Introduction
Neuroblastoma (NB) is the most common solid
extracranial tumor in children, accounting for 15% of all childhood
cancer-associated mortalities. In spite of significant advances in
detection and effort to reduce recurrence rates, the prognosis of
NB remains poor. The predominant obstacle of NB therapy is
recurrence, which is characterized by metastasis (1,2).
Therefore, it is urgent to identify the molecular mechanism
involved in NB metastasis.
MicroRNAs (miRNAs) regulate gene expression by
inhibiting gene translation or facilitating mRNA degradation
(3). In previous years,
accumulating evidence has revealed that miRNAs modulate a variety
of genes pivotal for tumor development (4). Deregulation of miRNAs contribute to
abnormal expression of oncogenes and tumor suppressors. Previously,
miRNA (miR)-203 has been reported to be involved in the development
of certain types of cancer. However, the detailed role of miR-203
in NB, as well as the underlying molecular mechanism, remain to be
elucidated.
Sam68 is a member of the K homology
domain-containing, RNA-binding, signal transduction-associated
protein family, and has been suggested to be involved in various
cellular processes, including RNA 3′-end formation, alternative
splicing, cell cycle regulation, adipogenesis, neuronal development
and tumorigenesis (5–7). Previously, Sam68 was identified to be
frequently upregulated in cancer and has an oncogenic role in
inflammatory carcinogenesis, tumor invasion and metastasis
(8,9). Sam68 is predicated to be a target
gene of miR-203, according to bioinformatic analysis, however,
whether miR-203 directly mediates the expression of Sam68 and their
roles in NB remain to be elucidated.
In the present study, the expression levels of Sam68
and miR-203 were determined in NB tissues and adjacent normal
tissues. The roles of Sam68 and miR-203 in the regulation of cell
proliferation, migration and invasion of human SK-N-SH and SH-SY5Y
NB cell lines were further assessed. In addition, the targeting
association between miR-203 and Sam68 was determined in SK-N-SH and
SH-SY5Y cells.
Materials and methods
Reagents
TRIzol reagent, Dulbecco's modified Eagle's medium
(DMEM), fetal bovine serum (FBS), Lipofectamine 2000, Opti-MEM
medium and an mirVana™ qRT-PCR microRNA Detection kit were
purchased from Life Technologies (Carlsbad, CA, USA). A standard
SYBR Green RT-PCR kit was purchased from Takara (Otsu, Japan).
Sam68 siRNA, miR-203 mimics and scramble miRNA were purchased from
Genecopoeia (Guangzhou, China). A Bradford DC protein assay kit was
purchased from Bio-Rad Laboratories, Inc. (Hercules, CA, USA).
Mouse anti-Sam68 (cat. no. sc-1238) and mouse anti-GAPDH (sc-59540)
antibodies, and rabbit anti-mouse secondary antibody (cat. no.
sc-358943) were purchased from Santa Cruz Biotechnology, Inc.
(Dallas, TX, USA). An enhanced chemiluminescence (ECL) kit was
purchased from Pierce Chemical (Rockford, IL, USA). A cell invasion
assay kit was purchased from Chemicon (Temecula, CA, USA) and a
Quick-Change Site-Directed Mutagenesis kit was purchased from
Stratagene (La Jolla, CA, USA). The PsiCHECK™2 vector and Dual-Glo
substrate system was purchased from Promega (Madison, WI, USA).
Tissue specimen collection and cell
culture
The present study was approved by the Ethics
Committee of Xinxiang Medical University (Xinxiang, China). A total
of 16 NB tissues and their matched normal adjacent tissues were
collected at the Department of Pediatrics, The First Affiliated
Hospital of Xinxiang Medical University (Weihui, China). Informed
consent was obtained from the parents/guardians of the patients.
Human SK-N-SH and SH-SY5Y NB cell lines were obtained from China
Center for Type Culture Collection (Wuhan, China). The cells were
cultured in DMEM, supplemented with 10% FBS, 100 IU/ml penicillin
and 100 μg/ml streptomycin sulfate (Life Technologies) at
37°C in a humidified incubator, containing 5% CO2.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
The total RNA was extracted from cells using TRIzol
reagent. The relative expression of miR-203 was determined by
RT-qPCR using a mirVana™ qRT-PCR microRNA Detection kit, according
to the manufacturer's instructions. The mRNA expression of Sam68
was detected by RT-qPCR using the standard SYBR Green RT-PCR kit,
according to the manufacturer's instructions. GAPDH was used as an
internal control. The specific primer pairs (Genecopoeia) were as
follows: Sam68, sense: 5′-ATTCTTGGACCACAAGGGAATAC-3′ and antisense:
5′-GCCATAAGAGCATAAGCCTCACA-3′; GAPDH, sense:
5′-GGAGCGAGATCCCTCCAAAAT-3′ and antisense:
5′-GGCTGTTGTCATACTTCTCATGG-3′. The relative mRNA expression levels
of Sam68 or miR-203 were quantified using the GraphPad Prism 4.0
software (GraphPad Software, San Diego, CA, USA) and the
2−ΔΔCt method (10).
Transfection
Transfection was performed using Lipofectamine 2000,
according to the manufacturer's instructions. At the time of
transfection, the cells were at least 70% confluent.
Oligonucleotides and plasmids were incubated in Opti-MEM medium at
37°C for 20 min. For miR-148a functional analysis, the cells were
transfected with 100 nM pre-miR-203, anti-miR-203, pre-miR-con and
anti-miR-con. For Sam68 functional analysis, the cells were
transfected with Sam68-specific siRNA.
Cell proliferation assay
For each group, 10,000 cells/well were seeded into a
96-well plate. Following transfection, the plates were incubated
for 48 h at 37°C with 5% CO2. To assess cell
proliferation, an MTT assay was performed, according to the
manufacturer's instructions. MTT reagent (10 μl; 5 mg/ml;
Life Technologies) in phosphate-buffered saline was added to each
well and incubated for 4 h at 37°C with 5% CO2. The
supernatant was subsequently removed and 100 μl dimethyl
sulfoxide was added. The absorbance was measured at 490 nm using a
Microplate Reader (Model 680; Bio-Rad Laboratories, Inc.). Each
assay was performed in triplicate wells and repeated three
times.
Cell migration assay
Cell migration was measured using a wound healing
assay. Briefly, the cells were cultured to confluence. Wounds of
~1 mm width were created with a plastic tip. Following
wounding, the cells were incubated for 24 h and were subsequently
fixed with 16% trichloroacetic acid and observed under a microscope
(IX83; Olympus Corporation, Tokyo, Japan).
Cell invasion assay
Cell invasion was measured using a Cell Invasion
Assay kit, according to the manufacturer's instruction. Briefly,
the cells in serum-free DMEM were seeded into the upper compartment
of the chambers and DMEM, containing 10% FBS, was added into the
lower chambers. Following incubation for 24 h at 37°C, the cells on
the upper face of the membrane were removed using a cotton swab and
t he cells on the lower face were fixed with 16% trichloroacetic
acid, stained with 0.1% crystal violet (Life Technologies) and
observed under a microscope (IX83; Olympus Corporation).
Western blotting
The total protein was extracted using the Tissue or
Cell Protein Extraction kit (Amresco, Beijing, China) and the
protein concentration was measured using the Bradford DC protein
assay kit. The proteins were separated by 10% SDS-PAGE (Pierce
Chemical) and blotted onto polyvinylidene difluoride membranes
(PVDF; Pierce Chemical), which were subsequently blocked with
Tris-buffered saline with 0.5% Tween-20, containing 50 g/l non-fat
milk, at room temperature for 4 h. Following incubation, the PVDF
membrane was incubated with the primary antibodies, anti-Sam68 and
anti-GAPDH (1:200 dilution), at 37°C for 1 h. The membranes were
rinsed with Dulbecco's phosphate-buffered saline (DPBS; Life
Technologies) and incubated for 1 h with the corresponding
peroxidase-conjugated rabbit anti-mouse secondary antibody
(1:20,000 dilution). The membranes were further washed with DPBS
and chemiluminescent detection was performed using the ECL kit.
Dual luciferase reporter assay
The SH-SY5Y cells were co-transfected with the
reporter constructs, Sam68-3′untranslated region (UTR)-psi-CHECK2
(containing the 3′-UTR of Sam68 and the miRNA-203 binding sites) or
the mutated (Mut)-Sam68-3′UTR-psi-CHECK2 (containing the
corresponding mutated sequence of 3′-UTR of Sam68), and the miR-203
mimics or scramble miRNA as a negative control, using Lipofectamine
2000. The luciferase activity was determined after 48 h using the
Dual-Glo substrate system and LD400 luminometer (Beckman Coulter,
Kraemer Boulevard Brea, CA, USA). The data are presented as the
ratio of Renilla luciferase to Firefly luciferase.
Statistical analysis
The data are expressed as the mean ± standard
deviation. Differences between the two groups were determined by
Student t-test. All analyses were performed using SPSS 17.0
software (IBM SPSS, Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Inverse expression levels of Sam68 and
miR-203 in NB tissues
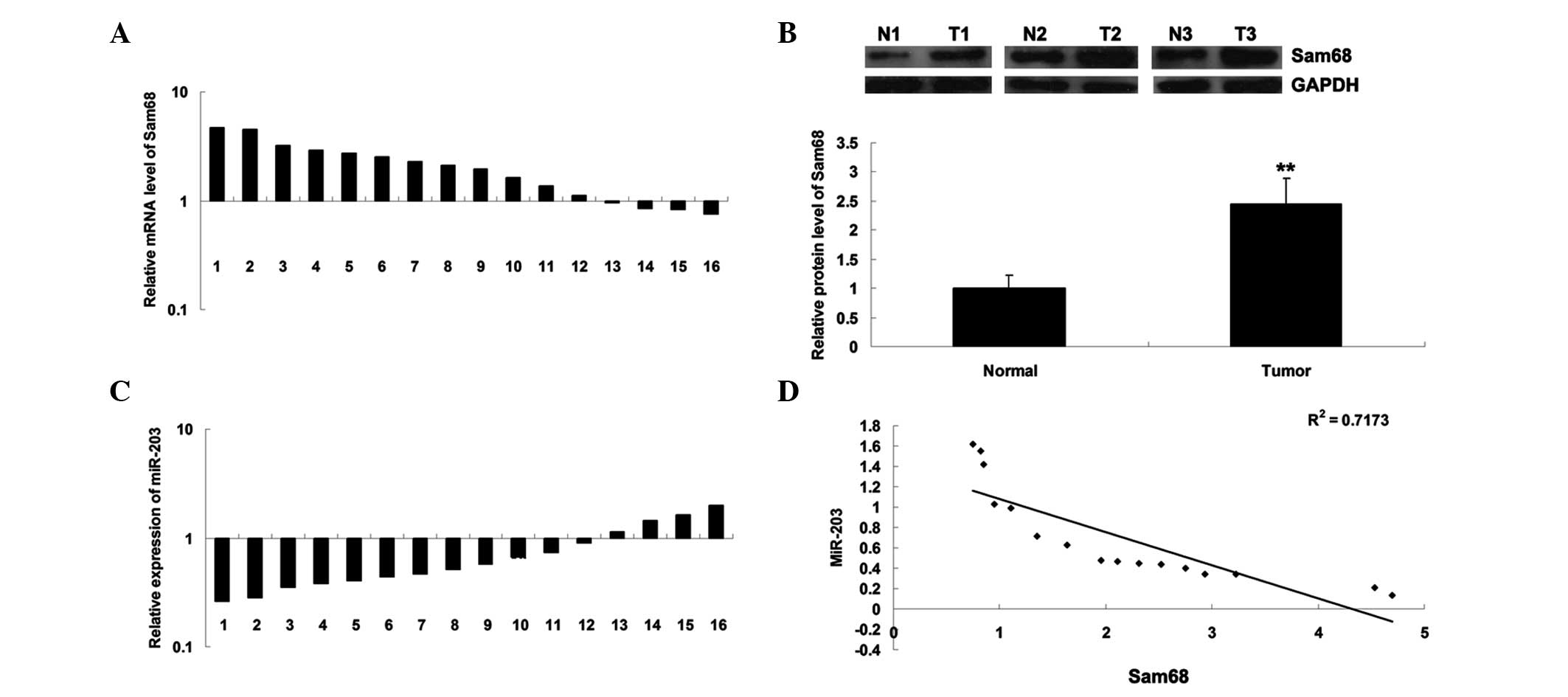
The present study investigated the expression of
Sam68 in NB tissues. As shown in Fig.
1A and B, the mRNA and protein expression levels of Sam68 were
frequently upregulated in the NB tissues compared with their
matched adjacent normal tissues. It was further demonstrated that
the expression level of miR-203 was frequently reduced in the NB
tissues compared with their matched normal adjacent tissues
(Fig. 1C). Notably, an inverse
correlation between the expression levels of miR-203 and Sam68 were
observed (R2=0.7173, Fig.
1D). Therefore, it was suggested that deregulation of Sam68 and
miR-203 may be important in NB.
Knockdown of Sam68 inhibits the
proliferation, migration and invasion of NB cells
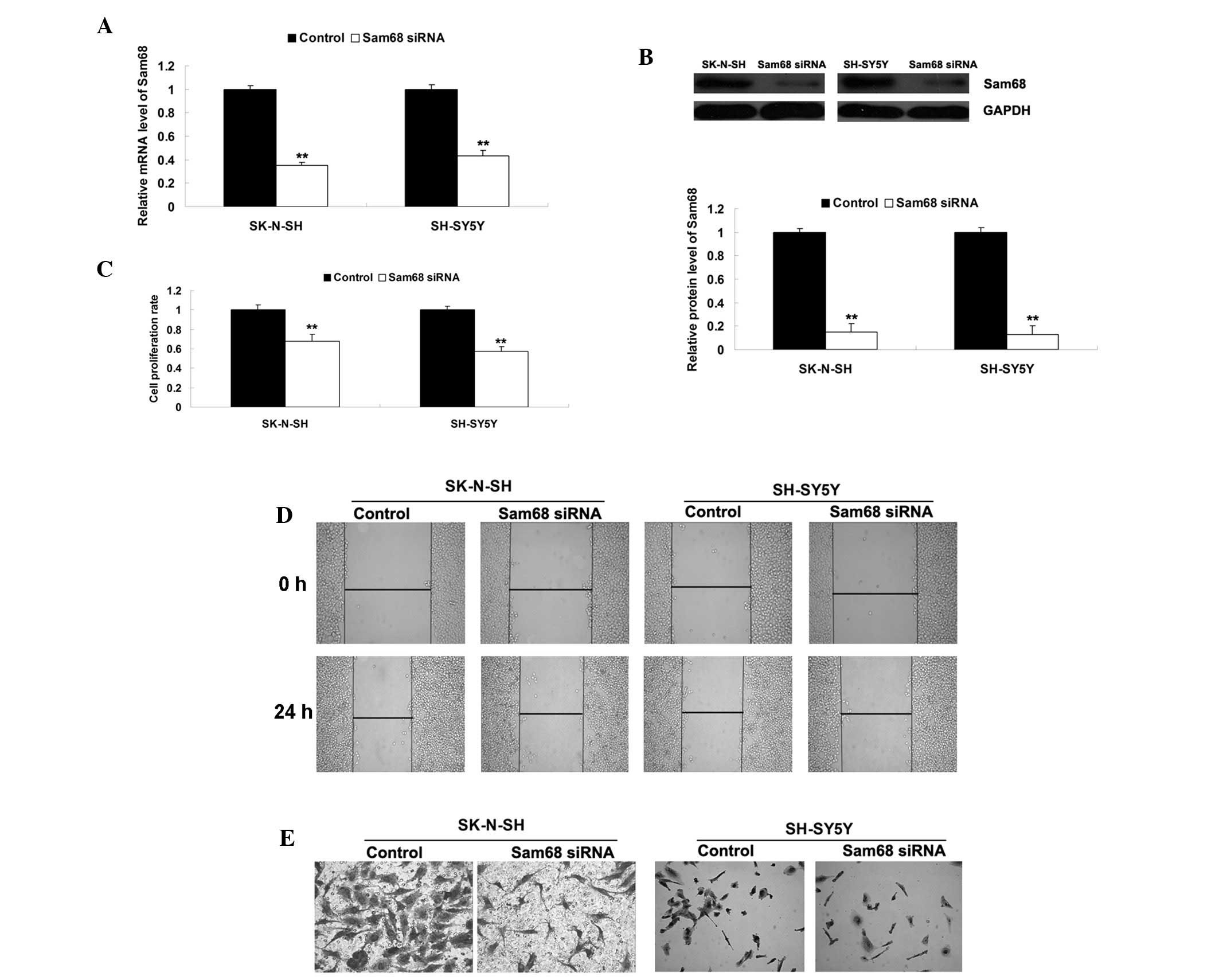
To investigate the role of Sam68 in NB, the SK-N-SH
and SH-SY5Y cells were transfected with Sam68-specific siRNA. As
shown in Fig. 2A and B, the mRNA
and protein expression levels of Sam68 were significantly decreased
in the SK-N-SH and SH-SY5Y cells following transfection with
Sam68-specific siRNA. It was also demonstrated that the
downregulation of Sam68 inhibited the proliferation, migration and
invasion of the SK-N-SH and SH-SY5Y cells (Fig. 2C–E). These findings suggested that
Sam68 may be involved in the progression of NB.
Overexpression of miR-203 suppresses the
proliferation, migration and invasion of NB cells
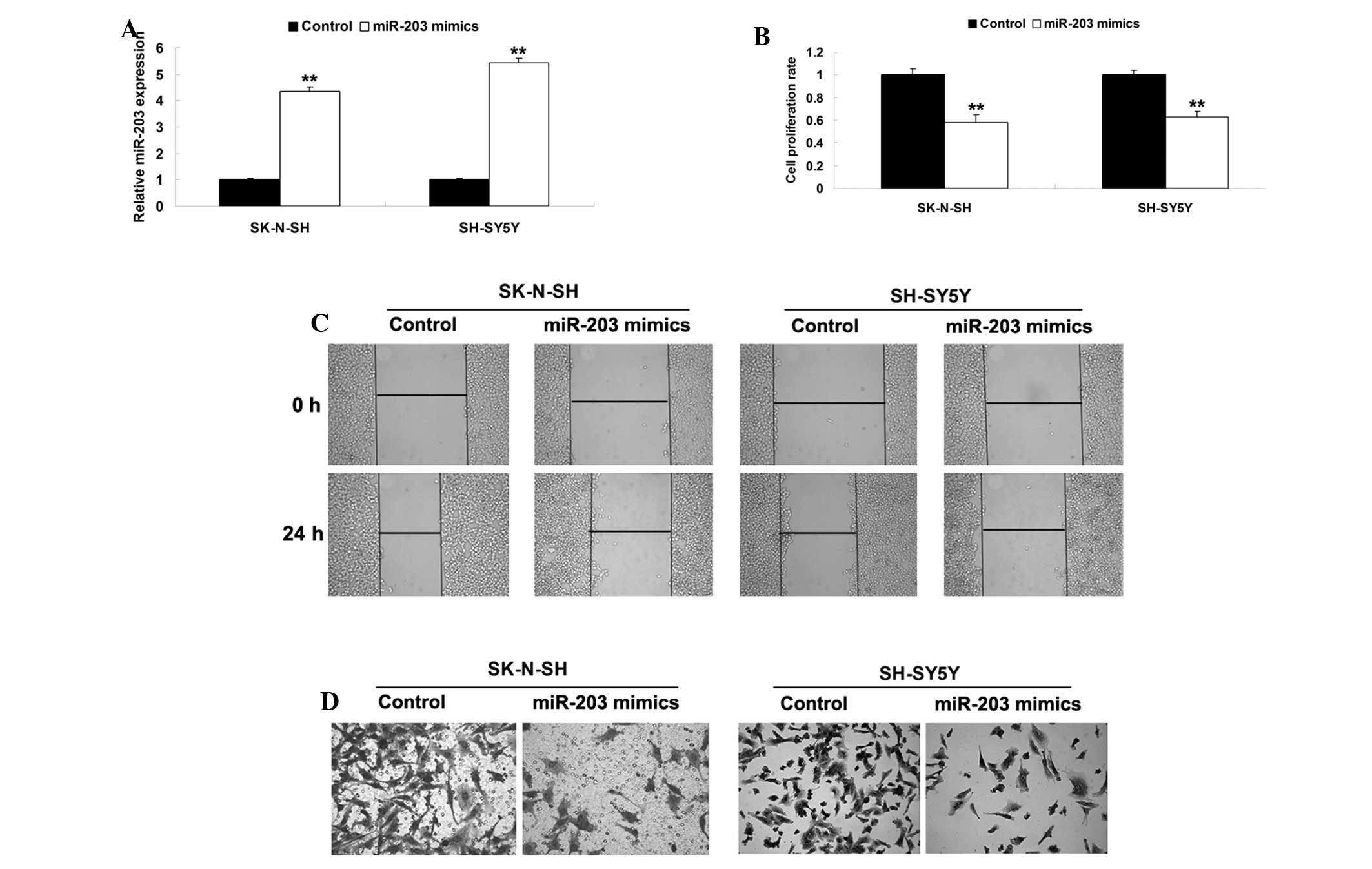
The present study further assessed the role of
miR-203 in NB. Since miR-203 was observed to be downregulated in
NB, the SK-N-SH and SH-SY5Y cells were transfected with pre-miR-203
plasmid. As shown in Fig. 3A,
transfection with pre-miR-203 significantly increased the
expression levels of miR-203 in the SK-N-SH and SH-SY5Y cells. It
was demonstrated that the upregulation of miR-203 markedly
suppressed proliferation, migration and invasion of the SK-N-SH and
SH-SY5Y cells (Fig. 3B–D). These
findings suggested that miR-203 functions as a tumor suppressor in
NB.
miR-203 negatively regulates the
expression of Sam68 by directly binding to its 3′-UTR
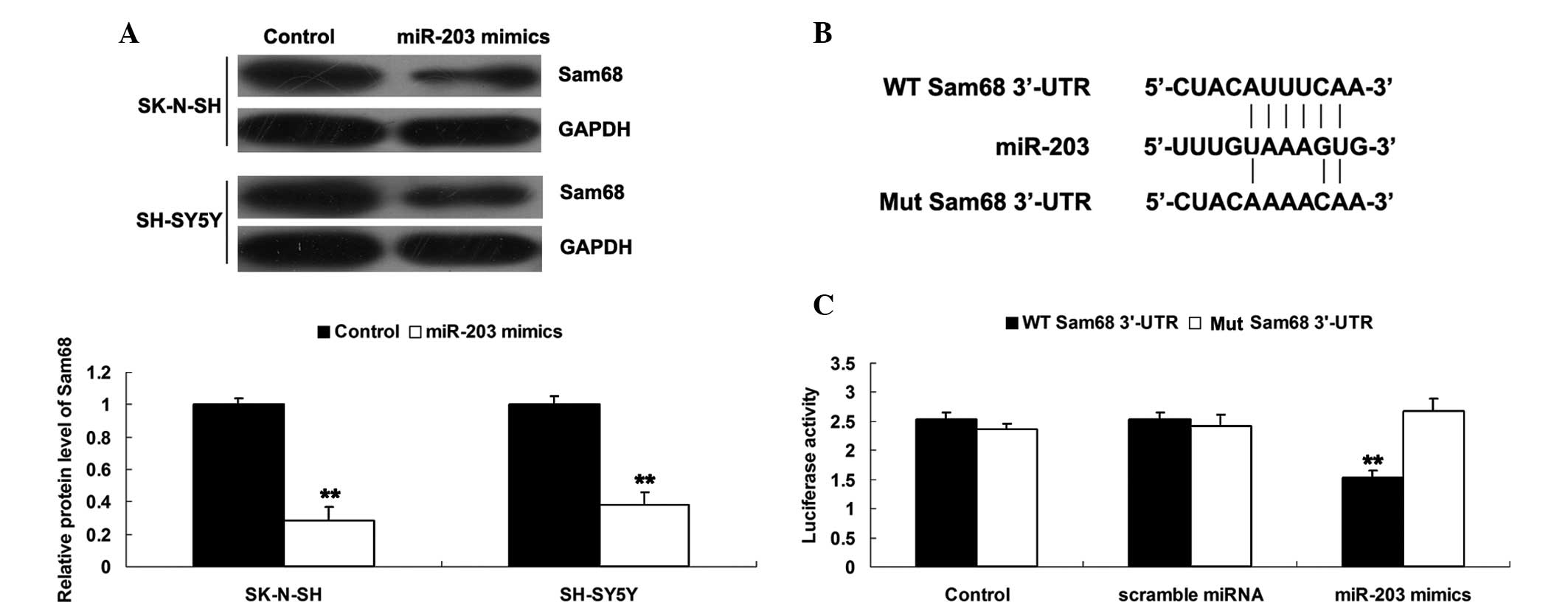
The effects of the upregulation of miR-203 on the
expression of Sam68 were further assessed. The SK-N-SH and SH-SY5Y
cells were transfected with miR-203 mimics and the protein
expression of Sam68 was determined by western blotting. As shown in
Fig. 4A, overexpression of miR-203
significantly inhibited the protein expression of Sam68 in the
SK-N-SH and SH-SY5Y cells. These data indicated that miR-203
negatively mediated the expression of Sam68 in NB cells.
Whether Sam68 is a direct target gene of miR-203 was
next investigated. The Sam68 3′-UTR, containing the wild type (WT)
or mutant type (Mut) of the miR-203 binding site, was subcloned
into psi-CHECK2 dual luciferase reporter vectors (Fig. 4B). As shown in Fig. 4C, overexpression of miR-203
markedly decreased the luciferase activity in the SH-SY5Y cells
transfected with the WT Sam68-3′UTR-psi-CHECK2 reporter vector,
however, the luciferase activity in the SH-SY5Y cells
co-transfected with the Mut-Sam68-3′UTR-psi-CHECK2 rep orter vector
and miR-203 exhibited no difference compared with the control group
(Fig. 4D). This indicated that
Sam68 is a novel target of miR-203. Based on these findings, the
upregulation of Sam68 may be induced partly by the downregulation
of miR-203 and miR-203 may act as a tumor suppressor in NB by
targeting Sam68.
Discussion
Sam68 localizes in the nucleus and is involved in
the formation of both nuclear and cytosolic multi-molecular
complexes, including stress granules and Sam68 nuclear bodies
(11). In addition, Sam68 mediates
gene expression by coupling with other proteins and RNA targets,
and therefore, is involved in the regulation of certain important
cellular functions, including cell proliferation, differentiation
and death (11). For instance,
Sam68 was recently identified to modulate the promoter specificity
of NF-κB and regulate the expression of CD25 in activated T cells
(12).
Deregulation of oncogenes or tumor suppressors is
critical in the initiation and progression of cancer. It has been
reported that high expression of Sam68 is a is a predictor of poor
prognosis in non-small cell lung cancer, oral tongue cancer,
early-stage cervical cancer and colorectal cancer (8,9,13,14).
In the present study, the expression level of Sam68 was
demonstrated to be higher in NB tissues compared with their matched
adjacent normal tissues. It was also demonstrated that the
knockdown of Sam68 effectively inhibited the proliferation,
migration and invasion of the SK-N-SH and SH-SY5Y NB cells.
Previously, Zhao et al (15) also reported that Sam68 was
upregulated in NB tissues and cell lines. Furthermore, the authors
revealed that the protein expression of Sam68 was positively
correlated with clinical stage, tumor histology and distant
metastasis, and that high expression of Sam68 was a predictor of
poor prognosis of patients with NB (15). Taken together, the present study
suggested that the deregulation of Sam68 may be involved in the
development and progression of NB.
The regulatory mechanisms of oncogenes, including
methylation, transcription factors and mutation, are complicated.
Previously, accumulating evidence has demonstrated that the
deregulation of oncogenes or tumor suppressors is due to the
aberrant expression of its regulatory miRNAs. However, the
molecular mechanism of the expression of Sam68 in NB remains to be
elucidated. In the present study, the expression of miR-203, a
theoretical regulatory miRNA of Sam68 predicted by algorithms, was
assessed in NB tissues and cell lines. The data revealed that the
expression level of miR-203 was significantly reduced in NB tissues
compared with their matched adjacent normal tissues, suggesting
that the aberrant downregulation of miR-203 may be involved in the
development and progression of NB. In fact, deregulation of miR-203
has been demonstrated to be associated with various cancer types,
including hepatocellular carcinoma, pancreatic cancer, glioma, lung
cancer, cervical cancer, prostate carcinoma, melanoma, esophageal
squamous cell carcinoma and colon cancer (16–25).
The present study further revealed that the
overexpression of miR-203 markedly suppressed the proliferation,
migration and invasion of the SK-N-SH and SH-SY5Y NB cells, similar
to the effects of Sam68 downregulation. In addition, the present
study demonstrated for the first time, to the best of our
knowledge, that Sam68 is a novel target of miR-203, and that
miR-203 and these miRNAs negatively regulated the expression of
Sam68 by targeting its 3′-UTR in NB cells. Therefore, it is
possible that high expression of Sam68 in NB is partly due to the
downregulation of the expression of miR-203. The suppressive role
of miR-203 in human cancer has been widely investigated. For
instance, Tian et al (26)
demonstrated that the expression of miR-203 was significantly
reduced in laryngeal squamous cell carcinoma tissues, and
downregulation of miR-203 was correlated with advanced clinical
stages, lymph node metastasis and poor prognosis (26).
An miRNA has various target genes and a gene can be
regulated by multiple miRNAs. It has been demonstrated that miR-203
can target survivin, leading to cell cycle arrest in laryngeal
carcinoma cells (27). It can also
target Hakai and inhibit the growth of human colon adenocarcinoma
(28). Additionally, those genes
involved in the regulation of cell proliferation, apoptosis, cell
cycle progression, migration and invasion, are also the targets of
miR-203, including Robo1, Kif5b, E2F3, LASP1, ASAP1, PKCα, BIRC5,
Bmi-1 and DeltaNp63 (23–26,29–33).
Whether these miR-203 targets are also involved in the development
and progression of NB remains to be elucidated.
In conclusion, the present study demonstrated that
Sam68 is upregulated in NB tissues and cell lines, which is likely
due to the downregulation of the expression of miR-203. In
addition, the overexpression of miR-203 effectively inhibited the
proliferation, migration and invasion of NB cell lines, partly at
least, by directly targeting Sam68.
References
|
1
|
Fulda S: The PI3K/Akt/mTOR pathway as
therapeutic target in neuroblastoma. Curr Cancer Drug Targets.
9:729–737. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jrebi NY, Iqbal CW, Joliat GR, Sebo TJ and
Farley DR: Review of our experience with neuroblastoma and
ganglioneuroblastoma in adults. World J Surg. 2871–2874. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baer C, Claus R and Plass C: Genome-wide
epigenetic regulation of miRNAs in cancer. Cancer Res. 73:473–477.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vogel G and Richard S: Emerging roles for
Sam68 in adipogenesis and neuronal development. RNA Biol.
9:1129–1133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Najib S, Martin-Romero C, Gonzalez-Yanes C
and Sanchez-Margalet V: Role of Sam68 as an adaptor protein in
signal transduction. Cell Mol Life Sci. 62:36–43. 2005. View Article : Google Scholar
|
|
7
|
Lukong KE and Richard S: Sam68, the KH
domain-containing superSTAR. Biochim Biophys Acta. 1653:73–86.
2003.PubMed/NCBI
|
|
8
|
Chen SW, Zhang Q, Yang AK, Li Z, Zhong Y,
Li H, Zeng Y, Zhuang SM, Wang LP, Song LB, et al: Overexpression
and cytoplasmic localization of Sam68 correlate with tumour
progression and poor prognosis in patients with clinically N0 oral
tongue cancer. Head Neck Oncol. 4:612012.
|
|
9
|
Zhang Z, Xu Y, Sun N, Zhang M, Xie J and
Jiang Z: High Sam68 expression predicts poor prognosis in non-small
cell lung cancer. Clin Transl Oncol. 16:886–891. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arocho A, Chen B, Ladanyi M and Pan Q:
Validation of the 2-DeltaDeltaCt calculation as an alternate method
of data analysis for quantitative PCR of BCR-ABL P210 transcripts.
Diagn mol pathol. 15:56–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sánchez-Jiménez F and Sánchez-Margalet V:
Role of Sam68 in post-transcriptional gene regulation. Int J Mol
Sci. 14:23402–23419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu K, Sun X, Zheng W, Wier EM, Hodgson A,
Tran DQ, Richard S and Wan F: Sam68 modulates the promoter
specificity of NF-kB and mediates expression of CD25 in activated T
cells. Nat Commun. 4:19092013. View Article : Google Scholar
|
|
13
|
Liao WT, Liu JL, Wang ZG, Cui YM, Shi L,
Li TT, Zhao XH, Chen XT, Ding YQ and Song LB: High expression level
and nuclear localization of Sam68 are associated with progression
and poor prognosis in colorectal cancer. BMC Gastroenterol.
14:1262014.
|
|
14
|
Li Z, Yu CP, Zhong Y, Liu TJ, Huang QD,
Zhao XH, Huang H, Tu H, Jiang S, Zhang Y, et al: Sam68 expression
and cytoplasmic localization is correlated with lymph node
metastasis as well as prognosis in patients with early-stage
cervical cancer. Ann Oncol. 23:638–646. 2012. View Article : Google Scholar
|
|
15
|
Zhao X, Li Z, He B, Liu J, Li S, Zhou L,
Pan C, Yu Z and Xu Z: Sam68 is a novel marker for aggressive
neuroblastoma. Onco Targets Ther. 6:1751–1760. 2013.PubMed/NCBI
|
|
16
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar
|
|
17
|
Greither T, Grochola LF, Udelnow A,
Lautenschlager C, Wurl P and Taubert H: Elevated expression of
microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated
with poorer survival. Int J Cancer. 126:73–80. 2010. View Article : Google Scholar
|
|
18
|
He J, Deng Y, Yang G and Xie W:
MicroRNA-203 down-regulation is associated with unfavorable
prognosis in human glioma. J Surg Oncol. 108:121–125. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheung TH, Man KN, Yu MY, Yim SF, Siu NS,
Lo KW, Doran G, Wong RR, Wang VW, Smith DI, et al: Dysregulated
microRNAs in the pathogenesis and progression of cervical neoplasm.
Cell Cycle. 11:2876–2884. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Boll K, Reiche K, Kasack K, Mörbt N,
Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn
F, et al: MiR-130a, miR-203 and miR-205 jointly repress key
oncogenic pathways and are downregulated in prostate carcinoma.
Oncogene. 32:277–285. 2013. View Article : Google Scholar
|
|
21
|
Kozubek J, Ma Z, Fleming E, Duggan T, Wu
R, Shin DG and Dadras SS: In-depth characterization of microRNA
transcriptome in melanoma. PLoS One. 8:e726992013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang C, Wang X, Liang H, Wang T, Yan X,
Cao M, Wang N, Zhang S, Zen K, Zhang C, et al: miR-203 Inhibits
cell proliferation and migration of lung cancer cells by targeting
PKCα. PLoS One. 8:e739852013. View Article : Google Scholar
|
|
24
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J,
Cheng HZ and Huang SD: MicroRNA-203 inhibits cell proliferation by
repressing ΔNp63 expression in human esophageal squamous cell
carcinoma. BMC Cancer. 11:572011. View Article : Google Scholar
|
|
26
|
Tian L, Li M, Ge J, Guo Y, Sun Y, Liu M
and Xiao H: MiR-203 is downregulated in laryngeal squamous cell
carcinoma and can suppress proliferation and induce apoptosis of
tumours. Tumour Biol. 35:5953–5963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bian K, Fan J, Zhang X, Yang XW, Zhu HY,
Wang L, Sun JY, Meng YL, Cui PC, Cheng SY, et al: MicroRNA-203
leads to G1 phase cell cycle arrest in laryngeal carcinoma cells by
directly targeting survivin. FEBS Lett. 586:804–809. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abella V, Valladares M, Rodriguez T, Haz
M, Blanco M, Tarrío N, Iglesias P, Aparicio LA and Figueroa A:
miR-203 regulates cell proliferation through its influence on Hakai
expression. PLoS One. 7:e525682012. View Article : Google Scholar
|
|
29
|
Dontula R, Dinasarapu A, Chetty C, Pannuru
P, Herbert E, Ozer H and Lakka SS: MicroRNA 203 modulates glioma
cell migration via Robo1/ERK/MMP-9 Signaling. Genes Cancer.
4:285–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noguchi S, Kumazaki M, Yasui Y, Mori T,
Yamada N and Akao Y: MicroRNA-203 regulates melanosome transport
and tyrosinase expression in melanoma cells by targeting kinesin
superfamily protein 5b. J Invest Dermatol. 134:461–469. 2014.
View Article : Google Scholar
|
|
31
|
Noguchi S, Mori T, Otsuka Y, Yamada N,
Yasui Y, Iwasaki J, Kumazaki M, Maruo K and Akao Y: Anti-oncogenic
microRNA-203 induces senescence by targeting E2F3 protein in human
melanoma cells. J Biol Chem. 287:11769–11777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takeshita N, Mori M, Kano M, Hoshino I,
Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, et
al: miR-203 inhibits the migration and invasion of esophageal
squamous cell carcinoma by regulating LASP1. Int J Oncol.
41:1653–1661. 2012.PubMed/NCBI
|
|
33
|
Yu X, Jiang X, Li H, Guo L, Jiang W and Lu
SH: MiR-203 inhibits the proliferation and self-renewal of
esophageal cancer stem-like cells by suppressing stem renewal
factor Bmi-1. Stem Cells Dev. 23:576–585. 2014. View Article : Google Scholar
|