Introduction
Glioma is the most common tumor type of the central
nervous system and they occur in 6.42/100,000 individuals (1). Despite the advancements in surgery,
radiotherapy and chemotherapy, the survival rate of patients with
high grade glioma has not significantly improved during the past
several decades, predominantly due to regional recurrence (2).
The kinesin superfamily proteins (KIFs) are a
conserved class of microtubule-dependent molecular motor proteins
exhibiting adenosine triphosphatase activity. KIFs are important in
mitosis, meiosis and macromolecular transport (3). The abnormal expression and
dysfunction of KIFs may lead to the development and progression of
various types of human cancer (4–8).
KIF2A, a member of the kinesin-13 family, is known to be involved
in mitosis and mitotic spindle assembly (9). Previously, the expression of KIF2A
was demonstrated to be upregulated in squamous cell carcinoma of
the oral tongue (SCCOT) and breast cancer, when compared with the
adjacent tissues (10,11). KIF2A promotes SCCOT progression and
metastasis, and breast cancer metastasis; however, little is known
about the expression and roles of KIF2A in glioma.
In the present study, the expression of KIF2A was
examined in 35 freshly isolated human glioma tissue samples, and
its prognostic value for glioma patients was evaluated. The
biological functions of KIF2A in glioma cells were also
analyzed.
Materials and methods
Patient specimens
Intracranial tissue specimens were collected from 35
patients [23 males (65.7%) and 12 females (34.3%)] and included 15
grade I–II cases (42.9%) and 20 grade III–IV cases (57.1%), ranging
in age from 4 to 70 years. The patients underwent primary and
curative resection for glioma, and were pathologically confirmed at
Qilu Hospital of Shandong University (Jinan, China) in 2014, for
reverse transcription quantitative-polymerase chain reaction
(RT-qPCR) and immunohistochemical (IHC) analyses. The patients were
diagnosed based on the World Health Organization (2007) standard
classification (12). The fresh
tumor tissues were immediately transferred to liquid nitrogen and
stored at −80°C for subsequent RT-qPCR. The remaining tissue was
immediately dipped in 10% formalin (Sangon Biotech Co., Ltd.,
Shanghai, China) for IHC analysis. The present study was approved
by the Institutional Review Board of Qilu Hospital of Shandong
University, and written informed consent was obtained from each
patient or the patient's family.
RT-qPCR
The total RNA was extracted from human intracranial
tissues or cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Single-stranded cDNA was
synthesized using oligo dT16 primers and Moloney Murine Leukemia
Virus Reverse Transcriptase, according to the manufacturer's
protocol (Invitrogen; Thermo Fisher Scientific, Inc.). RT-qPCR was
performed using a LightCycler 2.0 Instrument (Roche Diagnostics
GmbH, Penzberg, Germany) and the cycling conditions were as
follows: 30 sec at 95°C, and 40 cycles at 95°C for 5 sec, 60°C for
10 sec and 72°C for 15 sec for PCR amplification, according to the
instructions of the SYBR® Green Realtime PCR Master Mix
[Toyobo (Shanghai) Co., Ltd.], which was used as the detection dye.
Gene-specific amplifications were confirmed through a melting curve
analysis at the end of the RT-qPCR. Relative gene expression levels
were determined using the 2−ΔΔCq method with endogenous
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as the
reference gene. The following primers were used: KIF2A, sense:
5′-GCCTTTGATGACTCAGCTCC-3′ and antisense:
5′-TTCCTGAAAAGTCACCACCC-3′ (154 bp); GAPDH, sense:
5′-GGTGGTCTCCTCTGACTTCAACAG-3′ and antisense:
5′-GTTGCTGTAGCCAAATTCGTTGT (127 bp).
IHC analysis and evaluation of
immunostaining parameters
The IHC procedure was performed according to an
established protocol, with minor modifications (13). Briefly, all glioma tissues were
formalin-fixed, paraffin-embedded (Sangon Biotech Co., Ltd.),
sectioned at 4 µm thickness using a rotatory microtome
(Finesse™ 325; Thermo Shandon Ltd., Cheshire, UK) and then placed
on slides pretreated with 3-aminopropyltriethoxysilane (ZSGB-Bio,
Beijing, China). Following deparaffinization and rehydration, the
sections were microwaved for heat-induced epitope retrieval in
citrate buffer (ZSGB-Bio; 80°C). The sections were subsequently
washed with phosphate-buffered saline (PBS) and the endogenous
peroxidase activity was inhibited with 3% hydrogen peroxide for 10
min. Next, the specimens were blocked with PBS containing normal
sheep serum (ZSGB-Bio) at 37°C for 30 min. Subsequently, the
sections were incubated with rabbit anti-human anti-KIF2A antibody
(1:4,000; Abcam, Cambridge, UK; cat. no. ab37005) overnight at 4°C
in a humidified chamber. Following rinsing with PBS, the slides
were incubated for 30 min with secondary anti-rabbit antibody
conjugated with horseradish peroxidase, according to the
manufacturer's instructions for the PV-9000 2-step plus®
Poly-HRP Anti-Mouse/Rabbit IgG Detection System kit (ZSGB-Bio;
1:100). The slides were then exposed to diaminobenzidine for
visualization and hematoxylin (ZSGB-Bio) for nuclear
counterstaining.
The tissue sections were assessed by light
microscopy (Olympus IX51; Olympus Corp., Tokyo, Japan;
magnification, ×200) by two pathologists in a blinded manner.
Initially, a proportion score was assigned, representing the
estimated proportion of positive tumor cells (0, none; 1, 1/100; 2,
1/100-1/10; 3, 1/10-1/3; 4, 1/3-2/3; 5, >2/3). Next, an
intensity score was assigned, representing the average intensity of
the positive tumor cells (0, none; 1, weak; 2, intermediate; 3,
strong). The proportion and intensity scores were subsequently
added to obtain a total score ranging between 0 and 8, with 0–3
indicating negative and 4–8 indicating positive (14). The specimens were rescored if the
difference between the scores from the two pathologists was
>3.
Western blot analysis
Cultured cells (4×105) in a 6-well cell
plate were homogenized for 60 min in 150 µl ice-cold
radioimmunoprecipitation lysis buffer [50 mM Tris-HCl (pH 7.4), 150
mM NaCl,1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 2 mM
sodium pyrophosphate, 25 mM β-glycerophosphate, 1 mM EDTA, 1 mM
Na3VO4 and 0.5 mg/ml leupeptin] according to
the manufacturer's instructions (cat. no. P0013K; Beyotime
Institute of Biotechnology, Beijing, China), containing 1 mM
phenylmethylsulfonyl fluoride (Beyotime Institute of
Biotechnology). The samples were subsequently centrifuged at 15,000
× g for 10 min at 4°C. The protein concentration was measured using
a Bicinchoninic acid Protein Assay kit (Beyotime Institute of
Biotechnology). The protein samples were denatured by boiling for
10 min prior to electrophoresis. A total of 10 µg each
protein sample was separated by 10% sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE) and were subsequently
transferred onto a polyvinylidene fluoride membrane (EMD Millipore,
Darmstadt, Germany) at 100 V for 90 min. Following blocking with 5%
non-fat milk for 1 h at room temperature, each membrane was
incubated with primary antibodies, including rabbit polyclonal
anti-KIF2A (1:4,000), rabbit polyclonal anti-MMP-2 (1:1,000; Abcam;
cat. no. ab110186), rabbit polyclonal anti-MMP-9 (1:1,000; Cell
Signaling Technology, Danvers, MA, USA; cat. no. 3852) or rabbit
anti-GAPDH polyclonal GAPDH (1:2,000; Proteintech Group, Inc.,
Wuhan, China; cat. no. 10494-1-AP), overnight at 4°C, followed by
three 10 min washes with Tris-buffered saline, containing 0.1%
Tween-20 (TBST). Following incubation with peroxidase-conjugated
affinipure goat anti-rabbit IgG(H+L) (1:5,000; Santa Cruz
Biotechnology Inc., Santa Cruz, CA, USA; cat. no. SA00001-2) for 1
h at room temperature, the membranes were washed three times with
TBST for 10 min. The bands were subsequently detected by enhanced
chemiluminescence (EMD Millipore). The housekeeping protein GAPDH
served as a loading control. Positive immunoreactive bands and the
ratio of target proteins to GAPDH in optical density units were
obtained using Image Station 4000 MM software (Carestream Health,
Rochester, NY, USA).
Cell culture and RNA interference
The human malignant glioma cell lines, A172 and U251
were obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). The cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone;
Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5%
CO2 humidified incubator. For RNA interference, the A172
cells were transfected with 50 nM KIF2A small interfering (si)RNA
(5′-CACCGGCAAAGAGATTGACCTGGTTCAAGAGACCAGGTCAATCTCTTTGCCTTTTTTG-3′)
or scrambled siRNA (a universal negative control;
5′-CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG-3′)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol.
Cell proliferation assay
A172 cells were transfected with either KIF2A siRNA
or scrambled siRNA and seeded at a density of 1.0×104
cells/well in a 96-well plate. Following incubation for 0, 24, 48
and 72 h, a CCK-8 assay (Yiyuan Biotech Inc., Guangzhou, China) was
performed to measure cell proliferation, according to the
manufacturer's protocol. The optical densities were measured at 450
nm using a TECAN Infinite® M200 microplate reader (Tecan
US, Durham, NC, USA).
Matrigel invasion and Transwell migration
assays
The effects of KIF2A on the invasion and migration
of glioma cells were evaluated using a Matrigel invasion and a
transwell migration assay in a 24-well Transwell, containing
polycarbonate filters with 8 mm pores (Costar, Corning, NY, USA).
For the Matrigel cell invasion assay, the inserts were precoated
with 50 µl Matrigel matrix (dilution at 1:3; BD Bioscience,
Franklin Lakes, NJ, USA), according to the manufacturer's protocol.
At 48 h after the transfection with KIF2A siRNA or scrambled siRNA,
the A172 cells were trypsinized (Invitrogen; Thermo Fisher
Scientific, Inc.) and adjusted to 1×106 cells/ml in
DMEM. DMEM (600 µl), containing 10% FBS was loaded in the
lower chamber and 100 µl resuspended cell solution was
plated in the upper chamber. The plates were subsequently incubated
for 16 h under normal conditions, after which non-invading or
non-migrating cells on the upper surface of the membrane were
removed from the chamber. The cells, which had invaded the lower
surface of the membrane were fixed and stained with crystal violet
(Sangon Biotech Co., Ltd.). The cells, which had migrated to the
lower surface of the membrane were fixed and stained with eosin
(ZSGB-Bio). The number of invading and migrating cells was
calculated using a microscope (Olympus IX51; Olympus, Tokyo, Japan)
at a magnification of ×200 in five random fields. Three independent
experiments were performed.
Gelatin zymography
Gelatin zymography assays were performed, as
previously described (15), with
minor modifications. Briefly, equal quantities of protein in the
serum-free supernatant from transfected cells was diluted in 5X
sample buffer [10% SDS (w/v); 0.05% bromophenol blue (w/v) and 0.25
M Tris-HCl (pH 6.8)] and incubated at room temperature for 10 min.
Subsequently, the proteins were separated by 10% SDS-PAGE,
containing 1 mg/ml gelatin (Sigma-Aldrich, St. Louis, MO, USA).
Each gel was subsequently transferred onto a clean glass container
and washed in 2.5% Triton X-100 twice for 45 min. The gels were
incubated in a development buffer [50 mM Tris-HCl; 150 mM NaCl; 5
mM CaCl2; 1 µM ZnCl2 and 0.02% NaN3,
(pH 7.5)] for 16 h at 37°C. Next, the gels were stained with 0.1%
Coomassie brilliant blue R-250 (w/v; Sigma-Aldrich) in 45% methanol
and 10% acetic acid for 1 h and were subsequently destained in 10%
acetic acid for two 30 min washes. Clear bands were visualized in
the areas where the gelatin was degraded. The bands were analyzed
using densitometry with Image J software 1.46r (National Institute
of Health, Bethesda, MD, USA). Three independent experiments were
performed.
Flow cytometric analysis of
apoptosis
An Annexin V-fluorescein isothiocyanate (FITC) and
Propidium Iodide (PI) Apoptosis Detection kit (BestBio, Shanghai,
China) was used to assess the apoptotic rate of the A172 cells
pretreated with scrambled siRNA or KIF2A siRNA, according to the
manufacturer's protocol. Briefly, A172 cells transfected with
scrambled siRNA or KIF2A siRNA were seeded into 12-well plates and
cultured under normal conditions for 48 h. The cells were
subsequently collected with trypsin (no EDTA), washed twice with
cold PBS, resuspended with binding buffer and incubated with
Annexin V-FITC and PI staining solution in the dark for 10 min at
room temperature, and the apoptotic rate was immediately quantified
using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA,
USA). Three independent experiments were performed.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA) for Windows. The
χ2 test was used to examine the correlation between the
expression of KIF2A and various clinicopathological parameters.
Student's t-test or Mann-Whitney U test was used for statistical
analyses. P<0.05 was considered to indicate a statistically
significant difference.
Results
mRNA and protein expression levels of
KIF2A were significantly higher in grade III–IV compared with those
in grade I–II glioma tissues
A summary of the patient clinicopathological
features is provided in Table I.
The present study evaluated the mRNA and protein expression levels
of KIF2A in fresh glioma tissues by RT-qPCR and IHC, respectively.
The mRNA expression levels of KIF2A were demonstrated to be
significantly higher in grade III–IV glioma tissues compared with
those in grade I–II glioma tissues (Table I; P<0.05). IHC analysis results
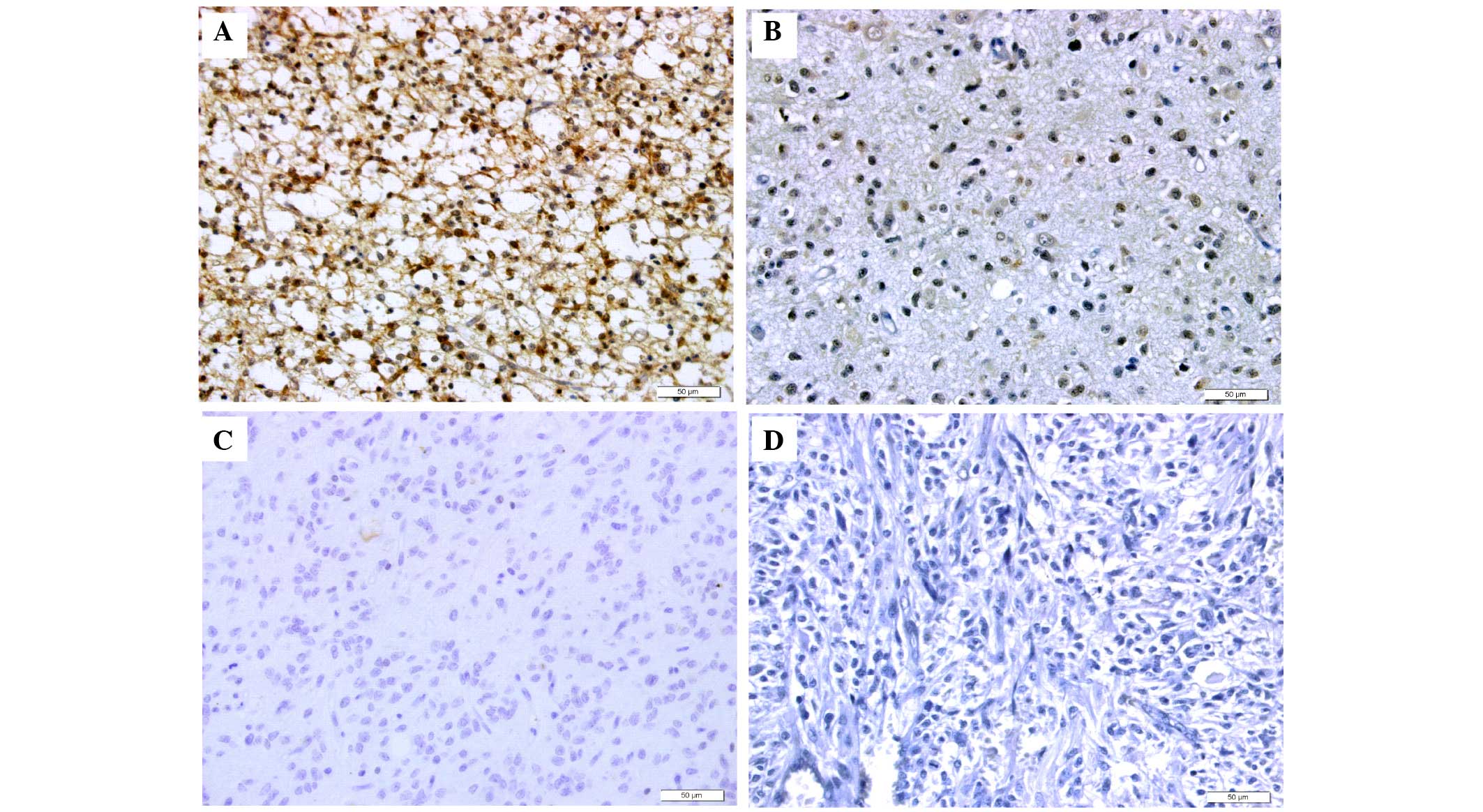
showed that KIF2A was predominantly localized in the nucleus and
cytoplasm of cancer cells (Fig.
1). Among the grade III–IV glioma specimens, 14 (40.0%) were
positively stained and 6 (17.14%) revealed no staining. Among the
grade I–II glioma specimens, 5 (14.29%) were positively stained and
10 (28.57%) revealed no staining. A significant increase in the
protein expression of KIF2A was observed in grade III–IV glioma
compared with grade I–II glioma (P<0.05; Table I). Representative images are shown
in Fig. 1.
 | Table ImRNA and protein expression levels of
KIF2A and its association with patient clinicopathological
features. |
Table I
mRNA and protein expression levels of
KIF2A and its association with patient clinicopathological
features.
| Variable | Patients, n | mRNA
expressiona
| Protein expression, n
(%)
|
|---|
| Median | IQR | P-value | Positive | Negative | P-value |
|---|
| Age, years | | | | 0.056b | | | 0.317d |
| ≤45 | 14 | 0.009 | 0.005–0.020 | | 6 (17.14) | 8 (22.88) | |
| >45 | 21 | 0.020 | 0.010–0.050 | | 13 (37.14) | 8 (22.88) | |
| Gender | | | | 0.263b | | | 1.000d |
| Male | 23 | 0.020 | 0.008–0.050 | | 12 (34.29) | 11 (31.43) | |
| Female | 12 | 0.020 | 0.004–0.028 | | 7 (20.00) | 5 (14.29) | |
| Tumor location | | | | 0.143c | | | 0.541d |
| Prefrontal | 14 | 0.020 | 0.005–0.050 | | 6 (17.14) | 8 (22.88) | |
| Temporal lobe | 13 | 0.020 | 0.009–0.055 | | 8 (22.88) | 5 (14.29) | |
| Other | 8 | 0.090 | 0.039–0.175 | | 5 (14.29) | 3 (8.57) | |
| Grade | | | | 0.033b | | | 0.044d |
| I–II | 15 | 0.009 | 0.005–0.020 | | 5 (14.29) | 10 (28.57) | |
| III–IV | 20 | 0.025 | 0.010–0.073 | | 14 (40.00) | 6 (17.14) | |
| Tumor size, cm | | | | 0.601b | | | 0.503d |
| ≥3 | 22 | 0.020 | 0.008–0.050 | | 13 (37.14) | 9 (25.71) | |
| <3 | 13 | 0.010 | 0.007–0.030 | | 6 (17.14) | 7 (20.00) | |
Correlation between the expression of
KIF2A and clinical parameters
The χ2 test results revealed that the
expression of KIF2A correlated closely with the World Health
Organization grade of glioma (Table
I; P<0.05). No significant correlation was observed between
the expression of KIF2A, and patient age, gender and tumor location
or size (Table I; P>0.05).
KIF2A knockdown significantly inhibits
proliferation and induces apoptosis in A172 cells
To identify the functions of KIF2A in malignant
glioma cells, the expression of KIF2A in the A172 and U251 glioma
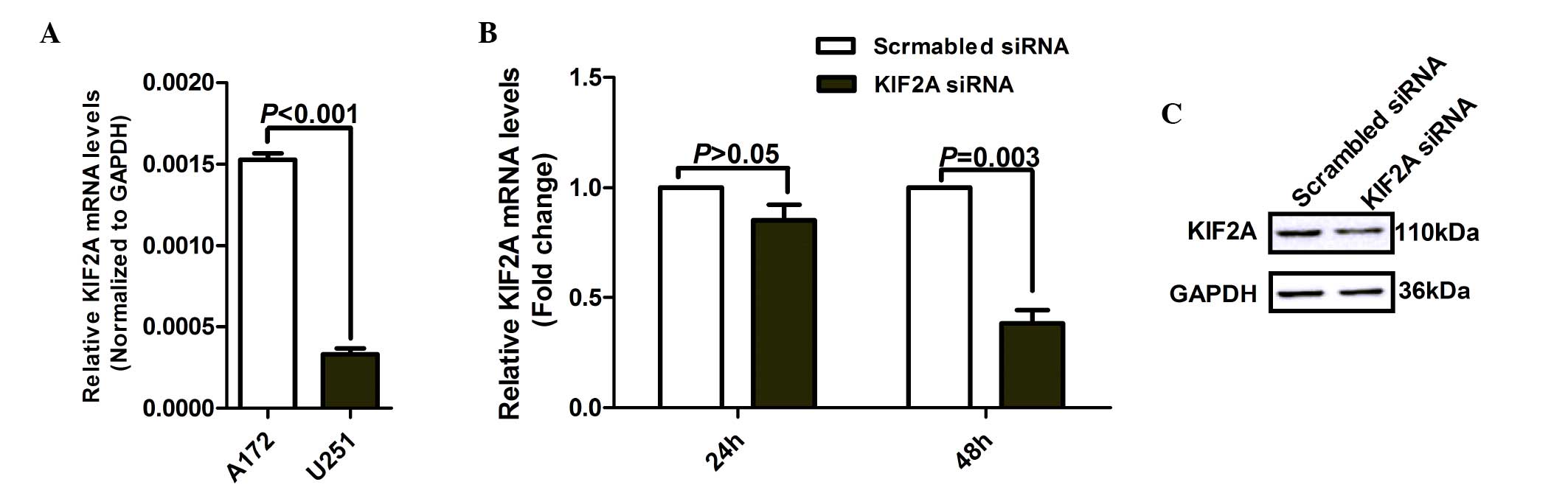
cell lines was analyzed. RT-qPCR results revealed that the mRNA
expression of KIF2A was significantly higher in A172 cells compared
with in U251 cells (Fig. 2A;
P<0.001). The A172 cells were therefore selected for the in
vitro experiments in the present study. Briefly, the human
glioma cell lines, A172 and U251, were passaged to obtain the
appropriate cells. The A172 and U251 cells were grown in DMEM
supplemented with 10% heat-inactivated FBS at 37°C in 5%
CO2. Total RNA was extracted from the A172 and U251
cells, then reverse transcribed into cDNA and amplified by RT-qPCR.
The A172 cell line then underwent further study. Compared with the
scrambled siRNA group, A172 cells transfected with KIF2A siRNA
exhibited significantly downregulated mRNA expression of KIF2A in a
time-dependent manner (Fig. 2B).
Consistent with the mRNA results, the protein expression of KIF2A
was also lower in A172 cells transfected with KIF2A siRNA for 48 h,
compared with the scrambled siRNA group (Fig. 2C). The above results suggested that
the specific siRNA targeting KIF2A significantly decreased the mRNA
and protein expression levels of KIF2A.
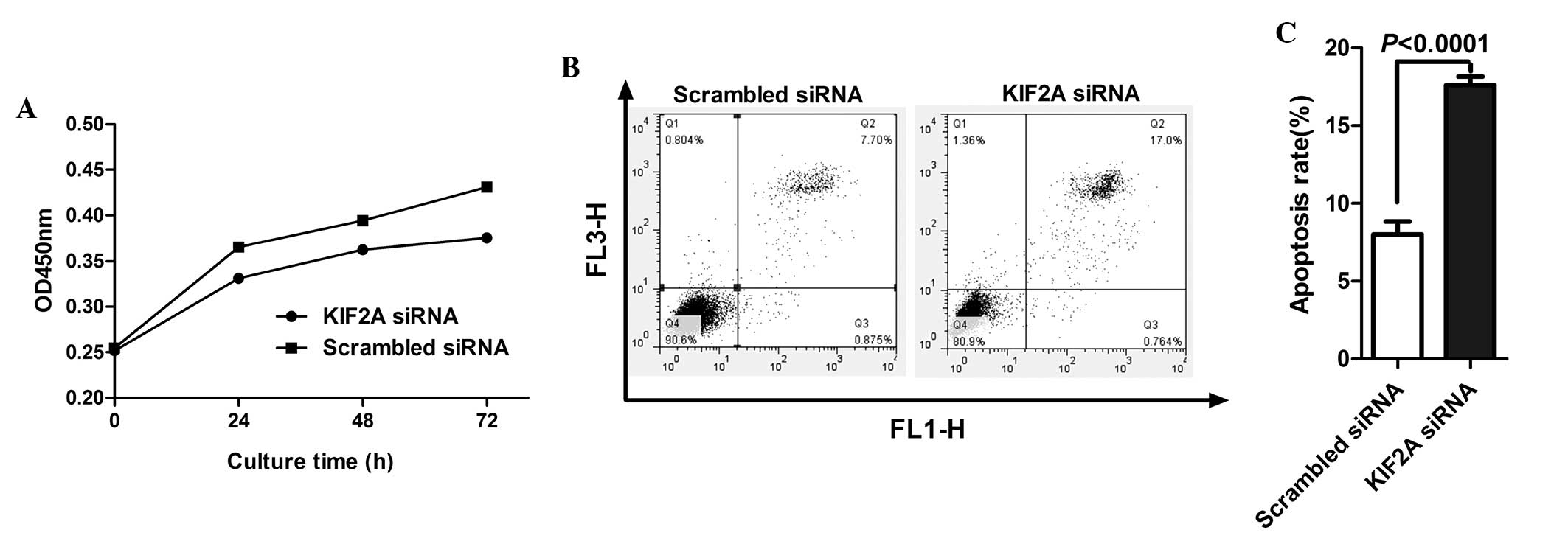
The CCK-8 assay was used to investigate the effect
of KIF2A on the proliferation of A172 cells. KIF2A knockdown
inhibited A172 cell proliferation in a time-dependent manner
(Fig. 3A). To assess whether the
decreased cell number was due to apoptosis, induced by KIF2A siRNA,
apoptosis in A172 cells following siRNA transfection was examined.
As shown in Fig. 3B and C, a
higher number of apoptotic cells were observed among the A172 cells
transfected with KIF2A siRNA, compared with the control cells
(P<0.0001). The above data suggested that KIF2A may be important
in the proliferation and apoptosis of glioma cells.
KIF2A knockdown significantly inhibited
the invasion and migration capacities of A172 cells by regulating
the activity and expression of MMP-2
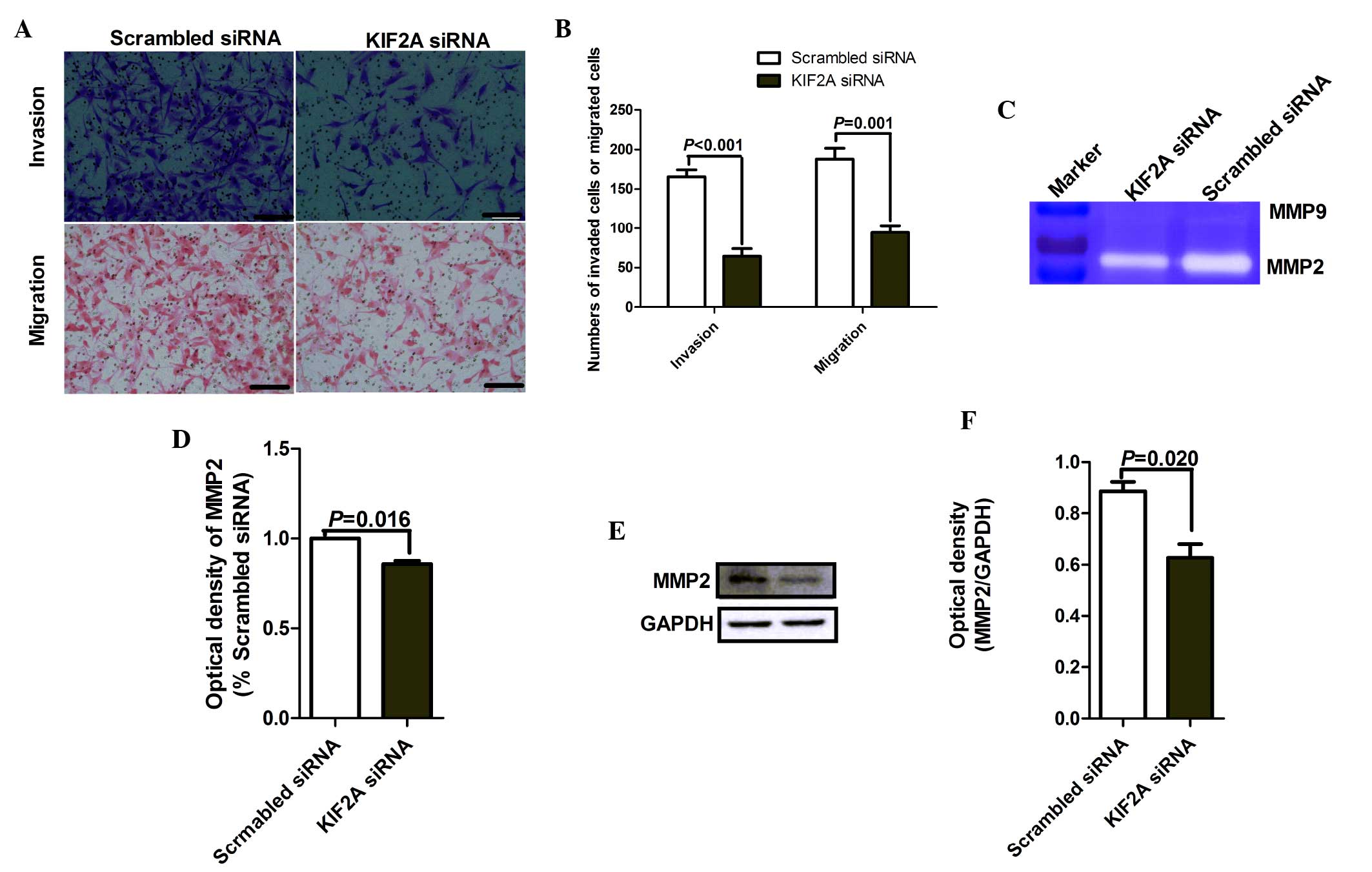
Matrigel cell invasion and Transwell cell migration
models were performed to analyze the effect of KIF2A expression on
the invasion and migration capacities of A172 cells. Compared with
the cells transfected with scrambled siRNA, the invasion and
migration capacities of A172 cells were significantly inhibited by
KIF2A knockdown (Fig. 4A and B;
P<0.001 and P=0.001, respectively).
Matrix metalloproteinases (MMPs) are a group of
peptidases involved in the degradation of the extracellular matrix,
and accumulated data have suggested that MMPs contribute to the
invasion and migration processes of glioma cells (16). Gelatin zymography was performed to
analyze the gelatinolytic activities of MMP-2 and MMP-9 in the
supernatants of A172 cells transfected with KIF2A siRNA or
scrambled siRNA. The results (Fig.
4C) and statistical analysis (Fig.
4D) demonstrated that KIF2A siRNA significantly decreased MMP-2
activity in the supernatant of A172 cells when compared with the
scrambled siRNA (P=0.016). By contrast, MMP-9 activity was barely
detected in the supernatant of A172 cells transfected with either
indicated siRNA. Western blot analysis further demonstrated that
KIF2A silencing significantly reduced the expression of MMP-2
(P=0.020; Fig. 4E and F), and
MMP-9 expression levels were below detection limits (data not
shown).
Discussion
The aim of the present study was to evaluate the
expression of KIF2A in glioma tissues, assess the association
between the expression of KIF2A and clinical parameters, as well as
identify the functions of KIF2A in malignant glioma cells.
KIF2A, a member of the kinesin-13 family, was first
cloned from the murine central nervous system by degenerate PCR in
1992 (17), and is said to act as
the key factor in human neuronal diseases (17,18).
KIF2A has been reported to specifically localize to centrosomes and
spindle poles (19), and is
important in both bipolar spindle assembly and chromosome movement
during mitosis (19,20). A previous study reported that
errors in the process of mitosis may result in numerous defects in
daughter cells, thereby leading to carcinogenesis (21). Other previous studies on malignant
tumor types have demonstrated that the overexpression of KIF2A is
associated with tumor progression, including that of SCCOT and
breast cancer (10,11); however, the expression of KIF2A and
its correlation with various clinical parameters in glioma remain
unclear.
In the present study, the findings of RT-qPCR and
IHC analysis demonstrated that the mRNA and protein expression
levels of KIF2A were significantly higher in grade III–IV glioma
tissues compared with those in grade I–II glioma tissues, and that
the expression of KIF2A was notably correlated with glioma grade
(Table I). This result was
consistent with previous studies performed on breast cancer and
SCCOT (10,11), and suggested that KIF2A may be
involved in the regulation of glioma tumor progression.
Previous studies have reported that the knockdown of
KIF2A in somatic or cancer cells can lead to a marked increase in
monopolar spindles (9,20). Therefore, KIF2A may be important in
the proliferation and apoptosis of those cells. In the present
study, it was revealed that KIF2A knockdown significantly inhibited
the proliferation of A172 cells (Fig.
3A), which supported the findings of previous studies on breast
cancer and SCCOT. It was further demonstrated that KIF2A knockdown
significantly induced apoptosis in A172 cells. A previous study
also reported that KIF2A has a central role in the apoptosis of
malignant cells (22). The precise
mechanisms by which KIF2A regulates the proliferation and apoptosis
of glioma cells remain to be elucidated.
Cellular morphology is supported by the
cytoskeleton, and cytoskeletal reorganizations have important
effects on the migration of neoplastic cells (12). Microtubules (MTs) are fundamental
components of the cytoskeleton in eukaryotic cells. Accumulating
evidence has revealed that decreases in MTs and MT depolymerization
are important in the invasion and migration capacities of malignant
tumors (23,24). Certain previous studies have
proposed that KIF2A has microtubule-depolymerizing activity
(9,25), which means that KIF2A may be
involved in the movement of cells. In the present study, it was
shown that KIF2A knockdown significantly inhibited the invasion and
migration capacities of A172 cells (Fig. 4A and B); similar results have also
been demonstrated in previous studies on breast cancer and SCCOT
(10,11). The present study further identified
why KIF2A knockdown regulates the invasion and migration capacities
of A172 cells. Due to the important role of MMPs in the invasion
and migration of glioma cells (16), the activity and expression of
MMP-2/9 in A172 cells trans-fected with KIF2A/scramble siRNA were
analyzed by gelatin zymography and western blot analysis. The
results revealed that KIF2A significantly inhibited the activity
and expression of MMP-2 in A172 cells, however, the activity and
expression of MMP-9 were below the detection limit (Fig. 4C–F). The precise mechanism by which
KIF2A regulates the expression and function of MMP-2 remains to be
elucidated.
In conclusion, the findings of the present study
demonstrated that the expression of KIF2A was upregulated in grade
III–IV glioma tissues compared with that in grade I–II glioma
tissues, and that it was markedly correlated with glioma grade.
Furthermore, it was revealed that KIF2A knockdown significantly
inhibited proliferation and induced apoptosis in A172 cells, as
well as significantly inhibited the invasion and migration
capacities of A172 cells by regulating the activity and expression
of MMP-2. These results suggested that KIF2A has a central role in
glioma development and that the inhibition of KIF2A expression may
prove to be a promising target in the control of glioma.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 31470885
and 31270971) and the Natural Science Foundation of Science and
Technology Department of Liaoning Province (grant no.
2014022020).
References
|
1
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maher EA, Furnari FB, Bachoo RM, Rowitch
DH, Louis DN, Cavenee WK and DePinho RA: Malignant glioma: Genetics
and biology of a grave matter. Genes Dev. 15:1311–1333. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miki H, Okada Y and Hirokawa N: Analysis
of the kinesin superfamily: Insights into structure and function.
Trends Cell Biol. 15:467–476. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lukong KE and Richard S: Breast tumor
kinase BRK requires kinesin-2 subunit KAP3A in modulation of cell
migration. Cell Signal. 20:432–442. 2008. View Article : Google Scholar
|
|
5
|
Corson TW and Gallie BL: KIF14 mRNA
expression is a predictor of grade and outcome in breast cancer.
Int J Cancer. 119:1088–1094. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Minakawa Y, Kasamatsu A, Koike H, Higo M,
Nakashima D, Kouzu Y, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H
and Uzawa K: Kinesin family member 4A: A potential predictor for
progression of human oral cancer. PLoS One. 8:e859512013.
View Article : Google Scholar
|
|
7
|
Bie L, Zhao G, Wang YP and Zhang B:
Kinesin family member 2C (KIF2C/MCAK) is a novel marker for
prognosis in human gliomas. Clin Neurol Neurosurg. 114:356–360.
2012. View Article : Google Scholar
|
|
8
|
Taniwaki M, Takano A, Ishikawa N, Yasui W,
Inai K, Nishimura H, Tsuchiya E, Kohno N, Nakamura Y and Daigo Y:
Activation of KIF4A as a prognostic biomarker and therapeutic
target for lung cancer. Clin Cancer Res. 13:6624–6631. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ganem NJ and Compton DA: The KinI kinesin
Kif2a is required for bipolar spindle assembly through a functional
relationship with MCAK. J Cell Biol. 166:473–478. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang CQ, Qu X, Zhang XY, Zhou CJ, Liu GX,
Dong ZQ, Wei FC and Sun SZ: Overexpression of Kif2a promotes the
progression and metastasis of squamous cell carcinoma of the oral
tongue. Oral Oncol. 46:65–69. 2010. View Article : Google Scholar
|
|
11
|
Wang J, Ma S, Ma R, Qu X, Liu W, Lv C,
Zhao S and Gong Y: KIF2A silencing inhibits the proliferation and
migration of breast cancer cells and correlates with unfavorable
prognosis in breast cancer. BMC Cancer. 14:4612014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Louis DN, Ohgaki H, Wiestler OD and
Cavenee WK: WHO classification of tumours of the central nervous
system. IV edition. IARC Press; Lyon: pp. 8–57. 2007
|
|
13
|
Wang N, Feng Y, Wang Q, Liu S, Xiang L,
Sun M, Zhang X, Liu G, Qu X and Wei F: Neutrophils Infiltration in
the tongue squamous cell carcinoma and its correlation with CEACAM1
expression on tumor cells. PLoS One. 9:e899912014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kawai H, Ishii A, Washiya K, Konno T, Kon
H, Yamaya C, Ono I, Minamiya Y and Ogawa J: Estrogen receptor alpha
and beta are prognostic factors in non-small cell lung cancer. Clin
Cancer Res. 11:5084–5089. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang H, Cheng H, Shao Q, Dong Z, Xie Q,
Zhao L, Wang Q, Kong B and Qu X: Leptin-promoted human extravillous
trophoblast invasion is MMP14 dependent and requires the cross talk
between Notch1 and PI3K/Akt signaling. Biol Reprod. 90:782014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakada M, Okada Y and Yamashita J: The
role of matrix metal-loproteinases in glioma invasion. Front
Biosci. 8:e261–e269. 2003. View
Article : Google Scholar
|
|
17
|
Aizawa H, Sekine Y, Takemura R, Zhang Z,
Nangaku M and Hirokawa N: Kinesin family in murine central nervous
system. J Cell Biol. 119:1287–1296. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Poirier K, Lebrun N, Broix L, Tian G,
Saillour Y, Boscheron C, Parrini E, Valence S, Pierre BS, Oger M,
et al: Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause
malformations of cortical development and microcephaly. Nat Genet.
45:639–647. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ganem NJ, Upton K and Compton DA:
Efficient mitosis in human cells lacking poleward microtubule flux.
Curr Biol. 15:1827–1832. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu C, Zhao J, Bibikova M, Leverson JD,
Bossy-Wetzel E, Fan JB, Abraham RT and Jiang W: Functional analysis
of human microtubule-based motor proteins, the kinesins and
dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol
Cell. 16:3187–3199. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weaver BA and Cleveland DW: Decoding the
links between mitosis, cancer and chemotherapy: The mitotic
checkpoint, adaptation and cell death. Cancer Cell. 8:7–12. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang K, Lin C, Wang C, Shao Q, Gao W, Song
B, Wang L, Song X, Qu X and Wei F: Silencing Kif2a induces
apoptosis in squamous cell carcinoma of the oral tongue through
inhibition of the PI3K/Akt signaling pathway. Mol Med Rep.
9:273–278. 2014.
|
|
23
|
Fink-Puches R, Hofmann-Wellenhof R, Smolle
J, Helige C and Kerl H: Cytoplasmic microtubules in two different
mouse melanoma cell lines: A qualitative and quantitative analysis
using confocal laser scanning microscopy and computer-assisted
image analysis. J Cutan Pathol. 24:350–355. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Watanabe T, Noritake J and Kaibuchi K:
Regulation of micro-tubules in cell migration. Trends Cell Biol.
15:76–83. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Niwa S: Kinesin superfamily proteins and
the regulation of microtubule dynamics in morphogenesis. Anat Sci
Int. 90:1–6. 2015. View Article : Google Scholar
|