Introduction
Relapsed acute myeloid leukemia (AML) is considered
to be the result of leukemic stem cell (LSC) survival following
chemotherapy (1). AML is a
heterogeneous clonal disorder, characterized by the accumulation of
immature myeloblasts (2).
Malignant cell proliferation is maintained by a small fraction of
LSCs, and similar to normal hematopoietic stem cells (HSCs), LSCs
exhibit certain stem cell properties, including self-renewal,
differentiation capacity and expression of cell surface phenotype
CD34+CD38− markers (3). LSCs also predominantly produce
colony-forming units (CFUs) in vitro, indicating their
potential for full differentiation (4). The CFU assay is traditionally used
for the detection of hematopoietic progenitor cells (HPCs) in the
blood (5). Despite the differences
in CFU formation between leukemic and normal progenitor cells, it
remains unclear whether colonies are derived from normal HPCs or
LSCs/HPCs expressing leukemia-associated genes.
Bone marrow (BM) microenvironments and
stem/progenitor cells communicate in order to sustain drug
resistance or differentiate into cell lineages; therefore
understanding the stromal condition against leukemic cells
expressing abnormal genes is required for the development of
advanced therapeutic strategies to prevent relapse. Since 1863,
when Rudolf Virchow highlighted the importance of the tumor
microenviroment for cell growth (6), studies have supported the existence
of an association between tumor cell fate and the microenvironment
(6–8).
Podoplanin, a 38 kDa integral membrane mucoprotein,
predominantly expressed in the lymphatic capillaries, has been
identified to be involved in tumor progression,
epithelial-to-mesenchymal transition and lymphatic function
(9,10). Its expression has also been
observed in intratumoral stromal cells, which can function as
normal stromal cells (11,12). Previous studies have been
demonstrated that podoplanin is a potent cancer-associated factor
in the microenvironments of various tumor types (11,13,14).
Podoplanin has been identified to be expressed in osteoblasts and
osteocytes in normal bone tissue, and highly expressed in
mesenchymal stromal cells, the main component of the BM
microenvironment, under conditions of abundant vascular endothelial
growth factor C (14,15). Despite the fact that the role of
podoplanin in tumor development has been extensively studied
(16–18), the role of podoplanin+
cells as tumor microenvironmental factors in leukemia remains to be
fully elucidated.
The present study examined the role of
podoplanin+ cells in leukemia, in addition to
investigating its protective role against apoptosis in leukemic
blasts, which are enriched by the fibromyalgia-like tyrosine
kinase-3 (FLT3) gene. These present study aimed to provide
insight into the role of podoplanin as a tumor microenvironmental
factor, and contribute to the development of targeted
therapies.
Materials and methods
Human primary cells and cell lines
All experiments were approved by the Institutional
Review Board of the Human Research at the Catholic University of
Korea (Seoul, South Korea). A total of 12 AML blood samples were
obtained from patients admitted to the Catholic Blood and Marrow
Transplantation Center at Seoul St. Mary's Hospital (Seoul, South
Korea). The patients were diagnosed with various subtypes of AML
using the World Health Organization (WHO) classification system
(19). A total of seven patients
had AML not otherwise specified, three had AML with an inversion in
chromosome 16, one had AML with myelodysplasia-related change, and
one had acute promyelocytic leukemia. BM and peripheral blood (PB)
samples were frozen in fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% dimethyl sulfoxide
(DMSO; Sigma-Aldrich, St. Louis, MO, USA) and stored in liquid
nitrogen. BM- and PB-derived mononuclear cells (MNCs) were
fractionated by density gradient centrifugation at 1,220 × g for 30
min at 4°C, using Ficoll-Paque™ (17-1440-03; GE Healthcare Life
Sciences, Shanghai, China). The clinical characteristics and
laboratory data of the patients with AML enrolled in the present
study are listed in Table I.
TIB152 human Jurkat cells (American Type Culture Collection,
Manassas, VA, USA), were grown in RPMI medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS in a humidified
atmosphere of 5% CO2 at 37°C. CellTrace™
carboxyfluorescein diacetate succinimidyl ester (CFSE; C34554;
Invitrogen, Thermo Fisher Scientific, Inc.) with 5 µM in
DMSO was used to stain the Jurkat cells.
 | Table IClinical and laboratory features of
patients with AML. |
Table I
Clinical and laboratory features of
patients with AML.
| Patient | WHO subtype | Cell | Age (years) | Gender | WBC/mm3
at diagnosis | Molecular
defects | Cytogenetic
anomalies |
|---|
| 1 | AML NOS | PB | 19 | M | 32240 | NEG | 46, XY [20] |
| 2 | AML NOS | PB | 64 | F | 127350 | FLT3 | 46, XX [20] |
| 3 | AML with MRC | BM | 65 | M | 260300 | MRC | 46, XY,
del(5)(q11.2q15)[4]/46,
XY[16] |
| 4 | APL | PB | 41 | M | 43010 | RARA | 46, XY,
t(15;17)(q22;q12)[20] |
| 5 | AML with
inv(16) | PB | 31 | M | 154500 | CBFB | 46, XY,
t(9;22)(q34;q11.2), inv(16)(p13.1q22) [13]/47, idem, +17[15]/48,
idem, +8, +17[2] |
| 6 | AML NOS | PB | 54 | F | 227830 | MLLT3 | 46, XX,
t(9;11)(p22;q23)[20] |
| 7 | AML NOS | PB | 41 | M | 248521 | NPM1 | 46, XY [20] |
| 8 | AML with
inv(16) | PB | 45 | M | 42234 | CBFB | 46, XY,
inv(16)(p13.1q22)[20] |
| 9 | AML NOS | PB | 54 | M | 195104 | NPM1 | 46, XY [20] |
| 10 | AML NOS | PB | 36 | F | 240640 | NPM1 | 46, XX [20] |
| 11 | AML with
inv(16) | PB | 46 | M | 108400 | CBFB | 46, XY,
inv(16)(p13.1q22)[20] |
| 12 | AML NOS | PB | 65 | F | 114510 | NEG | 46, XX [20] |
Magnetic-activated cell sorting and CFU
assay
Podoplanin+ cells (BAF3670; R&D
Systems, Inc., Minneapolis, MN, USA) were sorted and isolated from
AML primary cells using magnetic beads (130-056-701; Miltenyi
Biotec, Inc. Cambridge, MA, USA) in order to validate human
clonogenic hematopoietic progenitor properties. Anti-biotin
microbeads were used to isolate podoplanin (120-000-900; Miltenyi
Biotec, Inc.). Sorted cells were cultured in methylcellulose media
(H4434; STEMCELL Technologies, Inc., Vancouver, BC, Canada) for
7–10 days and colonies were counted using an inverted microscope
(Leica Microsystems, Inc., Buffalo Grove, IL, USA).
Jurkat proliferation assay
CFSE-labeled Jurkat cells (2.5×103) were
co-cultured with the sorted podoplanin+ and
podoplanin− cells (2.5×103) from BM-MNCs in
RPMI medium supplemented with 1% FBS. After 24 h, the cells were
stained with rabbit anti-human Ki67 antibody (cat no. ab15580;
Abcam, Cambridge, UK) and counterstained with
4′,6-diamidino-2-phenylindole (DAPI). Cells positive for green
fluorescent protein, CFSE-labeled Jurkat cells, Ki67 positive cells
and DAPI-stained cells were counted under the inverted
microscope.
Flow cytometry
Fluorescence activated cell sorting (FACS) staining
and analysis was performed as previously described (20). Briefly, the cells were resuspended
in 100 µl rinsing buffer and incubated with all antibodies
at 4°C for 20 min. These included phycoerythrin (PE)-conjugated
mouse anti-CD34 (1:20; cat no. 555822; BD Pharmingen, San Diego,
CA, USA) and PEcy™ 5-conjugated mouse anti-CD38 (1:20; cat no.
555461; BD Pharmingen) antibodies, which were used to label
leukemic stem cells (LSCs), and allophycocyanin (APC)-conjugated
anti-human podoplanin polyclonal antibody (1:20; cat no. FAB3670A;
R&D Systems, Inc.,), which was used for the detection of
podoplanin. Subsequently, the cells were incubated with PE-annexin
V (cat no. 556421; BD Pharminogen) for 20 min at room temperature
for the detection of apoptosis. Following washing with 1% bovine
serum albumin in phosphate-buffered saline (PBS; Thermo Fisher
Scientific, Inc.), the cells were analyzed using a FACSCalibur flow
cytometer equipped with CellQuest software, version 3.0 (BD
Biosciences, San Diego, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA isolation and DNA synthesis were performed
as previously described (21). PCR
reactions were performed in a 50 µl PCR reaction mixture
(Promega Corporation, Madison, WI, USA) containing 100 ng of each
primer, 1X Tris-ethylenediaminetetraacetic acid buffer, 100 ng
template DNA, 2.5 units HQ Taq polymerase, and 2.5 mM
deoxyribonucleotide triphosphate. PCR amplification was performed
using a conventional thermocycler (P×2 Thermal Cycler; Thermo
Fisher Scientific, Inc.) under the following cycling conditions:
94°C for 4 min; 30–36 cycles at 94°C for 1 min, 53°C for 1 min, and
72°C for 2 min; extension cycle was at 72°C for 7 min. The RT-qPCR
products were separated on a 2.0% agarose gel (Sigma-Aldrich) at 12
V/cm using a Tris-acetic acid-ethylenediaminetetraacetic acid
buffer, and were subsequently stained with ethidium bromide (Thermo
Fisher Scientific, Inc.), and visualized and photographed under an
ultra-violet transilluminator (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Information regarding the primer/probe sets
(TaqMan; Biosearch Technologies, Inc., Novato, CA, USA) and the
primers used in the present study is provided in Table II. The relative mRNA expression of
target genes was calculated using the comparative Cq method. All
target gene expression was normalized to the expression of
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in multiplexed
reactions performed in triplicate. Differences in Cq values were
calculated for each target mRNA by subtracting the mean value of
the GAPDH expression (relative expression = 2−ΔΔCq)
(22).
 | Table IIPrimers and probes for reverse
transcription-quantitative polymerase chain reaction. |
Table II
Primers and probes for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primers and probes
(5′-3′) |
|---|
| Mouse
GAPDH | F:
GGTGGTCTCCTCTGACTTCAACA
R: GTGGTCGTTGAGGGCAATG
P: CCACTCCTCCACCTTTGACGCTGG |
| Mouse
Wt1 | F:
AGCTGTCGGTGGCACAGTTGTCA
R: TGCCTGGGATGCTGGACTGTC
P: ACCCCTCAAAGCGCCAGCTGGAGTTT |
| Mouse
survivin | F:
TCTGCTTTAAGGAATTGGAAGG
R: CTCTGTCTGTCCAGTTTCAAG
P: ACGGTTAGTTCTTCCATCTGCTTCTTGAC |
| Human
GAPDH | F:
GGTGGTCTCCTCTGACTTCAACA
R: GTGGTCGTTGAGGGCAATG |
| Human
podoplanin | F:
CAGGTGCCGAAGATGATGTG
R: TGTTGCCACCAGAGTTGTCA |
| Human
FLT3 | F:
GCATGCCTGGTTCAAGAGAA
R: TGCCAGGGTAAGGATTCACA |
Immunostaining
Immunostaining was conducted as previously described
(14). Briefly, using the cytospin
method (4,23), cells were spun onto slides and
fixed with 2% paraformaldehyde (Sigma-Aldrich) for 10 min at 25°C.
Following washing with PBS, the cells were blocked with 5% horse
serum (Thermo Fisher Scientific, Inc.) and incubated with the
primary antibodies overnight at 4°C, followed by incubation with
the secondary antibody for 30 min at room temperature. The primary
antibodies used were as follows: Biotinylated anti-podoplanin (cat
no. BAF3670; R&D Systems, Inc.), rabbit anti-CD34 (cat no.
GWB-BBP214; GenWay Biotech, Inc., San Diego, CA, USA) and rabbit
anti-Ki67. The Cy3 affinipure goat anti-IgG (cat no. NC9771594;
Jackson Immuno-Research Laboratories, Inc., Inc., West Grove, PA,
USA) secondary antibody was used. The cells were incubated with
DAPI for 1 min at room temperature to stain the nuclei. Images were
captured using the Zeiss LSM 510 META confocal laser scanning
microscope and LSM 510 Imaging software, version 3.2 (Carl Zeiss,
Inc., Gottingen, Germany).
Statistical analysis
All results are presented as the mean ± standard
error. The comparison between groups was performed using the
Mann-Whitney U test. GraphPad Prism version 4 software (GraphPad
Software, Inc, La Jolla, CA, USA) was used for the statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
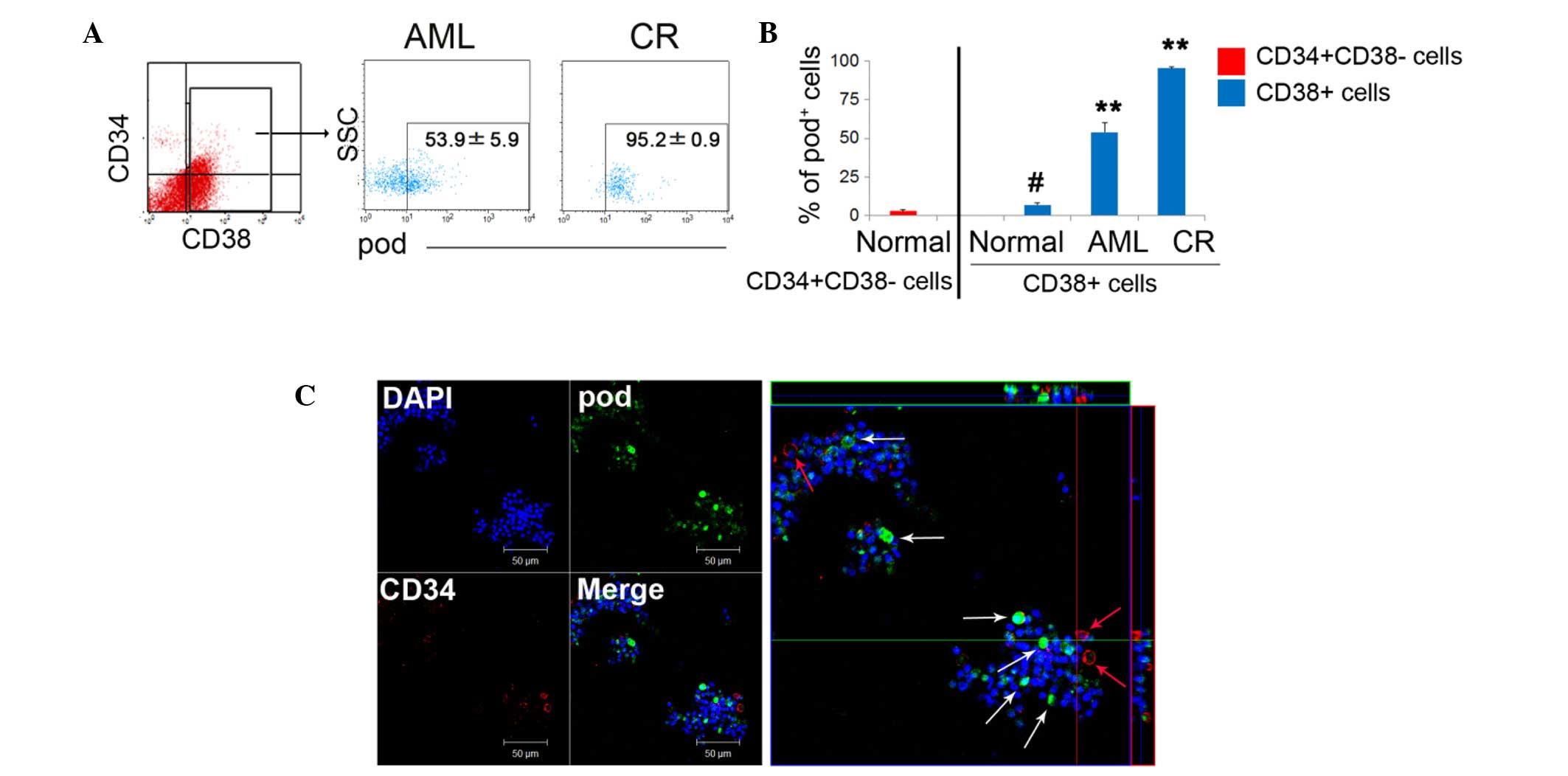
High podoplanin expression on
CD38+ differentiated cells in leukemia
To investigate the expression of podoplanin in
leukemic cells, FACS analysis was performed in AML patient-derived
cells. Under normal conditions, podoplanin is expressed in
CD45− stromal cells, including osteocytes and
osteoblasts; however, this protein is only expressed in
CD45+ hematopoietic cells under disease conditions
(14,21,24).
The results of the present study demonstrated that the expression
of podoplanin was markedly higher in mature CD38+ cells
in complete remission (CR) than in those cells in the de
novo AML state (AML, 53.9%; CR, 95.2%; Fig. 1A). Of note, under normal
conditions, podoplanin+ cells were significantly more
frequent in mature CD38+ cells (6.9%) than they were in
CD34+CD38− HSCs (1.7%) (Fig. 1B). In CD38+
differentiated cells, the expression of podoplanin was
significantly and gradually increased during the complete remission
(CR) state, compared with the AML and normal states. This suggests
that podoplanin-sustaining cells are required for BM reconstruction
or blast protection, and that most podoplanin+ cells
function as supportive cells rather than as LSCs. Due to the fact
that CD38+ cells consist of a number of immune cells
such as T, B, and nature killer cells, most CD38+
leukocytes that survive chemotherapy, may serve a role in blast
communication in the tumor environment. A low frequency of
CD34+ podoplanin+ cells was also detected in
flushed cells, whereas, podoplanin single positive cells exhibited
a high frequency (Fig. 1C), again
suggesting that podoplanin cells can potentially function as
supportive cells rather than as LSCs.
Enrichment of FLT3 in
podoplanin−, however not podoplanin+ cells
and high CFU-colony forming efficiency of podoplanin-cells
To further examine CFU potency, sorted cells were
cultured in Matrigel gel supplemented with cytokines, and CFUs were
observed after 10 days. Common myeloid progenitors were identified
to be able to differentiate into two cell lineages: i) Granulocyte,
erythrocyte, monocyte, megakaryocyte (GEMM), which includes
megakaryocytes and erythrocytes, and ii) granulocyte-macrophage
(GM) cells, which represent myeloblasts. Fig. 2 presents the colonies formed,
including GEMM, G, GM and M from podoplanin+ or
podoplanin− cells. The number of CFU-GM colonies
detected in CD34+ podoplanin− cells was
significantly higher than that of other colonies (Fig. 2). Colonies produced from normal
HSCs were characterized and enumerated by their distinct cell
morphology. Similarly, leukemic-derived colonies were also rapidly
formed by a progenitor population; however, leukemic-derived
colonies with atypical morphologies in CD34+
podoplanin− cells overwhelmingly produced abnormal HSCs.
The majority of formed colonies were small and condensed (<0.4
mm), which is consistent with previous studies (4,25,26),
suggesting a putative leukemic stem/progenitor cell function of
podoplanin− cells. To examine whether these CFUs
expressed leukemia-associated genes, and had a differential potency
based on podoplanin expression, CD34+
podoplanin+ or CD34+ podoplanin−
cells were isolated using a microbead system. Sorted cells were
immediately subjected to RT-qPCR to confirm the purity using
podoplanin-specific primers, and the cells were then measured for
FLT3, which is known to be overexpressed in patients with
leukemia (27,28). The RT-qPCR data demonstrated that
the podoplanin gene was exclusively expressed by the sorted
podo-planin+ cells, and that the FLT3 gene was
markedly increased in podoplanin− cells, however not in
podoplanin+ cells; however, the expression of these
genes was similar in both podoplanin+ and
podoplanin− cells during differentiation (Fig. 3A). Sorted cells exhibited
changeable expression of FLT3 and podoplanin at the
time of differentiation, implying that there is some flexibility in
the expression of AML genes.
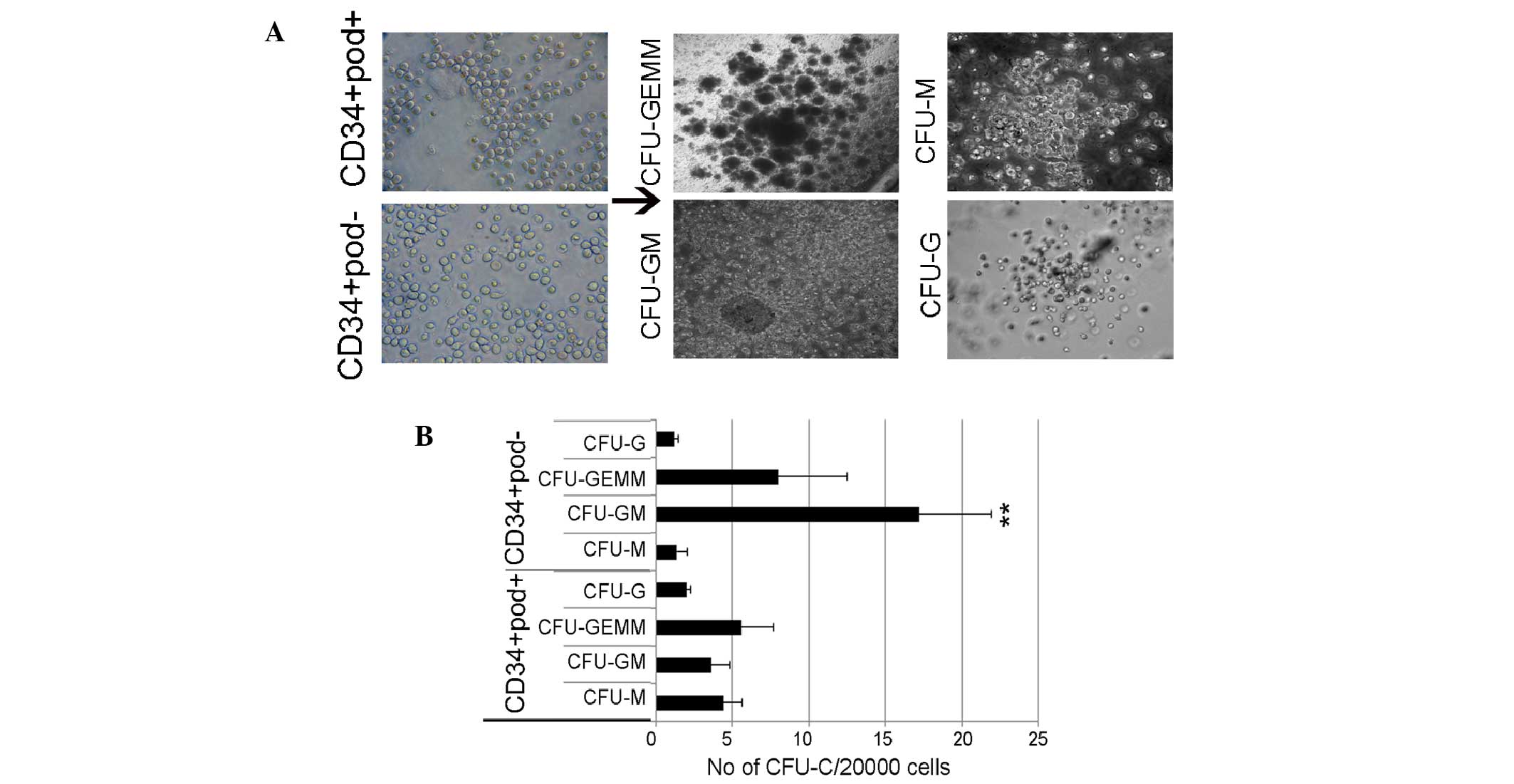 | Figure 2Leukemic-derived CFU-assay in
CD34+ podoplanin+ or CD34+
podoplanin− cells. (A) Morphologies of colonies. (B)
Podoplanin− cells produced high numbers of CFUs,
including CFU-GM and CFU-GEMM, compared with podoplanin+
cells. Values are expressed as the mean ± standard error.
**P<0.01 vs. CD34+ podoplanin+
cells. Scale bar, 100 µm. CFU, colony forming unit; GM,
granulocyte-macrophage; GEMM, granulocyte, erythrocyte, monocyte,
megakaryocyte; G, granulocyte; M, macrophage. |
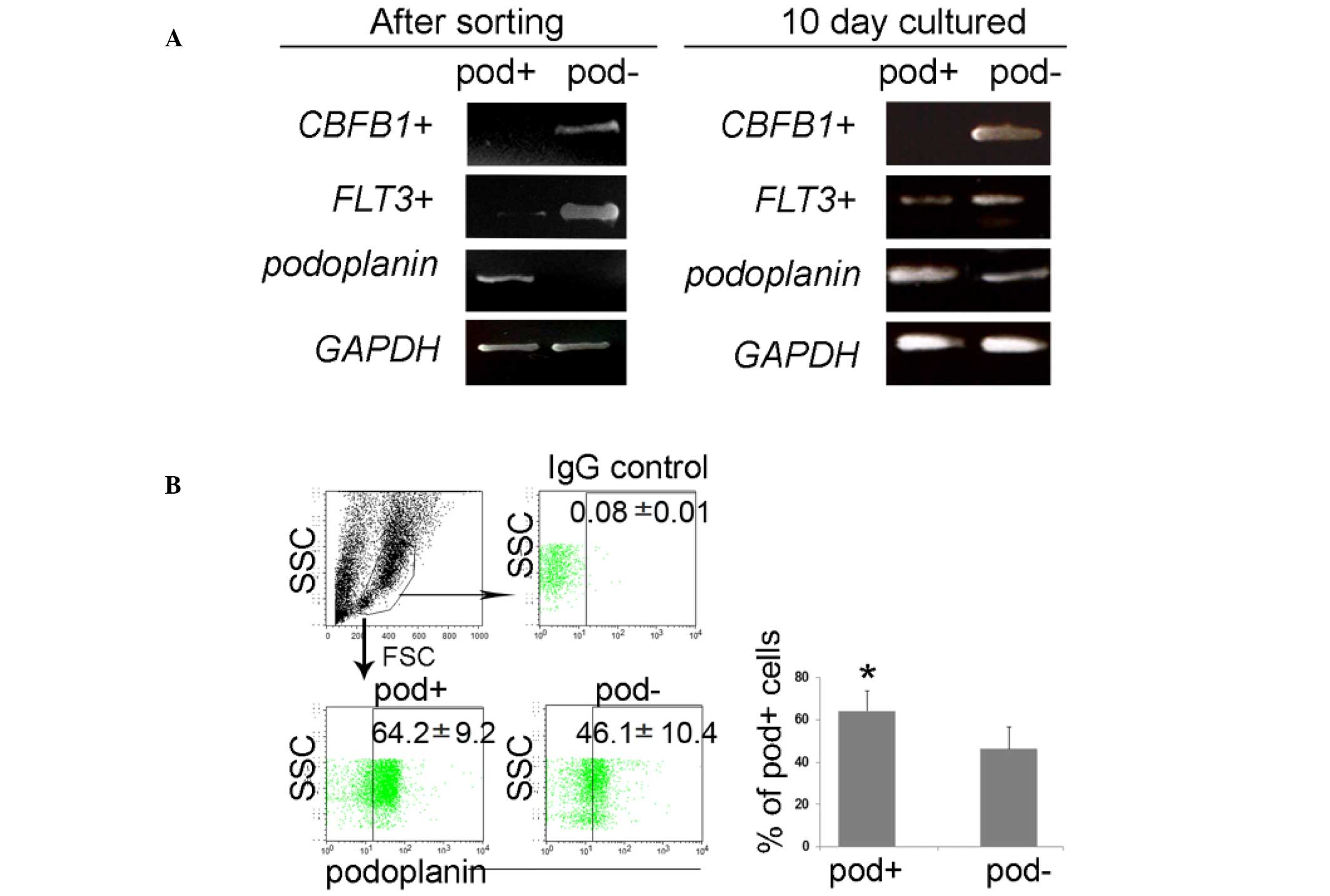 | Figure 3Enrichment of FLT3 in sorted
cells, and further differentiation from podoplanin+ or
podoplanin− cells. (A) Isolated podoplanin+
and podoplanin− cells maintained high purity following
magnetic-activated cell sorting, and FLT3 was exclusively
expressed in podoplanin− cells; however, their
expression was altered by differentiation. (B) At the protein
level, the podoplanin expression was also upregulated in the
podoplanin− cell population, implying flexibility in
leukemic status. Values are expressed as the mean ± standard error.
*P<0.05 vs. podoplanin− cells.
CBFB1, core-binding factor subunit beta 1; FLT3,
Fms-like tyrosine kinase-3; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase; IgG, immunoglobulin G; SSC, side scatter; FSC,
forward scatter; pod, podoplanin. |
These results suggested that leukemic properties are
enriched by podoplanin− rather than
podoplanin+ cells. FLT3 acts as a molecular
marker, and so it reflects a leukemic state (29,30);
however, podoplanin+ cells may not be directly
representative of leukemic cells. It has been reported that
translocation of the chromosome containing the core-binding factor
subunit beta 1 (CBFB1) gene results in AML (31). The expression of CBFB1 was
restricted in podoplanin− cells regardless of further
differentiation, suggesting that podoplanin+ cells may
function as stromal cells to podoplanin− cells (data not
shown), which contain leukemic stem cells expressing FLT3.
At a protein level, podoplanin is primarily sustained in
differentiated CFUs, and simultaneously detected in
podoplanin− cells (Fig.
3B), further suggesting its necessity in the maintenance of
leukemic cells.
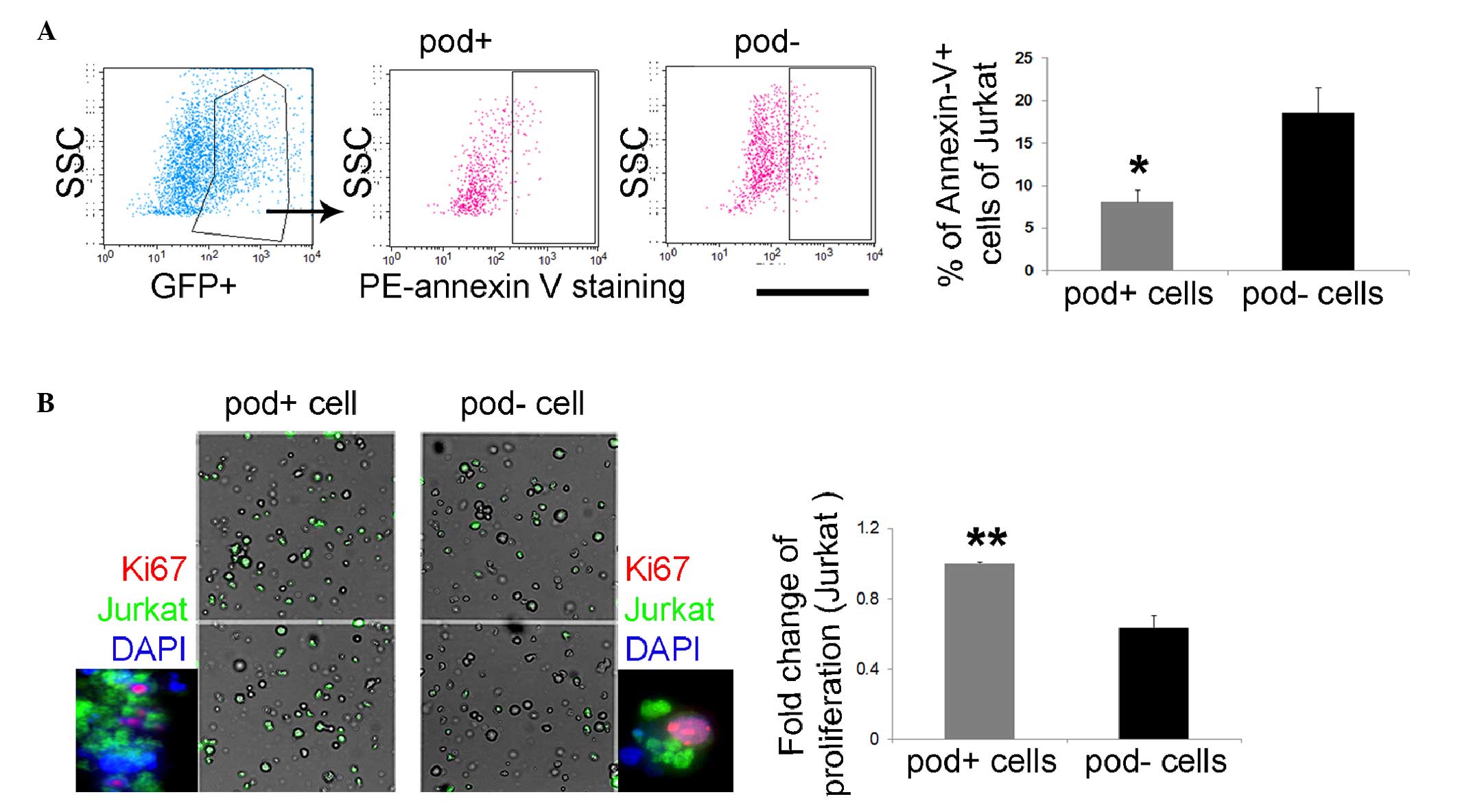
Leukemic cells can promote proliferative
and anti-apoptotic effects under co-culture with
podoplanin+ cells
To investigate the function of
podoplanin+ cells as stromal cells, CFSE-stained Jurkat
cells were cultured with podoplanin+ or
podoplanin− cells. After 24 h the
Jurkat/podoplanin+ co-cultured cells exhibited a lower
number of annexin-V+ cells (2.29-fold), compared with
the Jurkat/podoplanin− co-cultured cells (Fig. 4A), thus suggesting that
podoplanin+ cells can protect leukemic cells from
apoptosis. Additionally, Jurkat cells proliferated rapidly during
co-culture with podoplanin+ cells. There was a
significantly increased number of Ki67+ green
fluorescent protein+ Jurkat cells during co-culture with
podoplanin+ cells (1.47-fold), compared with the results
of co-culture with podoplanin− cells (Fig. 4B), suggesting the supportive role
of podoplanin+ cells in leukemic cell activity. These
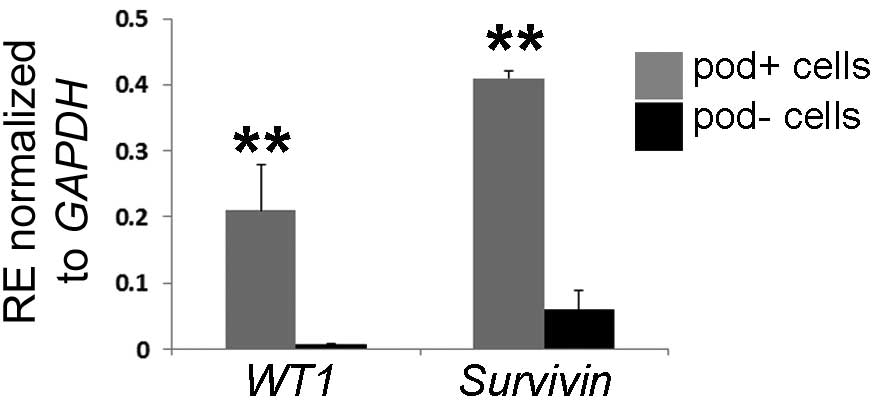
results raised the question of whether primary blasts are able to
upregulate their leukemic-associated genes in
podoplanin+ stromal cell. Wilms' tumor gene 1
(WT1) and survivin, an apoptosis inhibitor encoded by
survivin and expressed primarily in human blast cells, were
selected for co-culture with podoplanin+ or
podoplanin− cells. Both genes are commonly regarded as
leukemic-specific antigens and have been suggested to be
upregulated under leukemic conditions (32). It was identified that the
expression of WT1 and survivin was significantly
increased (27.4-fold and 6.2-fold, respectively) in the cells
co-cultured with podoplanin+ in vitro (Fig. 5), which supports a role of
podoplanin+ cells in the maintenance of leukemic
cells.
Discussion
Podoplanin was originally known as a protein marker
for lymphatic endothelium (10).
Previous studies have suggested a potential role of podoplanin in
sustaining tumor cells in the tumor microenvironment (33,34).
In addtition, podoplanin+ cells may function as
stem/progenitor cells under lymphan-giogenic or lymphavasculogenic
conditions in BM-derived cells (21) and regulate tumor metastasis
(35), suggesting a multifactorial
role of podoplanin in solid tumors. The role of podoplanin in
leukemia, however, remains unclear. Previous studies reported that
lymphangiogenic cytokines and markers, including podoplanin, are
involved in leukemia, and in the BM microenvironment in particular
(36,37).
Leukemic stem cells require stromal cells to survive
chemotherapy (38). In numerous
niches, stromal cells, including osteoblasts in normal BM, express
podoplanin; this expression has been demonstrated to increase
markedly under tumor conditions (24). In the present study, an increased
level of podoplanin was observed in leukemic cells, which is
consistent with previous studies of solid tumors (39–41).
Of note, CD38+ cells sustained a high podoplanin
expression in the de novo AML and CR states following
chemotherapy, and increased podo-planin is continuously required to
maintain BM reconstruction or blast survival. The high expression
of podoplanin in CD38+ cells, including leukocytes, may
be associated with the release of podoplanin-soluble mediators.
Cross-linkage between podoplanin-soluble mediator defensive action
and surviving leukemic stem cells should be investigated in order
to assist the development of targeted AML therapy.
Previously, Kim et al (42) reported that osteopontin (OPN)
production by tumor cells, however not by stromal cells, enhances
the propagation of tumor initiating cells in tumor environments,
and that OPN silencing can delay tumor growth and extramedullary
myelopoiesis. Like the diverse roles of OPN in tumor cells, the
effects of podoplanin may alter depending on the environment; thus
the present study investigated whether the inhibition of podoplanin
was able to suppress leukemic blasts. A protective effect of
podoplanin+ cells against apoptosis in blasts was
detected, and further studies are required to identify cell type
from podoplanin+ cells, which are associated with
leukemic blasts. Stromal cell impairment leads to deficient
hematopoiesis and chromosomal abnormalities, which may contribute
to leukemogenesis (43,44), indicating the importance of
micro-environment alteration in leukemia.
In the present study, leukemia-derived cells that
express leukemia-related genes were markedly increased on
podoplanin− CD34+ cells.
Podoplanin+ cells, which contain stromal cells, partly
expressed hematopoietic-associated genes during differentiation;
however, the mechanism through which this switching of podoplanin
expression occurs, and the way it evolves to the progression of
leukemic cells, remains unknown. Stromal cells appear to serve a
role in AML by preventing apoptosis (45). Boyerinas et al (7) suggested that dormant leukemic cells
are heavily regulated by the BM niche. By contrast, Flach et
al (46) and Schepers et
al (47) emphasized that DNA
damage is responsible for the conversion of normal HSCs into
malignant cells, and that LSC eventually leads to disruption of BM
niches. Despite the controversy, understanding the association
between LSCs and their surrounding environment is required for the
treatment of AML.
Chemotherapy-resistant leukemic stem cells are
typically observed in BM, and interact with stromal cells to
promote blast retention (47–50).
Since the development of leukemia leads to alterations in
microenvironmental factors, including immune and stromal cells,
these alterations may directly or indirectly affect leukemic cells
in a reciprocal manner (7,45,47).
In the present study, a marked reduction in blast cell apoptosis
was observed following co-culture with podoplanin+
cells, suggesting that blast cells rapidly promote cell
proliferation, and have a protective role.
Further studies on syngeneic mouse models are
required in order to gain insight into the function of podoplanin
cells in leukemia, as well as to fully elucidate the functional
properties of podoplanin+ stromal cells in the presence
of cytokines or trafficking leukemia-associated mutant genes. The
observations of the present study indicated that
podoplanin+ cells in patients with leukemia are able to
function as stromal cells, in order to protect against apoptosis
and leukemic propagation with increased leukemic antigens.
Acknowledgments
The current study was supported by the Basic Science
Research Program of the National Research Foundation of Korea
funded by the Ministry of Education (grant. nos. 2014R1A1A2053407
and 2015R1D1A1A01059819).
References
|
1
|
Pabst C, Krosl J, Fares I, Boucher G, Ruel
R, Marinier A, Lemieux S, Hébert J and Sauvageau G: Identification
of small molecules that support human leukemia stem cell activity
ex vivo. Nat Methods. 11:436–442. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shlush LI, Zandi S, Mitchell A, Chen WC,
Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW,
et al: Identification of pre-leukaemic haematopoietic stem cells in
acute leukaemia. Nature. 506:328–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang H, Mi JQ, Fang H, Wang Z, Wang C, Wu
L, Zhang B, Minden M, Yang WT, Wang HW, et al: Preferential
eradication of acute myelogenous leukemia stem cells by
fenretinide. Proc Natl Acad Sci USA. 110:5606–5611. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matsushita H, Nakajima H, Nakamura Y,
Tsukamoto H, Tanaka Y, Jin G, Yabe M, Asai S, Ono R, Nosaka T, et
al: C/EBPalpha and C/EBPvarepsilon induce the monocytic
differentiation of myelomonocytic cells with the MLL-chimeric
fusion gene. Oncogene. 27:6749–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wiley JM and Yeager AM: Predictive value
of colony-forming unit assays for engraftment and leukemia-free
survival after transplantation of chemopurged syngeneic bone marrow
in rats. Exp Hematol. 19:179–84. 1991.PubMed/NCBI
|
|
6
|
David H: Rudolf Virchow and modern aspects
of tumor pathology. Pathol Res Pract. 183:356–364. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boyerinas B, Zafrir M, Yesilkanal AE,
Price TT, Hyjek EM and Sipkins DA: Adhesion to osteopontin in the
bone marrow niche regulates lymphoblastic leukemia cell dormancy.
Blood. 121:4821–4831. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wiseman DH, Greystoke BF and Somervaille
TC: The variety of leukemic stem cells in myeloid malignancy.
Oncogene. 33:3091–3098. 2014. View Article : Google Scholar
|
|
9
|
Breiteneder-Geleff S, Soleiman A, Kowalski
H, Horvat R, Amann G, Kriehuber E, Diem K, Weninger W, Tschachler
E, Alitalo K and Kerjaschki D: Angiosarcomas express mixed
endothelial phenotypes of blood and lymphatic capillaries:
Podoplanin as a specific marker for lymphatic endothelium. Am J
Pathol. 154:385–394. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schacht V, Ramirez MI, Hong YK, Hirakawa
S, Feng D, Harvey N, Williams M, Dvorak AM, Dvorak HF, Oliver G and
Detmar M: T1alpha/podoplanin deficiency disrupts normal lymphatic
vasculature formation and causes lymphedema. EMBO J. 22:3546–3556.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawase A, Ishii G, Nagai K, Ito T, Nagano
T, Murata Y, Hishida T, Nishimura M, Yoshida J, Suzuki K and Ochiai
A: Podoplanin expression by cancer associated fibroblasts predicts
poor prognosis of lung adenocarcinoma. Int J Cancer. 123:1053–1059.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamanashi T, Nakanishi Y, Fujii G,
Akishima-Fukasawa Y, Moriya Y, Kanai Y, Watanabe M and Hirohashi S:
Podoplanin expression identified in stromal fibroblasts as a
favorable prognostic marker in patients with colorectal carcinoma.
Oncology. 77:53–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kadota K, Huang CL, Liu D, Nakashima N,
Yokomise H, Ueno M and Haba R: The clinical significance of the
tumor cell D2-40 immunoreactivity in non-small cell lung cancer.
Lung cancer. 70:88–93. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee JY, Park S, Kim DC, Yoon JH, Shin SH,
Min WS and Kim HJ: A VEGFR-3 antagonist increases IFN-γ expression
on low functioning NK cells in acute myeloid leukemia. J Clin
Immunol. 33:826–837. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Conrad C, Niess H, Huss R, Huber S, von
Luettichau I, Nelson PJ, Ott HC, Jauch KW and Bruns CJ: Multipotent
mesenchymal stem cells acquire a lymphendothelial phenotype and
enhance lymphatic regeneration in vivo. Circulation. 119:281–289.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raica M, Cimpean AM and Ribatti D: The
role of podoplanin in tumor progression and metastasis. Anticancer
Res. 28:2997–3006. 2008.PubMed/NCBI
|
|
17
|
Schacht V, Dadras SS, Johnson LA, Jackson
DG, Hong YK and Detmar M: Up-regulation of the lymphatic marker
podoplanin, a mucin-type transmembrane glycoprotein, in human
squamous cell carcinomas and germ cell tumors. Am J Pathol.
166:913–921. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wicki A and Christofori G: The potential
role of podoplanin in tumour invasion. Br J Cancer. 96:1–5. 2007.
View Article : Google Scholar
|
|
19
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A and Bloomfield CD: The 2008
revision of the World Health Organization (WHO) classification of
myeloid neoplasms and acute leukemia: Rationale and important
changes. Blood. 114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bae DS and Lee JK: Development of NK cell
expansion methods using feeder cells from human myelogenous
leukemia cell line. Blood Res. 49:154–161. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee JY, Park C, Cho YP, Lee E, Kim H, Kim
P, Yun SH and Yoon YS: Podoplanin-expressing cells derived from
bone marrow play a crucial role in postnatal lymphatic
neovascularization. Circulation. 122:1413–1425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Koh CM: Preparation of cells for
microscopy using cytospin. Methods Enzymol. 533:235–240. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ariizumi T, Ogose A, Kawashima H, Hotta T,
Li G, Xu Y, Umezu H, Sugai M and Endo N: Expression of podoplanin
in human bone and bone tumors: New marker of osteogenic and
chondrogenic bone tumors. Pathol Int. 60:193–202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gishizky ML and Witte ON: Initiation of
deregulated growth of multipotent progenitor cells by bcr-abl in
vitro. Science. 256:836–839. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng H, Hao S, Liu Y, Pang Y, Ma S, Dong
F, Xu J, Zheng G, Li S, Yuan W and Cheng T: Leukemic marrow
infiltration reveals a novel role for Egr3 as a potent inhibitor of
normal hematopoietic stem cell proliferation. Blood. 126:1302–1313.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Drexler HG: Expression of FLT3 receptor
and response to FLT3 ligand by leukemic cells. Leukemia.
10:588–599. 1996.PubMed/NCBI
|
|
28
|
Rosnet O, Bühring HJ, Marchetto S, Rappold
I, Lavagna C, Sainty D, Arnoulet C, Chabannon C, Kanz L, Hannum C
and Birnbaum D: Human FLT3/FLK2 receptor tyrosine kinase is
expressed at the surface of normal and malignant hematopoietic
cells. Leukemia. 10:238–248. 1996.PubMed/NCBI
|
|
29
|
Meshinchi S and Appelbaum FR: Structural
and functional alterations of FLT3 in acute myeloid leukemia. Clin
Cancer Res. 15:4263–4269. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Patel JP, Gonen M, Figueroa ME, Fernandez
H, Sun Z, Racevskis J, van Vlierberghe P, Dolgalev I, Thomas S,
Aminova O, et al: Prognostic relevance of integrated genetic
profiling in acute myeloid leukemia. N Engl J Med. 366:1079–1089.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miyoshi H, Shimizu K, Kozu T, Maseki N,
Kaneko Y and Ohki M: t(8;21) breakpoints on chromosome 21 in acute
myeloid leukemia are clustered within a limited region of a single
gene, AML1. Proc Natl Acad Sci USA. 88:10431–10434. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim HJ, Choi EJ, Sohn HJ, Park SH, Min WS
and Kim TG: Combinatorial molecular marker assays of WT1, survivin
and TERT at initial diagnosis of adult acute myeloid leukemia. Eur
J Haematol. 91:411–422. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Suzuki S, Ishii G, Matsuwaki R, Neri S,
Hashimoto H, Yamauchi C, Aokage K, Hishida T, Yoshida J, Kohno M,
et al: Ezrin-expressing lung adenocarcinoma cells and
podoplanin-positive fibroblasts form a malignant microenvironment.
J Cancer Res Clin Oncol. 141:475–484. 2015. View Article : Google Scholar
|
|
34
|
Chuang WY, Yeh CJ, Chao YK, Liu YH, Chang
YS, Tseng CK, Chang HK, Wan YL and Hsueh C: Concordant podoplanin
expression in cancer-associated fibroblasts and tumor cells is an
adverse prognostic factor in esophageal squamous cell carcinoma.
Int J Clin Exp Pathol. 7:4847–4856. 2014.PubMed/NCBI
|
|
35
|
Dang Q, Liu J, Li J and Sun Y: Podoplanin:
A novel regulator of tumor invasion and metastasis. Med Oncol.
31(24)2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JY and Kim HJ: (Lymph) angiogenic
influences on hematopoietic cells in acute myeloid leukemia. Exp
Mol Med. 46:e1222014. View Article : Google Scholar
|
|
37
|
Lee JY, Park S, Min WS and Kim HJ:
Restoration of natural killer cell cytotoxicity by VEGFR-3
inhibition in myelogenous leukemia. Cancer lett. 354:281–289. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Junttila MR and de Sauvage FJ: Influence
of tumour micro-environment heterogeneity on therapeutic response.
Nature. 501:346–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chang YW, Hsieh PW, Chang YT, Lu MH, Huang
TF, Chong KY, Liao HR, Cheng JC and Tseng CP: Identification of a
novel platelet antagonist that binds to CLEC-2 and suppresses
podoplanin-induced platelet aggregation and cancer metastasis.
Oncotarget. 6:42733–42748. 2015.PubMed/NCBI
|
|
40
|
Grau SJ, Trillsch F, Tonn JC, Goldbrunner
RH, Noessner E, Nelson PJ and von Luettichau I: Podoplanin
increases migration and angiogenesis in malignant glioma. Int J
Clin Exp Pathol. 8:8663–8670. 2015.PubMed/NCBI
|
|
41
|
Tanaka M, Kijima H, Shimada H, Makuuchi H,
Ozawa S and Inokuchi S: Expression of podoplanin and vimentin is
correlated with prognosis in esophageal squamous cell carcinoma.
Mol Med Rep. 12:4029–4036. 2015.PubMed/NCBI
|
|
42
|
Kim EK, Jeon I, Seo H, Park YJ, Song B,
Lee KA, Jang Y, Chung Y and Kang CY: Tumor-derived osteopontin
suppresses antitumor immunity by promoting extramedullary
myelopoiesis. Cancer Res. 74:6705–6716. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Blau O, Baldus CD, Hofmann WK, Thiel G,
Nolte F, Burmeister T, Türkmen S, Benlasfer O, Schümann E, Sindram
A, et al: Mesenchymal stromal cells of myelodysplastic syndrome and
acute myeloid leukemia patients have distinct genetic abnormalities
compared with leukemic blasts. Blood. 118:5583–5592. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Geyh S, Oz S, Cadeddu RP, Fröbel J,
Brückner B, Kündgen A, Fenk R, Bruns I, Zilkens C, Hermsen D, et
al: Insufficient stromal support in MDS results from molecular and
functional deficits of mesenchymal stromal cells. Leukemia.
27:1841–1851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Konopleva M, Konoplev S, Hu W, Zaritskey
AY, Afanasiev BV and Andreeff M: Stromal cells prevent apoptosis of
AML cells by up-regulation of anti-apoptotic proteins. Leukemia.
16:1713–1724. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Flach J, Bakker ST, Mohrin M, Conroy PC,
Pietras EM, Reynaud D, Alvarez S, Diolaiti ME, Ugarte F, Forsberg
EC, et al: Replication stress is a potent driver of functional
decline in ageing haematopoietic stem cells. Nature. 512:198–202.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schepers K, Pietras EM, Reynaud D, Flach
J, Binnewies M, Garg T, Wagers AJ, Hsiao EC and Passegué E:
Myeloproliferative neoplasia remodels the endosteal bone marrow
niche into a self-reinforcing leukemic niche. Cell Stem Cell.
13:285–299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ishikawa F, Yoshida S, Saito Y, Hijikata
A, Kitamura H, Tanaka S, Nakamura R, Tanaka T, Tomiyama H, Saito N,
et al: Chemotherapy-resistant human AML stem cells home to and
engraft within the bone-marrow endosteal region. Nat Biotechnol.
25:1315–1321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Saito Y, Uchida N, Tanaka S, Suzuki N,
Tomizawa-Murasawa M, Sone A, Najima Y, Takagi S, Aoki Y, Wake A, et
al: Induction of cell cycle entry eliminates human leukemia stem
cells in a mouse model of AML. Nat Biotechnol. 28:275–280.
2010.PubMed/NCBI
|
|
50
|
Bendall LJ, Kortlepel K and Gottlieb DJ:
Human acute myeloid leukemia cells bind to bone marrow stroma via a
combination of beta-1 and beta-2 integrin mechanisms. Blood.
82:3125–3132. 1993.PubMed/NCBI
|