Introduction
Deep vein thrombosis (DVT) is a serious disease,
which is prone to progress to post-thrombotic syndrome (PTS),
phlegmasia cerulea dolens, phlegmasia alba dolens and pulmonary
embolism (PE), which may lead to mortality if not treated properly.
Retrospective studies have shown that patients with venous
thromboembolism exhibit a high mortality rate between 5 and 23%
worldwide (1), which remains
between 1 and 2% even with anticoagulant treatment (2). It has been reported that one third of
patients with DVT develop PTS (3)
and 4–5% of patients with PE develop pulmonary hypertension
(4). DVT treatment is a clinical
problem; however, urokinase is a venous thrombolytic drug that is
commonly used in the clinical treatment of DVT. Urokinase-type
plasminogen activator (uPA) is a second-generation fibrin-selective
thrombolytic agent which is associated with a lower risk of
bleeding than urokinase due to its high selectivity to fibrin
(5–8). Adenovirus vectors are associated with
a high efficiency of gene transfer and protein expression in a
variety of cell types and are used in gene vaccines, gene therapy,
tissue engineering and other studies, and are becoming one of the
most widely used viral vectors. In the present study, the
pAD/CMV/V5-DEST™ Gateway® vector was used for the
construction of the adenovirus-uPA vector. The vector was then
transduced into human umbilical vein endothelial cells (HUVECs) to
investigate the effect of exogenous uPA on uPA expression and
fibrinolytic activity in HUVECs.
Materials and methods
uPA gene synthesis
In accordance with the sequencing results of the
complete uPA gene sequence, single-strand oligos were designed and
synthesized. The oligos were then spliced into a complete gene
using reverse-transcription polymerase chain reaction (RT-PCR) and
inserted into the pMD-18T vector (Takara Bio, Inc., Shiga, Japan)
for transformation into the competent DH5α cells. The gene
sequences in the recombinant clones were identified using DNA
sequencing. Mutations in the gene sequences were repaired using
overlapping PCR.
Construction of recombinant adenovirus
vectors
Construction of recombinant adenovirus vectors was
performed using the Gateway® Recombination Cloning
system (Invitrogen Life Technologies, St. Louis, MO, USA) in a
two-step process. The RT-PCR products of the complete uPA gene
sequence were isolated and purified from the gel and digested using
XhoI and SalI endonucleases to prepare the sequences
for insertion into the pIRES2-EGFP vector (Takara Bio, Inc.) and
the subsequent transformation into competent DH5α cells. Gene
sequencing was used to verify the recombinant plasmid. The plasmid
containing the gene sequences was puPA-IRES2-EGFP. It was then used
as a template to amplify a uPA-IRES2-EGFP fragment containing
restriction sites for attB1 and attB2. The following
gene-specific primers were used: Forward, 5′-GGG GAC AAG TTT GTA
CAA AAA AGC AGG CTT CGC CAC CAT GAG AGC CCT GC-3′ and reverse,
5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC TTA CTT GTA CAG CTC GTC
CAT GC-3′. The PCR product size was 2,600 bp.
The gene of interest was cloned into the entry
vector pDONR221 using BP Clonase™ (Invitrogen Life Technologies).
To produce the expression clone, the gene of interest from pDONR221
was subcloned into a destination vector pAD/CMV/V5-DEST containing
the sequence information necessary for expression using the LR
Reaction (Invitrogen Life Technologies). The recombinant plasmid
was then verified by gene sequencing using the nr database and
blastn program (http://www.ncbi.nlm.nih.gov; version 2.2.10).
Comparision and analysis of the detected sequences were then
performed.
Preparation and purification of the
adenovirus and determination of the viral titer
The recombinant plasmid was digested using the
PacI endonuclease and purified using a phenol-chloroform
extraction method. The purity and concentration of the purified
digestion product was determined using an ultraviolet
spectrophotometer prior to being transduced into HUVECs (Shanghai
Fuchun Zhongnan Biotechnology Co., Ltd., Shanghai, China) using
Lipofectamine® 2000 (Invitrogen Life Technologies)
according to the manufacturer’s instructions. The adenovirus was
released from the cells using repeated freeze-thawing and collected
by centrifugation. The viral solution was amplified primarily and
the viral titer was assessed using immunization.
Determination of the optimal multiplicity
of infection (MOI)
Subsequent to adenovirus transduction, HUVECs were
observed every 24 h and images were captured. The MOI that caused
the highest transduction efficiency and the least cell damage was
used for HUVEC transduction in the subsequent experiments. At 72 h
after transduction, HUVECs were harvested for quantitative (q)PCR
and western blot analyses. RNA was extracted by the TRIzol method
(Invitrogen Life Technologies).
qPCR analysis
The qPCR reaction conditions were as follows: 95°C
for 2 min followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec
and 72°C for 30 sec then 72°C for 10 min and 70 cycles beginning at
60°C and increasing by 0.5°C every 30 sec, following which the
temperature was increased to 95°C.
uPA mRNA expression was detected using qPCR
analysis. Relative quantification was calculated using the
2−ΔΔCt method
[ΔΔCt=(CtTarget−CtActin)Test−
(CtTarget−CtActin)Calibrator].
β-actin was used as an internal control. 1–2−ΔΔCt was
considered to be the silencing efficiency of the target gene. The
gene-specific uPA primer pair was as follows: Forward, 5′-CTA CTA
CGG CTC TGA AGT CAC CAC-3′ and reverse, 5′-GTA GAC GCC TGG CTT GTC
CT-3′.
Detection of uPA protein expression
Cells were harvested and uPA protein expression was
detected using western blot analysis (9). The supernatant of each group was
collected to determine uPA concentration using ELISA.
Detection of uPA activity using
colorimetric assay
The supernatants from the three HUVEC groups were
immediately placed into the microplate reader in order to detect
uPA activity using the spectrophotometer.
Statistical analysis
All data are presented as the mean ± standard
deviation and were analyzed using SAS v8.0 software (Statistical
Analysis System Institute Inc., Cary, NC, USA). Comparisons between
groups were performed using analysis of variance. P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of the adenovirus
expression vector
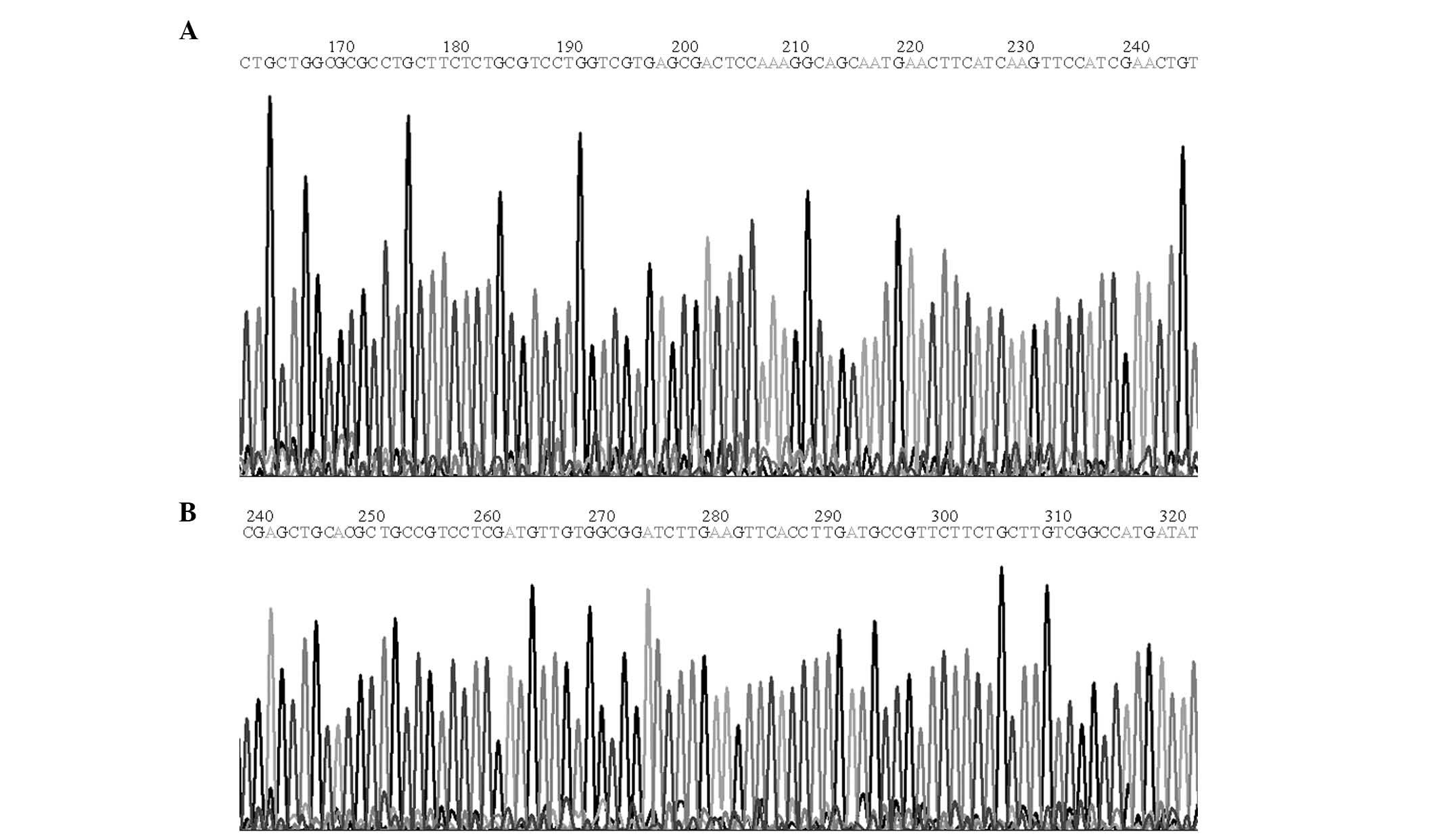
The constructed expression vector was identified
using DNA sequencing with an automatic sequencer (Fig. 1A and B). As verified using the
basic local alignment search tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the inserted
sequence in the positive recombinant plasmid demonstrated complete
homology to the human uPA cDNA sequence in the NCBI GenBank
(https://www.ncbi.nlm.nih.gov/genbank/) (Fig. 2). The viral titer obtained was
5.74×1010 ifu/ml following amplification. Thus, the
adenovirus gene expression vector was constructed correctly and the
virus was assembled successfully.
uPA mRNA expression in the transduced
HUVECs
Following transduction with the adenovirus gene
expression vector, HUVECs were harvested for uPA mRNA expression
detection using qPCR analysis. The relative expression of uPA mRNA
in the HUVECs in the ad/uPa group was significantly higher than
that in the HUVECs in the ad/neg or control groups (both P<0.01;
Table I).
 | Table IRelative uPA mRNA expression in the
human umbilical vein endothelial cells in the three groups
(n=3). |
Table I
Relative uPA mRNA expression in the
human umbilical vein endothelial cells in the three groups
(n=3).
| Group | Ct (target gene) | Ct (actin) | ΔCt | ΔΔCt |
2−ΔΔCt |
|---|
| Control | 25.31±0.10 | 19.86±0.16 | 5.45 | 0 | 1 |
| ad/neg | 14.57±0.11 | 11.31±0.29 | 3.26 | −2.19 | 4.5631 |
| ad/uPA | 27.55±0.14 | 26.03±0.53 | 1.52 | −3.93 | 15.2422a |
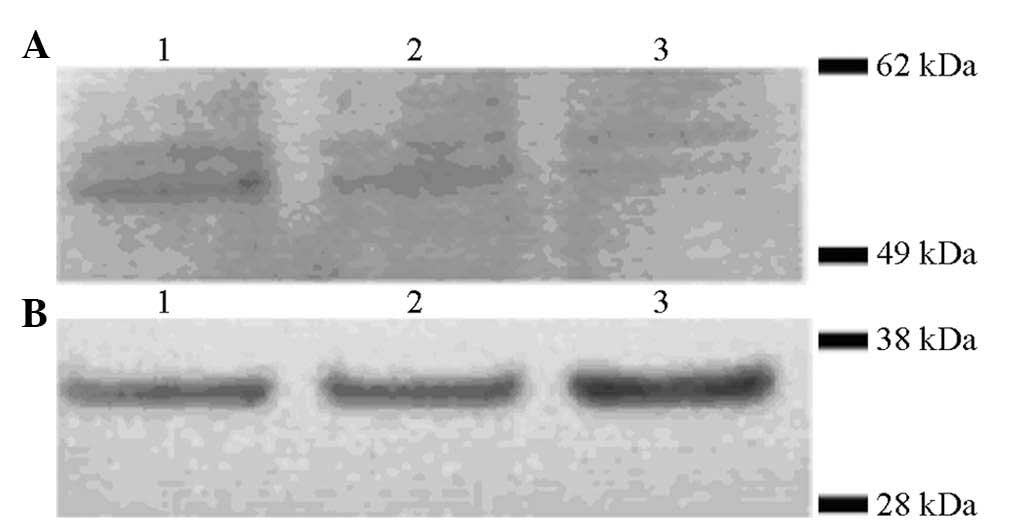
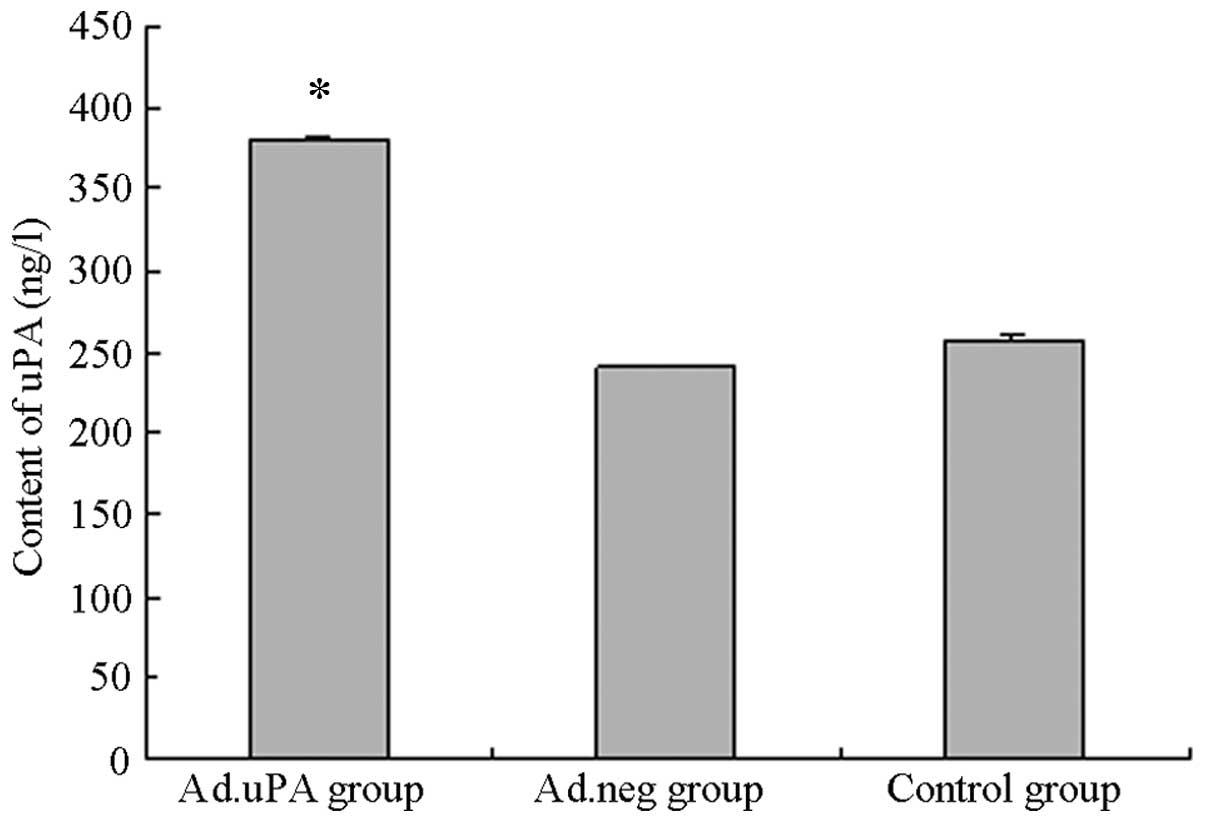
uPA protein expression in the transduced
HUVECs
uPA protein expression was analyzed in the HUVECs
and the HUVEC supernatant following transduction with the
adenovirus gene expression vector using western blot analysis
(Fig. 3) and ELISA (Fig. 4), respectively. Anti-Myc antibodies
were used to detect the expression of uPA as the uPA expression
vector contained a Myc tag. Western blot analysis revealed that uPA
expression in the HUVECs transfected with the adenovirus gene
expression vector (MOI: 800) was significantly higher than that in
the HUVECs transfected with negative vector or in the blank control
HUVECs. The uPA concentration in the supernatant of the ad/uPa
group was 379.40±2.46 ng/l, which was significantly higher than
that in the ad/neg (240.01±1.16 ng/l) or blank control
(256.1024±3.04 ng/l) groups (P<0.01; Fig. 4).
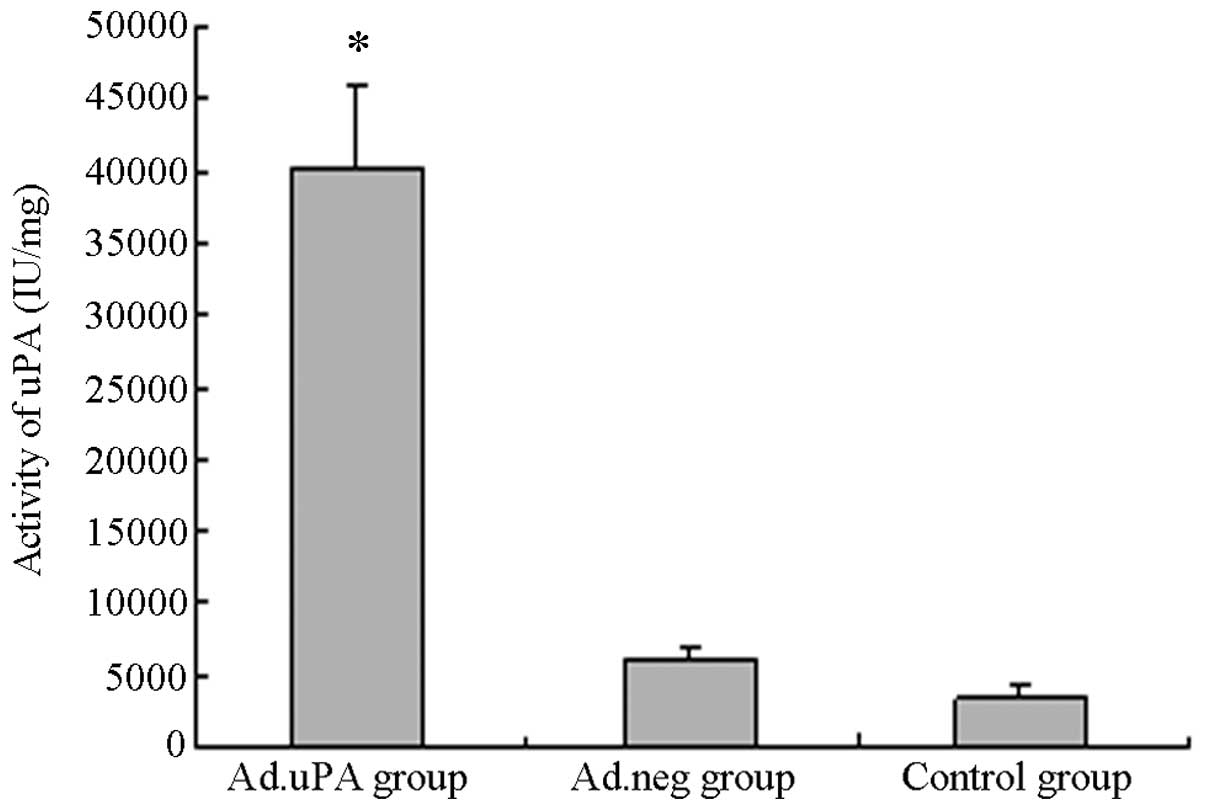
uPA activity in the transduced
HUVECs
uPA activity in the HUVECs in the ad/uPa group
(40238.49±5755 IU/mg) was found to be significantly increased
compared with that in the HUVECs in the ad/neg (6180.03±942.38
IU/mg) or control (3346.06±928.81 IU/mg) groups (P<0.01;
Fig. 5).
Discussion
DVT is a common disease, which is harmful to patient
health and quality of life. DVT has a poor clinical outcome and is
prone to progress to PE and PTS and may result in limb loss and
mortality. At present, the treatments for DVT primarily include
anticoagulation, thrombolysis, surgical treatment or intervention
therapy. However, to some extent, all of the above treatments are
associated with bleeding, mortality, disability, thrombotic
recurrence, as well as other disadvantages, which cause problems
for DVT treatment. Thus, alternative treatment methods, including
gene therapy, have attracted much research attention. Gene
therapies for vascular diseases are still in experimental and
subclinical stages (10–21). Appropriate genes and vectors are
key to gene therapy and uPA and tissue plasminogen activator (tPA)
are two thrombolytic agents, which have been proposed to have
potential (14–18).
The human body has two plasminogen activators: uPA
and tPA. uPA and tPA activate plasminogen into plasmin and thereby
have a thrombolytic effect. uPA was originally purified from urine.
Urogenital epithelial cells are the primary producers of uPA, while
uPA is not normally expressed in vascular endothelial cells (VECs).
However, under culture conditions, uPA is expressed if VECs are
stimulated with endotoxin or tumor necrosis factor (TNF) (22).
The human uPA gene is located on the long arm of
chromosome 10, at 10q24. The uPA gene consists of 11 exons and 10
introns with a size of 6.4 kb, which generates a mature mRNA of 2.4
kb and translates into pro-urokinase. uPA is a serine proteolytic
enzyme containing 411 amino acid residues. It is a glycoprotein
with a molecular weight of 50–60 kDa, constructed by two
polypeptide chains bridged by a disulfide bond. uPA activates
plasminogen directly without the cofactor fibrin.
Gene therapy involves introducing an exogenous gene
into somatic cells using genetic engineering technology, making the
cells express the corresponding protein in order to prevent or
treat a disease. As a major component of the vascular wall, VECs
are not only a natural barrier for the blood vessel wall, but also
an important endocrine organ that synthesizes a variety of
cytokines. Due to the location of VECs in the blood vessel wall,
the proteins secreted by VECs not only act in local blood vessels,
but also have a role in the systemic circulation. Thus, VECs are
particularly important target cells (23).
In vivo, VECs only express tPA; however, VECs
have been found to synthesize and secrete tPA and uPA in
vitro. Interleukin-1, TNFα, plasmin and α-tocopherol have been
reported to reduce the release of tPA from VECs. Plasminogen, uPA
and tPA receptors are expressed on the surface of endothelial cells
and promote the activation of plasminogen. Under pathological
conditions, the levels of uPA in the human body are not sufficient
to prevent and treat thrombosis. Therefore, introducing the
exogenous uPA gene into mammalian endothelial cells in order to
produce more uPA protein with higher biological activity is of
great importance for the prevention and treatment of
thrombosis.
qPCR is an assay which is used to analyze gene
transcription. qPCR quantitatively analyzes mRNA levels, prior to
colorimetric analysis, which saves time during clinical diagnosis
and treatment, particularly for predicting changes in the
fibrinolytic system (24,25). In addition, qPCR has a real-time
signal monitoring feature, which avoids the error caused by the
‘platform effect’. In the present study, qPCR analysis was used to
analyze uPA mRNA expression. During qPCR analysis, mRNA templates
are quantitatively analyzed using the Ct method and standard curves
are generated by analyzing the changes in the fluorescent signals
in each cycle of the PCR reaction. The Ct value refers to the
number of cycles required for the fluorescent signal in each
reaction to reach the set threshold. The Ct value of each mRNA
template has a linear negative correlation with its initial copy
number. A standard curve is generated using a proof sample with
known initial copy numbers. According to the standard curve, the
initial copy numbers of the test samples can be calculated using
their Ct values. The 2−ΔΔCt method is a convenient
method used to calculate the relative changes in gene expression in
qPCR analysis (26–30). qPCR analysis is more sensitive,
simple and specific than traditional northern blot analysis. In the
present study, uPA and an internal control were successfully
amplified using qPCR analysis. The uPA sequences amplified spanned
at least two exons in order to exclude the contamination of genomic
DNA. The housekeeping gene β-actin was used as an internal control,
which exhibits universal and constant expression in all tissues.
Thus, it can be used to reduce the error during qPCR analysis or
RNA quantitation.
In the present study, HUVECs transduced with the uPA
gene were observed to secrete more uPA protein than control HUVECs.
uPA activity was also found to be significantly increased in the
uPA-transduced HUVECs, indicating that the exogenous uPA gene is
transcribed and the uPA protein is synthesized and secreted in
HUVECs. An adenovirus containing the uPA gene was successfully
transduced, generating HUVECs stably expressing uPA and providing a
strong foundation for future research on direct or indirect uPA
gene therapy in vivo. However, prior to being used in
vivo, it must be verified that the uPA-adenovirus can be
successfully transfected into HUVECs and has an influence on the
biological characteristics of HUVECs. Whether such an exogenous
gene has biological effects in vivo requires further
investigation using animal experiments.
References
|
1
|
Goldhaber SZ: Pulmonary embolism. Lancet.
363:1295–1305. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spencer FA, Emery C, Lessard D, et al: The
Worcester Venous Thromboembolism Study: A population based study of
the clinical epidemiology of venous thromboembolism. J Gen Intern
Med. 21:722–727. 2006. View Article : Google Scholar
|
|
3
|
Chapman NH, Brighton T, Harris MF, Caplan
GA, Braithwaite J and Chong BH: Venous thromboembolism - management
in general practice. Aust Fam Physician. 38:36–40. 2009.PubMed/NCBI
|
|
4
|
Pengo V, Lensing AW, Prins MH, et al:
Thromboembolic Pulmonary Hypertension Study Group: Incidence of
chronic thromboembolic pulmonary hypertension after pulmonary
embolism. N Engl J Med. 350:2257–2264. 2004. View Article : Google Scholar
|
|
5
|
Vincenza Carriero M, Franco P, Vocca I, et
al: Structure, function and antagonists of urokinase-type
plasminogen activator. Front Biosci (Landmark Ed). 14:3782–3794.
2009.PubMed/NCBI
|
|
6
|
Radha KS, Madhyastha HK, Nakajima Y, Omura
S and Maruyama M: Emodin upregulates urokinase plasminogen
activator, plasminogen activator inhibitor-1 and promotes wound
healing in human fibroblasts. Vascul Pharmacol. 48:184–190. 2008.
View Article : Google Scholar
|
|
7
|
Nassar T, Yarovoi S, Fanne RA, et al:
Urokinase plasminogen activator regulates pulmonary arterial
contractility and vascular permeability in mice. Am J Respir Cell
Mol Biol. 45:1015–1021. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baldwin JF, Sood V, Elfline MA, et al: The
role of urokinase plasminogen activator and plasmin activator
inhibitor-1 on vein wall remodeling in experimental deep vein
thrombosis. J Vasc Surg. 56:1089–1097. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rao Gogineni V, Kumar Nalla A, Gupta R, et
al: Radiation-inducible silencing of uPA and uPAR in vitro and in
vivo in meningioma. Int J Oncol. 36:809–816. 2010.PubMed/NCBI
|
|
10
|
Ylä-Herttuala S: Cardiovascular gene
therapy with vascular endothelial growth factors. Gene.
525:217–219. 2013.
|
|
11
|
Zhao B, Li X, Dai X and Gong N:
Adenovirus-mediated anti-sense extracellular signal-regulated
kinase 2 gene therapy inhibits activation of vascular smooth muscle
cells and angiogenesis, and ameliorates transplant
arteriosclerosis. Transplant Proc. 45:639–642. 2013. View Article : Google Scholar
|
|
12
|
Dragneva G, Korpisalo P and Ylä-Herttuala
S: Promoting blood vessel growth in ischemic diseases: challenges
in translating preclinical potential into clinical success. Dis
Model Mech. 6:312–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kral BG and Kraitchman DL: From mice to
men: gene therapy’s future for treatment of myocardial infarction.
Circ Cardiovasc Imaging. 6:360–362. 2013.
|
|
14
|
Botkjaer KA, Fogh S, Bekes EC, et al:
Targeting the autolysis loop of urokinase-type plasminogen
activator with conformation-specific monoclonal antibodies. Biochem
J. 438:39–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sood V, Luke CE, Deatrick KB, et al:
Urokinase plasminogen activator independent early experimental
thrombus resolution: MMP2 as an alternative mechanism. Thromb
Haemost. 104:1174–1183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Humphries J, Gossage JA, Modarai B, et al:
Monocyte urokinase-type plasminogen activator up-regulation reduces
thrombus size in a model of venous thrombosis. J Vasc Surg.
50:1127–1134. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Madhyastha R, Madhyastha H, Nakajima Y,
Omura S and Maruyama M: Curcumin facilitates fibrinolysis and
cellular migration during wound healing by modulating urokinase
plasminogen activator expression. Pathophysiol Haemost Thromb.
37:59–66. 2010. View Article : Google Scholar
|
|
18
|
Shenkman B, Livnat T, Budnik I, Tamarin I,
Einav Y and Martinowitz U: Plasma tissue-type plasminogen activator
increases fibrinolytic activity of exogenous urokinase-type
plasminogen activator. Blood Coagul Fibrinolysis. 23:729–733. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lipskaia L, Hadri L, Lopez JJ, Hajjar RJ
and Bobe R: Benefit of SERCA2a gene transfer to vascular
endothelial and smooth muscle cells: a new aspect in therapy of
cardiovascular diseases. Curr Vasc Pharmacol. 11:465–479. 2013.
View Article : Google Scholar
|
|
20
|
Hennessy EJ and Moore KJ: Using microRNA
as an alternative treatment for hyperlipidemia and cardiovascular
disease: cardio-miRs in the pipeline. J Cardiovasc Pharmacol.
62:247–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Won YW, McGinn AN, Lee M, Nam K, Bull DA
and Kim SW: Post-translational regulation of a hypoxia-responsive
VEGF plasmid for the treatment of myocardial ischemia.
Biomaterials. 34:6229–6238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamaguchi Y, Yamada K, Suzuki T, et al:
Induction of uPA release in human peripheral blood lymphocytes by
[deamino-Cysl, D-Arg8]-vasopressin (dDAVP). Am J Physiol Endocrinol
Metab. 287:E970–E976. 2004.
|
|
23
|
De Meyer SF, Deckmyn H and Vanhoorelbeke
K: von Willebrand factor to the rescue. Blood. 113:5049–5057.
2009.PubMed/NCBI
|
|
24
|
Chey S, Claus C and Liebert UG: Validation
and application of normalization factors for gene expression
studies in rubella virus-infected cell lines with quantitative
real-time PCR. J Cell Biochem. 110:118–128. 2010.PubMed/NCBI
|
|
25
|
Gu Z, Pan J, Bankowski MJ and Hayden RT:
Quantitative real-time polymerase chain reaction detection of BK
virus using labeled primers. Arch Pathol Lab Med. 134:444–448.
2010.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
27
|
Oliveira MS, Skinner F, Arshadmansab MF,
et al: Altered expression and function of small-conductance (SK)
Ca(2+)-activated K+ channels in pilocarpine-treated
epileptic rats. Brain Res. 1348:187–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liang F, Arora N, Zhang KL, et al: A new,
multiplex, quantitative real-time polymerase chain reaction system
for nucleic Acid detection and quantification. Methods Mol Biol.
1039:51–68. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wagner EM: Monitoring gene expression:
quantitative real-time rt-PCR. Methods Mol Biol. 1027:19–45. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gentilini F and Turba ME: Two novel
real-time PCR methods for genotyping the von Willebrand disease
type I mutation in Doberman Pinscher dogs. Vet J. 197:457–460.
2013. View Article : Google Scholar : PubMed/NCBI
|