrated that SEPS1
protein expression in liver tissue decreased 12 and 24 h following
injection with SEPS1 siRNA. SEPS1 gene expression in liver tissue
also decreased significantly. These results demonstrated that the
SEPS1 gene was silenced.
Sepsis is defined as a systemic inflammatory
response to a microbial infection that results from excessive
stimulation of the host immune system by pathogen components,
producing various proinflammatory cytokines. The overproduction of
these cytokines then causes systemic inflammation that may lead to
the lethal multiple organ damage (21). In the present study, the influence
of important organ function and prognosis in mice with sepsis was
investigated following silencing of the SEPS1 gene with SEPS1
siRNA. Several biochemical parameters were selected in order to
reflect the important organ condition. The results demonstrated
that the levels of ALT, AST, BUN, CK-MB, CK and LDH all increased
in the SEPS1 siRNA group following SEPS1 siRNA silencing of the
SEPS1 gene, indicating that damage to the liver, kidney and heart
was aggravated. Thus, SEPS1 has an important role in protecting the
liver, kidney and heart from damage due to sepsis.
In order to investigate the protective role of SEPS1
in mice with sepsis, the effect of SEPS1 siRNA on circulating
levels of primary (TNF-α) and secondary (IL-6) proinflammatory
cytokines was assessed as an index of inflammation (22–24).
The TNF-α and IL-6 levels in the SEPS1 siRNA group significantly
increased compared with the control group. SEPS1 siRNA increased
the levels of TNF-α in liver tissue and may have subsequently
aggravated liver dysfunction.
The MAPK signaling pathways consist of a series of
kinases that are sequentially activated and consequently
phosphorylate the downstream kinases and transduce extracellular
stimuli into intracellular responses. The MAPK family includes
extracellular signal-regulated kinases, c-Jun N-terminal kinases
and p38 MAPK. One of the major functions of MAPK is the activation
of transcription factors, several of which bind to the promoters of
pro-inflammatory cytokines (25,26).
Furthermore, the MAPKs have been previously implicated in the
signaling pathways relevant to LPS-induced inflammation. LPS
activates all three MAPK type kinases in mouse macrophages
(27). p38 is activated by LPS
stimulation and has been theorized to have an important function in
the control of IL-6 and TNF-α gene expression. Numerous downstream
targets of the p38 MAPK pathway are transcription factors which
regulate transcription of proinflammatory mediators. In the present
study, the SEPS1 siRNA group enhanced the phospho-p38 expression at
12 and 24 h following LPS stimulation, suggesting that SEPS1 siRNA
treatment increased the phosphorylation of p38 MAPK. From this it
may be inferred that an increase in the levels of proinflammatory
cytokines is due to the activation of the p38 MAPK pathway.
The pathological results demonstrated that liver and
lung cell lesions markedly occurred in the SEPS1 siRNA group as
compared with the control group, which may correlate with the
decrease in SEPS1 gene expression. Therefore, it may be inferred
that the SEPS1 gene exhibited a protective effect on the livers and
lungs of mice with sepsis.
These data suggest that SEPS1 may have an important
role in regulating processes associated with sepsis, particularly
in protecting important organs from damage and decreasing the
production of inflammatory cytokines. Increasing SEPS1 gene
expression may be a new way to reduce sepsis, liver and other organ
damage.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 30972523) and the
Foundation of the ‘Twelfth Five-Year Plan’ for Medical Science
Development of the People’s Liberation Army (nos BWS11J048 and
CWS11J109).
References
|
1
|
Harrison C: Sepsis: calming the cytokine
storm. Nat Rev Drug Discov. 9:360–361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Delsesto D and Opal SM: Future
perspectives on regulating pro-and anti-inflammatory responses in
sepsis. Contrib Microbiol. 17:137–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shubin NJ, Monaghan SF and Ayala A:
Anti-inflammatory mechanisms of sepsis. Contrib Microbiol.
17:108–124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lewis DH, Chan DL, Pinheiro D,
Armitage-Chan E and Garden OA: The immunopathology of sepsis:
pathogen recognition, systemic inflammation, the compensatory
anti-inflammatory response, and regulatory T cells. J Vet Intern
Med. 26:457–482. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lorente JA, Delgado MA and Landin L:
Septic shock and nitric oxide. Enferm Infecc Microbiol Clin.
15:14–19. 1997.(In Spanish).
|
|
6
|
Lvovschi V, Arnaud L, Parizot C, et al:
Cytokine profiles in sepsis have limited relevance for stratifying
patients in the emergency department: a prospective observational
study. PLoS One. 6:e288702011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ando H, Takamura T, Ota T, Nagai Y and
Kobayashi K: Cerivastatin improves survival of mice with
lipopolysaccharide-induced sepsis. J Pharmacol Exp Ther.
294:1043–1046. 2000.PubMed/NCBI
|
|
8
|
Ye Y, Shibata Y, Yun C, Ron D and Rapoport
TA: A membrane protein complex mediates retro-translocation from
the ER lumen into the cytosol. Nature. 429:841–847. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye Y, Shibata Y, Kikkert M, van Voorden S,
Wiertz E and Rapoport TA: Recruitment of the p97 ATPase and
ubiquitin ligases to the site of retrotranslocation at the
endoplasmic reticulum membrane. Proc Natl Acad Sci USA.
102:14132–14138. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hart K, Landvik NE, Lind H, Skaug V,
Haugen A and Zienolddiny S: A combination of functional
polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk
of non-small cell lung cancer. Lung Cancer. 71:123–129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Curran JE, Jowett JB, Elliott KS, et al:
Genetic variation in selenoprotein S influences inflammatory
response. Nat Genet. 37:1234–1241. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rayman MP: Selenoproteins and human
health: insights from epidemiological data. Biochim Biophys Acta.
1790:1533–1540. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kryukov GV, Castellano S, Novoselov SV, et
al: Characterization of mammalian selenoproteomes. Science.
300:1439–1443. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blacker D, Bertram L, Saunders AJ, et al:
Results of a high-resolution genome screen of 437 Alzheimer’s
disease families. Hum Mol Genet. 12:23–32. 2003.PubMed/NCBI
|
|
15
|
Field LL, Tobias R and Magnus T: A locus
on chromosome 15q26 (IDDM3) produces susceptibility to
insulin-dependent diabetes mellitus. Nat Genet. 8:189–194. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zamani M, Pociot F, Raeymaekers P, Nerup J
and Cassiman JJ: Linkage of type I diabetes to 15q26 (IDDM3) in the
Danish population. Hum Genet. 98:491–496. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Susi M, Holopainen P, Mustalahti K, Maki M
and Partanen J: Candidate gene region 15q26 and genetic
susceptibility to coeliac disease in Finnish families. Scand J
Gastroenterol. 36:372–374. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fradejas N, Pastor MD, Mora-Lee S, Tranque
P and Calvo S: SEPS1 gene is activated during astrocyte ischemia
and shows prominent antiapoptotic effects. J Mol Neurosci.
35:259–265. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao Y, Hannan NR, Wanyonyi S, et al:
Activation of the selenoprotein SEPS1 gene expression by
pro-inflammatory cytokines in HepG2 cells. Cytokine. 33:246–251.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Paddison PJ, Caudy AA and Hannon GJ:
Stable suppression of gene expression by RNAi in mammalian cells.
Proc Natl Acad Sci USA. 99:1443–1448. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oberholzer A, Oberholzer C and Moldawer
LL: Sepsis syndromes: understanding the role of innate and acquired
immunity. Shock. 16:83–96. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saadeddin SM, Habbab MA and Ferns GA:
Markers of inflammation and coronary artery disease. Med Sci Monit.
8:RA5–RA12. 2002.PubMed/NCBI
|
|
23
|
Bickel C, Rupprecht HJ, Blankenberg S, et
al: Relation of markers of inflammation (C-reactive protein,
fibrinogen, von Willebrand factor, and leukocyte count) and statin
therapy to long-term mortality in patients with angiographically
proven coronary artery disease. Am J Cardiol. 89:901–908. 2002.
View Article : Google Scholar
|
|
24
|
de Maat MP and Kluft C: The association
between inflammation markers, coronary artery disease and smoking.
Vascul Pharmacol. 39:137–139. 2002.PubMed/NCBI
|
|
25
|
Lin YT, Chen YH, Yang YH, et al: Heme
oxygenase-1 suppresses the infiltration of neutrophils in rat liver
during sepsis through inactivation of p38 MAPK. Shock. 34:615–621.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qian F, Deng J, Gantner BN, et al: Map
kinase phosphatase 5 protects against sepsis-induced acute lung
injury. Am J Physiol Lung Cell Mol Physiol. 302:L866–L874. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ruland J and Mak TW: Transducing signals
from antigen receptors to nuclear factor kappaB. Immunol Rev.
193:93–100. 2003. View Article : Google Scholar : PubMed/NCBI
|