Introduction
Intervertebral discs lie between adjacent vertebrae
in the spine, forming a fibrocartilaginous joint to allow slight
movement of the vertebrae (1). The
layers of fibrocartilage contain the nucleus pulposus, which
functions as a shock absorber dissipating compressive forces
outward from its center to the surrounding annulus fibrosus
(2). However, the anulus fibrosus
becomes weaker with increasing age (3), resulting in intervertebral disc
degeneration (IDD). Beyond age 40, >60% of individuals show
symptoms of IDD; furthermore, IDD is the most common cause of
disability among workers aged 18–64 years (4). IDD is characterized by decreases in
intervertebral disc function and height due to cell loss through
apoptosis, increased breakdown of matrix or altered matrix
synthesis, with the underlying pathological processes being complex
(5–7).
Numerous studies have intended to explore the
molecular mechanisms involved in IDD. Vo et al (4) reported that IDD was a consequence of
increased catabolism of the extracellular matrix (ECM), since the
proteolytic degradation of ECM macromolecules led to marked
structural changes of the intervertebral disc. These catabolic
processes are mediated by a number of cytokines in the nucleus
pulposus, among which interleukin (IL)-1β and tumor necrosis factor
(TNF)-α have been suggested to have crucial roles in the
development of IDD. However, the involvement of IL-1β and TNF-α in
IDD has remained to be fully elucidated.
Markova et al (8) cultured rat intervertebral discs in
the presence of IL-1β, TNF-α and serum-limiting conditions to mimic
a degenerative insult to identify the differentially expressed
genes (DEGs) between experimental and control groups. Their
microarray data (no. GSE42611) have been deposited at the National
Center of Biotechnology Information Gene Expression Omnibus (GEO)
database (http://www.ncbi.nlm.nih.gov/geo/) and were used in the
present study for a bioinformatics analysis. DEGs between
experimental and control samples were identified, Gene Ontology
(GO) and pathway enrichment analyses were performed and a
protein-protein interaction (PPI) network was constructed. The
present study aimed to identify the DEGs and pathways associated
with IDD caused by the presence of IL-1β and TNF-α.
Materials and methods
Affymetrix microarray data
The microarray expression profile dataset GSE42611
(8) was downloaded from the GEO
(http://www.ncbi.nlm.nih.gov/geo/)
database based on the platform of Affymetrix Rat Gene 1.0 ST Array
[transcript (gene) version; Affymetrix Inc., Santa Clara, CA, USA].
The dataset contained four experimental nucleus pulposus (ENP)
samples and four control nucleus pulposus (CNP) samples. The
experimental lumbar discs had been cultured in Dulbecco's modified
Eagle's medium (DMEM; R&D Systems, Inc., Minneapolis, MN, USA)
containing 10 ng IL-1β (R&D Systems, Inc.), 100 ng/ml TNF-α
(R&D Systems, Inc.), 1% fetal bovine serum (FBS; R&D
Systems, Inc.), 50 µg/ml L-ascorbate (Cellgro; Corning
Incorporated, Corning, NY, USA), 40 mM NaCl (Cellgro; Corning
Incorporated), antibiotics and antimycotics (Cellgro; Corning
Incorporated), while the control discs were cultured in DMEM
containing 10% FBS, 50 µg/ml L-ascorbate, 40 mM NaCl,
antibiotics and antimycotics.
Data preprocessing and differential
expression analysis
The original array data were converted into gene
symbols and then subjected to background correction and quartile
data normalization using the robust multiarray average (9) algorithm in the oligo package
(10), which is available through
BioConductor (http://www.bioconductor.org).
The paired Student's t-test based on the Limma
package (11) in R was used to
identify DEGs between ENP and CNP samples. Multiple testing
correction was performed using the Benjamini-Hochberg method
(12) to obtain the adjusted
P-value. Subsequently, the log2-fold change
(log2FC) was calculated. Only genes with an adjusted
P<0.05 and a |log2FC|>1.0 were regarded as
DEGs.
GO and pathway enrichment analyses
GO (http://www.geneontology.org/) (13) is a database used for unification of
biological data, which comprises a structured, defined and
controlled vocabulary for large-scale gene annotation. The Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/) (14) database is a collection of online
databases of genomes, enzymatic pathways and biological
chemicals.
The present study performed GO and KEGG pathway
enrichment analyses to determine the function of DEGs using the
Database for Annotation, Visualization and Integrated Discovery
(http://david.abcc.ncifcrf.gov/)
(15) online tool, which is a
comprehensive functional annotation tool for associating functional
terms with gene lists using a clustering algorithm. P<0.05 and a
gene count >2 were set as thresholds.
PPI network construction
The Search Tool for the Retrieval of Interacting
Genes (STRING; http://string-db.org/) (16) database is a pre-computed global
resource, which was designed to predict functional associations
between proteins. In the present study, the STRING online tool was
applied to analyze the PPIs of DEGs with experimentally validated
interactions with a combined score of >0.4 considered
significant.
The previously obtained biological networks
indicated that the majority of the biological networks are
characterized as scale-free (17).
Thus, the connectivity degree was assessed by statistical analysis
of networks to identify important nodes, which are being referred
to as hub proteins (18).
Prediction of regulatory networks of
transcription factors (TFs)
The TRANSFAC® database (19), which is available at http://www.gene-regulation.com, consists of TFs,
their target genes and regulatory binding sites. In the present
study, the transcriptional regulatory network within the PPI
network was predicted based on the TRANSFAC®
database.
Network module identification and
functional enrichment analysis
After integrating the transcriptional regulatory
network into the PPI network, the modular complex detection plugin
(20) in cytoscape (http://www.cytoscape.org/) (21) was used to identify the network
modules. The selected modules with high node scores and
connectivity degrees were the subjected to GO and KEGG pathway
enrichment analyses.
Results
Identification of DEGs
After data pre-processing, a total of 536 DEGs were
obtained between ENP and CNP samples. Among these DEGs, 246 were
upregulated and 290 were downregulated.
GO and pathway enrichment analyses
The top two clustering groups obtained by GO
enrichment analysis are shown in Table
I. The upregulated DEGs were mainly enriched in biological
processes (BPs) associated with inflammatory response and responses
to organic substances. The downregulated DEGs were enriched in the
BP terms associated with cell adhesion and collagen fibril
organization.
 | Table IGO functional enrichment analysis for
the upregulated and downregulated differentially expressed
genes. |
Table I
GO functional enrichment analysis for
the upregulated and downregulated differentially expressed
genes.
| GO Term | Biological
process | Count | P-value |
|---|
| Enrichment score,
7.038 | | | |
| GO:0009611 | Response to
wounding | 24 |
2.10×10−8 |
| GO:0006954 | Inflammatory
response | 17 |
5.50×10−8 |
| GO:0006952 | Defense
response | 21 |
6.67×10−7 |
| Enrichment score,
5.439 | | | |
| GO:0010033 | Response to organic
substance | 34 |
4.24×10−7 |
| GO:0009719 | Response to
endogenous stimulus | 24 |
4.27×10−6 |
| GO:0009725 | Response to hormone
stimulus | 21 |
2.66×10−5 |
| Enrichment score,
6.414 | | | |
| GO:0007155 | Cell adhesion | 28 |
7.91×10−9 |
| GO:0022610 | Biological
adhesion | 28 |
7.91×10−9 |
| GO:0016337 | Cell-cell
adhesion | 12 |
9.22×10−4 |
| Enrichment score,
6.331 | | | |
| GO:0030199 | Collagen fibril
organization | 8 |
3.80×10−8 |
| GO:0030198 | Extracellular
matrix organization | 12 |
1.74×10−7 |
| GO:0043062 | Extracellular
structure organization | 12 |
1.54×10−5 |
The results of the pathway enrichment analysis are
shown in Table II. The
upregulated DEGs were enriched in nine pathways, which included
apoptotic pathways and the NOD-like receptor signaling pathway. The
downregulated DEGs were also enriched in nine pathways, including
pathways of ECM-receptor interaction and focal adhesion.
 | Table IIKEGG pathway enrichment analysis for
the upregulated and downregulated DEGs. |
Table II
KEGG pathway enrichment analysis for
the upregulated and downregulated DEGs.
| Term | Biological
process/pathway | Count | P-value |
|---|
| Upregulated
DEGs | | | |
| rno04210 | Apoptosis | 9 |
4.15×10−5 |
| rno04621 | NOD-like receptor
signaling pathway | 7 |
3.43×10−4 |
| rno04060 | Cytokine-cytokine
receptor interaction | 11 |
8.06×10−4 |
| rno04062 | Chemokine signaling
pathway | 9 |
4.53×10−3 |
| rno04630 | Jak-STAT signaling
pathway | 8 |
5.29×10−3 |
| rno05222 | Small cell lung
cancer | 6 |
8.70×10−3 |
| rno05200 | Pathways in
cancer | 12 |
8.81×10−3 |
| rno04620 | Toll-like receptor
signaling pathway | 6 |
1.21×10−2 |
| rno00230 | Purine
metabolism | 7 |
3.06×10−2 |
| Downregulated
DEGs | | | |
| rno04512 | Extracellular
matrix - receptor interaction | 16 |
7.56×10−12 |
| rno04510 | Focal adhesion | 20 |
1.13×10−9 |
| rno05200 | Pathways in
cancer | 14 |
4.34×10−3 |
| rno04640 | Hematopoietic cell
lineage | 6 |
1.30×10−2 |
| rno04670 | Leukocyte
transendothelial migration | 7 |
1.73×10−2 |
| rno05410 | Hypertrophic
cardiomyopathy | 6 |
1.75×10−2 |
| rno04110 | Cell cycle | 7 |
2.59×10−2 |
| rno05219 | Bladder cancer | 4 |
2.66×10−2 |
| rno04115 | p53 signaling
pathway | 5 |
3.08×10−2 |
PPI network construction
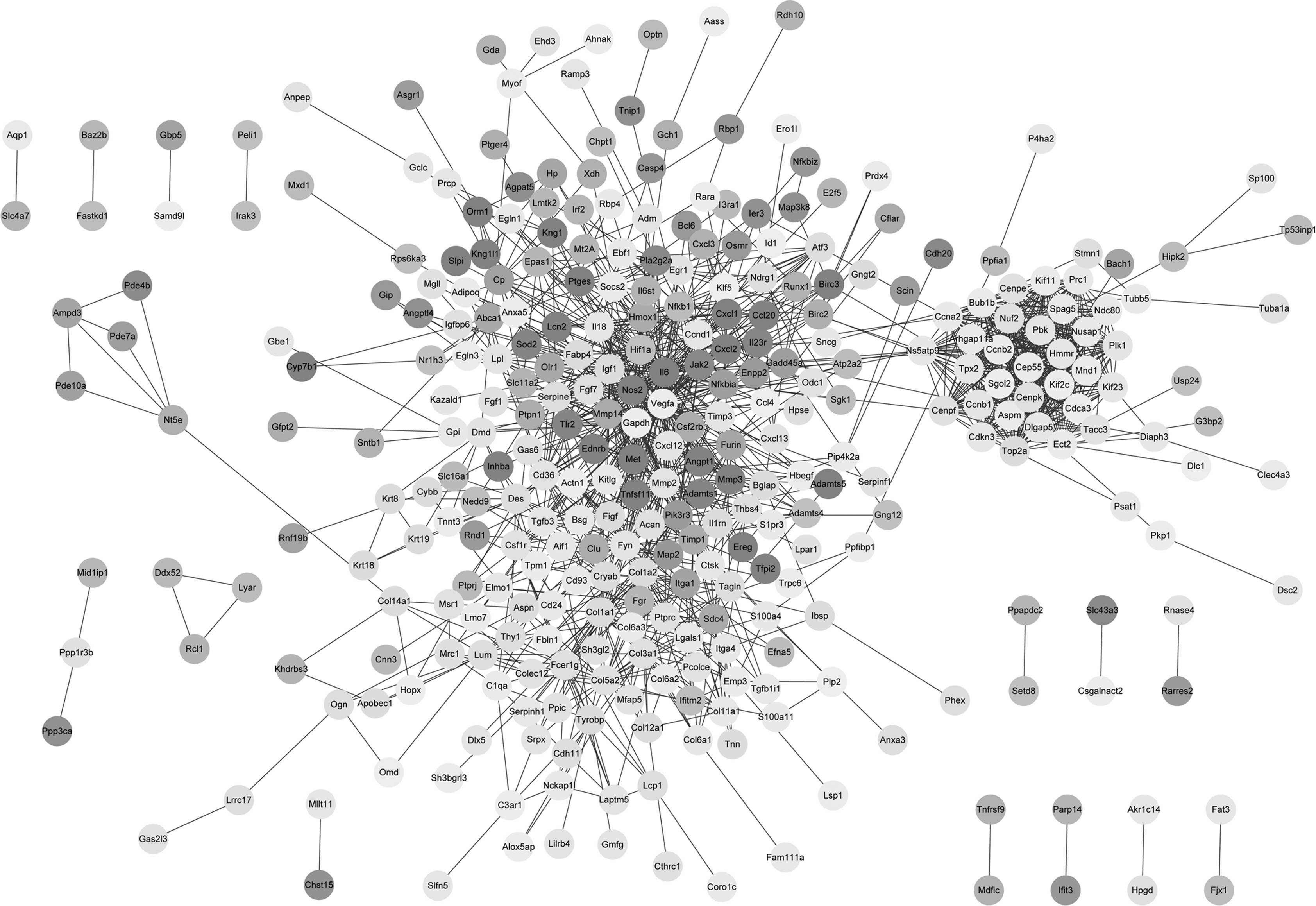
Based on the STRING database, 1,345 PPI pairs were
obtained and the PPI network was constructed (Fig. 1). The PPI network contained 36 DEGs
with a degree of >25, referred to as hub genes, which included
IL6 (degree, 67), COL1A1 (degree, 36), NFKB1
(degree, 31) and HIF1A (degree, 26).
Regulatory networks of TFs
Based on the PPI network and the
TRANSFAC® database, 81 PPI pairs of transcriptional
regulatory interactions were obtained. In addition, seven TFs were
obtained, namely RAR-α, ANPEP, ETS2,
ATF3, EGR1, HIF1A and NFKB1.
Network module identification and
functional enrichment analyses
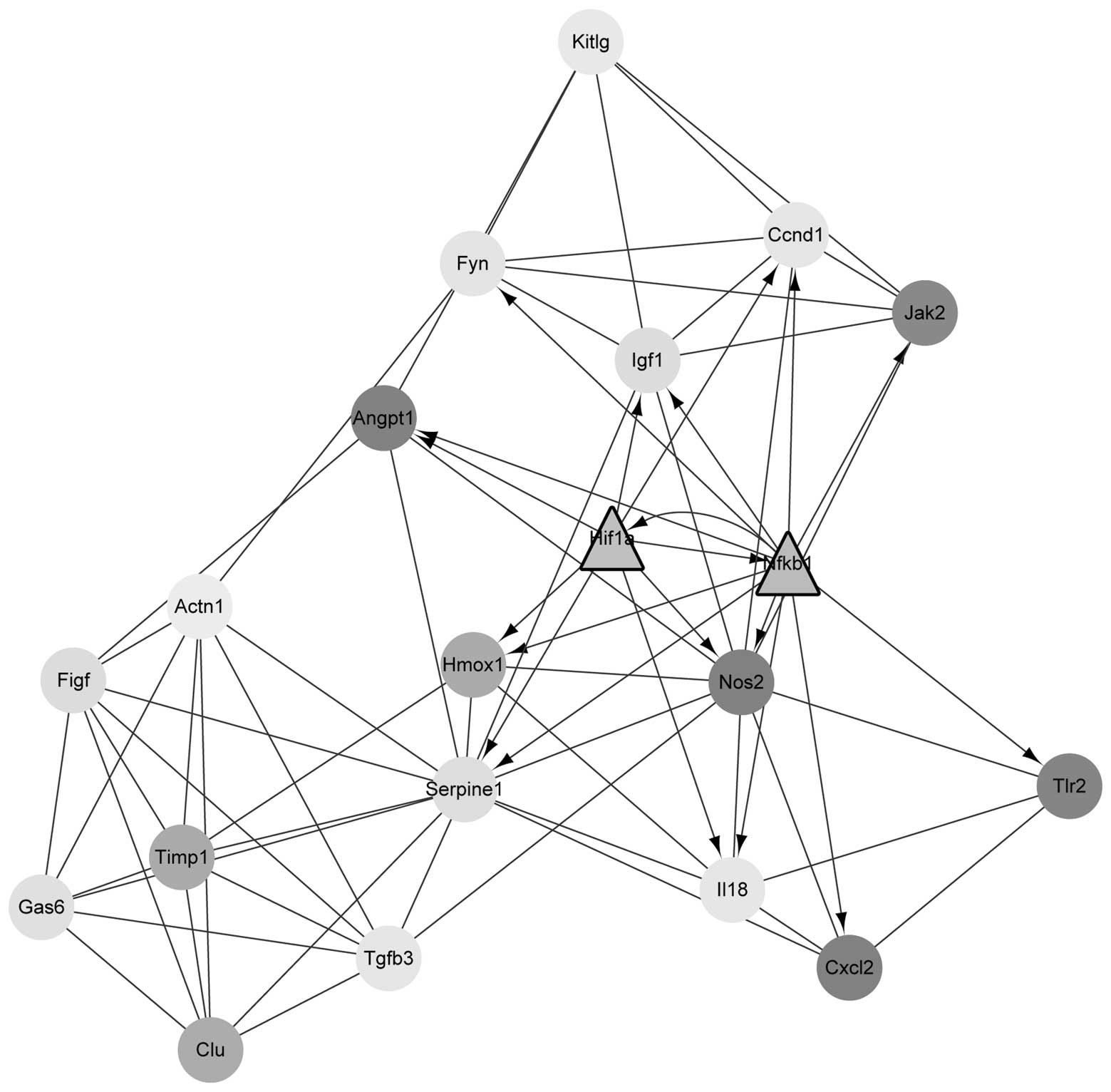
Using the MCODE plugin in cytoscape, four modules
were obtained. Module 1 contained 29 nodes and 376 edges; however,
it only contained downregulated DEGs and no TFs. Module 2 contained
20 nodes and 74 edges, and included the upregulated TFs
HIF1A and NFKB1 (Fig.
2). Module 3 contained 10 nodes and 33 edges while module 4
contained 14 nodes and 42 edges. Module 2 was further subjected to
GO and pathway enrichment analyses, and the results were shown in
Table III. The DEGs in module 2
were predominantly enriched in GO terms associated with the
regulation of cell proliferation and wound healing. In addition,
they were also enriched in pathways associated with cancer and
focal adhesion.
 | Table IIIGO and KEGG pathway enrichment
analyses for the differentially expressed genes in module 2. |
Table III
GO and KEGG pathway enrichment
analyses for the differentially expressed genes in module 2.
| Term | Biological
process/pathway | Count | P-value |
|---|
| GO | | | |
| GO:0042127 | Regulation of cell
proliferation | 12 |
6.47×10−10 |
| GO:0009611 | Response to
wounding | 10 |
4.98×10−9 |
| GO:0042060 | Wound healing | 7 |
1.89×10−7 |
| GO:0001666 | Response to
hypoxia | 7 |
3.28×10−7 |
| GO:0070482 | Response to oxygen
levels | 7 |
4.70×10−7 |
| GO:0009725 | Response to hormone
stimulus | 9 |
4.75×10−7 |
| GO:0031099 | Regeneration | 6 |
8.12×10−7 |
| GO:0009719 | Response to
endogenous stimulus | 9 |
1.15×10−6 |
| GO:0008284 | Positive regulation
of cell proliferation | 8 |
1.20×10−6 |
| GO:0001817 | Regulation of
cytokine production | 6 |
3.26×10−6 |
| KEGG | | | |
| rno05200 | Pathways in
cancer | 8 |
7.66×10−6 |
| rno05212 | Pancreatic
cancer | 4 |
7.37×10−5 |
| rno04510 | Focal adhesion | 5 |
1.45×10−3 |
| rno04150 | mTOR signaling
pathway | 3 |
8.56×10−3 |
| rno04621 | NOD-like receptor
signaling pathway | 3 |
1.16×10−2 |
| rno04115 | p53 signaling
pathway | 3 |
1.31×10−2 |
| rno05211 | Renal cell
carcinoma | 3 |
1.42×10−2 |
| rno04060 | Cytokine-cytokine
receptor interaction | 4 |
1.43×10−2 |
| rno05220 | Chronic myeloid
leukemia | 3 |
1.67×10−2 |
| rno05222 | Small cell lung
cancer | 3 |
2.02×10−2 |
| rno05215 | Prostate
cancer | 3 |
2.35×10−2 |
Discussion
IDD is present in adults with degenerative disc
disease and is considered a major source of back pain in
middle-aged adults, leading to a decrease of life quality or even
disability (22). To date, the
molecular mechanisms involved in the pathology of IDD have not been
fully elucidated. In the present study, a total of 246 upregulated
and 290 downregulated DEGs were identified between ENP and CNP
samples. Several upregulated DEGs, including NFKB1, were
enriched in apoptotic pathways, while downregulated DEGs, including
COL1A1, were enriched in the pathway of ECM-receptor
interaction. In addition, in the PPI network, IL6,
COL1A1, NFKB1 and HIF1A were hub genes with a
high connectivity degree. Furthermore, module analysis indicated
that the DEGs in module 2 were predominantly enriched in GO terms
associated with cell proliferation and pathways associated with
cancer. The results suggested that these genes and pathways may be
candidates for IDD diagnosis and treatment.
A central feature of IDD is loss of tissue
cellularity, which may be the result of programmed cell death
(23). The results of the pathway
enrichment analysis indicated that apoptosis was the most
significant pathway. Apoptosis is a type of programmed cell death
and is characterized by chromosomal concentration, DNA degradation
and cell shrinkage (24).
Apoptosis acts as a quality control mechanism for the maintenance
of tissue homeostasis by eliminating defective cells (25). Apoptosis not only exists in certain
physiological processes, but is also involved in numerous
pathological degenerative diseases, including neurodegeneration and
IDD (26,27). Boos et al (28) reported that the initiation of IDD
is associated with the changes of intervertebral disc cell
behavior, including increased cell death. Gruber and Hanley
(29) suggested that a large
proportion of intervertebral disc cells underwent apoptosis in
patients with degenerated intervertebral discs. Therefore, pathways
of apoptosis may be involved in the progression of IDD in the
ENP.
In addition, the present study found that the TF
NFKB1 was upregulated in the pathway of apoptosis. In
particular, NFKB1 was a hub gene in the PPI network.
NFKB1 encodes the nuclear factor kappa B p105/p50 isoforms
(30). NFKB proteins are a family
of transcription factors that have critical roles in the regulation
of various biological defense processes, including immune
responses, cell-growth control and apoptosis (31). Inappropriate activation of NFKB has
been associated with numerous inflammatory diseases (30). Wuertz et al (32) have reported that IDD is
characterized not only by an imbalance between anabolic and
catabolic processes, but also by inflammatory mechanisms. Of note,
IL-1β and TNF-α are pro-inflammatory cytokines, which have been
detected in the degenerated disc (33). In conclusion, the upregulation of
NFKB1 may have been stimulated by IL-1β and TNF-α, which
suggests that the TF NFKB1 may be an important biomarker for
IDD.
In the present study, several downregulated DEGs,
including COL1A1, were enriched in the pathway of ECM -
receptor interaction; furthermore, COL1A1 was a hub gene in
the PPI network. Over the previous decade, IDD research has focused
on elucidating the mechanisms of ECM degradation, as it causes
marked structural changes, including dehydration and fibrosis of
the nucleus pulposus, disorganization of the annulus fibrosus and
calcification of the cartilaginous end plates (28). These changes ultimately lead to
structural failure in IDD. Of note, the protein collagen type I,
alpha 1, encoded by COL1A1, is a major ECM component
(34). Col1a1 has been found in
most connective tissues and is abundant in bone (35). Several studies have demonstrated
that the expression of genes encoding connective tissue components,
particularly collagen, can be affected by numerous cytokines,
including IL-1 and TNF-α (36,37).
Mori et al (38) found that
TNF-α decreased Col1a1 expression through suppressing
promoter activity of Col1a1. Feng et al (39) suggested that Col1a1 can
provide tensile strength in the annulus fibrosus of the
intervertebral disc. Thus, the downregulated expression of
COL1A1 in the present study may be regulated by IL-1β and
TNF-α, resulting in the initiation of IDD.
In the PPI network, IL6 and HIF1A were
hub genes with a high connectivity degree, while HIF1A was
also a TF. IL6 together with IL-1α, IL-1β and TNF-α are among the
most potent catabolic cytokines and pro-inflammatory mediators as
mentioned above (40).
Noponen-Hietala et al (41)
reported that the features of pain, tissue destruction and
inflammation in IDD were linked with the functions of IL6, and IL6
was also associated with IDD-associated radiculopathy. Therefore,
IL6 is not the sole pro-inflammatory cytokine in the pathogenesis
of IDD and appears to be important in the mediation of pain in
IDD.
In the present study, the TF of HIF1A was
also found in module 2 and was involved in GO terms associated with
cell proliferation. HIF1 functions as a transcriptional regulator
of the adaptive response to hypoxia and activates transcription of
certain genes involved in energy metabolism, angiogenesis and
apoptosis (42). Wu et al
(43) found that Hif1a has an
important role in the metabolism and synthesis of ECM and is a
pivotal contributor to the survival of nucleus pulposus cells.
Therefore, HIF1A may be involved in the development of
IDD.
In conclusion, the present study provided a
comprehensive bioinformatics analysis of the molecular mechanisms
of IDD induced by IL-1β and TNF-α. Pathways of apoptosis and ECM -
receptor interaction as well as their enriched DEGs may have
important roles in the development and progression of IDD. In
addition, the TFs NFKB1 and HIF1A and the DEGs
COL1A1 and IL6 are hypothesized to interact with
IL1-β and TNF-α to participate in the development of IDD. However,
further bioinformatics and experimental studies with larger sample
sizes are required to confirm the results of the present study.
Acknowledgments
The present study was supported by the National
Natural Youth Science Foundation of China (grant no. 31300778).
References
|
1
|
Bolger C: Spine disorders (medical and
surgical management). The Surgeon. 8:1852010. View Article : Google Scholar
|
|
2
|
McCann MR, Tamplin OJ, Rossant J and
Séguin CA: Tracing notochord-derived cells using a Noto-cre mouse:
Implications for intervertebral disc development. Dis Model Mech.
5:73–82. 2012. View Article : Google Scholar :
|
|
3
|
Roberts S, Evans H, Trivedi J and Menage
J: Histology and pathology of the human intervertebral disc. J Bone
Joint Surg Am. 88(Suppl 2): 10–14. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors in intervertebral disc
aging and degeneration. Spine J. 13:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration and what causes it? Spine (Phila
Pa 1976). 31:2151–2161. 2006. View Article : Google Scholar
|
|
6
|
Johnson WE and Roberts S: 'Rumours of my
death may have been greatly exaggerated': A brief review of cell
death in human intervertebral disc disease and implications for
cell transplantation therapy. Biochem Soc Trans. 35:680–682. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Le Maitre C, Pockert A, Buttle D, Freemont
A and Hoyland J: Matrix synthesis and degradation in human
intervertebral disc degeneration. Biochem Soc Trans. 35:652–655.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Markova DZ, Kepler CK, Addya S, Murray HB,
Vaccaro AR, Shapiro IM, Anderson DG, Albert TJ and Risbud MV: An
organ culture system to model early degenerative changes of the
intervertebral disc II: Profiling global gene expression changes.
Arthritis Res Ther. 15:R1212013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc B (Methodological). 57:289–300.
1995.
|
|
13
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
15
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID Gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Albert R and Barabási AL: Statistical
mechanics of complex networks. Rev Mod Phys. 74:47–97. 2002.
View Article : Google Scholar
|
|
18
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Matys V, Fricke E, Geffers R, Gössling E,
Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis
OV, et al: TRANSFAC: Transcriptional regulation, from patterns to
profiles. Nucleic Acids Res. 31:374–378. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Akhatib B, Onnerfjord P, Gawri R, Ouellet
J, Jarzem P, Heinegård D, Mort J, Roughley P and Haglund L:
Chondroadherin fragmentation mediated by the protease HTRA1
distinguishes human intervertebral disc degeneration from normal
aging. J Biol Chem. 288:19280–19287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ariga K, Miyamoto S, Nakase T, Okuda S,
Meng W, Yonenobu K and Yoshikawa H: The relationship between
apoptosis of endplate chondrocytes and aging and degeneration of
the intervertebral disc. Spine (Phila Pa 1976). 26:2414–2420. 2001.
View Article : Google Scholar
|
|
24
|
Lockshin RA and Zakeri Z: Apoptosis,
autophagy and more. Int J Biochem Cell Biol. 36:2405–2419. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fuchs Y and Steller H: Programmed cell
death in animal development and disease. Cell. 147:742–758. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moreno S, Imbroglini V, Ferraro E,
Bernardi C, Romagnoli A, Berrebi AS and Cecconi F: Apoptosome
impairment during development results in activation of an autophagy
program in cerebral cortex. Apoptosis. 11:1595–1602. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye W, Xu K, Huang D, Liang A, Peng Y, Zhu
W and Li C: Age-related increases of macroautophagy and
chaperone-mediated autophagy in rat nucleus pulposus. Connect
Tissue Res. 52:472–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boos N, Weissbach S, Rohrbach H, Weiler C,
Spratt KF and Nerlich AG: Classification of age-related changes in
lumbar intervertebral discs: 2002 Volvo Award in basic science.
Spine (Phila Pa 1976). 27:2631–2644. 2002. View Article : Google Scholar
|
|
29
|
Gruber HE and Hanley EN Jr: Analysis of
aging and degeneration of the human intervertebral disc: Comparison
of surgical specimens with normal controls. Spine (Phila Pa 1976).
23:751–757. 1998. View Article : Google Scholar
|
|
30
|
Karban AS, Okazaki T, Panhuysen CI,
Gallegos T, Potter JJ, Bailey-Wilson JE, Silverberg MS, Duerr RH,
Cho JH, Gregersen PK, et al: Functional annotation of a novel NFKB1
promoter polymorphism that increases risk for ulcerative colitis.
Hum Mol Genet. 13:35–45. 2004. View Article : Google Scholar
|
|
31
|
Baldwin AS: Series introduction: The
transcription factor NF-kappaB and human disease. J Clin Invest.
107:3–6. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wuertz K, Vo N, Kletsas D and Boos N:
Inflammatory and catabolic signalling in intervertebral discs: The
roles of NF-κB and MAP kinases. Eur Cell Mater. 23:103–119.
2012.
|
|
33
|
Akeda K, An H, Gemba T, Okuma M, Miyamoto
K, Chujo T, Kitahara S and Masuda K: A new gene therapy approach:
In vivo transfection of naked NFkB decoy oligonucleotide restored
disc degeneration in the rabbit annular needle puncture model.
Trans Orthop Res Soc. 30:452005.
|
|
34
|
Wu F and Chakravarti S: Differential
expression of inflammatory and fibrogenic genes and their
regulation by NF-kappaB inhibition in a mouse model of chronic
colitis. J Immunol. 179:6988–7000. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Alberts B, Johnson A, Lewis J, Raff M,
Roberts K and Walter P: The extracellular matrix of animals.
2002
|
|
36
|
Postlethwaite AE, Raghow R, Stricklin GP,
Poppleton H, Seyer JM and Kang AH: Modulation of fibroblast
functions by interleukin 1: Increased steady-state accumulation of
type I procollagen messenger RNAs and stimulation of other
functions but not chemotaxis by human recombinant interleukin 1
alpha and beta. J Cell Biol. 106:311–318. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Scharffetter K, Heckmann M, Hatamochi A,
Mauch C, Stein B, Riethmüller G, Ziegler-Heitbrock HW and Krieg T:
Synergistic effect of tumor necrosis factor-alpha and
interferon-gamma on collagen synthesis of human skin fibroblasts in
vitro. Expe Cell Res. 181:409–419. 1989. View Article : Google Scholar
|
|
38
|
Mori K, Hatamochi A, Ueki H, Olsen A and
Jimenez S: The transcription of human alpha 1 (I) procollagen gene
(COL1A1) is suppressed by tumour necrosis factor-alpha through
proximal short promoter elements: Evidence for suppression
mechanisms mediated by two nuclear-factor binding sites. Biochem J.
319:811–816. 1996. View Article : Google Scholar
|
|
39
|
Feng H, Danfelter M, Strömqvist B and
Heinegård D: Extracellular matrix in disc degeneration. J Bone
Joint Surg Am. 88(Suppl 2): 25–29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Benoist M: The natural history of lumbar
disc herniation and radiculopathy. Joint Bone Spine. 69:155–160.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Noponen-Hietala N, Virtanen I, Karttunen
R, Schwenke S, Jakkula E, Li H, Merikivi R, Barral S, Ott J,
Karppinen J and Ala-Kokko L: Genetic variations in IL6 associate
with intervertebral disc disease characterized by sciatica. Pain.
114:186–194. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zou H, Kang X, Pang LJ, Hu W, Zhao J, Qi
Y, Hu J, Liu C, Li H, Liang W, et al: Xp11 translocation renal cell
carcinoma in adults: A clinicopathological and comparative genomic
hybridization study. Int J Clin Exp Pathol. 7:236–245, eCollection
2014. 2013.
|
|
43
|
Wu WJ, Zhang XK, Zheng XF, Yang YH, Jiang
SD and Jiang LS: SHH-dependent knockout of HIF-1 alpha accelerates
the degenerative process in mouse intervertebral disc. Int J
Immunopathol Pharmacol. 26:601–609. 2013.PubMed/NCBI
|