Introduction
The purpose of orthodontic treatment is to achieve
optimal occlusion for an individual. Orthodontic forces generate
mechanical stress and are a factor that brings about the remodeling
of the periodontal ligament (PDL) and alveolar bone (1,2). The
PDL contains ample vascular and cellular connective tissue,
including Sharpey's fibers, fibroblasts, osteoblasts, osteoclast
precursors and osteoclasts, and surrounds the roots of the teeth.
Osteoclast precursors and osteoclasts are present on the alveolar
bone.
Previous studies have revealed the response of
osteoblasts and fibroblasts to various types of mechanical
stimulation. Mechanical stimuli, including tensile force (3–5),
compressive force (6,7), fluid flow (8), hydrostatic pressure (9), rotation-associated stress (10) and other stimuli (11,12),
are influential factors in bone remodeling. Orthodontic forces
comprise tensile and compressive forces, and lead to a marked
compression of the region of the PDL which they are applied to
(13).
Tensile forces exerted using a Flexercell tension
system suppressed osteoclast differentiation and fusion (4,5).
Furthermore, when compressive forces were applied to osteoblasts
and PDL cells, essential factors that promote osteoclast
differentiation and activation, including prostaglandin
E2 (PGE2), interleukin (IL)-1β, IL-6, tumor
necrosis factor (TNF)-α, receptor activator of nuclear factor κB
ligand (RANKL) and macrophage colony-stimulating factor (M-CSF)
(11,12), were upregulated. Numerous studies
have reported that applying such forces leads to secretion of
cytokines associated with bone resorption and formation (14,15).
However, it has remained elusive whether direct compressive forces
regulate osteoclastogenesis.
Osteoclasts are derived from the monocyte/macrophage
lineage and are formed by the fusion of mononuclear precursors.
RANKL is an essential factor for osteoclast differentiation and
activation (16). RANKL is
secreted by osteoblasts and stromal cells, and is bound to RANK
(14). RANK is induced by M-CSF in
osteoclastogenesis, and is an essential factor for the formation of
mature osteoclasts (17). The
murine macrophage cell line RAW264.7 expresses RANK and can
differentiate into osteoclasts after RANKL stimulation without
M-CSF (5). RANKL signaling induces
the master transcription factor nuclear factor of activated T cells
c1 (NFATc1). NFATc1 is an essential regulator for numerous
osteoclast-specific genes associated with differentiation, fusion
and bone resorption (18).
Therefore, the aim of the present study was to
develop a method for applying compressive forces to RAW cells and
to investigate their osteoclastogenesis induced by applying various
magnitudes of force for various periods of up to 24 h.
Materials and methods
Cell culture
The murine monocyte/macrophage cell line RAW264.7
(TIB-71™; American Type Culture Collection, Manassas, VA, USA) was
used as osteoclast precursors. RAW264.7 cells have been shown to
differentiate into osteoclast-like cells in the presence of RANKL
(5). Cells were cultured in
Dulbecco's modified Eagle's medium (Wako, Osaka, Japan) containing
10% heat-inactivated fetal bovine serum (FBS; Invitrogen Life
Technologies, Carlsbad, CA, USA) and 66.7 μg/ml kanamycin sulfate
(Meiji Seika, Tokyo, Japan) at 37°C in a humidified atmosphere of
95% air and 5% CO2. Cells were seeded onto 100-mm
standard dishes (BD Biosciences, Franklin Lakes, NJ, USA) and
cultured overnight. 12-mm diameter glass cover slips (slips; Fisher
Microscope Cover Glass; Thermo Fisher Scientific, Waltham, MA, USA)
was placed into the wells of a 24-well culture plate (BD
Biosciences). For osteoclast differentiation, RAW264.7 cells were
transferred into the 24-well culture plates at 1.0×104
cells/well and were cultured in α-minimum essential medium (α-MEM;
Wako) supplemented with 10% heat-inactivated FBS, 2 μM
L-alanyl-L-glutamine (Wako), 284 μM L-ascorbic acid
2-phosphate (Sigma-Aldrich, St. Louis, MO, USA), 66.7 μg/ml
kanamycin sulfate and 50 ng/ml RANKL (Oriental Yeast Co., Ltd.,
Tokyo, Japan) at 37°C in a humidified atmosphere of 95% air and 5%
CO2. Medium containing these reagents was replaced every
other day.
Reversal of cells on glass cover
slips
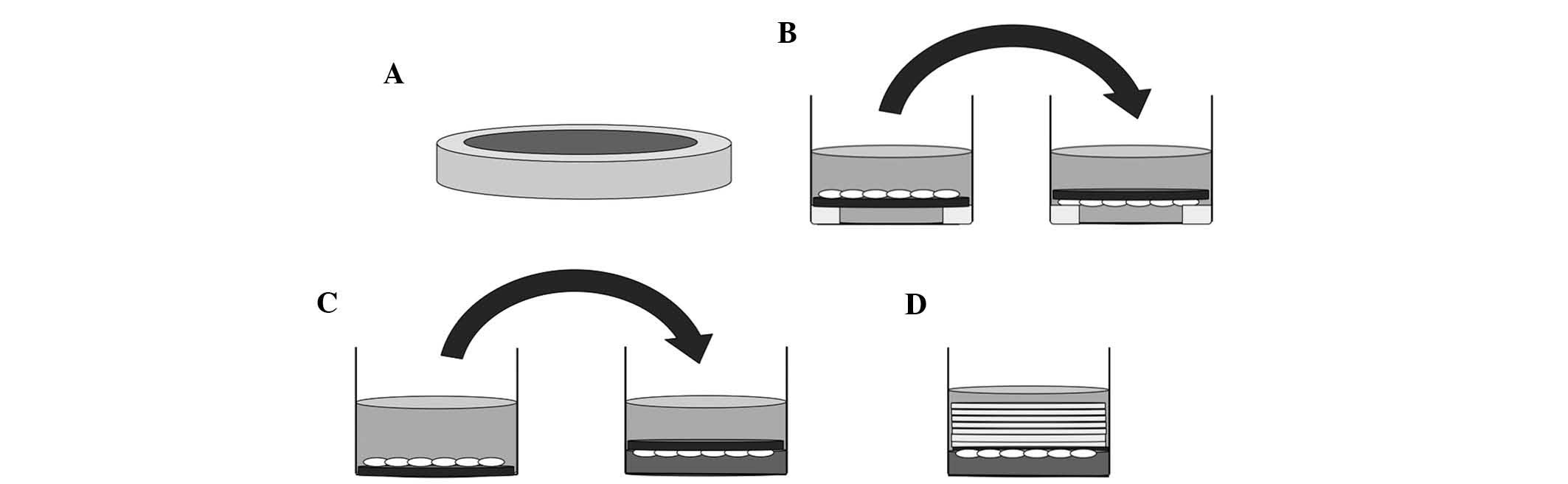
Prior to culture, 15 mm-diameter silicone tubes were
cut into 2-mm slices and sterilized in an autoclave (Fig. 1A). RAW264.7 cells were cultured on
slips which had been placed on sliced silicone tubes in 24-well
culture plates for five days, and the slips were reversed in the
same wells filled with culture medium after three days for 48 h, or
after four days for 24 h (Fig.
1B). The control group was cultured for the same period without
reversing the slips.
Preparation of collagen gels
Acid-soluble collagen solution (Cellmatrix; Nitta
Gelatin, Inc., Osaka, Japan), ten-fold concentrated α-MEM
(Invitrogen Life Technologies), and reconstruction buffer (2.2 g
NaHCO3 and 4.77 g
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid in 100 ml 0.05 N
NaOH; Nittal Gelatin, Inc.) were mixed at a volume ratio of 8:1:1,
and 10% heat-inactivated FBS, 284 µM l-ascorbic acid 2-phosphate
(Sigma-Aldrich) and 2 mM L-alanyl-L-gluta-mine (Wako) were added in
an ice-cold bath kept at <4°C. This prepared collagen mixture
was added to 24-well culture plates at a volume of 500
µl/well, and was subjected to gelation in a CO2
incubator at 37°C for 30 min. After the collagen mixture had
solidified, it was covered with 50 ng/ml RANKL in culture medium at
1 ml/well.
Application of compressive forces
Prior to the application of compressive forces,
RAW264.7 cells were incubated on the slip for three days (Fig. 1C, left). Collagen gel layers were
prepared in other 24-well culture plates. Subsequently, osteoclasts
on the slips were reversed and placed on top of these collagen gel
layers (Fig. 1C, right), and were
continuously compressed with layers of 3, 5, 7, 9 or 14 slips (~43
mg/slip) for 24 h, respectively (Fig.
1D). In the control group, the slips with cells attached were
reversed onto the collagen gel layer without application of any
additional force (Fig. 1C,
right).
Tartrate-resistant acid phosphatase
(TRAP) staining
After cells were compressed for the indicated times,
they were fixed in 10% neutral formalin. They were then washed with
distilled water and stained with Fast Red Violet LB Salt
(Sigma-Aldrich) in TRAP staining solution [acetate buffer (pH 5.0)
(Sigma-Aldrich), naphthol AS-MX phosphate (Sigma-Aldrich) as a
substrate, red violet LB (Sigma-Aldrich) as a stain, and 50 mM
sodium tartrate (Wako)]. TRAP-positive cells with 2–7 nuclei were
considered to be small osteoclasts, and those with >8 nuclei
were considered to be large osteoclasts (3,4).
Osteoclasts were counted under a light microscope (magnification,
×100; Olympus IMT-2; Olympus, Tokyo, Japan) in a 20-mm2
rectangle within the circular field.
Pit assay
Osteologic™ was used to characterize and quantify
osteoclast-mediated bone resorption in order to determine the
amount of osteoclastic differentiation. A disc (φ12-m m) made of
Osteologic™ (BD Biosciences) was coated with calcium phosphate.
Osteoclasts were cultured for three days on these discs under
normal cell culturing conditions as described above, and discs were
reversed onto the top of a collagen gel layer in 24-well culture
plates on the fourth day for 24 has a control group. Another group
of reversed osteoclasts was subjected to the compressive force of
seven layered slips. On the fifth day, the control and compressed
osteoclasts on the discs were rinsed twice with distilled water.
Subsequently, 1 ml bleach solution (6% NaOCl and 5.2% NaCl) was
added to each well, followed by pipetting up and down to remove
cells and incubation for five minutes at room temperature. Discs
were washed with 2 ml distilled water three times and were examined
by light microscopy (magnification, ×100; Olympus IMT-2; Olympus)
after drying (19,20). The resorption area was measured
using Image J software (v. 1.48a; National Institutes of Health,
Bethesda, MD, USA).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Cells were compressed for 1, 3, 6, 12 and 24 h in
24-well culture plates. Total RNA was extracted using TRIzol
(Invitrogen Life Technologies) according to the manufacturer's
instructions. Reverse transcription of 1 µg RNA was
performed to obtain cDNA. The PCR reaction mixture contained 50 mM
Tri-Hcl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 10 mM dithiothreitol,
0.01% Nonidet® P-40 and 50% glycerol. The thermocycling
protocol was as follows: Annealing at 30°C for 10 min, enzyme
reaction at 42°C for 20 min, denaturation at 99°C for 5 min and
cooling at 4°C. cDNA was amplified using Rever Tra Ace-α FSK-101
(Toyobo, Osaka Japan). Primers (Applied Biosystems, Thermo Fisher
Scientific) for the following genes were used: NFATc1 (Mm00479445_m
1), TRAP (Mm00475698_m1), RANK (Mm00437135_m matrix
metalloproteinase-9 (MMP-9; Mm00432271_m1), dendritic cell specific
trans membrane protein (DC-STAMP; Mm01168058_m1), osteoclast
stimulatory trans membrane protein (OC-STAMP; Mm00512445_m1),
integrin-av (Mm00434506_m1), integrin-β3 (Mm00443980_m1),
cathepsin-K (cath-K; Mm00484036_m1), chloride channel 7 (ClC-7;
Mm00442400_m1), adenosine triphosphatase H+ transporting
vacuolar proton pump member I (ATP6i; Mm00469395_g1) and GAPDH
(Mm99999915_g1). Real-time PCR was performed using the ABI Prism
7300 sequence detection system (Applied Biosystems). Data obtained
for each sample were standardized against the expression of GAPDH
and were calculated using the 2−ΔΔCt method (21, 22).
Statistical analysis
Values are expressed as the mean ± standard
deviation. Statistical analysis was performed using Microsoft Excel
for Mac 2011, version 14.4.8 (Redmont, WA, USA). Comparisons
between two groups were analyzed using the two-tailed unpaired
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Ostoclastogenesis increases with
culturing time
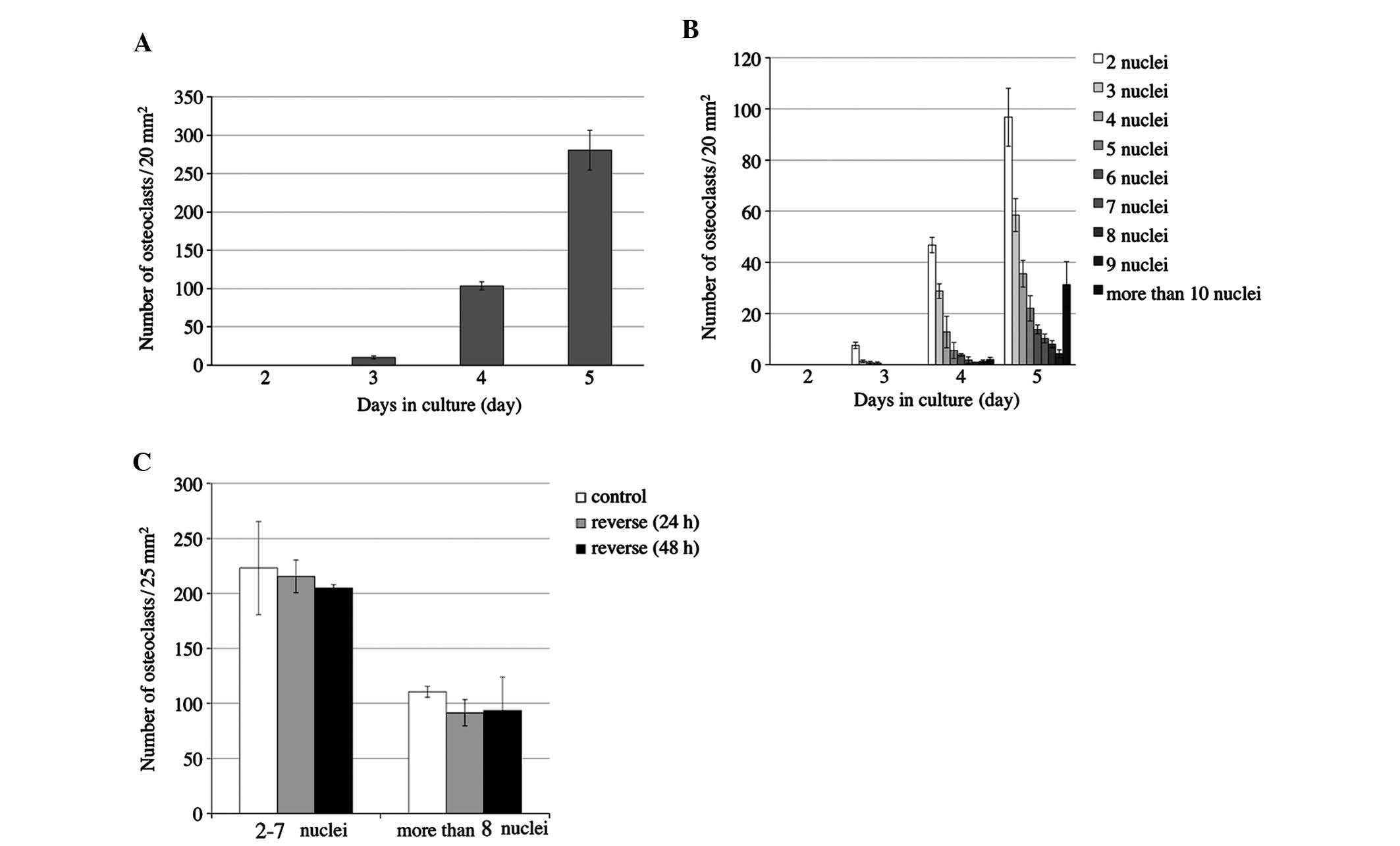
In order to determine at what time-point osteoclast
differentiation and fusion of RAW264.7 cells were activated, the
number of osteoclasts and the number of nuclei per cell were
monitored for five days (Fig. 2).
RAW264.7 cells were cultured with 50 ng/ml RANKL in 24-well culture
plates for five days to induce osteoclastonenesis. No osteoclasts
were identified on day two, while the number of osteoclasts was
slightly increased on days three and four, and was markedly
increased on day five (Fig. 2A).
Osteoclasts contained 2–4 nuclei on day three and began to appear
with >5 nuclei on day four. On day five, the number of
multinuclear cells further increased, and cells with >6 nuclei
were increasingly present (Fig.
2B). Only upon osteoclastic differentiation, RQW264.7 cells
begin to adhere to the cover slips. Day five was therefore
determined to be the ideal time-point for reversing the cover slips
in order to apply compressive forces on the cells.
Reversed culture without application of
compressive forces does not affect osteoclastogenesis
The present study investigated the effects of
reversing the slips on osteoclastogenesis on sliced silicone tubes
for 24 and 48 h (Fig. 1B).
Controls were cultured on slips without reversing and were fixed on
day 5. The number of TRAP-positive cells with 2–7 nuclei and those
with >8 nuclei was counted. The number of osteoclasts among the
reversed cells showed no significant difference when compared with
that in the control group at either 24 h or 48 h (Fig. 2C).
A compressive force exerted by loading
with seven cover slips for 24 h accelerates osteoclastogenesis
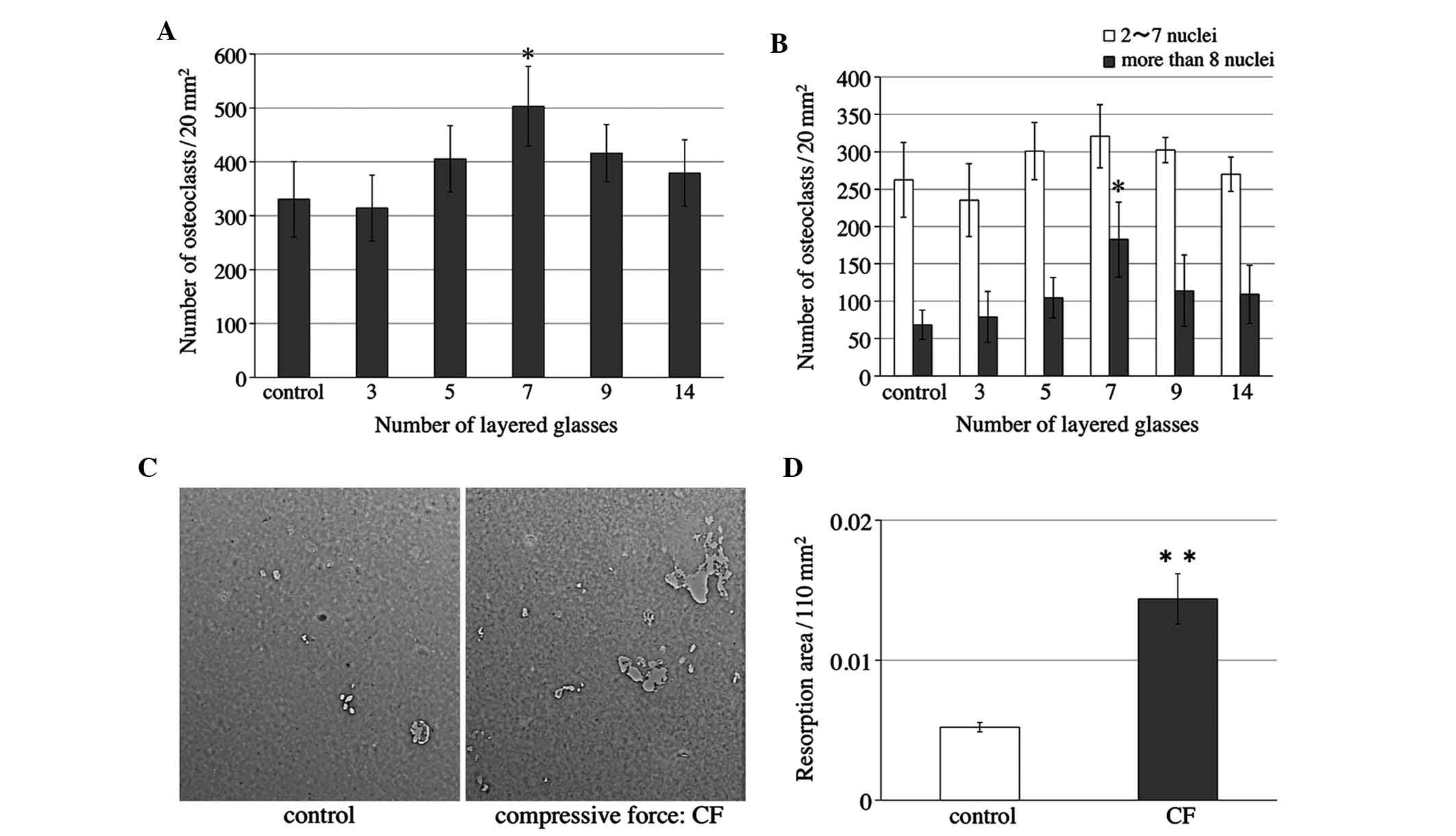
Various intensities of compressive force were
applied to osteoclasts for 24 h after 4–5 days of culture. RAW264.7
cells on slips were cultured as described above. Cells were
reversed onto the collagen gel layer in other plates on the 4th
day. Cells were then loaded with layers of 3–14 slips after being
reversed. The number of osteoclasts was significantly increased
when the cells were compressed with seven slips (Fig. 3A), and the number of osteoclasts
with >8 nuclei was significantly elevated (Fig. 3B).
Bone resorption activity of osteoclasts
subjected to compres-sive forces with 7 slips
The present study then examined the bone resorption
activity of osteoclasts when the cells were subjected to a
compressive force of seven slips. RAW264.7 cells were cultured on
Osteologic™ discs as described above. Light micrographs of the
Osteologic™ discs of the control and compressed (CF) groups were
captured (Fig. 3C). The areas in
which the calcium phosphate on the discs was completely resorbed
were measured. Quantification of the results showed that the
resorption areas in the compressed group were significantly larger
when compared with those in the control group (Fig. 3D).
Expression of osteoclast-associated
mRNA
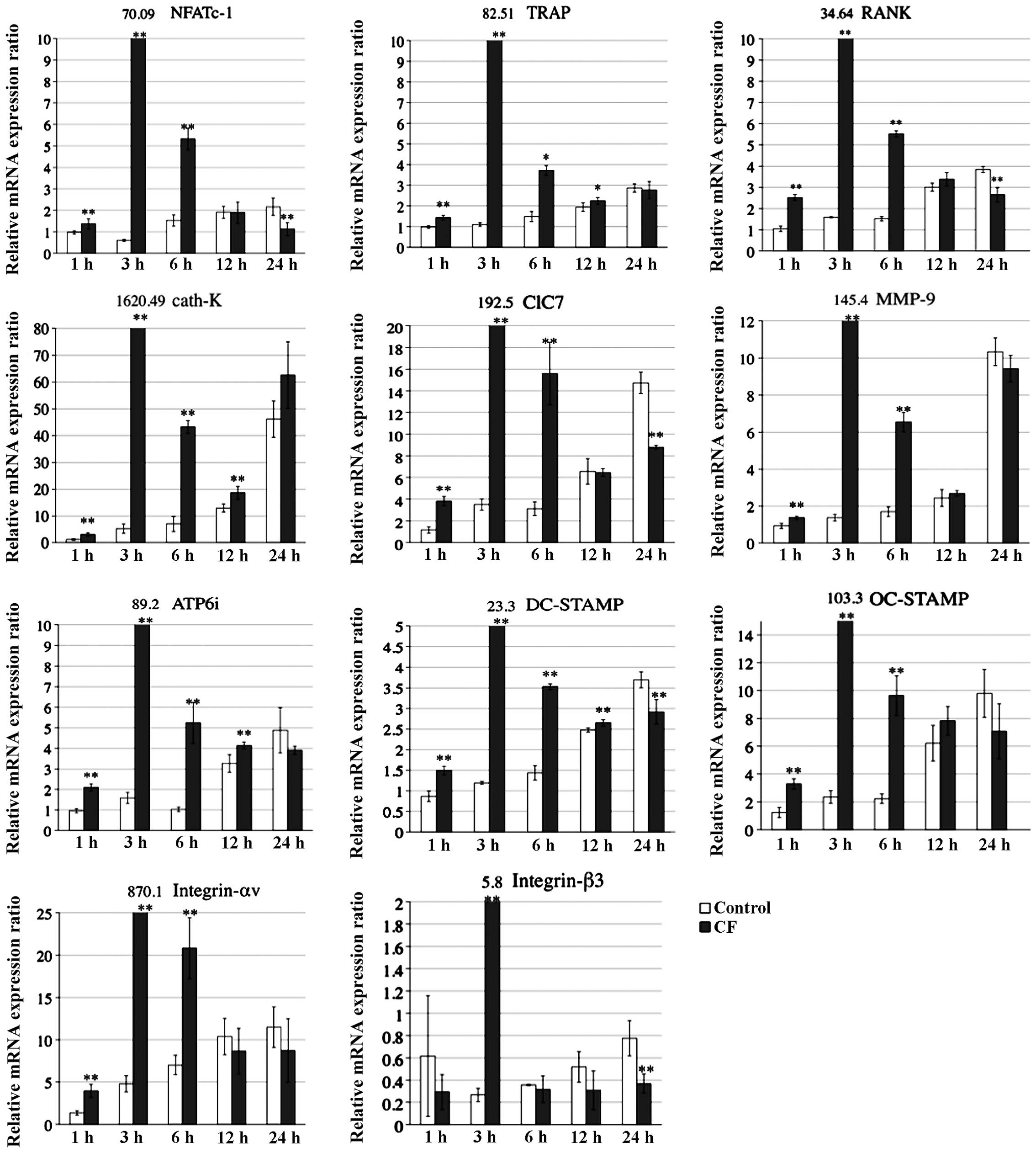
The present study examined the effects of
compressive forces on mRNA levels of osteoclast-specific genes
associated with differentiation, fusion and adherence using RT-qPCR
analysis (Fig. 4). The analysis
showed that the respective mRNA levels of osteoclast-associated
genes, including NFATc-1, TRAP, RANK, cath-K, ClC7, MMP-9, ATP6i,
DC-STAMP, OC-STAMP as well as Integrin-αv and -β3 (4), peaked at 3 h in the CF group, whereas
the mRNA levels peaked at 24 h in the control group.
Discussion
The present study was the first, to the best of our
knowledge, to directly apply compressive forces to
osteoclast-precursor cells using the abovementioned method. It was
shown that application of the optimal compressive force to RAW264.7
cells led to an upregulation of the expression of various
osteoclast-associated genes and promoted osteoclast differentiation
and fusion. Under normal culturing conditions and stimulation with
RANKL, osteoclastogenesis advanced rapidly on days four and five. A
previous study demonstrated that small osteoclasts with 2–7 nuclei
appeared on the third day (22).
Previous studies also showed that formation of osteoclasts
continued to increase up to the sixth day (22,23)
and cells began to undergo apoptosis from the seventh day (3). Therefore, the present study applied
compressive forces to the cells after 4–5 days of culture under
osteoclastic induction conditions.
In a pilot study, compressive force was applied
using layered glass cover slips on osteoclasts cultured on a
24-well culture plate. However, cell counts markedly decreased due
to the hindered supply of culture medium to the cells. Therefore, a
collagen gel (Cellmatrix) was used, against which cells were
pressed in subsequent experiments. The technical manual for
collagen gel noted that the gel was developed for tissue culture,
allowing it to be used for the culture of various cells.
Osteoclastogenesis was not promoted on the collagen gel, and cells
merely underwent cell division to form a mass of mononucleated
cells (results not shown). It was also observed that RAW264.7 cells
differentiated and fused on rigid surfaces. The impaired osteoclast
differentiation observed under these non-adherent conditions was
due to the absence of RANK-dependent signaling by osteoclast
precursors (24). When osteoclasts
were cultured on the bottom of a 24-well culture plate subjected to
compressive forces exerted by the collagen gel layer, it was
difficult to determine the cell number under the light microscope
and to measure mRNA expression in these osteoclasts.
The present study then cultured cells on slips and
reversed them onto the collagen gel layer in order to compress the
osteoclasts. Innutrition and cell death during application of
compression was thereby avoided, and the collagen gel layer was
able to act as a cushion material when compressive forces were
applied to the osteoclasts. Studies have shown that osteoclast
precursors and osteoclasts are present on alveolar bone and
adjacent to compressed PDL (13,25),
which suggested that compressive forces from orthodontic treatment
are transmitted to osteoclasts on the alveolar bone via the PDL.
Application of compressive forces to osteoclasts on slips via the
collagen gel therefore represents an in vitro model of the
application of orthodontic forces onto osteoclasts on the alveolar
bone surface via the PDL in vivo.
As a control experiment, the present study examined
the effects of reversing cover slips on osteoclast differentiation
when compared with osteoclasts on the same slips without reversal.
Differences in the numbers of osteoclasts with or without reversing
were not detected in the absence of compressive forces.
The present study examined the effects of various
weights (layers of 3–14 cover slips) on the number of osteoclasts.
The number of osteoclasts and multinuclear cells was greatest when
seven slips were used, which were therefore considered to exert the
optimal compressive force. Furthermore, the optimal compressive
force may have accelerated the fusion of osteoclasts. Application
of this compressive force also accelerated osteoclastogenesis of
RAW264.7 cells as compared with that in the control group. Bone
resorption areas of osteoclasts on Osteologic™ discs were
significantly elevated when the optimal compressive force was
applied.
There are several types of tooth movement in
orthodontic treatment, including tipping, bodily movement and
rotation. The optimal orthodontic forces are different for each
type of tooth movement and area of the root surface. Schwartz and
Austria (26) recommended that the
forces of orthodontic treatment should not exceed the capillary bed
blood pressure (~20–26 g/cm2 of the root surface) for
the most favorable outcomes. This is not in accordance with the
results of the present study, in which a force of ~300
mg/cm2 exerted by eight cover slips was found to be
optimal for accelerating osteoclastogenesis. Each tooth is attached
to and separated from the adjacent alveolar bone by heavy collagen
fibers known as Sharpey's fibers in the PDL (25). The optimal orthodontic force in
teeth may therefore be diminished by Sharpey's fibers in the
PDL.
In the present study, mRNA expression of
osteoclast-associated genes, including NFATc-1, TRAP, RANK, cath-K,
ClC7, MMP-9, ATP6i, DC-STAMP, OC-STAMP as well as Integrin-αv and
-β3, was shown to be upregulated and accelerated by compression at
the optimal force when compared with that in the controls. In the
compressed group, the expression of the respective mRNAs peaked at
3 h, whereas in the control group mRNA expression increased slowly
over 24 h. Among the genes assessed, NFATc1 is thought be an
essential transcriptional factor for autoamplification. These
results indicated that the optimal compressive force activates
osteoclastogenesis and upregulates osteoclast-associated mRNA
expression.
Numerous studies have suggested that applying
compressive forces to osteoblasts and PDL cells promotes
osteoclastogenesis. The present study succeeded in applying
compressive forces to osteoclasts and found that the optimal
compressive force accelerated osteoclastogenesis. These results
provided the basis for an alternative approach to the consideration
of optimal force, which focuses on osteoclast precursors and
osteoclasts.
Acknowledgments
The authors are grateful to Dr. S. Kameyama
(Departments of Orthodontics Dentistry, Hokkaido University
Graduate School of Dental Medicine, Sapporo, Japan) for technical
advice and support. This study was supported in part by the Japan
Society for the Promotion of Science Grant-in-Aid for Scientific
Research (nos. 25463161 to Y.Y., 24659906 to T.K. and 24593071 to
M.M.), and for Young Scientists (nos. 2586199403 to K.F. and
2586199303 to K.S).
References
|
1
|
Bourauel C, Vollmer D and Jäger A:
Application of bone remodeling theories in the simulation of
orthodontic tooth movements. J Orofac Orthop. 61:266–279. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Krisshnan V and Davidovitch Z: Cellular,
molecular and tissue-level reactions to orthodontic force. Am J
Orthod Dentofacial Orthop. 129:469.e1–e32. 2006. View Article : Google Scholar
|
|
3
|
Suzuki N, Yoshimura Y, Deyama Y, Suzuki K
and Kitagawa Y: Mechanical stress directly suppresses osteoclast
differentiation in RAW264.7 cells. Int J Mol Med. 21:291–296.
2008.PubMed/NCBI
|
|
4
|
Shibata K, Yoshimura Y, Kikuiri T,
Hasegawa T, Taniguchi Y, Deyama Y, Suzuki K and Iida J: Effect of
the release from mechanical stress on osteoclastogenesis in
RAW264.7 cells. Int J Mol Med. 28:73–79. 2011.PubMed/NCBI
|
|
5
|
Kameyama S, Yoshimura Y, Kameyama T,
Kikuiri T, Matsuno M, Deyama Y, Suzuki K and Iida J: Short-term
mechanical stress inhibits osteoclastogenesis via suppression of
DC-STAMP in RAW264.7 cells. Int J Mol Med. 31:292–298.
2013.PubMed/NCBI
|
|
6
|
Nishijima Y, Yamaguchi M, Kojima T, Aihara
N, Nakajima R and Kasai K: Levels of RANKL and OPG in gingival
crevicular fluid during orthodontic tooth movement and effect of
compression force on releases from periodontal ligament cells in
vitro. Orthod Craniofacial Res. 9:63–70. 2006. View Article : Google Scholar
|
|
7
|
Ichimiya H, Takahashi T, Ariyoshi W,
Takano H, Matayoshi T and Nishihara T: Compressive mechanical
stress promotes osteoclast formation through RANKL expression on
synovial cells. Oral Surg Oral Med Oral Pathol Oral Radiol Endod.
103:334–341. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tan SD, de Vries TJ, Kujipers-Jagtman AM,
Semeins CM, Everts V and Klein-Nulend J: Osteocytes subjected to
fluid flow inhibit osteoclast formation and bone resorption. Bone.
41:745–751. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rubin J, Biskobing D, Fan X, Rubin C,
McLeod K and Taylor WR: Pressure regulates osteoclast formation and
MCSF expression in marrow culture. J Cell Physiol. 170:81–87. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qing Hong Z, Meng Tao L, Yi Z, Wei L, Ju
Xiang S and Li L: The effect of rotative stress on CAII, FAS, FASL,
OSCAR and TRAP gene expression in osteoclasts. J Cell Biochem.
114:388–397. 2013. View Article : Google Scholar
|
|
11
|
Makihira S, Kawahara Y, Yuge L, Mine Y and
Nikawa H: Impact of the microgravity environment in a 3-dimentional
clinostat on osteoblast- and osteoclast-like cells. Cell Biol Int.
32:1176–1181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kadow-Romacker A, Hoffman JE, Duga G,
Wildemann B and Schmidmaier G: Effect of mechanical stimulation on
osteoblast and osteoclast-like cells in vitro. Cells Tissues
Organs. 190:61–68. 2009. View Article : Google Scholar
|
|
13
|
Graber LW, Vanarsdall RL and Vig KWL:
Orthodontics; current principles and techniques. Fifth edition.
Mosby & Co; Philadelphia, PA: pp. 247–286. 2012
|
|
14
|
Suda T, Takahashi N, Udagawa N, Jimi E,
Gillespie MT and Martin TJ: Modulation of osteoclast
differentiation and function by the new members of the tumor
necrosis factor receptor and ligand families. Endocr Rev.
20:345–357. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boyle WJ, Simonet WS and Lacey DL:
Osteoclast differentiation and activation. Nature. 423:337–342.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Väänänen HK and Laitala-Leinonen T:
Osteoclast lineage and function. Arch Biochem Biophys. 473:132–138.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshida H, Hayashi S, Kunusada T, Ogawa M
and Nishikawa S, Okamura H, Sudo T, Shultz LD and Nishikawa S: The
murine mutation osteopetrosis is in the coding region of the
macrophage colony stimulating factor gene. Nature. 345:442–444.
1990. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takayanagi H: The role of NFAT in
osteoclast formation. Ann N Y Acad Sci. 1116:227–237. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koshihara Y, Kodama S, Ishibashi H, Azuma
Y, Ohta T and Karube S: Reversibility of alendronate-induced
contraction in human osteoclast like cells formed from bone marrow
cells in culture. J Bone Miner Metab. 17:98–107. 1999. View Article : Google Scholar
|
|
20
|
Contractor T, Babiarz B, Kowalski A,
Rittling SR, Sørensen ES and Denhardt DT: Osteoclasts resorb
protein-free mineral (Osteologic discs) efficiently in the absence
of osteopontin. In Vivo. 19:335–342. 2005.PubMed/NCBI
|
|
21
|
Aguirre JI, Plotkin LI, Gortazar AR,
Millan MM, O'Brien CA, Manolagas SC and Bellido T: A novel ligand
independent function of the estrogen receptor is essential for
osteocyte and osteoblast mechanotransduction. J Biol Chem.
282:25501–25508. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nomura M, Yoshimura Y, Kikuiri T, Hasegawa
T, Taniguchi Y, Deyama Y, Koshiro K, Sano H, Suzuki K and Inoue N:
Platinum nanoparticles suppress osteoclatogenesis through
scavenging of reactive oxygen species produced in RAW264.7 cells. J
Pharmacol Sci. 117:243–252. 2011. View Article : Google Scholar
|
|
23
|
Abe K, Yoshimura Y, Deyama Y, Kikuiri T,
Hasegawa T, Tei K, Shinoda H, Suzuki K and Kitagawa Y: Effect of
bisphosphonates on osteoclastogenesis in RAW264.7 cells. Int J Mol
Med. 29:1007–1015. 2012.PubMed/NCBI
|
|
24
|
Mochizuki A, Takami M, Miyamoto Y,
Nakamaki T, Tomoyasu S, Kadono Y, Tanaka S, Inoue T and Kamijo R:
Cell adhesion signaling regulates RANK expression in osteoclast
precursors. PLoS One. 7:e487952012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Profit WR, Fields HW Jr and Sarver DM:
Contemporary Orthodontics. Fourth edition. Mosby; St Louis, MO: pp.
331–352. 2007
|
|
26
|
Schwartz AM and Austria MDV: Tissure
changes incidental to orthodontic tooth movement. Int J Orthod.
18:331–352. 1932.
|