Introduction
Hepatocellular carcinoma (HCC) is the third most
common cause of cancer-associated mortality, and affects
>500,000 individuals annually, worldwide (1). Hepatocarcinogenesis is a complicated,
multi-step process that is associated with various genetic and
epigenetic alterations (2,3). Despite recent advances in functional
genomics, the molecular pathogenesis of HCC remains to be
elucidated. Therefore, identifying novel molecules associated with
the development of HCC is required in order to combat this
aggressive malignancy.
MicroRNAs (miRNAs) are an abundant family of small
endogenous non-coding RNAs, with essential roles in the regulation
of gene expression. miRNAs bind to the 3′-untranslated region
(3′-UTR) of genes by imperfect base-pairing to complementary
sequences, and induce target mRNA degradation or translational
repression (4). Emerging evidence
has demonstrated the aberrant expression of miRNAs in numerous
types of cancer, including HCC (5–7).
Among them, miR-144 is aberrantly expressed in various
malignancies, including bladder (8), colorectal (CRC) (9) and lung cancer (10), as well as HCC (11). There are various signaling pathways
through which miR-144 plays a critical role in the development of
numerous cancers, by targeting different molecules involved in
those signalling pathways.
A number of these target molecules have been
experimentally identified, including enhancer of zeste homolog 2
(EZH2), mammalian target of rapamycin (mTOR) and Zinc finger
X-chromosomal protein (8–10).
In the present study, the effects of miR-144 on the
proliferation, migration and invasion of HCC cells were
investigated, and its downstream target was identified.
Materials and methods
Patient samples
A total of 29 HCC patients were enrolled in the
present study from the Third Affiliated Hospital (Changsha, China).
The present study was approved by the Ethics Committee of the Third
Affiliated Hospital, and informed consent was provided by each
patient. The samples were obtained during surgery, without
pre-surgical chemotherapy or radiation therapy. The tissues were
immediately snapfrozen and stored at −80°C
Cell culture and transfection
The HepG2, HuH7 and SMMC-7721 human HCC cell lines,
and the HL-7702 normal human hepatocyte cell line were obtained
from the Shanghai Institute of Cell Biology (Shanghai, China). The
cells were cultured in Dulbecco’s modified Eagle’s medium
(Invitrogen Life Technologies, Carlsbad, CA, USA) and were
incubated at 37°C in an atmosphere containing 5% CO2.
Transfections of the overexpression plasmids and the corresponding
control were performed using Lipofectamine 2000 (Invitrogen Life
Technologies), according to the manufacturer’s instructions.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
SYBR Green (Takara Bio, Inc., Otsu, Japan) reverse
transcription (RT)-qPCR (using RT kit; Toyobo Co., Ltd., Osaka,
Japan) was performed using an ABI StepOne machine (Applied
Biosystems Life Technologies, Foster City, CA, USA). Fold change
was calculated using the 2−ΔΔCt method. The primer
sequences used for RT-qPCR were as follows: AKT3 sense
5′-AAAACAGAACGACCAAAG-3′ and antisense 5′-TCTGCTACAGCCTGGATA-3′;
and GAPDH sense 5′-CCACTCCTCCACCTTTGAC-3′ and antisense
5′-ACCCTGTTGCTGTAGCCA-3′. The primers for miR-144 (cat.no
HmiRQP0190) and the control U6 (cat.no HmiRQP9001) were obtained
from GeneCopoeia, Inc. (Rockville, MD, USA). Tissue samples were
homogenized, and miRNA was extracted using an All-In-One microRNA
Extraction kit (GeneCopoeia, Inc.). RNA was extracted using TRIzol
(Invitrogen Life Technologies). The expression levels were
normalized to GAPDH or U6. Reactions were incubated for 10 min at
95°C, followed by 40 cycles of 95°C for 10 sec; 60°C for 20 sec and
72°C for 10 sec. The temperature was then increased from 68 to 95°C
to produce a melting curve.
Plasmid construction and luciferase
activity assay
miR-144 (pre-miR-144) and pre-miR negative control
(NC) were purchased from Ambion Life Technologies (Carlsbad, CA,
USA). pcDNA3-AKT3 was generated using the following primers: Sense:
5′-GGGGTACCCAAACCCTAAAGCTGATA-3′ and antisense:
5′-CCCTCGAGACAGTAGCAGCAACAGCA-3′. The PCR product was inserted into
pcDNA3.0 within the KpnI and XhoI restriction sites
(Invitrogen Life Technologies). The 3′-UTR of AKT3 mRNA was
amplified using the following primers: Sense:
5′-CCCTCGAGTCCCTCAGTGAAGGCTAA-3′ and antisense:
5′-TTGCGGCCGCTCTTCAGCCATCAGAGGT-3′. The PCR product and its mutant
were inserted into the dual luciferase reporter vector psiCHECK2
(Promega Corporation, Madison, Wi, USA), within the XhoI and
NotI restriction sites (Promega Corporation).
The SMMC-7721 cells were co-transfected with the
luciferase reporter vector, containing the 3′-UTR of AKT3 [wild
type (WT) or mutant (Mut)], and pre-miR-144 or pre-miR NC. The
luciferase activities were determined 48 h post-transfection using
the Dual-Luciferase Reporter Assay system (Promega
Corporation).
Analysis of cell viability and colony
formation
The cell viability was examined using a Cell
Counting kit-8 (CCK-8; Beyotime, Shanghai, China), according to the
manufacturer’s instructions, following transfection with
pre-miR-144 or pre-miR NC at the indicated time points. The
absorbance was measured at 450 nm by the absorbance reader (Thermo
Fisher Scientific, Waltham, MA, USA). For colony formation
analysis, 500 transfected SMMC-7721 cells were seeded in each well
of 6-well plates. The surviving colonies were stained with 0.5%
crystal violet and counted, following a two week incubation. The
experiments were performed in triplicate. For the observation of
cells a CK12 inverted microscope was used (Olympus Corporation,
Tokyo, Japan) according to the manufacturer’s instructions.
Analysis of cell invasion and
migration
Cell invasion and migration were examined by a
Transwell assay (EMD Millipore, Billerica, MA, USA). Filters in the
upper chamber were pre-coated with Matrigel (BD Biosciences, San
Jose, CA, USA) for the invasion assays. A total of 5×104
cells were seeded in the upper chamber, and the lower chamber was
filled with culture media supplemented with 10% fetal bovine serum
(Invitrogen, Foster City, CA, USA). The invaded and migrated cells
on the bottom surface were stained with 0.5% crystal violet and
counted, at the indicated time points. The experiments were
performed in triplicate, and four fields were counted per
filter.
Western blot analysis
The antibodies used were as follows: Rabbit
anti-human polyclonal AKT3 antibody and Rabbit anti-human
monoclonal GAPDH antibody (all 1:1,000; from Cell Signaling
Technology, Inc., Beverly, MA, USA). The protein concentrations
were determined by using BCA protein assay kit (Beyotime). The cell
lysates were prepared using a lysis buffer (pH 7.5, 50 mM Tris-HCl,
5 mM EDTA, 0.5% NP-40 and 150 mM NaCl). The protein samples were
separated by 10% SDS-PAGE and transferred to polyvinylidene
fluoride membranes (Millipore, MA, USA). The membranes were then
probed with primary antibodies, followed by an incubation with the
appropriate horseradish peroxidase-labeled secondary antibodies.
The blots were visualized using an Enhanced Chemiluminescence
reagent (GE Healthcare Life Sciences, Chalfont, UK). The protein
expression levels were normalized to GAPDH and quantified by
optical densitometry using ImageJ Software 1.46 (National Institute
of Health, Bethesda, MA, USA).
Statistical analysis
The data are presented as the mean ± standard
deviation. A one-way analysis of variance or Student’s
t-test was performed to determine statistical significance.
SPSS software, version 15.0 (SPSS Inc., Chicago, IL, USA) was used
for the statistical analyses. P<0.05 was considered to indicate
a statistically significant difference. Spearman’s correlation
analysis was used to analyze the correlation of miR-144 and
AKT3.
Results
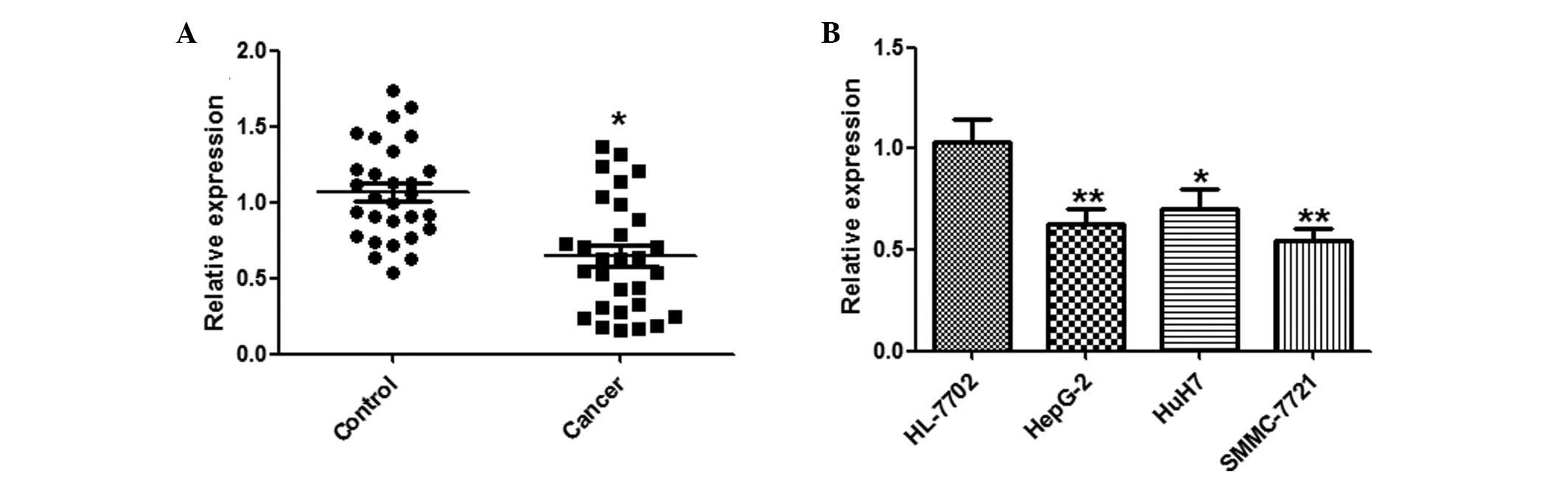
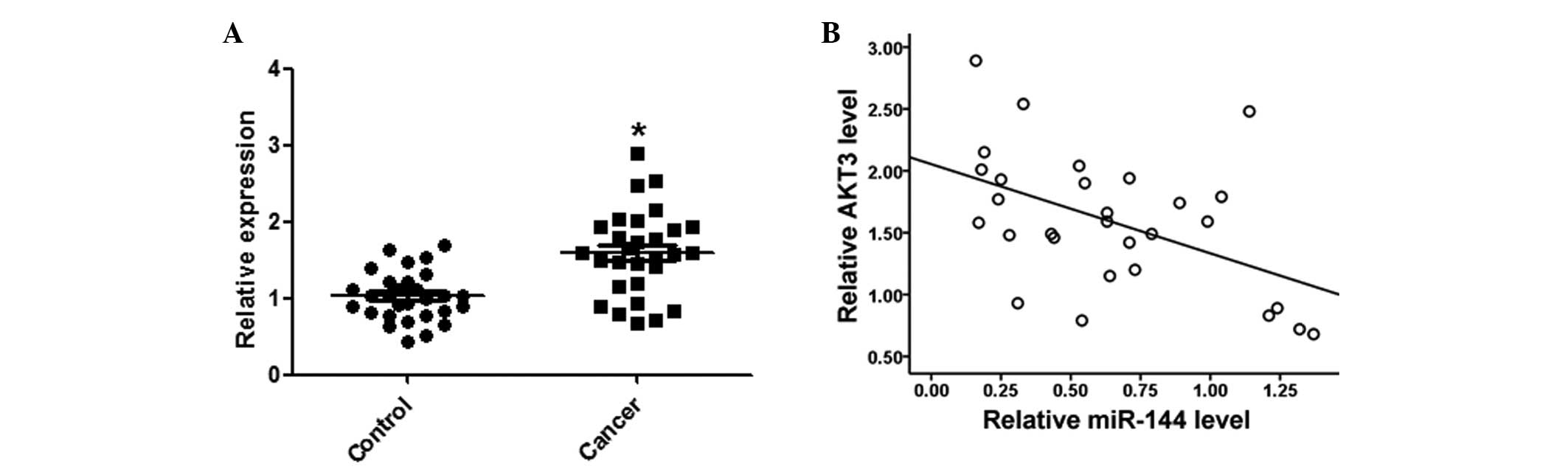
miR-144 is downregulated in HCC tissues
and cell lines
A qPCR analysis of miR-144 was conducted using
paired HCC and adjacent normal tissues from 29 HCC patients.
miR-144 was significantly downregulated in the HCC tissues, as
compared with the normal tissues (Fig.
1A). Furthermore, the expression levels of miR-144 in the three
human HCC cell lines: HepG2, HuH7 and SMMC-7721, and the HL-7702
normal human hepatocyte cell line, were examined by qPCR.
Concordant with the tissue results, miR-144 was markedly
downregulated in the HCC cell lines, as compared with the normal
hepatocytes (Fig. 1B).
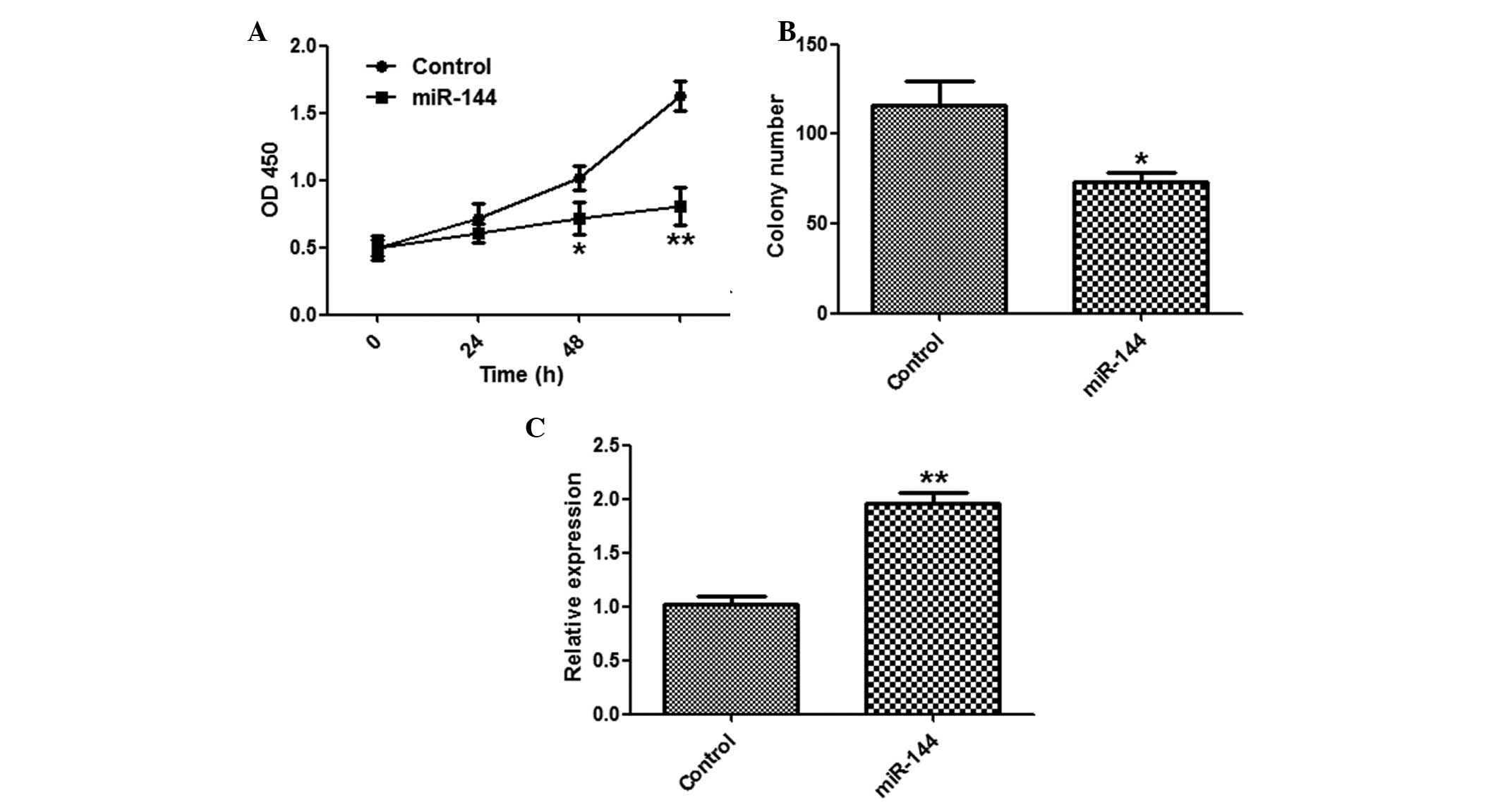
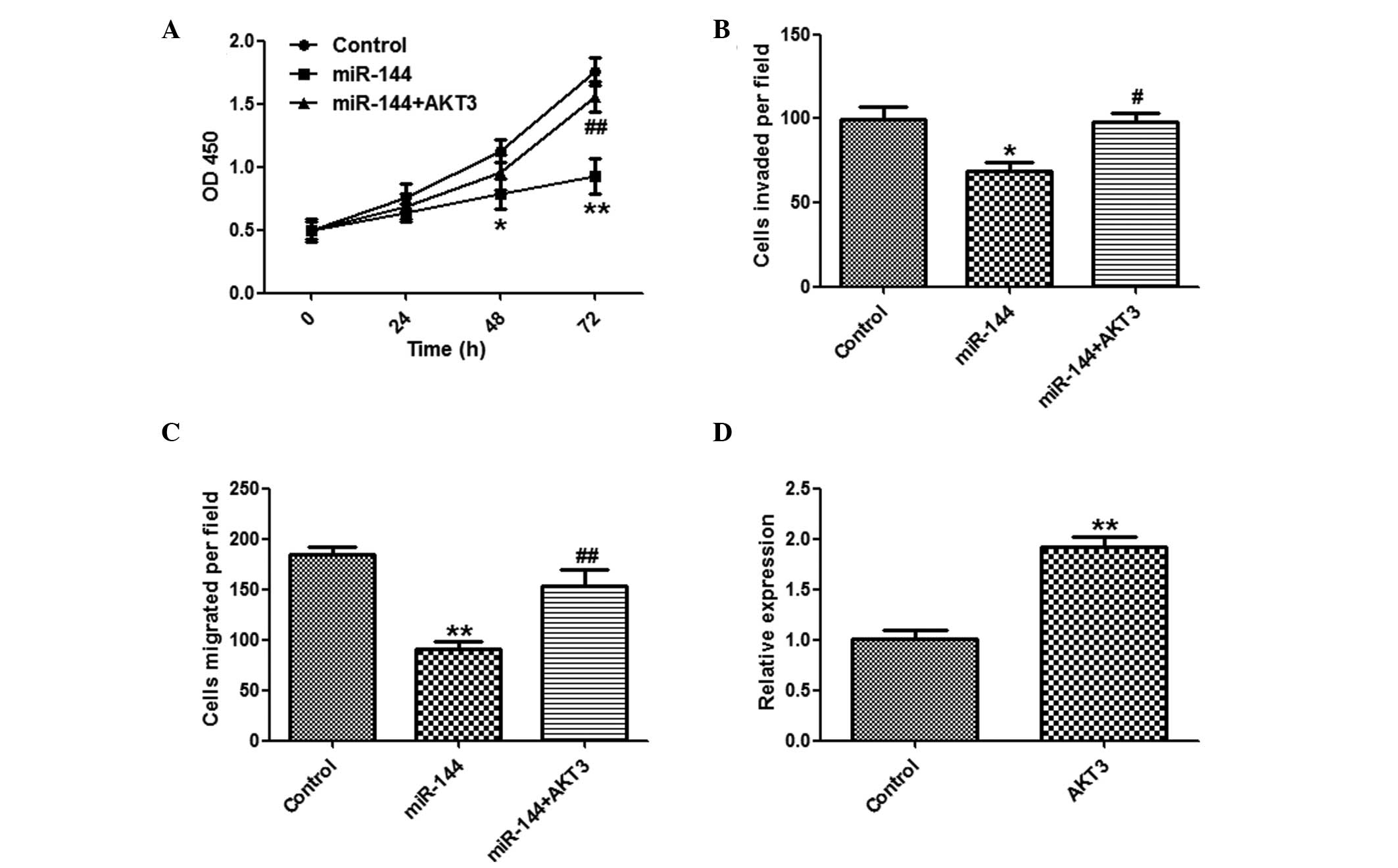
Suppression of HCC cell proliferation by
miR-144
To explore the function of miR-144 in tumor
proliferation, the SMMC-7721 cells were transfected with miR-144 or
a control miRNA. miR-144 overexpression significantly suppressed
the proliferation of SMMC-7721 cells, as determined by a CCK-8
assay (Fig. 2A). The cells
transfected with miR-144 formed fewer colonies, as compared with
the miR-NC group (Fig. 2B). The
effects of miR-144 overexpression were validated by qPCR (Fig. 2C).
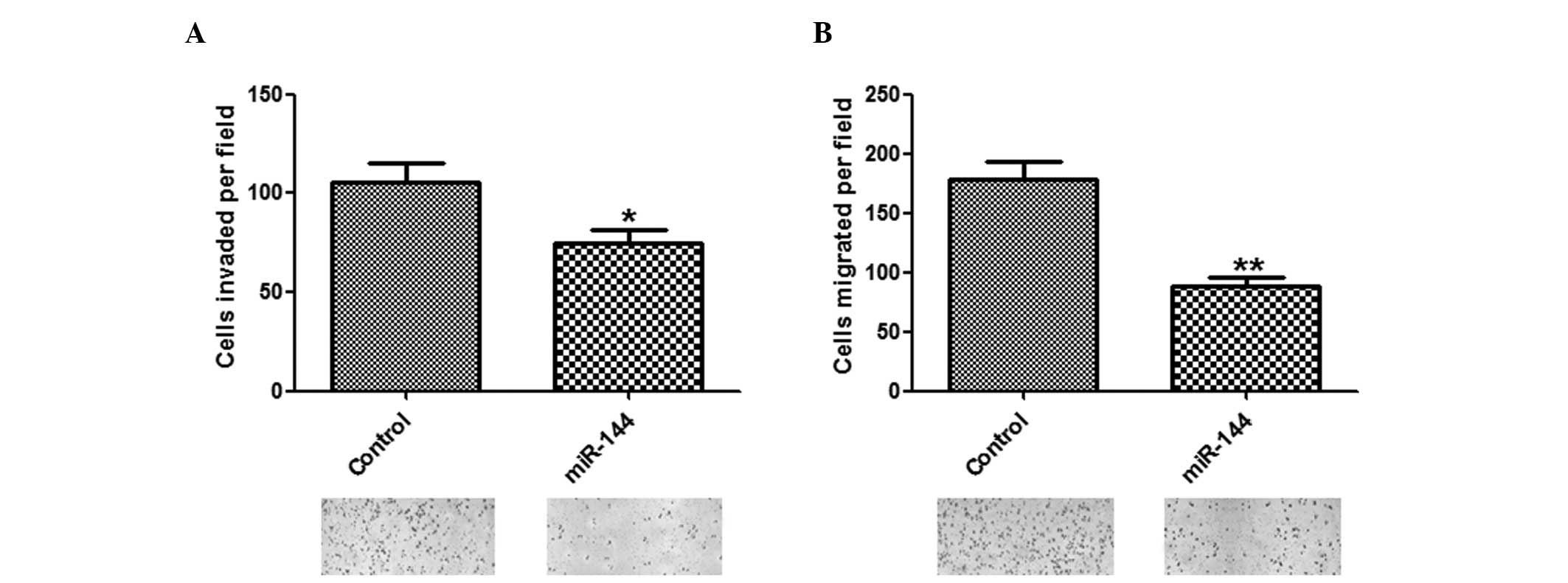
Inhibitory effects of miR-144 on invasion
and migration of HCC cells
To determine whether miR-144 has a crucial role in
cell invasion and migration, in vitro assays were performed.
The number of invasive and migratory cells were significantly
reduced in the group overexpressing miR-144, as compared with the
group transfected with control miRNA (Fig. 3A and B).
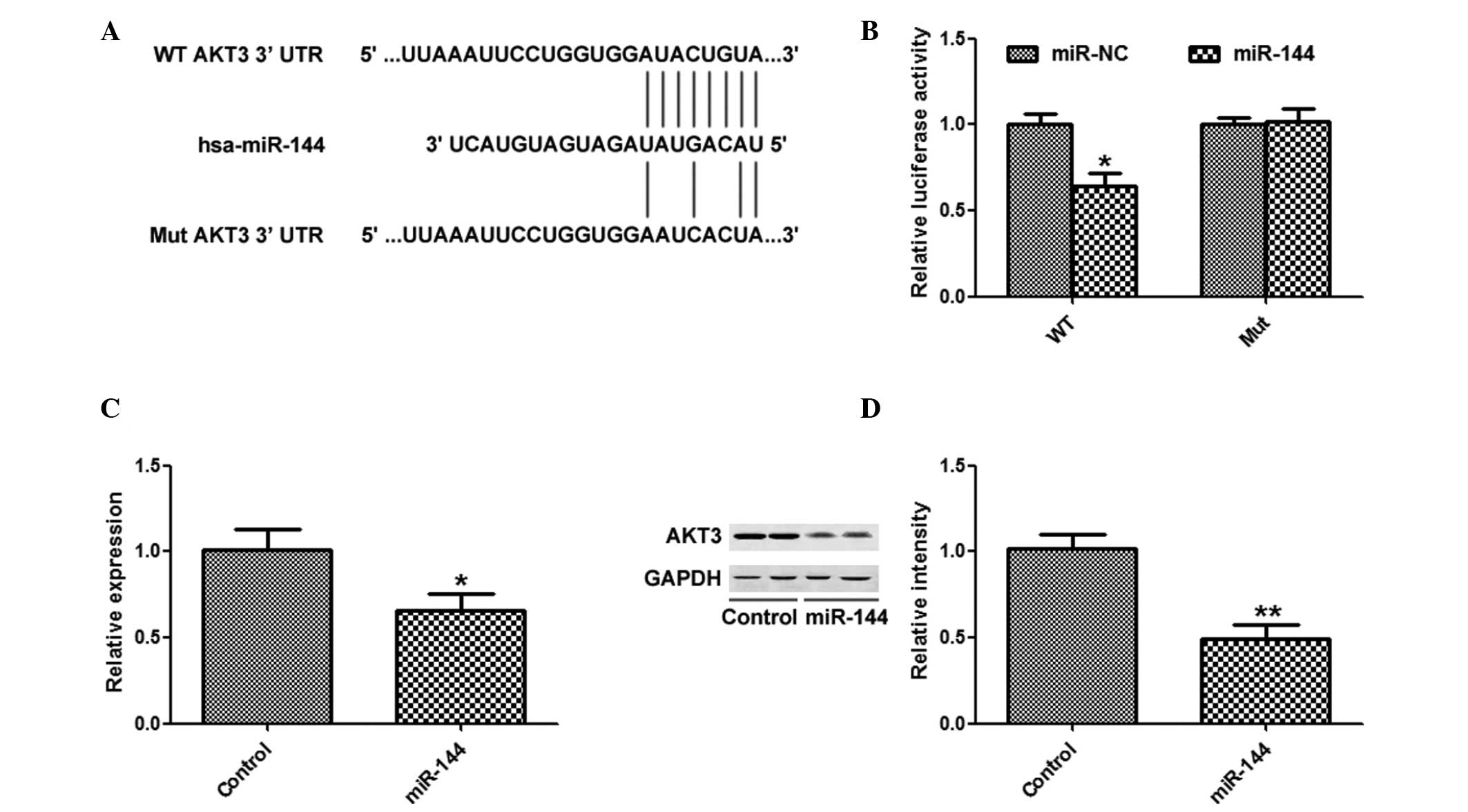
AKT3 is a target of miR-144 in HCC
cells
TargetScan was used to search for potential targets
of miR-144, and AKT3 was identified as a potential target (Fig. 4A). miR-144 overexpression
significantly inhibited the luciferase activity of the AKT3 WT
3′-UTR, but not the AKT3 Mut 3′-UTR (Fig. 4B). In addition, overexpression of
miR-144 significantly suppressed the mRNA and protein expression
levels of AKT3 (Fig. 4C and
D).
AKT3 overexpression attenuates the
suppressive effects of miR-144
It was further investigated whether overexpression
of AKT3 may reverse the suppressive effects of miR-144. A CCK8
assay (Fig. 5A), and invasion and
migration assays (Fig. 5B and C)
showed that overexpression of AKT3 markedly reversed the
suppressive effects of miR-144 on HCC cells. The effects of
pcDNA3-AKT3 were confirmed by qPCR (Fig. 5D).
AKT3 expression levels are inversely
correlated with miR-144 in HCC tissues
Expression levels of AKT3 in the HCC and matched
normal tissues were detected by qPCR. AKT3 expression levels were
significantly increased in the HCC tissues, as compared with the
matched normal tissues (Fig. 6A).
Furthermore, AKT3 was negatively correlated with miR-144 expression
levels in the same HCC tissues (Fig.
6B).
Discussion
The aim of the present study was to investigate the
role of miR-144 in hepatocarcinogenesis, and to determine the
underlying molecular mechanisms by which miR-144 exerts its
function. miR-144 was shown to be downregulated in the HCC tissues
and cell lines. Forced overexpression of miR-144 suppressed HCC
cell proliferation, colony formation, and the invasive and
migratory abilities of the cells. Theoretical targets of miR-144
were searched for using the target prediction software TargetScan.
AKT3 was identified as a potential target and was further tested.
Overexpression of AKT3 could partially attenuate the tumor
suppressive effects of miR-144 in HCC cells. Furthermore, AKT3 was
shown to be negatively correlated with miR-144 expression levels in
the HCC tissues. These results suggest that miR-144 may have a
potential role in the diagnosis and treatment of HCC.
The gene encoding miR-144 is located on chromosome
11 (9). Abnormal expression of
miR-144 has previously been detected in nasopharyngeal carcinoma
(NPC), CRC and HCC (9,11,12).
In NPC, miR-144 was shown to be elevated and act as an oncogene.
Repression of miR-144 substantially decreased cell proliferation,
colony formation, invasion, migration, and tumor formation in nude
mice. Furthermore, restoring miR-144 in miR-144-attenuated NPC
cells exerted a strong tumorigenic role (12). Conversely, in CRC, miR-144 has been
shown to be downregulated and miR-144 downregulation is associated
with poor CRC prognosis. Suppression of miR-144 facilitated
proliferation and increased rapamycin sensitivity of CRC cells,
through activation of the mTOR signaling pathway (9). Furthermore, in bladder cancer,
miR-144 served as a tumor suppressor by targeting EZH2 and
regulating Wnt signaling (8). The
present study expanded the role of miR-144 in HCC, and demonstrated
that miR-144 acts as a tumor suppressor in HCC.
The AKT family includes three isoforms: AKT1, AKT2
and AKT3, which share a highly conserved domain structure and have
similar roles in the regulation of cell proliferation, survival,
metabolism and numerous other cellular functions (13). The function of AKT isoforms in
mediating cancer development and progression is known to be
orchestrated in a tissue-specific manner (14). Nassirpour et al (15) reported that hepatitis B
virus-transformed cells had enhanced expression of AKT3, and
specific downregulation of AKT3 was able to suppress migration and
induce programmed cell death. They further demonstrated that AKT3,
but not AKT1 or AKT2, was required and sufficient to promote
migration and metastasis in certain HCCs. In concordance with these
findings, the present study demonstrated that supplementation with
AKT3 markedly attenuated the tumor suppressive effects of miR-144
on the proliferation, invasion and migration of HCC cells.
Restoration of miR-144 may induce antitumor activities by targeting
AKT3, thus suggesting that miR-144 could act as a tumor suppressor
in HCC, which harbors decreased miR-144 expression levels.
In conclusion, the results of the present study
showed that miR-144 bound to the 3′-UTR of AKT3, and overexpression
of miR-144 in HCC cells decreased AKT3 mRNA and protein expression
levels, resulting in the inhibition of cell proliferation, invasion
and migration. Ectopic overexpression of AKT3 was able to attenuate
the tumor suppressive characteristics induced by miR-144
overexpression, indicating that the regulation of tumorigenesis by
miR-144 was mediated by targeting AKT3 in HCC.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kong D, Chen H, Chen W, et al: Gene
expression profiling analysis of hepatocellular carcinoma. Eur J
Med Res. 18:442013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hauptman N and Glavac D: MicroRNAs and
long non-coding RNAs: prospects in diagnostics and therapy of
cancer. Radiol Oncol. 47:311–318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang W, Lin H, Zhou L, et al:
MicroRNA-30a-3p inhibits tumor proliferation, invasiveness and
metastasis and is downregulated in hepatocellular carcinoma. Eur J
Surg Oncol. Nov 19–2013.(Epub ahead of print).
|
|
6
|
Ke Y, Zhao W, Xiong J and Cao R:
Downregulation of miR-16 promotes growth and motility by targeting
HDGF in non-small cell lung cancer cells. FEBS Lett. 587:3153–3157.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang H, Li Q, Zhao W, Yuan D, Zhao H and
Zhou Y: miR-329 suppresses the growth and motility of neuroblastoma
by targeting KDM1A. FEBS Lett. 588:192–197. 2014. View Article : Google Scholar
|
|
8
|
Guo Y, Ying L, Tian Y, et al: miR-144
downregulation increases bladder cancer cell proliferation by
targeting EZH2 and regulating Wnt signaling. FEBS J. 280:4531–4538.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iwaya T, Yokobori T, Nishida N, et al:
Downregulation of miR-144 is associated with colorectal cancer
progression via activation of mTOR signaling pathway.
Carcinogenesis. 33:2391–2397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zha W, Cao L, Shen Y and Huang M: Roles of
Mir-144-ZFX pathway in growth regulation of non-small-cell lung
cancer. PLoS One. 8:e741752013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katayama Y, Maeda M, Miyaguchi K, et al:
Identification of pathogenesis-related microRNAs in hepatocellular
carcinoma by expression profiling. Oncol Lett. 4:817–823.
2012.PubMed/NCBI
|
|
12
|
Zhang LY, Ho-Fun Lee V, Wong AM, et al:
MicroRNA-144 promotes cell proliferation, migration and invasion in
nasopharyngeal carcinoma through repression of PTEN.
Carcinogenesis. 34:454–463. 2013. View Article : Google Scholar
|
|
13
|
Hers I, Vincent EE and Tavaré JM: Akt
signalling in health and disease. Cell Signal. 23:1515–1527. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stambolic V and Woodgett JR: Functional
distinctions of protein kinase B/Akt isoforms defined by their
influence on cell migration. Trends Cell Biol. 16:461–466. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nassirpour R, Mehta PP and Yin MJ: miR-122
regulates tumorigenesis in hepatocellular carcinoma by targeting
AKT3. PLoS One. 8:e796552013. View Article : Google Scholar : PubMed/NCBI
|