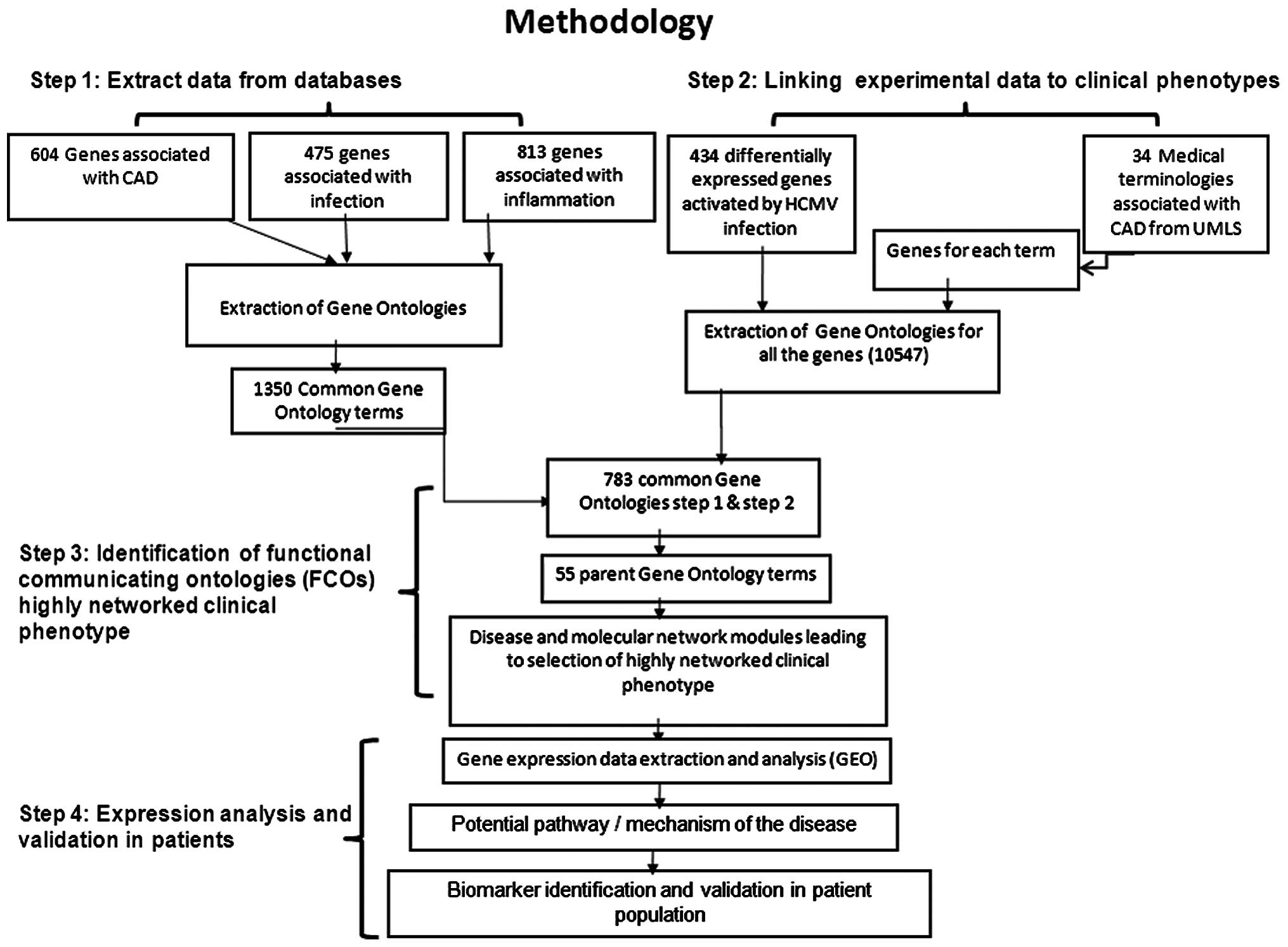
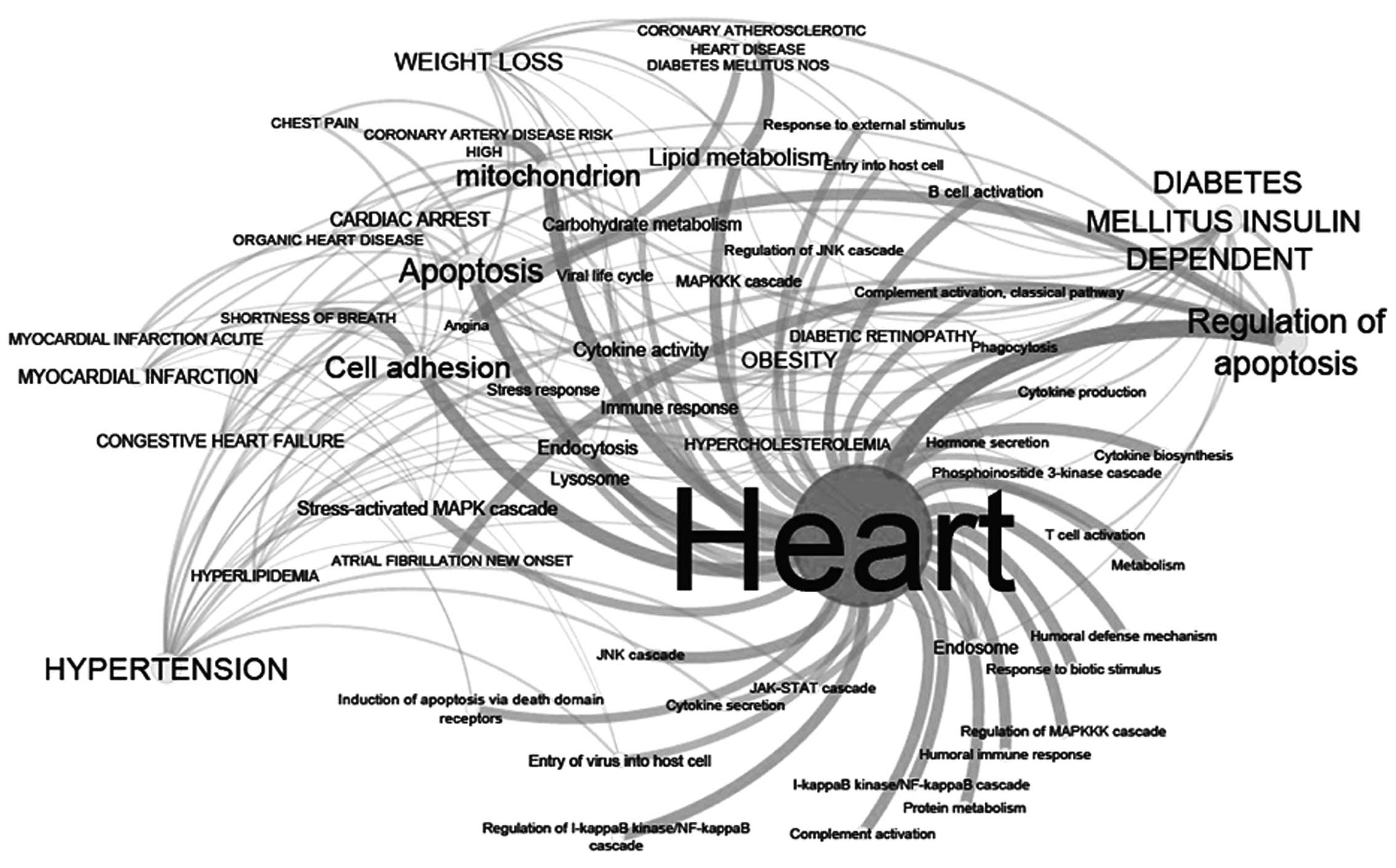
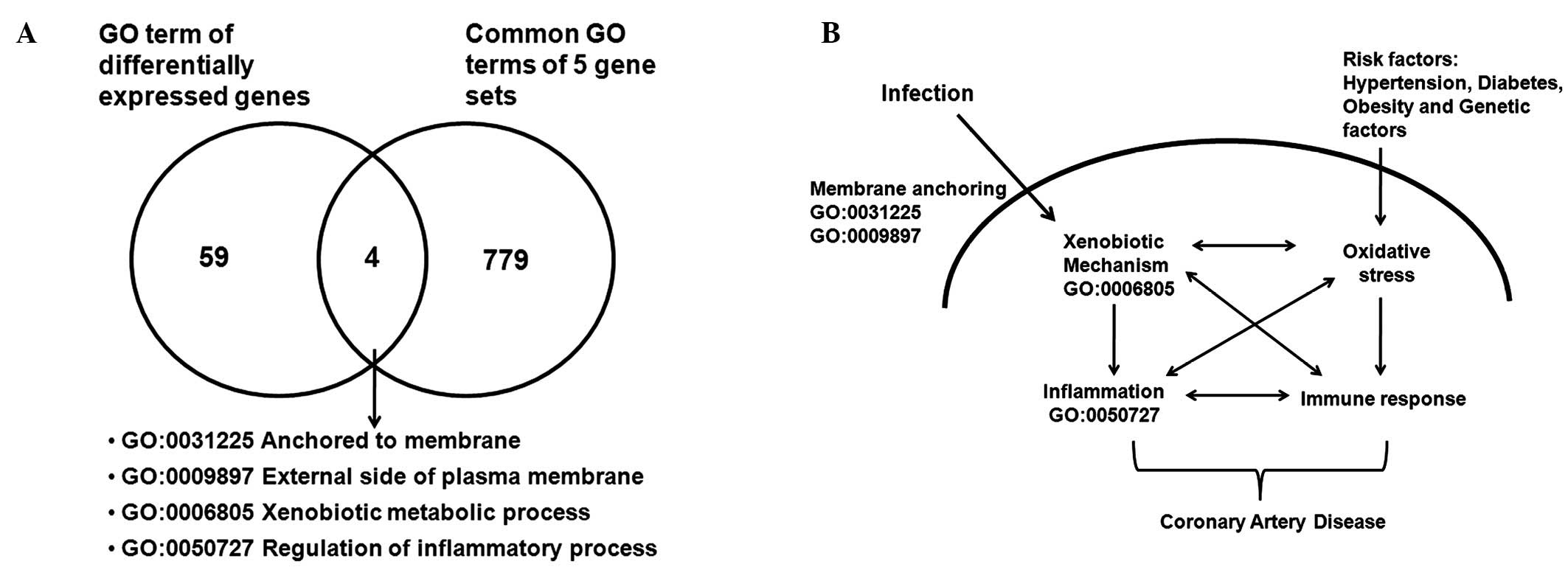
|
1
|
Cho DY, Kim YA and Przytycka TM: Chapter
5: Network biology approach to complex diseases. PLOS Comput Biol.
8:e10028202012. View Article : Google Scholar
|
|
2
|
Loscallzo J, Kohanne I and Barabasi AL:
Human disease classification in the post genomic era: A complex
systems approach to human pathobiology. Mol Sys Biol.
3:1242007.
|
|
3
|
Epstein SE, Zhu J, Najafi AH and Burnett
MS: Insights into the role of infection in atherogenesis and in
plaque rupture. Circulation. 119:3133–3141. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Epstein SE, Zhou YF and Zhu J: Infection
and atherosclerosis: Emerging mechanistic paradigms. Circulation.
100:e20–e28. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosenfield ME and Campbell LA:
Pathogenesis and atherosclerosis: Update on potential contribution
of multiple infectious organisms to pathogenesis of
atherosclerosis. Thromb Haemost. 106:858–867. 2011. View Article : Google Scholar
|
|
6
|
Shah PK: Link between infection and
atherosclerosis: Who are the culprits: Virises, bacteria, both, or
neither? Circulation. 103:5–6. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stassen FR, Vainas T and Bruggeman CA:
Infection and atherosclerosis. An alternative view on an outdated
hypothesis. Pharmacol Rep. 60:85–92. 2008.PubMed/NCBI
|
|
8
|
Mundkur LA, Rao VS, Hebbagudi S, Shanker
J, Shivanandan H, Nagaraj RK and Kakkar VV: Pathogen burden,
cytomegalovirus infection and inflammatory markers in the risk of
premature coronary artery disease in individuals of Indian origin.
Exp Clin Cardiol. 17:63–68. 2012.PubMed/NCBI
|
|
9
|
Gulbahce N, Yan H, Dricot A, Padi M,
Byrdsong D, Franchi R, Lee DS, Rozenblatt-Rosen O, Mar JC,
Calderwood MA, et al: Viral perturbations of host networks reflect
disease etiology. PLoS Comput Biol. 8:e10025312012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee DS, Park J, Kay KA, Christakis NA,
Oltvai ZN and Barabási AL: The implications of human metabolic
network topology for disease comorbidity. Proc Natl Acad Sci USA.
105:9880–9885. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goh KI, Cusick ME, Val le D, Childs B,
Vidal M and Barabási AL: The human disease network. Proc Natl Acad
Sci USA. 104:8685–8690. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Linghu B, Snitkin ES, Hu Z, Xia Y and
Delisi C: Genome-wide proirotization of disease genes and
identification of disease-disease associations from an integrated
human functional linkage network. Genome Biol. 10:R912009.
View Article : Google Scholar
|
|
13
|
Janić C and Pržulj N: Biological function
through network topology: A survey of the human diseasome. Brief
Funct Genomics. 11:522–532. 2012. View Article : Google Scholar
|
|
14
|
Emmert-Streib F, Tripathi S, de Matos
Simoes R, Hawwa AF and Dehmer M: The human disease network.
Opportunities for classification, diagnosis and prediction of
disorders and disease genes. Systems Biomedicine. 1:20–28. 2013.
View Article : Google Scholar
|
|
15
|
Piro RM and Di Cunto F: Computational
approached to disease-gene prediction: Rationale, classification
and successes. FEBS J. 279:678–696. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu S, Falck T, Daemen A, Tranchevent LC,
Suykens JA, De Moor B and Moreau Y: L2-norm multiple
kernel learning and its application to biomedical data fusion. BMC
Bioinformatics. 11:3092010. View Article : Google Scholar
|
|
17
|
Chen Y, Zhu J, Lum PY, Yang X, Pinto S,
MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al:
Variations in DNA molecular networks that cause disease. Nature.
452:429–435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu H, Liu W, Liao Y, Cheng L, Liu Q, Ren
X, Shi L, Tu X, Wang QK and Guo AY: CADgene: A comprehensive
database for coronary artery disease genes. Nucleic Acids Res.
39(Database issue): D991–D996. 2011. View Article : Google Scholar :
|
|
19
|
Wang A, Ren L and Li H: A systemic network
triggered by human cytomegalovirus entry. Adv Virol.
2011:2620802011. View Article : Google Scholar
|
|
20
|
Suresh R, Li X, Chiriac A, Goel K, Terzic
A, Perez-Terzic C and Nelson TJ: Transcriptome from circulating
cells suggests dysregulated pathways associated with long-term
recurrent events following frst-time myocardial infarction. J Mol
Cell Cardiol. 74:13–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shanker J, Mitra A, Rao VS, Mundkur L,
Dhanalakshmi B, Hebbagodi S and Kakkar VV: Rationale, design &
preliminary findings of the Indian atherosclerosis research study.
Indian Heart J. 62:286–295. 2010.
|
|
22
|
DeLong ER, DeLong DM and Clarke-Perason
DL: Comparing the areas under two or more correlated receiver
operating characteristics curves: A nonparametric approach.
Biometrics. 44:837–845. 1988. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ji YN, An L, Zhan P and Chen XH:
Cytomegalovirus infection and coronary heart disease risk: A
meta-analysis. Mol Biol Rep. 39:6537–6546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Span AH, Van Boven CP and Bruggeman CA:
The effect of cytomegalovirus infection on the adherence of
polymorphonuclear leucocytes to endothelial cells. Eur J Clin
Invest. 19:542–548. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Naghavi M, Wyde P, Litovsky S, Madjid M,
Akhtar A, Naguib S, Siadaty MS, Sanati S and Casscells W: Influenza
infection exerts prominent inflammatory and thrombotic effects on
the atherosclerotic plaques of apolipoprotein E-deficient mice.
Circulation. 107:762–768. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hsich E, Zhou YF, Paigen B, Johnson TM,
Burnett MS and Epstein SE: Cytomegalovirus infection increases
development of atherosclerosis in apolipoprotein-E knockout mice.
Atherosclerosis. 156:23–28. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Burnett MS, Gaydos CA, Madico GE, Glad SM,
Paigen B, Quinn TC and Epstein SE: Atherosclerosis in apoE knockout
mice infected with multiple pathogens. J Infect Dis. 183:226–231.
2001. View
Article : Google Scholar
|
|
28
|
Ezzahiri R, Neilssen-Vrancken HJ, Kurvers
HA, Stassen FR, Vliegen I, Grauls GE, van Pul MM, Kitslaar PJ and
Bruggeman CA: Chlamydophila pneumonia (Chlamidia pneumoniae)
accelerates the formation of complex atherosclerotic lesions in apo
E3-Leiden mice. Cardiovasc Res. 56:269–276. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ezzahiri R, Stassen FR, Kurvers HA, van
Pul MM, Kitslaar PJ and Bruggemann CA: Chlamydia pneumoniae
infection induces an uns- atherosclerotic plaque phenotype in
LDL-receptor, ApoE double knockout mice. Eur J Vasc Endovasc Surg.
26:88–95. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Janeway CA Jr, Travers P, Walport M and
Shlomchik MJ: The front line of host defense. Immunobiology: The
Immune System in Health and Disease. 5th edition. Garland Science;
New York: pp. 295–340. 2001
|
|
31
|
Smeeth L, Thomas SL, Hall AJ, Hubbard R,
Farrington P and Vallance P: Risk of myocardial infarction and
stroke after acute infection or vaccination. N Engl J Med.
351:2611–2618. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anderson JL and Muhlestein JB: Antiobiotic
trials for coronary heart disease. Tex Heart Inst J. 31:33–38.
2004.
|
|
33
|
Dhingra R, Gona P, Wang TJ, Fox CS,
D'Agostino RB Sr and Vasan RS: Serum gamma glutamyl transferase and
risk of heart failure in the community. Atheroscler Thromb Vasc
Biol. 30:1855–1860. 2010. View Article : Google Scholar
|
|
34
|
Gryaston JT: Antiobiotic treatment of
atherosclerotic cardiovascular disease. Circulation. 107:1228–1230.
2003. View Article : Google Scholar
|